Abstract
We report the characterization and cloning of the genes for an unusual type IV restriction–modification system, BspLU11III, from Bacillus sp. LU11. The system consists of two methyltransferases and one endonuclease, which also possesses methyltransferase activity. The three genes of the restriction–modification system, bsplu11IIIMa, bsplu11IIIMb and bsplu11IIIR, are closely linked and tandemly arranged. The corresponding enzymes recognize the dsDNA sequence 5′-GGGAC-3′/5′-GTCCC-3′, with M.BspLU11IIIa modifying the A (underlined) of one strand and M.BspLU11IIIb the inner C (underlined) of the other strand. R.BspLU11III has both endonuclease and adenine-specific methyltransferase activities and is able to protect the DNA against cleavage by itself. In contrast to all type IV restriction–modification systems described so far, which have only one adenine-specific methyltransferase, BspLU11III is the first type IV restriction–modification system that includes two methyltransferases, one of them being cytosine specific.
INTRODUCTION
Restriction–modification (R-M) systems are traditionally divided into three major classes: types I, II and III. However, not all of the R-M enzymes discovered at this time fit in these classes, hence, an additional class (type IV) has been proposed (1). There are at least six restriction enzymes that belong to type IV R-M systems: R.Eco57I (1), R.Bce83I (2), R.HaeIV (3), R.MmeI (4), R.BspLU11III (5) and BseMII (6). The common feature of these enzymes is that two enzymatic activities, methyltransferase (MTase) and endonuclease (ENase), are combined together in one polypeptide chain and the ENase activity is positively affected by S-adenosine-l-methionine (AdoMet) but ATP has no influence on activity of the enzymes. For all of these enzymes it has been shown that the associated MTase activity is m6A specific. The primary structures of R.Eco57I, R.BseMII and R.HaeIV have been determined (4,6,7). As for their recognition pattern, type IV enzymes usually recognize asymmetrical DNA sequences and ENase cleavage occurs at a defined distance from the recognition site, as in type IIS enzymes. An exception is R.HaeIV, where cleavage occurs on both sides of the recognition site. The suggestion was made that type IV enzymes are intermediate between type IIS and type III enzymes (1).
Screening for new thermostable R-M enzymes among thermophilic Bacillus strains identified a strain, Bacillus sp. LU11, which harbors at least three ENases: R.BspLU11I, R.BspLU11II (8) and R.BspLU11III (5). R.BspLU11I is an enzyme with a new sequence specificity, 5′-A↓CATGT-3′, and recently its isoschizomer R.PciI was found. R.BspLU11II is a thermostable isoschizomer of R.XbaI. R.BspLU11III recognizes the asymmetrical DNA sequence 5′-GGGAC-3′ and cuts the DNA at a distance of 14 bp from the recognition site, like IIS type ENases. However, unlike the IIS ENases, R.BspLU11III possesses m6A MTase activity resulting in methylation of the A residue in the upper strand of the recognition sequence. The ENase activity of R.BspLU11III is positively affected in vitro by low concentrations of AdoMet. These properties fit the definition of a type IV enzyme (5).
Here we describe cloning of the genes, primary structure analysis and characterization of the two MTases M.BspLU11IIIa and M.BspLU11IIIb and of the ENase R.BspLU11III from Bacillus sp. LU11.
MATERIALS AND METHODS
Bacterial strains and plasmids
Bacillus sp. LU11 was obtained from a soil isolate (8). Escherichia coli strains SURE [e14–(McrA–) Δ(mcrCB-hsdSMR-mrr)171 endA1 supE44 thi-1 gyrA96 relA1lac recB recJ sbcC umuC::Tn5 (Kanr) uvrC] and INVαF′ [F′ endA1 recA1 hsdR17(rk–, mk+) supE44 thi-1 gyrA96 relA1 φ80lacZΔM15 Δ(lacZYA-argF)U169] were used to generate the Bacillus sp. LU11 genomic DNA library and as hosts for subcloning procedures. Escherichia coli strain BL21(DE3) [B F– dcm ompT hsdS(rB– mB–) gal λ(DE3)] was used for protein expression. Bacillus subtilis strain YB886M was used for R.BspLU11III expression. For the determination of DNA MTase specificity plasmid pJRD184 (9), pUC18A5/A6 and pUC19A5/A6 (see below) DNAs and phage λ DNA were used. pET28b and Bacillus-specific shuttle vector pHP13 (10) were used for recombinant protein expression experiments.
DNA preparation and manipulation
Bacillus sp. LU11 genomic DNA was extracted and purified using a protein ‘salting out’ procedure (11). For competent E.coli cell preparation and transformation the protocol of Inoue et al. (12) was used. Bacillus subtilis YB886M competent cells were prepared and transformed according to the protocols described in Bott and Wilson (13).
For plasmid pUC18A5/A6 and pUC19A5/A6 construction oligonucleotides A5 (5′-CTAGGGGACGCGTG-3′) and A6 (5′-CTAGCACGCGTCCC-3′) were annealed to each other and the oligonucleotide duplex was ligated with plasmid pUC18 or pUC19 pre-cut with XbaI.
All restriction enzymes, T4 DNA ligase and DNA molecular weight markers were purchased from MBI Fermentas (Lithuania) or from New England Biolabs (USA).
DNA methylation assay
The ENase protection test was performed using phage λ DNA as substrate in MEB4 buffer (20 mM Tris–acetate pH 8.0, 0.1 mM EDTA, 50 mM KCH3COO) containing 100 µM AdoMet. After addition of DNA MTase, the reaction mixture was incubated at 50°C for 2 h and, after methylation, Mg(CH3COO)2 was added to a final concentration of 10 mM and incubation was continued in the presence of the restriction ENase R.BspLU11III at 50°C for 2 h.
The incorporation of radiolabeled methyl groups into DNA was performed in MEB4 buffer containing 1.5 µM [3H]AdoMet (15 Ci/mmol; Amersham, UK). Phage λ DNA and plasmid pJRD184 DNA were used as substrates. After methylation, the DNA was bound to glass filters, washed with 5% trichloroacetic acid and the radioactivity remaining on the filters was counted in a Beckman LS9800 scintillation counter (Beckman, USA).
Cloning of the genomic DNA fragment containing the m5C-specific MTase gene bsplu11IIIMb
Amplification of the bsplu11IIIMb gene fragment with degenerate primers 39 (5′-CGAATTCCARGGNTTYCCNGA-3′) and 40 (5′-GGGATCCGGNGGNACNGCRTTNCC-3′) was performed using a Mastercycler gradient PCR amplifier (Eppendorf, Germany) with 20 µl of a reaction mix containing 10 mM Tris–HCl pH 8.8, 50 mM KCl, 0.8% Nonidet P-40, 1.75 mM MgCl2, 100 ng Bacillus sp. LU11 genomic DNA, 3 µM each primer, 200 µM each dNTP and 1 U Taq DNA polymerase. The cycling conditions were as follows: an initial denaturation step of 5 min at 95°C, followed by 30 cycles of 30 s denaturation at 95°C, 30 s annealing over a temperature gradient of 42–62°C and 1 min elongation at 72°C. The last cycle was followed by extension for 7 min at 72°C. PCR amplification with degenerate primer 257 (5′-CCGAATTCAACGGYGGYCCYCCYTGYCA-3′) and specific primer LU1 (5′-CACTAGTATGCTTTCCTC-3′) was performed using the same conditions with the exception that the LU1 primer concentration was 0.3 µM. The PCR products were separated on an agarose gel and the PCR product of ∼800 bp in length was isolated and cloned into TA cloning vector pCR2.1 (Invitrogen, USA) and sequenced.
To construct the genomic DNA library, ClaI fragments of Bacillus sp. LU11 genomic DNA were ligated with ClaI pre-cut and dephosphorylated pBluescriptSK vector and the ligation mixture used to transform E.coli SURE competent cells. The transformed cells were plated on LB–agar plates containing 100 µg/ml ampicillin.
The colony hybridization procedure was performed as described (14). The 32P-labeled 780 bp long LU1 (5′-CACTAGTATGCTTTCCTC-3′)–LU3 (5′-AAGTGATGATGAGAGAGG-3′) PCR product was used as a probe for hybridization, which was carried out overnight at 55°C in a Biometra OV2 hybridization oven (Biometra, Germany) according to the protocol of Church and Gilbert (15).
Cloning of the ENase gene bspLU11IIIR
The flanking regions of the cloned ClaI 5.2 kb DNA fragment were amplified and cloned using an inverse PCR approach. Bacillus sp. LU11 genomic DNA was digested with EcoRI and the fragments obtained were circularized by self-ligation in a series of dilutions. The resulting ligation products were used as a template for PCR amplification with primers LU3 (5′-AAGTGATGATGAGAGAGG-3′) and LU10 (5′-CTTGTGCGTGCTCAGATC-3′). The first round PCR was followed by a second round PCR with nested primers LU6 (5′-AAGTCTTCAACAAAAAGAGG-3′) and LU9 (5′-GATCGTGCCATAGCTGTG-3′). The resulting PCR product from the second round PCR was cloned into TA cloning vector pCR2.1.
DNA sequencing
The cloned 5.2 kb DNA fragment of Bacillus sp. LU11 genomic DNA and the LU6–LU9 inverse PCR product were subcloned using ClaI, XbaI, EcoRV, SpeI and XhoI restriction sites which mapped within the fragment. The restriction fragments obtained were cloned into vector pBluescriptSK or pUC19. M13 universal (forward, reverse) sequencing primers or fragment-specific primers LU4 (5′-CTCTCTCATCATCACTTC-3′), LU6 (5′-AAGTCTTCAACAAAAAGAGG-3′), LU14 (5′-CTAATGAATCCACCATATG-3′) and LU18 (5′-AGTTAACGAGAGATATCCTG-3′) were used in an ABI-PRISM Ready Reaction DyeDeoxyterminator Cycle sequencing reaction (Applied BioSystems, USA). The sequencing reaction products were loaded into an Automated DNA Sequencer (Applied BioSystems, USA). The Wisconsin v.10.0 Unix software package (Genetics Computer Group, USA) and BLAST program (16) were used to compile and analyze the sequencing data.
Expression of the bsplu11IIIMa and bsplu11IIIMb genes in E.coli and protein purification
The bsplu11IIIMa gene was PCR amplified using primers LU7 (5′-GGAGGATCCAATGATGAAATTATCTAATC-3′) and LU11 (5′-GTTTCAAGCTTCCATACCTC-3′). The bsplu11IIIMb gene was amplified using primers LU5 (5′-CATGGATCCTTTGAAACCTACTGTAG-3′) and LU8 (5′-TTACAAGCTTCAACATAATATTG-3′). Plasmid pLUClaI was used as the DNA template. To reduce possible sequence errors a Taq/Pwo DNA polymerase mixture (15:1 U) was used. PCR products were gel purified, digested with BamHI and HindIII and then ligated with BamHI + HindIII-cut pET28b vector (Novagen, USA). The resulting plasmids, pET28bORF2 and pET28bORF3, were verified by restriction analysis and the region covering the vector–insert junction was sequenced. The recombinant plasmids pET28bORF2 and pET28bORF3 were used for transformation of E.coli strain BL21(DE3). Immediately after transformation cells were grown in 250 ml of TY medium in the presence of kanamycin (50 µg/ml) at 37°C. When the OD600 reached 0.8, IPTG was added to a final concentration of 0.3 mM and the cell culture was grown on for an additional 3 h at 37°C. Cells were harvested by centrifugation and disrupted using a French pressure cell (Aminco, USA). The recombinant proteins from cleared lysates were purified in two chromatographic steps: on Ni–NTA agarose (Qiagen, Germany) according to the manufacturer’s protocol, and then on heparin–Sepharose (Pharmacia, Sweden) using gradient elution with an increasing concentration of NaCl from 50 mM to 1 M. Elution fractions containing the protein of interest were pooled together and dialyzed against the storage buffer (20 mM Tris–HCl pH 7.5, 50 mM KCl, 1 mM DTT, 50% glycerol).
Expression of the bsplu11IIIR gene in B.subtilis and protein purification
The 4736 bp long BamHI–ClaI fragment from plasmid pLUClaI was ligated with the 1970 bp long ClaI–EcoRI fragment from the LU6–LU9 inverse PCR product (see Fig. 4) and the construct inserted into the pHP13 cloning vector pre-cut with BamHI and EcoRI. The resulting plasmid pHP13LUIII was used to transform B.subtilis strain YB886M cells. The transformed cells were plated on LB agar containing chloramphenicol (15 µg/ml) and a single colony was taken to inoculate 1 l of TY medium. Cells were grown in liquid culture until the OD600 reached 1.0. Then the cells were harvested by centrifugation and disrupted using a French pressure cell. R.BspLU11III from cleared lysate was purified by two step liquid chromatography on a S-Sepharose Fast Flow column (Pharmacia, Sweden) and on a heparin–Sepharose column using gradient elution, with an increasing concentration of NaCl from 50 mM to 1 M for both chromatography steps. Elution fractions from the heparin–Sepharose column containing ENase R.BspLU11III activity were pooled and dialyzed against storage buffer.
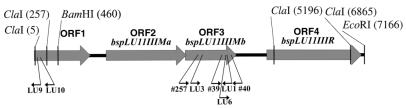
Figure 4.
Schematic map of the cloned region of Bacillus sp. LU11 genomic DNA. The cloned 5196 bp ClaI fragment and part of the inverse PCR product are combined together. The arrows indicate open reading frames (ORFs) which were found in the cloned region. Designations for genetic markers are: bspLU11IIIMa, gene encoding M.BspLU11IIIa m6A MTase; bspLU11IIIMb, gene encoding M.BspLU11IIIb m5C MTase; bspLU11IIIR, gene encoding R.BspLU11III ENase. Annealing sites for oligonucleotides 39, 40, 257, LU1, LU3, LU6, LU9 and LU10 are shown by small arrows.
Determination of the DNA recognition sequence and nature and position of methylated bases
Aliquots of 10 µg plasmid pJRD184 DNA were methylated with native M.BspLU11III, recombinant M.BspLU11IIIa or recombinant M.BspLU11IIIb MTase in buffer MEB4 supplemented with 1.5 µM [3H]AdoMet. Methylation was carried out at 50°C for 4 h, followed by ethanol precipitation of methylated DNA. Then the DNA was dissolved in H2O and cleaved with HpaII ENase in the recommended buffer. The restriction products obtained were separated on a 10% acrylamide gel, which was stained with ethidium bromide and photographed under UV light. Afterwards, the gel was soaked in Amplify scintillation liquid (Amersham, UK), dried and autoradiographed with Kodak BioMax X-Ray film (Kodak, UK) for 1 week at –70°C.
Thin layer chromatography was used to determine the methylated base within the recognition site. Aliquots of 25 µg phage λ DNA were methylated with M.BspLU11III, M.PvuII, M.EcoRII or M.EcoRI in 200 µl of MEB4 buffer in the presence of 1.5 µM [3H]AdoMet for 4 h at the appropriate temperature: 37°C for M.PvuII, M.EcoRII and M.EcoRI; 50°C for M.BspLU11III. MTases with known base specificities were taken as controls: M.EcoRII (m5C), M.EcoRI (m6A) and M.PvuII (m4C). Methylated DNA was enzymatically degraded to mononucleosides according to Gehrke et al. (17). The mononucleoside mixture obtained was lyophilized and dissolved in 50 µl of bidistilled water and applied to a Fixion 50 × 8 thin layer cation exchange chromatography plate (Chinoin, Hungary). Separation of mononucleosides was performed in butanol:H2O:NH4OH (60:10:0.1 v/v/v) (18). After separation plates were dried, treated with EN3HANCE spray (New England Nuclear, USA) and autoradiographed with Kodak BioMax X-Ray film at –70°C for 2 weeks.
Determination of the position of the m5C residue within the recognition site
Plasmid pUC18A5/A6 or pUC19A5/A6 DNA was methylated with either native M.BspLU11III or recombinant M.BspLU11IIIb MTase in buffer MEB4 supplemented with 100 µM AdoMet. To eliminate non-methylated DNA molecules, methylated plasmids were digested with R.BspLU11III. An aliquot of 100 ng methylated DNA was used for bisulfite/alkaline treatment, which was done according to Olek et al. (19). PCR amplification after bisulfite treatment was done using primers AC1 (5′-ATAATAATTCTAGATAGGAAATAGTTATGATT-3′) and AC2 (5′-AATCACAAGCTTATAAAACAACAACCAA-3′). The PCR product was cloned into TA cloning vector pCR2.1 and sequenced.
RESULTS
Isolation and biochemical characterization of a new m5C MTase from Bacillus sp. LU11
Methylation by R.BspLU11III of the only A in the upper strand of the recognition sequence 5′-GGGAC-3′/5′-GTCCC-3′ renders the site resistant to the restriction activity of this enzyme. During semi-conservative DNA replication, however, one of the replicated DNA molecules would become unmethylated and therefore sensitive to restriction, unless another MTase that could methylate the lower strand was available. Since there is no A residue in the lower strand of the recognition sequence, such an enzyme must be a C-specific MTase. To find such an activity we fractionated the proteins isolated from Bacillus sp. LU11 using chromatography on Sephadex G100, Blue Sepharose, HTP Biogel and heparin–agarose. After each chromatographic step aliquots from each fraction were tested for the presence of MTase activity protecting against R.BspLU11III restriction. As a result, a new MTase activity (M.BspLU11III) was detected after a second purification step on a Blue agarose column which was separable from the m6A-specific MTase activity of R.BspLU11III (data not shown). Additional purification steps on HTP Biogel and heparin–agarose allowed purification of the active protein without detectable contamination with other DNA MTases or ENases.
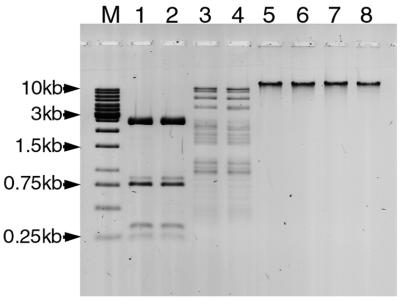
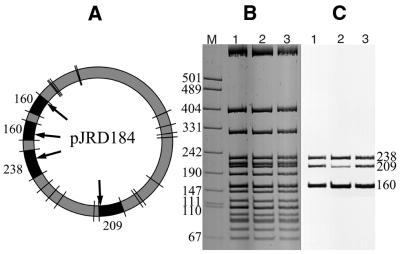
To determine whether M.BspLU11III recognizes the same DNA target 5′-GGGAC-3′/5′-GTCCC-3′ as does R.BspLU11III, two different assays were performed. In the first assay the ability of the MTase to protect phage λ DNA against restriction by R.BspLU11III was tested. M.BspLU11III completely protects phage λ DNA against digestion by R.BspLU11III (Fig. 1, lane 5). In another assay plasmid pJRD184 DNA containing four sites for R.BspLU11III was methylated by M.BspLU11III in the presence of [3H]AdoMet and then digested with HpaII. The cleavage of pJRD184 DNA by HpaII results in 24 DNA fragments ranging in size from 9 to 864 bp (Fig. 2). Four fragments of 238, 209, 160 and 160 bp contain the BspLU11III target sequence 5′-GGGAC-3′. Restriction fragments were separated on an acrylamide gel and fragments containing [3H]m5C residues were identified by autoradiography. Figure 2 shows that only the four fragments containing the sequence 5′-GGGAC-3′ were labeled by M.BspLU11III.
Figure 1.
Digestion and methylation assays on phage λ and plasmid pJRD184 DNA. Lane M, 1 kb DNA size marker ladder; lane 1, digestion of λ DNA with native R.BspLU11III; lane 2, digestion of λ DNA with recombinant R.BspLU11III encoded by ORF4; lane 3, digestion of pJRD184 with native R.BspLU11III; lane 4, digestion of pJRD184 with recombinant R.BspLU11III; lanes 5–8, protection of λ DNA against digestion with R.BspLU11III by methylation with native M.BspLU11III (lane 5), recombinant M.BspLU11IIIa (lane 6), recombinant M.BspLU11IIIb (lane 7) and recombinant R.BspLU11III in the absence of magnesium ions (lane 8).
Figure 2.
Sequence specificity determination of M.BspLU11III, M.BspLU11IIIa and M.BspLU11IIIb on pJRD184 plasmid DNA digested with HpaII. (A) Schematic map of plasmid pJRD184 with HpaII cut site locations. Fragments containing BspLU11III sites (shown by black arrows) are filled in black. (B) Ethidium bromide stained gel. (C) Autoradiograph of the gel showing HpaII-digested pJRD184 DNA methylated with M.BspLU11III (lane 1), M.BspLU11IIIa (lane 2) and M.BspLU11IIIb (lane 3).
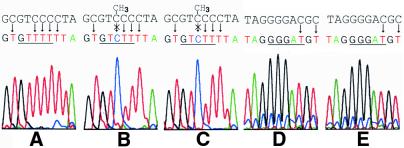
The base specificity of M.BspLU11III was determined by cation exchange thin layer chromatography. According to these data the enzyme methylates a C residue resulting in formation of m5C (data not shown). To determine the position of the methylated C within the recognition sequence we performed bisulfite mapping of m5C residues in DNA methylated by M.BspLU11III. The assay showed that the inner C residue (underlined) in DNA sequence 5′-GTCCC-3′ (Fig. 3) is modified and the only C residue in the complementary strand 5′-GGGAC-3′ is not methylated (Fig. 3).
Figure 3.
Determination of the position of the methylated cytosine residue. Shown are sequences and automated sequencing graphs of both strands of BspLU11III site 5′-GTCCC-3′/5′-GGGAC-3′ (underlined) after conversion of non-methylated cytosine residues to thymine residues by bisulfite/alkaline treatment. The upper string shows original sequences before bisulfite/alkaline treatment. Conversion of upper strand 5′-GTCCC-3′: non-methylated BspLU11III site (A) or methylated by native M.BspLU11III (B) or by recombinant M.BspLU11IIIb (C). Conversion of lower strand 5′-GGGAC-3′: BspLU11III site methylated by native M.BspLU11III (D) or by recombinant M.BspLU11IIIb (E).
Cloning of genes encoding the BspLU11III R-M system enzymes
The search for the gene encoding M.BspLU11III was based on the high similarity amongst C5 MTases. A comparison of the amino acid sequences of prokaryotic and eukaryotic m5C MTases identified at least 10 highly conserved motifs (I–X), which are arranged in canonical order (20,21). PCR technology with appropriate primers corresponding to the DNA coding some of these motifs was used to amplify segments of the m5C MTase gene. Here motifs IV, IX and X were selected to design specific degenerated oligonucleotides for PCR amplification of the corresponding parts of MTase genes from genomic DNA. At first we performed PCR amplification with degenerate oligonucleotides 39 and 40, which correspond to motifs IX (QGFPD) and X (GNAVPP), respectively. A 90 bp PCR product was extracted from an agarose gel, cloned into a TA vector and sequenced. Seven of eight positive clones contained a 93 bp insertion whose deduced amino acid sequence had substantial similarity to known m5C MTases. As a second step, PCR with primer LU1 specific for the 93 bp insertion was used in combination with degenerate primer 257, specific for the conserved motif IV (GGPPCQ). The resulting 830 bp product was cloned and sequenced. The 830 bp insertion contained an open reading frame corresponding to an amino acid sequence exhibiting high similarity to m5C MTases. Conserved m5C MTase motifs V–IX were present in the sequence in canonical order, which indicated that the cloned DNA fragment is part of a new m5C MTase gene. We then used primer LU3, specific for the amplified 830 bp fragment of the MTase, in combination with primer LU1 to generate a highly specific DNA probe of 780 bp for the given m5C MTase gene (Fig. 4).
This 32P-labeled 780 bp LU1–LU3 PCR product was used to probe the ClaI genomic DNA library of Bacillus sp. LU11 (Fig. 4). Three positive clones were found and the presence of the anticipated ClaI insert was verified by colony PCR using primers (LU1–LU3) specific for the putative m5C MTase gene. Plasmid pLUClaI with a 5.2 kb insert was isolated from one of the positive clones. To sequence the 5.2 kb insert in pLUClaI, the insert was subcloned using XbaI, EcoRV and SpeI restriction sites which mapped within the insert. Both strands of each subcloned fragment were sequenced and the resulting sequences were assembled using the GCG program package. Some parts of pLUClaI were sequenced using fragment-specific oligonucleotides. The exact size of the insert of pLUClaI, according to sequencing data, is 5196 bp. Two complete open reading frames, ORF2 and ORF3, and two incomplete open reading frames, ORF1 and ORF4, were found in the 5196 bp insert (Fig. 4). ORF1 starts outside and ends within the cloned fragment; ORF4 starts within and ends outside the cloned fragment.
The deduced amino acid sequence encoded by the 3′-part of ORF1 is 366 amino acids long and, according to BLAST analysis, has significant similarity to bacterial transposases (16).
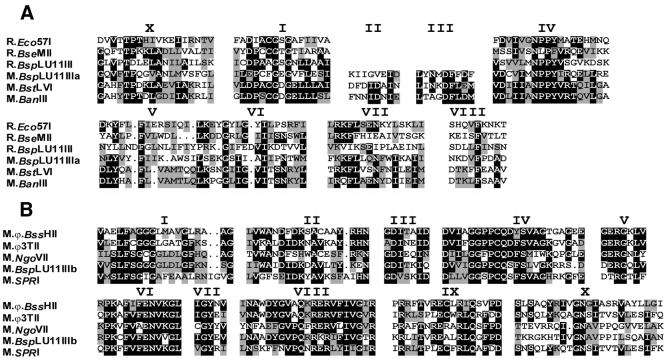
The deduced amino acid sequence encoded by ORF2, corresponding to the gene encoding M.BspLU11IIIa, is 569 amino acids long (66.5 kDa) and starts from the canonical ATG codon at position 1553 of the 5196 kb insert. DNA sequence upstream of this ATG contains pronounced –35 and –10 consensus DNA sequences and a potential ribosome-binding site (RBS) at position –9. Within the deduced amino acid sequence of the protein encoded by ORF2 nine conserved motifs characteristic for group γ m6A MTases could be identified (Fig. 5A) (22). A search in the GenBank database for similar proteins using the TBLAST algorithm (16) with the ORF2-encoded protein sequence showed that the protein has high similarity to type II m6A MTases M.BstLVI from Bacillus stearothermophilus LV (23) and M.BanIII from Bacillus aneurinolyticus (24) and other m6A MTases. The highest sequence similarity was limited to the nine conserved motifs (Fig. 5A).
Figure 5.
Amino acid sequence alignment of M.BspLU11IIIa and R.BspLU11III with known m6A-specific (A) and M.BspLU11IIIb with m5C-specific (B) MTases.
The translational start of ORF3, corresponding to the gene encoding M.BspLU11IIIb, appears to be a TTG codon at position 3259. The TTG codon corresponding to leucine can function as a translational start codon in Bacillus (25). –35, –10 and RBS DNA sequences could be found preceding the TTG codon. ORF3 encodes a protein that is 366 amino acids long with a molecular weight of 41 kDa. The initially cloned 830 bp fragment obtained by PCR with degenerate primers specific for conserved motifs of m5C MTases was a part of ORF3. Analysis of the deduced amino acid sequence of the protein encoded by ORF3 shows that the protein has the primary structure characteristic for prokaryotic m5C MTases. The 10 highly conserved motifs described for prokaryotic MTases can be identified (Fig. 5B) (20,21). TBLAST analysis of the ORF3-encoded protein showed that it exhibits very high similarity scores to prokaryotic type II m5C MTases M.NgoVII from Neisseria gonorrhoeae (26), bacillar phage encoded M.SPRI (27), M.ϕ3T (28) and M.BssHII (29). A potential target recognition domain (TRD) has been found between motifs VIII and XI. Amino acid sequence analysis of the TRD revealed homology to the TRDs of other m5C-specific MTases M.SinI, M.Eco47II, M.HgiDII and M.NgoVII (Fig. 6) (30).
Figure 6.
Amino acid sequence alignment of the M.BspLU11IIIb potential TRD region with those of M.SinI, M.Eco47I, M.HgiDII and M.NgoVII and their DNA recognition sequences.
Genes coding for R-M enzymes are usually linked to each other (31). Therefore, we proposed that the beginning of ORF4 found in the cloned ClaI fragment was the 5′-part of the ENase gene. In order to obtain the entire cloned ENase gene we applied the inverse PCR approach, which allows amplification of the unknown sequence flanking the known sequence (32). EcoRI fragments of Bacillus sp. LU11 genomic DNA were self-ligated in conditions favorable for circularization of the fragments. Two successive rounds of PCR, first with primers LU3 and LU10 and then a second round with nested primers LU6 and LU9, yielded a 9 kb long LU6–LU9 PCR product. Following digestion of the inverse PCR product with ClaI and EcoRI, we isolated the 1970 bp ClaI–EcoRI fragment, which covers the rest of the potential ENase gene. The fragment was subcloned using XhoI, ClaI and EcoRI restriction sites and sequenced. The sequences of both the ClaI fragment (5196 bp) and ClaI–EcoRI fragment (1970 bp) of the inverse PCR product were joined (Fig. 4) and the combined sequence was submitted to the GenBank database (accession no. AF169965).
ORF4, which begins in the ClaI 5196 bp fragment, continues in the combined sequence. This ORF is 2045 bp long and encodes a protein with a size of 681 amino acids and of predicted molecular weight 77 kDa. According to the computational analysis, ORF4 encodes a protein containing conserved motifs characteristic for group γ m6A MTases. These motifs are localized in the N-terminal part of the protein. Since R.BspLU11III has both ENase and MTase activities, we suggest that ORF4 corresponds to the gene encoding R.BspLU11III.
Expression and purification of the ORF2 and ORF3 encoded proteins
To construct vectors for inducible expression of both genes, ORF2 and ORF3 were cloned separately into vector pET28b under control of the T7 promoter, resulting in plasmids pET28bORF2 and pET28bORF3, respectively. To facilitate purification of the expressed proteins on Ni–NTA agarose, a DNA sequence encoding a 6×His tag was added in-frame to the 5′-ends of both ORFs. Escherichia coli strain BL21(DE3) lysogenic for phage λ with an IPTG-inducible phage T7 RNA polymerase gene was chosen as the host strain (33). Cells transformed with either pET28bORF2 or pET28bORF3 were grown until the cell density reached an OD600 of 0.8. After induction with IPTG, cells were grown for an additional 4 h. Cells expressing M.BspLU11IIIa reached an OD600 of 1.7 during this time. M.BspLU11IIIb-expressing cells grew only to an OD600 of 1.0. After this time point the cell density dropped because of extensive cell lysis.
To purify the expressed proteins, Ni–NTA agarose was used for affinity chromatography, followed by an additional purification step on heparin–Sepharose. The purity of the final enzyme preparations was ∼70% for M.BspLU11IIIa and 90% for M.BspLU11IIIb (data not shown.). No nuclease activity was detected in the final enzyme preparations.
Expression and purification of ORF4 encoded protein
Attempts to express the ORF4-encoded protein in E.coli were unsuccessful. To achieve expression of the putative ENase gene driven by its own promoter, the whole locus, bearing genes for both the MTase genes and the putative ENase gene (BamHI–EcoRI 460–7166 fragment, Fig. 4) was inserted into cloning vector pHP13, which is maintainable in both E.coli and B.subtilis. The resulting plasmid pHP13LUIII was used to transform B.subtilis YB886M. Transformed bacteria were checked for the presence of ENase activity in the lysate. ENase activity was detected in the lysates of cells bearing the plasmid pHP13LUIII, but not in those bearing the original plasmid pHP13. The expressed ENase was isolated from the cleared cell lysate by two-step column chromatography: the first step on Sepharose S Fast Flow, followed by a second chromatography step on heparin–Sepharose. One liter of liquid culture (OD600 = 1.0) of B.subtilis YB886M transformed with pHP13LUIII yielded ∼10 000 U ENase activity.
Enzymatic properties of the cloned MTases
The purified MTases were subjected to in vitro specificity tests. Both MTases fully protect phage λ and plasmid pJRD184 DNA against restriction with R.BspLU11III (Fig. 1, lanes 6 and 7) and they show the same methylation pattern on plasmid pJRD184 DNA (Fig. 2). Bisulfite sequencing shows that both recombinant M.BspLU11IIIb and native M.BspLU11III methylate the same C residue within the recognition site. M.BspLU11IIIa and M.BspLU11IIIb have maximum activity in a buffer containing 20 mM Tris–acetate pH 8.0, 50 mM KCH3COO, 100 µM AdoMet and 0.1 mM EDTA at 50°C.
Enzymatic properties of the recombinant ENase
The ENase encoded by ORF4 shows the same cleavage pattern on plasmid pJRD184 and phage λ DNA as native R.BspLU11III (Fig. 1, lanes 1–4), thus suggesting that both ENases recognize the same sequence in the DNA molecule and cleave it at the same point (14 bp from the recognition site). The methylation–restriction assay shows that the ORF4-encoded ENase is able to protect phage λ DNA against restriction by itself or by native R.BspLU11III (Fig. 1, lane 8). The ENase activity of the enzyme is greatly stimulated (up to 10-fold) by the presence of AdoMet in the reaction mixture at a concentration of 100 µM (data not shown). The optimal conditions for both cleavage and methylation were found to be: 50°C in a buffer containing 20 mM Tris–acetate pH 8.0, 50 mM KCH3COO, 100 µM AdoMet. An indispensable condition for ENase activity is the presence of 10 mM Mg(CH3COO)2 in the reaction buffer. Magnesium ions might be substituted by manganese ions, but ENase activity decreases ∼10-fold. Cobalt and calcium ions do not substitute for magnesium (data not shown).
DISCUSSION
Purification of a protein from strain Bacillus sp. LU11 protecting against restriction by R.BspLU11III resulted in the isolation of a C-specific MTase, M.BspLU11III. This MTase methylates only the inner C residue (underlined) within the sequence 5′-GTCCC-3′ in double-stranded DNA (Fig. 3C and E). An approach based on the conservation of the primary structure of m5C-specific MTases was used to clone the gene coding for M.BspLU11III. As a result, a 5196 bp ClaI fragment of Bacillus sp. LU11 chromosomal DNA containing two MTase genes was cloned. Both genes were expressed in E.coli. The recombinant enzymes were purified and were found to modify the underlined residues within the R.BspLU11III recognition sequence 5′-GGGAC-3′/5′-GTCCC-3′. Adenine methylation was not explicitly tested, but followed from the fact that there is only one A in the sequence. These data suggest that the cloned MTases belong to the BspLU11III R-M system and we designated them M.BspLU11IIIa for the m6A-specific MTase and M.BspLU11IIIb for the m5C-specific MTase, corresponding to MTase M.BspLU11III isolated from Bacillus sp. LU11. Methylation of one of the DNA strands within the recognition site by one of the MTases is sufficient to protect the DNA against restriction by R.BspLU11III. The ENase encoded by ORF4 shows the same enzymatic properties as R.BspLU11III; the identical cleavage patterns on different DNA substrates indicates that both enzymes recognize the same site in the DNA molecule. The recombinant ENase is also capable of protecting the substrate DNA against cleavage by R.BspLU11III (Fig. 1, lane 8). As for R.BspLU11III, it has been shown that both ENase and MTase activities are associated in one polypeptide chain (5) and, taking into account the similar enzymatic properties of R.BspLU11III and the ORF4-encoded ENase, we assume that ORF4 encodes the type IV ENase R.BspLU11III.
The three genes are closely linked and arranged in tandem. The upstream gene encodes a m6A-specific MTase and overlaps the m5C-specific MTase downstream gene by 1 bp (Fig. 4).
The conserved motifs characteristic for group γ m6A MTases are localized in the N-terminal part of the R.BspLU11III polypeptide except for motifs II and III, which are absent in all cloned type IV restriction enzymes (6).
Thus, each DNA strand within the dsDNA recognition sequence 5′-GGGAC-3′/5′-GTCCC-3′ is methylated by a different enzyme. Either M.BspLU11IIIa or R.BspLU11III methylates the only A residue in the recognition site. The biological significance of the ability of the ENase to methylate the same A residue within the recognition site as the corresponding MTase is not clear. It has been suggested that the binding of AdoMet as well as magnesium or manganese ions by type IV ENases is necessary for their restriction activity (6). If the latter is true, the main function of the MTase part within the type IV ENase would be just to bind AdoMet, while the ability to transfer methyl groups to the target DNA is a ‘side effect’. To answer this question, MTase knockout experiments need to be performed. R-M system BspLU11III is a most convenient object for such experiments because it is possible to partially switch off methylation of the recognition site by knocking out either M.BspLU11IIIa or M.BspLU11IIIb. Knockout of M.BspLU11IIIa would show the significance of the ability of R.BspLU11III to methylate the A residue within the recognition site. It has also been shown that both MTase and ENase methylate the same residue in the recognition sequence for other representatives of type IV R-M systems (Eco57I and BseMII) (1,6). However, in contrast to Eco57I and BseMII, two MTases are required to modify both DNA strands of the BspLU11III recognition site. The requirement for two different MTases to modify both DNA strands of the recognition site was also postulated for the HgaI R-M system (34) and BstF5I (35). We believe that modification of both DNA strands within the BspLU11III recognition site is crucial for bacteria to maintain the integrity of the chromosomal DNA after replication. However, for type III R-M systems it has been shown that modification of only one DNA strand is sufficient, because the necessary condition for DNA cleavage by type III restriction enzymes is the presence of two unmethylated recognition sites that are inversely oriented in the DNA double strand (36). We suppose that R.BspLU11III acts in a different way because it cleaves DNA efficiently even if only one recognition site is present on the substrate DNA pUC18A5/A6 (data not shown). M.BspLU11III is the first representative of type IV R-M systems described so far that includes a C-specific MTase.
Amino acid sequence analysis did not reveal any similarity between M.BspLU11IIIa, M.BspLU11IIIb and R.BspLU11III. All three genes have potential promoter regions, which means that they are unlikely to be organized in one operon. Based on these observations, we assume that the formation of this particular R-M system might be an example of convergent evolution. The presence of a transposase gene (ORF1) might indicate that the whole locus containing BspLU11III R-M genes is a part of a transposon.
Remarkably, we found that the potential TRD of M.BspLU11IIIb shows homology to TRDs of other m5C MTases whose recognition sites also have a G on the 5′-end and a C on the 3′-end (Fig. 6) and the homology region spans two potential successive glycine-rich recognition loops, which mediate interaction between the enzyme and the 5′-part of the recognition sequence (30). Prediction of the presence of these loops in TRDs is based on an X-ray crystal structure analysis of M.HhaI (37).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Brigitte Pawlek for her help with transformation of B.subtilis. This work was supported by the Russian Foundation for Fundamental Research under grant nos 99-0448098 and 99-0448272. K.L. was supported by fellowships from the European Molecular Biology Organization (ASTF9051) and the German Academic Exchange Service (DAAD).
DDBJ/EMBL/GenBank accession no. AF169965
REFERENCES
- 1.Janulaitis A., Petrusyte,M., Maneliene,Z., Klimasauskas,S. and Butkus,V. (1992) Purification and properties of the Eco57I restriction endonuclease and methylase—prototypes of a new class (type IV). Nucleic Acids Res., 20, 6043–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matvienko N.N., Kramarov,V.M., Ivanov,L.Y. and Matvienko,N.I. (1992) Bce83I, a restriction endonuclease from Bacillus cereus 83 which recognizes novel nonpalindromic sequence 5′-CTTGAG-3′ and is stimulated by S-adenosylmethionine. Nucleic Acids Res., 20, 1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piekarowicz A., Golaszewska,M., Sunday,A.O., Siwinska,M. and Stein,D.C. (1999) The HaeIV restriction modification system of Haemophilus aegyptius is encoded by a single polypeptide. J. Mol. Biol., 293, 1055–1065. [DOI] [PubMed] [Google Scholar]
- 4.Tucholski J., Zmijewski,J.W. and Podhajska,A.J. (1998) Two intertwined methylation activities of the MmeI restriction-modification class-IIS system from Methylophilus methylotrophus. Gene, 223, 293–302. [DOI] [PubMed] [Google Scholar]
- 5.Chernov A.V., Matvienko,N.N., Zheleznaya,L.A. and Matvienko,N.I. (1995) BspLUII III, a bifunctional restriction and modification enzyme from a thermohilic strain Bacillus species LUII. Nucleic Acids Res., 23, 1213–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jurenaite-Urbanaviciene S., Kazlauskiene,R., Urbelyte,V., Maneliene,Z., Petrusyte,M., Lubys,A. and Janulaitis,A. (2001) Characterization of BseMII, a new type IV restriction-modification system, which recognizes the pentanucleotide sequence 5′-CTCAG(N)10/8. Nucleic Acids Res., 29, 895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janulaitis A., Vaisvila,R., Timinskas,A., Klimasauskas,S. and Butkus,V. (1992) Cloning and sequence analysis of the genes coding for Eco57I type IV restriction-modification enzymes. Nucleic Acids Res., 20, 6051–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matvienko N.N., Zeleznaja,L.A. and Matvienko,N.I. (1993) BspLU11I, a novel site specific endonuclease which cleaves 5′-ACATGT-3′. Nucleic Acids Res., 21, 1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heusterspreute M., Ha Thi,V., Emery,S., Tournis-Gamble,S., Kennedy,N. and Davison,J. (1985) Vectors with restriction site banks. IV. pJRD184, a 3793-bp plasmid vector with 49 unique restriction sites. Gene, 39, 299–304. [DOI] [PubMed] [Google Scholar]
- 10.Haima P., Bron,S. and Venema,G. (1987) The effect of restriction on shotgun cloning and plasmid stability in Bacillus subtilis Marburg. Mol. Gen. Genet., 209, 335–342. [DOI] [PubMed] [Google Scholar]
- 11.Laitinen J., Samarut,J. and Holtta,E. (1994) A nontoxic and versatile protein salting-out method for isolation of DNA. Biotechniques, 17, 316, 318,, 320–322. [PubMed] [Google Scholar]
- 12.Inoue H., Nojima,H. and Okayama,H. (1990) High efficiency transformation of Escherichia coli with plasmids. Gene, 96, 23–28. [DOI] [PubMed] [Google Scholar]
- 13.Bott K.F. and Wilson,G.A. (1968) Metabolic and nutritional factors influencing the development of competence for transfection of Bacillus subtilis. Bacteriol. Rev., 32, 370–378. [PMC free article] [PubMed]
- 14.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Church G.M. and Gilbert,W. (1984) Genomic sequencing. Proc. Natl Acad. Sci. USA, 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 17.Gehrke C.W., McCune,R.A., Gama-Sosa,M.A., Ehrlich,M. and Kuo,K.C. (1984) Quantitative reversed-phase high-performance liquid chromatography of major and modified nucleosides in DNA. J. Chromatogr., 301, 199–219. [DOI] [PubMed] [Google Scholar]
- 18.Tomasz J. (1979) Separation of nucleic acid bases, nucleosides and nucleotides on strong cation-exchange thin layers. IX. Separation of cyclic nucleotides. J. Chromatogr., 169, 466–468. [DOI] [PubMed] [Google Scholar]
- 19.Olek A., Oswald,J. and Walter,J. (1996) A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res., 24, 5064–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauster R., Trautner,T.A. and Noyer-Weidner,M. (1989) Cytosine-specific type II DNA methyltransferases: a conserved enzyme core with variable target-recognizing domains. J. Mol. Biol., 206, 305–312. [DOI] [PubMed] [Google Scholar]
- 21.Posfai J., Bhagwat,A.S. and Roberts,R.J. (1988) Sequence motifs specific for cytosine methyltransferases. Gene, 74, 261–265. [DOI] [PubMed] [Google Scholar]
- 22.Malone T., Blumenthal,R.M. and Cheng,X. (1995) Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyl-transferases and suggests a catalytic mechanism for these enzymes. J. Mol. Biol., 253, 618–632. [DOI] [PubMed] [Google Scholar]
- 23.Vasquez C.C., Saavedra,C.P. and Pichuantes,S.E. (2000) Nucleotide sequence of the gene encoding the BstLVI DNA methyltransferase: comparison with other amino-DNA methyltransferases. Curr. Microbiol., 40, 114–118. [DOI] [PubMed] [Google Scholar]
- 24.Kawakami B., Sasaki,A., Oka,M. and Maekawa,Y. (1990) Nucleotide sequence of the gene coding for the BanIII DNA methyltransferase in Bacillus aneurinolyticus. Agric. Biol. Chem., 54, 3227–3233. [PubMed] [Google Scholar]
- 25.Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- 26.Stein D.C., Gunn,J.S., Radlinska,M. and Piekarowicz,A. (1995) Restriction and modification systems of Neisseria gonorrhoeae. Gene, 157, 19–22. [DOI] [PubMed] [Google Scholar]
- 27.Posfai G., Baldauf,F., Erdei,S., Posfai,J., Venetianer,P. and Kiss,A. (1984) Structure of the gene coding for the sequence-specific DNA-methyltransferase of the B.subtilis phage SPR. Nucleic Acids Res., 12, 9039–9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noyer-Weidner M., Walter,J., Terschuren,P.-A., Chai,S. and Trautner,T.A. (1994) M.Phi3TII: a new monospecific DNA (cytosine-C5) methyltransferase with pronounced amino acid sequence similarity to a family of adenine-N6-DNA-methyltransferases. Nucleic Acids Res., 22, 4066–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schumann J., Willert,J., Wild,C., Waler,J. and Trautner,T.A. (1995) M.BssHII: a new multispecific C5-DNA-methyltransferase. Gene, 157, 103–104. [DOI] [PubMed] [Google Scholar]
- 30.Lange C., Wild,C. and Trautner,T.A. (1996) Identification of a subdomain within DNA-(cytosine-C5)-methyltransferases responsible for the recognition of the 5′ part of their DNA target. EMBO J., 15, 1443–1450. [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson G.G. (1991) Organization of restriction-modification systems. Nucleic Acids Res., 19, 2539–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dieffenbach C.W. and Dveksler,G.S. (1995) PCR Primer: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 33.Studier F.W. and Moffatt,B.A. (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol., 189, 113–130. [DOI] [PubMed] [Google Scholar]
- 34.Sugisaki H., Yamamoto,K. and Takanami,M. (1991) The HgaI restriction-modification system contains two cytosine methylase genes responsible for modification of different DNA strands. J. Biol. Chem., 266, 13952–13957. [PubMed] [Google Scholar]
- 35.Degtyarev S.K., Netesova,N.A., Abdurashitov,M.A. and Shevchenko,A.V. (1997) Primary structure and strand specificity of BstF5I-1 DNA methyltransferase which recognizes 5′-GGATG-3′. Gene, 187, 217–219. [DOI] [PubMed] [Google Scholar]
- 36.Kruger D.H., Kupper,D., Meisel,A., Reuter,M. and Schroeder,C. (1995) The significance of distance and orientation of restriction endonuclease recognition sites in viral DNA genomes. FEMS Microbiol. Rev., 17, 177–184. [DOI] [PubMed] [Google Scholar]
- 37.Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) Hhal methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]