Abstract
Proprotein convertase subtilisin/kexin type 9 (PCSK9) is involved in the degradation of the low-density lipoprotein receptor. PCSK9 also targets proteins involved in lipid metabolism (very low–density lipoprotein receptor), immunity (major histocompatibility complex I), and viral infection (cluster of differentiation 81). Recent studies have also indicated that PCSK9 loss-of-function mutations are associated with an increased incidence of diabetes; however, the expression and function of PCSK9 in insulin-producing pancreatic beta cells remain unclear. Here, we studied PCSK9 regulation and function by performing loss- and gain-of-function experiments in the human beta cell line EndoC-βH1. We demonstrate that PCSK9 is expressed and secreted by EndoC-βH1 cells. We also found that PCSK9 expression is regulated by cholesterol and sterol regulatory element–binding protein transcription factors, as previously demonstrated in other cell types such as hepatocytes. Importantly, we show that PCSK9 knockdown using siRNA results in deregulation of various elements of the transcriptome, proteome, and secretome, and increases insulin secretion. We also observed that PCSK9 decreases low-density lipoprotein receptor and very low–density lipoprotein receptor levels via an extracellular signaling mechanism involving exogenous PCSK9, as well as levels of cluster of differentiation 36, a fatty acid transporter, through an intracellular signaling mechanism. Finally, we found that PCSK9 regulates the cell surface expression of PDL1 and HLA-ABC, proteins involved in cell–lymphocyte interaction, also via an intracellular mechanism. Collectively, these results highlight PCSK9 as a regulator of multiple cell surface receptors in pancreatic beta cells.
Keywords: human pancreatic beta cell, PCSK9, LDLR, CD36, PDL1, HLA-ABC
Abbreviations: BSA, bovine serum albumin; CD, cluster of differentiation; DMEM, Dulbecco’s modified Eagle’s medium; ER, endoplasmic reticulum; FACS, fluorescence-activated cell sorting; FCS, fetal calf serum; HBSS, Hanks' balanced salt solution; HEK293T, human embryonic kidney 293T cell line; LDL, low-density lipoprotein; LDLR, LDL receptor; LRP, LDL-related protein; MHC, major histocompatibility complex; PCSK9, proprotein convertase subtilisin/kexin type 9; qPCR, quantitative PCR; siCTRL, siControl; siPCSK9, siRNA targeting PCSK9; SRE, sterol regulatory element; SREBP, sterol regulatory element binding protein; TRL, triglyceride-rich lipoprotein; VLDLR, very low–density lipoprotein receptor
Proprotein convertase subtilisin/kexin type 9 (PCSK9) is the last discovered member of the proconvertase family (1). It is mainly expressed in liver, cerebellum, lung, and organs of the gastrointestinal tract including pancreas (https://gtexportal.org/home/gene/PCSK9) (2). Hepatocytes express the highest levels of PCSK9 and are the main source of circulating PCSK9 (3). PCSK9 is synthesized as an immature zymogen precursor and autocatalytically cleaved in the endoplasmic reticulum (ER) between its N-terminal prodomain and catalytic domain (4). The resulting mature PCSK9 is next released into the circulation, where it plays a key role in the regulation of cholesterol homeostasis (3, 4).
Regulation of PCSK9 expression has been deeply studied in hepatocytes (4), and it was found that PCSK9 promoter contains a sterol regulatory element (SRE) (5, 6). There, PCSK9 expression is dependent on both SRE binding protein (SREBP)-1 and SREBP-2, whereas its regulation by cholesterol is predominantly SREBP-2 dependent (5, 6). PCSK9 expression is also modulated by insulin in a SREBP-1c–dependent manner (7).
PCSK9 gain-of-function mutations are associated with familial hypercholesterolemia (8), whereas patients with loss-of-function PCSK9 mutations have reduced circulating low-density lipoprotein (LDL)-cholesterol levels (9). These discoveries led to validation of PCSK9 as a therapeutic target for the treatment of hypercholesterolemia. The first anti-PCSK9 monoclonal antibodies were approved for the treatment of hypercholesterolemia in 2015 (10) and recently, small interfering RNAs targeting intracellular PCSK9 were approved by the European Medicines Agency for the treatment of hypercholesterolemia or mixed dyslipidemia (11). Interestingly, PCSK9 loss-of-function mutations are associated with increased prevalence of type 2 diabetes (12). However, there is currently no evidence of an effect of anti-PCSK9 inhibitory antibodies on the transition to new onset diabetes, suggesting that reducing circulating PCSK9 does not impair pancreatic beta cell function (13, 14).
It is known that PCSK9 binds extracellularly to the LDL receptor (LDLR) and targets it for lysosomal degradation, preventing its recycling to the plasma membrane (4). In addition, a number of studies suggest that PCSK9 can also decrease LDLR abundance at the cell surface prior to its secretion via an intracellular route. Indeed, PCSK9 and LDLR interact early in the secretory pathway (15), and PCSK9 favors LDLR degradation even when its endocytosis is blocked (16). Moreover, inhibition of LDLR traffic between the trans-Golgi network and the lysosome abolishes PCSK9-induced LDLR degradation (17).
In addition to LDLR, PCSK9 enhances the degradation of other proteins: very low–density lipoprotein receptor (VLDLR), LDL-related protein-8 (LRP8; also called ApoER2) (18), cluster of differentiation 81 (CD81) (19), LDL-related protein-1 (LRP1) (20), CD36 (21), β-secretase 1 (BACE1) (22) as well as the epithelial sodium channel subunits (SCNN1A, SCNN1B, and SCNN1G, also called α-, β-, and γ-ENaC) (23). Very recently, it was also demonstrated that PCSK9 disrupts the recycling of major histocompatibility complex I (MHC-I) to the cell surface, thus positioning PCSK9 inhibitors as a new way to enhance immune checkpoint therapy for cancer (24). Some of the aforedescribed proteins including the LDLR have been shown to be regulated by PCSK9 both intracellularly and extracellularly (18, 19, 20, 21, 24). For other proteins such as BACE1, intracellular PCSK9-mediated protein degradation has been described as an important route (22). Moreover, it is the unique route for the epithelial sodium channel ENaC degradation by PCSK9 (23). Finally, for other proteins, their route of degradation by PCSK9 is far less studied.
The islets of Langerhans are micro-organs distributed throughout the pancreas. They contain beta, alpha, delta, PP, and epsilon endocrine cells that produce and release insulin, glucagon, somatostatin, pancreatic polypeptide, and ghrelin, respectively. PCSK9 was previously described as expressed in mouse and human pancreatic islets (25, 26). However, the precise cell type–expressing PCSK9 within islets remains unclear. Some studies demonstrate that PCSK9 is only expressed in somatostatin-producing delta cells (25, 27), whereas other studies indicate that beta cells express PCSK9 (26, 28, 29). The function of PCSK9 within islets is also debated.
The aim of this study was to clarify the role of PCSK9 in pancreatic beta cells. We hypothesized that PCSK9 is expressed in pancreatic beta cells and asked whether it regulates other proteins beyond LDLR.
We studied PCSK9 expression in alpha, beta, and delta cell populations purified by fluorescence-activated cell sorting (FACS) from mouse pancreatic islets. We demonstrate that PCSK9 mRNA is enriched in the beta cell population. By using the human pancreatic beta cell line EndoC-βH1 (30), we investigated PCSK9 expression, function, and its regulation. We show that PCSK9 is expressed and secreted by human pancreatic beta cell. In EndoC-βH1 cells, PCSK9 expression is regulated by cholesterol, lipoproteins, and SREBP transcription factors. We characterized transcriptome, proteome, and secretome of EndoC-βH1 cells following PCSK9 knockdown that revealed multiple deregulations. We also observed an increase in basal and glucose-stimulated insulin secretion. Finally, we performed gain- and loss-of-function experiments using siPCSK9 (siRNA targeting PCSK9) or recombinant PCSK9 treatments to search for additional proteins regulated by PCSK9 in human beta cells. We demonstrate that PCSK9 regulates LDLR and VLDLR, two lipoprotein receptors through extracellular mechanism, and CD36, a fatty acid transporter, through an intracellular mechanism. Finally, PCSK9 regulates through an intracellular mechanism cell surface expression of PDL1 and HLA-ABC that are involved in cell–lymphocytes interaction.
Taken together, our work identifies new cell surface proteins regulated by PCSK9 and provides new insights for the comprehension of the consequences of PCSK9 modulation on beta cell function.
Results
PCSK9 is expressed and secreted by pancreatic beta cells
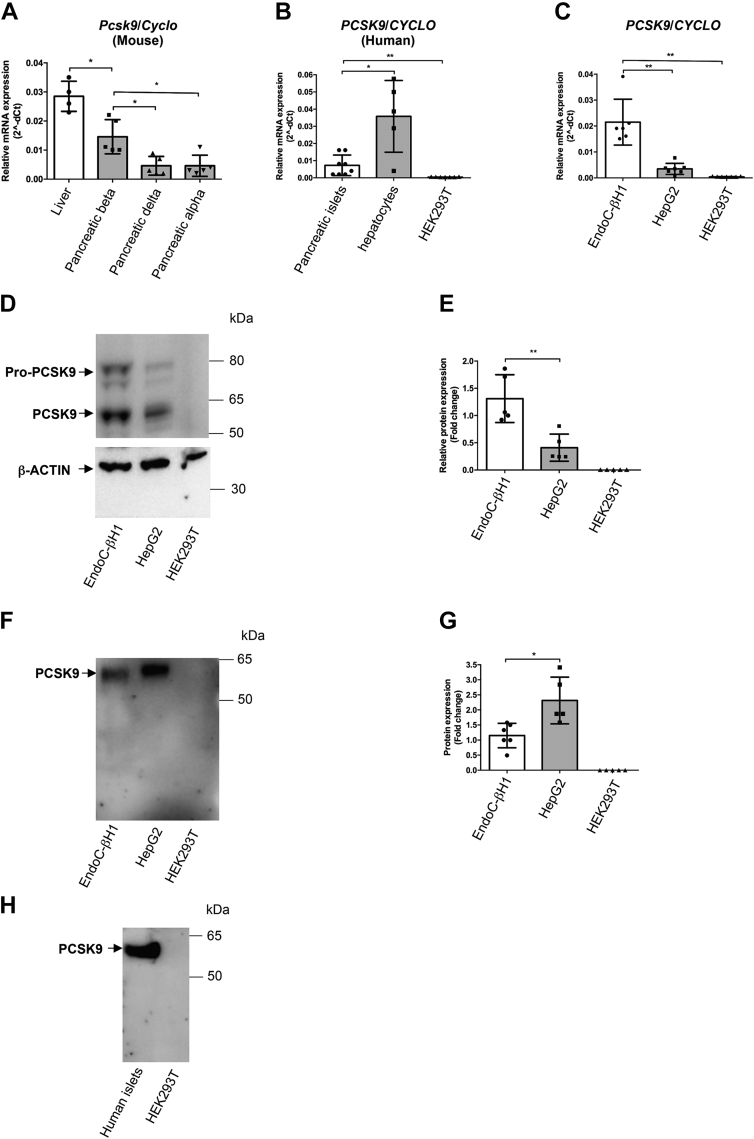
The cell types expressing PCSK9 in mouse and human pancreatic islets remain debated (25, 26, 27, 28). Here, we first analyzed PCSK9 expression in mouse pancreatic islet cell types by using a FACS-based strategy to purify beta, alpha, and delta cell populations (Fig. S1A) (30). RT–quantitative PCR (qPCR) analyses indicated that alpha (CD71−), beta (CD71+), and delta (CD24+) populations were highly enriched in glucagon, insulin-1, and somatostatin mRNA, respectively, validating the efficiency of the sorting (Fig. S1B). PCSK9 mRNA analyses indicated that its expression was threefold higher in beta than in the delta and alpha cell populations (Fig. 1A). PCSK9 mRNA level was also twofold lower in beta cells than in mouse liver (Fig. 1A). We then compared PCSK9 mRNA expression by RT–qPCR in human islet preparations versus human primary hepatocytes and human embryonic kidney 293T (HEK293T) cells (used as positive and negative controls, respectively). PCSK9 mRNAs were detected in human islets, although at a level fivefold lower than in human primary hepatocytes. As expected, PCSK9 was almost undetectable in HEK293T cells (Fig. 1B). We next investigated PCSK9 expression in human islet cell types. Since PCSK9 expression in bulk islets was low, it may be difficult to capture with single-cell sequencing because of limitations of the technology. Therefore, we used a combination of single cell RNA-Seq (E-MTAB-5061 (31), GSE81608 (32), and GSE86469 (33)) and RNA-Seq on highly purified FACS-sorted human beta and alpha cells (GSE67543 (34)) to analyze PCSK9 expression. PCSK9 transcripts were enriched in human adult beta cells (Fig. S1C). PCSK9 function was extensively studied in the human hepatoma cell line HepG2. We compared PCSK9 expression between EndoC-βH1, HepG2, and HEK293T cells. There, PCSK9 mRNA level was higher than in the human hepatoma cell line HepG2 (Fig. 1C). We next studied PCSK9 expression by Western blot (see Fig. S2, A–E for antibody validation). Western blot analysis revealed the presence of two major PCSK9 bands at ∼75 and ∼60 kDa. These bands corresponded to the expected sizes for pro-PCSK9 and mature PCSK9. Quantification of mature PCSK9 level showed a threefold higher expression in EndoC-βH1 compared with HepG2 cells and no expression in HEK293T cells (Fig. 1, D and E). Noteworthy, PCSK9 maturation rate (as measured by the mature PCSK9/total PCSK9 ratio) was higher in HepG2 cells than in the EndoC-βH1 (Fig. S1D). Given the fact that PCSK9 is a secreted protein, we also compared by Western blot PCSK9 levels in conditioned media from EndoC-βH1, HepG2, and HEK293T cells. In EndoC-βH1 and HepG2 cells, we detected a single band at approximately 60 kDa (as expected for the secreted mature form of PCSK9) and no signal in conditioned media from the HEK293T cells (Fig. 1F). PCSK9 secretion was twice lower in EndoC-βH1 when compared with HepG2 cells (Fig. 1, F and G). It was also the case when PCSK9 secretion was normalized to PCSK9 cell content (Fig. S1E). Finally, as PCSK9 transcripts were present in human islet preparations (Fig. 1B), we evaluated whether PCSK9 was secreted by human islets. By Western blot, we detected in conditioned media the presence of a band at ∼60 kDa, the expected size for the secreted form of PCSK9 (Fig. 1H). Omics data analysis confirmed that EndoC-βH1 cells are a representative in vitro model for human beta cells (35). Thus, mouse and human beta cells express PCSK9, and EndoC-βH1 cells can be used as a model to study PCSK9 regulation and function in human beta cells.
Figure 1.
PCSK9 is expressed and secreted by pancreatic beta cells. A–C, PCSK9 mRNA expression in (A) mouse liver compared with FACS-enriched pancreatic beta, delta, and alpha endocrine fractions (n = 4–5); (B) human islets, hepatocytes, and HEK293T cells (n = 5–8); (C) EndoC-βH1, HepG2, and HEK293T cells (n = 6–7). D and E, detection by Western blot and quantification of intracellular PCSK9 in EndoC-βH1, HepG2, and HEK293T cells (n = 5). F and G, detection by Western blot and quantification of secreted PCSK9 in EndoC-βH1, HepG2, and HEK293T cells (n = 5–6). H, Western blot analysis of PCSK9 secretion by human islets, compared with HEK293T cells (n = 7). Data represent the means ± SD. ∗p < 0.05 and ∗∗p < 0.01. FACS, fluorescence-activated cell sorting; HEK293T, human embryonic kidney 293T cell line; PCSK9, proprotein convertase subtilisin/kexin type 9.
Cholesterol, LDL and mevastatin modulate PCSK9 expression and secretion
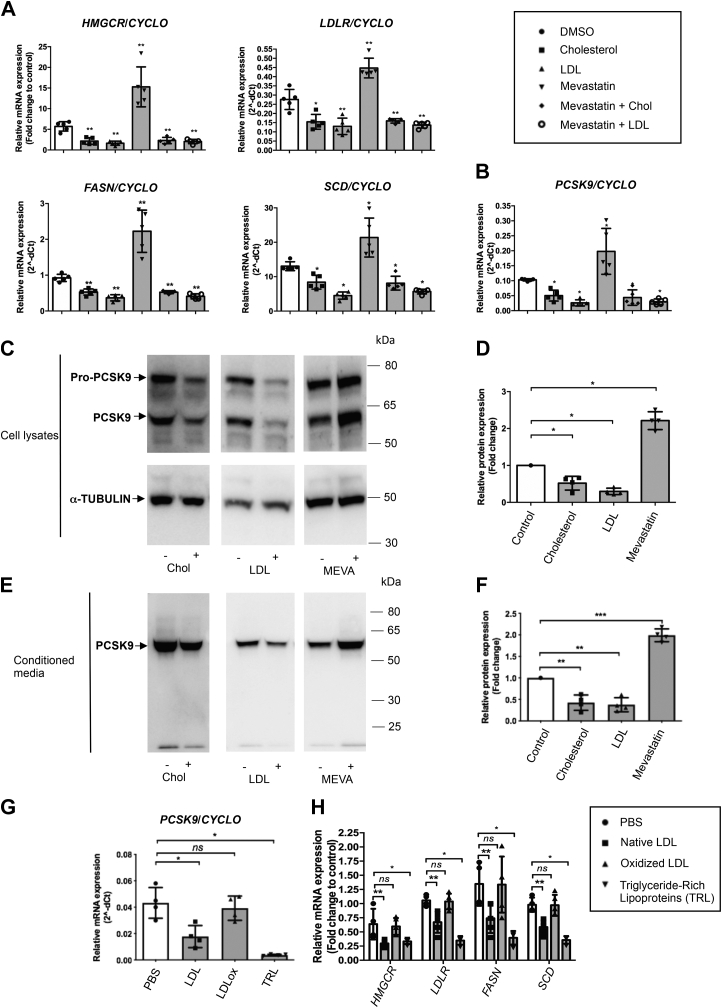
In mouse and human hepatocytes, PCSK9 expression is regulated by cholesterol and lipid-lowering drugs of the statin class (5, 36). Here, we evaluated the effects on PCSK9 expression of cholesterol, LDL, and mevastatin in EndoC-βH1 cells. Concentrations were chosen based on their effect on HMGCR gene expression, a SREBP-2 transcriptional target (Fig. S3, A–D). Cholesterol and LDL treatments reduced the mRNA expression of both SREBP-2 (HMGCR and LDLR) and SREBP-1 (FASN and SCD) transcriptional targets by ∼50%. Mevastatin treatment increased the expression of these genes by at least twofold, demonstrating the functionality of the LDL-cholesterol pathway in EndoC-βH1 cells (Fig. 2A). PCSK9 mRNA followed a similar pattern as expression was reduced by cholesterol and LDL treatment and increased following incubation with mevastatin (Fig. 2B). The effects of mevastatin were reverted by cholesterol or LDL treatments and were thus “cholesterol specific” (Fig. 2, A and B). Similar effects of cholesterol, LDL, and mevastatin were also observed at protein level. Indeed, Western blot analyses indicated that cholesterol and LDL treatments decreased, whereas mevastatin increased intracellular PCSK9 levels (Fig. 2, C and D). Similar results were also obtained when secreted PCSK9 was analyzed (Fig. 2, E and F). Finally, we tested whether other classes of human lipoproteins modulate PCSK9 expression in EndoC-βH1 cells. Treatment with triglyceride-rich lipoproteins (TRLs) mimicked the effects of LDL on PCSK9 mRNA expression and on SREBP-1 and SREBP-2 transcriptional targets, whereas oxidized LDL had no effect (Fig. 2, G and H).
Figure 2.
Cholesterol, LDL and mevastatin modulate PCSK9 expression and secretion.A–F, endoC-βH1 cells were exposed to 10 μg/ml human LDL, 0.25 mM cholesterol–methyl-β-cyclodextrin, and 10 μg/ml mevastatin for 24 h (A and B) SREBP-2 transcriptional targets (HMGCR and LDLR), SREBP-1 transcriptional targets (FASN and SCD), and PCSK9 mRNA were measured by RT–qPCR (n = 5). Intracellular (C and D) and secreted (E and F) PCSK9 were analyzed and quantified by Western blot (n = 4). G and H, endoC-βH1 cells were exposed to 10 μg/ml native LDL, oxidized LDL, or triglyceride-rich lipoproteins (TRLs) for 24 h. PCSK9, SREBP-1, and SREBP-2 targets were measured by RT–qPCR (n = 4). Data represent the means ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. LDL, low-density lipoprotein; PCSK9, proprotein convertase subtilisin/kexin type 9; qPCR, quantitative PCR; SREBP, sterol regulatory element binding protein.
PCSK9 is regulated by the SREBP-1 and SREBP-2 transcription factors
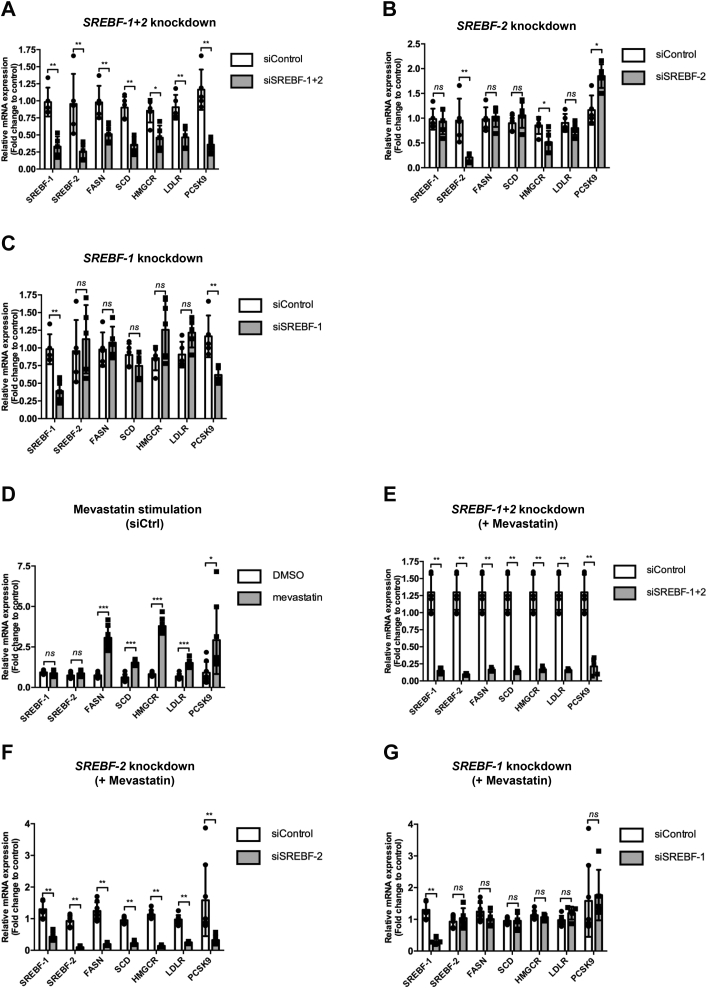
We first tested whether PCSK9 mRNA levels were dependent on SREBP transcription factors. Cotransfection of the EndoC-βH1 cells with siSREBF-1 + siSREBF-2 efficiently reduced mRNA expression of SREBF-1, SREBF-2, and their respective target genes: FASN, SCD (SREBP-1 targets) and HMGCR, LDLR (SREBP-2 targets). PCSK9 mRNA levels were downregulated indicating dependence of SREBP activity (Fig. 3A). Single siSREBF-2 downregulated SREBF-2 and its target HMGCR but not SREBF-1. It increased PCSK9 mRNA levels (Fig. 3B). Single siSREBF-1 transfection specifically reduced SREBF-1 expression. The expression of canonic SREBP-1 liver targets FASN and SCD was not modulated, indicating the existence of compensatory mechanism that maintain FASN, SCD, and LDLR expression when SREBF-1 or SREBF-2 is reduced following knockdown (compare Fig. 3, A–C). On the other hand, SREBF-1 knockdown was sufficient to decrease PCSK9 expression, indicating that SREBF-1 is essential to maintain PCSK9 expression (Fig. 3C).
Figure 3.
PCSK9 is regulated by the SREBP-1 and SREBP-2 transcription factors.A–C, endoC-βH1 cells were transfected with control nontarget siRNA (siCtrl), siRNA targeting SREBP-1 + SREBP-2 (siSREBP-1 + siSREBP-2), SREBP-2 (siSREBP-2), or SREBP-1 (siSREBP-1). RT–qPCR for SREBPs, and their targets were performed 3 days later (n = 5). D–G, endoC-βH1 cells were transfected with siRNA, cultured for 72 h, and next treated for 24 h with DMSO (control) or mevastatin before analyses by RT–qPCR (n = 5). Data represent the means ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. DMSO, dimethyl sulfoxide; PCSK9, proprotein convertase subtilisin/kexin type 9; qPCR, quantitative PCR; SREBP, sterol regulatory element binding protein.
We next assessed the SREBP dependence of PCSK9 upon mevastatin treatment. As expected, mevastatin treatment increased the mRNA expression of SREBP-1 targets, SREBP-2 targets, and also PCSK9 (Fig. 3D). This induction was reverted upon SREBF-1 + SREBF-2 double knockdown (Fig. 3E). Single SREBF-2 knockdown reduced the expression of SREBF-2 and its target genes HMGCR and LDLR in the presence of mevastatin. PCSK9 expression was also downregulated suggesting an SREBP-2 dependence (Fig. 3F). However, we noted that upon mevastatin treatment, siSREBF-2 decreased SREBF-1 levels (Fig. 3F). We thus performed single siSREBF1 knockdown that did not modulate PCSK9 expression (Fig. 3G). Taken together, our data indicate that SREBP-2, but not SREBP-1, is involved in the induction of PCSK9 by mevastatin in human beta cells.
Insights from hypothesis-generating omics analysis
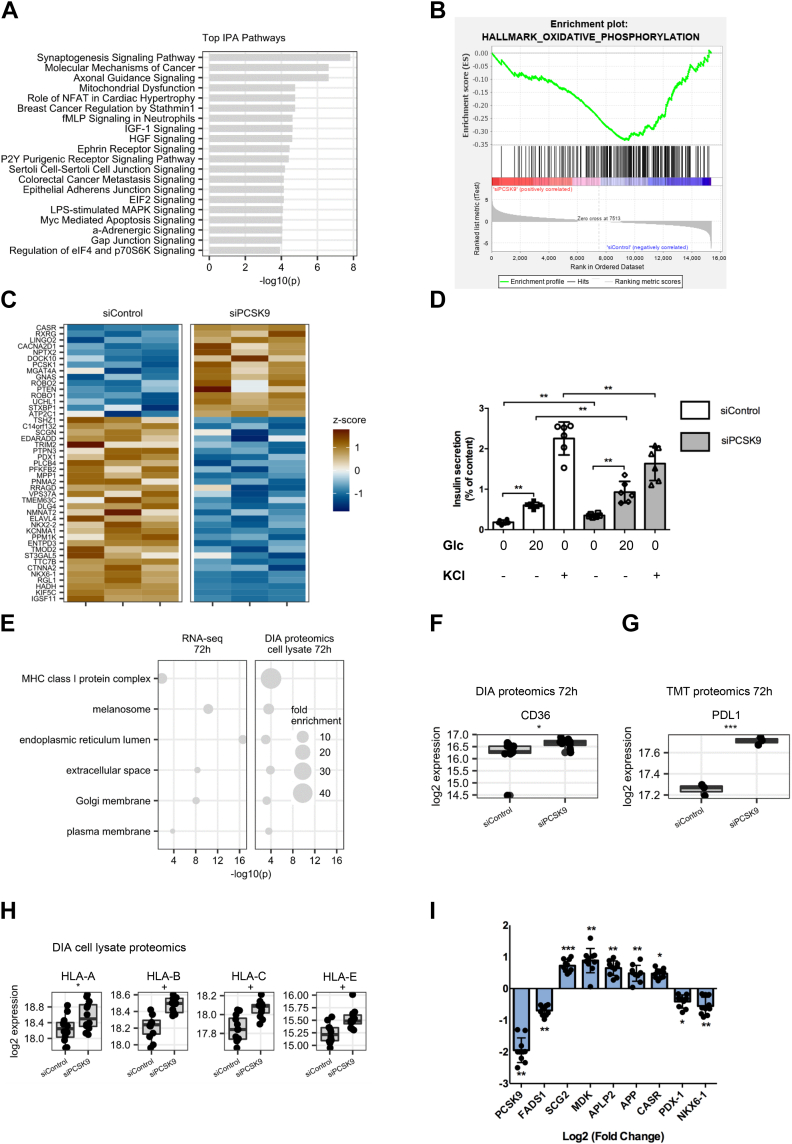
We investigated the effect of siRNA-mediated PCSK9 knockdown in EndoC-βH1 cells in transcriptomics and proteomics experiments. PCSK9 knockdown was confirmed in all omics experiments (Fig. S4). Changes in gene and protein expression levels were generally consistent and had the same directionality (60.3% for RNA-Seq versus data-independent acqusition cell lysate proteomics and 59.7% for RNA-Seq versus tandem mass tag cell lysate proteomics) (Fig. S5). Ingenuity pathway enrichment analysis indicated impaired mitochondrial function (Fig. 4A), which was confirmed by gene set enrichment analysis (Fig. 4B). Normal mitochondrial function is crucial for insulin secretion by beta cells (37). We investigated the expression of markers associated with glucose-stimulated insulin secretion (38). Of 147 markers, 45 (31%) were differentially expressed (Fig. 4C). We also observed consistent upregulation of PCSK1 levels, an enzyme involved in proinsulin to insulin processing, in the transcriptome and proteome experiments (Fig. S6). Interestingly, we observed an upregulation of insulin in data-independent acquisition secretome proteomics at 72 h after PCSK9 knockdown (Fig. S6D). Following PCSK9 knockdown, insulin secretion was impaired: basal and glucose-stimulated insulin secretion increased, whereas KCl-induced insulin secretion was reduced (Fig. 4D). Also note that insulin content decreased following PCSK9 knockdown (Fig. S7). Gene Ontology Cellular Component enrichment analysis indicated overrepresentation of MHC-I complex, and plasma membrane proteins among genes and proteins upregulated after PCSK9 knockdown (Fig. 4E). MHC-I complex enrichment was stronger on protein level than on mRNA level. Golgi and ER proteins were also upregulated, which is consistent with the reported role of PCSK9 in intracellular degradation of proteins in the ER-to-Golgi intermediate compartment (22). CD36, CD274 (PDL1), and MHC-I complex proteins (particularly HLA-A, B, C, and E) were top findings in proteomics experiments (Fig. 4, F–H). ENaC channel subunits were not expressed in EndoC-βH1 cells. LDLR, VLDL, and LRP8 levels were not altered (Fig. S8). CD81 and LRP1 were upregulated on mRNA but not protein level (Fig. S8). mRNA and whole-cell protein expression of APP and APLP2 was increased after PCSK9 knockdown (Fig. S8). Upregulation of APP and APLP2 mRNA was confirmed by RT–qPCR together with the other top differentially expressed genes (Fig. 4I).
Figure 4.
Insights from omics experiments. EndoC-βH1 cells were transfected with control nontarget siRNA (siControl) or siRNA targeting PCSK9 (siPCSK9). Three days later, cells were profiled by RNA-Seq and proteomics. RNA-Seq data were analyzed with (A) ingenuity pathway analysis (IPA) software, for the top enriched canonical pathways, and (B) Gene Set Enrichment Analysis (GSEA) software with the Hallmark_Oxidative_Phosphorylation gene set. C, GSIS markers deregulated upon PCSK9 knockdown with FDR <0.05 in RNA-Seq experiment. D, basal (0.5 mM), glucose-stimulated (20 mM), and KCl-stimulated (50 mM) insulin secretions were measured in EndoC-βH1 cells, 3 days following PCSK9 knockdown (n = 6). E, Gene Ontology analysis of cellular components terms enriched among genes and proteins upregulated following PCSK9 knockdown. F–H, Boxplot showing protein expression of top candidate PCSK9 degradation targets, measured on cell protein extracts by DIA and TMT proteomic. I, RT–qPCR validation of selected top differentially expressed genes (n = 10). For GSIS and RT–qPCR analyses, data are presented as means ± SD. For proteomics experiments, median expression and interquartile ranges are shown. + FDR < 0.05, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. DIA, data-independent acquisition; FDR, false discovery rate; GSIS, glucose-stimulated insulin secretion; PCSK9, proprotein convertase subtilisin/kexin type 9; qPCR, quantitative PCR; TMT, tandem mass tag.
PCSK9 knockdown does not affect LDLR levels
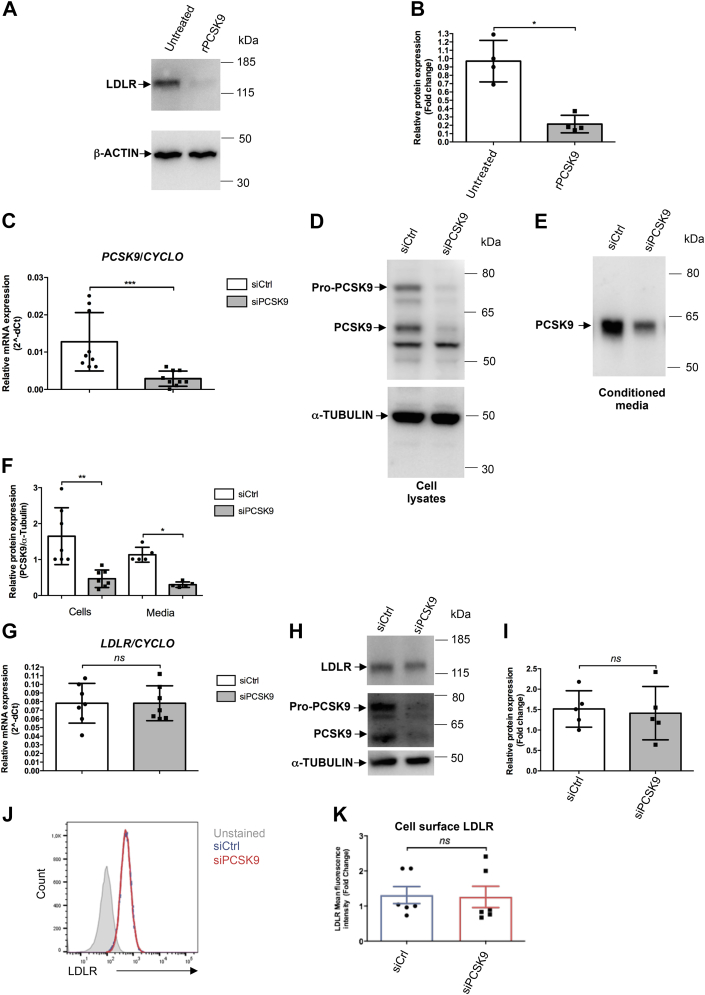
In hepatocytes, PCSK9 interacts with LDLR and favors its lysosomal degradation (4). Lack of LDLR dysregulation after PCSK9 knockdown in the omics experiments was unexpected, so we evaluated the effects of exogenously added human recombinant PCSK9 on LDLR expression. Incubating EndoC-βH1 cells with recombinant PCSK9 reduced LDLR protein levels (Fig. 5, A and B). We also performed a confirmatory siRNA experiment to test whether endogenously produced PCSK9 regulated LDLR expression. PCSK9 knockdown was efficient with a ∼80% decrease at the mRNA level (Fig. 5C), at the intracellular protein level (for both Pro-PCSK9 and mature PCSK9) (Fig. 5, D and F), and at the secreted protein level (Fig. 5, E and F). We next looked at LDLR levels. As expected, PCSK9 knockdown did not modulate LDLR mRNA levels (Fig. 5G). Moreover, despite the sharp decrease in PCSK9 protein expression and secretion, PCSK9 knockdown did not impair LDLR protein levels as measured by Western blot (Fig. 5,H and I). To determine whether PCSK9 specifically modulates LDLR at the cell surface, we performed FACS analyses. Again, PCSK9 knockdown did not modulate cell surface LDLR levels (Fig. 5, J and K). Taken together, these results indicate that LDLR can be targeted by extracellular PCSK9 but is not regulated by intracellular PCSK9 in EndoC-βH1 cells.
Figure 5.
PCSK9 knockdown does not affect LDLR levels. A and B, endoC-βH1 cells were exposed for 16 h to human recombinant PCSK9 (2.5 μg/ml). LDLR protein expression was studied by Western blot and quantified (n = 4). C–K, endoC-βH1 cells were transfected with control nontarget siRNA (siCtrl) or siRNA targeting PCSK9 (siPCSK9). Analyses were performed 3 days later. C–F, PCSK9 knockdown efficiency was analyzed by RT–qPCR (n = 9) (C) and Western blot for intracellular and secreted PCSK9 (n = 5–7) (D–F). G–K, the effect of PCSK9 knockdown on LDLR was measured by RT–qPCR (n = 7), Western blot (n = 5), and FACS for cell surface expression (n = 6). Data represent the means ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. FACS, fluorescence-activated cell sorting; LDLR, low-density lipoprotein receptor; PCSK9, proprotein convertase subtilisin/kexin type 9; qPCR, quantitative PCR.
PCSK9 knockdown potentiates the induction of PDL1 and HLA-ABC by interferon-γ
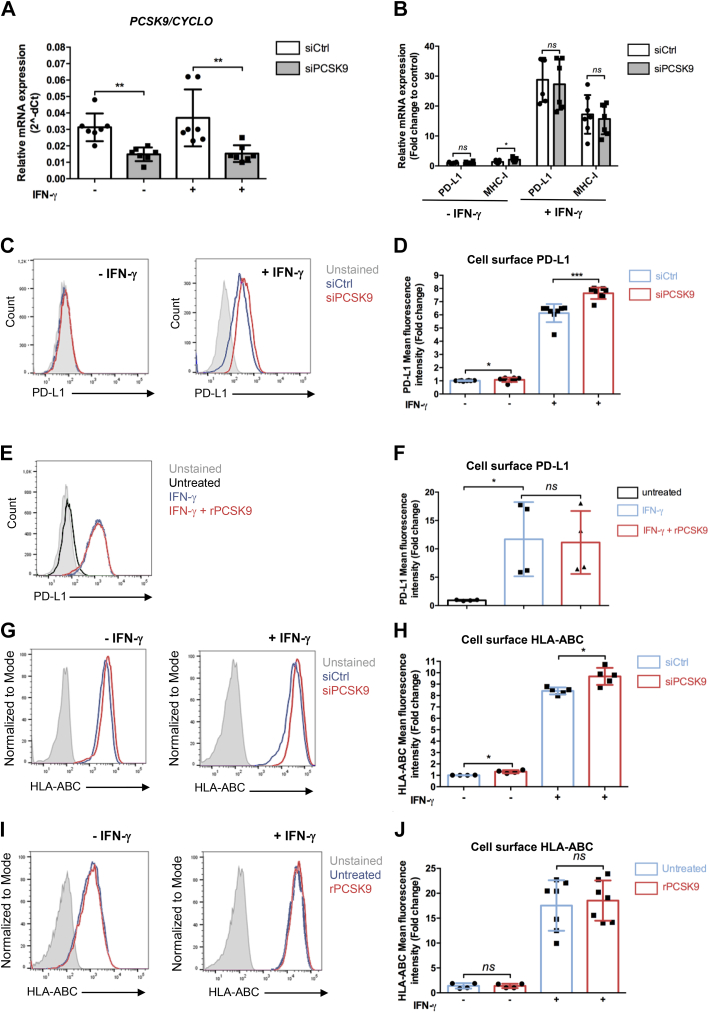
MHC-I and PDL1 are increased in pancreatic islets from type I diabetes and in response to interferon class I stimulation (39, 40). While characterizing the effect of PCSK9 knockdown on the proteome of EndoC-βH1, we observed upregulation of MHC-I (particularly HLA-A, B, C, and E) and PDL1 (Fig. 4, G and H). We thus asked whether PCSK9 regulates PDL1 and/or HLA-ABC cell surface expression in basal conditions and following interferon-γ treatment. PCSK9 knockdown consistently reduced PCSK9 mRNA levels in basal or interferon-γ stimulated conditions (Fig. 6A) with a slight increase of MHC-I mRNA under basal conditions (Fig. 6B). On the other hand, upon interferon-γ treatment, MHC-I and PDL1 mRNA levels were unchanged following PCSK9 knockdown (Fig. 6B). We next measured by FACS, cell surface expression of PDL1 following siRNA-mediated PCSK9 knockdown or following treatment with recombinant PCSK9. It revealed that while siRNA-mediated PCSK9 knockdown increased cell surface expression of PDL1 (Fig. 6, C and D), recombinant PCSK9 did not have any effects (Fig. 6, E and F). We performed a similar experiment, measuring cell surface expression of HLA-ABC. Again, while siRNA-mediated PCSK9 knockdown increased cell surface expression of HLA-ABC (Fig. 6, G and H), recombinant PCSK9 did not have any effects (Fig. 6,I and J). These data indicate that PDL1 and HLA-ABC cell surface expression is regulated by intracellular PCSK9.
Figure 6.
PCSK9 knockdown potentiates the induction of PDL1 and HLA-ABC by interferon-γ (IFN-γ).A–D, endoC-βH1 cells were transfected with control nontarget siRNA (siCtrl) or siRNA targeting PCSK9 (siPCSK9). Five days later, the cells were incubated for 24 h with 500 U/ml IFN-γ. A and B, RT–qPCR analysis of PCSK9, PDL1, and MHC-I (n = 6–7). C and D, cell surface PDL1 expression analyzed by FACS (n = 8–9). E and F, endoC-βH1 cells were treated with 500 U/ml IFN-γ. Twenty-four hours later, human recombinant PCSK9 (rPCSK9) (2.5 μg/ml) was added to the culture medium for 16 h. Cell surface PDL1 was studied by FACS (n = 4). G–J, cell surface HLA-ABC expression analyzed by FACS under the same experimental conditions as in (A–F) (n = 4–7). Data represent the means ± SD. ∗p < 0.05 and ∗∗p < 0.01. FACS, fluorescence-activated cell sorting; MHC-I, major histocompatibility complex; PCSK9, proprotein convertase subtilisin/kexin type 9; qPCR, quantitative PCR.
PCSK9 regulates the expression of VLDLR and CD36, two actors involved in fatty acid homeostasis
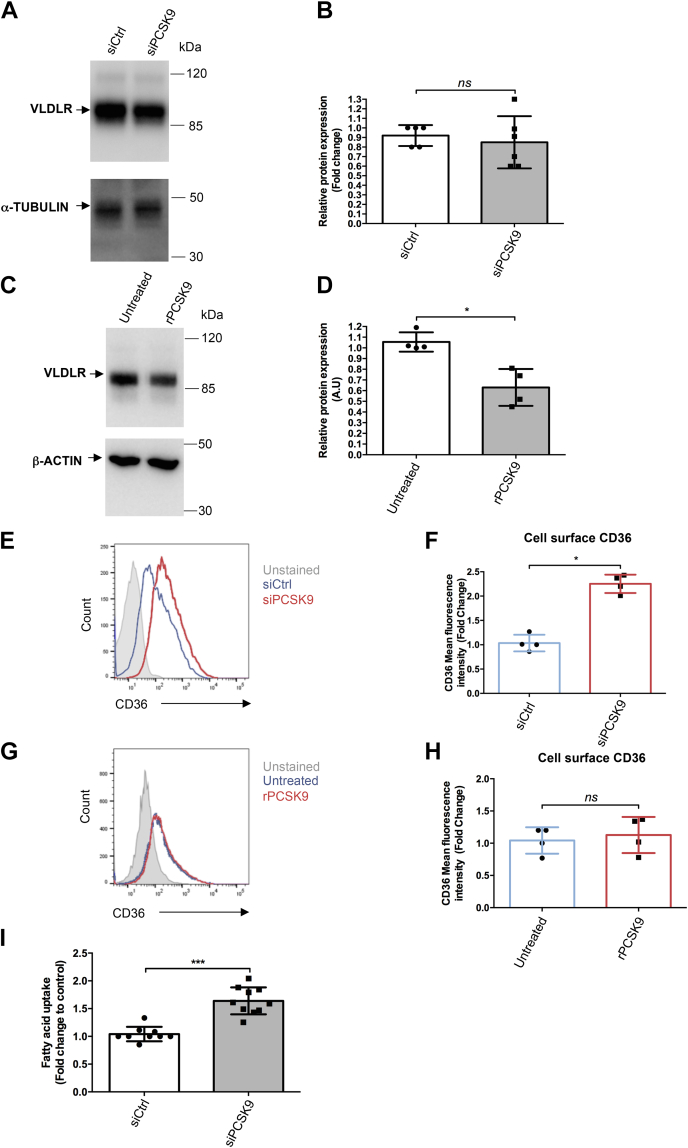
Fatty acid homeostasis is important for proper beta cell function (41). Interestingly, VLDLR and CD36 are involved in the uptake of TRLs and fatty acids, respectively, and are PCSK9 degradation targets in liver cells (18, 21). We further investigated their expression by RT–qPCR and Western blot. PCSK9 knockdown slightly decreased VLDLR mRNA (Fig. S9A) but did not impact VLDLR protein expression (Fig. 7, A and B). Incubating EndoC-βH1 cells with recombinant PCSK9 reduced VLDLR protein expression (Fig. 7, C and D). PCSK9 knockdown cells exhibited a clear reduction in PCSK9 mRNA level (Fig. S9B) and a slight but nonstatistically significant, increase in CD36 mRNA level (Fig. S9C). FACS analysis revealed a greater than twofold increase of CD36 cell surface expression (Fig. 7, E and F). On the other hand, incubating EndoC-βH1 cells with recombinant PCSK9 did not affect CD36 cell surface expression (Fig. 7, G and H). Finally, upon PCSK9 knockdown, fatty acid uptake increased by 50% (Fig. 7I).
Figure 7.
PCSK9 regulates the expression of VLDLR and CD36, two actors involved in fatty acid homeostasis. A and B, endoC-βH1 cells were transfected with control nontarget siRNA (siCtrl) or siRNA targeting PCSK9. Three days later, VLDLR expression was studied by Western blot and quantified (n = 5–6). C and D, endoC-βH1 cells were exposed for 16 h to human recombinant PCSK9 (2.5 μg/ml), and VLDLR protein expression was studied by Western blot (n = 4). E and F, endoC-βH1 cells were transfected with siRNA (siCtrl) or siRNA targeting PCSK9. Six days later, cell surface CD36 was analyzed by FACS (n = 4). G and H, endoC-βH1 cells were exposed for 16 h to 2.5 μg/ml human recombinant PCSK9, and cell surface CD36 was analyzed by FACS (n = 4). I, fatty acid uptake was evaluated 6 days following PCSK9 knockdown (n =10). Data represent the means ± SD. ∗p < 0.05 and ∗∗∗p < 0.001. CD36, cluster of differentiation; FACS, fluorescence-activated cell sorting; PCSK9, proprotein convertase subtilisin/kexin type 9; VLDLR, very low–density lipoprotein receptor.
In conclusion, both extracellular and intracellular PCSK9 can modulate beta cell fatty acid metabolism through the regulation of VLDLR and CD36 expression, respectively.
Discussion
Since its discovery in 2003, numerous studies have addressed PCSK9 expression and function in peripheral organs beyond liver including pancreas (4). PCSK9 mRNA was detected in human and mouse islets, but the cell type(s) expressing PCSK9 protein remained controversial. Moreover, most of the functional studies that address PCSK9 function in pancreatic beta cells focused on LDLR regulation without searching for additional targets (25, 26, 27, 28, 29). Here, we hypothesized that PCSK9 is expressed in pancreatic beta cells and that its role may not be restricted to the regulation of LDLR. We demonstrated that beta cells produce and secrete PCSK9. In beta cells, PCSK9 expression is regulated by cholesterol, lipoproteins, and the transcriptional factor SREBPs. In EndoC-βH1, PCSK9 knockdown resulted in transcriptome, proteome, and secretome deregulation and increased basal and glucose-stimulated insulin secretion. Finally, we observed that in EndoC-βH1, PCSK9 regulates the expression of multiple proteins that included LDLR and VLDLR through extracellular mechanisms, and CD36, HLA-ABC, and PDL1 through intracellular mechanisms.
In liver cells, PCSK9 is expressed as a proenzyme, processed intracellularly into its mature form, and finally secreted (1). By using EndoC-βH1 cells (42), we demonstrated here that it is also the case in human beta cells. Indeed, Western blot analyses detected both Pro-PCSK9 and mature PCSK9 in cell extracts, whereas only mature PCSK9 was detected in conditioned medium. We noted that the mature PCSK9/PCSK9 total ratio and secreted/intracellular PCSK9 ratio was lower in beta cells compared with liver cells. This suggests that Pro-PCSK9 processing is more efficient in liver cells compared with beta cells and that the intracellular PCSK9 pool is higher in beta cells. This may explain the preferential intracellular regulation of some proteins by PCSK9. The mechanisms that regulate Pro-PCSK9 processing and secretion have only been partially characterized (4). Our results also indicated that PCSK9 processing and secretion might be differentially regulated in liver and beta cells. Beta cells might thus be used as an additional model to study this complex processing.
In liver cells, the transcriptional regulation of PCSK9 has been studied in great detail (4). It has been shown that SREBP-2 regulates both basal and statin-induced expression of PCSK9 (6, 43, 44). Interestingly, we demonstrated that in beta cells, physiological PCSK9 levels are regulated by SREBP-1, whereas pharmacological regulation of PCSK9 by mevastatin is SREBP-2 dependent. This was intriguing as in the case of other SREBP-1 targets such as FASN and SCD, we observed an SREBP-2 mechanism that compensates for SREBP-1-mediated downregulation. This compensatory mechanism was not evident for PCSK9, indicating that physiological levels of PCSK9 in beta cells are highly SREBP-1 dependent.
The role of PCSK9 in pancreatic islet function is currently debated. Some studies indicated that PCSK9-deficient mice develop hyperglycemia and insulinopenia (26, 27), whereas others do not observe this phenotype (25, 28). The reasons for these discrepancies might be linked to the age and/or genetic background of the mice. Here, we observed a sharp decrease in several beta cell–enriched transcription factors upon PCSK9 knockdown in EndoC-βH1 cells. It was for example the case for PDX1 and NKX6-1 that are known to play major roles in beta cell development and function (45, 46, 47). We also detected some perturbation in glucose-stimulated insulin secretion with a specific increase in basal insulin secretion, which was not addressed by Ramin-Mangata et al. (29). Taken together, the phenotype observed upon PCSK9 knockdown (decreased expression of specific transcription factors and increased basal insulin secretion) was similar to that observed in immature fetal beta cells (48, 49) and insulin-producing beta cells differentiated in vitro from multipotent stem cells (50, 51). It suggests that PCSK9 might be important to maintain a mature phenotype in pancreatic beta cells.
In mouse islets, loss of beta-cell PCSK9 resulted in unchanged LDLR protein levels (28). Unexpectedly, data from a recently published study (29) indicated that PCSK9 knockdown in EndoC-βH1 cells increased LDLR cell surface expression by 20%. Our data do not support this last claim. We observed, as expected, that exogenously added PCSK9 decreased the expression of LDLR at the cell surface. On the other hand, efficient PCSK9 knockdown did not modify neither LDLR mRNA, nor the total amounts of LDLR protein measured either by Western blot or by global mass spectrometry, nor cell surface LDLR levels measured by FACS. Moreover, we observed similar results when looking at VLDLR, yet another PCSK9 target (18): VLDLR protein levels decreased upon treatment with exogenous PCSK9, without any effect upon PCSK9 knockdown. Our data are consistent with observations in mouse islets (28). The reasons for the discrepancy with the study by Ramin-Mangata et al. (29) are difficult to discuss, as this study presents FACS data in arbitrary units without supporting FACS plots. Taken together, we propose that in pancreatic beta cells, LDLR and VLDLR levels are regulated by PCSK9 extracellularly rather than through intracellular degradation.
CD36 has been described as another PCSK9 target both in mouse liver and adipocytes (21). There, PCSK9 increased CD36 degradation by a mechanism involving both lysosomes and proteasomes (21). Interestingly, in pancreatic beta cells, inhibiting CD36 has been described as a protective mechanism to prevent excessive lipid accumulation (52), whereas increased CD36 expression was found to be deleterious for proper beta-cell function (53). Here, we observe that PCSK9 knockdown in EndoC-βH1 increased cell surface CD36 expression and fatty acid uptake. Based on RNA-Seq and proteomics in PCSK9 knockdown cells and the insensitivity of cell surface CD36 expression to recombinant PCSK9, we postulate that PCSK9 regulates CD36 by an intracellular mechanism. The precise mechanism of this regulation was not identified but likely involves both proteasomal and lysosomal degradation (21). Further experiments, in particular by using proteasome or lysosome inhibitors, are needed to elucidate the nature of CD36 regulation by PCSK9 in the pancreatic beta cell.
In tumoral cells, MHC-I was recently described as a new PCSK9 target, as PCSK9 downregulates MHC-I cell surface expression by increasing its lysosomal degradation and disrupting its recycling (24). In this model, the authors observed marginal effects of PCSK9 on the immune regulatory protein PDL1. Interestingly, MHC-I and PDL1 play major roles in beta cells. Their expressions increase in pancreatic beta cell during inflammatory responses (39, 54) and are also induced in islets from type 1 diabetic patients (39, 54). There, it has been proposed that autoreactive CD8 T cells interact with beta cells and kill them through an MHC-I-mediated activation, whereas PDL1 would be protective against autoimmune injuries (55). Here, we found that PCSK9 knockdown increases both PDL1 and HLA-ABC cell surface expression, particularly upon interferon-γ stimulation. On the other hand, recombinant PCSK9 did not modulate HLA-ABC and PDL1 levels. We thus proposed that PCSK9 regulates the expression of PDL1 and HLA-ABC in human beta cells by an intracellular mechanism.
In conclusion, we found that PCSK9 is expressed and secreted by human pancreatic beta cells in a tightly regulated manner, which is altered in the presence of statins. In turn, we found that PCSK9 regulates cell surface expression of CD36, PDL1, and HLA-ABC through intracellular mechanisms but regulates LDLR and VLDLR expression through an extracellular mechanism in pancreatic beta cells. It would be interesting to determine if PCSK9 monoclonal antibodies or siRNA-based therapies against PCSK9 affect expression levels of LDLR and VLDLR, or CD36, PDL1, and HLA-ABC, respectively, in pancreatic beta cells, and whether cotreatment with statins interfered with the effect of PCSK9 siRNA in the pancreas.
Experimental procedures
Ethical statement
This study was performed according to the Declaration of Helsinki and the Declaration of Istanbul. No tissues were procured from prisoners. Human islets were provided by the Leiden University Medical Center and the human islet core facility of St-Louis Hospital. They were prepared from pancreata of brain-dead donors after informed consent was signed from next of kin and processed for islet isolation according to the procedures approved by the ethics committee of the French Agency of Biomedicine and the Dutch national law. All the animal studies complied with the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines and were conducted in strict accordance with the European Union Directive 2010/63/EU for animal experiments and with regard to specific national laws and INSERM (French National Institute of Health and Medical Research) guidelines.
Culture of EndoC-βH1 cells and treatment
The human pancreatic beta cell line EndoC-βH1 cells (Univercell Biosolutions [mycoplasma negative]) was cultured as previously described (42). Experiments were carried out 24 h after seeding the cells at a density of approximately 105 cells/cm2. The following compounds were used for treatment: cholesterol-methyl-β-cyclodextrin (catalog no.: C4951; Sigma–Aldrich), mevastatin (catalog no.: M2537; Sigma–Aldrich), human native LDL and copper-oxidized LDL or VLDL (see later for lipoprotein extraction protocol), human recombinant wildtype PCSK9 (CircuLex; catalog no.: CY-R2330).
Human hepatocytes, HepG2, and HEK293T culture
Human primary hepatocytes were isolated from liver fragments obtained from adult patients undergoing partial hepatectomy as previously described (56) and cultured in William’s Medium E (Life Technologies) supplemented with 10% fetal calf serum (FCS) (Perbio), 50 μM hydrocortisone hemisuccinate (SERB Laboratory), 5 μg/ml insulin (Sigma–Aldrich), 2 mM l-glutamine, 200 U/ml penicillin, and 200 μg/ml streptomycin. The human hepatocellular carcinoma cell line HepG2 and the HEK293T (mycoplasma free) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) and DMEM/F12 (Gibco), respectively, supplemented with 10 % FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Human islets
Islets were cultured in 12-well plates in CMRL medium supplemented with 10% FCS, Hepes, and penicillin/streptomycin (all from Thermo Fisher Scientific). Donor’s characteristic and islet details are presented in Table S1.
siRNA transfection
EndoC-βH1 cells were transfected as described (57) in OptiMEM using Lipofectamine RNAiMAX (Life Technologies) with siRNA SMARTpools (Horizon Discovery LTD). Medium was replaced 2.5 h later with fresh culture medium, and analyses were carried out 3 to 6 days following transfection. siRNA reagents used: control nontargeted siRNA (siControl [siCTRL]; catalog no.: D-001810-01-20); siRNA targeting: PCSK9 (siPCSK9) (catalog no.: M-005989-01-0005), SREBF-1 (siSREBF-1) (catalog no.: M-006891-00-0005), and SREBF-2 (siSREBF-2) (catalog no.: M-009549-00-0005) at the final concentration of 80 nM.
Cell sorting of mouse alpha, beta, and delta cell populations
Alpha, beta, and delta cell population were prepared from mouse pancreatic islets as previously described (30).
Lipoprotein preparation and LDL oxidation
LDL and VLDL were extracted from the plasma of healthy volunteers, by sequential ultracentrifugations, as previously described (58). Protein concentration of the lipoproteins was determined by using bicinchoninic acid assay (Pierce). Copper oxidation of LDL was done by incubating the native LDL with 5 μmol/l CuSO4 at 37 °C for 16 h followed by dialysis (three times) against PBS containing 0.1 mM ethylenediamine tetraacetic acid and storage at 4 °C.
RNA isolation, reverse transcription, and qPCR
An RNeasy Micro Kit (Qiagen) was used to extract total RNA from EndoC-βH1 cells (59). Genomic DNA was removed by DNAse treatment following the RNeasy Micro Kit protocol. RNAs were reverse transcribed by using the Maxima First Strand cDNA kit (Thermo Fisher Scientific). RT–qPCR was performed using Power SYBR Green mix (Applied Biosystems) with a QuantStudio 3 analyzer (Thermo Fisher Scientific). Custom primers were designed with Primer-Blast online, and their efficiency and specificity were determined for each pair by RT–qPCR on a serial dilution of complementary DNA samples. The list of primers is presented in Table S2. Relative quantification (2∧−dCt) was used to calculate the expression levels of each target gene, normalized to CYCLOPHILIN-A transcripts.
Western blot analysis
Western blot experiments were performed as described (59). For VLDLR immunoblot, cell extracts were loaded in 8% Bis–Tris polyacrylamide gels in the absence of heating and 2-beta-mercaptoethanol. The following antibodies were used: PCSK9 (1/200 dilution; catalog no.: AF3888; R&D Systems), diluted in Tris-buffered saline 0.1% Tween with 0.5% bovine serum albumin (BSA); LDLR (1/1000 dilution; catalog no.: ab52818; Abcam), VLDLR (1/1000 dilution; catalog no.: ab75591; Abcam), alpha-tubulin (1/2000 dilution; catalog no.: T9026; Sigma–Aldrich), and beta-actin (1/2000 dilution; catalog no.: A5441; Sigma–Aldrich). Species-specific horseradish peroxidase–linked secondary antibodies (1/1000 dilution; catalog nos.: 7074 and 7076; Cell Signaling Technology) were used. Antibodies were validated by knockdown experiments (PCSK9) or have passed application-specific testing standards. Densitometric quantification of Western blots was done using ImageJ software (U. S. National Institutes of Health) and normalized to α-tubulin or β-actin expression.
PCSK9 secretion
EndoC-βH1, HepG2, HEK293T, and human islets were cultivated for 1 h in their respective culture media free from BSA or FCS. These cells were then cultivated for 24 h in fresh BSA/FCS-free culture medium. Conditioned media were collected and centrifuged to eliminate cell debris. PCSK9 expression was studied by Western blot on a volume of conditioned medium normalized to cell count. In the case of human islets, secreted proteins were enriched following precipitation with acetone before Western blot analysis.
Flow cytometry
EndoC-βH1 cells were trypsinized, washed three times in Hanks' balanced salt solution (HBSS)/1% BSA and incubated at 4 °C for 15 min in the dark with the following antibodies: LDLR (1/100 dilution; catalog no.: MAB CL 472413; Fisher scientific), CD36 (1/10 dilution; catalog no.: 555455; BD Pharmingen), PDL1 (1/100 dilution; catalog no.: 329714; BioLegend), HLA-ABC (1/100 dilution; catalog no.: 3114410; BioLegend) diluted in HBSS medium/1% BSA (Gibco/Roche Diagnostics). Fluorochrome-conjugated primary antibodies were used with the exception of LDLR detection, for which a step of incubation with Alexa-Fluor 488 (1/400 dilution; Invitrogen) secondary antibody was performed. Following rinsing in HBSS/1% BSA and incubation with FACS medium containing propidium iodide (1/4000 dilution; Sigma–Aldrich), analysis was carried out using an FACS Aria III (BD Biosciences). Dead cells were excluded from analyses by using propidium iodide. Data were analyzed using FlowJo 10.7 software (Research Resource Identifier: SCR_008520; BD Life Sciences). Results are expressed in mean fluorescence intensity fold changes relative to the control condition.
Insulin secretion
EndoC-βH1 cells were transfected with siCTRL or siPCSK9. Two days later, cells were starved in DMEM containing 0.5 mM glucose for 24 h. Then, cells were washed twice and then preincubated in Krebs–Ringer bicarbonate Hepes buffer containing 0.2% fatty acid–free BSA in the absence of glucose for 1 h. Insulin secretion was measured following a 1 h incubation with Krebs–Ringer bicarbonate Hepes buffer containing 0.2% fatty acid–free BSA and 0 mM or 20 mM glucose or 50 mM KCl. Insulin secretion and intracellular insulin were measured by ELISA as previously described (42).
Fatty acid uptake
Fatty acid uptake in EndoC-βH1 was studied using the Fatty Acid Uptake Kit (catalog no.: MAK156; Sigma–Aldrich). Cells were transfected with siCTRL or siPCSK9. Six days later, cells were starved for 1 h in cell culture medium free from BSA. TF2-C12 fluorescent fatty acid was added to the culture medium for 1 h. Fluorescence signal was measured at λexcitation = 485 nm/λemission = 520 nm and normalized to cell count. Fatty acid uptake was calculated by subtracting the fluorescence intensity measured at 1 h incubation by that measured at time point 0, following the manufacturer’s instructions. Results were expressed in fatty acid uptake fold changes relative to control.
RNA-Seq and proteomics analyses
Acquisition of proteomics and RNA-Seq data and sample size calculation for omics experiments have been previously described (35). Benjamini–Yekutieli method (60) was used to adjust for multiple testing for ingenuity pathway analysis terms because of nonignorable overlap between genes underlying top pathways. Gene set enrichment analysis was conducted with GSEA, v4.0.3 software (Research Resource Identifier: SCR_003199; UC San Diego and Broad Institute).
Statistical analysis of RT–qPCR, Western blot, and FACS experiments
Each n represents an independent experiment. All the graphs show means ± SD, and the statistical analysis was conducted using Prism (GraphPad Software, Inc) either with t test if the normality condition was respected or with the nonparametric equivalent Mann–Whitney test. A p < 0.05 was considered significant, and symbols for indicating p values are ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.005.
Data availability
The datasets generated during and/or analyzed during the current study were previously published (35) and are available from the corresponding authors.
RNA-Seq data have been deposited to Gene Expression Omnibus database with accession number GSE182016. Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the following identifiers: PXD027921, PXD027911, and PXD027913.
Supporting information
This article contains supporting information..
Conflict of interest
K. Sengupta, M. R., G. M. H., A. F. J., C. R. U., and S. A. are employed by AstraZeneca. R. S. is a shareholder and consultant for Univercell Biosolutions. All other authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Natascha de Graaf (Leiden University Medical Center) for technical support. We also thank Latif Rachdi for helpful discussions.
Author contributions
K. Saitoski, M. R., G. M. H., A. F. J., K. Sengupta, S. A., and R. S. conceptualization; K. Saitoski, M. R., G. M. H., A. F. J., K. Sengupta, and R. S. methodology; M. R. and A. F. J. software; K. Saitoski, M. R., G. M. H., A. F. J., C. B., F. C., K. Sengupta, C. R. U., S. A., I. G., W. L. G., and R. S. validation; K. Saitoski, M. R., and A. F. J. formal analysis; K. Saitoski, G. M. H., C. B., F. C., I. G., and W. L. G. investigation; F. C., M. A., I. G., and W. L. G. resources; K. Saitoski, M. R., G. M. H., and A. F. J. data curation; K. Saitoski and R. S. Writing–original draft; K. Saitoski, M. R., G. M. H., A. F. J., K. Sengupta, C. R. U., S. A., I. G., W. L. G., and R. S. writing–review & editing; K. Saitoski, M. R., and R. S. visualization; K. Sengupta and R. S. supervision; K. Saitoski and R. S. project administration; R. S. funding acquisition.
Funding and additional information
The R. S. laboratory is supported by grants from the Fondation pour la Recherche Médicale (grant no.: EQU201903007793), the Dutch Diabetes Research Foundation, the DON Foundation, the Fondation Francophone pour la Recherche sur le Diabetes, the Agence Nationale de la Recherche (grant no.: ANR-19-CE15-0014-01), Laboratoire d’Excellence Consortium Revive, the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement nos. 115797 (INNODIA) and 945268 (INNODIA HARVEST). This joint undertaking receives support from the Union's Horizon 2020 Research and Innovation program EFPIA, JDRF, and the Leona M. and Harry B. Helmsley Charitable Trust.
Edited by Qi-Qun Tang
Supporting information
References
- 1.Seidah N.G., Benjannet S., Wickham L., Marcinkiewicz J., Jasmin S.B., Stifani S., et al. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. U. S. A. 2003;100:928–933. doi: 10.1073/pnas.0335507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaid A., Roubtsova A., Essalmani R., Marcinkiewicz J., Chamberland A., Hamelin J., et al. Proprotein convertase subtilisin/kexin type 9 (PCSK9): hepatocyte-specific low-density lipoprotein receptor degradation and critical role in mouse liver regeneration. Hepatol. Baltim. Md. 2008;48:646–654. doi: 10.1002/hep.22354. [DOI] [PubMed] [Google Scholar]
- 4.Seidah N.G., Awan Z., Chrétien M., Mbikay M. PCSK9: a key modulator of cardiovascular health. Circ. Res. 2014;114:1022–1036. doi: 10.1161/CIRCRESAHA.114.301621. [DOI] [PubMed] [Google Scholar]
- 5.Dubuc G., Chamberland A., Wassef H., Davignon J., Seidah N.G., Bernier L., et al. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 2004;24:1454–1459. doi: 10.1161/01.ATV.0000134621.14315.43. [DOI] [PubMed] [Google Scholar]
- 6.Jeong H.J., Lee H.-S., Kim K.-S., Kim Y.-K., Yoon D., Park S.W. Sterol-dependent regulation of proprotein convertase subtilisin/kexin type 9 expression by sterol-regulatory element binding protein-2. J. Lipid Res. 2008;49:399–409. doi: 10.1194/jlr.M700443-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Costet P., Cariou B., Lambert G., Lalanne F., Lardeux B., Jarnoux A.-L., et al. Hepatic PCSK9 expression is regulated by nutritional status via insulin and sterol regulatory element-binding protein 1c. J. Biol. Chem. 2006;281:6211–6218. doi: 10.1074/jbc.M508582200. [DOI] [PubMed] [Google Scholar]
- 8.Abifadel M., Varret M., Rabès J.-P., Allard D., Ouguerram K., Devillers M., et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 9.Cohen J., Pertsemlidis A., Kotowski I.K., Graham R., Garcia C.K., Hobbs H.H. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 10.Seidah N.G., Prat A., Pirillo A., Catapano A.L., Norata G.D. Novel strategies to target proprotein convertase subtilisin kexin 9: beyond monoclonal antibodies. Cardiovasc. Res. 2019;115:510–518. doi: 10.1093/cvr/cvz003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb Y.N. Inclisiran: first approval. Drugs. 2021;81:389–395. doi: 10.1007/s40265-021-01473-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt A.F., Swerdlow D.I., Holmes M.V., Patel R.S., Fairhurst-Hunter Z., Lyall D.M., et al. PCSK9 genetic variants and risk of type 2 diabetes: a mendelian randomisation study. Lancet Diabetes Endocrinol. 2017;5:97–105. doi: 10.1016/S2213-8587(16)30396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray K.K., Colhoun H.M., Szarek M., Baccara-Dinet M., Bhatt D.L., Bittner V.A., et al. Effects of alirocumab on cardiovascular and metabolic outcomes after acute coronary syndrome in patients with or without diabetes: a prespecified analysis of the ODYSSEY OUTCOMES randomised controlled trial. Lancet Diabetes Endocrinol. 2019;7:618–628. doi: 10.1016/S2213-8587(19)30158-5. [DOI] [PubMed] [Google Scholar]
- 14.Sattar N., Toth P.P., Blom D.J., Koren M.J., Soran H., Uhart M., et al. Effect of the proprotein convertase subtilisin/kexin type 9 inhibitor evolocumab on glycemia, body weight, and new-onset diabetes mellitus. Am. J. Cardiol. 2017;120:1521–1527. doi: 10.1016/j.amjcard.2017.07.047. [DOI] [PubMed] [Google Scholar]
- 15.Nassoury N., Blasiole D.A., Tebon Oler A., Benjannet S., Hamelin J., Poupon V., et al. The cellular trafficking of the secretory proprotein convertase PCSK9 and its dependence on the LDLR. Traffic Cph. Den. 2007;8:718–732. doi: 10.1111/j.1600-0854.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 16.Park S.W., Moon Y.-A., Horton J.D. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver. J. Biol. Chem. 2004;279:50630–50638. doi: 10.1074/jbc.M410077200. [DOI] [PubMed] [Google Scholar]
- 17.Poirier S., Mayer G., Poupon V., McPherson P.S., Desjardins R., Ly K., et al. Dissection of the endogenous cellular pathways of PCSK9-induced low density lipoprotein receptor degradation: evidence for an intracellular route. J. Biol. Chem. 2009;284:28856–28864. doi: 10.1074/jbc.M109.037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirier S., Mayer G., Benjannet S., Bergeron E., Marcinkiewicz J., Nassoury N., et al. The proprotein convertase PCSK9 induces the degradation of low density lipoprotein receptor (LDLR) and its closest family members VLDLR and ApoER2. J. Biol. Chem. 2008;283:2363–2372. doi: 10.1074/jbc.M708098200. [DOI] [PubMed] [Google Scholar]
- 19.Labonté P., Begley S., Guévin C., Asselin M.-C., Nassoury N., Mayer G., et al. PCSK9 impedes hepatitis C virus infection in vitro and modulates liver CD81 expression. Hepatol. Baltim. Md. 2009;50:17–24. doi: 10.1002/hep.22911. [DOI] [PubMed] [Google Scholar]
- 20.Canuel M., Sun X., Asselin M.-C., Paramithiotis E., Prat A., Seidah N.G. Proprotein convertase subtilisin/kexin type 9 (PCSK9) can mediate degradation of the low density lipoprotein receptor-related protein 1 (LRP-1) PLoS One. 2013;8:e64145. doi: 10.1371/journal.pone.0064145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demers A., Samami S., Lauzier B., Des Rosiers C., Ngo Sock E.T., Ong H., et al. PCSK9 induces CD36 degradation and affects long-chain fatty acid uptake and triglyceride metabolism in adipocytes and in mouse liver. Arterioscler. Thromb. Vasc. Biol. 2015;35:2517–2525. doi: 10.1161/ATVBAHA.115.306032. [DOI] [PubMed] [Google Scholar]
- 22.Jonas M.C., Costantini C., Puglielli L. PCSK9 is required for the disposal of non-acetylated intermediates of the nascent membrane protein BACE1. EMBO Rep. 2008;9:916–922. doi: 10.1038/embor.2008.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharotri V., Collier D.M., Olson D.R., Zhou R., Snyder P.M. Regulation of epithelial sodium channel trafficking by proprotein convertase subtilisin/kexin type 9 (PCSK9) J. Biol. Chem. 2012;287:19266–19274. doi: 10.1074/jbc.M112.363382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., Bao X., Hu M., Chang H., Jiao M., Cheng J., et al. Inhibition of PCSK9 potentiates immune checkpoint therapy for cancer. Nature. 2020;588:693–698. doi: 10.1038/s41586-020-2911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langhi C., Le May C., Gmyr V., Vandewalle B., Kerr-Conte J., Krempf M., et al. PCSK9 is expressed in pancreatic delta-cells and does not alter insulin secretion. Biochem. Biophys. Res. Commun. 2009;390:1288–1293. doi: 10.1016/j.bbrc.2009.10.138. [DOI] [PubMed] [Google Scholar]
- 26.Mbikay M., Sirois F., Mayne J., Wang G.-S., Chen A., Dewpura T., et al. PCSK9-deficient mice exhibit impaired glucose tolerance and pancreatic islet abnormalities. FEBS Lett. 2010;584:701–706. doi: 10.1016/j.febslet.2009.12.018. [DOI] [PubMed] [Google Scholar]
- 27.Da Dalt L., Ruscica M., Bonacina F., Balzarotti G., Dhyani A., Di Cairano E., et al. PCSK9 deficiency reduces insulin secretion and promotes glucose intolerance: the role of the low-density lipoprotein receptor. Eur. Heart J. 2019;40:357–368. doi: 10.1093/eurheartj/ehy357. [DOI] [PubMed] [Google Scholar]
- 28.Peyot M.-L., Roubtsova A., Lussier R., Chamberland A., Essalmani R., Murthy Madiraju S.R. Substantial PCSK9 inactivation in β-cells does not modify glucose homeostasis or insulin secretion in mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2021;1866:158968. doi: 10.1016/j.bbalip.2021.158968. [DOI] [PubMed] [Google Scholar]
- 29.Ramin-Mangata S., Thedrez A., Nativel B., Diotel N., Blanchard V., Wargny M., et al. Effects of proprotein convertase subtilisin kexin type 9 modulation in human pancreatic beta cells function. Atherosclerosis. 2021;326:47–55. doi: 10.1016/j.atherosclerosis.2021.03.044. [DOI] [PubMed] [Google Scholar]
- 30.Berthault C., Staels W., Scharfmann R. Purification of pancreatic endocrine subsets reveals increased iron metabolism in beta cells. Mol. Metab. 2020;42:101060. doi: 10.1016/j.molmet.2020.101060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segerstolpe Å., Palasantza A., Eliasson P., Andersson E.-M., Andréasson A.-C., Sun X., et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 2016;24:593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xin Y., Kim J., Okamoto H., Ni M., Wei Y., Adler C., et al. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metab. 2016;24:608–615. doi: 10.1016/j.cmet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 33.Lawlor N., George J., Bolisetty M., Kursawe R., Sun L., Sivakamasundari V., et al. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res. 2017;27:208–222. doi: 10.1101/gr.212720.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blodgett D.M., Nowosielska A., Afik S., Pechhold S., Cura A.J., Kennedy N.J., et al. Novel observations from next-generation RNA sequencing of highly purified human adult and fetal islet cell subsets. Diabetes. 2015;64:3172–3181. doi: 10.2337/db15-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryaboshapkina M., Saitoski K., Hamza G.M., Jarnuczak A.F., Pechberty S., Berthault C., et al. Characterization of the secretome, transcriptome, and proteome of human β cell line EndoC-βH1. Mol. Cell. Proteomics. 2022;21:100229. doi: 10.1016/j.mcpro.2022.100229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maxwell K.N., Soccio R.E., Duncan E.M., Sehayek E., Breslow J.L. Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J. Lipid Res. 2003;44:2109–2119. doi: 10.1194/jlr.M300203-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Maechler P. Mitochondrial function and insulin secretion. Mol. Cell. Endocrinol. 2013;379:12–18. doi: 10.1016/j.mce.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 38.Taneera J., Fadista J., Ahlqvist E., Atac D., Ottosson-Laakso E., Wollheim C.B., et al. Identification of novel genes for glucose metabolism based upon expression pattern in human islets and effect on insulin secretion and glycemia. Hum. Mol. Genet. 2015;24:1945–1955. doi: 10.1093/hmg/ddu610. [DOI] [PubMed] [Google Scholar]
- 39.Colli M.L., Hill J.L.E., Marroquí L., Chaffey J., Dos Santos R.S., Leete P., et al. PDL1 is expressed in the islets of people with type 1 diabetes and is up-regulated by interferons-α and-γ via IRF1 induction. EBioMedicine. 2018;36:367–375. doi: 10.1016/j.ebiom.2018.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foulis A.K., Farquharson M.A., Hardman R. Aberrant expression of class II major histocompatibility complex molecules by B cells and hyperexpression of class I major histocompatibility complex molecules by insulin containing islets in type 1 (insulin-dependent) diabetes mellitus. Diabetologia. 1987;30:333–343. doi: 10.1007/BF00299027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lytrivi M., Castell A.-L., Poitout V., Cnop M. Recent insights into mechanisms of β-cell lipo- and glucolipotoxicity in type 2 diabetes. J. Mol. Biol. 2020;432:1514–1534. doi: 10.1016/j.jmb.2019.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ravassard P., Hazhouz Y., Pechberty S., Bricout-Neveu E., Armanet M., Czernichow P., et al. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J. Clin. Invest. 2011;121:3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Dong B., Park S.W., Lee H.-S., Chen W., Liu J. Hepatocyte nuclear factor 1alpha plays a critical role in PCSK9 gene transcription and regulation by the natural hypocholesterolemic compound berberine. J. Biol. Chem. 2009;284:28885–28895. doi: 10.1074/jbc.M109.052407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyosawa K., Watanabe Y., Murakami K., Murakami T., Shibata H., Iwashita M., et al. New CETP inhibitor K-312 reduces PCSK9 expression: a potential effect on LDL cholesterol metabolism. Am. J. Physiol. Endocrinol. Metab. 2015;309:E177–190. doi: 10.1152/ajpendo.00528.2014. [DOI] [PubMed] [Google Scholar]
- 45.Jennings R.E., Scharfmann R., Staels W. Transcription factors that shape the mammalian pancreas. Diabetologia. 2020;63:1974–1980. doi: 10.1007/s00125-020-05161-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliver-Krasinski J.M., Stoffers D.A. On the origin of the beta cell. Genes Dev. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Servitja J.M., Ferrer J. Transcriptional networks controlling pancreatic development and beta cell function. Diabetologia. 2004;47:597–613. doi: 10.1007/s00125-004-1368-9. [DOI] [PubMed] [Google Scholar]
- 48.Blum B., Hrvatin S., Schuetz C., Bonal C., Rezania A., Melton D.A. Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat. Biotechnol. 2012;30:261–264. doi: 10.1038/nbt.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Meulen T., Huising M.O. Maturation of stem cell-derived beta-cells guided by the expression of urocortin 3. Rev. Diabet. Stud. 2014;11:115–132. doi: 10.1900/RDS.2014.11.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pagliuca F.W., Millman J.R., Gürtler M., Segel M., Van Dervort A., Ryu J.H., et al. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rezania A., Bruin J.E., Arora P., Rubin A., Batushansky I., Asadi A., et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 52.Khan S., Kowluru A. CD36 mediates lipid accumulation in pancreatic beta cells under the duress of glucolipotoxic conditions: novel roles of lysine deacetylases. Biochem. Biophys. Res. Commun. 2018;495:2221–2226. doi: 10.1016/j.bbrc.2017.12.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagao M., Esguerra J.L.S., Asai A., Ofori J.K., Edlund A., Wendt A., et al. Potential protection against type 2 diabetes in obesity through lower CD36 expression and improved exocytosis in β-cells. Diabetes. 2020;69:1193–1205. doi: 10.2337/db19-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas H.E., Parker J.L., Schreiber R.D., Kay T.W. IFN-gamma action on pancreatic beta cells causes class I MHC upregulation but not diabetes. J. Clin. Invest. 1998;102:1249–1257. doi: 10.1172/JCI2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roep B.O., Thomaidou S., van Tienhoven R., Zaldumbide A. Type 1 diabetes mellitus as a disease of the β-cell (do not blame the immune system?) Nat. Rev. Endocrinol. 2021;17:150–161. doi: 10.1038/s41574-020-00443-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silvie O., Rubinstein E., Franetich J.-F., Prenant M., Belnoue E., Rénia L., et al. Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity. Nat. Med. 2003;9:93–96. doi: 10.1038/nm808. [DOI] [PubMed] [Google Scholar]
- 57.Oshima M., Knoch K.-P., Diedisheim M., Petzold A., Cattan P., Bugliani M., et al. Virus-like infection induces human β cell dedifferentiation. JCI Insight. 2018;3:97732. doi: 10.1172/jci.insight.97732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Redgrave T.G., Roberts D.C., West C.E. Separation of plasma lipoproteins by density-gradient ultracentrifugation. Anal. Biochem. 1975;65:42–49. doi: 10.1016/0003-2697(75)90488-1. [DOI] [PubMed] [Google Scholar]
- 59.Rachdi L., Maugein A., Pechberty S., Armanet M., Hamroune J., Ravassard P., et al. Regulated expression and function of the GABAB receptor in human pancreatic beta cell line and islets. Sci. Rep. 2020;10:13469. doi: 10.1038/s41598-020-69758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study were previously published (35) and are available from the corresponding authors.
RNA-Seq data have been deposited to Gene Expression Omnibus database with accession number GSE182016. Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the following identifiers: PXD027921, PXD027911, and PXD027913.