Abstract
An aminooxy click chemistry (AOCC) strategy was used to synthesize nucleoside building blocks for incorporation during solid-support synthesis of oligonucleotides to enable bis-homo and bis-hetero conjugation of various biologically relevant ligands. The bis-homo aminooxy conjugation leads to bivalent ligand presentation, whereas the bis-hetero conjugation allows the placement of different ligands with either the same or different chemical linkages. This facile synthetic methodology allows introduction of two different ligands with different biological functions simultaneously.
Aminooxy or O-amino groups (-O-NH2) act as stronger nucleophiles than amines, due to the second lone-pair-bearing oxygen adjacent to the nucleophilic nitrogen. Moreover, their low basicity allows them to form more stable imines than Schiff bases.1 This phenomenon was first observed by Jencks and Carriuolo in 1960,2 and Edwards and Pearson called it the “alpha effect” in 1962.3 Aminooxy-functionalized sugars, nucleosides, and peptides have attracted interest as they can be easily synthesized and used for oxime ligation4−8 and N-oxyamide functionalization.9−11 A bis-functionalized aminooxy group was used for sugar modification in oligonucleotide chemistry,12 and nucleosides with a 5′-aminooxy group have been used in the synthesis of oligonucleotides with modified backbones;13 both syntheses involved very reactive formaldehyde.
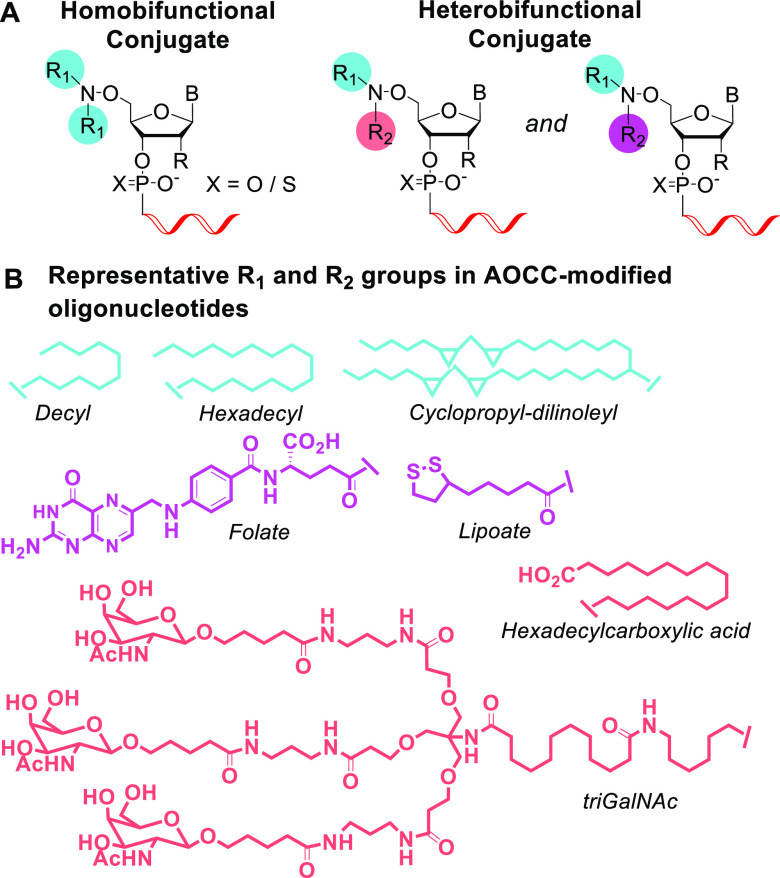
Conjugation chemistry has invigorated oligonucleotide therapeutics by enabling incorporation of moieties that modulate pharmacokinetics and that result in delivery into particular cell types, as demonstrated by the clinical success of small interfering RNAs (siRNAs) conjugated to triantennary N-acetylgalactosamine (tri-GalNAc), the ligand for the hepatocyte-specific receptor asialoglycoprotein.14 In other examples, conjugation to lipophilic molecules like cholesterol15 and fatty acids16 results in broad tissue distribution and cellular uptake in the liver and central nervous system;17,18 folic acid conjugation results in cancer cell targeting; and conjugation to cyclic RGD peptides results in uptake into cells that express integrins.19
The biocleavable oxime linkage has been used in oligonucleotide conjugation.1,5,6,8,20 Here, to facilitate bis-homo and bis-hetero conjugation, we developed a “click-type” aminooxy-based chemistry that we call aminooxy click chemistry (AOCC). The reported reaction meets the Sharpless definition of click chemistry:21 The reaction is modular and wide in scope and gives very high yields, and product isolation is simple. Two different strategies were envisioned: AOCC conjugate monomers could be synthesized as amidites and incorporated during solid-phase oligonucleotide synthesis, or precursors of aminooxy groups could be introduced into the oligonucleotide strands and functionalized postsynthetically. In the first approach, demonstrated in this report, a combination of two different aldehydes and a combination of an aldehyde with an acid were used for the bis-hetero ligation (Figure 1). Ligands with different biological functions were introduced, demonstrating the broad scope of the strategy.
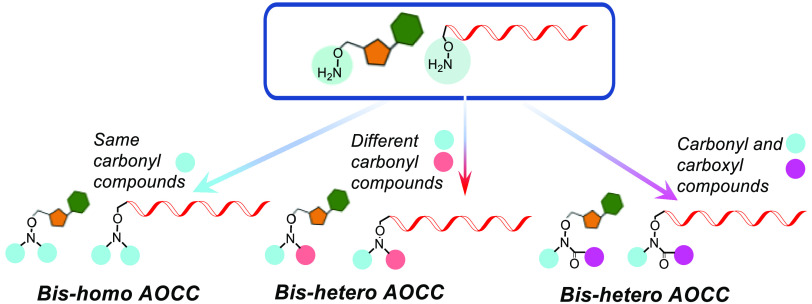
Figure 1.
(A) Schematic of 5′-end modification of oligonucleotides with AOCC-mediated bis-homo and bis-hetero conjugation. (B) Representative targeting and pharmacokinetic-modifier ligands.
For construction of building blocks to enable bis-homo and bis-hetero ligand conjugation, we synthesized nucleosides containing N-hydroxyphthalimido (OPhth), the precursors of O-amino. To generate 5′-OPhth-modified nucleosides, 2′-O-methyl (2′-OMe) and locked nucleic acid (LNA) nucleosides with protected 3′-OH were used. Mitsunobu reaction of compounds 1(22) and 8 afforded the OPhth-containing scaffolds 2 and 9, respectively.
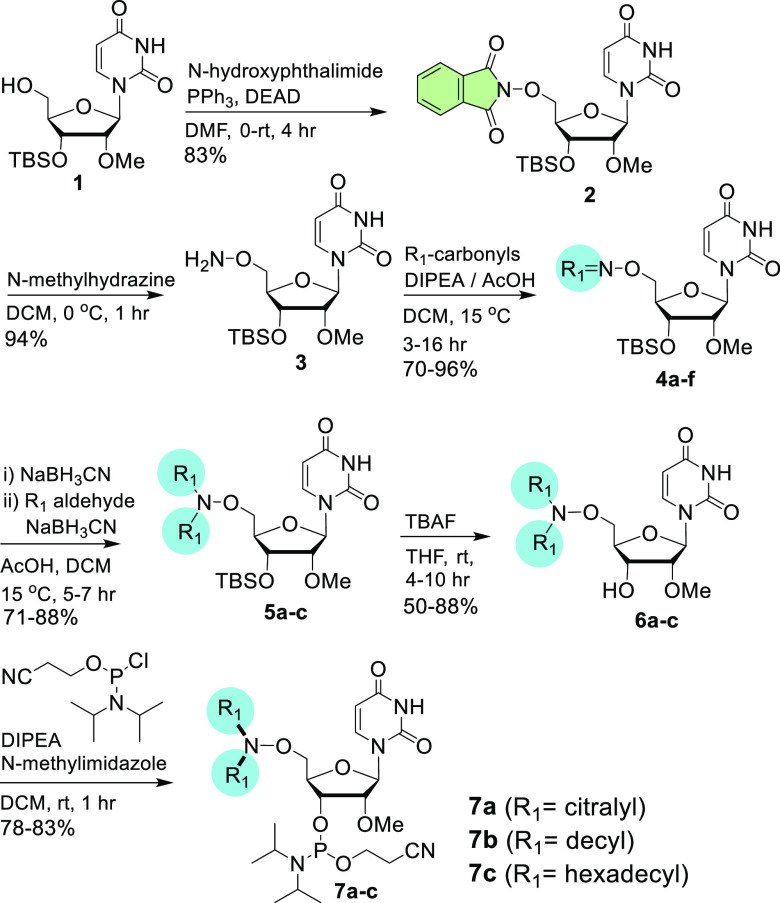
From these precursors, we first synthesized bis-homo ligand-containing modified nucleosides by generating an aminooxy linkage. Various aldehydes and ketones were used for facile oxime bond formation through a 5′-aminooxy functionality. For the oximes derived from aldehydes, an identical aldehyde was then attached to the aminooxy nitrogen atom through reductive amination. To illustrate this, 2 was deprotected in the presence of N-methylhydrazine to afford 3, which contains the aminooxy group. Reaction of 3 with carbonyl compounds [e.g., citral, decanal, hexadecanal, dilinoleyl aldehyde, cyclopropyl-dilinoleyl ketone, and protected tri-GalNAc aldehyde 3S (Scheme S1 and Table S1 in the Supporting Information)] resulted in oximes 4a–f either under basic or acidic conditions. The E- and Z-isomers of these oximes eluted together during column purification. The oxime was reduced with sodium cyanoborohydride under acidic conditions to afford the substituted aminooxy amine, which was reacted again with the same aldehyde. Thus, oximes 4a, 4b, and 4c were reacted with citral, decanal, and hexadecanal, respectively, under reductive amination conditions to produce compounds 5a, 5b, and 5c. The 3′-OTBS groups of 5a–c were removed by treatment with tetrabutylammonium fluoride (TBAF) to obtain 6a–c with free 3′-OH groups. Compounds 6a–c were then phosphitylated to afford bis-homo-AOCC-modified pyrimidine nucleoside phosphoramidite building blocks 7a–c (Scheme 1). Analogous purine compounds were synthesized using the same route.
Scheme 1. Synthesis of Bis-homo-aminooxy-modified 2′-OMe-uridine Building Blocks.
For 4a–7a, R1 is citralyl; for 4b–7b, R1 is decyl; for 4c–7c, R1 is hexadecyl; for 4d, R1 is dilinoleyl; for 4e, R1 is cyclopropyl-dilinoleyl; and for 4f, R1 is protected tri-GalNAc (structure shown in Scheme 3).
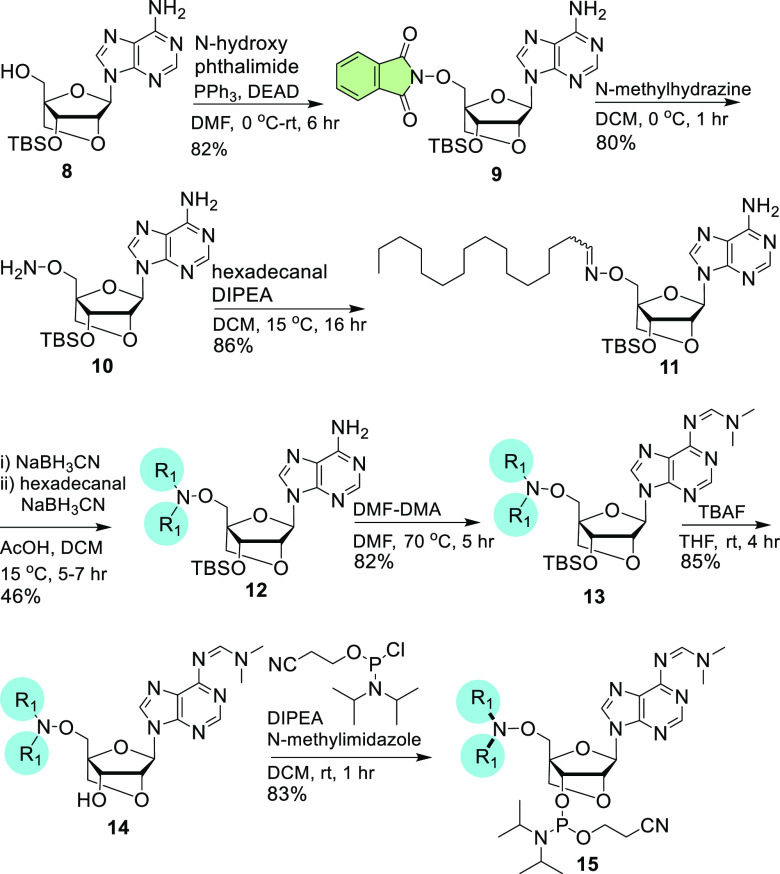
The adenosine LNA analogue 9 was prepared using an approach similar to that used for the synthesis of compound 2 from 3′-O-silyl-protected LNA-adenosine 8. The 5′-OPhth of compound 9 was deprotected to afford 10 in the presence of N-methylhydrazine. Compound 10 was reacted with hexadecanal to obtain oxime 11, which was subsequently converted to the bis-homoligated compound 12 using hexadecanal under reductive amination conditions. An additional step resulted in protection of the exocyclic N6-amine of adenosine with an amidine group to yield 13, and 3′-O-silyl deprotection of 13 produced 14. Phosphitylation of 14 produced the phosphoramidite 15 (Scheme 2).
Scheme 2. Synthesis of Bis-homo-Conjugated LNA-adenosine Building Blocks.
For 12–15, R1 is hexadecyl.
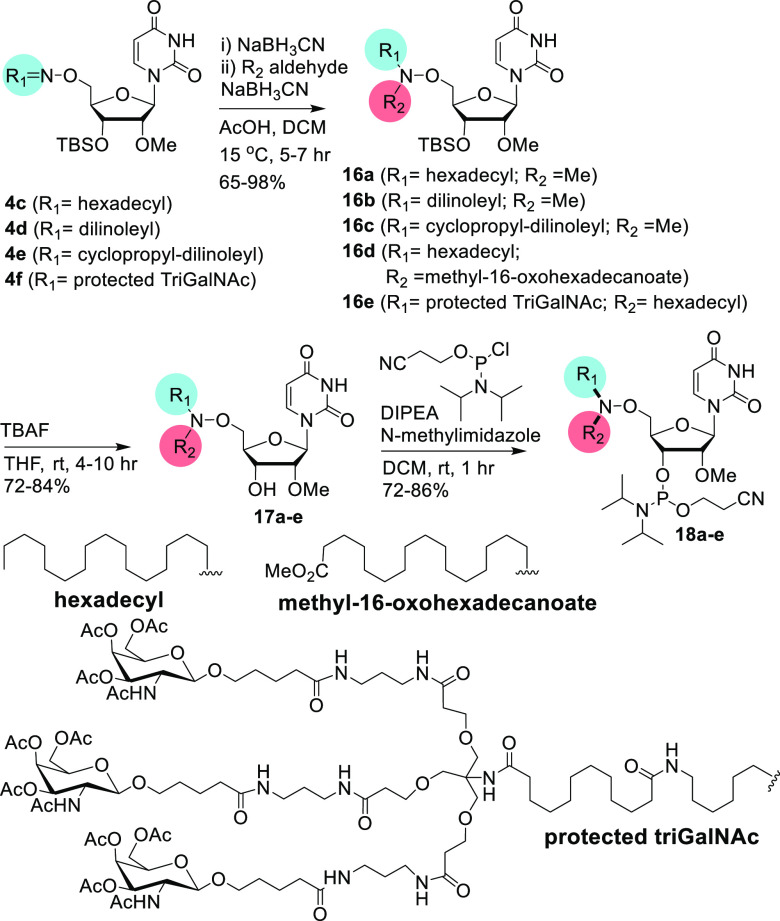
To synthesize the bis-hetero building blocks that allow introduction of two different ligands, we initially reacted three lipophilic oximes, 4c, 4d, and 4e, with formaldehyde to afford simple bis-hetero-AOCC analogues 16a, 16b, and 16c, respectively. Additionally, lipophilic oxime 4c and tri-GalNAc-conjugated oxime 4f were reacted with methyl-16-oxohexadecanoate and hexadecanal to afford 16d and 16e, respectively, under reductive amination conditions. These bis-hetero AOCC products were subsequently desilylated to obtain 17a–e, which were converted to the respective phosphoramidites 18a–e (Scheme 3). For the synthesis of bis-hetero building blocks, bulky ligands like tri-GalNAc or with keto-group-like dilinoleyl and cyclopropyl-dilinoleyl ketones were installed first for facile conversion and better yields.
Scheme 3. Synthesis of Bis-hetero-aminooxy-modified 2′-OMe-uridine Building Blocks.
For 16a–18a, R1 is hexadecyl, and R2 is CH3; for 16b–18b, R1 is dilinoleyl, and R2 is CH3; for 16c–18c, R1 is cyclopropyl-dilinoleyl, and R2 is CH3; for 16d–18d, R1 is hexadecyl, and R2 is methyl-16-oxohexadecanoate; for 16e–18e, R1 is protected tri-GalNAc, and R2 is hexadecyl.
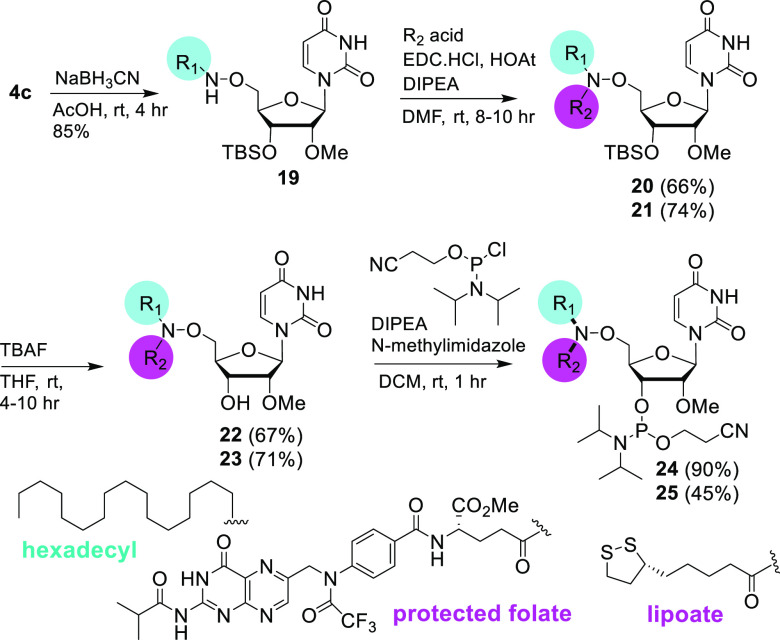
We also evaluated bis-heteroaminooxy-modified building blocks where the second ligand (R2) was an acid. Previously, this chemistry was used for conjugation to a neutral hydroxamate-modified DNA backbone.23 For this, 4c was reduced to afford the lipophilic aminooxy amine 19 and coupled with the protected folic acid 7S (Scheme S2 in Supporting Information) or lipoic acid to obtain the bis-hetero conjugates 20 and 21, respectively. The folate conjugation was achieved through the γ-carboxyl group.24 The amide coupling reaction was done using standard amide coupling conditions.25,26 The 3′-OTBS groups of 20 and 21 were deprotected to produce 22 and 23, respectively, which were converted to the phosphoramidites 24 and 25 in moderate to good yields (Scheme 4).
Scheme 4. Synthesis of Bis-hetero-aminooxy-modified 2′-OMe-uridine Building Blocks.
For 19, R1 is hexadecyl; for 20, 22, and 24, R1 is hexadecyl, and R2 is protected folate; and for 21, 23, and 25, R1 is hexadecyl, and R2 is lipoate.
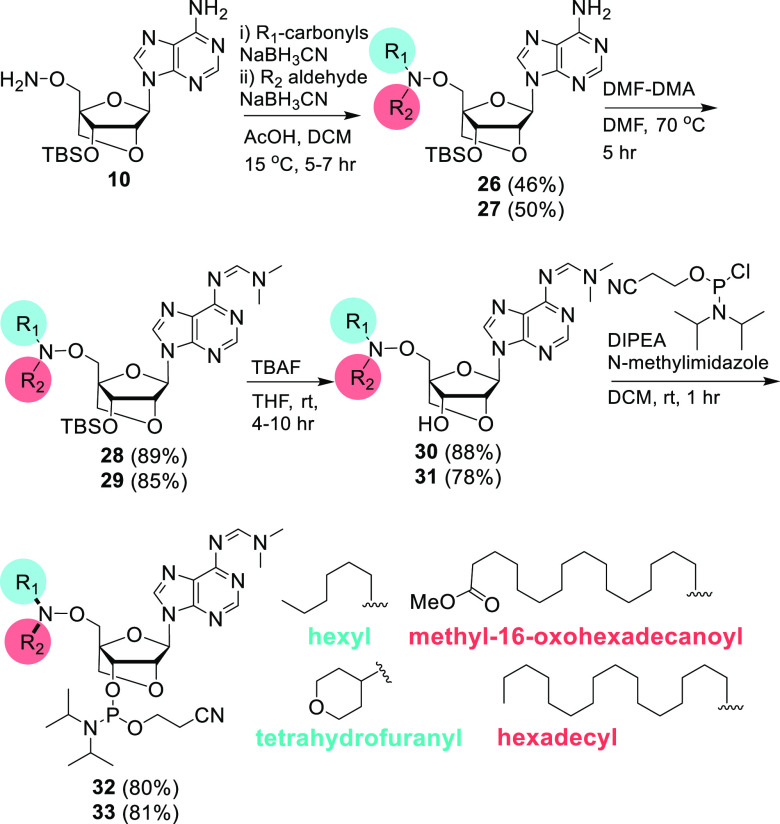
Finally, we tested the “click”-like behavior of the AOCC through a sequential one-pot reductive amination of 10 in the presence of two different carbonyl compounds. The LNA-A intermediate 10 with a 5′-aminooxy group was reacted first with hexanal. Then, without isolation of the oxime, the reductive amination step was performed with methyl-16-oxohexadecanoate to afford the bis-hetero product 26 in moderate yield. We did not observe the formation of the bis-homo-AOCC product upon incorporation of two units of hexanal/methyl-16-oxohexadecanoate. Similarly, a sequential one-pot reaction with tetrahydro-4H-pyran-4-one and hexadecanal produced the bis-hetero product 27 in moderate yield. Here also, the order of addition for the carbonyl compounds was important. For 27, addition of the ketone, tetrahydro-4H-pyran-4-one, followed by the aldehyde hexadecanal was more efficient than the reverse order. Amidine protection of 26 and 27 with DMF-DMA afforded 28 and 29, which were then desilylated with TBAF to yield 30 and 31, respectively. The phosphitylation reaction of 30 and 31 furnished the bis-heteroaminooxy-modified LNA-adenosine building blocks 32 and 33 in good yields (Scheme 5).
Scheme 5. Sequential One-Pot AOCC Reaction for the Synthesis of Bis-hetero-aminooxy-modified LNA-adenosine Building Blocks.
For 26, 28, 30, and 32, R1 is hexyl, and R2 is methyl-16-oxohexadecanoyl. For 27, 29, 31, and 33, R1 is tetrahydrofuranyl, and R2 is hexadecyl.
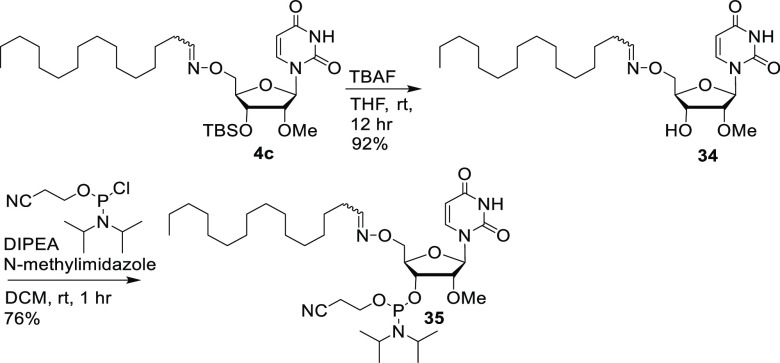
To demonstrate the representative first-generation oxime conjugates, we synthesized a lipophilic oxime building block. Intermediate 4c was desilylated with TBAF to afford compound 34 in good yield. Compound 34 was phosphitylated to obtain the starting monomer for oxime conjugation, the 5′-oxime-hexadecyl conjugate 35 (Scheme 6).
Scheme 6. Synthesis of the Oxime C16 Lipid Building Block (35).
The AOCC–conjugate building blocks were incorporated at the 5′ ends of oligonucleotides using standard oligonucleotide synthesis conditions. The oligonucleotides used were the sense strands of siRNAs targeting the SOD1, β-cat, and Apo-B mRNAs (Table 1). In these oligonucleotides, the modified monomers replaced the U nucleotide during solid-phase synthesis under standard conditions; the oligonucleotide conjugates were obtained with good coupling efficiencies and yields even with large ligands like folate and tri-GalNAc in combination with C16 (Figure 2). After deprotection and cleavage from the support under standard conditions, the conjugates were purified and characterized. As this was, to our knowledge, the first lipoic acid conjugation to oligonucleotides, we confirmed that there was no oxidation or degradation of disulfide-containing rings (Figure 2, ON3).
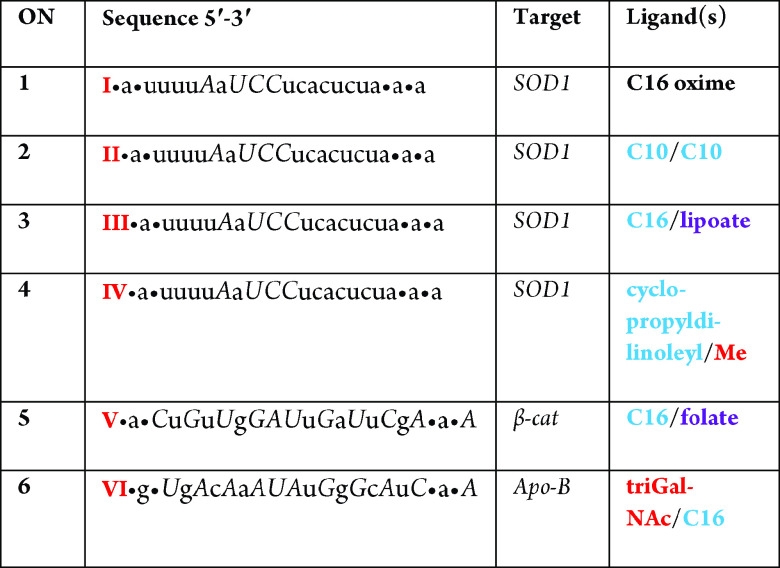
Table 1. Sequences, mRNA Targets, Chemistry, and Ligand Features of siRNA Sense Strand Oligonucleotides (ON) with AOCC Building Blocks at the 5′ Enda.
Figure 2.
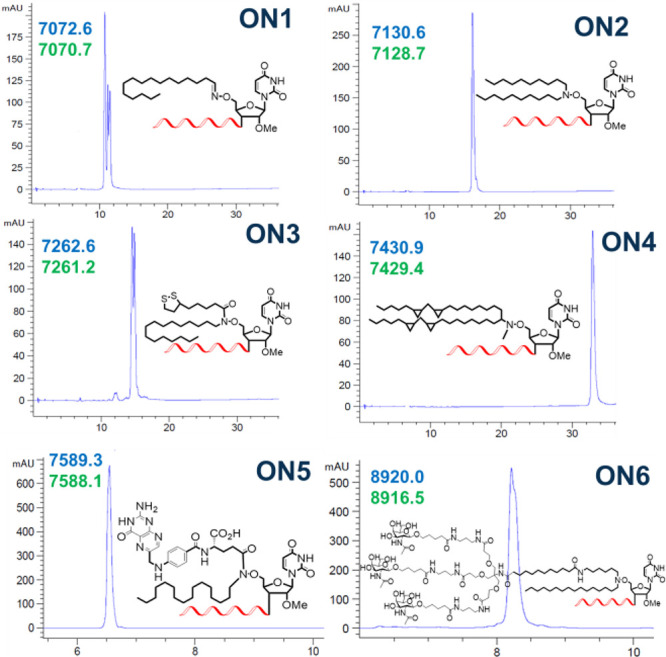
HPLC elution profiles (details in SI) of AOCC-modified oligonucleotides ON1–ON6 with calculated (blue) and observed (green) masses. The multiplicity of peaks is due to diastereomeric chiral phosphorothioates and/or oxime rotamers.
In summary, we have synthesized several nucleoside monomer building blocks equipped with the highly reactive nucleophilic aminooxy functionalities. These groups were converted into bis-homo and bis-hetero building blocks that were functionalized with two of the same or two different moieties, respectively. Both bis-homo- and bis-heterotype nucleoside conjugates were successfully incorporated at the 5′-ends of oligonucleotides. The ability to incorporate two different functionalities should prove especially useful in improving the therapeutic potential of all classes of therapeutic oligonucleotides. For example, siRNAs with single targeting agents like GalNAc, folate, and lipids enable tissue-type targeting. Use of these moieties as bis-hetero conjugates with the appropriate choice of a second ligand could improve tissue-specific delivery, cellular uptake, endosomal release, and pharmacodynamics.27,28 To date, conjugation of ligands has been performed mostly at the 3′ end14,15 of the sense strand of the siRNA. The strategy reported here will make 5′-end conjugation easier from the oligonucleotide synthesis and purification perspective. Conjugation at the 5′-position of the siRNA sense strand may improve antisense strand selection by the RNA interference machinery, thus eliminating sense-strand-mediated off-target effects,29 and should also enhance resistance to 5′-exonucleases.30 Work to test these possibilities is in progress.
Acknowledgments
We dedicate this publication to Professor John A. Gerlt for his broad contributions to science and for introducing oligonucleotide chemistry and enzymology of nucleic acids to the corresponding author at the JAG Laboratories.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.orglett.2c00988.
Experimental and compound characterization (PDF)
Author Contributions
The manuscript was written through contributions of all authors.
The authors declare the following competing financial interest(s): All authors are employees of Alnylam Pharmaceuticals.
Supplementary Material
References
- Kolmel D. K.; Kool E. T. Oximes and Hydrazones in Bioconjugation: Mechanism and Catalysis. Chem. Rev. 2017, 117, 10358–10376. 10.1021/acs.chemrev.7b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jencks W. P.; Carriuolo J. General Base Catalysis of the Aminolysis of Phenyl Acetate1. J. Am. Chem. Soc. 1960, 82, 675–681. 10.1021/ja01488a044. [DOI] [Google Scholar]
- Edwards J. O.; Pearson R. G. The Factors Determining Nucleophilic Reactivities. J. Am. Chem. Soc. 1962, 84, 16–24. 10.1021/ja00860a005. [DOI] [Google Scholar]
- Armstrong J. I.; Ge X.; Verdugo D. E.; Winans K. A.; Leary J. A.; Bertozzi C. R. A Library Approach to the Generation of Bisubstrate Analogue Sulfotransferase Inhibitors. Org. Lett. 2001, 3, 2657–2660. 10.1021/ol0162217. [DOI] [PubMed] [Google Scholar]
- Salo H.; Virta P.; Hakala H.; Prakash T. P.; Kawasaki A. M.; Manoharan M.; Lönnberg H. Aminooxy Functionalized Oligonucleotides: Preparation, On-Support Derivatization, and Postsynthetic Attachment to Polymer Support. Bioconjugate Chem. 1999, 10, 815–823. 10.1021/bc990021m. [DOI] [PubMed] [Google Scholar]
- Meyer A.; Vasseur J.-J.; Dumy P.; Morvan F. Phthalimide-Oxy Derivatives for 3′- or 5′-Conjugation of Oligonucleotides by Oxime Ligation and Circularization of DNA by ″Bis- or Tris-Click″ Oxime Ligation. Eur. J. Org. Chem. 2017, 2017, 6931–6941. 10.1002/ejoc.201701317. [DOI] [Google Scholar]
- Rodriguez E. C.; Marcaurelle L. A.; Bertozzi C. R. Aminooxy-, Hydrazide-, and Thiosemicarbazide-Functionalized Saccharides: Versatile Reagents for Glycoconjugate Synthesis. J. Org. Chem. 1998, 63, 7134–7135. 10.1021/jo981351n. [DOI] [PubMed] [Google Scholar]
- Noel M.; Clement-Blanc C.; Meyer A.; Vasseur J.-J.; Morvan F. Solid Supports for the Synthesis of 3′-Aminooxy Deoxy- or Ribo-oligonucleotides and Their 3′-Conjugation by Oxime Ligation. J. Org. Chem. 2019, 84, 14854–14860. 10.1021/acs.joc.9b00848. [DOI] [PubMed] [Google Scholar]
- Lee M.-r.; Lee J.; Baek B.-h.; Shin I. The First Solid-Phase Synthesis of Oligomeric α-Aminooxy Peptides. Synlett 2003, 2003, 0325–0328. 10.1055/s-2003-37127. [DOI] [Google Scholar]
- Xie J.; Peyrat S. Synthesis of Thymidine Dimers from 5′-O-Aminothymidine. Synthesis 2012, 44, 1718–1724. 10.1055/s-0031-1289759. [DOI] [Google Scholar]
- Yang D.; Chang X. W.; Zhang D. W.; Jiang Z. F.; Song K. S.; Zhang Y. H.; Zhu N. Y.; Weng L. H.; Chen M. Q. Chiral alpha-aminoxy acid/achiral cyclopropane alpha-aminoxy acid unit as a building block for constructing the alpha N-O helix. J. Org. Chem. 2010, 75, 4796–805. 10.1021/jo100810m. [DOI] [PubMed] [Google Scholar]
- Prakash T. P.; Manoharan M.; Kawasaki A. M.; Lesnik E. A.; Owens S. R.; Vasquez G. 2‘-O-{2-[N,N-(Dialkyl)aminooxy]ethyl}- Modified Antisense Oligonucleotides. Org. Lett. 2000, 2, 3995–3998. 10.1021/ol006555g. [DOI] [PubMed] [Google Scholar]
- Morvan F.; Sanghvi Y. S.; Perbost M.; Vasseur J.-J.; Bellon L. Oligonucleotide Mimics for Antisense Therapeutics: Solution Phase and Automated Solid-Support Synthesis of MMI Linked Oligomers. J. Am. Chem. Soc. 1996, 118, 255–6. 10.1021/ja9533959. [DOI] [Google Scholar]
- Nair J. K.; Willoughby J. L. S.; Chan A.; Charisse K.; Alam M. R.; Wang Q.; Hoekstra M.; Kandasamy P.; Kel’in A. V.; Milstein S.; Taneja N.; O’Shea J.; Shaikh S.; Zhang L.; van der Sluis R. J.; Jung M. E.; Akinc A.; Hutabarat R.; Kuchimanchi S.; Fitzgerald K.; Zimmermann T.; van Berkel T. J. C.; Maier M. A.; Rajeev K. G.; Manoharan M. Multivalent N-Acetylgalactosamine-Conjugated siRNA Localizes in Hepatocytes and Elicits Robust RNAi-Mediated Gene Silencing. J. Am. Chem. Soc. 2014, 136, 16958–16961. 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- Soutschek J.; Akinc A.; Bramlage B.; Charisse K.; Constien R.; Donoghue M.; Elbashir S.; Geick A.; Hadwiger P.; Harborth J.; John M.; Kesavan V.; Lavine G.; Pandey R. K.; Racie T.; Rajeev K. G.; Röhl I.; Toudjarska I.; Wang G.; Wuschko S.; Bumcrot D.; Koteliansky V.; Limmer S.; Manoharan M.; Vornlocher H.-P. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004, 432, 173–178. 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Prakash T. P.; Mullick A. E.; Lee R. G.; Yu J.; Yeh S. T.; Low A.; Chappell A. E.; Østergaard M. E.; Murray S.; Gaus H. J.; Swayze E. E.; Seth P. P. Fatty acid conjugation enhances potency of antisense oligonucleotides in muscle. Nucleic Acids Res. 2019, 47, 6029–6044. 10.1093/nar/gkz354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M.; Sena-Esteves M.; Chase K.; Sapp E.; Pfister E.; Sass M.; Yoder J.; Reeves P.; Pandey R. K.; Rajeev K. G.; Manoharan M.; Sah D. W. Y.; Zamore P. D.; Aronin N. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Nat. Acad. Sci. 2007, 104, 17204. 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn M. F.; Khvorova A. Improving siRNA Delivery In Vivo Through Lipid Conjugation. Nucleic Acid Ther. 2018, 28, 128–136. 10.1089/nat.2018.0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M. R.; Ming X.; Fisher M.; Lackey J. G.; Rajeev K. G.; Manoharan M.; Juliano R. L. Multivalent Cyclic RGD Conjugates for Targeted Delivery of Small Interfering RNA. Bioconjugate Chem. 2011, 22, 1673–1681. 10.1021/bc200235q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieslak J.; Grajkowski A.; Ausin C.; Gapeev A.; Beaucage S. L. Permanent or reversible conjugation of 2’-O- or 5′-O-aminooxymethylated nucleosides with functional groups as a convenient and efficient approach to the modification of RNA and DNA sequences. Nucleic Acids Res. 2012, 40, 2312–29. 10.1093/nar/gkr896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb H. C.; Finn M. G.; Sharpless K. B. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem., Int. Ed. 2001, 40, 2004–2021. . [DOI] [PubMed] [Google Scholar]
- Parmar R. G.; Brown C. R.; Matsuda S.; Willoughby J. L. S.; Theile C. S.; Charissé K.; Foster D. J.; Zlatev I.; Jadhav V.; Maier M. A.; Egli M.; Manoharan M.; Rajeev K. G. Facile Synthesis, Geometry, and 2′-Substituent-Dependent in Vivo Activity of 5′-(E)- and 5′-(Z)-Vinylphosphonate-Modified siRNA Conjugates. J. Med. Chem. 2018, 61, 734–744. 10.1021/acs.jmedchem.7b01147. [DOI] [PubMed] [Google Scholar]
- Ramasamy K. S.; He L.; Stoisavljevic V.; Harpham B.; Seifert W. Synthesis and biophysical studies of oligonucleotides containing hydroxamate linkages. Tetrahedron Lett. 2000, 41, 4317–4321. 10.1016/S0040-4039(00)00661-4. [DOI] [Google Scholar]
- Guzaev A. P.; Cook P. D.; Manoharan M.; Bhat B.. Preparation of nucleosidic and non-nucleosidic oligodeoxyribonucleotide-folate conjugates. US6335434B1, 2002.
- Valeur E.; Bradley M. Amide bond formation: beyond the myth of coupling reagents. Chem. Soc. Rev. 2009, 38, 606–631. 10.1039/B701677H. [DOI] [PubMed] [Google Scholar]
- El-Faham A.; Albericio F. Peptide Coupling Reagents, More than a Letter Soup. Chem. Rev. 2011, 111, 6557–6602. 10.1021/cr100048w. [DOI] [PubMed] [Google Scholar]
- Brown C. R.; Gupta S.; Qin J.; Racie T.; He G.; Lentini S.; Malone R.; Yu M.; Matsuda S.; Shulga-Morskaya S.; Nair A. V.; Theile C. S.; Schmidt K.; Shahraz A.; Goel V.; Parmar R. G.; Zlatev I.; Schlegel M. K.; Nair J. K.; Jayaraman M.; Manoharan M.; Brown D.; Maier M. A.; Jadhav V. Investigating the pharmacodynamic durability of GalNAc-siRNA conjugates. Nucleic Acids Res. 2020, 48, 11827–11844. 10.1093/nar/gkaa670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey D. R.; Damha M. J.; Manoharan M. Challenges and Opportunities for Nucleic Acid Therapeutics. Nucleic Acid Ther. 2022, 32, 8–13. 10.1089/nat.2021.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P.; Parmar R. G.; Brown C. R.; Willoughby J. L. S.; Foster D. J.; Babu I. R.; Schofield S.; Jadhav V.; Charisse K.; Nair J. K.; Rajeev K. G.; Maier M. A.; Egli M.; Manoharan M. 5′-Morpholino modification of the sense strand of an siRNA makes it a more effective passenger. Chem. Commun. 2019, 55, 5139–5142. 10.1039/C9CC00977A. [DOI] [PubMed] [Google Scholar]
- Mikami A.; Erande N.; Matsuda S.; Kel’in A.; Woods L. B.; Chickering T.; Pallan P. S.; Schlegel M. K.; Zlatev I.; Egli M.; Manoharan M. Synthesis, chirality-dependent conformational and biological properties of siRNAs containing 5′-(R)- and 5′-(S)-C-methyl-guanosine. Nucleic Acids Res. 2020, 48, 10101–10124. 10.1093/nar/gkaa750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.