Abstract
Background
Myasthenia gravis (MG) is a rare autoimmune disease with clinical symptoms of fluctuating muscle weakness. Due to the side effects of current therapies, there is an urgent need for a new medication for MG treatment. Nux vomica is a traditional Chinese medicine used in various diseases. However, the mechanism of action of Nux vomica against MG remains unclear.
Methods
Network pharmacology was used to explore the underlying mechanisms of Nux vomica in MG treatment, which was validated using molecular docking and in vivo experiments in mice.
Results
Twelve bioactive compounds and 72 targets in Nux vomica were screened. Seventy-nine myasthenia-related targets were obtained from the GENECARD and DisGeNET databases. PPI networks of Nux vomica- and myasthenia-related targets were constructed using Bisogenet, and these two networks were subsequently merged into an intersection to establish a core-target PPI network that consisted of 204 nodes and 4,668 edges. KEGG enrichment analysis indicated that 132 pathways were enriched in 204 core targets. In addition, we obtained 50 docking pairs via molecular docking. In vivo experiments revealed that Nux vomica can improve the symptoms of MG.
Conclusion
Nux vomica is involved in the pathogenesis of MG through the “multicomponent-target-pathway” mechanism.
1. Introduction
Myasthenia gravis (MG) is an autoantibody-mediated neuromuscular disease that causes fluctuating muscle weakness predominantly in the ocular, bulbar, and proximal limb muscles [1]. The prevalence rate is approximately one in 5,000 worldwide, and the incidence is higher in younger women and older men [2]. Currently, patients with MG are mainly treated with corticosteroids, long-term immunosuppressive drugs, plasma exchange, or thymectomy [3]. However, their clinical application is limited due to their high costs and side effects, such as cardiac rhythm disorders and hypotension [4]. Hence, alternative therapies with higher efficacy and fewer side effects are needed for MG treatment.
Traditional Chinese medicines (TCMs) have received increasing attention because of their high efficacy, safety, and few adverse effects. According to the TCM theory, MG is classified as “Weizheng” and diagnosed as “flaccidity syndrome” and “Qi-deficiency pattern” [5]. Thus, Qi-supplementation therapy is important for MG treatment. An increasing number of studies have shown that TCMs as an alternative or adjuvant of Western medicines could significantly improve the clinical efficacy of MG treatment [6–8]. Jianpiyiqi granules have been shown to remarkably alleviate the clinical symptoms in children with ocular MG accompanied by a decrease in acetylcholine receptor autoantibodies [6]. In addition, the Qiangji Jianli decoction has been shown to alleviate oxidative stress injury and facilitate mitochondrial biogenesis via the AMPK/PGC-1α-dependent signaling pathway, which restored muscle energy supply and ameliorated gastrocnemius muscle contractility in experimental autoimmune MG rats [7]. Another well-known TCM treatment for MG, Bu Zhong Yi Qi decoction, has been reported to decrease simulated weightlessness-induced muscle atrophy and promote myogenesis by reducing nuclear receptor corepressor 1 (NCoR1)-associated gene expression [8]. Nux vomica, also called Semen Strychni or Ma qianzi (Chinese: 马钱子), is the seed of Strychnos Nux-vomica L, belonging to the genus Strychnos of the family Loganiaceae. Studies have illustrated that Nux vomica has numerous biological activities, such as analgesic, anti-inflammatory, antitumor, and immunomodulatory effects [9]. A previous study found that Nux vomica could alleviate inflammatory response and thereby ameliorates MG in experimental autoimmune MG rats [10]. However, the molecular mechanisms of Nux vomica against MG remain unclear. Therefore, it is meaningful to explore the targets and further clarify the underlying mechanisms of action of Nux vomica on MG treatment based on network pharmacology.
Network pharmacology, first introduced by British pharmacologist Hopkins, has proven to be an effective strategy for revealing the complicated relationship between TCMs and diseases [11, 12]. TCMs exert their therapeutic functions through a multicompound-target-pathway network. Thus, the molecular mechanisms of TCMs can be clarified using network pharmacology systematically and integrally [13]. In this study, we screened the active components of Nux vomica and explored its underlying molecular mechanisms on MG based on network pharmacology. Our study provides a theoretical basis for the future clinical application of Nux vomica for treating MG.
2. Materials and Methods
2.1. Collection of Therapeutic Targets for MG
Therapeutic targets for MG were obtained from GeneCards [14] (https://www.genecards.org/) and DisGeNET [15] (https://www.disgenet.org/) databases. Candidate targets were collected after the deletion of duplicates.
2.2. Collection of Bioactive Ingredients and Therapeutic Targets for Nux Vomica
TCMSP (https://tcmspw.com/tcmsp.php) is an online platform for integrating pharmacokinetics, medicinal chemistry, therapeutic targets, and drug-target-disease networks [16]. Based on the TCMSP platform, bioactive ingredients and targets of Nux vomica were obtained using the screening criteria of oral bioavailability (OB) > 30% and drug-likeness (DL) > 0.18 [17]. In addition, strychnine was considered as a candidate bioactive ingredient based on a previous study. The pharmacokinetics and DL properties of strychnine were further assessed using the online SwissADME database (https://www.swissadme.ch/index.php). The therapeutic targets of strychnine were then predicted using the SwissTargetPrediction online tool (https://swisstargetprediction.ch/).
2.3. PPI Network Construction
A PPI network was constructed and visually analyzed using the Cytoscape plugin Bisogenet, which contains the following six PPI databases: the IntAct Molecular Interaction Database (IntAct), Biomolecular Interaction Network Database (BIND), Biological General Repository for Interaction Datasets (BioGRID), Database of Interacting Proteins (DIP), Molecular INTeraction Database (MINT), and Human Protein Reference Database (HPRD) [18]. PPI networks of the Nux vomica-related targets and MG-related targets were constructed using Cytoscape.
2.4. Central Network Evaluation
Central network evaluation is considered a primary method for core protein screening in PPI networks. First, the PPI networks of the Nux vomica-related targets and MG-related targets were merged into an intersection. Next, the Cytoscape plugin CytoNCA was applied to assess the intersection from the following six centrality measures: betweenness centrality (BC), closeness centrality (CC), degree centrality (DC), eigenvector centrality (EC), network centrality (NC), and local average connectivity-based method (LAC) [19]. In addition, CytoHubba, a Cytoscape plugin, was used to select the hub genes [20].
2.5. GO and KEGG Pathway Enrichment Analyses
Gene ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed based on the public database GO (https://geneontology.org/) and KEGG (https://www.genome.jp/kegg/).
A hypergeometric distribution model was used to evaluate whether the target gene set was significantly associated with a specific gene ontology or biological pathway using the following formula:
| (1) |
Here, N represents the total number of genes from the reference terms, M represents the number of genes annotated to specific GO terms or pathways, n represents the number of identified Nux vomica-target genes, and k represents the number of common genes between Nux vomica-target genes and the reference set. The clusterProfiler package in R (4.0) (https://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html) was adopted to perform the GO and KEGG pathway enrichment analyses with P values <0.01, adjusted by the Bonferroni correction.
2.6. Molecular Docking Analysis
The zinc database (https://zinc15.docking.org) was used to download two-dimensional (2D) structures of bioactive compounds [21]. AutoDockTools-1.5.6 was used to add nonpolar hydrogen, calculate charges, and set rotatable keys for the structure and save it as a “pdbqt” file [22]. The 3D structures of the hub targets were downloaded from the Protein Data Bank (PDB) database (https://www.rcsb.org/) [23]. The hub targets and the original ligands were separated and the water molecules were removed using PyMol 2.3.0 [24]. Next, the ligand and receptor were imported into AutoDockTools-1.5.6 to add nonpolar hydrogen, calculate charges, and save as a “pdbqt” file. The protein proligand was set as the center of the docking box; if no proligand was present in the target protein, the docking region was identified based on key amino acid residues near the active pocket of the target, as reported in previous studies, or the fpocket [25] online pocket parameter prediction website for analysis. The prepared protein files, ligand files, compound files, and scripts were placed in the appropriate folders, and molecular docking was performed using AutoDock Vina 1.1.2 [26]. Finally, the binding affinity was calculated using AutoDock Vina 1.1.2, and the docking results were visualized using the open-source version PyMol 2.3.0.
2.7. Animal Experiments
Female C57BL/6J mice (aged 7–8 weeks) were purchased from the Animal Resources Center (Murdoch, WA, Australia). All mice were housed in a controlled environment under a 12-h light/dark cycle with 21°C and 45–65% humidity and had free access to food and water.
After anaesthesia, mice in the model group were emulsified with 40 μg MuSK in 100 μL phosphate-buffered saline (PBS) and 100 μL complete Freund's adjuvant (CFA; Becton, Dickinson and Company, NJ, USA), which were intraperitoneally injected into the hind foot. The control group was injected with 100 μL of PBS and 100 μL of CFA. The mice in both groups were injected with cyclophosphamide monohydrate (300 mg/kg, dissolved in 0.9% saline to a final concentration of 10 mg/mL; Sigma-Aldrich, MO, USA) within 24 h to suppress immune resistance. The aforementioned operation was repeated after 30 days. The body weight of the mice was measured weekly, and a clinical examination was performed every 2 weeks to determine the model-building cycle. After the MG mouse model was successfully constructed, the mice were randomly divided into five groups (n = 8). The mice in the low-, mid-, and high-dose Nux vomica groups were administered low (4 mg/kg), mid (8 mg/kg), and high (16 mg/kg) doses of Nux vomica. The mice in the control and model groups were intragastrically administered equivalent doses of saline. The body weight of the mice was measured weekly, and a clinical examination was performed every 2 weeks. After 30 days, mice were sacrificed by cervical dislocation after injecting sodium pentobarbital (50 mg/kg), and tendon tissues were removed for subsequent experiments.
2.8. Clinical Evaluation
For clinical examination, mice were left for 3 min on a flat platform and observed for signs of MG. The clinical muscle weakness was graded as follows: Grade 0.5, the mouse exhibited slightly lower activity than normal after the standard exercise state, and the hanging time was >60 s; Grade 1, the mouse exhibited normal movement, lying prone, staying still, and/or hanging time <60 s; Grade 2, before and after exercise, the mouse was lying prone, staying still, and/or hanging test <30 s; Grade 3, severe weakness, dehydration and paralysis, weight loss >15%, and/or hanging time 0 s; Grade 4, the mouse died. The grip of the mice was observed every 2 weeks. First, the tail of each mouse was gently grabbed and the mouse was suspended above a metal grid. When the mouse grasped the metal grid, the mouse was lifted and the tension was measured 20 times. One end was fixed on the tension device, and the front end of the force measurement device was made into a grid shape. The tail of the mouse was grasped, the front end of the force measuring device was grasped, and the tail of the mouse was pulled forcefully. When the mouse was not strong enough to grasp the front end of the force measuring device, the force measuring device showed the grip force at this time. In addition, clinical progress was assessed using an inverted screening test every 2 weeks. In this test, the mouse was placed in the center of the grid and immediately rotated to the upside-down position, and stably maintained 50 cm above the soft cushion. The time when the mouse fell was recorded and the observation time was limited to 300 s.
2.9. Quantitative Real-Time PCR (qRT-PCR) Assay
Total RNA was extracted from the tendon tissues of the mice using TRIzol® Reagent (Ambion™) (Applied Biosystems, MA, USA), and 500 ng of RNA was reverse-transcribed into cDNA using TaqMan® Reverse Transcription Reagents (Thermo Fisher Scientific, MA, USA). The expression of target genes was examined using ABI Prism SDS 7000 (Thermo Fisher Scientific) at 95°C for 1 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min, and then calculated using the 2−ΔΔCt method. Primer sequences for qRT-PCR are listed in Table S1. GAPDH was used as an internal control.
2.10. Western Blot Analysis
The predicted target protein expression was detected by western blotting. Briefly, total proteins were extracted from tendon tissue using RIPA lysis buffer (Applied Biosystems, MA, USA). Then, the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Millipore, MA, USA). The membranes were blocked with 5% skimmed milk and incubated with primary antibodies anti-EGFR, anti-TP53, anti-AKT1, anti-MYC, and anti-GAPDH (1 : 1,000, Abcam, UK) at 4°C overnight. After washing with PBS, the membranes were incubated with a secondary antibody (1 : 1,000, Abcam). Enhanced chemiluminescent reagents (Millipore, MA, USA) were used to detect the protein bands. GAPDH was used as an internal control.
2.11. Statistical Analysis
Data are shown as the mean ± standard deviation (SD) and analyzed by one-way ANOVA followed by Tukey's test using SPSS 27.0 software (IBM, IL, USA). Differences were considered significant at P < 0.05.
3. Results
3.1. Nux Vomica Bioactive Compounds-Targets Network
The TCMSP database was used to search for the bioactive compounds of Nux vomica with the criteria of OB ≥ 30% and DL ≥ 0.18. The TCMSP database was also used to search for the targets of Nux vomica. As a result, 11 active compounds with the targets were found in Nux vomica. Previous studies have shown that strychnine is one of the major bioactive and principal toxic compounds of Nux vomica [9]. Therefore, strychnine was considered as a candidate in this study. The DL and pharmacokinetics of strychnine were subsequently evaluated using the online tool SwissADME. The results indicated that strychnine exhibited good gastrointestinal absorption and high DL properties, confirming that the strain was an active compound of Nux vomica. The targets of strychnine were predicted by SwissTargetPrediction, and the top 25 were viewed as key targets of strychnine. Collectively, we found 12 bioactive compounds and 72 targets in Nux vomica, and the details are shown in Tables 1 and 2. Subsequently, we constructed a Nux vomica bioactive compound-target network using Cytoscape (Figure 1).
Table 1.
Bioactive Compounds of Nux vomica.
| Chemical | Molecular Formula | Molecular Weight (g/mol) | OB (%) | DL |
|---|---|---|---|---|
| (+)-catechin | C15H14O6 | 290.27 | 54.83 | 0.24 |
| (2R)-5, 7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one | C15H12O5 | 272.25 | 42.36 | 0.21 |
| (S)-stylopine | C19H17NO4 | 323.37 | 51.15 | 0.85 |
| brucine-N-oxide | C23H26N2O5 | 410.51 | 49.17 | 0.38 |
| Icaride A | C22H28O7 | 404.5 | 48.74 | 0.43 |
| Isobrucine | C23H26N2O4 | 394.5 | 33.58 | 0.8 |
| Isostrychnine N-oxide (I) | C21H25N2O3 | 352.47 | 35.45 | 0.8 |
| Isostrychnine N-oxide (II) | C21H23N2O3 | 350.45 | 37.33 | 0.8 |
| Lokundjoside_qt | C29H44O10 | 552.65 | 32.82 | 0.76 |
| Stigmasterol | C29H48O | 412.77 | 43.83 | 0.76 |
| Vomicine | C22H24N2O4 | 380.437 | 47.56 | 0.65 |
| Strychnine | C21H22N2O2 | 334.4 | High | YES |
Table 2.
The Target Genes of Nux vomica.
| ID | Name |
|---|---|
| 126 | ADH1C |
| 148 | ADRA1A |
| 147 | ADRA1B |
| 146 | ADRA1D |
| 150 | ADRA2A |
| 153 | ADRB1 |
| 154 | ADRB2 |
| 367 | AR |
| 847 | CAT |
| 1128 | CHRM1 |
| 1129 | CHRM2 |
| 1131 | CHRM3 |
| 1132 | CHRM4 |
| 1133 | CHRM5 |
| 1135 | CHRNA2 |
| 1504 | CTRB1 |
| 2099 | ESR1 |
| 2100 | ESR2 |
| 3037 | HAS2 |
| 3359 | HTR3A |
| 3757 | KCNH2 |
| 4048 | LTA4H |
| 801 | CALM1 |
| 4129 | MAOB |
| 4128 | MAOA |
| 3356 | HTR2A |
| 2556 | GABRA3 |
| 1139 | CHRNA7 |
| 3363 | HTR7 |
| 2147 | F2 |
| 2159 | F10 |
| 5467 | PPARD |
| 5292 | PIM1 |
| 3778 | KCNMA1 |
| 8648 | NCOA1 |
| 10499 | NCOA2 |
| 4985 | OPRD1 |
| 4988 | OPRM1 |
| 5241 | PGR |
| 5328 | PLAU |
| 5644 | PRSS1 |
| 5742 | PTGS1 |
| 5743 | PTGS2 |
| 6256 | RXRA |
| 6331 | SCN5A |
| 6530 | SLC6A2 |
| 6531 | SLC6A3 |
| 6532 | SLC6A4 |
| 11255 | HRH3 |
| 3741 | KCNA5 |
| 1812 | DRD1 |
| 1803 | DPP4 |
| 3716 | JAK1 |
| 1595 | CYP51A1 |
| 1137 | CHRNA4 |
| 2837 | UTS2R |
| 54878 | DPP8 |
| 91039 | DPP9 |
| 3352 | HTR1D |
| 1230 | CCR1 |
| 9475 | ROCK2 |
| 120892 | LRRK2 |
| 3718 | JAK3 |
| 3717 | JAK2 |
| 1141 | CHRNB2 |
| 6093 | ROCK1 |
| 2155 | F7 |
| 2554 | GABRA1 |
| 4843 | NOS2 |
| 4306 | NR3C2 |
| 9475 | ROCK2 |
| 1432 | MAPK14 |
Figure 1.
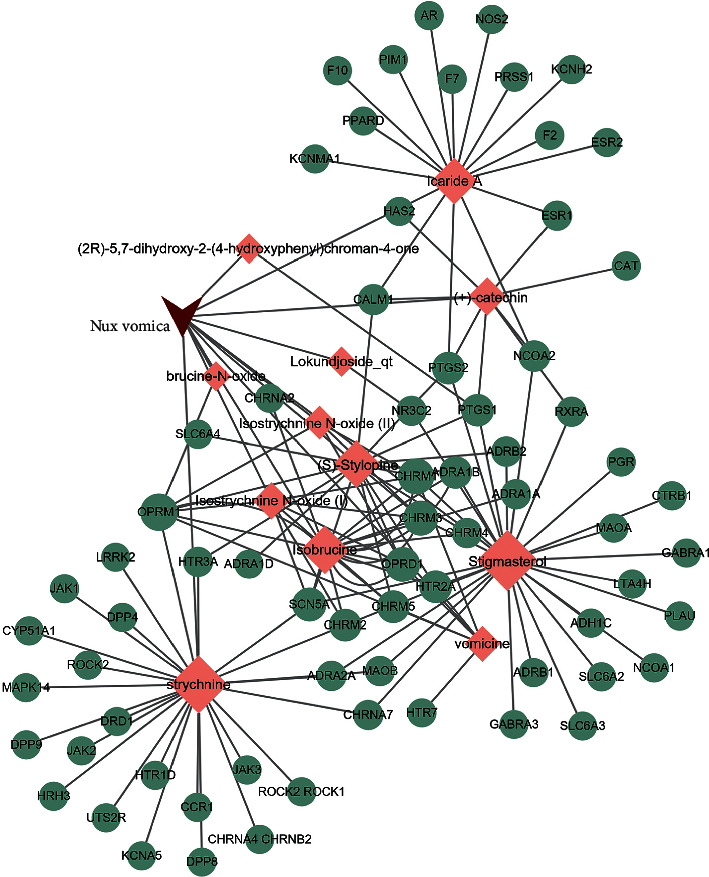
Nux vomica bioactive compounds-targets network. Red inverted triangle represents Nux vomica, pink rhombus represents bioactive compounds of Nux vomica, green circles represent the targets of Nux vomica, and the size represents the degree.
3.2. GO and KEGG Pathway Enrichment Analysis of Nux Vomica
To further investigate the 72 targets of the Nux vomica bioactive compounds, GO enrichment analysis was performed using biological processes (BP), molecular function (MF), and cellular component (CC) terms. A total of 103 MFs, 454 BPs, and 48 CCs were found to be enriched (P < 0.01). The GO analysis with the top 15 markedly enriched MF, BP, and CC terms is shown in Figures 2(a)–2(c). KEGG pathway enrichment analysis demonstrated that 72 targets of Nux vomica bioactive compounds were enriched in 17 associated pathways (P < 0.01). The top 15 KEGG pathways in which the targets of Nux vomica bioactive compounds were enriched are presented in Figure 2(d) and Table 3.
Figure 2.

Gene Ontology and KEGG pathway enrichment analysis of 72 targets of Nux vomica bioactive compounds. (a) Representative bubble plots of molecular function (MF) of identified targets. (b) Representative bubble plots of biological processes (BP) of identified targets. (c) Representative bubble plots of cellular components (CC) of identified targets. (d) Representative bubble plots of KEGG pathway of identified targets. Gene ratio = count/set size.
Table 3.
The Top 15 KEGG Pathways of Nux vomica-Related Target Genes.
| ID | Description | p.adjust | Count |
|---|---|---|---|
| hsa04080 | Neuroactive ligand-receptor interaction | 2.54E − 18 | 27 |
| hsa04020 | Calcium signaling pathway | 3.70E − 09 | 15 |
| hsa04022 | cGMP-PKG signaling pathway | 4.62E − 06 | 11 |
| hsa04725 | Cholinergic synapse | 1.07E − 05 | 9 |
| hsa04726 | Serotonergic synapse | 1.07E − 05 | 9 |
| hsa04970 | Salivary secretion | 2.05E − 05 | 8 |
| hsa05033 | Nicotine addiction | 0.000381 | 5 |
| hsa04261 | Adrenergic signaling in cardiomyocytes | 0.000549 | 8 |
| hsa04024 | cAMP signaling pathway | 0.001051 | 9 |
| hsa04270 | Vascular smooth muscle contraction | 0.001575 | 7 |
| hsa04915 | Estrogen signaling pathway | 0.001806 | 7 |
| hsa05031 | Amphetamine addiction | 0.003161 | 5 |
| hsa04810 | Regulation of actin cytoskeleton | 0.00453 | 8 |
| hsa05140 | Leishmaniasis | 0.00453 | 5 |
| hsa05030 | Cocaine addiction | 0.006586 | 4 |
3.3. Target Genes of MG
Thirty-seven MG-related targets were obtained from the GENECARD database, and 42 were from the DisGeNET database. Seventy-nine disease target genes were retrieved after combining the results of both databases, and the duplicates were deleted. The details are listed in Table 4.
Table 4.
The Target Genes of MG.
| ID | Name |
|---|---|
| 1756 | DMD |
| 5376 | PMP22 |
| 270 | AMPD1 |
| 2992 | GYG1 |
| 6904 | TBCD |
| 9499 | MYOT |
| 79823 | CAMKMT |
| 1437 | CSF2 |
| 1440 | CSF3 |
| 7273 | TTN |
| 6606 | SMN1 |
| 1674 | DES |
| 6647 | SOD1 |
| 7415 | VCP |
| 4000 | LMNA |
| 2110 | ETFDH |
| 2318 | FLNC |
| 64135 | IFIH1 |
| 4621 | MYH3 |
| 23345 | SYNE1 |
| 2200 | FBN1 |
| 773 | CACNA1A |
| 3123 | HLA-DRB1 |
| 1292 | COL6A2 |
| 55863 | TMEM126B |
| 55154 | MSTO1 |
| 7157 | TP53 |
| 1180 | CLCN1 |
| 4982 | TNFRSF11B |
| 2218 | FKTN |
| 28976 | ACAD9 |
| 8074 | FGF23 |
| 178 | AGL |
| 4038 | LRP4 |
| 1760 | DMPK |
| 4359 | MPZ |
| 26092 | TOR1AIP1 |
| 3767 | KCNJ11 |
| 57679 | ALS2 |
| 1605 | DAG1 |
| 4567 | TRNL1 |
| 7276 | TTR |
| 5373 | PMM2 |
| 9590 | AKAP12 |
| 1134 | CHRNA1 |
| 1146 | CHRNG |
| 4593 | MUSK |
| 5913 | RAPSN |
| 1145 | CHRNE |
| 43 | ACHE |
| 7124 | TNF |
| 1493 | CTLA4 |
| 6261 | RYR1 |
| 375790 | AGRN |
| 3586 | IL10 |
| 3558 | IL2 |
| 326 | AIRE |
| 3565 | IL4 |
| 3458 | IFNG |
| 4155 | MBP |
| 10673 | TNFSF13B |
| 7112 | TMPO |
| 4049 | LTA |
| 940 | CD28 |
| 941 | CD80 |
| 246750 | MYAS1 |
| 5354 | PLP1 |
| 3106 | HLA-B |
| 7293 | TNFRSF4 |
| 3115 | HLA-DPB1 |
| 942 | CD86 |
| 3119 | HLA-DQB1 |
| 1604 | CD55 |
| 1378 | CR1 |
| 154 | ADRB2 |
| 26191 | PTPN22 |
| 959 | CD40LG |
| 285489 | DOK7 |
3.4. Nux Vomica-MG PPI Network
The PPI networks of Nux vomica- and MG-related targets were constructed using Bisogenet. The PPI network of Nux vomica-related genes was constructed with 2,275 nodes and 92,252 edges (Figure 3(a)). In addition, the MG-related target network was constructed with 3,480 nodes and 121,919 edges (Figure 3(b)). Both networks were subsequently merged into an intersection with 1,517 nodes and 50,040 edges (Figure 3(c)). CytoNCA was used to assess the intersection of the PPI network by topological analysis. A Nux vomica on the myasthenia PPI network was first screened based on the criteria of “BC ≥ 902.385, CC ≥ 0.471, DC ≥ 52” (Figure 3(d)). A core-target PPI network was further screened using the criteria of “BC ≥ 1985.705, CC ≥ 0.500, DC ≥ 90,” which consists of 204 nodes and 4,668 edges (Figure 3(e)).
Figure 3.

Identification of core targets for Nux vomica against MG. (a) The PPI network of Nux vomica-related targets (2,275 nodes and 92,252 edges). (b) The PPI network of MG-related targets (3,480 nodes and 121,919 edges). (c) Intersection of PPI networks (1,517 nodes and 5,0040 edges). (d) PPI network by the screening criteria of BC ≥ 902.385, CC ≥ 0.471, DC ≥ 52 (567 nodes and 19,098 edges). (e) Core-target PPI network by the screening criteria of BC ≥ 1985.705, CC ≥ 0.500, DC ≥ 90 (204 nodes and 4,668 edges). BC, betweenness centrality; CC, closeness centrality; DC, degree centrality.
3.5. Enrichment Analysis of the Core-Target PPI Network
In the GO enrichment analysis, a total of 144 molecular functions, 1,570 biological processes, and 151 cell components were identified in 204 core targets of Nux vomica following MG treatment (P < 0.01). The top 15 significant GO terms, including MF, BP, and CC terms, are shown in Figures 4(a)–4(c). KEGG pathway enrichment analysis demonstrated that 132 pathways were enriched in 204 core targets (P < 0.01). The top 15 KEGG pathways enriched for the targets of bioactive compounds from Nux vomica are shown in Figure 4(d).
Figure 4.

Gene Ontology and KEGG pathway enrichment analysis of 204 core targets for Nux vomica against MG. (a) Representative bubble plots of molecular function (MF) of core targets. (b) Representative bubble plots of biological processes (BP) of core targets. (c) Representative bubble plots of cellular components (CC) of core targets. (d) Representative bubble plots of KEGG pathway of core targets. Gene ratio = count/set size.
3.6. Enrichment Analysis of the Hub Genes
Hub genes were selected using CytoHubba based on the core-target PPI network. The results showed that TP53, EGFR, UBC, MYC, HSP90AA1, EP300, HSPA8, RPS27A, AKT1, and GAPDH were the major hub nodes in the core-target PPI network, and the details are shown in Table 5. The PPI network of the hub genes is shown in Figure 5.
Table 5.
The Key Parameter Values of 10 Hub Genes.
| Genes | Betweenness | Closeness | Degree | Eigenvector | LAC | Network |
|---|---|---|---|---|---|---|
| TP53 | 78488.11 | 0.592188 | 492 | 0.123322 | 39.82724 | 226.9392 |
| EGFR | 29951.12 | 0.552277 | 302 | 0.069005 | 26.71523 | 98.37902 |
| UBC | 20784.32 | 0.547688 | 278 | 0.07858 | 32.83094 | 97.09218 |
| MYC | 17003.02 | 0.539502 | 246 | 0.060742 | 22.79675 | 63.83153 |
| HSP90AA1 | 12731.44 | 0.538544 | 245 | 0.075996 | 36.49796 | 85.84409 |
| EP300 | 10000.75 | 0.532117 | 215 | 0.060767 | 34.1907 | 75.79397 |
| HSPA8 | 7938.097 | 0.532303 | 200 | 0.065457 | 33.275 | 60.61926 |
| RPS27 A | 4139.276 | 0.51723 | 149 | 0.053001 | 29.77181 | 43.20848 |
| AKT1 | 5381.846 | 0.51776 | 143 | 0.038709 | 21.11189 | 32.85155 |
| GAPDH | 2874.597 | 0.515646 | 111 | 0.042723 | 24.31532 | 29.43539 |
Figure 5.

The PPI network of 10 hub genes.
The results of the most representative GO terms included regulation of protein modification by small protein conjugation or removal, cellular response to drugs, activation of the innate immune response, intrinsic apoptotic signaling pathway, regulation of protein ubiquitination, regulation of the innate immune response, positive regulation of cellular protein localization, regulation of protein stability, positive regulation of the innate immune response, regulation of the mRNA metabolic process, positive regulation of establishment of protein localization, regulation of binding, regulation of sequence-specific DNA binding transcription factor activity, regulation of apoptotic signaling pathway, and rhythmic process (Figure 6).
Figure 6.

The top 10 GO terms of hub genes.
In addition, the top 15 pathways in which the core targets were enriched were identified (Figure 7), mainly including the neurotrophin signaling pathway, PI3K-Akt signaling pathway, cell cycle, apoptosis, viral carcinogenesis, prostate cancer, hepatitis C, pancreatic cancer, Epstein-Barr (EB) virus infection, human cytomegalovirus infection, proteoglycans in cancer, hepatitis B, chronic myeloid leukemia, Kaposi sarcoma-associated herpes, and microRNAs (miRNAs) in cancer.
Figure 7.

The top 15 pathways of hub genes.
3.7. Molecular Docking Analysis
The five bioactive compounds (strychnine, stigmasterol, (S)-stylopine, brucine-N-oxide, and isostrychnineN-oxide (II)) are shown in Table 6. The top ten hug targets (TP53, EGFR, UBC, MYC, HSP90AA1, EP300, HSPA8, RPS27A, AKT1, and GAPDH) were selected as the core compounds and core targets for molecular docking. AutoDock Vina assesses the binding of small molecules mainly by binding energy (affinity) to assess the binding strength of a small molecule to a protein. If the affinity is <0, the ligand can spontaneously bind to the receptor, and the smaller the value, the easier it is for the active ingredient to bind to the receptor. Therefore, the interaction between the active compound and its corresponding target was analyzed by the binding energy, and a threshold affinity <–5 kcal/mol was set in this study. A total of 50 docking pairs were obtained, as shown in Table 7. Figure 8 shows the binding patterns of the top ten binding core targets and their corresponding compounds.
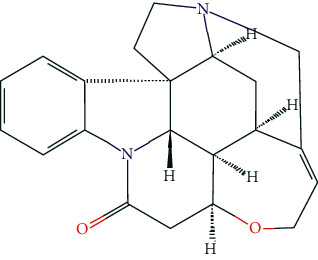
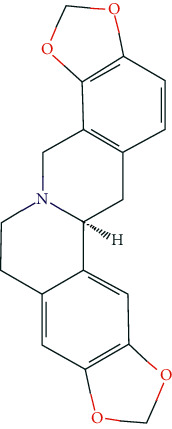
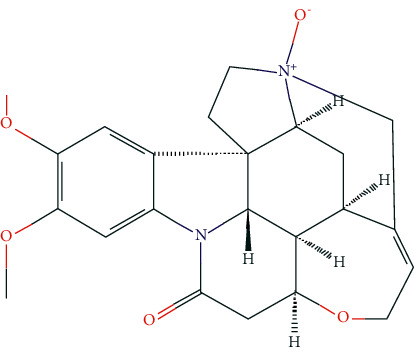
Table 6.
The chemical structure of five active compounds.
| Synonyms | Molecular Formula | 2D Structure |
|---|---|---|
| Strychnine | C21H22N2O2 |
|
|
| ||
| Stigmasterol | C29H48O |

|
|
| ||
| (S)-stylopine | C19H17NO4 |
|
|
| ||
| brucine-N-oxide | C23H26N2O5 |
|
|
| ||
| Isostrychnine N-oxide (II) | C21H23N2O3 |
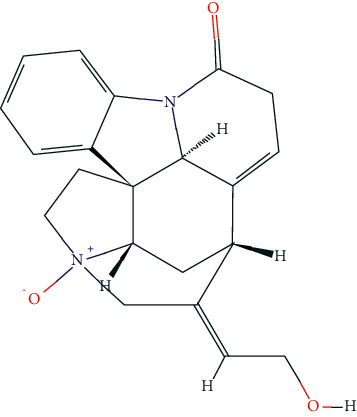
|
Table 7.
The result of molecular docking.
| Chem | PDB | Gene | Best affinity |
|---|---|---|---|
| Strychnine | 3poz | EGFR | −9.9 |
| Stigmasterol | 3biy | EP300 | −9.7 |
| (S)-stylopine | 3biy | EP300 | −9.6 |
| (S)-stylopine | 3poz | EGFR | −9.6 |
| Stigmasterol | 3poz | EGFR | −9.5 |
| brucine-N-oxide | 3poz | EGFR | −9.5 |
| Strychnine | 3biy | EP300 | −9.4 |
| (S)-stylopine | 1xqh | TP53 | −9.4 |
| IsostrychnineN-oxide (II) | 3poz | EGFR | −9 |
| Stigmasterol | 1u8f | GAPDH | −9 |
| (S)-stylopine | 1u8f | GAPDH | −8.9 |
| brucine-N-oxide | 3ldq | HSPA8 | −8.8 |
| IsostrychnineN-oxide (II) | 3biy | EP300 | −8.7 |
| brucine-N-oxide | 1u8f | GAPDH | −8.7 |
| IsostrychnineN-oxide (II) | 1xqh | TP53 | −8.6 |
| IsostrychnineN-oxide (II) | 1u8f | GAPDH | −8.6 |
| Strychnine | 1u8f | GAPDH | −8.6 |
| Strychnine | 1byq | HSP90AA1 | −8.5 |
| (S)-stylopine | 3ldq | HSPA8 | −8.4 |
| IsostrychnineN-oxide (II) | 1byq | HSP90AA1 | −8.3 |
| Strychnine | 1xqh | TP53 | −8.3 |
| Strychnine | 3ldq | HSPA8 | −8.3 |
| brucine-N-oxide | 1byq | HSP90AA1 | −8.3 |
| Strychnine | 5h07 | UBC | −8.2 |
| brucine-N-oxide | 3biy | EP300 | −8.2 |
| (S)-stylopine | 1byq | HSP90AA1 | −8.2 |
| IsostrychnineN-oxide (II) | 5h07 | UBC | −7.9 |
| IsostrychnineN-oxide (II) | 3ldq | HSPA8 | −7.7 |
| Stigmasterol | 1xqh | TP53 | −7.6 |
| brucine-N-oxide | 1xqh | TP53 | −7.6 |
| brucine-N-oxide | 5h07 | UBC | −7.5 |
| Stigmasterol | 1byq | HSP90AA1 | −7.4 |
| Stigmasterol | 3ldq | HSPA8 | −7.2 |
| brucine-N-oxide | 6sqs | RPS27A | −7.1 |
| (S)-stylopine | 5h07 | UBC | −6.8 |
| Stigmasterol | 6sqs | RPS27A | −6.6 |
| IsostrychnineN-oxide (II) | 6sqs | RPS27A | −6.5 |
| Stigmasterol | 5h07 | UBC | −6.5 |
| (S)-stylopine | 6sqs | RPS27A | −6.4 |
| (S)-stylopine | 1nkp | MYC | −6.4 |
| Strychnine | 6sqs | RPS27A | −6.3 |
| brucine-N-oxide | 1nkp | MYC | −6.3 |
| Stigmasterol | 1nkp | MYC | −6.1 |
| brucine-N-oxide | 1h10 | AKT1 | −5.9 |
| IsostrychnineN-oxide (II) | 1nkp | MYC | −5.8 |
| Strychnine | 1nkp | MYC | −5.7 |
| (S)-stylopine | 1h10 | AKT1 | −5.6 |
| Strychnine | 1h10 | AKT1 | −5.5 |
| Stigmasterol | 1h10 | AKT1 | −5.3 |
| IsostrychnineN-oxide (II) | 1h10 | AKT1 | −5.1 |
Figure 8.
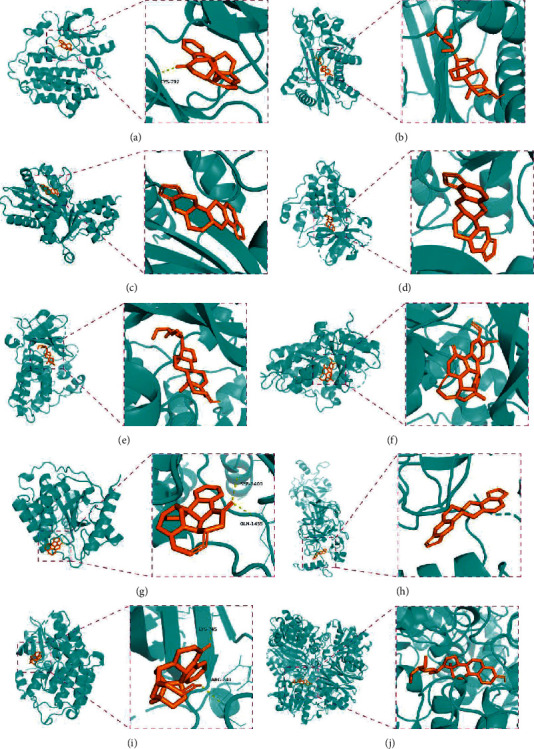
Molecular docking of hub genes with active compounds. (a) The binding poses of strychnine complexed with EGFR. (b) The binding poses of Stigmasterol complexed with EP300. (c) The binding poses of (S)-Stylopine complexed with EP300. (d) The binding poses of (S)-Stylopine complexed with EGFR. (e) The binding poses of Stigmasterol complexed with EGFR. (f) The binding poses of brucine-N-oxide complexed with EGFR. (g) The binding poses of strychnine complexed with EP300. (h) The binding poses of (S)-Stylopine complexed with TP53. (i) The binding poses of IsostrychnineN-oxide (II) complexed with EGFR. (j) The binding poses of Stigmasterol complexed with GAPDH.
3.8. MG Mouse Model Establishment
As shown in Figure 9, the body weight of the mice increased every week, and after 42 days, the body weight of the mice in the model group was significantly lower than that of the mice in the control group (P < 0.01). Clinical examinations were performed every 2 weeks. After 42 days, compared to the control group, the clinical scores were increased, the grip strengths were weakened, and the inverted screen times were shortened in the model group (P < 0.01).
Figure 9.
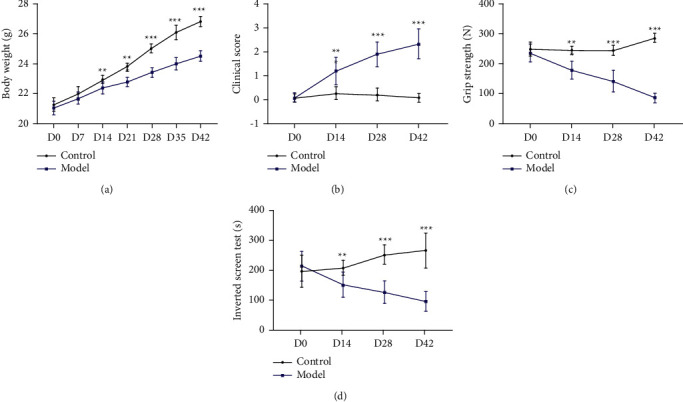
MG model constructs successful clinical examination indicators. (a) body weight; (b) clinical score; (c) grip strength; (d) inverted screen test.∗∗P < 0.01, and ∗∗∗P < 0.001vs. control group.
3.9. Nux Vomica Improves Clinical Examination Indexes of MG Model Mice
After treatment with Nux vomica, the clinical examination indicators of the MG model (weight, clinical score, grip strength, and inverted screen time) significantly improved. After 28 days of treatment, the weight of the MG mice increased significantly, clinical scores reduced, grip strength increased, and the inverted screen times were increased with the increased dose of Nux vomica (Figure 10).
Figure 10.
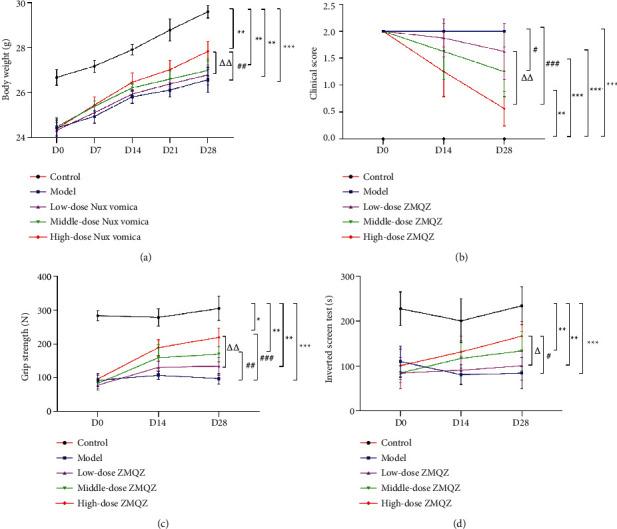
Nux vomica for MG clinical examination indicators. (a) Body weight; (b) clinical score; (c) grip strength; (d) inverted screen test. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001vs. control group; #P < 0.05, ##P < 0.01, and ### P < 0.001vs. model group; ΔP < 0.05, and ΔΔP < 0.01vs. low-dose Nux vomica group.
3.10. Expression of Hub Targets
To further verify the effect of Nux vomica against MG, we detected the expression of hub targets by qRT-PCR and western blotting. The results showed that compared to the model group, Nux vomica treatment significantly reduced EGFR and TP53 expression and increased MYC expression in a dose-dependent manner. Moreover, HSPA8 expression was not significantly changed, while AKT1 expression was significantly reduced, and the effect was particularly pronounced in the high-dose group. The qRT-PCR results also showed that the mRNA expression of RPS27, UBC, HSP90AA1, and EP300 was not significantly different between the groups (Figure 11).
Figure 11.
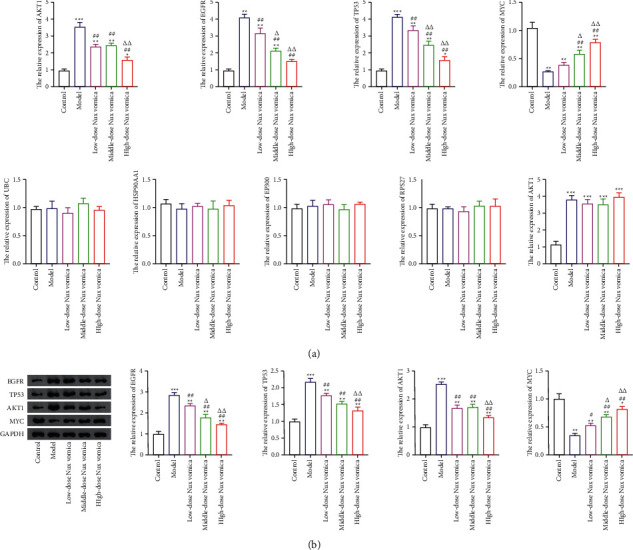
Hub targets expression. (a) The mRNA expression of EP300, EGFR, TP53, RPS27A, MYC, HSP90AA1, UBC, AKT1, GAPDH, and HSPA8 were detected by the qRT-PCR assay. (b) Western blot was used to detected the protein expression of MYC, EGFR, TP53, and AKT1 targets. ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001 vs. control group; #P < 0.05, and ##P < 0.01 vs. model group; ΔΔP < 0.01, and ΔP < 0.05 vs. low-dose Nux vomica group.
4. Discussion
MG is an autoimmune disease of the neuromuscular junction, which is mainly mediated by acetylcholine receptor (AChR) antibody and muscle-specific kinase (MuSK) [27, 28]. Presently, traditional therapies for long-term MG treatment have several adverse effects in some populations. Therefore, it is vital to explore new methods for treating MG. Owing to their high efficiency and few side effects, TCMs have been widely used in MG treatment as an alternative or adjuvant therapy. In addition, a previous meta-analysis indicated that compared to using Western medication alone, TCMs could function as adjuvant therapies to improve the efficacy and decrease the side effects and relapse rate in treating MG [29, 30]. As a TCM, Nux vomica is warm in nature, with a slightly bitter flavor due to its various pharmacological activities. However, the effects and mechanism of action of Nux vomica on MG treatment remain unclear. Therefore, our study systematically explored the underlying mechanism of Nux vomica treatment for MG based on network pharmacology and further validated the therapeutic effect of Nux vomica on MG in combination with in vivo experiments.
Based on the public databases, 12 bioactive compounds of Nux vomica were collected by setting OB ≥ 30% and DL ≥ 0.18 (Table 1). Studies have isolated many chemical compounds from Nux vomica, including alkaloids, glycosides, steroids, and organic acids [9]. Among them, alkaloids, including strychnine, brucine-N-oxide, isobrucine, vomicine, and brucine N-oxide, are the primary chemical components of Nux vomica and have been reported to have anticancer effects. Additionally, strychnine has been shown to exhibit significant anti-inflammatory activity [31], while brucine-N-oxide possesses a strong antinociceptive potential [32]. Moreover, stigmasterol can inhibit the activity of acetylcholinesterase (AChE) and has antioxidant, anti-inflammatory, and neuroprotective effects [33–35]. Thus, many compounds from Nux vomica possess favorable biological activity, including inhibiting AChE and immunomodulation and may exert a mitigative effect on MG.
MG occurrence is closely related to continuous viral infections, genetic factors, and abnormal immune responses. Growing evidence has demonstrated that EBV infection is correlated with the occurrence of MG [36, 37]. For example, EBV infection could affect TLR7/9-dependent innate immune responses, which enhance the inflammatory response, B-cell dysfunction, and autoimmunity in the thymus of patients with MG [36, 37]. Moreover, a recent pilot study showed that hepatitis E infection was relevant to MG, at least in China, and may contribute to MG by inducing replication in the thymus, bystander activation, and stimulation of antigen-specific T cells [38]. Target identification is a critical step for the mechanism investigation on Nux vomica treating MG. According to the PPI network of active compound targets, Nux vomica has the characteristics of multiple components and targets in the treatment of MG. Top 10 potential targets (EP300, EGFR, TP53, RPS27A, MYC, HSP90AA1, UBC, AKT1, GAPDH, HSPA8) of active compounds from Nux vomica were predicted to be associated with MG treatment. These 10 hub targets were validated in vivo, and results showed that Nux vomica treatment downregulated the expression of EGFR, TP53, and AKT1, while upregulated the MYC level in MG model mice. These findings indicated that EGFR, TP53, AKT1, and MYC may be the key targets for Nux vomica treating MG. In addition, the pathways of Nux vomica against MG were related to EBV infection, hepatitis B, hepatitis C, and other viral infections based on KEGG analysis of the core genes. We also identified the miRNA pathway in cancer. An increasing number of studies have suggested that miRNAs are involved in the occurrence and development of MG [39]. The miRNA profile of peripheral blood from MG patients was sequenced using a next-generation sequencing approach, and 41 miRNAs were found to be differentially expressed in nonresponders versus responders to immunosuppressive therapies. More importantly, miR-181d-5p, -323b-3p, -340-3p, -409-3p, and -485-3p can modulate the immune response and drug metabolism processes in patients with MG [40]. In another study, miR-612, miR-3651, and miR-3653 were found to be highly expressed in the peripheral blood mononuclear cells of patients with AChR-positive early-onset MG (AChR-EOMG), suggesting that these miRNAs may be involved in the pathogenesis of AChR-EOMG [41]. It was predicted that the active ingredients of Nux vomica may regulate the immune response by regulating the miRNA signaling pathway, thereby playing a role in the treatment of MG.
5. Conclusions
Our study systematically analyzed the potential effects of Nux vomica in the treatment of MG based on network pharmacology and validated its effects in combination with in vivo experiments. The findings showed that the bioactive compounds of Nux vomica exerted therapeutic effects on MG by acting on multiple targets and multiple pathways. Based on the network pharmacology, this study confirmed the therapeutic effects of Nux vomica on MG probably via downregulating the expression of EGFR, TP53, and AKT1 and upregulating MYC level. Our data may provide a valuable theoretical basis for future MG diagnosis and treatment and also serve as a reference for further mechanistic research on Nux vomica treating MG. However, we also found that the mechanisms of Nux vomica against are involved in the PI3K-Akt signaling pathway and miRNAs, so further experiments are needed to prove these predictions. Besides, the therapeutic efficacy of Nux vomica on MG is needed to be verified in a clinical trial.
Acknowledgments
This work was supported by the Medical and Health Research Projects of Zhejiang Province (number 2019RC058).
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Supplementary Materials
Table S1. The primer sequences for qRT-PCR.
References
- 1.Hehir M. K., Silvestri N. J. Generalized myasthenia gravis. Neurologic Clinics . 2018;36(2):253–260. doi: 10.1016/j.ncl.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Ciafaloni E. Myasthenia gravis and congenital myasthenic syndromes. Continuum: Lifelong Learning in Neurology . 2019;25(6):1767–1784. doi: 10.1212/con.0000000000000800. [DOI] [PubMed] [Google Scholar]
- 3.Mantegazza R., Antozzi C. From traditional to targeted immunotherapy in myasthenia gravis: prospects for research. Frontiers in Neurology . 2020;11:p. 981. doi: 10.3389/fneur.2020.00981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voinov V. Plasmapheresis in Treatment of Myasthenia Gravis . London, UK: IntecOpen; 2018. [Google Scholar]
- 5.Jiang X., Chen G., Huang J., et al. Modified Buzhong Yiqi decoction for myasthenia gravis. Medicine (Baltimore) . 2018;97(50) doi: 10.1097/md.0000000000013677.e13677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi G., Liu P., Gu S., Yang H., Dong H., Xue Y. Effect of Jianpiyiqi granule on ocular myasthenia gravis in children prepared with a formula from traditional Chinese medicine. Journal of Traditional Chinese Medicine . 2017;37:546–553. [PubMed] [Google Scholar]
- 7.Jiao W., Hu F., Li J., et al. Qiangji Jianli decoction promotes mitochondrial biogenesis in skeletal muscle of myasthenia gravis rats via AMPK/PGC-1α signaling pathway. Biomedicine & Pharmacotherapy . 2020;129 doi: 10.1016/j.biopha.2020.110482.110482 [DOI] [PubMed] [Google Scholar]
- 8.Zhu M., Liu Z., Gao M., et al. The effect of Bu Zhong Yi Qi decoction on simulated weightlessness-induced muscle atrophy and its mechanisms. Molecular Medicine Reports . 2017;16(4):5165–5174. doi: 10.3892/mmr.2017.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo R., Wang T., Zhou G., et al. Botany, phytochemistry, pharmacology and toxicity of strychnos nux-vomica L.: a review. The American Journal of Chinese Medicine . 2018;46:1–23. doi: 10.1142/s0192415x18500015. [DOI] [PubMed] [Google Scholar]
- 10.Jiang X. H., Chen Y., Ding Y. Y., Qiu H., Zhou D. Y., Qiu C. L. Effect of grilled nux vomica on differential RNA expression profile of gastrocnemius muscle and toll-like receptor 4 (tlr-4)/nuclear factor kappa b (nf-κb) signaling in experimental autoimmune myasthenia gravis rats. Medical Science Monitor . 2020;26 doi: 10.12659/msm.919150.e919150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopkins A. L. Network pharmacology: the next paradigm in drug discovery. Nature Chemical Biology . 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 12.Lee W. Y., Lee C. Y., Kim Y. S., Kim C. E. The methodological trends of traditional herbal medicine employing network pharmacology. Biomolecules . 2019;9 doi: 10.3390/biom9080362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu B., Song Z., Yu J., Li P., Tang Y., Ge J. The atherosclerosis-ameliorating effects and molecular mechanisms of BuYangHuanWu decoction. Biomedicine & Pharmacotherapy . 2020;123 doi: 10.1016/j.biopha.2019.109664.109664 [DOI] [PubMed] [Google Scholar]
- 14.Rappaport N., Twik M., Plaschkes I., et al. Malacards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Research . 2017;45:D877–D887. doi: 10.1093/nar/gkw1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piñero J., Bravo À., Queralt-Rosinach N., et al. Disgenet: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Research . 2017;45:D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J., Huang Q., Zhao R., Ma Z. A network pharmacology study on the Tripteryguim wilfordii hook for treatment of Crohn’s disease. BMC Complementary Medicine and Therapies . 2020;20(1):p. 95. doi: 10.1186/s12906-020-02885-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics . 2014;6(1):p. 13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin A., Ochagavia M. E., Rabasa L. C., Miranda J., Fernandez-de-Cossio J., Bringas R. BisoGenet: a new tool for gene network building, visualization and analysis. BMC Bioinformatics . 2010;11(1):p. 91. doi: 10.1186/1471-2105-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Y., Li M., Wang J., Pan Y., Wu F.-X. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems . 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Chin C. H., Chen S. H., Wu H. H., Ho C. W., Ko M. T., Lin C. Y. Cytohubba: identifying hub objects and sub-networks from complex interactome. BMC Systems Biology . 2014;8(4):p. S11. doi: 10.1186/1752-0509-8-s4-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sterling T., Irwin J. J. ZINC 15-ligand discovery for everyone. Journal of Chemical Information and Modeling . 2015;55(11):2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris G. M., Huey R., Lindstrom W., et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. Journal of Computational Chemistry . 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burley S. K., Berman H. M., Kleywegt G. J., Markley J. L., Nakamura H., Velankar S. Protein data bank (PDB): the single global macromolecular structure archive. Methods in Molecular Biology . 2017;1607:627–641. doi: 10.1007/978-1-4939-7000-1_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seeliger D., de Groot B. L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. Journal of Computer-Aided Molecular Design . 2010;24(5):417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidtke P., Le Guilloux V., Maupetit J., Tuffery P. fpocket: online tools for protein ensemble pocket detection and tracking. Nucleic Acids Research . 2010;38:W582–W589. doi: 10.1093/nar/gkq383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trott O., Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry . 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilhus N. E., Verschuuren J. J. Myasthenia gravis: subgroup classification and therapeutic strategies. The Lancet Neurology . 2015;14(10):1023–1036. doi: 10.1016/s1474-4422(15)00145-3. [DOI] [PubMed] [Google Scholar]
- 28.Sanders D. B., Wolfe G. I., Benatar M., et al. International consensus guidance for management of myasthenia gravis. Neurology . 2016;87(4):419–425. doi: 10.1212/wnl.0000000000002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S., Xu M.-B., Zhou X.-L., Rong P.-Q., Jin T.-Y., Zheng G.-Q. Chinese herbal medicine for myasthenia gravis: a systematic review and meta-analysis. Frontiers in Pharmacology . 2018;9:p. 969. doi: 10.3389/fphar.2018.00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie R., Liu L., Wang R., Huang C. Traditional Chinese medicine for myasthenia gravis. Medicine (Baltimore) . 2020;99(29) doi: 10.1097/md.0000000000021294.e21294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J., Wang X., Qu Y.-G., et al. Analgesic and anti-inflammatory activity and pharmacokinetics of alkaloids from seeds of Strychnos nux-vomica after transdermal administration: effect of changes in alkaloid composition. Journal of Ethnopharmacology . 2012;139(1):181–188. doi: 10.1016/j.jep.2011.10.038. [DOI] [PubMed] [Google Scholar]
- 32.Wu Y., Wang T.-S., Yin F.-Z., Cai B.-C. Analgesic and anti-inflammatory properties of brucine and brucine n-oxide extracted from seeds of Strychnos nux-vomica. Journal of Ethnopharmacology . 2003;88:205–214. doi: 10.1016/s0378-8741(03)00224-1. [DOI] [PubMed] [Google Scholar]
- 33.Panda S., Jafri M., Kar A., Meheta B. K. Thyroid inhibitory, antiperoxidative and hypoglycemic effects of stigmasterol isolated from butea monosperma. Fitoterapia . 2009;80(2):123–126. doi: 10.1016/j.fitote.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Walker C. I. B., Oliveira S. M., Tonello R., et al. Anti-nociceptive effect of stigmasterol in mouse models of acute and chronic pain. Naunyn-Schmiedeberg’s Archives of Pharmacology . 2017;390(11):1163–1172. doi: 10.1007/s00210-017-1416-x. [DOI] [PubMed] [Google Scholar]
- 35.Sun J., Li X., Liu J., Pan X., Zhao Q. Stigmasterol exerts neuro-protective effect against ischemic/reperfusion injury through reduction of oxidative stress and inactivation of autophagy. Neuropsychiatric Disease and Treatment . 2019;15:2991–3001. doi: 10.2147/ndt.s220224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavalcante P., Galbardi B., Franzi S., et al. Increased expression of Toll-like receptors 7 and 9 in myasthenia gravis thymus characterized by active Epstein–Barr virus infection. Immunobiology . 2016;221(4):516–527. doi: 10.1016/j.imbio.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Cavalcante P., Barzago C., Baggi F., et al. Toll-like receptors 7 and 9 in myasthenia gravis thymus: amplifiers of autoimmunity? Annals of the New York Academy of Sciences . 2018;1413(1):11–24. doi: 10.1111/nyas.13534. [DOI] [PubMed] [Google Scholar]
- 38.Wang L., Gao F., Lin G., et al. Association of hepatitis e virus infection and myasthenia gravis: a pilot study. Journal of Hepatology . 2018;68(6):1318–1320. doi: 10.1016/j.jhep.2018.01.040. [DOI] [PubMed] [Google Scholar]
- 39.Wang L., Zhang L. Emerging roles of dysregulated micrornas in myasthenia gravis. Frontiers in Neuroscience . 2020;14:p. 507. doi: 10.3389/fnins.2020.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cavalcante P., Mizrachi T., Barzago C., et al. Microrna signature associated with treatment response in myasthenia gravis: a2q further step towards precision medicine. Pharmacological Research . 2019;148:p. 104388. doi: 10.1016/j.phrs.2019.104388.104388 [DOI] [PubMed] [Google Scholar]
- 41.Barzago C., Lum J., Cavalcante P., et al. A novel infection- and inflammation-associated molecular signature in peripheral blood of myasthenia gravis patients. Immunobiology . 2016;221(11):1227–1236. doi: 10.1016/j.imbio.2016.06.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The primer sequences for qRT-PCR.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


