Abstract
Enrichment cultures of microbial consortia enable the diverse metabolic and catabolic activities of these populations to be studied on a molecular level and to be explored as potential sources for biotechnology processes. We have used a combined approach of enrichment culture and direct cloning to construct cosmid libraries with large (>30-kb) inserts from microbial consortia. Enrichment cultures were inoculated with samples from five environments, and high amounts of avidin were added to the cultures to favor growth of biotin-producing microbes. DNA was extracted from three of these enrichment cultures and used to construct cosmid libraries; each library consisted of between 6,000 and 35,000 clones, with an average insert size of 30 to 40 kb. The inserts contained a diverse population of genomic DNA fragments isolated from the consortia organisms. These three libraries were used to complement the Escherichia coli biotin auxotrophic strain ATCC 33767 Δ(bio-uvrB). Initial screens resulted in the isolation of seven different complementing cosmid clones, carrying biotin biosynthesis operons. Biotin biosynthesis capabilities and growth under defined conditions of four of these clones were studied. Biotin measured in the different culture supernatants ranged from 42 to 3,800 pg/ml/optical density unit. Sequencing the identified biotin synthesis genes revealed high similarities to bio operons from gram-negative bacteria. In addition, random sequencing identified other interesting open reading frames, as well as two operons, the histidine utilization operon (hut), and the cluster of genes involved in biosynthesis of molybdopterin cofactors in bacteria (moaABCDE).
The structure of microbial communities from many environmental samples is highly complex and diverse. A recent study has estimated that 1 g of soil may contain up to 4,000 different species (33). The complexity of these communities not only is intriguing but also presents a challenge to biotechnology. Current estimates indicate that less than 1% of the microorganisms present in many environments are readily culturable (1). In fact, most of the species in many environments have never been described, and this will not be possible until new culture technologies are developed. Many approaches currently used to explore the diversity and the potential of microbial communities are biased because of the limitations of cultivation methods (3). The classical cultivation techniques require that the different microorganisms derived from an environmental sample be cultured on an appropriate growth medium and then separated until individual clones are isolated. Separation of bacterial communities and growing them on different media, however, results in the loss of major portions of the microbial community, because of the different growth requirements of the many different microbes (3, 23). In addition, this approach is time-consuming and labor-intensive. To overcome difficulties and limitations associated with cultivation techniques, several methods that are DNA based and that bypass cultivation techniques have been developed (15, 25, 31–36). One technique has been very successfully used to construct different environmental DNA libraries and screen for enzymes which can be used in biotechnological processes (15). However, one of the drawbacks of this method is that only small DNA molecules can be cloned from environments. Thus, this technique is limited to the analysis of single genes and consequently precludes screening for metabolic activities encoded in operons or gene clusters. Also, while this paper was under review, Rondon et al. reported the cloning of large DNA fragments directly isolated from one soil type using bacterial artificial chromosomes (25). Although only DNA from one specific soil was isolated and cloned, the method might still cope with the difficulties associated with isolation of DNA from soil when different soils are tested.
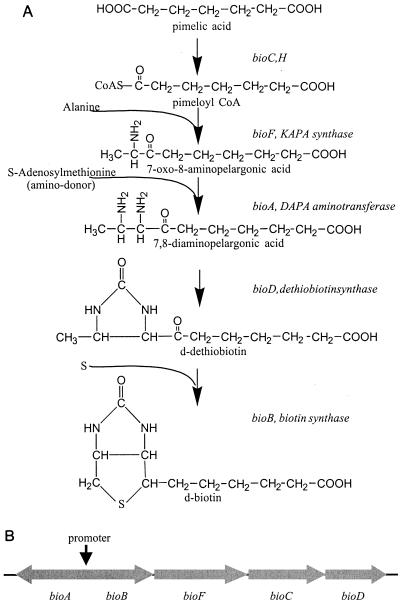
Because of the known difficulties associated with the construction of DNA libraries with DNA directly derived from environmental samples, we have chosen to isolate DNA for the construction of cosmid libraries from enrichment cultures. As an example, we used enrichment cultures to select for microorganisms producing high amounts of biotin. DNA was isolated from the enrichment cultures and used to construct cosmid libraries with inserts of >30 kb. The genetic diversity and potential of the different libraries were assessed by screening for biotin biosynthesis operons. This screening was possible because in Escherichia coli and many other gram-negative bacteria, the genes associated with biotin biosynthesis are located in two linked but divergently transcribed operons, which are controlled by a single operator (8). In E. coli, biotin is biosynthesized through the following intermediates: pimeloyl coenzyme A (pimeloyl-CoA) → 7-keto-8-aminopelargonic acid (KAPA) → 7, 8-diaminopelargonic acid (DAPA) → dethiobiotin → biotin (Fig. 1). Six structural genes and one regulatory element are involved in this biosynthetic pathway; five of the genes (bioA, bioB, bioC, and bio D) are located in a cluster at 17.5 min on the genetic map, forming a biotin operon (2, 8). The bioF, bioA, and bioD genes encode KAPA synthetase, DAPA aminotransferase, and dethiobiotin synthetase, respectively. An enzyme encoded by the bioB gene is involved in the conversion of dethiobiotin to biotin, and the bioC gene is thought to be involved in pimeloyl-CoA synthesis (5, 8, 9, 18, 22, 24).
FIG. 1.
(A) E. coli biotin biosynthesis pathway; (B) genetic organization of the E. coli biotin biosynthesis operon.
In this work we have isolated seven cosmid clones, each carrying a different biotin biosynthesis operon derived from environmental consortia. The cosmid clones were characterized on a molecular and physiological level. Although classical enrichment cultures have been widely used for the isolation of microbes with desirable traits, this is the first study to use this approach on a molecular basis, allowing the direct isolation, cloning, and comparative analysis of loci organized in operons or clusters, from diverse microbial niches.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and environmental samples used in this study.
Microbiological materials used in this work are listed in Table 1. E. coli was grown at 37°C on Luria-Bertani medium (28) supplemented with appropriate antibiotics. Enrichment cultures were inoculated with 2.0 g of the different environmental samples (Table 2) into 1-liter flasks containing 100 ml of M9 medium (28) supplemented with 6.5 U of avidin and glucose (10 mM) as the sole carbon source. Except for the enrichment culture derived from horse excrement (Table 2), all cultures were grown at 30°C. Enrichment cultures were incubated on a shaker (150 rpm) for 2 days, pelleted, and used for construction of the DNA libraries. Growth was monitored as optical density at 600 nm (OD 600). The enrichment culture inoculated with horse excrement was grown at 37°C for 24 h.
TABLE 1.
Strains and constructs used
| Strain or Construct | Relevant trait | Source or referencea |
|---|---|---|
| E. coli | ||
| VCS257 | DP50 derivative | Stratagene |
| JW1 | ΔlacZ thi ara pro | 21 |
| K12 | Wild-type isolate, thi | ATCC |
| ATCC33767 | K12ΔH1 derivative carrying a deletion in the bio region, Δ(bio-uvrB) | ATCC |
| L. plantarum | ATCC 8014 | DSMZ |
| Plasmids and cosmids | ||
| pBSK+ | pBluescript SK(+) multicopy cloning vector, Ampr | Stratagene |
| pHSG399 | Low-copy-number cloning vector, Cmr | 13 |
| pWE15 | Cosmid cloning vector for construction of DNA libraries, Ampr | Stratagene |
| pK18mobsacB | Multicopy cloning vector, sacB | 29 |
| pHE1-1 | Subclone of pCosHE1; in pHSG399, 1-kb fragment, containing parts of modC-bioA | This work |
| pHE1-2 | Subclone of pCosHE1; in pHSG399, 1.9-kb fragment, containing parts of hutH, complete orf-1, and partial bioA | This work |
| pHE1-3 | Subclone of pCosHE1; in pHSG399, 0.9-bp fragment, containing parts of bioA and bioB | This work |
| pHE1-4 | Subclone of pCosHE1; in pHSG399, 1.4-kb fragment, containing part of uvrB | This work |
| pHE1-5 | Subclone of pCosHE1; in pHSG399, 2.5-kb fragment, containing moaA–moaE | This work |
| pHE1-6 | Subclone of pCosHE1; in pHSG399, 2.8-kb fragment, containing moaE–rhlE region | This work |
| pHE2-1 | Subclone of pCosHE2; in pBSK+, 4.4-kb fragment, containing bioA–bioD | This work |
| pFS1-1 | Subclone of pCosFS1; in pHSG399, 0.9-kb fragment, containing parts of bioF and bioC | This work |
| pFS1-2 | Subclone of pCosFS1; in pHSG399, 3.6-kb fragment, containing moaE–rhlE | This work |
| pFS1-3 | Subclone of pCosFS1; in pHSG399, 1.8-kb fragment, containing parts of bioA and bioB | |
| pAS1-1 | Subclone of pCosFS1; in pHSG399, 1.6-kb fragment, containing bioD and elsA | This work |
| pAS1-2 | Subclone of pCosFS1; in pHSG399, 2.8-kb fragment, containing hutU, hutH, and orf-1 | This work |
| pAS1-3 | Subclone of pCosFS1; in pK18mobsacB, 0.8-kb fragment, containing partial bioF and bioC sequences | This work |
| pAS1-4 | Subclone of pCosFS1; in pHSG399, 1.3-kb fragment, containing part of hutU | This work |
| pAS1-5 | Subclone of pCosFS1; in pHSG399, 0.6-kb fragment, containing part of uvrB | This work |
| pAS1-6 | Subclone of pCosFS1; in pK18mobsacB, 0.4-kb fragment, containing partial hutG and hutC | This work |
| pCosHE1 | pWE15 containing a 30-kb insert with bio genes from microbial consortium | This work |
| pCosHE2 | pWE15 containing a 30-kb insert with bio genes from microbial consortium | This work |
| pCosAS1 | pWE15 containing a 30-kb insert with bio genes from microbial consortium | This work |
| pCosFS1 | pWE15 containing a 30-kb insert with bio genes from microbial consortium | This work |
ATCC, American Type Culture Collection, DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen.
TABLE 2.
Biotin measurements in supernatants and analysis of colony morphology of the first subculturesa
| Enrichment culture | Source of inoculum | Minimum no. of different colony types observed on plates | Biotin in supernatant of first subculture (pg/ml/OD) |
|---|---|---|---|
| HE | Horse excrement | 3 | 200 |
| AS | Agricultural soil | 4 | 875 |
| FS | Forest soil | 6 | 1,000 |
| VS | Volcanic soil | 5 | <50 |
| GS | Sandy soil | 7 | 1,040 |
Biotin in culture supernatants was measured by ELISA. For biotin measurements in supernatants, cells were removed as soon as no further increase in the (OD600) was observed. Biotin measurements represent values of three experiments; with the exception of HE, all subcultures were grown at 30°C. Subcultures of the enrichment cultures were grown in the absence of avidin.
Molecular techniques.
DNA isolation from the enrichment cultures was done as described previously (32), with minor modifications. After pelleting, bacteria were resuspended in Tris-EDTA (TE)-sucrose (20% [wt/vol]) buffer and lysed in DNA extraction buffer (100 mM Tris-HCl, 100 mM EDTA, 100 mM Na2HPO4 and 1.5 M NaCl, 1% [wt/vol] sodium dodecyl sulfate) for several hours. RNA was degraded with RNase A (10 mg/ml). The resulting DNA extracts were incubated with protease and sarcosyl (5%, [wt/vol]) in TE buffer overnight. Total genomic DNA was then purified repeatedly with chloroform-phenol (1:1, [vol/vol]) and then once with chloroform. DNA was then dialyzed against 2 liters of TE buffer at 4°C overnight. Finally, an aliquot of the DNA was analyzed on a 0.8% agarose gel to ensure that the DNA was not degraded.
Direct extraction of DNA from environmental samples was carried out as described by Henne et al. (15).
DNA cloning steps were performed with standard methods (30). DNA-modifying enzymes were used as specified by the manufacturer. For DNA hybridizations, restriction fragments were separated by electrophoresis in 0.8% agarose gels, transferred onto nylon membranes, and cross-linked with UV light. Hybridizations were performed overnight using digoxigenin-labeled DNA probes and high-stringency conditions (68°C). Hybridization signals were detected with colorimetric substances nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate and with CSPD using X-ray films.
Construction of cosmid libraries from the enrichment cultures.
Cosmid libraries containing DNA from the various cosmid libraries were prepared in pWE15 (Stratagene, La Jolla, Calif.) using standard protocols (28). DNA fragments (30 to 40 kb) obtained after partial Sau3A digestion were ligated into BamHI restriction sites of the cosmid vector. Phage packaging mixes were obtained from Stratagene, and infection of E. coli VCS257 was performed according to the manufacturer's protocol. Vectors used for subcloning are listed in Table 1; when required, gaps in the DNA sequences were filled by PCR. Automated DNA sequencing was performed using an ABI377 DNA sequencer and dye terminator chemistry following the manufacturer's instructions.
Biotin measurements in culture supernatants.
For biotin measurements, use either the Lactobacillus plantarum growth assay (7) or a competitive assay enzyme-linked immunosorbent (ELISA) (4), with some modifications. For this purpose, microtiter plates were coated with anti-rabbit immunoglobulin G at a dilution of 1:5,000 in phosphate-buffered saline (PBS) for 2 hours; washed and treated with blocking solution (vitamin free casein) for 1 h, and then washed with PBS. Extravidin-alkaline phosphatase conjugate (Sigma, Heidelberg, Germany) was added to the plates in a 1:20,000 dilution in PBS-Tween (0.025% [vol/vol]); 100-μl aliquots of the samples were added to the wells, and the microtiter plates were incubated at 37°C for 30 min. Serial dilutions of the various samples were performed prior to the tests, and a standard with known biotin concentrations was included in each microtiter plate. After repeated washing of the microtiter plates with PBS, substrate buffer (Tris-HCl, 100 mM; NaCl, 100 mM; MgCl2, 50 mM; p-nitrophenylphosphate, 2 mg/ml [pH 9.5]) was added, and color development was recorded at 405 nm in a microplate reader.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained have been deposited at GenBank; accession numbers are listed in Table 4.
TABLE 4.
Identified genes, GenBank accession numbers, and observed similarities
| Cosmid clone | Gene identified (accession no.) | Similar protein | Organism | % Identity | Region of identical amino acids | Score |
|---|---|---|---|---|---|---|
| pCosAS1 | hutH (AF248314) | Histidine ammonia lyase | Pseudomonas putida | 75 | 490 | 723 |
| Sinorhizobium meliloti | 50 | 480 | 420 | |||
| pCosAS1 | orf-1 (AF248314) | Hypothetical 17.1-kDa protein in modC–bioA intergenic region | Escherichia coli | 77 | 158 | 268 |
| Hypothetical protein Rv2140c | Mycobacterium tuberculosis | 42 | 154 | 116 | ||
| pCosAS1 | bioA (AF248314) | DAPA aminotransferase | E. coli | 85 | 427 | 767 |
| Erwinia herbicola | 85 | 427 | 757 | |||
| Xenorhabdus nematophilus | 70 | 421 | 628 | |||
| pCosAS1 | bioB (AF248314) | Biotin synthase | E. herbicola | 86 | 346 | 618 |
| E. coli | 84 | 346 | 602 | |||
| Serratia marcescens | 80 | 343 | 566 | |||
| pCosAS1 | bioF (AF248314) | 7-KAPA synthetase | E. herbicola | 67 | 384 | 509 |
| E. coli | 64 | 384 | 485 | |||
| S. marcescens | 55 | 378 | 386 | |||
| pCosAS1 | bioC (AF248314) | Biotin synthesis protein BioC | E. herbicola | 56 | 251 | 271 |
| E. coli | 53 | 251 | 261 | |||
| S. marcescens | 43 | 247 | 201 | |||
| pCosAS1 | bioD (AF248314) | Dethiobiotin synthetase | E. herbicola | 74 | 225 | 106 |
| E. coli | 76 | 251 | 271 | |||
| S. marcescens | 68 | 251 | 261 | |||
| pCosAS1 | elsA (AF248314) | Urea/short-chain amide ABC transporter, ATP-binding, putative branched-chain amino acid ABC transporter | Deinococcus radiodurans Anabaena sp. strain PCC 7120 Thermotoga maritima | 38 35 33 | 189 186 182 | 103 86 86 |
| Nodulation ATP-binding protein I | Rhizobium leguminosarum | 31 | 189 | 71 | ||
| pCosAS1 | hutG (AF250775) | Histidine utilization repressor G | Klebsiella aerogenes | 65 | 28 | 48 |
| pCosAS1 | hutC (AF250775) | Histidine utilisation repressor C | K. aerogenes | 76 | 70 | 109 |
| Yersinia pestis | 63 | 60 | 86 | |||
| P. putida | 65 | 60 | 83 | |||
| pCosAS1 | hutU (AF250766) | Urocanase | P. putida | 84 | 340 | 603 |
| pCosAS1 | uvrB (AF248315) | UvrB | E. coli | 89 | 297 | 543 |
| P. aeruginosa | 69 | 297 | 430 | |||
| Xanthomonas campestris | 65 | 296 | 406 | |||
| pCosHE1 | moaA (AF250774) | Molybdenum cofactor biosynthesis protein A | E. coli | 89 | 186 | 352 |
| Haemophilus influenzae | 61 | 185 | 245 | |||
| Synechococcus sp. | 41 | 185 | 164 | |||
| pCosHE1 | moaB (AF250774) | Molybdenum cofactor biosynthesis protein B | E. coli | 41 | 136 | 70 |
| pCosHE1 | moaC (AF250774) | Molybdenum cofactor biosynthesis protein C | E. coli | 75 | 59 | 85 |
| H. influenzae | 67 | 53 | 64 | |||
| Staphylococcus carnosus | 54 | 53 | 52 | |||
| pCosHE1 | moaD (AF250774) | Molybdenum cofactor biosynthesis protein D | E. coli | 85 | 46 | 86 |
| 65 | 35 | 46 | ||||
| H. influenzae | 61 | 46 | 66 | |||
| 45 | 41 | 41 | ||||
| pCosHE1 | moaE (AF250774) | Molybdopterin converting factor, subunit 2 | E. coli | 82 | 150 | 270 |
| H. influenzae | 56 | 150 | 184 | |||
| P. abyssi | 41 | 131 | 101 | |||
| pCosHE1 | hutH (AF250769) | Histidine ammonia lyase | Pyrococcus putida | 81 | 121 | 205 |
| S. meliloti | 54 | 128 | 119 | |||
| D. radiodurans | 48 | 115 | 104 | |||
| pCosHE1 | orf-1 (AF250769) | Hypothetical 17.1-kDa protein in modC-bioA intergenic region | E. coli | 81 | 158 | 278 |
| Hypothetical 19.5-kDa protein in emrE–rus intergenic region | E. coli | 46 | 165 | 152 | ||
| Hypothetical protein MTH273 | Methanobacterium thermoautotrophicum | 32 | 148 | 80 | ||
| pCosHE1 | bioA (AF250770) | DAPA aminotransferase | E. herbicola | 85 | 260 | 453 |
| E. coli | 83 | 260 | 453 | |||
| S. marcescens | 70 | 254 | 355 | |||
| X. nematophilus | 67 | 255 | 361 | |||
| pCosHE1 | bioB (AF250770) | Biotin synthase | E. herbicola | 84 | 127 | 228 |
| E. coli | 82 | 127 | 220 | |||
| S. marcescens | 72 | 142 | 200 | |||
| P. stutzeri | 64 | 119 | 168 | |||
| pCosHE1 | uvrB (AF250772) | Excision nuclease subunit B | E. coli | 81 | 323 | 511 |
| H. influenzae Rd | 69 | 323 | 459 | |||
| P. aeroginosa | 62 | 322 | 393 | |||
| pCosHE1 | elsB (AF250773) | Hypothetical 36.3-kDa protein in modC-bioA intergenic region | E. coli | 85 44 | 98 24 | 183 25 |
| (Hypothetical 30.2-kDa protein in modC 3′ region)-YbhE | X. nematophilus | 46 | 97 | 83 | ||
| pCosHE1 | elsC (AF250773) | Hypothetical 46.1-kDa protein in modC-bioA intergenic region | E. coli | 83 | 162 | 283 |
| Putative hypothetical protein in bioA 5′ region | E. coli | 46 | 152 | 123 | ||
| Pectinesterase B precursor | Erwinia chrysanthemi | 46 | 152 | 123 | ||
| pCosHE1 | elsG (AF250771) | Hypothetical 47.6-kDa protein in moaE-rhlE intergenic region | E. coli | 70 | 44 | 75 |
| pCosHE1 | elsH (AF2507771) | Hypothetical 28.8-kDa protein in moaE-rhlE intergenic region | E. coli | 85 | 99 | 185 |
| pCosFS1 | bioA (AF250768) | DAPA aminotransferase | S. marcescens | 86 | 393 | 695 |
| X. nematophilus | 74 | 387 | 617 | |||
| E. herbicola | 73 | 390 | 598 | |||
| pCosFS1 | bioB (AF250768) | Biotin synthase | S. marcescens | 95 | 342 | 665 |
| E. coli | 87 | 341 | 605 | |||
| E. herbicola | 85 | 341 | 605 | |||
| pCosFS1 | bioF (AF250768) | 7-KAPA synthetase | S. marcescens | 66 | 388 | 477 |
| E. herbicola | 56 | 382 | 412 | |||
| E. coli | 55 | 381 | 409 | |||
| pCosFS1 | bioC (AF250768) | Biotin synthesis protein BioC | S. marcescens | 84 | 186 | 313 |
| E. coli | 56 | 181 | 198 | |||
| E. herbicola | 51 | 175 | 177 | |||
| pCosFS1 | uvrB (AF303465) | Excision nuclease subunit B | E. coli | 86 | 38 | 71 |
| P. aeruginosa | 77 | 35 | 59 | |||
| H. influenzae Rd | 70 | 37 | 58 | |||
| pCosFS1 | elsD (AF250767) | Hypothetical 42.1-kDa protein in moaE-rhlE intergenic region YbhS | E. coli | 77 | 270 | 427 |
| Hypothetical 42.1-kDa protein in moaE-rhlE intergenic region, YbhR | E. coli | 27 | 231 | 125 | ||
| Hypothetical protein F648 | E. coli | 88 | 26 | 51 | ||
| ABC transporter, ATP-binding protein | Archaeoglobus fulgidus | 31 27 | 235 184 | 119 86 | ||
| pCosFS1 | elsF (AF250767) | ABC transporter, ATP-binding protein-YbhF | E. coli | 72 41 | 114 116 | 168 87 |
| ABC transporter, ATP-binding protein | M. thermoautotrophicum | 41 | 113 | 90 | ||
| A. fulgidus | 39 | 105 | 82 | |||
| pCosFS2 | elsF AF250767) | Hypothetical protein F355 | E. coli | 66 | 163 | 224 |
| pCosFS1 | rhlE (AF303464) | ATP-dependent RNA helicase RhlE | E. coli | 80 | 78 | 123 |
| Shewanella violacea | 73 | 65 | 100 | |||
| Vibrio cholerae | 70 | 65 | 98 | |||
| pCosHE2 | orf1 (AF250776) | Hypothetical 17.1-kDa protein in modC–bioA intergenic region hypothetical protein Rv2140c | E. coli M. tuberculosis | 99 47 | 158 150 | 339 138 |
| pCosHE2 | bioA (AF250776) | DAPA aminotransferase | E. coli | 96 | 429 | 860 |
| E. herbicola | 83 | 427 | 735 | |||
| X. nematophilus | 70 | 421 | 625 | |||
| pCosHE2 | bioB (AF250776) | Biotin synthase | E. coli | 99 | 346 | 697 |
| E. herbicola | 91 | 346 | 655 | |||
| S. marcescens | 86 | 344 | 611 | |||
| pCosHE2 | bioF (AF250776) | 7-KAPA synthetase | E. coli | 82 | 384 | 635 |
| E. herbicola | 62 | 384 | 471 | |||
| S. marcescens | 55 | 378 | 384 | |||
| pCosHE2 | bioC (AF250776) | Biotin synthesis protein BioC | E. coli | 93 | 251 | 490 |
| E. herbicola | 64 | 251 | 317 | |||
| S. marcescens | 48 | 248 | 235 | |||
| pCosHE2 | bioD (AF250776) | Dethiobiotin synthase | E. coli | 91 | 212 | 401 |
| E. herbicola | 75 | 207 | 330 | |||
| S. marcescens | 72 | 198 | 297 |
RESULTS
Enrichment cultures.
The enrichment cultures were inoculated with samples derived from five microbial environments (Table 2). The first sample was from a forest soil with an extremely high humic acid content, collected near Goettingen, Germany; the second sample was soil collected from an agricultural site near Goettingen. The third sample consisted of horse excrement collected from a meadow near Goettingen; this sample was likely already enriched with a dominant population of enteric bacteria. The fourth sample was collected from a beach in the north of Greece, near Kavalla, and the fifth sample was collected on Mount Hood, in Oregon (Table 2).
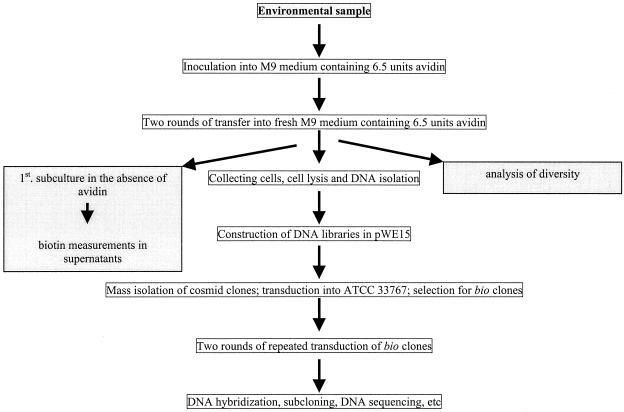
Attempts to directly extract the DNA from the environmental samples, in the high quality necessary for construction of cosmid libraries, failed. The isolated DNA appeared to be highly contaminated with humic acids, which could not be removed by repeated purification steps using the Wizard Plus Miniprep system or by employing a gel extraction kit; the DNA obtained by this method was not suitable for ligation and efficient packaging into phage heads. To avoid these difficulties and to keep the humic acid contaminations of the DNA to a minimum, small samples from the different environments were used to inoculate enrichment cultures. After two rounds of transfer into fresh medium and growth for 1 to 2 days, bacteria were pelleted and lysed to isolate total genomic DNA for construction of the cosmid libraries (Fig. 2). To verify a high degree of diversity within the different enrichment cultures, aliquots were microscopically examined. Different forms of bacteria were observed, including rods, cocci, spore-forming, and coryneform microbes. The presence of different colony types observed on agar plates, inoculated from the enrichment cultures, confirmed this finding, as did detailed electron microscope examinations of the same cultures (data not shown).
FIG. 2.
Scheme of the enrichment and cloning strategy used in this study. The samples were taken from five different microbial environments (an agriculture soil, a forest soil, horse excrement, volcanic soil, and sandy soil).
Two lines of evidence indicate that the avidin present in the enrichment cultures had a significant influence on the microbial consortia and that the avidin resulted in an increased selection for biotin-producing microbes. First, soil from the AS (agricultural soil) sample was used to inoculate a culture containing avidin (0.065 U /ml) and a control culture with no added avidin. Biotin contents of the supernatant was analyzed after 18 and 48 h of growth using the Lactobacillus growth assay. Interestingly, in the supernatant of the culture lacking the avidin, the biotin contents decreased significantly over time. While after 18 h of growth 15 pg of biotin/ml was detected, biotin was below the detection limit after 48 h of growth. This suggests that in the absence of added avidin, biotin was primarily taken up by the microorganisms present in the culture, and synthesis and release into the medium were of minor importance. In contrary, in the culture supernatants containing avidin, approximately 20 pg of free biotin was detected after 18 and 48 h of growth. A second line of evidence comes from additional DNA hybridization studies. In those studies, DNA directly isolated from the HE (horse excrement) sample and DNA obtained from the corresponding enrichment culture was spotted onto a nylon membrane and hybridized against the E. coli bio operon. While the E. coli biotin biosynthesis operon produced no detectable signal in the DNA extracted from the HE sample without enrichment, the probe used produced a strong hybridization signal when DNA was analyzed which was obtained from the enriched consortia (Fig. 3A). The E. coli bio operon was chosen as a DNA probe, because the E. coli K-12 wild-type isolate produces and releases significant amounts of biotin into the medium under defined conditions (Table 3). In further tests, DNA was extracted from enrichment cultures which had been inoculated with agricultural soil. The DNA was applied onto nylon membranes and hybridized with the bio operon encoded on pCosAS1 as a DNA probe. Results from these DNA-based studies also indicate that the DNA sequences hybridizing with the biotin biosynthesis genes from E. coli significantly increased during the enrichment process (Fig. 3B).
FIG. 3.
Dot blot hybridization of DNA extracted from environmental samples and enrichment cultures. (A) Nylon membrane with DNA directly isolated from the HE sample and DNA from a corresponding HE enrichment culture, which was grown in the presence of avidin; 780 ng of DNA was spotted per dot, and hybridizations were done with the E. coli biotin biosynthesis genes as a DNA probe. (B) Nylon membrane loaded with DNA which was isolated from two different enrichment cultures inoculated with soil from an agricultural site (AS); one of the cultures was grown in the presence of added avidin, while no avidin was added to the other culture. DNA was extracted after 48 h of growth, and 930 ng of DNA was spotted per dot onto the nylon membranes. Hybridizations were performed overnight under high-stringency conditions at 68°C employing the bio genes on pCosAS1 as a DNA probe.
TABLE 3.
Growth characteristics and biotin production of E. coli strain ATCC 33767 carrying cosmids containing bio operons derived from three different enrichment culturesa
| bio cosmid clone or control strain | Doubling time (h)
|
Lagging phase (h)
|
Biotin in culture supernatant (pg/ml)
|
|||
|---|---|---|---|---|---|---|
| Succinate | Glucose | Succinate | Glucose | Succinate | Glucose | |
| pCosHE1 | 6.0 | 2.9 | 6.3 | 3.6 | 414 | 100 |
| pCosHE2 | 12.2 | 3.0 | 6.3 | 4.3 | 3,800 | 42 |
| pCosAS1 | 5.0 | 2.9 | 5.3 | 4.1 | 360 | 1,200 |
| pCosFS1 | 5.1 | 2.9 | 9.5 | 5.0 | 1,740 | 3,600 |
| E. coli K-12 | 2.9 | 2.9 | 2.3 | 2.3 | 47 | 8 |
Biotin in culture supernatants was measured using the L. plantarum growth assay. Data are mean values of three independent experiments.
To obtain an estimate of the biotin biosynthesis capabilities of all the enriched consortia, subcultures were inoculated with aliquots derived from the different enrichment cultures and tested for biotin production (Fig. 2). Biotin amounts measured in the supernatants, obtained after pelleting the enriched consortia, ranged from 50 to 1,040 pg/ml/ OD (Table 2). Highest amounts of biotin were measured in culture supernatants derived from the GS (sandy soil) sample; the least amounts were found in supernatants of cultures derived from the volcanic soil collected near the top of Mount Hood (Table 2). The large differences in biotin levels observed clearly suggest that the samples contain widely different populations with a range of biotin synthesis capabilities. In subsequent studies, we focused on the isolation of bio genes associated with the HE, AS, and FS (forest soil) enrichment cultures (Table 2).
Construction of cosmid DNA libraries from enrichment cultures.
To identify biotin biosynthesis genes associated with the three enrichment cultures, DNA was isolated from bacterial pellets by direct lysis. The DNA was size fractionated to fragments ranging in size from 30 to 40 kb and ligated into the cosmid vector pWE15 (see Materials and Methods). After packaging of the DNA and transfection into the E. coli host strain, the three libraries (HE, AS, and FS) ranged in size from 6,000 to 35,000 clones. The quality of the libraries was ensured by estimating the insert sizes in 70 randomly picked clones; the average insert size was approximately 30 to 40 kb, and 98% of the clones analyzed contained inserts. In addition, restriction analysis of randomly chosen cosmid clones revealed high diversity of the DNA fragments cloned into pWE15.
Screening for DNA fragments containing bio operons.
To assess the biotin biosynthesis capabilities of the three environmentally derived cosmid libraries, cosmid DNA from the recombinant E. coli clones was extracted and used to transduce the E. coli biotin-auxotrophic strain ATCC 33767 (Fig. 2). This strain carries a deletion in the region of the bio operon, ranging from the lambda attachment site to the uvrB gene, and consequently is not able to synthesize biotin unless complemented by a complete biotin biosynthesis operon. After transduction, cells were transferred to M9 medium in the absence of biotin, allowing only growth of cells containing transduced DNA complementary to the host E. coli auxotrophic strain. Bacteria growing in liquid medium were spread onto agar plates and incubated overnight. From these possible bio+ clones, DNA was extracted and used to retransduce E. coli strain ATCC 33767. This procedure was repeated twice to avoid the isolation of false-positive clones. Only bio+ clones growing after the second transduction were subject to subcloning and further molecular analysis (Fig. 2). Employing this strategy, we isolated seven different cosmid clones which appeared to contain complete biotin synthesis operons and complemented the E. coli auxotrophic strain when grown on plates and in liquid medium. A DNA restriction analysis ensured that each clone was unique. Four of these seven cosmid clones carrying the different bio operons were subject to a detailed molecular and physiological analysis. The clones isolated, named pCosHE1 and pCosHE2, originated from the HE library. The cosmid clones pCosAS1 and pCosFS1 were isolated from the AS and FS libraries, respectively.
Biotin biosynthesis capabilities and growth characteristics of the bio cosmids.
To examine the biotin biosynthesis capabilities of the isolated cosmid clones, we measured growth and biotin synthesis capabilities of cosmid clones pCosHE1, pCosHE2, pCosAS1, and pCosFS1 under defined conditions. Because succinate and glucose as sole carbon sources are known to affect biotin synthesis, we chose to add these carbon sources to the defined medium. Data were compared to growth and biotin synthesis capabilities of the E. coli K-12 wild-type isolate. For the E. coli control strain, growth on succinate resulted in a 5.8-fold-higher biotin production than growth on glucose (Table 3). Growth on succinate also stimulated biotin biosynthesis for two of the clones, pCosHE1 and pCosHE2; the observed increase in biotin production was highest for pCosHE2 (90-fold), followed by pCosHE1 (4.1-fold). For pCosAS1 and pCosFS1, however, biotin production was highest after growth on glucose.
Clones grown on succinate showed a significant increase in both lag phase and doubling time compared to both their own growth on glucose and that of the E. coli K-12 control strain growing on succinate. Doubling times observed for growth on glucose as the sole carbon source were comparable to those observed for the E. coli control strain. Cosmid pCosHE2, which showed the overall highest biotin biosynthesis on succinate as a carbon source, also revealed a twofold-longer doubling time than any of the other cosmid clones when grown on this substrate. In addition, lag phases were significantly increased for all cosmid clones when grown on succinate. The largest increase was observed for pCosFS1 and pCosHE1, both showing an almost two-fold increase in lag phase when cultivated on succinate in comparison to lag phases observed for glucose-grown cells (Table 3). Altogether, these data indicated that diverse bio genes were associated with the different cosmid clones.
Molecular characterization of the different cosmids conferring biotin biosynthesis capability.
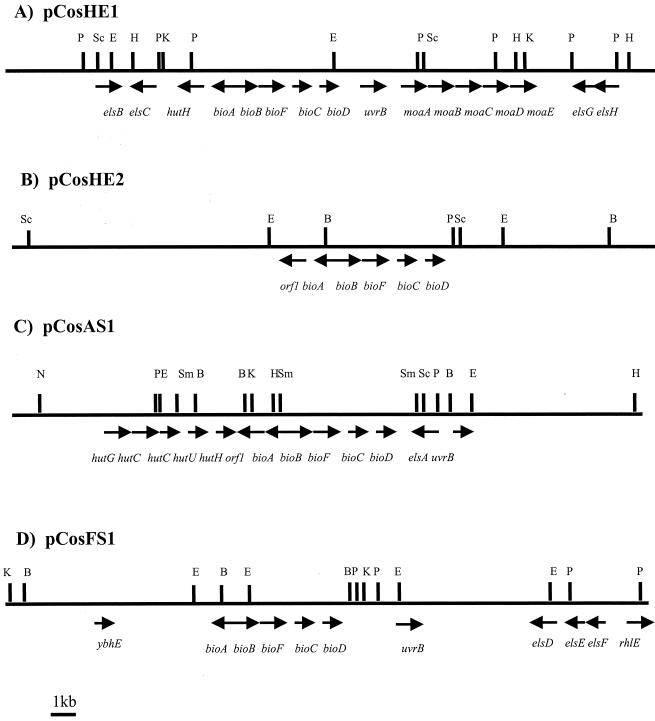
To further characterize the four bio clones, DNA was isolated, restricted with different enzymes, and subject to partial sequencing. Restriction analysis and shotgun sequencing confirmed that the insert sizes in pWE15 were approximately 30 to 40 kb and allowed a detailed mapping of the cosmids (Fig. 4). A partial sequencing of DNA derived from the cosmids in combination with DNA hybridizations identified and located four different biotin biosynthesis operons on the different cosmids. Figure 4 and Table 4 summarize the identified genes and similarities observed after comparison of the deduced amino acid sequences obtained with those available in the National Centre for Biotechnology Information databases. The organization of the bio genes identified on the four cosmids appeared to be the same as four for most gram-negative bacteria (e.g., E. coli). This finding is based on DNA sequencing and DNA hybridization studies. The deduced amino acid sequences of the bio genes identified by sequencing on pCosHE1 revealed highest similarities with the corresponding proteins in Erwinia herbicola. Identities observed for bioA and bioB were 84 and 85%, respectively. Downstream of the bio operon, the moaA-BCDE operon was identified. Comparison of the deduced amino acid sequences showed an overall 80% identity with MoaA-BCDE in E. coli. In addition, several other genes pCosHE1. (uvrB, elsB, elsC and hutH) were identified on. The deduced amino acid sequence of hutH was 81% identical to the deduced amino acid sequence of the Pseudomonas putida HutH. Also, we observed on this cosmid genes coding for proteins involved in DNA transfer and pilus assembly, as well two different antibiotic resistance markers (data not shown).
FIG. 4.
Restriction maps of the central parts of the different biotin cosmids isolated in this work; arrows indicate locations and directions of transcription of the identified ORFs on the different cosmids. (A) pCosHE1 restriction map of the bio cosmid derived from horse excrement showing highest homologies to genes of E. herbicola; (B) pCosHE2 bio cosmid isolated from an enrichment culture derived from the same source as in panel A but highly similar to E. coli; (C) pCosAS1 restriction map of a bio cosmid isolated from an agricultural soil; (D) pCosFS1 restriction map of a bio cosmid isolated from a forest soil enrichment culture. The bio genes identified in panels C and D show highest similarities to E. herbicola and S. marcescens, respectively. Observed similarities for selected ORFS are listed in Table 4, together with the assigned GenBank accession numbers. For genes of pCosHE1 and of pCosFS1, only partial sequences were obtained during the shotgun sequencing approach. DNA restriction enzymes: N, NcoI, B, BamHI; E, EcoRI; Sm, SmaI; P, PstI; K, KpnI; H, HindIII; Sc, SacI.
The biotin biosynthesis operon located on pCosHE2 was highly similar to the E. coli bio operon. The identified operon consists of 5,551 nucleotides, similar to the reported length of the classical E. coli bio operon, and the nucleotide sequence was 98% identical to the E. coli sequence. Direct comparison of the nucleotide sequence of the known E. coli biosynthesis operon with the biosynthesis genes located on pCosHE2 identified 67-b exchanges over the length of the entire operon, resulting in 21 amino acid substitutions (data not shown). In addition, the restriction pattern of the complete cosmid was highly similar to the restriction pattern of the DNA region flanking the classical E. coli bio genes (Fig. 4).
The complete nucleotide sequence of the biotin synthesis operon identified on pCosAS1 contained 5,521 nucleotides. With the exception of bioA and bioD, the deduced amino acid sequences for all other genes were highly similar to the corresponding proteins from E. herbicola. The amino acid sequences deduced from the bioB, bioF, and bioC genes showed, 86, 67, and 56% identity, respectively, to the corresponding E. herbicola bio genes. The deduced amino acid sequences of bioA and bioD were highly similar to the corresponding proteins in E. coli and E. herbicola (Table 4). Immediately upstream of open reading frame 1 (orf-1), we located a complete histidine utilization operon (hut) (Fig. 4C). Interestingly, the deduced amino acid sequences were highly similar to those known from P. putida and Klebsiella aerogenes; HutG and HutC were highly similar to the deduced amino acid sequences of the K. aerogenes proteins (65 and 76% identical amino acids, respectively), while HutU and HutH were highly similar to the corresponding P. putida proteins (84 and 75% identical amino acids, respectively).
The bio genes sequenced on the pCosFS1 were highly similar to those known from Serratia marcescens. The identity of the genes identified to the corresponding proteins in S. marcescens ranged from 73 to 92%. In addition, several other ORFs were identified on this cosmid, possibly linked to molybdopterin cofactor synthesis and similar to ATP-binding cassette (ABC) transporters (elsD, elsE, and elsF). Similarities observed were highest for similar proteins in E. coli or Archaeoglobus fulgidus. Further genes linked to the synthesis of aromatic amino acids, endonucleases, and antibiotic resistances were identified (Fig. 4).
In summary, the approach applied in this study led to the identification of DNA sequences of four different biotin biosynthesis operons. Sequence analysis indicates that all cosmids contained different DNA and were derived from different organisms. In addition to the biotin biosynthesis genes we have identified DNA sequences of many other genes including a complete histidine utilization operon and a molybdopterin cofactor synthesis operon. This further indicates that the use of enrichment cultures in combination with direct cloning allows a rapid screening for large DNA fragments derived from environmental consortia.
DISCUSSION
In this work we have developed an approach utilizing enrichment technology, which allows the isolation and comparative analysis of gene clusters or operons from diverse microbial consortia, as opposed to previous studies with the principal goal to obtain variants of one specific gene of interest. As a first step, our technique relies on the preparation of an enrichment. The technique of enrichment culture offers a powerful tool, with an intriguing potential to select for microbes with traits useful for biotechnological applications. In general, enrichment cultures have been successfully used to screen for single microbes or consortia with diverse catabolic capabilities. Specifically, enrichment cultures have been employed to select for microbes capable of degrading toluene (17) or phthalate (20), utilizing methane (6), or dechlorinating and degrading diverse aromatic compounds (10, 19, 26). The technique has also been successfully applied to enrich for microbial consortia or individual strains with enhanced capabilities for cellulose degradation (27) or scyllo-inosamine degradation (11). Nevertheless, while it is well known that enrichment cultures result in the loss of major portions of the microbial populations associated with any given environment (3), it is still possible for the enriched cultures to contain a highly diverse population of organisms (11, 19).
A major reason for the isolation of DNA from enrichment cultures arose from the difficulties associated with the isolation of high-molecular-mass DNA from microbial habitats. Although during the last decade different approaches for the isolation and purification of bacterial DNA from a variety of environments have been reported, the vast majority of the techniques described are not suitable for the efficient construction of libraries with large inserts. Most of the available techniques can be grouped in two major classes: first, the isolation of DNA after direct lysis of the microorganisms in the presence of the organic soil matrix; and second, the isolation of the DNA after separation of the bacteria from the soil matrix (16). However, one difficulty with both of these methods is that the purified DNA is often contaminated with polyphenolic compounds, which are copurified with the DNA. The contaminating compounds are difficult to remove, and it is well known that the polyphenols interfere with enzymatic modifications of the isolated DNA (16). As a result, the construction of environmentally derived DNA libraries with large inserts is almost impossible due to the poor quality of the isolated DNA. In two recently published model studies, the isolated DNA has been used for the construction of DNA libraries leading to the isolation of genes with desirable industrial traits (15, 25). Our attempts to construct DNA libraries with large inserts following the protocol published by Henne et al. (15), i.e., via extraction of the DNA directly from the different environmental samples failed. Therefore, we have developed a strategy that allows us to isolate highly pure DNA from microbial consortia after enrichment culture and that would allow construction of DNA libraries with insert sizes larger than 30 kb (Fig. 2). The construction of libraries with sufficiently large inserts was essential to allow screening for the presence of complete bacterial operons encompassing 5 to 6 kb in size.
In this work, we inoculated five enrichment cultures with environmental samples, four from soil and one from horse excrement (Fig. 2; Table 2). Because of the high selection pressure caused by the presence of the biotin-complexing agent avidin in the medium, bacteria able to synthesize high amounts of biotin were primarily enriched. Not much is known about the biotin biosynthesis rates of bacteria in a natural environment, and it can only be assumed that most microorganisms in microbial habitats are prototrophs. Only data on actual rates of biotin production of wild-type isolates in pure cultures are available. In our studies E. coli K-12 produced approximately 8 pg of biotin/ml in 24 h on a defined medium (Table 3); a Klebsiella pneumoniae isolate produced 350 pg/ml. Two typical soil bacteria, Rhizobium meliloti 1021 and Ralstonia eutropha H16, however, produced in the same time under the same conditions less than 10 pg of biotin/ml, which was below the detection limit.
DNA extracted from the enrichment cultures was sufficiently pure to allow the construction of cosmid libraries harboring large (>30-kb) inserts. Although the applied strategy likely resulted in the loss of major portions of the populations associated with each of the microbial communities (Table 2), we were able to use this method to isolate seven different cosmids complementing an E. coli bio mutant. Although we have not characterized all of the bio cosmids in detail on a molecular level, we can assume that each of the seven isolated cosmid clones carries a complete biotin biosynthesis operon; this is primarily because the E. coli auxotrophic mutant used can grow only if a complete bio operon is present. Four of the seven isolated cosmids conferring the ability to synthesize d-biotin were subject to a detailed study. Amounts of biotin produced were measured, and the results demonstrated that each of these bio clones produced significant amounts of biotin, indicating the successful expression of the identified operons.
DNA sequencing revealed that one of the isolated biotin biosynthesis operons was highly similar to the known E. coli bio operon (Table 4). The other identified biotin synthesis genes showed highest similarities to corresponding genes from E. herbicola and S. marcescens. Both microorganisms are closely related and belong to the family Enterobacteriaceae. Although the observed similarities were surprisingly high for several of the identified genes, we have no evidence indicating from which species the bio cosmids were derived. The isolated biotin synthesis operons appear to have the same bidirectional organization as the biotin biosynthesis operons of E. coli and other closely related organisms (Fig. 4) (2, 8, 12, 14). This suggests the possibility that the conserved organization of these genes is important for the synthesis of biotin or for its regulation. However, general rules for the organization of bio genes cannot be made. Finally, it must be kept in mind that our screen approach based on the complementation of an E. coli biotin auxotrophic mutant lacking the entire bio gene cluster allowed the isolation only of biotin biosynthesis operons whose promoters could be recognized by the host strain. This explains why the total number of bio+ clones isolated form an average of 6,000 to 30,000 screened clones is relatively low. Thus, we can only speculate that the use of a different screening system (e.g., a biotin-auxotrophic Bacillus strain) would have resulted in the isolation of bio genes closely related to gram-positive bacteria. Also the use of a mobilizable expression vector might have increased the number of resulting bio clones.
Although the isolated biotin biosynthesis operons were similar in structure, major differences were observed within the flanking regions (Fig. 4; Table 4). While in E. coli the galactose operon is upstream of the bio operon (2), we found the histidine utilization operon (hut) upstream of bioA in two of the characterized operons (Fig. 4). The presence of the hut operon upstream of the biotin biosynthesis genes has been described for Salmonella enterica serovar Typhimurium and K. aerogenes (12). However, none of the isolated bio cosmids shows a sufficient similarity to the bio genes isolated from S. enttrica serovar Typhimurium and K. aerogenes to warrant the claim that it was isolated from one of those organisms. In addition to the hut operon, we have identified the moaABCDE genes in pCosHE1, coding for genes involved in the synthesis of molybdopterin cofactors. Those genes are commonly found downstream of bio operons in gram-negative bacteria. The moa genes and the hut genes are two more examples of genes that are organized in a gene cluster and which we could isolate as a whole with the combined cloning and enrichment technique used in this work. Consequently, this technique will be a valuable tool for the isolation of operons or conserved gene clusters from environmentally derived microbial consortia.
ACKNOWLEDGMENTS
This work was supported by a grant from the Deutsche Bundesstiftung Umwelt and by the Fonds der chemischen Industrie.
W.R.S. thanks A. E. Shauger for correcting the English and for critical reading of the manuscript.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Caldwell D E, Wolfaardts G M, Korber D R, Lawrence J R. Cultivation of microbial consortia and communities. In: Hurst J H, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C.: ASM Press; 1997. pp. 79–90. [Google Scholar]
- 4.Chang Y S, Wu C H, Chang R J, Shiuan D. Determination of biotin concentration by a competitive enzyme-linked immunosorbent assay (ELISA) method. J Biochem, Biophys, Methods. 1994;29:321–329. doi: 10.1016/0165-022x(94)90042-6. [DOI] [PubMed] [Google Scholar]
- 5.Cheesman P, Pai C H. Partial purification and properties of d-dethiobiotin synthetase. J Bacteriol. 1970;104:726–733. doi: 10.1128/jb.104.2.726-733.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dedysh S N, Panikov N S, Tiedje J M. Acidophilic methanotrophic communities from Sphagnum peat bogs. Appl Environ Microbiol. 1998;64:922–929. doi: 10.1128/aem.64.3.922-929.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeMoll E, Shive W. Assay for biotin in the presence of dethiobiotin with Lactobacillus plantarum. Anal Biochem. 1986;158:55–58. doi: 10.1016/0003-2697(86)90587-7. [DOI] [PubMed] [Google Scholar]
- 8.Eisenberg M A. Regulation of the biotin operon in E. coli. Ann N Y Acad Sci. 1985;447:335–349. doi: 10.1111/j.1749-6632.1985.tb18449.x. [DOI] [PubMed] [Google Scholar]
- 9.Eisenberg M A, Star C. Synthesis of 7-oxo-8-aminopelargonic acid, a biotin vitamer, in cell-free extracts of Escherichia coli biotin auxotrophs. J Bacteriol. 1968;96:1291–1297. doi: 10.1128/jb.96.4.1291-1297.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fogel M M, Taddeo A R, Fogel S. Biodegradation of chlorinated ethenes by a methane-utilizing mixed culture. Appl Environ Microbiol. 1986;51:720–724. doi: 10.1128/aem.51.4.720-724.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardener B B M, de Bruijn F J. Detection and isolation of novel rhizopine-catabolizing bacteria from the environment. Appl Environ Microbiol. 1998;64:4944–4949. doi: 10.1128/aem.64.12.4944-4949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg R B, Magasanik B. Gene order of the histidine utilization (hut) operons in Klebsiella aerogenes. J Bacteriol. 1975;122:1025–1031. doi: 10.1128/jb.122.3.1025-1031.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto-Gotoh T, Kume A, Masahashi W, Takeshita S, Fukuda A. Improved vector, pHSG664, for direct streptomycin-resistance selection: cDNA cloning with G:C-tailing procedure and subcloning of double-digest DNA fragments. Gene. 1986;41:125–128. doi: 10.1016/0378-1119(86)90275-1. [DOI] [PubMed] [Google Scholar]
- 14.Hejazi A, Falkiner F R. Serratia marcescens. J Med Microbiol. 1997;46:903–912. doi: 10.1099/00222615-46-11-903. [DOI] [PubMed] [Google Scholar]
- 15.Henne A, Daniel R, Schmitz R A, Gottschalk G. Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl Environ Microbiol. 1999;65:3901–3907. doi: 10.1128/aem.65.9.3901-3907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holben W E. Isolation and purification of bacterial community DNA from environmental samples. In: Hurst J H, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C.: ASM Press; 1997. pp. 431–436. [Google Scholar]
- 17.Hubert C, Shen Y, Voordouw G. Composition of toluene-degrading microbial communities from soil at different concentrations of toluene. Appl Environ Microbiol. 1999;65:3064–3070. doi: 10.1128/aem.65.7.3064-3070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ifuku O, Kishimoto J, Haze S, Yanagi M, Fukushima S. Conversion of dethiobiotin to biotin in cell-free extracts of Escherichia coli. Biosci Biotechnol Biochem. 1992;56:1780–1785. doi: 10.1271/bbb.56.1780. [DOI] [PubMed] [Google Scholar]
- 19.Kengen S W, Breidenbach C G, Felske A, Stams A J, Schraa G, de Vos W M. Reductive dechlorination of tetrachloroethene to cis-1, 2-dichloroethene by a thermophilic anaerobic enrichment culture. Appl Environ Microbiol. 1999;65:2312–2316. doi: 10.1128/aem.65.6.2312-2316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleerebezem R, Hulshoff Pol L W, Lettinga G. Anaerobic degradation of phthalate isomers by methanogenic consortia. Appl Environ Microbiol. 1999;65:1152–1160. doi: 10.1128/aem.65.3.1152-1160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolmar H, Friedrich K, Pschorr J, Fritz H J. Hybrids of circular DNA single strands as intermediates in DNA cloning sequence analysis and directed mutagenesis. Technique. 1990;2:137–145. [Google Scholar]
- 22.Krell K, Gottesman M E, Parks J S, Eisenberg M A. Escape synthesis of the biotin operon in induced lambda b-2 lysogens. J Mol Biol. 1972;68:69–82. doi: 10.1016/0022-2836(72)90263-x. [DOI] [PubMed] [Google Scholar]
- 23.Ogram A, Feng X. Methods of soil microbial community analysis. In: Hurst J H, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C.: ASM Press; 1997. pp. 422–430. [Google Scholar]
- 24.Pai C H. Partial purification and properties of d-dethiobiotin synthetase. J Bacteriol. 1971;105:793–800. [Google Scholar]
- 25.Rondon M R, August P R, Bettermann A D, Brady S F, Grossman T H, Liles M R, Loiacono K A, Lynch B A, MacNeil I A, Minor C, Tiong C L, Gilman M, Osburne M S, Clardy J, Handelsman J, Goodman R M. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rooney-Varga J N, Anderson R T, Fraga J L, Ringelberg D, Lovley D R. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl Environ Microbiol. 1999;65:3056–3063. doi: 10.1128/aem.65.7.3056-3063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saddler J N, Khan A W. Cellulose degradation by a new isolate from sewage sludge, a member of the Bacteroidaceae family. Can J Microbiol. 1979;25:1427–1432. doi: 10.1139/m79-222. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 30.Staskawiez B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steffan R J, Goksoyr J, Bej A K, Atlas R M. Recovery of DNA from soils and sediments. Appl Environ Microbiol. 1988;54:2908–2915. doi: 10.1128/aem.54.12.2908-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Streit W, Bjourson A T, Cooper J E, Werner D. Application of subtraction hybridization for the development of a Rhizobium phaseoli and a Rhizobium tropici group-specific DNA probe for monitoring Rhizobium populations in soils. FEMS Microbiol Ecol. 1993;13:59–67. [Google Scholar]
- 33.Torsvik V, Goksoyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai Y L, Olson B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai Y L, Olson B H. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl Environ Microbiol. 1992;58:2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]