Abstract
Gene fusions involving the neurotrophic receptor tyrosine kinase genes NTRK1, NTRK2, and NTRK3, are well established oncogenic drivers in a broad range of pediatric and adult tumors. These fusions are also important actionable markers, predicting often dramatic response to FDA approved kinase inhibitors. Accurate interpretation of the clinical significance of NTRK fusions is a high priority for diagnostic laboratories, but remains challenging and time consuming given the rapid pace of new data accumulation, the diversity of fusion partners and tumor types, and heterogeneous and incomplete information in variant databases and knowledgebases. The ClinGen NTRK Fusions Somatic Cancer Variant Curation Expert Panel (SC-VCEP) was formed to systematically address these challenges and create an expert-curated resource to support clinicians, researchers, patients and their families in making accurate interpretations and informed treatment decisions for NTRK fusion-driven tumors. We describe a system for NTRK fusion interpretation (including compilation of key elements and annotations) developed by the NTRK fusions SC-VCEP. We illustrate this stepwise process on examples of LMNA∷NTRK1 and KANK1∷NTRK2 fusions. Finally, we provide detailed analysis of current representation of NTRK fusions in public fusion databases and the CIViC knowledgebase, performed by the NTRK fusions SC-VCEP to determine existing gaps and prioritize future curation activities.
Keywords: curation, pathogenicity, oncogenicity, reading-frame, actionable, biomarker
Introduction
Gene fusions involving the neurotrophic receptor tyrosine kinase (NTRK) genes, NTRK1, NTRK2 and NTRK3, have emerged as compelling diagnostic and therapeutic biomarkers in the care of children and adults with cancer. Early trials of NTRK-targeted inhibitors have demonstrated dramatic overall response rates of 75% in patients with solid tumors harboring NTRK gene fusions regardless of specific histologic diagnosis [1 – 3]. These data led to the FDA approval of two NTRK inhibitors, the first-in-class NTRK-specific inhibitor larotrectinib in 2018 and the kinase inhibitor entrectinib in 2019, for use in children and adults with NTRK fusion-positive solid tumors. This tissue-agnostic biomarker-centered approval is a pivotal event in personalized medicine, as a genetic alteration was identified as the key oncogenic therapeutic target in diverse tumor types [4, 5].
The process of variant curation makes use of literature, databases, and genomic expertise to evaluate the clinical relevance of individual genetic alterations. This process is particularly challenging when evaluating an NTRK fusion given the rapid emergence of new data, the multiplicity of fusion partners and databases, and different approaches deployed to evaluate fusions [6, 7]. Information about NTRK fusions is expanding rapidly. A search for “NTRK fusion” in PubMed reveals 399 unique articles over the last 10 years, with 188 articles (47.1%) published within the last year [8]. In addition to peer-reviewed articles, public fusion databases such as Mitelman, COSMIC fusions, and Quiver contain hundreds of NTRK fusion entries submitted from published literature and/or directly identified in patient samples [9 – 11]. With such an abundance of data, consolidating evidence into a concise interpretation that can be applied to a specific patient can be challenging for providers. Interpreting fusions individually also leads to extensive redundant effort on the part of busy healthcare providers and molecular pathologists and geneticists [12].
Expert crowdsourcing is an increasingly popular model for improving curation of genetic variants, with demonstrable benefits to both data quality and efficiency [13]. The National Institutes of Health (NIH) has funded the Clinical Genome Resource (ClinGen) to build a central resource that defines the clinical relevance of genes and variants for use in precision medicine and research [14]. Using the Clinical Interpretations of Variants in Cancer (CIViC) knowledgebase (civicdb.org), ClinGen has adopted crowdsourced clinical cancer somatic variant curation, providing a free public resource to the community of patients, caregivers, researchers, and healthcare providers to support education and patient care [15, 16]. CIViC is an open access, open source, community-driven web resource that releases data under a public domain dedication (CC0) enabling precision medicine through a sophisticated interface supporting structured curation, expert moderation, and dissemination of knowledge regarding the clinical significance of somatic cancer genome alterations.
Within ClinGen, a group of 13 researchers and clinicians was approved by the ClinGen Clinical Domain Oversight Committee to form an NTRK Fusions Somatic Cancer Variant Curation Expert Panel (SC-VCEP) to create structure and guidance for the SC-VCEP’s focused curation and interpretation of NTRK fusions. While other expert panels have been formed to address curation of germline alterations, the NTRK SC-VCEP represents the first panel dedicated to somatic alterations. NTRK alterations were prioritized by the ClinGen team due to their clinical significance, as described above. The goals of the group are to (1) develop NTRK -specific rules to evaluate evidence of oncogenicity and classify NTRK fusions based on a framework of the AMP/ASCO/CAP guidelines [17], (2) identify and prioritize specific NTRK alterations and associated cancer types for curation and (3) comprehensively curate NTRK fusion clinical evidence, apply aforementioned interpretation rules, and maintain a high quality corpus of publicly available NTRK clinical interpretations.
Here, we describe an evidence-based scoring framework for assessing oncogenicity and functional validity of NTRK fusions, which was developed by the NTRK SC-VCEP to supplement the 2017 AMP/ASCO/CAP guidelines for interpretation of clinical significance and actionability of somatic variants. We also describe key elements and annotations of NTRK genes and fusions, and illustrate the creation of Evidence Items for the LMNA∷NTRK1 and KANK1∷NTRK2 fusions in the CIViC knowledgebase using curation specifications developed to date by the NTRK SC-VCEP. Finally, we performed and described our detailed analysis of current representation of NTRK fusions in fusion databases and the CIViC knowledgebase to determine existing gaps and prioritize future curation activities.
Materials and methods
NTRK fusion SC-VCEP
The NTRK Fusions SC-VCEP was assembled in 2019 with members from nationally recognized academic centers consisting of clinical geneticists, molecular geneticists, cytogeneticists, and molecular pathologists who interpret and report NTRK fusions in cancers on a regular basis, oncologists involved in clinical trials for NTRK-targeted therapy, scientists who study the biology of NTRK fusion-driven neoplasms, and biocurators with specific knowledge in the field of variant curation and the use of clinical knowledgebases. The SC-VCEP defined the scope of curation to include known fusions involving NTRK1, NTRK2, and NTRK3 genes, including known kinase inhibitor resistance mutations in such genes. While activating point mutations in NTRK genes may emerge in the future as oncogenic [18], the initial scope of the VCEP is on fusions and subsequent resistance mutations. Tumors with high, intermediate, and low incidence of NTRK fusions, whether solid, hematological, adult, or pediatric cancers were all included within the scope of curation. The SC-VCEP meets at least monthly and develops consensus variant classification rules and curation protocols specific to NTRK fusions. The NTRK Fusions SC-VCEP is described at https://clinicalgenome.org/affiliation/50089/.
Fusion database content curation
NTRK fusion details, including gene fusion name, fusion partner orientation, fusion junction coordinates, cytogenetics data, and literature references were collected from six publicly accessible gene fusion databases that contain fusions curated from literature and/or patient submissions (Mitelman, Quiver, COSMIC, Tumor Fusion Portal, ChimerDB and Fusion GDB) [9, 10, 19 – 22]. To aid downstream comparisons, fusion names were reformatted to separate genes using a hyphen and references were standardized to PubMed IDs. The fusion list was deduplicated and events reported as NTRK-NTRK self fusions were removed. A list of 94 unique publicly available NTRK fusions was created along with documentation of their presence across each public database. References were collated and deduplicated on a per fusion basis. References listing NTRK as the 5′ partner were manually assessed for accuracy and fusions with incorrectly curated orientations were removed. Pilot curations were entered into CIViC based on CIViC’s standard operating procedure [23]. Finally, NTRK fusion inclusion in the CIViC knowledge base was compared to the NTRK public database list to determine curation coverage and future needs.
Specifications for NTRK fusions
The 2017 AMP/ASCO/CAP guidelines for variant interpretation were supplemented with specifications for assessing oncogenicity of NTRK fusions. Draft rules for assessing oncogenicity were developed based on known biological and clinical characteristics of NTRK fusions and were prepared with consensus input from members of the ClinGen NTRK Fusions SC-VCEP. These rules are reviewed and approved by the ClinGen Cancer Variant Interpretation Committee (CVI-C) followed by piloting the rules on prioritized variants.
Curation interface
NTRK fusion evidence was entered into CIViC as structured Evidence Items using the Add Evidence interface. A single Evidence Item is derived from a source publication and describes a single NTRK fusion event in terms of a particular cancer type and clinical significance (e.g., predicts sensitivity to larotrectinib). NTRK SC-VCEP curators enter an initial draft of each Evidence Item, which must then be reviewed by an approved editor in CIViC. Attaining editor status requires additional expertise, training, approval, and conflict of interest reporting (Editor requirements - https://docs.civicdb.org/en/latest/curating/editor.html). Comments for discussion and explanation of proposed revisions are documented in CIViC along with complete provenance surrounding the creator, revisors, and approvers of each Evidence Item. All activities of the NTRK SC-VCEP are summarized and associated with their defined organization within CIViC, so users can clearly differentiate contributions made by the expert panel as opposed to other members of the curation community. When a sufficient breadth and depth of Evidence Items have accumulated for a particular NTRK fusion, tumor type, and associated clinical significance, a CIViC Assertion is created to synthesize that knowledge into a definitive clinical interpretation.
Results
Annotation of NTRK gene features
For efficient curation of NTRK gene fusions, key annotation features for the representation of NTRK fusions were obtained and listed in Table 1. The NTRK gene family in humans consists of three members: NTRK1, NTRK2, and NTRK3. Important gene-level annotation features documented include gene symbol, gene name, stable HGNC and NCBI gene identifier, genomic and cytogenetic location, and reference sequences (Fig. 1).
Table 1.
Gene and protein annotation features for NTRK1–3.
| Gene | Alias symbols | HGNC Gene ID | NCBI Gene ID | RefSeq transcript | RefSeq exon count (TKD exons) | RefSeq Protein | ENSEMBL transcript (v75) | ENST exon count (TKD exons) | TKD exon CDS coordinates (GRCh37/hg19) | UniProt | AA (kinase domain) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NTRK1 | TRKA | 8031 | 4914 | NM_002529.4** | 17 (13–17) | NP_002520 | ENST00000524377.1 | 17 (13–17) | chr1:156,845,872–156,851,434(+) | P04629 | 796 (510–781) |
| NTRK2 | TRKB | 8032 | 4915 | NM_006180.6** | 19 (15–19) | NP_006171 | ENST00000376214.1 | 21 (17–21) | chr9:87,549,077–87,636,352(+) | Q16620 | 838 (554–823) |
| NTRK3 | TRKC | 8033 | 4916 | NM_001012338.3* | 20 (15–20) | NP_001012338 | ENST00000360948.2 | 19 (14–19) | chr15:88,420,166–88,483,984(−) | Q16288 | 839 (538–839) |
TKD, tyrosine kinase domain; CDS, coding sequences; AA, amino acids;
RefSeq Select, MANE Select;
RefSeq Select.
Fig. 1.
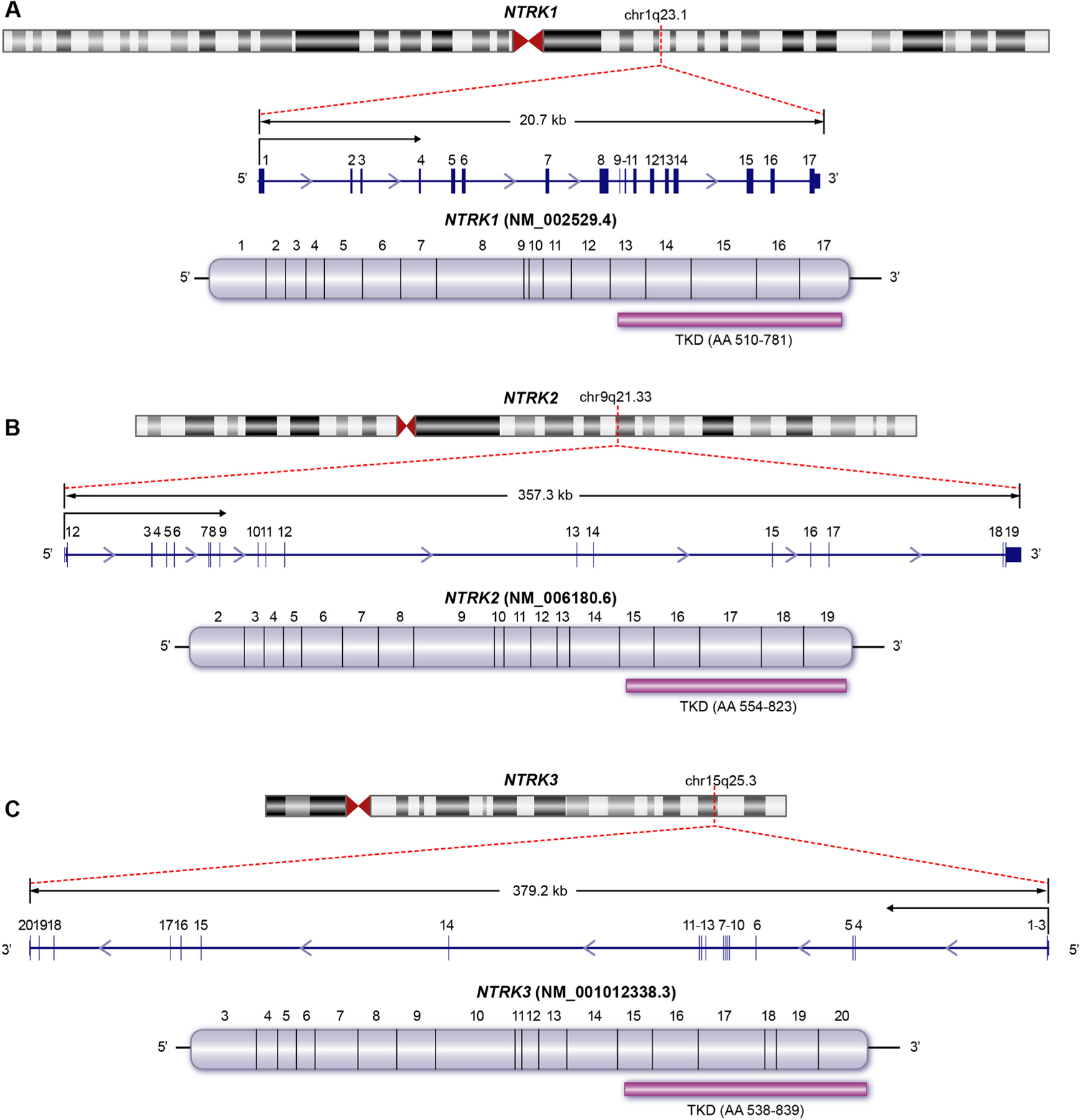
Key annotation features of NTRK genes, transcripts, and protein domains. The cytogenetic location, gene structure and size, exons (blue boxes), transcriptional orientation, and the open reading frame with coding exons (mauve) are shown for NTRK1 (A), NTRK2 (B) and NTRK3 (C). The 3′ exons encoding the C-terminal tyrosine kinase domains (TKD) of each gene are specifically illustrated along with the amino acid coordinates for the TKD (purple bar).
Transcript-level annotation can vary between reference genomic databases (e.g., NCBI RefSeq and Ensembl/Gencode ENST) and genome build versions (GRCh37 and GRCh38). In addition, variable use of transcript representation in published literature, the multiplicity of alternative transcripts, and ongoing updates to transcript versions in genomic databases pose further challenges to developing stable representation of transcripts for curation. Therefore, where available, a primary transcript for each NTRK gene was selected on the GRCh37/hg19 build following ongoing consensus efforts in transcript choice between the NCBI and Ensembl reference genomic databases (RefSeq Select and MANE Select) [24]. For NCBI RefSeq transcripts, the primary transcript selected for NTRK1 (NM_0 02529), NTRK2 (NM_0 06180) and NTRK3 (NM_001012338) all correspond to the designated transcript in the RefSeq Select transcript sets (Table 1); however, since the MANE transcript set from Ensembl/Gencode is currently unavailable for the hg19 assembly, and since primary transcript selection in the CIViC knowledgebase is based on GRCh37/hg19 build with Ensembl transcript version 75 (v75), the corresponding ENST v75 and RefSeq transcripts along with transcript versions, exon counts, and kinase domain encoding exons, and associated protein-level identifiers are also provided in Table 1.
For the purpose of curation, NTRK fusions are considered distinct based on the NTRK gene. For example, 5′ gene X∷NTRK1 fusion is considered distinct from 5′ gene X∷NTRK2 fusion. The critical genomic feature of NTRK fusions for curation is the location and integrity of the tyrosine kinase domain (TKD), which is encoded by the 3′ exons in all 3 NTRK genes (Fig. 1). As NTRK fusions can be represented in clinical genomic reports and the literature through several different annotation features such as exon number or junction coordinates, the location of each NTRK TKD is listed with encoding exons, genomic coordinates, and amino acid coordinates (Table 1).
The retention of the TKD regions is critical to maintaining a functional fusion protein capable of altering downstream signaling processes associated with tumorigenesis (Fig. 2). Providing genomic and protein level coordinates is essential for the standardized evaluation of the TKD status in NTRK fusions. Evidence to support the presence of the fusion typically comes from sequencing the fusion breakpoint, where stranding and directionality can help support the likelihood that a functional fusion product is produced. These considerations are particularly important for DNA sequencing, with highly variable intronic breakpoints and without the expression data available in RNA sequencing.
Fig. 2.
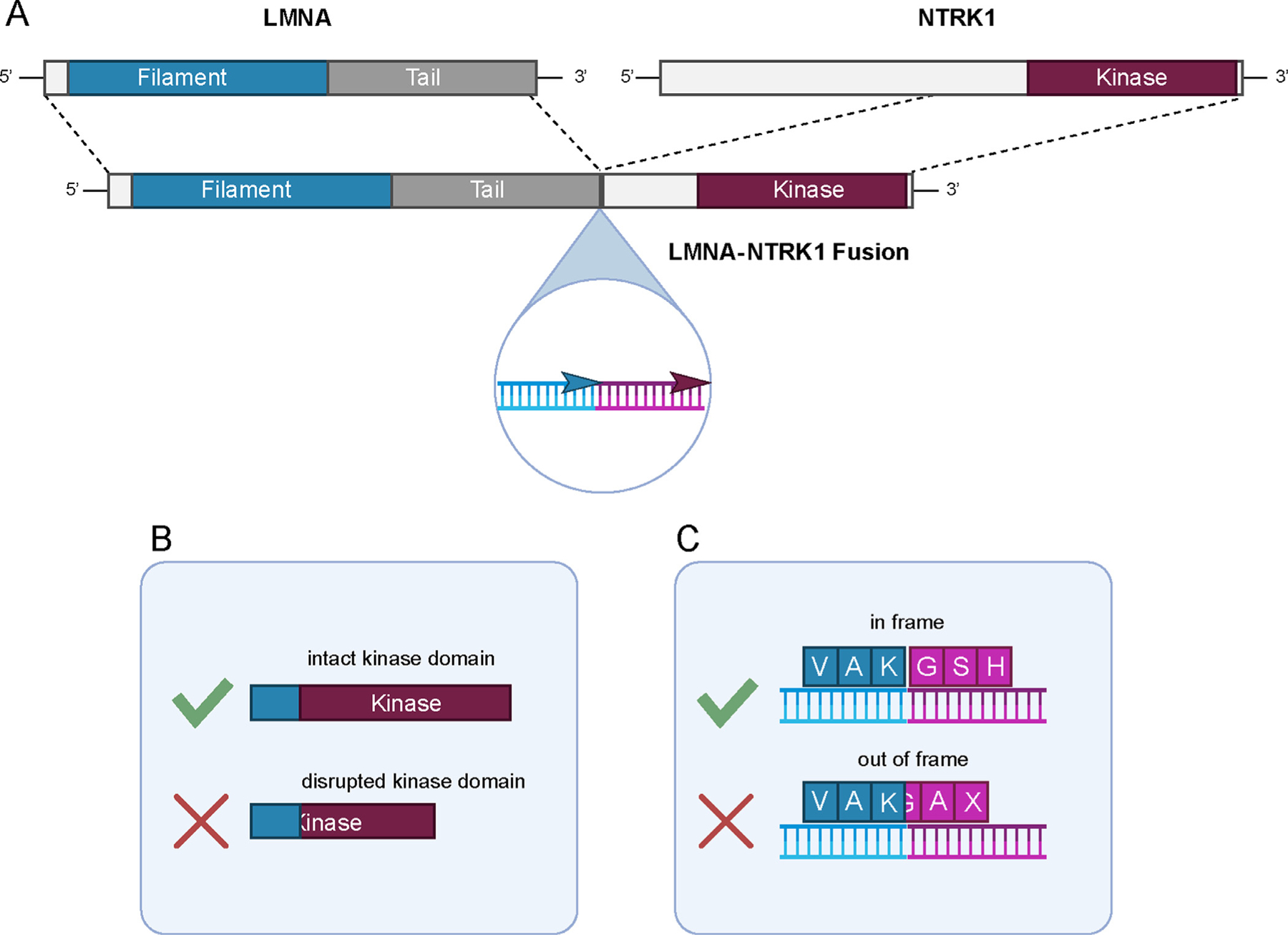
Representation of a canonical NTRK fusion and key features for assessing functionality. (A) The 5′ gene X - 3′ NTRK fusion gene structure is characteristic as shown here for LMNA∷NTRK1. The structure of the fusion gene should maintain transcriptional alignment between the two partner genes that can be assessed through transcript junction or genomic breakpoint evaluation (inset). Other key features in functional assessment include (B) evaluation of fusion transcript junction (or genomic breakpoint) to determine inclusion of a full non-disrupted NTRK kinase domain in the chimeric protein and (C) retention of the reading frame of the chimeric transcript downstream of the 5′ partner gene.
Accurate annotation of the 5′ gene partner, the 5′ fusion junction (or genomic breakpoint), the domain structure of the retained 5′ gene sequences, may also be important. A non-exhaustive list of 5′ gene partners curated from public databases is provided with this report (see later). While 5′ gene partners vary widely in prevalence, type, structural features such as inclusion of dimerization motifs, detailed annotation of the 5′ gene partner may in some circumstances provide evidence to distinguish a likely oncogenic fusion from one of uncertain significance.
In addition to annotations on fusion structure and protein domains, specific point mutations in the NTRK genes that have emerged as mechanisms of resistance to tyrosine kinase inhibitor (TKI) therapy, and informally referred to in the literature as gatekeeper mutations (NTRK1 p.F589L), solvent-front mutations (NTRK1 p.G595R and NTRK3 p.G623R) and xDFG motif mutations (NTRK1 p.G667C/S and NTRK3 p.G696A) were also annotated with the genomic coordinates for each codon (Supplemental Table 1).
NTRK fusion curation specifications
NTRK fusion evaluation involves two major steps: (1) Assessment of oncogenic validity (pathogenic/functional role), and (2) Assessment of clinical significance using supplemented AMP/ASCO/CAP rules (Fig. 3). While the AMP/ASCO/CAP guidelines provide a framework for assessing clinical significance of variants, additional supplementation has been proposed by the ClinGen NTRK fusion SC-VCEP for assessing oncogenicity and functional validity. The first of these specifications involves evaluation of NTRK fusion gene structure as follows. Canonical functional NTRK fusion proteins retain the NTRK TKD, which are encoded by the 3′ exons of all 3 NTRK genes and are located in the C-terminus downstream of the 5′ gene partner (Fig. 1). Therefore, the appropriate NTRK fusion gene structure and orientation should demonstrate a 5′ partner gene X fused to a 3′ NTRK gene. The fusion transcript junction assessed from fusion transcript/cDNA (or inferred from genomic breakpoint if assessed from genomic DNA) should be located such that the observed (or predicted) NTRK transcript junction is proximal to the TKD-encoding exons and the reading frame of the NTRK TKD is predicted to be retained. The second specification involves an assessment of cancer association, namely an evaluation of the recurrence of each NTRK fusion in cancers, regardless of the tumor type, as an indicator of oncogenicity. NTRK fusions are considered in general to be rare genetic events in cancers; however, certain tumor types are known to demonstrate a higher frequency. By this specification, the recurrence of a specific 5′ gene X∷NTRK fusion in cancers is considered a higher level of evidence for oncogenicity compared to a novel 5′ gene∷NTRK fusion. The third specification involves assessment of functional evidence of oncogenicity that may include evaluation of documented clinical response in patients (clinical trials or case reports) or of preclinical evidence (in vitro and in vivo studies e.g., signaling pathway activation or transformation and increased kinase partner expression). A schematic incorporating the above specifications into a general classification framework is shown in Fig. 3. Finally, the clinical significance of the fusions will be assessed based on AMP/ASCO/CAP rules customized for NTRK fusions to evaluate their diagnostic, prognostic, and therapeutic significance. These rules are under further development as part of the ClinGen Somatic Cancer expert panel process.
Fig. 3.
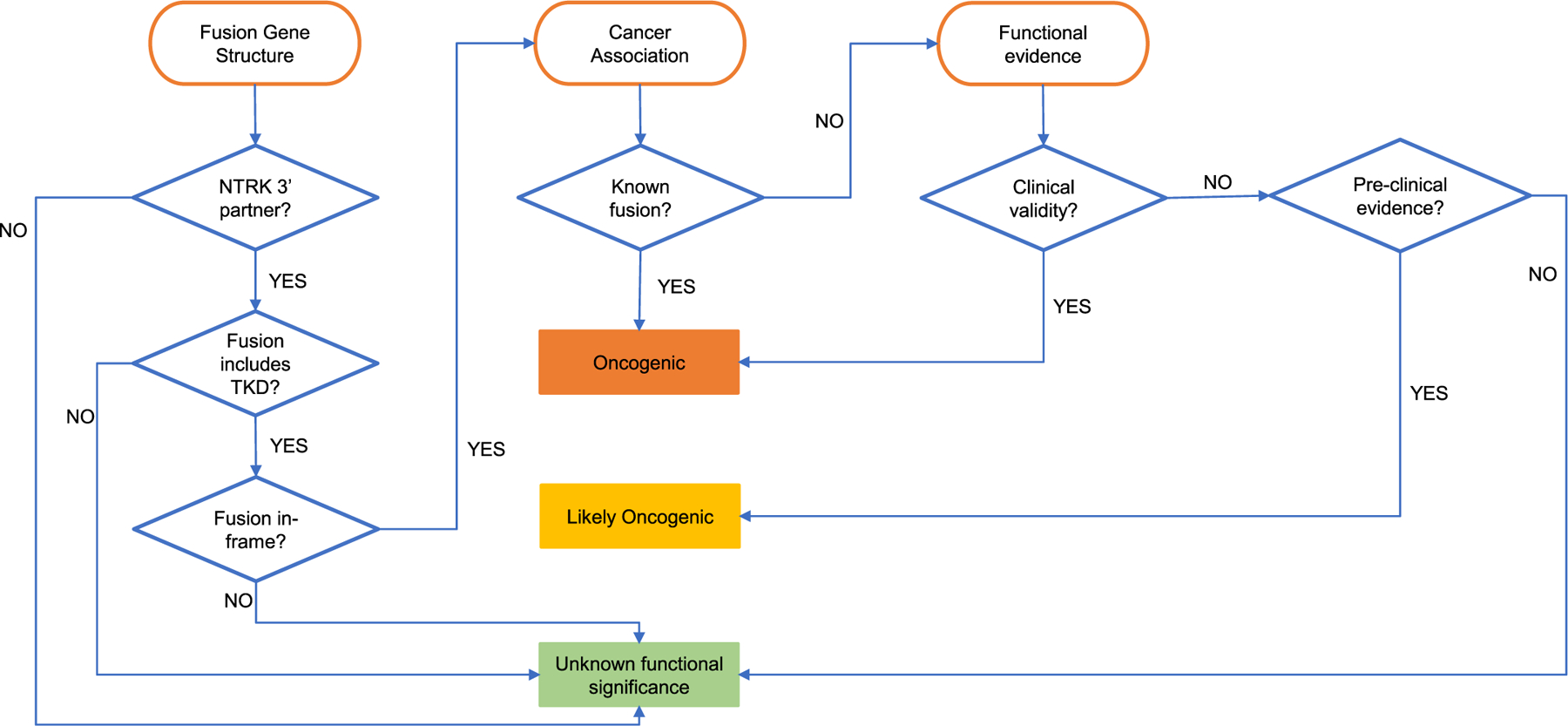
Framework for assessing oncogenicity of NTRK fusions. Three primary specification categories (fusion gene structure, cancer association, and functional evidence) and a decision support framework is presented to classify fusions based on varying strengths of evidence into three tiers of oncogenicity: oncogenic, likely oncogenic, and unknown functional significance.
NTRK fusion database mining
To better understand the breadth and depth of reported NTRK fusions, we conducted a query of six databases to evaluate accessibility and availability of details on fusions (fusion name, fusion junction information and/or genomic breakpoints, tumor type, clinical interpretation, and source information). Two of the databases (Mitelman & Quiver) source fusions from published literature, two (TCGA Fusion Portal & FusionGDB) from patient samples as part of large scale genomic projects, and two (COSMIC & ChimerDB) collect from both published literature and patient samples.
In total, 228 NTRK fusion entries were obtained from the six databases with Mitelman, Quiver and Chimerdb each containing over 40 entries (Supplemental Table 2). After removal of duplicate entries, 94 unique NTRK fusions were logged along with their presence across each database (Supplemental Table 3). Most fusions (n = 74) were reported with NTRK as the canonical 3′ partner 5′ X ∷ 3′ NTRK), whereas 20 were reported with NTRK as the 5′ partner (5′ NTRK ∷ 3′ X). We assessed the validity of the 20 5′ NTRK ∷ 3′ X fusions (Supplemental Figure 1) by reviewing their associated PubMed references and determined 8 to be spurious entries, of which 6 were the result of data entry errors where fusion orientation was listed with NTRK as the 3′ partner in the paper, but logged incorrectly in the database. For two others (NTRK1-CD5 and NTRK-Fc), careful review identified the absence of NTRK1-CD5 from the cited manuscript and NTRK-Fc was a representation of a recombinant fusion created for experimental purposes (Supplemental figure 1) [25, 26]. In the remaining twelve 5′ NTRK ∷ 3′ X fusions, 4 were found along with their reciprocal 5′ X ∷ 3′ NTRK fusion in patients (NTRK as the canonical 3′ partner), whereas 8 were documented in patients without mention of their reciprocal. Genomic database review suggests that the absence of a reported reciprocal for 3 of these 8 reported fusions (NTRK2∷RASEF, NTRK3∷PEAK1, and NTRK3∷SCAPER) may be explained by these likely resulting from intrachromosomal deletions. The lack of a reported reciprocal for the five other 5′ NTRK fusions may be due to the assays not designed for reciprocal fusion detection or the fusion calling pipelines having less sensitivity for such events. Further experimental data and analysis would be needed to determine the oncogenic potential of these 5′ NTRK fusions [27, 28]. For instance, NTRK3∷SCAPER [29] would be considered of unknown significance according to the specifications of oncogenicity proposed here since the NTRK TKD is absent in the fusion and any potential oncogenic impact would be outside of established mechanisms of downstream pathway activation.
Of the 74 unique fusions with NTRK reported as the 3′ partner, the majority were with NTRK1 (n = 32; 43.2%), followed by NTRK3 (n = 25; 33.8%), and NTRK2 (n = 17; 23.0%) (Fig. 4 a). Only 3/74 fusions (ETV6∷NTRK3, TFG∷NTRK1, and TPM3∷NTRK1) were found in all 6 databases (Fig. 4 b). Overall, 23 (24.5%) NTRK fusions were listed in at least 4 databases, 20 (27.0%) in 2 to 3 databases, leaving 31 (41.9%) found in only one database. Most of the entries recorded in just one database were curated from published literature (16 in Mitelman and 12 in Quiver) (Fig. 4 b). Across the 74 unique 3′ NTRK fusions, 68 unique 5′ partner genes were identified (Supplemental Table 4). While all of the NTRK genes displayed a degree of promiscuity regarding fusion with several different 5′ partner genes, only 5/68 of the 5′ partner genes (AFAP1, RBPMS, SQSTM1, STRN3, and TFG) were found to partner with more than one NTRK and of these only SQSTM1 was documented with all 3 NTRK s (Fig. 4 a, Supplemental Table 4). Overall 22 (32.4%) partners were found in at least 4 databases with 19 (27.9%) in 2 to 3 databases. Of the 27 5′ partner genes found in only one database, 24 were derived from the literature (13 in Mitelman and 11 in Quiver).
Fig. 4.
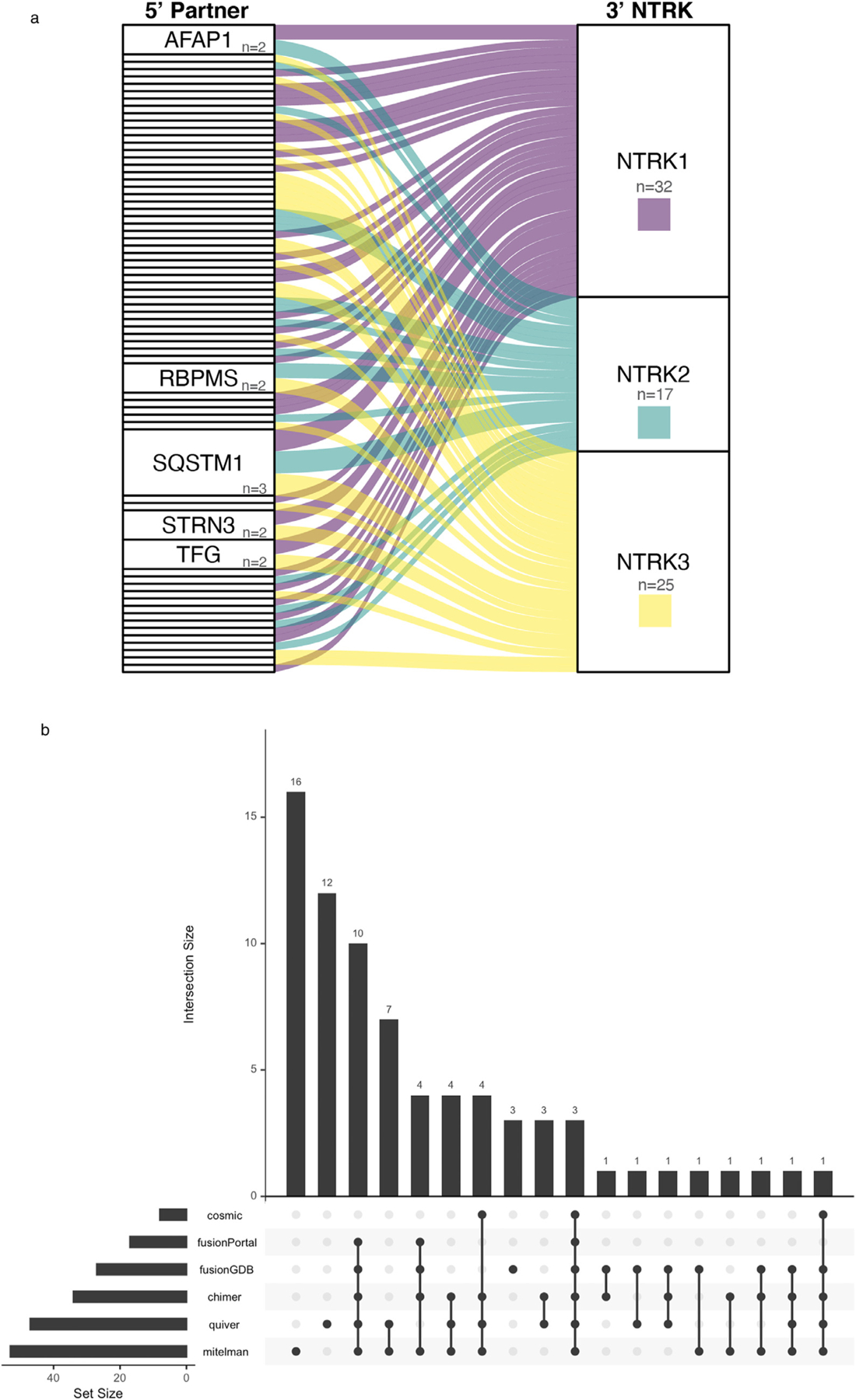
Representation of NTRK fusions in public databases. (A) Alluvial plot showing the multiplicity of 5′ partner genes for NTRK1 (n = 32), NTRK2 (n = 17), and NTRK3 (n = 25). Each rectangle in the 5′ partner column represents a single 5′ gene, and the height of the rectangle corresponds to frequency of the 5′ gene in partnering with one or more NTRK genes, e.g., AFAP1, RBPMS, STRN3, TFG (n = 2) have been reported to partner with 2 distinct NTRK genes, while only SQSTM1 (n = 3) has been reported with all three NTRK genes. Unlabeled rectangles only partner with a single NTRK gene. (B) Upset plot representing the overlap in the representation of NTRK fusions across commonly accessed public databases. The intersection among different database sets is shown as vertical bars, and the corresponding database sets with the overlap are represented below (filled circles). Only 3 fusions are represented in all 6 databases. The two largest database sets (Mitelman and Quiver) have 30 fusions in common, with n = 16 (Mitelman) and n = 12 (Quiver) as unique entries.
Fusion references were also collected and mapped to their PubMed IDs to create a table cataloging all references associated with individual fusions (n = 88 fusions) derived from published literature (Supplemental Table 5). ETV6∷NTRK3 was associated with the most references (173), TPM3∷NTRK1 had 45 references, and TPR∷NTRK1 had 28 (Supplemental Table 6). Alternatively, most fusions (n = 81) were associated with 5 or less references with the majority (n = 52) with only 1. In all, 257 unique PubMed articles were collected providing another useful resource to aid NTRK fusion curation.
NTRK fusion curation in CIViC
Having collected and harmonized the set of NTRK fusions, we next used the CIViC knowledgebase, which is the somatic variant curation platform of choice for the NTRK Fusions SC-VCEP, to apply detailed curation. To date, 46 total submitted or accepted Evidence Items have been entered into CIViC representing 12 NTRK fusions (Supplemental Table 7 e.g., ETV6∷NTRK3 most Evidence Items - 18). Only 7 overlap with the 74 3′ NTRK fusions list assembled from public databases and 4/7 are found across 5 or more databases. In addition, 5 NTRK fusions are unique to CIViC. Therefore, most NTRK fusions from the public database collection are not contained in CIViC and several CIViC fusions are not found in public databases. A similar non-overlap was seen also in the literature citations supporting the fusions: of the 33 PubMed papers used to create NTRK fusion Evidence Items in CIViC, 19 are not contained in the list of 257 papers indexed from the public databases. The incomplete overlap of NTRK fusions and papers between data and knowledgebases demonstrates the necessity for an organized and focused curation effort.
Below, we illustrate the NTRK fusion curation process in CIViC through evidence entries of LMNA∷NTRK1 and KANK1∷NTRK2 fusions that follow the rules defined to date by the NTRK fusion SC-VCEP. For LMNA∷NTRK1 e11-e10, evidence was curated into CIViC Evidence Item 8900 from a paper describing a patient with metastatic colorectal cancer harboring this fusion, whose tumor responded to the selective tyrosine kinase inhibitor entrectinib (Supplemental Figure 2) [30]. Through entry into structured fields, which allow for easily readable, searchable and comparable information, the orientation of the fusion (5′ LMNA −3′ NTRK1) and exons fused were recorded in the variant field. The disease (Colorectal Cancer, DOID:9256) was recorded in the Disease field and the reference manuscript was logged in the Source field. The clinical interpretation for the Evidence was described through the Evidence Type (Predictive). The Evidence Direction, Clinical Significance, and Drug fields combine to indicate the Evidence supports sensitivity of LMNA::NTRK1 expressing tumors to Entrectinib treatment. Patient information and response details were logged in the Evidence Statement. To aid fusion evaluation, special attention was applied to ensure the inclusion of specific study details related to fusion orientation, exons fused, preservation of the NTRK tyrosine kinase domain, and fusion frame status in the Evidence Statement. Fusion detection assays, validation method(s), and protein expression assessments were also noted to support the presence of the fusion. Finally, specific DNA breakpoints describing the fusion junction coordinates were noted in a Comment.
A second example curation (Evidence Item 8653) of a rare NTRK fusion, KANK1∷NTRK2, illustrates the collection of evidence from a single case report (Supplemental Figure 3). A 2 year-old with pilocytic astrocytoma was found to harbor a KANK1∷NTRK2 fusion in their initial and recurrent tumors [31]. Once again, the ontology-linked disease (tumor type) and source were entered into the appropriate fields and the clinical interpretation supporting the diagnosis of pilocytic astrocytoma was described through the Evidence Type, Direction, and Clinical Significance. Patient information and detection methods were logged into the Evidence Statement. However, multiple pieces of information such as fusion junction coordinates, exons fused, frame status, TKD preservation were not addressed in the paper. Their absence was noted in the comments to aid editor review and future evidence evaluation.
Discussion
Given the clinical implications of NTRK inhibitor therapy, evaluation of each identified NTRK structural alteration using a standardized curation process is critically important to provide patients and physicians with definitive data to guide treatment decisions. The relevance of having a well-established framework for curation and interpretation of the biological and clinical significance of NTRK fusions is underscored by the fact that not all NTRK fusions detected in tumor specimens represent functional oncogenic drivers [32].
Currently, no NTRK -specific curation rules or a centralized resource for documenting and sharing curated interpretations for NTRK fusions are available. Consequently, clinical interpretation of NTRK fusions relies on data from public databases and published literature to obtain supporting evidence for their oncogenic role and clinical actionability. This requires significant effort and poses many challenges for healthcare providers, due to abundance of data, inconsistencies in the published literature and databases, and lack of specialized, subject-matter expertise.
To address these unmet needs, the NTRK fusions SC-VCEP was established in 2019 to develop a systematic and sustainable process for the curation and interpretation of NTRK fusions and associated secondary alterations, and to provide high quality, publicly available NTRK clinical interpretations. Here, we present an evidence-based framework for the determination of functional validity and oncogenicity of NTRK structural rearrangements, as a supplementation to the AMP/ASCO/CAP guidelines [17]. The oncogenicity specifications focus on assessing the likelihood of a NTRK fusion to be a functionally valid activating alteration, independent of its clinical (diagnostic, prognostic, predictive) significance in any one given tumor type. As described here, systematic evaluation of fusion gene structure and reading frame, evaluation of cancer association, and functional evidence should be taken into account when assessing NTRK fusions. To assist with such evaluation, we also list key annotation features of NTRK genes and fusions, including gene-level, transcript-level (key exons, coordinates), and protein level (location of kinase domain) annotations.
Beyond annotation of the 3′ NTRK gene partner, the relative importance of the 5′ fusion partner to functional activation of the NTRK fusion remains uncertain. As reported elsewhere, over 65 unique 5′ partner genes have been documented for NTRK fusions [3, 6]. As shown here, these 5′ fusion partner genes appear to be fairly specific to a single 3′ NTRK member, with only 5 genes known to partner with more than 1 NTRK gene. Several 5′ gene partners have well-defined dimerization domains that have been considered important for fusion activation; however, the majority of 5′ partners either do not have a well-characterized dimerization domain, or activate NTRK fusions through unknown mechanisms [3]. The SC-VCEP is of the opinion that a novel 5′ gene partner by itself does not justify downgrading a NTRK fusion; however, a 5′ gene that is a known partner of NTRK or other kinase fusions constitutes a higher level of evidence to be considered along with the other specifications proposed.
When applied to different NTRK fusions reported in the literature, the curation specifications described here highlight the lack of adequate evidence for some fusions (e.g., 5′ NTRK fusions) while simultaneously supporting the validity of more established fusions (e.g., LMNA∷NTRK1). The SC-VCEP is proposing these specifications as a supplement to the AMP/ASCO/CAP somatic variant classification guidelines. Assessment of oncogenicity and assessment of clinical significance should be performed sequentially. Except for established NTRK fusions, for which all of the oncogenicity specifications have been demonstrated to be true, an uncommon or novel NTRK fusion should initially be assessed for oncogenicity. Fusions meeting classification criteria for ‘Oncogenic’ or ‘Likely Oncogenic’ will further be evaluated for clinical significance (diagnostic, prognostic and predictive). The clinical significance assessment and classification is contextual to the clinical presentation. For example, every ‘Oncogenic’ or ‘Likely Oncogenic’ NTRK fusion would be considered an AMP/ASCO/CAP tier I predictive marker in solid tumors, considering the tumor-type agnostic FDA approval for TRK inhibitors; however, an NTRK fusion in a childhood leukemia may be classified as a tier II (potential significance) predictive marker by AMP/ASCO/CAP criteria. If assessment of oncogenicity specifications results in inadequate evidence for oncogenicity, such fusions will be classified as fusions of uncertain functional/oncogenic significance, which would classify the fusion also as a ‘Variant of Unknown Clinical Significance (Tier III)’ category by the AMP/ASCO/CAP ‘clinical significance’ assessment. Fusions for which evidence shows they represent benign variants (well-described read-through transcripts) will be classified as ‘Benign’ in the ‘assessment of oncogenicity’, and Tier IV in the ‘clinical significance’ assessment. In combination with the customized AMP/ASCO/CAP variant classification rules under development by the NTRK fusions SC-VCEP, it is expected that when implemented, these specifications will serve as useful guidelines for assessment and classification of NTRK fusions.
Once finalized by the NTRK SC-VCEP, the oncogenicity framework and the NTRK fusion specifications of the AMP/ASCO/CAP rules will be evaluated and approved by the ClinGen CVI Committee, as part of the ClinGen SC-VCEP process. The SC-VCEP approval process will also include piloting the entire classification algorithm (oncogenicity evaluation and classification into tiers of clinical significance) on representative fusions, followed by formal implementation. Consensus classifications developed by the NTRK SC-VCEP will be added to CIViC as Assertions and submitted to ClinVar for public use. Specific somatic variants associated with resistance to NTRK inhibitors will also be curated by the NTRK SC-VCEP. This initiative will improve our ability to accurately classify NTRK somatic alterations and will thus play an essential role in clinical management.
Another NTRK fusion SC-VCEP initiative was to assess current representation of NTRK fusions in public fusion databases and the CIViC knowledgebase, in order to better prioritize future curation efforts. As demonstrated here, widely accessed fusion databases have several limitations in providing clinical-grade decision support: they lack standardized content and fusion nomenclature, may contain duplicate entries, may list fusion transcripts that are non-functional or contain data entry errors. In addition, database configuration, submission sources (e.g., literature or patient samples) and disease nomenclatures vary between databases. Importantly, we demonstrate that a significant proportion of NTRK fusions (41.9%) is only present in a single database, highlighting how fusion and reference citation lists in public databases are not exhaustive. The reference list of NTRK fusions compiled by our group from 6 different public databases (Supplemental Table 5) may be a useful resource to aid in NTRK fusion curation efforts; however, given the rapidly evolving landscape of NTRK fusions, a more dynamic system for ongoing assessments will be needed (e.g., enhancing the public fusion list with NTRK fusion instances called in our SC-VCEP member clinical laboratories).
Future efforts of the NTRK fusions SC-VCEP in collaboration with the CIViC team will include 1) creation of a detailed curation SOP for NTRK fusions, 2) improving searching methods and literature mining tools to find rare fusions, 3) collaboration with the Variant Interpretation for Cancer Consortium (VICC)/ClinGen/CGG/CAP initiative to standardize fusion nomenclature and representation in databases and knowledgebases and 4) improvement of CIViC infrastructure to capture fusion relevant data and structure text for easy machine readable parsing.
The ClinGen expert panels aim to use guided crowdsourcing to create a resource which will support clinicians, researchers, patients and their families in making informed decisions about choosing the right drug at the right time to support their cancer treatments. We invite readers to contribute their expertise to this effort through their publication of rare and common NTRK fusion cases and joining the curation community at https://civicdb.org – we are better together.
Supplementary Material
Acknowledgements
The authors wish to acknowledge Karen S. Prince for her assistance with graphical illustrations.
Funding
Jason Saliba, Shruti Rao, Arpad Danos, Gordana Raca, Malachi Griffith, Obi L Griffith and the CIViC project are supported by the National Institutes of Health (NIH) and National Cancer Institute (NCI) under Award Number U24CA237719 to Obi L Griffith (with Malachi Griffith as co-PI), including a funding supplement from the Childhood Cancer Data Initiative (CCDI). Malachi Griffith was also supported by the NIH NHGRI under award number R0 0HG0 07940. Funding for ClinGen with co-funding from the National Cancer Institute Grants are as follows: Baylor/Stanford - U24HG009649, Broad/Geisinger - U24HG006834, and UNC/Kaiser - U24HG009650.
COI
AC reports consulting and a member of the medical advisory board for Bayer. TL reports consulting for Bayer, Cellectis, Novartis, Deciphera, Jumo Health, and Y-mAbs Therapeutics and research support from Bayer, Pfizer, and Novartis. LZ reports that family members hold leadership positions and ownership interests of Decipher Medicine.
Footnotes
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi: 10.1016/j.cancergen.2022.03.001.
References
- [1].Drilon A, Siena S, Ou S-HI, Patel M, Ahn MJ, Lee J, et al. Safety and antitumor activity of the multitargeted Pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA-372–001 and STARTRK-1). Cancer Discov 2017;7:400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med 2018;378:731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 2018;15:731–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jørgensen JT. A paradigm shift in biomarker guided oncology drug development. Ann Transl Med 2019;7:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lemery S, Keegan P, Pazdur R. First FDA approval agnostic of cancer site - when a biomarker defines the indication. N Engl J Med 2017;377:1409–12. [DOI] [PubMed] [Google Scholar]
- [6].Hsiao SJ, Zehir A, Sireci AN, Aisner DL. Detection of tumor NTRK gene fusions to identify patients who may benefit from tyrosine kinase (TRK) inhibitor therapy. J Mol Diagn 2019;21:553–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Solomon JP, Benayed R, Hechtman JF, Ladanyi M. Identifying patients with NTRK fusion cancer. Ann Oncol 2019;30 viii16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].PubMed n.d. https://pubmed.ncbi.nlm.nih.gov/?term=ntrk+fusion (accessed August 12, 2021).
- [9].JBAMF Mitelman F (eds). Mitelman database of chromosome aberrations and gene fusions in cancer 2021. https://mitelmandatabase.isb-cgc.org (accessed August 12, 2021).
- [10].Archer Quiver Fusion Database n.d. http://quiver.archerdx.com/ (accessed August 12, 2021).
- [11].Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 2019;47:D941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ritter DI, Rao S, Kulkarni S, Madhavan S, Offit K, Plon SE. A case for expert curation: an overview of cancer curation in the clinical genome resource (ClinGen). Cold Spring Harb Mol Case Stud 2019;5. doi: 10.1101/mcs.a004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rödelsperger C, Athanasouli M, Lenuzzi M, Theska T, Sun S, Dardiry M, et al. Crowdsourcing and the feasibility of manual gene annotation: a pilot study in the nematode pristionchus pacificus. Sci Rep 2019;9:18789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, et al. ClinGen–the clinical genome resource. N Engl J Med 2015;372:2235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Griffith M, Spies NC, Krysiak K, McMichael JF, Coffman AC, Danos AM, et al. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat Genet 2017;49:170–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Danos AM, Ritter DI, Wagner AH, Krysiak K, Sonkin D, Micheel C, et al. Adapting crowdsourced clinical cancer curation in CIViC to the ClinGen minimum variant level data community-driven standards. Hum Mutat 2018;39:1721–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the association for molecular pathology, American society of clinical oncology, and college of American pathologists. J Mol Diagn 2017;19:4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Joshi SK, Qian K, Bisson WH, Watanabe-Smith K, Huang A, Bottomly D, et al. Discovery and characterization of targetable NTRK point mutations in hematologic neoplasms. Blood 2020;135:2159–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].COSMIC: The catalogue of somatic mutations in cancer - gene fusions n.d. https://cancer.sanger.ac.uk/cosmic/fusion (accessed August 12, 2021).
- [20].ChimerDB 4.0: an updated and expanded database of fusion genes n.d. https://www.kobic.re.kr/chimerdb_mirror/ (accessed August 12, 2021). [DOI] [PMC free article] [PubMed]
- [21].Kim P. And Zhou. FusionGDB: fusion gene annotation database n.d. https://ccsm.uth.edu/FusionGDB/index.html (accessed August 12, 2021). [DOI] [PMC free article] [PubMed]
- [22].Hu X, Wang Q, Tang M, Barthel F, Amin S, Yoshihara K, et al. TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res 2018;46:D1144–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Danos AM, Krysiak K, Barnell EK, Coffman AC, McMichael JF, Kiwala S, et al. Standard operating procedure for curation and clinical interpretation of variants in cancer. Genome Med 2019;11:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, et al. Ensembl 2020. Nucleic Acids Res 2020;4 8:D6 82–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shankar SL, O’Guin K, Cammer M, McMorris FA, Stitt TN, Basch RS, et al. The growth arrest-specific gene product Gas6 promotes the survival of human oligodendrocytes via a phosphatidylinositol 3-kinase-dependent pathway. J Neurosci 2003;23:4208–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lannon CL, Martin MJ, Tognon CE, Jin W, Kim S-J, Sorensen PHB. A highly conserved NTRK3 C-terminal sequence in the ETV6-NTRK3 oncoprotein binds the phosphotyrosine binding domain of insulin receptor substrate-1: an essential interaction for transformation. J Biol Chem 2004;279:6225–34. [DOI] [PubMed] [Google Scholar]
- [27].Kumar S, Vo AD, Qin F, Li H. Comparative assessment of methods for the fusion transcripts detection from RNA-Seq data. Sci Rep 2016;6:21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liu S, Tsai W-H, Ding Y, Chen R, Fang Z, Huo Z, et al. Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-seq data. Nucleic Acids Res 2016;44:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Friedman BJ, Hernandez S, Fidai C, Jiang A, Shwayder TA, Carskadon S, et al. A pediatric case of pigmented epithelioid melanocytoma with chromosomal copy number alterations in 15q and 17q and a novel NTRK3-SCAPER gene fusion. J Cutan Pathol 2020;47:70–5. [DOI] [PubMed] [Google Scholar]
- [30].Sartore-Bianchi A, Ardini E, Bosotti R, Amatu A, Valtorta E, Somaschini A, et al. Sensitivity to entrectinib associated with a novel LMNA-NTRK1 gene fusion in metastatic colorectal cancer. J Natl Cancer Inst 2016;108. doi: 10.1093/jnci/djv306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].López GY, Perry A, Harding B, Li M, Santi M. CDKN2A/B loss is associated with anaplastic transformation in a case of NTRK2 fusion-positive pilocytic astrocytoma. Neuropathol Appl Neurobiol 2019;45:174–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ameline B, Saba KH, Kovac M, Magnusson L, Witt O, Bielack S, et al. NTRK fusions in osteosarcoma are rare and non-functional events. Hip Int 2020;6:107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


