SUMMARY
G protein-coupled receptors (GPCRs) are critical mediators of cell signaling. Although capable of activating G proteins in a monomeric form, numerous studies reveal a possible association of class A GPCRs into dimers/oligomers. The relative location of individual protomers within these GPCR complexes remains a topic of intense debate. We previously reported that class A serotonin 5-HT2A receptor (5-HT2AR) and class C metabotropic glutamate 2 receptor (mGluR2) are able to form a GPCR heterocomplex. By introducing the photoactivatable unnatural amino acid p-azido-L-phenylalanine (azF) at selected individual positions along the transmembrane (TM) segments of mGluR2, we delineate the residues that physically interact at the heteromeric interface of the 5-HT2AR-mGluR2 complex. We show that 5-HT2AR crosslinked with azF incorporated at the intracellular end of mGluR2’s TM4, while no crosslinking was observed at other positions along TM1 and TM4. Together, these findings provide important insights into the structural arrangement of the 5-HT2AR-mGluR2 complex.
Graphical Abstract
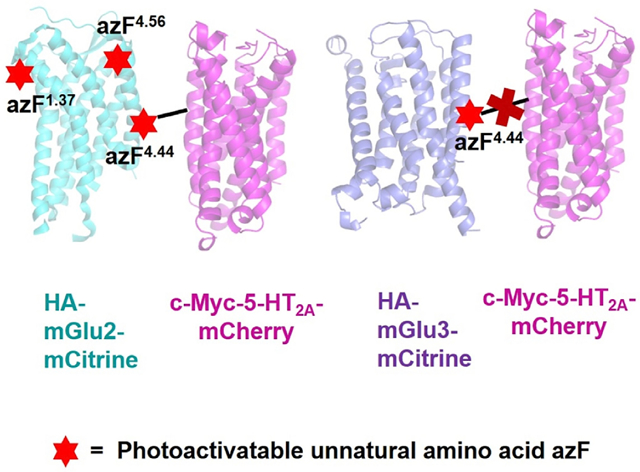
In Brief
Using site-specific incorporation of photoactivatable unnatural amino acids, Shah et al. identified that the intracellular end of TM4 of mGluR2 physically interacts with 5-HT2AR. These findings provide important insights into the structural arrangement of the 5-HT2AR-mGluR2 complex. This versatile method can be applied to study protein-protein complexes.
INTRODUCTION
G protein-coupled receptors (GPCRs), which share a common structural signature of seven hydrophobic transmembrane (TM) domains, represent the largest family of receptors in the human genome, and are a common target for therapeutic agents and drugs of abuse (Wacker et al., 2017; Hilger et al., 2018). Although most plasma membrane proteins, including enzyme-linked receptors and ligand-gated ion channels, exist and function as protein-protein complexes, GPCRs were traditionally assumed to recruit heterotrimeric G proteins and hence downstream signaling events as monomeric functional entities. This concept was challenged by the convincing demonstration that class C GPCRs, such as metabotropic glutamate receptors (mGluRs) and γ-aminobutyric acid B (GABAB) receptor, behave as obligate dimers (Niswender and Conn, 2010; Kniazeff et al., 2011; Pin and Bettler, 2016). Accordingly, follow-up studies provided further evidence that particular class A GPCRs, such as dopamine D2 and α1B-adrenergic receptors, can assemble as dimers or oligomers in living mammalian cells (Lopez-Gimenez et al., 2007; Guo et al., 2008). Based on these and other findings, it is now becoming clear that, whereas certain class A GPCRs signal as monomers, processes of trafficking and signaling of some class A GPCRs can only be explained by their homo/heteromerization (Milligan, 2009; González-Maeso, 2011; Ferre et al., 2014; Sleno and Hebert, 2019). Nevertheless, certain structural properties of these GPCR complexes, such as the inter-protomer interface within homo/heteromers, remain a topic of intense debate.
For example, the first X-ray and electron crystallographic studies of class A GPCRs identified TM1 and helix 8 as the possible GPCR dimer interface (Ruprecht et al., 2004; Cherezov et al., 2007), yet more recent ones revealed direct contacts through TM4, TM5, and TM6 (Wu et al., 2010; Manglik et al., 2012). The crystal structures of the mGluR1 7TM domain (i.e., mGluR1 construct without its relatively large amino-terminal extracellular domain, which is composed of a ligand-binding Venus flytrap [VFT] domain, and a cysteine-rich domain [Pin and Bettler, 2016]) showed a parallel dimer interface mediated mainly through TM1 (Wu et al., 2014). More recent findings based on crystal structures and cryoelectron microscopy (cryo-EM) of full-length mGluR5 reported that, for the inactive (apo) state, the two 7TM domains within an mGluR5 homodimer are well separated, whereas the presence of an orthosteric agonist leads to a physical interaction between TM6 and TM6 that initiates signaling (Koehl et al., 2019). These structural insights were obtained after protein purification or reconstitution into high-density lipoprotein particles.
In living mammalian cells, studies based on crosslinking of substituted cysteine residues suggest that the main mGluR2 homodimeric interface in its apo state is formed by TM4 and TM5 (Xue et al., 2015). As mGluR homodimers are already covalently linked by a disulfide bridge between their VFT domains (Pin and Bettler, 2016), this cysteine crosslinking approach was achieved with a mutant mGluR2 in which C121 was converted into alanine (C121A). However, previous results demonstrated that this particular mutation (C121A) alters the structure and dynamics of the mGluR2 homodimer (Levitz et al., 2016; Koehl et al., 2019). This raises questions regarding the relative orientation of the two mGluR2-C121A protomers as compared with those in the “wild-type” mGluR2 homodimer.
Within this context, we have previously shown that mGluR2 and the class A serotonin 5-HT2A receptor (5-HT2AR) are able to interact to form an inter-class GPCR heteromer (Gonzalez-Maeso et al., 2008; Fribourg et al., 2011). We also reported three residues located at the intracellular end of TM4 (A6774.40, A6814.44, and A6854.48) that are necessary for the mGluR2 to form a complex with 5-HT2AR in living HEK293 cells (Moreno et al., 2012, 2016) (superscripts in this form indicate [Ballesteros and Weinstein, 1995] numbering for conserved GPCR residues). These data provided the first evidence for specific protein domains responsible for inter-class GPCR heteromeric complex formation, yet they still did not yield direct information about the nature of the heteromeric interface. Thus, two alternative explanations can be put forward: (1) residues located at the TM4 of the mGluR2 are directly interacting with 5-HT2AR and (2) substitution of these residues distorts mGluR2 conformation and hence indirectly affects formation of the 5-HT2AR-mGluR2 complex through other protein domains. To solve this, here we incorporated photo-crosslinking amino acids at select individual positions along the TMs of mGluR2, and tested the ability of these constructs to photo-crosslink 5-HT2AR.
RESULTS
Incorporation of azF at Selected Individual Positions of mGluR2
The photo-crosslinking approach relies on the recognition of a nonsense codon, such as the low-abundance amber (TAG) stop codon, in the DNA construct of interest (Huber and Sakmar, 2014; Serfling and Coin, 2016). Using an orthogonal tRNA that is aminoacetylated with photoreactive unnatural amino acids (UAAs), such as p-azido-L-phenylalanine (azF) by an aminoacyl tRNA synthetase (aaRS), this method enables in cellulo incorporation of photo-crosslinking amino acids, which crosslink upon UV radiation (Figure S1A). Importantly, the technology does not require the introduction of the C121A mutation, which, as discussed above, may negatively affect mGluR2 homodimer structure— a concept that is further supported by our current findings (Figures S1B and S1C).
To incorporate azF into the mGluR2, the amber stop codon was inserted at three different sites located specifically at the extracellular half of TM1 (V5691.37), the intracellular half of TM4 (A6814.44), or the extracellular half of TM4 (I6934.56) (Figure 1A). The position A6814.44 was selected because our previous data demonstrated that substitution of any two of three residues located at the intracellular end of TM4 of mGluR2 (A6774.40, A6814.44, or A6854.48) with those at equivalent positions in the TM4 of the closely related mGluR3 is sufficient to disrupt its heteromeric association with 5-HT2AR (Moreno et al., 2012; Moreno et al., 2016). Position I6934.56 was chosen because previous findings suggested a parallel homomeric interface along the entire TM4 of class A GPCRs (Guo et al., 2003), whereas position V5691.37 was picked because previous cysteine crosslinking assays showed that TM1 facilitates the formation of class A GPCR dimer rows (Guo et al., 2008). In addition, these three residues (V5691.37, A6814.44, and I6934.56) are exposed to the lipid based on 3D models of monomeric mGluR2 (Figures 1A and 1B), and are different in the primary sequence between mGluR2 and mGluR3 to justify specificity of 5-HT2AR-mGluR2 complex formation. All the mGluR2 azF constructs were also tagged with the epitope HA at the N terminus, and sub-cloned in frame with the yellow fluorescent protein (mCitrine) at the C terminus (HA-mGluR2-TAG1.37-mCitrine, HA-mGluR2-TAG4.44-mCitrine, and HA-mGluR2-TAG4.56-mCitrine).
Figure 1. Introduction of the UAA azF into mGluR2.
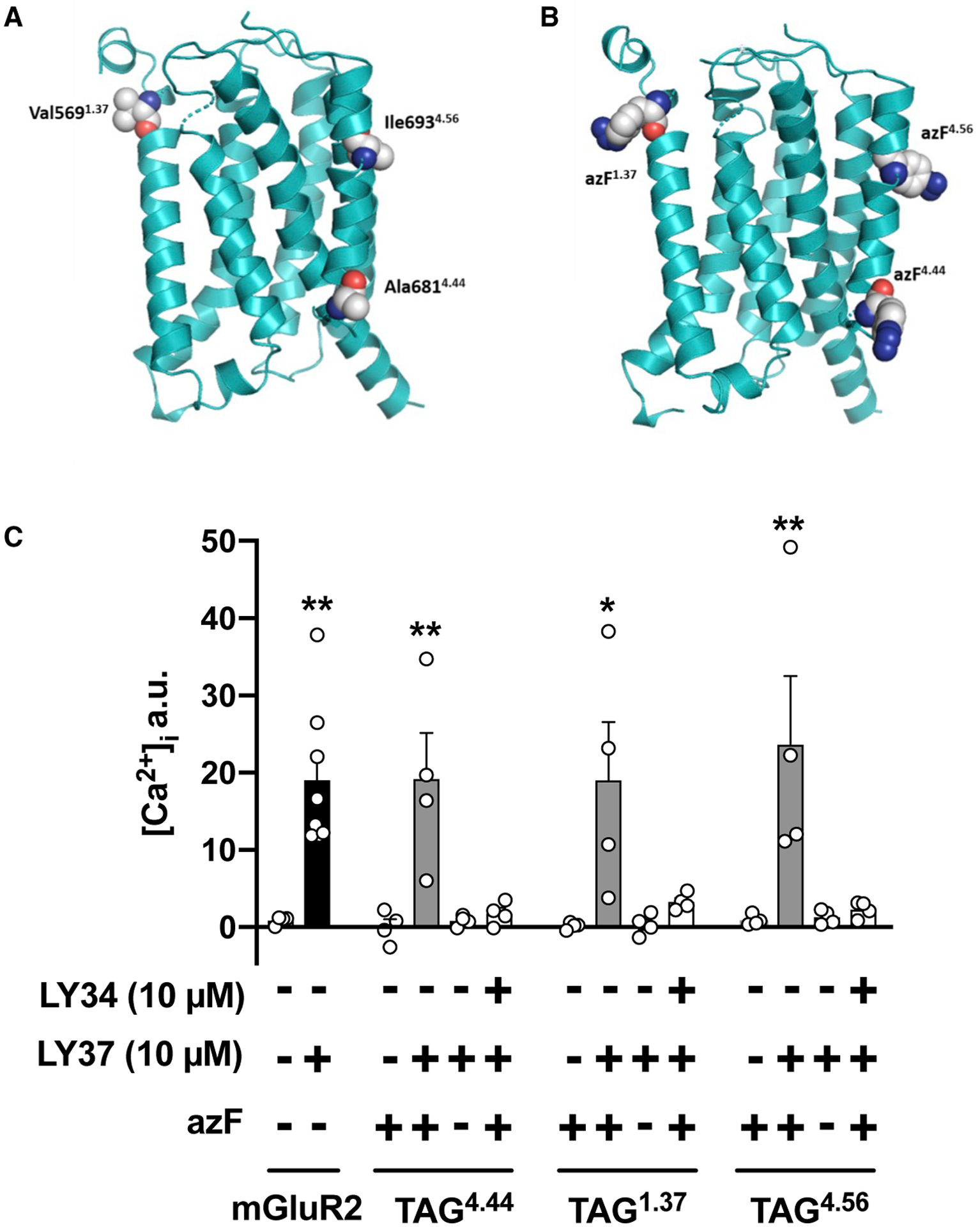
(A and B) Molecular model of the mGluR2 7TM domain. Residues V5691.37, A6814.44, and I6934.56 in mGluR2 (A) and the corresponding location of azF in the TAG HA-mGluR2 constructs (B) are shown as spheres
(C) Cells co-transfected with constructs encoding suppressor tRNA and azF aaRS, along with HA-mGluR2-TAG1.37-mCitrine, HA-mGluR2-TAG4.44-mCitrine, or HA-mGluR2-TAG4.56-mCitrine, were loaded with Fura-2 and monitored for intracellular calcium release after sequential administration of LY341495 (LY34) and/or LY379268 (LY37), or vehicle. Experiments were carried out with and without the unnatural amino acid azF. Controls included cells transfected with the wild-type HA-mGluR2-mCitrine construct (n = 4–8 independent experiments per experimental condition). Mean ± SEM. *p < 0.05 and **p < 0.01 by Student’s t test (mGluR2) or Dunnett’s post hoc test of one-way ANOVA (TAG mGluR2 constructs).
See also Figure S1
We first examined functional responses in HEK293 cells co-transfected with plasmids that encode the suppressor tRNA and azF aaRS, along with both the appropriate TAG HA-mGluR2-mCitrine construct (i.e., HA-mGluR2-TAG1.37-mCitrine, HA-mGluR2-TAG4.44-mCitrine, or HA-mGluR2-TAG4.56-mCitrine) and the chimeric Gα subunit Gαqi9, which allows activation of phospholipase C (PLC) and hence induction of a transient increase in the concentration of intracellular Ca2+ by an otherwise Gi/o-coupled GPCR (Xue et al., 2015), including the mGluR2. Upon exposure to the mGluR2/3 agonist LY379268 (10 μM), this increase in [Ca2+]i was corroborated in cells co-transfected with the wild-type HA-GluR2-mCitrine and Gαqi9 (Figure 1C). Importantly, LY379268 (10 μM) was able to recruit G protein-dependent signaling in cells co-transfected with suppressor tRNA, azF aaRS, and HA-mGluR2-TAG1.37- mCitrine, HA-mGluR2-TAG4.44-mCitrine, or HA-mGluR2-TAG4.56-mCitrine only when azF was added to the culture medium (Figure 1C). This effect of LY379268 was pharmacologically blocked by the mGluR2/3 antagonist LY341495 (Figure 1C). Additional controls that validate the specificity of this assay included absence of effect of LY379268 (10 and 100 μM) on G protein-dependent signaling in HEK293 cells co-transfected with the suppressor tRNA, azF aaRS, and the appropriate TAG HA-mGluR2-mCitrine construct, but not with Gαqi9 (Figure S2A). Furthermore, inappreciable effect of LY379268 on G protein-dependent signaling was detected in cells transfected with Gαqi9 only (Figure S2A).
Characterization of mGluR2 Containing azF along TM1 and TM4
We next explored the length of the translated constructs by immunoblot assays with anti-HA antibodies. In SDS-polyacryl-amide gels of membrane preparation material, upon addition of azF, anti-HA immunoreactivity was detected as a mixture of an ~70–75 kDa polypeptide and two bands at ~120 and ~240 kDa. These bands in the ~120- and ~240-kDa segments correspond to the molecular weights of wild-type HA-mGluR2-mCitrine monomer and dimer, respectively (Figure 2A). Absence of azF almost completely eliminated immunoblot signaling at ~ 120 and ~240 kDa, which confirms that incorporation of this UAA is required for translation of a full-length GPCR construct (Figure 2A). Site-specific incorporation of azF into the appropriate TAG HA-mGluR2-mCitrine construct was corroborated by immunoblot assays with the anti-HA antibody in membrane preparations of cells co-transfected with HA-mGluR2-TAG4.56- mCitrine, HA-mGluR2-TAG4.44-mCitrine, or HA-mGluR2-TAG1.37-mCitrine, along with suppressor tRNA and azF aaRS alone, or together (Figure 2B). Visualization of mCitrine and anti-HA staining of cells confirmed that the mCitrine signal for these constructs is observed only after azF treatment, which demonstrates detection of full-length HA-mGluR2-TAG4.44-mCitrine, HA-mGluR2-TAG4.56-mCitrine, or HA-mGluR2-TAG1.37-mCitrine (Figures 2C–2E). After addition of azF, radioligand binding saturation curves with the mGluR2/3 antagonist [3H] LY341495 in membrane preparations revealed that the level of expression of HA-mGluR2-TAG4.44-mCitrine, HA-mGluR2-TAG4.56-mCitrine, or HA-mGluR2-TAG1.37-mCitrine was relatively similar (Figures 2F–2H; Table S1). Most importantly, the capacity of HA-mGluR2-TAG4.44-mCitrine, HA-mGluR2-TAG4.56-mCitrine, or HA-mGluR2-TAG1.37-mCitrine to bind [3H] LY341495 was significantly reduced in HEK293 cells untreated with azF (Figures 2F–2H; Table S1). Density (Bmax values) of wild-type HA-mGluR2-mCitrine was significantly higher as compared with HA-mGluR2-TAG4.44- mCitrine, HA-mGluR2-TAG4.56-mCitrine, or HA-mGluR2-TAG1.37-mCitrine (Table S1), whereas affinities (KD values) were comparable (Table S1; see also Figure S2B for visualization of mCitrine and anti-HA staining in cells transfected with HA-mGluR2-mCitrine). Together with their functional properties (see Figure 1C, above), this absence of effect on KD values further suggests that introduction of azF at positions 4.44, 4.56, or 1.37 does not significantly disrupt the functional properties of the mGluR2 construct. Most importantly, these findings also demonstrate the feasibility of incorporating the photo-crosslinking amino acid azF into GPCRs upon transfection of HEK293 cells carrying the appropriate TAG HA-mGluR2-mCitrine construct, along with an orthogonal suppressor tRNA/aaRS pair for azF.
Figure 2. Characterization of mGluR2 Containing the Photo-Crosslinking UAA azF at Selected Positions.
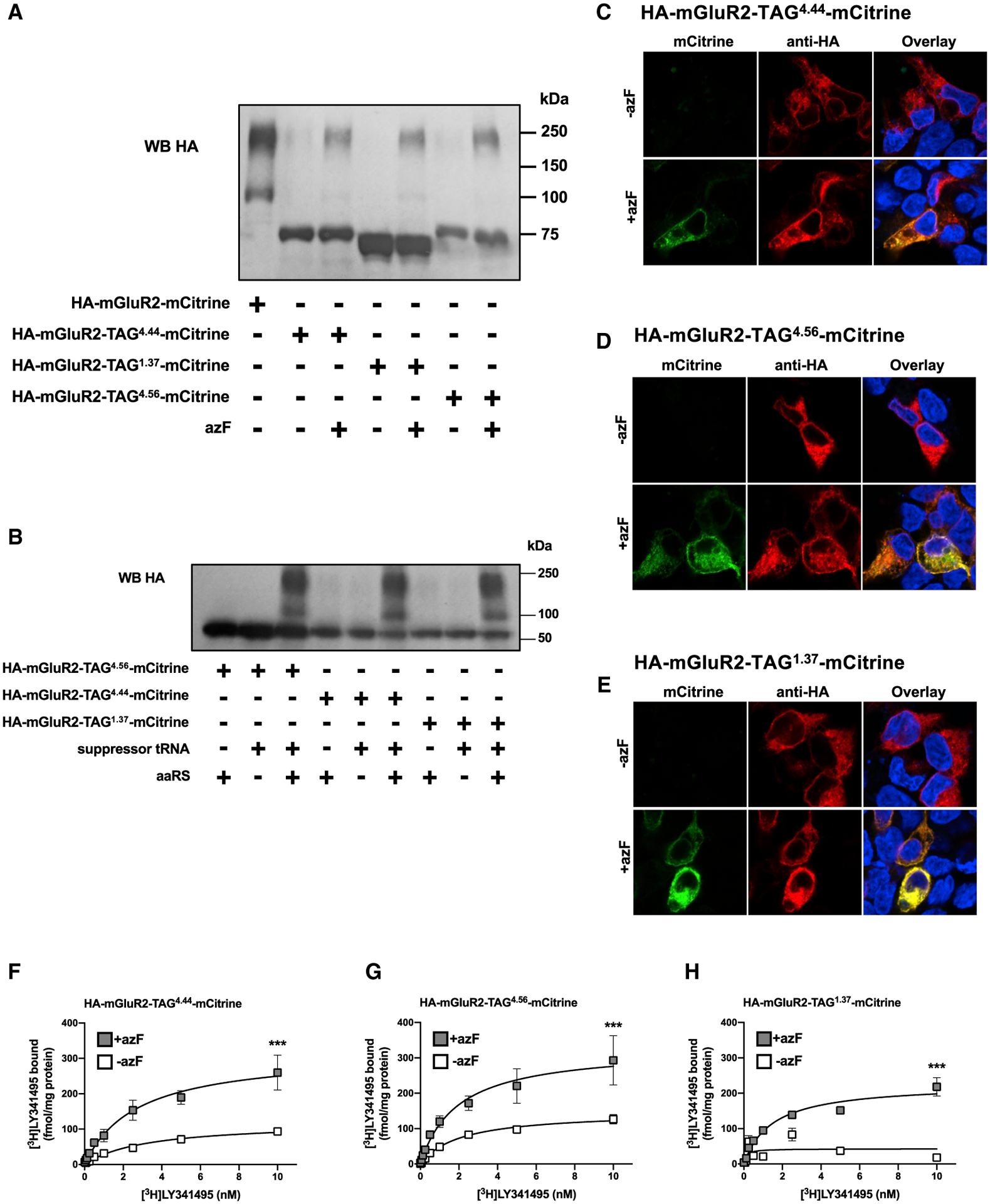
(A) Membrane preparations of cells exposed to azF or mock and co-transfected with constructs encoding suppressor tRNA and azF aaRS, along with HA-mGluR2-TAG1.37-mCitrine, HA-mGluR2-TAG4.44-mCitrine, or HA-mGluR2-TAG4.56-mCitrine were subjected to immunoblot assays with antibody against the HA tag. Bands in the ~120- and ~240-kDa segments correspond to the molecular weights of wild-type HA-mGluR2-mCitrine monomer and dimer, respectively; whereas bands in the ~70- to 75-kDa segments correspond to truncated TAG HA-mGluR2-mCitrine constructs where the amber codon is recognized as a stop codon rather than a codon for incorporation of the UAA azF.
(B) Membrane preparations of cells exposed to azF and co-transfected with constructs encoding suppressor tRNA and azF aaRS alone or together, along with HA-mGluR2-TAG1.37-mCitrine, HA-mGluR2-TAG4.44-mCitrine, or HA-mGluR2-TAG4.56-mCitrine were subjected to immunoblot assays with antibody against the HA tag. Western blots (A and B) are representative of three independent experiments.
(C–E) Cells exposed to azF (+azF) or mock (−azF) and co-transfected with constructs encoding suppressor tRNA and azF aaRS, along with HA-mGluR2-TAG4.44- mCitrine (C), HA-mGluR2-TAG4.56-mCitrine (D), or HA-mGluR2-TAG1.37-mCitrine (E) were then permeabilized to detect the HA epitope and imaged to detect mCitrine fluorescence. Nuclei were stained in blue with Hoechst.
(F–H) Radioligand binding saturation curves with [3H]LY341495 in membrane preparations of cells exposed to azF (+azF) or mock (−azF) and co-transfected with constructs encoding suppressor tRNA and azF aaRS, along with HA-mGluR2-TAG4.44-mCitrine (F), HA-mGluR2-TAG4.56-mCitrine (G), or HA-mGluR2-TAG1.37-mCitrine (H). Mean ± SEM. ***p < 0.001 by F test (representative results of four independent experiments, see also Table S1). See also Figure S2
Residues at the 5-HT2AR-mGluR2 Heteromeric Interface Using Amber Codon Suppression
In preliminary experiments, plasmids that encode the suppressor tRNA and azF aaRS, along with HA-mGluR2-TAG4.44-mCitrine and/or c-Myc-5-HT2AR-mCherry were used for transfections. Using immunoblot assays immediately after exposure to UV-A, or mock, we found that UV-induced crosslinking occurred only in cells co-transfected with HA-mGluR2-TAG4.44-mCitrine and c-Myc-5-HT2AR-mCherry, but not in cells expressing HA-mGluR2-TAG4.44-mCitrine alone (Figures S3A–S3C). This suggests the specificity of the UV-dependent effect. However, probably because of factors, such as the substantial amount of 5-HT2AR that migrates as dimer and higher-order oligomer (Gonzalez-Maeso et al., 2008; Moreno et al., 2012; Moreno et al., 2016), our ability for quantification of individual segments corresponding to the appropriate molecular weight was limited. To bypass this problem, we combined the UV-induced crosslinking approach with co-immunoprecipitation assays.
Upon incubation with azF, the same group of transfected cells with suppressor tRNA, azF aaRS, and c-Myc-5-HT2AR-mCherry, along with the appropriate TAG HA-mGluR2-mCitrine construct (i.e., HA-mGluR2-TAG4.44-mCitrine, HA-mGluR2-TAG4.56-mCitrine, or HA-mGluR2-TAG1.37-mCitrine) was then divided into two equal portions that were either exposed to UV-A, or mock. These two portions were then processed for membrane preparation followed by co-immunoprecipitation assays (Figure 3A). We found that, in mock-exposed cells, introduction of azF at positions 4.44, 4.56, or 1.37 of the appropriate TAG HA-mGluR2-mCitrine construct does not significantly affect co-immunoprecipitation with c-Myc-5-HT2AR-mCherry (Figures 3B and 3C). This shows the important finding that HA-mGluR2-TAG4.44-mCitrine, HA-mGluR2-TAG4.56-mCitrine, or HA-mGluR2-TAG1.37-mCitrine are still able to be part of the same protein complex with c-Myc-5-HT2AR-mCherry. Notably, UV-induced crosslinking augmented co-immunoprecipitation in cells co-transfected with c-Myc-5-HT2AR-mCherry and HA-mGluR2-TAG4.44-mCitrine, an effect that was not observed in cells co-transfected with c-Myc-5-HT2AR-mCherry and HA-mGluR2-TAG4.56-mCitrine or HA-mGluR2-TAG1.37-mCitrine (Figures 3B–3D). This UV-dependent effect was not observed in cells expressing HA-mGluR2-TAG4.44-mCitrine alone (Figures 3E and 3F), which supports the concept that augmentation of immunoprecipitation signal upon UV exposure requires co-expression of 5-HT2AR and not random collision with other signaling proteins. Together, our findings show that, under these experimental conditions, there is a direct physical interaction between 5-HT2AR and mGluR2 that occurs through the intracellular end of TM4 of the mGluR2 promoter.
Figure 3. Crosslinking Studies and Heteromeric Interface Determination of the 5-HT2AR-mGluR2 Heterocomplex.
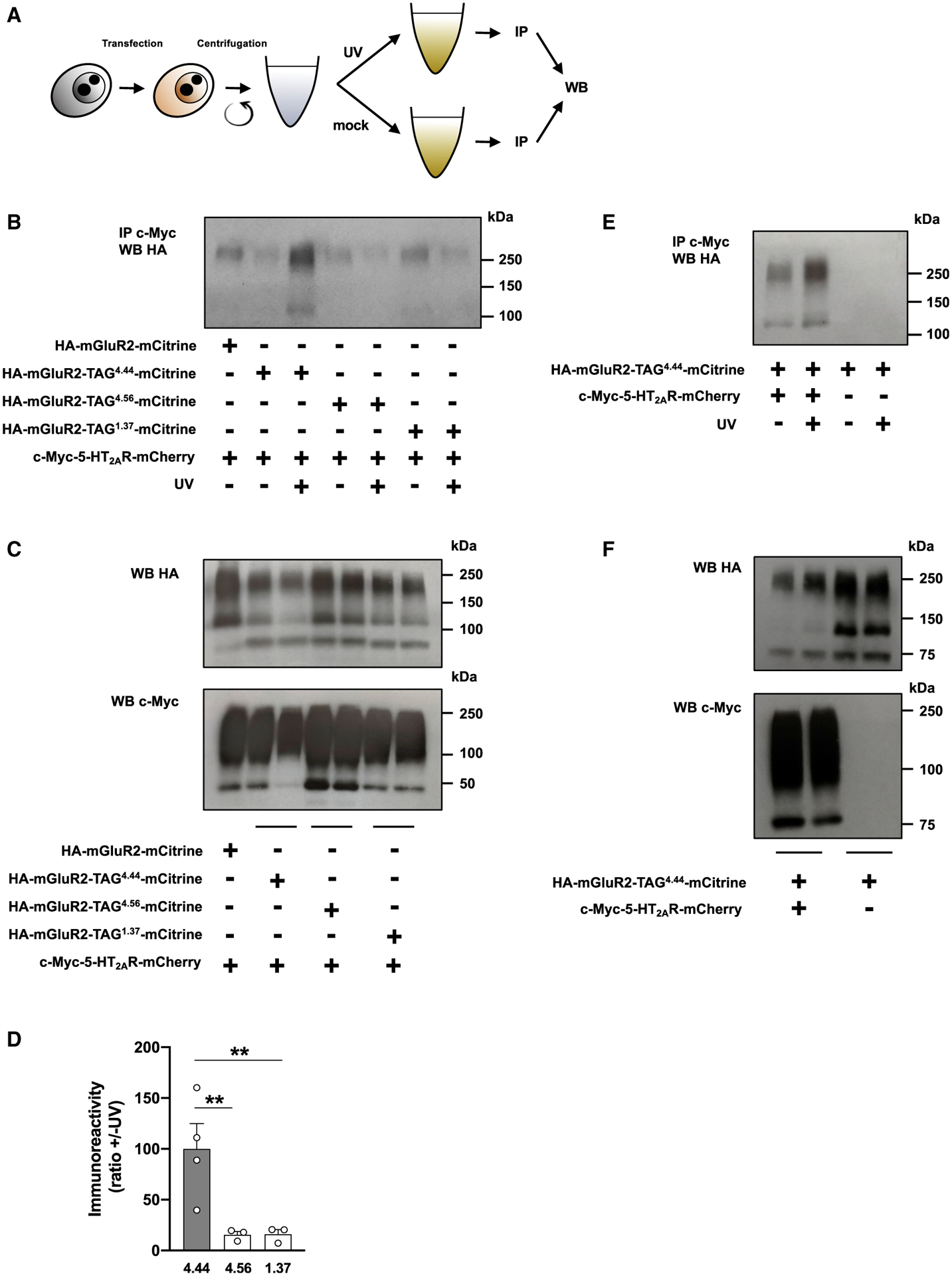
(A) Schematic illustration of the method of photo-crosslinking. Cells exposed to azF were co-transfected with constructs encoding suppressor tRNA and azF aaRS, along with c-Myc-5-HT2AR-mCherry and HA-mGluR2-TAG4.44-mCitrine, HA-mGluR2-TAG4.56-mCitrine, HA-mGluR2-TAG1.37-mCitrine, or HA-mGluR3-TAG4.44-mCitrine. After this manipulation, the same group of cells was divided into two equal portions that were either exposed to UV or mock. Cells (UV [+] and UV [−]) were afterward processed for membrane preparations and subjected to co-immunoprecipitation (IP) with an antibody against the c-Myc tag, and then analyzed by western blotting (WB) with an antibody against HA.
(B–D) Co-immunoprecipitation experiments of c-Myc-5-HT2AR-mCherry and HA-mGluR2-TAG4.44-mCitrine, HA-mGluR2-TAG4.56-mCitrine, or HA-mGluR2-TAG1.37-mCitrine in transfected cells (n = 3–4 separate experiments). For a control, co-immunoprecipitation experiments of c-Myc-5-HT2AR-mCherry and wild-type HA-mGluR2-mCitrine, where suppressor tRNA and azF aaRS were omitted, were assayed in parallel (B). Cell samples were also collected before UV or mock exposure, and analyzed by WB with antibodies against either HA or c-Myc (C). Representative immunoblots (B and C) and quantification of immunoreactivity (D). Mean ± SEM. **p < 0.01 by Dunnett’s post hoc test of one-way ANOVA.
(E and F) Absence of co-immunoprecipitation in cells co-transfected with constructs encoding suppressor tRNA, azF aaRS, and HA-mGluR2-TAG4.44-mCitrine, but not with the c-Myc-5-HT2AR-mCherry construct (E). Cell samples were also collected before UV or mock exposure, and analyzed by WB with antibodies against either HA or c-Myc (F).
See also Figures S1 and S3.
We previously reported that agonist activation of the Gi/o protein-coupled mGluR2 leads to Gq/11 protein-dependent increase in the concentration of intracellular Ca2+ (Moreno et al., 2016). We also demonstrated that the presence or absence of this crosstalk is profoundly affected by relative ratios and levels of expression of 5-HT2AR and mGluR2 (Gonzalez-Maeso et al., 2008; Baki et al., 2016; Moreno et al., 2016). Our data here showed that, even when the HA-mGluR2-TAG4.44-mCitrine construct was able to bind orthosteric antagonists and lead to functional outcomes upon agonist binding (see Figures 1C and 2F, above), the mGluR2/3 agonist LY379268 was not able to stimulate an increase in intracellular Ca2 release in cells cotransfected with HA-mGluR2-TAG4.44-mCitrine and c-Myc-5-HT2AR-mCherry (data not shown). This lack of crosstalk might be due to the significantly lower densities of the TAG HA-mGluR2-mCitrine constructs as compared with the wild-type HA-mGluR2-mCitrine in transfected cells (Table S1).
To further corroborate selectivity of the effect of UV exposure on co-immunoprecipitation signal, a similar group of experiments was designed using cells co-expressing HA-mGluR3-TAG4.44-mCitrine and c-Myc-5-HT2AR-mCherry. Importantly, although control assays validated the functional and biochemical properties of the HA-mGluR3-TAG4.44-mCitrine construct (Figures 4A–4D; Table S2), as well as the ability of HA-mGluR2-mCitrine, but not HA-mGluR3-mCitrine, to co-immunoprecipitate with c-Myc-5-HT2AR-mCherry (Figured S3D and S3E), our data showed that UV exposure does not affect the lack of co-immunoprecipitation of anti-HA immunoreactivity in cells co-expressing HA-mGluR3-TAG4.44- mCitrine and c-Myc-5-HT2AR-mCherry (Figures 4E and 4F).
Figure 4. Absence of UV-Induced Crosslinking in Cells Co-expressing 5-HT2AR and mGluR3.
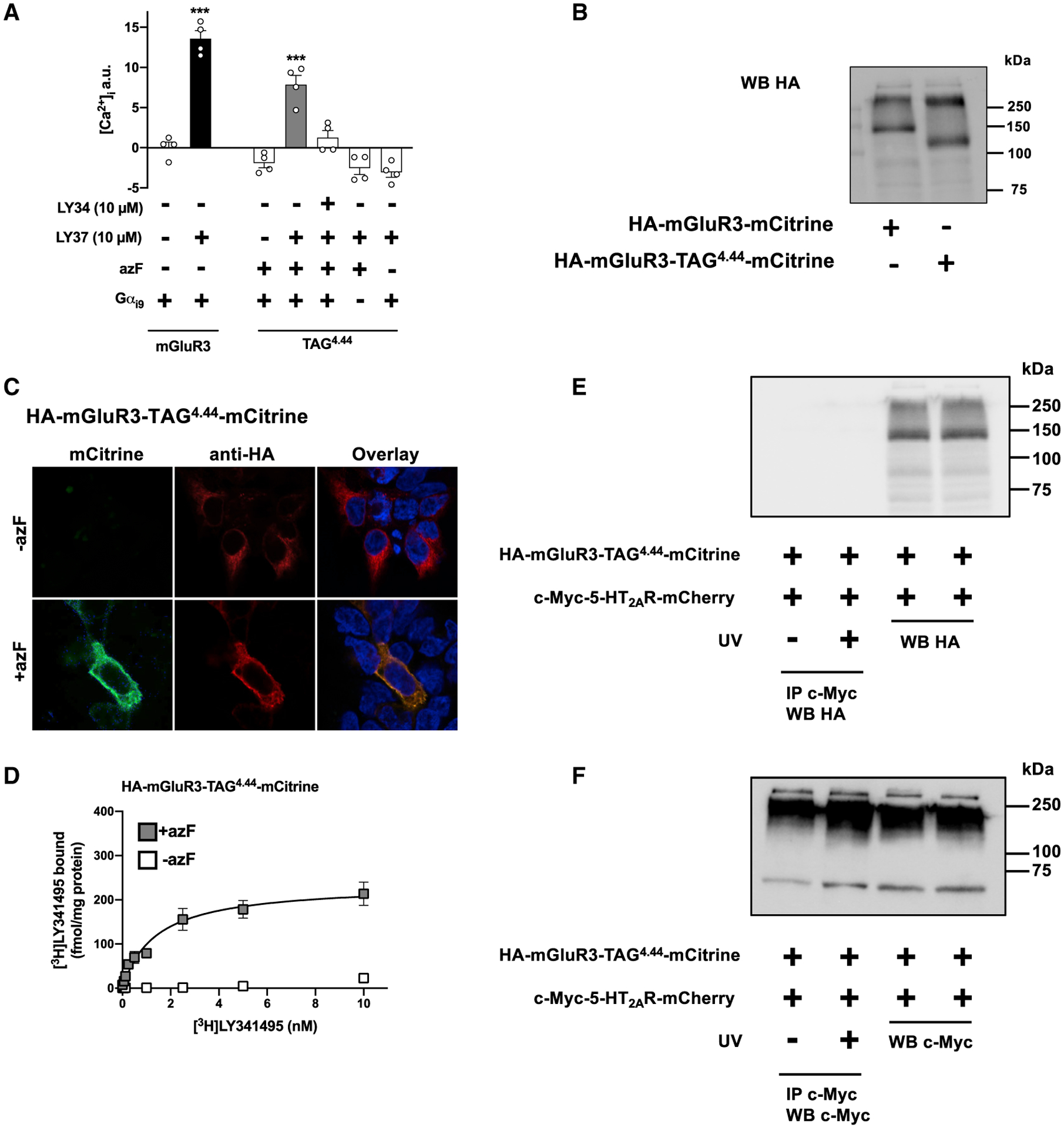
(A) Cells co-transfected with constructs encoding suppressor tRNA and azF aaRS, along with HA-mGluR3-TAG4.44-mCitrine, were loaded with Fura-2 and monitored for intracellular calcium concentration after sequential administration of LY341495 (LY34) and/or LY379268 (LY37), or vehicle. Controls included cells transfected with the wild-type HA-mGluR3-mCitrine construct, and cells untransfected with Gαi9 (n = 4 independent experiments per experimental condition). Mean ± SEM. ***p < 0.001 by Student’s t test (mGluR3) or Dunnett’s post hoc test of one-way ANOVA (TAG4.44).
(B) Membrane preparations of cells exposed to azF and co-transfected with constructs encoding suppressor tRNA and azF aaRS, along with HA-mGluR3-TAG4.44-mCitrine were subjected to immunoblot assays with antibody against the HA tag. The wild-type HA-mGluR3-mCitrine construct was used as a control. Bands in the ~130- and ~260-kDa segments correspond to the molecular weights of wild-type HA-mGluR3-mCitrine monomer and dimer, respectively; whereas the band in the ~110-kDa segment corresponds to the truncated HA-mGluR3-TAG4.44-mCitrine construct where the amber codon is recognized as a stop codon rather than a codon for incorporation of the UAA azF.
(C) Cells exposed to azF (+azF) or mock (−azF) and co-transfected with constructs encoding suppressor tRNA and azF aaRS, along with HA-mGluR3-TAG4.44- mCitrine were then permeabilized to detect the HA epitope and imaged to detect mCitrine fluorescence. Nuclei were stained in blue with Hoechst.
(D) Radioligand binding saturation curves with [3H]LY341495 in membrane preparations of cells exposed to azF (+azF) or mock (−azF) and co-transfected with constructs encoding suppressor tRNA and azF aaRS, along with HA-mGluR3-TAG4.44-mCitrine. Mean ± SEM. Non-linear regression analysis (binding saturation curve) was not applicable in mock-treated (−azF) cells (representative results of four independent experiments, see also Table S2).
(E and F) Co-immunoprecipitation experiments of c-Myc-5-HT2AR-mCherry and HA-mGluR3-TAG4.44-mCitrine. Cell samples were also collected before UV or mock exposure, and analyzed by WB with antibodies against either HA (E) or c-Myc (F). Western blots are representative of three independent experiments.).
See also Figure S3
DISCUSSION
Methods currently used to assess GPCR homo/heteromerization are mostly based on resonance energy transfer techniques, such as bioluminescence resonance energy transfer (BRET) and Förster resonance energy transfer (FRET). These are indirect approaches that rely on energy transfer between fluorescent and/or bioluminescent donor and acceptor proteins covalently linked to GPCR constructs (Wu and Brand, 1994). In addition, the distance needed to detect BRET or FRET signal between donor and acceptor is approximately 10–100Å, which does not fully demonstrate a direct physical proximity between the two GPCR protomers.
Protein crosslinking refers to the formation of one or more covalent bonds between amino acid residues, which results in the linking of distinct proteins and peptides and hence the formation of protein networks (Lossl and Sinz, 2016). Cysteine amino acids contain a highly reactive thiol group. Thus, when a pair of cysteines is exposed to each other in close molecular proximity, a disulfide bond can form between them, catalyzed by ambient oxygen or oxidizing agents, such as copper sulfate and 1,10-phenanthroline (Lossl and Sinz, 2016; Serfling and Coin, 2016). Cysteine is, however, one of the building blocks of virtually every protein, including GPCRs. Previous studies based on this cysteine crosslinking approach used GPCR constructs in which some of the highly conserved cysteine residues were substituted for nonpolar or polar amino acids, such as alanine or serine. This particular approach may, therefore, negatively impact structure and signaling properties of the tested cysteine-substituted GPCR construct. An important recent advance in structural biology has been the site-specific incorporation of UAAs through an amber codon (Suchanek et al., 2005; Huber and Sakmar, 2014). This site-specific incorporation of UAAs via genetic code expansion provides a powerful method to introduce synthetic moieties into specific positions along the protein of interest directly in live cells. Previous studies using mutational analysis and incorporation of photoreactive UAAs, such as azF and p-benzoyl-L-phenylalanine (BzF) into plasma membrane proteins have mapped out potential ligand-binding pockets within GPCRs (Ye et al., 2008; Grunbeck et al., 2011; Huber and Sakmar, 2014; Valentin-Hansen et al., 2014) or neurotransmitter transporters (Rannversson et al., 2016), as well as ligand-induced conformational dynamics of GPCRs (Park et al., 2015). However, this approach has so far not been used in studies of protein-protein interactions within GPCR heteromeric complexes. It should be noted that covalent crosslinking with the azido or benzophenone group of UAAs, such as azF or BzF, respectively, only occurs with properly oriented C-H bonds within a distance of about 2–4Å, and this approach can be used to prove a direct interaction between two proteins of a multiprotein complex in living mammalian cells (Sato et al., 2011; Ray-Saha et al., 2014). Using this approach, our data here precisely map the residues that physically interact at the heteromeric interface of the 5-HT2AR-mGluR2 complex by using amber codon suppression to introduce the photoreactive UAA azF at selected individual positions in the TMs of the mGluR2 promoter, which provides direct evidence for the structural arrangement among the protomers of an inter-class GPCR heterocomplex in living mammalian cells.
Our investigation of the 5-HT2AR-mGlu2R heteromeric interface was based on a combination of approaches that include photo-crosslinking followed by co-immunoprecipitation. We show that incorporation of the UAA azF at selected positions of the mGluR2 does not affect its predisposition to co-immunoprecipitate with the 5-HT2AR construct. Although our data showing that mGluR3 is not able to co-immunoprecipitate with 5-HT2AR support the selectivity and specificity of these findings, we are aware that the co-immunoprecipitation method uses cell lysates as a starting material, which raises concerns about the detection of false-positive interactions resulting from the loss of spatial organization during the lysis procedure. However, it is important to note that our photo-crosslinking method precedes cell membrane homogenization and co-immunoprecipitation protocols. Consequently, our data showing a significant increase in UV-induced crosslinking in cells co-transfected with 5-HT2AR and mGluR2-TAG4.44, but not in cells co-transfected with 5-HT2AR and mGluR2-TAG4.56 or mGluR2-TAG1.37 validate the specificity of the main conclusion of these findings that the TAG4.44 residue is located in close molecular proximity to the 5-HT2AR protomer and that there is a direct physical interaction between 5-HT2AR and mGluR2 via the TM4 of mGluR2. This concept is further supported by our findings showing lack of effect of UV exposure on the immunoblot signal of cells transfected with mGluR2-TAG4.44 alone, and absence of coimmunoprecipitation signal in UV-exposed cells co-expressing HA-mGluR3-TAG4.44-mCitrine and c-Myc-5-HT2AR-mCherry. Additional work, however, is necessary to solve the question related to individual positions along TMs of the 5-HT2AR that physically interact with the mGluR2 homodimer. Similarly, additional experimentation is required to fully characterize the dynamic changes within the quaternary structure of the 5-HT2AR-mGluR2 complex induced by orthosteric and allosteric agonists.
In summary, we successfully implemented a targeted photo-crosslinking approach with the genetically encoded UAA azF to delineate the amino acids that interact physically at the heteromeric interface of a GPCR complex. This method has wide applicability and will be important in future studies that may provide additional functional and structural insights into GPCR complexes as well as other large protein-protein complexes.
STAR⋆METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to the Lead Contact: Javier González-Maeso (javier.maeso@vcuhealth.org)
Materials Availability
Plasmids generated in this study (mGluR2 and mGluR3 constructs) are available upon request from the lead contact.
Plasmid pSVBpUC carrying the amber suppressor tRNA gene and the plasmid pcDNA3.1 carrying the azF aminoacyl-tRNA synthetase gene were donated by Thomas P. Sakmar (Ye et al., 2008).
The plasmid encoding the chimeric Gα subunit Gαqi9 was donated by Philippe Rondard (Xue et al., 2015). Requests for these plasmids should be directed to Thomas P. Sakmar and Philippe Rondard.
Data and Code Availability
The published article includes all datasets generated or analyzed during this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human (female) embryonic kidney (HEK293) cells (ATCC: CRL-1573) were used for this study (Shah et al., 2014) The cells were maintained at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) dialyzed fetal bovine serum (dFBS) and 1% (v/v) penicillin/streptomycin (Gibco) in a 5% CO2 humidified atmosphere.
METHOD DETAILS
Materials
(1R,4R,5S,6R)-4-Amino-2-oxabicyclo[3.1.0]hexane-4,6-dicarboxylic acid disodium salt (LY379268), (2S)-2-Amino-2-[(1S,2S)-2-carboxycycloprop-1-yl]-3-(xanth-9-yl) propanoic acid disodium salt (LY341495) and l-glutamic acid were purchased from Tocris Bioscience. [3H]LY341495 was purchased from American Radiolabeled Chemicals. p-Azido-L-phenylalanine (azF) was purchased from Chem-Impex International. All other chemicals were obtained from standard vendors.
Plasmid Construction
All assays were performed with PfuUltra Hotstart DNA Polymerase (Stratagene) in a Mastercycler EP Gradient Auto thermal cycler (Eppendorf). Cycling conditions were 30 cycles of 94°C for 30s, 55°C for 30 s, and 72°C for 1 min/kb of amplicon, with an initial denaturation/activation step of 94°C for 2 min and a final extension step of 72°C for 7 min. The constructs pcDNA3.1-c-Myc-5-HT2A-mCherry, pcDNA3.1-HA-mGluR2-mCitrine and pcDNA3.1-HA-mGluR3-mCitrine have previously been described (Moreno et al., 2016). Plasmid pSVBpUC carrying the amber suppressor tRNA gene and the plasmid pcDNA3.1 carrying the azF aminoacyl-tRNA synthetase gene were donated by Thomas P. Sakmar (Ye et al., 2008).
The plasmid encoding the chimeric Gα subunit Gαqi9 was donated by Philippe Rondard (Xue et al., 2015). Introduction of mutations Val569TAG, Ala681TAG, Ile693TAG or Cys121Ala into the pcDNA3.1-HA-mGluR2-mCitrine construct, and Phe690TAG into the pcDNA3.1-HA-mGluR3-mCitrine construct was performed with the QuikChange II Site Directed Mutagenesis Kit, according to the manufacturer’s protocol (Stratagene), (for primer pairs, see Table S3). All the constructs were confirmed by DNA sequencing.
Transient Transfection of HEK293 Cells
Human embryonic kidney (HEK293) cells (ATCC: CRL-1573) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) dialyzed fetal bovine serum (dFBS) and 1% (v/v) penicillin/streptomycin (Gibco) in a 5% CO2 humidified atmosphere. Transfection was performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocols. Plasmids encoding the appropriate TAG HA-mGluR2-mCitrine construct (i.e., HA-mGluR2-TAG1.37-mCitrine,
HA-mGluR2-TAG4.44-mCitrine, HA-mGluR2-TAG4.56-mCitrine HA-mGluR3-TAG4.44-mCitrine) along with amber suppressor tRNA suppressor tRNA and aminoacyl tRNA synthetase were transfected in a 1:1:0.1 ratio. For photo-crosslinking assays, TAG HA-mGluR2-mCitrine or TAG HA-mGluR3-mCitrine and c-Myc-5-HT2AR-mCherry constructs were transfected in a 2:1 ratio. The cell culture medium was supplemented with 0.5 mM azF.
Immunocytochemistry
Immunochemical assays were performed as previously reported (Moreno et al., 2012; Moreno et al., 2016), with minor modifications. Briefly, the media was removed and cells were fixed with 4% paraformaldehyde (Sigma) supplemented with 100 μM CaCl2 and 100 μM MgCl2 for 10 min at room temperature, rinsed with PBS, and washed twice with PBS supplemented with 20 mM glycine. Coverslips were then incubated with 0.2% Triton X-100 for 10 min at room temperature, and incubated for 60 min with PBS containing 3% goat serum (according to the secondary antibody). Primary antibody rabbit anti-HA (Cell Signaling Technology, catalog no. 2367; diluted at 1:800 was added (40 μl on coverslip) and incubated overnight at 4°C. After washing with blocking buffer, the cells were incubated with secondary antibodies Alexa 568-conjugated goat anti-rabbit (Invitrogen, A11011) for 60 min at room temperature. Coverslips were then mounted onto glass slides with ProLong Diamond Antifade mountant (ThermoFisher Scientific) and imaged 24 h later on a Carl Zeiss Axio Observer LSM 710 laser scanning confocal microscope (LSCM) with Plan-Apochromat 363/1.40 Oil DIC M27 or 340 Cal objective lens.
Immunoblot Assays
Western blot experiments were performed as previously reported (Moreno et al., 2012; Moreno et al., 2016), with minor modifications. Briefly, equal amounts of protein were resolved by 12% SDS-PAGE and transferred to nitrocellulose membranes by electroblotting. Detection of proteins by Western blotting with a mouse anti-c-Myc (Cell Signaling technology, catalog no. 2276), rabbit anti-HA (Cell Signaling technology, catalog no. 2367), rabbit anti-GFP (ThermoFisher Scientific, catalog no. A-11122), Amersham ECL Rabbit IgG, HRP-linked whole Ab (from donkey; cytiva catalog no. NA934–1mL) and Amersham ECL Mouse IgG, HRP-linked whole Ab (from sheep; cytiva catalog no. NA931–1mL) was performed with the enhanced chemiluminescence system according to the manufacturer’s instructions.
Radioligand Binding Assays
Saturation radioligand binding assays using [3H]LY341495 binding assays were performed as previously reported (Moreno et al., 2012; Moreno et al., 2016), with minor modifications. Briefly, harvested cell pellets were homogenized using a Teflon-glass grinder (50 up-and-down strokes) in 5 mL of binding buffer (1M K2HPO4, 1M KH2PO4, 100 mM KBr; pH 7.6). The volume was made up to 10 mL with binding buffer and the crude homogenate was centrifuged at 3,000 rpm for 5 min at 4°C. The supernatant was centrifuged at 18,000 rpm for 10 min at 4°C. The resultant pellet (P2) was washed with 10 mL of binding buffer and re-centrifuged at 18,000 rpm for 15 minutes. Aliquots were stored at –80°C until assay. Protein concentration was determined using the Bio-Rad protein estimation assay. Final volume in each well was 200 μL Saturation binding curves were carried out with 0, 0.0625, 0.125, 0.25, 0.5, 1, 2.5, 5, and 10 nM of [3H] LY341495. Nonspecific binding was determined in the presence of 1mM L-glutamic acid. The reaction mixtures were incubated at 4°C for 1 h. The free ligand was separated from bound ligand by rapid filtration under vacuum through GF/C glass fiber filters using a microbeta filtermat-96 harvester (PerkinElmer). These filters were then rinsed twice with 200 μL of ice-cold incubation buffer, and dried at 65°C for 1 h and counted for radioactivity by liquid scintillation spectrometry, using a MicroBeta2 detector (PerkinElmer).
[Ca2+]i Mobilization Assays
HEK293 cells were plated onto poly-D-lysine coated 96-well plates (Greiner Bio-One GmbH) and co-transfected with the appropriate plasmids, along with the chimeric Ga subunit (Gαqi9). On the day of the assay, cells were washed with Dulbecco’s phosphate-buffered saline (DPBS) and loaded with 3 μM Fura 2-AM in imaging solution (5 mM KCl, 0.4 mM KH2PO4, 138 mM NaCl, 0.3 mM Na2HPO4, 2 mM CaCl2, 1 mM MgCl2, 6 mM glucose, 20 mM HEPES, pH 7.4) supplemented with pluronic acid (10% solution in DMSO). Following incubation for 30 min at 37°C, cells were washed twice with imaging buffer before being placed on the FlexStation 3 microplate reader (Molecular Devices). For conditions that tested the effect of the mGluR2/3 antagonist LY341495 on the Ca2+ signal elicited by the mGluR2/3 agonist LY379268, cells were pre-treated with LY341495 for 30 min before the addition of LY379268. The Fura-2 signal was acquired at 510 nm by switching the excitation wavelength between 340/380 nm. Intracellular calcium concentration was expressed as a 340/380 nm ratio, and values were normalized to the basal 340/380 nm ratio recorded during 30 seconds before perfusion of the drug using Softmax Pro (Molecular Devices).
Photo-Crosslinking Assays
Assays were carried out in photo-crosslinking buffer (Hank’s Buffered Salt Solution [HBSS] containing 20 mM HEPES, pH 7.5 and 0.2% BSA). 48 h post-transfection, cells were harvested and cell pellets were collected. Cells were resuspended in the photo-crosslinking buffer, after which approximately half of the cells from each batch were pelleted down and stored as the [-UV] condition. The other half was photo-crosslinked at 4°C. To do this, the cell suspension was placed under a UV lamp at a distance of 5 inches and exposed to UV-A light for 15 min (ML-3500S MAXIM, Spectronics Corporation). The mixture was then resuspended and the cells were spun down, supernatant was removed and cell pellets were stored at –80°C until further use.
Co-immunoprecipitation Studies
Co-immunoprecipitation studies on membrane preparations of the cell pellets (photo-crosslinked and controls) were performed as described previously(Moreno et al., 2012; Moreno et al., 2016), with minor modifications. In brief, equal amounts of solubilized membrane fractions were incubated overnight with protein A/G beads and a mouse anti-c-Myc antibody at 4°C on a rotating wheel. A Western blot was performed on the co-immunoprecipitated samples using a anti-HA antibody as described above.
Molecular Modeling
Three-dimensional molecular models of the seven transmembrane domains of mGluR2 were built using the crystal structure of mGluR5 (PDB: 5CGC) as a structural template (Christopher et al., 2015). The amino acid sequences of the TM domains of the mGluR5 and the mGluR2 were aligned using Clustal X 2.1(Larkin et al., 2007). Homology models of the TM domains of the mGluR2 were generated using MODELLER v9.12 (Eswar et al., 2007). Hydrogen atoms were added to the homology models and disulfide bonds were built using SYBYL-X 2.1 (Tripos International). Modeling was obtained with the assistance of PyMOL (Molecular Graphics System, Version 1.7.4 Schrödinger, LLC).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed with GraphPad Prism software version 8. The statistical significance of experiments involving two groups was assessed by Student’s t test, whereas the statistical analysis involving three or more groups was assessed by one-way ANOVA followed by Dunnett’s post hoc test. An extra sum of square (F test) was used to determine the statistical difference for the simultaneous analysis of binding saturation curves. The level of significance was chosen at p = 0.05, and all assumptions (normality, equal variances) were met to perform parametric tests. All data are presented as means ± s.e.m. All statistical details of experiments can be found in the figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-c-Myc | Cell Signaling technology | Cat#2276; RRID: AB_331783 |
| Rabbit anti-HA | Cell Signaling technology | Cat# 2367; RRID: AB_10691311 |
| mouse anti-α-tubulin | Abcam | Cat#ab7291; RRID: AB_2241126 |
| rabbit anti-GFP | ThermoFisher Scientific | Cat# A-11122; RRID: AB_221569 |
| Alexa 568-conjugated goat anti-rabbit | Invitrogen | Cat# A11011: RRID: AB_143157 |
| Amersham ECL Rabbit IgG, HRP-linked whole Ab (from donkey) | Cytiva | Cat#NA934-1mL; RRID: AB_772206 |
| Amersham ECL Mouse IgG, HRP-linked whole Ab (from sheep) | Cytiva | Cat#NA931-1mL; RRID: AB_772210 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| p-Azido-L-phenylalanine (azF) | Chem-Impex International. | Cat#06162 |
| (1R,4R,5S,6R)-4-Amino-2-oxabicyclo[3.1.0] hexane-4,6-dicarboxylic acid disodium salt (LY379268) | Tocris Bioscience | Cat#5064 |
| (2S)-2-Amino-2-[(1S,2S)-2-carboxycycloprop-1-yl]-3-(xanth-9-yl) propanoic acid disodium salt (LY341495) | Tocris Bioscience | Cat#4062 |
| l-glutamic acid | Tocris Bioscience | Cat#0218 |
| [3H]]LY341495 | American Radiolabeled Chemicals | ART-1439 250 uCI |
| Experimental Models: Cell Lines | ||
| HEK293 cells (Female) | ATCC | Cat# CRL-1573 |
| Oligonucleotides | ||
| See Table S3 for sequences of primers used | Integrated DNA technologies (IDT) | N/A |
| Recombinant DNA | ||
| Plasmid: pSVBpUC carrying the amber suppressor tRNA gene were donated by | Thomas P. Sakmar (Ye et al., 2008) | N/A |
| Plasmid: pcDNA3.1 carrying the azF aminoacyl-tRNA synthetase gene | Thomas P. Sakmar (Ye et al., 2008) | N/A |
| Plasmid: chimeric Gα subunit Gαqi9 was donated by | Philippe Rondard (Xue et al., 2015) | N/A |
| Plasmid: pcDNA3.1-c-Myc-5-HT2A-mCherry and | Javier González-Maeso (Moreno et al., 2016) | N/A |
| Plasmid: pcDNA3.1-HA-mGluR2-mCitrine | Javier González-Maeso (Moreno et al., 2016) | N/A |
| Plasmid: pcDNA3.1-HA-mGluR3-mCitrine | Javier Gonzáez-Maeso (Moreno et al., 2016) | N/A |
| Plasmid: HA-mGluR2C121A-mCitrine | This paper | N/A |
| Plasmid: HA-mGluR2-TAG1.37-mCitrine | This paper | N/A |
| Plasmid: HA-mGluR2-TAG4.44-mCitrine | This paper | N/A |
| Plasmid: HA-mGluR2-TAG4.56-mCitrine | This paper | N/A |
| Plasmid: HA-mGluR3-TAG4.44-mCitrine | This paper | N/A |
| Software and Algorithms | ||
| Softmax Pro | Molecular Devices | https://www.moleculardevices.com/products/microplate-readers/acquisition-and-analysis-software/softmax-pro-software#gref |
| Prism software version 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Excel | Microsoft | https://www.microsoft.com/en-us/microsoft-365/excel |
| ImageJ | ImageJ developers | https://imagej.nih.gov/ij/ |
| Other | ||
| UV-A lamp | Spectronics Corporation | ML-3500S MAXIM |
Highlights.
Photoactivatable unnatural amino acids inform the structural interface of 5-HT2AR-mGluR2
TAG mGluR2 constructs were co-expressed with 5-HT2AR for photo-crosslinking
UV-induced crosslinking only in cells co-expressing 5-HT2AR and mGluR2-TAG4.44
5-HT2AR interacts with mGluR2 via the intracellular end of mGluR2’s TM4
SIGNIFICANCE.
GPCR homo/heteromerization is a highly controversial topic and the existence of GPCR oligomers has been questioned in multiple studies. Structural interface data of GPCR homo/heteromers as well as other plasma membrane proteins are limited by currently available methods. The methods that are available and commonly used to assess GPCR homo/heteromerization are mostly indirect approaches that include resonance energy transfer techniques, such as BRET and FRET, and cannot fully demonstrate a direct physical proximity between receptors. Cysteine crosslinking is one of the most commonly used methods to study protein-protein interactions and demonstrate direct physical proximity between GPCR protomers. However, this method suffers from certain disadvantages, such as the need to substitute some highly conserved cysteine residues, leading to a potential disruption of protein structure. Here, we use site-specific incorporation of the unnatural amino acid (UAA) azF by amber codon suppression to expand the genetic code coupled with photo-crosslinking and co-immunoprecipitation methods to study the structural interface of the inter-class 5-HT2AR-mGluR2 heteromeric complex. We introduced the photoactivatable UAA azF at selected individual positions in the TMs of the mGlu2R and tested the ability of the individual constructs to photo-crosslink with 5-HT2AR. Our data showed the residues that physically interact at the interface of this heteromeric complex. To further demonstrate the specificity of our approach, we also showed that the closely related mGluR3 could not photo-crosslink with 5-HT2AR. This method has wide applicability, and can be used in future studies to study structural interfaces as well as to demonstrate a direct physical interaction between protein-protein complexes.
ACKNOWLEDGMENTS
We thank Ashton Cropp and Scott Ramsey (Virginia Commonwealth University School of Medicine) for their critical review of the manuscript, Philippe Rondard (Montpellier University) for providing chimeric Gα subunit construct, and Thomas P. Sakmar (Rockefeller University) for providing amber suppressor tRNA and azF aaRS constructs. NIH R01 MH084894 and NIH R01 MH111940 (to J.G.-M.) participated in the funding of this study.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.chembiol.2020.07.006.
SUPPORTING CITATIONS
The following reference appears in the Supplemental Information: Preston and Wilson, 2013
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Baki L, Fribourg M, Younkin J, Eltit JM, Moreno JL, Park G, Vysotskaya Z, Narahari A, Sealfon SC, Gonzalez-Maeso J, et al. (2016). Cross-signaling in metabotropic glutamate 2 and serotonin 2A receptor heteromers in mammalian cells. Pflugers Arch. 468, 775–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros JA, and Weinstein H (1995). Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein coupled receptors. Methods Neurosci. 25, 366–428. [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, et al. (2007). High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318, 1258–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher JA, Aves SJ, Bennett KA, Dore AS, Errey JC, Jazayeri A, Marshall FH, Okrasa K, Serrano-Vega MJ, Tehan BG, et al. (2015). Fragment and structure-based drug discovery for a class C GPCR: discovery of the mGlu5 negative allosteric modulator HTL14242 (3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile). J. Med. Chem 58, 6653–6664. [DOI] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, and Sali A (2007). Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci Chapter 2, Unit 2.9. 10.1385/1-59259-890-0:831. [DOI] [PubMed] [Google Scholar]
- Ferre S, Casado V, Devi LA, Filizola M, Jockers R, Lohse MJ, Milligan G, Pin JP, and Guitart X (2014). G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol. Rev 66, 413–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourg M, Moreno JL, Holloway T, Provasi D, Baki L, Mahajan R, Park G, Adney SK, Hatcher C, Eltit JM, et al. (2011). Decoding the signaling of a GPCR heteromeric complex reveals a unifying mechanism of action of antipsychotic drugs. Cell 147, 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Maeso J (2011). GPCR oligomers in pharmacology and signaling. Mol. Brain 4, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Maeso J, Ang RL, Yuen T, Chan P, Weisstaub NV, Lopez-Gimenez JF, Zhou M, Okawa Y, Callado LF, Milligan G, et al. (2008). Identification of a serotonin/glutamate receptor complex implicated in psychosis. Nature 452, 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunbeck A, Huber T, Sachdev P, and Sakmar TP (2011). Mapping the ligand-binding site on a G protein-coupled receptor (GPCR) using genetically encoded photocrosslinkers. Biochemistry 50, 3411–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Shi L, and Javitch JA (2003). The fourth transmembrane segment forms the interface of the dopamine D2 receptor homodimer. J. Biol. Chem 278, 4385–4388. [DOI] [PubMed] [Google Scholar]
- Guo W, Urizar E, Kralikova M, Mobarec JC, Shi L, Filizola M, and Javitch JA (2008). Dopamine D2 receptors form higher order oligomers at physiological expression levels. EMBO J. 27, 2293–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilger D, Masureel M, and Kobilka BK (2018). Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol 25, 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber T, and Sakmar TP (2014). Chemical biology methods for investigating G protein-coupled receptor signaling. Chem. Biol 21, 1224–1237. [DOI] [PubMed] [Google Scholar]
- Kniazeff J, Prezeau L, Rondard P, Pin JP, and Goudet C (2011). Dimers and beyond: the functional puzzles of class C GPCRs. Pharmacol. Ther 130, 9–25. [DOI] [PubMed] [Google Scholar]
- Koehl A, Hu H, Feng D, Sun B, Zhang Y, Robertson MJ, Chu M, Kobilka TS, Laermans T, Steyaert J, et al. (2019). Structural insights into the activation of metabotropic glutamate receptors. Nature 566, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Levitz J, Habrian C, Bharill S, Fu Z, Vafabakhsh R, and Isacoff EY (2016). Mechanism of assembly and cooperativity of homomeric and heteromeric metabotropic glutamate receptors. Neuron 92, 143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gimenez JF, Canals M, Pediani JD, and Milligan G (2007). The alpha1b-adrenoceptor exists as a higher-order oligomer: effective oligomerization is required for receptor maturation, surface delivery, and function. Mol. Pharmacol 71, 1015–1029. [DOI] [PubMed] [Google Scholar]
- Lossl P, and Sinz A (2016). Combining amine-reactive cross-linkers and photo-reactive amino acids for 3D-structure analysis of proteins and protein complexes. Methods Mol. Biol 1394, 109–127. [DOI] [PubMed] [Google Scholar]
- Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, and Granier S (2012). Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature 485, 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan G (2009). G protein-coupled receptor hetero-dimerization: contribution to pharmacology and function. Br. J. Pharmacol 158, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JL, Miranda-Azpiazu P, Garcia-Bea A, Younkin J, Cui M, Kozlenkov A, Ben-Ezra A, Voloudakis G, Fakira AK, Baki L, et al. (2016). Allosteric signaling through an mGlu2 and 5-HT2A heteromeric receptor complex and its potential contribution to schizophrenia. Sci. Signal 9, ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JL, Muguruza C, Umali A, Mortillo S, Holloway T, Pilar-Cuellar F, Mocci G, Seto J, Callado LF, Neve RL, et al. (2012). Identification of three residues essential for 5-hydroxytryptamine 2A-metabotropic glutamate 2 (5-HT2A.mGlu2) receptor heteromerization and its psychoactive behavioral function. J. Biol. Chem 287, 44301–44319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender CM, and Conn PJ (2010). Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol 50, 295–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Sivertsen BB, Els-Heindl S, Huber T, Holst B, Beck-Sickinger AG, Schwartz TW, and Sakmar TP (2015). Bioorthogonal labeling of ghrelin receptor to facilitate studies of ligand-dependent conformational dynamics. Chem. Biol 22, 1431–1436. [DOI] [PubMed] [Google Scholar]
- Pin JP, and Bettler B (2016). Organization and functions of mGlu and GABAB receptor complexes. Nature 540, 60–68. [DOI] [PubMed] [Google Scholar]
- Preston GW, and Wilson AJ (2013). Photo-induced covalent cross-linking for the analysis of biomolecular interactions. Chem. Soc. Rev 42, 3289–3301. [DOI] [PubMed] [Google Scholar]
- Rannversson H, Andersen J, Sorensen L, Bang-Andersen B, Park M, Huber T, Sakmar TP, and Stromgaard K (2016). Genetically encoded photocrosslinkers locate the high-affinity binding site of antidepressant drugs in the human serotonin transporter. Nat. Commun 7, 11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Saha S, Huber T, and Sakmar TP (2014). Antibody epitopes on G protein-coupled receptors mapped with genetically encoded photoactivatable cross-linkers. Biochemistry 53, 1302–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht JJ, Mielke T, Vogel R, Villa C, and Schertler GF (2004). Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 23, 3609–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Mimasu S, Sato A, Hino N, Sakamoto K, Umehara T, and Yokoyama S (2011). Crystallographic study of a site-specifically cross-linked protein complex with a genetically incorporated photoreactive amino acid. Biochemistry 50, 250–257. [DOI] [PubMed] [Google Scholar]
- Serfling R, and Coin I (2016). Incorporation of unnatural amino acids into proteins expressed in mammalian cells. Methods Enzymol. 580, 89–107. [DOI] [PubMed] [Google Scholar]
- Shah K, McCormack CE, and Bradbury NA (2014). Do you know the sex of your cells? Am. J. Physiol. Cell Physiol 306, C3–C18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleno R, and Hebert TE (2019). Shaky ground—the nature of metastable GPCR signalling complexes. Neuropharmacology 152, 4–14. [DOI] [PubMed] [Google Scholar]
- Suchanek M, Radzikowska A, and Thiele C (2005). Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat. Methods 2, 261–267. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen L, Park M, Huber T, Grunbeck A, Naganathan S, Schwartz TW, and Sakmar TP (2014). Mapping substance P binding sites on the neurokinin-1 receptor using genetic incorporation of a photoreactive amino acid. J. Biol. Chem 289, 18045–18054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker D, Stevens RC, and Roth BL (2017). How ligands illuminate GPCR molecular pharmacology. Cell 170, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, et al. (2010). Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wang C, Gregory KJ, Han GW, Cho HP, Xia Y, Niswender CM, Katritch V, Meiler J, Cherezov V, et al. (2014). Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344, 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, and Brand L (1994). Resonance energy transfer: methods and applications. Anal. Biochem 218, 1–13. [DOI] [PubMed] [Google Scholar]
- Xue L, Rovira X, Scholler P, Zhao H, Liu J, Pin JP, and Rondard P (2015). Major ligand-induced rearrangement of the heptahelical domain interface in a GPCR dimer. Nat. Chem. Biol 11, 134–140. [DOI] [PubMed] [Google Scholar]
- Ye S, Kohrer C, Huber T, Kazmi M, Sachdev P, Yan EC, Bhagat A, RajBhandary UL, and Sakmar TP (2008). Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J. Biol. Chem 283, 1525–1533. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study.


