Abstract
The mammalian pocket protein family, which includes the Retinoblastoma protein (pRb) and Rb-like pocket proteins p107 and p130, regulates entry into and exit from the cell cycle by repressing cell cycle gene expression. Although pRb plays a dominant role in mammalian systems, p107 and p130 are the ancestral pocket proteins. The Rb-like pocket proteins interact with the highly conserved 5-subunit MuvB complex and an E2F-DP transcription factor heterodimer, forming the DREAM (for Dp, Rb-like, E2F, and MuvB) complex. DREAM complex assembly on chromatin culminates in repression of target genes mediated by the MuvB subcomplex. Here, we examined how the Rb-like pocket protein contributes to DREAM formation and function by disrupting the interaction between the sole Caenorhabditis elegans pocket protein LIN-35 and the MuvB subunit LIN-52 using CRISPR/Cas9 targeted mutagenesis. A triple alanine substitution of LIN-52’s LxCxE motif severed LIN-35-MuvB association and caused classical DREAM mutant phenotypes, including synthetic multiple vulvae, high-temperature arrest, and ectopic expression of germline genes in the soma. However, RNA-sequencing revealed limited upregulation of DREAM target genes when LIN-35-MuvB association was severed, as compared with gene upregulation following LIN-35 loss. Based on chromatin immunoprecipitation, disrupting LIN-35-MuvB association did not affect the chromatin localization of E2F-DP, LIN-35, or MuvB components. In a previous study, we showed that in worms lacking LIN-35, E2F-DP, and MuvB chromatin occupancy was reduced genome-wide. With LIN-35 present but unable to associate with MuvB, our study suggests that the E2F-DP-LIN-35 interaction promotes E2F-DP’s chromatin localization, which we hypothesize supports MuvB chromatin occupancy indirectly through DNA. Altogether, this study highlights how the pocket protein’s association with MuvB supports DREAM function but is not required for DREAM’s chromatin occupancy.
Keywords: DREAM, pocket protein, MuvB, CRISPR/Cas9 genome editing, transcriptional repression
Introduction
Members of the mammalian Retinoblastoma (Rb) protein family, pRb, p107, and p130, collectively called pocket proteins, serve key roles in regulating transcription during the cell cycle (Classon and Dyson 2001; Classon and Harlow 2002; Cobrinik 2005; Burkhart and Sage 2008; Dick and Rubin 2013). In mammalian cells, pRb interacts with activating E2F-DP transcription factor heterodimers (in mammals, E2F1/2/3-DP1/2), sequestering E2F-DP and preventing E2F-DP-mediated activation of early cell cycle genes (Helin et al. 1992; Lees et al. 1993; Liban et al. 2016). In contrast, the Rb-like proteins p107 and p130 interact with repressive E2F-DPs (in mammals, E2F4/5-DP1/2) and a highly conserved 5-subunit MuvB subcomplex (in mammals, LIN9, LIN37, LIN52, LIN54, and RBAP48), forming the 8-subunit DREAM (Dp, Rb-like, E2F, and MuvB) transcriptional repressor complex (Korenjak et al. 2004; Lewis et al. 2004; Harrison et al. 2006; Litovchick et al. 2007; Schmit et al. 2007). When associated with the DREAM complex, MuvB mediates transcriptional repression of early and late cell cycle genes (Litovchick et al. 2007; Goetsch et al. 2017; Muller et al. 2017). The transcriptional functions of the DREAM complex and pRb overlap, with each being sufficient to establish and maintain cellular quiescence (G0) if the other is inactive (Hurford et al. 1997; Litovchick et al. 2007; Muller et al. 2017). Upon progression into the cell cycle, pRb and the Rb-like pocket proteins are phosphorylated by CDK4/6-cyclin D, releasing their respective interaction partners and triggering activation of cell cycle genes (Tedesco et al. 2002; Pilkinton et al. 2007; Burke et al. 2010). Thus, the association and dissociation of pocket proteins from their respective transcriptional complexes governs the switch between cell cycle quiescence and cell cycle progression.
The Rb-like homologs p130 and p107 are the ancestral pocket proteins and likely the conserved components that mediate cell cycle control among eukaryotes (Cao et al. 2010; Liban et al. 2017). In Caenorhabditis elegans, LIN-35 is the sole pocket protein, most closely resembling p130/p107 (Lu and Horvitz 1998). The pocket protein-associated complex MuvB was isolated in Drosophila melanogaster (Korenjak et al. 2004; Lewis et al. 2004) and C. elegans (Harrison et al. 2006) before homologs were identified in mammals (Litovchick et al. 2007; Pilkinton et al. 2007; Schmit et al. 2007). The C. elegans complex, variably called DRM and DREAM, regulates cell cycle genes and requires MuvB to mediate gene repression (Boxem and van den Heuvel 2002; Goetsch et al. 2017). Caenorhabditis elegans DREAM also regulates cell-fate specification by antagonizing Ras signaling during vulval development (Myers and Greenwald 2005; Cui et al. 2006; Harrison et al. 2006) and by protecting somatic cells from expressing germline genes (Wang et al. 2005; Petrella et al. 2011).
Extensive biochemical analyses have demonstrated how the DREAM complex assembles on chromatin (Fig. 1a; Litovchick et al. 2007; Pilkinton et al. 2007; Schmit et al. 2007; Guiley et al. 2015; Asthana et al. 2022). E2F-DP and LIN54, a MuvB component, direct site-specific chromatin localization (Zwicker et al. 1995; Schmit et al. 2009; Muller and Engeland 2010; Müller et al. 2012; Marceau et al. 2016). The Rb-like pocket protein serves as a bridge between the 2 DNA-binding DREAM components (Guiley et al. 2015). LIN52 interacts with the pocket protein via an “LxCxE motif” in LIN52. In mammals, the LxCxE motif is instead a suboptimal LxSxExL sequence that is rendered optimal by phosphorylation of a nearby serine residue (S28) (Guiley et al. 2015; Fig. 1b). S28 phosphorylation by DYRK1A kinase induces formation of mammalian DREAM (Litovchick et al. 2011). In C. elegans, the conserved lin-52 gene encodes the optimal LxCxE sequence (Fig. 1b). Caenorhabditis elegans lacks a DYRK1A homolog and its corresponding consensus motif RxSP in LIN-52 (Fig. 1b), suggesting that in C. elegans a phospho-switch does not induce DREAM formation (Litovchick et al. 2011; Guiley et al. 2015). Importantly, the LxCxE binding motif mediates a high-affinity interaction that is employed by the human papillomavirus (HPV) viral oncoprotein E7 to prevent association of LIN52 with the mammalian pocket protein p130 (Guiley et al. 2015).
Fig. 1.
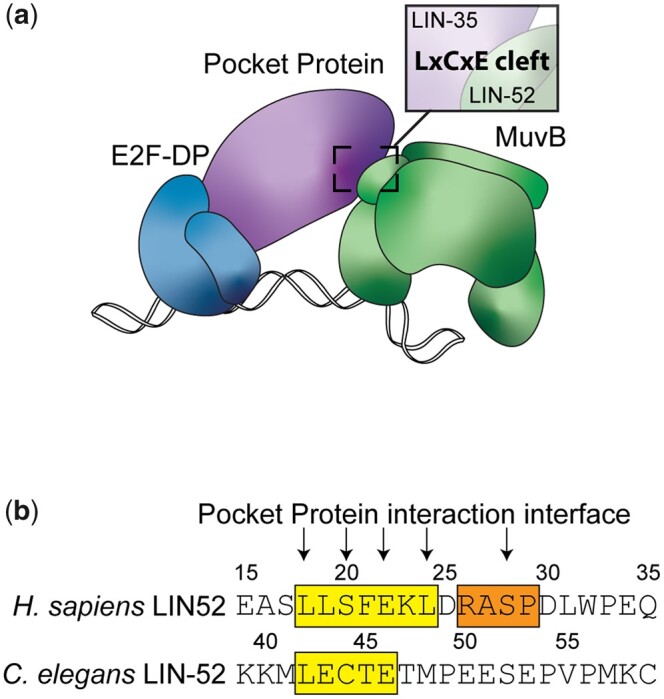
LIN-35 and MuvB associate via the LxCxE motif of LIN-52. a) Model of the C. elegans DREAM complex bound to DNA: E2F-DP, the pocket protein LIN-35, and the 5-subunit MuvB subcomplex. The highlighted region shows the target region for this study: an LxCxE binding motif in the MuvB subunit LIN-52 that interacts directly with the LIN-35 pocket protein. b) Alignment of Homo sapiens LIN52 and C. elegans LIN-52 sequences. The human LxSxExL and worm LxCxE sequences are highlighted in yellow, and the human DYRK1A consensus phosphorylation sequence is highlighted in orange. Arrows indicate residues involved in the interaction with the pocket protein and amino acid residues converted to alanine in this study.
Here, we assessed how the Rb-like pocket protein contributes to DREAM complex formation and function on chromatin. We previously reported that the absence of LIN-35 results in a genome-wide decrease in chromatin occupancy of both E2F-DP and MuvB, illustrating how DRM/DREAM disassembly likely proceeds during cell cycle progression (Goetsch et al. 2017). The model of DREAM complex assembly centers on reintroduction of the pocket protein associations with E2F-DP and MuvB as cells finish the cell cycle (Guiley et al. 2015). To test this model, we used CRISPR/Cas9-mediated genome editing of the C. elegans LIN-52 subunit of MuvB to sever the association of LIN-35 with the MuvB subcomplex. Disrupting that association caused phenotypes associated with impairment of DREAM function, including a highly penetrant synthetic multivulval (SynMuv) phenotype (Fay and Yochem 2007), high-temperature larval arrest (Petrella et al. 2011), and ectopic expression of germline genes in the soma (Wang et al. 2005; Petrella et al. 2011). However, our genome-wide transcript analysis revealed that severing the association of LIN-35 and LIN-52 led to upregulation of a relatively small set of DREAM target genes, which displayed a lower magnitude of upregulation than caused by loss of LIN-35. Moreover, chromatin immunoprecipitation (ChIP) revealed that the chromatin association of E2F-DP-LIN-35 and MuvB was not impaired by loss of LIN-35-MuvB association, even in gene promoters of upregulated DREAM target genes. Altogether, our results indicate that loss of the direct association between LIN-35 and MuvB causes limited upregulation of DREAM target genes, likely due to DREAM chromatin occupancy being relatively normal, but even those minimal effects lead to phenotypes consistent with impaired DREAM function in C. elegans.
Materials and methods
Worm strains
Strains were cultured on Nematode Growth Medium agarose plates with Escherichia. coli OP50 and incubated at 20°C. The following strains were used: Wild type (WT) N2 (Bristol), SS1240 lin-52(bn132(lin-52p::TagRFP-T^SEC^3xFLAG::lin-52 3' UTR)) III/hT2G [bli-4(e937) let-?(q782) qIs48] (I: III), SS1241 lin-52(bn133(lin-52p::TagRFP-T::3xFLAG::lin-52 3' UTR)) III/hT2G [bli-4(e937) let-?(q782) qIs48] (I: III), SS1325 lin-52(bn138(lin-52::GFP^SEC^3xFLAG)) III, SS1256 lin-52(bn139(lin-52::GFP::3xFLAG)) III, SS1273 lin-52(bn150(lin-52[C44A]::GFP::3xFLAG)) III, SS1276 lin-52(bn151(lin-52[L42A, C44A, E46A]::GFP: :3xFLAG)) III, SS1350 lin-52(bn139) III; lin-15A(n767) X, SS1351 lin-52(bn150) III; lin-15A(n767) X, SS1352 lin-52(bn151) III; lin-15A(n767) X, SS1406 lin-8(n2731) II; lin-52(bn139) III, SS1407 lin-8(n2731) II; lin-52(bn150) III, SS1408 lin-8(n2731) II; lin-52(bn151) III, PDG29 lin-52(bn139) III; pgl-1(sam52[pgl-1::mTagRFPT::3xFLAG]) IV, PDG30 lin-52(bn150) III; pgl-1(sam52) IV, and PDG31 lin-52(bn151) III; pgl-1(sam52) IV.
CRISPR/Cas9-mediated genome editing
For all genomic edits, 20 nucleotide crDNA targeting sequences were identified using the MIT CRISPR design tool (http://crispr.mit.edu). Single-guide RNA sequences were cloned into the PU6::unc119_sgRNA vector (Addgene plasmid no. 46169) using the overlapping PCR fragment method described in Friedland et al. (2013) or were cloned into pDD162 (Addgene plasmid no. 47549) using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs), as described in Dickinson et al. (2013). Homologous repair templates were cloned into pDD282 (Addgene plasmid no. 66823) or pDD284 (Addgene plasmid no. 66825) using Glibson Assembly (New England Biolabs; Gibson et al. 2009), as described in Dickinson et al. (2015). CRISPR/Cas9 component plasmids were coinjected with marker plasmids (Frøkjaer-Jensen et al. 2008) to identify strains with an extrachromosomal array instead of a mutated endogenous gene. For targeted mutagenesis, dpy-10(cn64) sgRNA (pJA58, Addgene plasmid no. 59933), and dpy-10(cn64) ssDNA template, dpy-10(cn64) guide and ssDNA template were coinjected to select for positive CRISPR activity in injectant progeny, as described in Arribere et al. (2014). Additional details are provided in Supplementary Materials and Methods.
Microscopy
L1 and L4 larvae were mounted on a 10% agarose pad and immobilized in a 1–2 µl suspension of 0.1 µm polystyrene beads (Polysciences), as described in Kim et al. (2013). Fluorescence images of L1 larvae were acquired using a Leica DM4B upright microscope with QImaging QIClick camera. Fluorescence images of L4 larvae were acquired using a Solamere spinning-disk confocal system with µManager software (Edelstein et al. 2014). The confocal microscope setup was as follows: Yokogawa CSUX-1 spinning disk scanner, Nikon TE2000-E inverted stand, Hamamatsu ImageEM X2 camera, solid-state 405-, 488-, and 561-nm laser lines, 435–485, 500–550, and 573–613 fluorescent filters, and Nikon Plan Fluor 40x air objective. Images were processed using Image J (Schneider et al. 2012).
Caenorhabditis elegans phenotype scoring
For brood size analyses, L4 individuals were cloned to fresh plates every 24 h and all progeny were counted. For SynMuv phenotype scoring, 3 replicate plates per strain were set up with 5–10 adults that were allowed to lay eggs for 6 h. Progeny were incubated at 20°C for 3 days, then scored for the presence or absence of pseudovulvae. The percentages of multivulva worms in each replicate population were averaged, and the standard deviation was calculated. For high-temperature arrest (HTA) phenotype scoring, L4 larvae of each strain were incubated at 24°C or 26°C overnight, then 3 plates per strain were set up with 9–12 adults that were allowed to lay eggs for 4–5 h. Progeny incubated at 24°C or 26°C were counted 2 days later and scored as arrested larvae (HTA) or adults (not HTA). Results from each plate were combined for each strain.
Immunoblotting and coimmunoprecipitation
For immunoblotting whole worm lysates, 200 adults from each strain were picked into SDS gel-loading buffer (50 mM pH 6.8 Tris-Cl, 2% sodium dodecyl sulfate, 0.1% bromophenol blue, 100 mM β-mercaptoethanol). For coimmunoprecipitation (co-IP), embryos collected after bleaching gravid worms were aged for 3.5 h and then frozen in liquid nitrogen, and lysates were prepared as described in Goetsch et al. (2017). For each IP, 8 mg of protein lysate was mixed with antibody-conjugated Dynabeads (ThermoFisher) and incubated for 2 h at 4°C. Proteins were separated by SDS/PAGE, and western blot analysis was performed using a 1:1,000–1:5,000 dilution of primary antibody and 1:2,000 dilution of an appropriate HRP-conjugated secondary antibody. Serial western blot analysis was performed by stripping the blot with buffer containing 0.2M pH 2.2 glycine, 0.1% SDS, and 1% Tween-20 between antibody probings. Additional details are provided in Supplementary Materials and Methods.
ChIP and sequential ChIP
Embryos collected after bleaching gravid worms were aged for 3.5 h and then frozen in liquid nitrogen. Lysates were prepared by grinding, crosslinking for 10 min in 1% formaldehyde, and sonicating to an average size of 250 base pairs in FA buffer (50 mM HEPES/KOH pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 150 mM NaCl) using a Bioruptor (Diagenode) on the high setting with 60 rounds of 30 seconds on and 1 min rest. Protein concentrations of lysates were determined using a Qubit fluorometer. ChIP and sequential ChIP experiments were performed as described in Goetsch et al. (2017) and in Supplementary Materials and Methods. Quantitative PCR was performed using SYBR green reagents on an Applied Biosystems QuantStudio 3 and ViiA 7 Real-Time PCR Systems (ThermoFisher).
Analysis of transcript levels by RT-qPCR and RNA-sequencing
Embryos collected after bleaching gravid worms were aged for 3.5 h and then frozen in Trizol for RNA isolation. A total of 1 μg RNA was treated with DNase and reverse transcribed using the High Capacity cDNA Kit (Applied Biosystems). qPCR was performed using SYBR green reagents on an Applied Biosystems QuantStudio 3 Real-Time PCR System (ThermoFisher). The relative quantity of experimental transcripts was calculated with act-2 as the control gene using the ΔCt method.
RNA-sequencing (RNA-seq) libraries were prepared using the NEBNext Poly(A) mRNA Magnetic Isolation Module and NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs). Samples were multiplexed using the NEBNext Multiplex Oligos for Illumina (NEB). Libraries were sequenced on an Illumina HiSeq X system by Novogene Corporation Inc. (Davis, CA, USA) to acquire 150-bp paired-end reads. Sequence reads were trimmed using FASTP (Chen et al. 2018) and mapped to transcriptome version WBcel235 using STAR (Dobin et al. 2013). Read counts per transcript were obtained using HTSeq (Anders et al. 2015) and differentially expressed genes were assessed using DEseq2 (Love et al. 2014).
Quantification and statistical analysis
For brood size analysis, significance was determined using a Wilcoxon–Mann–Whitney test comparing CRISPR/Cas9-genome-edited strains to WT (N2). For genome-wide differential expression analysis of lin-52(3A) vs lin-52(WT) late embryos, we used a 1.5-fold change and adjusted P-value < 0.05 cutoff as calculated by the Benjamini–Hochberg method performed by DEseq2. For expression analysis of lin-35(n745) vs N2 L1s, statistical analysis was performed using R using the Quantile normalization and robust multichip average algorithm in the affy package of Bioconductor using the 1.5-fold change and adjusted P-value < 0.05 cutoff (Bolstad et al. 2003; Irizarry et al. 2003). For transcript level analysis by RT-qPCR, significance was determined using a Student’s t-test between lin-35 vs WT (N2) or lin-52(1A) or lin-52(3A) vs lin-52(WT). For ChIP-qPCR experiments assessing individual subunit’s occupancy changes between strains, significance was determined using a Student’s t-test between lin-35 vs WT (N2) or lin-52(3A) vs lin-52(WT).
Results
Targeted mutagenesis to disrupt C. elegans DREAM complex formation
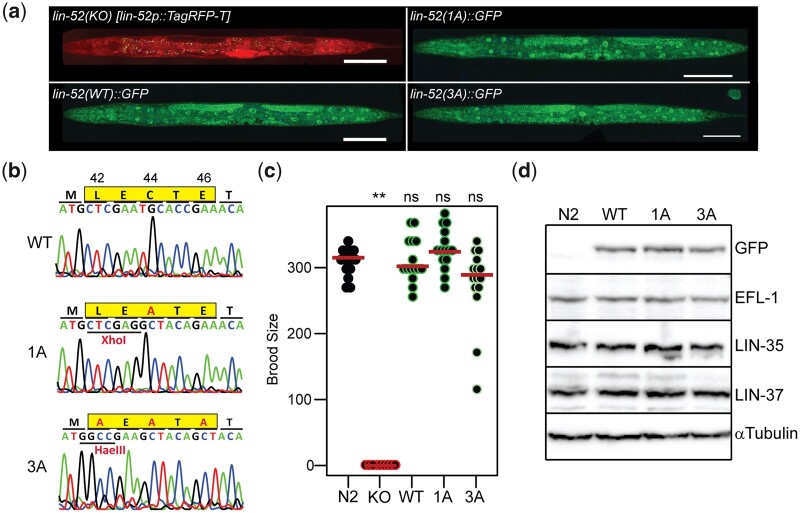
Structural studies previously demonstrated that MuvB interacts with the pocket protein via the LIN52 subunit (Fig. 1a;Guiley et al. 2015). Using the self-excising cassette method for C. elegans CRISPR/Cas9 genome editing (Dickinson et al. 2015), we generated a lin-52(KO) strain (lin52(bn133[lin-52p::TagRFP-T::3xFLAG]) by completely replacing lin-52 coding sequence with TagRFP-T coding sequence (Fig. 2a). We observed that lin-52(KO) rendered worms sterile (Fig. 2c), as previously observed in the lin-52(n3718) protein null strain (Ceol et al. 2006; Harrison et al. 2006). This resembles loss of other MuvB components, as loss of LIN-9, LIN-53 (C. elegans RBAP48), or LIN-54 in protein null strains also renders worms sterile and affects the levels of other MuvB subunits, suggesting that MuvB components require coexpression for assembly/stability of the complex (Harrison et al. 2006). Loss of LIN-37 does not cause sterility and does not affect assembly of the rest of MuvB in either C. elegans or mammalian cells (Harrison et al. 2006; Mages et al. 2017). We next replaced the TagRFP-T coding sequence with lin-52 tagged with a C-terminal GFP-3xFLAG coding sequence, generating the lin-52(WT) strain (lin-52(bn139[lin-52::GFP::3xFLAG]); Fig. 2a). lin-52(WT) completely rescued fertility (Fig. 2c), indicating that the GFP tag does not impair LIN-52 function.
Fig. 2.
Targeted mutagenesis to disrupt DREAM complex formation. a) Live worm fluorescence images of lin-52(KO), lin-52(WT), lin-52(1A), and lin-52(3A) L4 larvae. Composites were artificially straightened. Scale bars, 100 µM. b) Sanger sequencing of the lin-52 LxCxE coding region (highlighted in yellow) in lin-52(WT), lin-52(1A), and lin-52(3A). c) Swarm plots of the brood sizes of WT (N2) worms and lin-52(KO), lin-52(WT), lin-52(1A), and lin-52(3A) transgenic worms. Significance (**P-value < 0.01) was determined by a Wilcoxon-Mann-Whitney test comparing the indicated strains to WT (N2). d) Western blot analysis of DREAM subunits LIN-52 (via GFP tag), EFL-1, LIN-35, and LIN-37 using lysates from WT (N2) worms and lin-52(WT), lin-52(1A), and lin-52(3A) transgenic worms separated by SDS PAGE. Antibodies used are indicated on the right. Alpha-tubulin was used as a loading control. Full blots are shown in Supplementary Fig. 1.
Since LIN-52 is essential for C. elegans fertility, we sought to disrupt the LIN-35-LIN-52 interaction without affecting protein integrity. We directed targeted mutagenesis of the LIN-52 LxCxE sequence (Fig. 1b) using CRISPR/Cas9-mediated genomic editing. We generated 2 mutants of the LxCxE binding motif in lin-52(WT) using the dpy-10 co-CRISPR method of small oligo homology-directed repair (Arribere et al. 2014). We generated the lin-52(1A) single-alanine mutation strain (lin-52(bn150[lin-52[C44A]::GFP::3xFLAG)) and the lin-52(3A) triple-alanine mutation strain (lin-52(bn151(lin-52[L42A, C44A, E46A]::GFP::3xFLAG); Fig. 2b) with the intent to completely disrupt LIN-52’s interaction with the C. elegans pocket protein LIN-35. Additional silent mutations were included in the oligo repair templates to generate new restriction enzyme cut sites to aid in genotyping (Fig. 2b).
Full loss of C. elegans DREAM activity causes sterility, as observed in protein null mutants of worm E2F-DP (dpl-1 and efl-1) and worm MuvB (lin-9, lin-52, lin-53, and lin-54) (Beitel et al. 2000; Chi and Reinke 2006; Tabuchi et al. 2011). Since the C-terminally GFP-tagged lin-52 coding sequence completely rescued lin-52(KO) sterility, we were able to test whether lin-52(1A) and lin-52(3A) impair DREAM function. We observed that neither the 1A nor 3A mutation in the LIN-52 LxCxE sequence caused a significant reduction in brood size (Fig. 2c). Using western blot analysis of selected DREAM components from lin-52(WT) and mutant lysates, we observed that DREAM component protein levels were unaffected compared with WT (N2) (Fig. 2d and Supplementary Fig. 1). Similarly, using live-image analysis of lin-52(WT), lin-52(1A), and lin-52(3A) L4 larvae, we observed that LIN-52 level and localization appeared normal in mutants (Fig. 2a). Together, these results demonstrate that mutation of the LIN-52 LxCxE sequence does not cause a lin-52 null phenotype and does not alter the levels and tissue distribution of MuvB components.
The 3A substitution in the LIN-52 LxCxE sequence blocks DREAM assembly
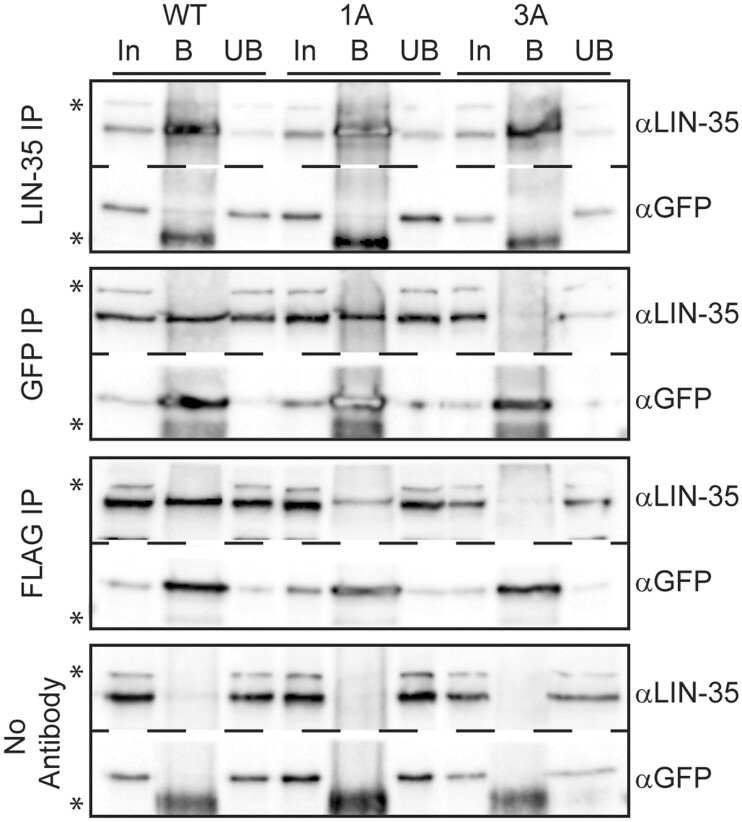
To test whether our CRISPR/Cas9-generated 1A and 3A substitutions disrupt the LIN-35-MuvB association, we performed co-IPs from protein extracts prepared from lin-52(WT), lin-52(1A), and lin-52(3A) late embryos. We pulled down LIN-35 and tested for LIN-52 association using the GFP epitope, and we pulled down LIN-52 using either the GFP or FLAG epitope and tested for LIN-35 association (Fig. 3 and Supplementary Fig. 2). In both co-IP experiments, we observed that LIN-52 association with LIN-35 was lost in lin-52(3A) extracts but not in lin-52(1A) extracts. In 1A extracts, we observed a weaker association between LIN-52 and LIN-35 when pulling down with an anti-FLAG antibody, but the effect was not replicated with the anti-GFP antibody. The inconsistent results suggest that the 1A substitution weakens but does not eliminate the LIN-52-LIN-35 interaction. Altogether, these results demonstrate that a 3A substitution in the LxCxE sequence is required to sever the protein–protein association between LIN-52 and LIN-35 in C. elegans.
Fig. 3.
lin-52 LxCxE binding motif mutants block DREAM formation. Late embryo extracts from lin-52(WT), lin-52(1A), and lin-52(3A) (each tagged with GFP and FLAG) were immunoprecipitated with anti-LIN-35, anti-GFP, and anti-FLAG antibodies, with no antibody serving as a negative control. Proteins bound (B) and unbound (UB) were separated by SDS PAGE, and western blot analysis was performed using the antibodies indicated on the right. 5% of Input (In) is shown on the left. Asterisks indicate nonspecific bands. Full blots are shown in Supplementary Fig. 2.
Blocking DREAM complex formation causes classic DREAM loss-of-function phenotypes
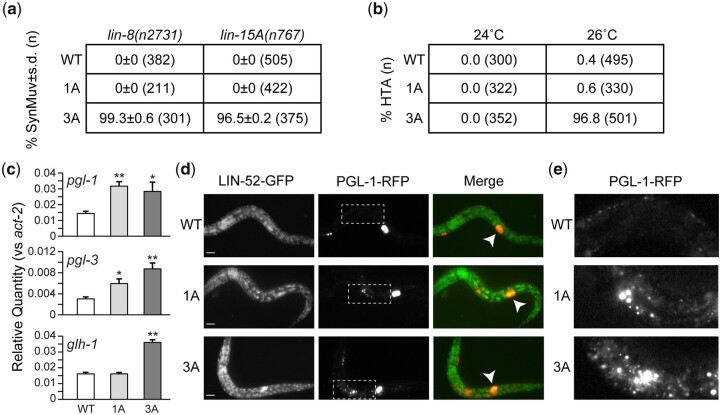
DREAM loss-of-function mutations when paired with SynMuv A gene mutations cause a synthetic multivulval (SynMuv) phenotype (Fay and Yochem 2007), high-temperature larval arrest (Petrella et al. 2011), and ectopic expression of germline genes in the soma (Wang et al. 2005; Petrella et al. 2011). We reasoned that if DREAM function was affected by severing the LIN-52-LIN-35 association, then we should observe each of the 3 phenotypes above. When paired with SynMuv A allele lin-8(n2731) (Harrison et al. 2007) or lin-15A(n767) (Huang et al. 1994), lin-52(3A) resulted in a SynMuv phenotype; lin-52(WT) and lin-52(1A) did not have this effect (Fig. 4a). When worms were grown at high temperature (26°C), lin-35(3A) displayed larval arrest; lin-52(WT) and lin-52(1A) did not cause larval arrest (Fig. 4b). When RT-qPCR was used to examine ectopic expression of the germline genes pgl-1, pgl-3, and glh-1, lin-52(3A) caused upregulation of all 3 genes; lin-52(1A) caused upregulaton of pgl-1 and pgl-3 but not glh-1 (Fig. 4c). To examine ectopic expression by another method, we crossed a germline-specific pgl-1::rfp reporter (Marnik et al. 2019) into our lin-52 mutant strains. We observed ectopic expression of pgl-1::rfp in lin-52(1A) and lin-52(3A) in the intestinal cells of larvae (Fig. 4, d and e), as expected (Petrella et al. 2011). These phenotypes along with our co-IP findings demonstrate that the 3A substitution has a greater impact on DREAM assembly and function than the 1A substitution.
Fig. 4.
Mutagenesis of the lin-52 LxCxE binding motif causes SynMuv B phenotypes. a) Table indicating the percentage Synthetic Multivulval (SynMuv) worms when lin-52(WT), lin-52(1A), and lin-52(3A) were combined with SynMuv A mutant alleles lin-8(n2731) or lin-15A(n767) with standard deviation indicated. The population size (n) is indicated in parentheses. b) Table indicating the percentage of HTA observed in lin-52(WT), lin-52(1A), and lin-52(3A) incubated at 24°C and 26°C. The population size (n) is indicated in parentheses. c) RT-qPCR analysis comparing transcript levels of 3 germline genes (pgl-1, pgl-3, and glh-1) in lin-52(WT) (white), lin-52(1A) (light grey), and lin-52(3A) (dark gray) late embryos. Expression values from 6 biological replicates were averaged and are presented as the relative quantity compared with act-2. Error bars indicate SEM, and significance was determined by a Student’s t-test between transcript levels in mutant (3A or 1A) vs WT (*P-value < 0.05, **P-value < 0.01). d, e) Live worm fluorescence images of lin-52(WT), lin-52(1A), and lin-52(3A) L1 larvae containing a PGL-1::RFP reporter gene. White arrowheads indicate the primordial germ cells Z2 and Z3. Scale bars, 10 µm. The white boxes in (d) indicate regions of ectopic expression of PGL-1::RFP in the intestine of lin-52(1A) and lin-52(3A) shown in (e).
Severing the LIN-35-MuvB connection has a minimal effect on repression of DREAM target genes
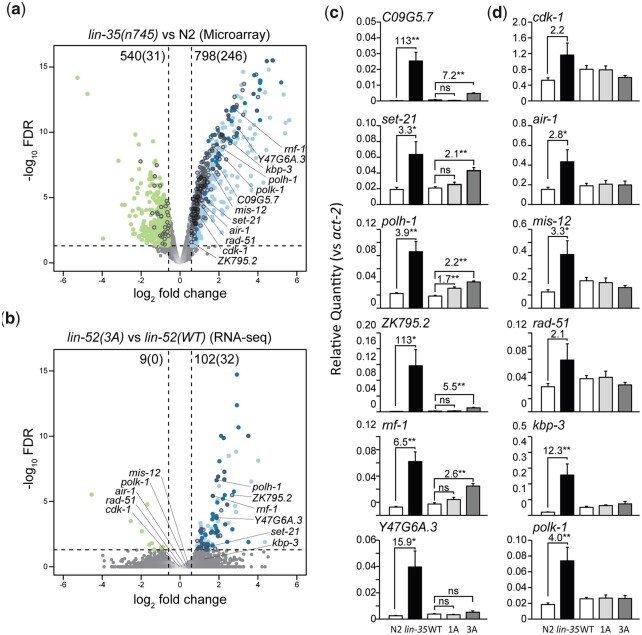
To investigate whether blocking DREAM assembly affects transcriptional repression of DREAM target genes, we performed whole-genome RNA-seq in lin-52(WT) and lin-52(3A) late embryos and compared our data to microarray-based expression data from WT and lin-35(n745) null mutant L1 larvae (Kirienko and Fay 2007). Our RNA-seq analysis detected 16,855 transcripts (>10 counts across samples), as compared with 16,810 transcripts tested in the microarray experiment (Supplementary Table 3). A total of 13,721 transcripts are shared between the 2 experiments, including 1,370 of the 1,515 genes previously reported to have DREAM bound to their promoters, which we categorize as DREAM targets (Goetsch et al. 2017). In lin-35(n745) vs WT (N2) L1 larvae, 798 genes (246 DREAM targets) were upregulated and 540 genes (31 DREAM targets) were downregulated (Fig. 5a). In contrast, in lin-52(3A) vs lin-52(WT) late embryos, 102 genes (32 DREAM targets) were upregulated and 9 genes (0 DREAM targets) were downregulated (Fig. 5b). Altogether, our transcript analyses reveal that the 3A substitution has a minimal effect on DREAM target regulation, with only ∼2% (32/1,370) of DREAM targets upregulated in lin-52(3A) late embryos compared with ∼18% (246/1,370) of DREAM targets upregulated in lin-35(n745) L1s.
Fig. 5.
Disruption of DREAM formation leads to upregulation of a small subset of target genes. a, b) Volcano plots of log2-fold change in transcript levels vs log10 false discovery rate (FDR) of 13,721 genes shared between (a) lin-35(n745) vs N2 L1 microarray expression analysis reported in (Kirienko and Fay 2007) and (b) lin-52(3A) vs lin-52(WT) late embryo RNA-seq. Genes downregulated or upregulated are highlighted, with dark circles indicating upregulated genes observed in both lin-35 and lin-52(3A) data sets. Black circle outlines indicate DREAM target genes, as reported in (Goetsch et al. 2017). The number of genes differentially expressed in each analysis is indicated at the top of each plot, with the number of DREAM target genes differentially expressed indicated in parentheses. Dashed lines indicate the significance cutoff of q = 0.05 (horizontal lines) and a 1.5-fold change in transcript level (vertical lines). Genes selected for RT-qPCR analysis are labeled. c, d) RT-qPCR analysis comparing transcript levels of 6 DREAM target genes that were upregulated in RNA-seq (c) and 6 DREAM target genes that were not upregulated in RNA-seq (d). Transcript levels in late embryos are from WT N2 (white), lin-35 (black), lin-52(WT) (white), lin-52(1A) (light gray), and lin-52(3A) (dark grey). Expression values from 6 biological replicates were averaged and are presented as the relative quantity compared with act-2. Error bars indicate SEM, fold change values > 1.5-fold are provided, and significance was determined by a Student’s t-test between transcript levels in mutant (3A or 1A) vs WT or lin-35 vs N2 (*P-value < 0.05, **P-value < 0.01).
To directly compare at 1 developmental stage the effect on transcription of loss of LIN-35 vs severing the LIN-35-MuvB association, we assessed transcript levels of a subset of DREAM target genes in N2, lin-35(n745), lin-52(WT), lin-52(1A), and lin-52(3A) late embryos using RT-qPCR analysis (Fig. 5, c and d). We tested 6 DREAM target genes that were upregulated (Fig. 5c) and 6 DREAM target genes that were not upregulated in lin-52(3A) late embryos (Fig. 5d), as identified from our RNA-seq analysis. For the 6 DREAM target genes that were upregulated in our RNA-seq analysis (C09G5.7, set-21, polh-1, ZK795.2, rnf-1, and Y47G6A.3), we consistently observed 1) modest but significant upregulation in lin-52(3A) vs lin-52(WT) and 2) a greater increase in transcript levels in lin-35 vs N2 than in lin-52(3A) vs lin-52(WT) (Fig. 5c). Consistent with our phenotype analysis, we observed that lin-52(1A) had a minimal effect on DREAM target gene levels (Fig. 5c). For the 6 DREAM target genes that were not upregulated in our RNA-seq analysis (cdk-1, air-1, mis-12, rad-51, kbp-3, and polk-1), we did not observe an increase in transcript levels in lin-52(3A) or lin-52(1A) compared with lin-52(WT) (Fig. 5d). Most (4 of 6) of these genes were significantly upregulated in lin-35 vs N2 late embryos. Altogether, our RNA-seq and RT-qPCR analyses demonstrate that blocking DREAM assembly has a limited impact on DREAM target gene repression in late embryos, in terms of both number of genes upregulated and the amplitude of upregulation, as compared with complete loss of LIN-35.
E2F-DP-LIN-35 and MuvB subcomplexes independently co-occupy chromatin sites
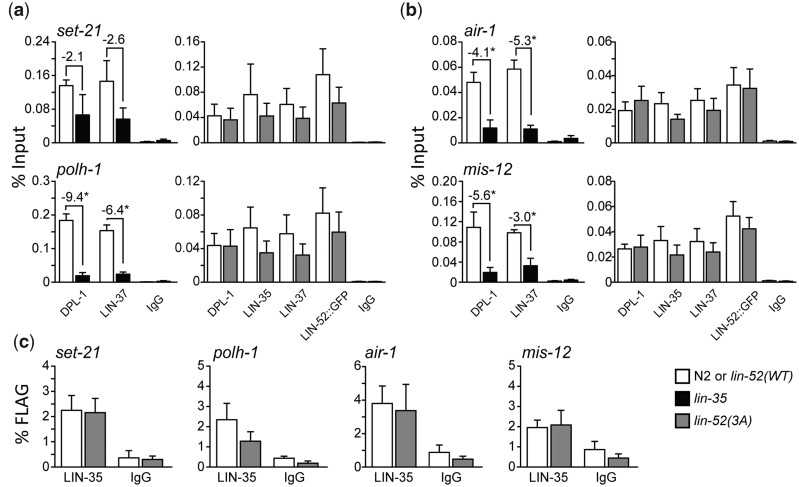
In the absence of LIN-35, E2F-DP, and MuvB do not associate with one another and their chromatin occupancy is reduced genome-wide (Goetsch et al. 2017). In our lin-52(3A) worm strain, LIN-35 is present, but its association with MuvB is severed. We tested the impact of this severing on the chromatin localization of DREAM components using ChIP. We chose 2 DREAM target genes that in our RNA-seq analysis were upregulated in lin-52(3A) (set-21 and polh-1; Fig. 6a), and 2 DREAM target genes that were not upregulated in lin-52(3A) (air-1 and mis-12; Fig. 6b). We performed ChIP-qPCR of DPL-1 and LIN-37 in lin-35 null compared with N2 embryos, and DPL-1, LIN-35, LIN-37, and LIN-52 via its GFP tag in lin-52(3A) compared with lin-52(WT) embryos. IgG was used as a negative control for both ChIP analyses. We observed that all tested DREAM components remained similarly enriched at the 4 selected promoters in lin-52(3A) as compared with lin-52(WT) (Fig. 6, a and b). In contrast, chromatin occupancy of both DPL-1 and LIN-37 were significantly reduced in 3 of the 4 selected promoters in lin-35(n745) as compared with N2. An additional 8 DREAM target gene promoters were tested and showed similar DREAM chromatin occupancy profiles (Supplementary Fig. 3, a and b). We did observe that LIN-37 and LIN-52 chromatin occupancy was reduced at some DREAM target gene promoters, but the observed decrease did not correspond to whether the target gene is misregulated (Supplementary Fig. 3, a and b). Notably, the key difference between lin-35(n745) and lin-52(3A) is the status of the pocket protein. Since DPL-1 and LIN-35 occupancy is not compromised by severing the LIN-52-LIN-35 association in lin-52(3A), whereas DPL-1 occupancy is compromised by loss of LIN-35, our results suggest that the chromatin association of the repressive E2F-DP transcription factor heterodimer is stabilized by its interaction with LIN-35.
Fig. 6.
Analysis of chromatin localization of DREAM subunits at target genes. a, b) ChIP-qPCR of DREAM subunits DPL-1 and LIN-37 in N2 (white) vs lin-35 (black) and DPL-1, LIN-35, LIN-37, and LIN-52 (via GFP tag) in lin-52(WT) (white) vs lin-52(3A) (dark gray) late embryo extracts at 2 DREAM target genes that were upregulated in RNA-seq (a) and 2 DREAM target genes that were not upregulated in RNA-seq (b). IgG was used as a negative control. Signals are presented as percentage of Input DNA, with negative fold-change values >2-fold noted. Error bars indicate SEM. Significance was determined by a Student’s t-test between subunit ChIP values in mutant (lin-35 or lin-52(3A)) vs WT control (N2 or lin-52(WT)) (*P-value < 0.05). c) Sequential ChIP-qPCR of LIN-52 (via FLAG tag) followed by LIN-35 or IgG from lin-52(WT) (white) and lin-52(3A) (dark gray) late embryo extracts at 4 DREAM target genes. Signals are presented as percentage of FLAG IP DNA. Error bars indicate SEM. Additional data are shown in Supplementary Fig. 3.
We tested whether MuvB and E2F-DP-LIN-35 co-occupy DREAM target regions by performing sequential ChIP analysis. We first ChIPed LIN-52 via its FLAG tag and then ChIPed LIN-35. We observed no significant difference in LIN-52 and LIN-35 co-occupancy in lin-52(3A) extracts vs lin-52(WT) extracts at 8 DREAM target gene promoters, as determined by a Student’s t-test (Fig. 6c and Supplementary Fig. 3c). Altogether, our results indicate that, although the interaction of LIN-35 and MuvB is disrupted, DREAM components can nevertheless colocalize at target promoters through their respective protein–DNA interactions.
Discussion
We previously postulated that DREAM assembly, initiated by DYRK1A phosphorylation of LIN52 in mammalian cells (Litovchick et al. 2011; Guiley et al. 2015), stabilizes MuvB-mediated repression of DREAM target genes (Goetsch et al. 2017). Using CRISPR/Cas9-mediated targeted mutagenesis, we generated a mutant C. elegans strain in which LIN-52 was rendered incapable of interacting with LIN-35, the sole C. elegans Rb-like pocket protein. We expected that, similar to loss of LIN-35 (Goetsch et al. 2017), MuvB chromatin occupancy would be destabilized and its repressive function would be impaired. Instead, we observed that chromatin occupancy was not destabilized at the genes tested and that relatively few genes were upregulated. We observed that ∼2% of all DREAM target genes were derepressed, indicating that for those genes MuvB chromatin occupancy alone is not sufficient for target gene repression. However, interrupting LIN-35-MuvB association did cause classic DREAM mutant phenotypes, including SynMuv, HTA, and ectopic expression of germline genes in the soma, consistent with impaired DREAM function. Altogether, we conclude that the LIN-35-MuvB association is not required to assemble DREAM at target sites but is necessary for full activity of the complex.
Our findings highlight that even minimal perturbation of DREAM-mediated transcriptional repression of target genes causes observable phenotypic consequences in C. elegans. Complete knockout of DREAM activity causes sterility, as demonstrated by our newly generated lin-52 null mutant and null mutants of other DREAM components (Harrison et al. 2006). In contrast, although our RNA-seq analysis revealed that ∼2% of DREAM target genes were upregulated in the lin-52(3A) mutant, that mutant caused highly penetrant SynMuv phenotypes with no corresponding effect on fertility. Thus, phenotype analysis provides a sensitive readout of DREAM function.
How the limited upregulation of some DREAM target genes in lin-52(3A) mutant embryos causes the observed phenotypes remains unknown. The SynMuv phenotype is caused by ectopic activation of LIN-3/EGF in the hyp7 hypodermal syncytium, which requires synthetic impairment of redundant regulatory pathways established by SynMuv A and SynMuv B class genes (Cui et al. 2006). Although there is some evidence that SynMuv A class proteins directly target lin-3’s promoter (Saffer et al. 2011), DREAM does not appear to regulate lin-3 directly (Goetsch et al. 2017). Similarly, we suspect that DREAM antagonism of HTA and ectopic expression of germline genes in the soma is also indirect (Petrella et al. 2011; Goetsch et al. 2017; Rechtsteiner et al. 2019). Instead, we speculate that SynMuv B class proteins including DREAM regulate chromatin compaction in somatic cells during development (Costello and Petrella 2019). Thus, even minimal perturbation of DREAM complex activity may delay embryonic chromatin compaction, providing a window for alternative regulatory pathways to trigger the developmental defects detected at later larval stages.
Our analysis provides important insight into how assembly of the mammalian DREAM complex maintains repression of cell cycle genes and suggests that the pocket protein’s primary function is to protect MuvB’s function as a transcriptional repressor. DREAM assembly is triggered by DYRK1A phosphorylation of LIN52, initiating MuvB association with p107/p130 (Litovchick et al. 2011; Guiley et al. 2015). Mammalian MuvB’s function switches from transcriptional repression in the DREAM complex during quiescence to transcriptional activation after associating with the B-Myb transcription factor and forming the Myb-MuvB (MMB) complex late in the cell cycle (Lewis et al. 2004; Osterloh et al. 2007; Schmit et al. 2007; Knight et al. 2009; Sadasivam et al. 2012). DYRK1A-mediated LIN52 phosphorylation also inhibits MuvB association with B-Myb (Litovchick et al. 2011), even though the 2 interaction interfaces do not physically overlap (Guiley et al. 2018). In Drosophila, Lin-52 protein is required for MMB to oppose repressive DREAM functions (Lewis et al. 2012), suggesting that Lin-52’s interaction with the pocket protein or B-Myb dictates MuvB’s transcriptional function (Guiley et al. 2018). Our data demonstrate that in worms MuvB localizes to chromatin sites and represses gene targets without direct association with the pocket protein. These findings suggest that the pocket protein’s association with LIN-52 serves a different purpose than just to direct assembly of DREAM. We propose that the pocket protein’s association with LIN-52 instead primarily functions to oppose B-Myb association with MuvB, preventing MuvB from converting to its transcriptionally active role. Since no B-Myb homolog exists in C. elegans (Vorster et al. 2020), loss of direct LIN-35-MuvB association had little effect on target gene repression. We predict that a similar 3A mutation in LIN-52 in Drosophila or mammalian cells would cause significantly more upregulation of DREAM target genes.
The switch between MuvB-associated cell cycle gene repression and activation is hijacked in cancer cells. All 3 mammalian pocket proteins are inactivated by the E7 viral oncoprotein present in high-risk HPV (Zhang et al. 2006; Huh et al. 2007). E7 interacts with the mammalian pocket proteins through its high-affinity LxCxE binding motif, disrupting MuvB association with the pocket protein in DREAM (Guiley et al. 2015). HPV E7 concurrently coaxes MuvB into its transcriptional activator function by stimulating MMB assembly (Pang et al. 2014). However, cancer cells resist cytotoxic chemotherapy by temporarily exiting the cell cycle (Boichuk et al. 2013), suggesting that MuvB’s capacity for transcriptional repression is retained. Based on our findings that MuvB does not require direct association with the pocket protein to repress target genes, MuvB’s function in cancer cells requires closer scrutiny.
We previously observed that E2F-DP and MuvB chromatin association is severely affected by loss of LIN-35 (Goetsch et al. 2017). By severing LIN-35-MuvB association, this study suggests a new model for DREAM complex formation where LIN-35 directly stabilizes E2F-DP chromatin occupancy. We also observed that MuvB chromatin occupancy is not disrupted even though MuvB no longer associates directly with E2F-DP-LIN-35. Importantly, in vitro analysis of the DNA-binding characteristics of heterodimeric mammalian E2F-DP identified a distinct induction of DNA bending, especially in the case of the homologs of C. elegans EFL-1-DPL-1 (E2F4-DP1/2; Tao et al. 1997). We propose that DNA-associated E2F-DP heterodimers promote MuvB co-occupancy through a DNA bending-dependent mechanism. Together, our results suggest a model in which the LIN-35 pocket protein promotes E2F-DP chromatin occupancy, which in turn promotes MuvB chromatin occupancy.
Our results support an exciting model for how local E2F-DP-mediated alterations to DNA shape enhanced by their interaction with a pocket protein promote MuvB co-occupancy. Even with evolutionary divergence from the ancestral pocket protein, this model may also apply to pRb function. Many histone deacetylases and chromatin remodeling complexes associate with pRb through the LxCxE binding cleft, although many of these associations have only limited support thus far from structural/biochemical interaction studies (Dyson 2016). Variation in pRb monophosphorylation events that can alter pRb structure and recognition of binding partners offers one explanation for how pRb can potentially interact with >300 individual protein partners (Rubin 2013; Narasimha et al. 2014). Our data provide an alternative, but not exclusive, possibility, namely that direct and stable pRb association with these myriad protein partners may be unnecessary. Perhaps pRb association with a few partners such as E2F-DPs promotes localization of multiprotein complexes to genomic sites. Additional dissection of DREAM and pRb structure and function will shed light on how the pocket proteins mediate their essential cellular roles.
Data availability
Requests for information, strains, and reagents should be directed to and will be fulfilled by PDG (pdgoetsc@mtu.edu). Supplemental Material is available at figshare: https://doi.org/10.25386/genetics.19491575. Supplementary information includes Supplementary Materials and Methods. Supplementary Fig 1 contains full western blots presented in Fig. 2d. Supplementary Fig. 2 contains full western blots presented in Fig. 3b. Supplementary Fig. 3 contains additional DREAM target genes tested using qPCR for ChIP and sequential ChIP analyses to complement Fig. 6. Antibodies used for western blot and ChIP analyses are available in Supplementary Table 1. Primers used for cloning and qPCR are available in Supplementary Table 2. DESeq2 differential gene expression analysis data are available in Supplementary Table 3. RNA-seq expression data used in this study are available from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) through GEO Series accession number GSE199287. Microarray expression analysis data used in this study are available through GEO Series accession number GSE6547 (Kirienko and Fay 2007).
Acknowledgments
We thank Seth Rubin and members of the Rubin and Strome labs for helpful discussions. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Funding
This work was supported by National Institutes of Health R01 grant GM034059 to S.S., American Cancer Society Postdoctoral Fellowship PF-16-106-01-DDC to P.D.G., and National Institutes of Health R15 grant GM137145 to P.D.G.
Conflicts of interest
None declared
Contributor Information
Paul D Goetsch, Department of Biological Sciences, Michigan Technological University, Houghton, MI 49931, USA; Department of Molecular, Cell and Developmental Biology, University of California, Santa Cruz, Santa Cruz, CA 95064, USA.
Susan Strome, Department of Molecular, Cell and Developmental Biology, University of California, Santa Cruz, Santa Cruz, CA 95064, USA.
Literature cited
- Anders S, Pyl PT, Huber W.. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BXH, Artiles KL, Hartman PS, Fire AZ.. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014;198(3):837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthana A, Ramanan P, Hirschi A, Guiley KZ, Wijeratne TU, Shelansky R, Doody MJ, Narasimhan H, Boeger H, Tripathi S, et al. The MuvB complex binds and stabilizes nucleosomes downstream of the transcription start site of cell-cycle dependent genes. Nat Commun. 2022;13(1):526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitel GJ, Lambie EJ, Horvitz HR.. The C. elegans gene lin-9, which acts in an Rb-related pathway, is required for gonadal sheath cell development and encodes a novel protein. Gene. 2000;254(1–2):253–263. [DOI] [PubMed] [Google Scholar]
- Boichuk S, Parry JA, Makielski KR, Litovchick L, Baron JL, Zewe JP, Wozniak A, Mehalek KR, Korzeniewski N, Seneviratne DS, et al. The DREAM complex mediates GIST cell quiescence and is a novel therapeutic target to enhance imatinib-induced apoptosis. Cancer Res. 2013;73(16):5120–5129. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP.. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. [DOI] [PubMed] [Google Scholar]
- Boxem M, van den Heuvel S.. C. elegans class B synthetic multivulva genes act in G(1) regulation. Curr Biol. 2002;12(11):906–911. [DOI] [PubMed] [Google Scholar]
- Burke JR, Deshong AJ, Pelton JG, Rubin SM.. Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J Biol Chem. 2010;285(21):16286–16293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart DL, Sage J.. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8(9):671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Peng B, Yao L, Zhang X, Sun K, Yang X, Yu L.. The ancient function of RB-E2F pathway: insights from its evolutionary history. Biol Direct. 2010;5:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol CJ, Stegmeier F, Harrison MM, Horvitz HR.. Identification and classification of genes that act antagonistically to let-60 Ras signaling in Caenorhabditis elegans vulval development. Genetics. 2006;173(2):709–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J.. FASTP: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Reinke V.. Promotion of oogenesis and embryogenesis in the C. elegans gonad by EFL-1/DPL-1 (E2F) does not require LIN-35 (pRB). Development. 2006;133(16):3147–3157. [DOI] [PubMed] [Google Scholar]
- Classon M, Dyson N.. p107 and p130: versatile proteins with interesting pockets. Exp Cell Res. 2001;264(1):135–147. [DOI] [PubMed] [Google Scholar]
- Classon M, Harlow E.. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer. 2002;2(12):910–917. [DOI] [PubMed] [Google Scholar]
- Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24(17):2796–2809. [DOI] [PubMed] [Google Scholar]
- Costello ME, Petrella LN.. C. elegans synMuv B proteins regulate spatial and temporal chromatin compaction during development. Development. 2019;146:dev174383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Chen J, Myers TR, Hwang BJ, Sternberg PW, Greenwald I, Han M.. SynMuv genes redundantly inhibit lin-3/EGF expression to prevent inappropriate vulval induction in C. elegans. Dev Cell. 2006;10(5):667–672. [DOI] [PubMed] [Google Scholar]
- Dick FA, Rubin SM.. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. 2013;14(5):297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B.. Streamlined genome engineering with a self-excising drug selection cassette. Genetics. 2015;200(4):1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B.. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10(10):1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson NJ. RB1: a prototype tumor suppressor and an enigma. Genes Dev. 2016;30(13):1492–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N.. Advanced methods of microscope control using muManager software. J Biol Methods. 2014;1(2):e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Yochem J.. The SynMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev Biol. 2007;306(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA.. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10(8):741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen S-P, Grunnet M, Jorgensen EM.. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40(11):1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. [DOI] [PubMed] [Google Scholar]
- Goetsch PD, Garrigues JM, Strome S.. Loss of the Caenorhabditis elegans pocket protein LIN-35 reveals MuvB's innate function as the repressor of DREAM target genes. PLoS Genet. 2017;13(11):e1007088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiley KZ, Iness AN, Saini S, Tripathi S, Lipsick JS, Litovchick L, Rubin SM.. Structural mechanism of Myb-MuvB assembly. Proc Natl Acad Sci U S A. 2018;115(40):10016–10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiley KZ, Liban TJ, Felthousen JG, Ramanan P, Litovchick L, Rubin SM.. Structural mechanisms of DREAM complex assembly and regulation. Genes Dev. 2015;29(9):961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Ceol CJ, Lu X, Horvitz HR.. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc Natl Acad Sci U S A. 2006;103(45):16782–16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Lu X, Horvitz HR.. LIN-61, one of two Caenorhabditis elegans malignant-brain-tumor-repeat-containing proteins, acts with the DRM and NuRD-like protein complexes in vulval development but not in certain other biological processes. Genetics. 2007;176(1):255–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K, Lees JA, Vidal M, Dyson N, Harlow E, Fattaey A.. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell. 1992;70(2):337–350. [DOI] [PubMed] [Google Scholar]
- Huang LS, Tzou P, Sternberg PW.. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol Biol Cell. 1994;5(4):395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh K, Zhou X, Hayakawa H, Cho J-Y, Libermann TA, Jin J, Harper JW, Munger K.. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J Virol. 2007;81(18):9737–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurford RK Jr, Cobrinik D, Lee MH, Dyson N.. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11(11):1447–1463. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP.. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. [DOI] [PubMed] [Google Scholar]
- Kim E, Sun L, Gabel CV, Fang-Yen C.. Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLoS One. 2013;8(1):e53419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirienko NV, Fay DS.. Transcriptome profiling of the C. elegans Rb ortholog reveals diverse developmental roles. Dev Biol. 2007;305(2):674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight AS, Notaridou M, Watson RJ.. A Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells. Oncogene. 2009;28(15):1737–1747. [DOI] [PubMed] [Google Scholar]
- Korenjak M, Taylor-Harding B, Binné UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A.. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119(2):181–193. [DOI] [PubMed] [Google Scholar]
- Lees JA, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, Helin K.. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol. 1993;13(12):7813–7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR.. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 2004;18(23):2929–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Sahoo D, Geng C, Bell M, Lipsick JS, Botchan MR.. Drosophila lin-52 acts in opposition to repressive components of the Myb-MuvB/dREAM complex. Mol Cell Biol. 2012;32(16):3218–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liban TJ, Medina EM, Tripathi S, Sengupta S, Henry RW, Buchler NE, Rubin SM.. Conservation and divergence of C-terminal domain structure in the retinoblastoma protein family. Proc Natl Acad Sci U S A. 2017;114(19):4942–4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liban TJ, Thwaites MJ, Dick FA, Rubin SM.. Structural conservation and E2F binding specificity within the retinoblastoma pocket protein family. J Mol Biol. 2016;428(20):3960–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Florens LA, Swanson SK, Washburn MP, DeCaprio JA.. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 2011;25(8):801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA, et al. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26(4):539–551. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Horvitz HR.. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95(7):981–991. [DOI] [PubMed] [Google Scholar]
- Mages CF, Wintsche A, Bernhart SH, Muller GA.. The DREAM complex through its subunit Lin37 cooperates with Rb to initiate quiescence. Elife. 2017;6:e26876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau AH, Felthousen JG, Goetsch PD, Iness AN, Lee H-W, Tripathi SM, Strome S, Litovchick L, Rubin SM.. Structural basis for LIN54 recognition of CHR elements in cell cycle-regulated promoters. Nat Commun. 2016;7:12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnik EA, Fuqua JH, Sharp CS, Rochester JD, Xu EL, Holbrook SE, Updike DL.. Germline maintenance through the multifaceted activities of GLH/Vasa in Caenorhabditis elegans P granules. Genetics. 2019;213(3):923–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller GA, Engeland K.. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 2010;277(4):877–893. [DOI] [PubMed] [Google Scholar]
- Müller GA, Quaas M, Schümann M, Krause E, Padi M, Fischer M, Litovchick L, DeCaprio JA, Engeland K.. The CHR promoter element controls cell cycle-dependent gene transcription and binds the DREAM and MMB complexes. Nucleic Acids Res. 2012;40(4):1561–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller GA, Stangner K, Schmitt T, Wintsche A, Engeland K.. Timing of transcription during the cell cycle: protein complexes binding to E2F, E2F/CLE, CDE/CHR, or CHR promoter elements define early and late cell cycle gene expression. Oncotarget. 2017;8(58):97736–97748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers TR, Greenwald I.. lin-35 Rb acts in the major hypodermis to oppose RAS-mediated vulval induction in C. elegans. Dev Cell. 2005;8(1):117–123. [DOI] [PubMed] [Google Scholar]
- Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, Dowdy SF.. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife. 2014;3:e02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterloh L, von Eyss B, Schmit F, Rein L, Hübner D, Samans B, Hauser S, Gaubatz S.. The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J. 2007;26(1):144–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang CL, Toh SY, He P, Teissier S, Ben Khalifa Y, Xue Y, Thierry F.. A functional interaction of E7 with B-Myb-MuvB complex promotes acute cooperative transcriptional activation of both S- and M-phase genes. (129 c). Oncogene. 2014;33(31):4039–4049. [DOI] [PubMed] [Google Scholar]
- Petrella LN, Wang W, Spike CA, Rechtsteiner A, Reinke V, Strome S.. synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development. 2011;138(6):1069–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilkinton M, Sandoval R, Colamonici OR.. Mammalian Mip/LIN-9 interacts with either the p107, p130/E2F4 repressor complex or B-Myb in a cell cycle-phase-dependent context distinct from the Drosophila dREAM complex. Oncogene. 2007;26(54):7535–7543. [DOI] [PubMed] [Google Scholar]
- Rechtsteiner A, Costello ME, Egelhofer TA, Garrigues JM, Strome S, Petrella LN.. Repression of germline genes in Caenorhabditis elegans somatic tissues by H3K9 dimethylation of their promoters. Genetics. 2019;212(1):125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin SM. Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem Sci. 2013;38(1):12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivam S, Duan S, DeCaprio JA.. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26(5):474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffer AM, Kim DH, van Oudenaarden A, Horvitz HR.. The Caenorhabditis elegans synthetic multivulva genes prevent ras pathway activation by tightly repressing global ectopic expression of lin-3 EGF. PLoS Genet. 2011;7(12):e1002418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit F, Cremer S, Gaubatz S.. LIN54 is an essential core subunit of the DREAM/LINC complex that binds to the cdc2 promoter in a sequence-specific manner. FEBS J. 2009;276(19):5703–5716. [DOI] [PubMed] [Google Scholar]
- Schmit F, Korenjak M, Mannefeld M, Schmitt K, Franke C, von Eyss B, Gagrica S, Hänel F, Brehm A, Gaubatz S, et al. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007;6(15):1903–1913. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW.. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi TM, Deplancke B, Osato N, Zhu LJ, Barrasa MI, Harrison MM, Horvitz HR, Walhout AJM, Hagstrom KA.. Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex. PLoS Genet. 2011;7(5):e1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Kassatly RF, Cress WD, Horowitz JM.. Subunit composition determines E2F DNA-binding site specificity. Mol Cell Biol. 1997;17(12):6994–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedesco D, Lukas J, Reed SI.. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev. 2002;16(22):2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorster PJ, Goetsch P, Wijeratne TU, Guiley KZ, Andrejka L, Tripathi S, Braden BJ, Rubin SM, Strome S, Lipstick J.. A long lost key opens an ancient lock: Drosophila Myb causes a synthetic multivulval phenotype in nematodes. Biol Open. 2020;9(5):bio051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Kennedy S, Conte D, Kim JK, Gabel HW, Kamath RS, Mello CC, Ruvkun G.. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436(7050):593–597. [DOI] [PubMed] [Google Scholar]
- Zhang B, Chen W, Roman A.. The E7 proteins of low- and high-risk human papillomaviruses share the ability to target the pRB family member p130 for degradation. Proc Natl Acad Sci U S A. 2006;103(2):437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwicker J, Lucibello FC, Wolfraim LA, Gross C, Truss M, Engeland K, Müller R.. Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 1995;14(18):4514–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Requests for information, strains, and reagents should be directed to and will be fulfilled by PDG (pdgoetsc@mtu.edu). Supplemental Material is available at figshare: https://doi.org/10.25386/genetics.19491575. Supplementary information includes Supplementary Materials and Methods. Supplementary Fig 1 contains full western blots presented in Fig. 2d. Supplementary Fig. 2 contains full western blots presented in Fig. 3b. Supplementary Fig. 3 contains additional DREAM target genes tested using qPCR for ChIP and sequential ChIP analyses to complement Fig. 6. Antibodies used for western blot and ChIP analyses are available in Supplementary Table 1. Primers used for cloning and qPCR are available in Supplementary Table 2. DESeq2 differential gene expression analysis data are available in Supplementary Table 3. RNA-seq expression data used in this study are available from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) through GEO Series accession number GSE199287. Microarray expression analysis data used in this study are available through GEO Series accession number GSE6547 (Kirienko and Fay 2007).