Abstract
Cryo-electron tomography (cryoET) and subtomogram averaging (STA) has had a rapid development in recent years. It provides structures of macromolecular complexes in situ and in cellular context at or below subnanometer resolution and has led to unprecedented insights into the inner working of molecular machines in their native environment, as well as their functional relevant conformations and spatial distribution within biological cells or tissues. Given the tremendous potential of cryoET STA in in situ structural cell biology, we previously developed emClarity, a GPU-accelerated image-processing software which offers subtomogram averaging and classification of macromolecular complexes at high resolution. However, the workflow remains challenging, especially for newcomers to the field. In this protocol, we describe a detailed workflow, processing and parameters associated with each step, from initial tomography tilt-series data to the final 3D density map, with several features unique to emClarity. We use four different samples, including HIV-1 Gag assemblies, ribosome and apoferritin, to illustrate the procedure and results of subtomogram averaging and classification. Following the processing steps described in this protocol, along with a comprehensive tutorial and guidelines for troubleshooting and parameter optimization, one can obtain density maps up to 2.8 Å resolution from six tilt series by cryoET STA.
INTRODUCTION:
Cryo-electron tomography (cryoET) has gained increasing importance to study molecular architectures of viruses, bacteria and cellular components in situ1–3. It can provide 3D reconstructions of pleomorphic objects such as organelles or cells in their close-to-native states, providing unique opportunities to capture the intermediate biological events in the cellular context. More importantly, the spatial relationship among macromolecules within a cellular tomogram can be determined4. In cryoET, a series of images from the same region of the specimen are recorded as the sample is tilted to various angles with respect to the incident electron beam. The images are subsequently aligned and reconstructed to generate a 3D tomogram. When there are many repeating objects, such as macromolecular complexes, in the tomogram, these objects can be aligned and averaged to improve the signal-to-noise ratio (SNR)5, a process referred to as cryoET subtomogram averaging (STA).
Compared with cryoEM single particle analysis (SPA), STA generally results in lower resolution. However, STA can resolve macromolecule structures in situ, unpurified, and in the cellular context, as well as provide a spatial relationship between molecules, which is important for interpreting their biological functions. Nonetheless, several studies have yielded high resolution density maps resolving secondary structural elements, including Coat Protein Complex I6, Nuclear Pore Complex4,7, polysomes8, chemotaxis signalling arrays9, retroviruses assembly10–14, bacteria surface layer15, and ribosomes16.
There are multiple additional challenges in STA compared to SPA1,17,18. Firstly, due to the physical limits of the goniometer as well as increasing sample thickness upon tilting, tilt-series are typically limited to tilt angles between −60° to 60°. The densities in a tomogram reconstructed from these tilt-series therefore suffers distortions, referred to as missing wedge effect. This distortion significantly affects the precision of subtomogram alignment and classification and must be considered for high-resolution STA. Secondly, biological samples are sensitive to radiation damage and the electron exposure applied to each tilted image is usually limited. As a result, the SNR of tilted image is much worse compared with images in SPA. Thirdly, specimens for cryoET are usually thick and the effective thickness of sample increases when sample tilts. The defocus gradient due to the thickness of sample and sample tilt also needs to be considered19. As many biological objects adopt multiple conformations or compositions, 3D classification is required to delineate these different variances. While STA has, in principle, an advantage in 3D classification over SPA since each particle exists as a unique 3D reconstruction thus allowing for direct analysis of the 3D variance, the low SNR and missing wedge effect often pose significant challenges20.
To deal with these challenges, a number of software packages has been developed for STA this far, including PEET21, EMAN222–24, RELION25,26, Dynamo27, Jsubtomo28, PyTom/AV329,30, Warp/M16, Protomo/i331 and emClarity32 (see review in Zhang, 2019 for a comparison1). We implemented several key features in emClarity. Firstly, an algorithm was implemented to estimate the defocus and astigmatism for each tilted image within the tilt series, to calculate the Contrast Transfer Function (CTF). The effect of CTF modulation of images is then corrected for during tomogram reconstruction, accounting for the depth-of-field32. Secondly, for accurate weighting during alignment, reconstruction and classification, emClarity computes 3D sampling functions (3DSF). The 3DSF of each subtomogram, which accounts for the missing wedge information, is updated during each step of processing and used as a weight. Thirdly, to address sample heterogeneity, emClarity implements a multiscale 3DSF-weighted, PCA based classification method, which allows the user to emphasise specific features of different length scales. Fourthly, local specimen motion and deformation place a major restriction on the quality of STA reconstructions. emClarity implemented Tomogram constrained projection refinement (tomoCPR) to refine local shifts, rotations, and magnification changes in the sample by using subtomograms as fiducial markers. This improves the tilt-series alignment, particularly for in situ cryoET dataset recorded from cryo-Focused Ion Beam milled lamellae, where gold bead fiducials are often not available.
Several high-resolution cryoEM maps have been successfully obtained by various research groups using emClarity1, including SARS2-Cov2 postfusion spikes33, in situ structure of Parkinson’s disease-linked LRRK234, cellular reovirus assembly intermediates35, Zika virus capsid protein36, nodaviral replication protein A crown complex37, native Leptospira spirochete flagellar filaments38 and bacterial chemotaxis signalling arrays39.
There are some major changes in the new version of emClarity (V1.5.3.10) since the original publication (V1.0)32. These include:
Per-tilt CTF refinement using embedded CTFFIND440;
Handedness check during CTF estimation;
Per-particle 3D sampling function is calculated;
3DSF calculation has been improved;
Switch to MATLAB 2019a;
Peak masks to limit translational search in alignment: the peak mask can be used to remove the cross correlation peaks from a given distance of the particle origin, i.e., it defines the maximum translation allowed.;
Reconstruction using the raw projection images using cisTEM.
Here, we describe a detailed workflow and processing steps using the new version of emClarity. The protocol has been tested by several novice users and the common issues that might arise during the procedure are detailed in the troubleshooting.
Overview of emClarity pipeline
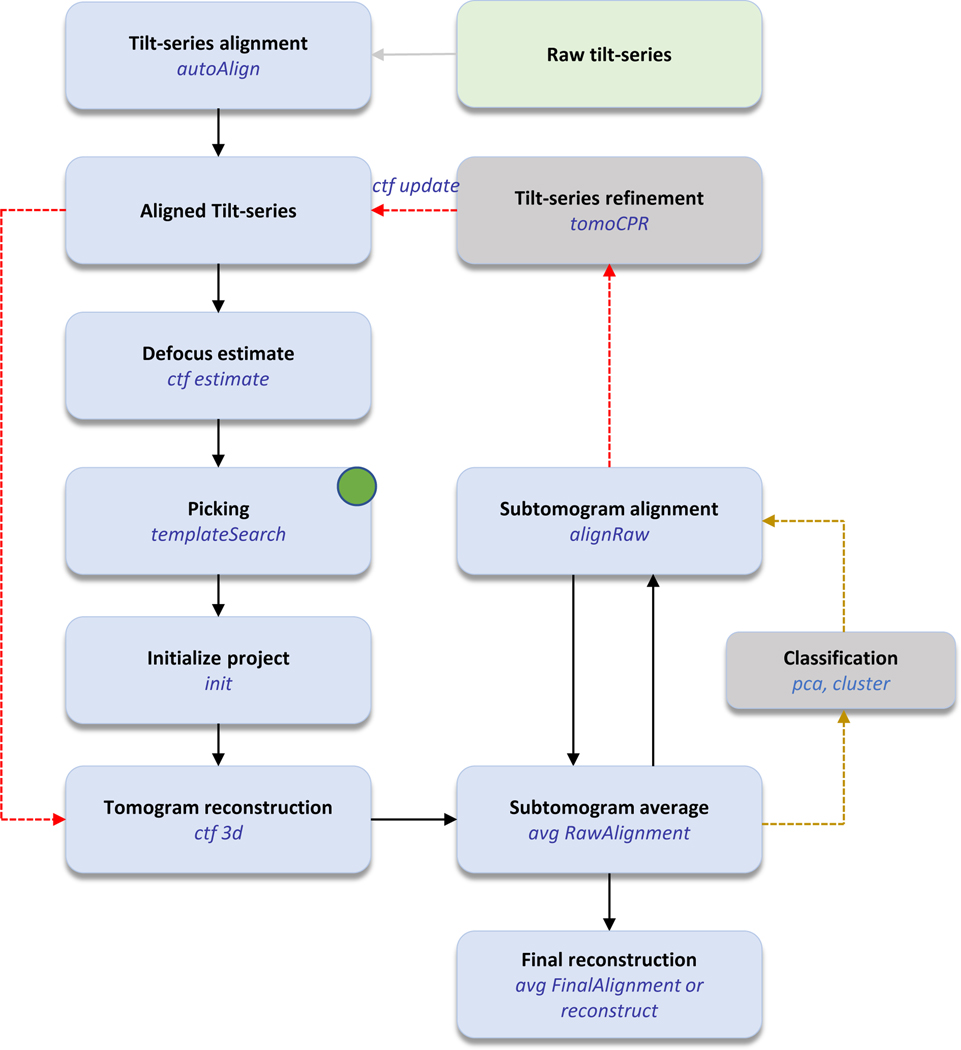
emClarity streamlines all steps in the pipeline (Figure 1). emClarity can align the raw tilt-series automatically using its “autoAlign” program. It can also import the aligned tilt-series from external software packages, as long as the file formats and naming conventions follow the requirement (Step 1). It then generates aligned tilt-series and estimates the CTF of each tilt-series (step 2–4). Users define the boundary of sub-region(s) in the tomogram for later reconstruction (step 5–7). The particles are then picked using template matching (step 8–12). emClarity manages the subtomogram associated metadata in a MATLAB database and updates the metadata after each processing step throughout the pipeline (step 13). The CTF corrected tomograms are then generated at the requested binning (step 14) and subtomogram averaging and alignment can be performed iteratively at each binning (step 15–18). TomoCPR can be performed (step 19–20) to refine tilt-series alignment, as well as subtomogram classification (step 22–30), both of which are optional steps. During the iterative alignment and averaging cycles, the data are kept in two fully separate half-sets following the “gold-standard” refinement procedure41. The half-sets are used to calculate an optimal filter for weighting the reconstructions, while reducing the risk of overfitting42. A final map can be generated combining the two half-sets with an additional B-factor sharpening optionally applied (step 31). A new feature is additionally implemented in emClarity, such that the raw projection images, instead of subtomograms, can be used for the final reconstruction using cisTEM. Table 1 lists cryoET data collection and processing details. emClarity processing run time for the main steps is illustrated in Table 2, along with specific GPU cards used for processing.
Figure 1 |. emClarity processing workflow.
Green boxes indicate data input. emClarity processing steps are in blue with optional steps in grey. Dashed red and gold lines are optional tomoCPR and classification processes, respectively. Subtomogram positions can also be imported from other software (indicated by the green dot). emClarity commands are written in blue text.
- I think that this is great.
- I think that it would be better if the colours in boxes were uniform.
- The arrow between “Aligned Tilt series” and “Tomogram reconstruction” is a little bit confusing. Does it mean that you can out the intermediate steps OR does it mean that you can “add” the “Aligned Tilt-series” to something in order to help with “Tomogram reconstruction”?
- I am okay that the meaning of the arrows is slightly ambiguous, but if it is easy make it more clear using language – please consider doing this. E.g. Sometimes that the text in the box implies an action, and sometimes it implies the result of doing an action.
Table 3 |.
Troubleshooting
| Steps | Problem | Possible reason | Solution |
|---|---|---|---|
| 3–4 | Gold fiducial beads are not removed correctly | The gold fiducial file fixedStacks/<prefix>.erase is not present or incorrect | Check Etomo alignment and recreate the <prefix>.erase and redo ctf estimate |
| 3–4 | The handedness is wrong | (i) Tilt-series rotation angle is not correct (incorrect by 180°) during Etomo alignment. (ii) Some detectors or software may save the raw image frames with additional rotation or flipping. |
(i) Redo Etomo alignment with correct tilt-series rotation angle (plus or minus 180 degrees). (ii) Flip or rotate the tilt-series to match the anticipated angle. |
| 3–4 | The estimated defocus is wrong | The anticipated defocus range defined in parameter defEstimate ± defWindow does not cover the real range. | Check whether the theoretical CTF estimate matches the radial average of the power spectrum of the tilt- series in fixedStacks/ctf/<prefix>_ali1_psRadial_1.pdf. Adjust the defocus range parameter defEstimate ± defWindow and rerun ctf estimate for the current tilt- series. The correct defocus peak can be found in fixedStacks/ctf/*_ccFIT.pdf |
| 9–11 | Convmap does not show clear local CC peaks | (i) The template pixel size is not calibrated. (ii) The tomogram is too noisy. For example, the tomogram is too thick. |
(i) The template matching is very sensitive to the correct pixel size. When starting with an external reference, it is probably best to process 10% or so of the data to generate a new reference for the full run. (ii) Use external software to improve the template. (iii) Optimize the Ali_mRadius and Ali_mType, especially for particles in lattice assembly. (iv) The sub-region is filtered at the spatial frequency of the first CTF at zero of each tilt-series by default. Overwrite it by including a different low-pass filter (parameter: lowResCut=40) in the parameter file. |
| 9–12 | Multiple points are picked on the same particle, or points are too close to each other | Ali_mRadius is too small | Increase particleRadius during template search, since it defines a region around a cross-correlation peak to remove from consideration after a particle is selected. |
| 13 | Init fails to generate database file | (i) convmap directory is not available. (ii) each <prefix>_<sub- region>_binX.mod should contain at least one point, i.e., one subtomogram. |
(i) Remember to rename convmap_wedgeType2_binX/ to convmap/ before running init. (ii) Make sure the number of convmap/<prefix>_<sub- region>_binX.mod matches the number of recon/<prefix> recon.coords files in the recon/ folder. |
| 15 | The average contrast is inverted | The non-CTF corrected tomograms are used for template search. The first cycle of average was performed at the same binning as template search, which uses tomograms generated from template search instead of ctf 3d. | Remove the previous cache/<prefix>_<sub- region>_binX.rec files generated during templateSearch before running ctf 3d if one wants to do the initial alignment at the same binning factor of template search. We generally start averaging and alignment at a different binning from the one used for template search. |
| 15–16 | Failure in average or alignment step ‘PEET error’ | The sub-region tomograms (*rec) or sampling function (*wgt) in cache/ directory are corrupted or not generated, which can happen when system disk is full during an emClarity step. | Check the integrity of the cache/<prefix>_<sub- region>_binX.rec and cache/<prefix>_binX.wgt using header or open with 3dmod command from IMOD. Remove the corrupted files and rerun ctf 3d. |
| 15–16 | ‘Reference to non-existent field cyclexxx” | The previous step (averaging, alignment, classification, etc.) did not finish successfully. | Rerun the previous step (averaging, alignment, classification, etc.) to update the <project>.mat. Check the logFile/emClarity.logfile to make sure it finishes properly. |
| 15–16 | Out of memory | emClarity exits when the required GPU memory is not available. | Reduce the number of parallel processing (set nCpuCores=2, for example). The requirement of GPU memory is related to the box size in average and alignment. In later stages of refinement with small binning factor, the required GPU memory for each process is substantially higher. Check usage of GPU memory (type nvidia-smi in the command line). |
| 19 | tomoCPR fails to run | (i) The average and alignment have not finished successfully. (ii) There is no particle in a subregion. |
(i) Rerun the average and alignment for the current cycle. (ii) Make sure there is at least one particle in a subregion. One can check the metadata <project>.mat or check the alignResume/cycleXXX_<project>/*.txt. Each text file should contain at least one line. Particle in a sub-region can be removed automatically if it drifts to the edge of sub-region. |
| 19 | ‘Error using BH_synthetic_ma pBack (line 978) mapBackRePrjSiz e = 4 is still too much for the Error in emClarity (line 366)’ | The amount of GPU memory needed for reconstruction depends on the size of the local shifts. Generally, this error only occurs if the local shifts are unrealistically large. |
Remove this tilt-series from your analysis. |
| 19 | tomoCPR results in misaligned tilt- series | (i) There are too few particles in the sub-region. (ii) All the particles are in one corner of field of view. |
To test whether tomoCPR helps, run average and alignment at one binning for several cycles until rotation and shifts are close to zero, then run tomoCPR and generate new tomogram in the same binning, redo averaging and alignment to see whether density map or FSC improves. tomoCPR may not improve tilt-series alignment equally for all tilt-series. |
| 25 | Cluster does not run | The Pca_coeffs file is not formatted correctly | Make sure that Pca_coeffs contains the same number for each pcaScaleSpace. The rows of Pca_coeffs should be equivalent to number of pcaScaleSpace. |
| 22–30 | Classification does not result in different classes | Suboptimal selection of eigenimages | Try a few different sets of Pca_coeffs and Pca_clusters and rerun cluster and average. |
| 31 | Out-of-memory in cisTEM reconstruct | CPU memory is not sufficient | Reduce the <max_exposure> to include less images |
Prerequisite for using the protocol
This protocol is broadly applicable to cryoET STA projects, but is focused on providing details needed for high-resolution refinement. emClarity uses GPU accelerations and parallelization tools to cope with large datasets. Since emClarity does not have a graphic user interface, users are expected to have basic knowledge of working with the command line on Unix/Linux-based systems. It is beneficial to have a good knowledge of fiducial based alignment as implemented in Etomo43. Familiarity with MATLAB scripting can be helpful, but is not required. Basic knowledge of principle component analysis (PCA) and commonly used clustering method (such as k-means clustering) is useful when carrying on emClarity subtomogram classification. Users could also refer to the associated emClarity Tutorial (Supplementary information 1) (https://github.com/ffyr2w/emClarity-tutorial) for in-depth understanding algorithms behind each step, as well as detailed step-by-step processes using a ribosome dataset (EMPIAR-10304).
Limitations:
Since emClarity uses a template-based particle picking method, it requires users to have a template for the object of interest. One should pay a close attention to the template search and be cautious to template bias. We recommend using a low-pass filtered template to minimize template bias. emClarity implement template matching with either non-CTF corrected or CTF-corrected tomograms, and comparison or combination of these two results can be informative for some challenging dataset. Small objects (<0.5MD), like SARS-COV2 spikes in cellular tomography dataset, can be identified through template search, albeit containing false positives. In this case, the existing prior information (like particle position and orientation relative to membrane) can be used to exclude these false positives. The number of desired particles during template search can be either determined automatically within emClarity or manually set by user. When templates are not available, one can use other software packages, such as Dynamo27 and PEET21, to generate an initial template. It is also possible to import particles (coordinates and angles) picked or refined from other software into emClarity (Figure 1, green dot). Although emClarity can refine tilt-series alignment by tomoCPR, we recommend aligning the initial tilt-series to a satisfactory level using emClarity autoAlign or other packages like Etomo43 or AreTomo (https://drive.google.com/drive/folders/1Z7pKVEdgMoNaUmd_cOFhlt-QCcfcwF3). In some cases, geometry refinement by tomoCPR might result in inadequate results.
MATERIALS
EQUIPMENT and SETUP
A computer or a computing cluster with NVIDIA GPU cards at least 12GB of memory, CUDA Version 7.5 or greater (Version 9 or newer preferred). An emClarity binary (Version 1.5.3.10) and installation procedure are available and detailed in emClarity wiki (https://github.com/bHimes/emClarity/wiki).
INPUT DATA
Data: raw tilt-series
Raw image movies need to be motion-corrected, but without exposure weighting, which is handled internally by emClarity. Motion-corrected images in a tilt-series should be ordered in the sequence of tilt angle, from −60 to 60, for example. Tilt-series can aligned using external software packages like Etomo and imported to emClarity. Users can also import the raw tilt-series and use emClarity to align it automatically. Details of required files and formats are listed in Step 1 in PROCEDURE.
Data: metadata
Microscope imaging conditions: voltage, pixel size, defocus range, amplitude contrast and Cs.
Data collection scheme (the order and exposure dose of image acquisition in a tilt-series).
emClarity currently uses a parameter file to manage inputs, usually named to reflect their function and cycle, such as param_ctf.m for CTF estimation and param1.m for cycle 1 alignment, averaging and classification. The parameters required for individual step are listed and explained in detail in the Tutorial (Supplementary information 1). A parameter file together with run commands for the processing of HIV-1 Gag dataset in this protocol is shown in Supplementary information 2 and a template is supplied with emClarity installation.
PROCEDURE
<CRITICAL> This protocol presents a stepwise working procedure for subtomogram averaging and classification using emClarity. Users run all the commands through a terminal shell inside the project directory. The entire iterative alignment, averaging and classification procedure can run to the end automatically through a runscript, as long as the parameter files are set properly for each cycle. Users should modify and optimize the key parameters relevant to their projects. In the following processing steps, step 1 to 31, we provide the individual run command with specific parameters and discuss the results, as well as troubleshoot potential issues. Novice users are recommended to follow the exact steps and check the outputs for each step and compare with the results described here. Users can refer to a more comprehensive tutorial (Supplementary information 1) (https://github.com/ffyr2w/emClarity-tutorial), which contains a detailed explanation of all parameters and basic algorithm for each processing step in emClarity.
Preparation: arrangement of input files and directories<Timing> ~30 min when using autoAlign
Tilt-series can be aligned automatically inside of emClarity, or externally using software like Etomo. In this protocol, some datasets were aligned using Etomo and imported to emClarity, and some were automatically aligned using the “emClarity autoAlign” program. The “autoAlign” function requires motion-corrected image stacks, tilt angle file and tilt axis rotation angle, and it prepares all the necessary files in fixedStacks/. Please refer to the supplementary tutorial for the parameters. If users align the tilt-series using external software like Etomo, please prepare the necessary files as indicated in Step 1.
1 | Make a project directory. Within the project directory, make a new directory called fixedStacks/. It is essential to strictly follow the naming conventions. Copy the following files into it.
<prefix>.fixed: the raw tilt-series corresponding to <prefix>.st.
<prefix>.xf: the transformation file generated from tiltalign in Etomo;
<prefix>.tlt: the refined tilt angle file;
(optional) <prefix>.local: the local alignment transformation file corresponding to <prefix>local.xf from tiltalign in Etomo.
(optional) <prefix>.erase: coordinates of the fiducial beads to erase, corresponding to <prefix>_erase.fid in Etomo.
(optional) <prefix>.order: refined tilt angles listed in the order of image acquisition. For example, if data collection starts from 0 degree and oscillate from 3, −3, 6, −6 … 60, −60, then the order file contains a single column listing these angles as 0, 3, −3, 6, 6 … 60, −60. However, we recommend generating the order file if the data acquisition scheme can not be represented by the exposure-weighting parameters (see Step 3).
If there are black images at high-angle in the tilt-series, we recommend removing these dark images during tilt-series alignment and make sure the corresponding .xf and .tlt are also updated. It is recommended to process the raw tilt-series with IMOD CCD eraser to remove hot and dead pixels.
2 | Set up appropriate working environment for emClarity (e.g. module load emClarity/1.5.3.10). Run emClarity using the provided command-list (Supplementary information 2). Users can run through the script entirely or run individual command separately as described below. If you have existing IMOD or UCSF Chimera in the environment, make sure there is no conflict. All the emClarity related logs are saved in logFile/emClarity.logfile.
Defocus estimate<Timing> ~25 min
3 | Estimate the defocus of the tilt-series. In this step, the raw tilt-series will be transformed into aligned tilt-series using the per-tilt transformation file; the gold-fiducials will be removed; and the aligned tilt-series will be used for per-tilt defocus and astigmatism estimation. The parameter file should contain the necessary imaging parameters. Copy a template parameter file to project directory and rename it as param_ctf.m.
| System parameters: | |
| nGPUs=4 | %% number of visible GPUs |
| nCpuCores=12 | %% maximum number of processes to run in parallel. |
| Microscope settings: | |
| PIXEL_SIZE=1.179e-10 | %% pixel size of raw tilt-series in meter |
| SuperResolution=0 | %% whether raw tilt-series pixel size corresponds to super-resolution image pixel size |
| Cs=2.7e-3 | %% in meter |
| VOLTAGE=300e3 | |
| AMPCONT=0.1 | %% amplitude contrast |
| beadDiameter=7e-9 | %% fiducial bead diameter in meter |
| Defocus range: | |
| defEstimate=2.3e-6 | %% in meter |
| defWindow=1.5e-6 | %% in meter |
| Exposure-weighting parameters: | |
| CUM_e_DOSE=123 | %% total exposure dose |
| doseAtMinTilt=3 | %% electron dose at minimum tilt |
| oneOverCosineDose=0 | %% whether Saxon scheme is used |
| startingAngle=0 | %% refined data collection starting angle |
| startingDirection=pos | %% data collection direction |
| doseSymmetricIncrement=1 | %% dose symmetric scheme group size |
The last three parameters in exposure-weighting are used to indicate the order of image acquisition for exposure weighting, which can also be specified by providing a <prefix>.order file in fixedStacks/. If a <prefix>.order is provided in the fixedStacks/, the exposure-weighting parameters will be ignored. For each tilt-series, run the following command:
emClarity ctf estimate <param> <prefix> emClarity ctf estimate param_ctf.m b2tilt20
A new directory aliStacks/ will be generated in the project directory and the aligned tilt-series aliStacks/<prefix>_ali1.fixed will be saved. For each tilt-series, per-tilt defocus and astigmatism estimation results are saved as fixedStacks/ctf/<prefix>_ali1_ctf.tlt, which contains the tilt geometry information, accumulated exposure dose and per-tilt defocus information. Repeat CTF estimation for all tilt-series:
#!/bin/bash
for stack in fixedStacks/*.fixed; do
prefix=${stack#fixedStacks/}
emClarity ctf estimate param_ctf.m ${prefix%.fixed}
done
4 | Inspect the results of CTF estimation for each tilt-series:
Open the transformed tilt-series in aliStacks/<prefix>_ali1.fixed in 3dmod and make sure they are correctly aligned and fiducial beads are removed properly;
emClarity also prints out the results of a tilt-series handedness check in the logfile/emClarity.logfile. The handedness check informs whether the expected defocus gradient matches the measured value. However, it should be noted that the handedness correctness does not necessarily indicate the biological handedness of density map is correct.
Open fixedStacks/ctf/<prefix>_ali1_psRadial_1.pdf and check that the theoretical CTF estimate matches the radial average of the power spectrum of the tilt-series.
<Troubleshooting> (Table 3)
Define sub-region boundaries<Timing> ~10 min
5 | In many cases, the regions of interest are in some local areas (sub-regions) in the whole tomogram. The boundary of a sub-region is defined in a binned tomogram with the entire field of view. Copy the recScript2.sh from emClarity installation directory to the project directory. Run the recScript2.sh script and a binned tomogram for each tilt-series will be generated in the bin10/ directory:
sh recScript2.sh −1
6 | Define the sub-region boundaries in the bin10 tomogram by defining 6 points (xmin, xmax, ymin, ymax, zmin and zmax) to enclose the sub-region. Inside the bin10/ directory, run:
3dmod <prefix>_bin10.rec
If you have 3 sub-regions in one tomogram, you will need to define 6 × 3 = 18 points. Save the model (File → Save model) with the same name as the tomogram but with the .mod extension in the bin10/ directory. One should generate one *.mod file per tilt-series. Leave at least a few pixels from the edge of the binned reconstruction for model boundary and sub-regions in a tomogram should not overlap. Sub-regions can be as big as the whole tomogram as long as the GPU cards have enough VRAM. In practice, splitting the tomogram into 2 sub-regions is supported for GPUs with >=12GB of memory. In this tutorial, we defined each virus-like particle as one sub-region so that multiple sub-regions can be processed in parallel to maximize computational throughput.
7 | Convert the <prefix>_bin10.mod file to an emClarity format. This generates a recon/ directory, within with <prefix>_recon.coords defines the boundary information of each sub-region of every tomogram. In the project directory, run:
./recScript2.sh <prefix>
To convert all the sub-regions of each tomogram, run:
#!/bin/bash
for stack in bin10/*.mod; do
prefix=${stack#bin10/}; ./recScript2.sh ${prefix%_bin10.mod};
donePick particles<Timing> ~ 1.5 h
<CRITICAL> emClarity uses a template-based particle picking method. A template is required (step 8) and template search for each sub-region is performed at designated binning (step 9 and 10). Check the template search result (step 11).
8 | Prepare the template for particle picking. The template used by emClarity need to have the same pixel size as that of the raw tilt-series (PIXEL_SIZE parameter). One may need to rescale the template from a source map to match the pixel size.
emClarity rescale <input> <output> <inputPixel> <outputPixel> cpu/GPU emClarity rescale EMD-8403.mrc emd_8403rescale.mrc 3.62 1.179 cpu
9 | Generate ctf corrected tomograms for template search. This step generates the binned tilt-series and CTF-corrected (i.e. CTF multiplied) tomograms for each sub-regions, and saves them as cache/<prefix>_<sub-region>_binX.rec.
| Parameters: | |
| Tmp_samplingRate=8 | %% binning factor for tomogram for template Search emClarity ctf 3d param_ts.m templateSearch |
10 |Run template search for each sub-region from each tomogram. One needs to decide the binning of tomogram for template search. Depending on the subtomogram size, we typically recommend running template search with tomograms at a final pixel size around 8–10Å/pixel. Ali_mRadius is the alignment mask radii. Test different Ali_mRadius and particleRadius to optimize particle picking, especially for subtomograms arranged in a lattice-like assembly. For the HIV Gag assembly, we set Ali_mRadius with the size of 7 Gag hexamers and particleRadius with size of one hexamer, so that the cross-correlation is calculated with a large molecular mass, while the individual hexamers positions can be picked. For the ribosome or apoferritin dataset, Ali_mRadius and particleRadius can be very close. Tmp_angleSearch defines the range and step of out-plane and in-plane angular search as [θout, Δout, θin, Δin] in degrees. For example, [180,9,35,7] specifies a ±180° out of plane search, with 9° each step, and ±35° in plane search with a 7° step. For subtomogram with cyclic symmetry, the in-plane search range can be limited to ±180/<symmetry>. Copy a template parameter file, rename it as param_ts.m and update the following parameters. The microscope parameters should remain constant as in ctf estimate.
| Parameters: | |
| Tmp_samplingRate=8 | %% binning factor for tomogram for template Search |
| particleRadius=[66,66,56] | %% in Angstrom. cross-correlation peak radius to remove from consideration after a particle in the current peak is selected. |
| Ali_mRadius=[116,116,72] | %% Radius of alignment mask in Angstrom |
| Tmp_angleSearch= [180,9,35,7] | %% in degrees |
| Tmp_threshold=1000 | %% estimate number of particles |
| symmetry=C6 | %% particle symmetry |
In the project directory, run:
emClarity templateSearch <param> <prefix> <sub-region> <template> <symmetry> <GPU_id> emClarity templateSearch param_ts.m b2tilt20 1 emd_8403rescale.mrc C6 1
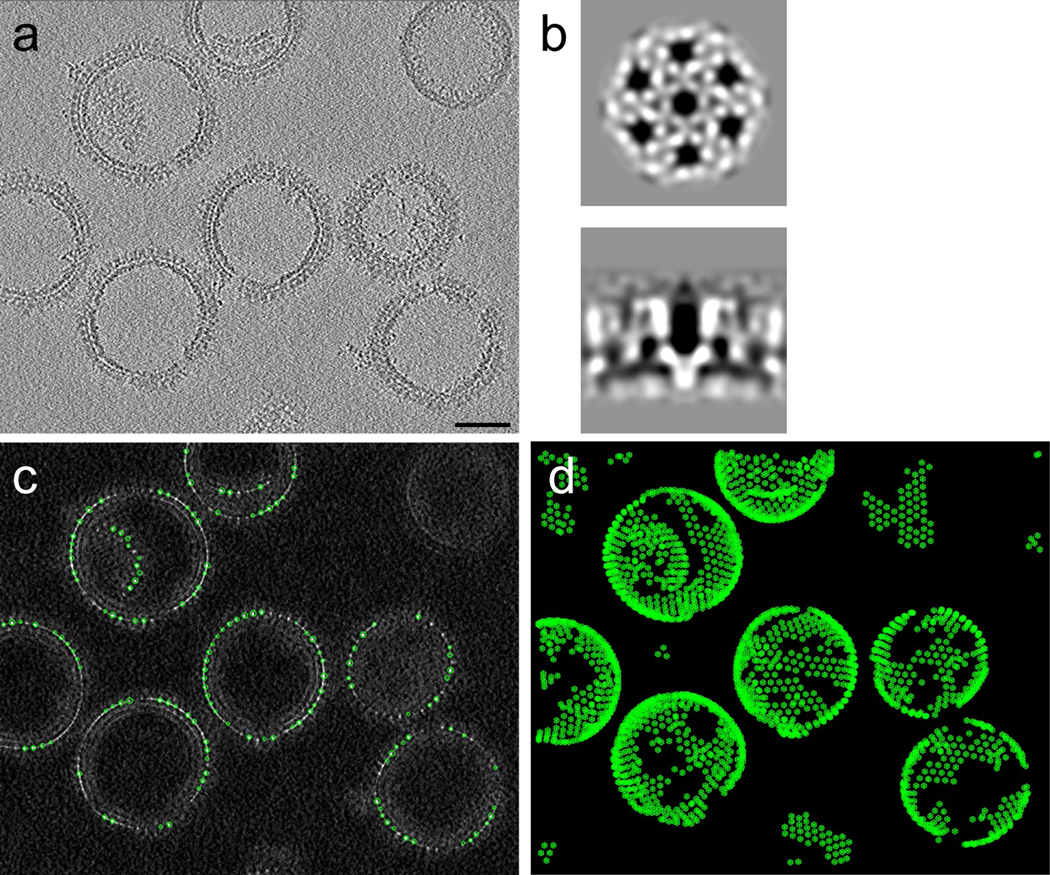
A new directory called convmap_wedge_Type2_binX/ contains the cross-correlation (CC) convolution map <prefix>_<region>_binX_convmap.mrc and model <prefix>_<region>_binX.mod, corresponding to the coordinates of picked particles. The resulting <prefix>_<region>_binX.csv file contains the unbinned coordinate and orientation information on all picked particles. Please refer to emClarity wiki for the convention and format of this file. A representative tomogram (bin8) and convolution map is shown in Figure 2.
Figure 2 |. Template matching.
(a) A typical tomographic slice (6 nm thick) depicting HIV-1 Gag T8I assemblies from the raw data. (b) The template used for particle picking, top and side views of HIV-1 Gag map (EMD-8403) low-pass filtered to 25 Å. (c) A tomographic slice of resulting convolution map overlaid with template matched model points of top cross-correlation peaks. (d) A projection view of model points through the tomogram volume. Scale bar is 50 nm.
- If found this confusing. What is the connection between the image in panel A and that shown in panel B?
- You mention “template search”. What template is used? If meaningful, could you add the template to the figure?
11 |Clean the false positive points using 3dmod. In the convmap_wedge_Type2_binX/ directory, run:
3dmod <prefix>_<sub-region>_binX_convmap.mrc <prefix>_<sub-region>_binX.mod
It is also useful to overlay the raw tomograms with convmap and model:
3dmod ../cache/<prefix>_<sub-region>_binX.rec <prefix>_<sub-region>_binX.mod
Check the <prefix>_<sub-region>_binX_convmap.mrc about the summed CC peaks to see whether they correspond to the desired subtomogram positions. Remove the false positive points, which are common in regions with strong features like ice contamination, carbon edges, gold beads residues, etc. Save the remaining points using the same model file name. Before averaging and alignment, one should ensure that the picked particles were mostly correct. It might be not necessary to clean all the false positive points as 3D classification usually can remove them.
12 |Rename the convmap_wedge_Type2_binX/ to convmap/, as emClarity will look into the convmap/ directory for subtomogram information in the next step.
Initialize the project<Timing> ~ 1 min
<CRITICAL> As mentioned above, emClarity stores all the project information in a MATLAB database. The database records information on the tilt-series and subtomograms including: sub-region boundary (recon/<prefix>.coords), per-tilt CTF estimate (fixedStacks/ctf/<prefix>_ali1.tlt) and information on each subtomogram (convmap/). This metadata will be used and updated throughout the emClarity data processing pipeline. A backup metadata will be saved as cycleXXX_<project>_backup.mat before a new cycle starts. Users can open the database in MATLAB to check the database structure.
13 | Generate an emClarity database <project>.mat. Copy param_ctf.m to param0.m and update the following parameters:
| Parameters: | |
| subTomoMeta=gag | %% <project> name |
| Tmp_samplingRate=8 | %% template matching binning |
| fscGoldSplitOnTomos=1 | %% whether or not the particles from the same sub-regions should be kept in the same half-set or distributed randomly |
Run the command as follows, which generates a metadata as gag.mat
emClarity init <param> emClarity init param0.m
Notes: fscGoldSplitOnTomos is typically set to 0 (randomly splitting subtomograms from each sub-region into ODD and EVEN datasets). However, if the particles within the alignment mask overlap substantially with their neighbour particles, such as in the Gag lattice, we used “1” to split sub-regions instead of subtomograms for ODD and EVEN datasets to avoid floating the FSC. For a small dataset with a limited number of tilt-series, we recommend defining more than two sub-regions for each tilt-series.
Reconstruct the tomograms for alignment and averaging<Timing> ~ 5 min
Reconstruct the sub-regions for all the tilt-series. This step generates the binned tilt-series and CTF-corrected (actually CTF multiplied) sub-regions tomograms, which are saved in the cache/ directory and are then used for the subtomograms extraction, averaging and alignment.
| Parameters: | |
| subTomoMeta=gag | |
| PIXEL_SIZE=1.179e-10 | |
| Ali_samplingRate=6 | %% Note this samplingRate is for alignment |
To generate tomogram at a binning factor of 6, run:
emClarity ctf 3d <param> emClarity ctf 3d param0.m
CTF corrected tomograms cache/<prefix>_<sub-region>_binX.rec will be generated and one can check the tomogram with 3dmod in IMOD.
Subtomogram averaging and alignment<Timing> variable, depending on subtomograms number, size and binning
<CRITICAL> Subtomogram averaging and alignment are performed iteratively using tomograms at a progressively reduced bin (e.g., from bin 6 to bin 1). The binned tomograms can enhance the SNR and help subtomogram alignment, at the cost of losing of high-resolution information. emClarity does not update alignment parameters automatically and allows the users to set the tomogram binning factor (Ali_samplingRate), angular search range and step (Raw_angleSearch) for each cycle and judge whether the refinement has converged. Each cycle starts by generating an average for each half map (step 15), which is then used as reference for alignment (step 16). For each binning, it is generally recommended to run several cycles (step 17). Similar to template search, for samples with lattice like structure, it is generally helpful to include several repetitive units (like Gag hexamers) during the averaging and alignment.
14 |emClarity does not extract the subtomograms onto disk by default, instead, the subtomograms will be extracted on the fly when needed, which can save large amounts of disk space for crowded samples.
| Parameters: | |
| subTomoMeta=gag | |
| PIXEL_SIZE=1.179e-10 | %% pixel size in meters |
| Ali_mRadius=[116,116,72] | %% in Å, enclosing 7 hexamers |
| Ali_mCenter=[0,0,0] | %% in Å |
| particleMass= 1 | %% in Megadalton |
| Ali_mType=sphere | %% sphere, cylinder, rectangle |
| particleRadius=[66,66,56] | %% corresponding to central hexamer size |
| Raw_className=0 | % class0 |
| FSC_bfactor=10 | %% b-factor applied to half maps |
| Ali_samplingRate=6 | %% binning factor |
| symmetry=C6 | %% symmetry |
Run the following command:
emClarity avg <param.m> <cycle_nb> RawAlignment emClarity avg param0.m 0 RawAlignment
This generates two half maps in the project directory: cycleXXX_< project>_class0_REF_EVE/ODD.mrc. The dimensions of maps are calculated based on Ali_mRadius with additional padding. Open these two maps in UCSF Chimera or 3dmod, or any software of your choice able to read MRC files, to check whether the maps match expectation. The corresponding (conical) Fourier shell correlation is available in FSC/cycleXXX_<project>_Raw-1-fsc_GLD.pdf, in which the dashed lines are conical FSC and the solid line is the overall FSC. The total sampling functions for both half maps cycleXXX_<project>_class0_REF_EVE/ODD_Wgt.mrc should be isotropic, if particles do not have preferred orientations in tomograms. Note that a molecular mask (FSC/cycleXXX_<project>_Raw-1-shapeMask_*mrc) is applied during FSC calculation. The overall sampling function and conical FSCs will indicate whether the subtomograms adopts preferred orientation. One can open the sampling function in 3dmod and look through the x-z plane to see whether the amplitude weight is isotropic.
15 |After the reference is generated with avg, emClarity can use this reference to align the particles. Similar to Tmp_angleSearch in template search, Raw_angleSearch in alignment step is also defined as [θout, Δout, θin, Δin]. Since most of the particles are picked correctly for the Gag dataset (step 9), the angular search ranges and step sizes for alignment are quite small.
| Parameters (other parameters are identical as avg): | |
| Raw_angleSearch=[0,0,20,5]; | %% Angular search, in degrees. |
emClarity alignRaw <param> <cycle_nb> emClarity alignRaw param0.m 0
The changes of rotation and translation for every subtomogram in each sub-region are saved in alignResume/cycleXXX_<project>/<prefix>_<sub-region>.txt. The number of lines in each file corresponds to the number of particles been aligned in the current cycle. After all the subtomograms are processed, the metadata <project>.mat will be updated.
16 |Copy param0.m to param1.m and param2.m, update Raw_angleSearch in these parameter files and repeat subtomogram averaging and alignment for a few cycles (step 14–15). For the speed of alignment, we usually alternate the in plane and out plane angular searches and perform a few cycles at each binning until the changes of rotation and shifts drop to around zero. In the same binning, one can repeat the same angular searches or gradually confine to finer angular searches. For the Gag dataset, two more cycles (cycle 1, 2) were run at bin6. Refer to supplementary information 2 for the list of commands and parameters at each cycle.
| Parameters | |
| Raw_angleSearch=[16,4,0,0]; | %% in param1.m |
| Raw_angleSearch=[0,0,9,3]; | %% in param2.m |
emClarity avg param1.m 1 RawAlignment emClarity alignRaw param1.m 1 emClarity avg param2.m 2 RawAlignment emClarity alignRaw param2.m 2
17 |Remove duplicated particles after alignment.
emClarity removeDuplicates param2.m 2
After these averaging and alignment cycles, one can run a tilt-series refinement by tomoCPR (steps 19–20, optional) and / or generate new tomograms and continue averaging and alignment (step 21).
(optional) Tilt-series refinement by tomoCPR
<Timing> variable, depending on subtomograms number, size and binning <CRITICAL> Tilt-series can be optionally refined by tomoCPR. subtomogram averaging provides accurate estimates of both particle positions and high SNR reconstructions, making them excellent fiducial markers. It is thus possible to leverage this information for improving the alignment of a tilt-series. In this protocol, we run tomoCPR for each binning.
18 | When using tomoCPR to refine the tilt-series geometry, the subtomograms are mapped back into raw tomograms to generate a synthetic tomogram containing an estimate of the background noise, plus the higher SNR particle, and projected into each view. A tile is cut out around each projected particle, convoluted with local CTF, and aligned to the corresponding particle in the raw data, to give rise the particle position in the tilt-series. These new positions of particles after local refinement will be used as new fiducial markers in tiltalign to refine the tilt-series alignment. Run the following command:
emClarity tomoCPR <param> <cycle_nb> emClarity tomoCPR param2.m 2
A temporary directory mapBack<n>/ is generated in cache/ and will be moved to project directory only after all the tilt-series are successfully processed. <n> indicates the current tomoCPR number. The overall and local transformation files will be written as mapBack<n>/<prefix>_ali<n>_ctf.tltxf and mapBack<n>/<prefix>_ali<n>_ctf.local for each tilt-series. The mapBack<n>/ directory should not be deleted since the local transformation file mapBack<n>/<prefix>_ali<n>_ctf.local will be used to generate new tomograms, although any of the image files can be deleted to save disk space. The metadata <project>.mat will be updated to record the current round of tomoCPR.
19 |Update the aligned tilt-series and geometry file. Copy param2.m to param3.m
| Parameters | |
| Ali_samplingRate=5; | %% tomogram binning |
emClarity ctf update <param> emClarity ctf update param3.m
A new geometry file fixedStacks/ctf/<prefix>_ali<n+1>_ctf.tlt and newly aligned tilt-series aliStacks/<prefix>_ali<n+1>.fixed will be created, which will be used to generate new tomograms. One can check whether the newly transformed tilt-series look well-aligned and do not deviate substantially from original aligned stacks.
20 |Generate the new tomogram at next binning (bin 5). Run the following command:
emClarity ctf 3d <param> emClarity ctf 3d param3.m
This is essentially repeating step 14 at a new binning, followed by the subtomogram averaging and alignment cycle (step 15–16), subtomogram duplicates removal (step 18) and tomoCPR (step 19–20). The cycle then continues as the binning reduces.
For the Gag dataset, we run three cycles of averaging and alignment using 6×, 5× and 4× binned subtomograms before 3D classification. Update the Ali_samplingRate and Raw_angleSearch in the parameter files at each cycle. Refer to the command-list in the Supplementary information 2.
(optional) Subtomogram Classification<Timing> ~40 min, depending on subtomograms number, size and binning
<CRITICAL> Subtomogram classification (steps 22–29) is optional in emClarity pipeline. In this protocol, we perform one cycle of 3D classification with bin 4 subtomograms after two rounds of tomoCPR and six cycles of subtomogram averaging and alignment (steps 14–21). emClarity uses PCA based classification method, with subtomograms band-pass filtered at various resolutions defined by users. It first computes an average map from all the subtomograms (step 22). emClarity will then analyse the heterogeneity of the dataset by comparing individual subtomograms with the current average map (the reference). Briefly, difference maps are calculated between each particle and the references, for each resolution bands that user defines. These maps are then analysed by Principal Component Analysis (PCA), using Singular Value Decomposition (SVD). This results in a decomposition revealing the major directions of variance (eigenimages) (step 23). Users will then select eigenimages corresponding to major direction of variance (step 24) and emClarity will project the whole dataset along each of these eigenvectors. The projected data, which is now denoised and much smaller in size, is then clustered (by default with k-means clustering algorithm, step 25). Then the class averages will be generated for each cluster as a montage (step 26) and particles from the undesired classes can be optionally removed from further analysis (step 27–28).
In principle, one can do classification at any binning and at any cycle. In practice, it is beneficial to have several rounds of alignment before classification and use an intermediate binning factor for a better SNR in tomograms (like bin4, bin3). It is generally not recommended to conduct classification at bin1 if it was already done at higher binning.
21 |Generate an average map for classification. Copy param7.m to param8.m and update flgClassify=1 to turn on classification flag in the parameter file. Besides the parameters inherited from previous alignment cycles, other parameters specific to classification includes:
| Parameters | |
| Ali_mRadius=[116,116,72] | %% in Å, enclosing 7 hexamers |
| Ali_mCenter=[0,0,0] | %% in Å |
| Ali_mType=sphere | |
| Ali_samplingRate=4 | %% binning factor for averaging |
| Raw_classes_odd=[0;1.*ones(2,1)] | %% C1 symmetry for half map 1 |
| Raw_classes_eve=[0;1.*ones(2,1)] | %% C1 symmetry for half map 2 |
| Cls_mRadius=[92,92,76] | %% classification mask radius |
| Cls_mCenter=[0,0,0] | |
| Cls_mType=sphere | %% classification mask |
| Cls_samplingRate=4 | %% binning factor for classification |
| flgClassify=1 | %% classification flag |
emClarity avg param8.m 8 RawAlignment
This will generate two half maps: cycleXXX_<project>_class0_Raw_EVE.mrc and cycleXXX_<project>_class0_Raw_ODD.mrc.
22 |Compute the difference map for each particle, with different band-pass filters. We set three band-pass filters at 10, 20 and 40 Å. The band-pass filters are selected according to the object one wishes to classify and typically below the maximum resolution of the current iteration. Most of variance is explained within the first 20 to 30 eigenimages and Pca_maxEigs is used to limit the number of eigenimages to save.
| Parameters: | |
| pcaScaleSpace=[10,20,40] | %% one can select as many band-pass filters as possible though three is typically sufficient. |
| Pca_maxEigs=25 | %% maximum number of eigenimages to save |
Run the following command:
emClarity pca <param> <cycle_nb> <subset> emClarity pca param8.m 8 0
It generates variance maps for each resolution band as cycleXXX_<project>_varianceMap25-STD-*.mrc and principle eigenimages as cycleXXX_<project>_eigenImage25-STD-*.mrc. To aid analysis, it is usually easier to look at cycleXXX_<project>_ eigenImage25-SUM-STD-mont_*.mrc, which add a common reference to the eigenimages.
23 |Select the main eigenimages by looking into each cycleXXX_<project>_ eigenImage25-SUM-STD-mont_*.mrc in 3dmod and save the eigenimages numbering into Pca_coeffs. The eigenimages are numbered from 1 to <Pca_maxEigs> counting from bottom left to top right by rows. For Gag dataset, eigenimages with hexagonal lattice feature can be selected, and eigenimages which display missing wedge effect are usually abandoned. Each resolution band requires the same number of eigenimages to be selected, which can be filled with zeros if there are not enough eigenimages in some resolution bands. Fill Pca_coeffs=[zeros(1,12);7:18;7:18] in param8.m.
24 |Cluster the PCA results according to the selected eigenimages; this step groups the subtomograms into different number of classes (Pca_clusters). Multiple classes can be generated.
| Parameters: | |
| Pca_clusters=[9 12 16] | %% different number of clusters |
emClarity cluster <param> <cycle_nb> emClarity cluster param8.m 8
This will use the Pca_coeffs and perform k-means clustering with 9, 12 and 16 target classes. The metadata will be updated and a text file <project>_cycleXXX_ClassIDX.txt listing the number of particles in each class will be generated.
25 |Generate the class averages as a 3d montage. For the Gag dataset, we generated 9 classes; the class average is numbered from 1 to <Cls_className> counting from bottom left to top right by rows (Figure 3). Set Cls_classes_odd=[1:9;1.*ones(1,9)], the first row specifying the class ID and the second row specifying the cyclic symmetry.
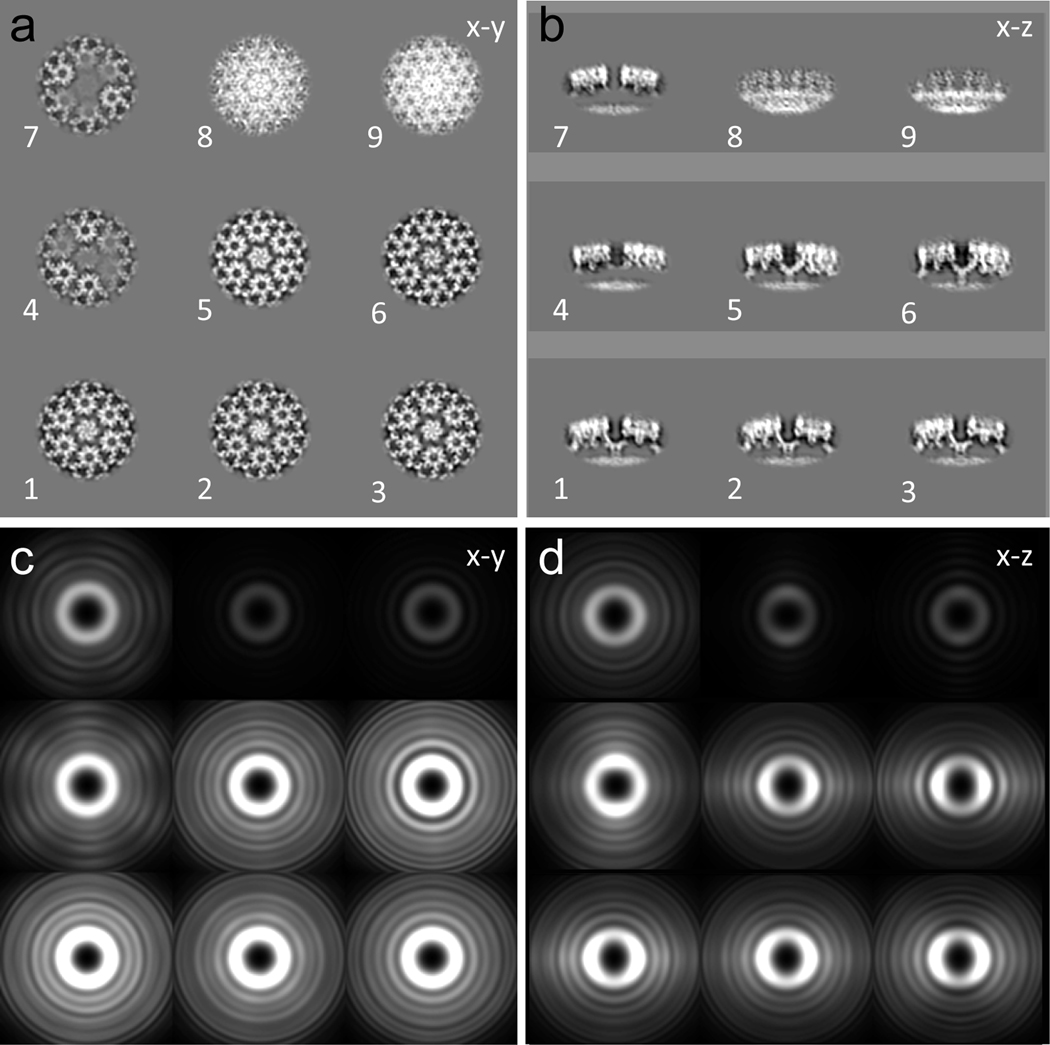
Figure 3 |. 3D classification and sampling function.
(a-b) A montage of nine 3D classes in x-y (a) and x-z slices (b). (c-d) 3D sampling functions of the corresponding classes in x-y (c) and x-z slices (d). Note 3D sampling function confirms that the classification is not biased by particle orientations, as different classes have similar sampling functions and are nearly isotropic in different orientations.
- My understanding is that I am looking at 9 different orientations of the complex – and that this means that the complex is resting on the surface at a different angles. Or does it mean that the complex has different conformations in the 9 different classes?
- I realise that I don’t understand what a “sampling function” is in this context.
| Parameters: | |
| Cls_className=9 | %% Name of classes |
| Cls_classes_odd=[1:9;1.*ones(1,9)] | %% C1 symmetry for half map 1 |
| Cls_classes_eve=[1:9;1.*ones(1,9)] | %% C1 symmetry for half map 2 symmetry=C1 |
emClarity avg <param> <cycle_nb> Cluster_cls emClarity avg param8.m 8 Cluster_cls
<Troubleshoot> (Table 3)
26 |Inspect the class averages in 3dmod or UCSF Chimera.
3dmod cycle008_gag_class9_Cls_EVE.mrc
We classified the particles into nine classes (Figure 3a–b). Seven of nine classes show clear hexagonal Gag lattice (class 1–7) and were merged together for further processing. It is generally informative to look at the sampling functions cycle008_gag_class9_Cls_EVE/ODD.Wgt to check whether the resulting classes have isotropic sampling function and proper coverage of defocus range (Figure 3). Depending on the selection of eigenimages, the missing-wedge effect may dominate the classification, resulting in stretched structures. Create a new model point for each class to remove and save the model file such as cycle008_remove.mod.
27 |Remove particles from the selected classes. STD refers to both the even and odd dataset.
emClarity geometry <param> <cycle_nb> RemoveClasses <remove.mod> STD emClarity geometry param8.m 8 Cluster_cls RemoveClasses remove_classes.mod STD
Subtomograms in these selected classes will be ignored for further analysis. The cycle008_ClassMods_STD.txt records the classes and number of subtomograms that have been removed. This should correspond exactly to the class populations from the clustering (step 27) listed in file <project>_cycleXXX_ClassIDX.txt. If it doesn’t, stop and make sure you followed the instructions from step 24.
28 | Skip the alignment for the current cycle, which prepares the metadata for the next cycle.
emClarity skip <param> <cycle_nb> emClarity skip param8.m 8
29 | Continue alignment and averaging cycles and tompCPR (optional) as in steps 15–21. Turn off the classification flag in these parameter files by setting flgClassify=0 and update the Ali_samplingRate and Raw_angleSearch for each cycle. We ran several cycles of alignment with each binned tomograms and ran tomoCPR in the end of alignment at each binning factor (bin3, bin2 and bin1). Refer to the command-list (Supplementary information 2) for the summary of all the cycles for the Gag project.
Final reconstruction<Timing> ~2.5 hrs
30 | For the final reconstruction, the two half datasets are combined. The updated versions of emClarity now offers two possibilities using either 3D subtomograms or their corresponding original 2D projections. To reconstruct through subtomograms, two half maps are reconstructed using avg as step 15 and the conical FSCs are calculated, as well as the transformation between the two maps. The subtomograms from the second group are reextracted and aligned to the first group using the aforementioned transformation. A final combined map is then generated averaging all aligned subtomograms from both halfsets and filtered using the FSC calculated, which is further sharpened with various b-factors.
| Parameters: | |
| Fsc_bfactor=[10,25,75,100,250] |
emClarity avg param19.m 19 RawAlignment emClarity avg param19.m 19 FinalAlignment
It generates the final reconstruction map cycleXXX_<project>_class0_final_<b-factor>.mrc. If one wants to use external software (like RELION44, cisTEM45, Bsoft46) to apply different b-factors, masks or FSC weighting, one can take the raw half maps in the final cycle without FSC weighting FSC/cycleXXX_<project>_Raw-*Ali.mrc.
Alternatively, the final reconstruction can also be calculated from the 2D particles using cisTEM, as implemented in the updated version of emClarity. In this case, emClarity reprojects the 3D coordinates of the particles. A cisTEM STAR file is created, containing parameters such as, for each particle and for each view of the tilt-series, its x and y position, rotation, defocus, pre- and post-exposure. cisTEM will then calculate an initial reconstruction using its reconstruct3d program, then refine it using refine3d (note that the angles are not refined) and then finally calculates the final reconstruction with reconstruct3d using this refinement. For this protocol, we set maximum exposure to 60 electrons to only include the images within this exposure and generated the final map as gag60e_refFilt_refined.mrc. The particleRadius is set to be equivalent to Ali_mRadius to reconstruct the final density map with the same area as alignment.
emClarity reconstruct <param> <cycle_nb> <prefix> <symmetry> <max_exposure> emClarity reconstruct param18recon.m 18 gag60e C6 60
TROUBLESHOOTING
Troubleshooting guidelines can be found in Table 3.
TIMING
The run time for each emClarity processing is listed in Table 2. Please note that the data processing time are for the Gag T8I dataset. The data processing time varies depending on the size of dataset, particle size, number of cycles, GPU models, etc.
Steps 1–2 Arrangement of input files and directories: ~30 min when using autoAlign
Steps 3–4 Defocus estimate: ~ 25 min
Steps 5–7 Define sub-region boundaries: ~ 10min
Steps 8–12 Pick particles: ~ 1.5 hrs
Step 13 Initialize the project: ~ 1min
Step 14 Reconstruct the tomograms for alignment and averaging: ~ 5 min, depending on the tomogram binning
Step 15–18 Subtomogram averaging and alignment: variable, depending on dataset size, particle size and binning
Step 19–21 Tilt-series refinement by tomoCPR: variable, depending on dataset size, particle size and binning, etc.
Step 22–30 Subtomogram classification: ~40 min, depending on dataset size, particle size, binning, etc.
Step 31 Final reconstruction: ~2.5 hrs, depending on dataset size, particle size and binning.
ANTICIPATED RESULTS
We illustrate the protocol using four different datasets: a wild-type Gag dataset (a subset of 5 tilt series) and a ribosome dataset (a subset of 12 tilt series) from EMPIAR (EMPIAR-10164, and EMPIAR-10304), a GagT8I assembly dataset (5 tilt series) from a previous study47 and a new apoferritin dataset (6 tilt series) collected in-house (see Table 1).
HIV-1 Gag T8I spherical assemblies
A challenging non-single particle dataset of HIV-1 Gag T8I immature spherical assemblies with overlapping densities, but no icosahedral symmetry is illustrated in detail in this protocol. These assemblies were produced in E. coli as part of a study aiming at resolving the extended 6-helix bundle of HIV-1 Gag hexamer.
The per-tilt CTF estimation of the tilt-series is consistent with expected values from experimental setting. After the template search, the convolution map reveals local peaks corresponding to each Gag hexamer. Most of the hexamers in the lattice are picked for further analysis; a small number of particles were found to be false positives (Figure 2). Subtomograms from each sub-region were assigned to the same half datasets to avoid mixing halfsets that had overlapping peripheral density (fscGoldSplitOnTomos=1). Subtomogram averaging and alignment was conducted using subtomograms binned at different factors (from 6× binned tomograms to 1× binned tomograms). After alignment is completed with each binned tomogram (except bin1), a tomoCPR tilt-series refinement was performed. Since tomoCPR is an optional step and requires tuning of some parameters, we recommend users working on a new STA project to run through iterative subtomogram averaging and alignment without tomoCPR for the first instance.
A 3D classification was performed using bin4, which gave 9 classes of images (Figure 3). The classes display different features as shown in x-y and x-z slices (Figure 3a–b), along with their corresponding overall 3d sampling functions in x-y and x-z slices (Figure 3c–d). Class 8 and 9 showed no clear Gag lattice (Figure 3a–b), therefore objects in these classes were removed from further processing. The sampling functions of the remaining classes reveal no preferential orientation, indicating that the 3D classification is not biased by the particle orientations in the raw tomogram
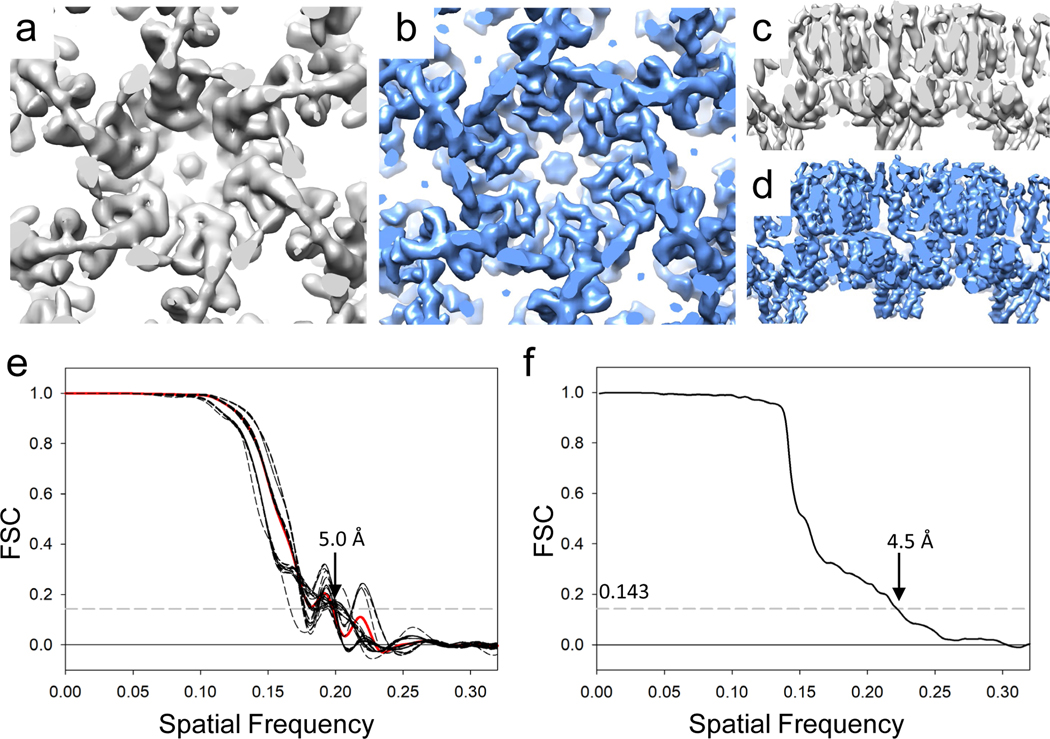
Further iterative cycles of subtomogram averaging, alignment and tomoCPR were carried out. The resulting final maps were generated using either subtomograms or 2D images with cisTEM, shown in Figure 4, along with its corresponding FSC plots. cisTEM reconstruction and refinement resulted in a higher resolution density map (4.5 Å) compared with averaging from subtomograms (5.0 Å) (Figure 4).
Figure 4 |. Subtomogram averages and conical FSC plots of HIV-1 Gag T8I assemblies (5 tilt series).
(a&c) A subtomogram averaged map of Gag T8I assemblies at 5.0 Å resolution, derived from seven classes (1–7) in Figure 3, viewed from top (a) and side (c). (b&d) Reconstruction of Gag T8I assemblies from projection images at 4.5 Å resolution using cisTEM with one cycle of additional translational refinement, viewed from top (b) and side (d). (e) Conical Fourier Shell Correlation plots of subtomogram averaged map shown in (a). The solid red curve represents the global FSC, while each dashed curve represents the FSC of a cone of 36° of half angle, with a 30° increment between each cone. (f) FSC plot of cisTEM reconstructed map shown in (b).
- I don’t know what a Fourier Shell Correlation plot is; and I don’t know what “spatial frequency” means.
- I don’t know what is special about 0.143
- I can see that there might be different information in the subtomogram and the cisTEM methods. But it is difficult to work what is important.
Wild-type Gag
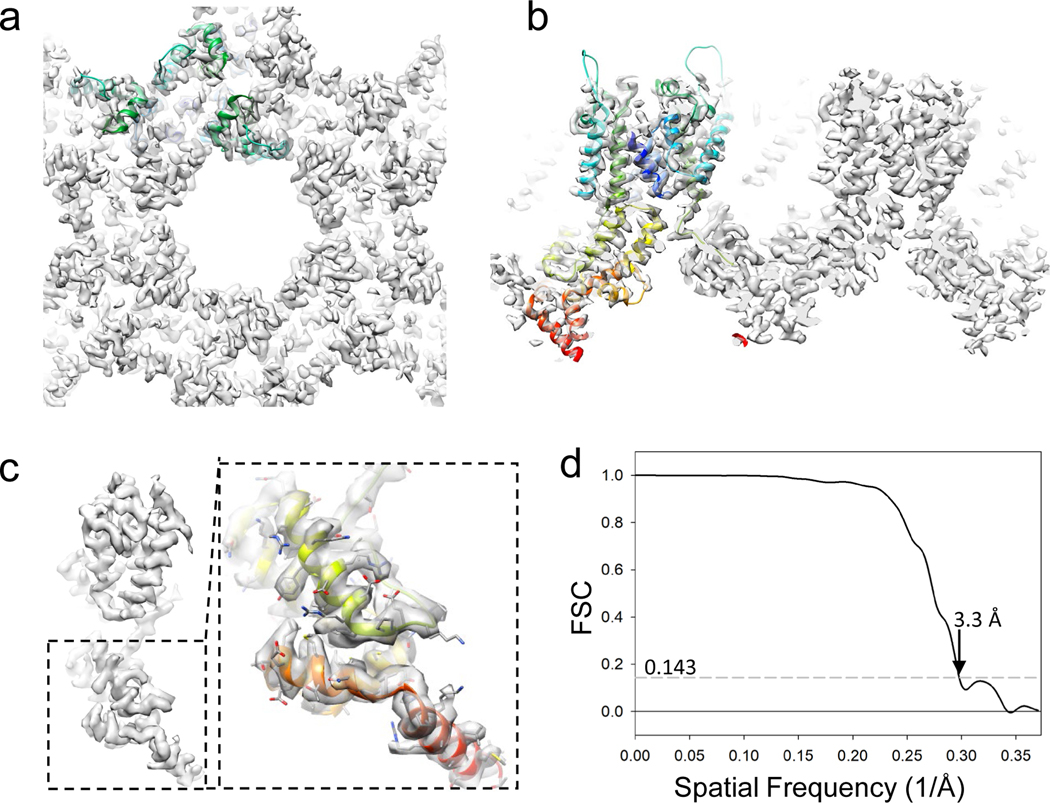
We also reprocessed a published 5 tilt-series of wild-type Gag (EMPIAR-10164, TS_001, 003, 043, 045 and 054), which yielded a subtomogram averaged map at 3.9 Å resolution previously19. The alignment procedure for this dataset is similar to that used for the Gag T8I dataset above, but does not include classification (Table 1 and Supplementary information 2). Given that the pixel size (1.35Å) is slightly larger in this dataset, the iterative alignment step used in emClarity starts from bin4 tomograms and 3 rounds of tomoCPR were conducted at bin4, bin3 and bin2, respectively. The same alignment mask size Ali_mRadius=[116,116,72] encompassing 7 hexamers as in the HIV-1 Gag T8I processing was used in the initial averaging/alignment steps. The size was changed to [88,88,72] in the last few iterations at bin1 to further improve the resolution. A final 6-fold symmetrized map at a resolution of 3.3 Å were obtained, revealing clear side chains of Gag domains (Figure 5).
Figure 5 |. Subtomogram averaging of WT Gag (5 tilt series, EMPIAR-10164).
(a-b) A subtomogram averaged map of Gag at 3.3 Å resolution. Top (a) and side (b) sectional views are shown. One asymmetric unit containing three Gag polypetides are fitted with a Gag structure model (PDB:5l93), colored in rainbow (blue to red) from N-terminal to C-terminal of each polypeptide. (c) A monomer extracted from the density map, with a close-up view of CTD region overlay with the atomic model (PDB:5l93). (d) Fourier Shell Correlation plot of Gag subtomogram averaged map.
Single ribosome particles
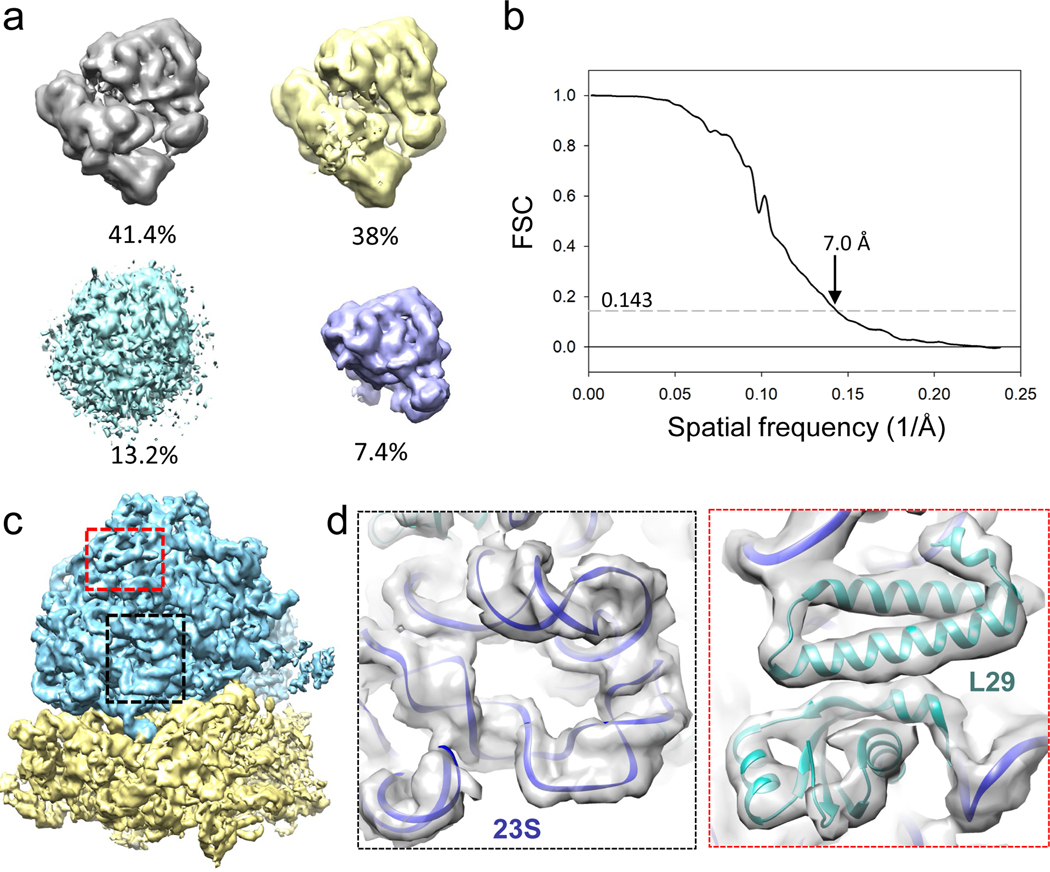
The emClarity processing of ribosome dataset of isolated single particles (EMPIAR-10304) is included in the software Tutorial (https://github.com/ffyr2w/emClarity-tutorial) along with emClarity installation. The tilt-series were aligned with emClarity autoAlign function and particles were picked through template search with bin6 tomograms. Subtomograms within the same sub-region were split into two random halves since there is no overlap among them (fscGoldSplitOnTomos=0). The alignment and averaging were iteratively performed from bin5 to bin1 with one round of tomoCPR before transition to lower binning. The classification was performed at bin3 to remove junk particles (Figure 6). Four resolution bands were used for 3D classification (pcaScaleSpace=[25,50,80,120]) and several different number of classes were tried (2, 3, 4, 6, 8, 14, 18), all of which resulted in classes with junk particles (~13.2%) and a small class (7.4%) containing only the large subunits (Figure 6a). The final reconstruction and refinement with cisTEM resulted a 7.0 Å resolution map, showing clear secondary structure elements like the RNA groves and α-helices (Figure 6 b–d).
Figure 6 |. Subtomogram classification and averaging of ribosome (12 tilt series, EMPIAR-10304).
(a) 3D classification of ribosome revealing 4 classes, with two major classes having both 50S and 30S subunits (grey and yellow), one class with junk particles (cyan) and one with 50S only (light blue). (b) Fourier Shell Correlation (FSC) of final map reconstructed from two major classes. (c) Density map of ribosome with 50S subunit colored in blue and 30S subunit in yellow. (d) Zoom-in view of two local regions (dashed black and red boxed) with model fitted in (PDB code: 5mdz), as boxed in c.
Apoferritin
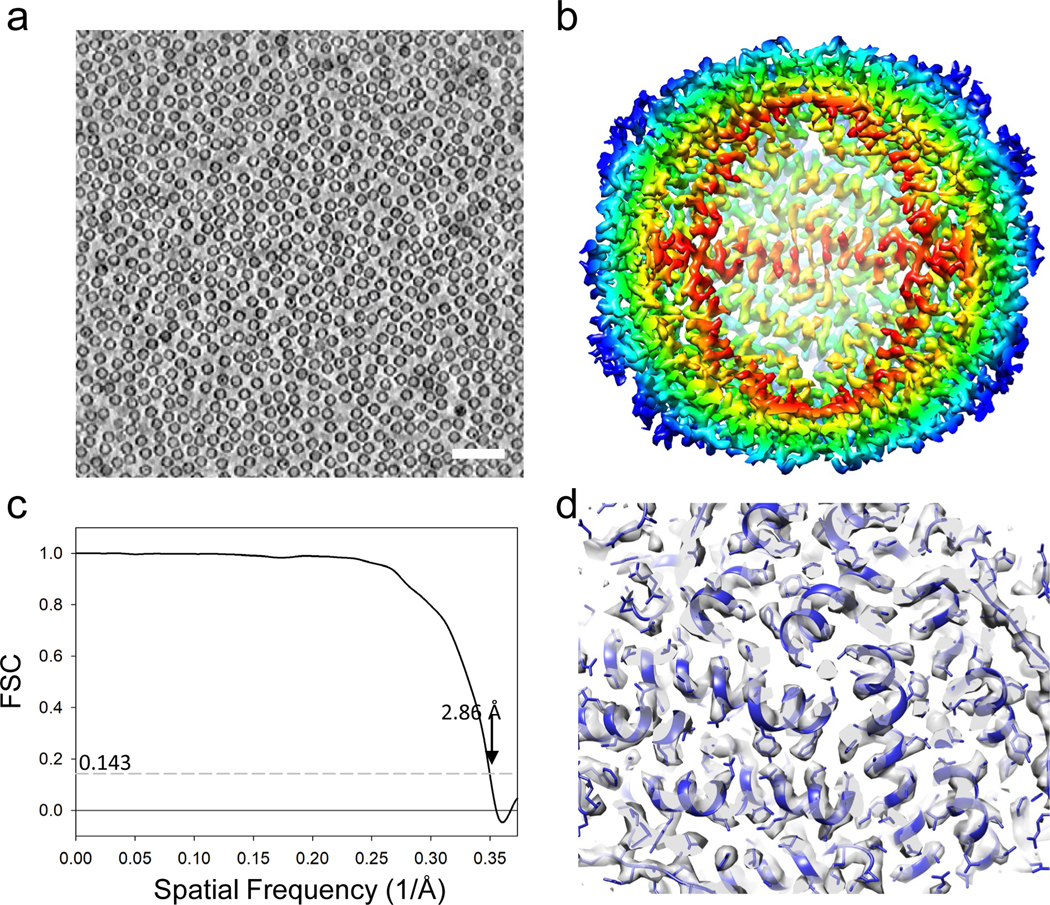
The final example is the apoferritin cryoEM sample, which was prepared using a graphene-coated EM grid, yielding a mono-dispersed thin layer of apoferritin (Figure 7a). Tilt-series were collected using the parameters shown in Table 1 and the emClarity commands are included in the Supplementary information 2. Six tilt-series were aligned with Etomo by patch tracking (no fiducial gold beads) and imported into emClarity. Octahedral symmetry was applied throughout alignment. The final subtomogram averaging map were obtained from less than 5000 subtomograms, with a 2.86 Å resolution, approaching the Nyquist frequency (2.68 Å) (Figure 7b–d).
Figure 7 |. Subtomogram averaging of apoferritin (6 tilt series).
(a) A tomographic slice of apoferritin on graphene grids. Scale bar: 50 nm. (b) Subtomogram averaged map of apoferritin at 2.86 Å resolution. The density map is coloured radially 40 Å (red) to 60 Å (blue) from the centre. (c) FSC plot of the averaged map. The resolution is 2.86 Å with an FSC cut-off of 0.143, approaching the Nyquist frequency which is the edge of the plot. (d) Representative density maps (fitted with PDB model 6s61).
Data availability
The Gag dataset (5 tilt-series) and apoferritin dataset (6 tilt-series) have been deposited in EMPIAR database under accession codes EMPIAR-10643 and EMPIAR-10787, respectively. The resulting final reconstructions have been deposited in EMDB under the following accession codes: Gag-T8I, EMD-13390; Gag-WT, EMD-13354; apoferritin, EMD-13271; and ribosome, EMD-13270.
Code availability
The software emClarity is freely available from https://github.com/bHimes/emClarity/wiki. The tutorial documentation is available at https://github.com/ffyr2w/emClarity-tutorial.
Supplementary Material
Supplementary information 1 | Supplementary emClarity tutorial. The tutorial provides in-depth understanding algorithms behind each step, as well as detailed step-by-step processes using a ribosome dataset, including explanation for each command and parameter.
Supplementary information 2 | Supplementary run command lists and parameter file. The commands are listed in sequence as they run, as well as associated parameter file and specific parameters chosen for each dataset.
- Would you please comment briefly about the list of comments at the end of the tables?
Table 1 |.
CryoET data collection and processing details
| Titan Krios† | GagT8I | Gag WT (EMPIAR-10164) | Ribosome (EMPIAR-10304) | Apoferritin |
|---|---|---|---|---|
|
| ||||
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Detector | Falcon 4 | Gatan K2 | Gatan K3 | Gatan K3 |
| Energy filter | Selectris X, 10 eV slit | Gatan bioquantum, 20 eV | Gatan bioquantum, 20 eV | Gatan bioquantum, 20 eV |
| Super-resolution mode | Yes | Yes | Yes | Yes |
| Pixel size (Å) | 1.18 | 1.35 | 2.1 | 1.34 |
| Total electron dose (e-/Å2) | 122 | ~120 | ~120 | 102 |
| Dose rate (e-/Å2/s ) | 3 | 3 | 4.2 | |
| Frame number | 10 | 10 | 10 | |
| Acquisition scheme | −60/60°, 3° | −60/60°, 3° | −60/60°, 3° | −60/60°, 3° |
| Defocus range (μm) | −1.36 ~ −3.11 | −1.5 ~ −3.96 | −2.2 ~ −4.3 | −1.5 ~ −3.5 |
| Number of tilt-series | 5 | 5 | 12 | 6 |
| Software | IMOD, emClarity | IMOD, emClarity | IMOD, emClarity | IMOD, emClarity |
| Number of tomograms | 5 | 5 | 12 | 6 |
| Number of initial subtomograms | 20010 | 15791 | 10441 | 5668 |
| Number of subtomograms after classification | 13844 | 15460 | 8131 | 4826 |
| Symmetry imposed | C6 | C6 | C1 | O |
| Resolution at 0.143 FSC | 5.0 Å/4.5 Å* | 3.3 | 7.0 | 2.8 |
| Data deposited | EMPIAR-10643 EMD-13390 | EMD-13354 | EMD-13270 | EMPIAR-10787 EMD-13271 |
Titan Krios is a 300kV electron microscope used for cryoEM data collection: https://www.thermofisher.com/uk/en/home/electron-microscopy/products/transmission-electronmicroscopes/krios-g4-cryo-tem.html
Density maps were calculated using subtomograms averaged in emClarity (5.0 Å) or using projection images (4.5 Å) reconstructed by cisTEM implemented in emClarity.
Table 2 |.
emClarity processing run time (5 tilt-series).
| emClarity processing steps | Binning | GPU card | # GPU units | Time |
|---|---|---|---|---|
|
| ||||
| ctf estimate | 1 | Tesla V100 | 1 | 25 m |
| template search | 8 | Tesla V100 | 1 | 1 h |
| init cycle 0–2 | 6 | Tesla V100 | 4 | 40 m |
| tomoCPR-1 cycle 3–5 | 5 | Tesla V100 | 4 | 2.5 h |
| tomoCPR-2 cycle 6–8 | 4 | Tesla V100 | 4 | 3 h |
| Classification | 4 | Tesla V100 | 4 | 40 m |
| cycle 9–10 | 4 | Tesla V100 | 4 | 1.5 h |
| tomoCPR-3 cycle 11–13 | 3 | Tesla V100 | 4 | 2 h |
| tomoCPR-4 cycle 14–16 | 2 | Tesla V100 | 4 | 3 h |
| tomoCPR-5 cycle 17–18 | 1 | Tesla V100 | 4 | 10 h |
| avg FinalAlignment | 1 | Tesla V100 | 4 | 1 h |
| cisTEM reconstruct/refine | 1 | Tesla V100 (cpu) | 12 cpu cores | 1.5 h |
Note: Each tomogram is divided into multiple sub-regions (one VLP/sub-region), which are processed in parallel.
Acknowledgements
We are grateful to Dr. Yanan Zhu for discussion and critical reading of manuscript. We acknowledge Diamond for access and support of the CryoEM facilities at the UK national electron bio-imaging centre (eBIC, proposal CM26464), funded by the Wellcome Trust, MRC and BBSRC. The computational aspects of this research were supported by the Wellcome Trust Core Award Grant Number 203141/Z/16/Z and the NIHR Oxford BRC. This work was supported by the National Institutes of Health grants AI150481, the UK Wellcome Trust Investigator Award 206422/Z/17/Z, the UK Biotechnology and Biological Sciences Research Council grant BB/S003339/1, and the ERC AdG grant 101021133.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Zhang P.Advances in cryo-electron tomography and subtomogram averaging and classification. Curr Opin Struct Biol 58, 249–258, doi: 10.1016/j.sbi.2019.05.021 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan M.et al. In Situ Imaging and Structure Determination of Biomolecular Complexes Using Electron Cryo-Tomography. Methods Mol Biol 2215, 83–111, doi: 10.1007/978-1-0716-0966-8_4 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Turk M.& Baumeister W.The promise and the challenges of cryo-electron tomography. FEBS Lett 594, 3243–3261, doi: 10.1002/1873-3468.13948 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Mahamid J.et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972, doi: 10.1126/science.aad8857 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Forster F.& Hegerl R.Structure determination in situ by averaging of tomograms. Methods Cell Biol 79, 741–767, doi: 10.1016/S0091-679X(06)79029-X (2007). [DOI] [PubMed] [Google Scholar]
- 6.Bykov YS et al. The structure of the COPI coat determined within the cell. Elife 6, doi: 10.7554/eLife.32493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y.et al. Molecular architecture of the luminal ring of the Xenopus laevis nuclear pore complex. Cell Res 30, 532–540, doi: 10.1038/s41422-020-0320-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeffer S.et al. Structure of the native Sec61 protein-conducting channel. Nat Commun 6, 8403, doi: 10.1038/ncomms9403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassidy CK et al. CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling. Elife 4, doi: 10.7554/eLife.08419 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodonova SO, Prinz S, Bilanchone V, Sandmeyer S.& Briggs JAG Structure of the Ty3/Gypsy retrotransposon capsid and the evolution of retroviruses. Proc Natl Acad Sci U S A 116, 10048–10057, doi: 10.1073/pnas.1900931116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattei S, Glass B, Hagen WJ, Krausslich HG & Briggs JA The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 354, 1434–1437, doi: 10.1126/science.aah4972 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Dick RA et al. Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly. PLoS Pathog 16, e1008277, doi: 10.1371/journal.ppat.1008277 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schur FK et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 353, 506–508, doi: 10.1126/science.aaf9620 (2016). https://www.ebi.ac.uk/empiar/EMPIAR-10164/ [DOI] [PubMed] [Google Scholar]
- 14.Qu K.et al. Structure and architecture of immature and mature murine leukemia virus capsids. Proc Natl Acad Sci U S A 115, E11751-E11760, doi: 10.1073/pnas.1811580115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Kugelgen A.et al. In Situ Structure of an Intact Lipopolysaccharide-Bound Bacterial Surface Layer. Cell 180, 348–358 e315, doi: 10.1016/j.cell.2019.12.006 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tegunov D, Xue L, Dienemann C, Cramer P.& Mahamid J.Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5 A in cells. Nat Methods 18, 186–193, doi: 10.1038/s41592-020-01054-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucic V, Rigort A.& Baumeister W.Cryo-electron tomography: the challenge of doing structural biology in situ. J Cell Biol 202, 407–419, doi: 10.1083/jcb.201304193 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan W.& Briggs JA Cryo-Electron Tomography and Subtomogram Averaging. Methods Enzymol 579, 329–367, doi: 10.1016/bs.mie.2016.04.014 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Turonova B, Schur FKM, Wan W.& Briggs JAG Efficient 3D-CTF correction for cryo-electron tomography using NovaCTF improves subtomogram averaging resolution to 3.4A. J Struct Biol 199, 187–195, doi: 10.1016/j.jsb.2017.07.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heumann JM, Hoenger A.& Mastronarde DN Clustering and variance maps for cryoelectron tomography using wedge-masked differences. J Struct Biol 175, 288–299, doi: 10.1016/j.jsb.2011.05.011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicastro D.et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944–948, doi: 10.1126/science.1128618 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Chen M.et al. Convolutional neural networks for automated annotation of cellular cryoelectron tomograms. Nat Methods 14, 983–985, doi: 10.1038/nmeth.4405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galaz-Montoya JG, Flanagan J, Schmid MF & Ludtke SJ Single particle tomography in EMAN2. J Struct Biol 190, 279–290, doi: 10.1016/j.jsb.2015.04.016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galaz-Montoya JG et al. Alignment algorithms and per-particle CTF correction for single particle cryo-electron tomography. J Struct Biol 194, 383–394, doi: 10.1016/j.jsb.2016.03.018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bharat TA & Scheres SH Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat Protoc 11, 2054–2065, doi: 10.1038/nprot.2016.124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bharat TAM, Russo CJ, Lowe J, Passmore LA & Scheres SHW Advances in Single-Particle Electron Cryomicroscopy Structure Determination applied to Sub-tomogram Averaging. Structure 23, 1743–1753, doi: 10.1016/j.str.2015.06.026 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castano-Diez D, Kudryashev M, Arheit M.& Stahlberg H.Dynamo: a flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J Struct Biol 178, 139–151, doi: 10.1016/j.jsb.2011.12.017 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Maurer UE et al. The structure of herpesvirus fusion glycoprotein B-bilayer complex reveals the protein-membrane and lateral protein-protein interaction. Structure 21, 1396–1405, doi: 10.1016/j.str.2013.05.018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forster F, Pruggnaller S, Seybert A.& Frangakis AS Classification of cryo-electron sub-tomograms using constrained correlation. J Struct Biol 161, 276–286, doi: 10.1016/j.jsb.2007.07.006 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Hrabe T.et al. PyTom: a python-based toolbox for localization of macromolecules in cryo-electron tomograms and subtomogram analysis. J Struct Biol 178, 177–188, doi: 10.1016/j.jsb.2011.12.003 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Winkler H.3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol 157, 126–137, doi: 10.1016/j.jsb.2006.07.014 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Himes BA & Zhang P.emClarity: software for high-resolution cryo-electron tomography and subtomogram averaging. Nat Methods 15, 955–961, doi: 10.1038/s41592-018-0167-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C.et al. The Architecture of Inactivated SARS-CoV-2 with Postfusion Spikes Revealed by Cryo-EM and Cryo-ET. Structure 28, 1218–1224 e1214, doi: 10.1016/j.str.2020.10.001 (2020). https://www.sciencedirect.com/science/article/pii/S0969212620303725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe R.et al. The In Situ Structure of Parkinson’s Disease-Linked LRRK2. Cell 182, 1508–1518 e1516, doi: 10.1016/j.cell.2020.08.004 (2020). https://www.sciencedirect.com/science/article/pii/S0092867420309958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutton G.et al. Assembly intermediates of orthoreovirus captured in the cell. Nat Commun 11, 4445, doi: 10.1038/s41467-020-18243-9 (2020). https://www.nature.com/articles/s41467-020-18243-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan TY et al. Capsid protein structure in Zika virus reveals the flavivirus assembly process. Nat Commun 11, 895, doi: 10.1038/s41467-020-14647-9 (2020). https://www.nature.com/articles/s41467-020-14647-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Unchwaniwala N.et al. Subdomain cryo-EM structure of nodaviral replication protein A crown complex provides mechanistic insights into RNA genome replication. Proc Natl Acad Sci U S A 117, 18680–18691, doi: 10.1073/pnas.2006165117 (2020). https://www.pnas.org/content/117/31/18680.short [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson KH et al. An asymmetric sheath controls flagellar supercoiling and motility in the leptospira spirochete. Elife 9, doi: 10.7554/eLife.53672 (2020). https://elifesciences.org/articles/53672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cassidy CK et al. Structure and dynamics of the E. coli chemotaxis core signaling complex by cryo-electron tomography and molecular simulations. Commun Biol 3, 24, doi: 10.1038/s42003-019-0748-0 (2020). https://www.nature.com/articles/s42003-019-0748-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohou A.& Grigorieff N.CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221, doi: 10.1016/j.jsb.2015.08.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheres SH & Chen S.Prevention of overfitting in cryo-EM structure determination. Nat Methods 9, 853–854, doi: 10.1038/nmeth.2115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenthal PB & Henderson R.Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333, 721–745, doi: 10.1016/j.jmb.2003.07.013 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Mastronarde DN & Held SR Automated tilt series alignment and tomographic reconstruction in IMOD. J Struct Biol 197, 102–113, doi: 10.1016/j.jsb.2016.07.011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530, doi: 10.1016/j.jsb.2012.09.006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant T, Rohou A.& Grigorieff N.cisTEM, user-friendly software for single-particle image processing. Elife 7, doi: 10.7554/eLife.35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heymann JB Guidelines for using Bsoft for high resolution reconstruction and validation of biomolecular structures from electron micrographs. Protein Sci 27, 159–171, doi: 10.1002/pro.3293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mendonca L.et al. CryoET structures of immature HIV Gag reveal six-helix bundle. Commun Biol 4, 481, doi: 10.1038/s42003-021-01999-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information 1 | Supplementary emClarity tutorial. The tutorial provides in-depth understanding algorithms behind each step, as well as detailed step-by-step processes using a ribosome dataset, including explanation for each command and parameter.
Supplementary information 2 | Supplementary run command lists and parameter file. The commands are listed in sequence as they run, as well as associated parameter file and specific parameters chosen for each dataset.
- Would you please comment briefly about the list of comments at the end of the tables?
Data Availability Statement
The Gag dataset (5 tilt-series) and apoferritin dataset (6 tilt-series) have been deposited in EMPIAR database under accession codes EMPIAR-10643 and EMPIAR-10787, respectively. The resulting final reconstructions have been deposited in EMDB under the following accession codes: Gag-T8I, EMD-13390; Gag-WT, EMD-13354; apoferritin, EMD-13271; and ribosome, EMD-13270.