Abstract
The sphingomyelin synthase (SMS) gene family has three members: SMS1 and SMS2 have SM synthase activity, while SMS-related protein (SMSr) has no SM synthase activity but has ceramide phosphorylethanolamine (CPE) synthase activity in vitro. Recently, we found that SMS family members are a group of phospholipase Cs (PLC). SMS1 and SMS2 are two phosphatidylcholine (PC)-PLCs and SMSr is a phosphatidylethanolamine (PE)-PLC. SMS family members not only influence SM levels but also influence the levels of diacylglycerol (DAG), PC, PE, and glycosphingolipids, thus influencing cell functions. In this chapter, we will discuss the recent progresses in the research field of SMS family and will focus on its impact on metabolic diseases.
Keywords: Sphingomyelin synthase, Phospholipase C, Metabolic diseases
Sphingomyelin Synthase 1
Sphingomyelin (SM) biosynthesis occurs via the actions of serine palmitoyltransferase (SPT), 3 ketosphinganine reductase, ceramide synthase, and dihydroceramide desaturase to produce ceramide, which is a substrate for the production of sphingomyelin (through sphingomyelin synthase, SMS) (Figure 1) (1,2). SM synthase has two isoforms, SMS1 and SMS2 (2,3). SMS1 is encoded by the SGMS1 gene which is located on human chromosome 11 (mouse chromosome 19), with 11 exons (4). Human SMS1 consist of 413 amino acids (2,3). Various alternative SMS1 mRNA transcripts, with different exon combination and length of 5′- or 3’-UTR, were found (5). Truncated SMS1 mRNA transcripts which do not yield full-length SMS1 protein were also discovered (6,7). It has been reported that certain circular noncoding RNAs (circRNAs), which contain sequences of 5′-UTR and/or exonic portions of the SGMS1 gene, were found in human, rat, and mouse tissues (8). These SMS1 circRNAs might play a role in the regulation of SMS1 expression, through binding certain microRNAs(8). Human SMS1 translation could be regulated through its mRNA 5′-untranslated region (4).
Fig. 1.
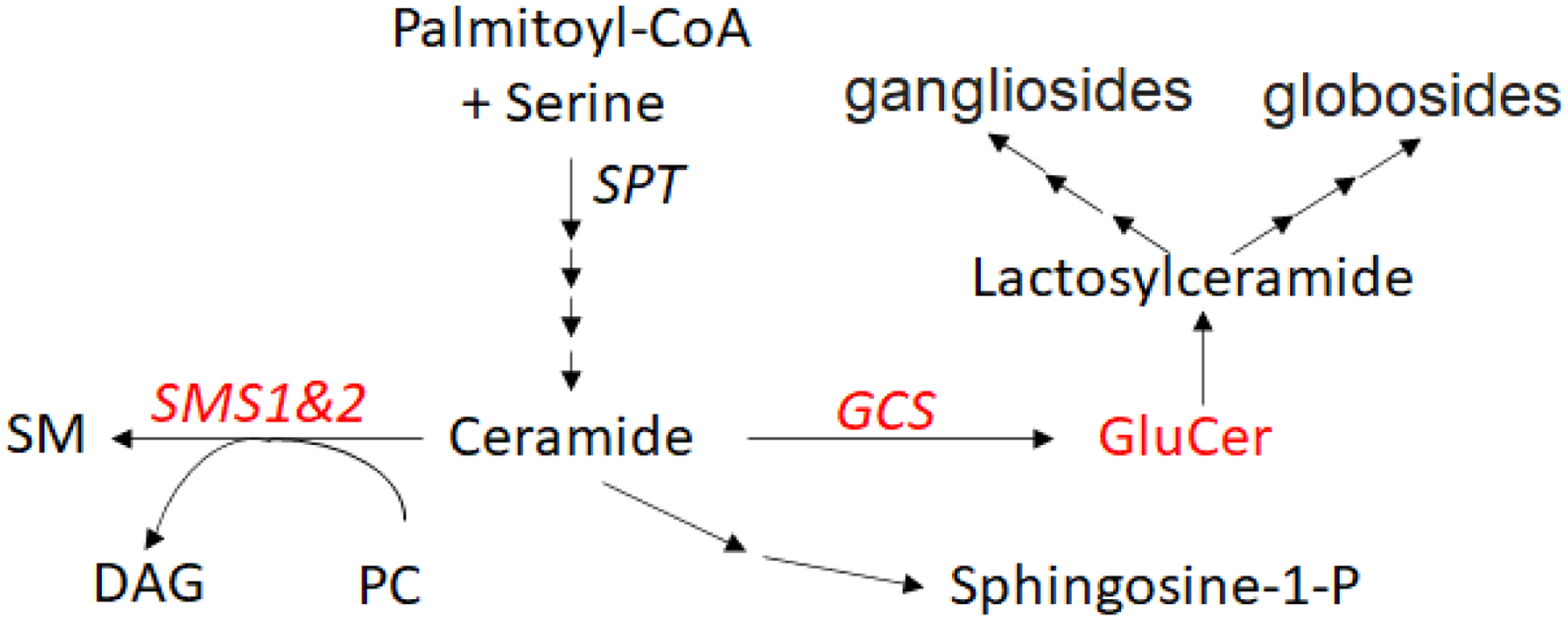
Scheme for ceramide, SM, and GluCer formation.
The three-dimensional structure of SMS remains unresolved so far. Thus, little is known about its overall folding and structure of catalytic site. However, there are two computer models have been reported. Using two proteins (with known structures) as templates and combined with homology and molecular dynamics, a computer model of human SMS1 was proposed by us 12-year ago (9). The conserved Histidine 285, Histidine 328, and Aspartate 332 along with the respective distances between residues are depicted (9). A recent study also proposed another human SMS1 computer model which showed that 1) the residence time of PC is shorter than SM; 2) SMS1 binding site is extremely conserved and three amino acids, Aspartate 101, Arginine 220, and Asparagine 358, are involved in SM synthesis; and 3) the hydroxylation of PC increases the rate of conversion from PC into SM (10).
SMS1-generated SM plays an important role in transferrin trafficking and cell proliferation (11) as well as involving in attachment and infection with Japanese encephalitis virus (12). SMS1 deficiency reduces bone formation due to impaired osteoblast differentiation (13). A very recent report indicated that SMS1 mediates hepatocyte pyroptosis to trigger nonalcoholic steatohepatitis (14). SMS1 is essential for male fertility in mice (15). We and another research group reported that global SMS1 deficiency exhibited moderate neonatal lethality (16,17), i.e., 25% of homozygotes die during the first 3 weeks (the remainders can grow to adulthood). Sms1 knockout (KO) mouse homozygous crosses did not yield viable progeny (17). It has been reported that Sms1 KO mice exhibited reduced body weight, loss of fat tissues mass, β cell mitochondrial dysfunction, insulin secretion inhibition, and insulin resistance (16). Sms1 KO mice have lipodystrophy which is related with an induction of oxidative stress in adipose tissues (18). SMS1 is the major isoform in macrophages, while SMS2 is the major one in the liver (17). The expression levels of both in the rest of tissues are comparable (17). To evaluate atherogenicity, we transplanted Sms1 KO mouse bone marrow into LDL receptor KO mice (Sms1−/−→Ldlr−/−). After 3 months with a Western type diet, these animals showed a significant decrease of atherosclerotic lesions in the root and the entire aorta, compared with WT→Ldlr−/− mice (17).
Based on hydrophobicity analysis, SMS1 contains six transmembrane α-helices connected by extramembrane loop domains with a sterile alpha motif (SAM) at its N-terminal (2). The SAM domain plays an important role in cell functions, such as development, signal transduction, and transcriptional regulation, through protein/lipid and protein/protein interactions (19). However, the proteins interacting with SAM domain of SMS1 have not been identified until recently. It is known that both SMS1 and glucosylceramide synthase (GCS) are involved in a sphingolipid metabolic branch point (Figure 1). SMS1 catalyzes the transfer of phosphocholine from phosphatidylcholine to ceramide to form SM, whereas GCS catalyzes the transfer of glucose from UDP-glucose to ceramide to form glucosylceramide (GluCer) (20). Similar to SMS1, GCS is located in the Golgi apparatus (21). There is an interaction between the SMS1 N-terminal SAM domain and GCS C-terminal domain to form a SMS1-GCS complex in the Golgi (Figure 2), controlling the metabolic fate of ceramide in the organelle (22). The formation of the SMS1-GCS complex increases SM synthesis and decreases GCS synthesis (22). In normal tissue, the formation of the SMS1-GCS complex can serve as a switch that controls the steady state levels of GluCer. However, the complex in the situation of SMS1 deficiency is depleted, thereby greatly releasing GCS activity, and promoting GluCer biosynthesis, which was observed in our study (17).
Fig. 2.
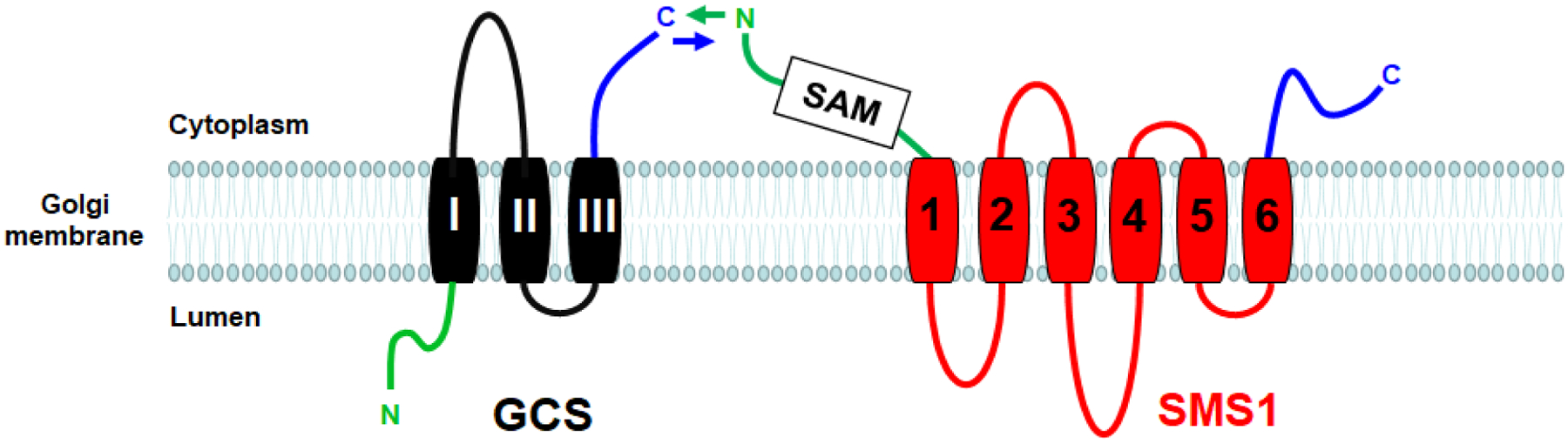
SMS1 and GCS complex formation on Golgi membrane.
Very recently, we studied the metabolic consequences of SMS1 deficiency-mediated GluCer accumulation. Liver-specific Sms1/global Sms2 double-knockout (dKO) exhibited severe liver steatosis under a high-fat diet. Further, old (more than 6-month-old) dKO mice had liver damage, inflammation, and fibrosis, compared with Sms2 KO and wild type mice. RNA sequencing analysis indicated the induction of various genes which are involved in lipogenesis, inflammation, and fibrosis. Furthermore, we found that GluCer administration promoted hepatocyte to secrete more activated TGFβ1, which could stimulate more collagen 1α1 production in hepatic stellate cells, thus promoting fibrosis. Additionally, GluCer accelerated more b-catenin translocation into the nucleus, thus promoting tumorigenesis. Significantly, human nonalcoholic steatohepatitis patients had higher levels of liver GCS and higher levels of serum GluCer. Our observation implicated that GluCer accumulation is one of triggers stimulating the development of nonalcoholic fatty liver disease into nonalcoholic steatohepatitis, then, fibrosis and tumorigenesis (23).
Sphingomyelin Synthase 2
SMS2 is encoded by the SGMS2 gene which is located on human chromosome 4 (mouse chromosome 3) with consist of 365 amino acids (2,3). SMS2 shares highly identical in protein sequence with SMS1 but have no SAM domain in its N-terminal. Same as SMS1, SMS2 is predicted as a six-pass transmembrane protein but predominantly localized on plasma membrane, with a minority portion found in Golgi (9). Thus, SMS2 contributes mainly to plasma membrane SMS activity (2,3). While SMS1 plays a role of SM producer, SMS2 was shown mostly a regulator of the SM in plasma membrane (2,3). We and other researchers have shown that SMS2 expression positively correlates SM levels in the lipid rafts of plasma membrane (24–26). SMS2 is required for the maintenance of plasma membrane microdomain fluidity (27) and is directly linked to cell membrane lipid messengers that play a role in cell survival and apoptosis (24,28). In contrast to the neonatal lethality found in Sms1 KO mice, the physiological consequence of SMS2 deficiency is relatively moderate. The Sms2 KO mice are health and viable (29,30).
SMS2 is one of the major determinants for plasma and liver SM levels in mice (29). We and other researchers have found that SMS2 deficiency prevented high fat diet-induced obesity and insulin resistance (30,31). Moreover, we found that liver SMS2 overexpression promoted fatty acid uptake and liver steatosis, while SMS2 deficiency had an opposite effect, in comparison with their respective controls. Importantly, the exogenous ceramide supplementation to Huh7 cells (a human hepatoma cell line reduced the expression of PPARγ2 and its target genes, CD36 and FSP27 (32). Thus, SMS2 deficiency-mediated ceramide induction suppressed lipogenesis in hepatocytes (32). We prepared Sms2 and Apoe double knockout (KO) mice. They showed a significant decrease in plasma lipoprotein SM levels. There was reduction of atherogenic lipoprotein aorta retention in the double KO mice. Importantly, the double KO mice showed a significant reduction in atherosclerosis, compared to controls (33).
G-protein coupled receptor family C group 5 member B (GPRC5B) recruitment of Src family kinases has been implicated in diet-induced insulin resistance (34). GPRC5B-mediated phosphorylation of sphingomyelin synthase 2 (SMS2) by Fyn is a crucial step in the development of insulin resistance (35). SMS2 deficiency inhibits osteoclastogenesis by decreasing RANKL expression in mouse primary osteoblasts (36). A recent paper reported that SMS2 deficiency suppresses steatosis but worsens fibrosis in the liver in a specific condition with choline-deficient, L-amino acid-defined, high-fat diet (37). In a very recent study, we indicated that SMS2 deficiency ameliorated cerebral ischemic reperfusion injury through reducing the recruitment of toll-like receptor 4 to lipid Rafts (38). We also prepared liver-specific Sms1/global Sms2 double KO mice to evaluate the effect of hepatocyte SM biosynthesis in lipoprotein metabolism. We found that 2 month-old the double KO mice significantly reduced their hepatocyte SM levels and reduced very low-density lipoprotein production (39). This phenomenon could be related with a reduction of atherogenicity.
In general, SMS2 deficiency decreases dietary induced obesity, insulin resistance (31), and atherosclerosis (33,40). We and others have suggested that SMS2 could be a therapeutic target for metabolic diseases, including NAFLD, type 2 diabetes, and atherosclerosis (30–33,41–43). In fact, there are some progresses in the development of SMS2 specific inhibitors. A selective SMS2 inhibitor ameliorates diet induced insulin resistance via the IRS-1/Akt/GSK-3β signaling pathway (44). The db/db mouse is an animal model for diabetic dyslipidemia (45). A specific SMS2 inhibitor reduced chronic inflammation in db/db mice(46). We have a specific Chapter to discuss this aspect.
It has been reported that the potent anticancer property of 2-hydroxy-oleic acid (2OHOA) is mediated by its activation of SMS which induced SM accumulation (47). Further, the same group researchers reported that 2OHOA had opposite effect towards SMS1 and SMS2 (48). However, we found that 2OHOA inhibited rather than activated purified rSMS1 and rSMS2 in a dose-dependent fashion (49). Thus, 2OHOA is not a SMS activator and that its anticancer property should be related other mechanisms.
Sphingomyelin Synthase-Related Protein
Sphingomyelin synthase related protein (SMSr) is the third member of the SMS family. SMSr is encoded by the SAMD8 gene which is located on human chromosome 10 (mouse chromosome 14), consisting 414 amino acids (2). SMSr is conserved throughout the animal kingdom (2,50) and ubiquitously expressed in all tested tissues (51). SMSr is located on endoplasmic reticulum (ER) (52). Like SMS1 and SMS2, SMSr also has six transmembrane domains and cytoplasmic N and C termini (2,50) as well as a conserved triad of two histidine and one aspartate residue (2,9,50). However, unlike SMS1 and SMS2, SMSr does not have SM synthase activity but instead catalyzes synthesis an SM analog, ceramide phosphoethanolamine (CPE), in test tubes (52). Interestingly, SMS2 (53) and SMS1 (54) are also capable to synthesize CPE in vitro.
It has been reported (52) that Smsr gene knockdown (by siRNA) in cultured HeLa and Drosophila S2 cells could lead to a significant increase in ER ceramide levels and a collapse of the early secretory pathway. Thus, the researchers hypothesized that SMSr regulates ceramide synthesis in ER. The same group researchers also reported that abnormal ceramide accumulation can lead to mislocalization of ceramide to mitochondria, triggering the mitochondrial apoptosis pathway, suggesting that SMSr might be a suppressor of apoptosis (55). However, another group researchers reported that Smsr knockdown in multiple cell lines did not alter sphingolipid biosynthesis, including ceramide (56).
To examine SMSr function in vivo, we generated Smsr KO mice which were fertile without obvious phenotypic alterations. Interestingly, quantitative MS analyses of plasma, liver, and macrophages from the KO mice revealed only marginal changes in CPE levels which were extremely low (54). Further, ceramide levels in the plasma, liver, and macrophages from the KO mice were not significantly different from those of WT controls (54). Similar phenomenon was also observed independently by another research group (57). Furthermore, since both SMS2 and SMSr have CPE synthase activity in test tubes (52,53), Smsr/Sms2 double KO mice were used to evaluate CPE-related biology. Unexpectedly, the double KO mice had no obvious impact on mouse development or fertility (57). While SMSr is widely expressed in all tested tissues, including the brain, blocking its catalytic activity did not affect ceramide levels or secretory pathway integrity in the brain or any other tissues (57). Then, two fundamental questions are asked: 1) is SMSr responsible for the production of the trace amount of CPE in tissues? and 2) is CPE synthesis a real biological function of SMSr? It could be possible that the trace amount of CPE in the circulation could come from microbiota which contains high levels of CPE (58) and is located in the lumen of intestine. Based on these unexpected results, we even raised a concern 6-year ago: “given the fact that tissue CPE levels are extremely low, it is not clear why SMSr is needed and so highly conserved in the animal kingdom” (54). Thus, SMSr’s real biological function was a big puzzle for more than 15 years.
Like SMS1, SMSr has a SAM domain (2). SMSr’s ER residency relies on homotypic oligomerization mediated by its SAM domain (59). SMSr functionally interact with diacylglycerol kinase δ through its SAM domains and such an interaction represent a new pathway independent of phosphatidylinositol turnover (60). These observations were linking the function of SMSr with lipid metabolism, other than sphingolipid metabolism. This will be the focus of the rest of this Chapter.
SMS family is a group of Phospholipase Cs
Phospholipase C cleaves the phospholipid into diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3) or phosphorylcholine (P-choline) or phosphorylethanolamine (P-ethanolamine). Phosphatidylinositol-specific Phospholipase C (PI-PLC) converts phosphatidylinositol 4,5-bisphosphate (PIP2) to inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG). DAG and IP3 each control diverse cellular processes and are also substrates for synthesis of other important signaling molecules. Mammals express six families of PI-PLCs each with both unique and overlapping controls over expression and subcellular distribution (61).
Phosphatidylcholine-specific phospholipase C (PC-PLC) or phosphatidylethanolamine-specific phospholipase C (PE-PLC) cleaves either PC or PE moiety from PC or PE, thereby generating and releasing DAG and P-choline (62) or P-ethanolamine. Considering the large abundance of PC and PE, it is possible that PC-PLC hydrolysis of PC or PE-PLC hydrolysis of PE can produce a more sustained DAG elevation than PI-PLC cleavage of phosphatidylinositol does (63). Moreover, PC and PE are the major lipid components on cell membrane (64), it is conceivable that PC-PLC and PE-PLC activities are important in maintaining cell membrane integrity and function.
Although bacterial PC-PLC was cloned (65), mammalian PC-PLC have not yet been cloned. Mechanistic PC-PLC studies in mammalian cells and in vivo have to rely on small molecule inhibitors of PC-PLC, such as tricyclodecan-9-yl-potassium xanthate (D609) (66). Similarly, bacterial PE-PLC has also been cloned (67), however, mammalian PE-PLC is still totally unknown. Interestingly, D609 actions are widely attributed to inhibiting PC-PLC and it also inhibits sphingomyelin synthase (SMS) (68).
As known from SM synthase reaction, SM formation can be separated into two steps: 1) PC-PLC step, where PC is hydrolyzed into P-choline and DAG; and 2) SM formation step, where P-choline is added onto ceramide to form SM. Similarly, CPE formation can also be separated into two steps: 1) PE-PLC step, where PE is hydrolyzed into P-ethanolamine and DAG; and 2) CPE formation step, where P-ethanolamine is added onto ceramide to form CPE (Figure 3). Besides six membrane-spanning domains, SMS1, SMS2, and SMSr contain four highly conserved sequence motifs, designated D1, D2, D3, and D4 (2). Motifs D3 (C-G-D-X3-S-G-H-T) and D4 (H-Y-X2-D-V-X2A-X-Y-I-T-T-R-L-F-X2-Y-H), containing conserved amino acids His-His-Asp, are similar to the C2 and C3 motifs in lipid phosphate phosphatase (LPPs) which form a catalytic triad mediating the nucleophilic attack on the lipid phosphate ester bond (69). In fact, the cloning of mammalian SMS gene family was based on the knowledge of LPP sequence (2). Thus, potentially, SMS family is a functional PLC family.
Fig. 3. SMS1/SMS2 mediated SM formation and SMSr mediated CPE formation.
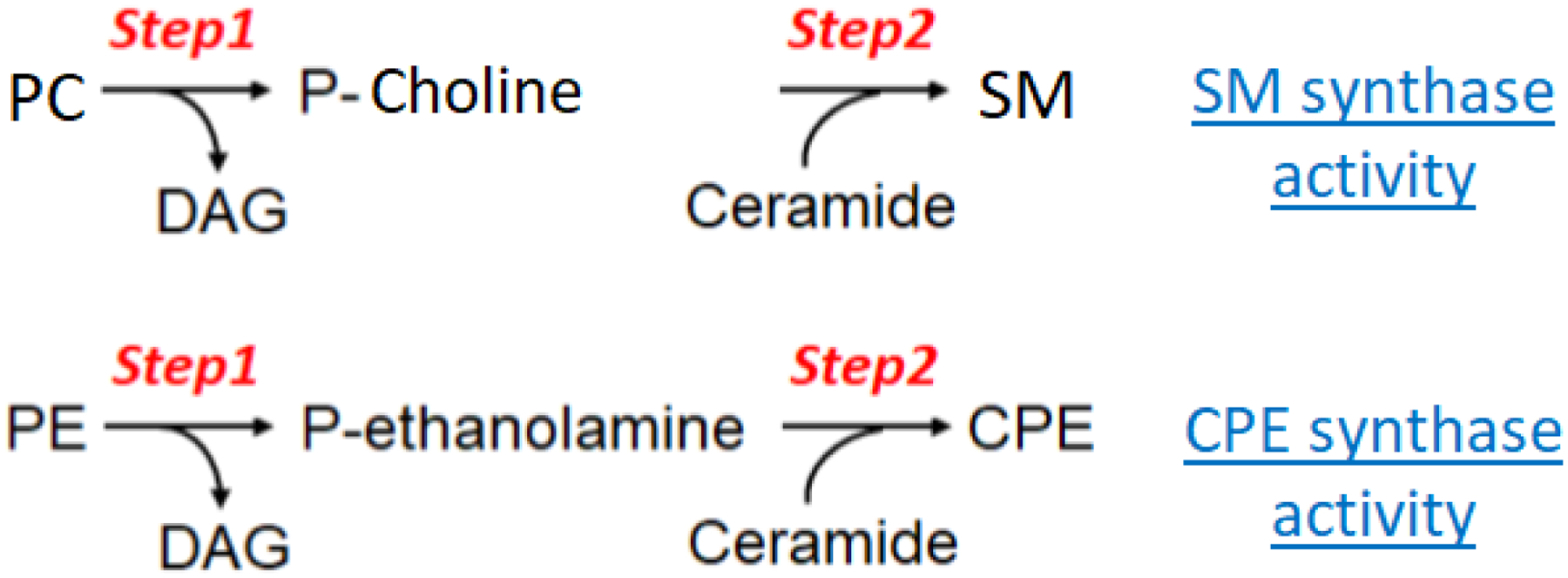
The first step of the reaction is hydrolysis of PC or PE (like PC-PLC or PE-PLC activity) to form P-choline or P-ethanolamine and diacylglycerol (DAG). The second step of the reaction is adding P-choline or P-ethanolamine onto ceramide to form sphingomyelin (SM) or ceramide phosphrylethanolamine (CPE).
Very recently, Murakami and Sakane (70) reported that SMSr, as a multi-glycerophospholipid PLC (or pan-PLC), generates diacylglycerol via the hydrolysis of glycerophospholipids in the absence of ceramide. Although we observed similar phenomenon that SMSr is a PLC, importantly, there is a significant difference between our results and theirs. Murakami and Sakane showed that rSMSr has an PC-PLC activity (70) and we found that rSMSr has no such an activity (71). Our observation is consistent with the knowledge that SMSr has no SM synthase activity, which consume PC and ceramide (2). Moreover, Murakami and Sakane (70) showed that SMSr can also hydrolyze phosphatidic acid (PA) to form DAG, but we did not find such an activity too (71). PA does not contain phosphodiester bone as PE and PC, thus, the rationale that SMSr has phosphatidic acid phosphatase (PAP) activity (which is different from PLC activity) is not that obvious. Given the fact that mammals express six families of PI-PLCs, with at least 13 members (61), and SMSr has no PS-PLC, PG-PLC, PC-PLC, and PAP activities (71), it is not likely that it has PI-PLC activity. Thus, we believe that SMSr is a specific PE-PLC but not a pan-PLC and its specificity is an important property of SMSr.
Interestingly, SMSr-mediated PE-PLC activity is not calcium dependent and can be inhibited by D609 in a dose-dependent fashion (71). Thus, our study together with the recent study (70) clearly solved a long-time puzzle, i.e., what is the real activity of SMSr in vivo? SMSr is a PE-PLC but not a CPE synthase in vivo. Importantly, SMSr can regulate steady state levels of PE in vivo (71), and it should be a new tool for PE-related biological study.
A question was asked more than 20-year ago: does SMS account for the putative PC-PLC? (72). Since SMS family was not cloned until 2004 (2), we had no clue for PC-PLC at that time. Given the fact that D609 can inhibit both PC-PLC and SM synthase activities (72), we hypothesize that both SMS1 and SMS2 have PC-PLC activity, i.e., producing DAG through hydrolysis of PC in the absence of ceramide. Indeed, so far, we have good evidence to show that both SMS1 and SMS2 have PC-PLC activity (Chiang et al. JBC in press). First, purified recombinant SMS1 (rSMS1) and rSMS2 have PC-PLC but not PE-PLC activity. Second, we prepared liver-specific Sms1/global Sms2 double knockout (dKO) mice. We found that liver PC-PLC activity was significantly reduced and steady state levels of DAG and PC in the liver were regulated by the deficiency, in comparison with wild type mice. Third, we respectively expressed Sms1 and Sms2 genes (using adenovirus) in the liver of the dKO mice and found that SMS1 and SMS2 expression can hydrolyze PC to produce phosphocholine and DAG. Thus, SMS1 and SMS2 exhibit PC-PLC activity in vitro and in vivo (Chiang et al JBC in press). Although DAG is known as an activator of certain protein kinases (PKCs) (73), DAG generated from different sources may have variable effects on PKCs. For example, DAG derived from phosphatidylinositol 4,5 biphosphate hydrolysis can activate PKCs whereas DAG produced from cannot activate PKCs(74). SMS/PC-PLC generated DAG pool may be different from PI-PLC generated DAG pool.
We concluded that SMS family is a PLC family. SMS1 and SMS2 are two specific PC-PLCs, while SMSr is a specific PE-PLC. The biological and pathological functions of SMS-mediated PLC activity deserve further investigation.
Abbreviation
- SMS
sphingomyelin synthase
- PLC
phospholipase C
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
References
- 1.Merrill AH Jr. (1983) Characterization of serine palmitoyltransferase activity in Chinese hamster ovary cells. Biochim Biophys Acta 754, 284–291 [DOI] [PubMed] [Google Scholar]
- 2.Huitema K, van den Dikkenberg J, Brouwers JF, and Holthuis JC (2004) Identification of a family of animal sphingomyelin synthases. EMBO J 23, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamaoka S, Miyaji M, Kitano T, Umehara H, and Okazaki T (2004) Expression cloning of a human cDNA restoring sphingomyelin synthesis and cell growth in sphingomyelin synthase-defective lymphoid cells. J Biol Chem 279, 18688–18693 [DOI] [PubMed] [Google Scholar]
- 4.Daian F, Esper BS, Ashrafi N, Yu GQ, Luciano G, Moorthi S, and Luberto C (2020) Regulation of human sphingomyelin synthase 1 translation through its 5’-untranslated region. FEBS Lett 594, 3751–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rozhkova AV, Dmitrieva VG, Zhapparova ON, Sudarkina OY, Nadezhdina ES, Limborska SA, and Dergunova LV (2011) Human sphingomyelin synthase 1 gene (SMS1): organization, multiple mRNA splice variants and expression in adult tissues. Gene 481, 65–75 [DOI] [PubMed] [Google Scholar]
- 6.Dergunova LV, Rozhkova AV, Sudarkina OY, and Limborska SA (2013) The use of alternative polyadenylation in the tissue-specific regulation of human SMS1 gene expression. Mol Biol Rep 40, 6685–6690 [DOI] [PubMed] [Google Scholar]
- 7.Sudarkina OY, Filippenkov IB, Brodsky IB, Limborska SA, and Dergunova LV (2015) Comparative analysis of sphingomyelin synthase 1 gene expression at the transcriptional and translational levels in human tissues. Mol Cell Biochem 406, 91–99 [DOI] [PubMed] [Google Scholar]
- 8.Filippenkov IB, Sudarkina OY, Limborska SA, and Dergunova LV (2015) Circular RNA of the human sphingomyelin synthase 1 gene: Multiple splice variants, evolutionary conservatism and expression in different tissues. RNA Biol 12, 1030–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeang C, Varshney S, Wang R, Zhang Y, Ye D, and Jiang XC (2008) The domain responsible for sphingomyelin synthase (SMS) activity. Biochim Biophys Acta 1781, 610–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piotto S, Sessa L, Iannelli P, and Concilio S (2017) Computational study on human sphingomyelin synthase 1 (hSMS1). Biochim Biophys Acta Biomembr 1859, 1517–1525 [DOI] [PubMed] [Google Scholar]
- 11.Shakor ABA, Taniguchi M, Kitatani K, Hashimoto M, Asano S, Hayashi A, Nomura K, Bielawski J, Bielawska A, Watanabe K, Kobayashi T, Igarashi Y, Umehara H, Takeya H, and Okazaki T (2011) Sphingomyelin synthase 1-generated sphingomyelin plays an important role in transferrin trafficking and cell proliferation. J Biol Chem 286, 36053–36062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumoto G, Hashizume C, Watanabe K, Taniguchi M, and Okazaki T (2019) Deficiency of sphingomyelin synthase 1 but not sphingomyelin synthase 2 reduces bone formation due to impaired osteoblast differentiation. Mol Med 25, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taniguchi M, Tasaki T, Ninomiya H, Ueda Y, Kuremoto KI, Mitsutake S, Igarashi Y, Okazaki T, and Takegami T (2016) Sphingomyelin generated by sphingomyelin synthase 1 is involved in attachment and infection with Japanese encephalitis virus. Sci Rep 6, 37829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh EH, Yoon JE, Ko MS, Leem J, Yun JY, Hong CH, Cho YK, Lee SE, Jang JE, Baek JY, Yoo HJ, Kim SJ, Sung CO, Lim JS, Jeong WI, Back SH, Baek IJ, Torres S, Solsona-Vilarrasa E, Conde de la Rosa L, Garcia-Ruiz C, Feldstein AE, Fernandez-Checa JC, and Lee KU (2020) Sphingomyelin synthase 1 mediates hepatocyte pyroptosis to trigger nonalcoholic steatohepatitis. Gut [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wittmann A, Grimm MO, Scherthan H, Horsch M, Beckers J, Fuchs H, Gailus-Durner V, Hrabe de Angelis M, Ford SJ, Burton NC, Razansky D, Trumbach D, Aichler M, Walch AK, Calzada-Wack J, Neff F, Wurst W, Hartmann T, and Floss T (2016) Sphingomyelin Synthase 1 Is Essential for Male Fertility in Mice. PLoS One 11, e0164298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yano M, Watanabe K, Yamamoto T, Ikeda K, Senokuchi T, Lu M, Kadomatsu T, Tsukano H, Ikawa M, Okabe M, Yamaoka S, Okazaki T, Umehara H, Gotoh T, Song WJ, Node K, Taguchi R, Yamagata K, and Oike Y (2011) Mitochondrial dysfunction and increased reactive oxygen species impair insulin secretion in sphingomyelin synthase 1-null mice. J Biol Chem 286, 3992–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Fan Y, Liu J, Li Y, Huan C, Bui HH, Kuo MS, Park TS, Cao G, and Jiang XC (2012) Impact of sphingomyelin synthase 1 deficiency on sphingolipid metabolism and atherosclerosis in mice. Arteriosclerosis, thrombosis, and vascular biology 32, 1577–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yano M, Yamamoto T, Nishimura N, Gotoh T, Watanabe K, Ikeda K, Garan Y, Taguchi R, Node K, Okazaki T, and Oike Y (2013) Increased oxidative stress impairs adipose tissue function in sphingomyelin synthase 1 null mice. PLoS One 8, e61380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiao F, and Bowie JU (2005) The many faces of SAM. Sci STKE 2005, re7. [DOI] [PubMed] [Google Scholar]
- 20.Ichikawa S, Sakiyama H, Suzuki G, Hidari KI, and Hirabayashi Y (1996) Expression cloning of a cDNA for human ceramide glucosyltransferase that catalyzes the first glycosylation step of glycosphingolipid synthesis. Proc Natl Acad Sci U S A 93, 4638–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Futerman AH, and Pagano RE (1991) Determination of the intracellular sites and topology of glucosylceramide synthesis in rat liver. Biochem J 280 (Pt 2), 295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi Y, Nemoto-Sasaki Y, Matsumoto N, Hama K, Tanikawa T, Oka S, Saeki T, Kumasaka T, Koizumi T, Arai S, Wada I, Yokoyama K, Sugiura T, and Yamashita A (2018) Complex formation of sphingomyelin synthase 1 with glucosylceramide synthase increases sphingomyelin and decreases glucosylceramide levels. J Biol Chem 293, 17505–17522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Chiang YP, He M, Worgall TS, Zhou H, and Jiang XC (2021) Liver sphingomyelin synthase 1 deficiency causes steatosis, steatohepatitis, fibrosis, and tumorigenesis: An effect of glucosylceramide accumulation. iScience 24, 103449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Hailemariam TK, Zhou H, Li Y, Duckworth DC, Peake DA, Zhang Y, Kuo MS, Cao G, and Jiang XC (2007) Inhibition of sphingomyelin synthase (SMS) affects intracellular sphingomyelin accumulation and plasma membrane lipid organization. Biochim Biophys Acta 1771, 1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyaji M, Jin ZX, Yamaoka S, Amakawa R, Fukuhara S, Sato SB, Kobayashi T, Domae N, Mimori T, Bloom ET, Okazaki T, and Umehara H (2005) Role of membrane sphingomyelin and ceramide in platform formation for Fas-mediated apoptosis. The Journal of experimental medicine 202, 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van der Luit AH, Budde M, Zerp S, Caan W, Klarenbeek JB, Verheij M, and Van Blitterswijk WJ (2007) Resistance to alkyl-lysophospholipid-induced apoptosis due to downregulated sphingomyelin synthase 1 expression with consequent sphingomyelin- and cholesterol-deficiency in lipid rafts. Biochem J 401, 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anjum F, Joshi K, Grinkina N, Gowda S, Cutaia M, and Wadgaonkar R (2012) Role of sphingomyelin synthesis in pulmonary endothelial cell cytoskeletal activation and endotoxin-induced lung injury. Am J Respir Cell Mol Biol 47, 94–103 [DOI] [PubMed] [Google Scholar]
- 28.Ding T, Li Z, Hailemariam T, Mukherjee S, Maxfield FR, Wu MP, and Jiang XC (2008) SMS overexpression and knockdown: impact on cellular sphingomyelin and diacylglycerol metabolism, and cell apoptosis. J Lipid Res 49, 376–385 [DOI] [PubMed] [Google Scholar]
- 29.Liu J, Zhang H, Li Z, Hailemariam TK, Chakraborty M, Jiang K, Qiu D, Bui HH, Peake DA, Kuo MS, Wadgaonkar R, Cao G, and Jiang XC (2009) Sphingomyelin synthase 2 is one of the determinants for plasma and liver sphingomyelin levels in mice. Arterioscler Thromb Vasc Biol 29, 850–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitsutake S, Zama K, Yokota H, Yoshida T, Tanaka M, Mitsui M, Ikawa M, Okabe M, Tanaka Y, Yamashita T, Takemoto H, Okazaki T, Watanabe K, and Igarashi Y (2011) Dynamic modification of sphingomyelin in lipid microdomains controls development of obesity, fatty liver, and type 2 diabetes. J Biol Chem 286, 28544–28555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Zhang H, Liu J, Liang CP, Li Y, Li Y, Teitelman G, Beyer T, Bui HH, Peake DA, Zhang Y, Sanders PE, Kuo MS, Park TS, Cao G, and Jiang XC (2011) Reducing plasma membrane sphingomyelin increases insulin sensitivity. Mol Cell Biol 31, 4205–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Dong J, Ding T, Kuo MS, Cao G, Jiang XC, and Li Z (2013) Sphingomyelin synthase 2 activity and liver steatosis: an effect of ceramide-mediated peroxisome proliferator-activated receptor gamma2 suppression. Arteriosclerosis, thrombosis, and vascular biology 33, 1513–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan Y, Shi F, Liu J, Dong J, Bui HH, Peake DA, Kuo MS, Cao G, and Jiang XC (2010) Selective reduction in the sphingomyelin content of atherogenic lipoproteins inhibits their retention in murine aortas and the subsequent development of atherosclerosis. Arterioscler Thromb Vasc Biol 30, 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim YJ, Sano T, Nabetani T, Asano Y, and Hirabayashi Y (2012) GPRC5B activates obesity-associated inflammatory signaling in adipocytes. Sci Signal 5, ra85. [DOI] [PubMed] [Google Scholar]
- 35.Kim YJ, Greimel P, and Hirabayashi Y (2018) GPRC5B-Mediated Sphingomyelin Synthase 2 Phosphorylation Plays a Critical Role in Insulin Resistance. iScience 8, 250–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshikawa Y, Yoshizawa T, Domae E, Hirai Y, Kamada A, Okazaki T, and Ikeo T (2019) Knockdown of sphingomyelin synthase 2 inhibits osteoclastogenesis by decreasing RANKL expression in mouse primary osteoblasts. Biomed Res 40, 189–196 [DOI] [PubMed] [Google Scholar]
- 37.Sugimoto M, Hamada T, Wakabayasi M, Yoshioka T, Kato H, Konishi H, Nagai R, Suzuki M, Numata Y, Igarashi Y, and Yukioka H (2020) Sphingomyelin synthase 2 loss suppresses steatosis but exacerbates fibrosis in the liver of mice fed with choline-deficient, L-amino acid-defined, high-fat diet. Biochem Biophys Res Commun 533, 1269–1275 [DOI] [PubMed] [Google Scholar]
- 38.Xue J, Yu Y, Zhang X, Zhang C, Zhao Y, Liu B, Zhang L, Wang L, Chen R, Gao X, Jiao P, Song G, Jiang XC, and Qin S (2019) Sphingomyelin Synthase 2 Inhibition Ameliorates Cerebral Ischemic Reperfusion Injury Through Reducing the Recruitment of Toll-Like Receptor 4 to Lipid Rafts. J Am Heart Assoc 8, e012885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Chiang YP, He M, Zhang K, Zheng J, Wu W, Cai J, Chen Y, Chen G, Chen Y, Dong J, Worgall TS, and Jiang XC (2021) Effect of Liver Total Sphingomyelin Synthase Deficiency on Plasma Lipid Metabolism. Biochim Biophys Acta Mol Cell Biol Lipids, 158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Huan C, Chakraborty M, Zhang H, Lu D, Kuo MS, Cao G, and Jiang XC (2009) Macrophage sphingomyelin synthase 2 deficiency decreases atherosclerosis in mice. Circ Res 105, 295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugimoto M, Shimizu Y, Zhao S, Ukon N, Nishijima K, Wakabayashi M, Yoshioka T, Higashino K, Numata Y, Okuda T, Tamaki N, Hanamatsu H, Igarashi Y, and Kuge Y (2016) Characterization of the role of sphingomyelin synthase 2 in glucose metabolism in whole-body and peripheral tissues in mice. Biochim Biophys Acta 1861, 688–702 [DOI] [PubMed] [Google Scholar]
- 42.Sakamoto H, Yoshida T, Sanaki T, Shigaki S, Morita H, Oyama M, Mitsui M, Tanaka Y, Nakano T, Mitsutake S, Igarashi Y, and Takemoto H (2017) Possible roles of long-chain sphingomyelines and sphingomyelin synthase 2 in mouse macrophage inflammatory response. Biochemical and biophysical research communications 482, 202–207 [DOI] [PubMed] [Google Scholar]
- 43.Lou B, Dong J, Li Y, Ding T, Bi T, Li Y, Deng X, Ye D, and Jiang XC (2014) Pharmacologic inhibition of sphingomyelin synthase (SMS) activity reduces apolipoprotein-B secretion from hepatocytes and attenuates endotoxin-mediated macrophage inflammation. PloS one 9, e102641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y, Huang T, Zhen X, Li Y, Mo M, Ye D, and Cheng N (2019) A selective sphingomyelin synthase 2 inhibitor ameliorates diet induced insulin resistance via the IRS-1/Akt/GSK-3beta signaling pathway. Pharmazie 74, 553–558 [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi K, Forte TM, Taniguchi S, Ishida BY, Oka K, and Chan L (2000) The db/db mouse, a model for diabetic dyslipidemia: molecular characterization and effects of Western diet feeding. Metabolism 49, 22–31 [DOI] [PubMed] [Google Scholar]
- 46.Mo M, Yang J, Jiang XC, Cao Y, Fei J, Chen Y, Qi X, Chu Y, Zhou L, and Ye D (2018) Discovery of 4-Benzyloxybenzo[d]isoxazole-3-amine Derivatives as Highly Selective and Orally Efficacious Human Sphingomyelin Synthase 2 Inhibitors that Reduce Chronic Inflammation in db/db Mice. J Med Chem 61, 8241–8254 [DOI] [PubMed] [Google Scholar]
- 47.Barcelo-Coblijn G, Martin ML, de Almeida RF, Noguera-Salva MA, Marcilla-Etxenike A, Guardiola-Serrano F, Luth A, Kleuser B, Halver JE, and Escriba PV (2011) Sphingomyelin and sphingomyelin synthase (SMS) in the malignant transformation of glioma cells and in 2-hydroxyoleic acid therapy. Proc Natl Acad Sci U S A 108, 19569–19574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandez-Garcia P, Rossello CA, Rodriguez-Lorca R, Beteta-Gobel R, Fernandez-Diaz J, Llado V, Busquets X, and Escriba PV (2019) The Opposing Contribution of SMS1 and SMS2 to Glioma Progression and Their Value in the Therapeutic Response to 2OHOA. Cancers (Basel) 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lou B, Liu Q, Hou J, Kabir I, Liu P, Ding T, Dong J, Mo M, Ye D, Chen Y, Bui HH, Roth K, Cao Y, and Jiang XC (2018) 2-Hydroxy-oleic acid does not activate sphingomyelin synthase activity. J Biol Chem 293, 18328–18336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tafesse FG, Ternes P, and Holthuis JC (2006) The multigenic sphingomyelin synthase family. J Biol Chem 281, 29421–29425 [DOI] [PubMed] [Google Scholar]
- 51.Ding T, Kabir I, Li Y, Lou C, Yazdanyar A, Xu J, Dong J, Zhou H, Park T, Boutjdir M, Li Z, and Jiang XC (2015) All members in the sphingomyelin synthase gene family have ceramide phosphoethanolamine synthase activity. J Lipid Res 56, 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vacaru AM, Tafesse FG, Ternes P, Kondylis V, Hermansson M, Brouwers JF, Somerharju P, Rabouille C, and Holthuis JC (2009) Sphingomyelin synthase-related protein SMSr controls ceramide homeostasis in the ER. J Cell Biol 185, 1013–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ternes P, Brouwers JF, van den Dikkenberg J, and Holthuis JC (2009) Sphingomyelin synthase SMS2 displays dual activity as ceramide phosphoethanolamine synthase. J Lipid Res 50, 2270–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding T, Kabir I, Li Y, Lou C, Yazdanyar A, Xu J, Dong J, Zhou H, Park T, Boutjdir M, Li Z, and Jiang XC (2015) All members in the sphingomyelin synthase gene family have ceramide phosphoethanolamine synthase activity. J Lipid Res 56, 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tafesse FG, Vacaru AM, Bosma EF, Hermansson M, Jain A, Hilderink A, Somerharju P, and Holthuis JC (2014) Sphingomyelin synthase-related protein SMSr is a suppressor of ceramide-induced mitochondrial apoptosis. J Cell Sci 127, 445–454 [DOI] [PubMed] [Google Scholar]
- 56.Siow DL, and Wattenberg BW (2012) Mammalian ORMDL proteins mediate the feedback response in ceramide biosynthesis. J Biol Chem 287, 40198–40204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bickert A, Ginkel C, Kol M, vom Dorp K, Jastrow H, Degen J, Jacobs RL, Vance DE, Winterhager E, Jiang XC, Dormann P, Somerharju P, Holthuis JC, and Willecke K (2015) Functional characterization of enzymes catalyzing ceramide phosphoethanolamine biosynthesis in mice. J Lipid Res 56, 821–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panevska A, Skocaj M, Krizaj I, Macek P, and Sepcic K (2019) Ceramide phosphoethanolamine, an enigmatic cellular membrane sphingolipid. Biochim Biophys Acta Biomembr 1861, 1284–1292 [DOI] [PubMed] [Google Scholar]
- 59.Cabukusta B, Kol M, Kneller L, Hilderink A, Bickert A, Mina JG, Korneev S, and Holthuis JC (2017) ER residency of the ceramide phosphoethanolamine synthase SMSr relies on homotypic oligomerization mediated by its SAM domain. Sci Rep 7, 41290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murakami C, Hoshino F, Sakai H, Hayashi Y, Yamashita A, and Sakane F (2020) Diacylglycerol kinase delta and sphingomyelin synthase-related protein functionally interact via their sterile alpha motif domains. J Biol Chem 295, 2932–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kadamur G, and Ross EM (2013) Mammalian phospholipase C. Annu Rev Physiol 75, 127–154 [DOI] [PubMed] [Google Scholar]
- 62.Cheng M, Bhujwalla ZM, and Glunde K (2016) Targeting Phospholipid Metabolism in Cancer. Front Oncol 6, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Exton JH (1994) Phosphatidylcholine breakdown and signal transduction. Biochim Biophys Acta 1212, 26–42 [DOI] [PubMed] [Google Scholar]
- 64.van Meer G, Voelker DR, and Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vazquez-Boland JA, Kocks C, Dramsi S, Ohayon H, Geoffroy C, Mengaud J, and Cossart P (1992) Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect Immun 60, 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Amtmann E (1996) The antiviral, antitumoural xanthate D609 is a competitive inhibitor of phosphatidylcholine-specific phospholipase C. Drugs Exp Clin Res 22, 287–294 [PubMed] [Google Scholar]
- 67.Barker AP, Vasil AI, Filloux A, Ball G, Wilderman PJ, and Vasil ML (2004) A novel extracellular phospholipase C of Pseudomonas aeruginosa is required for phospholipid chemotaxis. Mol Microbiol 53, 1089–1098 [DOI] [PubMed] [Google Scholar]
- 68.Adibhatla RM, Hatcher JF, and Gusain A (2012) Tricyclodecan-9-yl-xanthogenate (D609) mechanism of actions: a mini-review of literature. Neurochem Res 37, 671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neuwald AF (1997) An unexpected structural relationship between integral membrane phosphatases and soluble haloperoxidases. Protein Sci 6, 1764–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murakami C, and Sakane F (2021) Sphingomyelin synthase-related protein generates diacylglycerol via the hydrolysis of glycerophospholipids in the absence of ceramide. J Biol Chem, 100454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chiang YP, Li Z, Chen Y, Cao Y, and Jiang XC (2021) Sphingomyelin synthase related protein is a mammalian phosphatidylethanolamine phospholipase C. Biochim Biophys Acta Mol Cell Biol Lipids 1866, 159017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luberto C, and Hannun YA (1998) Sphingomyelin synthase, a potential regulator of intracellular levels of ceramide and diacylglycerol during SV40 transformation. Does sphingomyelin synthase account for the putative phosphatidylcholine-specific phospholipase C? J Biol Chem 273, 14550–14559 [DOI] [PubMed] [Google Scholar]
- 73.Huang KP (1989) The mechanism of protein kinase C activation. Trends Neurosci 12, 425–432 [DOI] [PubMed] [Google Scholar]
- 74.Pettitt TR, Martin A, Horton T, Liossis C, Lord JM, and Wakelam MJ (1997) Diacylglycerol and phosphatidate generated by phospholipases C and D, respectively, have distinct fatty acid compositions and functions. Phospholipase D-derived diacylglycerol does not activate protein kinase C in porcine aortic endothelial cells. J Biol Chem 272, 17354–17359 [DOI] [PubMed] [Google Scholar]


