Abstract
We have previously hypothesized that sulfide inhibits Hg methylation by decreasing its bioavailability to sulfate-reducing bacteria (SRB), the important methylators of Hg in natural sediments. With a view to designing a bioassay to test this hypothesis, we investigated a number of aspects of Hg methylation by the SRB Desulfobulbus propionicus, including (i) the relationship between cell density and methylmercury (MeHg) production, (ii) the time course of Hg methylation relative to growth stage, (iii) changes in the bioavailability of an added inorganic Hg (HgI) spike over time, and (iv) the dependence of methylation on the concentration of dissolved HgI present in the culture. We then tested the effect of sulfide on MeHg production by this microorganism. These experiments demonstrated that under conditions of equal bioavailability, per-cell MeHg production was constant through log-phase culture growth. However, the methylation rate of a new Hg spike dramatically decreased after the first 5 h. This result was seen whether methylation rate was expressed as a fraction of the total added Hg or the filtered HgI concentration, which suggests that Hg bioavailability decreased through both changes in Hg complexation and formation of solid phases. At low sulfide concentration, MeHg production was linearly related to the concentration of filtered HgI. The methylation of filtered HgI decreased about fourfold as sulfide concentration was increased from 10−6 to 10−3 M. This decline is consistent with a decrease in the bioavailability of HgI, possibly due to a decline in the dissolved neutral complex, HgS0.
The speciation of trace metals in aqueous systems can control their bioavailability to aquatic organisms. Active cellular uptake of many metals is dependent on either the concentration of free metal ion (e.g., Cu) or the concentration of specifically complexed forms (e.g., Fe) (30). In contrast, passive diffusion may be the important transport mechanism for Hg. The uptake of Hg by diatoms (27, 28) and diffusive transport of Hg across artificial membranes (20) were shown to be a function of the concentration of neutral HgCl20. Mason et al. (28) further showed that the octanol-water partitioning coefficient (Kow) of a Hg complex was proportional to its permeability to cell membranes, so Kow could be used as a measure of the bioavailability of Hg.
The uptake mechanism for inorganic Hg (HgI) by sulfate-reducing bacteria (SRB) prior to methylation is not known. Hg has no known physiological function in bacteria, and the ability to methylate Hg does not confer added resistance among SRB (21). In the one strain where methylation pathways have been examined (Desulfovibrio desulfuricans LS), methylmercury (MeHg) is produced in an accidental side reaction of a metabolic pathway (12, 13). Therefore, it is not likely that an active Hg transport pathway has evolved in SRB. A limited number of experiments with SRB suggest that Hg uptake is not an active process (21). For these reasons, passive diffusion across the cell membrane has been proposed as the important uptake mechanism for methylating bacteria (3, 4, 21). We have further hypothesized that under the sulfidic conditions typically surrounding SRB, HgS0 is the dominant neutral complex controlling HgI uptake (3, 5).
Support for this hypothesis comes from a model for Hg partitioning and speciation in sulfidic pore waters (3) that shows a strong relationship between predicted HgS0 concentration in the dissolved phase and MeHg concentration in bulk sediment. Further, determination of overall octanol-water partitioning coefficients (Dow) for Hg across a sulfide gradient showed a decline in Dow and experimentally supported the modeled decline in HgS0 concentration (5). We have suggested that this marked decrease in Dow of Hg at high (millimolar) sulfide concentrations leads to decreased bioavailability of HgI for methylation, and that it may explain the low levels of MeHg frequently observed in high-sulfide environments (4, 16, 19).
In the classic experiments where free ion Cu uptake by phytoplankton was demonstrated (e.g., references 1 and 35), ambient Cu speciation in culture medium was controlled with strong organic ligands, and toxicity was used as the endpoint indicating Cu transport into the cell. We wanted to devise a similar experimental system in which we manipulated the sulfide speciation of Hg in SRB cultures and used MeHg production as a surrogate for uptake. In this way, we could test whether our predicted (using chemical equilibrium calculations) HgS0 concentration could explain the changes in Hg methylation rates and MeHg concentrations that are observed across sulfide gradients in natural sediments.
Pursuant to this work, we have carried out a number of Hg methylation studies using pure cultures of Desulfobulbus propionicus (strain 1pr3). This microorganism was chosen because it is an efficient Hg methylator under sulfate-reducing and fermentative conditions, is well described, and is available in culture collections. In this paper we describe a set of experiments designed to investigate the effect of such variables as the concentration of Hg and sulfide on Hg bioavailability. They are not intended to address how pathways of carbon metabolism influence methylation, although this variable undoubtedly works in concert with bioavailability to control Hg methylation rates in natural sediments.
MATERIALS AND METHODS
Microorganism and culture conditions.
D. propionicus strain 1pr3 (DSM 2302 and ATCC 33891) was used in all experiments. Incomplete oxidation of propionate to acetate is characteristic (25, 34, 37). The genus Desulfobulbus along with the genera Desulfocapsa, Desulforhopalus, and Desulfofustis represent a deeply branching group within the family Desulfobacteriaceae, possibly a separate family (10). This organism is capable of Hg methylation under fermentative and sulfate-reducing conditions (2, 24; R. Devereux, M. R. Winfrey, C. C. Gilmour, and D. A. Stahl, unpublished data). Unless otherwise noted, experiments were carried out at 27°C in a medium that supports fermentative growth of D. propionicus. Because we wanted to be able to control the sulfide concentration in the cultures, S-containing organic substrates and reductants were excluded to prevent sulfide production from the breakdown of these compounds. Further, we wanted to limit the potential organic ligands for Hg. The medium contained pyruvate as an organic carbon source, no sulfate, and Ti-nitrilotriacetic acid (NTA) as a reductant. To make the medium, pyruvate (24 mM), salts (1.7 mM NaH2PO4, 4.7 mM NH4Cl, and 6.7 mM KCl), trace metal (0.5 ml liter−1) and vitamin (5 ml liter−1) stock solutions (20), 24 nM selenate, 25 nM tungstate, 3.6 μM FeCl2, and 48 mM carbonate (final pH 7.2) were mixed, boiled under O2-free 90% N2–10% CO2, dispensed into acid-washed bottles, sealed with rubber stoppers, and autoclaved. Resazurin at 1 mg liter−1 was used as a redox indicator. Just prior to use, MgCl2 and CaCl3 were injected into sealed bottles to final concentrations of 1.4 mM Ca and 3.1 mM Mg. Reductant (0.1 mM Ti-NTA [23]) was also added just prior to use. The Ti-NTA concentration was chosen based on previous tests of toxicity versus the ability to maintain adequately reduced medium for SRB growth (21), and it was used in all experiments except where noted.
Fermentatively grown cultures of D. propionicus were maintained between experiments on medium containing yeast extract, because we were unable to carry the cultures through successive transfers without it as an S source. Therefore, a small carryover of sulfide into test medium generally occurred, resulting in sulfide concentrations of about 1 μM without added sulfide. Sulfide concentrations were routinely monitored during experiments. Inoculation was at a ratio of 1 ml of inoculum to 50 ml of medium. Cell growth was monitored by measuring absorbance at 660 nm (optical density [OD]), and growth was terminated by freezing the cultures.
The relationship between OD and cell density (cells per milliliter) was determined directly in fermentatively grown cultures under conditions similar to those of the experiments. This relationship was examined to test its linearity and to provide a conversion factor from OD to cell density in other experiments. Cell counts were made using epifluorescence microscopy with acridine orange as the stain.
Subsampling.
Subsamples were taken from the cultures via degassed syringe. For analysis of filterable constituents, aliquots were passed through 0.2-μm-pore-size Acrodisc filter units. Acrodisc blanks for Hg contamination were determined by filtering deionized water through the units and found to be <1 ng of Hg liter−1. Sorption checks performed by filtering solutions containing 1,000 ng of Hg liter−1 in deionized water showed that less than 10% of the Hg was sorbed by the filters. Subsamples for total Hg or MeHg analysis were dispensed into Teflon vials, diluted with deionized water, and acidified to 0.5% with HCl as a preservative.
Analytical methods.
Total mercury (HgT) samples were digested overnight with BrCl and analyzed by SnCl2 reduction, gold amalgamation, and cold vapor atomic fluorescence detection (7, 18). MeHg concentration was determined by distillation (22, 23), followed by aqueous-phase derivitization and cold vapor atomic fluorescence detection (6). HgI concentration was calculated as the difference between HgT and MeHg. Dissolved gaseous Hg (DGM) was determined by purging and trapping undigested culture subsamples in the absence of SnCl2. Unfiltered HgT concentration in the medium was <10 ng liter−1 without added Hg spike. Unfiltered MeHg concentrations in uninoculated medium incubated with 1,000 ng of Hg liter−1 were indistinguishable from that in unspiked medium (<1 ng liter−1).
To prevent loss via oxidation, sulfide samples were not filtered. Sulfide samples were preserved in sulfide antioxidant buffer (9). Sulfide concentration was determined using a silver-sulfide ion-specific electrode.
Methylation in diluted cultures.
The effect of cell density on MeHg production was examined in diluted culture in order to keep other variables, including stage in life cycle, equal. A stationary-phase fermentative culture of D. propionicus was inoculated into fresh medium at 0.1 mM sulfide with dilution factors of 0.01, 0.02, 0.05, and 0.1 (volume of culture/volume of medium). Cultures were spiked to 800 ng/liter with Hg(II) standard (in 0.5% HNO3) and incubated for 4 h. Cultures were then frozen and saved for unfiltered MeHg analysis.
Methylation time course.
The methylation time course experiment was designed to monitor the production of MeHg by cultures over a growth cycle. The medium differed from that used in later experiments in that it contained 24 mM lactate instead of pyruvate as the organic carbon source, plus 0.5 g of yeast extract liter−1. Twelve bottles of medium were spiked with Hg(II) standard (in 0.5% HCl with an excess of BrCl) to a final concentration of 2,000 ng liter−1. After an equilibration period of 4 days, sulfide was added to six of the bottles to a final concentration of 1 mM (sulfide+ cultures). All bottles were inoculated 1 day later. In this experiment only, inoculation was from culture grown in medium with an initial sulfate concentration of 28 mM; consequently there was a carryover of 0.55 mM sulfate, which the cultures were able to respire to sulfide during the course of the experiment. Subsampling for sulfide and filtered Hg was performed 5, 7, and 13 days after inoculation. After subsampling, two each of the ambient and sulfide+ cultures were killed and saved for unfiltered MeHg determination. The conditions of this experiment are the same as those of a study of solid-phase mercury methylation reported elsewhere (2).
Mercury bioavailability time course.
Time course experiments were carried out to determine how the dissolved concentration and bioavailability of added Hg change over time in culture medium and to examine the effects of adding the Hg spike before and after inoculation. Medium with (Ti+) and without (Ti−) reductant was prepared. Two bottles of each type (called A cultures to denote that Hg was added after inoculation) were inoculated. At the same time, Hg(II) standard (in 0.5% HCl with an excess of BrCl) was added to 1,000 ng liter−1 and Na2S stock was added to 0.01 mM in two of each type of medium (called B cultures to denote that Hg was added before inoculation). Three days later, B cultures were inoculated, and A cultures were spiked with Hg and sulfide. After 5 h, filtered and unfiltered subsamples were taken from the A cultures. All cultures were allowed to grow for an additional 3 days before subsampling and freezing. Thus, the A cultures allowed determination of the short-term bioavailability of HgI to growing cultures within 5 h of addition and the long-term bioavailability of this spike over 3 days. The B cultures, on the other hand, were used to assess the long-term bioavailability of Hg that had been preequilibrated with medium prior to inoculation.
To check whether any DGM was produced in the cultures, either through reduction to Hg0 by Ti or via dimethylmercury production, DGM was determined on days 4 and 6 of the experiment. These times correspond to 4 and 6 days after the inoculation of A cultures and 1 and 3 days after inoculation of the B cultures.
Filtered controls were prepared by growing two cultures at 1 μM sulfide with no added Hg for 3 days, filtering through a 0.2-μm-pore-size Nalge filter unit in a glove box filled with O2-free N2, returning cell-free filtrates to sterile serum bottles, and incubating the bottles in the presence of HgI at 1,000 ng liter−1. These controls were used to ascertain that there was no significant MeHg production by spent culture medium in the absence of cells.
Concentration dependence.
The relationship between added HgI concentration and the amount of MeHg produced by cultures was examined across a concentration gradient of 0, 100, 400, 700, and 1,000 ng liter−1. Ten days prior to inoculation, medium was brought to 1 μM sulfide, and the concentration gradient was set up using Hg(II) standard (in 0.5% HNO3). Filtered and unfiltered subsamples were taken 1 h after inoculation (time zero [T = 0] for the experiment), and then cultures were incubated for 6 days (i.e., to T = 6). Previous chemical equilibrium modeling indicated that essentially all of the dissolved HgI should be present as HgS0 at this sulfide concentration (3). Average OD in the cultures at the end of the experiment was 0.166 ± 0.011 (2.9 × 107 cells ml−1).
This Hg gradient experiment was also designed to address the relationships between the Hg addition level and the resultant filtered HgI concentration at a constant sulfide concentration. Linearity between unfiltered HgI and filtered HgI would suggest that filtered HgI was controlled by sorption to solid phases (including cells), while a plateau in filtered HgI would suggest Hg precipitation at the higher Hg addition levels. Furthermore, a linear relationship between filtered HgI and methylation rate would be consistent with passive diffusion across the membrane, while a leveling-off of methylation would suggest saturation of an enzyme-mediated transport system.
Methylation across a sulfide gradient.
To investigate the effect of sulfide on filtered HgI concentration and MeHg production in cultures, medium was prepared with a gradient in sulfide concentration of 10−3, 10−4, 10−5, and 10−6 M (duplicates at each concentration), and Hg(II) standard (in 0.5% HCl with an excess of BrCl) was added to a concentration of 1,000 ng liter−1. Three days later, all media were inoculated, and cultures were incubated for 3 days. Filtered subsamples were taken for HgT and MeHg, and the remainder was frozen.
RESULTS
Methylation in diluted cultures.
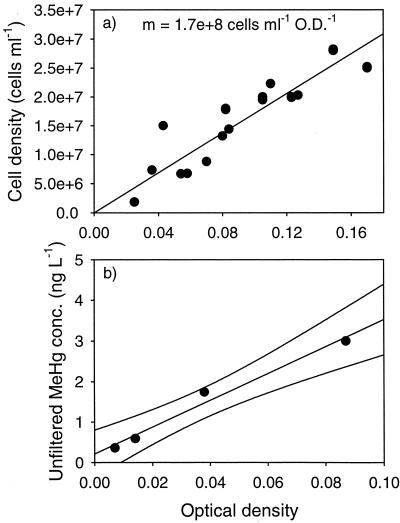
The cell density of cultures grown fermentatively under the conditions of these experiments was linearly related to measured OD, as shown in Fig. 1a, with a conversion factor of 1.7 × 108 cells ml−1 OD−1. Final MeHg concentration was also linearly related to OD in culture diluted up to 10 times (Fig. 1b); therefore, Hg methylation is also a linear function of cell density. Adsorption of Hg onto cell surfaces could deplete Hg from the dissolved phase, especially at higher cell densities, thereby lowering bioavailability of Hg for methylation. Within the range of OD examined in this experiment, this cell density effect was not observed. As discussed below, sorption onto cell surfaces appears to enhance, rather than hinder, diffusive uptake.
FIG. 1.
Relationship between cell density and MeHg production. (a) OD versus cell density; (b) OD in dilutions of a culture of D. propionicus versus final MeHg concentration. Spike concentration was 800 ng of Hg liter−1, and cultures were incubated for 4 h. The culture dilutions were not duplicated; lines represent the best fit determined by linear regression of the data, r2 = 0.98.
Methylation time course.
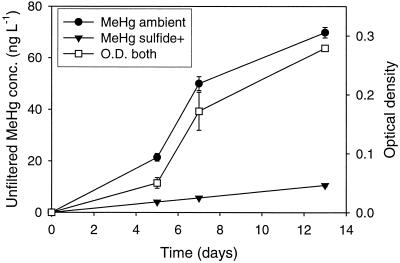
Increases in the concentration of MeHg in culture closely followed the growth curve of D. propionicus (Fig. 2). Similarly, Choi and Bartha (11) showed that MeHg production paralleled protein synthesis in D. desulfuricans LS cultures. Using the measured relationship between OD and cell density in the medium, we expressed MeHg production rates on a per-cell basis in order to examine potential changes in this rate during batch culture growth. The cellular MeHg production rate was 1.07 ag of cell−1 day−1 through day 5, 0.81 ag of cell−1 day−1 between days 5 and 7, and 0.10 ag of cell−1 day−1 between 7 and 13 days. These results suggest that per-cell methylation rate declined later in the growth cycle, when the growth rate decreased (Fig. 2). The time-averaged sulfide in these cultures was 0.55 mM, which resulted from the reduction of sulfate.
FIG. 2.
Total MeHg production and growth of D. propionicus in cultures with (sulfide+) and without (ambient) added sulfide during the methylation time course experiment. Averages for duplicate cultures are shown for MeHg concentration, averages for quadruplicate cultures (both types combined) are shown for OD, and error bars represent 1 standard deviation. Error bars that are not discernible fall within the symbol.
By comparison, overall and per-cell MeHg production was less in a parallel set of cultures grown in medium with 1.4 mM added sulfide (Fig. 2 and Table 1). Growth was similar at both sulfide concentrations, as evidenced by a final OD of 0.282 ± 0.004 in the ambient cultures and 0.276 ± 0.004 in the sulfide+ cultures; therefore, sulfide toxicity does not appear to be responsible for this difference. The magnitude of the difference in methylation in the ambient versus sulfide+ cultures depends on the manner in which it is expressed. Methylation can be expressed as turnover of the filtered HgI pool or as a fraction of the total Hg spike. The significance of each is discussed below.
TABLE 1.
Methylation of HgI spike by D. propionicus with and without added sulfidea
| Culture | Concn (ng liter−1)
|
Turnover | Fraction of spike methylated | |
|---|---|---|---|---|
| Unfiltered MeHg | Filtered HgI | |||
| Ambient A | 67.8 | 76.9 | 0.88 | 0.034 |
| Ambient B | 71.9 | 67.6 | 1.06 | 0.036 |
| Avg ± SE | 0.97 ± 0.09 | 0.035 ± 0.001 | ||
| Sulfide + A | 10.0 | 13.3 | 0.75 | 0.005 |
| Sulfide + B | 11.2 | 20.0 | 0.56 | 0.006 |
| Avg ± SE | 0.65 ± 0.10 | 0.005 ± 0.0003 | ||
Sulfide concentrations were 0.55 mM in ambient cultures and 1.4 mM in sulfide+ cultures; spike concentration was 2,000 ng Hg liter−1. Turnover = [unfiltered MeHg]/[filtered HgI]; fraction methylated = [unfiltered MeHg]/[spike HgI].
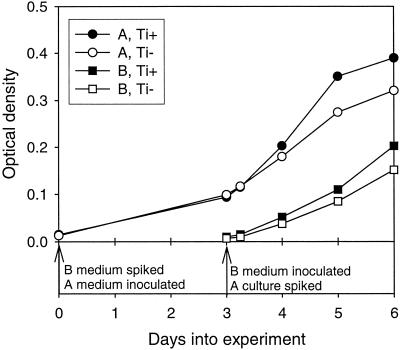
Mercury bioavailability time course.
The bioavailability of HgI added to medium before and after inoculation was determined, with MeHg production used as a surrogate for HgI bioavailability and uptake. The time line of this experiment is illustrated in Fig. 3. On day 0 of the experiment, Hg and sulfide were added to the B bottles, and the A bottles were inoculated. On day 3, B bottles were inoculated, and Hg and sulfide were added to A cultures. Five hours after the additions, subsamples were taken from A cultures for filtered and unfiltered HgT and MeHg in order to determine the short-term methylation rate. All cultures were further incubated until day 6, when the experiment was terminated.
FIG. 3.
Time line for Hg bioavailability time course experiment. Growth in the presence and absence of 0.1 M Ti-NTA is shown for A and B cultures.
The methylation rate (fraction of the total added HgI spike methylated per day) was much higher when Hg was added to growing cultures than when inoculation was into medium preequilibrated with Hg and sulfide, as indicated by the overall methylation rates given in Table 2. Further, the design of this experiment allowed comparison of methylation rates over the first 5 h versus the subsequent 3 days in the A cultures (Table 2). Clearly, the initial methylation rate was much greater even though the density of the culture increased throughout the experimental period (Fig. 3). The methylation rate in filtered, spent-medium controls was less than 1% of that in the A cultures, which shows that cells are required for Hg methylation (Table 2).
TABLE 2.
Methylation of HgI spike in time course experimentsa
| Culture | Methylation rate (day−1)
|
||
|---|---|---|---|
| First 5 h | 5 h–3 days | Overall (3 days) | |
| Filtered control | 0.008 | ||
| A + Ti-NTA | 1.11 | 0.047 | 0.13 |
| A − Ti-NTA | 1.14 | 0.065 | 0.15 |
| Avg ± SE | 1.13 ± 0.02 | 0.056 ± 0.013 | 0.14 ± 0.01 |
| B + Ti-NTA | 0.003 | ||
| B − Ti-NTA | 0.004 | ||
| Avg ± SE | 0.004 ± 0.001 | ||
Spike concentration was 1,000 ng of Hg liter−1.
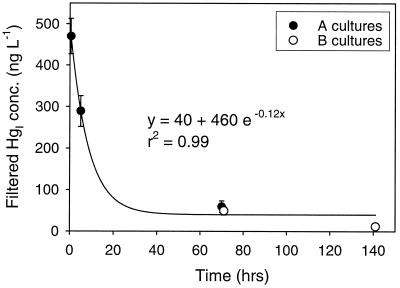
The concentration of filtered HgI declined rapidly after spike addition (Fig. 4). Therefore, it may be more meaningful to express methylation as turnover of the filtered substrate pool, where turnover = [unfiltered MeHg]/[filtered HgI], assuming that dissolved Hg species are the substrate for uptake and methylation. This way, effects of adsorption and/or precipitation on the availability of the spike are taken into account. Turnover of filtered HgI for the different time periods and culture types are summarized in Table 3.
FIG. 4.
Kinetics of the loss of added HgI from solution in the Hg bioavailability time course experiment. Filtered HgI is plotted for both experiments relative to the time of spike addition. Initial spike concentration was 1,000 ng liter−1. Averages for replicate cultures are shown; error bars represent 1 standard deviation. Error bars that are not discernible fall within the symbol.
TABLE 3.
Turnover by D. propionicusa
| Culture | First 5 h
|
5 h–3 days
|
||||
|---|---|---|---|---|---|---|
| Filtered HgI (ng liter−1) | Unfiltered MeHg (ng liter−1) | Turnover | Filtered HgI (ng liter−1) | Unfiltered MeHg (ng liter−1) | Turnover | |
| A + Ti-NTA | 420 | 233 | 3.3 | 88 | 129 | 0.26 |
| A − Ti-NTA | 340 | 240 | 4.1 | 102 | 180 | 0.37 |
| Avg ± SE | 3.7 ± 0.39 | 0.31 ± 0.054 | ||||
| B + Ti-NTA | 25 | 10 | 0.17 | |||
| B − Ti-NTA | 36 | 12 | 0.20 | |||
| Avg ± SE | 0.18 ± 0.013 | |||||
Turnover = [unfiltered MeHg]/[filtered HgI] per billion cells per day. Spike concentration was 1,000 ng of Hg liter−1.
As shown in Fig. 3, cultures grew better in the presence of Ti-NTA. However, Ti-NTA did not help preserve sulfide in the cultures, and methylation rates were not dramatically affected by the presence or absence of Ti-NTA (Tables 2 and 3). There were small and similar declines in sulfide concentration in all cultures over time (results not shown). The average DGM concentration in the cultures was 0.45 ng liter−1 on day 4 (detection limit [D.L.], 0.45 ng liter−1) and 0.66 ng liter−1 on day 6 (D.L. 0.51 ng liter−1), with no difference in DGM production in Ti+ compared to Ti− cultures. These values indicate that less than 0.07% of the added spike could have been lost via DGM production in the cultures. This can be compared to 40% (A cultures) and 1% (B cultures) of the spike converted to MeHg over the course of the experiment. The method used for DGM is nonspecific, and so this result indicates that neither dimethylmercury nor Hg0 was significantly formed.
Unfiltered HgT concentrations at the end of this experiment averaged 1,017 ± 126 ng liter−1, the same as the 1,000-ng liter−1 spike level, which shows that the loss of HgI from solution was not due to sorption to bottle walls. Instead, it appears that the spike partitioned onto suspended particles, including cells, over the course of the experiment. It is interesting that unfiltered HgI in the filtered, spent-medium controls averaged 829 ± 20 ng liter−1; thus, even in the absence of cells, there was significant formation of particles that precipitated or scavenged Hg from solution.
Concentration dependence.
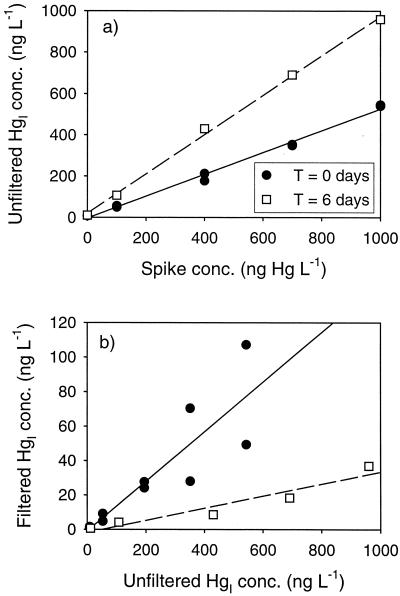
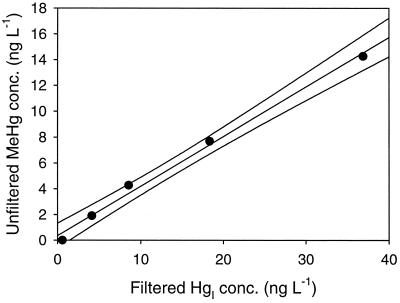
We examined the relationship between added Hg spike level, the resultant filtered and unfiltered HgI, and MeHg production. The unfiltered HgI concentration in the cultures was linearly related to the concentration of the Hg spike both shortly after inoculation (T = 0) and at the end of the experiment (T = 6). However, the slope of this relationship increased over time (Fig. 5a), which suggests a shift in Hg partitioning from the bottle walls to suspended solids, i.e., cells and precipitates, as the culture matured. The importance of adsorption as the mechanism controlling filtered HgI in these cultures is evidenced by the linear relationship between unfiltered and filtered HgI seen in Fig. 5b. The sharp decline in filtered HgI at T = 6 compared to T = 0 is consistent with production of particles in the form of cells and unknown precipitates over the course of the experiment. The final filtered HgI gradient in the experiment ranged from 0.9 to 41 ng liter−1. Over this concentration range, the MeHg produced was linearly correlated with filtered HgI concentration (Fig. 6).
FIG. 5.
Partitioning of the HgI spike in the concentration dependence experiment. (a) Addition level versus unfiltered HgI; (b) unfiltered HgI versus filtered HgI. Results from duplicate cultures are shown for T = 0.
FIG. 6.
Relationship between filtered HgI and MeHg in the concentration dependence experiment after 6 days of incubation. The individual Hg concentrations were not duplicated; lines represent the best fit determined by linear regression of the data, r2 = 0.99.
Sulfide gradient experiment.
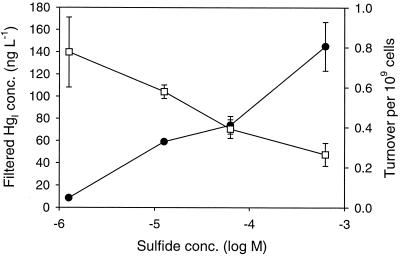
The filtered HgI concentration was found to increase with increasing sulfide, but turnover of the HgI pool into MeHg decreased (Fig. 7). The growth of the cultures in this experiment was uneven; in particular, growth was very poor in the cultures containing lower sulfide concentration. In order to make comparisons across the sulfide gradient, we expressed turnover on a cellular basis. Thus, the turnover in Fig. 7 is given per billion cells. All cultures were incubated for 3 days.
FIG. 7.
Methylation across a sulfide gradient. Plots show filtered HgI, (solid circles) and turnover ([unfiltered MeHg]/[filtered HgI]) per billion cells (open squares) across a range of sulfide concentrations. Spike concentration was 1,000 ng liter−1, and cultures were incubated for 3 days. Averages for duplicate cultures are shown; error bars represent 1 standard deviation.
DISCUSSION
These experiments demonstrate the ability of D. propionicus 1pr3 to methylate Hg under both sulfate-reducing conditions (methylation time course experiment, 0.5 mM sulfate) and fermentative conditions (other experiments). Due to the differences in medium chemistry, carbon substrate, and Hg addition levels, these experiments do not allow a direct comparison of relative Hg methylation rates during respiratory versus fermentative growth. Previous studies have shown Hg methylation by D. desulfuricans LS in the absence of sulfate during fermentation of pyruvate (11, 15) and during growth on lactate in coculture with a H2-utilizing methanogen (32). These results, along with those of the present study, indicate that sulfate reduction is not required for Hg methylation. These findings are in contrast to those of King et al. (24), who concluded of that Hg methylation is directly coupled to sulfate reduction based on the observation that pure cultures of D. desulfuricans and Desulfobacterium sp. strain BG-33 did not methylate Hg in the absence of sulfate. However, it is not clear whether these organisms were capable of fermentative growth under the culture conditions of their experiments.
A mercury methylation pathway has been described for only one SRB, D. desulfuricans LS. In this strain, a biosynthetic pathway donates formyl groups to tetrahydrofolate; then methylation proceeds through the acetyl coenzyme A (acetyl-CoA) pathway, with reduction to methyltetrahydrofolate followed by methyl group transfer from tetrahydrofolate to a corrinoid protein to Hg (13). The acetyl-CoA pathway has been shown to function in certain acetate-oxidizing SRB during growth with sulfate as the electron acceptor (33) and also during fermentation of propionate by Desulfotomaculum thermobenzoicum (36). It is possible that the same mechanism may be responsible for Hg methylation during both respiratory and fermentative growth. However, the biochemical pathway for the synthesis of MeHg by D. propionicus 1pr3 under any conditions is unknown. Support for involvement of the acetyl-CoA pathway in Hg methylation is provided by King et al. (24), who measured rates of MeHg production among a suite of SRB and found greatest Hg methylation activity by a Desulfobacterium strain that uses this pathway for complete acetate oxidation. Their measurement of lower but significant rates of methylation by a Desulfobacter strain, which oxidizes acetate via the citric acid cycle, may implicate transmethylation enzymes other than the corrinoid protein of the acetyl-CoA pathway. Further investigations in both pure cultures and natural sediments are needed to establish if the link between the acetyl-CoA pathway and Hg methylation by SRB is universal.
The first two experiments (Fig. 1 and 2) indicate that MeHg production by D. propionicus is related to cell density through log-phase growth (about 8 days, up to an OD of about 0.2). These results suggest that during active growth, SRB cultures methylate Hg at a fairly constant rate per cell. Therefore, in time course experiments, methylation rates can be compared if they are normalized to a per-cell-per-time basis. Alternatively, within an experiment where the same volume and incubation times were used for all cultures, relative turnover of the filtered HgI pool can be compared among treatments by simply scaling by the final OD of the cultures. In older cultures, the methylation rate declines, and relatively small amounts of MeHg were produced by the stationary-phase cells of the diluted culture experiment.
The MeHg concentration increased throughout the methylation time course experiment (Fig. 2), although the slope of the line decreased after 8 days, when cell growth declined. This declining net methylation rate may be a result of demethylation, which has been demonstrated for D. desulfuricans LS (31). We can estimate an upper limit on the effect of demethylation by assuming that the demethylation rate was comparable to the methylation rate. Comparison of the substrate concentrations (HgI was 2,000 ng of Hg liter−1 and MeHg was 50 ng of Hg liter−1 at day 7) suggests that demethylation would lower the net methylation rate only by about 25%. The nearly 10-fold decrease in per-cell methylation seen during the last 5 days of the experiment cannot be explained solely by demethylation. Instead, it was probably related to decreased metabolic activity of the cells.
The Hg bioavailability time course (Fig. 3) points out several important aspects of pure culture experiments used to study Hg methylation. First, the filtered HgI concentration declined rapidly (Fig. 4), and so cells were exposed to much higher ambient filtered HgI early in an experiment when the spike was first added to growing cultures. Presumably, it is some portion of the dissolved HgI pool that is bioavailable for uptake and methylation, as evidenced by studies that show inhibition of methylation with the addition of clays that sorb and remove HgI from solution (17). As a result of declining filtered HgI, the methylation rate (fraction of spike methylated per day) decreased over the course of the experiment (Table 2). The rate in A cultures was much higher in the first 5 h than over the next several days even though the culture density increased over the course of the experiment (Fig. 3).
After normalizing for cell density and ambient filtered HgI concentration, methylation expressed as turnover per billion cells was still about 10 times higher over the first 5 h of the incubation (Table 3) than over the next 3 days. The greater methylation rate early in the experiment was not only a result of higher initial filtered HgI concentration. The important implication of this finding is that Hg spiked into culture medium, and probably into other matrices, is initially highly bioavailable. This early pulse in methylation may result from the rapid scavenging of HgI by cell surfaces initially producing a steep concentration gradient across the membrane and enhancing uptake via passive diffusion. It appears that as the Hg distribution in the cultures reaches a steady state over time, the Hg concentration in the bulk medium comes to control uptake to a greater extent. Notice that the cellular turnover for A cultures after the first 5 h was much closer to the overall turnover in B cultures (Table 3). Alternatively, the dissolved speciation of HgI may change in the first few hours, with a shift toward less bioavailable complexes.
If one is interested in how the ambient concentration and speciation of Hg in culture media affect methylation, it is desirable to add the Hg spike prior to inoculation. In subsequent experiments, Hg and sulfide were added several days prior to inoculation so that Hg could equilibrate with suspended solids in the medium and the glass walls of the serum bottles. However, even when the Hg spike is preequilibrated with medium, on addition of inoculum, the partitioning of Hg is altered due to the high affinity of cell surfaces for Hg. Adding Hg with an associated solid phase that can compete with cells for Hg sorption should allow these methylation assays to mimic conditions in sediments more closely.
The concentration dependence experiment showed that at a set sulfide concentration (1 μM), Hg methylation is linearly related to filtered HgI up to 40 ng of Hg liter−1 (Fig. 6). In this experiment, all of the cultures grew quite similarly, and so no correction was applied. The relationship between concentration and methylation is consistent with passive uptake of HgI by SRB. The concentration dependence also shows that small differences in filtered HgI in cultures are reflected in the amount of MeHg produced. Again, to examine the effects of different treatments, e.g., sulfide concentration, on MeHg production it is useful to express methylation in terms of turnover of filtered HgI.
From the time course experiment (Fig. 2 and Table 1), it is evident that the fraction of the Hg spike methylated was about five times higher in the ambient than in the sulfide+ cultures, but the difference was less marked for turnover (Table 1). In other words, although the inhibition of methylation in the sulfide+ cultures was primarily brought about by a decrease in the filtered HgI concentration, turnover of HgI was still lower at the higher sulfide concentration. Thus, the inhibition by sulfide cannot be entirely explained by the loss of Hg from solution as has been previously suggested (8, 14, 38). This relatively small decrease in turnover in the sulfide+ culture is consistent with our model for Hg speciation (3), which predicts only a modest decline in HgS0 (the most bioavailable Hg complex) between 0.55 to 1.4 mM sulfide.
One explanation for the observed decrease in turnover is that sulfide changed the bioavailability of filtered HgI. We have previously suggested that a shift in Hg speciation with increasing sulfide concentration limits passive uptake and subsequent methylation of filtered HgI (4). In Benoit et al. (3), we modeled Hg speciation for (i) a system containing HgS(s) (cinnabar) in the presence of excess dissolved sulfide and (ii) sulfidic sediment pore waters. While the primary controls on filtered HgI were precipitation in the former case and adsorption to solids in latter, both systems exhibited a decline in the fraction of Hg present as HgS0 with increasing sulfide concentration. Furthermore, a measured decrease in Dow of filtered HgI across the same sulfide gradient supported the model (5). The changes in Dow were consistent with the predicted decline in HgS0 and suggest decreased diffusive transport of Hg across the cell membrane as this neutral complex is replaced with charged disulfide complexes.
The sulfide gradient experiment (Fig. 7) was designed to investigate the effect of sulfide on methylation across a broader sulfide concentration gradient. In this experiment, filtered HgI concentration increased with increasing sulfide. The dissolved concentration of Hg in cultures, as in natural sediments, is likely controlled by adsorption of Hg to solid phases rather than direct precipitation of HgS(s), and so the presence of solids with different adsorptive capacities can lead to different relationships between sulfide and filtered HgI (3). This point is illustrated by the observation that in the methylation time course, the filtered HgI concentration was lower in sulfide+ cultures than in ambient cultures (Table 1). This experiment was performed using medium that contained yeast extract, the inclusion of which may have led to the formation of organic solids with a high affinity for HgI. These experiments point out that the chemical composition of the medium strongly affects Hg distribution and speciation. This effect should be considered when pure cultures are used for methylation assays.
In the sulfide gradient experiment, turnover per billion cells decreased with sulfide concentration as expected (Fig. 7). The experimental trends are similar to what we observed in the Patuxent River estuary, where pore water-filtered HgI increased with sulfide concentration, but bulk sediment MeHg and modeled pore water HgS0 decreased (3, 4). Regression of predicted HgS0 against bulk sediment MeHg showed that the concentration of this neutral complex explained 70% of the variability in MeHg concentration in those sediments. Furthermore, the decline in pore water HgS0 concentration across a sulfide concentration gradient depended on the nature of the bulk solid and therefore differed in two different ecosystems (3). Application of the model to the sulfide gradient experiment is confounded by the differential growth of the cultures, which led to variable and unknown solid content. However, trends in turnover (Fig. 7) should reflect changes in the fraction of filtered HgI that was present as HgS0. Therefore, it appears that this fraction was about three times greater at 1 μM than at 1 mM sulfide.
Since methylation rates and MeHg concentrations vary over a much larger range in aquatic sediments (19), this experimental design fell short of adequately simulating field conditions. The lack of a major solid-phase substrate to compete with cell surfaces for Hg binding appears to be an important shortcoming of these methylation assays. Since the concentration of dissolved Hg in pore waters is controlled through adsorption, the inclusion of known Hg-containing solids to culture experiments might more realistically mimic natural conditions. Diatom culture medium closely approximates the natural chemistry of surface sea or fresh waters, and pure cultures of diatoms have been successfully used to investigate the role of Hg speciation on Hg bioavailability (27, 28). However, pure cultures of SRB fail to duplicate the components of the sediment-pore water system, thereby limiting their usefulness as microcosms to test hypotheses about the effects of sulfide, and other ligands, on Hg speciation and methylation in natural sediments and soils.
Although we have not explicitly determined the nature of particles in cultures, it is likely that precipitates including Ca and Mg carbonates, elemental sulfur, and metal sulfides are formed. In growing cultures, bacterial cells also provide sites for Hg adsorption, as these surfaces have a strong affinity for Hg (21). The undefined nature of Hg interaction with cells and precipitates makes modeling Hg speciation in culture extremely difficult. Our pore water model (3) suggests that adsorption to sediment solids strongly influences dissolved HgI concentration and speciation in aquatic sediments. Therefore, to better simulate natural conditions, and to facilitate estimation of HgS0 concentration in pure culture experiments, it is desirable to provide a solid-phase source of Hg. This approach has been investigated in our laboratory (2).
In summary, these experiments with D. propionicus demonstrated that under conditions of equal bioavailability, per-cell MeHg production was constant through log-phase culture growth. In long-term methylation assays, methylation rate expressed either as the fraction of the added spike or as turnover of filtered HgI dramatically decreased after the first 5 h. The bioavailability of added Hg in cultures, therefore, decreases over time due to loss of HgI from solution and changes in the partitioning and/or complexation of the dissolved pool. At low sulfide concentration, MeHg production depended on the concentration of filtered HgI. At higher sulfide concentration the production of MeHg from filtered HgI decreased, which is consistent with a decrease in the bioavailability of HgI, and consistent with our model that HgS0 is the dominant complex taken up prior to methylation. Taken together, these experiments underscore the importance of considering how medium composition and assay conditions affect Hg distribution and speciation when interpreting the results of pure-culture methylation studies. In particular, they point out the difficulty in recreating in situ Hg bioavailability in the laboratory.
ACKNOWLEDGMENTS
We thank Georgia Riedel for assistance with anaerobic culture technique.
This research was supported by an EPA STAR grant, South Florida Water Management District contract C-7690, and Florida DEP contract SP-434 and by the Electric Power Research Institute. J. Benoit was supported by an EPA STAR graduate fellowship.
REFERENCES
- 1.Anderson D M, Morel F M M. Copper sensitivity of Gonyaulax tamarensis. Limnol Oceanogr. 1978;23:283–295. [Google Scholar]
- 2.Benoit, J. M., C. C. Gilmour, and R. P. Mason. The influence of sulfide on solid-phase mercury bioavailability for methylation by pure cultures of Desulfobulbus propionicus (1pr3). Environ. Sci. Technol., in press. [DOI] [PubMed]
- 3.Benoit J M, Gilmour C C, Mason R P, Heyes A. Sulfide controls on mercury speciation and bioavailability to methylating bacteria in sediments pore waters. Environ Sci Technol. 1999;33:951–957. [Google Scholar]
- 4.Benoit J M, Gilmour C C, Mason R P, Riedel G F, Riedel G S. Behavior of mercury in the Patuxent River estuary. Biogeochemistry. 1998;40:249–265. [Google Scholar]
- 5.Benoit J M, Mason R P, Gilmour C C. Estimation of mercury-sulfide speciation in sediments pore waters using octanol-water partitioning and implications for availability to methylating bacteria. Environ Toxicol Chem. 1999;18:2138–2141. doi: 10.1002/etc.5620181004. [DOI] [PubMed] [Google Scholar]
- 6.Bloom N S. Determination of picogram levels of methylmercury by aqueous phase ethylation followed by cryogenic gas chromatography with cold vapour atomic fluorescence detection. Can J Fish Aquat Sci. 1989;46:1131–1140. [Google Scholar]
- 7.Bloom N S, Fitzgerald W F. Determination of volatile mercury species at the picogram level by low temperature gas chromatography with cold vapour atomic fluorescence detection. Anal Chim Acta. 1988;208:151–161. [Google Scholar]
- 8.Blum J E, Bartha R. Effect of salinity on methylation of mercury. Bull Environ Contam Toxicol. 1980;25:404–408. doi: 10.1007/BF01985546. [DOI] [PubMed] [Google Scholar]
- 9.Brouwer H, Murphy T P. Diffusion method for the determination of acid-volatile sulfides (AVS) in sediments. Environ Sci Technol. 1994;13:1273–1275. [Google Scholar]
- 10.Castro H F, Williams N H, Ogram A. Phylogeny of sulfate-reducing bacteria. FEMS Microb Ecol. 2000;31:1–9. doi: 10.1111/j.1574-6941.2000.tb00665.x. [DOI] [PubMed] [Google Scholar]
- 11.Choi S-C, Bartha R. Cobalamin-mediated mercury methylation by Desulfovibrio desulfuricans LS. Appl Environ Microbiol. 1993;59:290–295. doi: 10.1128/aem.59.1.290-295.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi S-C, Chase T, Jr, Bartha R. Enzymatic catalysis of mercury methylation by Desulfovibrio desulfuricans LS. Appl Environ Microbiol. 1994;60:1342–1346. doi: 10.1128/aem.60.4.1342-1346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi S-C, Chase T, Jr, Bartha R. Metabolic pathways leading to mercury methylation in Desulfovibrio desulfuricans LS. Appl Environ Microbiol. 1994;60:4072–4077. doi: 10.1128/aem.60.11.4072-4077.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Compeau G, Bartha R. Effects of sea-salt anions on the formation and stability of methyl mercury. Bull Environ Contam Toxicol. 1983;31:486–493. doi: 10.1007/BF01622282. [DOI] [PubMed] [Google Scholar]
- 15.Compeau G, Bartha R. Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediments. Appl Environ Microbiol. 1985;50:498–502. doi: 10.1128/aem.50.2.498-502.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig P J, Moreton P A. Total mercury, methyl mercury and sulphide in River Carron sediments. Mar Pollut Bull. 1983;14:408–411. [Google Scholar]
- 17.Farrell R E, Huang P M, Germida J J. Biomethylation of mercury(II) adsorbed on mineral colloids common in freshwater sediments. Appl Organomet Chem. 1998;12:613–620. [Google Scholar]
- 18.Gill G A, Fitzgerald W F. Picomolar mercury measurements in seawater and other materials using stannous chloride reduction and two-stage gold amalgamation with gas phase detection. Mar Chem. 1987;20:227–243. [Google Scholar]
- 19.Gilmour C C, Riedel G S, Ederington M C, Bell J T, Benoit J M, Gill G A, Stordal M C. Methymercury concentrations and production rates across a trophic gradient in the northern Everglades. Biogeochemistry. 1998;40:327–345. [Google Scholar]
- 20.Gutknecht J J. Inorganic mercury (Hg2+) transport through lipid bilayer membranes. J Membr Biol. 1981;61:61–66. [Google Scholar]
- 21.Henry E A. Ph.D. dissertation. 1992. The role of sulfate-reducing bacteria in environmental mercury methylation. Harvard University, Cambridge, Mass. [Google Scholar]
- 22.Horvat M, Bloom N S, Liang L. Comparison of distillation with other current isolation methods for the determination of MeHg compounds in low level environmental samples. Part I. Sediment. Anal Chim Acta. 1993;282:135–152. [Google Scholar]
- 23.Horvat M, Liang L, Bloom N S. Comparison of distillation with other current isolation methods for the determination of MeHg compounds in low level environmental samples. Part II. Water. Anal Chim Acta. 1993b;282:153–168. [Google Scholar]
- 24.King J K, Kostka J E, Frischer M E, Saunders F M. Sulfate-reducing bacteria methylate mercury at variable rates in pure culture and in marine sediments. Appl Environ Microbiol. 2000;66:2430–2437. doi: 10.1128/aem.66.6.2430-2437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kremer D R, Hansen T A. Pathway of propionate degradation in Desulfobulbus propionicus. FEMS Microbiol Lett. 1988;49:273–277. [Google Scholar]
- 26.Laanbroek H J, Pfennig N. Oxidation of short-chain fatty acids by sulfate-reducing bacteria in freshwater and in marine sediments. Arch Microbiol. 1981;128:330–335. doi: 10.1007/BF00422540. [DOI] [PubMed] [Google Scholar]
- 27.Mason R P, Reinfelder J R, Morel F M M. Bioaccumulation of mercury and methylmercury. Water Air Soil Pollut. 1995;80:915–921. [Google Scholar]
- 28.Mason R P, Reinfelder J R, Morel F M M. Uptake, toxicity, and trophic transfer of mercury in a coastal diatom. Environ Sci Technol. 1996;30:1835–1845. [Google Scholar]
- 29.Moench T T, Zeikus J G. An improved method for a titanium(III) media reductant. J Microbiol Methods. 1983;1:199–202. [Google Scholar]
- 30.Morel F M M, Hering J G. Principles and applications of aquatic chemistry. New York, N.Y: John Wiley & Sons; 1993. [Google Scholar]
- 31.Pak K-R, Bartha R. Mercury methylation and demethylation in anoxic lake sediments and by strictly anaerobic bacteria. Appl Environ Microbiol. 1998;64:1013–1017. doi: 10.1128/aem.64.3.1013-1017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pak K-R, Bartha R. Mercury methylation by interspecies hydrogen and acetate transfer between sulfidogens and methanogens. Appl Environ Microbiol. 1998;64:1987–1990. doi: 10.1128/aem.64.6.1987-1990.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schauder R, Eikmanns B, Thauer R K, Widdel F, Fuchs G. Acetate oxidation to CO2 in anaerobic bacteria via a novel pathway not involving reactions of the citric acid cycle. Arch Microbiol. 1986;145:162–172. [Google Scholar]
- 34.Stams A J M, Kreer D R, Nicolay K, Weenk G H, Hansen T A. Pathway of propionate formation in Desulfobulbus propionicus. Arch Microbiol. 1984;139:167–173. [Google Scholar]
- 35.Sunda W G, Guillard R R. Relationship between cupric ion activity and the toxicity of copper to phytoplankton. J Mar Res. 1976;34:511–529. [Google Scholar]
- 36.Tasaki M, Kamagata Y, Nakamura K, Okamura K, Minami K. Acetogenesis from pyruvate by Desulfotomaculum thermobenzoicum and differences in pyruvate metabolism among three sulfate-reducing bacteria in the absence of sulfate. FEMS Microbiol Lett. 1993;106:259–264. [Google Scholar]
- 37.Widdel F, Pfennig N. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. II. Incomplete oxidation of propionate by Desulfobulbus propionicus gen. nov., sp. nov. Arch Microbiol. 1982;131:360–365. doi: 10.1007/BF00406470. [DOI] [PubMed] [Google Scholar]
- 38.Winfrey M R, Rudd J W M. Environmental factors affecting the formation of methylmercury in low pH lakes: a review. Environ Toxicol Chem. 1990;9:853–869. [Google Scholar]