Abstract
Benzoxazinoids are a class of protective and allelopathic plant secondary metabolites that have been identified in multiple grass species and are encoded by the Bx biosynthetic gene cluster (BGC) in maize. Data mining of 41 high-quality grass genomes identified complete Bx clusters (containing genes Bx1–Bx5 and Bx8) in three genera (Zea, Echinochloa, and Dichanthelium) of Panicoideae and partial clusters in Triticeae. The Bx cluster probably originated from gene duplication and chromosomal translocation of native homologs of Bx genes. An ancient Bx cluster that included additional Bx genes (e.g., Bx6) is presumed to have been present in ancestral Panicoideae. The ancient Bx cluster was putatively gained by the Triticeae ancestor via horizontal transfer (HT) from the ancestral Panicoideae and later separated into multiple segments on different chromosomes. Bx6 appears to have been under less constrained selection compared with the Bx cluster during the evolution of Panicoideae, as evidenced by the fact that it was translocated away from the Bx cluster in Zea mays, moved to other chromosomes in Echinochloa, and even lost in Dichanthelium. Further investigations indicate that purifying selection and polyploidization have shaped the evolutionary trajectory of Bx clusters in the grass family. This study provides the first candidate case of HT of a BGC between plants and sheds new light on the evolution of BGCs.
Keywords: biosynthetic gene cluster, horizontal transfer, benzoxazinoid, grass, purifying selection
Biosynthetic gene clustering and horizontal gene transfer are two evolutionary inventions that enable rapid adaptation by organisms. In this manuscript, by mining 41 plant genomes, Wu et al. deciphered the organization of the Bx cluster, a biosynthetic gene cluster for benzoxazinoids in grasses, and identified horizontal transfer of the Bx cluster from Panicoideae to Triticeae. This study provides the first candidate case of the horizontal transfer of a biosynthetic gene cluster in plants.
Introduction
Biosynthetic gene clusters (BGCs) are specialized genomic organizations composed of a cluster of non-homologous genes that contribute to the biosynthesis of chemical defensive metabolites (Nützmann and Osbourn, 2014; Nützmann et al., 2018). The selective advantages of clustering, such as gene co-regulation and co-inheritance, may promote the formation of BGCs (Nützmann and Osbourn, 2014; Nützmann et al., 2016; Rokas et al., 2018). Natural selection, including long-term purifying selection, positive selection, and balancing selection, has also driven the establishment and maintenance of BGCs (Carbone et al., 2007; Takos and Rook, 2012; Rokas et al., 2018; Liu et al., 2020). The formation and evolution of BGCs have been studied extensively in fungi (Rokas et al., 2018). Approximately 30 examples of BGCs in plants have been identified in recent years (Boycheva et al., 2014; Guo et al., 2018). The Bx cluster for the biosynthesis of benzoxazinoids was the first BGC identified in plants (Frey et al., 1997).
Gene duplication, neofunctionalization, and relocation have been suggested as the origins of BGCs in most fungi and plants (Nützmann et al., 2018; Rokas et al., 2018). The DAL gene cluster involved in allantoin metabolism originated from duplication of native genes and relocation in the yeast Saccharomyces cerevisiae (Wong and Wolfe, 2005). The GAL cluster in Candida yeasts originated from the relocation of native unclustered genes (Slot and Rokas, 2010). Horizontal transfer (HT) also leads to the emergence and spread of BGCs and is an important source of genomic innovation (Khaldi et al., 2008; Slot and Rokas, 2011; Reynolds et al., 2018; Kominek et al., 2019; Tralamazza et al., 2019; Li et al., 2020). In the fungus Aspergillus clavatus, the ACE1 gene cluster originated by HT from a donor closely related to the rice blast fungus Magnaporthe grisea (Khaldi et al., 2008). The GAL cluster of Schizosaccharomyces yeasts was acquired from a Candida yeast (Slot and Rokas, 2010). A full operon encoding siderophore biosynthesis genes was horizontally transferred from bacteria to a group of budding yeasts (Kominek et al., 2019). In animals, bdelloid rotifers, small freshwater invertebrates, appear to have acquired a BGC for cell wall peptidoglycan biosynthesis composed of a racemase and a ligase from bacteria (Gladyshev et al., 2008). In plants, BGCs are unlikely to have been derived from microbes via HT (Nützmann et al., 2018), and no BGCs acquired by HT have been identified.
Benzoxazinoids are a class of indole-derived protective and allelopathic secondary metabolites that function in plant defense against insect herbivores, microbial pathogens, and neighboring competing plants (reviewed in Frey et al., 2009). 2,4-Dihydroxy-1,4-benzoxazin-3-one (DIBOA) and its 7-methoxy analog DIMBOA are the predominant representatives of benzoxazinoids in plants (Frey et al., 1997, 2009), and these compounds have been identified in many plants, including maize (Zea mays), wheat (Triticum aestivum), and barnyardgrass (Echinochloa crus-galli) (Frey et al., 2009; Guo et al., 2017). In the weed species Echinochloa, DIBOA functions as an allelopathic compound against rice in paddy fields (Guo et al., 2017).
The pathway of benzoxazinoid biosynthesis has been elucidated extensively in Z. mays (Figure 1A). The first step is the biosynthesis of indole from indole-3-glycerolphosphate in the chloroplast by Bx1, a homolog of the α-subunit of tryptophan synthase. Four P450 monooxygenases from the CYP71C subfamily (Bx2–Bx5) add four oxygen atoms at the four positions of the indole to synthesize DIBOA, the simplest benzoxazinoid (Frey et al., 1997). Two uridine diphosphate (UDP)-glucosyltransferases (UGTs), Bx8 and Bx9, attach a glucose moiety to DIBOA to produce DIBOA-Glc (Von Rad et al., 2002). Bx6, a 2-oxoglutarate-dependent dioxygenase (2-ODD), oxidizes DIBOA-Glc to TRIBOA-Glc, and Bx7 (O-methyltransferase [OMT]) subsequently methylates TRIBOA-Glc to produce DIMBOA-Glc (Jonczyk et al., 2008). Four functionally redundant OMTs (Bx10–Bx12 and Bx14) catalyze the conversion of DIMBOA-Glc to HDMBOA-Glc (Meihls et al., 2013). Bx13, a Bx6-like 2-ODD, converts DIMBOA-Glc to TRIMBOA-Glc, and TRIMBOA-Glc is further methylated by Bx7 to produce DIM2BOA-Glc (Handrick et al., 2016). Bx14 catalyzes the reaction from DIM2BOA-Glc to HDIM2BOA-Glc by methylation (Handrick et al., 2016).
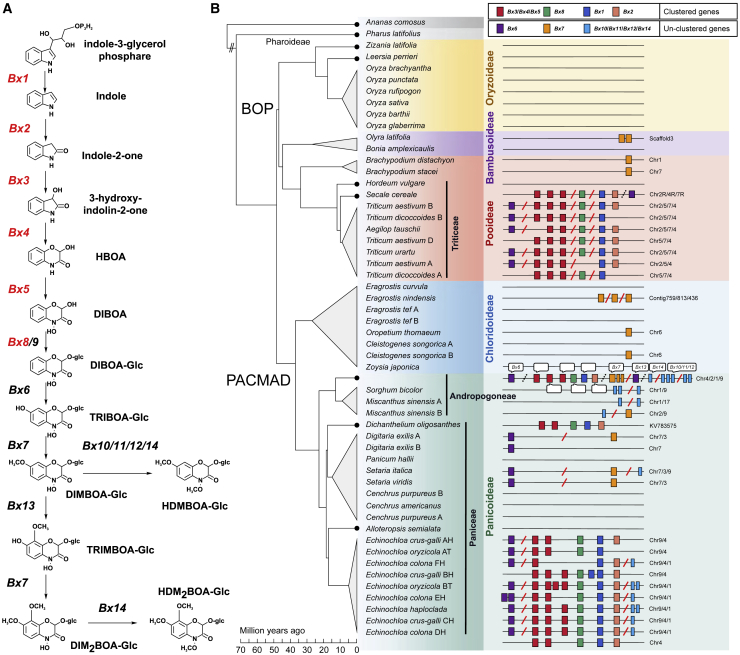
Figure 1.
Benzoxazinoid biosynthesis pathway and distribution of Bx genes in grass.es
(A) Biosynthetic pathway of benzoxazinoid secondary metabolites in maize. The pathway (Bx)-related genes in the Bx cluster are marked in red.
(B) Phylogeny and Bx gene distribution of grass species. Background colors represent different subfamilies in Poaceae. The lineage divergence time is adopted from the TimeTree database (www.timetree.org). Each rectangle represents one gene element. Genes flanking the red slash are located on two different chromosomes, and genes flanking the black dashed slash are on the same chromosome but not clustered.
In maize, six Bx genes (Bx1–Bx5 and Bx8) that encode enzymes for the first few steps of DIMBOA biosynthesis form a well-defined BGC (the Bx cluster) at the tip region of chromosome 4 (Frey et al., 1997, 2009). Bx genes have been identified in an intact cluster in barnyardgrass and in dispersed subclusters in wheat and rye (Sue et al., 2011; Guo et al., 2017). Previous studies have supported a monophyletic origin of Bx genes for benzoxazinoid biosynthesis (Frey et al., 2009; Sue et al., 2011; Dutartre et al., 2012; Nützmann and Osbourn, 2014). The progenitors evolved Bx genes before the divergence of the Triticeae and the Panicoideae (Sue et al., 2011; Dutartre et al., 2012). However, it should be noted that limited sampling may lead to overinterpretation of gene phylogeny. Frequent gene loss and rearrangements and patchy distribution across divergent species have complicated our understanding of BGC evolution (Lind et al., 2017). The broad availability of high-quality genomes of important crops and wild grasses has facilitated the discovery of more BGCs (Guo et al., 2018), enabling us to trace the organization and evolution of BGCs more comprehensively and reliably.
Here, we identified homologs of Bx genes in the grass family using 41 high-quality monocot genomes, explored the origin of the Bx cluster, and reconstructed its evolutionary trajectory. Through the analysis of sequence similarities, phylogeny, and genomic synteny, we provide evidence that the Bx clusters currently observed in grasses have originated from a complex evolutionary process that includes HT. The HT event and further natural selection have shaped the presence of Bx clusters in the grass family.
Results
Identification and distribution of Bx genes in the grass family
Key genes in the benzoxazinoid biosynthesis pathway of Z. mays, including those in the Bx cluster (Bx1–Bx5 and Bx8) and Bx genes dispersed in the genome (Bx6, Bx7, and Bx9–Bx14) were used as bait to search for Bx genes and homologs in the genomes of 41 monocot species covering five subfamilies of core grasses (Bambusoideae, Oryzoideae, and Pooideae [BOP] from the BOP lineage, and Chloridoideae and Panicoideae from the Panicoideae, Arundinoideae, Chloridoideae, Micrairoideae, Aristidoideae, and Danthonioideae [PACMAD] lineage) and the basal group of Poaceae (Pharus latifolius) (Figure 1B; Supplemental Table 1; Saarela et al., 2018). Homologs of Bx genes were identified in grass genomes based on their sequence similarities, phylogeny, and genomic physical positions (Figure 1B; Supplemental Table 2).
In addition to the Bx clusters previously reported in Z. mays and Echinochloa (Frey et al., 1997; Guo et al., 2017), a Bx cluster was also found in Dichanthelium oligosanthes, Scribner’s rosette grass, a C3 panicoid grass (Figure 1B). In Triticeae, the Bx cluster is split into three subclusters located on three different chromosomes. In total, 12 clusters were found in 6 grass species, 10 of which are in the Echinochloa genus, with 1 cluster in each monoploid genome (except 1 subgenome in Echinochloa colona with 2 copies) (Wu et al., 2022). The Bx gene orders in the clusters are entirely consistent among Z. mays, D. oligosanthes, and Echinochloa, implying a single origin of the Bx clusters (Figure 1B). Although the Bx cluster is split in Triticeae, the order of Bx3–Bx5 is the same as the Bx cluster in Panicoideae, showing the potentially close relationship between Bx genes in the BOP and PACMAD lineages. Bx6 is a distant 1.31 Mb away from the Bx cluster in the Z. mays genome, although both the gene and the cluster are located on chromosome 4. Bx6 genes are located on chromosome 9 in Echinochloa, rather than on chromosome 4 where the Bx clusters are located. Bx6 was also identified in Digitaria and Setaria from Panicoideae. In Triticeae, Bx6 genes are located on chromosome 2 in Triticum and Aegilops but on chromosome 7R in rye (Secale cereale), where Bx1 and Bx2 in rye are also located (Figure 1B; Supplemental Table 2). Bx7 genes are found in both BOP and PACMAD lineages, in spite of massive loss.
Formation and HT of the Bx cluster (Bx1–Bx5 and Bx8)
The phylogeny of Bx1 homologs across the grass family was constructed, and the phylogenetic tree is clearly separated into three clades: Igl1 (indole-3-glycerol phosphate lyase 1), TSA1 (tryptophan synthase α subunit 1), and TSAh1 (tryptophan synthase A homolog 1) (Figure 2A and Supplemental Figure 1). Igl1, TSA1, and TSAh1 are paralogs of Bx1 in maize. In the phylogeny, the Bx1 clade is nested within the TSAh1 clade, implying that Bx1 genes originated from the duplication of TSAh1.
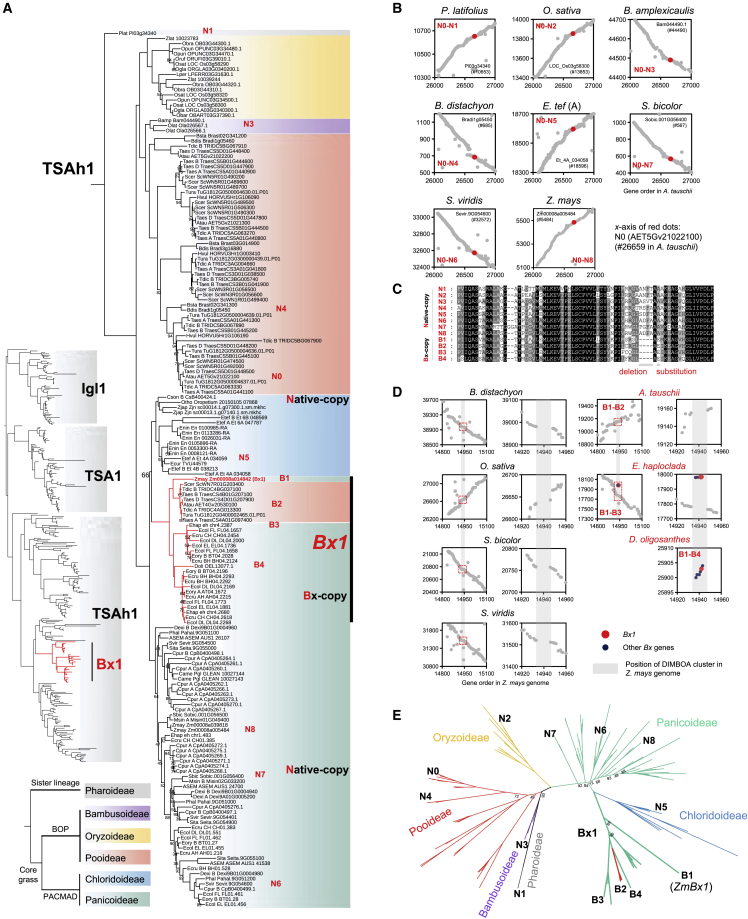
Figure 2.
Phylogeny and genomic synteny of Bx1 in grass.
(A) Phylogeny of Igl1, TSA1, and TSAh1 gene clades and a maximum-likelihood phylogenetic tree of Bx1 (TSAh1 clade) using protein sequences from grasses with P. latifolius as an outgroup species. Bootstrap values less than 95 are labeled at branches. The node label is composed of the genome abbreviation and gene ID. Background fill colors represent subfamilies. The Bx1 clade is highlighted as Bx-copy (e.g., B1–B4), and the native homologs of Bx1 (TSAh1 genes) are labeled as native-copy (e.g., N0–N7). The left bottom tree shows the phylogenetic relationships of five subfamilies.
(B) Genomic synteny of native Bx1 homologs (TSAh1 genes) among species. Red dots representing the native Bx1 homologs (TSAh1 genes) are syntenic.
(C) Local protein sequence alignments of Bx1 genes and their native homologs (TSAh1 genes). Bx-copy specific deletion and amino acid substitution are marked in gray rectangles.
(D) Genomic synteny between Z. mays and other species around the position of Bx1. For each species, the synteny around Bx1 is magnified locally in the right panel.
(E) Phylogenetic tree of Bx homologs using nucleotide sequences from the third codon position. Bootstrap values less than 95 are indicated on branches.
With P. latifolius from Pharoideae (N1 Pl03g34340 in Figure 2A) serving as an outgroup, the TSAh1 clade is divided into the BOP and PACMAD lineages, in line with the species tree (Figure 2A). Bx1 genes form a monoclade, composed of Bx1 copies from previously identified species with Bx clusters. To distinguish other Bx1 homologs from Bx1 copies in the monoclade, we referred to the other Bx1 homologs in the subtree as TSA1h homologs. The TSA1h homologs are native and extraordinarily conserved across the grass family, and they show good synteny among the genomes (Figure 2B). The TSA1h homolog AET5Gv21022100 (N0 in Figure 2A) in Aegilops tauschii from Pooideae is syntenic to the TSA1h homolog Pl3g34340 (N1) in P. latifolius from the basal lineage of Poaceae, as well as the TSA1h homologs LOC_Os3g58300 (N2) in Oryza sativa from Oryzoideae, Et_4A_034058 (N5) in Eragrostis tef from Chloridoideae, Sevir.9G054600 (N6) in Setaria viridis from Paniceae, Panicoideae, and Zm00008a005484 (N8) in Z. mays from Andropogoneae, Panicoideae. Sequence alignments show that they are conserved with the tryptophan synthase domain (Figure 2C and Supplemental Figure 2). In contrast to the native TSA1h homologs, the clade of Bx1 copies, which is nested between native TSA1h homologs of Chloridoideae and Panicoideae and is sister to native homologs of Panicoideae, is an extra lineage-specific copy duplicated in the ancestor of Panicoideae (Figure 2A). Therefore, based on the position of the Bx1 clade in the phylogeny and the conserved evolution of TSAh1 homologs across the whole family, the hypothesis that Bx1 originated before the divergence between Panicoideae and Triticeae should be rejected. To confirm the lineage-specific duplication event, local synteny of Bx1 was scanned between Z. mays and other genomes (Figure 2D). The two flanking genomic regions of the Bx cluster in Z. mays show high synteny to Brachypodium distachyon and A. tauschii from Pooideae, O. sativa from Oryzoideae, Sorghum bicolor from Andropogoneae, Panicoideae, and Setaria viridis from Paniceae, Panicoideae. However, the Bx cluster is entirely absent in these genomes. Comparing the gene positions of Bx clusters between Z. mays and Echinochloa haploclada from Echinochloa, their Bx clusters are in a large syntenic block, and the order of the Bx genes is consistent, implying a common origin of the Bx cluster in their common ancestor before the divergence of Andropogoneae and Paniceae, although there is a translocation between them. Although the scaffold that harbors the Bx cluster in D. oligosanthes is short, the sequences of five Bx genes were successfully assembled, and their orders are consistent with those in Z. mays, further supporting the origin of the Bx cluster in ancestral Panicoideae. Sequence alignment of Bx1 genes and their native homologs shows Bx1 lineage-specific deletion and substitution, confirming a single origin of Bx1 genes (Figure 2C).
Within the Bx1 clade, Bx1 genes from the Triticeae form a monoclade nested among Bx1 genes from Panicoideae, indicating a single origin of these genes in Triticeae. Because the divergence between the Pooideae and Panicoideae is ancient, estimated at more than 50 mya (Ma et al., 2021), and native TSA1h homologs are present, the positional congruence of the Triticeae Bx1 clade is unlikely to be derived from sexual hybridization, incomplete lineage sorting (ILS), or convergent evolution, but rather from HT from the Panicoideae (Figure 2A). To further confirm the robustness of the Bx1 phylogeny based on protein sequences, phylogenetic trees of Bx1 were constructed from coding sequence (CDS), codon12 (first and second codon positions), and codon3 (third codon position, whose evolution can serve as a proxy for synonymous substitution), and their topologies confirmed the gene duplication and HT of Bx1 (Figure 2E and Supplemental Figure 3).
We built a phylogeny and scanned the genomic synteny of Bx2–Bx5 and Bx8 across the whole Poaceae (Supplemental Figures 4 and 5). Native homologs of Bx2 could be traced and are highly conserved (Supplemental Figure 4). Bx3, Bx4, and Bx5 are three tandem duplicated CYP71C genes from the cytochrome P450 superfamily. The native ancestral homologs of Bx3–Bx5 are massively lost, but the retained homologs show high genomic synteny among subfamilies (Supplemental Figure4). Based on the Bx8 phylogeny, Bx8 genes are the duplicated products of native homologs, and Bx8 genes in the Triticeae are nested within those of Panicoideae. Bx9 is a maize-specific duplicate of Bx8 (Supplemental Figure 5). In brief, the topologies of the five Bx genes (Bx2–Bx5 and Bx8) are similar to those observed in the Bx1 phylogeny, implying that Bx genes in the cluster may be derived from a single origin and that Bx genes in Triticeae have probably been acquired via HT of an intact Bx cluster from Panicoideae.
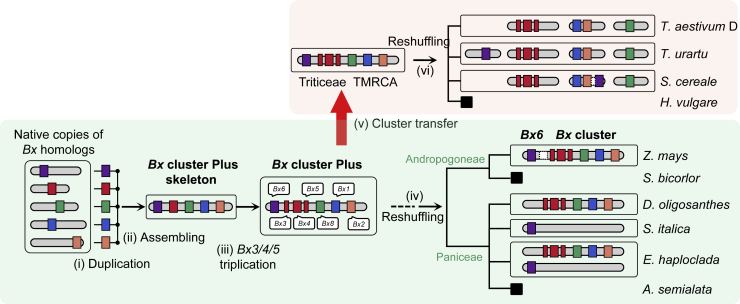
Figure 5.
A hypothetical scenario for the origin and evolution of the Bx cluster in grasses. TMRCA, the most recent common ancestor.
To formally test the hypothesis of a Panicoideae origin of the Bx genes in Triticeae, we reconstructed phylogenies under constraints that the Bx genes in Triticeae had a Panicoideae Bx clade origin (PO) or originated outside of that clade (Non-PO). To determine whether the PO phylogenies were statistically better explanations than the non-PO phylogenies, we used the approximately unbiased (AU) test, the resampling estimated log-likelihood method (RELL), and the Shimodaira-Hasegawa (SH) test. All tests of all Bx genes in the cluster (Bx1–Bx5 and Bx8) strongly rejected the alternative hypothesis that Bx genes in Triticeae are not derived from Panicoideae (all p < 0.001 for AU tests) (Supplemental Table 3). These results indicate that the obtained tree topologies of all Bx genes are highly robust and reflect HT of the Bx genes from Panicoideae to Triticeae.
Evolution of Bx6 and other Bx genes
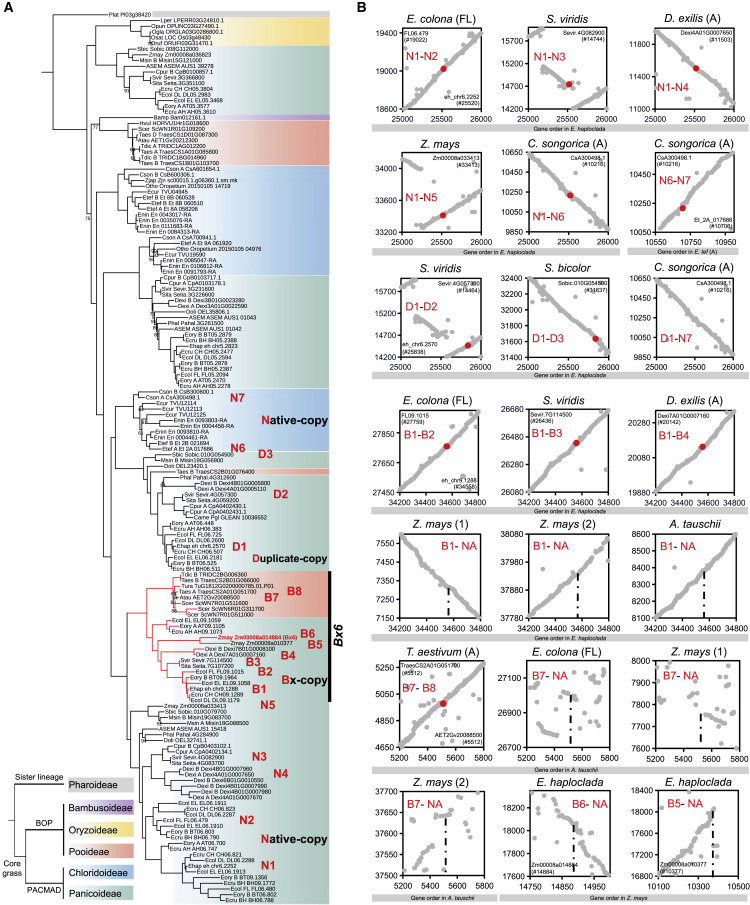
The Bx6 gene whose encoded product is responsible for oxidizing DIBOA-Glc to TRIBOA-Glc, the subsequent enzymatic step that follows the activity of the Bx cluster genes in maize, is located away from the Bx cluster (Figure 1A). The phylogeny of Bx6 shows a pattern similar to that of Bx1, in that the Bx clade is duplicated from native Bx6 homologs and HT from Panicoideae is probably responsible for the inheritance of the Bx6 genes in Triticeae (Figure 3A). Multi-species genome synteny analyses support the above results (Figure 3B). Topology tests confirm the robustness of the Bx6 phylogeny, and Bx6 genes in Triticeae are nested within the Panicoideae Bx6 clade (p < 0.001 for AU test) (Supplemental Table 3). Notably, in addition to species harboring the Bx cluster, Setaria and Digitaria from Panicoideae also contained Bx6 genes (Figures 1B and 3A). We also observed that Bx13 is a maize-specific duplicate of Bx6 (Figure 3A).
Figure 3.
Phylogeny and genomic synteny of Bx6 in grasses.
(A) A maximum-likelihood phylogenetic tree of Bx6 using protein sequences from grasses. Bootstrap values less than 95 are labeled at branches. Background fill colors represent subfamilies. The Bx6 clade is highlighted as Bx-copy (e.g., B1–B8) and the native homologs of Bx6 are labeled as native-copy (e.g., N1–N7). The other duplicates of Bx6 native homologs are labeled as duplicate-copy (e.g., D1–D3). The left bottom tree shows the phylogenetic relationships of five subfamilies.
(B) Genomic synteny among Bx6 genes and their homologs between species based on gene order in each genome. Red dots representing the Bx6 genes or homologs are syntenic.
We identified the presence of other dispersed Bx genes and built phylogenetic trees to trace their evolutionary histories. Bx7 catalyzes the conversion of TRIBOA-Glc to DIMBOA-Glc (Figure 1A). Only limited homologs could be identified in grasses, and their phylogenetic tree reveals that Bx7 has been conserved in evolution; the tree is consistent with the species phylogeny, although massive losses have occurred (Supplemental Figure 6). Bx10/Bx11/Bx12/Bx14 encode OMTs that act as metabolic switches between caterpillar and aphid resistance by transforming DIMBOA-Glc to HDMBOA-Glc (Li et al., 2018). From the phylogenetic analyses, the clade Bx10/Bx11/Bx12/Bx14 contains maize-specific duplicates and is nested in a well-defined Panicoideae-specific clade (Supplemental Figure 7). No Triticeae homologs are found within this clade. In wheat (T. aestivum), two OMT genes are characterized as functional DIMBOA-Glc OMTs; both are designated as TaBx10 but are phylogenetically close to Bx7 rather than Bx10 in Z. mays, indicating the functional convergence of OMT genes in grasses during the process of O-methylation (Li et al., 2018). This case implies that other paralogs of OMTs could function as Bx10/Bx11/Bx12/Bx14 in the process of O-methylation and that Bx10/Bx11/Bx12/Bx14 are not required for benzoxazinoid biosynthesis. Taken together, this evidence suggests that Bx7 and Bx10/Bx11/Bx12/Bx14 are alternative and to some extent dispensable in the Bx pathway. Hence, in the following analyses, we focused on the Bx cluster and Bx6.
Constrained purifying selection on the Bx cluster
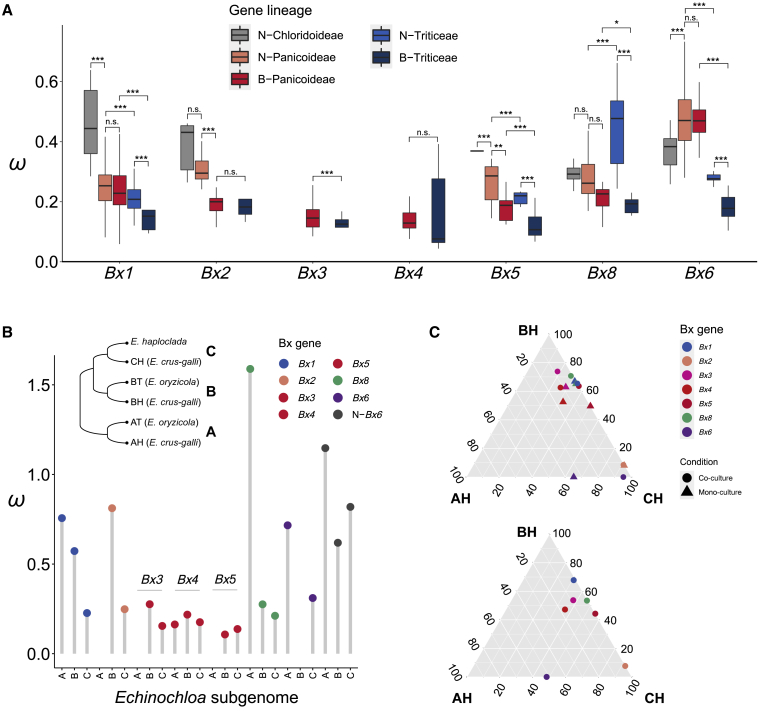
Natural selection shapes the evolutionary dynamics of BGCs (Slot and Rokas, 2010; Rokas et al., 2018; Liu et al., 2020). Selection pressure was measured by ω (dN/dS, the ratio between non-synonymous site substitution and synonymous site substitution) in each lineage of the individual Bx genes. In general, both the Bx genes and their native homologs are under purifying selection (ω < 1). Compared with the outgroup lineage N-Chloridoideae (native Bx homologs in Chloridoideae), constrained purifying selection was detected in all of the native Bx genes of Panicoideae, with the exception of Bx6. The native homologs of Bx6 in Panicoideae undergo relaxed selection with a higher ω value relative to other Bx native genes. Compared with the native homologs, ω values are lower for Bx genes in the cluster in Panicoideae (B-Panicoideae), whereas no difference in selection was found for Bx6, which is separate from the Bx cluster. This selection bias in Panicoideae corresponds to the presence and absence (PAV) of Bx genes and their homologs (Supplemental Figure 8). The loss of native homologs of Bx genes in the cluster is more frequent than the loss of Bx genes in the cluster, which mirrors the relaxed selection, especially for Bx2, Bx5, and Bx8. The presence of Bx6 native homologs is highly conserved, with one copy within one analyzed genome, corresponding to unbiased selection pressure compared with Bx6 genes (Supplemental Figure 8). In Triticeae, all of the Bx genes exhibit constrained selection, despite the conserved presence of Bx native homologs (Figures 4A and Supplemental Figure 8). Although Bx genes in Triticeae are inferred to have been gained from Panicoideae, stronger selection was detected in Triticeae Bx genes than in those of Panicoideae, especially for Bx1 and Bx6. To eliminate the effects of biases from species sampling and PAV of Bx genes or native homologs, selection pressure was measured focusing on Echinochloa and Triticeae. The results further confirmed the selection profiling of Bx genes (Supplemental Figure 9).
Figure 4.
Selection and polyploidization effects on the Bx genes.
(A) Selection pressure estimated by ω of Bx genes and native homologs. N-Chloridoideae, native homologs of Bx genes in Chloridoideae; N-Panicoideae, native homologs of Bx genes in Panicoideae; B-Panicoideae, Bx genes in Panicoideae; N-Triticeae, native homologs of Bx genes in Triticeae; and B-Triticeae, Bx genes in Triticeae. In the boxplots, the horizontal line shows the median value, and the whiskers show the 25% and 75% quartile values of ω. Pairwise t tests were performed to evaluate significance. n.s., not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.0001.
(B) Pairwise ω of Bx genes and native homologs of Bx6 (N-Bx6) in subgenomes A, B, and C between E. crus-galli and its progenitors (E. haploclada and E. oryzicola). The topology shows the phylogenetic relationships among subgenomes in the three Echinochloa species, where AT and AH belong to subgenome A, BT and BH belong to subgenome B, and E. haploclada and CH belong to subgenome C.
(C) Relative expression (upper ternary diagram) and relative response contribution (lower ternary diagram) of multi-copy homologous Bx genes in E. crus-galli subgenomes (AH, BH, and CH) under the control and allelopathy treatments.
Dominance of Bx cluster genes in polyploids
Polyploids are commonly seen in species whose genomes contain Bx genes (hexaploid T. aestivum, E. crus-galli, and E. colona and tetraploid Triticum dicoccoides and Echinochloa oryzicola in this study). We investigated the effects of polyploidization on Bx clusters or genes from three different viewpoints: PAV, selection, and gene expression. Duplicated genes tend to be lost in polyploids owing to gene redundancy or dosage effects (Soltis and Soltis, 2009; Van de Peer et al., 2017). Not unexpectedly, Bx genes tend to be lost in polyploids, especially in Echinochloa (Figures 1B and Supplemental Figure 8). The core Bx gene set is intact in diploid Echinochloa haploclada, whereas Bx losses are found in three polyploid Echinochloa species. In this case, only one intact copy of the core Bx gene set is retained in one subgenome of each species (e.g., BT in E. oryzicola, CH in E. crus-galli, DH2 in E. colona).
The selection strength on homologous duplicates usually varies in polyploids (Ye et al., 2020). The genomes of E. crus-galli and its progenitors (E. oryzicola and E. haploclada) provide a model in which to study the selection dominance of multi-copy homologous Bx genes, and we calculated the ω values of Bx genes in each subgenome between E. crus-galli and its parents (Figure 4B). Bx genes in subgenome A are generally under relaxed purifying selection, with higher ω values compared with those in subgenomes B and C (e.g., Bx1, Bx8). For native homologs, selection on the subgenome A copy is relaxed in the example of Bx6. In general, biased selection is observed for Bx genes in Echinochloa, and Bx genes in subgenome A are under less constrained selection post-hexaploidization.
Expression dominance has been commonly observed in polyploids (Van de Peer et al., 2017; Ye et al., 2020). The response contribution (relative change in expressed transcripts from one subgenome compared with the total expression change) is also biased among subgenomes (Ye et al., 2020). To explore the effect of polyploidization on the gene expression of multi-copy Bx genes, we investigated the expression levels of Bx genes in E. crus-galli under allelopathy treatment (i.e., co-culture with rice) (Guo et al., 2017). Expression and response contribution were both suppressed for Bx genes in subgenome AH (Figure 4C). The dominance of selection and gene expression or response are associated, such that Bx genes in subgenome A under less constrained selection show suppression of expression and response contribution (Supplemental Figure 9).
Discussion
Evolutionary trajectory of the Bx cluster in grass
Given that the Bx cluster and Bx6 catalyze the first seven steps in benzoxazinoid biosynthesis and are sufficient for the synthesis of benzoxazinoid compounds without other Bx genes (e.g., in wheat), we consider the Bx cluster (Bx1–Bx5 and Bx8) and Bx6 to be the core set of Bx genes in the pathway (Figure 1A). Based on the results from all phylogenetic analyses of the core Bx genes, the evolutionary trajectory of the Bx genes can be inferred. Native Bx homologs are found in all phylogenetic trees of core Bx genes; they are evolutionarily conserved and show good genomic synteny among subfamilies. Therefore, the Bx genes in the Bx cluster and Bx6 appear to originate from duplication of native Bx homologs. Previous studies proposed that Bx1 evolved from duplication and modification of the alpha subunit of tryptophan synthase (TSA) (Grün et al., 2005; Frey et al., 2009). Here, we comprehensively identified the native homologs of the Bx1 genes and found that TSAh1 was the ancestor of Bx1 genes (Figure 2A). Gene duplication, followed by neofunctionalization and/or subfunctionalization, and recurrent genomic translocation may have gathered Bx genes together to form the Bx cluster. The processes of gene duplication and translocation may be induced by the activities of retrotransposon elements.
Previous studies proposed that the Bx genes in grasses were of monophyletic origin before the divergence of the Triticeae and Panicoideae (Frey et al., 2009; Grün et al., 2005). Here, our integrated evidence strongly indicates that the Bx genes in Triticeae originated from Panicoideae via HT. Triticeae and Panicoideae diverged more than 50 mya, ruling out the possibility of natural hybridization between them and ILS. Previous studies found no benzoxazinoid biosynthesis in Brachypodium (basal genus in Pooideae) (Frey et al., 2009), consistent with the absence of identifiable Bx genes in two Brachypodium genomes (Figure 1B). Benzoxazinoids were produced in wild but not cultivated Hordeum (H. vulgare in Triticeae), indicating that Bx genes may have been lost in cultivated Hordeum (Grün et al., 2005; Sue et al., 2011). Therefore, we speculate that the transfers occurred in the common ancestor of Triticeae after the divergence with Brachypodium. To trace the potential donor of the Bx genes, we considered the topologies of Bx genes from Triticeae, Andropogoneae (e.g., Z. mays) and Paniceae (e.g., Echinochloa, D. oligosanthes). Bx2 and Bx6 genes support the common ancestor of Andropogoneae and Paniceae as the donor of Bx genes in Triticeae. However, five other Bx genes show discordant topologies, implying that the transfer event may have taken place at a time close to the divergence of Andropogoneae and Paniceae, which would result in an ILS-like phylogeny. It is noteworthy that phylogenies of individual genes based on different sequence types (e.g., amino acid or nucleotide sequences), different substitution models, and even different parameters are sometimes misleading. For example, in the phylogenies of Bx1, Triticeae and Andropogoneae (Z. mays) Bx1 genes formed a monoclade in the protein sequence tree, whereas Bx1 genes from Triticeae and Paniceae formed a monoclade in the nucleotide sequence trees (CDS, codon12, and codon3) (Figure 2A and Supplemental Figure 3).
Overall consistency in Bx gene order and orientation in Panicoideae implies that the Bx cluster was assembled before the divergence of Andropogoneae and Paniceae, although we could not detect sequence collinearity in Bx intergenic regions among clusters, reflecting the high evolutionary rate and weak selection pressure in intergenic regions (Figure 1B and Supplemental Figure 11). The dispersal distributions of Bx genes in Triticeae could result from HT of an intact Bx cluster and subsequent recurrent chromosomal rearrangements or from multiple independent HT events when Bx genes were not gathered together as an intact cluster (e.g., individual HT events for segments Bx1/Bx2, Bx3/Bx4/Bx5, and Bx8) or more complicated routes. Not unexpectedly, we found no conservation in intergenic regions of Bx genes from Bx clusters or subclusters in Panicoideae and Triticeae (Supplemental Figure 11). Based on the principle of parsimony in evolution and on empirical evidence for the occurrence frequency and possibility of HT and chromosomal rearrangement, we hypothesize that an intact Bx cluster has been transferred to Triticeae (Figure 5). With subsequent massive genome reshuffling in Triticeae, the intact ancient Bx cluster was split into segments and scattered on multiple chromosomes (Frey et al., 2009). Gene loss resulted in the partial loss of Bx genes (e.g., T. urartu) and the complete loss (e.g., H. vulgare) in Triticeae.
The positional relationship between the Bx cluster and Bx6 in grass appears to be dynamic. In Panicoideae, Bx6 and the Bx cluster are both located on chromosome 4 in Z. mays, whereas they are separated on different chromosomes in Echinochloa (Figure 1B). In Triticeae, Bx6 genes are located on chromosome 2 in Triticum/Aegilops, and Bx1 and Bx2 genes are on chromosome 4, whereas in Secale, these three Bx genes are on chromosome 7R, despite the fact that Bx1 and Bx2 are over 700 Mb away from the two tandemly duplicated Bx6 genes. In contrast to the lack of synteny between Bx6 genes from maize and Echinochloa, Bx6 genes are syntenic between Triticum/Aegilops and Secale. Comparison between genomes indicates that chromosome 7R in rye is syntenic to chromosomes 5, 4, 7, and 2 in Triticum/Aegilops (Supplemental Figure 12). Therefore, in brief, Secale-specific chromosome fusions led to the location of Bx1, Bx2, and Bx6 on chromosome 7R in rye, a pattern not seen in the genomes of Triticum/Aegilops and the outgroup, Hordeum (Supplemental Figure 12).
In terms of the challenge of determining whether Bx genes were transferred to Triticeae as an intact cluster or through multiple HT events, it is difficult to trace the route by which Bx6 was transferred, with or without the Bx cluster. However, given that the genes in the Bx cluster and Bx6 show almost the same evolutionary phylogenies and that Bx6 catalyzes the reaction that follows those catalyzed by the gene products of the Bx cluster, it is reasonable to speculate that Bx6 co-evolved with the Bx cluster and may have been located in an ancient Bx cluster with the other Bx genes (Figure 5). In this hypothesis, the common ancestor of Panicoideae had a cluster of Bx genes, including Bx6, and the intact ancient Bx cluster was transferred to Triticeae via a single HT event. After the divergence of Andropogoneae and Paniceae, different genomic rearrangements occurred in the two tribes (Figure 5). In Andropogoneae, the Bx cluster and Bx6 were retained in Z. mays but lost completely in other species (e.g., S. bicolor and Miscanthus sinensis). Furthermore, Bx6 was separated from the Bx cluster by translocation in Z. mays, although it is still on the short arm of chromosome 4. In Paniceae, massive losses are found in the Bx genes. The Bx cluster is retained in D. oligosanthes, but Bx6 is lost. By contrast, Bx6 has been retained in Setaria and Digitaria, but the Bx clusters are missing. Both the Bx cluster and Bx6 are absent in Panicum, Cenchrus, and Alloteropsis. Echinochloa is the only genus in which the Bx cluster and Bx6 are on two chromosomes (Figure 1B). Nonetheless, we cannot exclude the possibility that Bx6 was transferred independently and has never been gathered together with the other Bx genes in a cluster, based on current genomic evidence.
Horizontal gene transfer in plants
HT is an important driving force of trait innovation in various levels of organisms (Soucy et al., 2015). In plants, HT is commonly seen between parasites and corresponding host species and between grafting rootstock and scion, owing to their intimate physical cell-to-cell contacts (Kim et al., 2014; Fuentes et al., 2014). HT can also emerge without direct contact, a phenomenon that has been studied somewhat in grasses (Hibdige et al., 2021; Dunning et al., 2019; Park et al., 2021). In addition to gene elements, transposon elements have also been transferred among divergent grass species, as in the Echinochloa genus and the Oryza punctata lineage (Park et al., 2021). Among these reported HT events, a few have involved large genomic segments. A block containing 10 protein-coding genes was transmitted from Iseilema membranaceum (Andropopgoneae) to Alloteropsis semialata (Panicoideae) (Dunning et al., 2019). Here, we provide strong and unambiguous evidence that seven Bx biosynthetic genes in Triticeae are derived from an ancestral Panicoideae donor via HT (Figure 5). HT occurs more frequently between closely related species (Soucy et al., 2015; Hibdige et al., 2021), whereas Triticeae and Panicoideae were split more than 50 mya. DNA transfer events from Panicoideae to Triticeae have been reported before. Several nuclear ribosomal DNA (rDNA) sequences in wild Hordeum and Elymus species are Panicum-like, indicating their foreign origins (Mahelka and Kopecký, 2010; Mahelka et al., 2017). Recently, a large chromosomal segment (∼68 kb) harboring five stress-related protein-coding genes was reported to have been transferred from Panicum to wild Hordeum species (Mahelka et al., 2021; Verhage, 2021). Some of these genes remain functional in the recipient Hordeum genomes. These cases suggest that the transfer of exotic DNA is not as rare among plants as previously supposed (Mahelka et al., 2021), at least from Panicoideae to Triticeae in grasses. It is reasonable to infer that more HT events from Panicoideae to Triticeae may be detected in future studies and that this unidirectional and biased HT pathway has accelerated the capacity to respond to environmental stress in Triticeae.
Compared with previously reported plant-to-plant transfers, here, we provide the first candidate case of HT of an intact gene cluster that functions in the biosynthesis of multi-effect chemical compounds in plants. The clustering of a series of biosynthetic genes facilitates inheritance and stress response by co-inheritance and co-expression in organisms, making it an ingenious invention in long-term adaptive evolution. The combination of HT and gene clustering constitutes a rapid strategy for acquiring highly efficient weapons to defend against external stress. This phenomenon appears to be rare but universal in the kingdom of life, as transfers of BGCs have also been detected in fungi (Khaldi et al., 2008; Slot and Rokas, 2011; Reynolds et al., 2018). As for how the transfer between phylogenetically distant plant species occurs, a possible explanation is that it takes place because of occasional contact (e.g., natural grafting) or is facilitated by vector transfer (e.g., insects, fungi, viruses) (Wang et al., 2020; Xia et al., 2021). The transfer of DNA between insect vectors and plants has been reported recently. For example, whitefly has acquired the plant-derived phenolic glucoside malonyltransferase gene BtPMaT1 from a plant host, enabling it to neutralize plant toxin phenolic glucosides (Xia et al., 2021). Similarly, the transfer of Fhb7 from the fungus Epichloë to Thinopyrum wheatgrass (Triticeae) provides broad resistance to both Fusarium head blight and crown rot in wheat (Wang et al., 2020).
Natural selection on gene clusters
The driving forces for the organization and maintenance of BGCs remain in debate. Nevertheless, it is widely accepted that natural selection must inevitably shape their evolution. Selection analysis of BGCs is rare, owing to the limited identification of BGCs and comparable sequences. In Saccharomycetes, the galactose BGCs are widely conserved in terms of sequence and function, suggesting the influence of long-term purifying selection (Slot and Rokas, 2010). Balancing selection also plays roles in maintaining the diversity of BGCs, as in the case of the aflatoxin gene cluster in the fungus Aspergillus parasiticus (Carbone et al., 2007). In Arabidopsis, the thalianol BGC appears to be under relaxed selection compared with genes in the phytosterol biosynthetic pathway, but it is still under strong purifying selection (Liu et al., 2020). In this study, we used multiple copies of Bx genes and their corresponding native homologs across a broad range of grass species to profile the selection landscapes of Bx clusters. Similar to findings in the thalianol BGC, Bx genes in both Panicoideae and Triticeae show purifying selection. Compared with that on native homologs, selection on Bx genes in clusters is more constrained (Figure 4A). The strength of selection pressure is similar for Bx6 and its native homologs in Panicoideae, perhaps as a result of the dispersal of Bx6 away from other core Bx genes in the cluster. It has been suggested that lateral pathway genes are less constrained by selection pressure than early pathway genes in the biosynthesis of thalianol in Arabidopsis (Liu et al., 2020). Here, we noticed that Bx6, whose product functions after reactions catalyzed by enzymes encoded in the Bx cluster, exhibited the highest ω value among the seven core Bx genes (Figure 4A). Bx8, which is within the Bx cluster, is less constrained than other Bx genes in the cluster. All of the identified Bx clusters or genes are transcribed in the various genomes and function in stress response, further indicating a role for purifying selection in conserving the functions of the Bx clusters.
Subgenome dominance of gene clusters in polyploids
We found that several species identified as having Bx clusters or whole-set core Bx genes are polyploids (Figure 1B). In most cases, polyploidization confers stronger growth and higher tolerance to environmental stress relative to the original diploid (Soltis and Soltis, 2009; Van de Peer et al., 2017). On this basis, biosynthetic gene clustering further offers these species a powerful weapon with which to respond to external stimuli. To some extent, the existence of BGCs in these polyploids has assisted in allowing them to become major crops under artificial selection (e.g., hexaploid and tetraploid wheat, paleo-tetraploid maize) or successful agricultural weeds (hexaploid and tetraploid barnyardgrass). In polyploids, subgenome dominance usually exists in selection and gene expression. The dominance of BGCs in polyploids has not been well studied. Differential expression of Bx genes in hexaploid wheat has been detected (Nomura et al., 2005). The main contribution in hexaploid and tetraploid wheat is by subgenome B. In the hexaploid barnyardgrass E. crus-galli, we found obvious suppression of the expression of Bx genes from subgenome AH compared with the other two subgenomes (Figure 4C). The dominance pattern of Bx genes is consistent with the overall profiling across whole subgenomes, with a significantly higher proportion of suppressed genes in subgenome AH (Ye et al., 2020). Highly expressed metabolic genes tend to be preferentially retained after polyploidization owing to selection pressure (Gout et al., 2009). The selection on Bx genes from subgenome A is indeed less constrained than that on the other two Bx homologs (Figure 4B and Supplemental Figure 10). Furthermore, three out of four Bx gene losses in the E. crus-galli pedigree are from subgenome A (Supplemental Figure 8). More transposon elements on subgenome A have somewhat increased the degree of methylation, which could inactivate gene expression (Ye et al., 2020). As seen in the cases of wheat (Nomura et al., 2005) and barnyardgrass, genomic bias in the expression of Bx genes in polyploids is putatively derived from their tetraploid progenitors. Subsequent selection then shapes the PAV of Bx genes in each genome. Clearly, additional studies are needed to decipher the mechanism of dominance of BGCs in polyploids.
Materials and methods
Datasets
Amino acid sequences of whole-genome protein and coding nucleotide sequences of 40 grass genomes (including grass basal group: Pharus latifolius; Oryzoideae: Zizania latifolia, Leersia perrieri, Oryza brachyantha, O. punctata, O. rufipogon, O. sativa, O. barthii, and O. glaberrima; Bambusoideae: Olyra latifolia and Bonia amplexicaulis; Pooideae: Brachypodium distachyon, B. stacei, Hordeum vulgare, Secale cereale, Triticum aestivum, T. dicoccoides, Aegilops tauschii, and T. urartu; Chloridoideae: Eragrostis curvula, E. nindensis, E. tef, Oropetium thomaeum, Cleistogenes songorica, and Zoysia japonica; Panicoideae: Zea mays, Sorghum bicolor, Miscanthus sinensis, Dichanthelium oligosanthes, Digitaria exilis, Panicum hallii, Setaria italica, S. viridis, Cenchrus purpureus, C. americanus, Alloteropsis semialata, Echinochloa crus-galli, E. oryzicola, E. colona, and E. haploclada) and the outgroup species Ananas comosus were downloaded from Phytozome (https://phytozome-next.jgi.doe.gov) and the National Genomics Data Center (NGDC) (https://ngdc.cncb.ac.cn) (Supplemental Table 1). Polyploids with chromosome-level assemblies (hexaploid T. aestivum, E. crus-galli, and E. colona and tetraploid T. dicoccoides, E. tef, C. songorica, M. sinensis, D. exilis, C. purpureus, and E. oryzicola) were split into subgenomes (Supplemental Table 1). A total of 54 diploid genomes or subgenomes were used to construct the grass phylogeny. OrthoFinder was used to identify single-copy orthologs in the 41 species genomes (Emms and Kelly, 2019). Individual phylogenetic trees of 45 single-copy genes were constructed using IQ-TREE (version 1.6.12) with the best substitution model from ModelFinder (Nguyen et al., 2015) and integrated into a species tree using ASTRAL (version 5.7.4) (Zhang et al., 2018). The divergence time was adopted from the TimeTree database (www.timetree.org) (Kumar et al., 2017).
Identification of Bx genes in grass
The protein sequences of Bx genes in Z. mays (PH207) (Bx1, Zm00008a014942; Bx2, Zm00008a014943; Bx3, Zm00008a014937; Bx4, Zm00008a014938; Bx5, Zm00008a014940; Bx6, Zm00008a014884; Bx7, Zm00008a015292; Bx8, Zm00008a014941; Bx9, Zm00008a003056; Bx10, Zm00008a001636; Bx11, Zm00008a001638; Bx12, Zm00008a001639; Bx13, Zm00008a010377; and Bx14, Zm00008a008314) were used as bait to search Bx genes in grass species by BLASTP. The homologs of individual Bx genes were filtered by parameters of e-value less than 1e−30 and identity greater than 50%. Homologs were then aligned using MAFFT (version 7.310) (Katoh and Standley, 2013), and phylogenetic trees were built using IQ-TREE with the substitution model parameters from ModelFinder with 1000 bootstrap replicates (Nguyen et al., 2015). Using the homologs in A. comosus or P. latifolius as the outgroup, we kept only the closest homologous copies of Bx genes across the grass family as native homologs. In Bx trees for which Bx homologs could not be found in the outgroup species A. comosus and P. latifolius, we referred to the topological relationships among homologs in the five subfamilies to identify Bx genes and their native homologs.
Phylogenetic analysis
Bx homologs (Bx genes and native homolog Bx-like genes) were re-aligned using MAFFT (Katoh and Standley, 2013). Substitution models were selected using ModelFinder, and the maximum-likelihood phylogenetic trees were reconstructed by IQ-TREE using ultrafast bootstrap approximation (1000 replicates) for branch support (Nguyen et al., 2015). To examine the robustness of the phylogeny, especially the location of Bx clades and candidate HT events from Panicoideae to Triticeae, we compared the real and constraint trees for each Bx gene. Three tests on tree topologies, the RELL approximation, the SH test, and the AU test, were performed using IQ-TREE with 10 000 bootstrap replicates (Nguyen et al., 2015). AU and SH tests return p values; thus, a tree is rejected if its p value is less than 0.05. The RELL approximation test returns posterior weights. To eliminate the effects of protein sequence alignment gaps, we also used Gblocks (Castresana, 2000) to remove gaps from alignments with the parameter “-b4 = 5 -b5 = h.” The trimmed alignments of conserved regions were used in topology tests. Phylogenies of Bx genes based on CDS, codon12 (first and second positions within a codon), and codon3 (third position within a codon) were constructed using MAFFT for alignment and IQ-TREE with the best substitution model (ModelFinder) and 1000-replicate ultrafast bootstrap analysis (Nguyen et al., 2015).
Genome synteny analysis
Whole-genome protein sequences were compared pairwise among the 40 grass species using BLASTP. The best hit for each blast search was retained. We also required that the e-value should be less than 1e−30 and the identity greater than 50%. According to the physical positions of the genes on each chromosome of each species, the genes or proteins were ordered. We performed gene-to-gene synteny analysis among grass species based on the gene orders within each genome. Synteny or collinearity analysis between Bx clusters or gene regions based on nucleotide sequences was performed using MUMmer (version 4.0) (Marçais et al., 2018). The alignment parameter in the nucmer module was set as “-l 12 -g 200 -c 20.” Three randomly selected conserved blocks between E. haploclada and Z. mays were determined by whole-genome protein sequence blast and integration by DAGchainer (Haas et al., 2004).
Selection analysis
Selection pressure was measured by the indicator ω, the ratio between non-synonymous substitution rate (dN) and synonymous substitution rate (dS), with ω = 1 usually indicative of neutral mutations, ω < 1 of purifying selection, and ω > 1 of diversifying positive selection. Bx homologs whose CDS or protein sequence lengths were greater than twice or less than half of the lengths of Bx genes or proteins in Z. mays were removed. Within each clade in the phylogenetic tree of each Bx gene, only one copy was retained in the following analysis within one (sub)genome for tandem duplicates, and duplicate copies with abnormal sequence lengths (usually much shorter) were removed. The CDS and protein sequences were aligned using MAFFT and PAL2NAL (Suyama et al., 2006). The dN and dS values were calculated using KaKs_calculator with the NG model for all pairs of genes within each clade (Bx clade or native homolog clade) (Zhang et al., 2006).
Gene expression analysis
RNA sequencing (RNA-seq) data from an analysis of E. crus-galli seedlings grown in a monoculture or co-cultured with rice were downloaded from NCBI (BioProject PRJNA268892) (Guo et al., 2017), and low-quality reads were removed using the NGS QC toolkit (version 2.3.348) (Patel and Jain, 2012). The clean reads were mapped to the chromosome-level reference genome of E. crus-galli (STB08) using TopHat (version 2.1.1) (Trapnell et al., 2012). Relative gene expression levels were quantified and normalized to fragments per kilobase of exon per million mapped fragments (FPKM) values using Cufflinks (version 2.2.1) (Trapnell et al., 2012). The determination of expression dominance and response contributions of Bx genes in subgenomes of E. crus-galli followed a previously described approach (Ye et al., 2020).
Funding
This work was supported by grants from the Zhejiang Natural Science Foundation (LZ17C130001), and the Jiangsu Collaborative Innovation Center for Modern Crop Production, 111 Project (B17039).
Author contributions
L.F. and D.W. conceived and designed the research. L.F. and C.-Y.Y. supervised the research. D.W. and B.J. carried out the data analysis. D.W., L.F., M.P.T., and C.-Y.Y. analyzed the findings and wrote the manuscript.
Acknowledgments
We are grateful to Jie Qiu (Shanghai Normal University) for insightful suggestions on the manuscript. The authors declare no competing interests.
Published: May 9, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- Boycheva S., Daviet L., Wolfender J., Fitzpatrick T.B. The rise of operon-like gene clusters in plants. Trends Plant Sci. 2014;19:447–459. doi: 10.1016/j.tplants.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Carbone I., Jakobek J.L., Ramirez-Prado J.H., Horn B.W. Recombination, balancing selection and adaptive evolution in the aflatoxin gene cluster of Aspergillus parasiticus. Mol. Ecol. 2007;16:4401–4417. doi: 10.1111/j.1365-294X.2007.03464.x. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Dunning L.T., Olofsson J.K., Parisod C., Choudhury R.R., Moreno-Villena J.J., Yang Y., Dionora J., Quick W.P., Park M., Bennetzen J.L., et al. Lateral transfers of large DNA fragments spread functional genes among grasses. Proc. Natl. Acad. Sci. U S A. 2019;116:4416–4425. doi: 10.1073/pnas.1810031116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutartre L., Hilliou F., Feyereisen R. Phylogenomics of the benzoxazinoid biosynthetic pathway of Poaceae: gene duplications and origin of the Bx cluster. BMC Evol. Biol. 2012;12:64. doi: 10.1186/1471-2148-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M., Chomet P., Glawischnig E., Stettner C., Grun S., Winklmair A., Eisenreich W., Bacher A., Meeley R.B., Briggs S.P., et al. Analysis of a chemical plant defense mechanism in grasses. Science. 1997;277:696–699. doi: 10.1126/science.277.5326.696. [DOI] [PubMed] [Google Scholar]
- Frey M., Schullehner K., Dick R., Fiesselmann A., Gierl A. Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry. 2009;70:1645–1651. doi: 10.1016/j.phytochem.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Fuentes I., Stegemann S., Golczyk H., Karcher D., Bock R. Horizontal genome transfer as an asexual path to the formation of new species. Nature. 2014;511:232–235. doi: 10.1038/nature13291. [DOI] [PubMed] [Google Scholar]
- Gladyshev E.A., Meselson M., Arkhipova I.R. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320:1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- Gout J.-F., Duret L., Kahn D. Differential retention of metabolic genes following whole-genome duplication. Mol. Biol. Evol. 2009;26:1067–1072. doi: 10.1093/molbev/msp026. [DOI] [PubMed] [Google Scholar]
- Grün S., Frey M., Gierl A. Evolution of the indole alkaloid biosynthesis in the genus Hordeum: distribution of gramine and DIBOA and isolation of the benzoxazinoid biosynthesis genes from Hordeum lechleri. Phytochemistry. 2005;66:1264–1272. doi: 10.1016/j.phytochem.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Guo L., Qiu J., Li L., Lu B., Olsen K., Fan L. Genomic clues for crop–weed interactions and evolution. Trends Plant Sci. 2018;23:1102–1115. doi: 10.1016/j.tplants.2018.09.009. [DOI] [PubMed] [Google Scholar]
- Guo L., Qiu J., Ye C., Jin G., Mao L., Zhang H., Yang X., Peng Q., Wang Y., Jia L., et al. Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat. Commun. 2017;8:1031. doi: 10.1038/s41467-017-01067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Delcher A.L., Wortman J.R., Salzberg S.L. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 2004;20:3643–3646. doi: 10.1093/bioinformatics/bth397. [DOI] [PubMed] [Google Scholar]
- Handrick V., Robert C.A.M., Ahern K.R., Zhou S., Machado R.A.R., Maag D., Glauser G., Fernandez-Penny F.E., Chandran J.N., Rodgers-Melnick E., et al. Biosynthesis of 8-O-methylated benzoxazinoid defense compounds in maize. Plant Cell. 2016;28:1682–1700. doi: 10.1105/tpc.16.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibdige S.G.S., Raimondeau P., Christin P., Dunning L.T. Widespread lateral gene transfer among grasses. New Phytol. 2021;230:2474–2486. doi: 10.1111/nph.17328. [DOI] [PubMed] [Google Scholar]
- Jonczyk R., Schmidt H., Osterrieder A., Fiesselmann A., Schullehner K., Haslbeck M., Sicker D., Hofmann D., Yalpani N., Simmons C., et al. Elucidation of the final reactions of DIMBOA-glucoside biosynthesis in maize: characterization of Bx6 and Bx7. Plant Physiol. 2008;146:1053–1063. doi: 10.1104/pp.107.111237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaldi N., Collemare J., Lebrun M., Wolfe K.H. Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biol. 2008;9:R18. doi: 10.1186/gb-2008-9-1-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G., LeBlanc M.L., Wafula E.K., DePamphilis C.W., Westwood J.H. Genomic-scale exchange of mRNA between a parasitic plant and its hosts. Science. 2014;345:808–811. doi: 10.1126/science.1253122. [DOI] [PubMed] [Google Scholar]
- Kominek J., Doering D.T., Opulente D.A., Shen X., Zhou X., DeVirgilio J., Hulfachor A.B., Groenewald M., Mcgee M.A., Karlen S.D., et al. Eukaryotic acquisition of a bacterial operon. Cell. 2019;176:1356–1366.e10. doi: 10.1016/j.cell.2019.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Suleski M., Hedges S.B. TimeTree: a resource for timelines, timetrees, and divergence Times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Li B., Förster C., Robert C.A.M., Züst T., Hu L., Machado R.A.R., Berset J.-D., Handrick V., Knauer T., Hensel G., et al. Convergent evolution of a metabolic switch between aphid and caterpillar resistance in cereals. Sci. Adv. 2018;4:1–15. doi: 10.1126/sciadv.aat6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Cheng J., Liu X., Guo X., Liu Y., Fan W., Lu L., Ma Y., Liu T., Tao S., et al. Origin and evolution of fusidane-type antibiotics biosynthetic pathway through multiple horizontal gene transfers. Genome Biol. Evol. 2020;12:1830–1840. doi: 10.1093/gbe/evaa163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind A.L., Wisecaver J.H., Lameiras C., Wiemann P., Palmer J.M., Keller N.P., Rodrigues F., Goldman G.H., Rokas A. Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLoS Biol. 2017;15:e2003583. doi: 10.1371/journal.pbio.2003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Cheema J., Vigouroux M., Hill L., Reed J., Paajanen P., Yant L., Osbourn A. Formation and diversification of a paradigm biosynthetic gene cluster in plants. Nat. Commun. 2020;11:5354. doi: 10.1038/s41467-020-19153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P.-F., et al. The Pharus latifolius genome bridges the gap of early grass evolution. Plant Cell. 2021;33(4):846–864. doi: 10.1093/plcell/koab015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahelka V., Kopecký D. Gene capture from across the grass family in the allohexaploid Elymus repens (L.) Gould (Poaceae, Triticeae) as evidenced by ITS, GBSSI, and molecular cytogenetics. Mol. Biol. Evol. 2010;27:1370–1390. doi: 10.1093/molbev/msq021. [DOI] [PubMed] [Google Scholar]
- Mahelka V., Krak K., Fehrer J., Caklová P., Nagy Nejedlá M., Čegan R., Kopecký D., Šafář J. A Panicum-derived chromosomal segment captured by Hordeum a few million years ago preserves a set of stress-related genes. Plant J. 2021;105:1141–1164. doi: 10.1111/tpj.15167. [DOI] [PubMed] [Google Scholar]
- Mahelka V., Krak K., Kopecký D., Fehrer J., Šafář J., Bartoš J., Hobza R., Blavet N., Blattner F.R. Multiple horizontal transfers of nuclear ribosomal genes between phylogenetically distinct grass lineages. Proc. Natl. Acad. Sci. U S A. 2017;114:1726–1731. doi: 10.1073/pnas.1613375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Delcher A.L., Phillippy A.M., Coston R., Salzberg S.L., Zimin A. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 2018;14:e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meihls L.N., Handrick V., Glauser G., Barbier H., Kaur H., Haribal M.M., Lipka A.E., Gershenzon J., Buckler E.S., Erb M., et al. Natural variation in maize aphid resistance is associated with 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell. 2013;25:2341–2355. doi: 10.1105/tpc.113.112409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Ishihara A., Yanagita R.C., Endo T.R., Iwamura H. Three genomes differentially contribute to the biosynthesis of benzoxazinones in hexaploid wheat. Proc. Natl. Acad. Sci. U S A. 2005;102:16490–16495. doi: 10.1073/pnas.0505156102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann H., Huang A., Osbourn A. Plant metabolic clusters – from genetics to genomics. New Phytol. 2016;211:771–789. doi: 10.1111/nph.13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann H.-W., Osbourn A. Gene clustering in plant specialized metabolism. Curr. Opin. Biotechnol. 2014;26:91–99. doi: 10.1016/j.copbio.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Nützmann H.-W., Scazzocchio C., Osbourn A. Metabolic gene clusters in eukaryotes. Annu. Rev. Genet. 2018;52:159–183. doi: 10.1146/annurev-genet-120417-031237. [DOI] [PubMed] [Google Scholar]
- Park M., Christin P., Bennetzen J.L. Sample sequence analysis uncovers recurrent horizontal transfers of transposable elements among grasses. Mol. Biol. Evol. 2021;38:3664–3675. doi: 10.1093/molbev/msab133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R.K., Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds H.T., Vijayakumar V., Gluck-Thaler E., Korotkin H.B., Matheny P.B., Slot J.C. Horizontal gene cluster transfer increased hallucinogenic mushroom diversity. Evol. Lett. 2018;2:88–101. doi: 10.1002/evl3.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A., Wisecaver J.H., Lind A.L. The birth, evolution and death of metabolic gene clusters in fungi. Nat. Rev. Microbiol. 2018;16:731–744. doi: 10.1038/s41579-018-0075-3. [DOI] [PubMed] [Google Scholar]
- Saarela J.M., Burke S.V., Wysocki W.P., Barrett M.D., Clark L.G., Craine J.M., Peterson P.M., Soreng R.J., Vorontsova M.S., Duvall M.R. A 250 plastome phylogeny of the grass family (Poaceae): topological support under different data partitions. PeerJ. 2018;6:e4299. doi: 10.7717/peerj.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot J.C., Rokas A. Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Curr. Biol. 2011;21:134–139. doi: 10.1016/j.cub.2010.12.020. [DOI] [PubMed] [Google Scholar]
- Slot J.C., Rokas A. Multiple GAL pathway gene clusters evolved independently and by different mechanisms in fungi. Proc. Natl. Acad. Sci. U S A. 2010;107:10136–10141. doi: 10.1073/pnas.0914418107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis P.S., Soltis D.E. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 2009;60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- Soucy S.M., Huang J., Gogarten J.P. Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 2015;16:472–482. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- Sue M., Nakamura C., Nomura T. Dispersed benzoxazinone gene cluster: molecular characterization and chromosomal localization of glucosyltransferase and glucosidase genes in wheat and rye. Plant Physiol. 2011;157:985–997. doi: 10.1104/pp.111.182378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos A.M., Rook F. Why biosynthetic genes for chemical defense compounds cluster. Trends Plant Sci. 2012;17:383–388. doi: 10.1016/j.tplants.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Tralamazza S.M., Rocha L.O., Oggenfuss U., Corrêa B., Croll D. Complex evolutionary origins of specialized metabolite gene cluster diversity among the plant pathogenic fungi of the Fusarium graminearum species complex. Genome Biol. Evol. 2019;11:3106–3122. doi: 10.1093/gbe/evz225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y., Mizrachi E., Marchal K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017;18:411–424. doi: 10.1038/nrg.2017.26. [DOI] [PubMed] [Google Scholar]
- Verhage L. A hitchhiker’s guide to foreign genomes. Plant J. 2021;105:1139–1140. doi: 10.1111/tpj.15192. [DOI] [PubMed] [Google Scholar]
- Von Rad U., Hüttl R., Lottspeich F., Gierl A., Frey M. Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize. Plant J. 2002;28:633–642. doi: 10.1046/j.1365-313x.2001.01161.x. [DOI] [PubMed] [Google Scholar]
- Wang H., Sun S., Ge W., Zhao L., Hou B., Wang K., Lyu Z., Chen L., Xu S., Guo J., et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science. 2020;80:368. doi: 10.1126/science.aba5435. [DOI] [PubMed] [Google Scholar]
- Wong S., Wolfe K.H. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat. Genet. 2005;37:777–782. doi: 10.1038/ng1584. [DOI] [PubMed] [Google Scholar]
- Wu D., Shen E., Jiang B., Feng Y., Tang W., Lao S., Jia L., Lin H.-Y., Xie L., Weng X., et al. Genomic insights into the evolution of Echinochloa species as weed and orphan crop. Nat. Commun. 2022;13:689. doi: 10.1038/s41467-022-28359-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Guo Z., Yang Z., Han H., Wang S., Xu H., Yang X., Yang F., Wu Q., Xie W., et al. Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell. 2021;184:1693–1705.e17. doi: 10.1016/j.cell.2021.02.014. [DOI] [PubMed] [Google Scholar]
- Ye C., Wu D., Mao L., Jia L., Qiu J., Lao S., Chen M., Jiang B., Tang W., Peng Q., et al. The genomes of the allohexaploid Echinochloa crus-galli and its progenitors provide insights into polyploidization-driven adaptation. Mol. Plant. 2020;13:1298–1310. doi: 10.1016/j.molp.2020.07.001. [DOI] [PubMed] [Google Scholar]
- Zhang C., Rabiee M., Sayyari E., Mirarab S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinf. 2018;19:153. doi: 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Li J., Zhao X., Wang J., Wong G.K., Yu J. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics. Proteomics Bioinformatics. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.