Abstract
Background
To assess the prevalence and evolution of transmitted drug resistance (TDR) in Belgium, a total of 3708 baseline human immunodeficiency virus (HIV)-1 polymerase sequences from patients diagnosed between 2013 and 2019 were analyzed.
Methods
Protease and reverse-transcriptase HIV-1 sequences were collected from the 7 national Aids Reference Laboratories. Subtype determination and drug resistance scoring were performed using the Stanford HIV Drug Resistance Database. Trends over time were assessed using linear regression, and the maximum likelihood approach was used for phylogenetic analysis.
Results
A total of 17.9% of the patients showed evidence of TDR resulting in at least low-level resistance to 1 drug (Stanford score ≥15). If only the high-level mutations (Stanford score ≥60) were considered, TDR prevalence dropped to 6.3%. The majority of observed resistance mutations impacted the sensitivity for nonnucleoside reverse-transcriptase inhibitors (NNRTIs) (11.4%), followed by nucleoside reverse-transcriptase inhibitors (6.2%) and protease inhibitors (2.4%). Multiclass resistance was observed in 2.4%. Clustered onward transmission was evidenced for 257 of 635 patients (40.5%), spread over 25 phylogenetic clusters.
Conclusions
The TDR prevalence remained stable between 2013 and 2019 and is comparable to the prevalence in other Western European countries. The high frequency of NNRTI mutations requires special attention and follow-up. Phylogenetic analysis provided evidence for local clustered onward transmission of some frequently detected mutations.
Keywords: HIV resistance, HIV transmission, prevalence, TDR
Thanks to the availability of antiretroviral drugs (ART) from different drug classes, human immunodeficiency virus (HIV) has evolved from a life-threatening disease to a chronic infection [1]. As of 2019, 6 classes of ART are available for clinical use: nucleoside reverse-transcriptase inhibitors (NRTIs), nonnucleoside reverse-transcriptase inhibitors (NNRTIs), protease inhibitors (PIs), integrase strand transfer inhibitors (INSTIs), entry inhibitors, and postattachment inhibitors. To guide the choice of the first-line ART, sequencing of the reverse-transcriptase (RT), protease (PR), and integrase genes of the virus at diagnosis is now standard of care in most high-income countries [2, 3]. Human immunodeficiency virus drug resistance may emerge when suppression of HIV replication is incomplete and the virus is allowed to replicate in the presence of suboptimal drug concentrations. Onward transmission of acquired resistant variants will lead to presence of transmitted drug resistance (TDR) in treatment-naive individuals [4, 5]. Transmitted drug resistance was initially observed in high-income countries, and overall TDR levels in these regions remain amongst the highest of the world (>10%) [3, 6]. Recently, however, increasing TDR levels are reported for low- and middle-income countries [6–8], while a steady-state level or a decrease is reported for Europe [6–10]. Depending on the patients selected, opposing trends in TDR prevalence have been reported for North America [6, 11, 12]. Subtype B infection and Asian race were associated with increased TDR, whereas African American race was associated with a reduced TDR risk [12].
In many studies, NNRTI mutations are the most frequently detected TDR, questioning the use of NNRTIs in first-line regimens [6, 13]. In addition, in general, high figures are reported for NRTI mutations while the number of PI mutations remains low. Because routine examination of the integrase gene was introduced only recently, limited information is available about the evolution of TDR to this class of drugs, but it appears that baseline INSTI resistance overall remains very low [14]. A retrospective study to assess the presence of INSTI mutations in 313 patients diagnosed in Belgium between 2010 and 2016 and initiating an INSTI-based first-line regimen showed that 7 (2.2%) harbored a single INSTI mutation with expected low-level impact on INSTI susceptibility: T66I (1 patient), E138K (2 patients), S147G (2 patients), G163K (1 patient), and S230R (1 patient) (unpublished data, EV 2018).
Transmitted drug resistance may result from transmission from an HIV-positive person experiencing therapy failure. If the mutant strain is fit enough to persist in the absence of drugs, however, ongoing transmission between treatment-naive individuals can take place [15, 16]. There is growing evidence that this onward transmission of resistant variants accounts for a large part of the observed TDR [17, 18]. In Belgium for instance, the importance of ongoing transmission on the spread of the K103N mutation has been documented [16].
In this multilaboratory retrospective study, we analyzed the prevalence and evolution of TDR in a joint collaboration among the 7 Belgian Aids Reference laboratories, covering all patients diagnosed in Belgium. All ART-naive individuals newly diagnosed between 2013 and 2019 for whom a baseline sequence was available were included (n = 3708). Partial HIV-1 pol sequences covering the complete protease (amino acids 9 to 99) and the first part of the reverse transcriptase (amino acids 41 to 239) gene were collected. To assess presence of INSTI mutations, the whole integrase gene (amino acids 1 to 288) was sequenced, but this analysis was limited to the patients diagnosed in 2019 (n = 533) because routine monitoring of baseline integrase resistance was only introduced then. Selection of resistance mutations was performed using the Stanford HIV Drug Resistance Database (HIVdb algorithm, version 8.9-1). Phylogenetic cluster analysis was conducted to gain more insight on the origin of the TDR and to look for indications of local transmission.
METHODS
Ethics Statement
Ethics approval was obtained from the ethical committee of Ghent University Hospital (EC Ref. BC11065). All data were collected as part of routine clinical testing. Pseudonymized identifiers were used to assemble the sequences and remove duplicates. Full anonymization was then implemented before initiating the resistance and phylogenetic analyses.
Resistance Analysis
All baseline sequences generated in 1 of the 7 Belgian Reference Laboratories between 2013 and 2019 from newly diagnosed, treatment-naive patients were included (GenBank accession numbers OL674561–OL675195). The sequences covered part of the HIV-1 pol gene (amino acids 9 to 99 of the protease and amino acids 41 to 239 of the reverse transcriptase). Sequences were generated from a blood plasma sample collected as soon as possible after diagnosis using either an in-house method, the ViroSeq HIV-1 Genotyping System (Abbott Molecular, Des Plaines, IL), the TRUGENE HIV-1 Genotyping Assay (Siemens Diagnostics, Tarrytown, NY), or the Sentosa SQ HIV-1 genotyping assay (Vela Diagnostics, Hamburg, Germany).
Subtype determination and drug resistance scoring were performed using the Stanford HIV Drug Resistance Database (HIVdb algorithm version 8.9-1) [19, 20]. Stanford defines 5 drug resistance levels: scores <10 indicate susceptible, scores between 10 and 14 potential low-level resistance, scores between 15 and 29 low-level resistance, scores between 30 and 59 intermediate resistance, and scores of ≥60 high-level resistance. In our analysis, TDR was defined as the presence of 1 or more mutations with a score of at least 15 for at least 1 drug. To analyze only mutations with a high impact on drug resistance, a score of ≥60 was applied.
In addition, the prevalence of resistant mutations was assessed using the HIV mutation lists provided by the World Health Organization (WHO) [21] and the International Antiviral Society–USA (IAS-USA) [22].
Phylogenetic Analysis
For phylogenetic analysis, all sequences with TDR were selected and aligned. From the 664 sequences selected, 29 were removed because they missed a small part of the 864 nucleotides fragment covered by the majority of the sequences. To avoid bias, in addition, the nucleotides at positions involved in resistance were removed, leading to final fragments of 756 nucleotides long. Alignments were composed in BioEdit [23]. The maximum likelihood approach implemented in PhyML 3.0 with automatic selection of the best-fit, evolutionary model of deoxyribonucleic acid substitution was used for phylogenetic analysis. For cluster identification, a bootstrap value of ≥0.97 was applied. Only clusters of 3 or more patients were considered. Pairs (clusters of 2 patients) were analyzed separately.
Statistical Analysis
We used generalized binomial logistic regression models to assess the relationship between sample year and TDR and calculated the odds ratio for yearly increases in TDR prevalence. The χ2 tests were used for subtype analysis. The level of significance was set at P ≤ .05. All data were analyzed using SPSS Statistics version 26 (SPSS Inc., Chicago, IL).
RESULTS
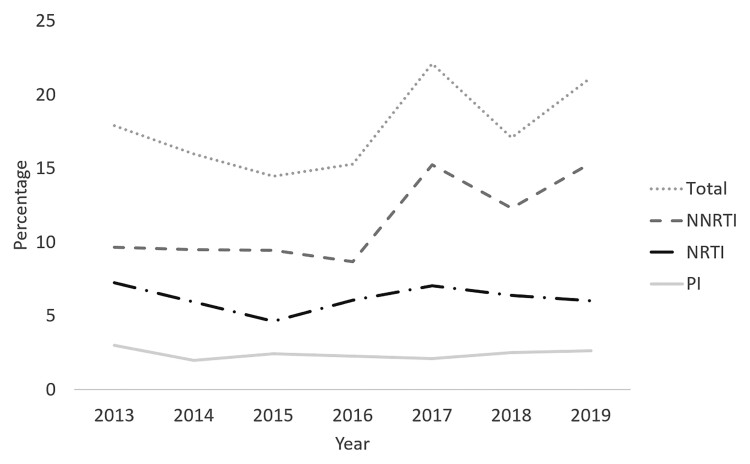
Of the 3708 patients included, 2801 (75.5%) were men. A subtype B virus was present in 1762 (47.5%). The overall prevalence of TDR (Stanford score ≥15) was 17.9% (664 of 3708 patients). Minor fluctuations in yearly TDR prevalence were observed (odds ratio, 1.04 per year; 95% confidence interval, 1.00–1.08; P = .034) (Figure 1). The majority of detected resistance mutations impacted susceptibility for NNRTI (423 patients; 11.4%), followed by NRTI mutations (230 patients; 6.2%) and PI mutations (89 patients; 2.4%) (Figure 1). Multiclass resistance was observed in 88 patients (2.4%). Three of 533 patients (0.56%) analyzed for INSTI resistance had a single INSTI resistance mutation.
Figure 1.
Evolution of transmitted drug resistance prevalence between 2013 and 2019 when considering all patients with a Stanford score of at least 15 for at least 1 of the nucleoside reverse-transcriptase inhibitors (NRTI), nonnucleoside reverse-transcriptase inhibitors (NNRTI), and/or protease inhibitors (PI). Patients with more than 1 resistance mutation were counted only once in the overall calculations.
For comparative purposes, the overall prevalence of NNRTI, NRTI, and PI TDR was also defined using the WHO and the IAS-USA lists of TDR mutations. This resulted in a respective TDR prevalence of 10.9% and 19.0%. When only the IAS-USA list of mutations labeled as “major” were considered, the prevalence was 13.8%. When only the mutations with a Stanford score of ≥60 were considered, the prevalence dropped to 6.3%. Supplementary Table 1 gives an overview of the mutations taken into consideration by the different algorithms.
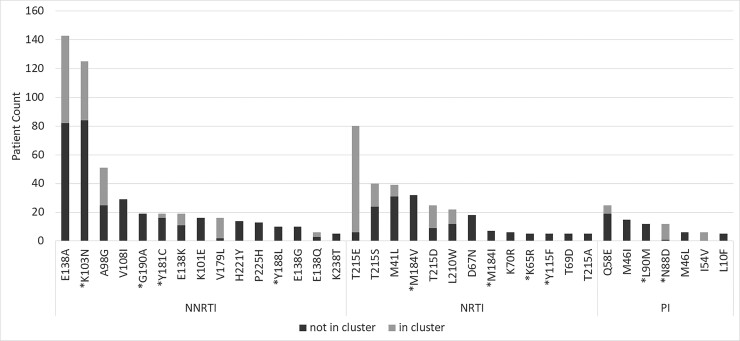
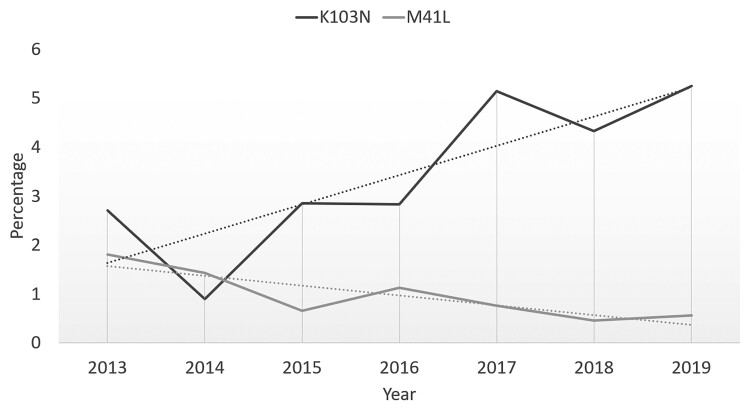
Of the mutations with a Stanford score ≥60, the most frequently observed (≥5 patients; ≥0.13%) were the NNRTI mutations K103N (123 patients; 3.37%), Y181C (18 patients; 0.51%), G190A (18 patients; 0.51%), and Y188L (10 patients; 0.27%) (Figure 2). The NRTI mutation M184V was detected in 32 patients (0.86%), followed by M184I (7 patients; 0.19%), K65R (5 patients; 0.13%), and Y115F (5 patients; 0.13%). The most prevalent PI mutations were N88D (12 patients; 0.32%) and L90M (12 patients; 0.32%). A trend over time was observed for 2 mutations, with a significant decrease in prevalence of M41L (P = .022) and a slight but not significant increase in the prevalence of K103N (P = .082) (Figure 3).
Figure 2.
Overview of the most frequently observed mutations (cut off prevalence: ≥5 times; ≥0.13%). The number of patients with transmitted drug resistance that are part of a transmission cluster is indicated in light gray. Mutations with a Stanford score of ≥60 are marked with a star. Abbreviations: NNRTI, nonnucleoside reverse-transcriptase inhibitors; NRTI, nucleoside reverse-transcriptase inhibitors; PI, protease inhibitors.
Figure 3.
Evolution of the prevalence of M41L and K103N.
Phylogenetic analysis of sequences with at least 1 resistant mutation revealed the presence of 25 clusters with an average cluster size of 10 (minimum–maximum: 3-64 patients) (Table 1; Supplementary Figure 1). These 25 clusters comprised 257 (40.5%) of all patients with TDR. In 18 clusters, all members had the same drug-resistant mutation or combination of mutations. The mutations most prevalent in clusters were K103N (41 patients; 5 clusters), E138A (45 patients; 8 clusters), E138K (8 patients; 1 cluster), and 215rev (T215D, T215E, T215L, or T215S; 113 patients; 9 clusters) (Table 1). There were 6 clusters with transmission of a variant combining at least 2 mutations including either the polymorphic mutation 138A or Q or a revertant amino acid at position 215. In the 7 clusters in which not all members had the same mutational pattern (clusters 19 to 25) (Table 1), the differences were mainly due to inconsistent presence of 215 revertants. A clear example thereof is a large cluster of 12 patients (cluster 22) with both PR and RT resistance but with small differences in mutation pattern.
Table 1.
Overview of Clusters Based on Phylogenetic Analysis of Viral Sequences Harboring at Least 1 Transmitted Drug Resistancea
| Cluster Number | Mutations | No. of Patients With Mutation | Cluster Size | Subtype |
|---|---|---|---|---|
| 1 | NNRTI_K103N | 26 | 26 | B |
| 2 | NNRTI_E138A | 12 | 12 | B |
| 3 | NNRTI_E138A | 10 | 10 | B |
| 4 | NNRTI_E138K | 8 | 8 | B |
| 5 | PI_Q58E | 6 | 6 | B |
| 6 | NNRTI_E138A | 6 | 6 | B |
| 7 | NRTI_M41L | 5 | 5 | B |
| 8 | NNRTI_K103N | 5 | 5 | B |
| 9 | NNRTI_K103N | 4 | 4 | F |
| 10 | NNRTI_E138A | 4 | 4 | B |
| 11 | NRTI_T215E | 4 | 4 | B |
| 12 | NNRTI_K103N | 3 | 3 | A |
| 13 | NRTI_M41L | 3 | 3 | A |
| 14 | NNRTI_E138A, NRTI_T215D | 3 | 3 | CRF02_AG |
| 15 | NNRTI_E138Q, NNRTI_V179L | 3 | 3 | CRF02_AG |
| 16 | NNRTI_Y181C, NRTI_T215D | 3 | 3 | B |
| 17 | NRTI_T215L | 3 | 3 | B |
| 18 | NNRTI_E138A | 3 | 3 | B |
| 19 | NRTI_T215E, NNRTI_V179L | 11 | 64 | B |
| NRTI_T215E | 53 | |||
| 20 | NNRTI_A98G | 26 | 33 | F |
| 21 | NRTI_T215D | 2 | 14 | B |
| NRTI_T215E | 6 | |||
| NRTI_T215S | 6 | |||
| 22 | PI_I54V, PI_N88D | 1 | 12 | B |
| PI_I54V, PI_N88D, NRTI_L210W, NRTI_T215S | 5 | |||
| PI_N88D, NRTI_L210W, NRTI_T215S | 5 | |||
| 23 | NRTI_T215D | 7 | 10 | B |
| NNRTI_E138A | 2 | |||
| NNRTI_E138A, NRTI_T215D | 1 | |||
| 24 | NNRTI_E138A | 5 | 9 | A |
| 25 | NNRTI_K103N | 3 | 4 | CRF-other |
Abbreviations: CRF, circulating recombinant form; NNRTI, nonnucleoside reverse-transcriptase inhibitors; NRTI, nucleoside reverse-transcriptase inhibitors; PI, protease inhibitors.
Codons associated with the resistance mutations were removed. For each cluster, mutation details, number of patients with the specified mutation, cluster size, and viral subtype are listed.
More than 75% of the patients harboring the PI mutation N88D, the NRTI mutation T215E, or the NNRTI mutation V179L were members of a transmission clusters. Apart from the clustered transmission, phylogenetic analysis also provided evidence for 21 paired transmissions of resistant variants. Here too the most frequently observed mutations were E138A (4 pairs) and K103N (3 pairs).
High-level resistant mutations (Stanford score ≥60) were less frequently observed in subtype B infections than in non-B infections (5.7% vs 9.3%) (P < .001). Mutations with a Stanford score ≥60 in non-B infections were most prevalent in patients infected with a circulating recombinant form (CRF) other than CRF_02AG or CRF01_AE (17.8%; P < .001), followed by subtype A (11.9%; P = .002) and C (11.8%; P = .013) infections. Mutations that were significantly more prevalent in subtype B infections than in non-B infections were M41L (1.38% vs 0.71%; P = .043), I54V (0.35% vs 0.00%; P = .009), N88D (0.69% vs 0.00%; P < .001), V179L (0.69% vs 0.20%; P = .024), L210W (1.04% vs 0.20%; P = .001), T215D (1.10% vs 0.30%; P = .003), T215E (4.61% vs 0.00%; P < .001), and T215S (1.67% vs 0.56%; P = .001) (Table 2). Mutations more prevalent in the non-B subtypes were K65R (0.25% vs 0.00%; P = .036), K70R (0.30% vs 0.00%; P = .022), A98G (2.38% vs 0.23%; P < .001), V108I (1.22% vs 0.29%; P = .001), M184V (1.27% vs 0.40%; P = .005), G190A (0.86% vs 0.12%; P = .002), and P225H (0.56% vs 0.12%; P = .023) (Table 2).
Table 2.
List of Mutations With Significantly Different Prevalence in Subtype B and Non-B Infections
| Subtype | Drug Class | Mutation | Distribution Subtype B vs Non-Ba | P Value | Stanford Score |
|---|---|---|---|---|---|
| B | NNRTI | V179L | 0.69% vs 0.20% | .024 | 15 |
| NRTI | M41L | 1.38% vs 0.71% | .043 | 15 | |
| L210W | 1.04% vs 0.20% | .001 | 15 | ||
| T215D | 1.10% vs 0.30% | .003 | 20 | ||
| T215E | 4.61% vs 0.00% | <.001 | 20 | ||
| T215S | 1.67% vs 0.56% | .001 | 20 | ||
| PI | I54V | 0.35% vs 0.00% | .009 | 20 | |
| N88D | 0.69% vs 0.00% | <.001 | 60 | ||
| Non-B | NNRTI | A98G | 0.23% vs 2.38% | <.001 | 30 |
| V108I | 0.29% vs 1.22% | .001 | 15 | ||
| G190A | 0.12% vs 0.86% | .002 | 60 | ||
| P225H | 0.12% vs 0.56% | .023 | 45 | ||
| NRTI | K65R | 0.00% vs 0.25% | .036 | 60 | |
| K70R | 0.00% vs 0.30% | .022 | 30 | ||
| M184V | 0.40% vs 1.27% | .005 | 60 |
Abbreviations: NNRTI, nonnucleoside reverse-transcriptase inhibitors; NRTI, nucleoside reverse-transcriptase inhibitors; PI, protease inhibitors.
Percentage of patients infected with a virus of subtype B with the specified mutation vs the percentage of patients infected with a virus of subtype non-B carrying that same mutation.
In the 533 available integrase sequences (only samples from 2019), INSTI resistance was found in 3 (0.56%). The detected mutations were E92Q, G163K, and G163R. The E92Q is a nonpolymorphic mutation known to reduce susceptibility to elvitegravir and raltegravir [24], whereas G163K/R are nonpolymorphic accessory mutations [25].
DISCUSSION
To assess the prevalence of TDR in Belgium, HIV-1 pol sequences from 3708 treatment-naive patients newly diagnosed between 2013 and 2019 were analyzed. The results revealed an overall TDR prevalence of 17.9% with minor yearly fluctuations over the studied period. These figures are in line with those reported by others for comparable populations [12, 26–28]. However, comparison of prevalence rates is complicated by the use of different algorithms to identify TDR mutations. The 3 most frequently used algorithms are the WHO list for TDR surveillance, the IAS-USA drug resistance mutations list, and the Stanford HIV Drug Resistance Database. The first 2 are specifically designed for the purpose of TDR monitoring. The Stanford HIV Drug Resistance Database on the other hand provides a complete list of all mutations associated with drug resistance, with each mutation scored according to its presumed impact on the susceptibility of individual drugs. Important assets of this database are the completeness and the fact that it is regularly updated (last update March 2021). This is not the case for the WHO and IAS-USA lists that were last updated in 2009 and 2019, respectively. These still frequently used mutation lists do not take into account some of the key mutations implicating the activity of currently prescribed drugs and overestimate the importance of polymorphisms, revertant mutations, and mutations only affecting the activity of drugs that are no longer prescribed. We compared the outcome of the 3 algorithms for our study population and found significant differences, with a TDR prevalence of 10.9% using the WHO list, 17.9% using the Stanford database, and 19.0% using the IAS-USA list. We choose to continue the analyses based on the Stanford HIV Drug Resistance Database because this is the only one that is regularly updated, and we set our score cut off at ≥15 to identify mutations with at least a low-level impact on drug susceptibility or ≥60 to identify the mutations with high impact on drug susceptibility. The overall TDR prevalence of 17.9% using the ≥15 cut off is almost identical to the 17.2% reported for the German seroconverter study cohort using the same algorithm [9]. The level of 10.9% using the WHO list was similar to what has been described for a German (10.6%) and Hungarian (10.7%) population [9, 18].
Transmitted drug resistance prevalence is mainly driven by the presence of NNRTI mutations as has been observed before in other patient populations from high-income countries [7, 12, 26, 29]. A recent meta-analysis showed that presence of NNRTI mutations before initiation of ART resulted in poorer treatment outcomes in patients initiating an efavirenz-based regimen [30]. However, the impact of the NNRTI mutations that we observed most frequently, E138A (3.86%) and K103N (3.37%), on the currently most prescribed second-generation NNRTIs, rilpivirine, etravirine, and doravirine, is expected to be very low. Mutations with a considerable impact (Stanford score ≥60) on 1 or more of these second-generation NNRTIs were nevertheless detected, although at low frequency, for doravirine (Y188L, 0.27%; G190E, 0.05%; and M230L, 0.05%) and for rilpivirine (L100I, 0.05%; E138K, 0.51%; Y188L, 0.27%; and M230L, 0.05).
Nevirapine- and efavirenz-based regimens have been used extensively in the past. When these regimens fail, the K103N mutation is easily selected. The low impact of this mutation on viral fitness allowed its persistence and swift spread [27, 31, 32]. Ruelle et al [16] first described the presence of a large K103N transmission cluster in Belgium. Our results revealed not only the further expansion of this cluster but also the presence of 3 additional K103N clusters. Together these 4 clusters comprised 41 patients. We also found indications for clustered spread of the rilpivirine-resistant E138K variants as well as the E138A containing variants. The polymorphic E138A only slightly reduces rilpivirine activity, but initiation of rilpivirine in the presence of this mutation is not recommended [33, 34]. No association was observed between presence of E138A mutation and the subtype, with a prevalence varying between 4.86% and 2.27% in subtype A, B, C, CRF02_AG, and F (results not shown). Considering the recent launch of the long-acting injectable drug combination of the integrase inhibitor cabotegravir with rilpivirine, the frequent observation of mutations reducing the rilpivirine sensitivity necessitates close monitoring. The prevalence of transmitted PI mutations was extremely low (2.4%) in line with reports from other comparable studies, with none of the observed mutations expected to reduce the activity of the most frequently prescribed PI, darunavir [35, 36]. However, NRTI resistance mutations were present at higher frequency (6.2%) with the majority of them being thymidine analog resistance mutations (TAMs) (M41L, D67N/G, K70R, L210W, T215Y/F, T215rev, and K219Q/E/R/N). These TAMs have little or no impact on the currently most used NRTIs [27, 37]. They likely originate from long time ago and have spread steadily through onward transmission. For the T215rev and M41L, we found clear indications for clustered spread. Onward transmission of NRTI-associated mutations has been reported before, especially for the 215rev variants [27, 38, 39]. The question can be asked whether it remains justified to still count T215rev as resistant mutations. Failure to identify the mutants T215Y/T as minority variants in persons with T215rev argues against the original belief that presence of T215rev may indicate a hidden presence of the true resistance mutants T215Y/T [38]. Although there have been reports showing some impact of T215rev on the risk of virological failure when initiating a thymidine analog-containing regimen [40], there are no indications that T215rev confers resistance to today’s most commonly used NRTI, tenofovir [27, 41]. The NRTI mutations with probably the highest clinical impact on today’s regimen are M184V/I and K65R. M184V/I reduces susceptibility to lamivudine and emtricitabine, K65R reduces the activity of tenofovir, but all 3 are important components of recommended first-line regimens. M184V/I was detected in 1.05% of the samples. This may be an underestimation of the true presence of M184V/I because it is known that fitness disadvantages may lead to the banishment of viral strains carrying M184V/I to the latent reservoir, whereas the more fit wild-type strains predominate the plasma [31, 32]. It remains to be defined to what extend the more sensitive next-generation sequencing techniques and/or analysis of the viral reservoir will increase the frequency of identification of M184V/I. In a recent study using next-generation sequencing on a large international cohort of patients, the overall M184V/I prevalence remained low [14].
Only 5 cases of K65R presence were observed; however, in 4 of these 5 cases, the K65R was present in combination with M184V. This combination is sufficient to abrogate the activity of the backbone of regimens comprising abacavir or tenofovir and lamivudine or emtricitabine [42].
As we previously stated, we found multiple indications for clustered transmission of drug-resistant variants, which are not limited to variants with single mutations. Comparable findings were also reported by Oroz et al [39]. In 6 clusters, transmission of a variant combining at least 2 resistance mutations was evidenced. The majority of the clusters with TDR involved subtype B infections; however, 2 subtype A, 2 subtype CRF02_AG, and 2 subtype F clusters with TDR were also identified. These are the subtypes that overall contribute the most to local HIV transmission in our region [43, 44]. Similar observations were made when analyzing HIV epidemics in the United Kingdom and Switzerland [29, 45, 46]. The multiple indications for clustered transmission of drug-resistant variants highlight the importance of early diagnosis and treatment as a way to reduce the circulation of these resistant variants.
The HIV-infected population in Belgium is highly heterogeneous with only half of the infections attributed to subtype B and presence of a wide variety of non-B subtypes. Resistance mutations with a Stanford score ≥60 were more frequently detected in non-B infections than in subtype B infections. Higher prevalence of TDR has previously been associated with subtypes present in people with a migration background [47, 48]. In Italy and the United Kingdom, however, higher prevalence of TDR in patients with subtype B compared with non-B was reported [10, 28]. Non-B subtypes were associated with presence of the high-level resistance mutations K65R, M184V, and G190A, confirming the findings of others [49–51]. These mutations highly impact the activity of currently prescribed regimen. The only high-level resistance mutation associated with subtype B was the infrequently observed N88D (0.69% vs 0.00%), a non-polymorphic PI mutation reducing susceptibility to nelfinavir with low-level cross resistance to atazanavir and saquinavir, drugs that are irrelevant for contemporary regimens.
The strength of this study is its nationwide investigation conducted over a relatively long but recent time period in a country with a heterogeneous HIV epidemic. However, there were also some limitations. Only treatment-naive patients newly diagnosed in Belgium were included, but for patients arriving in Belgium from abroad and especially for those from African countries, it is not always clear whether they were truly unaware of their serostatus and treatment naive. This may have resulted in a slight overestimation of TDR in non-B subtype infections. Another weak point is the lack of data on transmitted resistance to integrase inhibitors. However, subsequent analysis of the 2020 data (not presented) and previously performed analyses on a selection of patients who initiated an integrase inhibitor-based, first-line regimen illustrated that, in agreement with other studies in comparable populations [52, 53], baseline presence of integrase mutations remains rare.
CONCLUSIONS
In conclusion, the currently used algorithms for TDR detection may overestimate the clinical importance of TDR because they take into account mutations with no or only low impact on the currently subscribed drugs. However, our findings clearly show that viral variants with a significantly reduced sensitivity for today’s recommended and most used drug regimens can be identified in treatment-naive individuals and are transmitted locally. Efforts to define TDR as a way to prevent failure of first-line ART thereby remains important to avoid mortgaging future treatment options. Moreover, having a baseline sequence available will facilitate the interpretation of mutations in sequences generated later, during episodes of virological treatment failure. This is particularly important when new drugs or drug combinations are approved for clinical use.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Acknowledgments
Financial support. The AIDS Reference Laboratory is supported by the Belgian Ministry of Social Affairs through a fund within the Health Insurance System.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
Contributor Information
Virginie Mortier, Aids Reference Laboratory, Department of Diagnostic Sciences, Ghent University, Ghent, Belgium.
Laurent Debaisieux, Aids Reference Laboratory, Université Libre de Bruxelles, Cliniques universitaires de Bruxelles Hôpital Erasme, Brussels, Belgium.
Géraldine Dessilly, Aids Reference Laboratory, Medical Microbiology Unit, Université Catholique de Louvain, Brussels, Belgium.
Karolien Stoffels, Aids Reference Laboratory, Centre Hospitalier Universitaire St. Pierre, Brussels, Belgium.
Dolores Vaira, Aids Reference Laboratory, Centre Hospitalier Universitaire de Liège, Liège, Belgium.
Ellen Vancutsem, Aids Reference Laboratory, Vrije Universiteit Brussel VUB, Brussels, Belgium.
Kristel Van Laethem, Department of Microbiology and Immunology, Rega Institute for Medical Research, University of Leuven, Leuven, Belgium; Aids Reference Laboratory, University Hospitals Leuven, Leuven, Belgium.
Fien Vanroye, Aids Reference Laboratory, Clinical Reference Laboratory, Department of Clinical Sciences, Institute of Tropical Medicine, Antwerp, Belgium.
Chris Verhofstede, Aids Reference Laboratory, Department of Diagnostic Sciences, Ghent University, Ghent, Belgium.
References
- 1. Murphy EL, Collier AC, Kalish LA, et al. Highly active antiretroviral therapy decreases mortality and morbidity in patients with advanced HIV disease. Ann Intern Med 2001; 135:17–26. [DOI] [PubMed] [Google Scholar]
- 2. European AIDS Clinical Society . EACS Guidelines version 10.1. Available at: https://www.eacsociety.org/guidelines/eacs-guidelines/eacs-guidelines.html. Accessed March 23, 2021.
- 3. Günthard HF, Calvez V, Paredes R, et al. Human immunodeficiency virus drug resistance: 2018 recommendations of the International Antiviral Society-USA Panel. Clin Infect Dis 2019; 68:177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Paredes R, Lalama CM, Ribaudo HJ, et al. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis 2010; 201:662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Simen BB, Simons JF, Hullsiek KH, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis 2009; 199:693–701. [DOI] [PubMed] [Google Scholar]
- 6. Rhee SY, Kassaye SG, Barrow G, Sundaramurthi JC, Jordan MR, Shafer RW. HIV-1 transmitted drug resistance surveillance: shifting trends in study design and prevalence estimates. J Int AIDS Soc 2020; 23:e25611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hofstra LM, Sauvageot N, Albert J, et al. Transmission of HIV drug resistance and the predicted effect on current first-line regimens in Europe. Clin Infect Dis 2016; 62:655–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tostevin A, White E, Dunn D, et al. Recent trends and patterns in HIV-1 transmitted drug resistance in the United Kingdom. HIV Med 2017; 18:204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Machnowska P, Meixenberger K, Schmidt D, et al. Prevalence and persistence of transmitted drug resistance mutations in the German HIV-1 Seroconverter Study Cohort. PLoS One 2019; 14:e0209605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calza L, Tamburello M, Borderi M, et al. Prevalence of transmitted drug resistance mutations among newly diagnosed HIV-1-infected patients in a large teaching hospital of the Northern Italy. J Med Virol 2020; 92:929–31. [DOI] [PubMed] [Google Scholar]
- 11. Kagan RM, Dunn KJ, Snell GP, Nettles RE, Kaufman HW. Trends in HIV-1 drug resistance mutations from a U.S. reference laboratory from 2006 to 2017. AIDS Res Hum Retroviruses 2019; 35:698–709. [DOI] [PubMed] [Google Scholar]
- 12. Rhee SY, Clutter D, Fessel WJ, et al. Trends in the molecular epidemiology and genetic mechanisms of transmitted human immunodeficiency virus type 1 drug resistance in a large US clinic population. Clin Infect Dis 2019; 68:213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Avila-Rios S, Sued O, Rhee SY, Shafer RW, Reyes-Teran G, Ravasi G. Surveillance of HIV transmitted drug resistance in Latin America and the Caribbean: a systematic review and meta-analysis. PLoS One 2016; 11:e0158560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baxter JD, Dunn D, Tostevin A, et al. Transmitted HIV-1 drug resistance in a large international cohort using next-generation sequencing: results from the Strategic Timing of Antiretroviral Treatment (START) study. HIV Med 2021; 22:360–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yerly S, Junier T, Gayet-Ageron A, et al. The impact of transmission clusters on primary drug resistance in newly diagnosed HIV-1 infection. AIDS 2009; 23:1415–23. [DOI] [PubMed] [Google Scholar]
- 16. Ruelle J, Ingels MG, Jnaoui K, et al. Transmission network of an HIV type 1 strain with K103N in young Belgian patients from different risk groups. AIDS Res Hum Retroviruses 2013; 29:1306–9. [DOI] [PubMed] [Google Scholar]
- 17. Cope AB, Powers KA, Kuruc JD, et al. Ongoing HIV transmission and the HIV care continuum in North Carolina. PLoS One 2015; 10:e0127950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Áy E, Müller V, Mezei M, et al. Transmitted drug resistance in newly diagnosed and treatment-naive HIV type 1-infected patients in Hungary. J Glob Antimicrob Resist 2020; 20:124–30. [DOI] [PubMed] [Google Scholar]
- 19. Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res 2003; 31:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis 2006; 42:1608–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bennett DE, Camacho RJ, Otelea D, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 2009; 4:e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wensing AM, Calvez V, Ceccherini-Silberstein F, et al. 2019 update of the drug resistance mutations in HIV-1. Top Antivir Med 2019; 27:111–21. [PMC free article] [PubMed] [Google Scholar]
- 23. Hall TA. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/INT. Nucleic Acids SympSer 1999; 41:95–8. [Google Scholar]
- 24. Hatano H, Lampiris H, Fransen S, et al. Evolution of integrase resistance during failure of integrase inhibitor-based antiretroviral therapy. J Acquir Immune Defic Syndr 2010; 54:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rhee SY, Sankaran K, Varghese V, et al. HIV-1 protease, reverse transcriptase, and integrase variation. J Virol 2016; 90:6058–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Skoura L, Metallidis S, Buckton AJ, et al. Molecular and epidemiological characterization of HIV-1 infection networks involving transmitted drug resistance mutations in Northern Greece. J Antimicrob Chemother 2011; 66:2831–7. [DOI] [PubMed] [Google Scholar]
- 27. Margot NA, Wong P, Kulkarni R, et al. Commonly transmitted HIV-1 drug resistance mutations in reverse-transcriptase and protease in antiretroviral treatment-naive patients and response to regimens containing tenofovir disoproxil fumarate or tenofovir alafenamide. J Infect Dis 2017; 215:920–7. [DOI] [PubMed] [Google Scholar]
- 28. Mazzuti L, Melengu T, Falasca F, et al. Transmitted drug resistance mutations and trends of HIV-1 subtypes in treatment-naive patients: a single-centre experience. J Glob Antimicrob Resist 2020; 20:298–303. [DOI] [PubMed] [Google Scholar]
- 29. Yang WL, Kouyos R, Scherrer AU, et al. Assessing the paradox between transmitted and acquired HIV type 1 drug resistance mutations in the Swiss HIV Cohort Study from 1998 to 2012. J Infect Dis 2015; 212:28–38. [DOI] [PubMed] [Google Scholar]
- 30. Bertagnolio S, Hermans L, Jordan MR, et al. Clinical impact of pretreatment human immunodeficiency virus drug resistance in people initiating nonnucleoside reverse transcriptase inhibitor-containing antiretroviral therapy: a systematic review and meta-analysis. J Infect Dis 2021; 224:377–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pingen M, Wensing AM, Fransen K, et al. Persistence of frequently transmitted drug-resistant HIV-1 variants can be explained by high viral replication capacity. Retrovirology 2014; 11:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang WL, Kouyos RD, Böni J, et al. Persistence of transmitted HIV-1 drug resistance mutations associated with fitness costs and viral genetic backgrounds. PLoS Pathog 2015; 11:e1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu HT, Colby-Germinario SP, Asahchop EL, et al. Effect of mutations at position E138 in HIV-1 reverse transcriptase and their interactions with the M184I mutation on defining patterns of resistance to nonnucleoside reverse transcriptase inhibitors rilpivirine and etravirine. Antimicrob Agents Chemother 2013; 57:3100–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Picchio GR, Rimsky LT, Van Eygen V, Haddad M, Napolitano LA, Vingerhoets J. Prevalence in the USA of rilpivirine resistance-associated mutations in clinical samples and effects on phenotypic susceptibility to rilpivirine and etravirine. Antivir Ther 2014; 19:819–23. [DOI] [PubMed] [Google Scholar]
- 35. McKeage K, Perry CM, Keam SJ. Darunavir: a review of its use in the management of HIV infection in adults. Drugs 2009; 69:477–503. [DOI] [PubMed] [Google Scholar]
- 36. De Luca A, Dunn D, Zazzi M, et al. Declining prevalence of HIV-1 drug resistance in antiretroviral treatment-exposed individuals in Western Europe. J Infect Dis 2013; 207:1216–20. [DOI] [PubMed] [Google Scholar]
- 37. Vercauteren J, Wensing AM, van de Vijver DA, et al. Transmission of drug-resistant HIV-1 is stabilizing in Europe. J Infect Dis 2009; 200:1503–8. [DOI] [PubMed] [Google Scholar]
- 38. Mitsuya Y, Varghese V, Wang C, et al. Minority human immunodeficiency virus type 1 variants in antiretroviral-naive persons with reverse transcriptase codon 215 revertant mutations. J Virol 2008; 82:10747–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oroz M, Begovac J, Planinić A, et al. Analysis of HIV-1 diversity, primary drug resistance and transmission networks in Croatia. Sci Rep 2019; 9:17307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Violin M, Cozzi-Lepri A, Velleca R, et al. Risk of failure in patients with 215 HIV-1 revertants starting their first thymidine analog-containing highly active antiretroviral therapy. AIDS 2004; 18:227–35. [DOI] [PubMed] [Google Scholar]
- 41. Margot N, Ram R, Abram M, Haubrich R, Callebaut C. Antiviral activity of tenofovir alafenamide against HIV-1 with thymidine analog-associated mutations and M184V. Antimicrob Agents Chemother 2020; 64:e02557-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Borroto-Esoda K, Parkin N, Miller MD. A comparison of the phenotypic susceptibility profiles of emtricitabine and lamivudine. Antivir Chem Chemother 2007; 18:297–300. [DOI] [PubMed] [Google Scholar]
- 43. Verhofstede C, Dauwe K, Fransen K, et al. Phylogenetic analysis of the Belgian HIV-1 epidemic reveals that local transmission is almost exclusively driven by men having sex with men despite presence of large African migrant communities. Infect Genet Evol 2018; 61:36–44. [DOI] [PubMed] [Google Scholar]
- 44. Vinken L, Fransen K, Cuypers L, et al. Earlier initiation of antiretroviral treatment coincides with an initial control of the HIV-1 sub-subtype F1 outbreak among men-having-sex-with-men in Flanders, Belgium. Front Microbiol 2019; 10:613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mourad R, Chevennet F, Dunn DT, et al. A phylotype-based analysis highlights the role of drug-naive HIV-positive individuals in the transmission of antiretroviral resistance in the UK. AIDS 2015; 29:1917–25. [DOI] [PubMed] [Google Scholar]
- 46. Ragonnet-Cronin ML, Shilaih M, Günthard HF, et al. A direct comparison of two densely sampled HIV epidemics: the UK and Switzerland. Sci Rep 2016; 6:32251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sluis-Cremer N, Jordan MR, Huber K, et al. E138A in HIV-1 reverse transcriptase is more common in subtype C than B: implications for rilpivirine use in resource-limited settings. Antiviral Res 2014; 107:31–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Theys K, Van Laethem K, Gomes P, et al. Sub-epidemics explain localized high prevalence of reduced susceptibility to rilpivirine in treatment-naive HIV-1-infected patients: subtype and geographic compartmentalization of baseline resistance mutations. AIDS Res Hum Retroviruses 2016; 32:427–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lessells RJ, Katzenstein DK, de Oliveira T. Are subtype differences important in HIV drug resistance? Curr Opin Virol 2012; 2:636–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Singh K, Flores JA, Kirby KA, et al. Drug resistance in non-B subtype HIV-1: impact of HIV-1 reverse transcriptase inhibitors. Viruses 2014; 6:3535–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu HT, Colby-Germinario SP, Quashie PK, Bethell R, Wainberg MA. Subtype-specific analysis of the K65R substitution in HIV-1 that confers hypersusceptibility to a novel nucleotide-competing reverse transcriptase inhibitor. Antimicrob Agents Chemother 2015; 59:3189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mesplède T, Quashie PK, Wainberg MA. Resistance to HIV integrase inhibitors. Curr Opin HIV AIDS 2012; 7:401–8. [DOI] [PubMed] [Google Scholar]
- 53. Anstett K, Brenner B, Mesplede T, Wainberg MA. HIV drug resistance against strand transfer integrase inhibitors. Retrovirology 2017; 14:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.