Abstract
Purpose
Segmentation and evaluation of in vivo confocal microscopy (IVCM) images requires manual intervention, which is time consuming, laborious, and non-reproducible. The aim of this research was to develop and validate deep learning–based methods that could automatically segment and evaluate corneal nerve fibers (CNFs) and dendritic cells (DCs) in IVCM images, thereby reducing processing time to analyze larger volumes of clinical images.
Methods
CNF and DC segmentation models were developed based on U-Net and Mask R-CNN architectures, respectively; 10-fold cross-validation was used to evaluate both models. The CNF model was trained and tested using 1097 and 122 images, and the DC model was trained and tested using 679 and 75 images, respectively, at each fold. The CNF morphology, number of nerves, number of branching points, nerve length, and tortuosity were analyzed; for DCs, number, size, and immature–mature cells were analyzed. Python-based software was written for model training, testing, and automatic morphometric parameters evaluation.
Results
The CNF model achieved on average 86.1% sensitivity and 90.1% specificity, and the DC model achieved on average 89.37% precision, 94.43% recall, and 91.83% F1 score. The interclass correlation coefficient (ICC) between manual annotation and automatic segmentation were 0.85, 0.87, 0.95, and 0.88 for CNF number, length, branching points, and tortuosity, respectively, and the ICC for DC number and size were 0.95 and 0.92, respectively.
Conclusions
Our proposed methods demonstrated reliable consistency between manual annotation and automatic segmentation of CNF and DC with rapid speed. The results showed that these approaches have the potential to be implemented into clinical practice in IVCM images.
Translational Relevance
The deep learning–based automatic segmentation and quantification algorithm significantly increases the efficiency of evaluating IVCM images, thereby supporting and potentially improving the diagnosis and treatment of ocular surface disease associated with corneal nerves and dendritic cells.
Keywords: deep learning, confocal microscopy, segmentation, corneal nerves, dendritic cells
Introduction
Dry eye disease (DED) is a multifactorial, immune-based inflammatory disease of the ocular surface and tears that includes ocular discomfort, dryness, pain, and alteration of tear composition, resulting in disturbance of tear production and tear evaporation. DED currently impacts 5% to 35% of the world's population, with variation in prevalence due to geographic location, age, and gender.1–3 Currently, DED is considered a major health problem due to its significant impact on the patient's quality of vision and life, which leads to socioeconomic burdens.4,5
Pain is defined as an unpleasant sensory and emotional experience that might exist or occur over a short or a prolonged period.6 A wide range of ocular disorders can cause ocular pain; however, the most common denominator of ocular pain is ocular surface and corneal disorders,7–9 including dry eye.10–12 In DED patients, this ocular pain not only is creating irritation but also indicates the severity of DED. Moein et al.7 and Galor et al.13 have shown that, compared with patients without ocular pain, patients who suffer from DED with ocular pain have more severe signs (such as decreased corneal nerve fibers [CNFs] number and density and increased DCs density) and symptoms (such as burning, hypersensitivity to wind, and sensitivity to light and temperature). Diabetic peripheral neuropathy (DPN) affects roughly 50% of the patients that suffer from diabetes and is the main driving factor for painful diabetic neuropathy. A major challenge is identifying early neuropathy, which predominantly affects small nerve fibers first, rather than advanced neuropathy, which later affects the large nerve fibers. Furthermore, DPN showed a progressive reduction in corneal nerve fiber density, length, and branching points in patients with increasing severity of diabetic neuropathy.14
In vivo confocal microscopy (IVCM) is a non-invasive optical imaging modality that enables histological visualization of the CNFs and dendritic cells (DCs). IVCM is clinically widely accepted and commonly used in the diagnosis of various ocular surface disorders because it provides high resolution and detailed morphometric information regarding CNFs and DCs.15,16 Previous work has utilized IVCM to demonstrate that the density of CNF is significantly reduced and the number of DCs is significantly higher in patients with dry eye.7,12,17–19 Furthermore, CNFs demonstrate early and progressive pathology in a range of peripheral and central neurodegenerative conditions.20–23 Previous studies have demonstrated analytical validation by showing that IVCM reliably quantifies early axonal damage in diabetic peripheral24,25 neuropathy with high sensitivity and specificity.26,27
For accurate quantification of CNF morphology and detection of DC, the nerves and DCs must be accurately segmented in the IVCM images. Traditionally, the methods used to segment CNFs and DCs have been manual or semi-automated techniques that require experienced personnel and are laborious, cost ineffective, and potentially subject to user bias. To address these issues, several automatic CNF segmentation and quantification software tools have been developed.28–31 For example, Dabbah et al.28,32 developed a dual-model automated CNF detection method that showed excellent correlation with manual grading (r = 0.92); further extension of this method used a dual-model property in a multi-scale framework to generate feature vectors at every pixel. The authors achieved high correlation with manual grading (r = 0.95).
In contrast to traditional image processing or machine learning–based image segmentation techniques, a deep learning–based method offers the advantage of learning useful features and representations from raw images automatically. This method is preferable, as manually extracted features selected for a specific corneal disease may not be generally transferable to other corneal diseases.33,34 The implementation of deep learning in the field of ophthalmology has increased in recent years.35–37 Using a convolutional neural network (CNN) and a large volume of image data, a neural network can learn to segment and detect specific objects from ophthalmic images such as retinal blood vessels,38 retinal layers,39 optic disc,40 anterior chamber angle,41 and meibomian glands.42 Several CNN-based CNF segmentation models have been proposed. Williams et al.43 demonstrated the efficacy of deep learning models to identify DPN with high interclass correlation with manual ground-truth annotation. Wei et al.44 and Colonna et al.45 developed a U-Net–based CNN model to segment CNF that provides high sensitivity and specificity in the segmentation task. Oakley et al.46 developed a U-Net–based model to analyze macaque corneal sub-basal nerve fibers which achieved high correlation between readers and CNN segmentation. Yıldız et al.47 proposed CNF segmentation using a generative adversarial network (GAN), and they achieved correlation and Bland–Altman analysis results similar to those for U-Net; however, the GAN showed higher accuracy compared to U-Net in receiver operating characteristic (ROC) curves. Most of the previous studies, however, have provided only nerve segmentation without further quantification, with the exception of Williams et al.,43 and all CNN-based models focus on CNFs, not DCs. Therefore, there is a need for fully automatic segmentation and quantification of both CNFs and DCs.
The aim of this research study was to develop and validate deep learning methods for automatic CNF and DC segmentation and the morphometric evaluation of IVCM images.
Materials and Methods
Datasets
Corneal Nerve Fiber Dataset
In total, 1578 images were acquired from both normal (n = 90) and pathological (n = 105, including 52 patients with diabetes) subjects using the Heidelberg Retina Tomograph 3 (HRT3) with Rostock Cornea Module (RCM) (Heidelberg Engineering GmbH, Heidelberg, Germany) at the Peking University Third Hospital, Beijing, China. This dataset is available for further research purposes on request.48,49 After removing the 359 duplicate images, the remaining 1219 images were used for our model training and testing. All images cover an area of 400 × 400 µm (384 × 384 pixels). On average, eight images were obtained per subject. The images were from both central and inferior whorl regions.
Dendritic Cells Dataset
A total of 754 clinical IVCM images of both males and females from patients with dry eye and neuropathic corneal pain (n = 54) were collected using the HRT3-RCM at the Eye Hospital, University Hospital Cologne, Germany. These images have the same dimensions as the cornea nerve dataset (400 × 400 µm; 384 × 384 pixels). On average, 14 images were obtained per subject. The image collection adhered to the tenets of the Declaration of Helsinki and was approved by the ethics and institutional review board (IRB) at the University of Cologne, Germany (#16-405). As this study was conducted retrospectively using completely anonymized non-biometric image data, the ethics committee and IRB waived the need to obtain informed consent from the participants.
Image Annotation
Corneal Nerve Fiber Annotation
The CNF dataset from Peking University Third Hospital, Beijing, China, was manually annotated using ImageJ (National Institutes of Health, Bethesda, MD) with the NeuronJ (Biomedical Imaging Group, Lausanne, Switzerland) plug-in. All ground-truth images were verified and corrected by experienced ophthalmologists, thus providing validated ground-truth masks images along with raw IVCM images for further research.48,49
Dendritic Cells Annotation
All collected DC images were manually annotated using VGG Image Annotator (VIA),50 an open-source, web-based, image annotation software. The user draws a polygon region around the DC body and hyperreflective (dendrite) area. The VIA software tool saves all polygons in a JSON file format, which forms the ground-truth mask for the DC image dataset. One author (MAKS) created all of the initial ground-truth masks. The initial ground-truth masks were then verified and corrected by an experienced senior ophthalmologist (PS) from the University Hospital Cologne, Germany, before they were used in deep learning model training, validation, and testing.
Data Allocation
Corneal Nerve Fiber Data Allocation
The CNF image dataset, comprised of a total of 1219 images, was divided into training and testing datasets. Among 1219 images, ∼1097 were used for training, and ∼122 were used to test the model at each fold during the cross-validation.
Dendritic Cells Data Allocation
Images of DCs were also divided into training, and testing datasets. Among 754 images, ∼679 were used for model training and ∼75 images were used to test the model at each fold during cross-validation. To increase the number of training images and reduce the risk of model overfitting during the model training phase, data augmentation techniques were applied. The applied augmentations were horizontal flip, vertical flip, rotation (90°), gamma contrast (±30%), and random crop (25%) with subsequent resizing to original image size (Supplementary Fig. S1). Data augmentation increased the total number of training images to around 4753 (679 × 7).
Deep Learning Model Design and Training
Corneal Nerve Fiber Segmentation Model
The CNF segmentation model was based on the U-Net architecture.51 A U-Net CNN architecture establishes a pixel-wise segmentation map to attain full-image resolution segmentation which makes it an ideal choice for medical image segmentation.52–54 The detailed U-Net network architecture is shown in Figure 1. The ability of U-Net to provide pixel-level segmentation is a feature of the two sides of the “U” shape which form an encoding and a decoding path, respectively. The general pattern of encoding path includes a repeating group of convolution, dropout, convolution, batch normalization, and max pooling layer; the pattern of decoding path includes a repeating group of transpose convolution, concatenation, convolution, dropout, convolution, and batch normalization. The direct connections between the encoding path and the decoding path allow reuse of the extracted features and strengthen feature propagation. At the end of the network architecture, the fully connected layer used a sigmoid activation function to produce the probabilistic segmentation map. This probabilistic segmentation map was converted into a binary image using a cut-off threshold value of 0.1.
Figure 1.
Detailed U-Net architecture. Each dark blue rectangular block represents a multi-channel feature map passing through 3 × 3 convolution followed by rectified linear unit (ReLU) operations. Dark gray and light blue blocks denote dropout with a rate of 0.2 and batch normalization. Orange and dark yellow blocks denote 2 × 2 max pooling and 3 × 3 transpose convolution, respectively. Green blocks denote the concatenation of feature maps. The light gray block denotes a 1 × 1 convolution operation followed by sigmoid activation. The number of convolution filters is indicated at the top of each column.
A topology-preserving loss function, clDice,55 was used during the CNF segmentation model training. Briefly, clDice preserves connectivity while segmenting tubular-like structures. One of the most efficient gradient-based stochastic optimization algorithms, Adam,56 was used to optimize the CNF segmentation model during training. It optimizes individual learning rates for individual parameters used in the model training. The CNF segmentation model was trained for 50 epochs with an initial learning rate of 0.0001, a momentum of 0.9, and a batch size of 32 per fold during cross-validation.
Dendritic Cells Detection Model
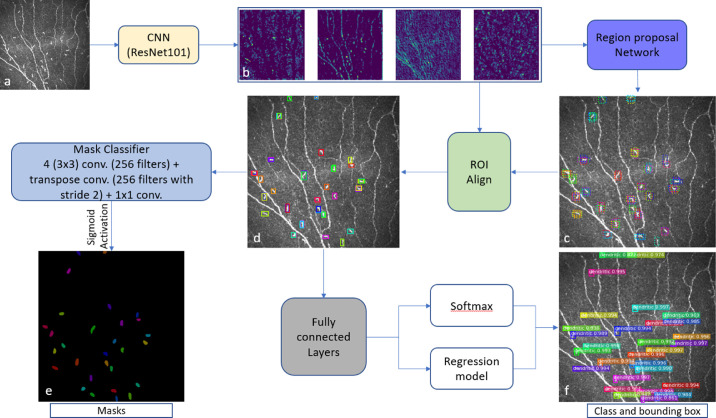
The DC detection model was adopted from the Mask R-CNN architecture.57 Mask R-CNN is divided into two stages: a Region Proposal Network (RPN), which proposes bounding boxes and objects, followed by a binary mask classifier to generate segmentation masks for each detected object inside the bounding box. The detailed Mask R-CNN network architecture is presented in Figure 2. First, the CNN (light yellow box in Fig. 2), based on the ResNet10158 backbone that was pretrained with the MS COCO dataset,59 generates a feature map (Fig. 2b) from the input image (Fig. 2a). Then, the RPN network (denoted by the purple box in Fig. 2) generates multiple regions of interest (ROIs) (dotted bounding box in Fig. 2c) using predefined bounding boxes referred to as anchors. Then, the ROI align network (green box in Fig. 2) takes both the proposed bounding boxes from the RPN network and the feature maps from the ResNet101 CNN as inputs and uses this information to find the best-fitting bounding box (Fig. 2d) for each proposed dendritic cell.
Figure 2.
Mask R-CNN architecture. The pretrained ResNet101 (light yellow box) generates feature maps (b) from the input image (a). From the feature maps, the RPN (purple box; 3 × 3 convolution with 512 filters, padding the same) generates multiple ROIs (dotted bounding box) with the help of predefined bounding boxes referred to as anchors (c). The green box denotes the ROI Align network, which takes both the proposed bounding boxes from the RPN network and the feature maps as inputs and uses this information to find the best-fitting bounding box (d) for each proposed DC. These aligned box maps are fed into fully connected layers (7 × 7 convolution with 1024 filters + 1 × 1 convolution with 1024 filters), denoted by the gray box, and then generates a class and bounding box for each object using softmax and a regression model, respectively (f). Finally, the aligned box maps are fed into the Mask classifier (4 3 × 3 convolution with 256 filters + transpose convolution with 256 filters and stride = 2 + 1 × 1 convolution + sigmoid activation), denoted by the light blue box, to generate binary masks for each object (e).
These aligned bounding box maps are then fed into fully connected layers (gray box in Fig. 2) to predict object class and bounding boxes (Fig. 2f) using softmax and regression models, respectively. Finally, the aligned bounding box maps are also fed into another CNN (light blue box in Fig. 2) consisting of four convolutional layers, transpose convolution, and sigmoid activation. This CNN is named the mask classifier, and it generates binary masks (Fig. 2e) for every detected DC. The complete Mask R-CNN uses a multi-task loss function that combines object class, bounding box, and segmentation mask. The loss function is
| (1) |
where Lloss is the total loss, and Lclass, Lbox, and Lmask are the loss of object class, bounding box, and segmented masks, respectively. The model training occurred in two steps with the Adam56 optimizer. First, the model was trained for 25 epochs with a learning rate of 0.0001 and a momentum of 0.9 and without data augmentations. In the second stage, we trained our model for another 25 epochs with a reduced initial learning rate of 0.00001, with momentum of 0.9, and with data augmentations per fold during cross-validation. We removed all DC detections with less than 90% confidence.
Both deep learning models training and testing were conducted on a laptop computer running Windows 10 Professional on a 64-bit Intel Core i7-9750H processor at 2.6 GHz with 12 MB of cache memory; Samsung 970 PRO NVMe Series SSD 512 GB M.2; Samsung 970 PRO PCIe 3.0 × 4 NVMe, RAM 32 GB DDR4 at 2666 MHz; and NVIDIA GeForce RTX 2070 Max-Q with 8 GB of GDDR6 memory. Data preparation, deep learning model design, training, evaluation, and testing were written in Python 3.6.6 using Keras 2.3.1 with TensorFlow 1.14.0 (CUDA 10.0, cuDNN 7.6.2) as the backend.
Cross-Validation Study
To evaluate our deep learning models, k-fold cross-validation was used. In this study, k = 10 was applied. The entire dataset of images (1219 for CNFs and 754 for DCs) was randomly split into 10 subgroups, and each time nine groups were selected as a training dataset and one group was selected as a test dataset. In total ∼1097 and ∼122 images were randomly selected as training and testing datasets, and ∼679 and ∼75 images were randomly selected as training and testing datasets, respectively, for the CNF and DC segmentation models on every fold for training. To improve the accuracy of the CNF and DC models, an ensemble network of 10 trained models obtained using 10-fold cross-validation was used. The final segmentation was computed by a majority vote over the segmentation of the ensemble network.
Evaluation Metrics
For the CNF segmentation and DC detection tasks, sensitivity (Sen) and specificity (Spe) were used for CNFs, and precision (P), recall (R), and F1 score were used for DC detection. The CNF or DC pixels (e.g., white pixels in the binary predicted masks) are considered as positive instances. Based on the combination of the ground-truth masks and predicted masks, these pixels are categorized into four categories: true positive (TP), true negative (TN), false positive (FP), and false negative (FN). Sensitivity, specificity, precision, and recall are defined by the following equations, respectively:
| (2) |
| (3) |
| (4) |
| (5) |
The F1 score is a weighted average of precision and recall and is defined by the following equation:
| (6) |
Morphometric Parameter Assessment
To better analyze the CNF morphology, the number of nerves (number/frame), number of branching points (number/frame), length (mm), and tortuosity were measured; for the DC morphology, total cell number, cell size (µm2), and mature–immature cell number were determined. All of these morphometric parameters were directly computed from the binary segmented image.
Nerve Length
To compute the nerve length (mm), the binary segmented image was first skeletonized. The branching points were then found to break up the total nerve into branches segments. Finally, the total CNF length was calculated by summing up the distance between two consecutive pixels in the nerve branch segments using the following equation:
| (7) |
where N is the total number of pixels of a nerve segment, and (xi, yi) is the coordinate of the corresponding pixel; the inter-pixel distance is 1.0416 µm.
Nerve Tortuosity
For a curvilinear structure such as corneal nerves, tortuosity is a useful metric to calculate the curvature changes of the nerves. In this study, the average tortuosity of all detected CNFs in the image was calculated. First, the nerve length was calculated for each nerve segments. Then, the tortuosity60 τ was calculated for each nerve segment (n) dividing the path length by the straight distance (Euclidean distance) between the start and end points of that nerve segment as follows:
| (8) |
Finally, the average tortuosity was calculated for the whole image by calculating the arithmetic average of the tortuosity values derived from each nerve segment. To calculate the average tortuosity, both main nerves (red) and branch nerves (orange) (Fig. 3c) were considered.
Figure 3.
Calculation of number of corneal nerves. First, the corneal nerves were divided into clusters (A, B, and C) and then each start and end point was provided with a cluster point number (A1, A2, …, B1, B2, …, C1, C2, …). After that, an abstract graph was created for each cluster using the associated nodes and edges. Then, all possible paths (if branch nerves exist in a cluster such as clusters A and C) within the abstract graph were created, and the tortuosity was calculated for each path. The main nerve was selected with the lowest tortuosity for all clusters (a). All nodes creating this main nerve were removed and the process was run again to find another main nerve if one exists (cluster C); otherwise, the remaining nerve was considered the branch nerve (cluster A) (b). If there was no additional main nerve, then the remaining nodes and edges were considered branch nerves (orange and green in cluster C) (c). Finally, the total number of corneal nerves was calculated by summing the red and orange nerves while discarding the branch nerves (green) with length less than 20% (80 µm) of the image.
Branching Points
To calculate the branching points, we first automatically defined the start and end points for each nerve segment. All of these points are stored and evaluated. Then, if the same point is both a start and an end point for one nerve segment and a start and en end point for a different nerve segment, then this point is classified as a branch point. For example, in cluster A (Fig. 3a), there are three nerve segments (A1–A3, A2–A3, and A3–A4) and four points. A1–A2 is the starting point, and A4 is the end point. However, A3 is both an end point for the A1–A3 and the A2–A3 nerve segments and a start point for the A3–A4 nerve segments. So, A3 is considered a branch point.
Number of Corneal Nerves
To calculate the total number of nerves presented in a frame, we first created an abstract graph using the nodes (corresponding to the start or end points of nerve segments) and edges (corresponding to the nerve segments between the start and end points) for each separated CNF cluster. Then, we extracted all of the possible paths in the graph between the isolated nodes (the start/end points that are not branching points), thus ensuring that all of the paths were “long”—more specifically, that they extended through the entire CNF cluster. Next, the tortuosities for each path were calculated. The main nerve was selected as the path with the lowest tortuosity (depicted as the red nerves in Fig. 3a). We then removed the main nerve and the branch nerves connected to it from the abstract graph and iteratively re-ran the process to find other main nerves on that CNF cluster, if existing (cluster C in Fig. 3b), until there are only two nodes left. Otherwise, the remaining edges were considered branch nerves (cluster A in Fig. 3b). Finally, we calculated the total number of nerves in a frame by summing the number of main nerves (red) and branch nerves (orange) that were longer than a predetermined threshold length. Here, we discarded nerves whose length was less than 20% (80 µm) of the image height (green). However, these discarded nerves were still considered for total nerve length, branching points, and tortuosity calculations.
Comparison with ACCMetrics
We compared corneal nerve fiber length (CNFL) using the widely used automated CNF analysis software, ACCMetrics (Early Neuropathy Assessment Group, University of Manchester, Manchester, UK).29
DC Size
The DC size, representing the total area surrounding the cell body and hyperreflective (dendrites) area,61,62 was measured by calculating the segmented polygon area around the DC and reported as square micrometers.
Immature and Mature DCs
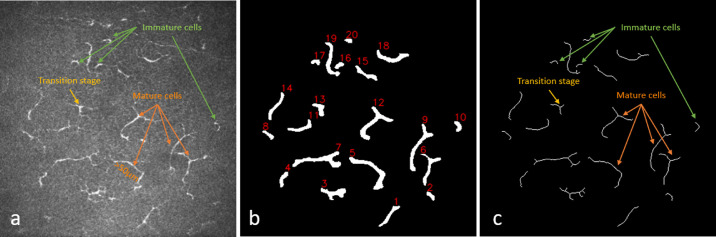
Hamrah et al.63 differentiated immature DCs from mature DCs by their location and the absence or presence of dendrites; immature cells were located at the center of the cornea and had no dendrites, and mature cells were located in the peripheral cornea and had dendrites. Building on this classification system, Zhivov et al.64 proposed three types of DCs: (1) individual cell without dendrites (immature cell), (2) cell with small dendrites in the transition stage (starting dendrite processes), and (3) cell with long dendrites (mature cell). To differentiate among the three types of DCs, a total cell length threshold and the presence of dendrites were used as parameters. Mature cells were defined as greater than 50 µm in length with or without the presence of dendrites; immature cells were defined as cells without the presence of dendrites and less than 50 µm in length.65,66 Finally, dendritic cells in the transition stage were defined as less than 50 µm in length with the presence of dendrites (Fig. 4a). To calculate the cell length and number of dendrites, first the binary segmented image (Fig. 4b) was skeletonized into one pixel width (Fig. 4c). Then, total cell length including dendrites and the presence of dendrites (branch numbers) were calculated for each individual cell presented in the skeletonized image using the Python-based library FilFinder 1.7.
Figure 4.
Immature and mature cell number calculation. (a) Original image, (b) binary segmented image with individual cell identification number, and (c) skeletonize image. The green arrows indicate immature cells (<50 µm) without dendrites, yellow arrows indicate transition-stage cells (<50 µm) with dendrites, and orange arrows indicate mature cells (>50 µm) with or without dendrites.
Statistics
Our proposed CNF and DC models were compared with manual segmentation and detection. The performance of both deep learning models was measured using the Bland–Altman method.67 Interclass correlation coefficient (ICC)68 was used to measure agreement between manual annotation and deep learning segmentation. Python 3.6.6–based SciPy 1.5.2 and NumPy 1.18.1 libraries were used for the statistical analysis.
Results
CNF Segmentation
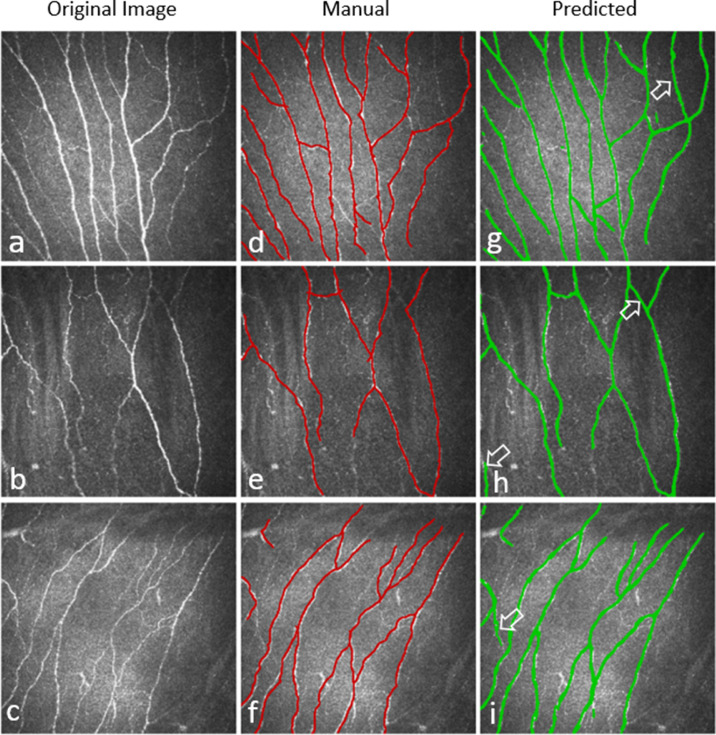
Training the CNF segmentation model for 50 epochs with a batch size of 32 per epoch on the full training dataset took approximately 10 hours and 28 minutes to complete, and segmenting the 122 testing images took around 9 minutes per fold on the described hardware and software. Figure 5 presents an example of testing images, along with their respective manual annotations, and automated segmentation obtained by our developed model. In general, the CNF segmentation model reliably segmented all testing images in each fold. The model achieved on average 86.1% ± 0.008% sensitivity and 90.1% ± 0.005% specificity with an average area under the ROC curve (AUC) value of 0.88 ± 0.01 (Supplementary Fig. S2) during 10-fold cross-validation.
Figure 5.
Three examples of CNF segmentation. (a–c) Original image, (d–f) manually segmented CNFs, and (g–i) predicted CNFs by the deep learning model. White marked arrows in the predicted images indicate the thin CNFs that were predicted by the deep learning model but were not annotated in the ground-truth images.
Morphometric Parameter Assessment
The morphometric parameters of CNF number, CNF length (mm), number of branching points, and tortuosity were computed from the binary segmented image. These are important clinical parameters to analyze the CNF health status. Our software provides all of these morphometric parameters automatically from the binary segmented image. Figure 6 shows an example image where the total CNFs number (sum of red and orange) is 11, total nerve length is 3.59 mm (density, 22.43 mm/mm2), number of branching points is 9, and average tortuosity of the nerves present in the frame is 1.18.
Figure 6.
Example of an automatic CNF quantification from the segmented binary image of the deep learning model. (a) Original image, and (b) binary segmented image with automatic quantification.
Morphological parameters from the automatically segmented images from the test dataset of 10-fold cross-validation were compared with the parameters from the manually annotated images with the P values (paired t-test) presented in the Table. The ICC of CNF number, length, branching points, and tortuosity were compared between automatic segmentation and manual annotation, and the values were 0.85, 0.87, 0.95, and 0.88, respectively, for 1219 (∼122 × 10) testing images from 10-fold cross-validation.
Table.
Morphometric Parameter Analysis of 10-Fold CNF Testing Dataset
| Mean ± SD | |||
|---|---|---|---|
| Parameter | Manual Annotation | Automatic Segmentation | P |
| CNF number | 10.14 ± 3.37 | 9.97 ± 3.26 | 0.06 |
| CNF length (mm) | 4.29 ± 3.81 | 4.30 ± 3.85 | 0.97 |
| Branching points, n | 6.45 ± 5.99 | 6.36 ± 5.85 | 0.31 |
| Tortuosity | 2.05 ± 1.34 | 2.00 ± 1.32 | 0.40 |
P values are between manual annotation and automatic segmentation.
To determine the consistency between automatic and manual segmentation, Bland–Altman analysis was performed for CNF number, length (mm), branching points, and tortuosity. The results are presented in Figure 7. A total of 1219 (∼122 × 10) test images from 10-fold cross-validation were used for this analysis.
Figure 7.
Bland–Altman plots present the consistency of CNF number, CNF length (mm), number of branching points, and tortuosity between manual annotation and deep learning segmentation methods. The middle solid line indicates the mean value of the two methods, and the two dotted lines indicate the limits of agreement (±1.96 SD). The gray bands indicate a confidence interval of 95%.
Comparison With ACCMetrics
The total mean CNFL of 1219 (∼122 × 10) test images from 10-fold cross-validation were 26.81 mm/mm2, 26.87 mm/mm2, and 13.94 mm/mm2 for segmentation of manual ground truth, our proposed method, and ACCMetrics, respectively. The remaining quantification parameters from ACCMetrics software are tabulated in the Supplementary Table.
To better understand the difference between the proposed method and ACCMetrics, we performed further in-depth analyses by grouping the image dataset of 1219 images according to image quality: (1) high-/average-quality images (∼691), defined by uniform illumination and high contrast; (2) low-quality images (∼428), defined by non-uniform illumination, contrast variations, and artifacts; and (3) images of the endothelial layer, epithelial layer, and stromal keratocytes (∼100). For the high-/average-quality images, there was better concordance between ACCMetrics (20.03 mm/mm2) and the deep learning–based method (29.84 mm/mm2). Also, there was concordance between both methods in images of the endothelial layer, epithelial layer, and stromal keratocytes (where there were no nerves present). However, there was clear discordance between ACCMetrics (7.36 mm/mm2) and the deep learning–based method (22.98 mm/mm2) for low-quality images containing artifacts such as nerves slightly out of focus, strong specular reflection, faint nerves, nerves that are not continuous (interrupted), speckle noise, and low contrast. Comparisons between the ACCMetrics and deep learning–based segmentation methods for low-quality images are presented in Supplementary Figure S3.
Dendritic Cell Detection
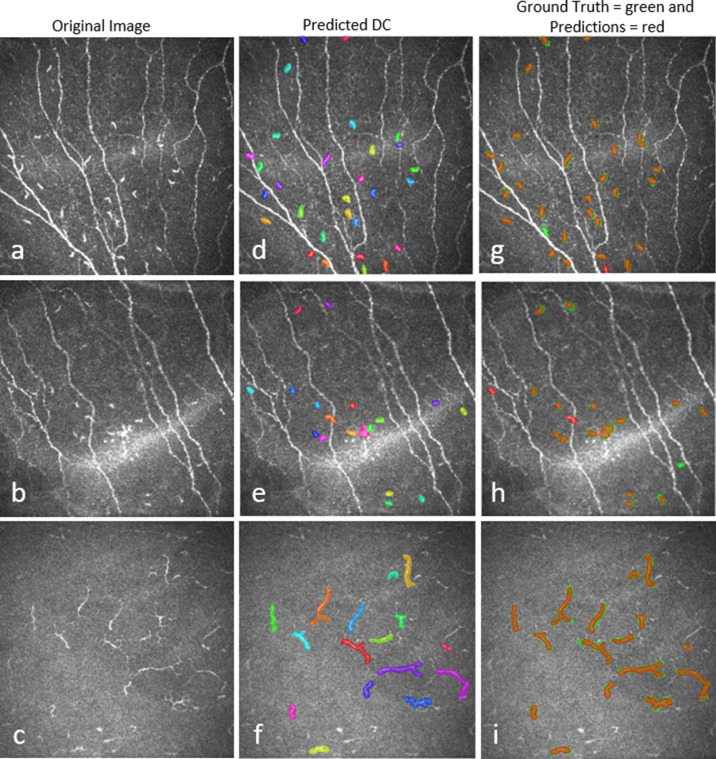
Training the DC detection model for 50 epochs, in two stages, on the full training dataset with data augmentation took approximately 3 hours per fold, and DC detection from 75 test images took around 3 minutes 45 seconds on the described hardware and software. A comparison between manual and automated DC detection of the test images is presented in Figure 8. The model achieved on average 89.37% ± 0.12% precision, 94.43% ± 0.07% recall, and 91.83% ± 0.09% F1 score. The means of the total number of DCs and DC size were 9.74 ± 7.74 and 11.41 ± 7.75 (P = 0.38) and 1269.40 ± 939.65 µm2 and 1296.84 ± 832.37 µm2 (P = 0.11), respectively, for 754 (∼75 × 10) testing images from 10-fold cross-validation of manual annotation and automatic segmentation.
Figure 8.
Three examples of DC prediction by the deep learning model. (a–c) Original image, d–f) predicted DC regions, and (g–i) overlay of manual annotation and predicted DCs.
Morphometric Parameter Assessment
The morphometric parameters of DC number, size (µm2), and number of immature, transition-stage, and mature cells were computed directly from the binary segmented image. These are important clinical parameters for analyzing corneal health status. Our software provides all of these morphometric parameters automatically from binary segmented images. Figure 9 shows an example image of automatic DC segmentation with morphometric evaluation where the total number of DCs was 20, with seven immature cells (cell identification numbers 2, 4, 8, 10, 16, 17, and 20), one transition-stage cell (cell identification number 13), and 12 mature cells (cell identification numbers 1, 3, 5, 6, 7, 9, 11, 12, 14, 15, 18, and 19); DC density was 125.0 cells/mm2, and total cell size was 7555.34 µm2.
Figure 9.
DC segmentation and quantification. (a) Original image, (b) segmentation overlay with quantification parameters, and (c) skeletonized binary segmented image with cell identification numbers. The immature cell identification numbers are 2, 4, 8, 10, 16, 17, and 20; the transition-stage cell identification number is 13; and the mature cell identification numbers are 1, 3, 5, 6, 7, 9, 11, 12, 14, 15, 18, 19.
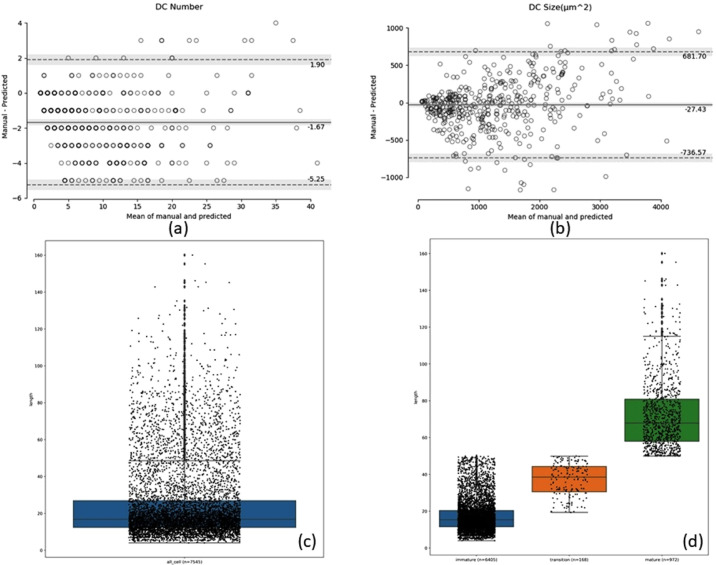
The Bland–Altman analysis was also performed on 754 (∼75 × 10) images from 10-fold cross-validation to determine the consistency of manual annotation and automatic segmentation of total DC number and size (µm2), and the results are shown in Figures 10a and 10b. Furthermore, the total number of segmented cells without branches (dendrites) is shown in a scatterbox plot (Fig. 10c), and the immature, transition-stage, and mature dendritic cells are shown in a scatterbox plot (Fig. 10d) where the length of most of the immature cells (n = 6405) was between 10 and 20 µm (dense data point cloud). The lengths of the mature cells (n = 972) and transition-stage cells (n = 168) were evenly distributed. All 754 (∼75 × 10) test images from 10-fold cross-validation were used to analyze the results for immature, transition-stage, and mature cells.
Figure 10.
Bland–Altman plots indicate the consistency of the total DC number (a) and size (µm2) (b) between manual annotation and automatic segmentation. The middle solid line indicates the mean value of the two methods, and the two dotted lines indicate the limits of agreement (±1.96 SD). The gray bands indicate a confidence interval of 95%. (c) Scatterbox plot of all segmented cells from the test images, and (d) scatterbox plot of the number of immature, transition-stage, and mature cells identified in the test images.
Discussion
In this study, fully automatic CNF and DC segmentation and quantification methods have been developed for corneal IVCM images. To the best of our knowledge, this is the first deep neural network–based DC segmentation and evaluation method to be proposed. This research study validates our deep learning methods and demonstrates segmentation performance that is comparable with manual annotation while reducing the amount of time required for analysis. In particular, the methods we developed show high interclass correlation between manual annotation and automatic segmentation in quantification metrics. In addition to automatic segmentation, our developed methods provide fully automatic, quantitative, clinical variables that can have utility in the diagnosis of dry eye disease, neuropathic corneal pain, and other corneal diseases.
IVCM images play an important role in the diagnosis of many corneal diseases in clinical practice. Clinicians must analyze the images multiple times to ensure accurate results for disease diagnosis or scientific research. During their manual analyses, clinicians may quantify the CNFs or DCs differently at different patient appointments which could lead to within-observer variability. In addition, two different clinicians could quantify the same image of CNFs or DCs differently, leading to between-observer variability. Therefore, a fully automatic quantitative evaluation of IVCM images is needed to obtain stable, constant, fast, and reproducible results. Unlike traditional feature engineering, deep neural network–based automatic segmentation is comparatively easier and faster than the conventional image processing approaches where various types of filters and graphs have been used.28–31 In contrast to conventional machine learning methods such as support vector machine,29 the deep learning–based method eliminates the need for manual feature selection and extraction and allows the machine to learn complex features using hundreds of filters.
We developed a new method of automatically characterizing corneal nerves into main trunk nerves and branching nerves for further morphometric analysis after segmentation using a deep neural network. Our trained model achieved on average 86.1% sensitivity and 90.1% specificity and AUC of 0.88 on the test dataset during cross-validation. We automatically calculated average nerve tortuosity, total nerve density, and total number of branch points, which have been shown to be useful clinical parameters to measure the severity of DED and ocular pain.7,8,12 Our work builds upon the first deep learning–based CNF segmentation and evaluation method based on U-Net architecture, which was proposed by Williams et al.43 This approach achieved ICC of 0.933 for total CNF length, 0.891 for branching points, 0.878 for number of nerve segments, and 0.927 for fractals between manual annotation and automatic segmentation (Liverpool Deep Learning Algorithm (LDLA) method) but had a different method for corneal nerve characterization. Williams et al.43 calculated the total number of nerve segments by calculating the nerve segments between two branching points, two end points, or an end and a branching point, and they used fractal dimensions to describe the nerve curvature. Supplementary Figure S4 illustrates the difference between the method of Williams et al.43 and our new proposal for detecting the trunk and branch nerves, highlighting the difference in total nerve count. Our work also differs in the loss function used. Williams et al.43 used the Dice similarity coefficient as a loss function, whereas we used clDice,55 a state-of-the-art topology preserving loss function that preserves connectivity among the segmented nerves. Another recent deep learning–based CNF segmentation based on U-Net was proposed by Wei et al.44 This method achieved 96% sensitivity and 75% specificity for segmenting CNFs from IVCM images. In contrast, our U-Net–based model has achieved 86.1% sensitivity and 90.1% specificity for segmenting CNFs and provides validated automatic morphometric evaluation parameters such as the number of nerves, nerve density, nerve length, branching points, and tortuosity. Furthermore, our proposed method demonstrates better performance compared to ACCMetrics for low-quality images. In particular, average CNFL per segment obtained by our proposed method are closer to those obtained by manual annotation than the values obtained by ACCMetrics. By comparing both methods with regard to the image quality of the dataset used, we found that there is a higher requirement for image quality when using ACCMetrics to calculate CNFL, as out-of-focus, faint, or thinner nerves on the images cannot be detected properly. This is in agreement with previous studies using ACCMetrics in which only images with high optical quality with regard to brightness, contrast, and sharpness were selected for the analysis.69,70 In this context, Williams et al.43 proposed further research on interrupted CNF segments. Our proposed deep learning model was trained using low-quality images; thus, it seems able to segment more nerves in the category of low-quality images.
We have proposed a new method of automatically differentiating among immature, transition-stage, and mature DCs for different pathological (dry eye, diabetic, and neuropathic corneal pain) patient data, along with cell density and cell size after the segmentation, using a deep neural network. This method could be a potential image-based biomarker to differentiate the severity of patients among the different pathological patient groups. However, in our future work, we will correlate these findings with clinical information for various patient groups. To segment DCs, we used the instance segmentation algorithm Mask R-CNN,57 which is different from the semantic segmentation algorithm U-Net.51 Mask R-CNN first predicts the bounding boxes that contain a DC, then segments the DC inside the bounding box. Therefore, the Mask R-CNN has the potential to detect objects more accurately than U-Net71,72; however, it struggles to predict good segmentation masks inside the bounding box.71 A previous study71 in microscopy image segmentation found that the two-step CNN process of the Mask R-CNN enables more precise localization compared with the single-step CNN used in typical U-Net architectures. For DCs, determining accurate localization and number is important; therefore, we compromised on accurate segmentation of dendritic cell border in favor of focusing on precise localization and determining accurate numbers.
In this research study, our results demonstrate that the methods we have developed can reliably and automatically segment and quantify CNFs and DCs with rapid speed. The average individual image segmentation and quantification time for CNFs was approximately 4.5 seconds, whereas for DCs it was approximately 3 seconds (based on the mentioned software and hardware). Our developed deep neural network–based methods significantly reduce image analysis time when applied to a large volume of clinical images. Manual annotation and automated segmentation of the same images appear similar, with important features segmented in both methods. Overall, the newly developed CNN models significantly reduce computational processing time while providing an objective approach to segmenting and evaluating CNFs and DCs.
Further studies are needed to identify the feasibility of implementing these methods in clinical practice and diagnostic devices. The CNF images that were acquired from Peking University Third Hospital in China and the DC images obtained from University Hospital Cologne in Germany were based on a small group of patients. Therefore, a larger patient cohort data in other racial populations could potentially enhance the strength and generalizability of the developed methods. Finally, the developed software should be assessed using different types of IVCM devices.
Conclusions
Deep neural network–based fully automatic CNF and DC segmentation and quantification methods have been proposed in this work. Automatic and objective analysis of IVCM images can assist clinicians in the diagnosis of several corneal diseases, thereby reducing user variability and time required to analyze a large volume of clinical images. Our results demonstrate that the deep learning–based approaches provide automatic quantification of CNFs and DCs and have the potential to be implemented in clinical practice for IVCM imaging.
Supplementary Material
Acknowledgments
The authors thank Yitian Zhao (Cixi Institute of Biomedical Engineering, Chinese Academy of Sciences) for providing the CNF images along with verified ground-truth mask images to support our research.
Supported by the European Union's Horizon 2020 research and innovation program (under Marie Skłodowska-Curie grant agreement number 765608 for Integrated Training in Dry Eye Disease Drug Development [IT-DED3]) and Deutsche Forschungsgemeinschaft Research Unit FOR2240 (DFG STE1928/7-1, DFG GE3108/1-1). The Division of Dry-Eye and Ocular GvHD received donations from Novaliq, Ursapharm, and Juergen and Monika Ziehm.
Author contributions: MAKS designed and developed the deep learning algorithm and morphology assessment software, processed, and anonymized the DC data, generated ground truth mask images, and wrote the main manuscript text. SS and GM provided technical knowledge and reviewed the manuscript. MES participated in study design and reviewed and commented on the manuscript. PS generated the main idea of this research, provided clinical background knowledge, collected and verified the clinical IVCM data, and wrote and reviewed the manuscript.
Disclosure: M.A.K. Setu, None; S. Schmidt, Heidelberg Engineering (E); G. Musial, None; M.E. Stern, ImmunEyez (E), Novaliq (C); P. Steven, Novaliq (R), Roche (R), Bausch & Lomb (R), Ursapharm (R)
References
- 1. Craig JP, Nelson JD, Azar DT, et al.. TFOS DEWS II Report Executive Summary. Ocul Surf. 2017; 15(4): 802–812. [DOI] [PubMed] [Google Scholar]
- 2. Stapleton F, Alves M, Bunya VY, et al.. TFOS DEWS II Epidemiology Report. Ocul Surf. 2017; 15(3): 334–365. [DOI] [PubMed] [Google Scholar]
- 3. Craig JP, Nichols KK, Akpek EK, et al.. TFOS DEWS II Definition and Classification Report. Ocul Surf. 2017; 15(3): 276–283. [DOI] [PubMed] [Google Scholar]
- 4. McDonald M, Patel DA, Keith MS, Snedecor SJ.. Economic and humanistic burden of dry eye disease in Europe, North America, and Asia: a systematic literature review. Ocul Surf. 2016; 14(2): 144–167. [DOI] [PubMed] [Google Scholar]
- 5. Uchino M, Schaumberg DA.. Dry eye disease: impact on quality of life and vision. Curr Ophthalmol Rep. 2013; 1(2): 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belmonte C, Acosta MC, Merayo-Lloves J, Gallar J.. What causes eye pain? Curr Ophthalmol Rep. 2015; 3(2): 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moein HR, Akhlaq A, Dieckmann G, et al.. Visualization of microneuromas by using in vivo confocal microscopy: an objective biomarker for the diagnosis of neuropathic corneal pain? Ocul Surf. 2020; 18(4): 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hamrah P, Cruzat A, Dastjerdi MH, et al.. Corneal sensation and subbasal nerve alterations in patients with herpes simplex keratitis: an in vivo confocal microscopy study. Ophthalmology. 2010; 117(10): 1930–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dieckmann G, Goyal S, Hamrah P.. Neuropathic corneal pain: approaches for management. Ophthalmology. 2017; 124(11): S34–S47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pflipsen M, Massaquoi M, Wolf S.. Evaluation of the painful eye. Am Fam Physician. 2016; 93: 991–998. [PubMed] [Google Scholar]
- 11. Cruzat A, Qazi Y, Hamrah P.. In vivo confocal microscopy of corneal nerves in health and disease. Ocul Surf. 2017; 15(1): 15–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Benítez Del Castillo JM, Wasfy MAS, Fernandez C, Garcia-Sanchez J.. An in vivo confocal masked study on corneal epithelium and subbasal nerves in patients with dry eye. Invest Ophthalmol Vis Sci. 2004; 45(9): 3030–3035. [DOI] [PubMed] [Google Scholar]
- 13. Galor A, Zlotcavitch L, Walter SD, et al.. Dry eye symptom severity and persistence are associated with symptoms of neuropathic pain. Br J Ophthalmol. 2015; 99(5): 665–668. [DOI] [PubMed] [Google Scholar]
- 14. Petropoulos IN, Ponirakis G, Khan A, et al.. Corneal confocal microscopy: ready for prime time. Clin Exp Optom. 2020; 103(3): 265–277. [DOI] [PubMed] [Google Scholar]
- 15. Villani E, Baudouin C, Efron N, et al.. In vivo confocal microscopy of the ocular surface: from bench to bedside. Curr Eye Res. 2014; 39(3): 213–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Niederer RL, McGhee CNJ.. Clinical in vivo confocal microscopy of the human cornea in health and disease. Prog Retin Eye Res. 2010; 29(1): 30–58. [DOI] [PubMed] [Google Scholar]
- 17. Benítez-Del-Castillo JM, Acosta MC, Wassfi MA, et al.. Relation between corneal innervation with confocal microscopy and corneal sensitivity with noncontact esthesiometry in patients with dry eye. Invest Ophthalmol Vis Sci. 2007; 48(1): 173–181. [DOI] [PubMed] [Google Scholar]
- 18. Erdélyi B, Kraak R, Zhivov A, Guthoff R, Németh J.. In vivo confocal laser scanning microscopy of the cornea in dry eye. Graefes Arch Clin Exp Ophthalmol. 2007; 245(1): 39–44. [DOI] [PubMed] [Google Scholar]
- 19. Khamar P, Nair AP, Shetty R, et al.. Dysregulated tear fluid nociception-associated factors, corneal dendritic cell density, and vitamin D levels in evaporative dry eye. Invest Ophthalmol Vis Sci. 2019; 60(7): 2532–2542. [DOI] [PubMed] [Google Scholar]
- 20. Sivaskandarajah GA, Halpern EM, Lovblom LE, et al.. Structure-function relationship between corneal nerves and conventional small-fiber tests in type 1 diabetes. Diabetes Care. 2013; 36(9): 2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tavakoli M, Quattrini C, Abbott C, et al.. Corneal confocal microscopy: a novel noninvasive test to diagnose and stratify the severity of human diabetic neuropathy. Diabetes Care. 2010; 33(8): 1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kemp HI, Petropoulos IN, Rice ASC, et al.. Use of corneal confocal microscopy to evaluate small nerve fibers in patients with human immunodeficiency virus. JAMA Ophthalmol. 2017; 135(7): 795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ferdousi M, Azmi S, Petropoulos IN, et al.. Corneal confocal microscopy detects small fibre neuropathy in patients with upper gastrointestinal cancer and nerve regeneration in chemotherapy induced peripheral neuropathy. PLoS One. 2015; 10(10): e0139394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Petropoulos IN, Alam U, Fadavi H, et al.. Corneal nerve loss detected with corneal confocal microscopy is symmetrical and related to the severity of diabetic polyneuropathy. Diabetes Care. 2013; 36(11): 3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petropoulos IN, Manzoor T, Morgan P, et al.. Repeatability of in vivo corneal confocal microscopy to quantify corneal nerve morphology. Cornea. 2013; 32(5): 83–89. [DOI] [PubMed] [Google Scholar]
- 26. Petropoulos IN, Alam U, Fadavi H, et al.. Rapid automated diagnosis of diabetic peripheral neuropathy with in vivo corneal confocal microscopy. Invest Ophthalmol Vis Sci. 2014; 55(4): 2071–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alam U, Jeziorska M, Petropoulos IN, et al.. Diagnostic utility of corneal confocal microscopy and intra-epidermal nerve fibre density in diabetic neuropathy. PLoS One. 2017; 12(7): e0180175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dabbah MA, Graham J, Petropoulos IN, Tavakoli M, Malik RA.. Automatic analysis of diabetic peripheral neuropathy using multi-scale quantitative morphology of nerve fibres in corneal confocal microscopy imaging. Med Image Anal. 2011; 15(5): 738–747. [DOI] [PubMed] [Google Scholar]
- 29. Chen X, Graham J, Dabbah MA, Petropoulos IN, Tavakoli M, Malik RA.. An automatic tool for quantification of nerve fibers in corneal confocal microscopy images. IEEE Trans Biomed Eng. 2017; 64(4): 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scarpa F, Grisan E, Ruggeri A.. Automatic recognition of corneal nerve structures in images from confocal microscopy. Invest Ophthalmol Vis Sci. 2008; 49(11): 4801–4807. [DOI] [PubMed] [Google Scholar]
- 31. Kim J, Markoulli M.. Automatic analysis of corneal nerves imaged using in vivo confocal microscopy. Clin Exp Optom. 2018; 101(2): 147–161. [DOI] [PubMed] [Google Scholar]
- 32. Dabbah MA, Graham J, Petropoulos I, Tavakoli M, Malik RA.. Dual-model automatic detection of nerve-fibres in corneal confocal microscopy images. Med Image Comput Comput Assist Interv. 2010; 13(pt 1): 300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lecun Y, Bengio Y, Hinton G.. Deep learning. Nature. 2015; 521(7553): 436–444. [DOI] [PubMed] [Google Scholar]
- 34. Shen D, Wu G, Suk H II. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017; 19: 221–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ting DSW, Peng L, Varadarajan AV, et al.. Deep learning in ophthalmology: the technical and clinical considerations. Prog Retin Eye Res. 2019; 72: 100759. [DOI] [PubMed] [Google Scholar]
- 36. Ting DSW, Pasquale LR, Peng L, et al.. Artificial intelligence and deep learning in ophthalmology. Br J Ophthalmol. 2019; 103(2): 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grewal PS, Oloumi F, Rubin U, Tennant MTS.. Deep learning in ophthalmology: a review. Can J Ophthalmol. 2018; 53(4): 309–313. [DOI] [PubMed] [Google Scholar]
- 38. Jiang Z, Zhang H, Wang Y, Ko SB.. Retinal blood vessel segmentation using fully convolutional network with transfer learning. Comput Med Imaging Graph. 2018; 68: 1–15. [DOI] [PubMed] [Google Scholar]
- 39. Mishra Z, Ganegoda A, Selicha J, Wang Z, Sadda SVR, Hu Z.. Automated retinal layer segmentation using graph-based algorithm incorporating deep-learning-derived information. Sci Rep. 2020; 10(1): 9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sreng S, Maneerat N, Hamamoto K, Win KY.. Deep learning for optic disc segmentation and glaucoma diagnosis on retinal images. Appl Sci. 2020; 10(14): 4916. [Google Scholar]
- 41. Fu H, Baskaran M, Xu Y, et al.. A deep learning system for automated angle-closure detection in anterior segment optical coherence tomography images. Am J Ophthalmol. 2019; 203: 37–45. [DOI] [PubMed] [Google Scholar]
- 42. Setu MAK, Horstmann J, Schmidt S, Stern ME, Steven P.. Deep learning-based automatic meibomian gland segmentation and morphology assessment in infrared meibography. Sci Rep. 2021; 11(1): 7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Williams BM, Borroni D, Liu R, et al.. An artificial intelligence-based deep learning algorithm for the diagnosis of diabetic neuropathy using corneal confocal microscopy: a development and validation study. Diabetologia. 2020; 63(2): 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wei S, Shi F, Wang Y, Chou Y, Li X.. A deep learning model for automated sub-basal corneal nerve segmentation and evaluation using in vivo confocal microscopy. Transl Vis Sci Technol. 2020; 9(2): 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Colonna A, Scarpa F, Ruggeri A.. Segmentation of corneal nerves using a U-Net-based convolutional neural network. In: Stoyanov D, Taylor Z, Ciompi F, et al., eds. Computational Pathology and Ophthalmic Medical Image Analysis. Cham: Springer; 2018: 185–192. [Google Scholar]
- 46. Oakley JD, Russakoff DB, McCarron ME, et al.. Deep learning-based analysis of macaque corneal sub-basal nerve fibers in confocal microscopy images. Eye Vis (Lond). 2020; 7(1): 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yıldız E, Arslan AT, Taş AY, et al.. Generative adversarial network based automatic segmentation of corneal subbasal nerves on in vivo confocal microscopy images. Transl Vis Sci Technol. 2021; 10(6): 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mou L, Zhao Y, Chen L, et al.. CS-Net: channel and spatial attention network for curvilinear structure segmentation. In: Shen D, Liu T, Peters TM, et al., eds. Medical Image Computing and Computer-Assisted Intervention—MICCAI 2019, LNCS Vol. 11764. Cham: Springer; 2019: 721–730. [Google Scholar]
- 49. Zhao YY, Zhang J, Pereira E, et al.. Automated tortuosity analysis of nerve fibers in corneal confocal microscopy. IEEE Trans Med Imaging. 2020; 39(9): 2725–2737. [DOI] [PubMed] [Google Scholar]
- 50. Dutta A, Zisserman A.. The VIA annotation software for images, audio and video. In: MM ’19: Proceedings of the 27th ACM International Conference on Multimedia. New York: Association for Computing Machinery; 2019: 2276–2279. [Google Scholar]
- 51. Ronneberger O, Fischer P, Brox T.. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, eds. Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, LNCS Vol. 9351. Cham: Springer; 2015: 234–241. [Google Scholar]
- 52. Zhu W, Huang Y, Zeng L, et al.. AnatomyNet: deep learning for fast and fully automated whole-volume segmentation of head and neck anatomy. Med Phys. 2019; 46(2): 576–589. [DOI] [PubMed] [Google Scholar]
- 53. Nasrullah N, Sang J, Alam MS, Mateen M, Cai B, Hu H.. Automated lung nodule detection and classification using deep learning combined with multiple strategies. Sensors (Basel). 2019; 19(17): 3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Norman B, Pedoia V, Majumdar S.. Use of 2D U-net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology. 2018; 288(1): 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shit S, Paetzold JC, Sekuboyina A, et al.. clDice – a topology-preserving loss function for tubular structure segmentation. arXiv. 2021, 10.48550/arXiv.2003.07311. [DOI]
- 56. Kingma DP, Ba J.. Adam: a method for stochastic optimization. arXiv. 2017, 10.48550/arXiv.1412.6980. [DOI]
- 57. He K, Gkioxari G, Dollár P, Girshick R.. Mask R-CNN. In: Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV). Piscataway, NJ: Institute of Electrical and Electronics Engineers; 2017: 2980–2988. [Google Scholar]
- 58. He K, Zhang X, Ren S, Sun J.. Deep residual learning for image recognition. In: Proceedings of IEEE Computer Society Conference on Computer Vision and Pattern Recognition. Piscataway, NJ: Institute of Electrical and Electronics Engineers; 2016: 770–778. [Google Scholar]
- 59. Lin TY, Maire M, Belongie S, et al.. Microsoft COCO: common objects in context. In: Fleet D, Pajdla T, Schiele B, Tuytelaars T, eds. European Conference on Computer Vision—ECCV 2014, LNCS Vol. 8693. Cham: Springer; 2014: 740–755. [Google Scholar]
- 60. Smedbyörjan, Högman N, Nilsson S, Erikson U, Olssori AG, Walldius G. Two-dimensional tortuosity of the superficial femoral artery in early atherosclerosis. J Vasc Res. 1993; 30(4): 181–191. [DOI] [PubMed] [Google Scholar]
- 61. Cavalcanti BM, Cruzat A, Sahin A, Pavan-Langston D, Samayoa E, Hamrah P.. In vivo confocal microscopy detects bilateral changes of corneal immune cells and nerves in unilateral herpes zoster ophthalmicus. Ocul Surf. 2018; 16(1): 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kheirkhah A, Rahimi Darabad R, Cruzat A, et al.. Corneal epithelial immune dendritic cell alterations in subtypes of dry eye disease: a pilot in vivo confocal microscopic study. Invest Ophthalmol Vis Sci. 2015; 56(12): 7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hamrah P, Huq SO, Liu Y, Zhang Q, Dana MR.. Corneal immunity is mediated by heterogeneous population of antigen-presenting cells. J Leukoc Biol. 2003; 74(2): 172–178. [DOI] [PubMed] [Google Scholar]
- 64. Zhivov A, Stave J, Vollmar B, Guthoff R.. In vivo confocal microscopic evaluation of Langerhans cell density and distribution in the normal human corneal epithelium. Graefes Arch Clin Exp Ophthalmol. 2005; 243(10): 1056–1061. [DOI] [PubMed] [Google Scholar]
- 65. Ferdousi M, Romanchuk K, Mah JK, et al.. Early corneal nerve fibre damage and increased Langerhans cell density in children with type 1 diabetes mellitus. Sci Rep. 2019; 9(1): 8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Khan A, Li Y, Ponirakis G, et al.. Corneal immune cells are increased in patients with multiple sclerosis. Transl Vis Sci Technol. 2021; 10(4): 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Martin Bland J, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986; 327(8476): 307–310. [PubMed] [Google Scholar]
- 68. Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016; 15(2): 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chin JY, Yang LWY, Ji AJS, et al.. Validation of the use of automated and manual quantitative analysis of corneal nerve plexus following refractive surgery. Diagnostics. 2020; 10(7): 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wu PY, Wu JH, Hsieh YT, et al.. Comparing the results of manual and automated quantitative corneal neuroanalysing modules for beginners. Sci Rep. 2021; 11(1): 18208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vuola AO, Akram SU, Kannala J.. Mask-RCNN and U-Net ensembled for nuclei segmentation. In: Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging. Piscataway, NJ: Institute of Electrical and Electronics Engineers; 2019: 208–212. [Google Scholar]
- 72. Zhao T, Yang Y, Niu H, Chen Y, Wang D.. Comparing U-Net convolutional network with Mask R-CNN in the performances of pomegranate tree canopy segmentation. Proc SPIE. 2018; 10780: J-1–J-9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.