Abstract
Bread wheat (Triticum aestivum, genome BBAADD) is a young hexaploid species formed only 8,500–9,000 years ago through hybridization between a domesticated free-threshing tetraploid progenitor, genome BBAA, and Aegilops tauschii, the diploid donor of the D subgenome. Very soon after its formation, it spread globally from its cradle in the fertile crescent into new habitats and climates, to become a staple food of humanity. This extraordinary global expansion was probably enabled by allopolyploidy that accelerated genetic novelty through the acquisition of new traits, new intergenomic interactions, and buffering of mutations, and by the attractiveness of bread wheat’s large, tasty, and nutritious grain with high baking quality. New genome sequences suggest that the elusive donor of the B subgenome is a distinct (unknown or extinct) species rather than a mosaic genome. We discuss the origin of the diploid and tetraploid progenitors of bread wheat and the conflicting genetic and archaeological evidence on where it was formed and which species was its free-threshing tetraploid progenitor. Wheat experienced many environmental changes throughout its evolution, therefore, while it might adapt to current climatic changes, efforts are needed to better use and conserve the vast gene pool of wheat biodiversity on which our food security depends.
We describe the evolution of bread wheat in nature and under human selection with an emphasis on the donors of its subgenomes, evolution under polyploidy, and the “where when and how” of its domestication.
Introduction
Bread wheat (Triticum aestivum L.), also known as common wheat, is an annual, predominantly autogamous species belonging to the Triticeae tribe of the grasses (Poaceae) family. It is an allohexaploid species, composed of 21 chromosome pairs organized in three subgenomes, A, B, and D, Genome BBAADD, 2n = 6x = 42 (Sears, 1952). Its nutritious grain contributes an important fraction of the calorific daily intake in many parts of the world, for example, 40%–50% in Egypt and Turkey and ∼20% in the UK (Shewry and Hey, 2015). It is also an important source of protein, dietary fibers, B vitamins, minerals, and other phytochemicals in the human diet. Starting with domestication in the fertile crescent, its cultivation spread worldwide in an unrivaled range of locations as far north as 67° N in Norway, Finland, and Russia, and south to 45° S in Argentina, being mostly grown in temperate regions and to a lesser extent also in sub-tropical and tropical habitats (Feldman, 2001). In 2020 and 2021, worldwide annual wheat production reached a record of ∼770 mt (FAO, 2021) with China and India as the top two producers. Once considered the food of western civilizations, wheat became one of the most important staple foods of humanity. Therefore, the question of wheat origin and evolution has fascinated humans since ancient times. Mac Key classified T. aestivum as having five subspecies, two hulled, ssp. spelta, ssp. macha, and three free threshing, ssp. aestivum, ssp. Compactum, and ssp. sphaerococcum (Mac Key, 1954). All subspecies are domesticated, except an additional one, for one semi-wild form, ssp. tibetanum, presumably a feral wheat that escaped cultivation in Tibet (Shao et al., 1983). Here, we focus on T. aestivum ssp. aestivum, which is of global importance, the other subspecies being only locally cultivated, and we refer to it as bread wheat. In this review, we follow Mac Key’s classification, yet additional classifications were proposed as summarized by Sharma et al. (2021).
Despite the sequencing of its large approximately 16,000 Mbp genome (IWGS, 2018) and recent advances, major questions regarding bread wheat evolution remain: Which species are its progenitors? What was the impact of hybridization and allopolyploidization on the genome structure and stability? What were the main genetic changes during recent evolution under cultivation, domestication, and modern breeding? We present our current understanding of wheat evolution, starting from early discoveries to recent achievements and we discuss open questions.
The diploid and tetraploid donors of wheat subgenomes and cytoplasm
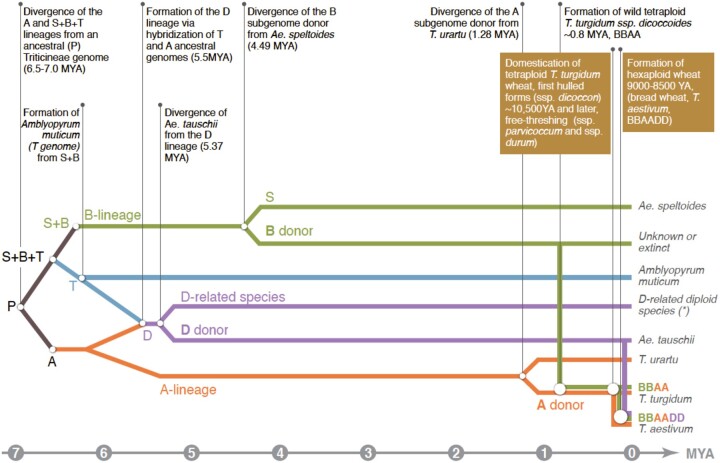
A major milestone in the study of wheat evolution was the discovery of the chromosome number of the wheat species (Sakamura, 1918; Sax, 1918), showing that the wheats comprise a polyploid series consisting of diploids (2n = 2x = 14), tetraploids (2n = 4x = 28), and hexaploids including bread wheat (2n = 6x = 42). Soon after this discovery, Kihara (1919, 1924), Sax (1921, 1922), and others (see tribute to Elisabeth Schiemann [Kilian et al., 2014; and references therein]) crossed representatives of the different ploidy levels and found that polyploid wheats are allopolyploids, that is, having subgenomes (chromosome sets) that derived from different species. When hexaploid bread wheat was crossed with domesticated emmer tetraploid wheat, T. turgidum ssp. dicoccon (genome BBAA 2n = 4x = 28), analysis of chromosomal pairing at meiosis of the F1 pentaploid hybrids indicated that hexaploid wheat, T. aestivum, contains the B and A subgenomes of tetraploid wheat with an additional diploid subgenome (Kihara, 1919; McFadden and Sears, 1944, 1946). Morphological (Kihara, 1944) and cytogenetic studies (McFadden and Sears, 1944, 1946) showed that the third subgenome of hexaploid wheat, designated D, was derived from Aegilops tauschii. McFadden and Sears (1946) produced a synthetic hexaploid from the cross of tetraploid emmer wheat with Ae. tauschii, that resembled the hulled form of T. aestivum, ssp. spelta in most morphological traits, had the hexaploid number of chromosomes, and produced fertile F1 hybrids with T. aestivum exhibiting complete chromosomal pairing at meiosis. This demonstrated unequivocally that Ae. tauschii (formerly Ae. squarrosa), is the donor of the third subgenome of T. aestivum. When the genome of Ae. tauschii was sequenced, first as a draft (Jia et al., 2013), then as a high-quality sequence (Luo et al., 2017; Zhao et al., 2017), it became apparent that the origin of the D genome, derived 5.37 million years ago (MYA) from the D-lineage, resulted from hybridization ∼5.5 MYA (Figure 1) between the ancestral donors of the A and B subgenomes (Marcussen et al., 2014).
Figure 1.
Phylogenetic representation of wheat evolution. Wheat evolution is shown starting ∼7 MYA from a progenitor that gave rise to the (A), (B), and (D) lineages that merged to form bread wheat. The relative timing of the major speciation events is shown in the horizontal axis and described in the boxes above. The tree is adapted from Glémin et al. (2019); Li et al. (2022); Avni et al. (2021). The diploid species related to the D genome (asterisks) are Aegilops bicornis, Ae. longissima, Ae. searsii, Ae. sharonensis, Ae. caudata, Ae. comosa, Ae. umbellulata, and Ae. uniaristata.
The wild progenitor of tetraploid wheat that contributed to the A and B subgenomes of T. aestivum had been discovered in nature in 1906 by Aaronson (Aaronsohn, 1910). The fertile F1 hybrids between ssp. durum, ssp. Dicoccon, and wild emmer wheat (T. turgidum ssp. dicoccoides) (Tschermak, 1914), as well as the subsequent demonstration of the full pairing of their chromosomes (Sax, 1921, 1922; Percival, 1921; Kihara, 1924) indicated that wild emmer was the wild progenitor of domesticated tetraploid wheat (Figure 1) and thus of the A and B subgenomes of T. aestivum. Whole-genome sequencing confirmed the high synteny between ssp. dicoccoides (Avni et al., 2017) and ssp. durum (Maccaferri et al., 2019).
Thus, bread wheat evolved through two rounds of allopolyploidization (Figure 1). The first event led to the formation of tetraploid wild emmer wheat approximately 800,000 years ago following hybridization between two diploid species: the male donor of the A subgenome, a species very similar to Triticum urartu, but that diverged from it ∼1.3 MYA, and the female donor of the B subgenome, a species related to Ae. speltoides, but that diverged from it ∼4.5 MYA (Marcussen et al., 2014; Li et al., 2022). The second allopolyploidization event that produced hexaploid wheat involved allotetraploid wheat as the female donor of the B and A subgenomes and Ae. tauschii as the male donor of the D subgenome. This event first formed hulled allohexaploid wheat from which the more advanced free-threshing forms originated by mutations (Feldman, 2001). The cytoplasm of T. aestivum, designated B, is identical to that of its maternal parent, T. turgidum (Tsunewaki, 2009, and references therein).
While the identity of the diploid donors of two out of the three subgenomes of T. aestivum, namely, T. urartu (A subgenome) and Ae. tauschii (D subgenome), is well established, that of the B subgenome, the cytoplasm donor of bread wheat, remains elusive. There is no extant diploid species whose chromosomes have a high affinity for the B subgenome, suggesting that the progenitor is either extinct or remains to be discovered, or that the B subgenome has a polyphyletic origin and evolved at the polyploid level through multiple hybridizations with close extant species, for example, the five species from the Sitopsis section: Ae. speltoides; Ae. bicornis; Ae. sharonensis; Ae. Longissima; and Ae. searsii. Recent insight from whole-genome sequences suggests that Ae. speltoides diverged from the B subgenome ∼4.5 MYA (Li et al., 2022). The estimate that wild emmer wheat (genome BBAA) was formed ∼0.8 MYA (Marcussen et al., 2014) rules out the possibility that Ae. speltoides is the direct progenitor of the B subgenome. The results of Li et al. (2022) support the hypothesis that the donor of the bread wheat B subgenome is a diploid species closely related to, but distinct from extant Ae. speltoides. Moreover, despite evidence for introgressions from other Sitopsis species, the B subgenome appears to have a mainly monophyletic origin. According to this scenario, the B subgenome donor is either extinct or remains to be discovered. Considering that morphological differences are sometimes very subtle among Sitopsis species, it cannot be ruled out that the B subgenome donor is still present in the wild. The B subgenome donor and Ae. speltoides thus belong to the B lineage, and both diverged from Amblyopyrum muticum (2n = 2x = 14, T genome) ∼6.4 MYA (Figure1), making Am. muticum the most ancestral extant representative of the B lineage (Glémin et al., 2019; Bernhardt et al., 2020; Avni et al., 2021; Li et al., 2022).
In summary, one could say that bread wheat was formed from its diploid and tetraploid progenitors through a cycle of “diverge-and-merge” speciation events. Indeed, after their divergence ∼7 MYA, the A and B genomes merged to form a new homoploid hybrid species, Ae. tauschii (genome DD) 5.5 MYA, then, 800,000 YA, A and B reunited again, but this time to form an allotetraploid species (ssp. dicoccoides, genome BBAA). And to close the cycle of “diverge-and-merge,” the D genome, which is itself an ancient homoploid hybrid between B and A, merged with the BBAA genome approximately 9,000 YA to form allohexaploid bread wheat (Figure 1). Another insight from recent studies points to the seminal role of A. muticum as an ancestral contributor to both the B and D lineages (Glémin et al., 2019; Avni et al., 2021; Li et al., 2022).
Evolution under allopolyploidy
Speciation
Allopolyploidization is a biological process that played a central role in plant speciation and evolution (Soltis and Soltis, 2009), including in wheat, which is an archetype of evolution via polyploidization (Feldman and Levy, 2012). It constitutes a radical and rapid mode of speciation that produces a new species by means of inter-specific or inter-generic hybridization of two diverging species, followed by chromosome doubling of the F1 hybrids. There has been much debate on the fitness of polyploids (Stebbins, 1950, 1971; Soltis and Soltis, 1999; Mayrose et al., 2011). A recent meta-analysis of tens of thousands of species (Rice et al., 2019) suggests that the prominence of polyploidy depends on several factors, for example, polyploids are relatively more abundant in cold or dry climates with less species richness, presumably thanks to facilitated competition with endemic diploids. Such ability to outcompete diploid relatives can be achieved through a diversity of mechanisms, including novel intergenomic interaction, rewiring of gene expression, new dosage effects, new protein complexes, and fixed heterosis (Birchler and Veitia, 2021). Domestication, a recent event in plant evolution, seems to have favored allopolyploids in wheat and in several other crop species, possibly as it allowed for rapid adaptation to the new cultivation environment (Dubcovsky and Dvorak, 2007). Diploid einkorn wheat is grown on a smaller scale than tetraploid durum, which in turn represents only ∼10% of the area and production of hexaploid bread wheat.
All diploid grass species are thought to be paleopolyploids that underwent at least one event of whole-genome duplication (Salse et al., 2008; Wang et al., 2015; Murat et al., 2017), and on the evolutionary scale, polyploidization was followed by cytological and genetic diploidization (Bolot et al., 2009). Genetic diploidization can be considered as proposed by Ohno (1970), as a regulatory process that brings redundant or unbalanced gene systems in allopolyploids toward a diploid-like mode of expression. This can be achieved through multiple mechanisms (Conant et al., 2014), such as gene mutation, elimination, pseudogenization, or gene dosage compensation (Galili et al., 1986). Likewise, cytological diploidization is essential for the success of the new allopolyploid species to achieve full fertility and to provide a stable disomic inheritance for new beneficial interactions between subgenomes (Feldman and Levy, 2012).
Karyotypic evolution
The ancestral Triticeae karyotype has been proposed to have derived from the n = 12 ancestral grass karyotype as seen in rice (Salse et al., 2008; Luo et al., 2009). Comparative genetics showed that the reduction from n = 12 to n = 7, without the loss of genes, was accomplished through four centromeric ancestral chromosome fusions (leading to functional monocentric neochromosomes), one fission, and two telomeric ancestral chromosome fusions (Salse et al., 2008; Luo et al., 2009). The Triticeae basic chromosome number of n = 7 evolved via the loss of five functional centromeres, four of which correspond to those of rice chromosomes Os4, Os5, Os6, and Os9, and the fifth to either that of Os3 or Os11 (Luo et al., 2009).
The genome of common wheat, T. aestivum, shares the ancestral reciprocal translocation (between A4 and A5 on the A subgenome) as well as an additional lineage-specific translocation (between chromosomes 4A and 7B) to reach the modern 21 chromosomes (Salse et al., 2008; Luo et al., 2009; Murat et al., 2014). One of the most prominent large-scale karyotypic variants that occurred in wheat is a translocation between chromosomes 5B and 7B. This translocation was found in the majority of the lines tested by Walkowiak et al. (2020). Sequence data showed that the translocation between chromosomes 5B and 7B, which are ∼737 and 762 Mb long, respectively, gave rise to translocated chromosomes of 488 Mb (5BS/7BS) and 993 Mb (7BL/5BL) in length. The translocation breakpoint was mapped to a ∼5-kb GAA microsatellite when comparing cv. ArinaLrFor and SY Mattis.
In addition to the series of rearrangements that shaped the present-day bread wheat chromosomes, functional units, such as centromeres or nucleolar organizers, have also rapidly evolved under allopolyploidy in wheat. The Triticeae centromeres are no exception to other plants, forming through the assembly of the CENH3 proteins with arrays of satellite repeats and retroelements (Cheng and Murata, 2003). The primary constrictions of barley, wheat, Aegilops, and rye chromosomes, that is, the physical locations of centromeres, harbor retroelement-like sequences (Presting et al., 1998; Fukui et al., 2001; Cheng and Murata, 2003). In wheat, a detailed chromatin immunoprecipitation analysis, using a CENH3 antibody followed by sequencing of the precipitated DNA fraction, enabled the identification of a distinct dynamic structure for wheat centromeres, possibly due to evolution through frequent hybridization and polyploidization (Su et al., 2019). Unlike typical plant centromeres, which carry a satellite of tandem repeats ranging from 150 to 180 bp, the wheat CENH3 nucleosomes were associated with two different types of repeats much larger than other plant centromeric repeats, namely 550 and 566 bp long, respectively (Su et al., 2019). Moreover, different subgenomes tended to contain different repeat types, and some chromosomes lacked satellite repeats altogether. Phylogenetic analyses indicate that the repeat signals were stronger in diploids than in polyploids and that centromere structure evolved rapidly at the polyploid level (Su et al., 2019).
The nucleolar organizing regions (NORs) where nucleoli form is seen as secondary constrictions on chromosomes (McClintock, 1934). NORs contain ribosomal (rDNA) genes that code for the ribosomal RNA (rRNA) and are organized in a gene array of thousands of tandemly arranged copies per NOR. The total number of rDNA units in the fully sequenced genome of cv. Chinese Spring was estimated at 11,160 copies corresponding to 100 Mb, 30.5% of which are on the Nor-B1 locus (Chr. 1B), 60.9% on Nor-B2 (Chr. 6B), and 8.6% in other NORs (Handa et al., 2018), which is consistent with earlier estimates (Flavell and O’Dell, 1979; Appels and Honeycutt, 1986). In hybrids and polyploids, the rDNA genes of one parental set are transcribed, while most or all rDNA genes inherited from the other parent remain silent. This phenomenon, known as nucleolar dominance (Pikaard, 2000), is common in the allopolyploid species of the genera Aegilops and Triticum (Feldman et al., 2012). The diploid species of wheat, Triticum monococcum and T. urartu, contain two nucleolar organizer regions, one on chromosome arm 1AS and the second on 5AS (Miller et al., 1983). In the allopolyploid wheat species, the NOR of 1AS is inactive, while that of 5AS was lost (Miller et al., 1983; Jiang and Gill, 1994). Newly synthesized allopolyploids exhibit genetic and epigenetic changes in their rRNA-encoding genes similar to those occurring in natural allopolyploids, indicating that these changes are rapid and reproducible and were generated early during allopolyploid formation (Baum and Feldman, 2010). Guo and Han (2014) conducted a detailed molecular analysis of the fate of wheat NORs in several allopolyploids, including the synthetic and natural BBAADD genomes. They concluded that the NORs from the B subgenome are dominant in several genomic combinations and that the elimination of the other NORs proceeds in two steps—first through silencing and hypermethylation in the first generations, then through progressive elimination of the non-B rDNA copies, starting in the fourth and ending by the seventh generation after polyploidization (Guo and Han, 2014).
Evolution of gene order and content
Allopolyploidy involves genetic redundancy, even in an allohexaploid where the different subgenomes diverged from each other millions of years ago. This was shown in a milestone study by Ernie Sears with the demonstration that each of the seven pairs of chromosomes of a subgenome could compensate for the absence of a specific pair of another subgenome (Sears, 1954, 1959). The compensating pair was called homoeologous (with partially homology). The wheat genome could thus be arranged into seven homoeologous groups and three subgenomes (Sears, 1959). From an evolutionary point of view, this study also implied that, despite their divergence, the subgenomes maintained a relatively high degree of similarity.
Pioneering molecular studies revealed a high level of gene synteny and collinearity in the homoeologous chromosomes of bread wheat (Moore et al., 1995). The nature of the divergence/similarity between the hexaploid wheat subgenomes was fully understood only after the publication of an annotated high-quality sequence of bread wheat cv. Chinese Spring, the same cultivar used by Ernie Sears in his analysis of wheat chromosomes homoeology (IWGSC, 2018). Annotation of CS genes identified 107,891 “high confidence” genes distributed almost equally between the A, B, and D subgenomes. Synteny was highly conserved in the three subgenomes. Synteny was more prominent, with larger syntenic blocks, in interstitial regions compared to sub-telomeric regions or pericentric regions (IWGS, 2018). As shown by Akhunov et al. (2003), a higher rate of gene duplication and deletion in the sub-telomeric regions was in part responsible for the reduction in synteny. Only 55% of the genes had a syntenic homoeolog in all three subgenomes, whereas 15% had at least one missing homoeolog but had a paralog. The percent of missing homoeologs (gene loss) was similar (∼10%) in each subgenome (IWGS, 2018).
Gene organization within the chromosome analyzed through sequencing of single wheat BAC clones and BAC clone contigs suggested that genes are clustered into many small islands of 3–4 genes interspersed by retroelements (Feuillet and Keller, 1999; Wicker et al. 2005; Choulet et al. 2010). Gene density in the distal regions of chromosome 3B of common wheat was higher than in the rest of the genome due to a higher incidence of duplicated genes in these regions (Dvorak and Akhunov, 2005; Luo et al. 2013). For example, when comparing the Ae. tauschii and bread wheat D subgenome, only 87.4% of the genes were present at the expected orthologous position, highlighting a dynamic gene copy-number variation that is not related to whole-genome duplication (Zhou et al., 2021). Some gene families, such as prolamine genes (including glutenins and gliadins) and disease-resistance genes were among those most prone to be nonorthologous with other grasses (Zhou et al., 2021).
In bread wheat, gene family expansion generally occurred in the wild progenitor or the common ancestor of the subgenomes: out of 8,592 expanded families, 6,216 expanded in all three A, B, and D subgenomes while 1,109 expanded in only one of the subgenomes, and only 78 gene families contracted (IWGS, 2018). Interestingly, there were significant differences between subgenomes when functions were assigned to the families that expanded: there was an over-representation of seed-related genes (embryo and endosperm) in the A subgenome and of vegetative growth and development in the B subgenome. Families that expanded in all three genomes were enriched in genes that were hypothesized to play an important role in wheat breeding for trait related to yield or biotic and abiotic stress resistance (IWGS, 2018); however, a causal relationship has not been shown to date.
Walkowiak et al. (2020) analyzed the genomic sequence of different bread wheat lines and found high collinearity of gene order on homologous chromosomes and overall similar genome sizes. Nevertheless, ∼12% of the genes showed a “Presence/Absence Variation” between the different cultivars. Deletions in the polyploid background can be 10 times faster than in the diploid background (Dvorak and Akhunov, 2005). On the other hand, the repeat-rich intergenic environment of genes can accelerate gene duplication by 20-fold (Akhunov et al., 2007). Indeed, repeats, often TEs, can promote gene duplication by unequal (Molinier et al., 2004) or ectopic crossover (Shalev and Levy, 1997), or by gene-flanking helitrons (Morgante et al., 2005), or by capturing genes in mobile structures (Jiang et al., 2004). This dynamic gene copy number variation might be made possible by the buffering effect of whole-genome duplication enabling toleration of a broad range of genetic changes over time.
Rapid genetic and genomic changes leading to genetic and cytological diploidization
Genome merging in hybrids or allopolyploids generates both cytological and genetic challenges to the nascent species. For example, homoeologous pairing may lead to partial sterility and multisomic inheritance, and genomic clashes between regulatory elements of different subgenomes can lead to hybrid necrosis (Bomblies and Weigel, 2007). Rewiring of gene expression (Tirosh et al., 2009) may also reduce fitness of the nascent species. Presumably, a new allopolyploid species unable to rapidly overcome such challenges would not be able to establish itself and survive in nature.
Upon allopolyploidization, the immediate triggering of a variety of cardinal genetic and epigenetic changes that affect genome structure and gene expression might be essential to bring about genetic and cytological diploidization. Such rapid changes have been reported in newly formed allopolyploids, including elimination of coding and noncoding DNA sequences, transposable element (TE) and tandem repeat elimination or amplification, and gene expression modifications (Feldman et al., 1997; Liu et al., 1998; Ozkan et al., 2001; Shaked et al., 2001; Han et al., 2003, 2005; Salina et al., 2004; Ma and Gustafson, 2006; Baum and Feldman, 2010; Guo and Han, 2014). Remarkably, sequence elimination is often reproducible, as is evident from the elimination of the same sequences in synthetic and natural allopolyploids bearing the same genomic combinations (Feldman et al., 1997; Ozkan et al., 2001; Han et al., 2005). In many instances, sequence elimination leaves homologous-specific sequences in the genome, instead of two or three originally present in the parental genomes. Such differential elimination leads to rapid divergence of the homoeologous chromosomes, contributing a physical basis to facilitate chromosome homology recognition, and enabling hexaploid wheat to exhibit exclusively intra-subgenomic pairing in the form of bivalents between fully homologous chromosomes (diploid-like meiotic behavior). Allopolyploid species of the wheat group contain 2%–10% less DNA than the additive sum of their parental species, and synthetic allopolyploids exhibit a similar loss, indicating again that DNA elimination occurs soon after allopolyploidization (Furuta et al., 1974; Eilam et al., 2008).
TEs activity can be triggered by allopolyploidization, affecting gene expression and inducing many structural rearrangements, including deletions or duplications (Kashkush et al., 2002, 2003; Yaakov and Kashkush, 2011; Senerchia et al., 2015; Bariah et al., 2020). Due to their high copy number, TEs have the highest potential to differentiate between subgenomes. In bread wheat, TEs constitute ∼80%–85% of the total genome with ∼70% long terminal-repeat retrotransposons (LTRs) and ∼12% DNA transposons (Walkowiak et al., 2020). LTR elements that are “young,” that is, that have transposed recently, tend to be located in the gene-rich recombinogenic distal part of chromosomal arms, while “old” LTR elements tend to be conserved among wheat lines and to be located in the pericentric heterochromatic regions of the chromosomes (Walkowiak et al., 2020). Recent transpositions, therefore, contribute to subgenome divergence in regions important for promoting homologous pairing. TEs also have the potential to affect genome structure and function through transposition, induction of homologous recombination, and epigenetic re-patterning (Shalev and Levy, 1997; Bennetzen, 2005; Slotkin and Martienssen, 2007; Fedoroff, 2012). Since the activity of TEs is governed by epigenetic regulation, their activation might be induced by genetic and environmental perturbations (Fedoroff, 2012). Hence, TEs may mutate genes, alter gene regulation, and generate new genes, in response to environmental challenges, thus providing fuel for evolution (Kidwell and Lisch, 1997). TEs associated with genes can affect the transcription of neighboring genes, as shown for retroelements, through readout activity of their LTRs (Kashkush et al., 2002, 2003). Interestingly, inspection of the expression of homoeologs for which all three subgenomes were represented (triads) showed that for ∼70% of the triads, the level of expression was balanced, that is, homoeoalleles were generally expressed at the same level. For the ∼30% remaining triads, differential expression was often associated with the diversity of TEs in proximity to the promoter region (Ramírez-González et al., 2018). Wheat TEs can affect splicing and intron retention as shown for SINEs that are enriched in introns (Keidar et al., 2018). Miniature inverted-repeat transposable elements (MITEs) also show a strong association with wheat genes, being near genes, or within the transcriptome (Keidar-Friedman et al., 2018). All this suggests that TEs are drivers of dynamic genomic changes and modulation of gene expression and contribute massively toward shaping genome structure, gene expression, and evolution. However, these dynamic changes, which can lead to deleterious genome instability, are mitigated by several epigenetic genome stabilizing factors that suppress TE activities such as cytosine methylation (Fedoroff, 2012) or small RNAs (Kenan-Eichler et al., 2011). Nevertheless, TEs have retained some extent of mobility throughout wheat evolution as suggested from their high diversity between the subgenomes of bread wheat and between varieties in the same subgenome (Wicker et al., 2018) and as shown for MITEs in a newly synthesized wheat allohexaploid (Yaakov and Kashkush, 2012).
Introgression was an important mechanism to increase genetic diversity during wheat evolution. Indeed, there has been extensive natural introgression from wild emmer wheat into the background of domesticated emmer wheat or bread wheat, leading to higher variability in these genomes (He et al., 2019; Zhou et al., 2020). This was facilitated by the ease of crossing between wild emmer and domesticated subspecies of T. turgidum, having the same BBAA genome. Moreover, the initial process of domestication might have taken approximately 2–3,000 years (Kislev, 1984; Tanno and Willcox, 2006) as seen from the archeological records where fragile, partly fragile, and nonfragile spikes can be found in the same farming sites, providing high chances to hybridize. Consistent with these biological and archeological data, the variation in the domesticated-emmer genome includes ∼73% of that of wild emmer (Zhou et al., 2020; Keilwagen et al., 2022; Sharma et al., 2021). Genotyping of T. aestivum landraces has shown that a significant part of the variation in wild or domesticated tetraploid wheat has also found its way into the A and B subgenomes of the bread wheat genome suggesting that there was only a limited genetic barrier between tetraploid and hexaploid wheat (Dvorak et al., 2006). The barrier between diploid Ae. tauschii and hexaploid wheat is harder to cross because the resulting genome ABDD tetraploid hybrid is highly sterile, which is consistent with the finding that the variability of the D subgenome is 2–5 times lower than that of the A and B subgenomes (Caldwell et al., 2004). Nevertheless, some degree of introgression took place as seen from whole-genome sequencing (Gaurav et al., 2021; Zhou et al., 2021). Introgressions were found between nonhomologous genomes, seen as an incongruent block of single-nucleotide polymorphisms (SNPs) that are specific to another species and do not follow the phylogeny of the species, mostly from the B to D lineage and to a lesser extent from D to B lineage. However, these introgressions were relatively rare and took place prior to the formation of tetraploid wheat (Li et al., 2022).
Homologous and homoeologous recombination
Meiotic recombination between homologous chromosomes is a major engine of diversity in wheat evolution and breeding. Introgressions via recombination between homologous genomes of bread wheat and wild relatives have been relatively frequent for the A and B genomes as discussed above. Dvorak (2009) reviewed data indicating that Triticeae genomes have a steep recombination gradient along the centromere–telomere axis following gene content which increases from the proximal toward the distal region. In their detailed study of the pattern of crossovers along the sequenced chromosome 3B of bread wheat, Saintenac et al. (2009) also show that crossover frequency increases gradually from the centromeres to the telomeres. Moreover, they show that small chromosome segments of high gene density and frequent recombination interspersed with relatively large regions of low gene density and infrequent recombination (Saintenac et al., 2009). Multiple crossovers occurred within these gene-dense regions where the degree of recombination is at least 11-fold greater than the genomic average (Faris et al., 2000). Fine mapping of crossover in chromosome 3B showed that 82% of the crossovers occurred in 19% of the chromosome length in the sub-telomeric regions which carry 60%–70% of the genes (Valenzuela et al., 2013; Darrier et al., 2017). The remaining ∼35% of the genes are located in recombinationally poor chromosomal regions (Erayman et al., 2004; Darrier et al., 2017), hindering the elimination of deleterious mutations or introgression of beneficial ones in these regions. Recombination tends to occur in or near genes, often in promoter regions (Darrier et al., 2017). Most recombination events take place in hotspots that are characterized by specific sequence motifs, such as CCN or CTT repeats, or A-rich regions (Darrier et al., 2017) and are associated with typical chromatin modifications (Liu et al., 2021). A new member of the RecQ helicase gene family was recently shown to be associated with high crossover frequency in hexaploid wheat (Gardiner et al., 2019).
Superimposed on the divergence of the homoeologous chromosomes is a genetic system that contributes to the maintenance and reinforcement of the exclusive bivalent pairing in the allopolyploid Triticum species, ensuring that chromosome pairing is wholly restricted to homologous chromosomes (reviewed by Sears, 1976). The most potent suppressor of homoeologous pairing between wheat subgenomes and in interspecific or intergeneric hybrids is a gene designated Pairing homoeologues1 (Ph1). First described by Okamoto (1957), Sears and Okamoto (1958), and Riley and Chapman (1958), Ph1 is located on chromosome arm 5BL and suppresses homoeologous chromosome pairing without affecting homologous chromosome pairing. A deletion of Ph1 was induced by Sears (1977) via X-irradiation and shown by Gill and Gill (1991) to encompass 73.5 Mbp (the ph1b locus). In the absence of Ph1, pairing occurs between the homoeologues of the A, B, and D subgenomes of bread wheat or between homoeologous chromosomes in hybrids between hexaploid wheat and wild relatives (Sears, 1976; Feldman, 1988). Later analysis of deletions in 5B enabled restricting the size of the Ph1 locus to a 2.5-Mb region (Griffiths et al., 2006). This region still contains several candidate genes that may affect chromosome synapsis and recombination: C-Ph1 (Bhullar et al., 2014), CDK2-like genes, methyl transferase genes and Hyp3, now called TaZIP4-B2, and it is possible that the Ph1 phenotypes are due to the combined effect of several genes as proposed by Griffiths et al. (2006). Recently, two mutants in TaZIP4-B2, a point mutation induced by ethyl methanesulfonate (EMS) and a small deletion induced by clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9 yielded high levels of homoeologous pairing (Rey et al., 2017) making it a strong Ph1 candidate. Nevertheless, recovering the whole complexity of the Ph1 phenotype might require mutations in additional genes at the same locus. In addition to Ph1, there are at least two other suppressors of homoeologous pairing in T. aestivum, the 3DS and 3AS genes, designated Ph2 and Ph3, respectively, that are less potent than Ph1 (Sears, 1982, 1984). The 3DS Ph2 locus was recently isolated (Serra et al., 2021) and found to encode the wheat homolog of DNA mismatch repair gene MSH7, TaMSH7.
How homoeologous suppression evolved is still poorly understood. It seems to have existed at the diploid levels (Attia et al., 1979). However, diploid activity seems to have been too weak to explain homoeologous suppression as seen in bread wheat. Riley (1960) and Sears (1976) assumed that Ph1 evolved at the allotetraploid level, in parallel to, or soon after, the allopolyploidization process. The evolution of Ph1 seems to have been gradual with activity increasing with polyploidy with diploid < tetraploid < hexaploid (Ozkan and Feldman, 2001). Tracing the evolution of homoeologous pairing suppression will be possible only when the molecular mode of action of Ph1 is more fully understood but will remain limited in the absence of an extant B donor species.
In nature, the number of introgressions that took place by homoeologous recombination in the bread wheat lineage seems to have been relatively infrequent (Li et al., 2022). Nevertheless, when it happens, as shown in a synthetic allotetraploid lacking Ph1, homoeologous recombination events were shown to occur preferentially within exons, generating novel hybrid transcripts and proteins (Zhang et al., 2020a), possibly due to the higher homology in these regions. This contrasts with homologous recombination that preferentially occurs in promoters (Darrier et al., 2017).
In summary, allopolyploidy in the wheat genome has enabled, and in some cases induced, rapid changes at many levels, starting from providing a rapid speciation mode that triggered massive changes in repetitive DNA such as centromeres, NORs, and TEs, as well as gene elimination and duplication, thus leading to divergence of homoeologous genomes. Despite these rapid alterations, stability has been maintained thanks to epigenetic regulation and to a genetic system superimposed on the diverged homoeologs, namely the Ph1 locus, that was present in diploids but further evolved in polyploids, controlling the preferential pairing of homologous rather than homoeologous chromosomes. It is assumed that all these mechanisms have contributed to the rapid genetic and cytological diploidization of the wheat genome, ensuring disomic inheritance and fertility of the nascent species, eliminating redundancies, and maintaining heterotic interactions between subgenomes.
Wheat domestication
Wheat is a staple food of humanity and its evolution under human selection is part of a never-ending endeavor to reach food security. Proto-humans likely fed on foraged wheat since the Paleolithic. What induced hunter–gathers in the Near East to start cultivation and then to domesticate plants and become farmers? This question has inspired myths and religions (e.g. Genesis 3: 17–23) and has fascinated archeologists, anthropologists, plant geneticists, and evolutionary biologists, including Darwin (1868). Despite various hypotheses, such as pressure on food resources due to an increase in human population and to climate change (see Figure 2), the “why” of wheat cultivation and domestication remains a mystery. Likewise, an important and unanswered question is whether domestication was an intentional process, namely due to active selection by early farmers (as modern breeders would do) of mutants adapted to cultivation, such as nonfragile spike and free threshing, or whether it was a genetic drift due to the increased fitness under cultivation of these mutants that became fixed after several generations. Both are plausible and not mutually exclusive scenarios.
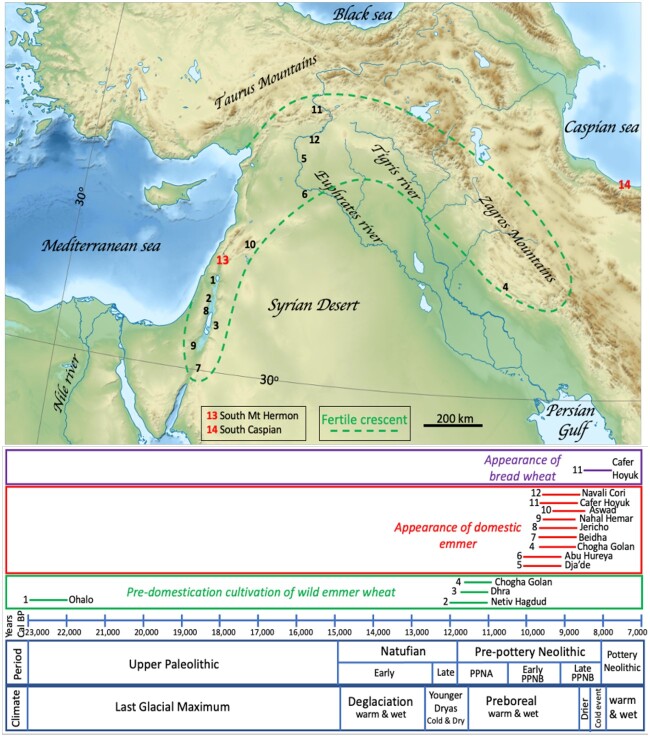
Figure 2.
Archaeological evidence for wheat cultivation and domestication in the near-east. The location of the fertile crescent is shown as dashed green lines. Its boundaries correspond to the distribution of wild progenitors of wheat, barley, and several legumes as well as to early domestication of these crops. The western part, called the Levant or levantine corridor (Bar-Yosef, 1998), goes south, around the Jordan valley between the dashed line on the side of the Syrian desert and the Mediterranean Sea. The south Levant is the region between Beidha (#6) and Aswad (#9) and the north Levant is north of Aswad (#10), for example, in Dja’de (#5) and Abu Hureya (#6). The northern area of the fertile crescent is also referred to as the upper Euphrates (e.g. Cafer Hoyuk, #11), and the east of the fertile crescent, in the Zagros mountain is represented by sites such as Chogha Golan (#4). The years on the blue horizontal axis correspond to “Calibrated years before present” (Cal BP). The bottom boxes represent the climatic and the archeological periods when cultivation and domestication took place. Horizontal lines flanked by the location number (see map) and names indicate the relevant period when archeological evidence of cultivation or domestication was found. Numbers in red indicate two regions where evidence came from genomic data rather than archaeological data for the putative progenitors of domestic emmer (#13) and for the donor of the D subgenome of bread wheat (#14). Archaeological data were gathered from Nesbitt (2001); Willcox (2012); Zeder (2011); Riehl, et al. (2013). A blank topographic map from Wikipedia (Middle East topographic map-blank 3000bc.svg, by Fulvio314, CCBY 3.0) served as the background on which text and data were added.
Wheat domestication: where and when?
The phases of wheat evolution under human selection are: (1) the gatherers (also called foraging) period when wheat was harvested from wild stands; (2) a predomestication cultivation period when wild wheat was grown in small plots; (3) the domestication of wild emmer wheat, namely, the period when mutations in genes affecting spike morphology appeared and became fixed. These mutations, which hindered seed dispersal and facilitated threshing, suited the farmer rather than the wild plant and turned wheat into an organism dependent on humans for its dissemination; (4) the formation of bread wheat under domestication; (5) the spread of bread wheat and the accumulation of landraces; and (6) the green revolution and modern breeding.
We know little of the predomestication period, when human were gatherers and brought grains near their dwellings, due to a limited amount of available data. The earliest data relevant to wheat comes from archeological records from the upper paleolithic Last Glacial Maximum period approximately 23,000 years ago in the hunter–gatherer sedentary camp of Ohalo II on the shores of the lake of Galilee (Figure 2), suggesting that there might have been a period of cultivation of wheat that was not yet domesticated (Snir et al., 2015). The evidence is that extensive farming-related activity was detected at this site, as deduced from various flints, sickle blades and stone grinding tools, fauna remains, and large amounts of seeds (approximately 10,000 seeds from cereals) including wild emmer wheat, wild barley, and wild oats, in what was a human-disturbed environment containing seeds from weedy species (presumably a field). If this interpretation is correct, this would be the earliest known site of wild wheat cultivation. Alternatively, it might be a site where wild emmer wheat, harvested from nearby wheat stands, was brought and processed (Piperno et al., 2004). Ohalo II is a singular case as there is a gap of approximately 10,000 years before other human sedentary settlements were found during the Younger Dryas in the Late Natufian and early Pre-pottery Neolithic-A period throughout the fertile crescent (Figure 2). There is a consensus among archaeobotanists (not considering Ohalo II) that the cultivation of wild cereals predated morphological domestication by more than 1,000 years with wild einkorn in the north Levant as soon as 13,000 Cal-Y BP (Abu Hureyat I and Mureybit I-III) and 11–12,000 Cal-Y BP, wild emmer in the south Levant in Netiv Hagdud and Zaharat adh-Dhra (see reviews by Nesbitt, 2001 and Willcox, 2012) and wild barley and to a lesser extent also wild emmer in the eastern part of the fertile crescent Chogha Golan site in the Zagros mountains (Riehl et al., 2013). During these periods, there was no clear morphological evidence for the presence of domesticated wheat.
The morphological hallmark of domesticated cereals is the nonbrittle rachis, marking the transition from seed dispersal through spike fragility to dispersal by farmers (Figure 3). Domesticated emmer wheat appears first in the archeological record of the northern Levant (Figure 2) at Dja’de and Abu Hureya II in the early PPNB (starting approximately 10,000 Cal-Y BP), and briefly after that in the southern Levant at Beidha, Jericho, Nahal Hemar, in the central fertile crescent at Cafer Hoyuk and Navali Cori (reviewed by Nesbitt, 2001 and Willcox, 2012) and east of the fertile crescent at Chogha Golan (Riehl et al., 2013). Early genetic studies searching for the location of emmer wheat domestication did not provide clear answers and did not always match archaeological data, with evidence pointing to either the upper Euphrates or the Levant (Ozkan et al., 2005; Luo et al., 2007; Civan et al., 2013). Confusion on these points was exacerbated by the mobility of Neolithic farmers, rapidly spreading seeds and cultivation technology and enabling multiple and diverse introgressions among wild in domesticated wheat (Civan et al., 2013). Part of this confusion can be clarified by looking at the fragility loci which are more diagnostic than the rest of the genome, being the first hallmarks of domestication (Avni et al., 2017; Nave et al., 2019). Recent genetic evidence shows that a haplotype surrounding a major gene controlling spike fragility (further discussed below) that is found in all the modern tetraploid and hexaploid wheats, originates from wild emmer wheat from the south foothills of Mt. Hermon in the south Levant (Nave et al., 2019). The “where” origin is thus fuzzy, spanning possible sites from the south Levant all the way to the North of the fertile crescent in the upper Euphrates (Figure 2). Nonfragile spikes were initially found together with fragile spikes in early PPNB sites and it took 2–3 millennia for nonbrittleness to be fully dominant among cultivated forms of wheat suggesting that domestication was a gradual process as proposed by Kislev (1984) for emmer wheat and by Tanno and Willcox (2006) for einkorn. The loss of fragility gave rise to the first known domesticated wheat, T. turgidum ssp. dicoccon which had a hulled grain and a nonbrittle rachis. Free-threshing tetraploid wheat appeared soon following the nonfragile types, in the Near east, late Pre-Pottery Neolithic B (PPNB) (Kislev, 1980; Nesbitt, 2001; Schultze-Motel, 2019).
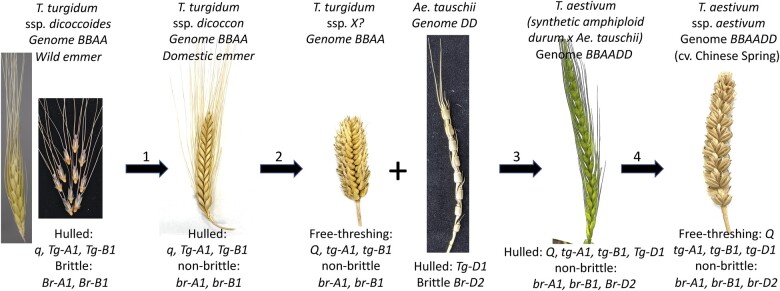
Figure 3.
Major mutations and morphological changes during wheat domestication. 1, The transition from ssp. dicoccoides to ssp. dicoccon involved mutation in the Brittle rachis loci. Some modern emmer wheat lines might also contain mutations in some but not all loci affecting free threshing. 2, Free-threshing tetraploid wheat, named ssp. parvicoccum, appears in the archeological record approximately 2,000 years before ssp. durum. It is now extinct but might have resembled the tetraploid wheat (Genome BBAA) shown here as ssp. X that was extracted from hexaploid wheat and has a compact spike and small grains. Its genotype must have been similar to durum, namely free threshing with soft glumes, with mutants Q and tg-A1, tg-B1. 3, The hybridization of this free-threshing tetraploid wheat with the DD subgenome donor, Ae. tauschii, gave rise to a primitive hulled hexaploid wheat, different from spelt wheat due to the Q factor, and absent from the archaeological record. It likely resembled the picture shown from a synthetic hexaploid between ssp. durum and Ae. tauschii shown here. 4, Soon after its formation, hexaploid wheat became free threshing thanks to a mutation in Tg-D1 and its rachis became thicker thanks to a mutation in Br-D2.
By the middle PPNB, approximately 9,800 Cal-Y BP, domesticated tetraploid wheat was already found east of the fertile crescent (Riehl et al., 2013), and it is assumed that it came into contact with Ae. tauschii to form hexaploid wheat (Feldman, 2001; Zohary et al., 2012). Various biochemical and molecular studies indicated that the most likely area of origin of T. aestivum is the south-western corner of the Caspian belt (Nakai, 1979; Jaaska, 1981; Dvorak, 1998). A recent whole-genome analysis of a core collection of 278 accessions covering the eco-geographic distribution of Ae. tauschii, in arid and semi-arid habitats from central Asia, Transcaucasia to China, confirmed earlier studies suggesting that the wheat D subgenome is mostly derived from the strangulata subgroup originating from the south Caspian Sea and further narrowing down the origin to accessions from the Mazandaran province (Zhou et al., 2021). A recent analysis of 242 Ae. tauschii accessions showed that a rare and distinct lineage (different from strangulata) from Transcaucasia also contributed ∼1% on average to the current wheat D subgenome (Gaurav et al., 2021), in accordance with earlier studies that analyzed allelic variation of high molecular weight (HMW) glutenins (Giles and Brown, 2006). HMW glutenins from the D subgenome have contributed to wheat baking quality (Orth and Bushuk, 1973; Shewry, 2009). Likewise, the Transcaucasian D lineage contains an HMW glutenin subunit contributing to superior baking quality (Delorean et al., 2021). Hence, a likely scenario is that bread wheat was formed in the south Caspian under domestication, that a few additional introgressions with strangulata lines took place such as the Ae. tauschii lineage from Transcaucasia, and that there is no true wild T. aestivum. Note that there is one semi-wild form of hexaploid wheat, ssp. tibetanum, growing in Tibet, whose spike is fragile, hulled and breeds freely with bread wheat (Shao and Li, 1983), but it is generally considered a feral form of bread wheat T. aestivum ssp. aestivum. A recent study showed that Tibetan wheat became feral through mutations in a local landrace that involved a Btr1/2 homolog on 3D and Q on 5A (Guo et al., 2020). Dvorak et al. (2012) suggested, on the basis of the mapping of the tenacious glume (Tg) locus in several wheat species, that the BBAA subgenomes of hexaploid wheat originated from a free-threshing tetraploid form and therefore it could not be T. turgidum ssp. dicoccon, the most ancient domesticated hulled grain tetraploid wheat. This leaves us with the question of the identity of the domesticated tetraploid parent of hexaploid wheat. The free-threshing tetraploid macaroni wheat T. turgidum ssp. durum is also an unlikely donor since there is no evidence that it existed approximately 8,500 YA when T. aestivum was formed. The domesticated free-threshing tetraploid wheat that was cultivated approximately 9,000 YA, starting in the Levant and spreading throughout the fertile crescent, when T. aestivum was formed is the primitive tetraploid T. turgidum ssp. parvicoccum (1980, Kislev, 2009; Schultze-Motel, 2019), making it a candidate as donor of the BBAA genomes. This is reinforced by the fact that the extracted tetraploids produced by Kerber (1964), having the BBAA subgenomes from hexaploid wheat and lacking the D subgenome (see Figure 3), were free threshing and had similar small and oval grain shape as ssp. parvicoccum (Kislev, 1980, 2009). While ssp. parvicoccum was found in several archaeological sites beyond the fertile crescent, in the Balkans and in Transcaucasia, and as late as approximately 1,000 years ago in Georgia (Schultze-Motel, 2019), it is now extinct as it was replaced during the Roman period by the more prolific and large-grain ssp. durum (Kislev, 2009). Therefore, the precise identity of the free-threshing domesticated tetraploid that hybridized with Ae. tauschii to form T. aestivum remains uncertain. Another puzzling fact about hexaploid wheat origin is that there is no archeological record of the hulled, spelt-like type that should have emerged from a T. turgidum × Ae. tauschii allohexaploid (see below). The earliest archeological findings of hexaploid wheat are from an 8,400-year-old, free-threshing type in Cafer Hoyuk, upper Euphrates (Bilgic et al., 2016) rather than from the location of origin predicted from the genomic data (Zhou et al., 2021) in the south Caspian (Figure 2). The lack of hulled, spelt-like types in the archeological record suggests that the free-threshing mutation occurred very soon after hexaploid wheat formation and was preferred, in most regions, over the original Spelt-like type. The free-threshing hexaploid wheat settled in central Anatolia and spread rapidly to Europe (Bogaard, 2016).
Domestication: how?
The brittle rachis phenotype in ssp. dicoccoides is controlled by two major loci mapped on the short arm of chromosomes 3A and 3B (Levy and Feldman, 1989; Nalam et al., 2006; Watanabe et al., 2006; Millet et al., 2013). Avni et al. (2017) isolated these two genes, homologous to the barley Brittle rachis Btr1 and Btr2 genes (Pourkheirandish et al., 2015). They show that, while a Btr2 homolog exists in wheat, only a mutation in the Btr1 orthologs in the wild emmer wheat loci (br-A1 and br-B1) leads to nonbrittleness (Figure 3). The mutations are likely a loss of function: br-A1 has a 2 bp deletion 290 bases from the start codon and br-B1 contains a 4-kb TE insertion in its coding region. A mutation in a single homoeoallele, (br-A1 or br-B1) led to partial fragility while the combination of both was needed for the full nonbrittle rachis phenotype. A panel of 113 wild emmer, 85 domestic emmer, and 9 durum lines showed that all domesticated forms carried both mutations (Avni et al., 2017). The accumulation of one mutation after another was thus necessary to reach full nonbrittleness which is consistent with the gradual evolution of this phenotype as seen in the archeological record (Kislev, 1984; Tanno and Willcox, 2006). Moreover, additional QTLs with weaker effects were mapped to other loci (Tzarfati et al., 2014) and might have been selected to further strengthen the rachis. Remarkably, the function and mode of action of the barley Btr1 and Btr2 genes or of their wheat homologs remain unknown. In barley, mutations in these genes affect cell wall thickness in the spikelet abscission zone, but not cell wall composition (Pourkheirandish et al., 2015). Nave et al. (2019) investigated the geographical birthplace of the recessive mutations in the Brittle Rachis genes br-A1 and br-B1; mutations that determine spike nonbrittleness. They analyzed a core collection of more than 400 lines of wild and domesticated wheat, including hexaploid wheat. They show that mutations in br-A1 and br-B1 are of monophyletic origin in the domestic germplasm. They also analyzed the precursor of the domesticated haplotype of br-A1 in wild accessions and found that it is widespread throughout the Levant, from central Israel to central Turkey. In contrast, the precursor of the domesticated haplotype of br-B1 was found only in the wild accessions, all from the southern Levant. Moreover, one particular accession from the southern foothills of Mt Hermon carried the haplotype around both br-A1 and br-B1 that is found in all domesticated polyploid wheat with the AA and BB genomes (Nave et al., 2019). This suggests a central role for the southern Levant in the domestication of wheat, where one or both mutations might have occurred as opposed to the earlier proposal for the upper Euphrates (Ozkan et al., 2005). In this regard, genetic and archeological evidence do not fully match because the most ancient nonfragile spikes were found in the north Levant at Abu Hureyra and Mureybit (Figure 2). These conflicting conclusions could be reconciled if the eco-geographic distribution of wild wheat drifted during the past 10,000 years since domestication started, or if the archeological record is incomplete.
Concerning hexaploid wheat, one might assume that the hybrid between domesticated free-threshing tetraploid wheat and Ae. tauschii that gave rise to bread wheat would be fragile due to the wild-type (WT) fragility locus in 3D, Br-D2 (Figure 3). Fragility in Ae. tauschii is not taking place at the rachis node, as in ssp. dicoccoides, but the breakage occurs below the rachis node (Zohary and Hopf, 2000) so that the spikelet falls with the rachis internode beside it and not below it, as in wild emmer. This fragility trait is controlled by a gene mapping to the long arm of chromosome 3D (Li and Gill, 2006) and designated Br-D2, while Br-A1 and Br-B1 are located on the short arm, and its mode of action is not clear. At any rate, the spike of the amphiploids between domesticated tetraploid wheat and Ae. tauschii turned out to be hulled but nonfragile (Figure 3) even though it becomes brittle upon slight pressure indicating co-dominance of the Br1 and Br2 genes (McFadden and Sears, 1946). Had it been fragile it might never have been domesticated.
Free threshing is another important domestication trait that is beneficial to the farmer but reduces fitness in the wild. The WT grain is tightly enclosed within the glumes which are large and hard and protect it from birds and grazers while the grain of free-threshing wheat can be easily separated from the glumes. Muramatsu (1986) showed that free threshing emerged at the tetraploid level in an already domesticated nonfragile crop, ssp. dicoccon (Figure 3), and that all the free-threshing (or naked) forms of tetraploid wheat carry a dominant allele, the Q factor, located on the long arm of chromosome 5A (Sears, 1954). The Q allele has numerous pleiotropic effects, changing the shape and thickness of the glumes, and leading to a more compact spike and to several other phenotypes (Mac Key, 2005; Simons et al., 2006). Cloning of the Q factor showed that it encodes an AP2-like transcription factor, which is consistent with its effect on multiple pathways (Simons et al., 2006). The q homoeoalleles underwent pseudogenization on 5B and neofunctionalization on 5D (Zhang et al., 2011). The Q mutation is not a duplication as previously thought (Muramatsu, 1963) but a gain-of-function mutation that is found in all the domesticated wheat forms, tetraploid or hexaploid (Simons et al., 2006). Although Simons et al. (2006) considered a SNP, leading to the substitution of valine by isoleucine at position 329, as a possible cause for the Q mutation, they also noted a conserved SNP in the miRNA 172 binding site. Subsequent work by Debernardi et al. (2017) showed that in fact, the SNP within the miRNA binding site is the causal polymorphism for the functional difference between the Q and q alleles. Moreover, Q is more abundantly transcribed than WT q, which is consistent with its dominant nature, as well as with the disruption of the miRNA 172 suppressive effect (Debernardi et al., 2017) and with the positive dosage effects shown by Muramatsu (1963) who found that extra doses of q mimic the effect of Q in common wheat. Recently, Zhang et al. (2020b) reported new insights into Q’s mode of action using transcriptomics and phenotypic analyses. They show that modification of cell wall thickness and composition of glumes, for example, lignin versus cellulose ratio, correlates with the expression of genes involved in secondary cell wall biosynthesis.
When free-threshing domesticated tetraploid wheat hybridized with the D donor to form bread wheat (Dvorak et al., 2012), we might have expected that the resulting allohexaploid would be free threshing because the Q factor is dominant. However, all crosses of either hulled or free-threshing tetraploid wheat with all lines of Ae. tauschii yielded only hulled forms resembling ssp. spelta, indicating that this subspecies is the prototype of hexaploid wheat (McFadden and Sears, 1946; Kerber and Rowland, 1974). Further analyses showed that the free-threshing Q-phenotype was suppressed by the Tg gene from Ae. tauschii, located on chromosome 2D (Tg-D1, Figure 3), and that a single mutation from Tg to tg produced free-threshing forms (Kerber and Rowland, 1974). The identity of the Tg homoeoalleles (Tg-A1, Tg-B1, and Tg-D1) remains unknown. Simonetti et al. (1999), Faris et al. (2014), as well as Sharma et al. (2019), performed fine mapping of the Tg loci and showed that dominant homoeoalleles on chromosomes 2A and 2B of emmer wheat (Tg-A1 and Tg-B1) contribute together with the q WT allele, to the nonfree-threshing phenotype (Figure 3). They also showed that fully free threshing is achieved by mutations in all three loci and that each mutation has an additive effect on threshability with Q having the most profound effect. Free-threshing tetraploid wheats such as T. turgidum ssp. durum, and possibly ssp. parvicoccum, were formed through mutations in all three loci, leading to a tg-A1tg-A1/tg-B1tg-B1/Q-A1Q-A1 genotype. However, when, where, and in what order these mutations occurred remains unknown. The hulled hexaploid wheat formed through hybridization between free-threshing tetraploid wheat and Ae. tauschii was thus expected to have the Q/tg-A1tg-A1/tg-B1tg-B1/Tg-D1Tg-D1 genotype (Figure 3). Such genotypes have not been found among extant hulled wheats. Indeed, most hulled hexaploid spelt wheat lines (T. aestivum ssp. spelta) examined so far did not have this genotype but rather had a recessive tg-D1 allele and a dominant Tg-B1 and q allele and are thus thought to be derived from hybridization between free threshing hexaploid wheat with tetraploid hulled emmer wheat (Dvorak et al., 2012). The free-threshing bread wheat, ssp. aestivum, with the Q/tg-A1tg-A1/tg-B1tg-B1/tg-D1tg-D1 genotype (Figure 3) must have been preferred by the early farmers of the region and the single mutation in Tg-D1 enabled the quick replacement of the hulled forms. The isolation of the still elusive Tg homoeoalleles will facilitate a better understanding of the evolution of free-threshing tetraploid and hexaploid wheat and the origin of spelt wheat.
In addition to the above-mentioned classical domestication traits, several other traits were selected during the process of domestication. These include plant erectness versus the prostrate grassy types, increased number of seeds per spikelet, increased grain size, and reduced seed dormancy (Feldman, 2001). Overall, the number of domestication-related QTLs mapped to the A subgenome was two-fold higher than those found on the B subgenome, that is, 24 QTL effects for domestication and domestication-related traits in the A subgenome, versus only 11 such QTLs in the B subgenome (Tzarfati et al., 2014). This is in accordance with the concept of “genome asymmetry,” implying that the A subgenome is dedicated to the control of morphological traits, housekeeping metabolic reactions, and yield components (Peng et al., 2003; Feldman et al., 2012).
All of the major evolutionary processes required to produce domesticated tetraploid and hexaploid wheat were completed by the end of the Pre-Pottery Neolithic period in the Near East, about 7,500 BP (Kislev, 1984), and wheat spread away from its Near-East cradle. The appearance and fixation of the nonfragility and free-threshing traits happened mostly within a millennium, between 9,500 and 8,500 years BP for tetraploid and hexaploid wheat. The spread of wheat westward was relatively swift. A surprising finding was the report of wheat DNA in 8,000 years old sediments in a submarine archeological site off the isle of Wight, in the southern UK (Smith et al., 2015). The remarkable transformation and rapid evolution of wheat were probably due to several factors. The allopolyploidy in wheat likely contributed to a highly evolvable nature (Dubcovsky and Dvorak, 2007). While some diploid species such as barley, which has a similar domestication history as wheat, can grow abundantly in a broad range of habitats (see Haas et al. (2019) for a comparison of wheat and Barley domestication and evolution), the wheat diploids grow in limited habitats where they are not the dominant species. In fact, diploid einkorn, which was the first domesticated wheat, did not have the same success as tetraploid wheat, which itself is cultivated approximately 10 times less than hexaploid bread wheat. Intergenomic interactions, described above, together with the impressive capacity of wheat to sustain high rates of mutations (Dvorak and Akhunov, 2005; Akhunov et al., 2007) might have enabled rapid evolution in the allopolyploid background. Moreover, the spread of domesticated tetraploid wheat to new regions facilitated contact between the domesticated and related species or genera, with which they could hybridize and exchange genes. One such encounter is of course the hybridization between domesticated tetraploid wheat and Ae. Tauschii, which led to hexaploid wheat formation. While domesticated tetraploid wheats, in keeping with their Near Eastern origin, are adapted to the Mediterranean-type environments (with mild winters and warm, rainless summers), addition of the D subgenome from the central Asiatic Ae. Tauschii greatly extended the range of adaptation of hexaploid wheats to a more continental climate over the continental plateaus of Asia and the colder temperate areas in eastern, central, and northern Europe (Feldman, 2001; Zohary et al., 2012). The D genome also contributed resistance to several fungal diseases (Appels and Lagudah, 1990; Yildirim et al., 1995) as well as many desirable grain qualities (Orth and Bushuk, 1973; Delorean et al., 2021) that promoted bread wheat spread and global adoption as the preferred wheat type and the preferred cereal compared to barley.
Major milestones in wheat breeding during the past century, including the green revolution, were recently reviewed (Haas et al., 2019; Venske et al., 2019). The phylogeny of a broad collection of landraces and modern bread wheat lines shows clades that reflect the early east versus west spread of wheat and the adaptation to different climates and in more recent breeding the separation of two major gene pools, one from the western block and one from the Warsaw pact (Pont et al., 2019). Selection for mutations in vernalization (VRN genes) that enabled growth under harsh winter conditions or photoperiod sensitivity (e.g. PPD-D1) that enabled growth under a broad range of latitude, enabled the expansion of wheat to new areas. The selection of reduced height (Rht genes) enabled the green revolution through reduction of lodging under chemical fertilization and high yield.
The future of wheat evolution
Breeders will continue to rely on wheat biodiversity and wild wheat will keep evolving in nature as they did for the past million years, surviving multiple climatic changes. However, it is not certain that food security can be ensured in the face of anthropogenic activities, nor that biodiversity won’t be affected. Indeed, climate changes, population growth, urbanization, arable land expansion, overgrazing, soil deterioration, etc., might lead to the extinction of several wild wheat populations, to a reduction in biodiversity, and to the emergence of new diseases. Biodiversity conservation in seeds banks and in situ protection are thus essential. Conservation of landraces is mostly taking place in seeds banks, with notable efforts to characterize them genetically (Kilian et al., 2011, 2021; Cavanagh et al., 2013; Dempewolf et al., 2017; Frankin et al., 2020) and to maintain their culture, in situ in farmer’s fields, for niche markets (Negri, 2003). In situ conservation of wild wheat is much more limited. An analysis of a long-term study of in situ conservation of wild emmer wheat showed the resilience, for now, of the genetic structure of a population during the past 36 years in a protected area where temperature raised by ∼1.5°C and CO2 concentration by ∼70 ppm (Dahan-Meir et al., 2022). Establishing natural reserves for in situ wild wheat species conservation is critical to ensure their survival and evolution in the wild and to protect biodiversity. Wild emmer wheat (Huang et al., 2016; Klymiuk et al., 2018; Faris et al., 2020; Li et al., 2020) and Ae. Tauschii (Bhatta et al., 2018; Gaurav et al., 2021; Zhou et al., 2021) whose chromosomes readily pair with their bread wheat homologs are an invaluable resource for breeders in particular when it comes to biotic and abiotic resistance genes not found in the domestic wheat gene pool. Also, the A subgenome of wild Triticum Timopheevii and the A genome of T. rartu are homologous to the A subgenome of bread wheat and can be used as a source of useful genes (Badaeva et al., 2021; Zeibig et al., 2021). The gene pool for wheat breeding can be extended to more distant relatives whose homoeologous chromosomes can recombine with wheat subgenomes in the absence of Ph1. In this respect, sequencing the genome of several wheat relatives would provide an invaluable source to facilitate broad gene transfers. The ability to increase the rate of homoeologous recombination in a ph1 mutant background has been widely used during wheat breeding for the transfer of genes from wild relatives of wheat to the wheat background (Feldman and Sears, 1981). This might be further enhanced by a premeiotic treatment with magnesium in the ZIP4 (Ph1 candidate) mutant (Rey et al., 2018) or in the WT background by a virus-induced gene silencing treatment of Ph1 candidates (Bhullar et al., 2014). During the 20th century, wheat geneticists and breeders have transferred genes from at least 52 species whose chromosomes do not pair regularly with domesticated wheat chromosomes (Aegilops, Agropyron, Ambylopyrum, Dasypyrum, Elymus, Hordeum, Leymus, Lophopyrum, Psathyrostachys, Pseudoroegneria, Secale, Thinopyrum, and Triticum; Wulff and Moscou, 2014). Several contemporary introgressions from wild relatives were precisely mapped following de novo sequencing of multiple wheat lines (Walkowiak et al., 2020; Keilwagen et al., 2022). The main motivation for these wide transfers was for both resistance to diseases and tolerance to various abiotic stresses. Improving our ability to perform wide gene transfer precisely and effectively is thus critical. The new technologies of genome editing, shown to work well in wheat (Gao, 2021; Li et al., 2021), offer the promise of inducing new targeted genetic variation in wheat in the near future. The mapping of important QTLs in a broad germplasm, rich datasets of genomes and transcriptome sequencing, future data on proteomics and metabolomics, computational tools including Artificial Intelligence, together with speed-breeding (Watson et al., 2018), will enable breeding wheat more efficiently, including locally adapted varieties, to face the challenges of climate change (Xiong et al., 2021). In summary, we can expect that breeding will accelerate to create new varieties that will contain new genetic variants, either induced or from wild wheat relatives.
Acknowledgments
We thank Tal Dahan and Bao Liu for their comments on the manuscript.
Funding
Our research on wheat was supported by the Yeda-Sela grant from the Weizmann Institute and the Israel Science Foundation-China National Natural Science Foundation collaborative grant to Bao Liu and A.A.L.
Conflict of interest statement. None declared.
Contributor Information
Avraham A Levy, Department of Plant and Environmental Sciences, Weizmann Institute of Science, Rehovot, 76100 Israel.
Moshe Feldman, Department of Plant and Environmental Sciences, Weizmann Institute of Science, Rehovot, 76100 Israel.
Both authors contributed to writing this article.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Avraham A. Levy(avi.levy@weizmann.ac.il) and Moshe Feldman (moshe.feldman@weizmann.ac.il).
References
- Aaronsohn A (1910) Agricultural and botanical explorations in Palestine. Bull Plant Ind 180: 1–63 [Google Scholar]
- Akhunov ED, Akhunova AR, Dvorak J (2007) Mechanisms and rates of birth and death of dispersed duplicated genes during the evolution of a multigene family in diploid and tetraploid wheats. Mol Biol Evol 24: 539–550 [DOI] [PubMed] [Google Scholar]
- Akhunov ED, Akhunova AR, Linkiewicz AM, Dubcovsky J, Hummel D, Lazo G, Chao SM, Anderson OD, David J, Qi LL, et al (2003) Synteny perturbations between wheat homoeologous chromosomes caused by locus duplications and deletions correlate with recombination rates. Proc Natl Acad Sci USA 100: 10836–10841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appels R, Honeycutt RL (1986) rDNA: evolution over a billion years. In Dutta SK, ed, DNA Systematics. CRC Press, Boca Raton, FL, pp 81–135 [Google Scholar]
- Appels R, Lagudah ES (1990) Manipulation of chromosomal segments from wild wheat for the improvement of bread wheat. Austral J Plant Physiol 17: 253–266 [Google Scholar]
- Attia T, Ekingen H, Robbelen G (1979) origin of 3D-suppressor of homoeologous pairing in hexaploid wheat. Z. Pflanzenzücht 83: 131–126 [Google Scholar]
- Avni R, Lux T, Minz-Dub A, Millet E, Sela H, Distelfeld A, Deek J, Yu G, Steuernagel B, Pozniak C, et al. (2021) Genome sequences of Aegilops species of section Sitopsis reveal phylogenetic relationships and provide resources for wheat improvement. Plant J 110: 179–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni R, Nave M, Barad O, Baruch K, Twardziok SO, Gundlach H, Hale I, Mascher M, Spannagl M, Wiebe K, et al (2017) Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357: 93–97 [DOI] [PubMed] [Google Scholar]
- Badaeva E, Konovalov F, Knuepffer H, Fricano A, Ruban A, Kehel Z, Zoshchuk S, Surzhikov S, Neumann K, Graner A, et al. (2021) Genetic diversity, distribution and domestication history of the neglected GGAtAt genepool of wheat. Theor Appl Genet 134: 3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariah I, Keidar-Friedman D, Kashkush K (2020) Where the wild things are: transposable elements as drivers of structural and functional variations in the wheat genome. Front Plant Sci 11: 585515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Yosef O (1998) On the nature of transitions: the middle to upper Palaeolithic and the Neolithic revolution. Cambridge Archaeol J 8: 141–163 [Google Scholar]
- Baum BR, Feldman M (2010) Elimination of 5S DNA unit classes in newly formed allopolyploids of the genera Aegilops and Triticum. Genome 53: 430–438 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL (2005) Transposable elements, gene creation and genome rearrangement in flowering plants. Curr Opin Genet Dev 15: 621–627 [DOI] [PubMed] [Google Scholar]
- Bernhardt N, Brassac J, Dong X, Willing E, Poskar C, Kilian B, Blattner F (2020) Genome‐wide sequence information reveals recurrent hybridization among diploid wheat wild relatives. Plant J 102: 493–506 [DOI] [PubMed] [Google Scholar]
- Bhatta M, Morgounov A, Belamkar V, Baenziger PL, Poland J (2018) Unlocking the novel genetic diversity and population structure of synthetic Hexaploid wheat. BMC Genomics 19: 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhullar R, Nagarajan R, Bennypaul H, Sidhu GK, Sidhu G, Rustgi S, Von Wettstein D, Gill KS (2014) Silencing of a metaphase I-specific gene results in a phenotype similar to that of the pairing homeologous 1 (Ph1) gene mutations. Proc Natl Acad Sci USA 111: 14187–14192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgic H, Hakki EE, Pandey A, Khan MK, Akkaya MS (2016) Ancient DNA from 8400 year-old catalhoyuk wheat: implications for the origin of neolithic agriculture. PLoS One 11: e0151974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA (2021) One hundred years of gene balance: how stoichiometric issues affect gene expression, genome evolution, and quantitative traits. Cytogenet Genome Res 161: 1–22 [DOI] [PubMed] [Google Scholar]
- Bogaard A (2016) Archaeobotany: the wheat and the chaff. Nat Plants 2: 16079. [DOI] [PubMed] [Google Scholar]
- Bolot S, Abrouk M, Masood-Quraishi1 U, Stein N, Messing J, Feuillet C, Salse J (2009) The ‘inner circle’ of the cereal genomes. Curr Opin Plant Biol 12: 119–125 [DOI] [PubMed] [Google Scholar]
- Bomblies K, Weigel D (2007) Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet 8: 382–393 [DOI] [PubMed] [Google Scholar]
- Caldwell KS, Dvorak J, Lagudah ES, Akhunov E, Luo MC, Wolters P, Powell W (2004) Sequence polymorphism in polyploid wheat and their D-genome diploid ancestor. Genetics 167: 941–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh CR, Chao S, Wang S, Huang BE, Stephen S, Kiani S, Forrest K, Saintenac C, Brown-Guedira GL, Akhunova A, et al (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci USA 110: 8057–8062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZJ, Murata M (2003) A centromeric tandem repeat family originating from a part of Ty3/gypsy retroelement in wheat and its relatives. Genetics 164: 665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulet F, Wicker T, Rustenholz C, Paux E, Salse J, Leroy P, Schlub S, Le Paslier MC, Magdelenat G, Gonthier C, et al (2010) Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell 22: 1686–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civan P, Ivanicova Z, Brown TA (2013) Reticulated origin of domesticated emmer wheat supports a dynamic model for the emergence of agriculture in the Fertile Crescent. PLoS One 8: e81955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Birchler JA, Pires JC (2014) Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Curr Opin Plant Biol 19: 91–98 [DOI] [PubMed] [Google Scholar]
- Dahan Meir T, Ellis TJ, Mafessoni F, Sela H, Manisterski J, Avivi-Ragolsky N, Raz A, Feldman M, Anikster Y, Nordborg M, et al (2022) The genetic structure of a wild wheat population has remained associated with microhabitats over 36 years bioRxiv 2022.01.10.475641; 10.1101/2022.01.10.475641 [DOI]
- Darrier B, Rimbert H, Balfourier F, Pingault L, Josselin AA, Servin B, Navarro J, Choulet F, Paux E, Sourdille P (2017) High-resolution mapping of crossover events in the hexaploid wheat genome suggests a universal recombination mechanism. Genetics 206: 1373–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin CR (1868). The Variation of Animals and Plants Under Domestication. John Murray, London [Google Scholar]
- Debernardi JM, Lin H, Chuck G, Faris JD, Dubcovsky J (2017) microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development 144: 1966–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorean E, Gao L, Lopez JFC, Open Wild Wheat Consortium, Wulff BBH, Ibba MI, Poland J (2021) High molecular weight glutenin gene diversity in Aegilops tauschii demonstrates unique origin of superior wheat quality. Commun Biol 4: 1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempewolf H, Baute G, Anderson J, Kilian B, Smith C, Guarino L (2017) Past and Future Use of Wild Relatives in Crop Breeding. Crop Sci 57: 1070–1082 [Google Scholar]
- Dubcovsky J, Dvorak J (2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316: 1862–1866 10.1126/science.1143986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J (1998) Genome analysis in the Triticum-Aegilops alliance. In Slinkard AE, ed, Proceedings of the 9th International Wheat Genetics Symposium. University Extension Press, University of Saskatoon, Saskatoon, Saskatchewan, Canada, pp 8–11 [Google Scholar]
- Dvorak J (2009) Triticeae genome structure and evolution. In Feuiller C, Muehlbauer GJ, eds, Genetics and Genomics of the Triticeae, Plant Genetics and Genomics: Crops and Models, Vol 7. Springer, Berlin, Germany, pp 685–711 [Google Scholar]
- Dvorak J, Akhunov ED (2005) Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics 171: 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J, Akhunov ED, Akhunov AR, Deal KR, Luo MC (2006) Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Mol Biol Evol 23: 1386–1396 [DOI] [PubMed] [Google Scholar]
- Dvorak J, Deal KR, Luo M-C, You FM, von Borstel K, Dehghani H (2012) The origin of spelt and free-threshing hexaploid wheat. J Hered 103: 426–441 [DOI] [PubMed] [Google Scholar]
- Eilam T, Anikster Y, Millet E, Manisterski J, Feldman M (2008) Nuclear DNA amount and genome downsizing in natural and synthetic allopolyploids of the genera Aegilops and Triticum. Genome 51: 616–627 [DOI] [PubMed] [Google Scholar]
- Erayman M, Sandhu D, Sidhu D, Dilbirligi M, Baenziger PS, Gill KS (2004) Demarcating the gene-rich regions of the wheat genome. Nucleic Acids Res 32: 3546–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2021) FAO cereal supply and demand brief. http://www.fao.org/worldfoodsituation/csdb/en/
- Faris JD, Haen KM, Gill BS (2000) Saturation mapping of a gene-rich recombination hot spot region in wheat. Genetics 154: 823–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris JD, Overlander ME, Kariyawasam GK, Carter A, Xu SS, Liu Z (2020) Identification of a major dominant gene for race-nonspecific tan spot resistance in wild emmer wheat. Theor Appl Genet 133: 829–841 [DOI] [PubMed] [Google Scholar]
- Faris JD, Zhang Z, Chao S (2014) Map-based analysis of the tenacious glume gene Tg-B1 of wild emmer and its role in wheat domestication. Gene 542: 198–208 [DOI] [PubMed] [Google Scholar]
- Fedoroff NV (2012) Presidential address. Transposable elements, epigenetics, and genome evolution. Science 338: 758–767 [DOI] [PubMed] [Google Scholar]
- Feldman M (1988) Cytogenetic and molecular approaches to alien gene transfer in wheat. In TE Miller, RMD Koebner, eds, Proceedings of the 7th International Wheat Genetics Symposium, Vol. 1. Cambridge, England, pp 23–32 [Google Scholar]
- Feldman M (2001) The origin of cultivated wheat. In Bonjean A, Angus W, eds, The World Wheat Book. Lavoisier Tech. & Doc., Paris [Google Scholar]
- Feldman M, Levy AA (2012) Genome evolution due to allopolyploidization in wheat. Genetics 192: 763–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Levy AA, Fahima T, Korol A (2012) Genome asymmetry in allopolyploid plants - wheat as a model. J Exp Bot 63: 5045–5059 [DOI] [PubMed] [Google Scholar]
- Feldman M, Liu B, Segal G, Abbo S, Levy AA, Vega JM (1997) Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147: 1381–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Sears ER (1981) The wild gene resources of wheat. Sci Am 244: 102–112 [Google Scholar]
- Feuillet C, Keller B (1999) High gene density is conserved at syntenic loci of small and large grass genomes. Proc Natl Acad Sci USA 96: 8265–8270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell RB, O’Dell M (1979) The genetic control of nucleolus formation in wheat. Chromosoma 71: 135–152 [Google Scholar]
- Frankin S, Kunta S, Abbo S, Sela H, Goldberg BZ, Bonfil DJ, Levy AA, Avivi-Ragolsky N, Nashef K, Roychowdhury R, et al (2020) The Israeli-Palestinian wheat landraces collection: restoration and characterization of lost genetic diversity. J Sci Food Agric 100: 4083–4092 [DOI] [PubMed] [Google Scholar]
- Fukui KN, Suzuki G, Lagudah ES, Rahman S, Appels R, Yamamoto M, Mukai Y (2001) Physical arrangement of retrotransposon-related repeats in centromeric regions of wheat. Plant Cell Physiol 42: 189–119 [DOI] [PubMed] [Google Scholar]
- Galili G, Levy AA, Feldman M (1986) Gene-dosage compensation of endosperm proteins in hexaploid wheat Triticum aestivum. Proc Natl Acad Sci USA 83: 6524–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C (2021) Genome engineering for crop improvement and future agriculture. Cell 184: 1621–1635 [DOI] [PubMed] [Google Scholar]
- Gardiner LJ, Wingen LU, Bailey P, Sánchez-Martín J, Horsnell R, Gao L, Brar GS, Widrig V, Raupp WJ, Singh N, et al. (2019) Analysis of the recombination landscape of hexaploid bread wheat reveals genes controlling recombination and gene conversion frequency. Genome Biol 20: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaurav K, Arora S, Silva P, Sanchez-Martin J, Horsnell R, Gao L, Brar GS, Widrig V, John Raupp W, Singh N, et al (2021) Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nat Biotechnol 10: 422–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles RJ, Brown TA (2006) GluDy allele variations in Aegilops tauschii and Triticum aestivum: implications for the origins of hexaploid wheats. Theor Appl Genet 112: 1563–1572 [DOI] [PubMed] [Google Scholar]
- Gill KS, Gill BS (1991) A DNA fragment mapped within the submicroscopic deletion of ph1, a chromosome pairing regulator gene in polyploid wheat. Genetics 129: 257–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S, Scornavacca C, Dainat J, Burgarella C, Viader V, Ardisson M, Sarah G, Santoni S, David J, Ranwez V (2019) Pervasive hybridizations in the history of wheat relatives. Sci Adv 5: eaav9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S, Colas I, Moore G (2006) Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752 [DOI] [PubMed] [Google Scholar]
- Guo X, Han F (2014) Asymmetric epigenetic modification and elimination of rDNA sequences by polyploidization in wheat. Plant Cell 26: 4311–4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Xin M, Wang Z, Yao Y, Hu Z, Song W, Yu K, Chen Y, Wang X, Guan P, et al (2020) Origin and adaptation to high altitude of Tibetan semi-wild wheat. Nat Commun 11: 5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas M, Schreiber M, Mascher M (2019) Domestication and crop evolution of wheat and barley: genes, genomics, and future directions. J Integr Plant Biol 61: 204–225 [DOI] [PubMed] [Google Scholar]
- Han FP, Fedak G, Guo WL, Liu B (2005) Rapid and repeatable elimination of a parental genome specific DNA repeat (pGcIR-1a) in newly synthesized wheat allopolyploids. Genetics 170: 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han FP, Fedak G, Ouellet T, Liu B (2003) Rapid genomic changes in interspecific and intergeneric hybrids and allopolyploids of Triticeae. Genome 46: 716–723 [DOI] [PubMed] [Google Scholar]
- Handa H, Kanamori H, Tanaka T, Murata K, Kobayashi F, Robinson SJ, Koh CS, Pozniak CJ, Sharpe AG, Paux E, et al (2018) Structural features of two major nucleolar organizer regions (NORs), Nor-B1 and Nor-B2, and chromosome-specific rRNA gene expression in wheat. Plant J 96: 1148–1159 [DOI] [PubMed] [Google Scholar]
- He F, Pasam R, Shi F, Kant S, Keeble-Gagnere G, Kay P, Forrest K, Fritz A, Hucl P, Wiebe K, et al. (2019) Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat Genet 51: 896–904 [DOI] [PubMed] [Google Scholar]
- Huang L, Raats D, Sela H, Klymiuk V, Lidzbarsky G, Feng L, Krugman T, Fahima T (2016) Evolution and adaptation of wild emmer wheat populations to biotic and abiotic stresses. Annu Rev Phytopathol 54: 279–301 [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (IWGSC) (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: eaar7191. [DOI] [PubMed] [Google Scholar]
- Jaaska V (1981) Aspartate aminotransferase and alcohol dehydrogenase isoenzyme: intraspecific differentiation in Aegilops tauschii the origin of the D genome polyploids in the wheat group. Plant Syst Evol 137: 259–273 [Google Scholar]
- Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X, et al (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496: 91–95 [DOI] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR (2004) Pack-MULE transposable elements mediate gene evolution in plants. Nature 431: 569–573 [DOI] [PubMed] [Google Scholar]
- Jiang J, Gill BS (1994) New 18S-26S ribosomal RNA gene loci: chromosomal landmarks for the evolution of polyploid wheats. Chromosoma 103: 179–185 [DOI] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA (2002) Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160: 1651–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33: 102–106 [DOI] [PubMed] [Google Scholar]
- Keidar D, Doron C, Kashkush K (2018) Genome-wide analysis of a recently active retrotransposon, Au SINE, in wheat: content, distribution within subgenomes and chromosomes, and gene associations. Plant Cell Rep 37: 193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keidar-Friedman D, Bariah I, Kashkush K (2018) Genome-wide analyses of miniature inverted-repeat transposable elements reveals new insights into the evolution of the Triticum-Aegilops group. PLoS One 13: e0204972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilwagen J, Lehnert H, Berner T, Badaeva E, Himmelbach A, Börner A, Kilian B (2022) Detecting major introgressions in wheat and their putative origins using coverage analysis. Sci Rep 12: 1908. 10.21203/rs.3.rs-910879/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenan-Eichler M, Leshkowitz D, Tal L, Noor E, Melamed-Bessudo C, Feldman M, Levy AA (2011) Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 188: 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber ER (1964) Wheat: reconstitution of the tetraploid component (AABB) of hexaploids. Science 143: 253–255 [DOI] [PubMed] [Google Scholar]
- Kerber ER, Rowland GG (1974) Origin of the free threshing character in hexaploid wheat. Can J Genet Cytol 16: 145–154 [Google Scholar]
- Kidwell MG, Lisch D (1997) Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci USA 94: 7704–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara H (1919) Uber cytologische Studien bei einigen Getreidiarten. I. Species-bastarde des weizens und weizenroggen-bastarde. Bot Mag Tokyo 32: 17–38 [Google Scholar]
- Kihara H (1924) Cytologische und Genetische Studien bei Wichtigcn Getreidearten mit Besonderer Rücksicht ouf das Verhalten der Chromosomen und die Sterilitat in den Bastarden. Memoirs of the College of Science, Kyoto imperial University, Kyoto, Japan, pp 1–200 [Google Scholar]
- Kihara H (1944) Discovery of the DD-analyser, one of the ancestors of Triticum vulgare. Agric Hortic 19: 13–14 [Google Scholar]
- Kilian B, Dempewolf H, Guarino L, Werner P, Coyne C, Warburton ML (2021) Crop Science special issue: adapting agriculture to climate change: a walk on the wild side. Crop Sci 61: 32–36. [Google Scholar]
- Kilian B, Knupffer H, Hammer K (2014) Elisabeth Schiemann (1881–1972): a pioneer of crop plant research, with special reference to cereal phylogeny. Genet Resour Crop Ev 61: 89–106 [Google Scholar]
- Kilian B, Mammen K, Millet E, Sharma R, Graner A, Salamini F, Hammer K, Özkan H (2011) Aegilops L. In Kole C, ed, Wild Crop Relatives: Genomic and Breeding Resources Cereals. Springer, Berlin, Germany, pp 1–76. [Google Scholar]
- Kislev ME (1980) Triticum parvicoccum sp. nov, the oldest naked wheat. Isr J Bot 28: 95–107 [Google Scholar]
- Kislev ME (1984) Emergence of wheat agriculture. Paleorient 10: 61–70 [Google Scholar]
- Kislev ME (2009) Reconstructing the ear morphology of ancient small-grain wheat (Triticum turgidum ssp. parvicoccum). In Fairbairn A, Weiss E, eds, From Foragers to Farmers: Papers in Honour of Gordon C Hillman. Oxbow Books, England, pp 235–238 [Google Scholar]
- Klymiuk V, Yaniv E, Huang L, Raats D, Fatiukha A, Chen S, Feng L, Frenkel Z, Krugman T, Lidzbarsky G, et al (2018) Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat Commun 9: 3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy AA, Feldman M (1989) Genetics of morphological traits in wild wheat, Triticum turgidum var. dicoccoides. Euphytica 40: 275–281 [Google Scholar]
- Li M, Dong L, Li B, Wang Z, Xie J, Qiu D, Li Y, Shi W, Yang L, Wu Q, et al (2020) A CNL protein in wild emmer wheat confers powdery mildew resistance. New Phytol 228: 1027–1037 [DOI] [PubMed] [Google Scholar]
- Li W, Gill B (2006) Multiple genetic pathways for seed shattering in the grasses. Funct Integr Genomics 6: 300–309 [DOI] [PubMed] [Google Scholar]
- Li T, Hu J, Sun Y, Li B, Zhang D, Li W, Liu J, Li D, Gao C, Zhang Y, et al (2021) Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture. Mol Plant 14: 1787–1798 [DOI] [PubMed] [Google Scholar]
- Li LF, Zhang ZB, Wang ZH, Li N, Sha Y, Wang XF, Ding N, Li Y, Zhao J, Wu Y, et al. (2022) Genome sequences of the five Sitopsis species of Aegilops and the origin of polyploid wheat B subgenome. Mol Plant 15: 488–503 [DOI] [PubMed] [Google Scholar]
- Liu B, Vega JM, Feldman M (1998) Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome 41: 535–542 [DOI] [PubMed] [Google Scholar]
- Liu Y, Yuan J, Jia G, Ye W, Chen ZJ, Song Q (2021) Histone H3K27 dimethylation landscapes contribute to genome stability and genetic recombination during wheat polyploidization. Plant J 105: 678–690 [DOI] [PubMed] [Google Scholar]
- Luo MC, Deal KR, Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, Blake N, Clegg MT, Coleman-Derr D, Conley EJ, et al (2009) Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc Natl Acad Sci USA 106: 15780–15785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Gu YQ, Puiu D, Wang H, Twardziok SO, Deal KR, Huo N, Zhu T, Wang L, Wang Y, et al. (2017) Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551: 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Gu YQ, You FM, Deal KR, Ma Y, Hu Y, Huo N, Wang Y, Wang J, Chen S, et al. (2013) A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc Natl Acad Sci USA 110: 7940–7945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Yang ZL, You FM, Kawahara T, Waines JG, Dvorak J (2007) The structure of wild and domesticated emmer wheat populations, gene flow between them, and the site of emmer domestication. Theor Appl Genet 114: 947–959 [DOI] [PubMed] [Google Scholar]
- Ma XF, Gustafson JP (2006) Timing and rate of genome variation in triticale following allopolyploidization. Genome 49: 950–958 [DOI] [PubMed] [Google Scholar]
- Mac Key J (1954) The taxonomy of hexaploid wheat. Svensk Bot Tidskr 48: 579–590 [Google Scholar]
- Mac Key J (2005) Wheat: its concept, evolution and taxonomy. In Royo C, Di Fonzo N, eds, Durum Wheat, Current Approaches, Future Strategies, Vol. 1. CRC Press, Boca Raton, FL, pp 3–61 [Google Scholar]
- Maccaferri M, Harris NS, Twardziok SO, Pasam RK, Gundlach H, Spannagl M, Ormanbekova D, Lux T, Prade VM, Milner SG, et al (2019) Durum wheat genome highlights past domestication signatures and future improvement targets. Nat Genet 51: 885–895 [DOI] [PubMed] [Google Scholar]
- Marcussen T, Sandve SR, Heier L, Spannagl M, Pfeifer M, Jakobsen KS, Wulff BBH, Steuernagel B, Klaus FX, Mayer KFX, et al (2014) Ancient hybridizations among the ancestral genomes of bread wheat. Science 345: 288–291 [DOI] [PubMed] [Google Scholar]
- Mayrose I, Zhan SH, Rothfels CJ, Magnuson-Ford K, Barker MS, Rieseberg LH, Otto SP (2011) Recently formed polyploid plants diversify at lower rates. Science 333: 1257. [DOI] [PubMed] [Google Scholar]
- McClintock B (1934) The relation of a particular chromosomal element to the development of the nucleoli in Zea mays. Zeit Zellforsch Mik Anat 21: 294–328 [Google Scholar]
- McFadden ES, Sears ER (1944) The artificial synthesis of Triticum spelta. Records Genet Soc Amer 13: 26–27 [Google Scholar]
- McFadden ES, Sears ER (1946) The origin of Triticum spelta and its free-threshing hexaploid relatives. J Hered 37: 81–89, 107–116 [DOI] [PubMed] [Google Scholar]
- Miller TE, Hutchinson J, Reader SM (1983) The identification of the nucleolus organizer chromosomes of diploid wheat. Theor Appl Genet 65: 145–147 [DOI] [PubMed] [Google Scholar]
- Millet E, Rong JK, Qualset CO, McGuire PE, Bernard M, Sourdille P, Feldman M (2013) Production of chromosome-arm substitution lines of wild emmer in common wheat. Euphytica 190: 1–17 [Google Scholar]
- Molinier J, Ries G, Bonhoeffer S, Hohn B (2004) Interchromatid and Interhomolog Recombination in Arabidopsis thaliana. Plant Cell 16: 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G, Devos KM, Wang Z, Gale MD (1995) Cereal genome evolution. Grasses, line up and form a circle. Curr Biol 5: 737–739 [DOI] [PubMed] [Google Scholar]
- Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A (2005) Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet 37: 997–1002 [DOI] [PubMed] [Google Scholar]
- Muramatsu M (1963) Dosage effect of the spelta gene q of hexaploid wheat. Genetics 48: 469–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M (1986) The vulgare super gene, Q: its universality in durum wheat and its phenotypic effects in tetraploid and hexaploid wheats. Can J Genet Cytol 28: 30–41 [Google Scholar]
- Murat F, Armero A, Pont C, Klopp C, Salse J (2017) Reconstructing the genome of the most recent common ancestor of flowering plants. Nat Genet 49: 490–496 [DOI] [PubMed] [Google Scholar]
- Murat F, Pont C, Salse J (2014) Paleogenomics in Triticeae for translational research. Curr Plant Biol 1: 34–39 [Google Scholar]
- Nakai Y (1979) Isozyme variation in Aegilops and Triticum. IV. The origin of the common wheats revealed from the study on esterase isozymes in synthesized wheats. Jpn J Genet 54: 175–189 [Google Scholar]
- Nalam VJ, Vales MI, Watson CJW, Kianian SF, Riera-Lizarazu O (2006) Map-based analysis of genes affecting the brittle rachis character in tetraploid wheat (Triticum turgidum L.). Theor Appl Gen 112: 373–381 [DOI] [PubMed] [Google Scholar]
- Nave M, Avni R, Çakır E, Portnoy V, Sela H, Pourkheirandish M, Ozkan H, Hale I, Takao Komatsuda T, et al (2019) Wheat domestication in light of haplotype analyses of the Brittle rachis 1 genes (BTR1-A and BTR1-B). Plant Sci 285 193–199 [DOI] [PubMed] [Google Scholar]
- Negri V (2003) Landraces in central Italy: where and why they are conserved and perspectives for their on-farm conservation. Genet Resourc Crop Evol 50: 871–885 [Google Scholar]
- Nesbitt M (2001) Wheat evolution: integrating archaeological and biological evidence. In Caligari P.D.S., Brandham PE eds, Wheat Taxonomy: The Legacy of John Percival, Linnean Society, London, pp 37–59 [Google Scholar]
- Furuta Y, Nishikawa K, Tanino T (1974) Stability in DNA content of AB genome component of common wheat during the past seven thousand years. Jpn J Genet 49: 179–187 [Google Scholar]
- Ohno S (1970) Evolution by Gene Duplication. Springer, New York, NY [Google Scholar]
- Okamoto M (1957) Asynaptic effect of chromosome V. Wheat Inf Serv 5: 6 [Google Scholar]
- Orth RA, Bushuk W (1973) Studies of glutenin: III. Identification of subunits coded by the D-genome and their relation to breadmaking quality. Cereal Chem 50: 680–687 [Google Scholar]
- Ozkan H, Brandolini A, Pozzi C, Effgen S, Wunder J, Salamini F (2005) A reconsideration of the domestication geography of tetraploid wheats. Theor Appl Genet 110: 1052–1060 [DOI] [PubMed] [Google Scholar]
- Ozkan H, Feldman M (2001) Genotypic variation in tetraploid wheat affecting homoeologous pairing in hybrids with Aegilops peregrina. Genome 44: 1000–1006 [PubMed] [Google Scholar]
- Ozkan H, Levy AA, Feldman M (2001) Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13: 1735–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Ronin Y, Fahima T, Röder MS, Nevo E, Korol A (2003) Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc Natl Acad Sci USA 100: 2489–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival J (1921) The Wheat Plant: A Monograph. Duckworth, London, pp 1–46 [Google Scholar]
- Pikaard CS (2000) The epigenetics of nucleolar dominance. Trend Genet 16: 495–500 [DOI] [PubMed] [Google Scholar]
- Piperno DR, Weiss E, Holst I, Nadel D (2004) Processing of wild cereal grains in the upper Palaeolithic revealed by starch grain analysis. Nature 430: 670–673 [DOI] [PubMed] [Google Scholar]
- Pont C, Leroy T, Seidel M, Tondelli A, Duchemin W, Armisen D, Lang D, Bustos-Korts D, Goue N, Balfourier F, et al (2019) Tracing the ancestry of modern bread wheats. Nat Genet 51 905–911 [DOI] [PubMed] [Google Scholar]
- Pourkheirandish M, Hensel G, Kilian B, Senthil N, Chen G, Sameri M, Azhaguvel P, Sakuma S, Dhanagond S, Sharma R, et al (2015) Evolution of the grain dispersal system in barley. Cell 162: 527–539 [DOI] [PubMed] [Google Scholar]
- Presting GG, Malysheva L, Fuchs J, Schubert IZ (1998) A TY3/GYPSY retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J 16: 721–728 [DOI] [PubMed] [Google Scholar]
- Ramírez-González RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, Davey M, Jacobs J, van Ex F, Pasha A, et al. (2018) The transcriptional landscape of polyploid wheat. Science 361: eaar6089. [DOI] [PubMed] [Google Scholar]
- Rey MD, Martín AC, Higgins J, Swarbreck D, Uauy C, Shaw P, Moore G (2017) Exploiting the ZIP4 homologue within the wheat Ph1 locus has identified two lines exhibiting homoeologous crossover in wheat-wild relative hybrids. Mol Breed 37: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey MD, Martín AC, Smedley M, Hayta S, Harwood W, Shaw P, Moore G (2018) Magnesium increases homoeologous crossover frequency during meiosis in ZIP4 (Ph1 Gene) mutant wheat-wild relative hybrids. Front Plant Sci 9: 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice A, Šmarda P, Novosolov M, Drori M, Glick L, Sabath N, Meiri S, Belmaker J, Mayrose I (2019) The global biogeography of polyploid plants. Nat Ecol Evol 3: 265–273 [DOI] [PubMed] [Google Scholar]
- Riehl S, Zeidi M, Conard NJ (2013) Emergence of agriculture in the foothills of the Zagros Mountains of Iran. Science 341: 65–67 [DOI] [PubMed] [Google Scholar]
- Riley R (1960) The diploidization of polyploid wheat. Heredity 15: 407–429 [Google Scholar]
- Riley R, Chapman V (1958) Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182: 713–715 [Google Scholar]
- Saintenac C, Falque M, Martin OC, Paux E, Feuillet C, Sourdille P (2009) Detailed recombination studies along chromosome 3B provide new insights on crossover distribution in wheat (Triticum aestivum L.). Genetics 181: 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamura T (1918) Kurze Mitteilung uber die chromosomenzahalen und die Verwandtschaftsverhaltnisse der Triticum Arten. Bot Mag Tokyo 32: 151–154 [Google Scholar]
- Salina EA, Numerova OM, Ozkan H, Feldman M (2004) Alterations in subtelomeric tandem repeats during early stages of allopolyploidy in wheat. Genome 47: 860–867 [DOI] [PubMed] [Google Scholar]
- Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, Calcagno T, Cooke R, Delseny M, et al. (2008) Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sax K (1918) The behavior of chromosomes in fertilization. Genetics 3: 309–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sax K (1921) Chromosome relationships in wheat. Science 54: 413–415 [DOI] [PubMed] [Google Scholar]
- Sax K (1922) Sterility in wheat hybrids. II. Chromosome behavior in partially sterile hybrids. Genetics 7: 513–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze-Motel J (2019) Triticum parvicoccum Kislev in Transcaucasia. Genet Resour Crop Ev 66: 1363–1366 [Google Scholar]
- Sears ER (1952) Homoeologous chromosomes in Triticum aestivum. Genetics 37: 624 [Google Scholar]
- Sears ER (1954) The aneuploids of common wheat. Missouri Agric Exp Stn Res Bull 572: 1–58 [Google Scholar]
- Sears ER (1959) Wheat cytogenetics. Ann Rev Genet 3: 451–468 [Google Scholar]
- Sears ER (1976) Genetic control of chromosome pairing in wheat. Ann Rev Genet 10: 31–51 [DOI] [PubMed] [Google Scholar]
- Sears ER (1977) An induced mutant with homoeologous chromosome pairing. Can J Genet Cytol 19: 585–593 [Google Scholar]
- Sears ER (1982) A wheat mutation conditioning an intermediate level of homoeologous chromosome pairing. Can J Genet Cytol 24: 715–719 [Google Scholar]
- Sears ER (1984) Mutations in wheat that raise the level of meiotic chromosome pairing. In Gustafson JP (ed) Gene Manipulation in Plant Improvement. Stadler Genetics Symposia Series. Springer, Boston, MA, pp 295–300 [Google Scholar]
- Sears ER, Okamoto M (1958) Intergenomic chromosome relationships in hexaploid wheat. Proc 10th Intern Congr Genet, Montreal, Quebec 2: 258–259 [Google Scholar]
- Senerchia N, Felber F, Parisod C (2015) Genome reorganization in F1 hybrids uncovers the role of retrotransposons in reproductive isolation. Proc Biol Sci Royal Soc 282: 20142874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra H, Svacina R, Baumann U, Whitford R, Sutton T, Bartos J, Sourdille P (2021) Ph2 encodes the mismatch repair protein MSH7-3D that inhibits wheat homoeologous recombination. Nat Commun 12: 803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA (2001) Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev G, Levy AA (1997) The maize transposable element Ac induces recombination between the donor site and an homologous ectopic sequence. Genetics 146: 1143–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Q, Li C (1983) Basang C semi-wild wheat from Xizang (Tibet). In S Sakamoto, ed, 6th International Wheat Genetics Symposium, Kyoto, Japan, pp 111–114
- Sharma JS, Running KLD, Xu SS, Zhang Q, Peters Haugrud AR, Sharma S, McClean PE, Faris JD (2019) Genetic analysis of threshability and other spike traits in the evolution of cultivated emmer to fully domesticated durum wheat. Mol Genet Genomics 294: 757–771 [DOI] [PubMed] [Google Scholar]
- Sharma S, Schulthess A, Bassi F, Badaeva E, Neumann K, Graner A, Özkan H, Werner P, Knüpffer H, Kilian B (2021) Introducing beneficial alleles from plant genetic resources into the wheat germplasm. Biology 10: 982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR (2009) Wheat. J Exp Bot 60: 1537–1553 [DOI] [PubMed] [Google Scholar]
- Shewry PR, Hey SJ (2015) The contribution of wheat to human diet and health. Food Energy Secur 4: 178–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti MC, Bellomo MP, Laghetti G, Perrino P, Simeone R, Blanco A (1999) Quantitative trait loci influencing free-threshing habit in tetraploid wheats. Genet Resource Crop Evol 46: 267–271 [Google Scholar]
- Simons KJ, Fellers JP, Trick HN, Zhang Z, Tai YS, Gill BS, Faris JD (2006) Molecular characterization of the major wheat domestication gene Q. Genetics 172: 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Smith O, Momber G, Bates R, Garwood P, Fitch S, Pallen M, Gaffney V, Allaby RG (2015) Archaeology. Sedimentary DNA from a submerged site reveals wheat in the British Isles 8000 years ago. Science 347: 998–1001 [DOI] [PubMed] [Google Scholar]
- Snir A, Nadel D, Groman-Yaroslavski I, Melamed Y, Sternberg M, Bar-Yosef O, Weiss E (2015) The origin of cultivation and proto-weeds, long before neolithic farming. PLoS One 10: e0131422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS (1999) Polyploidy: recurrent formation and genome evolution. Trends Ecol Evol 14: 348–352 [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE (2009) The role of hybridization in plant speciation. Annu Rev Plant Biol 60: 561–588 [DOI] [PubMed] [Google Scholar]
- Stebbins GL (1950) Variation and Evolution in Plants. Columbia University Press, New York, NY [Google Scholar]
- Stebbins GL (1971) Chromosomal Evolution in Higher Plants. Edward Arnold Ltd, London [Google Scholar]
- Su H, Liu Y, Liu C, Shi Q, Huang Y, Han F (2019) Centromere satellite repeats have undergone rapid changes in polyploid wheat subgenomes. Plant Cell 31: 2035–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno K, Willcox G (2006) How fast was wild wheat domesticated? Science 311: 1886. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N (2009) A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324: 659–662 [DOI] [PubMed] [Google Scholar]
- Tschermak E (1914) Die Verwertung der Bastardierung für phylogenetische Fragen in der Getreidegruppe. Zeitschr Pflanzenzucht 2: 291–312 [Google Scholar]
- Tsunewaki K (2009) Plasmon analysis in the Triticum-Aegilops complex. Breed Sci 59: 455–470 [Google Scholar]
- Tzarfati RV., Barak V, Krugman T, Fahima T, Abbo A, Saranga Y, Korol AB (2014) Novel quantitative trait loci underlying major domestication traits in tetraploid wheat. Mol Breed 34: 1613–1628 [Google Scholar]
- Valenzuela NT, Perera E, Naranjo T (2013) Identifying crossover-rich regions and their effect on meiotic homologous interactions by partitioning chromosome arms of wheat and rye. Chromosome Res 21: 433–445 [DOI] [PubMed] [Google Scholar]
- Venske E, Dos Santos RS, Busanello C, Gustafson P, Costa de Oliveira A (2019) Bread wheat: a role model for plant domestication and breeding. Hereditas 156: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkowiak S, Gao L, Monat C, Haberer G, Kassa MT, Brinton J, Ramirez-Gonzalez RH, Kolodziej MC, Delorean E, Thambugala D, et al. (2020) Multiple wheat genomes reveal global variation in modern breeding. Nature 588: 277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang J, Jin D, Guo H, Lee TH, Liu T, Paterson AH (2015) Genome alignment spanning major Poaceae lineages reveals heterogeneous evolutionary rates and alters inferred dates for key evolutionary events. Mol Plant 8: 885–898 [DOI] [PubMed] [Google Scholar]
- Watanabe N, Fujii Y, Kato N, Ban T, Martinek P (2006) Microsatellite mapping of the genes for brittle rachis on homoeologous group 3 chromosomes in tetraploid and hexaploid wheats. J Appl Genet 47: 93–98 [DOI] [PubMed] [Google Scholar]
- Watson A, Ghosh S, Williams MJ, Cuddy WS, Simmonds J, Rey MD, Asyraf Md Hatta M, Hinchliffe A, Steed A, Reynolds D, et al (2018) Speed breeding is a powerful tool to accelerate crop research and breeding. Nat Plants 4: 23–29 [DOI] [PubMed] [Google Scholar]
- Wicker T, Gundlach H, Spannagl M, Uauy C, Borrill P, Ramirez-Gonzalez RH, De Oliveira R, Mayer KFX, Paux E, Frederic C (2018) Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol 19: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Zimmermann W, Perovic D, Paterson AH, Ganal M, Graner A, Stein N (2005) A detailed look at 7 million years of genome evolution in a 439 kb contiguous sequence at the barley Hv-eIF4E locus: recombination, rearrangements and repeats. Plant J 41: 184–194 [DOI] [PubMed] [Google Scholar]
- Willcox G (2012). Pre-domestic cultivation during the late pleistocene and early Holocene in the Northern Levant. In Gepts P, Famula T, Bettinger R, Brush S, Damania A, McGuire P, Qualset CO, eds, Biodiversity in Agriculture: Domestication, Evolution, and Sustainability. Cambridge University Press, Cambridge, pp 92–109 [Google Scholar]
- Wulff BBH, Moscou MJ (2014) Strategies for transferring resistance into wheat: from wide crosses to GM cassettes. Front Plant Sci 5: 692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Reynolds MP, Crossa J, Schulthess U, Sonder K, Montes C, Addimando N, Singh RP, Ammar K, Gerard B, et al (2021) Increased ranking change in wheat breeding under climate change. Nat Plants 7: 1207–1212 [DOI] [PubMed] [Google Scholar]
- Yaakov B, Kashkush K (2011) Methylation, transcription, and rearrangements of transposable elements in synthetic allopolyploids. Int J Plant Genomics 2011: 569826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaakov B, Kashkush K (2012) Mobilization of Stowaway-like MITEs in newly formed allohexaploid wheat species. Plant Mol Biol 80: 419–427 [DOI] [PubMed] [Google Scholar]
- Yildirim A, Jones SS, Murray TD, Cox TS, Line RP (1995) Resistance to stripe rust and eyespot diseases of wheat in Triticum tauschii. Plant Disease 79: 1230–1236 [Google Scholar]
- Zeder MA (2011) The origins of agriculture in the Near East. Curr Anthropol 52: S221–S235 [Google Scholar]
- Zeibig F, Kilian B, Frei M (2021) The grain quality of wheat wild relatives in the evolutionary context. Theor Appl Genet 10.1007/s00122-021-04013-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Belcram H, Gornicki P, Charles M, Just J, Huneau C, Magdelenat G, Couloux A, Samain S, Gill BS, et al (2011) Duplication and partitioning in evolution and function of homoeologous Q loci governing domestication characters in polyploid wheat. Proc Natl Acad Sci USA 108: 18737–18742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gou X, Kun H, Bian Y, Ma XL, Li M, Gong L, Feldman M, Liu B, Levy AA (2020a) Homoeologous exchanges occur through intragenic recombination generating novel transcripts and proteins in wheat and other polyploids. Proc Natl Acad Sci USA 117: 14561–14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li A, Song G, Geng S, Gill BS, Faris JD, Mao L (2020b) Comprehensive analysis of Q gene near-isogenic lines reveals key molecular pathways for wheat domestication and improvement. Plant J 102: 299–310 [DOI] [PubMed] [Google Scholar]
- Zhao G, Zou C, Li K, Wang K, Li T, Gao L, Zhang X, Wang H, Yang Z, Liu X, et al (2017) The Aegilops tauschii genome reveals multiple impacts of transposons. Nat Plants 3: 946–955. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Bai S, Li H, Sun G, Zhang D, Ma F, Zhao X, Nie F, Li J, Chen L, et al (2021) Introgressing the Aegilops tauschii genome into wheat as a basis for cereal improvement. Nat Plants 7: 774–786 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhao X, Li Y, Xu J, Bi A, Kang L, Xu D, Chen H, Wang Y, Wang YG, et al. (2020) Triticum population sequencing provides insights into wheat adaptation. Nat Genet 52: 1412–1422 [DOI] [PubMed] [Google Scholar]
- Zohary D, Hopf M (2000) Domestication of Plants in the Old World, Ed 3. Oxford Science Publications, Oxford [Google Scholar]
- Zohary D, Hopf M, Weiss E (2012) Domestication of Plants in the Old World-The origin and spread of domesticated plants in south-west Asia, Europe and the Mediterranean Basin, Ed 4. Oxford University Press, Oxford, pp 1–243 [Google Scholar]