Abstract
UV absorption and CD spectroscopy, along with polyacrylamide gel electrophoresis, were used to study conformational properties of DNA fragments containing the trinucleotide repeat (GCC)n (n = 4, 8 or 16), whose expansion is correlated with the fragile X chromosome syndrome. We have found that the conformational spectrum of the (GCC)n strand is wider than has been shown so far. (GCC)n strands adopt the hairpin described in the literature under a wide range of salt concentrations, but only at alkaline (>7.5) pH values. However, at neutral and slightly acid pH (GCC)4 and (GCC)8 strands homodimerize. Our data suggest that the homodimer is a bimolecular tetraplex formed by two parallel-oriented hairpins held together by hemi-protonated intermolecular C·C+ pairs. The (GCC)16 strand forms the same tetraplex intramolecularly. We further show that below pH 5 (GCC)n strands generate intercalated cytosine tetraplexes, whose molecularity depends on DNA strand length. They are tetramolecular with (GCC)4, bimolecular with (GCC)8 and monomolecular with (GCC)16. i-Tetraplex formation is a complex and slow process. The neutral tetraplex, on the other hand, arises with fast kinetics under physiological conditions. Thus it is a conformational alternative of the (GCC)n strand duplex with a complementary (GGC)n strand.
INTRODUCTION
Fragile X chromosome syndrome, the most common form of inherited mental retardation in humans (reviewed in 1), is associated with expansion of DNA triplet microsatellites (CCG)n·(CGG)n (reviewed in 2,3). Single strands of (CCG)n, as well as (CGG)n, adopt non-canonical DNA structures (reviewed in 4–6) that may be the cause of the expansions (7–9). The unusual structures include various kinds of single-stranded hairpins (10–16) that have been reported (11,13–15) to be dominant structures of the C-rich strand. The main structural feature shared by all reported hairpins is the presence of classical Watson–Crick G·C pairs in the hairpin stem. The hairpins, however, differ in the number of nucleotides in the hairpin loop, the conformation (intrahelical or extrahelical) of cytosines not involved in G·C pairs, the protonation of unpaired cytosines and, especially, whether the first (GpC) or second (CpG) cytosine of the CCG motif is involved in G·C pairs. The first possibility is called frame 1, the latter frame 2 (5).
The strands of (GCC)5–11 have been reported (13,14) to adopt hairpins exclusively in frame 1 (in 10–150 mM NaCl, pH 7.5–6) (i.e. the G·C pairs are in GpC steps). It has been concluded (11,13,14) that the mispaired cytosine residues (at the CpG steps) are intrahelical, well stacked on the neighboring G·C pairs and probably connected by a single hydrogen bond. On the other hand, Yu et al. (15) have shown that (CCG)15 is a frame 2 hairpin under a wide range of solvent conditions (1–600 mM NaCl, pH 8.5–6) in which the GpC step cytosines are protonated at physiological pH values. The protonated cytosines are stacked on each other in the minor groove of the distorted hairpin helix, forming the so-called extended e-motif (15). This motif was first observed (17) in the antiparallel homoduplex d(CCG)2·d(CCG)2. Long (CCG)18 and (CCG)20 strands have also been reported (15) to form frame 2 hairpins. Their unpaired cytosines have remarkably high (>8) pK values.
In this report we show that DNA strands containing (GCC)n repeats (n = 4, 8 or 16) can adopt a broader spectrum of conformations than has so far been demonstrated. Apart from the above mentioned hairpins, (GCC)4 and (GCC)8 adopt a bimolecular structure under close to physiological conditions. We suggest that the structure is a tetraplex formed by two parallel oriented hairpins. The 48mer (GCC)16 adopts this tetraplex intramolecularly. We further show that the (GCC)n strands form intercalated cytosine tetraplexes (i-tetraplexes) below pH 5.
MATERIALS AND METHODS
The oligonucleotides (GCC)4, (GCC)8, (GGC)4 and (GGC)8 were bought from the Laboratory of Plant Molecular Physiology, Faculty of Science, Masaryk University (Brno, Czech Republic). The oligonucleotides (GCC)16 and (GGC)16 were synthesized and purified by VBC Genomics Bioscence Research (Vienna, Austria). The lyophilized oligonucleotides were dissolved in 1 mM sodium phosphate, 0.3 mM EDTA, pH 7, to give a stock solution concentration of ∼100 OD/ml. The sample concentrations were determined from their absorption measured at room temperature in the same buffer (the solution pH was 8.5 for the C-rich strands) on a Unicam 5625 UV/Vis spectrometer. The following molar extinction coefficients were used at the absorption maximum: ɛ259 = 8490 M–1 cm–1 for (GCC)4, ɛ259 = 8180 M–1 cm–1 for (GCC)8, ɛ258 = 7850 M–1 cm–1 for (GCC)16, ɛ254 = 9460 M–1 cm–1 for (GGC)4, ɛ253 = 8790 M–1 cm–1 for (GGC)8 and ɛ253 = 8440 M–1 cm–1 for (GGC)16. The molar extinction coefficients were assessed from their single-strand extinction coefficients calculated according to Gray et al. (18) and from their hypochromic effects.
Required volumes of concentrated Robinson–Britton buffer and concentrated NaCl were added to the oligonucleotide samples to obtain the conditions shown in the figure captions. The pH was adjusted by adding dilute HCl or NaOH directly to the cells. The pH values were measured using a Sentron Titan pH meter and Sentron Red-Line Probe electrode. The salt and DNA concentrations were corrected for the sample volume increase.
CD spectra were measured using the Jobin-Yvon Mark VI dichrograph in 0.1 and 0.01 cm path length Hellma cells placed in a thermostatted holder. The DNA concentration was chosen to give an absorption of ∼0.8 at the absorption maximum, which gives the optimum signal-to-noise ratio. Δɛ was expressed as per M–1 cm–1, the molarity M being related to the number of nucleoside residues in the DNA samples.
Non-denaturing PAGE was performed in a thermostatted submersible apparatus (SE-600; Hoefer Scientific, San Francisco, CA). Gels (16%, 29:1 acrylamide:bisacrylamide), 14 × 16 × 0.1 cm in size, were run for 20 h at 70 V (∼5 V/cm) and 0°C. The gels were stained with Stains-All (Sigma). Densitometry was performed using a Personal Densitometer SI, Model 375-A (Molecular Dynamics, Sunnyvale, CA).
RESULTS
The (GCC)n strand hairpin
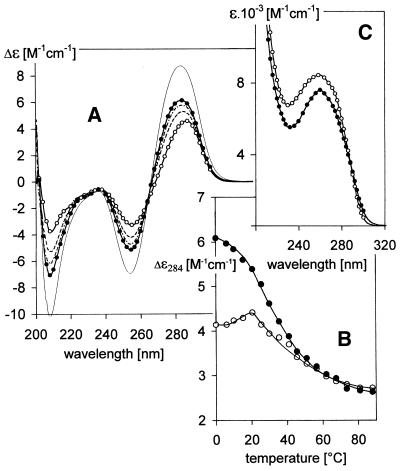
(GCC)4, (GCC)8 and (GCC)16 run as single strands in slightly alkaline gels (Fig. 1A). Up to 0.35 M Na+ (the highest salt concentration allowing electrophoretic migration) neither of the (GCC)n fragments formed a bimolecular structure (not shown). The spectroscopic measurements, however, reveal (Fig. 2A and B) that the single strands are ordered. This fact, along with the B-like shape (see for example 19–22) of the circular dichroism (CD) spectra (Fig. 2A), agree with NMR results demonstrating (13,14) that this motif folds back into hairpins stabilized by classic G·C pairs. Similar to B-DNA duplexes (see for example 23,24), the temperature-induced CD spectral changes comprise two steps (Fig. 2B). Before denaturation, an increase in temperature, similarly to a decrease in ionic strength (not shown), leads to opposite changes in the CD spectra to denaturation (Fig. 2B). The denaturation of (GCC)n displays a low cooperativity characteristic (25,26) of hairpins. Both regions of the temperature changes shift towards higher temperatures with increasing (GCC)n length (not shown). Ionic strengths >0.15 M Na+ induce no noticeable changes in the CD spectra of (GCC)n (not shown), suggesting that the hairpins remain stable even under high salt concentrations.
Figure 1.
The pH dependence of migration in polyacrylamide gels of (GCC)4, (GCC)8 and (GCC)16 and of heteroduplexes with their complementary strands (which serve as markers). The gels were run at 0°C in 0.15 M Na+ (Robinson–Britton buffer + NaCl) at (A) pH 8.5, (B) pH 7, (C) pH 6.5 and (D) pH 6.
Figure 2.
pH-induced changes in the (A) CD spectra and (C) UV absorption spectra of (GCC)4 dissolved in 0.15 M Na+ (Robinson–Britton buffer + NaCl) measured at 0°C. The pH values were: pH 8.5 (open circles), pH 7.2 (dashes), pH 6.9 (dash-dot) and pH 6 (filled circles). These CD and absorption spectra were measured in a 0.1 cm path length cell with a DNA concentration of 0.7 mM; thin line in (A), (GCC)4 in the same solvent at pH 6 measured in a 0.01 cm path length cell with a DNA concentration of 7 mM. (B) Temperature-induced changes in CD spectra of (open circles) the (GCC)4 hairpin in 0.15 M Na+, pH 8.5, and (full circles) of (GCC)4 homodimer in 0.15 M Na+, pH 6.
Homodimers of (GCC)4 and (GCC)8
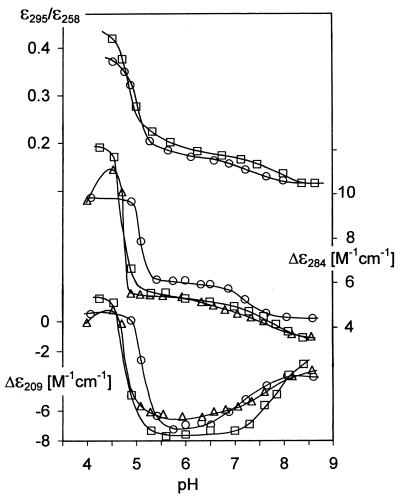
The electrophoretic results (Fig. 1A–D) show that (GCC)4 and (GCC)8 homodimerize at pH < 7. (GCC)n·(GGC)n heteroduplexes serve as markers in the gel to show retardation resulting from the doubled sample molecular weight. Due to fast exchange between the hairpin and homodimer, the electrophoretic trace of (GCC)4 gradually shifts towards the trace of the (GCC)4·(GGC)4 duplex between pH 7 and 6 (compare lanes 1 and 4 in Fig. 1A–D). The trace of (GCC)8 has already started to shift towards larger molecules and becomes hazy at pH 7 [when migration of (GCC)4 has not yet shown distinct changes]. The homodimer becomes the dominant conformer of (GCC)8 at pH 6.5 (compare lanes 2 and 5 in Fig. 1A–D). Homodimers of (GCC)4, as well as of (GCC)8, migrate slightly faster than the corresponding heteroduplexes with their complementary strands (Fig. 1C and D). Similarly, the hairpins migrate faster than their corresponding unstructured single strands (27,28) or than half-length duplexes (Fig. 1A). The 48mer (GCC)16 remains monomolecular under conditions under which (GCC)4 and (GCC)8 undergo a hairpin-to-homodimer transition (Fig. 1A–D, compare lanes 3 and 6). At pH 6.5 a slight trace in (GCC)16 dimers appears which disappears at more acid pH values (Fig. 1C and D). A slight band corresponding to the monomolecular fraction also becomes evident with (GCC)8, the population of which increases when going from pH 6.5 to 6.0 (Fig. 1C and D). Decreasing pH is accompanied by an increase in all amplitudes of the CD spectrum of (GCC)4, especially by a deepening of the negative short wavelength band at 210 nm (Fig. 2A). Simultaneously, its UV absorption spectrum becomes markedly hypochromic at its maximum, while a slight increase in absorption is observed in the vicinity of 300 nm (Figs 2C and 3). Although the global shape of the CD spectrum of (GCC)4 does not change substantially, it is obvious that the hairpins transform into a more stable arrangement on acidification. This transition is an all-or-none process (isoelliptical point at 263 nm) lasting seconds, at most, and it is immediately reversible. The CD and absorption changes (Fig. 3) are the same with all (GCC)n studied. The CD spectra have only moderately lower amplitudes and the transition shifts slightly towards more alkaline pH with increasing length of the molecule (Fig. 3).
Figure 3.
pH-induced changes in the CD and absorption spectra of (GCC)4 (circles), (GCC)8 (triangles) and (GCC)16 (squares) dissolved in 0.15 M Na+ (Robinson–Britton buffer + NaCl) measured at 0°C in a 0.1 cm path length cell (conditions as for Fig. 2): (upper) the ratio of absorption at 295 and 258 nm; (middle and bottom) CD changes monitored at 284 and 209 nm, respectively. All points in the dependences correspond to equilibrium states.
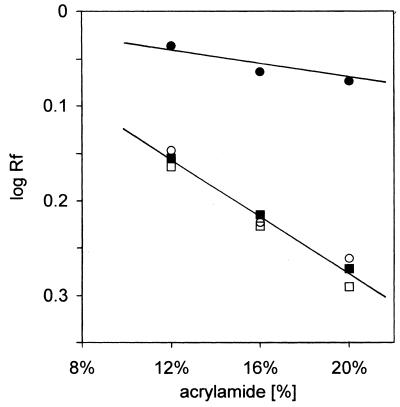
To unambiguously demonstrate dimerization of (GCC)n molecules on going from pH 8.5 to 6, we have undertaken a further electrophoretic characterization of the samples by studying their mobility as a function of gel concentration. The results are shown in Figure 4 in the form of a Ferguson plot (29). The slope of the linear dependence enables estimation of the retardation coefficient, which is related to the size and shape of the molecule studied (29–31). In Figure 4 we show Ferguson plots for (GCC)8 at pH 8.5 and 6 and also for a heteroduplex with (GGC)8 at both pH values. It is evident that the slope of the (GCC)8 single hairpin at pH 8.5 is different from that of the remaining three dependences. In contrast, the dependence of (GCC)8 at pH 6 is nearly the same as those of the heteroduplex at both pH 6 and 8. The retardation coefficient of (GCC)8 at pH 6 represents homodimerization of the original module. The same result was obtained for (GCC)4 (not shown).
Figure 4.
Ferguson plots of the (GCC)8 hairpin (full circles) at pH 8.5 and of the hairpin dimer (open circles) at pH 6 compared with the heteroduplex (GCC)8·(GGC)8 at pH 8.5 (full squares) and pH 6 (opens squares), respectively. The logarithm of relative mobility [with respect to that of the (CAG)4 hairpin] is plotted against the concentration of acrylamide in the gel. The gels were run at 0°C in 0.15 M Na+ (Robinson–Britton buffer + NaCl).
In line with the electrophoretic migration, suggesting a bimolecular structure (Figs 1C and D and 4), the CD spectrum amplitudes of (GCC)4 [as well as of (GCC)8; not shown] depend on DNA concentration at slightly acidic pH. The CD spectrum of 7 mM nucleoside concentration (GCC)4 (Fig. 2A) displays high amplitudes, but its global shape remains B-like. It shares the isoelliptic point of the CD spectra reflecting the two-state, pH-induced hairpin–homodimer transition of (GCC)4 (measured at an order of magnitude lower DNA concentration). Thus the DNA concentration-dependent CD spectra reflect the same process, i.e. stabilization of the homodimer. The homodimer melts cooperatively (Fig. 2B). Melting is a two-state process up to ∼30°C (not shown) and it has fast kinetics (not measurable by CD). At pH 6 and higher temperatures the temperature dependence of (GCC)4 follows the melting curve of the (GCC)4 hairpin, which is stable at pH 8.5 (Fig. 2B), indicating dissociation of the homodimer of (GCC)4 into hairpins. This conclusion is confirmed by electrophoresis at pH 6 at ∼30°C (not shown). The dimer–hairpin dissociation is fully reversible and fast.
The longer (GCC)n fragments behave like (GCC)4, according to the CD results (Fig. 3). The electrophoretic results (Fig. 1), however, show that (GCC)4 and (GCC)8 are predominantly bimolecular at slightly acidic pH, but (GCC)16 remains predominantly monomolecular. Thus the monomolecular structure of (GCC)16 is similar to the bimolecular structures of (GCC)4 and (GCC)8. The 24mer (GCC)8 also partly adopts the monomolecular arrangement. Exchange between the bi- and monomolecular arrangements is slow.
Acidic tetraplexes of (GCC)n strands
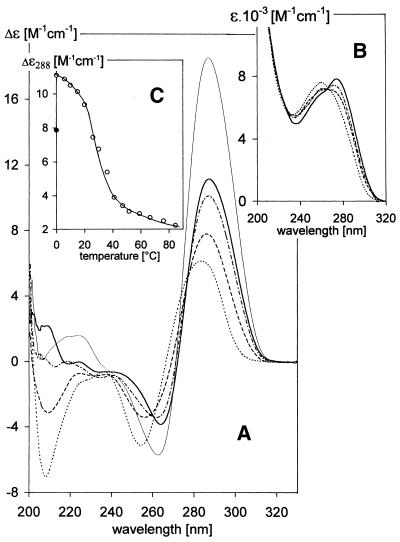
As shown in Figure 3, the bimolecular structures of (GCC)4 and (GCC)8 and the intramolecular structure of (GCC)16 are destabilized below pH 5.5–5, when spectral changes indicate another conformational transition (Fig. 5). The positive long wavelength CD band increases, the negative CD band at 250 nm (in contrast to the preceding process) diminishes, the short wavelength negative band completely disappears and the whole CD spectrum shifts towards the red (Fig. 5A). Thus a distinct type of CD spectrum appears. The UV absorption spectrum maximum at 258 nm decreases, while at longer wavelengths it increases and finally dominates (Fig. 5B). These spectral changes are characteristic of extensive protonation of cytosines and formation of hemi-protonated C·C+ pairs (32). The changes have very slow kinetics, which testify to an awkward transition between two ordered structures. It takes days to reach the equilibrium state near the transition midpoint. The structure again melts cooperatively with temperature (Fig. 5C). Melting is a slow, two-state process between 20 and 40°C. The CD spectra do not change with time at temperatures >40°C and they become similar in this temperature region to the spectra of (GCC)4 hairpins at pH 6 and 8.5 (Fig. 2B).
Figure 5.
pH-induced changes in the (A) CD spectra and (B) UV absorption spectra of (GCC)4. The sample was dissolved in 0.15 M Na+ (Robinson–Britton buffer + NaCl), pH 6 (dots). The latter three spectra were measured 4 days after adjusting the pH to: pH 5.1 (dashes), pH 4.9 (dash-dot) and pH 4.4 (solid trace). The spectra were measured at 0°C in a 0.1 cm path length cell at a DNA concentration of 0.7 mM; (thin line) (GCC)4 in the same solvent, pH 5, measured in a 0.01 cm path length cell at a DNA concentration of 6 mM, after 4 days equilibration at pH 5 and 0°C. (C) Temperature-induced changes in the CD spectra of (GCC)4 dissolved in 0.15 M Na+, pH 4.4 [conditions as for solid trace in (A) and (B)]. The filled circle corresponds to the CD value obtained immediately after cooling the sample from 85 to 0°C. No time-dependent changes in CD spectra were observed in the temperature interval 0–20°C; the spectra changed with time in the temperature interval 20–40°C, so the spectra were taken after 70 min equilibration at the particular temperatures at which all the samples reached equilibrium. No changes in CD spectra with time were again observed at temperatures >40°C.
The shape of the CD spectrum of (GCC)4 is identical at pH < 5 to the CD spectra of intercalated cytosine tetraplexes (i-tetraplexes) (33–35). The band wavelengths precisely correspond to the i-tetraplex CD spectra, only the amplitudes are lower. However, the CD spectrum of (GCC)4 completely corresponds to the i-tetraplex spectrum (including the large amplitude of the positive band approaching a value of 20 M–1 cm–1) (Fig. 5A) when measured at a DNA concentration an order of magnitude higher than for the other samples. Besides the noticeable increase in CD amplitude, higher DNA concentrations shift formation of the i-tetraplex towards less acidic pH values. The acid pH-induced transition is the same with all (GCC)n studied. It becomes slower and requires a lower pH (Fig. 3) with increasing length of the DNA molecule.
The electrophoretic results (Fig. 6) clearly show that (GCC)n generate tetraplexes at very acidic pH. We prepared two sets of (GCC)n samples. The first set (lanes 1, 3 and 5) was loaded onto the gel immediately after dilution of the samples in electrophoretic buffer of pH 5. The second set (lanes 2, 4 and 6) was loaded onto the gel after 2 days equilibration at pH 4.3 and 0°C. The oligonucleotides in the first set did not form (or did not have sufficient time to form) tetraplexes, so that they yielded marker bands corresponding to structures stable at pH > 5. (GCC)4 that was equilibrated at pH 4.3 provided two new bands (Fig. 6, compare lanes 1 and 2) migrating slower than the (GCC)8·(GGC)8 heteroduplex. The bands thus reflect two types of four-stranded (GCC)4 tetraplexes. (GCC)8 yielded a new band (possibly again two) corresponding to a bimolecular tetraplex (Fig. 6, compare lanes 3 and 4). (GCC)16 (Fig. 6, lane 6) yielded an additional slowly migrating monomolecular tetraplex. This population prevailed with a (GCC)16 sample that was equilibrated at still more acidic pH (Fig. 6, lane 7). In addition, another slowly migrating bimolecular tetraplex of (GCC)16 appeared at this very acid pH. (GCC)n·(GGC)n heteroduplexes were not stable at acid pH (Fig. 6, lanes 8–13) and their strands partially generated ordered homostructures. Electrophoreses run at pH < 5 yielded largely the same results as shown in Figure 6 (lanes 2, 4 and 6). However, we never reached conditions that solely stabilized tetraplexes.
Figure 6.
Acid PAGE of (GCC)4, (GCC)8 and (GCC)16 and of heteroduplexes with their complementary strands. The gel was run at 0°C in 0.15 M Na+ (Robinson–Britton buffer + NaCl), pH 5. The samples were loaded onto the gel in two sets: at pH 5, immediately after dissolving the oligonucleotides in electrophoresis buffer (lanes 1, 3 and 5); at pH 4.3, after 2 days equilibration at 0°C (lanes 2, 4 and 6); (GCC)16 loaded onto the gel after 5 days equilibration at pH 4.2 and 0°C (lane 7). Duplexes (GCC)4·(GGC)4, (GCC)8·(GGC)8 and (GCC)16·(GGC)16 were loaded immediately after mixing at pH 5 (lanes 8, 10 and 12) or pH 4.3 (lanes 9, 11 and 13) after 2 days equilibration of particular strands at 0°C.
DISCUSSION
Hairpins of (GCC)n
All the literature data, though differing in detail, are consistent with the conclusion that trinucleotide repeats of one G and two C residues form hairpins over a wide range of experimental conditions. It was shown that (GCC)5–11 folded back in a frame 1 hairpin with an overhanging 3′-end C (13,14), which is in line with the results we obtained independently with (GCC)2 and (GCC)4 (unpublished). The hairpin (GCC)n, n = 5–11, is thus stabilized by couples of G·C pairs formed in the GpC steps. The remaining cytosines have been reported to remain intrahelical (13,14), i.e. stacked between the G·C pairs. Our CD and UV absorption spectra, as well as the electrophoretic results, are in line with the literature data at slightly alkaline pH. The CD spectrum of (GCC)n corresponds to B-form DNA (see for example 19–22) containing classical G·C pairs while the intervening cytosines do not disturb the B-type arrangement of the hairpin. Even molar salt concentrations do not transform (GCC)n hairpins to a bimolecular duplex.
(GCC)n in neutral and slightly acidic solutions
Our observations are in line with the literature data only at slightly alkaline pH. pH values <7.5 cause evident homodimerization of (GCC)4 and (GCC)8 (Figs 1 and 4).
Homodimerization increases all amplitudes of the CD spectra, but the global shapes of the spectra remain B-like (Fig. 2A). Nevertheless, the dimer adopts a more stable arrangement compared to hairpins (Fig. 2C). What is the homodimer? Why had this homodimer escaped detection in previous studies?
We observed a strong hypochromism at the absorption maximum and a slight increase in absorption at ∼300 nm when the solution pH of (GCC)n was decreased from alkaline to neutral and then to slightly acid (Fig. 2C). The ratio of absorption at 295 nm to that at the absorption maximum reflected a partial protonation of cytosines (Fig. 3). Identical changes accompanied formation of acid pH-induced parallel duplexes of oligonucleotides containing (CGA)n (28,36,37) or (GAC)n (38). In contrast, UV absorption spectra of (CAG)4 and (CTG)4, which remained as hairpins up to acid denaturation (16,28), exhibited a hyperchromic effect at acid pH. The pK of cytosine is <5 if cytosine occurs in a hairpin, while it shifts towards 8 in parallel homoduplexes of (CGA)n and (GAC)n, where protonation stabilizes C·C+ pairs (28,38). C·C+ pairs are also formed by the (GCC)n fragments in this study around neutral pH, as judged by the completely analogous changes in UV absorption spectra. We therefore conclude that (GCC)4 as well as (GCC)8 hairpins homodimerize in a parallel-stranded fashion and that homodimerization is induced and stabilized by C·C+ pairs. The transition is fast, so that it can hardly involve unpairing of G·C pairs in the hairpin stem. In contrast, hairpin–duplex isomerization is a very slow process (25,26), lasting days (28). Moreover, the hairpin–duplex (antiparallel as well as parallel) transition becomes more complicated with increasing DNA strand length (38), which is the opposite behavior to that we observed for (GCC)n homodimerization (Figs 1 and 3). Furthermore, the CD spectrum hardly changes during this transition. Hence the bimolecular structure is obviously a homodimer of two hairpins that associate through intermolecular C·C+ pairing of the cytosines which do not pair to the complementary guanines in the hairpin stem.
The tetraplex of (GCC)n stabilized by intermolecular C·C+ pairs at neutral pH
The model of the structure formed by two parallel-oriented hairpins is sketched in Figure 7. The CpG cytosines in the hairpin stem that do not bind to the complementary guanines can remain intrahelical, but they can also ‘flip out’ (14) to generate C·C+ pairs connecting the hairpins. Hairpin homodimerization is promoted by increasing molecule length, which supports the model with hairpins associating side-by-side (Fig. 7), rather than a kissing dimer held together by a loop–loop interaction (39). We thus conclude that (GCC)4 and (GCC)8 form a bimolecular tetraplex under physiological conditions. The tetraplex is peculiar because it contains classical G·C pairs as well as hemi-protonated C·C+ pairs (Fig. 7B). That it is indeed a tetraplex is nicely demonstrated by the observation that the 48mer (GCC)16 [and partly also (GCC)8] adopts the same structure within a single molecule (Fig. 7B, bottom). Interestingly, this intramolecular tetraplex is stable at low ionic strength (1 mM sodium phosphate) even at alkaline pH. Only pH > 9 (at 0°C) denatures this tetraplex, as suggested by changes in the CD spectra corresponding to the presence of single hairpins (not shown).
Figure 7.
Schematic models of (GCC)n structures. (A) A hairpin of (GCC)4 existing at pH > 8. (B) (Top) Neutral tetraplex of (GCC)4 arising in the course of decreasing the pH from 8 to ∼6.5, as a result of homodimerization of two parallel oriented hairpins; (bottom) model of the intramolecular neutral tetraplex of (GCC)16. (C) (Top) i-Tetraplex of (GGC)4 formed by four strands at pH < 5; (bottom) model of the intramolecular acidic tetraplex of (GCC)16.
Qualitatively, the same pH-dependent changes in CD spectra as those we observed with (GCC)n were reported for (CCG)15 (15). The authors ascribed the changes around neutral pH to increased stacking and/or base pairing in single-stranded (CCG)15+. Our experiments indicate that too low a DNA concentration, a high temperature and, especially, a long molecule were the reasons why these authors did not detect the neutral tetraplex. In fact, we noticed the monomolecular neutral tetraplex of (GCC)16 only because of the parallel analysis of (GCC)4, (GCC)8 and (GCC)16.
Intercalated tetraplexes of (GCC)n
In this paper we have, furthermore, demonstrated that (GCC)n forms another tetraplex. Its CD spectrum (Fig. 5A) precisely corresponds to cytosine i-tetraplexes (33–35,40–42). The tetraplex is tetramolecular with (GCC)4, bimolecular with (GCC)8 and monomolecular with (GCC)16. Surprisingly, the presence of guanines does not noticeably influence the intercalated tetraplex structure. The i-tetraplexes, formed at acidic pH, migrate more slowly on electrophoresis than neutral tetraplexes formed by the same number of strands. Electrophoretic migration reflects DNA structure differences, including orientation of the interacting strands (28,43). The two electrophoretic bands observed with the i-tetraplexes of (GCC)n (Fig. 6, lanes 2, 4 and 7) most probably indicate two kinds of duplex intercalation in the i-tetraplex, where either the 3′- or 5′-end C·C+ pairs are located on the tetraplex edges. In the latter case the 5′-guanines would completely extrude, which would definitely influence the electrophoretic mobility of the molecule. The two types of i-tetraplexes have already been described with another molecule (44).
Conclusions
Trinucleotide repeats probably expand during replication due to an unusual conformation of their DNA (7,8,45,46). In this paper we demonstrate that the (GCC) trinucleotide repeat, associated with fragile X chromosome syndrome, can adopt not only the hairpins described in the literature (5,6,11,13,14), but also various kinds of tetraplexes. These include i-tetraplexes at acid pH and, especially, another type of tetraplex at neutral pH, which contains Watson–Crick G·C pairs as well as hemi-protonated C·C+ pairs. The neutral tetraplex arises with fast kinetics under physiological conditions. If the (GCC)n repeat is sufficiently long, then the neutral tetraplex is intramolecular. The threshold length falls somewhere between (GCC)8 and (GCC)16. The intramolecular tetraplex may arise as a result of two subsequent coils of the (GCC)n strand resembling a paperclip, or by crossing and folding back of a hairpin. Stable hairpins of expanded (CNG)n repeats exist in vivo (47). The intramolecular tetraplex of (GCC)16 described above is a conformational alternative to the Watson–Crick duplex with the complementary (GGC)16 strand, as well as to the hairpin of (GCC)16. The tetraplex is more compact and more thermostable than the hairpin. Both of these properties are of interest not only in studies of DNA conformation but also in considerations of the pathological consequences of (GCC)n strand expansions.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to Dr Jaroslav Kypr for kind discussions and advice. Mrs Marcela Muchová is thanked for expertly conducting the electrophoretic experiments. The results were obtained within the framework of research aim Z 5004920 of the Academy of Sciences of the Czech Republic. The study was supported by grant 204/01/0561 awarded to M.V. by the Grant Agency of the Czech Republic.
REFERENCES
- 1.Ashley C.T.J. and Warren,S.T. (1995) Trinucleotide repeat expansion and human disease. Annu. Rev. Genet., 29, 703–728. [DOI] [PubMed] [Google Scholar]
- 2.Oostra B.A. and Willems,P.J. (1995) A fragile gene. Bioessays, 17, 941–947. [DOI] [PubMed] [Google Scholar]
- 3.Jin P. and Warren,S.T. (2000) Understanding the molecular basis of fragile X syndrome. Hum. Mol. Genet., 9, 901–908. [DOI] [PubMed] [Google Scholar]
- 4.Mitas M. (1997) Trinucleotide repeats associated with human disease. Nucleic Acids Res., 25, 2245–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darlow J.M. and Leach,D.R.F. (1998) Secondary structures in d(CGG) and d(CCG) repeat tracts. J. Mol. Biol., 275, 3–16. [DOI] [PubMed] [Google Scholar]
- 6.Pearson C.E. and Sinden,R.R. (1998) Trinucleotide repeat DNA structures: dynamic mutations from dynamic DNA. Curr. Opin. Struct. Biol., 8, 321–330. [DOI] [PubMed] [Google Scholar]
- 7.Wells R.D. (1996) Molecular basis of genetic instability of triplet repeats. J. Biol. Chem., 271, 2875–2878. [DOI] [PubMed] [Google Scholar]
- 8.Gordenin D.A., Kunkel,T.A. and Resnick,M.A. (1997) Repeat expansion–all in a flap? Nature Genet., 16, 116–118. [DOI] [PubMed] [Google Scholar]
- 9.Usdin K. and Grabczyk,E. (2000) DNA repeat expansions and human disease. Cell. Mol. Life Sci., 57, 914–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith S.S., Laayoun,A., Lingeman,R.G., Baker,D.J. and Riley,J. (1994) Hypermethylation of telomere-like foldbacks at codon 12 of the human c-ha-ras gene and the trinucleotide repeat of the FMR-1 gene of fragile X. J. Mol. Biol., 243, 143–151. [DOI] [PubMed] [Google Scholar]
- 11.Chen X., Mariappan,S.V.S., Catasti,P., Ratliff,R., Moyzis,R.K., Laayoun,A., Smith,S.S., Bradbury,E.M. and Gupta,G. (1995) Hairpins are formed by the single DNA strands of the fragile X triplet repeats: structure and biological implications. Proc. Natl Acad. Sci. USA, 92, 5199–5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng M., Huang,X., Smith,G.K., Yang,X. and Gao,X. (1996) Genetically unstable CXG repeats are structurally dynamic and have a high propensity for folding. An NMR and UV spectroscopic study. J. Mol. Biol., 264, 323–336. [DOI] [PubMed] [Google Scholar]
- 13.Mariappan S.V.S., Catasti,P., Chen,X., Ratliff,R., Moyzis,R.K., Bradbury,E.M. and Gupta,G. (1996) Solution structures of the individual single strands of the fragile X DNA triplets (GCC)n.(GGC)n. Nucleic Acids Res., 24, 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariappan S.V.S., Silks,L.A., Bradbury,E.M. and Gupta,G. (1998) Fragile X DNA triplet repeats, (GCC)n, form hairpins with single hydrogen-bonded cytosine.cytosine mispairs at the CpG sites: isotope-edited nuclear magnetic resonance spectroscopy on (GCC)n with selective N4-labeled cytosine bases. J. Mol. Biol., 283, 111–120. [DOI] [PubMed] [Google Scholar]
- 15.Yu A., Barron,M.D., Romero,R.M., Christz,M., Gold,B., Dai,J., Gray,D.M., Haworth,I.S. and Mitas,M. (1997) At physiological pH, d(CCG)15 forms a hairpin containing protonated cytosines and a distorted helix. Biochemistry, 36, 3687–3699. [DOI] [PubMed] [Google Scholar]
- 16.Vorlícková M., Zimulová,M., Kovanda,J., Fojtík,P. and Kypr,J. (1998) Conformational properties of DNA dodecamers containing four tandem repeats of the CNG triplets. Nucleic Acids Res., 26, 2679–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao X., Huang,X., Smith,G.K., Zheng,M. and Liu,H. (1995) New antiparallel duplex motif of DNA CCG repeats that is stabilized by extrahelical bases symmetrically located in the minor groove. J. Am. Chem. Soc., 117, 8883–8884. [Google Scholar]
- 18.Gray D.M., Hung,S.-H. and Johnson,K.H. (1995) Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol., 246, 19–34. [DOI] [PubMed] [Google Scholar]
- 19.Gray D.M., Hamilton,F.D. and Vaughan,M.R. (1978) The analysis of circular dichroism spectra of natural DNAs using spectral components from synthetic DNAs. Biopolymers, 17, 85–106. [DOI] [PubMed] [Google Scholar]
- 20.Allen F.S., Gray,D.M., Roberts,G.P. and Tinoco,I.J. (1972) The ultraviolet circular dichroism of some natural DNAs and an analysis of the spectra for sequence information. Biopolymers, 11, 853–879. [DOI] [PubMed] [Google Scholar]
- 21.Kypr J., Chládková,J., Zimulová,M. and Vorlícková,M. (1999) Aqueous trifluorethanol solutions simulate the environment of DNA in the crystalline state. Nucleic Acids Res., 27, 3466–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vorlícková M. (1995) Conformational transitions of alternating purine-pyrimidine DNAs in perchlorate ethanol solutions. Biophys. J., 69, 2033–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Studdert D.S., Patroni,M. and Davis,R.C. (1972) Circular dichroism of DNA: temperature and salt dependence. Biopolymers, 11, 761–779. [DOI] [PubMed] [Google Scholar]
- 24.Kypr J., tepán,J., Chládková,J. and Vorlícková,M. (1999) Circular dichroism spectroscopy analysis of conformational transitions of a 54 base pair DNA duplex composed of alternating CGCGCG and TATATA blocks. Biospectroscopy, 5, 253–262. [DOI] [PubMed] [Google Scholar]
- 25.Xodo L.E., Manzini,G., Quadrifoglio,F., van der Marel,G.A. and van Boom,J.H. (1988) Oligodeoxynucleotide folding in solution: loop size and stability of B-hairpins. Biochemistry, 27, 6321–6326. [DOI] [PubMed] [Google Scholar]
- 26.Xodo L.E., Manzini,G., Quadrifoglio,F., van der Marel,G. and van Boom,J.H. (1989) Hairpin structures in synthetic oligodeoxynucleotides: sequence effects on the duplex-to-hairpin transition. Biochimie, 71, 793–803. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell J.E., Newbury,S.F. and McClellan,J.A. (1995) Compact structures of d(CNG)n oligonucleotides in solution and their possible relevance to Fragile X and related human genetic diseases. Nucleic Acids Res., 23, 1876–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kejnovská I., Tumová,M. and Vorlícková,M. (2001) (CGA)4: parallel, anti-parallel, right-handed and left-handed homoduplexes of a trinucleotide repeat DNA. Biochim. Biophys. Acta, 1527, 73–80. [DOI] [PubMed] [Google Scholar]
- 29.Rodbard D. and Chrambach,A. (1971) Estimation of molecular radius, free mobility and valence using polyacrylamide gel electrophoresis. Anal. Biochem., 40, 95–134. [DOI] [PubMed] [Google Scholar]
- 30.Manzini G., Xodo,L.E., Gasparotto,D., Quadrifoglio,F., van der Marel,G.A. and van Boom,J.H. (1990) Triple helix formation by oligopurine-oligopyrimidine DNA fragments. Electrophoretic and thermodynamic behavior. J. Mol. Biol., 213, 833–843. [DOI] [PubMed] [Google Scholar]
- 31.Bullock B.P. and Habener,J.F. (1998) Phosphorylation of the cAMP response element binding protein CREB by cAMP-dependent protein kinase A and glycogen synthase kinase-3 alters DNA-binding affinity, conformation and increases net charge. Biochemistry, 3, 3795–3809. [DOI] [PubMed] [Google Scholar]
- 32.Antao V.P. and Gray,D.M. (1993) CD spectral comparison of the acid-induced structures of poly[d(A)], poly[r(A)], poly[d(C)] and poly[r(C)]. J. Biomol. Struct. Dyn., 10, 819–839. [DOI] [PubMed] [Google Scholar]
- 33.Manzini G., Yathindra,N. and Xodo,L.E. (1994) Evidence for intramolecularly folded i-DNA structures in biologically relevant CCC-repeat sequences. Nucleic Acids Res., 22, 4634–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehara H., Mizuguchi,M., Tajima,K., Kanaori,K. and Makino,K. (1997) Spectroscopic evidence for the formation of four-stranded solution structure of oligodeoxycytidine phosphorothioate. Biochemistry, 36, 1790–1797. [DOI] [PubMed] [Google Scholar]
- 35.Simonsson T., Pribylová,M. and Vorlícková,M. (2000) A nuclease hypersensitive element in the human c-myc promoter adopts several distinct i-tetraplex structures. Biochem. Biophys. Res. Commun., 278, 158–166. [DOI] [PubMed] [Google Scholar]
- 36.Robinson H., van der Marel,G.A., van Boom,J.H. and Wang,A.H.-J. (1992) Unusual DNA conformation at low pH revealed by NMR: parallel-stranded DNA duplex with homo base pairs. Biochemistry, 31, 10510–10517. [DOI] [PubMed] [Google Scholar]
- 37.Robinson H. and Wang,A.H.-J. (1993) 5′-CGA sequence is a strong motif for homo base-paired parallel-stranded DNA duplex as revealed by NMR analysis. Proc. Natl Acad. Sci. USA, 90, 5224–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vorlícková M., Kejnovská,I., Tumová,M. and Kypr,J. (2001) Conformational properties of DNA fragments containing (GAC) trinucleotide repeats associated with skeletal displasias. Eur. Biophy. J., 30, 179–185. [DOI] [PubMed] [Google Scholar]
- 39.Comolli L.R., Pelton,J.G. and Tinoco,I.,Jr (1998) Mapping of a protein-RNA kissing hairpin interface: Rom and Tar-Tar*. Nucleic Acids Res., 26, 4688–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gehring K., Leroy,J.L. and Gueron,M. (1993) A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature, 363, 561–565. [DOI] [PubMed] [Google Scholar]
- 41.Leroy J.L., Gehring,K., Kettani,A. and Guéron,M. (1993) Acid multimers of oligodeoxycytidine strands: stoichiometry, base-pair characterization and proton exchange properties. Biochemistry, 32, 6019–6031. [DOI] [PubMed] [Google Scholar]
- 42.Leroy J.-L., Guéron,M., Mergny,J.-L. and Hélene,C. (1994) Intramolecular folding of a fragment of the cytosine-rich strand of telomeric DNA into an i-motif. Nucleic Acids Res., 22, 1600–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van de Sande J.H., Ramsing,N.B., Germann,M.W., Elhorst,W., Kalisch,B.W., Kitzing,E., Pon,R.T., Clegg,R.C. and Jovin,T.M. (1988) Parallel stranded DNA. Science, 241, 551–557. [DOI] [PubMed] [Google Scholar]
- 44.Kanaori K., Maeda,A., Kanehara,H., Tajima,K. and Makino,K. (1998) 1H nuclear magnetic resonance study on equilibrium between two four-stranded solution conformations of short d(CnT). Biochemistry, 37, 12979–12986. [DOI] [PubMed] [Google Scholar]
- 45.McMurray C.T. (1995) Mechanisms of DNA expansion. Chromosoma, 104, 2–13. [DOI] [PubMed] [Google Scholar]
- 46.McMurray C.T. (1999) DNA secondary structure: a common and causative factor for expansion in human disease. Proc. Natl Acad. Sci. USA, 96, 1823–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore H., Greenwell,P.W., Liu,C.P., Arnheim,N. and Petes,T.D. (1999) Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl Acad. Sci. USA, 96, 1504–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]