Abstract
The evolution of eukaryotic genomes is accompanied by fluctuations in chromosome number, reflecting cycles of chromosome number increase (polyploidy and centric fissions) and decrease (chromosome fusions). Although all chromosome fusions result from DNA recombination between two or more nonhomologous chromosomes, several mechanisms of descending dysploidy are exploited by eukaryotes to reduce their chromosome number. Genome sequencing and comparative genomics have accelerated the identification of inter-genome chromosome collinearity and gross chromosomal rearrangements and have shown that end-to-end chromosome fusions (EEFs) and nested chromosome fusions (NCFs) may have played a more important role in the evolution of eukaryotic karyotypes than previously thought. The present review aims to summarize the limited knowledge on the origin, frequency, and evolutionary implications of EEF and NCF events in eukaryotes and especially in land plants. The interactions between nonhomologous chromosomes in interphase nuclei and chromosome (mis)pairing during meiosis are examined for their potential importance in the origin of EEFs and NCFs. The remaining open questions that need to be addressed are discussed.
Comparative (cyto)genomics have shown that end-to-end chromosome fusions and nested chromosome fusions played a more important role in the evolution of eukaryotic genomes than previously thought.
Introduction
Why should we be aware of, and celebrate, scientific anniversaries? One reason is that anniversaries give us the opportunity to pause, look back over the decades since the event, and reflect on the brilliant achievements of great scientists. By traveling back in time, we can compare past accomplishments with our own work, identify questions that were not answered at the time, and revisit decades-old problems in the context of advanced knowledge and methodologies. And, perhaps most importantly, we pay tribute to our heroes of science! This year marks the bicentennial of the birth of Johann Gregor Mendel (1822–1884), a man ahead of his time who laid the foundations of genetics and breeding. We also commemorate 120 years since the birth of two distinguished chromosomal geneticists of the 20th century, Barbara McClintock (1902–1992) and Cyril D. Darlington (1903–1981), and reflect on the 90 years that have passed since Darlington published his influential synthesis, Recent Advances in Cytology (1932).
Heinzendorf (now Hynčice) was once a small village in the former Austrian Silesia, a region that became part of the newly formed Czechoslovakia (1918) and later the Czech Republic (1993). It was here 200 years ago, on July 20, 1822, that a boy was born who would, much later, become famous as the “father of genetics” (Figure 1). In 1843, 21-year-old Johann Mendel entered the Augustinian Order in the St Thomas Monastery on Klosterplatz (since 1910, Mendel Square) in Altbrünn (Old Brno) and was given the name Gregor. It was here, in the monastery garden and a newly built greenhouse, that Gregor Mendel became fascinated with the inheritance of various plant characters and conducted his extensive breeding experiments with garden peas (and other plant species), from which he drew general conclusions about the inheritance of morphological traits (for a detailed account of Mendel’s life, see, e.g. Edelson, 2001; Iltis, 2018). Mendel published his conclusions in 1866 (Versuche über Pflanzen-Hybriden), but 35 more years passed before the now-familiar Mendel’s Laws were rediscovered by Hugo de Vries (1848–1935), Carl Correns (1864–1933), and Erich von Tschermak (1871–1962); the year 1900 is, therefore, considered by many to be the year that the new discipline—genetics—was born.
Figure 1.
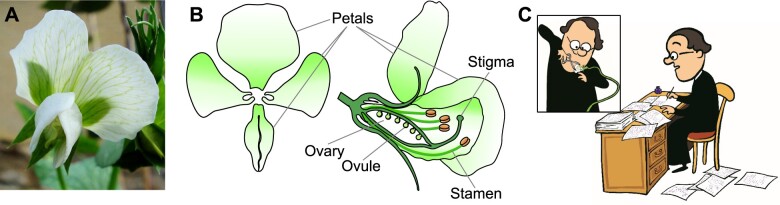
Johann Gregor Mendel (1822–1884). Mendel cultivated as many as 27,000 garden pea plants during his famous crossing experiments (1854–1863) at the St Thomas monastery in Brno (Orel, 1971). Since peas are self-pollinating, Mendel had to emasculate the flowers before manual cross-pollination. In the winter of 1885, Mendel reported on his pea experiments and presented the general law of inheritance in two lectures in Brno. A year later, the edited lectures were published as his famous article “Experiments on Plant Hybrids” (1866). A, Pea flower, original photograph from Flickr (https://www.flickr.com/photos/wheatfields/2670660145/), by allispossible.org.uk, CC BY 4.0. B, diagram of pea floral structure. C, Mendel at work, images courtesy of Mendel University in Brno.
During Mendel’s lifetime, the connection between inheritance of morphological characters and the physical units that mediate their transmission from generation to generation was unknown to science. Several now-famous cytologists, such as Walther Flemming (1843–905), Emil Heuser (1851–1928), Carl Rabl (1853–1917), Eduard Strasburger (1844–1912), and Wilhelm Waldeyer (1836–1921), made significant discoveries in cell biology by describing the structure and dynamics of darkly stained threads, now known as chromosomes, in the cell nucleus (see, e.g. Cremer and Cremer, 1988, 2006 for detailed historical accounts). In the same year that Mendel died in Brno (January 6, 1884), Emil Heuser described, for the first time, two chromatids of each mitotic chromosome that separated and moved to opposite poles of the cell (Heuser, 1884). Only a year later, Carl Rabl observed the continuity of mitotic chromosomes in resting (interphase) nuclei and their polarized centromere–telomere arrangement (Rabl, 1885), now known as the Rabl configuration. However, the term chromosome was not used until after 1888, when it was coined by Wilhelm Waldeyer.
After the independent rediscovery of Mendel’s rules in 1900, chromosomes began to be considered as possible physical carriers of Mendelian factors (see Martins, 1999, on the history of the chromosome theory of heredity). About 120 years ago, a 25-year-old student at Columbia University, Walter Sutton (1877–1916), recognized that the segregation of Mendelian factors during the production of germ cells was consistent with the segregation of chromosomes in meiosis, and concluded, “I may finally call attention to the probability that the association of paternal and maternal chromosomes in pairs and their subsequent separation during the reducing division … may constitute the physical basis of the Mendelian law of heredity” (Sutton, 1902). The year Sutton published his treatise on the role of chromosomes in heredity, Barbara McClintock was born (for a detailed account of McClintock’s life, see Keller, 1983; Campbell, 1993; Comfort, 2001). She made her first significant discovery 29 years later, when she and her student Harriet Creighton described the relationship between meiotic recombination and crossovers between homologous chromosomes (Creighton and McClintock, 1931). But there were still many mysteries about chromosomes that Barbara and her contemporary Cyril Darlington would challenge in the forthcoming years.
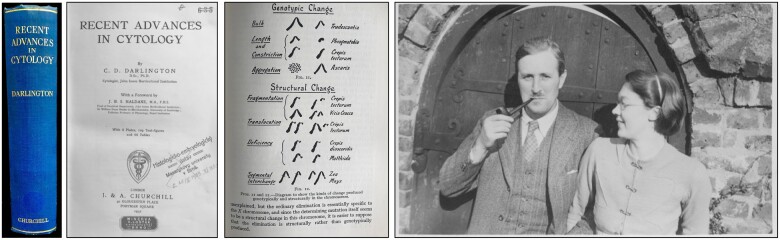
In 1923, Cyril Darlington joined the John Innes Horticultural Institute in Merton, England, then headed by William Bateson (1861–1926), who was an active supporter of Mendel’s work and the first to call research on heredity “genetics” (Bateson, 1906; Lipshitz, 2021). Within just 10 years, Darlington familiarized himself with the modern techniques of chromosome analysis and captured most of what was known about chromosomes at that time by publishing his famous Recent Advances in Cytology in 1932 (second edition 1937; Figure 2). The book was not so much concerned with the biology of the cell, but almost exclusively with the structure and behavior of chromosomes. Advances has become the “chromosome bible” for some scientists, but at the same time, the book and its author stirred controversy from day one (e.g. Belling, 1933; Lewis, 1983; Harman, 2003, 2004, 2006). Why did Darlington’s book provoke such a fierce polemic? It was his unconventional working method and courage to shake up the calm waters of descriptive karyology by making bold and sometimes daring predictions and generalizations. As Darlington’s senior colleague J.B.S. Haldane wrote in the preface to his book, “it [the book] marks the beginning of a new epoch, the transition from an essentially descriptive to a largely deductive science.” The value of the book lay in the author’s attempt to collate scattered individual chromosomal studies into a synthetic account, which in turn enabled him to deduce common patterns across phylogenetic phyla. The significance of Darlington’s synthesis, including reservations about his working methods, was aptly described by McClintock in her book review entitled “Cyto-genetics in 1932”: “There are too many unproven postulates. By such a procedure, however, a clearer view of the present state of the subject is made possible and a definite direction for future investigation is indicated. The theoretical postulates in the field in which the author has carried on his investigations and centered his thoughts are admirable and provocative. Unfortunately, the author has a tendency to dismiss evidence which, due to his lack of acquaintance with the material, appears inconsequential to him” (McClintock, 1932). Despite the natural shortcomings of Darlington’s compendium, his book was a milestone in the integration of hitherto descriptive karyology with genetics and evolutionary views of genetic variation.
Figure 2.
Cyril D. Darlington (1903–1981). Darlington’s Recent Advances in Cytology, his first both famous and controversial book, was published in 1932. A year later, Margaret Upcott began her Ph.D. at the John Innes under Darlington’s supervision and they were married in 1937 (photo from this period). In September 1939, Darlington became director of the John Innes until 1953, when he accepted the Sherardian Professorship of Botany at the University of Oxford (Harman, 2004). Photo of CDD and MU courtesy of Clare Passingham.
At the time of the first edition of Darlington’s Advances, the reduction of chromosome number by reciprocal translocations (i.e. the so-called chromosome fusions) was well known, including the type of translocation in which two telocentric or acrocentric chromosomes recombine to form a metacentric (V-shaped) chromosome (Robertson, 1916), still called a Robertsonian (Rb) translocation today (see Darlington’s Translocation in Figure 2). However, not all types of translocations between nonhomologous chromosomes, which (in addition to polyploidy) have been held responsible for the observed chromosome number variation (Darlington, 1932), could be reconstructed as Rb fusions using the then standard methods of comparative cytogenetics. Mather and Stone (1933), working alongside Darlington at the John Innes Institute, observed dicentric chromosomes in Crocus corm root tips after irradiation with X-rays: “In these nuclei one less than the normal somatic number of chromosomes appear on the metaphase plate, but in each case one chromosome has an unusual appearance. These chromosomes appear to possess two attachment constrictions [i.e. centromeres].” In cases like this, even conventional chromosome staining and chromosome morphology were sufficient to identify a nascent fusion chromosome with two centromeres (Mather and Stone, 1933). However, as evolutionary time since chromosome fusion increases, so too does the likelihood of centromere inactivation, deletion or repositioning, inversions, reciprocal translocations, and other changes. Therefore, some dysploidal chromosomal rearrangements (i.e. rearrangements that reduce chromosome number), such as end-to-end chromosome fusions (EEFs) or nested chromosome fusions (NCFs), went unnoticed until they were (re)discovered in the first two decades of this century, thanks to sequence-based comparative genomics.
In this review, I pay tribute to the great discoveries and achievements of Mendel, McClintock, and Darlington, to name just three of our scientific predecessors, by discussing the recent findings on the (re)discovered and still not well-known mechanisms of chromosomal descending dysploidy (i.e. reduction of chromosome number), namely EEFs and NCFs. With the increasing number of analyzed eukaryotic genomes containing fusion chromosomes, these two dysploid mechanisms are gaining increasing prominence in the literature.
End-to-end fusions
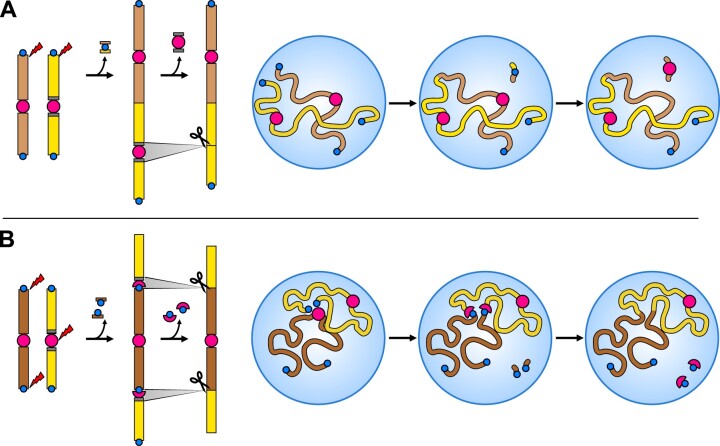
Recombination between the ends of two nonhomologous chromosomes is described in scientific literature by various terms, such as EEF, end-to-end translocation, end-to-end joining, terminal chromosome translocation, telomere fusion, or head-to-head fusion. As these rearrangements are commonly referred to as EEF, this term is used here, although it is factually incorrect, since telomeric chromatin normally precludes simple “fusion” of chromosome ends. In EEF, two different chromosomes are tandemly fused by recombination between their “sticky” ends (Figure 3A), with both chromosomes retained in their entirety within the fusion chromosome; the possible deletion at the junction is minimal and limited to repeats. In eukaryotes with monocentric chromosomes, an end-to-end fusion makes the new chromosome dicentric initially. If such a chromosome should be stably transmitted, one of the centromeres must be inactivated or eliminated (Figure 3A). Organisms with holokinetic chromosomes by default avoid the problem of dicentricity (Mandrioli and Manicardi, 2020).
Figure 3.
Chromosome number reduction by EEF and NCF. A, EEF. Two nonhomologous chromosomes recombine at their (sub)telomeres. The resulting dicentric chromosome is stabilized by epigenetic inactivation and/or recombinational removal (scissors) of one of the two centromeres. The figure on the right shows an EEF event occurring in an interphase nucleus with non-Rabl configuration (Figure 4B) or during meiotic prophase I (postbouquet, Figure 4E). B, NCF. Two nonhomologous chromosomes recombine within the (peri)centromere of the recipient chromosome (yellow) and the (sub)telomeres of the inserted chromosome (brown). The resulting di- or tricentric chromosome is stabilized by epigenetic inactivation and/or recombinational removal (scissors) of the recipient chromosome (peri)centromere. The figure on the right shows an NCF event occurring in an interphase nucleus with non-Rabl configuration (Figure 4B) or during meiotic prophase I (postbouquet, Figure 4E). Centromeres and telomeres are shown as red circles and blue dots, respectively.
There is an important difference between EEF types (Figure 4). End fusions between centric ends/short arms of two telocentric chromosomes (i.e. centric fusions) or a telocentric and an acrocentric chromosome, and probably sometimes between two acrocentric chromosomes, are called Rb translocations/fusions. Some Rb translocations can be dicentric (Page et al., 1996; Stimpson et al., 2010), with both centromeres adjacent or in close proximity to each other, and possibly both acting as one functional centromere. However, the focus of the present review is on dicentric end-to-end fusions involving both short and long arms of acrocentric and telocentric chromosomes, and both arms of metacentrics. Unlike Rb translocations, these EEFs do not result in two centromeres being adjacent or in close proximity to each other (Figures 3A and 4).
Figure 4.
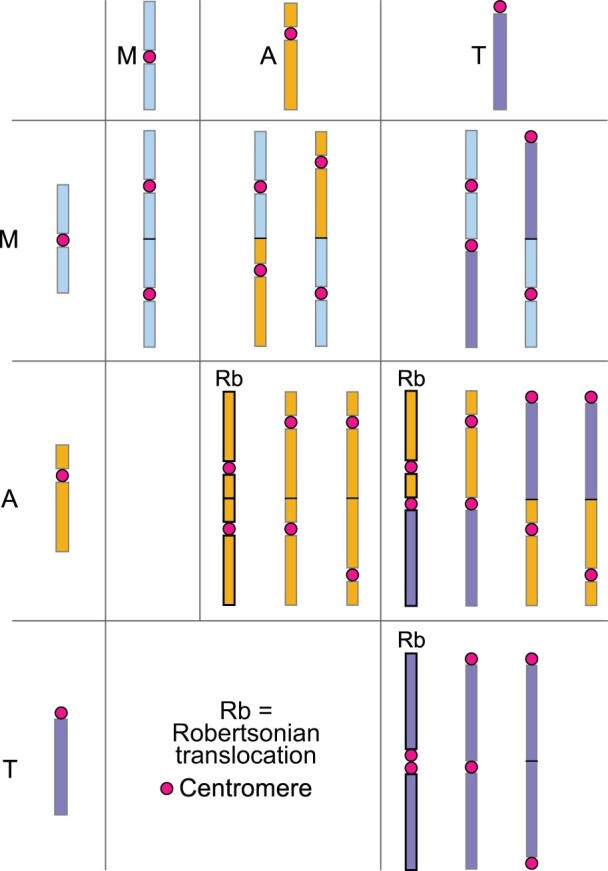
Fusion chromosomes resulting from all theoretically possible end-to-end fusions between different types of monocentric chromosomes. Only three chromosome types are shown: metacentric (M), acrocentric (A), and telocentric (T). With the exception of centric fusion (T/T translocation), all fusion chromosomes are primarily dicentric, with the two centromeres separated by a fluctuating distance. Fusions involving telocentric and acrocentric chromosomes, and probably sometimes between two acrocentric chromosomes, are referred to as Rb translocations/fusions.
When expressed sequence tag (EST) -based genetic maps and whole-genome sequences enabled comparative paleogenomics studies in plants, the inferred mergers of homoeologous chromosomes into a smaller number of linkage groups were often referred to as “chromosome fusions” or “fusions” (e.g. Murat et al., 2010; Salse, 2016; see Table 1 for additional references). Insufficient resolution of comparative genome maps and/or rearrangements shuffling the primary structure of fusion chromosomes, unknown ancestral karyotypes, or more than one equally parsimonious reconstruction of descending dysploidy do not always allow inference of the type of the translocation that mediated the inferred chromosome fusions. In other words, despite the fact that some chromosomes are graphically represented as resulting from end-to-end fusions, in fact, only some were formed by true EEFs.
Table 1.
Examples of end-to-end chromosome fusions reported in plant species
| Taxon | 2n | Family | Estimated no. of EEFs | Method: references |
|---|---|---|---|---|
| Eudicots | ||||
| Daucus carota | 18 | Apiaceae | 2 | G: Song et al. (2021) |
| Betulaceae | 16, 22 | Betulaceae | 1–7 | G: Li et al. (2021b) |
| Arabidopsis thaliana | 10 | Brassicaceae | 1 | C/G: Hu et al. (2011), Lysak (2014), Wang et al. (2015b) |
| Biscutella laevigata | 18 | Brassicaceae | ND | C/G: Geiser et al. (2016) |
| Boechereae (min. 5 genera) | 14 (21, 28) | Brassicaceae | 1 | C: Mandáková et al. (2015) |
| Brassica rapa | 20 | Brassicaceae | ND | G: Cheng et al. (2013) |
| Camelina neglecta | 12 | Brassicaceae | 1 | C: Mandáková et al. (2019) |
| Pugionium spp. | 22 | Brassicaceae | ND | C/G: Hu et al. (2021) |
| Origin of ancestral PCK (n = 7) genome from ancPCK (n = 8) | 14 | Brassicaceae | 1 | C: Mandáková et al. (2018) |
| Origin of ancestral karyotype of clade E (n = 7) from n = 8 genome | 14 | Brassicaceae | 1 | C: Mandáková et al. (2017a) |
| Microlepidieae (many genera) | 8–48 | Brassicaceae | 3 (Arabidella trisecta) 4 (Pachycladon spp.) ND (mult. spp.) |
C: Mandáková et al. (2010a, 2010b, 2017b) |
| Ricotia aucheri | 28 | Brassicaceae | 1 | C: Mandáková et al. (2018) |
| Cardamine cordifolia | 24 | Brassicaceae | 4 | C: Mandáková et al. (2016) |
| Megadenia pygmaea | 12 | Brassicaceae | 1 | C: Guo et al. (2021); G: Yang et al. (2021) |
| Cucumis sativus | 14 | Cucurbitaceae | 1 | C: Zhao et al. (2021) |
| Monocots | ||||
| Ancestral n = 8 genome, Cocos nucifera | 32 | Arecaceae | 2 + 5 | G: Wang et al. (2021c) |
| Spirodela intermedia | 36 | Lemnaceae | 1 | C: Hoang and Schubert (2017) |
| Aegilops tauschii | 14 | Poaceae | 1 | G: Luo et al. (2017) |
Listed are only EEF events documented by comparative (cyto)genomic analyses as of 2010 (author’s compilation). Note that the number of EEFs detected is actually higher. However, many of the published EEF events are referred to as chromosome fusions without specifying whether they resulted from EEF or Robertsonian translocation. The last column shows the method used to infer EEF: C (cytogenomics), G (genome sequence assembly).
Dicentric chromosomes and their stabilized variants, pseudodicentrics (Barra and Fachinetti, 2018), were reported in fungi (Schotanus et al., 2021), yeast (Pennaneach and Kolodner, 2009; Pobiega and Marcand, 2010; Gordon et al., 2011; Sato et al., 2012; Sankaranarayanan et al., 2020), insect (Imai and Taylor, 1989; Agudo et al., 2000), rodents (Matveevsky et al., 2020), fish (Liu et al., 2022), humans (Ijdo et al., 1991; Stimpson et al. 2010, 2012), and plants (reviewed by Lysak, 2014 and see Table 1).
Historical reminiscence
Probable end fusions started to be more frequently documented in the 1930s as dicentric chromosomes and corresponding anaphase bridges occurring spontaneously (Upcott, 1937; Darlington and Upcott, 1941) or after X-ray irradiation (Mather and Stone, 1933; McClintock, 1941) were observed. Some of the dicentric chromosomes arose by recombination between sister chromatids (or between homologous chromosomes) and entered the breakage–fusion–bridge (BFB) cycle (McClintock, 1939, 1941), some were dicentric Rb translocations producing a dicentric fusion chromosome and acentric fragment, and some may have been dicentrics arisen from end-to-end fusions. The uncertainty associated with the detection of EEFs was due to the methodological limitations that existed at the time and for many decades thereafter. Only in some favorable cases could EEFs be inferred on the basis of different morphological features of the chromosomes involved, such as centromere (Upcott, 1937) or heterochromatic knob positions (McClintock, 1941). When chromosomes lacked chromatin landmarks and/or the position of their centromeres was not apparent, or when one of them was eliminated immediately after the fusion, inference of (prior) EEFs was impossible. It was not until the advent of whole-genome sequence-based analyses and modern cytogenomic methods that EEFs were discovered to be an important mechanism of descending dysploidy (Lysak, 2014; Wang et al., 2015b; Birchler and Han, 2018).
I well remember the feeling of uncertainty of stepping into terra incognita when we concluded that not one but several chromosomes in species of the tribe Microlepidieae (Brassicaceae, the mustard family or crucifers) must have originated by EEFs and not by a double pericentric inversion and subsequent Rb-like translocations (Mandáková et al., 2010a, 2010b). At that time, overlooking McClintock’s X-ray-induced dicentric chromosome in maize (McClintock, 1941; see also Birchler and Han, 2018), this type of chromosomal rearrangement was apparently not reported for plants on such a scale. Due to the massive occurrence of EEFs in Microlepidieae and using newly available whole-genome sequences (Hu et al., 2011), the previously reconstructed origin of chromosome 2 in Arabidopsis thaliana (Lysak et al., 2006) was parsimoniously reinterpreted as an EEF between ancestral chromosomes 3 and 4 (Wang et al., 2015b). In animals, EEF was purported to mediate the origin of the single metacentric chromosome (n = 1) in the ant Myrmecia pilosula (jack jumper ant; Imai and Taylor, 1989) and for the origin of human chromosome 2, reducing the 48 chromosomes to 2n = 46 in hominoids (Ijdo et al., 1991).
In the last decade, several EEF events have been discovered in yeast, animal, and plant species (Table 1) as the number of sequenced and cytogenetically investigated genomes has rapidly increased, allowing comparisons of chromosomal collinearity between genomes with different chromosome numbers. An EEF event is assumed when two homoeologs in one genome are tandemly merged (end-to-end) within a single homoeolog in another genome.
How EEFs originate
Double-strand breaks (DSBs) introduced during meiosis are essential for maintaining the genetic variation, correct bivalent alignment on the metaphase I spindle, and faithful segregation of homologous chromosomes. Meiotic DSBs at telomeres or subtelomeres are repaired by homologous recombination. However, if DSBs occur in telomeres of nonhomologous chromosomes associated accidentally or clustered in the telomere bouquet (Figure 4), their repair by nonhomologous end joining (NHEJ) or nonallelic homologous recombination (NAHR), using a nonsister chromatid (sub)telomere as a template, may result in EEF.
Telomeres are nucleoprotein complexes that protect the ends of eukaryotic chromosomes that would be otherwise recognized by the cell as DSBs and processed by the activated DNA damage response (e.g. Muraki and Murnane, 2017; Lee et al., 2021). The shelterin complex, formed of six telomere-specific proteins, associates with and protects telomere ends from degradation and the formation of EEFs. Dysfunctional shelterin-based telomere capping is one of the factors promoting illegitimate telomere–telomere recombination. For example, transient removal of TRF2, one of the shelterin subunits important for the formation of telomeric t-loops, results in a high frequency of EEFs between nonhomologous human chromosomes (Stimpson et al., 2010). Similarly, EEFs were observed in Trf1-deficient mouse spermatocytes (Wang et al., 2018a, 2018b).
EEFs can also result from telomere shortening due to a telomerase deficiency or DNA polymerase failing to replicate the lagging telomeric strand, making telomeres progressively shorter and prone to fusions with nonhomologous telomeric sequences. EEFs were induced in telomerase-deficient A. thaliana mutant plants (e.g. Riha et al., 2001) and budding yeast mutants (Pennaneach and Kolodner, 2009). In naturally occurring wild house mice, Rb translocations were found to be more frequent in mice with shortened telomeres (Sanchez-Guillen et al., 2015). As the repetitive (sub)telomeric sequences are difficult to replicate, telomere fusions may originate more often from replication fork stalling and collapse than from shelterin uncapping or the formation of spontaneous DSBs (Stroik and Hendrickson, 2020). When telomerase is not expressed, some cells and organisms elongate their telomeres by NAHR, known as alternative lengthening of telomeres, which often leads to EEFs (Kim et al., 2021; Lee et al., 2021).
Overall, telomere shortening, dysfunctional telomere capping, aberrant replication of (sub)telomeric sequences, and meiotic and somatic DSBs facilitate EEFs by misrepairing these breaks through NHEJ or NAHR.
Where and when EEFs occur
For EEF to occur, the ends of two different chromosomes should be in close physical proximity, which can be promoted, for example, by increased mobility of dysfunctional telomeres and DSB ends (Lottersberger et al., 2015 and references therein). The ends of any two nonhomologous chromosomes can be accidentally juxtapositioned throughout the cell cycle. However, the spatial arrangements of chromosomes during interphase and meiosis are probably most relevant for the formation of chromosome translocations including EEFs.
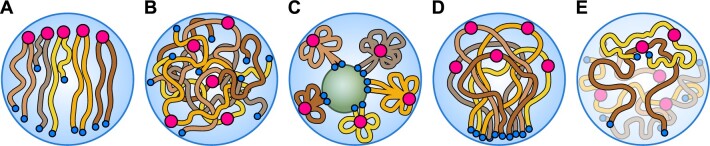
Since the seminal work of Dong and Jiang (1998), the organization of chromosomes during interphase in plants remains largely unexplored. Here we recognize three main patterns of interphase chromosome configuration: (1) Rabl, (2) non-Rabl, and (3) rosette-like organization (i.e. nucleolus-associated clustering of telomeres) (Figure 5). The Rabl configuration (Rabl, 1885) refers to the lateral (horseshoe-like) arrangement of chromosome arms distributing the centromeres and telomeres approximately to the opposite nuclear poles (Figure 5A). The non-Rabl pattern (Figure 5B) is the least well-known (see Oko et al., 2020) and refers to nuclei where chromosome arms are not arranged along one axis. Instead, the centromeres are located at the nuclear periphery, and (some) telomeres are anchored in the nuclear interior or at the nuclear periphery, or the centromeres, and telomeres are distributed throughout the nuclear interior (Zhou et al., 2019; Shan et al., 2021). The rosette-like pattern (Figure 5C; Fransz et al., 2002) is known in the genus Arabidopsis and other Brassicaceae species with small genomes (Shan et al., 2021). In these nuclei, the centromeres (chromocenters) are located at the nuclear periphery, while the telomeres are attached to the nucleolus. The “rosette” (Tiang and Pawlowski, 2012) alludes to chromosome-arm loops being anchored to the chromocenters and the chromosome ends associated with the nucleolus (Fransz et al., 2002).
Figure 5.
Interphase chromosome configurations and interactions between nonhomologous chromosomes during meiotic prophase I. A, Rabl configuration. B, Non-Rabl configuration. C, Rosette-like configuration (nucleolus-associated telomere clustering). D, Telomere bouquet in prophase I. E, Prebouquet and postbouquet chromosome dynamics allowing centromere–telomere and telomere–telomere interaction. Centromeres and telomeres are shown as red circles and blue dots, respectively.
Although centromeres and telomeres in an interphase nucleus are expected to be positioned to avoid their undesirable association and chromosomal rearrangements (see e.g. Pouokam et al., 2019), all types of interphase organization allow interactions between nonhomologous chromosomes (Figure 5). In Rabl and rosette-like configurations (Figure 5A and C), the ends of nonhomologous chromosomes are brought into close physical proximity permitting interactions between nonhomologous telomeres (e.g. Ashley, 1979). In nuclei with a non-Rabl organization (Figures 3A and 5B), some nonhomologous telomeres are likely to come into contact on the nuclear envelope or inside the nucleus. Numerous studies localizing telomeres within interphase nuclei by fluorescence in situ hybridization have reported lower than expected (2n × 2) numbers of telomeric signals (e.g. Joachimiak, 1987; Armstrong et al., 2001; Shan et al., 2021), suggesting that the (sub)telomeric chromatin of homologous and nonhomologous chromosomes may be closely linked and possibly recombine by NAHR. In all types of interphase organization, nucleolar organizer regions (NORs) positioned on chromosomes terminally may facilitate interactions between nonhomologous telomeres in the nucleolus. Such a fusion (primarily dicentric) chromosome would have a hybrid NOR positioned interstitially within one of its arms.
The closest physical proximity of telomeres is achieved during meiotic prophase I, when telomeres cluster to form a telomeric bouquet (Figure 5D). The bouquet is formed by telomere tethering to the inner nuclear membrane (INM) of the nuclear envelope in leptotene/zygotene (e.g. Scherthan, 2001), followed by homologous chromosome pairing initiated at telomeres (most likely at subtelomeric sequences), toward the fully synapsed homologs (bivalents) in zygotene and pachytene. In genomes with the Rabl configuration, the bouquet is formed when chromosome ends positioned at one nuclear pole move and cluster in a small area on the INM (Carlton et al., 2003; Sheehan and Pawlowski, 2009; Richards et al., 2012; Alleva and Smolikove, 2017). However, the bouquet is also formed in plants with non-Rabl configuration (e.g. in rice, Zhang et al., 2017; Zhou et al., 2019) or telomeres associated with the nucleolus during premeiotic interphase (e.g. Arabidopsis: Varas et al., 2015; Hurel et al., 2018). The proximity of nonhomologous telomeres in the bouquet (Figure 5D), in combination with meiotic DSBs at the chromosome termini or aberrant telomeric chromatin (see above), can lead to EEFs. Because telomeres are bound to the INM in most eukaryotes from leptotene to pachytene, their movements along the INM may mediate accidental inter-chromosomal contacts and recombination before and after the bouquet stage (Figure 5E). The origin of end-to-end (tandem) fusions and other inter-chromosomal rearrangements on the INM during meiosis has been advocated primarily by Hirotami Imai with his minimum interaction theory (Imai, 1986, Imai et al., 2001). The formation of dicentric pachytene bivalents resulting from recombination between telomeres on short arms of acrocentric chromosomes has been observed in rodents (Matveevsky et al., 2020).
As telomeric repeats are highly conserved and not chromosome-specific, homologous recognition, and pairing in meiotic prophase I, which is mostly initiated at terminal chromosomal regions, might actually rely on chromatin similarity at subtelomeres (e.g. in a review by Aguilar and Prieto, 2021). On the other hand, subtelomeres could be extremely variable due to amplification and mutation of satellite repeats and transposable elements, segmental duplications, and increased meiotic recombination in terminal chromosomal regions (Linardopoulou et al., 2005; Louis and Vershinin, 2005; Muraki and Murnane, 2017; Aguilar and Prieto, 2021). As a result, the subtelomeric repeat-rich regions can be implicated as hotspots of DSBs and inter-chromosomal NAHR in some species (Linardopoulou et al., 2005; Aguilar and Prieto, 2021), provided that at least some repeats on nonhomologous chromosomes are in opposite directions, allowing chromosome fusion (Louis and Vershinin, 2005; Birchler and Han, 2018; Oizumi et al., 2021). It can be assumed that some chromosomal fusion events thought to be telomere-to-telomere fusions, in fact, arise from recombination in subtelomeric regions.
Dicentric chromosome and centromere elimination
Probably, the most puzzling questions related to EEFs are (1) how the cell copes with two centromeres after an end fusion between two monocentric chromosomes and (2) what are the consequences of chromosome elongation by adding an entire chromosome, that changes the morphology, and overall length and weight of the fusion chromosome?
In eukaryotes with localized centromeres, a single functional centromere is the preferred evolutionary solution, whereas chromosomal dicentricity usually has deleterious consequences. In dicentrics, the magnitude of this problem is likely to increase with increasing distance between the two centromeres. When the short-arm ends of two acrocentric chromosomes recombine (e.g. Ijdo et al., 1991; Stimpson et al., 2010), the resulting distance between two centromeres is shorter than, for example, the distance after joining short and long arms of two acrocentrics (Figure 4). It is assumed that the shorter the distance between the two centromeres, the greater the chance that both centromeres will bind kinetochore microtubules from only one cell pole. Some dicentrics can probably segregate regularly for a time, but it is more likely that the two centromeres will be pulled to opposite poles, an anaphase bridge will form, and the chromosome will break between its two centromeres. In sporophytic tissues, the broken chromosomes can be rescued by de novo telomere addition (telomere healing) or enter the BFB cycle through recombination with another chromosome with compromised telomere(s). In gametophytic tissues, the uncapped chromosomes may enter either the chromatid- or chromosome-type BFB cycle (McClintock, 1939, 1941; see also Birchler and Han, 2018). It is possible that some dicentric anaphase bridges are resolved by a break at the fusion breakpoint and the two original monocentric chromosomes are restored. Indeed, this was observed in 40% of dicentrics in the budding yeast (Saccharomyces cerevisiae) and suggested as a rescue pathway to avoid dicentric chromosome fusions (Pobiega and Marcand, 2010).
To avoid dicentricity and stabilize the newly formed fusion chromosome, one centromere must be suppressed and/or eliminated (Figure 3A). Based on current paradigms in cell biology, recombination and/or epigenetic silencing have been proposed as mechanisms of centromere elimination. However, the exact mechanism of centromere elimination on dicentric chromosomes is not entirely clear (Barra and Fachinetti, 2018), and the process may well have different features in different cells and organisms.
A dicentric chromosome can be stabilized by deletion of chromatin containing CENP-A/CENH3 nucleosomes (reviewed in MacKinnon and Campbell, 2011; Lysak, 2014; Barra and Fachinetti, 2018). Probably, the best-studied centromere elimination is that on human chromosome 2, where one of the two ancestral centromeres was most likely removed by a one-step excision of higher-ordered centromeric satellite DNA, rather than by stepwise recombinational removal or epigenetic inactivation (Chiatante et al., 2017). Centromere deletions can occur through a single DSB, followed by end resection and NHEJ-based repair (Pennaneach and Kolodner, 2009, and references therein). Two DSBs within homologous (peri)centromeric sequences (e.g. between centromeric tandem repeats or between LTRs of two different retrotransposons) can also lead to deletion of the centromere (Ma and Bennetzen, 2006; Stimpson et al., 2010). Nevertheless, available data on dicentrics in yeast, animals, and plants suggest that epigenetic mechanisms are more commonly involved in centromere inactivation (for reviews, see, e.g. MacKinnon and Campbell, 2011; Fu et al., 2012; Stimpson et al., 2012; Liu et al., 2020). The functional and temporal relationships between centromere excision and epigenetic (functional) centromere inactivation remain to be elucidated.
Many inactive centromeres on pseudodicentric chromosomes lack the centromere-specific CENP-A/CENH3 nucleosomes and essential kinetochore proteins (e.g. CENP-C), whereas the corresponding DNA sequence is either (1) intact, (2) partially excised, or (3) completely eliminated (e.g. Han et al., 2009; Stimpson et al., 2010). Because the identity and function of the centromere are epigenetically determined, primarily by CENH3 replacing the canonical histone H3 in centromeric nucleosomes, the process of centromere inactivation should ensure that CENH3 is loaded into only one of the two centromeric regions (Dawicki-McKenna and Black, 2019). One possibility is to modify centromeric chromatin by DNA hypermethylation, as observed in maize pseudodicentrics (Koo et al., 2011). In a rye-wheat translocation chromosome with a hybrid centromere, hypermethylation of wheat centromeric repeats was purported to be responsible for the binding of CENH3-containing nucleosomes only to rye repeats (Karimi-Ashtiyani et al., 2021). Centromere inactivation is often associated with enhanced levels of methylation of histone 3 (H3K27me2 and H3K27me3, Zhang et al., 2010). This has also been observed in artificially generated dicentric chromosomes in fission yeast, where the central centromere domain was heterochromatinized by dimethylation of histone 3 on lysine 9 (H3K9me2) and hypoacetylation (H3K14 and H3K9) (Sato et al., 2012). In general, based on the limited data, epigenetic marks specific for euchromatin are thought to be modified to marks specifying (inactive) heterochromatin. However, in the long term, heterochromatin of the inactive centromere may be reduced, as shown during centromere decay after centromere repositioning on cucumber chromosomes (Han et al., 2009), followed by recombinational removal of (peri)centromeric repeats. In some cases, the inactive and likely heterochromatinized centromere can be epigenetically reactivated (Han et al., 2009).
If a dicentric chromosome should become stable, centromere elimination must be rapid to minimize the risk of defective spindle attachment, anaphase bridge formation, and entry into the BFB cycle. Pulling both centromeres to the same pole can rescue the dicentric chromosome shortly after its formation, and it is likely that faithful segregation can occur repeatedly in subsequent divisions (e.g. Stimpson et al. 2010). Recently, the time frame of centromere inactivation on dicentric chromosomes in maize was investigated using gamma-irradiated pollen in plants with B chromosomes. Some of the generated dicentrics showed centromere inactivation within one or only a few cell cycles (Liu et al., 2020), confirming previous observations of centromere inactivation on maize dicentric chromosomes (Han et al., 2006; Gao et al., 2011). The number of EEFs detected in various eukaryotes (see above and Table 1) indirectly suggests that centromere inactivation may occur early enough to ensure stabilization and regular segregation of some pseudodicentric chromosomes.
Longer chromosomes need a stronger centromere
When a new pseudodicentric chromosome is stabilized by centromere elimination, its “old kinetochore” may not be able to bind a sufficient number of spindle microtubules to ensure optimal mobility and segregation of sister chromatids or homologous chromosomes of increased length and weight. There is a lower limit to the number of kinetochore microtubules (e.g. McEwen et al., 1998), and centromere size appears to be positively correlated with chromosome and genome size in eukaryotes (Plačková et al., 2021), particularly in plants (e.g. Jenkins and Bennett, 1981; Wang et al., 2021a). It has been demonstrated experimentally that centromeric CENH3 binding domains can expand significantly when chromosomes with small(er) centromeres are introgressed into a genome with predominant large(er) centromeres (Wang et al., 2014; Wang et al., 2021a). Consequently, the functional centromere of the pseudodicentric chromosome is expected to become “stronger” as the CENH3-binding domain expands, increasing the size of the kinetochore and the number of kinetochore microtubules. It could be investigated whether fusion chromosomes formed by EEFs have larger (“stronger”) centromeres than (1) other chromosomes in the karyotype and (2) the corresponding homoeologous (nonfused) chromosomes in related genomes. Alternatively, chromosomes with larger centromeres might be more tolerant to EEFs because their centromeres would be more compatible with increased chromosome size (Plačková et al., 2021).
Fusion chromosome becomes longer
Another mechanistic problem of pseudodicentric chromosomes is their altered morphology and length due to elongation of one arm by joining an entire chromosome (Figures 3A and 4). If a chromosome arm becomes too long, movement and segregation of the fusion chromosome during mitosis and meiosis could be negatively affected. Eukaryotic chromosomes have an upper size limit as demonstrated experimentally using engineered chromosomes in field bean lines (Schubert and Oud, 1997). The authors showed that “the longest chromosome arm must not exceed half of the average length of the spindle axis at telophase.” If the sister chromatid arms are too long, they do not separate completely in mitosis and their distal parts could be broken off during the formation of a new cell wall. The consequences can be fatal for the cell progeny containing the chromosome with the terminal deletion. Congruently, by analyzing 68 eukaryotic genomes, Li et al. (2011) showed that there is an upper limit to chromosome size variation in diploid eukaryotes and that chromosome lengths in most genomes tend to center around a species-specific average chromosome length. A testable assumption might be that in karyotypes containing chromosomes of different size, EEFs among short chromosomes or among short and long chromosomes will occur more frequently than end fusions between long chromosomes, analogously to fusions/fissions among microchromosome and macrochromosomes during vertebrate genome evolution (Waters et al., 2021). Similarly, EEF-mediated elongation of the long arm of acrocentric and telocentric fusion chromosomes may exceed the length limit for chromosome arms, whereas pseudodicentrics with arms of approximately equal size are more likely to be regularly transmitted (Figure 4). The upper limit to chromosome size (besides other constraints) may also explain why most documented EEFs involved two but not three or more chromosomes.
EEFs in organisms with holocentric chromosomes
EEFs should be more common in organisms with holocentric chromosomes because segregation of fusion chromosomes is not hindered by two centromeres (Mandrioli and Manicardi, 2020; Kim et al., 2021). Indeed, eukaryotic taxa with holocentric chromosomes are often characterized by long rows of chromosome numbers, as has been reported for sedges (Carex) and butterflies (Guerra, 2016; Hill et al., 2019; Mandrioli and Manicardi, 2020). Although chromosome fusions and fissions were readily inferred from altered chromosome number and size (Hill et al., 2019), only recent advances in comparative genomics, based on whole-genome sequencing, have allowed a reassessment of the role of EEFs (and chromosome fissions) in the evolution of karyotypes with holocentric chromosomes.
EEF events have been implicated to drive descending dysploidies in butterflies (e.g. Hill et al., 2019; Cicconardi et al., 2021), with the caveat that some chromosome fusions may be Rb translocations (Hill et al., 2019). Recently, comparisons of whole-genome assemblies in three beak-sedge species (Rhynchospora) identified EEFs as the predominant mechanism of descending dysploidy in this genus with holocentric chromosomes (P. G. Hofstatter et al., 2022, manuscript under review). A 172-bp centromeric repeat was present at most fusion breakpoints, whereas an interstitial telomeric repeat was detected at a single fusion site. In Rhynchospora, numerous EEFs mediated structural genome diploidization after whole-genome duplications (WGDs), analogous to postpolyploid chromosomal diploidization documented in angiosperms with monocentric chromosomes (Mandáková and Lysak, 2018).
Nested chromosome fusions
NCF, also termed nested chromosome insertion (NCI), is a mechanism of descending dysploidy that combines two nonhomologous chromosomes by the apparent insertion of one chromosome between the chromosome arms of another one (Figure 3B). The centromere of the inserted chromosome remains the functional centromere of the fusion chromosome, while the centromere on the recipient chromosome loses its function (Schubert and Lysak, 2011). A minimum of three DSBs are required for NCF to occur: one at the (peri)centromere of the recipient chromosome and two at both (sub)telomeres of the inserted chromosome (Luo et al., 2009; Schubert and Lysak, 2011; Wang et al., 2015a, 2015b). If telomere length and/or protein capping of the two chromosome ends are compromised, only one DSB at the recipient chromosome (peri)centromere is required (Birchler and Han, 2018).
Historical reminiscence
NCFs were first described by Luo et al. (2009) in the tribe Triticeae (grasses, Poaceae) as the dominant mechanism of descending dysploidy, reducing the number of chromosomes from n = 12 to n = 7. Further studies confirmed NCFs as a common mechanism of descending dysploidy in grasses (International Brachypodium Initiative, 2010; Murat et al., 2010 and references in Table 2). The discovery of NCFs has been driven by methodological advances. Luo et al. (2009) compared the EST-based genetic map of Aegilops tauschii (n = 7) with similar orthologous sequences in sorghum (n = 10) and rice (n = 12) genome. At a later stage, chromosome-level genome assemblies (International Brachypodium Initiative, 2010; Luo et al., 2018; Wang et al., 2019; see Table 2 for more examples) and chromosome-specific painting probes (e.g. Mandáková et al., 2010b) accelerated the identification of NCF events primarily in grass genomes and, to a lesser extent, in eudicots (Table 2).
Table 2.
NCFs in plants
| Taxon | 2n | Family/subfamily | Estimated no. of NCFs | Method: references |
|---|---|---|---|---|
| Eudicots | ||||
| Betulaceae | 16, 22 | Betulaceae | 2, 2 | G: Li et al. (2021b) |
| Hornungia alpina | 12 | Brassicaceae | 1 | C: Lysak et al. (2006) |
| Pachycladon spp. | 20 | Brassicaceae | 1 | C: Mandáková et al. (2010b) |
| Cardamine pratensis | 30 | Brassicaceae | 1 | C: Mandáková et al. (2013) |
| Cucumis sativus | 14 | Cucurbitaceae | 1 | C: Yang et al. (2014) |
| Phaseolus leptostachyus | 20 | Fabaceae | 1 | C: Fonsêca et al. (2016) |
| Malus × domestica | 34 | Rosaceae | 1 | G: Velasco et al. (2010) |
| Monocots | ||||
| Aegilops tauschii | 14 | Poaceae/Pooideae | 4 | G: Luo et al. (2009, 2017) |
| Brachypodium distachyon | 10 | Poaceae/Pooideae | 7 | G: International Brachypodium Initiative (2010); C: Lusinska et al. (2019) |
| Cynodon dactylon | 36 | Poaceae/Chloridoideae | 2 | G: Fang et al. (2020) |
| Eremochloa ophiuroides | 18 | Poaceae/Panicoideae | 3 | G: Wang et al. (,2021b) |
| Secale cereale | 14 | Poaceae/Pooideae | 2 | G: Li et al. (2021a) |
| Setaria italica | 18 | Poaceae/ Panicoideae | 3 | G: Zhang et al. (2012) |
| Sorghum bicolor | 20 | Poaceae/ Panicoideae | 2 | G: Luo et al. (2009) |
| Zea mays | 20 | Poaceae/ Panicoideae | 17 | G: Wang and Bennetzen (2012) |
| Zoysia japonica | 40 | Poaceae/Chloridoideae | 2 | G: Wang et al. (2015a) |
| Grasses (summary articles) | 10–40 | Poaceae | 1–17 | G: Murat et al. (2010), Salse (2016), Wang et al. (2019); Ma et al. (2021), Wang et al. (2021a, 2021b) |
Only NCF events documented by comparative (cyto)genomic analyses from 2009 onwards are listed (author’s compilation). The last column shows the method used to infer NCF: C (cytogenomics), G (genome sequence assembly).
How NCFs originate
The origin of NCFs is associated with several questions that have not yet been satisfactorily answered. First, the nature of the rearrangement requires that two telomeric ends of the inserted chromosome become “sticky” simultaneously with a break in the (peri)centromere of the recipient chromosome. It can be assumed that intra-chromosomal NAHR between the (sub)telomeric repeats on the inserted chromosome makes its two ends “sticky” to either form a ring chromosome (e.g. Schubert, 2021) or to be prone to recombination with the (peri)centromeric “sticky” ends of the recipient chromosome (Wang et al. 2015a, 2015b). Second, there must be a mechanism to ensure that in the case of multiple DSBs, only the four free ends of only two chromosomes recombine to form the tripartite structure typical of NCF events (Figure 3B). In other words, without such a constraint, the two free (peri)centromeric ends could recombine with the free telomeric ends of more than one chromosome and, conversely, two “sticky” telomeric ends could recombine with free (peri)centromeric ends of two (or more) different chromosomes. Third, if the (peri)centromeric breakage per se and/or subsequent end resection of free centromeric ends do not remove the CENH3-containing nucleosomes, a NCF would generate a dicentric or tricentric fusion chromosome with one strong centromere from the inserted chromosome and one or two weaker centromere(s) of the recipient chromosome. Thus, the centromere of the recipient chromosome or its segments must be inactivated or deleted to stabilize the fusion chromosome (Figure 3B).
Centromere and telomeres after NCF
Several chromosome-level genome assemblies of grass species (Table 2) should be sufficient to provide information on the fate of the (sub)telomeric repeats of the inserted chromosome and the (peri)centromere of the recipient chromosome after NCF events. Nevertheless, as these repeats have usually not been identified on fusion chromosomes in grass species (cf. references in Table 2), the exact molecular mechanism of NCFs and subsequent evolution at fusion breakpoints are unknown. When a break occurs in the centromere, or perhaps more likely within the repeat-rich pericentromere on the recipient chromosome (Luo et al., 2009, 2017; Wang and Bennetzen, 2012; Wang et al., 2019), the megabase-sized recipient centromere is disrupted and its two segments shifted toward the middle position on both arms of the nascent fusion chromosome. Thus, both arms of the fusion chromosome should contain adjacently arranged (peri)centromeric and telomeric repeats (Figure 3B). However, as peaks of pericentromeric repeats at the fusion breakpoints have only rarely been reported, it is assumed that the repeats are deleted during or after the resealing. In Brachypodium distachyon (n = 5), footprints of the recipient chromosome centromere in the form of centromeric (Bd_CENT) repeats and retrotransposons were detected on three chromosomes (International Brachypodium Initiative, 2010). The footprint of a 141-bp array of telomeric repeats remained on chromosome 2D in Ae. tauschii (Luo et al., 2017). However, when detecting sequence footprints of NCFs, it should be taken into account that despite conserved chromosomal collinearity, centromeres may have migrated along chromosomes (Schneider et al., 2016; Mandáková et al., 2020), and therefore centromeres on ancestral, nonfused chromosomes in related genomes may have changed their positions since the time of the NCF.
Where and when NCFs originate
Because the repeat-rich (peri)centromeres of various eukaryotic organisms are frequently implicated in chromosomal rearrangements, these chromosomal regions are considered prone to breakage and (mis)repair by NHEJ or NAHR (reviewed by Barra and Fachinetti, 2018). Centromere misdivision, that is, centromere rupture in a univalent after the kinetochore has been attached to the spindle microtubules from both cell poles (Sears, 1952; Birchler and Han, 2018), could be one of the mechanisms for centromere breakage. Although meiotic DSBs and crossovers are suppressed within centromeres (Nambiar and Smith, 2016), DSBs can occur within (peri)centromeres (Fernandez et al., 2019). Therefore, breakage within the recipient chromosome pericentromere due to meiotic DSBs, centromere misdivision, or the tension-driven resolution of interlocked chromosomes (Wang et al., 2009) are plausible events. However, the tripartite structure of fusion chromosomes implies that some spatial (and temporal) constraint must be imposed to ensure that the “sticky” ends of only one chromosome are involved in a (peri)centromere break repair on just one recipient chromosome. Thus, where can centromeres and telomeres come into close physical contact to generate an NCF? Because chromosomes are not static chromatin structures during interphase (e.g. Tortora et al., 2020; Arpon et al., 2021) and meiosis (e.g. Carlton et al., 2003; Sheehan and Pawlowski, 2009; Alleva and Smolikove, 2017; Aguilar and Prieto, 2021), centromeric and telomeric chromatin can co-localize and recombine, particularly in nuclei with a non-Rabl configuration (Figures 3B and 5B). In contrast, Rabl and rosette-like interphase configurations support the spatial separation of centromeres and telomeres (Figure 5A and C). The observation that fusion chromosomes formed by NCFs appear to be rare during plant genome evolution (Table 2) may indirectly support the notion that centromere–telomere recombination à la NCF may be infrequent and/or that the nascent fusion chromosomes are not stable. However, many NCF events were probably “overwritten” by other chromosomal rearrangements, so their actual numbers may be underestimated in comparative genomics analyzes.
In contrast to the different organization of centromeres and telomeres in interphase, the telomere bouquet is a universal hallmark of meiosis in most eukaryotes (Scherthan, 2001). Here, I explore the possibility that NCFs may (also) form during the meiotic prophase I. As discussed above for the origin of end-to-end fusion chromosomes, DSBs start to occur from leptotene, coinciding with tethering of telomeres on the INM during the leptotene/zygotene transition. The formation of the telomere bouquet (Figure 5D) is associated with chromosome movement along the INM (Carlton et al., 2003; Sheehan and Pawlowski, 2009) and possible contacts between telomeric and centromeric regions of unsynapsed nonhomologous chromosomes. Similarly, the transition from the bouquet to fully synapsed pachytene bivalents is marked by the dispersal of telomeres at the INM in the late zygotene (Wang et al., 2018a, 2018b). At this stage, some centromeres and telomeres are brought into close contact (Figure 5E; Sepsi et al., 2018). There is evidence that some nonhomologous, unsynapsed, or synapsed chromosomes may be entangled or interlocked from the prebouquet stage to early/mid pachytene (Wang et al., 2009; Sepsi et al., 2017; Martinez-Garcia et al., 2018). Interlocks are resolved by chromosome movement and/or breakage and resealing, up to their full resolution in the late pachytene (Wang et al., 2009; Martinez-Garcia et al., 2018); however, centromere–telomere interlocks have not been reported in the plant species analyzed (Wang et al., 2009).
NCFs in grasses
Because of the economic importance of cereal grasses, comparative genomics in Poaceae is ahead of comparable analyses in other land plant families. This partly explains why NCF events have been documented so frequently in grasses. However, several lines of evidence suggest that NCF is indeed an important mechanism of chromosome number reduction in Poaceae (Luo et al. 2009, 2017; Murat et al., 2010; Wang et al., 2019, Ma et al., 2021; Table 2). Not surprisingly, the seminal work of Dong and Jiang (1998), showing that not all plants have the Rabl interphase configuration, was based on a comparative analysis of grass genomes. Despite major advances in comparative genomics of grasses, only a tiny fraction of the more than 11,500 species (Soreng et al., 2017) have been examined for the presence of NCFs, and even less is known about potentially relevant intrinsic genomic traits underlying NCFs, such as interphase chromosome organization, repeat diversity and abundances, or meiotic chromosome pairing.
NCF events mediated the reduction in chromosome number during genome diploidization after several clade-specific WGDs in grasses (e.g. Murat et al., 2010; Salse, 2016; Ma et al., 2021). After the last clade-specific ρ-WGD followed by descending dysploidy, the 12 pairs of ancestral chromosomes were retained in Pharus latifolius (Pharoideae; Ma et al., 2021) and in the rice genome (Oryzoideae). In other grass subfamilies (Chloridoideae, Panicoideae, and Pooideae), the 12 ancestral chromosomes were reduced to n = 10, 9, 7, 5, and 2 (Saunders and Houben, 2001; Kotseruba et al., 2005; International Brachypodium Initiative, 2010; Betekhtin et al., 2014; Salse, 2016; Luo et al., 2017; Lusinska et al., 2019; Wang et al., 2019; Ma et al., 2021). Interestingly, NCF was a dominant mechanism mediating the independent descending dysploidies in all three grass subfamilies. Independent NCFs involving different progenitor chromosomes over more than 40 million years of grass genome evolution (Ma et al., 2021) strongly suggest that some intrinsic genomic features predispose grass chromosomes to NCFs. To elucidate why NCFs are common in grasses, more comprehensive comparative studies to identify NCF-triggering factors in combination with artificially induced DSBs that would mimic the onset of NCF events are needed.
Evolutionary impacts of EEF and NCF events
The effects of fusion chromosomes on their carriers and their long-term adaptive significance including their role in speciation are poorly understood (Guerrero and Kirkpatrick, 2014). While attention has been paid to impacts of Rb fusions, end-to-end fusions have been studied only marginally, and virtually nothing is known about the evolutionary impacts of NCFs.
As the formation of fusion chromosomes reduces the total number of chromosomes in a complement by one (n − 1), the number of centromeres (−1) and telomeres (−2) is also reduced, and thus the number of potential substrates for ectopic recombination (NAHR) between nonhomologous chromosomes. It could be modeled whether an initial wave of EEF and NCF events, for example after genome duplications (Mandáková and Lysak, 2018; Cheng et al., 2020), slows the rate of chromosome fusion events. However, the reduction in the number of centromeres/telomeres can be compensated by amplification of repeats in pericentromeric and subtelomeric regions, which increases the probability of NAHR, at least in plants. While EEF and NCF events may reduce the probability of the usually deleterious telomere–telomere and telomere–centromere recombination, the increasing length of fusion chromosomes may affect their own organization and the organization of nonfused chromosomes in interphase and during meiosis. This was demonstrated by Vara et al. (2021) by comparing mouse individuals with 40 acrocentric chromosomes to those with lower chromosome numbers (2n = 39−28) resulting from Rb translocations (centric fusions). In the studied spermatocytes, the lower number of longer metacentric chromosomes altered the nuclear occupancy of the fused, but also nonfused chromosomes and their mutual physical interactions.
Both EEFs and NCFs decrease per chromosome recombination rates (e.g. Cicconardi et al., 2021; Vara et al., 2021), which can have both positive (adaptive) and negative (maladaptive) effects on carriers of such dysploid karyotypes. In eukaryotes, recombination rates are usually suppressed or low near (peri)centromeres and increase toward the chromosome ends. Therefore, in the case of EEF events (Figure 3A), the high recombination rate in the distal regions of two merging chromosome arms is likely to decrease in these regions after the end-to-end fusion. Similarly, after NCF (Figure 3B), the recombination landscapes of all four chromosome arms involved are altered: the highly recombining distal regions of the inserted chromosome are positioned more proximally within the fusion chromosome (lower recombination rate on average), whereas genes that were originally located at or adjacent to the pericentromere of the recipient chromosome are shifted more distally within the fusion chromosome (higher recombination rate on average). Recombination landscapes were analyzed on fusion chromosomes formed by EEFs and NCIs in Ae. tauschii (Luo et al., 2017). However, the expected peaks of altered recombination rates along fusion chromosomes were not detected, likely due to recombination conforming to rates typical of different regions on grass chromosomes (Luo et al., 2017).
All dysploid events can lead to the establishment of reproductive barriers and eventually to speciation due to the compromised fertility of hybrids from crosses between individuals with different chromosome numbers. Genome comparison of three stickleback fish species showed that two independent chromosome fusions occurred in two species, reducing chromosome number from 23 to 21 pairs (Liu et al., 2022). The fusion chromosomes differed from nonfused ones by an enrichment of quantitative trait loci facilitating freshwater adaptation, likely due to low recombination rates on the fusion chromosomes. Luo et al. (2018) used clustered regularly interspaced short palindromic repeats-Cas9 (CRISPR-Cas9)-mediated genome editing to produce lines of S. cerevisiae with chromosome numbers ranging from n = 16 (the wild-type) to n = 2. They showed that eight fusion events (n = 16 → n = 8) resulted in almost complete reproductive isolation between the n = 16 and n = 8 strains. Future experimental research based on engineered fusion chromosomes (cf. Luo et al., 2018) will provide more precise evidence on the immediate and long-term effects of chromosome fusions.
Outlook and outstanding questions
As mentioned, the emerging evolutionary significance of EEF and NCF events in the evolution of eukaryotic genomes has been driven by methodological advances. However, the triggering factors and evolutionary significance of these mechanisms of descending dysploidy are still not fully understood. Genome-wide analyses across a broader range of eukaryotic clades and phyla will likely reveal instances of nascent or evolutionarily young fusion chromosomes and karyotypes formed by EEFs/NCFs. This should be key to better understanding the origin of such fusion chromosomes and their immediate and long-term consequences for their carriers, including their role in speciation. An attractive alternative to naturally occurring EEF and NCF events in status nascendi is genome editing applied (1) to simultaneously generate DSBs at (sub)telomeres and (peri)centromeres and (2) to remove centromeric and/or telomeric sequences on engineered fusion chromosomes (e.g. Adikusuma et al., 2017; Luo et al., 2018). The engineered fusion chromosomes would also allow analysis of recombination rate, epigenetic changes, and potentially altered gene expression along the newly formed fusion chromosomes.
The currently known EEF and NCF events raise several research questions. Are some chromosomes more likely to be involved in fusions than others, and what factors determine a chromosome’s susceptibility to “fusions”? How do chromosome organization in interphase and meiotic chromosome pairing affect EEF- and NCF-mediated chromosome fusions, and what are the usual DNA sequence and/or epigenetic features of fusion breakpoints? Does the cell impose any mechanistic (or other) constraints on the length of fusion chromosomes?
The fate of the two centromeres after EEF and NCF needs further investigation. When an end fusion occurs between two monocentric chromosomes, which of the two centromeres remain functional? How does the cell achieve inactivation and eventual deletion of the nonfunctional centromere, and, importantly, what is the time window in which this process can be accomplished? What are the molecular mechanisms of elimination of the (peri)centromeric and (sub)telomeric sequences after NCF?
Acknowledgments
I dedicate this article to Professor Ingo Schubert on the occasion of his 75th birthday. I thank Adel Sepsi, Terezie Mandáková, Jim Birchler and Ingo Schubert for their comments on the manuscript. I acknowledge Jiří Fajkus and Karel Říha for discussing telomeres, and Clare Pansigham and Oren Harman for their help with photographs of C. D. Darlington. I thank Patrice Salomé, Science Editor of Plant Cell, for his assistance in preparing the figures for this article.
Funding
This work was supported by a Czech Science Foundation research grant (21-07748L) focused on elucidating the mechanisms of descending dysploidy in land plants.
Conflict of interest statement. None declared.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is Martin A. Lysak (martin.lysak@ceitec.muni.cz).
References
- Adikusuma F, Williams N, Grutzner F, Hughes J, Thomas P (2017) Targeted deletion of an entire chromosome using CRISPR/Cas9. Mol Ther 25: 1736–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agudo M, Abad JP, Molina I, Losada A, Ripoll P, Villasante A (2000) A dicentric chromosome of Drosophila melanogaster showing alternate centromere inactivation. Chromosoma 109: 190–196 [DOI] [PubMed] [Google Scholar]
- Aguilar M, Prieto P (2021) Telomeres and subtelomeres dynamics in the context of early chromosome interactions during meiosis and their implications in plant breeding. Front Plant Sci 12: 672489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alleva B, Smolikove S (2017) Moving and stopping: regulation of chromosome movement to promote meiotic chromosome pairing and synapsis. Nucleus 8: 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong SJ, Franklin FCH, Jones GH (2001) Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J Cell Sci 114: 4207–4217 [DOI] [PubMed] [Google Scholar]
- Arpòn J, Sakai K, Gaudin V, Andrey P (2021) Spatial modeling of biological patterns shows multiscale organization of Arabidopsis thaliana heterochromatin. Sci Rep 11: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley T (1979) Specific end-to-end attachment of chromosomes in Ornithogalum virens. J Cell Sci 38: 357–367 [DOI] [PubMed] [Google Scholar]
- Barra V, Fachinetti D (2018) The dark side of centromeres: types, causes and consequences of structural abnormalities implicating centromeric DNA. Nat Commun 9: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson W (1906) The progress of genetic research. In Wilks W, ed, Report of the Third International Conference on Genetics. Spottiswoode & Co., London, pp 90–97 [Google Scholar]
- Belling J (1933) Critical notes on Darlington’s recent advances in cytology. Univ Cal Pubs Bot 17: 75–110 [Google Scholar]
- Betekhtin A, Jenkins G, Hasterok R (2014) Reconstructing the evolution of Brachypodium genomes using comparative chromosome painting. PLoS One 9: e115108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Han F (2018) Barbara McClintock’s unsolved chromosomal mysteries: parallels to common rearrangements and karyotype evolution. Plant Cell 30: 771–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A (1993) Barbara McClintock. Ann Rev Genet 27: 1–32 [DOI] [PubMed] [Google Scholar]
- Carlton PM, Cowan CR, Cande WZ (2003) Directed motion of telomeres in the formation of the meiotic bouquet revealed by time course and simulation analysis. Mol Biol Cell 14: 2832–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Mandáková T, Wu J, Xie Q, Lysak MA, Wang X (2013) Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell 25: 1541–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Shang D, Luo M, Huang C, Lai F, Wang X, Zhao Y, Zhang L, Long M, Zhou R, et al. (2020) Whole genome-wide chromosome fusion and new gene birth in the Monopterus albus genome. Cell Biosci 10: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiatante G, Giannuzzi G, Calabrese FM, Eichler EE, Ventura M (2017) Centromere destiny in dicentric chromosomes: new insights from the evolution of human chromosome 2 ancestral centromeric region. Mol Biol Evol 34: 1669–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicconardi F, Lewis JJ, Martin SH, Reed RD, Danko CG, Montgomery SH (2021) Chromosome fusion affects genetic diversity and evolutionary turnover of functional loci but consistently depends on chromosome size. Mol Biol Evol 38: 4449–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comfort N (2001) The Tangled Field: Barbara McClintock’s Search for the Patterns of Genetic Control. Harvard University, Cambridge [Google Scholar]
- Creighton HB, McClintock B (1931) A correlation of cytological and genetical crossing-over in zea mays. Proc Natl Acad Sci USA 17: 492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C (1988) Centennial of Wilhelm Waldeyer's introduction of the term “chromosome” in 1888. Cytogenet Cell Genet 48: 66–67 [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C (2006) Rise, fall and resurrection of ch0romosome territories: a historical perspective Part I. The rise of chromosome territories. Eur J Histochem 50: 161–176 [PubMed] [Google Scholar]
- Darlington CD (1932) Recent Advances in Cytology, Churchill. P. Blakiston’s Son and Co., Philadelphia, PA
- Darlington CD, Upcott MB (1941) Spontaneous chromosome change. J Genet 41: 297–338 [Google Scholar]
- Dawicki-McKenna JM, Black BE (2019) Chromosomes: keeping centromeric chromatin tidy through S phase. Curr Biol 29: R35–R37 [DOI] [PubMed] [Google Scholar]
- Dong F, Jiang J (1998) Non-Rabl patterns of centromere and telomere distribution in the interphase nuclei of plant cells. Chromosome Res 6: 551–558 [DOI] [PubMed] [Google Scholar]
- Edelson E (2001) Gregor Mendel: And the Roots of Genetics. Oxford University Press, Oxford [Google Scholar]
- Fang T, Dong H, Yu S, Moss JQ, Fontanier CH, Martin DL, Fu J, Wu Y (2020) Sequence-based genetic mapping of Cynodon dactylon Pers. reveals new insights into genome evolution in poaceae. Commun Biol 3: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J, Bloomer H, Kellam N, LaRocque JR (2019) Chromosome preference during homologous recombination Repair of DNA double-strand breaks in Drosophila melanogaster. G3: Genes Genom Genet 9: 3773–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonsêca A, Ferraz ME, Pedrosa-Harand A (2016) Speeding up chromosome evolution in Phaseolus: multiple rearrangements associated with a one-step descending dysploidy. Chromosoma 125: 413–421 [DOI] [PubMed] [Google Scholar]
- Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I (2002) Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci 99: 14584–14589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S, Gao Z, Birchler J, Han F (2012) Dicentric chromosome formation and epigenetics of centromere formation in plants. J Genet Genomics 39: 125–130 [DOI] [PubMed] [Google Scholar]
- Gao Z, Fu S, Dong Q, Han F, Birchler JA (2011) Inactivation of a centromere during the formation of a translocation in maize. Chromosome Res 19: 755–761 [DOI] [PubMed] [Google Scholar]
- Geiser C, Mandáková T, Arrigo N, Lysak MA, Parisod C (2016) Repeated whole-genome duplication, karyotype reshuffling, and biased retention of stress-responding genes in Buckler mustard. Plant Cell 28: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JL, Byrne KP, Wolfe KH (2011) Mechanisms of chromosome number evolution in yeast. PLoS Genet 7: e1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra M (2016) Agmatoploidy and symploidy: a critical review. Genet Mol Biol 39: 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero RF, Kirkpatrick M (2014) Local adaptation and the evolution of chromosome fusions. Evolution 68: 2747–2756 [DOI] [PubMed] [Google Scholar]
- Guo X, Mandáková T, Trachtová K, Özüdoğru B, Liu J, Lysak MA (2021) Linked by ancestral bonds: multiple whole-genome duplications and reticulate evolution in a Brassicaceae tribe. Mol Biol Evol 38: 1695–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Lamb JC, Birchler JA (2006) High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci USA 103: 3238–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Zhang Z, Liu C, Liu J, Huang S, Jiang J, Jin W (2009) Centromere repositioning in cucurbit species: implication of the genomic impact from centromere activation and inactivation. Proc Natl Acad Sci USA 106: 14937–14941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman OS (2003) Darlington and the ‘invention’of the chromosome. Endeavour 27: 69–74 [DOI] [PubMed] [Google Scholar]
- Harman OS (2004) The Man who Invented the Chromosome: a Life of Cyril Darlington. Harvard University Press, CA, MA, USA [Google Scholar]
- Harman OS (2006) Method as a function of “Disciplinary Landscape”: C.D. Darlington and cytology, genetics and evolution, 1932–1950. J History Biol 39: 165–197 [DOI] [PubMed] [Google Scholar]
- Heuser E (1884) Beobachtungen über Zellkerntheilung. Botanisches Zentralblatt 17: 2711 [Google Scholar]
- Hill J, Rastas P, Hornett EA, Neethiraj R, Clark N, Morehouse N, de la Paz Celorio-Mancera M, Cols JC, Dircksen H, Wheat CW, et al. (2019) Unprecedented reorganization of holocentric chromosomes provides insights into the enigma of lepidopteran chromosome evolution. Sci Adv 5: eaau3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang PT, Schubert I (2017) Reconstruction of chromosome rearrangements between the two most ancestral duckweed species Spirodela polyrhiza and S. intermedia. Chromosoma 126: 729–739 [DOI] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Guo YL, et al. (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Ma Y, Mandáková T, Shi S, Chen C, Sun P, Zhang L, Feng L, Zheng Y, Feng X, Yang W, et al. (2021) Genome evolution of the psammophyte Pugionium for desert adaptation and further speciation. Proc Natl Acad Sci USA 118: e2025711118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurel A, Phillips D, Vrielynck N, Mézard C, Grelon M, Christophorou N (2018) A cytological approach to studying meiotic recombination and chromosome dynamics in Arabidopsis thaliana male meiocytes in three dimensions. Plant J 95: 385–396 [DOI] [PubMed] [Google Scholar]
- Ijdo JW, Baldini A, Ward DC, Reeders ST, Wells RA (1991) Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc Natl Acad Sci USA 88: 9051–9055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iltis H (2018) Life of Mendel. Routledge, Abingdon [Google Scholar]
- Imai HT (1986) Modes of species differentiation and karyotype alteration in ants and mammals. In Iwatsuki K, Raven PH, Bock WJ, eds, Modern Aspect of Species. University of Tokyo Press, Tokyo, Japan, pp 87–105 [Google Scholar]
- Imai HT, Taylor RW (1989) Chromosomal polymorphisms involving telomere fusion, centromeric inactivation and centromere shift in the ant Myrmecia (pilosula) n = 1. Chromosoma 98: 456–460 [Google Scholar]
- Imai HT, Satta Y, Takahata N (2001) Integrative study on chromosome evolution of mammals, ants and wasps based on the minimum interaction theory. J Theor Biol 210: 475–497 [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Jenkins G, Bennett MD (1981) The intranuclear relationship between centromere volume and chromosome size in Festuca scariosa × drymeja. J Cell Sci 47: 117–125 [DOI] [PubMed] [Google Scholar]
- Joachimiak A (1987) Telomere arrangement in interphase nuclei of Allium cepa L. Acta Biol Cracov Ser Bot 29: 64–70 [Google Scholar]
- Karimi-Ashtiyani R, Schubert V, Houben A (2021) Only the rye derived part of the 1BL/1RS hybrid centromere incorporates CENH3 of wheat. Front Plant Sci 12: 802222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller EF (1983) A Feeling for the Organism: The Life and Work of Barbara McClintock. W.H. Freeman and Company, New York, NY [Google Scholar]
- Kim E, Kim J, Kim C, Lee J (2021) Long-read sequencing and de novo genome assemblies reveal complex chromosome end structures caused by telomere dysfunction at the single nucleotide level. Nucleic Acids Res 49: 338–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo DH, Han F, Birchler JA, Jiang J (2011) Distinct DNA methylation patterns associated with active and inactive centromeres of the maize B chromosome. Genome Res 21: 908–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotseruba V, Pistrick K, Gernand D, Meister A, Ghukasyan A, Gabrielyan I, Houben A (2005) Characterisation of the low-chromosome number grass Colpodium versicolor (Stev.) Schmalh.(2n = 4) by molecular cytogenetics. Caryologia 58: 241–245 [Google Scholar]
- Lee JJ, Lee J, Lee H (2021) Alternative paths to telomere elongation. Semin Cell Dev Biol 113: 88–96 [DOI] [PubMed] [Google Scholar]
- Lewis D (1983) Cyril Dean Darlington 1903–1981. Biographical Memoirs of the Royal Society, Vol. 29. Royal Society, London [Google Scholar]
- Li G, Wang L, Yang J, He H, Jin H, Li X, Han X, Zhao X, Dong L, Wang D, et al. (2021a) A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nat Genet 53: 574–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chengsong Z, Zhongwei L, Yun W, Dabao Z, Guihua B, Weixing S, Ma J, Muehlbauer GJ, Scanlon MJ, Jianming Y, et al. (2011) Chromosome size in diploid eukaryotic species centers on the average length with a conserved boundary. Mol Biol Evol 28: 1901–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun P, Lu Z, Chen J, Wang Z, Du X, Zheng Z, Wu Y, Hu H, Yang J, Ma J (2021b) The Corylus mandshurica genome provides insights into the evolution of Betulaceae genomes and hazelnut breeding. Hortic Res 8: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou EV, Williams EM, Fan Y, Friedman C, Young JM, Trask BJ (2005) Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 437: 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshitz HD (2021) The origin of GENETICS. Genetics 217: iyaa024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Su H, Zhang J, Shi L, Liu Y, Zhang B, Bai H, Liang S, Gao Z, Han F, et al. (2020) Rapid birth or death of centromeres on fragmented chromosomes in maize. Plant Cell 32: 3113–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Roesti M, Marques D, Hiltbrunner M, Saladin V, Peichel CL (2022) Chromosomal fusions facilitate adaptation to divergent environments in threespine stickleback. Mol Biol Evol 39: msab358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottersberger F, Karssemeijer RA, Dimitrova N, de Lange T (2015) 53BP1 and the LINC complex promote microtubule-dependent DSB mobility and DNA repair. Cell 163: 880–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis EJ, Vershinin AV (2005) Chromosome ends: different sequences may provide conserved functions. Bioessays 27: 685–697 [DOI] [PubMed] [Google Scholar]
- Luo J, Sun X, Cormack BP, Boeke JD (2018) Karyotype engineering by chromosome fusion leads to reproductive isolation in yeast. Nature 560: 392–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Deal KR, Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, Blake N, Clegg MT, Coleman-Derr D, Dvorak J, et al. (2009) Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc Natl Acad Sci USA 106: 15780–15785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Gu YQ, Puiu D, Wang H, Wardziok SO, Deal KR, Huo N, Zhu T, Wang L, Dvořák J (2017) Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551: 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusinska J, Betekhtin A, Lopez-Alvarez D, Catalan P, Jenkins G, Wolny E, Hasterok R (2019) Comparatively barcoded chromosomes of Brachypodium perennials tell the story of their karyotype structure and evolution. Int J Mol Sci 20: 5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA (2014) Live and let die: centromere loss during evolution of plant chromosomes. New Phytologist 203: 1082–1089 [Google Scholar]
- Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci USA 103: 5224–5229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL (2006) Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc Natl Acad Sci USA 103: 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma PF, Liu YL, Jin GH, Liu JX, Wu H, He J, Guo ZH, Li DZ (2021) The Pharus latifolius genome bridges the gap of early grass evolution. Plant Cell 33: 846–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon RN, Campbell LJ (2011) The role of dicentric chromosome formation and secondary centromere deletion in the evolution of myeloid malignancy. Genet Res Int 2011: 643628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Lysak MA (2018) Post-polyploid diploidization and diversification through dysploid changes. Curr Opin Plant Biol 42: 55–65 [DOI] [PubMed] [Google Scholar]
- Mandáková T, Joly S, Krzywinski M, Mummenhoff K, Lysak MA (2010a) Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant Cell 22: 2277–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Heenan PB, Lysak MA (2010b) Island species radiation and karyotypic stasis in Pachycladon allopolyploids. BMC Evol Biol 10: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Kovařík A, Zozomová-Lihová J, Shimizu-Inatsugi R, Shimizu KK, Mummenhoff K, Marhold K, Lysak MA (2013) The more the merrier: recent hybridization and polyploidy in Cardamine. Plant Cell 25: 3280–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Schranz ME, Sharbel TF, de Jong H, Lysak MA (2015) Karyotype evolution in apomictic Boechera and the origin of the aberrant chromosomes. Plant J 82: 785–793 [DOI] [PubMed] [Google Scholar]
- Mandáková T, Gloss AD, Whiteman NK, Lysak MA (2016) How diploidization turned a tetraploid into a pseudotriploid. Am J Bot 103: 1187–1196 [DOI] [PubMed] [Google Scholar]
- Mandáková T, Hloušková P, German D, Lysak MA (2017a) Monophyletic origin and evolution of the largest crucifer genomes. Plant Physiol 174: 2062–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Pouch M, Harmanová K, Zhan SH, Mayrose I, Lysak MA (2017b) Multispeed genome diploidization and diversification after an ancient allopolyploidization. Mol Ecol 26: 6445–6462 [DOI] [PubMed] [Google Scholar]
- Mandáková T, Guo X, Özüdoğru B, Mummenhoff K, Lysak MA (2018) Hybridization‐facilitated genome merger and repeated chromosome fusion after 8 million years. Plant J 96: 748–760 [DOI] [PubMed] [Google Scholar]
- Mandáková T, Pouch M, Brock JR, Al-Shehbaz IA, Lysak MA (2019) Origin and evolution of diploid and allopolyploid Camelina genomes were accompanied by chromosome shattering. Plant Cell 31: 2596–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Hloušková P, Koch MA, Lysak MA (2020) Genome evolution in Arabideae was marked by frequent centromere repositioning. Plant Cell 32: 650–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrioli M, Manicardi GC (2020) Holocentric chromosomes. PLoS Genet 16: e1008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins LC (1999) Did Sutton and Boveri propose the so-called Sutton-Boveri chromosome hypothesis? Genetics Mol Biol 22: 261–272 [Google Scholar]
- Martinez-Garcia M, Schubert V, Osman K, Darbyshire A, Sanchez-Moran E, Franklin FCH (2018) TOPII and chromosome movement help remove interlocks between entangled chromosomes during meiosis. J Cell Biol 217: 4070–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather K, Stone LHA (1933) The effect of X-radiation upon somatic chromosomes. J Genet 28: 1–24 [Google Scholar]
- Matveevsky S, Kolomiets O, Bogdanov A, Alpeeva E, Bakloushinskaya I (2020) Meiotic chromosome contacts as a plausible prelude for Robertsonian translocations. Genes 11: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B (1932) Cyto-Genetics in 1932. J Hered 23: 497–498 [Google Scholar]
- McClintock B (1939) The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc Natl Acad Sci USA 25: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B (1941) The stability of broken ends of chromosomes in Zea Mays. Genetics 26: 234–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Ding Y, Heagle AB (1998) Relevance of kinetochore size and microtubule-binding capacity for stable chromosome attachment during mitosis in PtK1 cells. Chromosome Res 6: 123–132 [DOI] [PubMed] [Google Scholar]
- Muraki K, Murnane JP (2017) The DNA damage response at dysfunctional telomeres, and at interstitial and subtelomeric DNA double-strand breaks. Genes Genet Syst 92: 135–152 [DOI] [PubMed] [Google Scholar]
- Murat F, Xu JH, Tannier E, Abrouk M, Guilhot N, Pont C, Messing J, Salse J (2010) Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res 20: 1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar M, Smith GR (2016) Repression of harmful meiotic recombination in centromeric regions. Seminars in Cell & Developmental Biology, Vol. 54. Academic Press, Cambridge, MA, pp 188–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oizumi Y, Kaji T, Tashiro S, Takeshita Y, Kanoh J (2021) Complete sequences of Schizosaccharomyces pombe subtelomeres reveal multiple patterns of genome variation. Nat Commun 12: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oko Y, Ito N, Sakamoto T (2020) The mechanisms and significance of the positional control of centromeres and telomeres in plants. J Plant Res 133: 471–478 [DOI] [PubMed] [Google Scholar]
- Orel V (1971) A reconstruction of Mendel’s Pisum experiments and an attempt at an explanation of Mendel’s way of presentation. Folia Mendel 6: 41–60 [Google Scholar]
- Page SL, Shin JC, Han JY, Andy Choo KH, Shaffer LG (1996) Breakpoint diversity illustrates distinct mechanisms for Robertsonian translocation formation. Hum Mol Genet 5: 1279–1288 [DOI] [PubMed] [Google Scholar]
- Pennaneach V, Kolodner RD (2009) Stabilization of dicentric translocations through secondary rearrangements mediated by multiple mechanisms in S. cerevisiae. PLoS One 4: e6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plačková K, Bureš P, Zedek F (2021) Centromere size scales with genome size across Eukaryotes. Sci Rep 11: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobiega S, Marcand S (2010) Dicentric breakage at telomere fusions. Genes Dev 24: 720–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouokam M, Cruz B, Burgess S, Segal MR, Vazquez M, Arsuaga J (2019) The Rabl configuration limits topological entanglement of chromosomes in budding yeast. Sci Rep 9: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl C (1885) Über Zelltheilung. Morphol Jahrbuch 10: 214–330 [Google Scholar]
- Richards DM, Greer E, Martin AC, Moore G, Shaw PJ, Howard M (2012) Quantitative dynamics of telomere bouquet formation. PLoS Comput Biol 8: e1002812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, McKnight TD, Griffing LR, Shippen DE (2001) Living with genome instability: plant responses to telomere dysfunction. Science 291: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Robertson WR (1916) Chromosome studies: I. Taxonomic relationships shown in the chromosomes of Tettigidae and Acrididae: V-shaped chromosomes and their significance in Acrididae, Locustidae, and Gryllidae: chromosomes and variation. J Morphol 27: 179–331 [Google Scholar]
- Salse J (2016) Deciphering the evolutionary interplay between subgenomes following polyploidy: a paleogenomics approach in grasses. Am J Botany 103: 1167–1174 [DOI] [PubMed] [Google Scholar]
- Sánchez-Guillén RA, Capilla L, Reig-Viader R, Martínez-Plana M, Pardo-Camacho C, Andrés-Nieto M, Ventura J, Ruiz-Herrera A (2015) On the origin of Robertsonian fusions in nature: evidence of telomere shortening in wild house mice. J Evol Biol 28: 241–249 [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan SR, Ianiri G, Coelho MA, Reza MH, Thimmappa BC, Ganguly P, Vadnala RN, Sun S, Siddharthan R, Sanyal K, et al. (2020) Loss of centromere function drives karyotype evolution in closely related Malassezia species. Elife 9: e53944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Masuda F, Takayama Y, Takahashi K, Saitoh S (2012) Epigenetic inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Curr Biol 22: 658–667 [DOI] [PubMed] [Google Scholar]
- Saunders VA, Houben A (2001) The pericentromeric heterochromatin of the grass Zingeria biebersteiniana (2n = 4) is composed of Zbcen1-type tandem repeats that are intermingled with accumulated dispersedly organized sequences. Genome 44: 955–961 [DOI] [PubMed] [Google Scholar]
- Scherthan H (2001) A bouquet makes ends meet. Nat Rev Mol Cell Biol 2: 621–627 [DOI] [PubMed] [Google Scholar]
- Schneider KL, Xie Z, Wolfgruber TK, Presting GG (2016) Inbreeding drives maize centromere evolution. Proc Natl Acad Sci USA 113: E987–E996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotanus K, Yadav V, Heitman J (2021) Epigenetic dynamics of centromeres and neocentromeres in Cryptococcus deuterogattii. PLoS Genet 17: e1009743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I (2021) Boon and bane of DNA double-strand breaks. Int J Mol Sci 22: 5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I, Lysak MA (2011) Interpretation of karyotype evolution should consider chromosome structural constraints. Trend Genet 27: 207–216 [DOI] [PubMed] [Google Scholar]
- Schubert I, Oud JL (1997) There is an upper limit of chromosome size for normal development of an organism. Cell 88: 515–520 [DOI] [PubMed] [Google Scholar]
- Sears ER (1952) Misdivision of univalents in common wheat. Chromosoma 4: 535–550 [DOI] [PubMed] [Google Scholar]
- Sepsi A, Higgins JD, Heslop‐Harrison JS, Schwarzacher T (2017) CENH3 morphogenesis reveals dynamic centromere associations during synaptonemal complex formation and the progression through male meiosis in hexaploid wheat. Plant J 89: 235–249 [DOI] [PubMed] [Google Scholar]
- Sepsi A, Fábián A, Jäger K, Heslop-Harrison JS, Schwarzacher T (2018) ImmunoFISH: simultaneous visualisation of proteins and DNA sequences gives insight into meiotic processes in nuclei of grasses. Front Plant Sci 9: 1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W, Kubová M, Mandáková T, Lysak MA (2021) Nuclear organization in crucifer genomes: nucleolus‐associated telomere clustering is not a universal interphase configuration in Brassicaceae. Plant J 108: 528–540 [DOI] [PubMed] [Google Scholar]
- Sheehan MJ, Pawlowski WP (2009) Live imaging of rapid chromosome movements in meiotic prophase I in maize. Proc Natl Acad Sci USA 106: 20989–20994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Sun P, Yuan J, Gong K, Li N, Meng F, Zhang Z, Li X, Hu J, Wang X, et al. (2021) The celery genome sequence reveals sequential paleo‐polyploidizations, karyotype evolution and resistance gene reduction in Apiales. Plant Biotechnol J 19: 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreng RJ, Peterson PM, Romaschenko K, Davidse G, Teisher JK, Clark LG, Barberá P, Gillespie LJ, Zuloaga FO (2017) A worldwide phylogenetic classification of the Poaceae (Gramineae) II: An update and a comparison of two 2015 classifications. J Syst Evol 55: 259–290 [Google Scholar]
- Stimpson KM, Song IY, Jauch A, Holtgreve-Grez H, Hayden KE, Bridger JM, Sullivan BA (2010) Telomere disruption results in non-random formation of de novo dicentric chromosomes involving acrocentric human chromosomes. PLoS Genet 6: e1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson KM, Matheny JE, Sullivan BA (2012) Dicentric chromosomes: unique models to study centromere function and inactivation. Chromosome Res 20: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroik S, Hendrickson EA (2020) Telomere fusions and translocations: a bridge too far? Curr Opin Genet Dev 60: 85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton WS (1902) On the morphology of the chromosome group in Brachystola magna. Biol Bull 4: 24–39 [Google Scholar]
- Tiang CL, He Y, Pawlowski WP (2012) Chromosome organization and dynamics during interphase, mitosis, and meiosis in plants. Plant Physiol 158: 26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortora MM, Salari H, Jost D (2020) Chromosome dynamics during interphase: a biophysical perspective. Curr Opin Genet Dev 61: 37–43 [DOI] [PubMed] [Google Scholar]
- Upcott M (1937) Spontaneous chromosome changes in pollen grains. Nature 139: 153–153 [Google Scholar]
- Vara C, Paytuví-Gallart A, Cuartero Y, Álvarez-González L, Marín-Gual L, Garcia F, Ruiz-Herrera A, et al. (2021) The impact of chromosomal fusions on 3D genome folding and recombination in the germ line. Nat Commun 12: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varas J, Graumann K, Osman K, Pradillo M, Evans DE, Santos JL, Armstrong SJ (2015) Absence of SUN 1 and SUN 2 proteins in Arabidopsis thaliana leads to a delay in meiotic progression and defects in synapsis and recombination. Plant J 81: 329–346 [DOI] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42: 833–839 [DOI] [PubMed] [Google Scholar]
- Wang H, Bennetzen JL (2012) Centromere retention and loss during the descent of maize from a tetraploid ancestor. Proc Natl Acad Sci USA 109: 21004–21009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CJR, Carlton PM, Golubovskaya IN, Cande WZ (2009) Interlock formation and coiling of meiotic chromosome axes during synapsis. Genetics 183: 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Singh R, Genovesi AD, Wai CM, Huang X, Chandra A, Yu Q (2015a) Sequence‐tagged high‐density genetic maps of Zoysia japonica provide insights into genome evolution in Chloridoideae. Plant J 82: 744–757 [DOI] [PubMed] [Google Scholar]
- Wang N, Liu J, Ricci WA, Gent JI, Dawe RK (2021a) Maize centromeric chromatin scales with changes in genome size. Genetics 217: iyab020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zi H, Wang R, Liu J, Wang H, Chen R, Li L, Guo H, Chen J, Li J, Zong J (2021b) A high-quality chromosome-scale assembly of the centipedegrass [Eremochloa ophiuroides (Munro) Hack.] genome provides insights into chromosomal structural evolution and prostrate growth habit. Horticu Res 8: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wu Y, Zhang W, Dawe RK, Jiang J (2014) Maize centromeres expand and adopt a uniform size in the genetic background of oat. Genome Res 24: 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tu Z, Liu C, Liu H, Kaldis P, Chen Z, Li W (2018a) Dual roles of TRF1 in tethering telomeres to the nuclear envelope and protecting them from fusion during meiosis. Cell Death Differ 25: 1174–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Xiao Y, Zhou ZW, Yuan J, Guo H, Yang Z, Wang X, Fan H, Chen LL, Luo J, et al. (2021c) High-quality reference genome sequences of two coconut cultivars provide insights into evolution of monocot chromosomes and differentiation of fiber content and plant height. Genome Biol 22: 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhai B, Yang X, Zhang L (2018b) Protect chromosomes from end-to-end fusion during meiotic bouquet. Sci China Life Sci 61: 736–738 [DOI] [PubMed] [Google Scholar]
- Wang X, Jin D, Wang Z, Guo H, Zhang L, Wang L, Li J, Paterson AH (2015b) Telomere‐centric genome repatterning determines recurring chromosome number reductions during the evolution of eukaryotes. New Phytologist 205: 378–389 [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang J, Pan Y, Lei T, Ge W, Wang L, Zhang L, Li Y, Zhao K, Liu T, Song X, et al. (2019) Reconstruction of evolutionary trajectories of chromosomes unraveled independent genomic repatterning between Triticeae and Brachypodium. BMC Genom 20: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters PD, Patel HR, Ruiz-Herrera A, Álvarez-González L, Lister NC, Simakov O, Ezaz T, Kaur P, Frere C, Graves JAM, et al. (2021) Microchromosomes are building blocks of bird, reptile, and mammal chromosomes. Proc Natl Acad Sci USA 118: e2112494118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Koo DH, Li D, Zhang T, Jiang J, Luan F, Renner SS, Henaff E, Sanseverino W, Garcia-Mas J, Casacuberta J (2014) Next‐generation sequencing, FISH mapping and synteny‐based modeling reveal mechanisms of decreasing dysploidy in Cucumis. Plant J 77: 16–30 [DOI] [PubMed] [Google Scholar]
- Yang W, Zhang L, Mandáková T, Huang L, Li T, Jiang J, Yang Y, Lysak MA, Liu J, Hu Q (2021) The chromosome‐level genome sequence and karyotypic evolution of Megadenia pygmaea (Brassicaceae). Mol Ecol Resource 21: 871–879 [DOI] [PubMed] [Google Scholar]
- Zhang F, Tang D, Shen Y, Xue Z, Shi W, Ren L, Cheng Z, et al. (2017) The F-box protein ZYGO1 mediates bouquet formation to promote homologous pairing, synapsis, and recombination in rice meiosis. Plant Cell 29: 2597–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Liu X, Quan Z, Cheng S, Xu X, Pan S, Zeng P, Yue Z, Wang W, Wang J, et al. (2012) Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotechnol 30: 549–554 [DOI] [PubMed] [Google Scholar]
- Zhang W, Friebe B, Gill BS, Jiang J (2010) Centromere inactivation and epigenetic modifications of a plant chromosome with three functional centromeres. Chromosoma 119: 553–563 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Meng Y, Wang P, Qin X, Cheng C, Zhou J, Yu X, Li J, Lou Q, Jahn M, Chen J (2021) Reconstruction of ancestral karyotype illuminates chromosome evolution in the genus Cucumis. Plant J 107: 1243–1259 [DOI] [PubMed] [Google Scholar]
- Zhou S, Jiang W, Zhao Y, Zhou DX (2019) Single-cell three-dimensional genome structures of rice gametes and unicellular zygotes. Nat Plants 5: 795–800 [DOI] [PubMed] [Google Scholar]