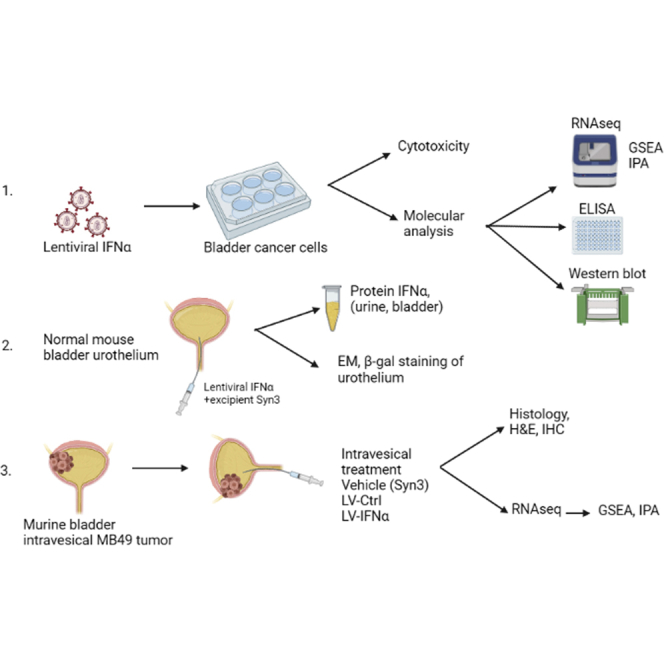
Abstract
Interferon alpha (IFNα) gene therapy is emerging as a new treatment option for patients with non-muscle invasive bladder cancer (NMIBC). Adenoviral vectors expressing IFNα have shown clinical efficacy treating bacillus Calmette-Guerin (BCG)-unresponsive bladder cancer (BLCA). However, transient transgene expression and adenoviral immunogenicity may limit therapeutic activity. Lentiviral vectors can achieve stable transgene expression and are less immunogenic. In this study, we evaluated lentiviral vectors expressing murine IFNα (LV-IFNα) and demonstrate IFNα expression by transduced murine BLCA cell lines, bladder urothelium, and within the urine following intravesical instillation. Murine BLCA cell lines (MB49 and UPPL1541) were sensitive to IFN-mediated cell death after LV-IFNα, whereas BBN975 was inherently resistant. Upregulation of interleukin-6 (IL-6) predicted sensitivity to IFN-mediated cell death mediated by caspase signaling, which when inhibited abrogated IFN-mediated cell killing. Intravesical therapy with LV-IFNα/Syn3 in a syngeneic BLCA model significantly improved survival, and molecular analysis of treated tumors revealed upregulation of apoptotic and immune-cell-mediated death pathways. In particular, biomarker discovery analysis identified three clinically actionable targets, PD-L1, epidermal growth factor receptor (EGFR), and ALDHA1A, in murine tumors treated with LV-IFNα/Syn3. Our findings warrant the comparison of adenoviral and LV-IFNα and the study of novel combination strategies with IFNα gene therapy for the BLCA treatment.
Keywords: interferon alpha, gene therapy, bladder cancer, IL-6, lentiviral vector, intravesical, MB49
Graphical abstract
Interferon alpha gene therapy is emerging as a promising alternative to BCG in bladder cancer. We describe here the development of lentiviral vectors that effectively transduce bladder cancer cell lines and urothelium and also resulted in improved survival in BLCA syngeneic tumor model.
Introduction
Approximately 70% of newly diagnosed bladder cancers (BLCAs) are non-muscle invasive (NMIBC). Conventional treatment of these tumors includes transurethral resection followed by intravesical therapy. Bacillus Calmette-Guerin (BCG) has been the mainstay of intravesical therapy for patients with high-risk disease for decades.1 Although this treatment achieves initial clinical responses in the majority of patients, disease recurrence and progression to a BCG-unresponsive state are common.2 Currently, the optimal management of BCG-unresponsive disease is radical cystectomy, an invasive procedure associated with significant morbidity and potential mortality.2
Several agents have and continue to be tested in the BCG-unresponsive setting.3 To date, few have provided durable disease responses. Immunostimulatory gene therapy is a novel therapeutic strategy with promise to improve the durability of disease responses after intravesical therapy through immune-mediated mechanisms of tumor killing. Since the first successful therapeutic gene transfer over 30 years ago, gene therapy has evolved into an asset in our fight against cancer.4,5
Adenoviral interferon-alpha 2b (Ad-IFNα2b) combined with a novel excipient (Syn-3), an excipient that facilitates gene transfer across the urothelium,6 has shown promising clinical activity in phase 1, 2, and 3 trials.7, 8, 9 In these studies, Ad-IFNα2b/Syn3 appeared to be well tolerated, provided sustained urinary IFNα levels, and demonstrated a 35% 1-year recurrence-free survival (RFS) in patients enrolled in a phase 2 trial. Accordingly, the recently concluded phase 3 trial showed a 60% response rate at 3 months with maintenance of complete response in 51% of patients at 12 months.9 These findings represent favorable improvement in the management of BCG-unresponsive disease compared with currently approved agents.10,11
The anti-tumor efficacy of Ad-IFNα is the result of IFNα’s pleiotropic anti-tumor effects. Preclinical studies demonstrated that IFNα directly induced apoptosis in some human BLCA cells by inducing autocrine tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) production.12 Furthermore, Ad-IFNα treatment inhibited angiogenesis in human xenografts,13,14 and studies conducted in immune-competent syngeneic murine models demonstrated that the immune response to IFN was mediated by activation of both innate and adaptive immunity.15 Also, type 1 IFN signaling is important for initiating anti-tumor responses in dendritic cells.16
While Ad-IFNα gene therapy is clinically efficacious, IFN transgene expression from this DNA viral vector is transient.17 In contrast, lentiviral vectors can integrate their genomes into the DNA of host cells and achieve stable immunostimulatory transgene expression from both dividing and non-dividing tumor cells.18 In addition, whereas adenoviral vectors are highly immunogenic,19 lentiviral vectors are less immunostimulatory and hosts carry lower levels of pre-existing humoral immunity. Lentiviruses have been used as vectors for transgene delivery in several clinical trials.20, 21, 22 The studies presented here are the first to evaluate lentiviral IFNα gene therapy (LV-IFNα) for BLCA.
Results
Expression of IFNα protein and its target genes in syngeneic mouse BLCA cell lines after LV-IFNα transduction
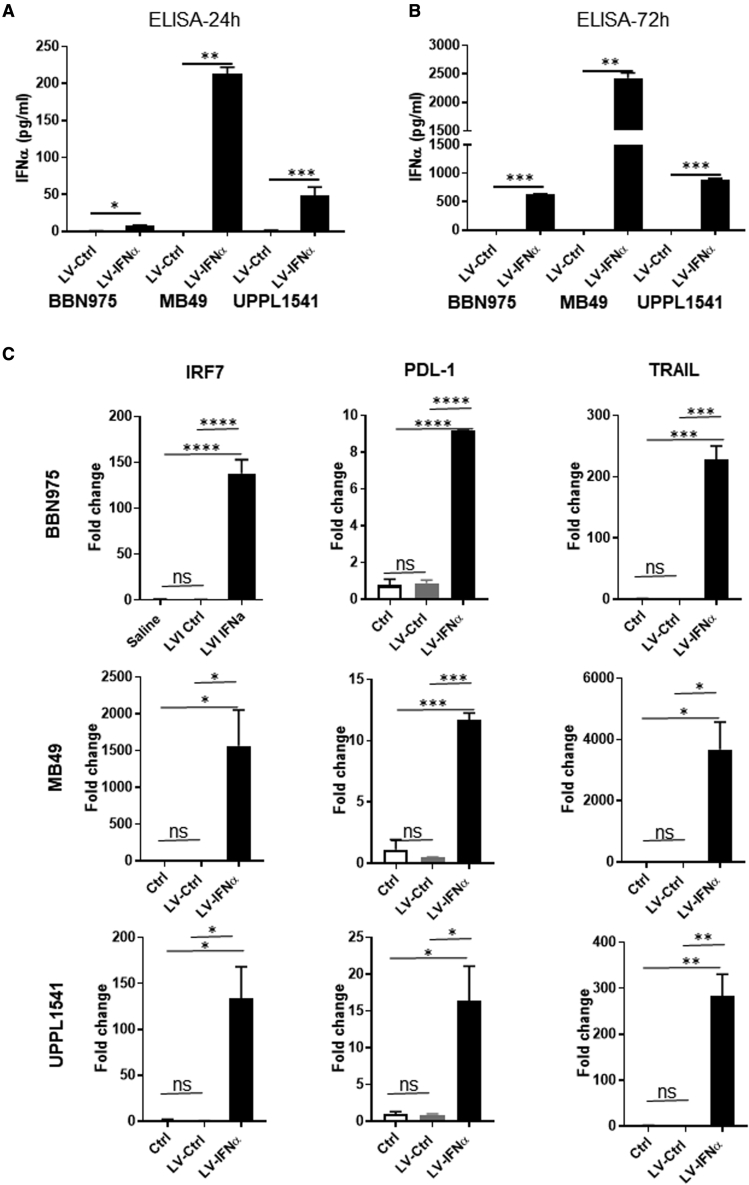
The anti-tumor efficacy of LV-IFNα was tested in the murine BLCA cell lines BBN975 and UPPL1541, which recapitulate NMIBC, along with the carcinogen-induced cell line MB49 that is frequently used in preclinical studies.23 GFP expression confirmed complete transduction of all cells by 48–72 h after treatment using increasing multiplicities of infection (MOIs) (2, 6, and 8; Figure S1). Cell lines were then treated with lentiviral control vector (LV-Ctrl) or LV-IFNα at a MOI of 2. IFNα-specific ELISA performed on cell-free supernatants collected 24 and 72 h after transduction with LV-IFNα showed robust and significantly higher IFNα protein expression in all three cell lines compared with LV-Ctrl (Figures 1A and 1B). Quantitative real-time PCR using RNA from transduced cells showed upregulation of the IFNα target genes IRF7, PD-L1, and TRAIL (Figure 1C). IRF7 expression was significantly induced in all the three cell lines but was highest in MB49 (1,564-fold), followed by BBN975 (138-fold) and UPPL1541 cells (134-fold). PD-L1 was comparably upregulated in all the three cell lines (9.165-fold in BBN975, 11.7-fold in MB49, and 16.35-fold in UPPL1541). TRAIL expression was highest in MB49 (3,677-fold), followed by UPPL1541 (282.5-fold) and BBN975 (228.3-fold).
Figure 1.
Expression of murine IFNα protein and its target genes in syngeneic BLCA cell lines
(A and B) BLCA cell lines were transduced with LV-Ctrl or LV-IFNα vectors at an MOI of 2 and murine IFNα was measured in the cell-free supernatants at 24 h (A) and 72 h (B) after viral transduction. All the three cell lines showed significant expression of IFNα protein in LV-IFNα-transduced cells compared with controls. (C) qPCR analysis of IFNα target genes IRF7, PDL-1, and TRAIL in BLCA cell lines show increased expression following treatment with LV-IFNα virus when compared with control. Not significant (ns), p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Cytotoxic effects of IFNα protein on BLCA cell lines after transduction with LV-IFNα
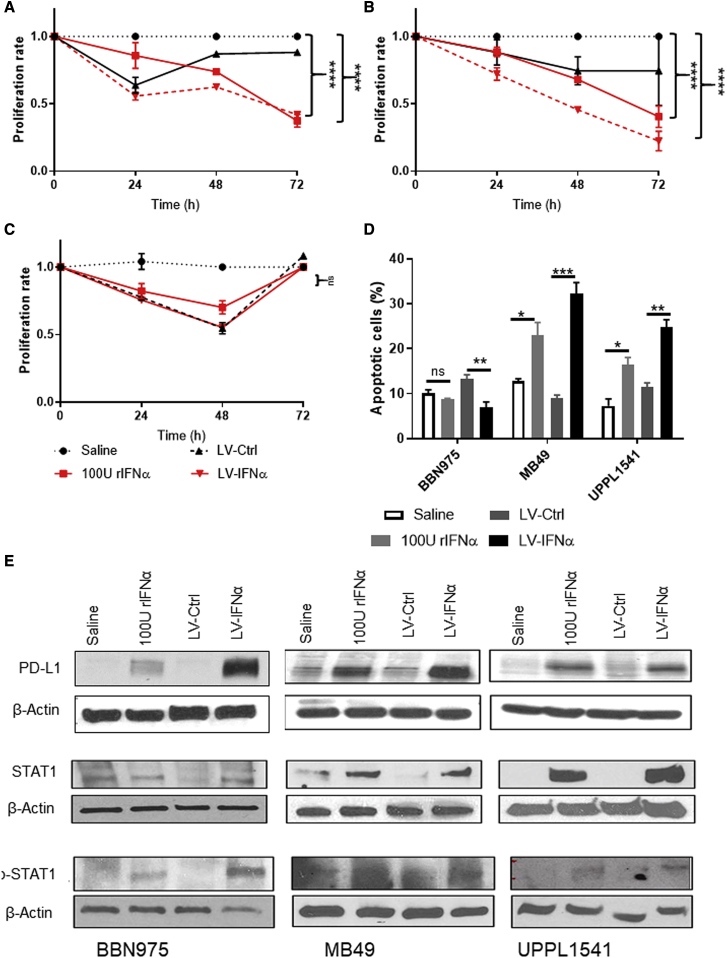
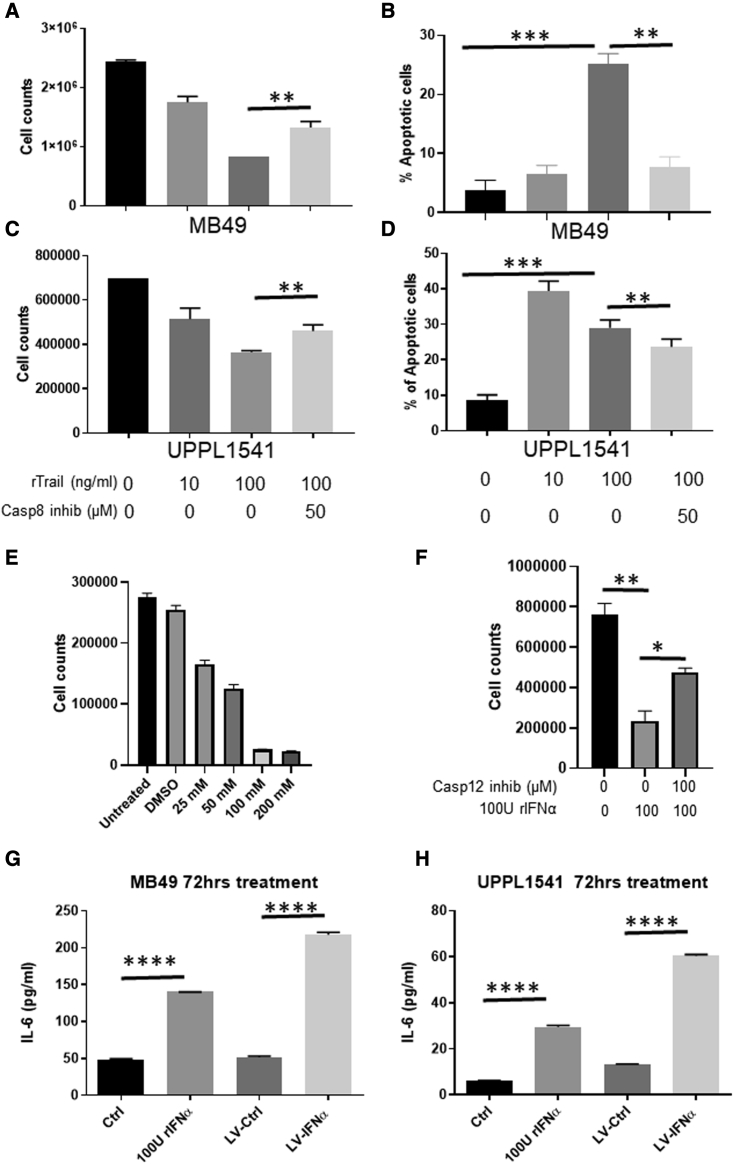
To determine whether IFNα gene therapy induced direct cytotoxic effects, we assessed cell viability of BLCA cell lines by Trypan blue dye exclusion after LV-IFNα transduction. In two of the three cell lines (MB49 and UPPL1541), viable cell counts decreased after LV-IFNα and 100 U of recombinant IFNα (rIFNα) treatment in a time-dependent manner (Figures 2A and 2B). This result was confirmed by MTT assay at 72 h after treatment, which demonstrated that MB49 cell proliferation was significantly lower after LV-IFNα treatment compared with LV-Ctrl (Figure S2A). BBN975 cells showed an initial response to LV-IFNα and 100 U rIFNa, but by 72 h, the viable cell count was comparable to controls (saline or LV-Ctrl; Figure 2C). Recombinant IFNα dose-response curves illustrated that BBN975 cells were resistant to rIFNα (up to 600 U/mL conc.; Figure S2B), whereas MB49 and UPPL1541 were sensitive at all dose levels tested (Figures S2C and S2D). The cytotoxicity of IFNα was mediated by cell-death pathways, as demonstrated by increased annexin V staining in MB49 and UPPL1541 cells following treatment with LV-IFNα and 100 U of rIFNα compared with controls (Figure 2D). Following the observation that IFNα gene therapy promoted PD-L1 expression through IFNAR1/STAT1 signaling, we performed western blot analysis on lysates from our three cell lines after treatment with LV-IFNα, rIFNα, LV-Ctrl, or vehicle, confirming that PD-L1, STAT1, and p-STAT1 were all upregulated following LV-IFNα of MB49, UPPL1541, and BBN975 (Figure 2E).
Figure 2.
Cytotoxic effect of LV-IFNα on BLCA cell lines
(A–C) Murine cell lines were exposed to murine recombinant IFNα (rIFNα) or transduced with LV-Ctrl or LV-IFNα and cell counts were measured using Trypan blue dye exclusion method. At 72 h, MB49 (A) and UPPL1541 (B) showed significant reduction in cell counts, while BBN975 (C) showed no change in cell numbers when compared with the saline treatment. (D) At 72 h, annexin V and propidium iodide staining showed increased apoptotic cells after 100 U rIFNα or LV-IFNα treatment in MB49 and UPPL1541 cells. Percentage of apoptotic cells in BBN975 remained unchanged. ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. (E) Western blot for IFNα target genes shows increased expression of PD-L1, STAT1, and p-STAT1 in cell lines treated with 100 U rIFNα or LV-IFNα.
Cellular pathway discovery
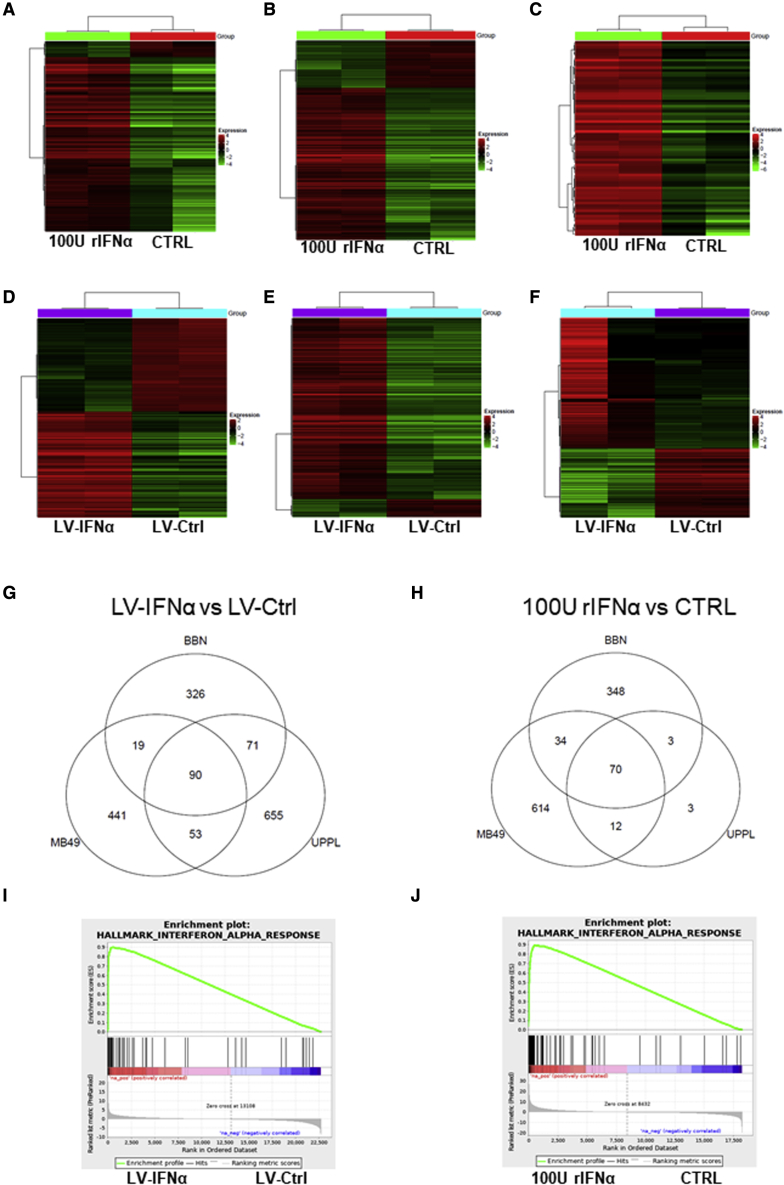
To better understand how LV-IFNα affects global gene expression, we performed RNA sequencing (RNA-seq) on cell lines treated with rIFNα, LV-Ctrl, or LV-IFNα and compared them with untreated control cells (Ctrl). Hierarchical clustering and principal-component analyses showed distinct gene expression patterns for the four groups (Figures S3A and S3B). To identify candidate genes contributing to IFNα-mediated effects on cell viability, we compared differentially expressed genes between the rIFNα/Ctrl and LV-IFNα/LV-Ctrl pairs and generated heatmaps using a false discovery rate (FDR) cutoff of 0.05 and fold change of 2 (Figures 3A–3F). The gene lists are provided in Table S1.
Figure 3.
RNA-seq analysis of mouse BLCA cell lines treated with 100 U rIFNα or LV-IFNα
(A–F) Heatmaps of significant genes (FDR cutoff 0.05 and fold change of 2) between 100 U IFNα and CTRL samples for BBN975 (A), MB49 (B), and UPPL1541 (C) and between LV-IFNα and LV-Ctrl samples for BBN975 (D), MB49 (E), and UPPL1541 (F). (G and H) Venn diagrams show significantly altered genes (FDR cutoff 0.05 and fold change of 1.5) common or unique to all the three cell lines after treatment with LV-IFNα and rIFNα. (I and J) GSEA shows enrichment of interferon alpha response pathway in MB49 cells treated with LV-IFNα and rIFNα.
We then used a more inclusive fold change cutoff of 1.5 to identify genes that are commonly altered among the three cell lines tested. We generated Venn diagrams that identified 506, 603, and 869 differentially expressed genes between LV-IFNα and LV-Ctrl treatment of BBN975, MB49, and UPPL1541, respectively, with 90 genes common to all cell lines (Figure 3G). Similarly, 455, 730, and 88 genes were differentially expressed across the respective cell lines between 100 U rIFNα-treated and untreated controls (Ctrl), with 70 genes common to all cell lines (Figure 3H). The complete gene list for the three cell lines is included in Table S2.
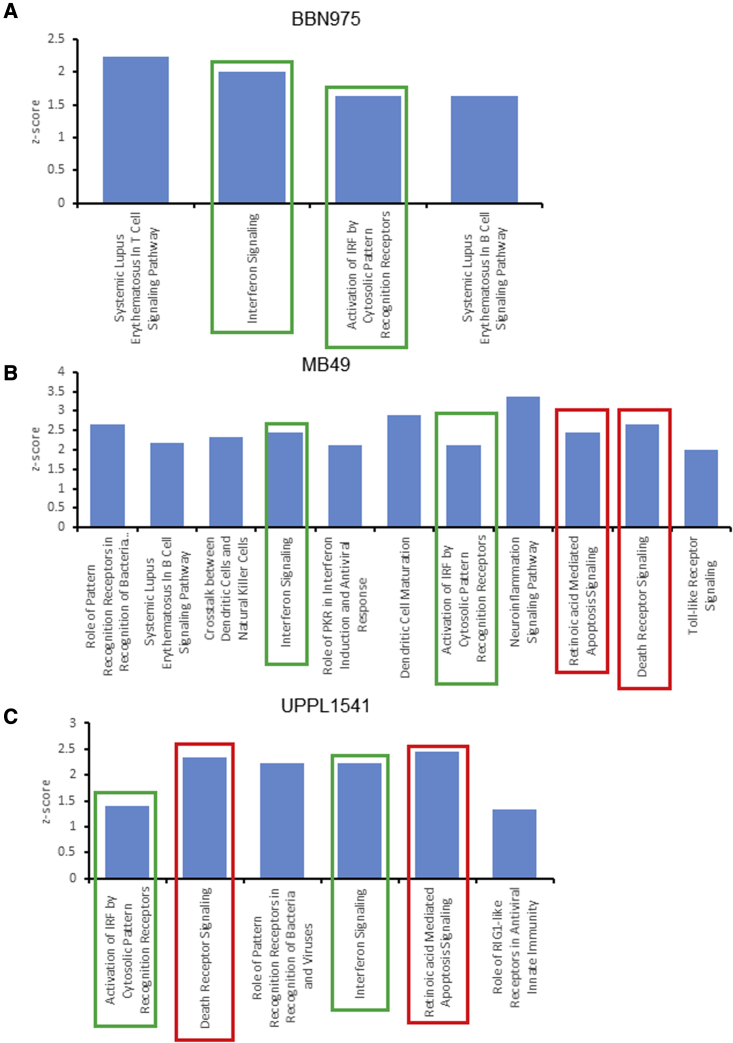
To identify major pathways activated in these cells, we used the sequencing data to perform gene set enrichment analysis (GSEA). Top GSEA hallmark pathways are shown in Table S3. As expected, GSEA showed enrichment of the IFNα response pathway in both LV-IFNα- and rIFNα-treated MB49 cells when compared with respective controls (Figures 3I and 3J). GSEA of top pathways for BBN975 and UPPL1541 is shown in Figure S3C. We also performed ingenuity pathway analysis (IPA) on differentially expressed genes between LV-IFNα- and LV-Ctrl-treated cells in all three cell lines. IFN signaling and activation of IRF by cytosolic pattern recognition receptors were commonly enriched in all three cell lines with a positive Z score whereas retinoic-acid-mediated apoptosis signaling pathway and death receptor signaling were enriched only in the IFNα-sensitive MB49 and UPPL1541 (Figures 4A–4C) lines. PARP proteins and TRAIL (Tnfsf10) were commonly enriched among the two apoptotic pathways, whereas DAXX was specific to the death receptor signaling pathway and was upregulated >2-fold in both MB49 and UPPL1541 cells. The complete gene list for the apoptotic pathways is provided in Table S4.
Figure 4.
Ingenuity pathway analysis (IPA) of canonical pathways altered after treatment with LV-IFNα
IPA analysis of significant genes (FDR cutoff 0.05 and fold change of 2) was performed to identify top candidate canonical pathways with a positive Z score in BBN975 (A), MB49 (B), and UPPL1541 (C). Pathways that are common to all three cell lines are indicated by green bars, and pathways common between MB49 and UPPL1541 are shown in red.
Mechanism underlying IFNα-mediated cell death
Given that our sequencing studies confirmed increased expression of TRAIL in all three cell lines (Figure 1C), we next evaluated caspase 8 expression, which has been shown to mediate TRAIL-dependent cell death in human BLCA cell lines.12 Caspase 8 inhibition in MB49 (Figures 5A and 5B) and UPPL1541 (Figures 5C and 5D) partially rescued TRAIL-mediated cell death. In contrast, TRAIL-mediated cell death was not observed in BBN975, which is resistant to IFNα treatment (Figures S4A and S4B). Cell-surface expression of TRAIL was also increased in MB49 following treatment with LV-IFNα, which was rescued following caspase 8 inhibition (Figure S4C). In addition to caspase 8, in MB49 cells, we detected caspase 4 and 12 overexpression (2.0- and 2.2-fold, respectively), genes involved in endoplasmic reticulum (ER) stress response and whose expression has been linked to cell death (Table S1). Treatment of MB49 with tunicamycin to induce ER stress also resulted in cell death in a dose-dependent manner (Figure 5E), suggesting that MB49 cells are sensitive to ER stress induction. Treatment with IFNα together with caspase 12 inhibition partially rescued cell death in MB49 cells (Figure 5F). Interestingly, caspase 12 expression was downregulated in UPPL1541 (2.2-fold), suggesting that this caspase may not be involved in death of these cells (Table S1).
Figure 5.
Caspase-mediated cell death in IFNα-sensitive cells
(A and C) Cell counts by Trypan blue dye exclusion method in MB49 (A) and UPPL1541 (C) cells treated with recombinant TRAIL (rTRAIL, 100 ng/mL) in the presence or absence of caspase 8 inhibitor (50 μM). rTRAIL reduced cell counts in a dose-dependent manner and was partially rescued by addition of caspase 8 inhibitor. (B and D) Percentage of apoptotic cells measured by annexin V/propidium iodide (PI) staining showed increased apoptosis in MB49 (B) and UPPL1541 (D) cells treated with rTRAIL and was rescued in cells treated with rTRAIL and caspase 8 inhibitor. (E) MB49 cells treated with increasing concentrations of tunicamycin, ER stress inducer, showed dose-dependent reduction in cell counts. (F) Caspase 12 inhibitor rescues cell death induced by rIFNα. (G and H) IL-6 protein expression measured by ELISA in cell-free supernatants of MB49 (G) and UPPL1541 (H) cells is shown. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
We recently reported that interleukin-6 (IL-6) was important for IFN-mediated death in MB49 cells and that upregulation of IL-6 in clinical samples collected within the phase 2 trial correlated with response to IFNα gene therapy.15 We therefore evaluated the relative expression of IL-6 in all cell lines following treatment with LV-IFNα by ELISA. IL-6 protein expression was detected at 72 h after LV-IFNα treatment in MB49 and UPPL1541 (Figures 5G and 5H) cells, whereas its expression was undetectable in BBN975 (data not shown). This underscores the link between the induction of IL-6 and response to IFNα.
Transduction of the bladder urothelium with LV-IFNα
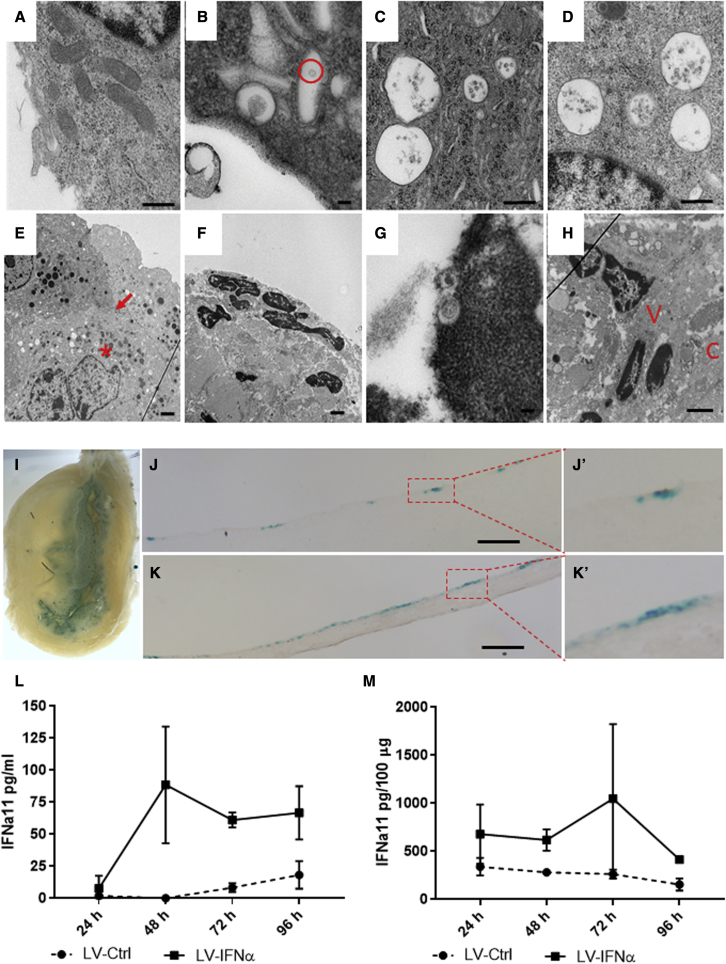
We next tested the transduction efficiency of LV-IFNα after murine intravesical instillation. Following pre-administration of the excipient Syn3,24 LV-IFNα transduction of murine urothelium was confirmed by electron microscopy (EM), with viral particles detectable inside vacuoles as early as 4 h post-instillation and persisting for up to 96 h post-instillation (Figures 6A–6D). Sequential instillations resulted in deeper vector penetration of the urothelium (Figures 6E and 6H) and apoptotic urothelial cells with shrunken cytoplasm and viral particles detected in the nuclei (Figures 6F and 6G). In addition to EM, we verified urothelial transduction using a lentivirus-beta-galactosidase (LV-βgal) vector (Figure 6I). Histological sections showed multifocal regions of βgal staining throughout the urothelium (Figures 6J and 6K). Finally, expression of IFNα in urine from treated mice measured by ELISA confirmed LV-IFNα transduction of and IFNα transgene expression from murine urothelium first detectable at 48 h and maintained for up to 96 h post-treatment, with protein expression of IFNα in bladder lysates peaking at 72 h post-transduction (Figures 6L and 6M).
Figure 6.
Electron microscopy and transduction of bladder urothelium treated with LV vectors
(A) Normal mouse urothelium at 50,000×. (B) Mouse urothelium 4 h post-LV-IFNα instillation demonstrating one vacuole containing a virus particle inside the cytoplasm (red circle) at 100,000× is shown. (C) Twenty-four hours post-instillation of LV-IFNα demonstrates four vacuoles with LV vectors within them at 50,000×. (D) Ninety-six hours post-LV-IFNα instillation, four vacuoles containing LV vectors adjacent to the nucleus at 75,000× are shown. (E) After five weekly instillations, at 10,000×, there are numerous vector-filled vacuoles outside the nucleus in a second cell layer (∗ denotes vacuoles and arrow denotes cell layer). (F) Mouse urothelium after 3 months of BBN followed by monthly instillations of LV-IFNα for 3 months is shown. At 10,000×, a dark nucleus with shrunken cytoplasm is visible within an apoptotic cell. (G) In the same mouse as in image (F) at 100,000×, viral vectors are seen within a darkened nucleus, apoptotic. (H) At 10,000×, viral vectors (marked by V) are seen near nuclei deep to the connective tissue (marked by C). (I) Whole mounts of mouse bladders instilled with LV-βgal vector showing β-gal-positive blue cells in the urothelium are shown. (J and K) Tissue sections showing β-gal-positive cells in the urothelium are shown. Insets (J′) and (K′) show higher magnifications. Scale bars, 100 μm. (L and M) Measurement of IFNα11 levels by ELISA in urine of mice treated with LV-Ctrl or LV-IFNα (L) or tissue (M) at 24, 48, 72, and 96 h post-instillation is shown.
Efficacy of LV-IFNα in murine model for BLCA
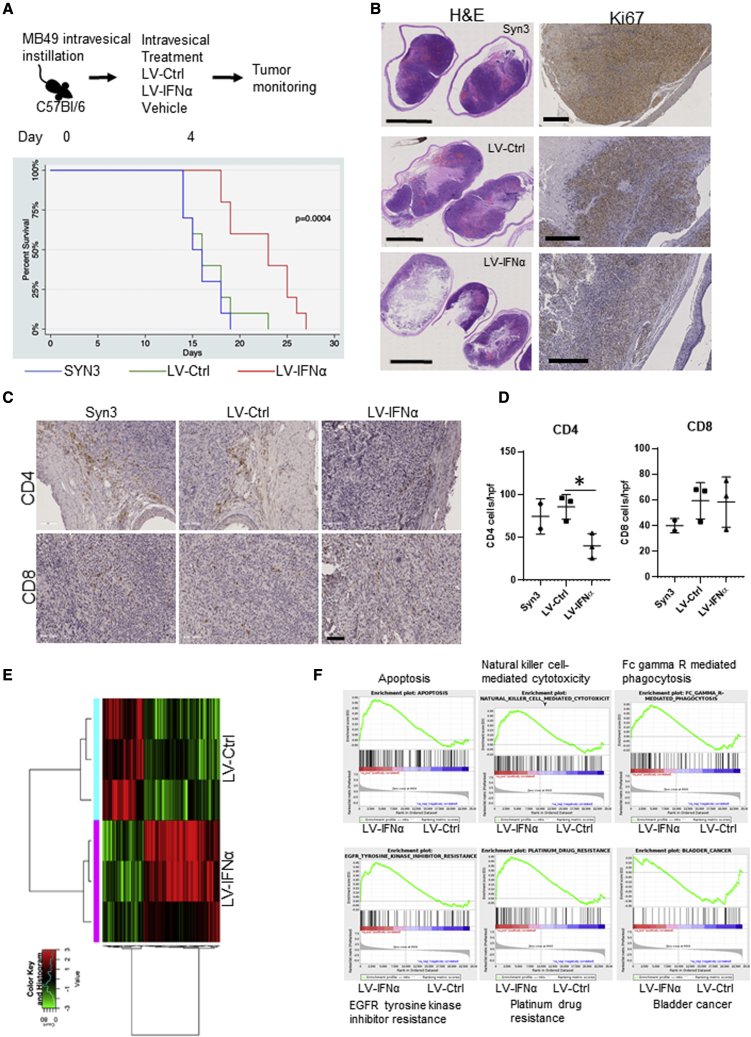
The anti-tumor efficacy of LV-IFNα was tested in the syngeneic MB49 murine BLCA model after intravesical administration. LV-IFNα treatment resulted in a significant improvement in median survival compared with LV-Ctrl (26 and 19 days, respectively; p < 0.001; Figure 7A). Histological examination of murine bladders treated with LV-IFNα revealed reduced tumor burden and decreased tumor proliferation (Figure 7B). To investigate intratumoral immune responses induced by LV-IFNα, we performed immunohistochemistry (IHC) on treated tumors and confirmed decreased numbers of tumor-infiltrating CD4 cells following LV-IFNα treatment compared with LV-Ctrl treatment. CD8 cells were not significantly altered (Figures 7C and 7D).
Figure 7.
Efficacy of LV-IFNα in murine MB49 intravesical model
(A) Kaplan-Meier plot showing percent survival in C57Bl/6, MB49 intravesical disease model treated with vehicle (Syn3), LV-Ctrl, and LV-IFNα (p = 0.0004). (B) H&E staining of whole-mount bladder tissues with corresponding Ki67 staining in MB49 is shown. (C) IHC for CD4 and CD8 T cells on MB49 intravesical tumors treated with Syn3 (vehicle), LV-Ctrl, and LV-IFNα vectors is shown. (D) Quantification of CD4 and CD8 cells in the MB49 intravesical model is shown. (E) Heatmap of 190 significantly altered genes at an FDR cutoff 0.05 and log2 fold change of one between LV-IFNα and LV-Ctrl groups. (F) GSEA analysis shows enrichment of apoptosis, natural-killer-cell-mediated cytotoxicity, and Fc-gamma-receptor-mediated cytotoxicity (top panel) and EGFR tyrosine kinase inhibitor resistance, platinum drug resistance in LV-IFNα-treated tumors, and enrichment of bladder cancer gene set in LV-Ctrl group.
In vivo gene expression profiles induced by LV-IFNα treatment
Finally, we sequenced murine MB49 bladder tumors treated with LV-IFNα and compared their expression profile with those from LV-Ctrl and vehicle-treated bladder tumors. Hierarchical clustering analysis was performed using log-transformed normalized count data. The clustered dendrogram is presented with a heatmap of variably expressed genes (top 1,500) in Figure S5. Using an FDR cutoff of 0.05 and log2 fold change of 1, this analysis identified 190 genes differentially expressed between LV-IFNα and LV-Ctrl treatment groups (Figure 7E), with the complete gene list provided in Table S4. Specifically, we found upregulation of several granzyme genes that are positive regulators of apoptosis in LV-IFNα-treated tumors.25 Consistent with our in vitro results (Table S4), we also identified upregulation of caspase 12 in LV-IFNα-treated tumors. GSEA identified greater expression of apoptotic pathways in LV-IFNα-treated tumors when compared with LV-Ctrl tumors (Figure 7F). Immune-related pathways, such as natural-killer-cell-mediated cytotoxicity and Fc-gamma-receptor-mediated phagocytosis pathways, were also enriched in LV-IFNα-treated tumors compared with LV-Ctrl.
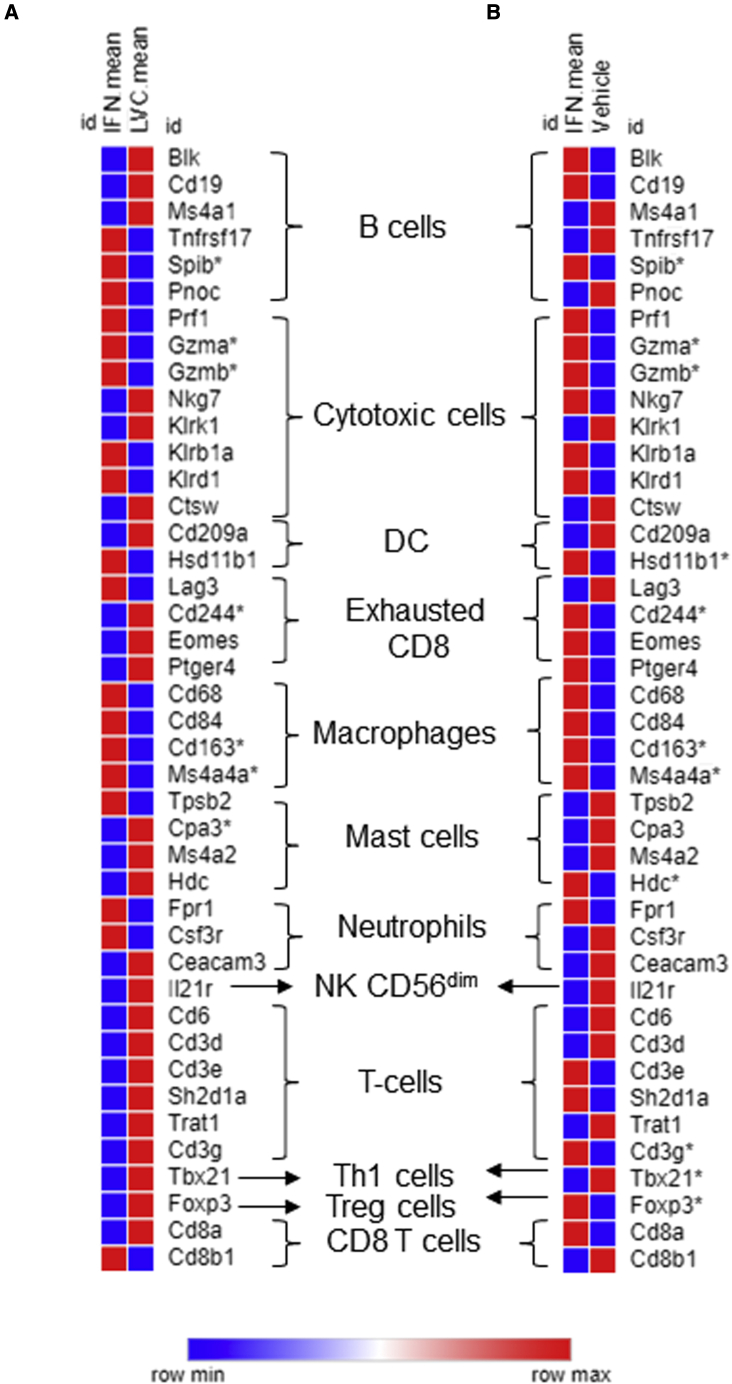
In order to identify biomarkers associated with IFNα sensitivity, we used IPA with sequencing results from LV-IFNα-treated tumors using an FDR cutoff of 0.05 and fold change of 2. We identified 11 genes associated with cancer, inflammatory disease, and renal and urological disease (Table 1). The biomarker application for each of these genes, fold change (expr log ratio), and p value is listed (Table 1). We also evaluated expression of genes involved in cytotoxicity and immune cell recruitment. Directionality of expression of most genes involved in cytotoxicity and immune cell recruitment was consistent with their previously described functions (Table 2). We also evaluated the differential expression of candidate genes associated with immune cell subtypes following LV-IFNα versus LV-Ctrl or vehicle treatment (Figures 8A and 8B; Table S5). The most differentially expressed genes included SPIB (B cell marker), granzyme A and B (cytotoxic cell marker), and CD163 and MS4A4A (macrophage markers), suggesting involvement of these immune cell subtypes in IFNα-mediated cytotoxicity.
Table 1.
Biomarkers identified by IPA in mouse tumors
| Symbol | Entrez gene name | Family | Drug(s) | Expr log ratio | Expr p value | Biomarker application(s) | Diseases (relevant) |
|---|---|---|---|---|---|---|---|
| ALDH1A1 | aldehyde dehydrogenase 1 family member A1 | enzyme | disulfiram, chlorpropamide |
3.437 | 0.041 | diagnosis and disease progression | cancer, immunological disease, inflammatory disease, inflammatory response, and renal and urological disease |
| ALDH3A1 | aldehyde dehydrogenase 3 family member A1 | enzyme | 2.577 | 0.012 | diagnosis | cancer, inflammatory disease, and renal and urological disease | |
| CARD9 | caspase recruitment domain family member 9 | other | 3.979 | 0.000 | diagnosis | cancer, inflammatory disease, inflammatory response | |
| CAV1 | caveolin 1 | receptor | 1.839 | 0.047 | diagnosis | cancer | |
| EGFR | epidermal growth factor receptor | kinase | hemay020, cetuximab and erlotinib, CK-101, etc. | 2.162 | 0.043 | diagnosis, disease progression, efficacy, prognosis, response to therapy, safety | cancer, immunological disease, inflammatory disease, inflammatory response, and renal and urological disease |
| ERCC5 | ERCC excision repair 5, endonuclease | enzyme | 2.105 | 0.016 | diagnosis and response to therapy | cancer, immunological disease, inflammatory disease, inflammatory response, and renal and urological disease | |
| HP | haptoglobin | peptidase | 4.224 | 0.004 | diagnosis and efficacy | cancer, immunological disease, inflammatory disease, inflammatory response, and renal and urological disease | |
| IL-23A | interleukin-23 subunit alpha | cytokine | LY3074828, tildrakizumab, and guselkumab | −3.196 | 0.016 | diagnosis | cancer |
| LGALS7/LGALS7B | galectin 7 | other | −5.229 | 0.005 | disease progression | cancer and renal and urological disease | |
| PPL | periplakin | other | 2.072 | 0.049 | diagnosis | cancer and renal and urological disease | |
| VEGFD | vascular endothelial growth factor D | growth factor | 2.867 | 0.014 | diagnosis, disease progression, efficacy, and prognosis | cancer, hypersensitivity response, immunological disease, inflammatory disease, and inflammatory response |
Table 2.
IPA of gene sets involved in cytotoxicity and recruitment of neutrophils and granulocytes
| Gene ID | Prediction (based on measurement direction) | Log2fold change | Findings from other studies |
|---|---|---|---|
| Cytotoxicity of cells | |||
| Klra3 | increased | 5.374 | increased |
| Gzma | increased | 4.611 | increased |
| Sgk1 | decreased | 3.628 | decreased |
| Kitl | increased | 2.536 | increased |
| Lamp1 | increased | 2.027 | increased |
| Il23a | decreased | −3.196 | increased |
| Recruitment of neutrophils | |||
| Il21r | affected | −4.028 | affected |
| Lcn2 | increased | 3.693 | increased |
| Adrb2 | increased | 3.284 | increased |
| Cxcl1 | increased | 3.165 | increased |
| Xdh | increased | 2.688 | increased |
| P2rx1 | decreased | −3.127 | increased |
| Recruitment of granulocytes | |||
| Agt | increased | 7.097 | increased |
| Card9 | decreased | 3.979 | decreased |
| Lcn2 | increased | 3.693 | increased |
| Adrb2 | increased | 3.284 | increased |
| Cxcl1 | increased | 3.165 | increased |
| Xdh | increased | 2.688 | increased |
| Kitl | increased | 2.536 | increased |
| P2rx1 | decreased | −3.127 | increased |
| Il21r | decreased | −4.028 | increased |
Figure 8.
Expression of cell-type-specific immune cell markers in MB49 tumors
Heatmap of expression comparing LV-IFNα versus LV-Ctrl (A) and LV-IFNα versus vehicle (B) in MB49 tumors. Asterisk indicates significantly altered genes.
Discussion
Adenoviral-based IFNα2b gene therapy has shown promising efficacy in clinical trials. In addition to being well tolerated with a convenient dosing schedule, it achieved an RFS rate of 30.5% at 12 months.9 Although this represents a significant improvement over the agents currently approved for BCG-unresponsive disease,2 there remains a need to understand and improve upon intravesical IFNα gene therapy in clinical non-responders. This can be accomplished by identifying biomarkers predictive of sensitivity or resistance, by better understanding its mechanisms of action, and through the development of more efficient gene therapy vectors. We report here the initial study evaluating LV-IFNα therapy against BLCA, identifying mechanisms driving IFNα’s direct anti-tumor activity and its role(s) in immune modulation of the tumor microenvironment. Given the established clinical efficacy of IFNα gene therapy, improved intensity and duration of IFNα transgene expression following lentiviral transduction has the potential to improve the rate and durability of clinical responses.17
Unlike adenoviruses, lentiviruses can efficiently transduce both dividing and non-dividing cells and achieve stable transgene expression, as shown in several clinical trials.20, 21, 22 Additional advantages to lentiviral vectors include reduced baseline seropositivity and decreased immunogenicity following sequential administrations.19,26,27 One significant practical limitation to the development of lentiviral vectors for clinical use was the challenge for large-scale manufacturing and purification. This limitation has been overcome using bioreactors for clinical vector production.28 The lentiviral vector appears to be more efficient than adenoviral vectors, as higher IFN levels were generated at a much lower MOI than required with the adenovirus (data not shown). We observed little to no toxicity in normal cells infected with the adenoviral-IFNα29 and similarly have not observed any obvious evidence of toxicity in animals treated with lenti-IFNα. However, if lenti-IFNα is developed for clinical use, more formal toxicity studies will be performed. One concern with lentiviral gene therapy is the potential risk for integration into the host’s genome and the theoretical risk for insertional mutagenesis and development of a secondary malignancy. However, the benefits of long-term IFNα production may outweigh this theoretical risk. Using the lentiviral vector, we observed robust expression of the IFN transgene in multiple cell lines in vitro, from murine urothelium, and from syngeneic murine bladder tumors after intravesical therapy. LV-IFNα treatment resulted in significantly improved survival in our BLCA murine models, which also correlated with upregulation of IFN-target genes, including PD-L1 and TRAIL, and multiple cellular pathways involved in cytotoxicity and immune cell recruitment.
Interestingly, despite comparable expression levels of IFNα following LV-IFNα transduction in BBN975 cells, only MB49 and UPPL1541 cells lines were sensitive to IFNα-mediated cell death. We found that this sensitivity correlated with TRAIL expression and activation of death receptor signaling and retinoic-acid-mediated apoptotic signaling pathways. Our group has previously reported TRAIL-mediated cell death in human BLCA cells mediated by caspase 8.12 Consistent with this mechanism, we observed that caspase 8 inhibition rescued TRAIL-mediated cell death in MB49 and UPPL1541 cells after exposure to rIFNα, whereas in the resistant BBN975 cells, recombinant TRAIL failed to induce cell death. We acknowledge that this is indirect evidence, and more detailed studies are being carrying out to molecularly tease out the pathways contributing to cell death.
Molecular analyses of IFNα-treated cells clarified some of the mechanisms driving the therapeutic efficacy of and cellular resistance to LV-IFNα. Ninety IFNα response genes were upregulated in all three cell lines and included CD274 (PD-L1); chemokines, such as CXCL10 and CXCL5; GBP family proteins; IRF7; USP18; and the SLFN family known to be activated by IFN treatment.30 When restricting the comparison to the cell lines sensitive to LV-IFNα (MB49 and UPPL1541), Tlr3, IL-15, CXCL11, TNFSF10 (TRAIL), and caspase 12 were differentially expressed. Of these, only caspase 12, which is involved in ER stress response,31,32 was differentially upregulated in MB49 and downregulated in UPPL1541. Caspase 12 inhibition rescued cell death in MB49, suggesting that at least two caspases, caspase 8 and caspase 12, are involved in cell death pathways in at least a subset of BLCAs. Notably, MB49 cells were also sensitive to cell death by tunicamycin, an ER-stress-inducing compound, suggesting ER-mediated stress may be a novel mechanism of direct toxicity associated with IFNα gene therapy. It remains to be determined whether ER-mediated cellular stress responses are specific to IFNα gene therapy or mediated by lentiviral transduction.
In earlier work, we identified IL-6 as an important mediator of anti-tumor effects of IFNα in our murine MB49 subcutaneous model.15 Here, we found that IL-6 was detectable at the protein level upon treatment with LV-IFNα in only the two sensitive cell lines MB49 and UPPL1541 and not in the resistant BBN975 line. This further supports the observation that IL-6 induction predicts response to IFNα.15 This result was confirmed by MTT assay at 72 h after treatment, which demonstrated that MB49 cell proliferation was significantly lower in LV-IFNα-treated cells compared with LV-Ctrl (Figure S2A). This is supported by other studies showing STAT1-mediated upregulation of IL-6 results in apoptosis in different cell types.33,34 STAT1 was upregulated and phosphorylated in all three cell lines; however, IL-6 was detectable only in the IFNα-sensitive cell lines. Taken together, IFNα-mediated cell death in culture may therefore be occurring by at least three mechanisms: (1) TRAIL-mediated cell death through caspase 8 in MB49 and UPPL1541, (2) ER-stress-induced cell death mediated by caspase 4 and 12 in MB49, and (3) IL-6 induction that induces cell death via STAT1 activation in MB49 and UPPL1541 cells.
GSEA of mouse tumors confirmed that in vivo efficacy resulted from activation of apoptotic pathways, natural-killer-cell-mediated cytotoxicity, and Fc-gamma-receptor-mediated phagocytosis in LV-IFNα-treated tumors. Accordingly, IPA showed increased expression of cytotoxicity genes, such as Klra3, Gzma, Kitl, and Lamp1.35, 36, 37, 38 In addition, when immune cell populations were analyzed by whole-tumor RNA-seq, we identified enrichment of genes identifying cytotoxic immune cell populations, suggesting multiple distinct mechanisms of tumor killing (apoptosis, ER stress, and cytotoxic immune cell recruitment) may be driving the observed in vivo efficacy of LV-IFNα. Future studies utilizing single-cell sequencing techniques and fluorescence-activated cell sorting (FACS) analysis will be required to conclusively identify the tumor-associated immune cell populations necessary for IFNα efficacy.
Finally, we identified three biomarkers that are part of clinically targetable molecular pathways using the biomarker filter of IPA. Aldha1, an enzyme that has been associated with stem cell properties in cancer and resistance to drugs, is upregulated and can be targeted by disulfiram and chloropropamide.39,40 Epidermal growth factor receptor (EGFR) has been targeted in BLCA, with both cetuximab and erlotinib demonstrating effective anti-tumor activity in preclinical models.41 Finally, PD-L1 targeting has revolutionized cancer treatment in multiple disease settings, including BLCA, and there is great clinical potential for synergistic gene therapy and checkpoint inhibitor regimens currently being evaluated in clinical trials. Biomarker-driven combination strategies may provide therapeutic benefit for patients who would otherwise fail to respond or develop resistance to IFNα monotherapy.
One concern with LV-IFNα treatment includes the potential for local toxicity. We did not identify morphological changes to the urothelium or clinical symptoms in mice treated with LV-IFNα, suggesting good local tolerance. Additional areas for future investigation include identifying optimal dosing regimens, the long-term safety and durability of lentiviral-mediated IFNα expression by transduced urothelium, and the systemic immune responses elicited against both the lentiviral vector and tumor neoantigens following intravesical LV-IFNα therapy.
In summary, our investigation demonstrates the feasibility of using lentiviral vectors for the treatment of BLCA. We also improved our mechanistic understanding of LV-IFNα-mediated cytotoxicity within BLCA cell lines and murine preclinical disease models. Future studies will directly compare the efficacy of adeno- and lentiviral-based IFNα gene therapy and the therapeutic potential of combination therapy regimens.
Materials and methods
Cell lines
Mouse urothelial cell line MB49/GFP-luciferase was a generous gift from Dr. Robert Svatek (The University of Texas Health, San Antonio), and UPPL1541 (derived from UPII PTEN/p53 null) and BBN975 (p53+/− mice treated with BBN) lines were a generous gift from Dr. William Kim (University of North Carolina). Cells were grown in minimum essential medium (MB49) or Dulbecco’s modified Eagle medium (BBN975 and UPPL1541) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin). For viral transduction, cells were seeded in 6-well culture dishes at varying densities (MB49 50,000; UPPL1541 75,000; and BBN975 75,000). After overnight attachment, media were replaced with polybrene-containing media (4 μg/mL) and viral particles were added at desired MOI. Recombinant proteins murine IFNα (100 U/mL), TRAIL (100 ng/mL), caspase 8 inhibitor (50 mM), and pancaspase inhibitor (50 mM) were also added to seeded cells. Trypan blue dye exclusion method (ViCell, Beckman Coulter Genomics) was used to determine cytotoxicity. Annexin V allophycocyanin and propidium iodide staining (Thermo Fisher Scientific, 88-8102-72) were used to determine percentage of apoptotic cells.
Viral vectors
FKD Therapies, University of Finland, provided lentiviral vectors (LV-GFP, LV-Ctrl, and LV-IFN). Briefly, Invitrogen’s Gateway cloning technology was used for cloning murine IFNα11 gene into pDONR211 vector to obtain pENTRY mIFNα11 and then subcloned into pSWOP shuttle vector to generate lentiviral murine IFNα11 vector. Viral particles with the vector were used to transduce HeLa cells to check efficiency of IFNα11 production. After confirmation of protein expression, the LV-IFNα was used to transduce BLCA cell lines.
qPCR analysis
RNA was extracted using Ambion miRVANA kit (AM1561) and quantified using NanoDrop ND-1000 spectrophotometer. Twenty nanograms of RNA was used along with AgPAth-ID One-Step RT-PCR kit (Thermo Fisher Scientific, 4387391) with Taqman probes to detect IRF7 (Mm00516788_m1), PD-L1 (Mm00452054), TRAIL (Mm01283606_m1), and GAPDH (Mm99999915_g1) on a StepOnePlus Real-Time PCR instrument. Comparative Ct method was used to determine relative gene expression.42
ELISA for murine IFNα
Cell-free supernatants from cells treated with the lentiviral vectors were collected, and levels of murine IFN were measured by ELISA (PBL, 42115–1). Urine IFNα levels were measured by Cloud Clone Corp murine IFNa11 kit (SE5090Mu).
Gene expression profiling
Whole transcriptome RNA-seq was performed on Ion Gene Studio S5 (Thermo Fisher Scientific). RNA was isolated from cell lines using miRVANA miRNA isolation kit (Thermo Fisher Scientific) using manufacturer’s instructions. Quality of RNA was assessed using Nanodrop ND-1000 spectrophotometer and by 4200 Tapestation (Agilent Technologies). Twenty nanograms of RNA was transcribed into cDNA using Ion AmpliSeq transcriptome mouse gene expression chef-ready kit (A36412). cDNA was amplified and subsequently ligated with adapters and barcode. Purified libraries were quantified using Ion library quantitation kit (Thermo Fisher Scientific) and pooled in a set of eight followed by enrichment on IonChef (Thermo Fisher Scientific). Enriched samples were then loaded onto Ion 540 chips and run on the Ion Gene Studio S5. Primary analysis of RNA-seq data was performed using AmpliseqRNA analysis plugin in the Torrent Suite software. Raw reads were aligned with mouse reference genome Ampliseq_Mouse_Transcriptome_V1_Reference. Raw counts and normalized reads per gene per million mapped reads (rpm) were generated, which was used for subsequent analysis. Unsupervised analysis was performed to identify outliers and assess overall similarities and differences among the samples. Differential expression analysis was performed using t tests, and FDR was estimated using the beta-uniform mixture method.43
Electron microscopy
Samples were fixed with a solution containing 3% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer at pH 7.3 and then washed in 0.1 M sodium cacodylate buffer and treated with 0.1% Millipore-filtered cacodylate buffered tannic acid, post-fixed with 1% buffered osmium, and stained en bloc with 1% Millipore-filtered uranyl acetate. The samples were dehydrated in increasing concentrations of ethanol, infiltrated, and embedded in LX-112 medium. The samples were polymerized in a 60°C oven for approximately 3 days. Ultrathin sections were made using a Leica Ultracut microtome (Leica, Deerfield, IL), stained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined in a JEM 1010 transmission electron microscope (JEOL, USA, Peabody, MA) at an accelerating voltage of 80 kV. Digital images were obtained using AMT Imaging System (Advanced Microscopy Techniques, Danvers, MA)
Western blotting
Cells were grown in monolayer and treated with lentiviral vectors. After 48 h, cells were washed once with phosphate-buffered saline (PBS) and scraped off the plates into whole-cell lysis buffer (50 mM Tris-HCl [pH 7.4]; 150 mM NaCl; 5 mM EDTA; 25 mM NaF; 1% Triton X-100; 1% NP-40; 0.1 mM Na3VO4; 12.5 mM β-glycerophosphate; 1 mM PMSF) containing protease and phosphatase inhibitors. Cell lysates were prepared by incubation on ice for 30–40 min with intermittent vortexing every 10 min. Protein concentration was measured (Micro BCA protein assay kit, Thermo Fisher Scientific), and 30 μg of protein was resolved on 4%–15% gradient gel (Bio-Rad) and transferred to a nitrocellulose membrane. The blots were blocked in 10% mild powder in PBS and probed with antibodies against PD-L1 (AF1019, Bio-Techne), STAT1 (33–1400, Invitrogen), pSTAT1 (Cell Signaling Technology, no. 9171), and β-actin (AC-74). Species-specific secondary antibodies were used to detect proteins bands using an enhanced chemiluminescence detection kit (GE Healthcare).
In vivo experiments
All animal experiments were conducted in compliance with the Institutional Animal Care and Use Committee guidelines at MD Anderson Cancer Center (Houston, TX). For checking gene transfer efficiency in murine bladder, 6- to 8-week-old female C57Bl/6 mice were anesthetized using isoflurane and the urethra was catheterized using a 20G angiocatheter. Bladders were flushed with PBS and instilled with lentiviral vectors/Syn3 (1 mg/mL) for 40 min. Mice were allowed to recover and returned to their cages. Urine was collected after instillations, and mouse bladders were collected for βgal staining or IFN ELISA.
Superficial bladder tumors were established using previously published protocols.44 Briefly, after anesthetizing mice with isoflurane, mouse urethra was catheterized and instilled with 100 μL of poly(l-lysine) (PLL) (0.01 μg/mL) for 15 min. After emptying the bladder, 25,000 MB49 cells in 100 μL of HBSS were instilled into mouse bladders for 30 min. Mice were allowed to recover, and tumor burden was assessed by IVIS Spectrum In vivo Imaging System and Living Image Software (PerkinElmer). Mice were treated intravesically with either Syn3 (vehicle), LV-Ctrl (3 × 107 virus particles), or LV-IFN (3 × 107 virus particles, treatment), and tumor growth was monitored. Mice with significant reduction in body weight, lethargy, and hematuria were deemed as moribund and were humanely euthanized. All experiments were repeated thrice.
Histology and immunostaining
Mouse tissues were fixed in buffered formalin, embedded in paraffin, and sectioned at the Research Histology core laboratory at MD Anderson Cancer Center. Immunostaining was performed with the specified antibodies, and species-specific horseradish-peroxidase-conjugated secondary antibodies were used to detect proteins using the 3,3-diaminobenzidine substrate kit (Vector Laboratories). Sections were counterstained with hematoxylin and mounted in Permount. Images were captured using Nanozoomer image scanner (Hamamatsu). For βgal staining, mouse bladders were fixed and stained as described earlier.45
Statistical methods
Statistical analysis was performed using GraphPad Prism 7 software. ANOVA was used to make multiple group comparisons with Tukey’s test, and results were considered significant when p < 0.05. Log rank test was used to perform the survival analysis.
Data availability statement
The RNA-seq data are available through NCBI Gene Expression Omnibus (GEO): GSE205493 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE205493).
Acknowledgments
We are grateful to Dr. William Kim (University of North Carolina, Chapel Hill) for donating BBN975 and UPPL1541 cells and Dr. Robert Svatek (University of Texas Health Science Center, San Antonio) for donating MB49-luc-tagged cells. We thank I-Ling Lee at MD Anderson for all the help with sequencing experiments. Research is supported in part by A.I. Virtanen Institute for Molecular Sciences (Kuopio, Finland) and MD Anderson CCSG program (P30 016672).
Author contributions
C.P.D., D.J.M., N.R.P., S.Y.-H., and S.M. conceived the project and wrote the manuscript. S.M., V.M.N., A.H.L., J.J.D., A.K., T.S.M., A.P.M., D.P., S.S.A., M.J.M., K.D., and T.N. performed experiments, developed reagents, and analyzed data. B.J. and K.S.S. helped in data interpretation.
Declaration of interests
C.P.D. received personal compensation from FKD Therapies, Oy for consulting and advisory services, including serving as the Independent Chairman of steering committee for the phase 3 nadofaragene firadenovec (rAd-IFNα/Syn3) trial.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2022.06.005.
Contributor Information
Sharada Mokkapati, Email: smokkapa@mdanderson.org.
Colin P. Dinney, Email: cdinney@mdanderson.org.
Supplemental information
References
- 1.Chang S.S., Boorjian S.A., Chou R., Clark P.E., Daneshmand S., Konety B.R., Pruthi R., Quale D.Z., Ritch C.R., Seigne J.D., et al. Diagnosis and treatment of non-muscle invasive bladder cancer: AUA/SUO guideline. J. Urol. 2016;196:1021–1029. doi: 10.1016/j.juro.2016.06.049. [DOI] [PubMed] [Google Scholar]
- 2.Kamat A.M., Colombel M., Sundi D., Lamm D., Boehle A., Brausi M., Buckley R., Persad R., Palou J., Soloway M., Witjes J.A. BCG-unresponsive non-muscle-invasive bladder cancer: recommendations from the IBCG. Nat. Rev. Urol. 2017;14:244–255. doi: 10.1038/nrurol.2017.16. [DOI] [PubMed] [Google Scholar]
- 3.Siddiqui M.R., Grant C., Sanford T., Agarwal P.K. Current clinical trials in non-muscle invasive bladder cancer. Urol. Oncol. 2017;35:516–527. doi: 10.1016/j.urolonc.2017.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wysocki P.J., Karczewska-Dzionk A., Mackiewicz-Wysocka M., Mackiewicz A., KArczewskA-Dzionk A., MAckiewicz A. Human cancer gene therapy with cytokine gene-modified cells. Expert Opin. Biol. Ther. 2004;4:1595–1607. doi: 10.1517/14712598.4.10.1595. [DOI] [PubMed] [Google Scholar]
- 5.Leick M.B., Maus M.V., Frigault M.J. Clinical perspective: treatment of aggressive B cell lymphomas with FDA-approved CAR-T cell Therapies. Mol. Ther. 2020;29:433–441. doi: 10.1016/j.ymthe.2020.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connor R.J., Engler H., Machemer T., Philopena J.M., Horn M.T., Sutjipto S., Maneval D.C., Youngster S., Chan T.M., Bausch J., et al. Identification of polyamides that enhance adenovirus-mediated gene expression in the urothelium. Gene Ther. 2001;8:41–48. doi: 10.1038/sj.gt.3301348. [DOI] [PubMed] [Google Scholar]
- 7.Dinney C.P., Fisher M.B., Navai N., O'Donnell M.A., Cutler D., Abraham A., Young S., Hutchins B., Caceres M., Kishnani N., et al. Phase I trial of intravesical recombinant adenovirus mediated interferon-α2b formulated in Syn3 for Bacillus calmette-guérin failures in nonmuscle invasive bladder cancer. J. Urol. 2013;190:850–856. doi: 10.1016/j.juro.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shore N.D., Boorjian S.A., Canter D.J., Ogan K., Karsh L.I., Downs T.M., Gomella L.G., Kamat A.M., Lotan Y., Svatek R.S., et al. Intravesical rAd–IFNα/syn3 for patients with high-grade, Bacillus calmette-guerin–refractory or relapsed non–muscle-invasive bladder cancer: a phase II randomized study. J. Clin. Oncol. 2017;35:3410–3416. doi: 10.1200/JCO.2017.72.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boorjian S.A., Alemozaffar M., Konety B.R., Shore N.D., Gomella L.G., Kamat A.M., Bivalacqua T.J., Montgomery J.S., Lerner S.P., Busby J.E., et al. Intravesical nadofaragene firadenovec gene therapy for BCG-unresponsive non-muscle-invasive bladder cancer: a single-arm, open-label, repeat-dose clinical trial. Lancet Oncol. 2020;22:107–117. doi: 10.1016/S1470-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinney C.P., Greenberg R.E., Steinberg G.D. Intravesical valrubicin in patients with bladder carcinoma in situ and contraindication to or failure after bacillus Calmette-Guérin. Urol. Oncol. 2013;31:1635–1642. doi: 10.1016/j.urolonc.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Balar A.V., Kamat A.M., Kulkarni G.S., Uchio E.M., Boormans J.L., Roumiguié M., Krieger L.E.M., Singer E.A., Bajorin D.F., Grivas P., et al. Pembrolizumab monotherapy for the treatment of high-risk non-muscle-invasive bladder cancer unresponsive to BCG (KEYNOTE-057): an open-label, single-arm, multicentre, phase 2 study. Lancet Oncol. 2021;22:919–930. doi: 10.1016/S1470-2045(21)00147-9. [DOI] [PubMed] [Google Scholar]
- 12.Papageorgiou A., Dinney C.P., McConkey D.J. Interferon-alpha induces TRAIL expression and cell death via an IRF-1-dependent mechanism in human bladder cancer cells. Cancer Biol. Ther. 2007;6:872–879. doi: 10.4161/cbt.6.6.4088. [DOI] [PubMed] [Google Scholar]
- 13.Dinney C.P., Bielenberg D.R., Perrotte P., Reich R., Eve B.Y., Bucana C.D., Fidler I.J. Inhibition of basic fibroblast growth factor expression, angiogenesis, and growth of human bladder carcinoma in mice by systemic interferon-alpha administration. Cancer Res. 1998;58:808–814. [PubMed] [Google Scholar]
- 14.Izawa J.I., Sweeney P., Perrotte P., Kedar D., Dong Z., Slaton J.W., Karashima T., Inoue K., Benedict W.F., Dinney C.P.N. Inhibition of tumorigenicity and metastasis of human bladder cancer growing in athymic mice by interferon-beta gene therapy results partially from various antiangiogenic effects including endothelial cell apoptosis. Clin. Cancer Res. 2002;8:1258–1270. [PubMed] [Google Scholar]
- 15.Plote D., Choi W., Mokkapati S., Sundi D., Ferguson J.E., 3rd, Duplisea J., Parker N.R., Yla-Herttuala S., Committee S.C.B., McConkey D., et al. Inhibition of urothelial carcinoma through targeted type I interferon-mediated immune activation. Oncoimmunology. 2019;8:e1577125. doi: 10.1080/2162402X.2019.1577125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diamond M.S., Kinder M., Matsushita H., Mashayekhi M., Dunn G.P., Archambault J.M., Lee H., Arthur C.D., White J.M., Kalinke U., et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J. Exp. Med. 2011;208:1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vorburger S.A., Hunt K.K. Adenoviral gene therapy. Oncologist. 2002;7:46–59. doi: 10.1634/theoncologist.7-1-46. [DOI] [PubMed] [Google Scholar]
- 18.Park F. Lentiviral vectors: are they the future of animal transgenesis? Physiol. Genom. 2007;31:159–173. doi: 10.1152/physiolgenomics.00069.2007. [DOI] [PubMed] [Google Scholar]
- 19.Vannucci L., Lai M., Chiuppesi F., Ceccherini-Nelli L., Pistello M. Viral vectors: a look back and ahead on gene transfer technology. New Microbiol. 2013;36:1–22. [PubMed] [Google Scholar]
- 20.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C., et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S., et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 22.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K., et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito R., Smith C.C., Utsumi T., Bixby L.M., Kardos J., Wobker S.E., Stewart K.G., Chai S., Manocha U., Byrd K.M., et al. Molecular subtype-specific immunocompetent models of high-grade urothelial carcinoma reveal differential neoantigen expression and response to immunotherapy. Cancer Res. 2018;78:3954–3968. doi: 10.1158/0008-5472.CAN-18-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamashita M., Rosser C.J., Zhou J.H., Zhang X.Q., Connor R.J., Engler H., Maneval D.C., Karashima T., Czerniak B.A., Dinney C.P.N., Benedict W.F. Syn3 provides high levels of intravesical adenoviral-mediated gene transfer for gene therapy of genetically altered urothelium and superficial bladder cancer. Cancer Gene Ther. 2002;9:687–691. doi: 10.1038/sj.cgt.7700488. [DOI] [PubMed] [Google Scholar]
- 25.Voskoboinik I., Whisstock J.C., Trapani J.A. Perforin and granzymes: function, dysfunction and human pathology. Nat. Rev. Immunol. 2015;15:388–400. doi: 10.1038/nri3839. [DOI] [PubMed] [Google Scholar]
- 26.Kajon A.E., Lamson D.M., Bair C.R., Lu X., Landry M.L., Menegus M., Erdman D.D., St George K. Adenovirus type 4 respiratory infections among civilian adults, northeastern United States, 2011-2015(1) Emerg. Infect. Dis. 2018;24:201–209. doi: 10.3201/eid2402.171407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mingozzi F., High K.A. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood. 2013;122:23–36. doi: 10.1182/blood-2013-01-306647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valkama A.J., Oruetxebarria I., Lipponen E.M., Leinonen H.M., Käyhty P., Hynynen H., Turkki V., Malinen J., Miinalainen T., Heikura T., et al. Development of large-scale downstream processing for lentiviral vectors. Mol. Ther. Methods Clin. Dev. 2020;17:717–730. doi: 10.1016/j.omtm.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benedict W.F., Tao Z., Kim C.S., Zhang X., Zhou J.H., Adam L., McConkey D.J., Papageorgiou A., Munsell M., Philopena J., et al. Intravesical Ad-IFNα causes marked regression of human bladder cancer growing orthotopically in nude mice and overcomes resistance to IFN-α protein. Mol. Ther. 2004;10:525–532. doi: 10.1016/j.ymthe.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 30.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakagawa T., Yuan J. Cross-talk between two cysteine protease families. J. Cell Biol. 2000;150:887–894. doi: 10.1083/jcb.150.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakagawa T., Zhu H., Morishima N., Li E., Xu J., Yankner B.A., Yuan J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 33.Borsini A., Cattaneo A., Malpighi C., Thuret S., Harrison N.A., Consortium M.R.C.I., Zunszain P.A., Pariante C.M. Interferon-alpha reduces human hippocampal neurogenesis and increases apoptosis via activation of distinct STAT1-dependent mechanisms. Int. J. Neuro. psycho. pharmacol. 2018;21:187–200. doi: 10.1093/ijnp/pyx083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regis G., Icardi L., Conti L., Chiarle R., Piva R., Giovarelli M., Poli V., Novelli F. IL-6, but not IFN-gamma, triggers apoptosis and inhibits in vivo growth of human malignant T cells on STAT3 silencing. Leukemia. 2009;23:2102–2108. doi: 10.1038/leu.2009.139. [DOI] [PubMed] [Google Scholar]
- 35.Fehrenbacher N., Bastholm L., Kirkegaard-Sørensen T., Rafn B., Bøttzauw T., Nielsen C., Weber E., Shirasawa S., Kallunki T., Jäättelä M. Sensitization to the lysosomal cell death pathway by oncogene-induced down-regulation of lysosome-associated membrane proteins 1 and 2. Cancer Res. 2008;68:6623–6633. doi: 10.1158/0008-5472.CAN-08-0463. [DOI] [PubMed] [Google Scholar]
- 36.Roger J., Chalifour A., Lemieux S., Duplay P. Cutting edge: ly49A inhibits TCR/CD3-induced apoptosis and IL-2 secretion. J. Immunol. 2001;167:6–10. doi: 10.4049/jimmunol.167.1.6. [DOI] [PubMed] [Google Scholar]
- 37.Shi L., Chen G., MacDonald G., Bergeron L., Li H., Miura M., Rotello R.J., Miller D.K., Li P., Seshadri T., et al. Activation of an interleukin 1 converting enzyme-dependent apoptosis pathway by granzyme B. Proc. Natl. Acad. Sci. U S A. 1996;93:11002–11007. doi: 10.1073/pnas.93.20.11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van de Wetering D., de Paus R.A., van Dissel J.T., van de Vosse E. IL-23 modulates CD56+/CD3- NK cell and CD56+/CD3+ NK-like T cell function differentially from IL-12. Int. Immunol. 2009;21:145–153. doi: 10.1093/intimm/dxn132. [DOI] [PubMed] [Google Scholar]
- 39.Clark D.W., Palle K. Aldehyde dehydrogenases in cancer stem cells: potential as therapeutic targets. Ann. Transl Med. 2016;4:518. doi: 10.21037/atm.2016.11.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Januchowski R., Wojtowicz K., Zabel M. The role of aldehyde dehydrogenase (ALDH) in cancer drug resistance. Biomed. Pharmacother. 2013;67:669–680. doi: 10.1016/j.biopha.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Massari F., Ciccarese C., Santoni M., Brunelli M., Conti A., Modena A., Montironi R., Santini D., Cheng L., Martignoni G., et al. The route to personalized medicine in bladder cancer: where do we stand? Target. Oncol. 2015;10:325–336. doi: 10.1007/s11523-015-0357-x. [DOI] [PubMed] [Google Scholar]
- 42.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Pounds S., Morris S.W. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19:1236–1242. doi: 10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- 44.Ang W.X., Zhao Y., Kwang T., Wu C., Chen C., Toh H.C., Mahendran R., Esuvaranathan K., Wang S. Local immune stimulation by intravesical instillation of baculovirus to enable bladder cancer therapy. Sci. Rep. 2016;6:27455. doi: 10.1038/srep27455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mokkapati S., Niopek K., Huang L., Cunniff K.J., Ruteshouser E.C., deCaestecker M., Finegold M.J., Huff V. β-Catenin activation in a novel liver progenitor cell type is sufficient to cause hepatocellular carcinoma and hepatoblastoma. Cancer Res. 2014;74:4515–4525. doi: 10.1158/0008-5472.CAN-13-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data are available through NCBI Gene Expression Omnibus (GEO): GSE205493 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE205493).