Abstract
The sea urchin mitochondrial displacement (D)-loop binding protein mtDBP has been previously identified and cloned. The polypeptide (348 amino acids) displays a significant homology with the human mitochondrial transcription termination factor mTERF. This similarity, and the observation that the 3′ ends of mitochondrial RNAs coded by opposite strands mapped in correspondence of mtDBP-binding sites, suggested that mtDBP could function as transcription termination factor in sea urchin mitochondria. To investigate such a role we tested the capability of mtDBP bound to its target sequence in the main non-coding region to affect RNA elongation by mitochondrial and bacteriophage T3 and T7 RNA polymerases. We show that mtDBP was able to terminate transcription bidirectionally when initiated by human mitochondrial RNA polymerase but only unidirectionally when initiated by T3 or T7 RNA polymerases. Time-course experiments indicated that mtDBP promotes true transcription termination rather than transcription pausing. These results indicate that mtDBP is able to function as a bipolar transcription termination factor in sea urchin mitochondria. The functional significance of such an activity could be linked to the previously proposed dual role of the protein in modulating mitochondrial DNA transcription and replication.
INTRODUCTION
Sea urchin mitochondrial DNA (mtDNA) is a circular 15.7 kb double-stranded molecule which codes for the same genes found in vertebrate mitochondrial genomes (1–3). However, the gene order and distribution among the two strands are strikingly different; the major differences concern the separation of the two rRNA genes, the clustering of 15 tRNA genes and the reduced size of the main non-coding region (NCR) that is ∼130 bp long. A heavy (H)-strand replication origin, associated with a displacement (D)-loop triplex of ∼80 nucleotides (nt), was mapped in the NCR of Strongylocentrotus purpuratus mtDNA (4). The light (L)-strand might initiate from multiple points, one of which could be located in proximity of the main H-strand replication pause site, at the boundary of the ATPase 6 and COIII genes (5). Studies on the transcription of sea urchin mtDNA suggested a mechanism based on the existence of multiple and partially overlapping transcription units starting from the AT-rich consensus sequences, which are located in five different regions of the genome (6,7).
Recent work on proteins assisting mitochondrial replication and transcription processes has provided a consistent amount of information on these factors. A single-stranded DNA-binding protein (mtSSB) and an ATP-dependent DNA helicase were identified in mitochondria from Paracentrotus lividus eggs (8,9). Moreover, four sequence-specific mtDNA-binding proteins, mtPBP-1, mtPBP-2, mtAT1-BP and mtDBP, were found in P.lividus and S.purpuratus. MtPBP-1 and mtPBP-2 are two polypeptides of 18 and 15 kDa, respectively, which interact with two sequences located at the boundary of ATPase 6 and COIII genes. A possible role in the regulation of L-strand replication has been proposed for them (10,11). MtAT1-BP is a DNA-binding activity, probably involved in transcription initiation, which interacts with the AT-consensus sequence located in the NCR of P.lividus mtDNA (12). MtDBP is a 40 kDa protein which binds specifically two homologous sequences: one is located in the NCR and contains the H-strand replication origin, the other encompasses the adjacent 3′ ends of the oppositely transcribed ND5 and ND6 genes (12–14). Cloning of mtDBP cDNA revealed that the protein is a 348 amino acid polypeptide containing two leucine zipper-like domains and two small N- and C-terminal basic domains (15). The protein binds DNA as a monomer rather than as a dimer, as expected from the presence of the leucine zippers. This suggests that the heptad repeats could be involved in forming intramolecular coiled-coil interactions.
MtDBP displays as the most significant homology a match with the human mitochondrial transcription termination factor mTERF. This protein has been found to promote specific termination of the H-strand ribosomal transcription unit at the 16S rRNA-tRNALeu (UUR) boundary by binding to a tridecameric sequence within the tRNALeu (UUR) gene (16). Similarly to mtDBP, mTERF binds DNA as a monomer and contains three leucine zippers probably establishing intramolecular interactions needed to expose the two basic binding domains to the target DNA sequences (17). These similarities and the observation that the 3′ ends of mitochondrial RNAs coded by opposite strands mapped in correspondence with mtDBP-binding sites (4; P.Cantatore et al., unpublished results), pointed to the role of mtDBP as transcription termination factor. In this paper we show that mtDBP bound to its target site in the NCR of sea urchin mtDNA is able to terminate transcription bidirectionally when initiated by a heterologous mtRNA polymerase but only unidirectionally when initiated by T3 or T7 RNA polymerases.
MATERIALS AND METHODS
Plasmid constructs
The templates used for the transcription termination assay were obtained by inserting HindIII–EcoRI chimeric fragments into pBluescript KS(+) vector. The chimeric fragments were obtained as follows. For template pDBP-term1(F), a HindIII–BamHI fragment of 883 bp, comprising nt 1–739 and 3063–3195 of human mtDNA, was obtained by polymerase chain reaction (PCR) on pTER clone (16); it was then ligated to a 432 bp BamHI–EcoRI fragment (nt 1061–1492) prepared by PCR on P.lividus mtDNA (see Fig. 1A). For the plasmid pDBP-term2(F), the HindIII–EcoRI chimeric insert was constructed as follows. A HindIII–BamHI fragment of 394 bp, comprising nt 490–739 and 3063–3195 of human mtDNA, was obtained by PCR on pTER clone (16); it was then ligated to a 77 bp BamHI–XhoI fragment (nt 1090–1166) obtained by PCR on P.lividus mtDNA. The resulting HindIII–XhoI fragment was ligated to a 320 bp XhoI–EcoRI fragment (nt 3275–3594) prepared by PCR on pTER clone. For the version pDBP-term2(R), the chimeric insert was obtained as for pDBP-term2(F), except that the ends of the PCR-amplified sea urchin mtDNA fragment were inverted (XhoI–BamHI instead of BamHI–XhoI). In this way the mtDBP-binding sequence was placed in the reverse polarity with respect to H-strand promoter region (HSP) promoter (see Fig. 2A).
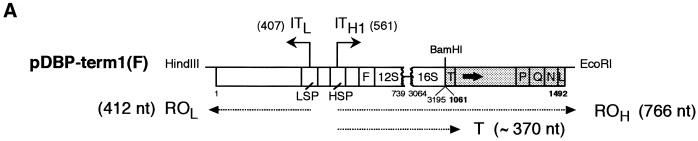
Figure 1.
MtDBP promotes termination of transcription by human mtRNA polymerase. (A) Schematic diagram of the DNA template used. Open and filled boxes indicate the human and sea urchin mtDNA part, respectively. Numbers give the position of human mtDNA nucleotides (thin numbers) (22) and sea urchin mtDNA nucleotides (bold numbers) (1). LSP and HSP, human HSP and LSP, respectively; ITH1 and ITL, two major transcription initiation sites. The enclosed arrow in the sea urchin mtDNA portion marks the orientation of mtDBP target site with respect to HSP. ROH and ROL, run-off transcripts proceeding from HSP and LSP, respectively; T: terminated transcript. Transcripts are indicated by a dotted arrowed line; sizes are in brackets. The DNA templates were digested with HindIII and EcoRI. (B) Autoradiogram of a 5% polyacrylamide/7 M urea gel showing products of transcription assays, indicated by arrow, obtained in the presence and absence of in vivo affinity-purified or purified bacterially expressed mtDBP (bmtDBP). Reactions contained 0.5 µg of the indicated template DNA, 5 µl of S-100 extract and either no mtDBP (–) or the indicated amounts of mtDBP. The reactions with no protein contained always column elution buffer corresponding to the highest volume of eluted protein used in the assay. Size marker (M) positions are shown to the left.
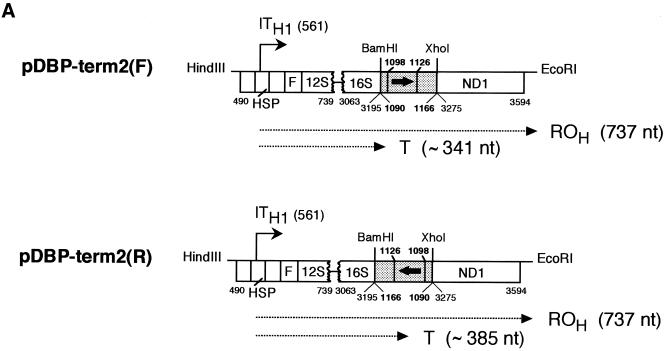
Figure 2.
Transcription termination assays by recombinant mtDBP (bmtDBP). (A) Scheme of the templates used in the transcription assays (see legend of Fig. 1A for description of the templates). Numbering is as in Figure 1A; bold numbers above the filled box mark the borders of the region footprinted by mtDBP (15). Run-off (ROH) and terminated (T) transcripts are indicated by a dotted arrowed line; the calculated sizes are in brackets. (B) Autoradiogram of a 5% polyacrylamide/7 M urea gel showing the position of the transcripts. All reactions were performed using 0.5 µg of the indicated template, 5 µl of S-100 extract and the indicated amounts of recombinant mtDBP (bmtDBP).
S-100 fraction preparation
For the preparation of the mitochondrial S100 fraction, HeLa cells were grown on a solid substrate in Dulbecco’s modified Eagle’s medium with 5% fetal calf serum until they were 70–80% confluent. After trypsinisation, the cells were collected by centrifugation at 370 g for 7 min and washed twice with 10 vol of packed cells (v.p.c.) of NKM buffer (1 mM Tris–HCl, pH 7.4, 0.13 M NaCl, 5 mM KCl, 7.5 mM MgCl2). The cells were resuspended in 6 v.p.c. of homogenisation buffer (10 mM Tris–HCl, pH 6.7, 10 mM KCl, 0.15 mM MgCl2), mixed gently and kept for 2 min on ice. The homogenisation was performed using six to eight strokes with a motor-driven Teflon pestle using a glass homogeniser and the homogenate was immediately poured into a conical centrifuge tube containing 1 v.p.c. of 2 M sucrose. The unbroken cells, nuclei and large debris were pelleted at 1200 g for 3 min, and the supernatant was centrifuged once more under the same conditions, discarding again the pellet. The mitochondria were pelleted at 7000 g for 10 min, resuspended in 3 v.p.c. of mitochondria suspension buffer (10 mM Tris–HCl, pH 6.7, 0.25 M sucrose, 0.15 mM MgCl2) and washed twice under the same conditions. The pellet was then resuspended in 1/3 v.p.c. of lysis buffer [10% glycerol, 25 mM HEPES–KOH, pH 7.6, 5 mM MgCl2, 0.5 mM EDTA, 1 mM dithiothreitol (DTT) and 1 mM freshly added phenylmethylsulfonyl fluoride (PMSF)]. The suspension was homogenised by five slow strokes, and then Tween-20 and KCl were added to final concentrations of 0.5% and 0.5 M, respectively. The homogenisation was repeated 10 times, and the mitochondrial lysate was spun at 39 000 r.p.m. (100 000 gav) for 60 min at 4°C. The clear supernatant was carefully collected avoiding the fluffy layer over the pellet to yield the final S-100 fraction of the mitochondrial lysate. It was aliquoted, frozen in liquid nitrogen and stored at –80°C.
In vitro transcription termination assays
Standard reactions for termination of transcription by human mtRNA polymerase were carried out in 50 µl of a mixture containing 10 mM Tris–HCl, pH 8.0, 10% glycerol, 10 mM MgCl2, 1 mM DTT, 100 µg/ml bovine serum albumin, 1 mM ATP, 0.1 mM CTP and GTP, 0.01 mM UTP, 1 µl of [α-32P]UTP (400 Ci/mmol, 10 mCi/ml), 5 µl of a HeLa S-100 fraction, the template DNA at a final concentration of 10 µg/ml and the desired volume of the purified mtDBP fraction. Control reactions always received the column elution buffer corresponding to the highest volume of eluted protein used in the assay, and the final volume was made 50 µl with diethyl pyrocarbonate (DEPC)-treated water. The components were mixed by vortexing, the mixture was briefly spun and then incubated at 30°C for 30 min. Termination of transcription by T3 or T7 RNA polymerases was performed under the same reaction conditions except that the S-100 fraction was omitted, the nucleotide concentration was 0.5 mM for ATP, CTP and GTP and 0.05 mM for UTP, and 5 U of RNA polymerase were added. For time-course experiments with T7 RNA polymerase, 100 µl reactions were set up in the same conditions as described before but containing 10 µl of [α-32P]UTP. The reactions were incubated for 5 min at 30°C and then 2.5 µl of 100 mM cold UTP were added. Fractions (20 µl) of the reactions were taken at 0, 30, 100, 200 and 250 min after addition of cold UTP, and processed as described below. Reactions were stopped by adding 150 µl of transcription stop buffer (10 mM Tris–HCl pH 7.4, 0.5% SDS, 0.2 M NaCl, 10 mM EDTA) and 20 µg of yeast tRNA; they were then phenol extracted and the nucleic acids were ethanol precipitated. The pellet, dissolved in 150 µl of transcription stop buffer, was phenol extracted again and ethanol precipitated. The final pellet was resuspended in 15 µl of DEPC-treated water and 1 vol of urea-dye was added. Samples were then denatured at 80°C for 10 min, loaded onto 5% polyacrylamide/7 M urea gels in TBE and run at 300 V. Gels were washed twice with water, stained with ethidium bromide for gel documentation and vacuum-dried at 80°C for 1 h, before exposure for autoradiography.
RESULTS
MtDBP mediates termination of transcription by human mitochondrial RNA polymerase in an orientation-independent manner
The sequence and structural similarities between mtDBP and mTERF prompted us to set up an assay to test whether the sea urchin protein could function as a transcription termination factor. The termination activity of human mTERF in HeLa cell mitochondria was characterised by using a submitochondrial transcription system consisting of the S-100 fraction of a mitochondrial lysate programmed by an exogenous template derived from human mtDNA (16,17). As an analogous system from sea urchin mitochondria was not available, we decided to test the effect of mtDBP on in vitro transcription termination by using the human S-100 extract, as a source of RNA polymerase activity, and a chimeric template containing the human transcription promoters and the sea urchin mtDBP-binding site. We reasoned that the sea urchin protein, by virtue of its structural and putative functional homology to mTERF, could productively interact with the human mitochondrial transcription apparatus, when bound to its target site, and possibly arrest RNA chain elongation. The templates used in the transcription assays all included the HSP region and the sea urchin mtDBP target sequence in the NCR, placed downstream of HSP, in either orientation. Templates were designated as ‘forward’ or F when the orientation of the mtDBP-binding site relative to HSP was the same as it is with respect to the direction of transcription of sea urchin ribosomal genes (6,7), and ‘reverse’ or R when mtDBP-binding site was placed in the opposite orientation.
The first evidence of the capability of mtDBP to support transcription termination was obtained with the template pDBP-term1(F) (Fig. 1A). This template contained an 883 bp fragment which includes the human L-strand promoter (LSP) and HSP regions followed by a 432 bp portion of sea urchin mtDNA (nt 1061–1492) encompassing part of the tRNAThr gene, the entire NCR with the mtDBP target site, the genes for tRNAPro, tRNAGln and tRNAAsn and part of the tRNALeu(CUN) gene. After digestion with HindIII and EcoRI (Fig. 1A), the template was transcribed in the absence and presence of a DNA-affinity fraction containing mtDBP. As shown in Figure 1B, lane 1, in the absence of mtDBP two run-off transcripts of 766 and 412 nt were detected; the longer molecules (ROH) correspond to transcripts initiating at the HSP promoter and ending at the EcoRI-generated end of the digested template, whereas the shorter molecules (ROL) represent transcripts initiating at the LSP promoter and ending at the HindIII-generated end. Addition of the affinity-purified mtDBP (Fig. 1B, lanes 2 and 3) caused the appearance of a shorter transcript of ∼370 nt whose size was consistent with a transcription event starting at the HSP promoter and ending at the mtDBP-binding site. A marked reduction in the total transcription activity was also observed, which could be due to protein factors present in the chromatography fraction that partially inhibit the transcription initiation complex. The results suggest that binding of purified mtDBP to its cognate target site likely stops the progression of human mitochondrial RNA polymerase. Experiments with the purified bacterially expressed mtDBP gave similar results (Fig. 1B, lanes 4–6) thus confirming that formation of the shorter transcript was actually due to mtDBP by itself rather than to contaminants present in the affinity-purified fraction. As the recombinant protein produced a stronger transcription termination and a less overall reduction in the transcription activity, probably due to the high purity of the protein fraction, further transcription assays were performed with the recombinant mtDBP. Moreover we decided to simplify the template by eliminating the LSP-containing tract as well as the sea urchin sequences flanking mtDBP-binding site, such as a guanosine stretch (nt 1177–1202) and the tRNAs genes (Fig. 1A) that could affect RNA chain elongation. Thus, a shorter fragment (nt 1090–1166) containing almost only the mtDBP-binding site was placed in either orientation downstream of the HSP promoter, followed by 320 bp of the human mitochondrial ND1 gene. These two new templates, named pDBP-term2(F) and pDBP-term2(R) (Fig. 2A), were transcribed with the same transcription system used above. As shown in Figure 2B, lanes 1 and 5, in the absence of mtDBP both templates generated only a full-length run-off transcript of 737 nt. The addition of increasing amounts of recombinant mtDBP caused, with both templates, a decrease in the intensity of the run-off band and the appearance of transcripts whose size corresponded to that of molecules arrested at the protein-binding site. A minor slower-migrating band was observed with the pDBP-term2(R) template, which was probably caused by nucleases present in the extract and acting preferentially on the terminated transcripts. The efficiency of the termination promoting activity was similar with both templates although slightly higher for the reverse than for the forward orientation, reaching 97 and 86%, respectively (average of two experiments), with the maximum amount of added protein which corresponds to a ratio of 20 molecules of protein per DNA molecule.
In order to determine more precisely the 3′ ends of the terminated species, their size was compared with that of the run-off transcripts generated by templates pDBP-term2(F) and pDBP-term2(R) digested with HindIII–BamHI and HindIII–XhoI, respectively (Fig. 2A). This allowed us to locate the 3′ end of the terminated RNA chain around nt 1101, that is, at the promoter-proximal border of the region contacted by mtDBP (15) when the F template was used, and around nt 1111, that is, inside the sequence bound by mtDBP (15) in the case of the R template.
MtDBP arrests T3 and T7 RNA polymerases in a polar way
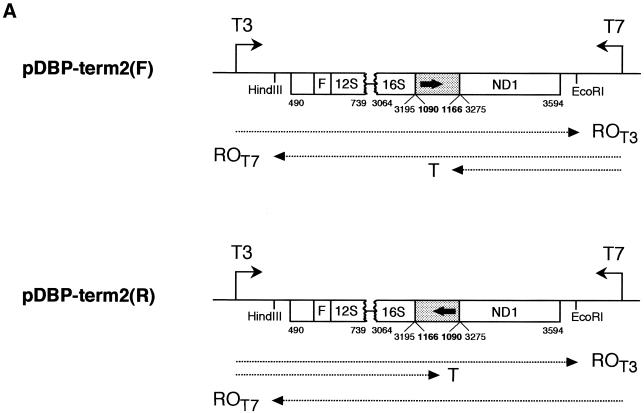
The transcriptional arrest operated by mtDBP raises the question of whether the transcription termination is the result of a specific protein–protein interaction between mtDBP and the polymerase or whether it may be caused by a physical blockage of RNA polymerase progression by the DNA-bound protein. To investigate this point, we analysed the ability of mtDBP to interfere with RNA chain elongation catalysed by bacteriophage RNA polymerases. As the constructs were subcloned in pBluescript vector, the templates already carried the standard promoters for T3 and T7 RNA polymerases located upstream of their EcoRI and HindIII ends, respectively. Each plasmid construct, carrying the mtDBP-binding site in either orientation relative to the bacteriophage promoter (Fig. 3A), was linearised downstream of the selected promoter, and then transcribed with the proper RNA polymerase, in the absence and presence of mtDBP. Figure 3B, lanes 1–3, shows that when the termination site was placed in the F orientation with respect to T3 promoter, transcription with T3 RNA polymerase yielded only run-off transcripts. Their length corresponded to the distance between the promoter and the end of the template, indicating that the transcribing enzyme bypassed the termination site even when the protein was bound to it. When the orientation of the protein-binding site was reversed (template R), elongation of the RNA chain by the bacteriophage enzyme was clearly affected by the presence of mtDBP in the assay (Fig. 3B, lanes 4–6). In fact ∼80% of the transcripts were terminated in the presence of mtDBP. Their size corresponded to molecules starting at the T3 promoter and ending upstream of the termination site. Transcription with T7 RNA polymerase on the properly linearised template F generated 85–90% of the terminated transcript in the presence of mtDBP (Fig. 3C, lanes 1–3); however, no such an arrest of transcription but almost only run-off transcripts were detected when the template R was employed (Fig. 3C, lanes 4–6).
Figure 3.
MtDBP acts as a polar blocker of transcription elongation by T3 and T7 RNA polymerases. (A) Schematic diagram of the DNA templates used (see legend of Fig. 1A for description) showing the position of T3 and T7 promoters and the mtDBP-binding site (filled box with enclosed arrow) placed in either orientation with respect to the promoters. DNA was linearised with HindIII or EcoRI. Run-off (ROT3, ROT7) and terminated (T) transcripts are indicated by a dotted arrowed line. (B and C) Autoradiogram of a 5% polyacrylamide/7 M urea gel showing the run-off (ROT3, ROT7) and terminated (T) transcripts obtained with T3 RNA polymerase (B) and T7 RNA polymerase (C). Reactions contained 0.5 µg of properly linearised template DNA, 5 U of the bacteriophage polymerase and either no protein (–) or the indicated amounts of bmtDBP.
MtDBP promotes true termination of transcription and not only polymerase pausing
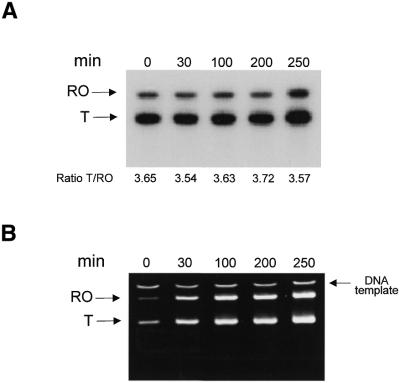
The shorter truncated transcripts could derive from mtDBP-mediated transcription termination or from transcription pausing. To prove one of these two hypotheses we performed a time-course experiment. The transcription reaction was carried out in the presence of T7 RNA polymerase on pDBP-term2 template in the forward orientation (Fig. 3C). After a 5 min pulse of radiolabelled UTP it followed a chase of 250 min with a 1000-fold excess of cold UTP. If pausing of RNA polymerase occurred, it is expected that the ratio of terminated to run-off transcripts would decrease progressively with time, due to resumption of elongation by the stalled enzyme. On the contrary, in the case of transcription termination, which involves dissociation of RNA polymerase from the DNA template, the above transcript ratio should not change. Figure 4A shows that the shorter transcript persisted for up to 250 min of incubation, a time longer than mtDBP half-life on its target site (150 min) (13); during this time the ratio between terminated and run-off transcripts did not change appreciably in spite of the fact that transcription activity persisted as it can be seen from the ethidium bromide stained gel (Fig. 4B). These results imply that in these conditions the terminated transcripts are likely formed by a transcription termination event rather than by pausing.
Figure 4.
Time-course of transcription elongation by T7 RNA polymerase. (A) Autoradiogram of a 5% polyacrylamide/7 M urea gel showing the run-off (RO) and terminated (T) transcripts obtained with T7 RNA polymerase. The reaction contained 1 µg of template pDBP-term2(F) linearised with HindIII, 160 ng of recombinant mtDBP and 5 U of T7 RNA polymerase. After a pulse-label of 5 min with [α-32P]UTP, the reaction was chased with an excess amount of unlabelled UTP and samples were taken at the time points indicated. The ratio of labeling of the terminated to the run-off transcript is indicated for each time point. (B) Image of the same gel as in (A) stained with ethidium bromide.
DISCUSSION
Sea urchin mtDBP is a mitochondrial DNA-binding protein, which binds specifically two homologous sequences located in the NCR and at the junction of ND5/ND6 genes (12–15). Sequence and structural similarities between mtDBP and mTERF, the human mitochondrial transcription termination factor, suggested that the sea urchin protein might function as a transcription termination factor. In this paper, to demonstrate such a role, in vitro transcription assays using the S-100 fraction of a human mitochondrial lysate were performed on chimeric templates containing a combination of the human HSP promoter and mtDBP-binding site in the NCR, in both orientations. We observed consistently that the addition of mtDBP to the assay caused the arrest of HSP-initiated transcription in correspondence of the protein-binding site. MtDBP-mediated transcriptional arrest appeared to be independent of the orientation of the protein-binding site; moreover, termination occurred with approximately the same efficiency with both orientations. This mode of transcription termination could be interpreted by assuming that mtDBP works simply as non-specific roadblock to elongating RNA polymerase. If so, the protein would block the progression of any RNA polymerase sliding on DNA. However, transcription experiments with T3 and T7 bacteriophage polymerases demonstrated that arrest of mitochondrial RNA polymerase by mtDBP must be more than a physical blockage imposed by a tightly DNA-bound protein. In fact, we observed that mtDBP exerted a polar termination effect on both T3 and T7 RNA polymerases: it arrested the passage of the enzymes that were moving in the same direction as that of transcription of sea urchin mtDNA L-strand, whereas the DNA-bound protein was bypassed when the enzymes travelled in the opposite sense, that is, in the direction of H-strand transcription (6,7) (Fig. 5).
Figure 5.
Diagram summarising the polar transcription termination activity of mtDBP. The region of sea urchin mtDNA contacted by mtDBP and additional flanking sequences are shown. OH, DNA replication origin; AT, AT-rich region. Thick arrows indicate the direction of progression and the points of arrest of the RNA polymerases used in the transcription assays. Thin arrows mark the suggested direction of movement and the points of arrest of sea urchin mtRNA polymerase, according to the proposed functional model of mtDBP; the dashed arrow refers to the direction of the DNA replication block. A possible asymmetric structure of mtDBP is also shown: the blunt and concave sides are suggested to function as roadblock and specific domain, respectively.
The fact that mtDBP blocks the progression of the bacteriophage enzymes in a polar fashion whereas it functions in an orientation-independent manner with the human mitochondrial RNA polymerase could suggest that the protein–DNA complex is structurally asymmetrical in that it exposes two distinct ‘surfaces’ to RNA polymerases moving on opposite directions. In this scenario (Fig. 5), one domain would be required by the protein to establish specific contacts with the mitochondrial RNA polymerase that is approaching the bound protein in the same direction as that of H-strand transcription. The specificity of this domain is confirmed by the observation that when it is encountered by the bacteriophage polymerase no such specific protein–protein interactions take place; as a consequence, the DNA-bound protein is ‘ignored’ and no termination event occurs. The opposite domain, on the contrary, seems to function in a less specific fashion as it not only arrests the mitochondrial RNA polymerase, but also stops T3 and T7 RNA polymerases. In this case a mechanism of physical blockage to elongation caused by the tight binding of the protein can be proposed.
Concerning the termination mode of mTERF, the interpretation of early studies suggested a simple model of transcriptional arrest for the protein based primarily on a physical road block to RNA chain elongation (18); however, recent findings (17) indicated that the recombinant protein fails to terminate transcription, even though it is able to bind DNA. This observation suggests that mTERF cannot simply act as a roadblock and adds a further common feature shared by mtDBP and mTERF.
Another interesting feature of mtDBP-mediated transcription termination concerns the finding that once elongation by T7 RNA polymerase has been stopped by mtDBP, it cannot resume after a period that extends over the half-life (∼250 min) of the association between mtDBP and its binding site. This implies that the mtDBP block causes a release of the RNA from the stalled RNA polymerase and/or dissociation of the RNA polymerase from the DNA template. This behaviour is similar to that shown by mTERF (18) and different from that of the termination factor of either Escherichia coli or T7 and SP6 RNA polymerases. In fact, in the latter case once the transcription block is eliminated, the paused polymerases are fully capable of resuming RNA elongation (19,20).
The proposed role for mtDBP as bidirectional transcription termination factor allows refinement of the transcription model of sea urchin mtDNA. Data from our and other laboratories showed that the sea urchin mitochondrial genome is transcribed via multiple and partially overlapping transcription units (6,7) initiating at the conserved AT-rich non-coding sequences located in five regions of the genome. The bidirectional termination function of mtDBP at the NCR and at the ND5/ND6 border could be used to control the overlapping of the large number of transcripts generated by the different transcription units. The existence of two binding sites as opposed to the single mTERF target site in mammals might be due to the multiplicity of transcription units in sea urchin mitochondria that requires more than one checkpoint to regulate RNA synthesis.
It was inferred previously that mtDBP had a role in regulating mtDNA replication, as the mtDBP-binding site in the NCR contains the 3′ end of the (D-loop) structure, where D-loop formation or H-strand extension take place (14). The functional model presented here supports the view that the same protein, besides acting as transcription termination factor, could also play a role in controlling the replication cycle. By promoting H-strand transcription arrest with the specific surface, mtDBP would produce the 12S RNA precursor transcript and also protect the replication origin from possible inactivation by transcript invasion. On the other hand, the roadblock activity exerted by the opposite face could serve two functions: creating the 3′ end of the replication primer by arresting L-strand transcription and/or forming the D-loop structure by blocking DNA polymerase progression. The use of the same protein to perform both roles is justified by the compact organisation of the sea urchin D-loop region (∼130 nt as opposed to ∼1000 nt in mammals) and by the observation that the 3′ end of the primer is very close (20–30 bp) to the 3′ end of the newly synthesised DNA (4). There is at least one precedent for a DNA-binding protein serving such two functions. In mammals it was shown that the nuclear transcription termination factor TTF-1 bound to its target sequence blocks the progression of both elongating RNA polymerase I and DNA polymerase, as both transcriptional and replicative elements are co-localised within a short region of DNA (21).
It is tempting to speculate that mtDBP function could be modulated on the basis of changes in leucine zipper interactions, which could result in exposing or ‘hiding’ the blocking surface to polymerases, depending on the physiological conditions. Precise mapping of the domains that come to close contact with the enzyme, isolation of mutants of mtDBP that are able to bind DNA but fail to terminate transcription and, particularly, solving the crystal structure of the protein will contribute to prove the proposed model, leading to a better understanding of the molecular mechanisms that control gene expression in sea urchin mitochondria.
Acknowledgments
ACKNOWLEDGEMENTS
We are very grateful to A. Sagliano for collaboration in the initial stages of this investigation. The technical assistance of V. Cataldo is gratefully acknowledged. This work was supported in part by Ministero della Ricerca Scientifica e Tecnologica (Progetto PRIN 2000-MURST), in part by University of Bari (Progetto di Ricerca di Ateneo, ex 60%) and in part by the Spanish Dirección General de Enseñanza Superior e Investigación Científica (PB971019).
REFERENCES
- 1.Cantatore P., Roberti,M., Rainaldi,G., Gadaleta,M.N. and Saccone,C. (1989) The complete nucleotide sequence, gene organization, and genetic code of the mitochondrial genome of Paracentrotus lividus. J. Biol. Chem., 264, 10965–10975. [PubMed] [Google Scholar]
- 2.Jacobs H.T., Elliott,D.J., Math,V.B. and Farquharson,A. (1988) Nucleotide sequence and gene organization of sea urchin mitochondrial DNA. J. Mol. Biol., 202, 185–217. [DOI] [PubMed] [Google Scholar]
- 3.De Giorgi C., Martiradonna,A., Lanave,C. and Saccone,C. (1996) Complete sequence of the mitochondrial DNA in the sea urchin Arbacia lixula: conserved features of the echinoid mitochondrial genome. Mol. Phylogenet. Evol., 5, 323–332. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs H.T., Herbert,E.R. and Rankine,J. (1989) Sea urchin egg mitochondrial DNA contains a short displacement loop (D-loop) in the replication origin region. Nucleic Acids Res., 17, 8949–8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayhook A.G., Rinaldi,A.M. and Jacobs,H.T. (1992) Replication origins and pause sites in sea urchin mitochondrial DNA. Proc. R. Soc. Lond. B, 248, 85–94. [DOI] [PubMed] [Google Scholar]
- 6.Cantatore P., Roberti,M., Loguercio Polosa,P., Mustich,A. and Gadaleta,M.N. (1990) Mapping and characterization of Paracentrotus lividus mitochondrial transcripts: multiple and overlapping transcription units. Curr. Genet., 17, 235–245. [DOI] [PubMed] [Google Scholar]
- 7.Elliott D.J. and Jacobs,H.T. (1989) Mutually exclusive synthetic pathways for sea urchin mitochondrial rRNA and mRNA. Mol. Cell. Biol., 9, 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberti M., Musicco,C., Loguercio Polosa,P., Gadaleta,M.N., Quagliariello,E. and Cantatore,P. (1997) Purification and characterization of a mitochondrial, single-stranded-DNA-binding protein from Paracentrotus lividus eggs. Eur. J. Biochem., 247, 52–58. [DOI] [PubMed] [Google Scholar]
- 9.Roberti M., Musicco,C., Loguercio Polosa,P., Gadaleta,M.N. and Cantatore,P. (1996) DNA-helicase activity from sea urchin mitochondria. Biochem. Biophys. Res. Commun., 219, 134–139. [DOI] [PubMed] [Google Scholar]
- 10.Qureshi S.A. and Jacobs,H.T. (1993) Characterization of a high-affinity binding site for a DNA-binding protein from sea urchin embryo mitochondria. Nucleic Acids Res., 21, 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qureshi S.A. and Jacobs,H.T. (1993) Two distinct, sequence-specific DNA-binding proteins interact independently with the major replication pause region of sea urchin mtDNA. Nucleic Acids Res., 21, 2802–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberti M., Loguercio Polosa,P., Musicco,C., Milella,F., Qureshi,S., Gadaleta,M.N., Jacobs,H.T. and Cantatore,P. (1999) In vivo mitochondrial DNA–protein interactions in sea urchin eggs and embryos. Curr. Genet., 34, 449–458. [DOI] [PubMed] [Google Scholar]
- 13.Roberti M., Mustich,A., Gadaleta,M.N. and Cantatore,P. (1991) Identification of two homologous mitochondrial DNA sequences, which bind strongly and specifically to a mitochondrial protein of Paracentrotus lividus. Nucleic Acids Res., 19, 6249–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loguercio Polosa P., Roberti,M., Mustich,A., Gadaleta,M.N. and Cantatore,P. (1994) Purification and characterization of a mitochondrial DNA-binding protein that binds to double-stranded and single-stranded sequences of Paracentrotus lividus mitochondrial DNA. Curr. Genet., 25, 350–356. [DOI] [PubMed] [Google Scholar]
- 15.Loguercio Polosa P., Roberti,M., Musicco,C., Gadaleta,M.N., Quagliariello,E. and Cantatore,P. (1999) Cloning and characterisation of mtDBP, a DNA-binding protein which binds two distinct regions of sea urchin mitochondrial DNA. Nucleic Acids Res., 27, 1890–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kruse B., Narasimhan,N. and Attardi,G. (1989) Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell, 58, 391–397. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Silva P., Martinez-Azorin,F., Micol,V. and Attardi,G. (1997) The human mitochondrial transcription termination factor (mTERF) is a multizipper protein but binds to DNA as a monomer, with evidence pointing to intramolecular leucine zipper interactions. EMBO J., 5, 1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang J. and Clayton,D.A. (1994) Human mitochondrial transcription termination exhibits RNA polymerase independence and biased bipolarity in vitro. J. Biol. Chem. 269, 29112–29120. [PubMed] [Google Scholar]
- 19.Pavco P.A. and Steege,D.A. (1990) Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J. Biol. Chem. 265, 9960–9969. [PubMed] [Google Scholar]
- 20.Pavco P.A. and Steege,D.A. (1991) Characterization of elongating T7 and SP6 RNA polymerases and their response to a roadblock generated by a site-specific DNA binding protein. Nucleic Acids Res., 19, 4639–4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerber J.K., Goegel,E., Berger,C., Wallisch,M., Mueller,F., Grummt,I. and Grummt,F. (1997) Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell, 90, 559–567. [DOI] [PubMed] [Google Scholar]
- 22.Anderson S., Bankier,A.T., Barrell,B.G., De Bruijn,M.H.L., Coulson,A.R., Drouin,J., Eperon,I.C., Nierlich,D.P., Roe,B.A., Sanger,F., Schreier,P.H., Smith,A.J.H., Staden,R. and Young,I.G. (1981) Sequence and organization of the human mitochondrial genome. Nature, 290, 457–465. [DOI] [PubMed] [Google Scholar]