Abstract
Testing hypothesis about the biogeography of genes using large data resources such as Tara Oceans marine metagenomes and metatranscriptomes requires significant hardware resources and programming skills. The new release of the ‘Ocean Gene Atlas’ (OGA2) is a freely available intuitive online service to mine large and complex marine environmental genomic databases. OGA2 datasets available have been extended and now include, from the Tara Oceans portfolio: (i) eukaryotic Metagenome-Assembled-Genomes (MAGs) and Single-cell Assembled Genomes (SAGs) (10.2E+6 coding genes), (ii) version 2 of Ocean Microbial Reference Gene Catalogue (46.8E+6 non-redundant genes), (iii) 924 MetaGenomic Transcriptomes (7E+6 unigenes), (iv) 530 MAGs from an Arctic MAG catalogue (1E+6 genes) and (v) 1888 Bacterial and Archaeal Genomes (4.5E+6 genes), and an additional dataset from the Malaspina 2010 global circumnavigation: (vi) 317 Malaspina Deep Metagenome Assembled Genomes (0.9E+6 genes). Novel analyses enabled by OGA2 include phylogenetic tree inference to visualize user queries within their context of sequence homologues from both the marine environmental dataset and the RefSeq database. An Application Programming Interface (API) now allows users to query OGA2 using command-line tools, hence providing local workflow integration. Finally, gene abundance can be interactively filtered directly on map displays using any of the available environmental variables. Ocean Gene Atlas v2.0 is freely-available at: https://tara-oceans.mio.osupytheas.fr/ocean-gene-atlas/.
Graphical Abstract
Graphical Abstract.
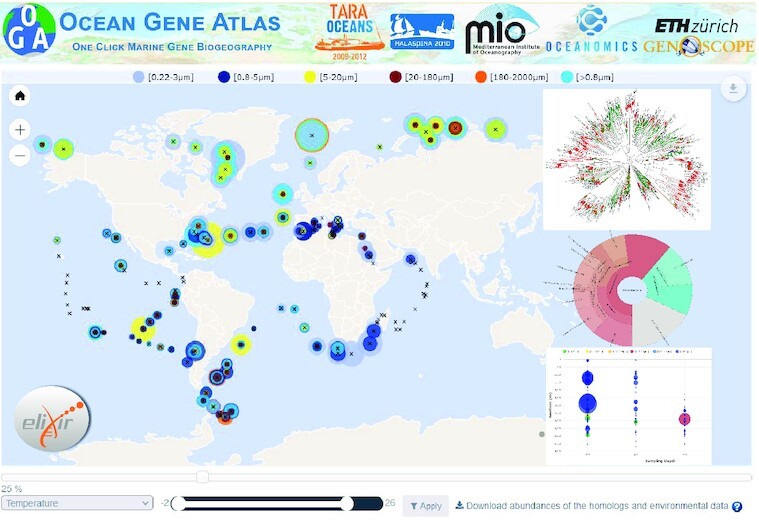
The Ocean Gene Atlas v2.0: online exploration of the biogeography and phylogeny of plankton genes.
INTRODUCTION
Marine plankton ecosystems represent main actors in global climate regulation (1). Marine microorganisms export photosynthetically fixed carbon to the deep ocean and contribute about half of global primary production (2). Their role in biogeochemical cycles such as the biological carbon pump in the ocean is crucial in the context of climate change (3). Intense large scale oceanographic sampling campaigns are providing precious observational data from the planet's largest but still underexplored continuous biome. Such samples subjected to high throughput DNA and RNA sequencing have in turn provided increasingly comprehensive and insightful environmental genomics resources, mostly from uncultivated organisms. In the wake of the Global Ocean Sampling (GOS) expedition, which produced a 6.1 million gene catalogue mostly from marine prokaryotes (4), the Tara Oceans pan-oceanic expedition applied a holistic sampling of plankton from viruses to fish larvae coupled with comprehensive in situ biogeochemical measurements, albeit with sampling bias towards the epipelagic sunlit layer (5,6). Marine gene catalogues released from the Tara Oceans sequencing effort include datasets specific to prokaryotes (7) as well as eukaryotes (8). The Malaspina 2010 global circumnavigation (9) used a similar sampling approach applied to the tropical and subtropical deep oceans from surface down to 4000 m depth.
Resulting environmental genomics resources have been made available via a variety of modes, including the MAR databases (10), MGnify (11), Planet Microbe (12) and the Ocean Microbiomics Database (13). The updated Ocean Gene Atlas (14) presented here is unique in presenting 8 trillion of marine environmental read sequences in their environmental context, hence allowing marine biologists to explore the biogeography and phylogeny of plankton genes among a total of 228 millions. Indeed, the Ocean Gene Atlas v2.0 (OGA2) provides an integrated interactive interface to mine all major Tara Oceans and Malaspina gene datasets characterized as of early 2022 without any requirement for programming or dedicated hardware. Moreover, no account or identification is necessary to run queries, and results visualization occurs on-the-fly.
OGA v2.0: NEW FEATURES AND UPDATES
The Ocean Gene Atlas v2.0 (OGA2) web service provides a user-friendly interface to identify and geolocate marine environmental homologous sequences using a nucleic acid or protein sequence query.
The web service update consists on the one hand in the integration of six datasets from Tara Oceans and Malaspina consortium sequencing efforts, and on the other hand new tools to quantitatively explore contextualized genes of interest in the global ocean ecosystem. An updated user manual is provided online from the OGA2 service web pages.
New resources
The first version of the Ocean Gene Atlas deployed its analyses based on two datasets: (i) the Ocean Microbial Reference Gene Catalogue (OM-RGC) comprising 40 million non-redundant mostly prokaryotic gene sequences associated with both Tara Oceans and Global Ocean Sampling (GOS) gene abundances (7) and (ii) the Marine Atlas of Tara Ocean Unigenes (MATOU) composed of >116 million eukaryotic unigenes (8).
The OGA2 includes the following new Tara Oceans and Malaspina datasets:
713 non-redundant and manually curated eukaryotic MAGs and SAGs containing 10 million genes (15). This EUK_SMAGs dataset was built from 280 billion Tara Oceans metagenomic reads from polar, temperate, and tropical sunlit oceans and covers eukaryotic environmental genomes ranging from 10 Mb to 1.3 Gb.
1888 non-redundant and manually curated bacterial and archaeal MAGs containing 4.5 million genes (16). This BAC_ARC_MAGs dataset was built using the same 280 billion Tara Oceans metagenomic reads.
924 non-redundant MetaGenomic Transcriptomes (MGTs) containing 7 million unigenes (17). This MGT database is mostly eukaryotic and was built based on the MATOU catalogue (Tara Oceans).
530 bacterial and archaeal MAGs containing 1 million genes (18). This Arctic_MAGs dataset was built using the Tara Oceans Polar Circle expedition.
317 bacterial and archaeal MAGs containing 0.9 million genes (19). This MDeep-MAGs dataset was built using the Malaspina metagenomes.
version 2 of Ocean Microbial Reference Gene Catalogue (OM-RGCv2) with additional data from the Arctic Ocean comprising a total of 47 million non-redundant gene sequences from 370 marine metagenomes and 187 metatranscriptomes (20).
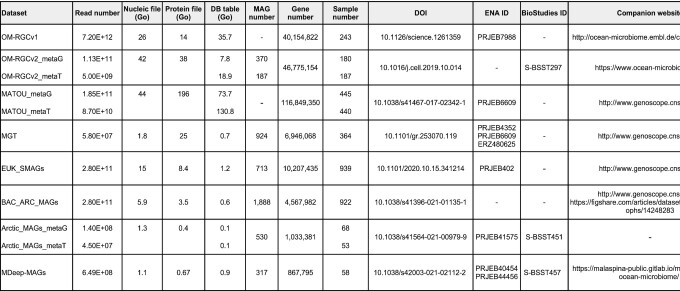
Together with the gene sequence catalogues, two additional complementary data objects were also included in OGA2: gene abundances for each sample, and sample biogeochemical environmental context (see Data availability section and Table 1).
Table 1.
Dataset information
|
New implementations
Application Programming Interface (API)
The Application Programming Interface (API) offers researchers the option of command line to facilitate access to OGA2 and ensure the datasets are explored to their fullest. The API uses standard protocols and readily available programming languages, allowing for instance full control of OGA2 through a simple bash script. A tutorial with examples of codes is available at the following address: https://tara-oceans.mio.osupytheas.fr/ocean-gene-atlas/build/script/API_tutorial.pdf. Three types of API commands are possible as described in Figure 1. The first type is to ‘Submit a request’ using a JSON file with search parameters, such as a FASTA sequence or Pfam identifier, as well as the dataset to be mined. The Laravel application server (detailed in the ‘Data integration and framework’ paragraph) from OGA2 sends a response in JSON format with the request identifier and an estimation of the computation time. The second type of command is to ‘Check results’ accompanied with the request identifier provided after the initial query submission above. Once the computation is completed, the server will return the URL of the results web page. The third API command is the ‘Fetch results’ request using the request identifier and the results file of interest. Three files can be provided containing the alignment results, the homologues sequences or the homologues abundances together with the associated contextual environmental data. A throttling limits users to 200 jobs per 24 h, and we advise users to submit no more than one request every 30 seconds. In order to provide the best possible interactive experience, queries launched from the web interface have priority over API requests.
Figure 1.
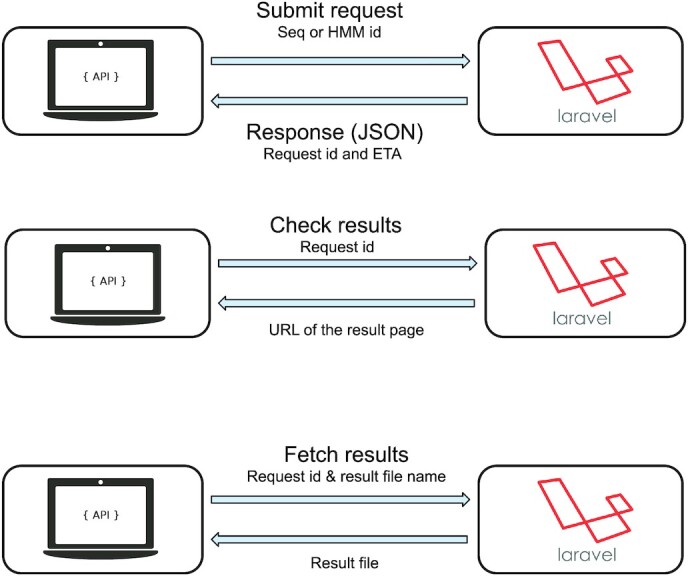
The three types of API request. The first type of API request, ‘Submit a request’, uses a JSON file with parameters (such as FASTA sequence, Hidden Markov Model profile or Pfam identifier). Then the Laravel application server from OGA2 sends a JSON formatted response with the request identifier and an estimation of the time of arrival (ETA) or computation time. The second type of command ‘Check results’ can be ran accompanied with the request identifier provided after the initial query submission. The OGA2 server then returns the URL of the results web page when the computation is over. The last command, ‘Fetch results’, uses the request identifier and the resulting file name.
Phylogenetic analysis
A new feature of OGA2 is the phylogenetic analysis of the user query sequence together with its closest (applying the user defined E-value threshold, or the maximum number of aligned sequences) BLAST hits in both marine metagenomic homologues and reference databases. To do so, the ‘Phylogenetic tree’ option should be selected on the website submission form. An additional panel section will then display a phylogenetic tree in the results page.
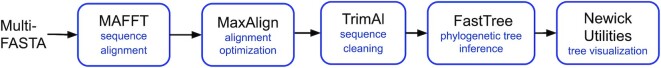
For this purpose, the sequence query is used to search homologues in the RefSeq database (21). If the number of RefSeq homologues is greater than the number of metagenomic homologues, the RefSeq homologous sequences are progressively clustered with CD-HIT (22) until the sequence number is equal or less than that of metagenomic homologues sequences (to avoid some cases we observed where RefSeq homologues could clutter the resulting tree, such as queries close to over-represented enterobacteria). This clustering step is done iteratively by gradually decreasing the threshold of clustering from 100% to a minimum of 60%. The sequences in the resulting combined dataset, consisting of the user query sequence, the metagenomic homologues, and the reference RefSeq homologues, are then aligned with MAFFT (23). This alignment is cleaned with MaxAlign (24) and trimAl (25) before submission to FastTree (26) for phylogenetic tree inference (Figure 2). To visualize the resulting tree, the Newick Utilities (27) tools suite is used.
Figure 2.
Phylogenetic pipeline. All sequences identified as homologous to the user query sequence are first aligned with MAFFT (23), the sequence alignment is treated with MaxAlign (24) to maximize the number of amino acid symbols in the alignment area and cleaned with an automated alignment trimming tool named trimAl (25). FastTree (26), with the default settings, allows to infer approximately-maximum-likelihood phylogenetic tree from the resulting alignment with the JTT (Jones-Taylor-Thornton 1992) model of amino acid evolution, and computes local support values with the Shimodaira-Hasegawa test. The tree visualization is done with Newick Utilities tools suite (27).
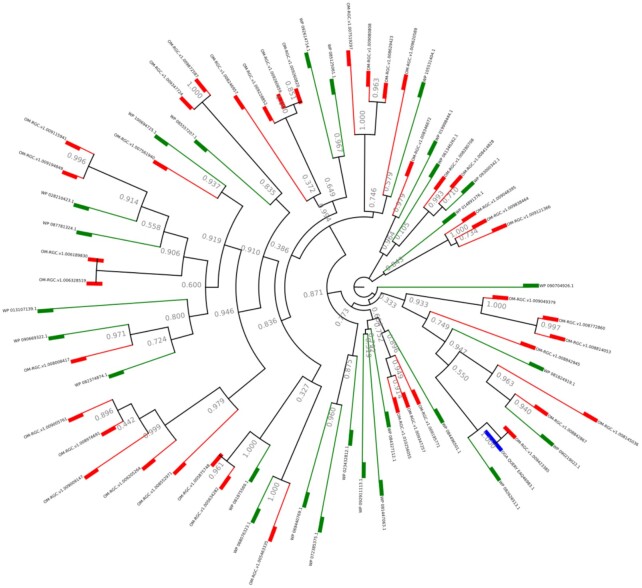
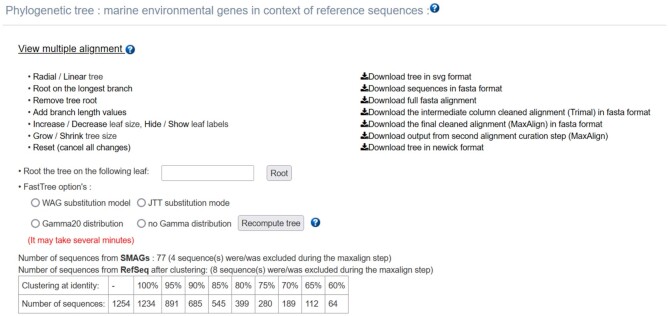
Once the phylogeny workflow has completed successfully, the resulting phylogenetic tree is rendered in the results interface in a new panel with several phylogenetic tree formatting options. The user query sequence is represented in blue, the metagenomic homologues appear in red, and the RefSeq reference homologues are labelled in green (Figure 3). One can download the tree in SVG format as well as all intermediate files used in the workflow (multi-FASTA homologues, multiple alignment before and after trimming, Newick formatted tree) (Figure 4). It is also possible to interact with the tree (change from radial to linear), change the substitution mode or tree inference (gamma law), but also to root the tree (with the longest branch or branch specified by the user) and zoom in or out. The colored multiple sequence alignment with selected positions (as output by trimAl) can also be displayed.
Figure 3.
An example of phylogenetic tree. In the phylogenetic tree, the user query sequence is colored in blue, the metagenomic homologues in red, and the RefSeq reference homologues in green.
Figure 4.
Phylogenetic analysis options. Several options allow user to download the tree in SVG format and intermediate files used in the phylogeny workflow (multi-FASTA homologues, multiple alignment before and after trimming and newick formatted tree). The link ‘view multiple alignment’ shows the HTML file generated by trimAl. The tree can be changed from radial to linear, the substitution mode or tree inference (gamma law) can be modified, and it is possible to root the tree (with the longest branch or branch specified by the user) and zoom in or out. The colored multiple sequence alignment can also be displayed.
Selection of homologous sequences using an environmental parameter range
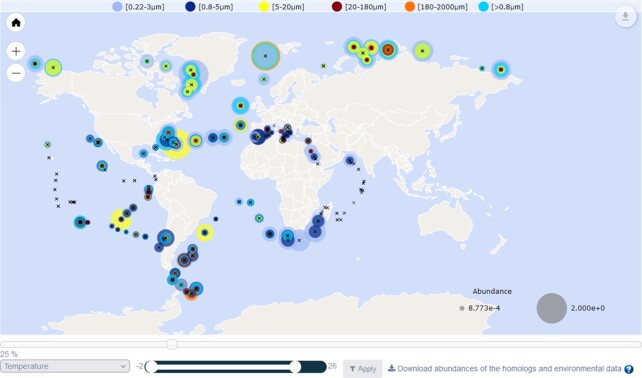
Below the map showing geographic distribution of homologues abundances (see Figure 5), users can select a sequence subset defined by an environmental parameter range: users first choose an environmental variable from the drop-down list (e.g. temperature), and then define the desired range using the associated slider (e.g. 4–12°C). When the ‘Apply’ button is clicked, only samples corresponding to the selected range are displayed on the map. It is then possible to download the abundance files and environmental variables corresponding to the subset selection.
Figure 5.
Interactive world map. The geographic distribution of homologues abundances are represented on a map and an environmental parameter can be selected from the drop-down list (e.g. temperature). Using the associated slider (e.g. 4–12°C) and the ‘Apply’ button, only the sequence subset corresponding to the selected range are displayed on the map. The abundance files and environmental variables corresponding to the subset selection can be downloaded.
Abundance normalization
The abundance of each catalogue gene (for OM-RGCv1 and MATOU) in specific biosamples was estimated by evaluating the coverage of raw sequencing reads mapped to the gene's nucleotide sequence as described earlier (14). Briefly, depending on the database queried (Table 2), abundance estimates may be expressed in one of three available normalization schemes: (i) the gene's read coverage is divided by the sum of the total gene coverages for the sample (‘percent of total coverage’), (ii) the gene's read coverage is divided by the total number of reads for the sample (‘percent of total reads’), (iii) the gene's read coverage is divided by the median of the coverages of a set of 10 universal single copy marker genes (‘average copies per cell’) that were previously benchmarked for their suitability for prokaryotes metagenomics data analysis (28).
Table 2.
Dataset abundance normalization methods
| Datasets | Percent of total coverage RPKM or RPKG | Percent of total reads | Average copies per cell |
|---|---|---|---|
| OM-RGCv1 | x | x | x |
| OM-RGVv2 | x | ||
| MATOU | x | x | |
| MGT | x | ||
| EUK_SMAGs | x | ||
| BAC_ARC_MAGs | x | ||
| Arctic_MAGs | x | ||
| MDeep-MAGs | x |
In order to estimate the abundance and expression of each MGT unigene in each sample, cleaned reads (from metagenomes and metatranscriptomes) were mapped against the reference catalog as described in (17). Reads covering at least 80% of read length with at least 95% of identity were retained for further analysis. Unigene expression values and genomic occurrences were computed in RPKM (reads per kilo base covered per million of mapped reads).
Gene abundance from MAG catalogues was computed using reads per genomic kilobase and metagenomic gigabase (RPKG). For Euk_SMAGs, BAC_ARC_MAGs and Arctic MAGs gene abundance, we attributed to gene its MAGs abundance computed as described in (15,16,18).
For the MDeep-MAGs dataset, the abundance of each MAG was expressed by the number of mapped reads per genomic kilobase and sample gigabase as described in (19). And each gene abundance is expressed as mean read coverage (best read map, with at least 95% identity over at least 90% of the read length).
Data integration and framework
All data objects (sample gene abundance tables, environmental context and gene catalogues) were downloaded from ENA, Pangaea or companion websites (Table 1) and preprocessed using bash, perl or R (version 4.03) scripts to generate files for database integration (Figure 6). Figure 7 represents the MariaDB version 10.3.27 managed relational database schema (note that MAG datasets use a dedicated table). These datasets are queried by Laravel 5.4 PHP application server that uses a classical Model-View-Controller pattern architecture to create web interfaces. Hosted on dedicated Linux hardware, the application server communicates with the user through an Apache2 HTTP server using HTML5, CSS3, Javascript and AJAX to retrieve user requests and display results. As per FAIR principles (29), a database dump is done every week to save the data to a remote backup server and the scripts are hosted under bitbucket and gitlab (see Data accessibility section) in order to facilitate updates and collaborative work.
Figure 6.
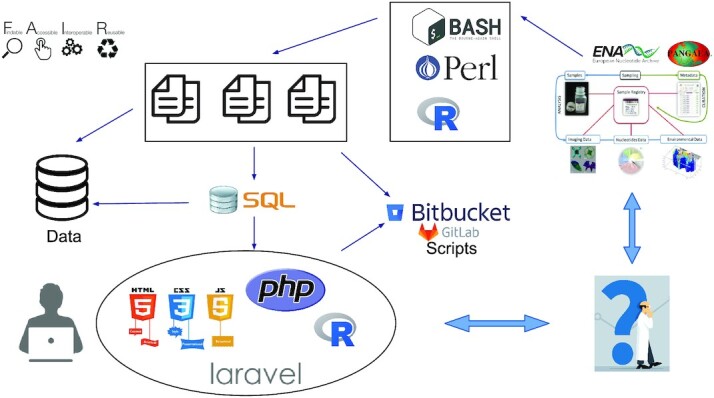
OGA2 processing. To answer to the FAIR principles (Findability, Accessibility, Interoperability, and Reusability), the processing of OGA2 server is the following: the metadata (sample gene abundance tables, environmental context and gene catalogues) are collected from a data warehouse such as ENA, PANGAEA or companion websites. All data objects are preprocessed using bash, perl or R scripts to generate the files for database integration and BLAST databases are generated from sequence files. Laravel requests allow to query the different datasets and the application server communicates with the user through an Apache 2 HTTP server to display results. Every week an OGA2 database dump is done to save the data to a remote backup server. In order to facilitate updates and collaborative work, the scripts are hosted under bitbucket and gitlab.
Figure 7.
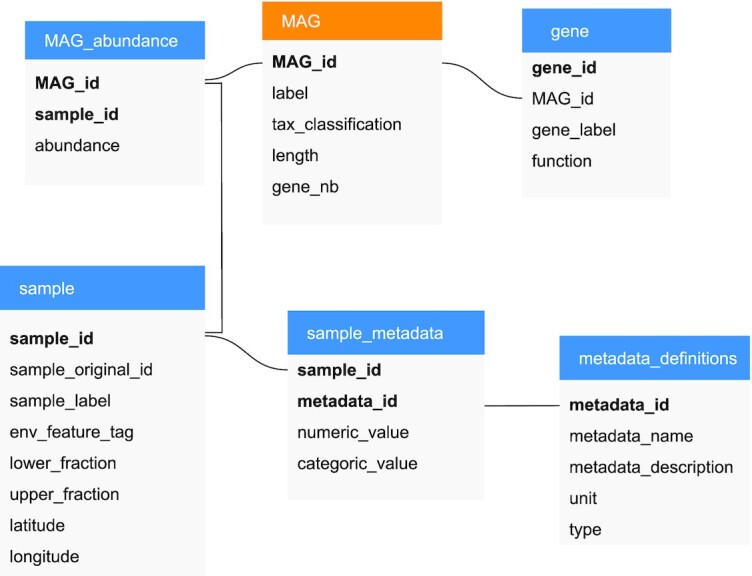
Relational schema of the OGA2 database. For the gene catalogue dataset, five tables are used and a sixth table is needed for the MAGs dataset. The primary key is in bold in each table. Relation between tables are represented with solid lines.
CONCLUSIONS
OGA 2.0 is a web service for biogeographical analysis of large scale marine environmental genomics datasets. The additional datasets presented here now offer users access to a comprehensive set of environmental sequences, including metagenomes, metatranscriptomes, MAGs and SAGs. The API allows users to run several requests using a command line to facilitate access and ensure the datasets are explored to their fullest. Moreover programmatic execution allows a better documentation of the requests (with the trace of the script) and increased repeatability of results. The automated phylogenetic tree option provides an initial view of the homologues neighborhood that can be valuable for evolutionary studies (30).
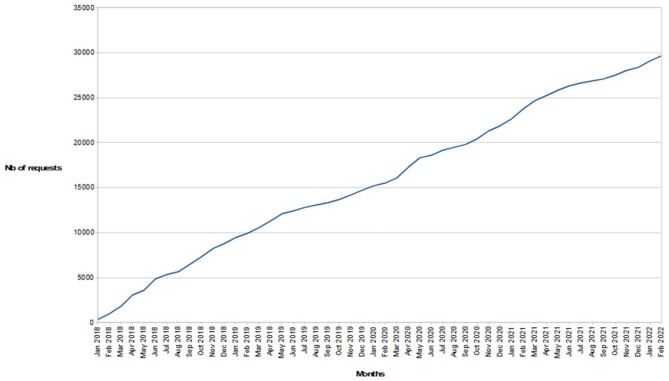
OGA 2.0 has recently been awarded ELIXIR-FR accreditation for its Service Delivery Plan and we maintain our commitment to high performance and stability. The increasing number of users since 2018 illustrated in Figure 8 and the appreciable number of citations (74) since the initial OGA paper (14) underline community interest in the services offered by the Ocean Gene Atlas.
Figure 8.
Request number on OGA2 webserver. Since the first publication of OGA in January 2018, the number of webserver requests is increasing.
In terms of future development, we plan to explore further available dataset annotation such as MAG ecological niches (15) (https://end.mio.osupytheas.fr/Ecological_Niche_database/) as well as Gene Ontology to allow users to query MAG contig sequences for a particular gene but also gene environment to address genome plasticity and evolution (e.g. collinearity and synteny).We encourage scientists to solicit our help in order to integrate additional datasets into OGA2, to which end we can provide a user-friendly data preparation and integration tool.
DATA AVAILABILITY
Ocean Gene Atlas 2.0 is freely available and can be accessed via the following link: https://tara-oceans.mio.osupytheas.fr/ocean-gene-atlas/.
Source code is available at GitLab repository: https://gitlab.osupytheas.fr/ocean_atlas/oga.
Shotgun sequences are available at the European Nucleotide Archive (ENA, https://www.ebi.ac.uk/ena) under accession number PRJEB7988 (OM-RGCv1 and v2), PRJEB6609 (MATOU), PRJEB41575 (Tara Arctic metagenome co-assemblies) and PRJEB402 (EUK_SMAGs) (see Table 1).
The predicted genes from the OM-RGC are available at ENA under the accession numbers ERZ094224 and ERZ096909 to ERZ097151, and the protein sequences are available at: ftp://ftp.genome.jp/pub/db/mgenes/Environmental/Tara.pep.gz.
For OM-RGCv2, all data files can be found through BioStudies with the accession S-BSST297 and for the 530 Tara Arctic metagenome co-assemblies with S-BSST451.
All MATOU, EUK_SMAGs, MGT and BAC_ARC_MAGs resources are available at http://www.genoscope.cns.fr/tara/.
Registry of all the samples from the Tara Oceans Expedition (2009–2013) with environmental metadata are available at PANGAEA: https://doi.org/10.1594/PANGAEA.875582.
For the Global Malaspina 2010 Expedition, all raw sequences are publicly available at both DOE's JGI Integrated Microbial Genomes and Microbiomes (IMG/MER) and the European Nucleotide Archive (ENA). Individual metagenome assemblies, annotation files, and alignment files can be accessed at IMG/MER. All accession numbers are listed at https://www.nature.com/articles/s42003-021-02112–2#MOESM4 in Supplementary Data 1. The metagenomic data can be found through ENA with accession number PRJEB44456 and the co-assembly for the MAG dataset construction with accession number PRJEB40454, the nucleotide sequence for each MAG and their annotation files can be found through BioStudies with accession S-BSST457 and also in the companion publication website at: https://malaspina-public.gitlab.io/malaspina-deep-ocean-microbiome/.
The user manual is available at https://tara-oceans.mio.osupytheas.fr/ocean-gene-atlas/build/pdf/Ocean-Gene-Atlas_User_Manual.pdf.
ACKNOWLEDGEMENTS
This article is contribution number 134 of Tara Oceans.
The web server is hosted by the OSU Pythéas cluster with the help of Cyrille Blanpain and SIP members. Adrien Malgoyre from SIP is thanked for the development of the OSU Pythéas gitlab. We are grateful to the Institut Français de Bioinformatique for providing help and computing resources. Tara Oceans (which includes both the Tara Oceans and Tara Oceans Polar Circle expeditions) would not exist without the leadership of the Tara Ocean Foundation and the continuous support of Tara Oceans consortium members. We further thank the commitment of the following sponsors: CNRS (in particular Groupement de Recherche GDR3280 and the Research Federation for the study of Global Ocean Systems Ecology and Evolution, FR2022/Tara Oceans-GOSEE), European Molecular Biology Laboratory (EMBL), Genoscope/CEA, the French Ministry of Research, and the French Government ‘Investissements d'Avenir’ programmes, FRANCE GENOMIQUE, MEMO LIFE and PSL* Research University. We also thank the support and commitment of agnès b. and Etienne Bourgois, the Prince Albert II de Monaco Foundation, the Veolia Foundation, Region Bretagne, Lorient Agglomeration, Serge Ferrari, Worldcourier, and KAUST. The global sampling effort was enabled by countless scientists and crew who sampled aboard the Tara from 2009–2013, and we thank MERCATOR-CORIOLIS and ACRI-ST for providing daily satellite data during the expeditions. We are also grateful to the countries who graciously granted sampling permissions. The authors declare that all data reported herein are fully and freely available from the date of publication, with no restrictions, and that all of the analyses, publications, and ownership of data are free from legal entanglement or restriction by the various nations whose waters were sampled by the Tara Oceans expedition.
Author contributions: C.V. designed the database schema, the associated query system, and implemented the production server. C.V. and P.H. conceived the overall analysis pipeline strategy, tested the platform and contributed to the writing of the manuscript. J.L. implemented the server system architecture (virtual machine, storage and backups). P.S., S.S., T.O.D., S.G.A., E.P. provided datasets. M.L. contributed to the coordination of the scientific project and supervised developments at the M.I.O., tested the platform and wrote the manuscript. TOC enabled the unprecedented Tara Oceans dataset to be generated. All authors read and approved the final manuscript.
APPENDIX
The Tara Oceans Consortium, Coordinators and Affiliations
Silvia G. Acinas1, Marcel Babin2, Peer Bork3,4,5, Emmanuel Boss6, Chris Bowler7, Guy Cochrane8, Colomban de Vargas9, Gabriel Gorsky10, Lionel Guidi10,11, Nigel Grimsley12,13, Pascal Hingamp14, Daniele Iudicone15, Olivier Jaillon16,17,18, Stefanie Kandels-Lewis3,19, Lee Karp-Boss6, Eric Karsenti7,19, Fabrice Not20, Hiroyuki Ogata21, Nicole Poulton22, Stéphane Pesant23,24, Christian Sardet10,25, Sabrina Speich26,27, Lars Stemmann10, Matthew B. Sullivan28,29, Shinichi Sunagawa30, and Patrick Wincker16,17,18.
1Department of Marine Biology and Oceanography, Institut de Ciències del Mar (CSIC), Barcelona, Catalonia, Spain.
2Département de biologie, Québec Océan and Takuvik Joint International Laboratory (UMI3376), Université Laval (Canada) - CNRS (France), Université Laval, Québec, QC, G1V 0A6, Canada.
3Structural and Computational Biology, European Molecular Biology Laboratory, Meyerhofstrasse 1, 69117 Heidelberg, Germany.
4Max Delbrück Centre for Molecular Medicine, 13125 Berlin, Germany.
5Department of Bioinformatics, Biocenter, University of Würzburg, 97074 Würzburg, Germany.
6School of Marine Sciences, University of Maine, Orono, Maine 04469, USA.
7Ecole Normale Supérieure, PSL Research University, Institut de Biologie de l'Ecole Normale Supérieure (IBENS), CNRS UMR 8197, INSERM U1024, 46 rue d'Ulm, F-75005 Paris, France.
8European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Welcome Trust Genome Campus, Hinxton, Cambridge, UK.
9CNRS, UMR 7144, EPEP & Sorbonne Universités, UPMC Université Paris 06, Station Biologique de Roscoff, 29680 Roscoff, France.
10Sorbonne Universités, UPMC Université Paris 06, CNRS, Laboratoire d'oceanographie de Villefranche (LOV), Observatoire Océanologique, 06230 Villefranche-sur-Mer, France.
11Department of Oceanography, University of Hawaii, Honolulu, HI 96822, USA.
12CNRS, UMR 7232, BIOM, Avenue du Fontaulé, 66650 Banyuls-sur-Mer, France.
13Sorbonne Universités Paris 06, OOB UPMC, Avenue du Fontaulé, 66650 Banyuls-sur-Mer, France.
14Aix Marseille Univ, Université de Toulon, CNRS, IRD, MIO, Marseille, France.
15Stazione Zoologica Anton Dohrn, Villa Comunale, 80121 Naples, Italy.
16CEA - Institut de Génomique, Genoscope, 2 rue Gaston Crémieux, Evry France.
17CNRS, UMR 8030, 2 rue Gaston Crémieux, Evry France.
18Université d'Evry, UMR 8030, CP5706, Evry France.
19Directors’ Research European Molecular Biology Laboratory Meyerhofstr. 1 69117 Heidelberg Germany.
20CNRS, UMR 7144, Sorbonne Universités, UPMC Université Paris 06, Station Biologique de Roscoff, 29680 Roscoff, France.
21Institute for Chemical Research, Kyoto University, Gokasho, Uji, Kyoto, 611-001, Japan.
22Bigelow Laboratory for Ocean Sciences, East Boothbay, ME 04544, USA.
23MARUM, Center for Marine Environmental Sciences, University of Bremen, Bremen, Germany.
24PANGAEA, Data Publisher for Earth and Environmental Science, University of Bremen, Bremen, Germany.
25CNRS, UMR 7009 Biodev, Observatoire Océanologique, F-06230 Villefranche-sur-mer, France.
26Laboratoire de Physique des Océans, UBO-IUEM, Place Copernic, 29820 Plouzané, France.
27Department of Geosciences, Laboratoire de Météorologie Dynamique (LMD), Ecole Normale Supérieure, 24 rue Lhomond, 75231 Paris Cedex 05, France.
28Department of Microbiology, The Ohio State University, Columbus, OH 43214, USA.
29Department of Civil, Environmental and Geodetic Engineering, The Ohio State University, Columbus, OH 43214, USA.
30Department of Biology, Institute of Microbiology and Swiss Institute of Bioinformatics, ETH Zurich, Vladimir-Prelog-Weg 4, 8093 Zurich, Switzerland.
Contributor Information
Caroline Vernette, Aix-Marseille Université, Université de Toulon, IRD, CNRS, Mediterranean Institute of Oceanography (MIO) UM 110, Marseille, France; Research Federation for the study of Global Ocean systems ecology and evolution, FR2022/Tara Oceans-GOSEE, Paris, France.
Julien Lecubin, SIP, OSU PYTHEAS, Marseille, France.
Pablo Sánchez, Department of Marine Biology and Oceanography, Institute of Marine Sciences (ICM), CSIC, Barcelona, Spain.
Shinichi Sunagawa, Department of Biology, Institute of Microbiology and Swiss Institute of Bioinformatics, ETH Zurich, Zurich, Switzerland.
Tom O Delmont, Research Federation for the study of Global Ocean systems ecology and evolution, FR2022/Tara Oceans-GOSEE, Paris, France; Génomique Métabolique, Genoscope, Institut de Biologie François-Jacob, CEA, CNRS, Univ Evry, Univ Paris-Saclay, 91057 Evry, France.
Silvia G Acinas, Department of Marine Biology and Oceanography, Institute of Marine Sciences (ICM), CSIC, Barcelona, Spain.
Eric Pelletier, Research Federation for the study of Global Ocean systems ecology and evolution, FR2022/Tara Oceans-GOSEE, Paris, France; Génomique Métabolique, Genoscope, Institut de Biologie François-Jacob, CEA, CNRS, Univ Evry, Univ Paris-Saclay, 91057 Evry, France.
Pascal Hingamp, Aix-Marseille Université, Université de Toulon, IRD, CNRS, Mediterranean Institute of Oceanography (MIO) UM 110, Marseille, France.
Magali Lescot, Aix-Marseille Université, Université de Toulon, IRD, CNRS, Mediterranean Institute of Oceanography (MIO) UM 110, Marseille, France; Research Federation for the study of Global Ocean systems ecology and evolution, FR2022/Tara Oceans-GOSEE, Paris, France.
Tara Oceans Coordinators:
Silvia G Acinas, Marcel Babin, Peer Bork, Emmanuel Boss, Chris Bowler, Guy Cochrane, Colomban de Vargas, Gabriel Gorsky, Lionel Guidi, Nigel Grimsley, Pascal Hingamp1, Daniele Iudicone, Olivier Jaillon, Stefanie Kandels-Lewis, Lee Karp-Boss, Eric Karsenti, Fabrice Not, Hiroyuki Ogata, Nicole Poulton, Stéphane Pesant, Christian Sardet, Sabrina Speich, Lars Stemmann, Matthew B Sullivan, Shinichi Sunagawa, and Patrick Wincker
FUNDING
French Government ‘Investissements d'Avenir’ programmes OCEANOMICS [ANR-11-BTBR-0008]; FRANCE GENOMIQUE [ANR-10-INBS-09-08]; Institut Français de Bioinformatique (IFB) [ANR-11-INBS-0013]; SeqDigger [ANR-19-CE45-0008]; AO-EMBRC [ANR-21-ESRE-0038]. Funding for open access charge: ANR [ANR-19-CE45-0008].
Conflict of interest statement. None declared.
REFERENCES
- 1. Falkowski P. Ocean science: the power of plankton. Nature. 2012; 483:S17–S20. [DOI] [PubMed] [Google Scholar]
- 2. Field C.B., Behrenfeld M.J., Randerson J.T., Falkowski P.. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 1998; 281:237–240. [DOI] [PubMed] [Google Scholar]
- 3. Guidi L., Chaffron S., Bittner L., Eveillard D., Larhlimi A., Roux S., Darzi Y., Audic S., Berline L., Brum J.R.et al.. Plankton networks driving carbon export in the oligotrophic ocean. Nature. 2016; 532:465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rusch D.B., Halpern A.L., Sutton G., Heidelberg K.B., Williamson S., Yooseph S., Wu D., Eisen J.A., Hoffman J.M., Remington K.et al.. The sorcerer II global ocean sampling expedition: northwest atlantic through eastern tropical pacific. PLoS Biol. 2007; 5:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karsenti E., Acinas S.G., Bork P., Bowler C., De Vargas C., Raes J., Sullivan M., Arendt D., Benzoni F., Claverie J.-M.et al.. A holistic approach to marine eco-systems biology. PLoS Biol. 2011; 9:e1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pesant S., Not F., Picheral M., Kandels-Lewis S., Bescot N.L., Gorsky G., Iudicone D., Karsenti E., Speich S., Troublé R.et al.. Open science resources for the discovery and analysis of tara oceans data. Scientific Data. 2015; 2:150023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sunagawa S., Coelho L.P., Chaffron S., Kultima J.R., Labadie K., Salazar G., Djahanschiri B., Zeller G., Mende D.R., Alberti A.et al.. Structure and function of the global ocean microbiome. Science. 2015; 348:1261359. [DOI] [PubMed] [Google Scholar]
- 8. Carradec Q., Pelletier E., Silva C.D., Alberti A., Seeleuthner Y., Blanc-Mathieu R., Lima-Mendez G., Rocha F., Tirichine L., Labadie K.et al.. A global ocean atlas of eukaryotic genes. Nat. Commun. 2018; 9:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duarte C.M. Seafaring in the 21St century: the malaspina 2010 circumnavigation expedition. Limnol. Oceanogr. Bull. 2015; 24:11–14. [Google Scholar]
- 10. Klemetsen T., Raknes I.A., Fu J., Agafonov A., Balasundaram S.V., Tartari G., Robertsen E., Willassen N.P.. The MAR databases: development and implementation of databases specific for marine metagenomics. Nucleic Acids Res. 2018; 46:D692–D699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mitchell A.L., Almeida A., Beracochea M., Boland M., Burgin J., Cochrane G., Crusoe M.R., Kale V., Potter S.C., Richardson L.J.et al.. MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res. 2020; 8:D570–D578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ponsero A.J., Bomhoff M., Blumberg K., Youens-Clark K., Herz N.M., Wood-Charlson E.M., Delong E.F., Hurwitz B.L.. Planet microbe: a platform for marine microbiology to discover and analyze interconnected ‘omics and environmental data. Nucleic Acids Res. 2021; 49:D792–D802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paoli L., Ruscheweyh H.-J., Forneris C.C., Kautsar S., Clayssen Q., Salazar G., Milanese A., Gehrig D., Larralde M., Carroll L.M.et al.. Uncharted biosynthetic potential of the ocean microbiome microbiology. 2021); bioRxiv doi:24 March 2021, preprint: not peer reviewed 10.1101/2021.03.24.436479. [DOI]
- 14. Villar E., Vannier T., Vernette C., Lescot M., Cuenca M., Alexandre A., Bachelerie P., Rosnet T., Pelletier E., Sunagawa S.et al.. The ocean gene atlas: exploring the biogeography of plankton genes online. Nucleic Acids Res. 2018; 46:W289–W295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delmont T.O., Pierella Karlusich J.J., Veseli I., Fuessel J., Eren A.M., Foster R.A., Bowler C., Wincker P., Pelletier E.. Heterotrophic bacterial diazotrophs are more abundant than their cyanobacterial counterparts in metagenomes covering most of the sunlit ocean. ISME J. 2022; 16:927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Delmont T.O., Gaia M., Hinsinger D.D., Fremont P., Vanni C., Fernandez-Guerra A., Eren A.M., Kourlaiev A., d'Agata L., Clayssen Q.et al.. Functional repertoire convergence of distantly related eukaryotic plankton lineages abundant in the sunlit ocean. Cell Genomics. 2022; 2:100123. 10.1016/j.xgen.2022.100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vorobev A., Dupouy M., Carradec Q., Delmont T.O., Annamalé A., Wincker P., Pelletier E.. Transcriptome reconstruction and functional analysis of eukaryotic marine plankton communities via high-throughput metagenomics and metatranscriptomics. Genome Res. 2020; 30:647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Royo-Llonch M., Sánchez P., Ruiz-González C., Salazar G., Pedrós-Alió C., Sebastián M., Labadie K., Paoli L.M., Ibarbalz F., Zinger L.et al.. Compendium of 530 metagenome-assembled bacterial and archaeal genomes from the polar arctic ocean. Nat. Microbiol. 2021; 6:1561–1574. [DOI] [PubMed] [Google Scholar]
- 19. Acinas S.G., Sánchez P., Salazar G., Cornejo-Castillo F.M., Sebastián M., Logares R., Royo-Llonch M., Paoli L., Sunagawa S., Hingamp P.et al.. Deep ocean metagenomes provide insight into the metabolic architecture of bathypelagic microbial communities. Commun. Biol. 2021; 4:604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salazar G., Paoli L., Alberti A., Huerta-Cepas J., Ruscheweyh H.-J., Cuenca M., Field C.M., Coelho L.P., Cruaud C., Engelen S.et al.. Gene expression changes and community turnover differentially shape the global ocean metatranscriptome. Cell. 2019; 179:1068–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li W., O'Neill K.R., Haft D.H., DiCuccio M., Chetvernin V., Badretdin A., Coulouris G., Chitsaz F., Derbyshire M.K., Durkin A.S.et al.. RefSeq: expanding the prokaryotic genome annotation pipeline reach with protein family model curation. Nucleic Acids Res. 2021; 49:D1020–D1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H., Durbin R.. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katoh K., Standley D.M.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gouveia-Oliveira R., Sackett P.W., Pedersen A.G.. MaxAlign: maximizing usable data in an alignment. BMC Bioinf. 2007; 8:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T.. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009; 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Price M.N., Dehal P.S., Arkin A.P.. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010; 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Junier T., Zdobnov E.M.. The newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics. 2010; 26:1669–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sunagawa S., Mende D.R., Zeller G., Izquierdo-Carrasco F., Berger S.A., Kultima J.R., Coelho L.P., Arumugam M., Tap J., Nielsen H.B.et al.. Metagenomic species profiling using universal phylogenetic marker genes. Nat. Methods. 2013; 10:1196. [DOI] [PubMed] [Google Scholar]
- 29. Wilkinson M.D., Dumontier M., Aalbersberg Ij.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E.et al.. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016; 3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vannier T., Hingamp P., Turrel F., Tanet L., Lescot M., Timsit Y.. Diversity and evolution of bacterial bioluminescence genes in the global ocean. NAR Genomics Bioinformatics. 2020; 2:lqaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Ocean Gene Atlas 2.0 is freely available and can be accessed via the following link: https://tara-oceans.mio.osupytheas.fr/ocean-gene-atlas/.
Source code is available at GitLab repository: https://gitlab.osupytheas.fr/ocean_atlas/oga.
Shotgun sequences are available at the European Nucleotide Archive (ENA, https://www.ebi.ac.uk/ena) under accession number PRJEB7988 (OM-RGCv1 and v2), PRJEB6609 (MATOU), PRJEB41575 (Tara Arctic metagenome co-assemblies) and PRJEB402 (EUK_SMAGs) (see Table 1).
The predicted genes from the OM-RGC are available at ENA under the accession numbers ERZ094224 and ERZ096909 to ERZ097151, and the protein sequences are available at: ftp://ftp.genome.jp/pub/db/mgenes/Environmental/Tara.pep.gz.
For OM-RGCv2, all data files can be found through BioStudies with the accession S-BSST297 and for the 530 Tara Arctic metagenome co-assemblies with S-BSST451.
All MATOU, EUK_SMAGs, MGT and BAC_ARC_MAGs resources are available at http://www.genoscope.cns.fr/tara/.
Registry of all the samples from the Tara Oceans Expedition (2009–2013) with environmental metadata are available at PANGAEA: https://doi.org/10.1594/PANGAEA.875582.
For the Global Malaspina 2010 Expedition, all raw sequences are publicly available at both DOE's JGI Integrated Microbial Genomes and Microbiomes (IMG/MER) and the European Nucleotide Archive (ENA). Individual metagenome assemblies, annotation files, and alignment files can be accessed at IMG/MER. All accession numbers are listed at https://www.nature.com/articles/s42003-021-02112–2#MOESM4 in Supplementary Data 1. The metagenomic data can be found through ENA with accession number PRJEB44456 and the co-assembly for the MAG dataset construction with accession number PRJEB40454, the nucleotide sequence for each MAG and their annotation files can be found through BioStudies with accession S-BSST457 and also in the companion publication website at: https://malaspina-public.gitlab.io/malaspina-deep-ocean-microbiome/.
The user manual is available at https://tara-oceans.mio.osupytheas.fr/ocean-gene-atlas/build/pdf/Ocean-Gene-Atlas_User_Manual.pdf.