Abstract
Paraquat (PQ) is the third most used broad-spectrum nonselective herbicide around the globe after glyphosate and glufosinate. Repeated usage and overreliance on this herbicide have resulted in the emergence of PQ-resistant weeds that are a potential hazard to agriculture. It is generally believed that PQ resistance in weeds is due to increased sequestration of the herbicide and its decreased translocation to the target site, as well as an enhanced ability to scavenge reactive oxygen species. However, little is known about the genetic bases and molecular mechanisms of PQ resistance in weeds, and hence no PQ-resistant crops have been developed to date. Forward genetics of the model plant Arabidopsis thaliana has advanced our understanding of the molecular mechanisms of PQ resistance. This review focuses on PQ resistance loci and resistance mechanisms revealed in Arabidopsis and examines the possibility of developing PQ-resistant crops using the elucidated mechanisms.
Keywords: herbicide, paraquat, paraquat resistance, Arabidopsis thaliana, weed
This article reviews our current understanding of paraquat resistance mechanisms in weeds and Arabidopsis thaliana and discusses their potential application to the development of paraquat-resistant crops.
Introduction
Paraquat (1,1-dimethyl-4,4′-bipyridinium dichloride, PQ) is a post-emergence, broad-spectrum, foliar-applied, and nonselective herbicide. It has been used in agriculture for weed control since 1960 (Preston, 1994). The herbicidal potential of PQ was first discovered at Jealott’s Hill Research Center in 1955 (Hawkes, 2014). This divalent cation is highly soluble in water and kills plants within a few hours under sunlight (Fuerst and Vaughn, 1990). PQ is being used in more than 100 countries and is the third most extensively used herbicide in the world after glyphosate and glufosinate. In addition, PQ is frequently used as a free radical initiator for studying oxidative stress response in the laboratory.
PQ is quickly adsorbed to soil colloids, making it inactive; the sowing of crops immediately after herbicide treatment is possible without the fear of phytotoxicity, and PQ is therefore widely used in non-tillage farming. Owing to soil adsorption, its limited leaching and reduced runoff by rainwater reduce the ecotoxicity of this herbicide. Its rapid uptake by plant tissues makes it an optimal choice in areas where rainfall is frequent. Moreover, its non-penetration of woody tissues ensures its safe use in orchards.
Repeated use of PQ has given rise to PQ-resistant weeds. In 1981, the first case of PQ resistance was reported in Conyza bonariensis (hairy fleabane) in Egypt (Youngman, 1981). To date, at least 28 weed species with PQ resistance have been reported in 20 countries (Heap, 2021). The evolution of PQ resistance in weeds is a relatively slow process. Even after use of this herbicide for 50 years, PQ resistance in weeds is not a major economic concern (Hawkes, 2014). The evolution of PQ resistance is generally observed to be a slow phenomenon, resulting mostly from immense exposure to this herbicide over a duration of 10 years (or longer) (Soar et al., 2003). For example, PQ resistance in capeweed (Arctotheca calendula) appeared after one application/year of a diquat and PQ mixture for a period of 23 years (Powles et al., 1989). Hence, PQ-resistant weeds are not yet a major problem, but they are a potential threat to agriculture. Studies on PQ-resistant weeds provide important clues to hypothesize the mechanisms of PQ resistance in weeds. However, little is known about their genetic bases and molecular mechanisms. Although PQ-resistant weeds are not a major problem at present, the development of PQ-resistant crops may provide a novel tool for weed management. However, the limited understanding of PQ resistance mechanisms is a barrier to the creation of PQ-resistant crops. With the power of genetics, the model plant Arabidopsis thaliana offers a unique opportunity to dissect the molecular mechanisms of PQ resistance.
Mechanism of PQ action
In plants, PQ is absorbed and transported from the external environment to the chloroplasts, the major site of PQ action. The plant cuticle does not impede the absorption of this herbicide (Bishop et al., 1987). The uptake of PQ by plant cells is mediated by plasma membrane-localized transporters such as the polyamine transporters (Hart et al., 1992b). However, it is not yet clear how PQ crosses the chloroplast membranes. Once it has arrived at the site of action, under conditions of illumination, the PQ dication (PQ2+) accepts a free electron from ferredoxin in photosystem I (PSI) to form a stable monocation radical (PQ·+), which quickly reacts with divalent oxygen to produce superoxide (Farrington et al., 1973). PQ is then recycled to continuously generate superoxide. Superoxide is decomposed into oxygen and hydrogen peroxide through the activity of superoxide dismutase (SOD). Then, hydrogen peroxide can oxidize Fe2+ to Fe3+ through the Fenton reaction and generate hydroxyl free radicals (Gutteridge, 1984), thus causing redox reaction chains to generate various forms of reactive oxygen species (ROS) (Babbs et al., 1989). Excessive ROS (e.g., superoxide radical, hydroxyl radical, singlet oxygen, hydrogen peroxide) quickly destroy the cell membranes, leading to leaf chlorosis and wilting and ultimately to desiccation (Hawkes, 2014).
Proposed mechanisms of PQ resistance in weeds
In general, herbicide resistance involves two broad mechanisms: resistance produced by target site mutations, known as target site resistance (TSR), and resistance produced by non-target site mutations, known as non-target site resistance (NTSR). In TSR, a lethal dose of herbicide reaches the target, but its effect is mitigated because of mutations in the target site. NTSR includes mechanisms that reduce the quantity of herbicide that approaches the target site or protect the plant against herbicide-mediated damage (Powles and Yu, 2010; Délye et al., 2013, 2015).
The target of PQ is presumably ferredoxin in PSI. However, there is currently no evidence of TSR in PQ-resistant plants. PQ is thought to accept electrons from ferredoxin rather than from PSI (Hawkes, 2014). However, there is no clear evidence of PQ accepting electrons from ferredoxins, PSI, or both. As a redox cycler, PQ has a more negative reduction potential (E0 = −446 mV) than ferredoxin. The midpoint reduction potential exhibited by leaf ferredoxins of Arabidopsis and maize (E0 = ∼ −425 mV) is more positive than that of PQ (Hanke et al., 2004), making it possible that PQ accepts electrons from the donor by simple proximity but without binding to the “target site.” Alternatively, mutations in the target site may be lethal, so that no TSR mutants have been isolated. All PQ-resistant weeds are the NTSR type. Based on studies of PQ-resistant weeds, several mechanisms of NTSR have been proposed, involving disrupted PQ transport to the target organelle (chloroplast), enhanced PQ sequestration in the vacuole, enhanced ability to scavenge ROS, and PQ detoxification via metabolism.
Disrupted transport of PQ to its target organelle
PQ is taken up into the plant cell by plasma membrane-localized transporters (Hart et al., 1992a, 1992b). Thus, impaired uptake of PQ may result in enhanced resistance to PQ. For example, leaf sections of resistant C. bonariensis exhibited 81 times more resistance to PQ than leaf sections of the sensitive genotype. Lateral 14C-PQ movement from the point of herbicide application was restricted in leaf sections of the resistant plants relative to the sensitive ones (Norman et al., 1993, 1994), suggesting a resistance mechanism involving reduced translocation of PQ (perhaps a transporter) in the resistant plants. Interestingly, polyamines can competitively reduce PQ translocation. For example, exogenous application of polyamines on leaf discs of a sensitive biotype of Arctotheca calendula reduced the translocation of PQ, thereby increasing PQ tolerance (Soar et al., 2004), which suggests that polyamine transporters play a role in PQ translocation.
The plant homolog of Escherichia coli EmrE, which is involved in the transport of various xenobiotic compounds in bacteria (Yerushalmi et al., 1995), was proposed to be a potent transporter of PQ in C. canadensis (Jóri et al., 2007). Cationic amino acid transporters, usually involved in polyamine transport (Moretti et al., 2017), may be able to transport PQ. For instance, cationic amino acid transporter 4 was found to be involved in enhanced PQ resistance of C. canadensis (Soós, 2005; Jóri et al., 2007).
Once PQ is taken up into the cytosol, it can be transported to its target organelle, the chloroplast. Impaired transport to the chloroplast would also result in enhanced resistance to PQ. For instance, the increased expression of PqST2 in a resistant biotype of Eleusine indica (goosegrass) relative to a sensitive biotype after PQ treatment may be associated with reduced PQ transport to the target organelle in the resistant biotype (Luo et al., 2019). PqST2 is a homolog of ABCB1 that encodes a PQ efflux transporter in human and mice (Wen et al., 2014). Furthermore, enhanced expression of PqST1, a homolog of SYP121 that encodes a protein involved in vesicle trafficking, increased secretion of PQ out of the cells of resistant goosegrass plants compared with sensitive plants after PQ treatment (Luo et al., 2019), thereby reducing cellular PQ level.
Enhanced sequestration of PQ
PQ sequestration is one of the major proposed mechanisms by which weeds have acquired resistance to PQ. The PQ level can be reduced in cytoplasm and target organelles by enhanced sequestration to metabolically inactive compartments such as the cell wall and vacuole. For example, the enhanced PQ resistance of C. bonariensis was considered to be related to PQ exclusion from its site of action in the chloroplast via an unknown sequestration mechanism (Fuerst et al., 1985). The PQ resistance of leaf sections was dramatically higher in a resistant biotype than in a sensitive one, but illuminated preparations of protoplasts and chloroplasts did not show different sensitivities to PQ treatment, strongly suggesting that the resistance was related to restricted diffusion of the herbicide to its site of action in the chloroplast, possibly because of a sequestration mechanism. As PQ is a polar dication and the plant cell wall has cation-exchange ability (Baydoun and Brett, 1988), the hypothesis of PQ binding to the cell wall seems significant. Reduced translocation of PQ in resistant versus sensitive Hordeum glaucum was attributed to its sequestration, possibly in the apoplast (Bishop et al., 1987) or vacuole (Lasat et al., 1997). Similarly, in Lolium rigidum (ryegrass), decreased translocation of PQ in resistant plants was attributed to elevated sequestration of this herbicide in the vacuoles (Yu et al., 2007). In C. canadensis (horseweed), experiments on uptake, distribution, and localization of PQ into cell organelles revealed that a significant proportion of applied herbicide was retained in the vacuole plus cytosol fraction. The PQ-resistant horseweed plants with reduced PQ in chloroplasts after PQ treatment showed that the PQ resistance was caused by its sequestration in the vacuole (Jóri et al., 2007). The leaf protoplasts of PQ-treated resistant plants of L. rigidum exhibited two to three times more PQ than those of PQ-treated susceptible plants, and the mechanism of resistance was probably attributed to enhanced sequestration of PQ in the vacuoles of the resistant plants (Yu et al., 2010). The movement of 14C-labeled PQ (under light conditions) was confined to the PQ-treated leaf in the resistant biotype of L. multiflorum (Italian ryegrass) compared with the sensitive one, from whose treated leaf the herbicide transport rate was approximately 20 times higher (Brunharo and Hanson, 2017). Vacuolar PQ sequestration, possibly by tonoplast-localized polyamine transporters, contributed to the enhanced PQ tolerance of the resistant biotype of Italian ryegrass (Brunharo and Hanson, 2017). These studies strongly suggest that there are efflux transporters that localize in the plasma membrane and tonoplast and export PQ into the vacuole and apoplast from the cytosol, although this hypothesis nevertheless awaits molecular genetic proof.
Enhanced ROS scavenging ability
The enhanced capability of ROS scavenging helps weeds to minimize PQ-generated ROS in resistant biotypes. The extensively studied enzymes in this regard are those related to the ascorbate-glutathione (Halliwell–Asada) cycle that protects cells from oxidative damage (Délye, 2013). These enzymes include glutathione reductase (GR), SOD, monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR), glutathione peroxidase (GPX), and ascorbate peroxidase (APX) (Foyer and Noctor, 2011; Noctor et al., 2012, 2016). Nevertheless, the generators of ascorbate and glutathione (reducing equivalents) that are responsible for driving the ascorbate-glutathione cycle come from PSI (the same site from which PQ effectively diverts electrons). Therefore, compared to the reduced transport of PQ to its target site, the capability of this cycle to provide a high level of PQ resistance is a bit doubtful (Hawkes, 2014).
Increased ROS scavenging ability has been found in PQ-resistant weeds. For instance, enhanced PQ resistance was attributed to enhanced activities of antioxidant enzymes in various PQ-resistant weeds compared with sensitive ones (Shaaltiel et al., 1988a, 1988b; Shaaltiel and Gressel, 1986). Similarly, the increased activity of SOD after PQ treatment in resistant biotypes of Mazus pumilus (Japanese mazus) was found to be responsible for PQ resistance in the resistant plants (Tsuji et al., 2013).
PQ detoxification by plant metabolism
PQ detoxification is a plausible mechanism, but little is known about it. To date, there are no reports of PQ degradation by plant metabolism. It is generally thought that PQ is stable in plant cells (Hawkes, 2014). The metabolic detoxification of PQ was not found to be associated with enhanced PQ resistance in the R biotype of L. perenne (Harvey et al., 1978) and C. bonariensis (Norman et al., 1993). However, PQ can be degraded by microorganisms in the soil (Bromilow, 2004; Huang et al., 2019). By contrast, the metabolic degradation of other types of herbicides has been reported (Busi et al., 2011).
Arabidopsis mutants with enhanced PQ resistance confirm the proposed mechanisms in weeds
Due to technical difficulties, especially the lack of genetic analyses and genome sequence information for weeds, none of the mechanisms proposed above have been confirmed at the molecular level. Thanks to the model plant Arabidopsis, PQ resistance mutants provide key molecular genetic evidence for PQ resistance mechanisms.
It should be noted that PQ application as an herbicide for weed control differs from that as a selection reagent in an Arabidopsis mutant screen. PQ is usually sprayed on leaves in the former case, whereas PQ is applied to the medium to select PQ-resistant Arabidopsis mutants during germination and early seedling growth. In addition, the dose of PQ used for mutant selection is significantly lower. Therefore, the mechanisms of PQ resistance revealed in Arabidopsis may have some differences from those in weeds and should be cautiously assessed when used for PQ-resistant crop development.
Nevertheless, the PQ resistance genetic loci revealed in Arabidopsis to date have basically confirmed all of the proposed mechanisms of PQ resistance in weeds.
Impaired uptake and transport of PQ
Mutations in plasma membrane transporters can lead to reduced uptake of PQ into the cell, increased cellular efflux, and impaired intracellular transport. Arabidopsis mutants with defects in PQ uptake have paved the way to understanding the molecular bases of PQ transporters. For example, study of the PQ-tolerant mutant pqt24-1 revealed that PQT24/AtPDR11 (pleiotropic drug resistance 11) acts as a PQ transporter (Xi et al., 2012). AtPDR11 belongs to the ATP binding cassette (ABC) family, which is one of the largest protein families and is found in all living organisms (Henikoff et al., 1997). Some members of this family are involved in ATP-driven transport activities (Higgins, 1992). A loss-of-function mutation in this transporter contributed to two-fold enhancement of PQ tolerance, as shown by seedlings of the null mutant pqt24-1 compared with the wild type (Xi et al., 2012). Consistent with its function, the PQT24/AtPDR11 protein is localized in the plasmalemma, supporting the idea that PQ resistance was due mainly to its reduced influx into the cell from the extracellular environment. Considering that PQT24/AtPDR11 is a PQ importer, one might expect that PQ treatment would lead to an increased accumulation of intracellular PQ in the wild type with the passage of time. In fact, an equilibrium was evident after 2 h of PQ treatment, indicating that a PQ efflux mechanism must exist to counterbalance PQ import and maintain equilibrium. PQ is an opportunistic substrate of this transporter, whose PQ transport activity is competitively inhibited by putrescine (Xi et al., 2012), like that observed in maize roots (Hart et al., 1992a, 1992b). As PQ and polyamines have similar uptake characteristics (Hart et al., 1992b), it is possible that polyamines and PQ share common transporters.
Another transporter capable of PQ uptake in Arabidopsis is the L-type amino acid transporter1 (AtLAT1) revealed by the rmv1 (resistant to methyl viologen1) mutant (Fujita et al., 2012). PQ uptake capacity of the knockout mutant rmv1 was decreased approximately two- to four-fold compared with the wild type. Transgenic plants overexpressing RMV1 displayed higher PQ uptake, indicating that RMV1 acts as a PQ transporter. RMV1 is a plasma membrane-localized transporter, consistent with its role in importing PQ from the external environment. The transport activity of RMV1 is competitively inhibited by polyamines, suggesting that RMV1 is a polyamine transporter (Fujita and Shinozaki, 2014).
The enhanced PQ resistance of par1 (paraquat resistant1) mutants (at least 10 times higher PQ resistance than the wild type in germination) and the elevated sensitivity of PAR1-overexpressing plants compared with the wild type suggest that PAR1 is involved in the intracellular transport of PQ (Li et al., 2013). PAR1 encodes AtLAT4, another L-type amino acid transporter. PAR1/AtLAT4 is Golgi localized, which is why the rate of PQ uptake is not significantly different between the mutant and the wild type; rather its concentration is reduced in the chloroplasts of mutant plants, suggesting that PAR1 is involved in the transport of PQ to the chloroplasts. Reduced import of PQ to the chloroplast by brefeldin-A-sensitive cycling was proposed as a possible mechanism of PQ resistance in par1 (Li et al., 2013). However, vesicle-based trafficking has not yet been confirmed from the Golgi apparatus to the chloroplast; hence, direct transport of PQ from the Golgi apparatus to the chloroplast remains unresolved. Li et al. (2013) proposed that PAR1 may be involved in the endocytosis-mediated transfer of PQ from the Golgi apparatus to the cytosol or into another intracellular organelle, followed by transport into the chloroplast via unidentified transporters. They also speculated that a small proportion of PAR1 may be localized in the chloroplast and may transport PQ into the chloroplast from the cytosol (Li et al., 2013). A nonsense mutation in the coding region of PAR1 resulting in enhanced PQ tolerance in the Arabidopsis pqr2 mutant compared with the wild type (Dong et al., 2016) and more than 20 par1 alleles isolated by Xia et al. (2021) reinforce the idea that PAR1 is a major locus of PQ resistance in Arabidopsis. It would be interesting to further explore the molecular mechanism by which PQ is specifically transported to the chloroplast from the cytosol.
Enhanced sequestration of PQ
Cellular efflux of the herbicide is an attractive mechanism, but no membrane efflux protein related to PQ transport has yet been reported in plants. During genetic screening for PQ-tolerant mutants from a near-saturated ethyl methanesulfonate-mutagenized M2 library of Arabidopsis, Xia et al. identified the mutant pqt15-D (paraquat tolerance 15-D) that is highly tolerant to PQ (approximately 80 times more tolerant than the wild type). A dominant mutation in PQT15/DETOXIFICATION EFFLUX CARRIER 6 (DTX6) causes a G311E substitution and increases the negative charge at the substrate tunnel of DTX6 and the binding of positively charged PQ, probably enhancing the substrate (PQ) binding affinity and efflux activity of DTX6. The reduced PQ tolerance of the knockout mutant dtx6 and the enhanced tolerance of transgenic plants overexpressing DTX6 support the notion that PQT15/DTX6 is a PQ efflux transporter. The DTX6 protein is mainly localized in the plasma membrane, consistent with the role of DTX6 as an efflux transporter (Xia et al., 2021). DTX6 belongs to the MATE (multidrug and toxic compound extrusion) family of transporters that use the membrane electrochemical gradient to drive transport activities (Kuroda and Tsuchiya, 2009). Members of this family function as efflux pumps to export xenobiotic compounds out of the cell. Recent studies have shown that these proteins function as antiporters and transport substrates via H+ exchange in plants (He et al., 2010; Lu et al., 2013). DTX6 is indeed involved in the cellular efflux of PQ, as demonstrated by mesophyll protoplasts of DTX6-overexpressing plants that export approximately 1.25 times more PQ than those of the wild type, further confirming the efflux nature of this transporter. Similar findings for DTX6 were obtained independently by another research group and published back to back (Lv et al., 2021). Lv et al. also presented evidence that DTX6 localizes in the tonoplast of leaf cells and transports PQ into the vacuole. These two recent reports support the well-known proposed mechanism of PQ sequestration in the vacuole and cell wall.
DTX6 seems to work in a manner similar to that of the bacterial membrane protein PqrA from Ochrobactrum anthropi that is responsible for the efflux of toxic compounds. It confers resistance against PQ when expressed in E. coli, providing evidence for its transporter nature and increasing efflux of the herbicide (Won et al., 2001). The PqrA gene also confers PQ resistance when heterologously expressed in tobacco (Jo et al., 2004).
Enhanced ROS scavenging ability
While exerting its herbicidal mode of action, PQ2+ accepts electrons from PSI and is converted into PQ·+. PQ·+ can immediately react with molecular oxygen present in chloroplasts and is re-oxidized to PQ2+, generating ROS (Vicente et al., 2001; Wang et al., 2021). Given an adequate supply of electrons and molecular oxygen, the constant redox cycling of PQ results in dose-dependent ROS production (Reczek et al., 2017). The reservoir of antioxidants must be maintained at a level that corresponds to the rate of electron transfer. Hence, detoxification of PQ-generated ROS by cellular antioxidant mechanisms has some limitations in providing adequate PQ resistance, as PQ is recycled during ROS production, allowing it to produce new ROS until something else happens to PQ.
Increased antioxidant activities have been observed in a number of Arabidopsis mutants. For example, the Arabidopsis mutant gigantea is tolerant to PQ and H2O2 and flowers later than the wild type, suggesting that GIGANTEA is involved in oxidative stress tolerance and delayed flowering (Kurepa et al., 1998).
While screening a mutagenized Arabidopsis library to find mutants with stable photoautotrophic growth under salt stress, Tsugane et al. (1999) discovered a mutant called photoautotrophic salt tolerance1 (pst1). The recessive mutation in pst1 was responsible for enhanced tolerance to salt and higher light in the mutant compared with the wild type. In addition, the PQ tolerance of pst1 seedlings was 10 times higher than that of wild-type seedlings. The dramatically upregulated transcripts of ROS scavengers (APX and SOD) in the mutant were considered to be responsible for the enhanced PQ tolerance of the pst1 plants. Hence, the PST1 gene is thought to regulate a number of genes that are involved in the detoxification of ROS (Tsugane et al., 1999).
Fujibe et al. isolated a PQ-resistant mutant that was found to be allelic to ozone-sensitive radical-induced cell death1 (rcd1) (Overmyer et al., 2000) and called it rcd1-2 (Fujibe et al., 2004). The PQ tolerance of rcd1-2 was enhanced approximately four-fold compared with that of the wild type. The rcd1-2 mutant was also tolerant of freezing and UVB light. In contrast to the wild type, rcd1-2 showed an increase in the expression of chloroplastic antioxidant enzymes such as Cu/Zn SOD, thylakoid APX, and stromal APX under PQ treatment, and its enhanced oxidation tolerance was credited to the elevated levels of these antioxidant enzymes (Fujibe et al., 2004).
The recessive mutant par2-1 (paraquat resistant2-1) exhibits reduced cell death compared with the wild type under PQ treatment (Chen et al., 2009). PAR2 encodes S-nitrosoglutathione reductase1. S-Nitrosoglutathione is an active species of nitric oxide (NO), and par2-1 therefore exhibits an enhanced level of NO compared with the wild type. Upon PQ treatment, superoxide levels in par2-1 and the wild type were similar, suggesting that PAR2 was not involved in superoxide generation or turnover. Thus, PAR2 regulates cell death downstream of superoxide. Protein modification by nitrosylation, resulting in increased availability of NO, may be involved in mitigating events downstream of superoxide, thus reducing PQ-mediated cell death in par2-1 (Chen et al., 2009).
The mutant paraquat tolerance3 (pqt3) was isolated from the same transfer DNA (T-DNA) insertion library as pqt24-1 (Xi et al., 2012). The recessive mutation in pqt3 enhanced the PQ resistance of this mutant 30-fold relative to the wild type. PQT3 encodes a ubiquitin E3 ligase that acts as a molecular switch to turn off plant antioxidant mechanisms when oxidative stress is absent (Luo et al., 2016). PQT3 negatively regulates the oxidative stress response by interacting with protein methyltransferase 4b (PRMT4b), which upregulates the expression of antioxidant enzymes such as GPX and APX by histone methylation under stress conditions, thereby protecting the plant from oxidative stress. PQT3 ubiquitinates PRMT4b for 26S proteasome-mediated degradation. Thus, elevated activities of GPX and APX and decreased expression of PQT3 were responsible for the enhanced PQ resistance of pqt3 (Luo et al., 2016).
It is conceivable that any mutations that improve antioxidant ability will help the mutants to survive PQ stress.
Energy-related metabolism also contributes to PQ resistance, as shown by the mutant pqr-216, an activation-tagged line with increased tolerance to PQ, in which the T-DNA insertion has activated the expression of AtNUDX2 (At5g47650). AtNUDX2 encodes ADP-ribose pyrophosphatase, which affects the maintenance of ATP and NAD+ homeostasis by nucleotide recycling from free ADP-ribose molecules (Ogawa et al., 2009). Likewise, the decreased activities of poly(ADP-ribose) polymerases enhanced stress tolerance (De Block et al., 2005).
PQ detoxification by plant metabolism: myth or reality?
Apart from transporters and antioxidant systems, a more desirable strategy for PQ resistance is the degradation of PQ by metabolism. The cytochrome P450 superfamily plays significant roles in plant metabolism and the detoxification of xenobiotics (Hansen et al., 2021). There are many reports of P450-mediated herbicide degradation (Höfer et al., 2014; Azab et al., 2018; Yamada et al., 2000; Didierjean et al., 2002; Jang et al., 2020). However, no evidence for PQ metabolism has been found in plants to date.
In a genetic screen for PQ resistance mutants of Arabidopsis from the same T-DNA insertion library as pqt24-1 (Xi et al., 2012), Huang et al. (2021) isolated the gain-of-function mutant paraquat tolerance11D (pqt11D) in which the T-DNA insertion with 35S enhancers activated PQT11/CYP86A4, which encodes a P450 protein. The PQ resistance phenotype can be recapitulated by overexpressing PQT11 in the wild type, and the resulting overexpression lines were twice as resistant to PQ as the wild type in the presence of a high concentration of PQ (10 μM). Huang et al. (2021) demonstrated that PQT11/CYP86A4 could demethylate PQ to N-demethyl PQ, which was found to be nontoxic to Arabidopsis. Therefore, Huang et al. clearly revealed a novel PQ resistance mechanism by which PQ is detoxified via plant metabolism. However, PQ is not a natural substrate of PQT11/CYP86A4, and the natural substrate of this P450 remains unknown. More importantly, it should be explored whether other plant enzymes such as glutathione S-transferases and glycosyltransferases participate in PQ degradation. The fate of N-demethyl PQ in the cell also needs to be investigated in the future.
Potential strategies for developing PQ-resistant crops
Sustainable crop production is crucial for world food security, as arable land is continually decreasing. The infestation and intrusion of weeds on crops is a major threat to world crop production, thus necessitating the use of herbicides to control weeds. The most common strategy to control weeds in the field is the combination of herbicide application and herbicide-resistant crops. Crops resistant to major commercial herbicides have been developed by overexpressing herbicide-resistance genes (Mazur and Falco, 1989; Duke and Cerdeira, 2010; Guo et al., 2015; Fartyal et al., 2018; Achary et al., 2020) or by genome editing (Sun et al., 2016; Zhang et al., 2019; Kuang et al., 2020; Wu et al., 2020; Liu et al., 2021). However, no PQ-resistant crops have yet been developed owing to a lack of understanding of the molecular mechanisms of PQ resistance.
The idea of developing PQ-resistant crops is interesting, as PQ is the third most commonly used herbicide in the world because of its cost-effectiveness, labor savings, and rapid mode of action (Mercado and Caleño, 2021). Despite being banned in a few countries (Li et al., 2019), PQ is currently used in more than 120 countries for approximately 100 crops (Mercado and Caleño, 2021). Although China banned the aqueous solution of this herbicide in 2016, it is still being marketed under different formulations (Huang et al., 2019). However, owing to its rapid mode of action and the production of enormous amounts of ROS (Novaes et al., 2016), PQ can also damage the crop species, limiting its application mainly to no-tillage farms and orchards. Clearly, it will be important to develop PQ-resistant crops to enable confident use of this herbicide without damage to agricultural crops; this would not only expand the application acreage of PQ but also boost its production. More importantly, we may be forced to resort to PQ to control weeds that have evolved resistance to other herbicides in crop fields.
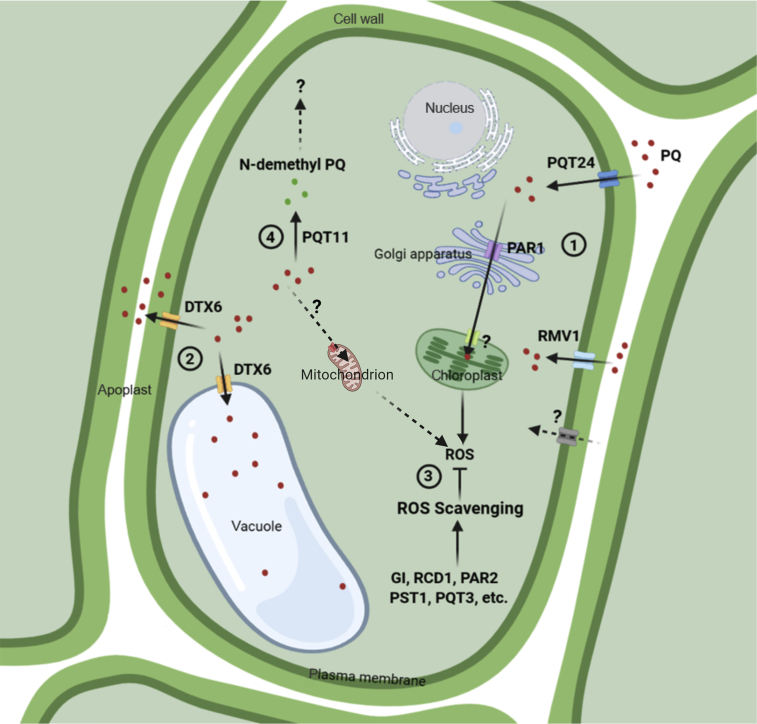
As more and more information on the molecular mechanisms of PQ resistance is revealed, more genetic loci will be available for the development of PQ-resistant crops. With the model plant Arabidopsis, we have been able to dissect the molecular mechanisms of PQ resistance. Over the past decade, several key molecular mechanisms have been unraveled and their pertinent genes identified (Figure 1); these findings can serve as guidelines and tools for the development of PQ-resistant crops (Figure 2).
Figure 1.
Molecular mechanisms of PQ resistance elucidated in Arabidopsis thaliana
PQ is transported across the plasma membrane into the cytosol from the external environment by ABC transporters such as PDR11/PQT24 and L-amino acid transporters such as RMV1/LAT1, as well as other unidentified transporters that recognize PQ as their mimic substrate. Once inside the cell, PQ faces several fates. First, PQ is transported by PAR1/LAT4 and other unidentified transporters to its site of action, the chloroplast, where it competes for electrons from PSI and generates large amounts of ROS that are scavenged by antioxidant enzymes. Any mutations that enhance ROS-scavenging ability, such as pqt3 and pst1, would help plants to tolerate PQ. Second, PQ is exported to the vacuole and apoplast by efflux transporters such as DTX6/PQT15/RTP1. Third, PQ is catabolized to nontoxic products by plant enzymes such as PQT11/CYP86A4. Therefore, PQ resistance mechanisms revealed by Arabidopsis mutants to date include (1) impaired PQ transport (rvm1, pqt24, and par1); (2) enhanced PQ export to the vacuole and apoplast (dtx6D); (3) enhanced ROS-scavenging capability (e.g., pst1 and pqt3); and (4) enhanced metabolic detoxification of PQ (pqt11D). Unconfirmed PQ transporters and transport routes are indicated by question marks and dashed lines, respectively. Red dots represent PQ, and green dots represent N-demethyl PQ.
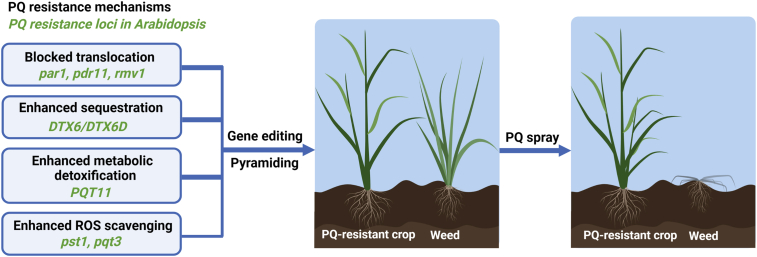
Figure 2.
Potential strategies for the development of PQ-resistant crops
Thanks to extensive studies in weeds and Arabidopsis, major PQ resistance mechanisms have been elucidated and the pertinent genes identified; these can serve as candidate genes for the development of PQ-resistant crops. Multiple genes contribute to PQ resistance, making the pyramiding of different PQ resistance genes a powerful strategy for PQ-resistant crop development. Because PQ translocation and sequestration are the major PQ resistance mechanisms in weeds, these mechanisms may be prioritized for PQ-resistant crops, in addition to metabolic detoxification. The genes that confer PQ resistance in Arabidopsis could be pyramided in crops by gene editing to knock out the crop allelic PDR11, RMV1, PAR1, PQT3, and PST1 and by increasing the expression of the crop allelic DTX6, DTX6D, and PQT11. The development of crops that are resistant to commercial-level PQ applications by pyramiding is a promising strategy.
First, plant cells use plasma membrane-localized transporters such as PDR11 and RMV1 to take up PQ (as a natural substrate mimic) (Fujita et al., 2012; Xi et al., 2012). Disruption of these PQ importers will reduce PQ intake and accumulation in cells, thus making plants more resistant to PQ. Once PQ is taken up into a plant cell, it can be transported to the chloroplast, its site of action. PAR1 plays a predominant role in this transport process, as clearly shown by the knockout mutant par1 (Li et al., 2013). Therefore, par1, pdr11, and rmv1 may constitute the basis of PQ resistance conferred by impaired PQ transport, and this information can be easily applied to crops by editing the corresponding homologous genes in the crop genomes. Indeed, OsPAR1 RNAi knockdown rice lines show strong PQ resistance to a 140-μM PQ spray under field conditions (Li et al., 2013). Second, PQ inside the plant cell can be exported out of the cytosol and into the apoplast and vacuole. Enhanced activities of the relevant exporters would increase PQ resistance. The recently identified MATE efflux transporter DTX6 demonstrated this molecular mechanism (Lv et al., 2021; Xia et al., 2021). The overexpression of DTX6D in Arabidopsis confers strong resistance to a near-commercial PQ dose (Xia et al., 2021). Third, PQ inside plant cells could be catabolized to nontoxic metabolites, a much-welcomed mechanism of herbicide resistance that also alleviates the food safety concern of herbicide residues. Developing crops with an enhanced ability to break down PQ is the most desirable solution. Recent progress on PQ catabolism in plants sheds light on this strategy (Huang et al., 2021). Fourth, an enhanced capacity for ROS scavenging would help plants tolerate PQ to some extent. There are many genes that can be used to enhance antioxidation, such as PST1 (Tsugane et al., 1999) and PQT3 (Luo et al., 2016). Knockout of OsPQT3 improves abiotic stress tolerance and increases grain yield in rice (Alfatih et al., 2020).
As multiple genes contribute to PQ resistance (Gaines et al., 2020), pyramiding different mechanisms of PQ resistance offers a potent strategy for the development of PQ-resistant crops. The genes described in this review that confer PQ resistance in Arabidopsis could be pyramided in crops by gene editing (crop allelic PDR11, RMV1, PAR1, PQT3) and overexpression (DTX6, DTX6D, and PQT11). It is promising to attempt to develop crops that are resistant to commercial-level PQ applications by combining genes for nonoverlapping major mechanisms of PQ resistance. However, economic traits such as yield and disease resistance should not be compromised in the future development of PQ-resistant crops by pyramiding different mechanisms.
The extensive study of PQ resistance in weeds provides instructive clues for the development of PQ-resistant crops. PQ translocation and sequestration appear to be the major resistance mechanisms in weeds, and they are likely to produce the resistance to a commercial PQ dose that is required for PQ-resistant crops. The mechanisms of PQ resistance and the genes identified in Arabidopsis, especially those involved in translocation, sequestration, and metabolic detoxification of PQ, greatly increase the likelihood that commercial PQ-resistant crops will be developed successfully. However, most studies of the molecular mechanisms of PQ resistance in Arabidopsis have been conducted in the laboratory. Whether the future development of PQ-resistant crops based on these molecular mechanisms will provide PQ resistance sufficient to enable survival under commercial doses of PQ remains to be tested in field trials.
To complete the picture of PQ resistance mechanisms in plants, more and more genetic components involved in the process must be investigated. Special focus should be given to chloroplast and vacuolar transporters of PQ, as well as PQ-detoxifying enzymes. The natural substrates of these transporters and the P450 enzyme also need to be elucidated in the future. Expanding our knowledge of PQ resistance mechanisms will open an avenue for the confident use of this herbicide on crops. It would also be quite interesting to investigate whether mutations similar to those that helped to identify PQ resistance loci in Arabidopsis have also occurred in various PQ-resistant weeds in nature. Likewise, the extremely short evolutionary history of PQ resistance mechanisms may be quite attractive to evolutionary biologists. It is worth noting that the chloroplast may not be the only organelle targeted by PQ. Because PQ is toxic to non-photosynthetic organisms (Mercado and Caleño, 2021), it is reasonable to believe that PQ may attack other organelles with electron transfer chains, such as the mitochondrion, or even enzymes and protein complexes from which PQ can obtain electrons. Perhaps we should expand our study beyond the current territory of PQ resistance.
Pros and cons of developing PQ-resistant crops
Apart from providing protection against weeds, PQ-resistant crops are expected to have a major impact on crop rotation, tillage farming, yield, and the ecosystem. PQ-resistant crops will allow us to confidently cultivate formerly PQ-sensitive crops, providing advantages in terms of managing the timing and dosage of herbicide application. Tillage farming results in long-term environmental damage owing to loss of the upper fertile soil layer by water or wind, a loss that may take many years to be reversed. Moreover, tillage leads to the loss of carbon and moisture from the soil, thereby reducing its fertility (Duke, 2015). As PQ is a broad-spectrum, post-emergence, soil-inactivated, and foliar-applied herbicide that promotes no-tillage farming (Baldwin et al., 1968; Hawkes, 2014), reductions in tillage through the use of PQ-resistant crops will significantly prevent soil loss from cultivated land and help to preserve soil carbon and moisture content. PQ-resistant crops will reduce labor costs in the areas where hand-picking of weeds is a common practice, making them economical for growers.
If PQ-resistant crops are commercialized, use of this herbicide is expected to increase every year, with increasing resistance in already-resistant weed species. Monoculture practices will intensify the situation. Therefore, crops with enhanced resistance to PQ must be developed, and stacking different mechanisms of PQ resistance in a single crop offers the best solution in this regime. An alternative solution will be crop rotation, which ensures high agricultural productivity by reducing herbicide use, weed infestation, and weed resistance to the relevant herbicide due to selection pressure (Schütte et al., 2017). Transgene flow from PQ-resistant crops to feral relatives must be given special consideration when developing transgenic crops. Gene flow from glyphosate-resistant crops to their non-transgenic counterparts has already been documented (Mallory-Smith and Zapiola, 2008). However, gene flow is affected by a number of factors, such as crop species, genotype and pollination system, hybridization frequency, compatible relative species, weather circumstances, pollinator activities, and distance from transgenic crop species to their concurrently flowering wild relatives (Schütte et al., 2017). Therefore, it is difficult to predict the extent and frequency with which transgene flow will occur from PQ-resistant crops to feral biotypes.
PQ-resistant crops are expected to offer remarkable benefits in terms of efficient weed management and cost-effectiveness compared with conventional crops, although they will be linked with some potential risks. In our opinion, the benefits of PQ-resistant crops will outweigh the potential risks, and such risks can be managed with cautious agricultural practices.
Author contributions
X.-T.C., P.X., and C.-B.X. conceived the review. T.N. and Y.-J.H. prepared the first draft. J.Z. prepared the figures. All of the authors contributed input and revisions.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (grant nos. 31770273, 31270302, and 30770189). The authors declare no competing interests.
Published: May 9, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Contributor Information
Xiao-Teng Cai, Email: xtcai@mail.ustc.edu.cn.
Ping Xu, Email: pingxu@mail.ustc.edu.cn.
Cheng-Bin Xiang, Email: xiangcb@ustc.edu.cn.
References
- Achary V.M.M., Sheri V., Manna M., Panditi V., Borphukan B., Ram B., Agarwal A., Fartyal D., Teotia D., Masakapalli S.K. Overexpression of improved EPSPS gene results in field level glyphosate tolerance and higher grain yield in rice. Plant Biotechnol. J. 2020;18:2504–2519. doi: 10.1111/pbi.13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfatih A., Wu J., Jan S.U., Zhang Z.S., Xia J.Q., Xiang C.B. Loss of rice PARAQUAT TOLERANCE 3 confers enhanced resistance to abiotic stresses and increases grain yield in field. Plant Cell Environ. 2020;43:2743–2754. doi: 10.1111/pce.13856. [DOI] [PubMed] [Google Scholar]
- Azab E., Kebeish R., Hegazy A. Expression of the human gene CYP1A2 enhances tolerance and detoxification of the phenylurea herbicide linuron in Arabidopsis thaliana plants and Escherichia coli. Environ. Pollut. 2018;238:281–290. doi: 10.1016/j.envpol.2018.03.025. [DOI] [PubMed] [Google Scholar]
- Babbs C.F., Pham J.A., Coolbaugh R.C. Lethal hydroxyl radical production in paraquat-treated plants. Plant Physiol. 1989;90:1267–1270. doi: 10.1104/pp.90.4.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin B., Clarke C., Wilson I. Paraquat in chloroplasts. Biochim. Biophys. Acta -Bioenerg. 1968;162:614–617. doi: 10.1016/0005-2728(68)90069-8. [DOI] [PubMed] [Google Scholar]
- Baydoun E.A., Brett C.T. Properties and possible physiological significance of cell wall calcium binding in etiolated pea epicotyls. J. Exp. Bot. 1988;39:199–208. [Google Scholar]
- Bishop T., Powles S., Cornic G. Mechanism of paraquat resistance in Hordeum glaucum. 11.∗ paraquat uptake and translocation. Funct. Plant Biol. 1987;14:539–547. [Google Scholar]
- Bromilow R.H. Paraquat and sustainable agriculture. Pest Manag. Sci. 2004;60:340–349. doi: 10.1002/ps.823. [DOI] [PubMed] [Google Scholar]
- Brunharo C.A., Hanson B.D. Vacuolar sequestration of paraquat is involved in the resistance mechanism in Lolium perenne L. spp. multiflorum. Front. Plant Sci. 2017;8:1485. doi: 10.3389/fpls.2017.01485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busi R., Vila-Aiub M.M., Powles S.B. Genetic control of a cytochrome P450 metabolism-based herbicide resistance mechanism in Lolium rigidum. Heredity. 2011;106:817–824. doi: 10.1038/hdy.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Sun S., Wang C., Li Y., Liang Y., An F., Li C., Dong H., Yang X., Zhang J. The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 2009;19:1377–1387. doi: 10.1038/cr.2009.117. [DOI] [PubMed] [Google Scholar]
- De Block M., Verduyn C., De Brouwer D., Cornelissen M. Poly(ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J. 2005;41:95–106. doi: 10.1111/j.1365-313X.2004.02277.x. [DOI] [PubMed] [Google Scholar]
- Délye C. Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: a major challenge for weed science in the forthcoming decade. Pest Manag. Sci. 2013;69:176–187. doi: 10.1002/ps.3318. [DOI] [PubMed] [Google Scholar]
- Délye C., Jasieniuk M., Le Corre V. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 2013;29:649–658. doi: 10.1016/j.tig.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Délye C., Duhoux A., Pernin F., Riggins C.W., Tranel P.J. Molecular mechanisms of herbicide resistance. Weed Sci. 2015;63:91–115. [Google Scholar]
- Didierjean L., Gondet L., Perkins R., Lau S.-M.C., Schaller H., O'Keefe D.P., Werck-Reichhart D. Engineering herbicide metabolism in tobacco and Arabidopsis with CYP76B1, a cytochrome P450 enzyme from Jerusalem artichoke. Plant Physiol. 2002;130:179–189. doi: 10.1104/pp.005801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Hu H., Wang Y., Xu Z., Zha Y., Cai X., Peng L., Feng S. A pqr2 mutant encodes a defective polyamine transporter and is negatively affected by ABA for paraquat resistance in Arabidopsis thaliana. J. Plant Res. 2016;129:899–907. doi: 10.1007/s10265-016-0819-y. [DOI] [PubMed] [Google Scholar]
- Duke S.O. Perspectives on transgenic, herbicide-resistant crops in the United States almost 20 years after introduction. Pest Manag. Sci. 2015;71:652–657. doi: 10.1002/ps.3863. [DOI] [PubMed] [Google Scholar]
- Duke S.O., Cerdeira A.L. In: Transgenic Crop Plants. Kole C., Michler C.H., Abbott A.G., Hall T.C., editors. Springer Berlin Heidelberg; 2010. Transgenic crops for herbicide resistance; pp. 133–166. [DOI] [Google Scholar]
- Farrington J., Ebert M., Land E., Fletcher K. Pulse radiolysis studies of the reaction of paraquat radical with oxygen. Implication for the mode of action of bipuridyl herbicides. Biochim. Biophys. Acta. 1973;314:372–381. doi: 10.1016/0005-2728(73)90121-7. [DOI] [PubMed] [Google Scholar]
- Fartyal D., Agarwal A., James D., Borphukan B., Ram B., Sheri V., Yadav R., Manna M., Varakumar P., Achary V.M.M. Co-expression of P173S mutant rice EPSPS and igrA genes results in higher glyphosate tolerance in transgenic rice. Front. Plant Sci. 2018;9:144. doi: 10.3389/fpls.2018.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 2011;155:2–18. doi: 10.1104/pp.110.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst E.P., Vaughn K.C. Mechanisms of paraquat resistance. Weed Technol. 1990;4:150–156. [Google Scholar]
- Fuerst E.P., Nakatani H.Y., Dodge A.D., Penner D., Arntzen C.J. Paraquat resistance in Conyza. Plant Physiol. 1985;77:984–989. doi: 10.1104/pp.77.4.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujibe T., Saji H., Arakawa K., Yabe N., Takeuchi Y., Yamamoto K.T. A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol. 2004;134:275–285. doi: 10.1104/pp.103.033480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Shinozaki K. Identification of polyamine transporters in plants: paraquat transport provides crucial clues. Plant Cell Physiol. 2014;55:855–861. doi: 10.1093/pcp/pcu032. [DOI] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Iuchi S., Yamada K., Kobayashi Y., Urano K., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. Natural variation in a polyamine transporter determines paraquat tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2012;109:6343–6347. doi: 10.1073/pnas.1121406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines T.A., Duke S.O., Morran S., Rigon C.A., Tranel P.J., Küpper A., Dayan F.E. Mechanisms of evolved herbicide resistance. J. Biol. Chem. 2020;295:10307–10330. doi: 10.1074/jbc.REV120.013572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Guo Y., Hong H., Jin L., Zhang L., Chang R.-Z., Lu W., Lin M., Qiu L.-J. Co-expression of G2-EPSPS and glyphosate acetyltransferase GAT genes conferring high tolerance to glyphosate in soybean. Front. Plant Sci. 2015;6:847. doi: 10.3389/fpls.2015.00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge J.M. Lipid peroxidation initiated by superoxide-dependent hydroxyl radicals using complexed iron and hydrogen peroxide. FEBS Lett. 1984;172:245–249. doi: 10.1016/0014-5793(84)81134-5. [DOI] [PubMed] [Google Scholar]
- Hanke G.T., Kimata-Ariga Y., Taniguchi I., Hase T. A post genomic characterization of Arabidopsis ferredoxins. Plant Physiol. 2004;134:255–264. doi: 10.1104/pp.103.032755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C.C., Nelson D.R., Moller B.L., Werck-Reichhart D. Plant cytochrome P450 plasticity and evolution. Mol. Plant. 2021;14:1772. doi: 10.1016/j.molp.2021.09.013. [DOI] [PubMed] [Google Scholar]
- Hart J.J., Di Tomaso J.M., Linscott D.L., Kochian L.V. Characterization of the transport and cellular compartmentation of paraquat in roots of intact maize seedlings. Pestic. Biochem. Physiol. 1992;43:212–222. [Google Scholar]
- Hart J.J., DiTomaso J.M., Linscott D.L., Kochian L.V. Transport interactions between paraquat and polyamines in roots of intact maize seedlings. Plant Physiol. 1992;99:1400–1405. doi: 10.1104/pp.99.4.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey B., Muldoon J., Harper D. Mechanism of paraquat tolerance in perennial ryegrass: I. Uptake, metabolism and translocation of paraquat. Plant Cell Environ. 1978;1:203–209. [Google Scholar]
- Hawkes T.R. Mechanisms of resistance to paraquat in plants. Pest Manag. Sci. 2014;70:1316–1323. doi: 10.1002/ps.3699. [DOI] [PubMed] [Google Scholar]
- He X., Szewczyk P., Karyakin A., Evin M., Hong W.-X., Zhang Q., Chang G. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature. 2010;467:991–994. doi: 10.1038/nature09408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap I. The international herbicide-resistant weed database. 2021. www.weedscience.org
- Henikoff S., Greene E.A., Pietrokovski S., Bork P., Attwood T.K., Hood L. Gene families: the taxonomy of protein paralogs and chimeras. Science. 1997;278:609–614. doi: 10.1126/science.278.5338.609. [DOI] [PubMed] [Google Scholar]
- Higgins C.F. ABC transporters: from microorganisms to man. Annu. Rev. Cel. Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Höfer R., Boachon B., Renault H., Gavira C., Miesch L., Iglesias J., Ginglinger J.-F., Allouche L., Miesch M., Grec S. Dual function of the cytochrome P450 CYP76 family from Arabidopsis thaliana in the metabolism of monoterpenols and phenylurea herbicides. Plant Physiol. 2014;166:1149–1161. doi: 10.1104/pp.114.244814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-J., Huang Y.-P., Xia J.-Q., Fu Z.-P., Chen Y.-F., Huang Y.-P., Ma A., Hou W.-T., Chen Y.-X., Qi X. AtPQT11, a P450 Enzyme, Detoxifies Paraquat via N-Demethylation. bioRxiv. 2021 doi: 10.1101/2021.06.23.449549. Preprint at. [DOI] [PubMed] [Google Scholar]
- Huang Y., Zhan H., Bhatt P., Chen S. Paraquat degradation from Contaminated environments: current achievements and perspectives. Front. Microbiol. 2019;10:1754. doi: 10.3389/fmicb.2019.01754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J., Khanom S., Moon Y., Shin S., Lee O.R. PgCYP76B93 docks on phenylurea herbicides and its expression enhances chlorotoluron tolerance in Arabidopsis. Appl. Biol. Chem. 2020;63:1–11. [Google Scholar]
- Jo J., Won S.-H., Son D., Lee B.-H. Paraquat resistance of transgenic tobacco plants over-expressing the Ochrobactrum anthropipqrA gene. Biotechnol. Lett. 2004;26:1391–1396. doi: 10.1023/B:BILE.0000045638.82348.7a. [DOI] [PubMed] [Google Scholar]
- Jóri B., Soos V., Szegő D., Páldi E., Szigeti Z., Rácz I., Lásztity D. Role of transporters in paraquat resistance of horseweed Conyza canadensis (L.) Cronq. Pestic. Biochem. Physiol. 2007;88:57–65. [Google Scholar]
- Kuang Y., Li S., Ren B., Yan F., Spetz C., Li X., Zhou X., Zhou H. Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms. Mol. Plant. 2020;13:565–572. doi: 10.1016/j.molp.2020.01.010. [DOI] [PubMed] [Google Scholar]
- Kurepa J., Smalle J., Va M., Montagu N., Inzé D. Oxidative stress tolerance and longevity in Arabidopsis: the late-flowering mutant gigantea is tolerant to paraquat. Plant J. 1998;14:759–764. doi: 10.1046/j.1365-313x.1998.00168.x. [DOI] [PubMed] [Google Scholar]
- Kuroda T., Tsuchiya T. Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta Proteins Proteomics. 2009;1794:763–768. doi: 10.1016/j.bbapap.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Lasat M.M., DiTomaso J.M., Hart J.J., Kochian L.V. Evidence for vacuolar sequestration of paraquat in roots of a paraquat-resistant Hordeum glaucum biotype. Physiol. Plant. 1997;99:255–262. [Google Scholar]
- Li H., Zhu Q., Wang S., Huang T., Li X., Ni C., Fang Y., Li L., Lian Q., Ge R.-S. Paraquat exposure delays stem/progenitor Leydig cell regeneration in the adult rat testis. Chemosphere. 2019;231:60–71. doi: 10.1016/j.chemosphere.2019.05.104. [DOI] [PubMed] [Google Scholar]
- Li J., Mu J., Bai J., Fu F., Zou T., An F., Zhang J., Jing H., Wang Q., Li Z., et al. Paraquat Resistant1, a Golgi-localized putative transporter protein, is involved in intracellular transport of paraquat. Plant Physiol. 2013;162:470–483. doi: 10.1104/pp.113.213892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Kuang Y., Yan F., Li S., Ren B., Gosavi G., Spetz C., Li X., Wang X., Zhou X. Developing a novel artificial rice germplasm for dinitroaniline herbicide resistance by base editing of OsTubA2. Plant Biotechnol. J. 2021;19:5. doi: 10.1111/pbi.13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Symersky J., Radchenko M., Koide A., Guo Y., Nie R., Koide S. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc. Natl. Acad. Sci. U S A. 2013;110:2099–2104. doi: 10.1073/pnas.1219901110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Cai X.-T., Du J., Zhao T.-L., Wang P.-F., Zhao P.-X., Liu R., Xie Q., Cao X.-F., Xiang C.-B. PARAQUAT TOLERANCE3 is an E3 ligase that switches off activated oxidative response by targeting histone-modifying PROTEIN METHYLTRANSFERASE4b. PLoS Genet. 2016;12:e1006332. doi: 10.1371/journal.pgen.1006332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q., Wei J., Dong Z., Shen X., Chen Y. Differences of endogenous polyamines and putative genes associated with paraquat resistance in goosegrass (Eleusine indica L.) PLoS One. 2019;14:e0216513. doi: 10.1371/journal.pone.0216513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Z., Zhao M., Wang W., Wang Q., Huang M., Li C., Lian Q., Xia J., Qi J., Xiang C., et al. Changing Gly311 to acidic amino acid in the MATE family protein DTX6 leads to enhanced resistance of Arabidopsis to the dihydropyridine herbicides. Mol. Plant. 2021;14:2115–2125. doi: 10.1016/j.molp.2021.09.002. [DOI] [PubMed] [Google Scholar]
- Mallory-Smith C., Zapiola M. Gene flow from glyphosate-resistant crops. Pest Manag. Sci. 2008;64:428–440. doi: 10.1002/ps.1517. [DOI] [PubMed] [Google Scholar]
- Mazur B.J., Falco S.C. The development of herbicide resistant crops. Annu. Rev. Plant Phys. 1989;40:441–470. doi: 10.1146/annurev.pp.40.060189.002301. [DOI] [Google Scholar]
- Mercado S.A.S., Caleño J.D.Q. Use of Lens culinaris Med test as environmental bioindicator to identify the cytogenotoxic effect of paraquat pesticide. Environ. Sci. Pollut. Res. 2021:1–8. doi: 10.1007/s11356-021-14352-0. [DOI] [PubMed] [Google Scholar]
- Moretti M.L., Alarcon-Reverte R., Pearce S., Morran S., Hanson B.D. Transcription of putative tonoplast transporters in response to glyphosate and paraquat stress in Conyza bonariensis and Conyza canadensis and selection of reference genes for qRT-PCR. PLoS One. 2017;12:e0180794. doi: 10.1371/journal.pone.0180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Mhamdi A., Foyer C.H. Oxidative stress and antioxidative systems: recipes for successful data collection and interpretation. Plant Cell Environ. 2016;39:1140–1160. doi: 10.1111/pce.12726. [DOI] [PubMed] [Google Scholar]
- Noctor G., Mhamdi A., Chaouch S., Han Y., Neukermans J., Marquez-Garcia B., Queval G., Foyer C.H. Glutathione in plants: an integrated overview. Plant Cell Environ. 2012;35:454–484. doi: 10.1111/j.1365-3040.2011.02400.x. [DOI] [PubMed] [Google Scholar]
- Norman M.A., Fuerst E.P., Smeda R.J., Vaughn K. Evaluation of paraquat resistance mechanisms in Conyza. Pestic. Biochem. Physiol. 1993;46:236–249. [Google Scholar]
- Norman M.A., Smeda R.J., Vaughn K.C., Fuerst E.P. Differential movement of paraquat in resistant and sensitive biotypes of Conyza. Pestic. Biochem. Physiol. 1994;50:31–42. [Google Scholar]
- Novaes R.D., Gonçalves R.V., Cupertino M.C., Santos E.C., Bigonha S.M., Fernandes G.J., Maldonado I.R., Natali A.J. Acute paraquat exposure determines dose-dependent oxidative injury of multiple organs and metabolic dysfunction in rats: impact on exercise tolerance. Int. J. Exp. Pathol. 2016;97:114–124. doi: 10.1111/iep.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Ishikawa K., Harada K., Fukusaki E., Yoshimura K., Shigeoka S. Overexpression of an ADP-ribose pyrophosphatase, AtNUDX2, confers enhanced tolerance to oxidative stress in Arabidopsis plants. Plant J. 2009;57:289–301. doi: 10.1111/j.1365-313X.2008.03686.x. [DOI] [PubMed] [Google Scholar]
- Overmyer K., Tuominen H., Kettunen R., Betz C., Langebartels C., Sandermann H., Jr., Kangasjarvi J. Ozone-sensitive arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell. 2000;12:1849–1862. doi: 10.1105/tpc.12.10.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powles S.B., Yu Q. Evolution in action: plants resistant to herbicides. Annu. Rev. Plant Biol. 2010;61:317–347. doi: 10.1146/annurev-arplant-042809-112119. [DOI] [PubMed] [Google Scholar]
- Powles S.B., Tucker E.S., Morgan T.R. A capeweed (Arctotheca calendula) biotype in Australia resistant to bipyridyl herbicides. Weed Sci. 1989;37:60–62. [Google Scholar]
- Preston C. Herbicide Resistance in Plants: Biology and Biochemistry. Lewis Publ.; 1994. Resistance to photosystem I disrupting herbicides; pp. 61–82. [Google Scholar]
- Reczek C.R., Birsoy K., Kong H., Martínez-Reyes I., Wang T., Gao P., Sabatini D.M., Chandel N.S. A CRISPR screen identifies a pathway required for paraquat-induced cell death. Nat. Chem. Biol. 2017;13:1274–1279. doi: 10.1038/nchembio.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütte G., Eckerstorfer M., Rastelli V., Reichenbecher W., Restrepo-Vassalli S., Ruohonen-Lehto M., Saucy A.-G.W., Mertens M. Herbicide resistance and biodiversity: agronomic and environmental aspects of genetically modified herbicide-resistant plants. Environ. Sci. Europe. 2017;29:1–12. doi: 10.1186/s12302-016-0100-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaaltiel Y., Gressel J. Multienzyme oxygen radical detoxifying system correlated with paraquat resistance in Conyza bonariensis. Pestic. Biochem. Physiol. 1986;26:22–28. [Google Scholar]
- Shaaltiel Y., Chua N.-H., Gepstein S., Gressel J. Dominant pleiotropy controls enzymes co-segregating with paraquat resistance in Conyza bonariensis. Theor. Appl. Genet. 1988;75:850–856. [Google Scholar]
- Shaaltiel Y., Glazer A., Bocion P., Gressel J. Cross tolerance to herbicidal and environmental oxidants of plant biotypes tolerant to paraquat, sulfur dioxide, and ozone. Pestic. Biochem. Physiol. 1988;31:13–23. [Google Scholar]
- Soar C., Karotam J., Preston C., Powles S. Reduced paraquat translocation in paraquat resistant Arctotheca calendula (L.) Levyns is a consequence of the primary resistance mechanism, not the cause. Pestic. Biochem. Physiol. 2003;76:91–98. [Google Scholar]
- Soar C., Preston C., Karotam J., Powles S. Polyamines can inhibit paraquat toxicity and translocation in the broadleaf weed Arctotheca calendula. Pestic. Biochem. Physiol. 2004;80:94–105. [Google Scholar]
- Soós V. Role of transporters in the mechanism of paraquat resistance of horseweed (Conyza canadensis (L.) Cronq.) Acta Biol. Szeged. 2005;49:191–193. [Google Scholar]
- Sun Y., Zhang X., Wu C., He Y., Ma Y., Hou H., Guo X., Du W., Zhao Y., Xia L. Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol. Plant. 2016;9:628–631. doi: 10.1016/j.molp.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Tsugane K., Kobayashi K., Niwa Y., Ohba Y., Wada K., Kobayashi H. A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell. 1999;11:1195–1206. doi: 10.1105/tpc.11.7.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji K., Hosokawa M., Morita S., Miura R., Tominaga T. Resistance to paraquat in M azus pumilus. Weed Res. 2013;53:176–182. [Google Scholar]
- Vicente J.A., Peixoto F., Lopes M.L., Madeira V.M. Differential sensitivities of plant and animal mitochondria to the herbicide paraquat. J. Biochem. Mol. Toxicol. 2001;15:322–330. doi: 10.1002/jbt.10010. [DOI] [PubMed] [Google Scholar]
- Wang H., Xu D., Zhu X., Wang M., Xia Z. The maize SUMO conjugating enzyme ZmSCE1b protects plants from paraquat toxicity. Ecotoxicol. Environ. Saf. 2021;211:111909. doi: 10.1016/j.ecoenv.2021.111909. [DOI] [PubMed] [Google Scholar]
- Wen X., Gibson C.J., Yang I., Buckley B., Goedken M.J., Richardson J.R., Aleksunes L.M. MDR1 transporter protects against paraquat-induced toxicity in human and mouse proximal tubule cells. Toxicol. Sci. 2014;141:475–483. doi: 10.1093/toxsci/kfu141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S.-H., Lee B.-H., Lee H.-S., Jo J. An Ochrobactrum anthropi gene conferring paraquat resistance to the heterologous host Escherichia coli. Biochem. Biophys. Res. Commun. 2001;285:885–890. doi: 10.1006/bbrc.2001.5268. [DOI] [PubMed] [Google Scholar]
- Wu J., Chen C., Xian G., Liu D., Lin L., Yin S., Sun Q., Fang Y., Zhang H., Wang Y. Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing. Plant Biotechnol. J. 2020;18:1857. doi: 10.1111/pbi.13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi J., Xu P., Xiang C.B. Loss of AtPDR11, a plasma membrane-localized ABC transporter, confers paraquat tolerance in Arabidopsis thaliana. Plant J. 2012;69:782–791. doi: 10.1111/j.1365-313X.2011.04830.x. [DOI] [PubMed] [Google Scholar]
- Xia J.Q., Nazish T., Javaid A., Ali M., Liu Q.Q., Wang L., Zhang Z.Y., Zhang Z.S., Huang Y.J., Wu J., et al. A gain-of-function mutation of the MATE family transporter DTX6 confers paraquat resistance in Arabidopsis. Mol. Plant. 2021;14:2126–2133. doi: 10.1016/j.molp.2021.09.004. [DOI] [PubMed] [Google Scholar]
- Yamada T., Kambara Y., Imaishi H., Ohkawa H. Molecular cloning of novel cytochrome P450 species induced by chemical treatments in cultured tobacco cells. Pestic. Biochem. Physiol. 2000;68:11–25. [Google Scholar]
- Yerushalmi H., Lebendiker M., Schuldiner S. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J. Biol. Chem. 1995;270:6856–6863. doi: 10.1074/jbc.270.12.6856. [DOI] [PubMed] [Google Scholar]
- Youngman R. In: Akoyunoglou G., editor. Balaban International Science Services; 1981. On the Mechanism of Paraquat Resistance in Conyza Sp; pp. 537–544. (Photosynthesis and Plant Productivity, Photosynthesis and Environment). [Google Scholar]
- Yu Q., Cairns A., Powles S. Glyphosate, paraquat and ACCase multiple herbicide resistance evolved in a Lolium rigidum biotype. Planta. 2007;225:499–513. doi: 10.1007/s00425-006-0364-3. [DOI] [PubMed] [Google Scholar]
- Yu Q., Huang S., Powles S. Direct measurement of paraquat in leaf protoplasts indicates vacuolar paraquat sequestration as a resistance mechanism in Lolium rigidum. Pestic. Biochem. Physiol. 2010;98:104–109. [Google Scholar]
- Zhang R., Liu J., Chai Z., Chen S., Bai Y., Zong Y., Chen K., Li J., Jiang L., Gao C. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat. Plants. 2019;5:480–485. doi: 10.1038/s41477-019-0405-0. [DOI] [PubMed] [Google Scholar]