Abstract
Sclerotinia sclerotiorum acidifies its ambient environment by producing oxalic acid. This production of oxalic acid during plant infection has been implicated as a primary determinant of pathogenicity in this and other phytopathogenic fungi. We found that ambient pH conditions affect multiple processes in S. sclerotiorum. Exposure to increasing alkaline ambient pH increased the oxalic acid accumulation independent of carbon source, sclerotial development was favored by acidic ambient pH conditions but inhibited by neutral ambient pH, and transcripts encoding the endopolygalacturonase gene pg1 accumulated maximally under acidic culture conditions. We cloned a putative transcription factor-encoding gene, pac1, that may participate in a molecular signaling pathway for regulating gene expression in response to ambient pH. The three zinc finger domains of the predicted Pac1 protein are similar in sequence and organization to the zinc finger domains of the A. nidulans pH-responsive transcription factor PacC. The promoter of pac1 contains eight PacC consensus binding sites, suggesting that this gene, like its homologs, is autoregulated. Consistent with this suggestion, the accumulation of pac1 transcripts paralleled increases in ambient pH. Pac1 was determined to be a functional homolog of PacC by complementation of an A. nidulans pacC-null strain with pac1. Our results suggest that ambient pH is a regulatory cue for processes linked to pathogenicity, development, and virulence and that these processes may be under the molecular regulation of a conserved pH-dependent signaling pathway analogous to that in the nonpathogenic fungus A. nidulans.
Sclerotinia sclerotiorum is a filamentous ascomycete phytopathogen that produces oxalic acid during growth in vitro and in planta. Mutants deficient in the ability to produce oxalic acid are nonpathogenic and fail to produce sclerotia (14). Multiple synergistic roles have been proposed for oxalic acid during pathogenesis (29, 32). During plant infection, the secretion of oxalic acid creates an acidic pH environment necessary for the activities of many hydrolytic enzymes (3), including polygalacturonases (24). Polygalacturonase enzymes have been implicated as colonization and virulence factors in other plant-infecting fungi (44, 49). Furthermore, oxalic acid chelates calcium, resulting in a destabilization of pectate polymers allowing increased access and sensitivity to pathogen-produced pectolytic enzymes (32). Oxalic acid also suppresses the plant oxidative burst (5) and inhibits the activities of plant-produced polyphenol oxidase (27, 29), suggesting that oxalic acid plays more subtle roles in pathogenesis as well.
Secretion of oxalic acid by S. sclerotiorum results in the acidification of the growth environment. The pH of in vitro-grown liquid cultures and infected host tissues can be as low as 2 and 4, respectively (29, 32). Numerous carbon sources, including components of plant cell walls, can support oxalic acid accumulation when provided as the sole carbon source (30, 32, 51). Culture pH also is a strong regulator of oxalic acid biosynthesis (32, 51). Oxalic acid production increases with the ambient pH of the growth medium, as does oxaloacetase activity, the enzyme proposed to catalyze oxalic acid production by hydrolysis of oxaloacetate (31). Since S. sclerotiorum acidifies the extracellular environment and yet cytosolic pH is assumed to remain relatively stable, any effects of external pH change on intracellular enzyme activities should be transient. These findings suggest that a signaling pathway responsive to external pH conditions regulates the expression of a gene(s) for oxalic acid biosynthesis.
In Aspergillus nidulans, an ambient pH-sensing signal transduction pathway affects expression of genes encoding several secreted and outer membrane bound proteins, as well as enzymes that synthesize metabolites destined for export (4, 8, 9, 26, 43, 50). The gene product of pacC is the terminal component of the pH signaling pathway and the regulator of pH-dependent gene expression (4). This protein has a zinc finger DNA-binding domain with a core DNA consensus binding site of 5′-GCCARG-3′ (50). pacCc mutations result in constitutive activation of the pH-responsive gene expression pathway and mimic gene expression patterns observed in the wild type under alkaline growth conditions (4). pacC-null mutations result in severe defects in growth and conidiation, and gene expression is similar to that observed in the wild type at acidic pH (50).
Our objectives in this study were (i) to determine if ambient pH was a major regulator of oxalic acid biosynthesis and sclerotial development. (ii) to identify ambient pH-responsive gene expression, and (iii) to determine if a homolog of the ambient pH transcriptional regulator pacC was present in S. sclerotiorum. We hypothesize that ambient pH is a signal for the transcriptional regulation of genes necessary for the disease process and developmental life cycle of this organism. We found that diverse processes in S. sclerotiorum are influenced by ambient pH and that pac1 is a functional homolog of the pacC pH-responsive transcription factor. Our results suggest that pH-responsive gene regulation plays a role in S. sclerotiorum pathogenicity and development.
MATERIALS AND METHODS
S. sclerotiorum growth conditions.
We utilized wild-type S. sclerotiorum isolate 1980 obtained from J. R. Steadman at the University of Nebraska–Lincoln. This isolate was originally obtained from dry bean culls in western Nebraska (14) and was routinely cultured on potato dextrose agar (PDA; Difco, Detroit, Mich.). To analyze the effects of ambient pH and carbon source on oxalic acid accumulation, we inoculated 500 ml of YPSu medium (containing, per liter, 4 g of yeast extract [Difco], 15 g of sucrose, 1 g of K2HPO4, and 0.5 g of MgSO4) in a 1-liter flask with 20 ∼5-mm3 agar-mycelium plugs of S. sclerotiorum obtained from the advancing margin of a PDA culture. This liquid culture was incubated at room temperature, with shaking at 150 rpm, for 5 days and then blended in a Waring blender and incubated for an additional 24 h. The entire culture was harvested by vacuum filtration onto filter paper and washed three times by resuspension in 600 ml of distilled water. Then, 10-ml aliquots of the washed mycelia (average mycelial wet weight, 200 mg) were harvested by vacuum filtration and resuspended in 50 ml of fresh medium containing, per liter, 4 g of yeast extract, 1 g of K2HPO4, 0.5 g of MgSO4, 0.5 M 3-(N-morpholino)propanesulfonic acid (MOPS) buffer, and either one of four carbon sources—sucrose (7.5 g), glucose (15 g), mannitol (15 g), or soluble starch (15 g)—or no carbon. Preliminary results indicated that the need for greatest control of pH would be at pH 7; therefore, although MOPS provides little buffering capacity below pH 7, it was used for all pH treatments to control for buffer effects. Nine flasks were inoculated for each carbon source, three each with initial pH values of 3.0, 5.0, or 7.0. These cultures were incubated for an additional 24 h. These secondary cultures were harvested by vacuum filtration, and supernatants were saved for oxalic acid quantification.
To determine the effect of ambient pH on sclerotial development, 50 ml of YPSu liquid medium cultures were inoculated with four ∼5-mm3 agar-mycelium plugs of S. sclerotiorum. Cultures were grown for 4 days and then blended with a surface-sterilized hand mixer, and the incubation was continued. At 24 h after blending, cultures were harvested by vacuum filtration onto filter paper (Whatman no. 1) and washed with four volumes of sterile distilled water. The filter paper containing the mycelial mat was transferred to a 9-cm glass petri dish containing a single layer of 5-mm-diameter glass beads and 20 ml of 0.5 M MOPS solution at pH 5.0 or pH 7.0. The buffer was replaced every 24 h for 4 days to ensure stable pH conditions for the duration of the experiment. Cultures were monitored for sclerotial development and then photographed after 4 days. Three cultures were evaluated at each pH, and the experiment was repeated twice.
Oxalic acid quantification.
Supernatants from cultures grown on different carbon sources and at various pH levels were analyzed for oxalic acid content with an enzymatic assay kit (Sigma, St. Louis, Mo.) according to the manufacturer's instructions. The oxalic acid concentration was calculated by extrapolation from a standard curve and adjusted for the dilution factors. Values represent the mean and standard deviation of three replications and are adjusted by subtraction of background obtained by incubating in buffer alone at the three different pH values.
Library construction and pac1 cloning.
A 145-bp sequence from the A. nidulans pacC gene was obtained by PCR amplification of A. nidulans genomic DNA with forward primer 5′-AACCTCAACCTGAACTTGTCAATGGG-3′ and reverse primer 5′-AAATCCTGGGGACGCTT GAA-3′ followed TOPO TA Cloning into pCRII (Invitrogen, Carlsbad, Calif.) to generate pCRPacC. Total genomic DNA was isolated from the mycelia of S. sclerotiorum using the protocol of Panaccione et al. (35). Southern hybridization of S. sclerotiorum DNA digested with PstI and fractionated on a 0.8% agarose gel was carried out by standard protocols (2) using a 32P-labeled probe prepared by PCR labeling (33) of the pCRPacC insert. Low-stringency hybridization was performed in 1% bovine serum albumin (BSA), 1 mM EDTA, 0.5 M NaHPO4, and 7% sodium dodecyl sulfate (SDS) at 55°C, followed by two room temperature washes and two 55°C washes in 0.5% BSA, 1 mM EDTA, 40 mM NaHPO4, and 5% SDS.
We constructed a library from total genomic DNA of S. sclerotiorum isolate 1980 by ligating partially Sau3AI-digested, size-selected- (14 to 23-kb) DNA fragments into the BamHI site of the λ replacement vector λEMBL3 (Promega, Madison, Wis.). This library was plated and screened using standard protocols (2) under the same conditions and with the same 32P-labeled pacC probe as those described above for the Southern hybridization. Positive plaques were identified by autoradiography and purified by three successive rounds of screening. Sequences hybridizing to the pacC probe were localized to a 9.5-kb PstI fragment on clone λPacS7.2 and to a 6.7-kb XbaI fragment on clone λPacS4.2. These fragments were cloned into the pBluescript II KS(−) vector (Stratagene, La Jolla, Calif.) and were designated pBSPacS7.2 and pBSPacS4.2, respectively. The pacC-related sequence was further localized to a 543-bp HindIII-Sau3AI fragment on pBSPacS7.2, and a subclone, pBSPacS7.2HS, containing this fragment was made.
We prepared a cDNA library in the λZipLox vector (Gibco-BRL, Rockville, Md.) with a SuperScript λcloning System (Gibco-BRL) from poly (A)+ RNA obtained from developing sclerotia of S. sclerotiorum isolate 1980. S. sclerotiorum was cultured on PDA medium in 9-cm-diameter petri plates by inoculating a single mycelium-agar plug onto the center of each plate. After 4 days of growth at room temperature, nonmelanized, developing sclerotia with liquid exudate on the surface were harvested with forceps, frozen in liquid nitrogen, and stored at −80°C. These immature sclerotia were frozen in liquid nitrogen and ground to a fine powder with a mortar and pestle. Total RNA was isolated with Trizol reagent (Gibco-BRL) according to the manufacturer's instructions. Poly(A)+ RNA was purified with a Dynabead mRNA detection kit (Dynal, Oslo, Norway) according to the manufacturer's instructions. This poly(A)+ RNA was used to construct a unidirectional cDNA library. This library was screened with the pacC-hybridizing HindIII-Sau3AI fragment from pBSPacS7.2HS according to standard protocols (2).
Hybridization was done in 1% BSA, 1 mM EDTA, 0.5 M NaHPO4, and 7% SDS at 65°C, followed by a 10-min room temperature wash in 0.5% BSA, 1 mM EDTA, 40 mM NaHPO4, and 5% SDS and then three 10-min washes, the first at room temperature and the last two at 65°C, in 1 mM EDTA, 40 mM NaHPO4, and 1% SDS. cDNA clones were recovered as autonomously replicating pZL plasmids by using the in vivo excision protocol supplied with the cloning kit (Gibco-BRL). One of these clones with a 2.4-kb insert, designated pZLPacS1, was chosen for further analysis.
pac1 sequence analysis.
Plasmids pBSPacS4.2 and pZLPacS1, containing the genomic and cDNA sequences of pac1, respectively, were used as DNA sequencing templates. Both strands of each insert were sequenced using universal, reverse, and custom-made oligonucleotide primers (DNA Sequencing Core Research Facility, University of Nebraska–Lincoln). Sequence contigs were assembled and analyzed with DNASIS (Hitachi Software Engineering Co., Yokohama, Japan). Homology searches were conducted with the BLAST algorithm (1). Multiple amino acid sequence alignment with S. sclerotiorum Pac1 (accession no. AY005467), A. niger PacC (accession no. X98417), A. nidulans PacC (accession no. Z47081), Penicillium chrysogenum PacC (accession no. U44726), Yarrowia lipolytica YIRIM101 (accession no. X99616), Candida albicans HRM101 (accession no. AF173841), and Saccharomyces cerevisiae RIM1 (accession no. X72960) were conducted with the Wisconsin Sequence Analysis Program 9.1 (Genetics Computer Group, Madison, Wis.). Shading of conserved residues on the multiple amino acid sequence analysis output was conducted with MacBoxShade v2.01 (http://ulrec3.unil.ch/software/BOX_form.html).
Northern analysis.
Primary cultures were grown and harvested as described above for experiments to determine the effect of ambient pH on sclerotial development. For ambient pH series experiments, harvested cultures were transferred to fresh YPSu medium buffered with citric acid-sodium phosphate buffer to achieve initial pH values between 3.0 and 7.0; the actual initial pH value for each culture was determined after autoclaving. These secondary cultures were incubated for 6 h, harvested by vacuum filtration, and quick frozen in liquid nitrogen. For expression kinetics experiments, primary cultures were incubated for 0, 10, and 30 min and for 1, 2, 3, 4, 5, and 6 h in YPSu medium buffered with citric acid-sodium phosphate buffer at pH 7.9. Total RNA was prepared with Trizol reagent as described above. For Northern blot analysis, 15 μg of RNA and RNA size standards was electrophoresed in a 0.8% agarose–0.66 M formaldehyde gel and transferred to MagnaGraph nylon membranes (Micron Separations, Inc., Westborough, Mass.) according to standard protocols (2). Hybridization probes were generated by 32P-random primer labeling (11). The sequences used as hybridization probes were the pac1 cDNA sequence from pZLPacS1; the endopolygalacturonase-encoding pg1 coding sequence (39), obtained by PCR amplification of S. sclerotiorum DNA with primers derived from the published sequence (39) and subsequent TOPO TA cloning into the pCRII vector (Invitrogen); and the rDNA repeat sequence from Neurospora crassa from pMF2 (13). Hybridization and washing conditions were the same as for cDNA library screening. The washed blots were autoradiographed and exposed to a phosphor screen that was scanned by a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Hybridization signals were quantified using ImageQuant software (Molecular Dynamics), and the reported relative level of signal in each lane was corrected based on the hybridization signal from the rDNA probe. All pH-transfer experiments and Northern blots were conducted a minimum of three times.
A. nidulans strains and transformation.
A. nidulans strain RDP2 (pabaA1 argB2 pacC veA1) (37) carrying the null pacC allele (50) was provided by Nancy Keller. This strain was transformed with the argB-containing plasmid pPK1 (53) and pPKPacS, created by ligating a 5.9-kb XbaI-XhoI fragment from pBSPacS4.2 containing the pac1 coding sequence plus 3.3 kb of the upstream sequence and 666 bp of the downstream sequence into pPK1. To obtain RDP2 protoplasts, the strain was cultured on solid pH 4-buffered minimal medium (7) supplemented with 200 mg of l-arginine hydrochloride, 85 μg of p-aminobenzoate (PABA), and 5 g of yeast extract (Difco) per liter. Conidia were collected from the surface of plates by scraping into sterile water. Conidia and hyphae collected from all 10 plates were inoculated into 400 ml of liquid, pH 4-buffered minimal medium (7), supplemented as necessary, and incubated at 37°C and at 150 rpm for 16 h. Protoplasts were prepared and transformed essentially as described by Yelton et al. (55). Transformants were selected on solid, pH 4-buffered minimal medium containing 1.2 M sorbitol and 85 μg of PABA per liter. Transformants were evaluated for rate of growth, conidiation, and morphology on pH 4- and pH 8-buffered minimal medium supplemented with PABA. Photomicrographs of conidiophores were taken on a Zeiss (Jena, Germany) Axioskop compound light microscope with differential interference contrast optics. Comparisons were made between strains RKS1 (pabaA1 yA2 veA1), containing the wild-type pacC allele, and the recipient strain RDP2, RDP2 transformed with pPK1, and RDP2 transformed with pPKPacS.
Nucleotide sequence number.
The pac1 genomic sequence has been deposited in the GenBank, EMBL, and DDBJ DNA data banks under accession no. AY005467.
RESULTS
Ambient pH and carbon source regulation of oxalic acid accumulation.
Total oxalic acid accumulation in media buffered at an initial pH of 7.0 ranged from an average of 1,500 mg of oxalic acid per liter of culture supernatant with glucose, sucrose, and starch as the carbon source to 150 mg of oxalic acid per liter of culture supernatant in media with mannitol as the carbon source (Table 1). When oxalic acid levels in media were compared at three different initial pH values, pH 3, 5, and 7, the highest levels of oxalic acid were observed at pH 7, with lower levels at pH 5 and the lowest levels at pH 3 (Table 1). If the initial culture pH was 5 or 3, then no difference in oxalic acid accumulation between mannitol media and media prepared from other carbon sources was observed.
TABLE 1.
Influence of pH and carbon source on oxalic acid accumulation
| Carbon source | pHia | pHffb | Mean oxalic acid concn (mg/liter) ± SD |
|---|---|---|---|
| Glucose | 3.0 | 2.5 | 3.9 ± 2.0 |
| 5.0 | 3.3 | 90 ± 7.4 | |
| 7.0 | 6.6 | 1,500 ± 400 | |
| Sucrose | 3.0 | 2.6 | 3.2 ± 1.4 |
| 5.0 | 3.5 | 79 ± 23 | |
| 7.0 | 6.7 | 1,500 ± 173 | |
| Starch | 3.0 | 2.8 | 2.0 ± 1.2 |
| 5.0 | 3.7 | 21 ± 5.8 | |
| 7.0 | 6.9 | 1,400 ± 140 | |
| Mannitol | 3.0 | 2.8 | 1.7 ± 0.8 |
| 5.0 | 3.7 | 74 ± 20 | |
| 7.0 | 6.9 | 150 ± 44 |
pH of the culture before mycelial inoculation.
pH of the culture 24 h after mycelial inoculation. Within treatments, the final pH values varied by 0.1 or less.
Ambient pH and sclerotial development.
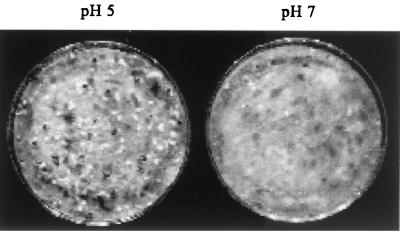
Sclerotia do not develop from mycelia in liquid submerged culture but do develop synchronously when mycelia are transferred to aerial conditions. After transfer to conditions conducive for sclerotial development at pH 5, sclerotial initials were evident by 24 h, melanized sclerotia were evident by 48 h, and nearly mature sclerotia were evident by 4 days (Fig. 1). In contrast, sclerotial initiation and development were almost completely inhibited under the same conditions when the pH was held constant at pH 7 for 4 days (Fig. 1). This pH effect on sclerotial development was independent of nutrient requirements since sclerotia developed normally and with the same kinetics when YPSu medium buffered at pH 5 was used and since development was inhibited in buffered YPSu medium at pH 7 (data not shown). In these same YPSu cultures 24 h after transfer to sclerotium-inducing conditions, nearly five times as much oxalic acid (1,600 ± 30 mg/liter) accumulates at pH 7 relative to pH 5 (330 ± 45 mg/liter).
FIG. 1.
pH regulation of sclerotial development. Mycelial cultures were harvested and transferred to stationary, sclerotium-inducing conditions in 9-cm-diameter petri dishes with the liquid underlay buffered with 0.5 M MOPS at pH 5 or pH 7. Buffer was replaced every 24 h to maintain constant pH, and treatments were performed in triplicate with three replications. Representative cultures from each treatment are shown.
pg1 expression.
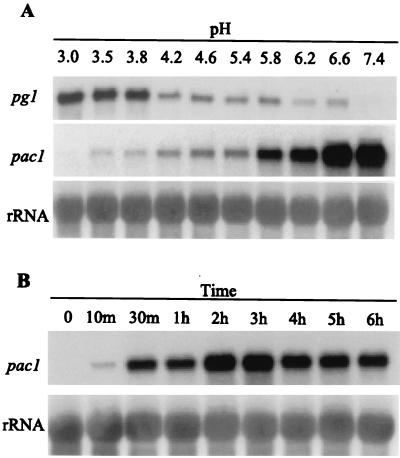
The influence of ambient culture pH on the accumulation of transcripts encoding the endopolygalacturonase enzyme PG1 was examined by Northern blot analysis. Mycelia were transferred from primary cultures with an average pH of 3.2 to fresh cultures with a series of pH values from 3.0 to 7.4. pg1 expression decreases as ambient pH increases (Fig. 2A). Transcript levels of pg1 were highest between pH 3.0 and 3.8 and decreased dramatically at pH 4.2 and above (Fig. 2A).
FIG. 2.
pg1 and pac1 transcript accumulation in response to different ambient pH conditions. (A) Lanes 1 to 10 show a Northern analysis of total RNA isolated from S. sclerotiorum mycelia 6 h after transfer to fresh YPSu medium buffered with citrate-phosphate buffer at the indicated pH values. Blots were probed with pac1 and then sequentially stripped and reprobed with pg1 and ribosomal DNA (rDNA) probes as indicated. (B) Northern analysis of total RNA isolated from S. sclerotiorum mycelia at various time points after transfer to fresh YPSu medium buffered with citrate-phosphate buffer at pH 7.9. Lane 1, total RNA from mycelia of the primary culture (final pH, 3.0); lanes 2 to 9, total RNA from cultures at 10 min to 6 h after transfer to pH 7.9. Blots were probed with pac1, stripped, and reprobed with the rDNA probe. To facilitate comparisons of transcript levels between various time points, the level of pac1 signal detected at the 10-min time point is set at 1X.
Isolation and characterization of a pacC homolog.
We probed a Southern blot of restriction enzyme-digested S. sclerotiorum genomic DNA at low stringency with a fragment of the pacC gene and observed a single, weakly hybridizing band. From the S. sclerotiorum genomic library in λEMBL3, we identified six hybridizing clones. We localized the pacC-related sequences to an approximately 6.7-kb XbaI fragment from clone λPacS4.2. This fragment was subcloned into pBluescript II KS(−) to create pBSPacS4.2.
Sequences from pBSPacS4.2 hybridized to nine clones in the λZipLox library that we had constructed from S. sclerotiorum cDNA. The largest insert size, 2.4 kb, was present in three of these clones. We sequenced one of these cDNA clones designated pZLPacS1. We identified a single open reading frame of 1,878 bp, predicted to encode a 626-amino-acid, 68-kDa protein (GenBank accession no. AY005467). This predicted protein shares high sequence similarity with the PacC/RIM1 family of transcription factors which have been identified from A. nidulans (50), A. niger (25), P. chrysogenum (47), S. cerevisae (46), Y. lipolytica (20), and C. albicans (38). Over its entire sequence, the predicted peptide shares greatest identity (44%) and similarity (51%) to the A. nidulans and A. niger PacC peptides. We refer to this gene as pac1 and the encoded peptide as Pac1.
Several of the sequence features described for the A. nidulans PacC peptide (50) are conserved in Pac1. These include a tyrosine-rich region and two proline-glycine-rich regions downstream of the zinc finger. Additionally, the carboxy-terminal half of Pac1 is serine- and threonine-rich, and the S/TPXX motif that is frequently found in transcription factors (48), including PacC (50), occurs 13 times in Pac1. Two putative nuclear localization signals (NLS) were identified in the Pac1 sequence using PSORT II (34). The first, KRNFDNLNDFFAGAAKRR, begins at residue 236 and fits the pattern of a bipartite NLS (40). These Pac1 residues align with a putative bipartite NLS in the PacC homologs (data not shown). A second potential NLS of the SV40 large T antigen type (16) was identified, PRRR, beginning at residue 475.
The amino terminus of Pac1, comprising 44 residues upstream of the zinc finger domain, is glutamine-rich (11 of 44 residues), including a stretch of six contiguous glutamine residues, serine-rich (10 of 44 residues), and threonine-rich (6 of 44 residues) but, in contrast to the N termini of the aspergilli and Penicillium PacC homologs, is not particularly alanine-rich (4 of 44 residues in Pac1).
The most highly conserved sequence feature among the PacC/RIM1 homologs is the zinc finger region. Pac1 contains three Cys2His2 zinc finger domains designated Zf1, Zf2, and Zf3 (Fig. 3). The Cys-Cys and His-His spacing (Zf1: C4C, H4H; Zf2: C4C, H3H; and Zf3: C2C, H3H) present in the other homologs is conserved in Pac1. The greatest sequence conservation is observed in Zf3, in which Pac1 Zf3 contains one conservative substitution, Glu (E) for Asp (D) at position c2 (following the conventional notation established by Jacobs [17]), relative to Zf3, c2 residues in aspergilli, and Penicillium PacC homologs. Compared to Zf2 of the aspergilli PacC peptides, Pac1 Zf2 contains two nonconservative amino acid substitutions: Gly (G) for Gln (Q) at position c2 and Asn (N) for Gly (G) at position c4. The amino acid substitutions observed in Zf2 and Zf3 of Pac1 relative to PacC are in the predicted b2 strand and not in residues of the alpha helix involved with specific DNA base recognition by PacC (10). Among the zinc fingers of all the PacC/RIM1 homologs, the weakest sequence conservation is within Zf1. In the Pac1 Zf1, there are four conservative and six nonconservative substitutions relative to the Zf1 of A. nidulans PacC. However, a pair of Trp (W) residues that are thought to be necessary for Zf1-Zf2 interaction are conserved at the c3 position within the Cys knuckles of Zf1 and Zf2.
FIG. 3.
Amino acid sequence alignment of zinc finger domains from Pac1 and PacC/RIM1 homologs. Residues conserved in at least four of the seven sequences are shaded with black boxes, and conservative amino acid substitutions are shaded with gray boxes. The three zinc finger domains conforming to the consensus of Y/FCX2,4CX3FX5LX2HX3,5H (17) are indicated with an overline and are labeled Zf1, Zf2, and Zf3. Sequences are aligned from greatest to least homology to the derived S. sclerotiorum Pac1 sequence. GenBank accession numbers: S.scl-Pac, AY005467; A.nig-Pac, X98417; A.nid-Pac, Z47081; P.chr-Pac, U44726; Y.lip-Rim, X99616; C.alb-Rim, AF173841; S.cer-Rim, X72960.
The 6.7-kb insert of the genomic pac1 clone pBSPacS4.2 was sequenced (GenBank accession no. AY005467) and contains the entire pac1 gene. No significant open reading frame other than pac1 was found. We compared the sequences of the cDNA and genomic clones and found that the pac1 genomic sequence contains three introns. The codon position of the first pac1 intron is the same as that for pacC from A. nidulans, A. niger, and P. chrysogenum, and the codon position of the third pac1 intron is conserved in A. nidulans and A. niger. The second intron is unique to pac1. The 5′ limit of the pac1 cDNA sequence is 240 bp upstream of the putative ATG translation start site. The 5′ cytosine in this position occurs in a pyrimidine-rich environment (CACTTTTT) and thus is a putative transcriptional start site for pac1. The 240-base transcribed, untranslated sequence is AT-rich (67% A+T), as is the entire 1,100-bp sequence (60% A+T) upstream of the coding region. Eight copies of the PacC core consensus binding sequence (5′-GCCARG-3′) were found between the −339 and −599 bases upstream of the ATG start codon. Seven of the eight copies were found in the plus orientation, and all conformed to the more strictly defined consensus of 5′-GCCAAG-3′.
Effect of ambient pH on pac1 expression.
Primary mycelial cultures, average pH 3.2, were transferred to fresh medium buffered with citrate phosphate buffer to maintain a pH between 3.0 and 7.4. After a 6-h incubation, cultures were harvested and examined by Northern analysis (Fig. 2A). The pac1 gene hybridizes with a single mRNA transcript of approximately 2.4 kb. The steady-state levels of pac1 are influenced by and increase with ambient pH. The difference between pac1 transcript accumulations at pH 3.0 and at pH 7.4 is approximately 300-fold. pac1 transcripts accumulate following transfer from acidic (pH 3.0) to alkaline (pH 7.9) growth conditions. pac1 transcripts are barely detectable in total RNA from primary cultures (Fig 2B, time zero), but after 10 min at pH 7.9 the pac1 transcripts are easily detected (Fig. 2B). The steady-state level of pac1 transcripts increases under alkaline conditions, peaking at between 2 and 3 h after transfer with levels 12- to 13-fold higher than those found at the 10-min time point (Fig. 2B). By 6 h at alkaline pH, pac1 levels had dropped to approximately one-half of their maximal levels.
Complementation of a null pacC A. nidulans strain with pac1.
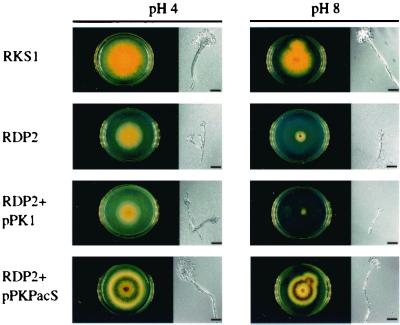
We used vectors pPKPacS and pPK1 to independently transform a null pacC argB A. nidulans strain (RDP2). Transformants were selected on pH4 minimal medium, and argB+ prototrophs were retained for further analysis. Several hundred pPKPacS and pPK1 transformants were collected. All pPK1 transformants were arginine prototrophs but otherwise retained RDP2 growth and conidiation phenotypes (Fig. 4). Single-spore strains of pPKPacS transformants all had the same phenotypes: arginine prototrophy and wild-type growth and conidiation at both alkaline and acidic ambient pH levels (Fig. 4).
FIG. 4.
pac1 complementation of an A. nidulans pacC-null strain. The colonial character and conidiation of RKS1, a yellow-spored A. nidulans pacC wild-type strain; RDP2, an A. nidulans pacC-null strain; RDP2+pPK1, and A. nidulans pacC-null strain transformed with the argB vector pPK1; and RDP2+pPKPacS, an A. nidulans pacC-null strain transformed with the argB, pac1 vector pPKPacS are shown. All were grown in 6-cm diameter petri dishes on minimal medium buffered with citrate-phosphate buffer and supplemented with PABA. The RDP2 culture also was supplemented with arginine. Scale bars represent 30 μm.
DISCUSSION
S. sclerotiorum is a broad-host-range phytopathogenic fungus that produces oxalic acid in high concentrations, thus creating an acidic environment in which it grows and causes disease. The carbon source and the pH of the growth medium influence the accumulation of oxalic acid in culture. Previous studies on the effect of pH on oxalic acid production in Sclerotinia spp. have focused on single carbon sources (32, 51), and most studies of carbon source effects on oxalic acid accumulation do not examine a range of pH values (30, 36, 51). We grew S. sclerotiorum on various carbon sources that support different levels of oxalic acid accumulation at a neutral ambient pH and compared these levels with the levels of oxalic acid accumulation at lower pH. Carbon source plays a substantial role in the ability to synthesize oxalic acid, but an alkaline environment increases oxalic acid biosynthesis independent of carbon source. Determining the hierarchial positioning of carbon and ambient pH regulation of oxalic acid biosynthesis awaits the identification of molecular components of this pathway.
The synthesis of oxalic acid in S. sclerotiorum is proposed to be catalyzed by oxaloacetate acetylhydrolase. This enzyme activity increases as the pH of the ambient environment increases, paralleling oxalic acid accumulation (19, 21, 31, 42). A second enzymatic activity, oxalate decarboxylase, catalyzes the breakdown of oxalic acid. This enzyme activity is higher in mycelial extracts from cultures grown under acidic growth conditions (28). In Aspergillus spp., genes encoding enzymes in the penicillin, sterigmatocystin, and aflatoxin biosynthetic pathways are regulated by ambient pH (6, 18, 43). Based on these findings, a differential pH expression strategy would appear to be a plausible approach for cloning genes involved in oxalic acid biosynthesis and degradation from S. sclerotiorum.
Oxalic acid synthesis and degradation may be tightly regulated to provide an optimal pH environment for lytic enzyme activities. Numerous pectinolytic, proteolytic, cellulytic, and other hydrolytic enzymes from S. sclerotiorum with acidic pH optima have been described (15, 22, 23, 24). Our Northern analysis with the endopolygalacturonase-encoding gene pg1 demonstrate that ambient pH can also regulate enzyme levels through gene transcription. pg1 is a member of a multigene family whose members share a high degree (97 to 98%) of sequence identity (12). At least one member of this family displayed acidic pH-specific expression. Specifically determining which member(s) of this family was regulated by ambient pH was not possible with the probe we used. Whether genes encoding other lytic enzymes also are pH regulated has yet to be determined. In Colletotrichum gloeosporioides, endopolygalacturonase gene expression and pectate lyase gene transcription and enzyme secretion were recently demonstrated to be regulated by ambient pH (54). Transcriptional regulation of host-degrading enzymes by ambient pH also has been demonstrated in the entomopathogenic fungus Metarhizium anisopliae (45). In this fungus, the kinetics of extracellular protease and chitinase transcript accumulation is pH dependent, with expression patterns closely paralleling the pH optima of enzyme activities. Thus, a dynamic system of gene regulation based on ambient pH sensing and modification of the ambient pH environment may have a central role in determining the pathogenic success of fungal pathogens such as C. gloeosporioides, M. anisopliae, S. sclerotiorum, and possibly other phytopathogenic fungi.
In addition to its role in pathogenesis, environmental pH may regulate sclerotial development in S. sclerotiorum (51). Sclerotia are compact, melanized, multihyphal structures that can survive long periods of time under adverse environmental conditions and carpogenically germinate to produce apothecia and ascospores. These properties of sclerotia make them essential for the long-term survival and dissemination of Sclerotinia spp. In the present study we found that ambient pH affects sclerotial development. At a neutral or alkaline ambient pH, sclerotial development is inhibited. If ambient conditions are maintained at a neutral pH by buffering, this inhibition of sclerotial development occurs despite the accumulation of high concentrations of oxalic acid. Thus, under normal developmental conditions, it is the lowering of ambient pH due to oxalic acid accumulation, rather than oxalic acid accumulation per se, that appears to be the important regulator of sclerotial development.
The relationship between oxalic acid production and sclerotial development is likely more complex than that of pH alone. Sclerotial development in mutants that can neither synthesize oxalic acid nor develop sclerotia (14) is not restored by simply lowering ambient pH (unpublished observations). Furthermore, cyclic AMP (cAMP)-dependent signaling is known to regulate sclerotial development and oxalic acid biosynthesis in S. sclerotiorum (41), and the effects of high exogenous cAMP levels parallel the effects of elevated ambient pH reported here, i.e., increased oxalic acid production and inhibition of sclerotial development. We are interested in determining whether cAMP-dependent and pH-dependent signaling pathways are components of a common signaling circuit or whether they operate through independent pathways but regulate common components.
The finding that ambient pH plays a major regulatory role in oxalic acid biosynthesis, sclerotial development, and pg1 expression suggests that an ambient pH signal transduction pathway exists in S. sclerotiorum. Such a pathway has been characterized in A. nidulans, and several components of this pathway, including the pH-dependent transcriptional regulator pacC, have been cloned and characterized (50). pacC homologs also have been identified in closely related filamentous fungi (25, 47) and in yeasts (20, 38, 46, 52). The conservation of the zinc finger region among the various fungal homologs and the central role that PacC plays in mediating pH-dependent signaling made the pacC gene the first choice for determining if homologs of a pH-dependent signaling pathway existed in S. sclerotiorum. Pac1 shares the greatest amino acid sequence similarity (51%) with the aspergillus PacC proteins. The most convincing sequence characteristic suggesting that pac1 is a structural homolog of pacC is the existence of the three zinc finger domains that are 85 to 86% identical to the aspergillus PacC zinc finger domains. Furthermore, the spacing, number, and organization of these domains are conserved, and the sequences are 100% homologous for residues proposed to be involved in base specific contacts with the PacC consensus recognition sequences (10). Like pacC, pac1 displays pH-regulated expression with steady-state levels increasing as the pH of the growth medium is increased. Additionally, pacC consensus binding sites found 5′ upstream of the pac1 coding sequence suggest that pac1 also is autogenously regulated. The eight perfect 5′-GCCAAG-3′ hexamer sequence matches 340 to 600 bp upstream of the putative translational start site represent the largest number of consensus sequences upstream of any pacC homolog.
We showed that the pac1 gene product can serve as a functional homolog of the A. nidulans PacC protein by heterologous complementation of an A. nidulans pacC-null strain. Although sequence-specific DNA binding and regulation by proteolytic processing have not been demonstrated for Pac1, the ability to complement a pacC-null mutant demonstrates that pac1 and pacC are functional homologs and suggests that the pH-responsive pathway regulating pacC and pac1 in both A. nidulans and S. sclerotiorum will contain additional structurally and functionally homologous components.
Our results demonstrate that ambient pH is a major regulator of a pathogenicity determinant, oxalic acid; a possible virulence factor, pg1; and a process necessary for long-term survival, sclerotial development. At least one component of a conserved regulatory pathway mediating pH-regulated gene expression, Pac1, exists in this fungus. These findings suggest that S. sclerotiorum uses the ambient pH environment as a regulatory cue for disease and morphological development. Oxalic acid appears to play a central role in pH responsiveness. Its production is a pH-regulated process and, in turn, its accumulation, by virtue of environment acidification, may serve as a regulator of acid pH-regulated processes such as pg1 expression and sclerotial development. Interfering with ambient pH sensing and gene regulation may be viable strategies for blocking disease development in this broad-host-range plant pathogen.
ACKNOWLEDGMENTS
This work was supported by in part by USDA/NRICGP grant 9802312 and by grant BARD 2473-94 from the United States-Israel Binational Agricultural Research and Development Fund.
We thank Young-Sil Ha for providing technical assistance and Nancy Keller, Robert Butchko, and David Piñero for providing A. nidulans strains, techniques, and helpful discussion during the course of this work.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1991. [Google Scholar]
- 3.Bateman D F, Beer S V. Simultaneous production and synergistic action of oxalic acid and polygalacturonase during pathogenesis by Sclerotium rolfsii. Phytopathology. 1965;58:204–211. [PubMed] [Google Scholar]
- 4.Caddick M X, Brownlee A G, Arst H N., Jr Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol Gen Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 5.Cessna S, Sears V, Dickman M, Low P. Oxalic acid, a pathogenicity factor of Sclerotinia sclerotiorum, supresses the host oxidative burst. Plant Cell. 2000;12:2191–2199. doi: 10.1105/tpc.12.11.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotty P J. Aflatoxin and sclerotial production by Aspergillus flavus: Influence of pH. Phytopathology. 1988;78:1250–1253. [Google Scholar]
- 7.Cove D J. Chlorate toxicity in Aspergillus nidulans. Mol Gen Genet. 1976;146:147–159. doi: 10.1007/BF00268083. [DOI] [PubMed] [Google Scholar]
- 8.Espeso E A, Tilburn J, Arst H N, Jr, Peñalva M A. pH regulation is a major determinant in expression of a fungal penicillin biosynthetic gene. EMBO J. 1993;12:3947–3956. doi: 10.1002/j.1460-2075.1993.tb06072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espeso E A, Peñalva M A. Three binding sites for the Aspergillus nidulans PacC zinc-finger transcription factor are necessary and sufficient for regulation by ambient pH of the isopenicillin N gene promoter. J Biol Chem. 1996;271:28825–28830. doi: 10.1074/jbc.271.46.28825. [DOI] [PubMed] [Google Scholar]
- 10.Espeso E A, Tilburn J, Sánchez-Pulido L, Brown C V, Valencia A, Arst H N, Jr, Peñalva M A. Specific DNA recognition by the Aspergillus nidulans three zinc finger transcription factor PacC. J Mol Biol. 1999;274:466–480. doi: 10.1006/jmbi.1997.1428. [DOI] [PubMed] [Google Scholar]
- 11.Feinberg B P, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 12.Fraissninet-Thachet L, Reymond-Cotton P, Févre M. Characterization of a multigene family encoding an endopolygalacturonase in Sclerotinia sclerotiorum. Curr Genet. 1995;29:96–99. doi: 10.1007/BF00313199. [DOI] [PubMed] [Google Scholar]
- 13.Free S J, Rice P W, Metzenberg R L. Arrangement of genes coding for ribosomal ribonucleic acids in Neurospora crassa. J Bacteriol. 1979;137:1219–1226. doi: 10.1128/jb.137.3.1219-1226.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godoy G, Steadman J R, Dickman M B, Dam R. Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol Mol Plant Pathol. 1990;37:179–191. [Google Scholar]
- 15.Hancock J G. Degradation of pectic substances associated with pathogenesis by Sclerotinia sclerotiorum in sunflower and tomato stems. Phytopathology. 1966;56:975–979. [Google Scholar]
- 16.Hicks G R, Raikhel N V. Protein import into the nucleus: an integrated view. Annu Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs G H. Determination of the base recognition positions of zinc fingers from sequence analysis. EMBO J. 1992;11:4507–4517. doi: 10.1002/j.1460-2075.1992.tb05552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller N P, Nesbitt C, Sarr B, Phillips T D, Burow G B. pH regulation of sterigmatocystin and aflatoxin biosynthesis in Aspergillus spp. Phytopathology. 1997;87:643–648. doi: 10.1094/PHYTO.1997.87.6.643. [DOI] [PubMed] [Google Scholar]
- 19.Kubicek C P, Schreferl-Kunar G, Wöhrer W, Röhr M. Evidence for a cytoplasmic pathway of oxalate biosynthesis in Aspergillus niger. Appl Environ Microbiol. 1988;54:633–637. doi: 10.1128/aem.54.3.633-637.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert M, Blanchin-Roland S, Le Louedec F, Lépingle A, Gaillardin C. Genetic analysis of regulatory mutants affecting synthesis of extracellular proteinases in the yeast Yarrowia lipolytica: identification of a RIM101/pacC homolog. Mol Cell Biol. 1997;17:3966–3976. doi: 10.1128/mcb.17.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenz H, Wunderwald P, Eggerer H. Partial purification and some properties of oxalacetase from Aspergillus niger. Eur J Biochem. 1976;63:225–236. doi: 10.1111/j.1432-1033.1976.tb10409.x. [DOI] [PubMed] [Google Scholar]
- 22.Lumsden R D. Sclerotinia sclerotiorum infection of bean and the production of cellulase. Phytopathology. 1969;59:653–657. [Google Scholar]
- 23.Lumsden R D. Phosphotidase of Sclerotinia sclerotiorum produced in culture and in infected bean. Phytopathology. 1970;60:1106–1110. [Google Scholar]
- 24.Lumsden R D. Pectolytic enzymes of Sclerotinia sclerotiorum and their localization in infected bean. Can J Bot. 1976;54:2630–2641. [Google Scholar]
- 25.MacCabe A P, van den Hombergh J P T W, Tilburn J, Arst H N, Jr, Visser J. Identification, cloning and analysis of the Aspergillus niger gene pacC, a wide domain regulatory gene responsive to ambient pH. Mol Gen Genet. 1996;250:367–374. doi: 10.1007/BF02174395. [DOI] [PubMed] [Google Scholar]
- 26.MacCabe A P, Orejas M, Perez-Gonzalez J A, Ramon D. Opposite patterns of expression of two Aspergillus nidulans xylanase genes with respect to ambient pH. J Bacteriol. 1998;180:1331–1333. doi: 10.1128/jb.180.5.1331-1333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magro P, Marciano P, Di Lenna P. Oxalic acid production and its role in pathogenesis of Sclerotinia sclerotiorum. FEMS Microbiol Lett. 1984;24:9–12. [Google Scholar]
- 28.Magro P, Marciano P, Di Lenna P. Enzymatic oxalate decarboxylation in isolates of Sclerotinia sclerotiorum. FEMS Microbiol Lett. 1988;49:49–52. [Google Scholar]
- 29.Marciano P, Di Lenna P, Magro P. Oxalic acid, cell wall-degrading enzymes and pH in pathogenesis and their significance in the virulence of two Sclerotinia sclerotiorum isolates on sunflower. Physiol Plant Pathol. 1983;22:339–345. [Google Scholar]
- 30.Marciano P, Magro P, Favaron F. Sclerotinia sclerotiorum growth and oxalic acid production on selected culture media. FEMS Microbiol Lett. 1989;61:57–60. [Google Scholar]
- 31.Maxwell D P. Oxalate formation in Whetzelinia sclerotiorum by oxaloacetate acetylhydrolase. Physiol Mol Plant Pathol. 1973;3:279–288. [Google Scholar]
- 32.Maxwell D P, Lumsden R D. Oxalic acid production by Sclerotinia sclerotiorum in infected bean and in culture. Phytopathology. 1970;60:1395–1398. [Google Scholar]
- 33.Mertz L M, Rashtchian A. Nucleotide imbalance and polymerase chain reaction: effects on DNA amplification and synthesis of high specific activity radiolabeled DNA probes. Anal Biochem. 1994;221:160–165. doi: 10.1006/abio.1994.1392. [DOI] [PubMed] [Google Scholar]
- 34.Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panaccione D G, McKiernan M, Hanau R M. Colletotrichum graminicola transformed with homologous and heterologous benomyl-resistance genes retains expected pathogenicity to corn. Mol Plant-Microbe Interact. 1988;1:113–120. [Google Scholar]
- 36.Pierson P E, Rhodes L H. Effect of culture medium on the production of oxalic acid by Sclerotinia trifoliorum. Mycologia. 1992;84:467–469. [Google Scholar]
- 37.Piñero D. M.S. thesis. Isolation and characterization of the Aspergillus parasiticus pacC gene. College Station: Texas A&M University; 1999. [Google Scholar]
- 38.Ramon A M, Porta A, Fonzi W A. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J Bacteriol. 1999;181:7524–7530. doi: 10.1128/jb.181.24.7524-7530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reymond P, Deleage G, Rascle C, Févre M. Cloning and sequence analysis of a polygalacturonase-encoding gene from the phytopathogenic fungus Sclerotinia sclerotiorum. Gene. 1994;146:233–237. doi: 10.1016/0378-1119(94)90298-4. [DOI] [PubMed] [Google Scholar]
- 40.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequences: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 41.Rollins J A, Dickman M B. Increase in endogenous and exogenous cyclic AMP level inhibits sclerotial development in Sclerotinia sclerotiorum. Appl Environ Microbiol. 1998;64:2539–2544. doi: 10.1128/aem.64.7.2539-2544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruijter G J G, van de Vondervoort P J I, Visser J. Oxalic acid production by Aspergillus niger: an oxalate-non-producing mutant produces citric acid at pH 5 and in the presence of manganese. Microbiology. 1999;145:2569–2576. doi: 10.1099/00221287-145-9-2569. [DOI] [PubMed] [Google Scholar]
- 43.Shah A J, Tilburn J, Adlard M W, Arst H N., Jr pH regulation of penicillin production in Aspergillus nidulans. FEMS Microbiol Lett. 1991;77:209–212. doi: 10.1016/0378-1097(91)90553-m. [DOI] [PubMed] [Google Scholar]
- 44.Shieh M, Brown R L, Whitehead M P, Carey J W, Cotty P J, Cleveland T E, Dean R A. Molecular genetic evidence for the involvement of a specific polygalacturonase, P2c, in the invasion and spread of Aspergillus flavus in cotton bolls. Appl Environ Microbiol. 1997;63:3548–3552. doi: 10.1128/aem.63.9.3548-3552.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.St. Leger R A, Joshi L, Roberts D. Ambient pH is a major determinant in the expression of cuticle-degrading enzymes and hydrophobin by Metarhiziyum anisopliae. Appl Environ Microbiol. 1998;64:709–713. doi: 10.1128/aem.64.2.709-713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su S S Y, Mitchell A. Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Res. 1993;21:3789–3797. doi: 10.1093/nar/21.16.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suárez T, Peñalva M A. Characterization of a Penicillium chrysogenum gene encoding a PacC transcription factor and its binding sites in the divergent pcbAB-pcbC promoter of the penicillin biosynthetic cluster. Mol Microbiol. 1996;20:529–540. doi: 10.1046/j.1365-2958.1996.5421065.x. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki M. SPXX, a frequent sequence motif in gene regulatory proteins. J Mol Biol. 1989;207:61–84. doi: 10.1016/0022-2836(89)90441-5. [DOI] [PubMed] [Google Scholar]
- 49.ten Have A, Mulder W, Visser J, van Kan J A L. The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol Plant-Microbe Interact. 1998;11:1009–1016. doi: 10.1094/MPMI.1998.11.10.1009. [DOI] [PubMed] [Google Scholar]
- 50.Tilburn J, Sarkar S, Widdick D A, Espeso E A, Orejas M, Mungroo J, Peñalva M A, Arst H N., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acidic- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vega R R, Corsini D, Tourneau D L. Nonvolatile organic acids produced by Sclerotinia sclerotiorum in synthetic liquid media. Mycologia. 1970;62:332–338. [PubMed] [Google Scholar]
- 52.Wilson R B, Davis D, Mitchell A P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yager L N, Lee H-O, Nagle D L, Zimmerman J E. Analysis of fluG mutation that affect light-dependent conidiation in Aspergillus nidulans. Genetics. 1998;149:1777–1786. doi: 10.1093/genetics/149.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yakoby N, Kobiler I, Dinoor A, Prusky D. pH regulation of pectate lyase secretion modulates the attack of Colletotrichum gloeosporioides on avocado fruits. Appl Environ Microbiol. 2000;66:1026–1030. doi: 10.1128/aem.66.3.1026-1030.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yelton M M, Hamer J E, Timberlake W E. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc Natl Acad Sci USA. 1984;81:1470–1474. doi: 10.1073/pnas.81.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]