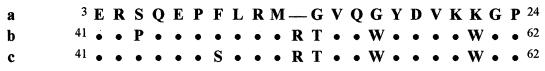Abstract
A 41-kDa protein of Nitrosomonas eutropha was purified, and the N-terminal amino acid sequence was found to be nearly identical with the sequence of AmoB, a subunit of ammonia monooxygenase. This protein was used to develop polyclonal antibodies, which were highly specific for the detection of the four genera of ammonia oxidizers of the β-subclass of Proteobacteria (Nitrosomonas, including Nitrosococcus mobilis, which belongs phylogenetically to Nitrosomonas; Nitrosospira; Nitrosolobus; and Nitrosovibrio). In contrast, the antibodies did not react with ammonia oxidizers affiliated with the γ-subclass of Proteobacteria (Nitrosococcus oceani and Nitrosococcus halophilus). Moreover, methane oxidizers (Methylococcus capsulatus, Methylocystis parvus, and Methylomonas methanica) containing the related particulate methane monooxygenase were not detected. Quantitative immunoblot analysis revealed that total cell protein of N. eutropha consisted of approximately 6% AmoB, when cells were grown using standard conditions (mineral medium containing 10 mM ammonium). This AmoB amount was shown to depend on the ammonium concentration in the medium. About 14% AmoB of total protein was found when N. eutropha was grown with 1 mM ammonium, whereas 4% AmoB was detected when 100 mM ammonium were used. In addition, the cellular amount of AmoB was influenced by the absence of the substrate. Cells starved for more than 2 months contained nearly twice as much AmoB as actively growing cells, although these cells possessed low ammonia-oxidizing activity. AmoB was always present and could even be detected in cells of Nitrosomonas after 1 year of ammonia starvation.
Nitrification, the microbial oxidation of ammonia to nitrate, is an essential part of the microbial nitrogen cycle in marine, freshwater, and soil environments. Two physiologically different groups of chemolithoautotrophic bacteria, the ammonia and nitrite oxidizers, are involved in this oxidation. The ammonia oxidizers derive their energy from the oxidation of ammonia to nitrite. The first step, the oxidation of ammonia to hydroxylamine is catalyzed by ammonia monooxygenase (AMO) (18, 60). Hydroxylamine is further oxidized to nitrite by hydroxylamine oxidoreductase (HAO) (5, 53). Since the AMO is an important key enzyme of nitrification, many efforts have been initiated to isolate the enzyme. However, it has not been purified thus far since the enzyme is not stable once isolated from the cells (16, 51, 52). Therefore, little is known about its structure and enzymatic mechanism. Information on the molecular properties of AMO has been deduced from studies with intact cells. It was demonstrated that the AMO has a broad substrate specificity (4, 22, 28, 54) and is irreversibly inhibited by C2H2 (25, 26). A similar substrate range and inhibitor profiles including C2H2 effects were found for the biochemically related particulate methane monooxygenase (pMMO) of methane oxidizing bacteria (7, 14, 20, 42). Moreover, the AMO and the pMMO may be evolutionary related, since their encoding genes share high sequence similarities (3, 19).
Inactivation of AMO by 14C2H2 labels a membrane bound 27-kDa polypeptide, which is called AmoA (21). This protein seems to be the active-site-containing subunit of the enzyme (21, 24). A corresponding gene amoA has been identified and, within the same operon, another gene called amoB was sequenced (39). The amoB codes for a 41-kDa polypeptide (AmoB), which could be copurified with the 27-kDa AmoA (10, 39). Upstream of the amoA-amoB tandem, a third gene was identified, amoC (3, 30). The numbers of copies of the amo operon seem to be genus specific. Two nearly identical copies are present in strains of Nitrosomonas and Nitrosovibrio, and three copies were found in strains of Nitrosospira and Nitrosolobus (29, 31, 39, 41, 45), whereas only a single copy could be detected in marine Nitrosococcus strains of the γ-subclass of Proteobacteria (3). However, neither the expressed polypeptides of amoA and amoB nor the purified proteins from Nitrosomonas cell homogenates showed ammonia-oxidizing activity (21, 23).
In previous studies antibodies were developed using whole cells of ammonia oxidizers, which recognize epitopes of the cell wall (8, 43, 47, 55, 56). These antibodies were applied in ecological studies to detect and count ammonia oxidizers in bacterial communities by using fluorescence microscopy. Their application was supposed to overcome the disadvantages of traditional counting methods such as the most-probable-number technique (38), which often underestimates the number of ammonia oxidizers in the natural environment (9). However, the application of these antibodies was limited since ammonia oxidizers show high serological diversity even within one genus. Therefore, ecologically relevant strains had to be isolated prior to antibody development. In the case of nitrite-oxidizing bacteria, it was shown that antibodies recognizing the conserved key enzyme can be used for the detection of all known genera of these organisms (6). They can be applied for studies of the key enzyme, as well as for the detection of as-yet-nonisolated strains in the environment.
In this study, the AmoB subunit of the AMO of N. eutropha N904 was used for the development of polyclonal antibodies. Evidence is given that these antibodies are highly specific for all genera of ammonia oxidizers affiliated with the β-subclass of Proteobacteria. Quantitative immunoblot analysis was used to measure the cellular amount of AmoB of Nitrosomonas eutropha N904 under different growth and starvation conditions.
(This study is based in part on the doctoral study of C. Pinck at the University of Hamburg.)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains of ammonia oxidizers isolated from soil and used in this study were Nitrosomonas communis Nm 2 (34), Nitrosomonas oligotropha Nm 45 (34), Nitrosomonas ureae Nm 10 (34), Nitrosospira sp. strain Nsp 1, and Nitrosolobus multiformis Nl 13 (ATCC 25196). Nitrosomonas europaea Freitag and Nitrosomonas nitrosa Nm 90 (34) were obtained from sewage. The strains Nitrosomonas sp. strain Dave and N. eutropha N904 originated from a biowaste fermenter and from cattle manure, respectively. Nitrosomonas sp. strain Nm R1.24, Nitrosospira sp. strain Nsp G1.6, Nitrosospira sp. strain Nsp M1.3, Nitrosospira sp. strain Nsp R6.2, Nitrosovibrio sp. strain Nv G1.3, and Nitrosovibrio sp. strain Nv K7.1 (49) were isolated from the sandstone of historical buildings. The marine ammonia oxidizers of the β-subclass of the Proteobacteria isolates Nitrosomonas aestuarii Nm 36 (34), Nitrosomonas cryotolerans Nm 55, Nitrosomonas halophila Nm 1 (34), Nitrosomonas marina Nm 22 (34), Nitrosococcus mobilis Nc 2 (32), and the two marine ammonia oxidizers belonging to the γ-subclass of Proteobacteria isolates Nitrosococcus halophilus Nc 4 (33) and Nitrosococcus oceani Nc 1 (ATCC 19707) were obtained from seawater.
The nitrite oxidizers Nitrobacter hamburgensis X14 (11) and Nitrobacter winogradskyi Engel (12) originated from soil. Nitrospira moscoviensis M-1 (15) was obtained from a heating system. Nitrobacter vulgaris K48 (12) originated from the sandstone of historical buildings; Nitrospina gracilis 3(211) (57), Nitrospina sp. strain 347, and Nitrococcus mobilis 231 (57) were isolated from seawater.
All ammonia oxidizers, nitrite oxidizers, the methane oxidizers Methylococcus capsulatus Bath (NCIMB 11132), Methylocystis parvus 4a, Methylomonas methanica Oo52006, and Bacillus subtilis 019, Escherichia coli K-12/067 (ATCC 23716), Methylobacterium radiotolerans, Paracoccus denitrificans 001 (ATCC 19367), and Pseudomonas sp. strain AM1 are stored in the culture collection of the Institut für Allgemeine Botanik, Abteilung Mikrobiologie, Universität Hamburg. The strains Achromobacter cycloclastes, Agrobacterium tumefaciens GM 19023, Alcaligenes faecalis (ATCC 8750), Azorhizobium sp. strain 24, Azospirillum lipoferum (ATCC 29707), Bacillus azotoformans (ATCC 29788), Bradyrhizobium denitrificans, Chromobacterium violaceum, and Pseudomonas sp. strain AK 15 were obtained from C. Coeur (University of Lyon I, Villeurbanne, France).
Terrestrial and freshwater ammonia oxidizers were grown at 28°C in mineral salt medium (34) in the presence of 10 mM ammonium. N. cryotolerans Nm 55, N. aestuarii Nm 36, N. halophila Nm 1, and Nitrosococcus mobilis Nc 2 were grown in the same medium containing 10 g of NaCl liter−1. N. marina Nm 22, Nitrosococcus oceani Nc 1, and Nitrosococcus halophilus Nc 4 were cultivated in seawater medium, with the following composition: 10 mM NH4Cl, 0.4 mM KH2PO4, 3 g of HEPES, and 1 ml of 0.05% (wt/vol) cresol red solution per liter of 40% seawater.
For quantitative immunoblots, N. eutropha N904 was grown lithoautotrophically in mineral salt medium containing different concentrations of NH4Cl (1, 10, or 100 mM). After cells of N. eutropha N904 were grown for 10 days with different substrate concentrations, the AmoB amount was determined. For mixotrophic growth the mineral salt medium was supplemented with 5 mM pyruvate, 1.5 g of yeast extract (Difco) liter−1, and 1.5 g of peptone (Difco) liter−1, and either 9.1 mM pyruvate or 9.1 mM alanine.
Batch cultures of N. eutropha N904, Nitrosomonas sp. strain Dave and N. europaea Freitag were starved of ammonia at 16°C in the dark. To test the AmoB amount by immunoblotting, samples were obtained after 1, 29, 58, 129, and 360 days of ammonia starvation.
Nitrobacter hamburgensis X14, Nitrobacter winogradskyi Engel, and Nitrobacter vulgaris K48 were grown mixotrophically in the presence of 2 g of NaNO2 liter−1 (12). Nitrospira moscoviensis M-1 was cultivated in mineral medium with 0.2 g of NaNO2 liter−1 (15). Nitrospina gracilis 3(211), Nitrospina sp. strain 347, and Nitrococcus mobilis 231 were cultivated in seawater media according to the method of Watson and Waterbury (57). The cultures were incubated at 28°C, expect for Nitrospira moscoviensis M-1, which was incubated at 37°C.
The methane oxidizers were cultivated in nitrate mineral salt medium (58) including 0.25 μM CuSO4 at 3% methane synthetic air atmosphere. The methylotrophs were grown in mineral medium containing 0.15% (wt/vol) methanol (17). All other bacterial strains were cultivated according to the American Type Culture Collection and National Collection of Industrial and Marine Bacteria instructions.
AmoB isolation, sequencing, and production of antibodies.
Cells of N. eutropha N904 were harvested by centrifugation, washed twice, and suspended in 0.9% NaCl. Cell homogenates were prepared by passing the cell suspension (1010 cells ml−1) through a French pressure cell at 140 MPa or by sonification on ice for 15 to 30 min by using a Biorupter apparatus. The protein concentrations of the crude extracts were determined colorimetrically according to the method of Bradford (13) as modified by Spector (48). Crude extracts were adjusted to 3.0 mg of protein ml−1. The samples were diluted (1:1) with 10 mM Tris-HCl buffer (pH 6.8) containing 2% sodium dodecyl sulfate (SDS), 20% glycerol, 1% 2-mercaptoethanol, and 0.001% bromophenol blue and then solubilized for 15 min at room temperature. Samples (75 μl) were loaded onto lanes of 0.75-mm-thick SDS-polyacrylamide gels, prepared as described by Laemmli (36). The stacking and separating gels contained 4 and 12% polyacrylamide, respectively. Electrophoresis was performed at 80 V and 10°C by using a PROTEAN II Slab Cell (20 by 16 cm; Bio-Rad). After electrophoresis the gels were reversibly stained by using the Zinc Stain & Destain Kit (Bio-Rad) to determine the position of the 41-kDa protein. The protein bands were cut out of the gels. The staining of the slices was removed, and the protein was electroeluted from the gel at 60 mA for 6 h in an Electro-Eluter Model 422 (Bio-Rad). The protein was concentrated by lyophilization (Freezemobile 12; Virtis). Antiserum against this polypeptide was produced by Valbex (Villeurbanne, France) in chickens. Injections of 50 μg of protein were given at prescribed intervals.
For protein sequencing by Edman degradation, the 41-kDa protein was isolated by a modified SDS-polyacrylamide gel electrophoresis (PAGE). In order to avoid N-terminal blockage the gel had been pre-electrophoresed for 2 h, adding 0.07% sodium thioglycolate to the Laemmli running buffer. The separated proteins were electroblotted (Pegasus; PHASE) onto a 0.2-μm-pore-size polyvinylidene difluoride membrane filter (Schleicher & Schuell), using the procedure described by Matsudaira (37). The amino acid sequencing of the isolated protein was done by the Institute of Biology and Chemistry of Proteins (University of Lyon I, Villeurbanne, France). Protein sequences were used to search for homologous proteins in the EMBL and SwissProt data banks (http://www.ncbi.nlm.nih.gov/BLAST/) (2).
Immunoblotting.
Cells were harvested, and crude extracts were prepared as described above. SDS-PAGE analyses were performed at 40 mA by using a Mini-PROTEAN II Cell (8 by 7.3 cm; Bio-Rad). The separated proteins were electroblotted (Pegasus; PHASE) for 2 h at 0.8 mA per cm2 onto a cellulose nitrate membrane (pore size, 0.2 μm; Schleicher & Schuell) using a discontinuous buffer system (35). The membrane was then blocked for 1.5 h in a phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA). The proteins on the nitrocellulose membrane were incubated with antiserum (diluted 1:32,000 in PBS containing 0.05% BSA and 0.025% Tween 20) overnight at room temperature. After two washes with PBS, the proteins were incubated with biotin-conjugated secondary antibodies (Biotrend, Cologne, Germany) diluted 1:30,000 in PBS containing 0.05% BSA–0.025% Tween 20 for 1.5 h at room temperature. The membrane was washed twice with PBS, and the proteins were incubated with a streptavidin-biotinylated alkaline phosphatase complex (diluted 1:3,000 in PBS containing 0.05% BSA and 0.025% Tween 20) for another 1.5 h. After two washes with 10 mM Tris-HCl (pH 8.6) containing 0.02% BSA and 0.05% Tween 20, the cellulose nitrate membrane was incubated with a substrate solution containing 0.005% 5-bromo-4-chloro-3-indolyl-phosphate (BCIP), 0.001% 4-nitroblue tetrazolium, 0.1 M NaHCO3, 0.05 M Na2CO3, and 0.004 M MgCl2. The enzymatic reaction was stopped by adding distilled water. A dense blue color indicated a positive reaction. Each immunoblotting experiment was reproduced at least three times. The membrane was scanned (ScanMagic 9636 S; Mustek), a densitogram of the lanes was performed, and the AmoB was quantified using software program origin 4.0 (Microcal).
Determination of ammonia oxidation activity.
Exponentially grown cells of N. eutropha N904 cultivated in 1-liter Erlenmeyer flasks were harvested by centrifugation, washed, and resuspended in mineral medium. The ammonia oxidation activity of the cells was reflected by the decrease in ammonia as well as by the increase in nitrite concentration in suspensions of 5 × 107 cells per ml. The activities were determined as mean values of three experiments. Ammonia and nitrite levels were measured by high-pressure liquid chromatography (50).
RESULTS
Purification and identification of AmoB protein.
The N terminus of the 41-kDa protein of N. eutropha N904, separated by SDS-PAGE, was sequenced by Edman degradation (positions 3 to 24). The derived amino acid sequence was compared with the amino acid sequences deduced from the amoB genes of N. eutropha Nm 57, since an amoB gene sequence of N. eutropha N904 is not available yet. The AmoB subunit of N. eutropha Nm 57 is encoded by two nearly identical gene copies (amoB1 and amoB2, GenBank accession numbers U51630 and U72670). The N terminus of the isolated 41-kDa protein of N. eutropha N904 showed 82% amino acid sequence identity to the deduced AmoB sequences of N. eutropha Nm 57 from positions 41 to 62 onwards (Fig. 1). The sequences AmoB1 and AmoB2 from N. eutropha Nm 57 differ in positions 43 and 47, respectively, compared to the N-terminal sequence from N. eutropha N904 (positions 5 and 9). As a consequence, the AmoB sequence of N. eutropha N904 tallied only with one amino acid of AmoB1 or AmoB2 of N. eutropha Nm 57, respectively. Mismatches were often represented by the amino acid glycine (positions 14 and 17 of N. eutropha N904).
FIG. 1.
Comparison of amino acid sequences of the isolated 41-kDa protein of N. eutropha N904 (a) and the deduced amino acid sequences of AmoB1 (b) and AmoB2 (c) of N. eutropha Nm 57 encoded by the amoB genes. (The SwissProt accession numbers for N. eutropha Nm 57 amoB1 and amoB2 are U51630 and U72670, respectively.) The periods represent an identical match to the sequence of the isolated 41-kDa protein of N. eutropha N904. The space represents a gap among the sequences. The isolated 41-kDa protein of N. eutropha N904 showed 82% (18 of 22) amino acid sequence identity compared to both AmoB proteins of N. eutropha Nm 57.
Specificity of the antibodies.
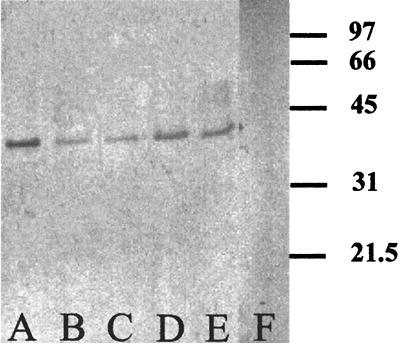
Polyclonal antibodies were produced against the purified AmoB of N. eutropha N904. The reactivity of the antibodies was tested by immunoblotting of crude extracts of numerous ammonia oxidizers, including all described species of the genus Nitrosomonas. All tested strains are listed in Materials and Methods. The antibodies were highly specific for the detection of 41-kDa proteins in cell extracts of the four genera of ammonia oxidizers of the β-subclass of Proteobacteria (Nitrosomonas, including Nitrosococcus mobilis, which belongs phylogenetically to Nitrosomonas; Nitrosospira; Nitrosolobus; and Nitrosovibrio). In six strains of ammonia oxidizers isolated from building stones, strains which have been characterized as yet only by their morphology, 41-kDa proteins were recognized as well. The antibodies did not show any unspecific reactions with other proteins of these crude extracts (Fig. 2). No proteins were recognized in crude extracts of Nitrosococcus oceani Nc 1 (Fig. 2) and Nitrosococcus halophilus Nc 4 (data not shown), which belong to the γ-subclass of Proteobacteria. Hence, the antiserum could be used to detect AmoB in ammonia oxidizers of the β-subclass of Proteobacteria including not-yet-described isolates, but not for the detection of AmoB in ammonia oxidizers of the γ-subclass of Proteobacteria.
FIG. 2.
Immunoblot using antiserum recognizing the AmoB of different ammonia-oxidizing bacteria. Lane A, N. eutropha N904; lane B, Nitrosovibrio sp. strain K7.1; lane C, Nitrosospira sp. strain R6.2; lane D, Nitrosococcus mobilis Nc 2; lane E, Nitrosolobus multiformis, Nl 13; lane F, Nitrosococcus oceani Nc 4. Standard protein molecular masses are indicated on the right (in kilodaltons). Crude extracts of cells were added to the gel at protein amounts of 10 μg per lane.
In order to prove the specificity of the antiserum, control experiments were performed with pure cultures of the methane-oxidizing bacteria Methylococcus capsulatus Bath, Methylomonas methanica Oo52006, and Methylocystis parvus 4a. The key enzyme in CH4 oxidation of these organisms, the pMMO, was shown to possess high sequence similarities to the AMO. Furthermore, the heterotrophic nitrifier Paracoccus denitrificans 001 was analyzed, since this organism also contains an ammonia-oxidizing system. In addition, immunoblot analysis was carried out with several other bacteria, such as nitrite oxidizers and methylotrophic and denitrifying bacteria. The antiserum did not react with any of these organisms.
Quantitative immunoblot analysis of the AmoB amount.
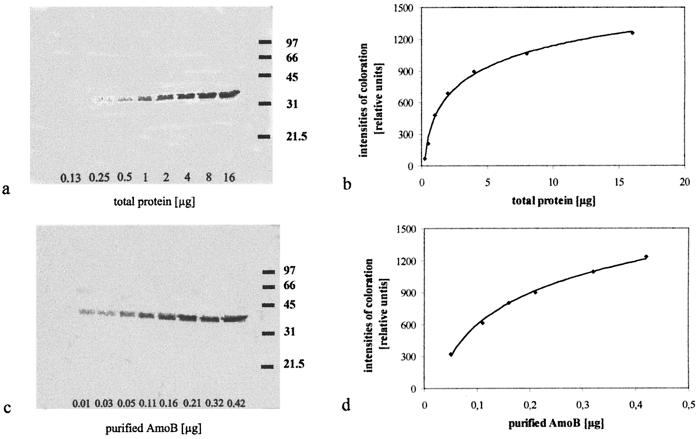
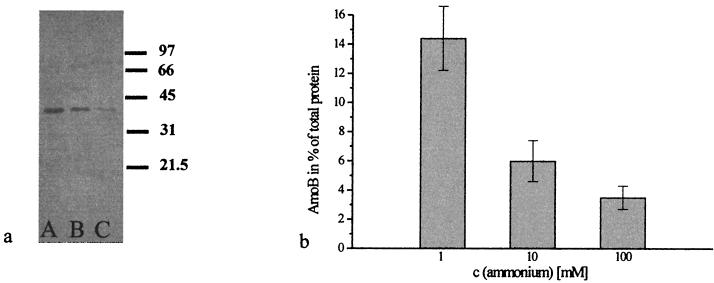
The AmoB amount of total cellular protein of N. eutropha N904 was measured using immunoblot analysis of crude extracts with protein amounts ranging from 0.25 to 16 μg (Fig. 3a and b), which corresponded to 2.9 × 106 cells and 1.9 × 108 cells, respectively. The protein coloration increased with the protein amount, and a saturation curve was obtained. These data could be used to determine the specific cellular amount of AmoB (Table 1). Purified AmoB ranging from 0.03 to 0.42 μg served as standards (Fig. 3c and d). By using standard growth conditions (mineral medium with 10 mM ammonium), a cellular amount of 5.9% ± 1.8% AmoB was found in the total cell protein of N. eutropha N904. As shown in Fig. 4, this cellular AmoB amount depended on the ammonium concentration in the mineral medium. The amount of AmoB increased to 14% ± 1.4% when the substrate was reduced to 1 mM. When cells were grown with 100 mM ammonium, only a low AmoB amount of 4% ± 0.8% was found.
FIG. 3.
Coloration intensities of the AmoB after immunostaining in correlation to the amount of total protein in N. eutropha N904 grown with 10 mM ammonium (a and b) and in correlation to the amount of purified AmoB (c and d). The coloration intensities obtained by immunoblotting (a and c) are plotted against the amounts of total protein (b) and against the amounts of purified AmoB (d). The values on the right of the immunoblots are molecular masses (in kilodaltons).
TABLE 1.
Specific AmoB amount in cells of N. eutropha N904 grown with 10 mM ammonium as measured by using immunoblot analysisa
| Total protein amt (μg) | Specific AmoB amtb
|
|
|---|---|---|
| μg | % Total protein | |
| 16 | ND | ND |
| 8 | 0.3 | 3.8 |
| 4 | 0.21 | 5.3 |
| 2 | 0.13 | 6.5 |
| 1 | 0.08 | 8.0 |
| 0.5 | ND | ND |
| 0.25 | ND | ND |
Cell extracts of different total protein amounts were analyzed to obtain statistically meaningful data. Purified AmoB ranging from 0.01 to 0.42 μg of protein were used as standards. The mean value of the specific AmoB amount was calculated with 5.9% ± 1.8% of the total protein (100%). The standard deviation for three replicated experiments was ±9%.
ND, not detectable.
FIG. 4.
Quantification of AmoB in crude extract of N. eutropha N904 grown with different ammonium concentrations. (a) An immunoblot was performed using crude extracts containing 4 μg of total protein. Lane A, cells grown with 1 mM; lane B, cells grown with 10 mM; lane C, cells grown with 100 mM ammonium. The values on the right of the immunoblot are molecular masses (in kilodaltons). (b) Decrease of specific AmoB amount of the total protein by increasing ammonium concentrations in the mineral media. The error bars represent the standard deviations for the average of three replicate experiments.
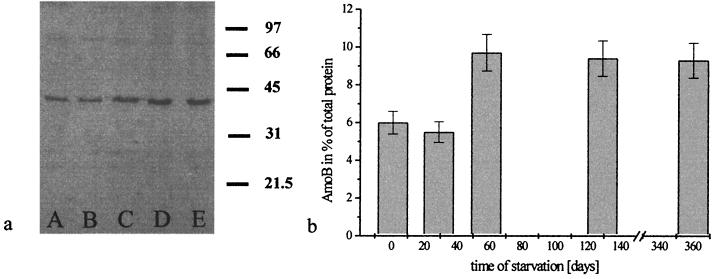
The AmoB amount increased when N. eutropha N904 was starved of ammonia. Cells, which did not receive ammonium for more than 60 days, contained 9.7% ± 0.9% AmoB (Fig. 5). That is nearly twice the amount compared to that found in active growing cells or cells that had been starved of ammonia for 20 days. The AmoB could even be detected after 1 year of ammonia starvation. These results did not correlate with the ammonia oxidation activity. The highest activity was found within cells in the exponential growth phase using standard cultivation conditions (575 μmol of NH4+ g of protein−1 h−1). The specific activity was reduced to 255 μmol of NH4+ g of protein−1 h−1 when cells were starved of ammonia for 1 month, a level which remained nearly constant during further starvation. Experiments with cells of Nitrosomonas sp. strain Dave and N. europaea Freitag showed similar results (data not shown).
FIG. 5.
Quantification of AmoB in crude extract of starved cells of N. eutropha N904. (a) Immunoblot was performed with crude extracts containing 4 μg of total protein. Lane A, cells ammonia starved for 1 day; lane B, cells ammonia starved for 29 days; lane C, cells ammonia starved for 58 days; lane D, cells ammonia starved for 129 days, and lane E, cells ammonia starved for 360 days. The values on the right of the immunoblot are molecular masses (in kilodaltons). (b) After different times of ammonia starvation the specific AmoB percentage of the total protein amount increased. The error bars represent the standard deviation for the average of three replicate experiments.
Mixotrophic growth of the bacteria with ammonia and organic N compounds had no influence on the AmoB amount of the cells. Cells of N. eutropha N904, which were grown mixotrophically in the presence of pyruvate, yeast extract, and peptone, as well as pyruvate or alanine, showed nearly the same AmoB amount as lithoautotrophically grown cells.
DISCUSSION
The isolated 41-kDa protein of N. eutropha N904 shows high sequence similarity to the sequences AmoB1 and AmoB2 of N. eutropha Nm 57. The mismatches in positions 14 and 17 of the partially sequenced AmoB protein of N. eutropha N904 (Fig. 1) might be a contamination of glycine used in the transfer buffer for blotting the protein on the membrane. The amoB genes of N. eutropha Nm 57 encode hydrophobic 37-amino-acid leader sequences (39) that was not present in the isolated AmoB of N. eutropha N904. Apparently, this part of the N terminus was removed during protein processing. Nevertheless, the purified 41-kDa protein can be regarded as the AmoB of N. eutropha N904. Sequence differences in the AmoB peptides between N. eutropha N904 and N. eutropha Nm 57 may be due to strain differences.
In this study, an antiserum against the AmoB was developed which recognized the 41-kDa subunit of the AMO in crude extracts of N. eutropha N904. It could be demonstrated that this antiserum has a broad serological specificity for all tested ammonia oxidizers of the β-subclass of Proteobacteria. Thus far, only antibodies recognizing specific epitopes of the cell wall of ammonia oxidizers have been described (43, 47, 55), and these were limited in the application to specific serological groups (8, 56). The AMO is a highly conserved enzyme in ammonia oxidizers affiliated with the β-subclass of Proteobacteria. Therefore, the broad serological specificity of the antibodies against AmoB may be due to their 73% AmoB sequence similarity (3). Thus, it seems likely that the antibodies can be used for in situ detection of ammonia oxidizers of the β-subclass in natural bacterial communities. First, evidence is given as the antibodies reacted with the AmoB in new strains of ammonia oxidizers isolated from building stones. Recent studies also proved in a similar approach that monoclonal antibodies recognizing the key enzyme of nitrite oxidizers are a useful tool for microbial ecological studies (1, 6).
In contrast, the antiserum did not react with the ammonia oxidizers belonging to the γ-subclass of Proteobacteria. This might be due to the low similarities between the AmoB of these bacteria to that of ammonia oxidizers belonging to the β-subclass of Proteobacteria. The phylogenetically related pMMO of methanotrophic bacteria also did not react with the antibodies. Indeed, the amino acid sequence of the AmoB of Nitrosococcus oceani (γ-proteobacteria) shows higher similarity (50 to 52%) to the pMMO sequence of Methylococcus capsulatus (γ-proteobacteria) than to the AmoB sequence (38 to 39%) of the ammonia oxidizers of the β-subclass of Proteobacteria (3).
The antibodies could be used to determine for the first time the amount of AmoB in cells of N. eutropha N904. It was shown that the total cell protein consisted of approximately 6% AmoB when cells were grown using standard substrate conditions (mineral medium containing 10 mM ammonium). During cell growth, the specific cellular amount of the AmoB was regulated by the ammonium concentration in the medium. When ammonium was limited, higher amounts of AmoB could be detected in cells of N. eutropha N904 in comparison to cells grown with standard concentrations. At ammonia concentrations below the Km value of the AMO (1.8 mM) (46), the low activity of the enzyme seemed to be compensated for by high amounts of the key enzyme. At high substrate concentrations the activity of the enzyme is maximal. Therefore, the cells were able to grow, although the enzyme concentration was reduced.
Organic compounds had no influence on the AmoB amount in cells of N. eutropha N904 as it was found for the nitrite oxidizer Nitrobacter. In Nitrobacter spp. a higher level of the nitrite oxidoreductase was observed in mixotrophically growing cells compared to cells growing in mineral medium (1).
Starved cells contained higher amounts of AmoB than actively growing cells, although they possessed far less ammonia oxidation activity. Previous studies also found considerable amounts of active AMO in starved cells of N. europaea (40, 59). Hence, our studies and these investigations indicate that the amount of AMO does not correlate with the activity of ammonia oxidation in Nitrosomonas. Although the AmoB was detected in high concentrations in cells of N. eutropha N904, N. europaea Freitag, and Nitrosomonas strain Dave after 1 year of ammonia starvation and their AMO remained active, the AMO seems not to be a constitutive enzyme. Sayavedra-Soto et al. (44) found that the mRNA of the AMO in cells of N. europaea was totally degraded a few hours after the depletion of ammonia. Moreover, it was shown that the endogenous respiration of N. cryotolerans cells decreased to undetectable levels under starvation conditions (27). Accordingly, the AMO seems to be strongly protected from degradation so that the energy supply is ensured as soon as ammonia is available. The increase of the AmoB amount in the cells during ammonia starvation might be due to the decline of unprotected proteins.
Similar results were also reported for the second key enzyme of the ammonia oxidation, the HAO. Using immunoblot analysis, the HAO level remained constant within 81 days of ammonia starvation in cells of N. europaea and a high amount of active HAO was detected (40, 59), while the mRNA was degraded during ammonia starvation (44).
This study demonstrated that the antibodies recognizing the AmoB could be applied successfully for physiological studies. Cytological analyses are in progress that will provide information about the localization of the enzyme by immunogold and immunofluorescence labeling. If the antibodies can be employed for isolation of the enzyme, there is even a good prospect for a detailed biochemical characterization of the AmoB protein.
ACKNOWLEDGMENTS
This work was supported by a grant from the Deutscher Akademischer Austauschdienst (DAAD, PROCOPE) and by the European Union (ENV4-CT98-0707).
We thank H.-P. Koops for contributing pure cultures of ammonia oxidizers, S. Bartosch for technical assistance and comments on the manuscript, and E. Spieck for the initiation of antibody development and for scientific discussions.
REFERENCES
- 1.Aamand J, Ahl T, Spieck E. Monoclonal antibodies recognizing nitrite oxidoreductase of Nitrobacter hamburgensis, N. winogradskyi, and N. vulgaris. Appl Environ Microbiol. 1996;62:2352–2355. doi: 10.1128/aem.62.7.2352-2355.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Alzerreca J J, Norton J M, Klotz M G. The amo operon in marine, ammonia-oxidizing γ-proteobacteria. FEMS Microbiol Lett. 1999;180:21–29. doi: 10.1111/j.1574-6968.1999.tb08773.x. [DOI] [PubMed] [Google Scholar]
- 4.Arciero D M, Vanneli T, Logan M, Hooper A B. Degradation of trichloroethylene by the ammonia-oxidizing bacterium Nitrosomonas europaea. Biochem Biophys Res Commun. 1989;159:640–643. doi: 10.1016/0006-291x(89)90042-9. [DOI] [PubMed] [Google Scholar]
- 5.Arciero D M, Hooper A B. Hydroxylamine oxidoreductase from Nitrosomonas europaea is a multimer of an octa-heme subunit. J Biol Chem. 1993;268:14645–14654. [PubMed] [Google Scholar]
- 6.Bartosch S, Wolgast I, Spieck E, Bock E. Identification of nitrite-oxidizing bacteria with monoclonal antibodies recognizing the nitrite oxidoreductase. Appl Environ Microbiol. 1999;65:4126–4133. doi: 10.1128/aem.65.9.4126-4133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bédard C, Knowles R. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol Rev. 1989;53:68–84. doi: 10.1128/mr.53.1.68-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belser L W, Schmidt E L. Serological diversity within a terrestrial ammonia-oxidizing population. Appl Environ Microbiol. 1978;36:589–593. doi: 10.1128/aem.36.4.589-593.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belser L W. Population ecology of nitrifying bacteria. Annu Rev Microbiol. 1979;16:309–333. doi: 10.1146/annurev.mi.33.100179.001521. [DOI] [PubMed] [Google Scholar]
- 10.Bergmann D J, Hopper A B. Sequence of the gene, amo B, for the 43 kDa polypeptide of ammonia monooxygenase of Nitrosomonas europaea. Biochim Biophys Res Commun. 1994;204:759–762. doi: 10.1006/bbrc.1994.2524. [DOI] [PubMed] [Google Scholar]
- 11.Bock E, Sundermeyer-Klinger H, Stackebrandt E. New facultative lithoautotrophic nitrite-oxidizing bacteria. Arch Microbiol. 1983;136:281–284. [Google Scholar]
- 12.Bock E, Koops H-P, Möller U C, Rudert M. A new facultatively nitrite oxidizing bacterium, Nitrobacter vulgaris sp. nov. Arch Microbiol. 1990;153:105–110. [Google Scholar]
- 13.Bradford M. A rapid and sensitive method for the quantification of microgram of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 14.Colby J, Stirling D I, Dalton H. The soluble methane monooxygenase of Methylococcus capsulatus (Bath): its ability to oxygenate n-alkanes, n-alkenes, ethers, and alicyclic, aromatic, and heterocyclic compounds. Biochem J. 1977;165:395–407. doi: 10.1042/bj1650395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol. 1995;164:16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- 16.Ensign S A, Hyman M R, Arp D J. In vitro activation of ammonia monooxygenase from Nitrosomonas by copper. J Bacteriol. 1993;175:1971–1998. doi: 10.1128/jb.175.7.1971-1980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green P N, Bousfield I J, Hood D. Three new Methylobacterium species: M. rhodesianum sp. nov., M. zatmani sp. nov., M. fuyisawanease sp. nov. Int J Syst Bacteriol. 1988;38:124–127. [Google Scholar]
- 18.Hollocher T C, Tate M E, Nicholas D J D. Oxidation of ammonia by Nitrosomonas europaea: definitive 18O-tracer evidence that hydroxylamine formation involves a monooxygenase. J Biol Chem. 1981;256:10834–10836. [PubMed] [Google Scholar]
- 19.Holmes A J, Costello A, Listrom M E, Murrell J C. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 20.Hubley J H, Thomson A W, Wilkinson J F. Specific inhibitors of methane oxidation in Methylosinus trichosporium. Arch Microbiol. 1975;102:199–202. [Google Scholar]
- 21.Hyman M R, Wood P M. Suicidal inactivation and labelling of ammonia mono-oxygenase by acetylene. Biochem J. 1985;227:719–725. doi: 10.1042/bj2270719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyman M R, Murton I B, Arp D J. Interaction of ammonia monooxygenase from Nitrosomonas europaea with alkanes, alkenes, and alkynes. Appl Environ Microbiol. 1988;54:3187–3190. doi: 10.1128/aem.54.12.3187-3190.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyman M R, Kim C Y, Arp D J. Inhibition of ammonia monooxygenase from Nitrosomonas europaea by carbon disulfide. Biochem J. 1990;172:4775–4782. doi: 10.1128/jb.172.9.4775-4782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hyman M R, Arp D J. 14C2H2- and 14CO2-labelling studies of the de novo synthesis of polypeptides by Nitrosomonas europaea during recovery from acetylene and light inactivation of ammonia monooxygenase. J Biol Chem. 1992;267:1534–1545. [PubMed] [Google Scholar]
- 25.Hynes R K, Knowles R. Inhibition of acetylene of ammonia oxidation in Nitrosomonas europaea. FEMS Microbiol Lett. 1978;4:319–321. doi: 10.1093/femsle/fnx068. [DOI] [PubMed] [Google Scholar]
- 26.Hynes R K, Knowles R. Effect of acetylene on autotrophic and heterotrophic nitrification. Can J Microbiol. 1982;28:334–340. [Google Scholar]
- 27.Johnstone B H, Jones R D. Physiological effects of long-term energy-source deprivation on the survival of a marine chemolithotrophic ammonia-oxidizing bacterium. Mar Ecol Prog Ser. 1988;49:295–303. [Google Scholar]
- 28.Jones R D, Morita R Y. Methane oxidation by Nitrosococcus oceanus and Nitrosomonas europaea. Appl Environ Microbiol. 1983;45:401–410. doi: 10.1128/aem.45.2.401-410.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klotz M G, Norton J M. Sequence of an ammonia monooxygenase subunit A-encoding gene from Nitrosospira sp. NpAV Gene. 1995;163:159–160. doi: 10.1016/0378-1119(95)00392-j. [DOI] [PubMed] [Google Scholar]
- 30.Klotz M G, Alzerreca J, Norton J M. A gene encoding a membrane protein exists upstream of the amo A/amo B genes in ammonia oxidizing bacteria: a third member of the amo operon? FEMS Microbiol Lett. 1997;150:65–73. doi: 10.1111/j.1574-6968.1997.tb10351.x. [DOI] [PubMed] [Google Scholar]
- 31.Klotz M G, Norton J M. Multiple copies of ammonia monooxygenase (amo) operons have evolved under biased AT/GC mutational pressure in ammonia-oxidizing autotrophic bacteria. FEMS Microbiol Lett. 1998;168:303–311. doi: 10.1111/j.1574-6968.1998.tb13288.x. [DOI] [PubMed] [Google Scholar]
- 32.Koops H-P, Harms H, Wehrmann H. Isolation of a moderate halophilic ammonia-oxidizing bacterium, Nitrosococcus mobilis nov. sp. Arch Microbiol. 1976;107:277–282. doi: 10.1007/BF00425339. [DOI] [PubMed] [Google Scholar]
- 33.Koops H-P, Böttcher B, Möller U C, Pommerening-Röser A, Stehr G. Description of a new species of Nitrosococcus. Arch Microbiol. 1990;154:244–248. [Google Scholar]
- 34.Koops H-P, Böttcher B, Möller U C, Pommerening-Röser A, Stehr G. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov. Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. J Gen Microbiol. 1991;137:1689–1699. [Google Scholar]
- 35.Kyse-Anderson J. Electroblotting of multiple gels: simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- 36.Laemmli U K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 38.Matulevich V A, Strom P F, Finstein M S. Length of incubation for enumerating nitrifying bacteria present in various environments. Appl Microbiol. 1975;29:265–268. doi: 10.1128/am.29.2.265-268.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McTavish H, Fuchs J A, Hooper A B. Sequence of the gene coding for ammonia-monooxygenase in Nitrosomonas europaea. J Bacteriol. 1993;175:2436–2444. doi: 10.1128/jb.175.8.2436-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nejidat A, Shmuely H, Abeliovich A. Effect of ammonia starvation on hydroxylamine oxidoreductase activity of Nitrosomonas europaea. J Biochem. 1997;121:957–960. doi: 10.1093/oxfordjournals.jbchem.a021679. [DOI] [PubMed] [Google Scholar]
- 41.Norton J M, Low J M, Klotz M G. The gene encoding ammonia monooxygenase subunit A exists in three nearly identical copies in Nitrosospira sp. NpAV FEMS Microbiol Lett. 1996;139:181–188. doi: 10.1111/j.1574-6968.1996.tb08200.x. [DOI] [PubMed] [Google Scholar]
- 42.Prior S D, Dalton H. Acetylene as a suicide substrate and active site probe for methane monooxygenase from Methylococcus capsulatus (Bath) FEMS Microbiol Lett. 1985;29:105–109. [Google Scholar]
- 43.Sanden B, Grunditz C, Hansson Y, Dalhammar G. Quantification and characterisation of Nitrosomonas and Nitrobacter using monoclonal antibodies. Water Sci Tech. 1994;29:1–6. [Google Scholar]
- 44.Sayavedra-Soto L A, Hommes N G, Russell S A, Arp D J. Induction of ammonia monooxygenase and hydroxylamine oxidoreductase mRNAs by ammonium in Nitrosomonas europaea. Mol Microbiol. 1996;20:541–548. doi: 10.1046/j.1365-2958.1996.5391062.x. [DOI] [PubMed] [Google Scholar]
- 45.Sayavedra-Soto L A, Hommes N G, Alzerreca J J, Arp D J, Norton J M, Klotz M G. Transcription of the amoC, amoA and amoB genes in Nitrosomonas europaea and Nitrosospira sp. NpAV FEMS Microbiol Lett. 1998;167:81–88. doi: 10.1111/j.1574-6968.1998.tb13211.x. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt I, Bock E. Anaerobic ammonia oxidation by cell-free extracts of Nitrosomonas eutropha. Antonie Leeuwenhoek. 1998;73:271–278. doi: 10.1023/a:1001572121053. [DOI] [PubMed] [Google Scholar]
- 47.Smorczewski W T, Schmidt E L. Numbers, activities, and diversity of autotrophic ammonia-oxidizing bacteria in a freshwater, eutrophic lake sediment. Can J Microbiol. 1991;37:828–833. [Google Scholar]
- 48.Spector T. Refinement of Coomassie-blue method of protein quantification. Ann Biochem. 1978;86:142–146. doi: 10.1016/0003-2697(78)90327-5. [DOI] [PubMed] [Google Scholar]
- 49.Spieck E, Meincke M, Bock E. Taxonomic diversity of Nitrosovibrio strains isolated from building sandstones. FEMS Microbiol Ecol. 1992;102:21–26. [Google Scholar]
- 50.Stüven R, Vollmer M, Bock E. The impact of organic matter on nitric oxide formation by Nitrosomonas europaea. Arch Microbiol. 1992;158:439–443. [Google Scholar]
- 51.Suzuki I, Kwok S-C. Cell-free ammonia oxidation by Nitrosomonas europaea extracts: effects of polyamines, Mg2+ and albumin. Biochem Biophys Res Commun. 1970;39:950–955. doi: 10.1016/0006-291x(70)90416-x. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki I, Kwok S-C, Dular U, Tsang D C Y. Cell-free ammonia-oxidizing system of Nitrosomonas europaea: general conditions and properties. Can J Biochem. 1981;59:477–483. doi: 10.1139/o81-066. [DOI] [PubMed] [Google Scholar]
- 53.Terry K R, Hooper A B. Hydroxylamine oxidoreductase: a 20-heme, 200,000 molecular weight cytochrome c with unusual denaturation properties which forms a 63,000 molecular weight monomer after heme removal. Biochemistry. 1981;20:7026–7032. doi: 10.1021/bi00527a039. [DOI] [PubMed] [Google Scholar]
- 54.Vanneli T, Logan M, Arciero D M, Hooper A B. Degradation of halogenated aliphatic compounds by the ammonia-oxidizing bacterium Nitrosomonas europaea. Appl Environ Microbiol. 1990;56:1169–1171. doi: 10.1128/aem.56.4.1169-1171.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Völsch A, Nader W F, Geiss H K, Nebe G, Birr C. Detection and analysis of two serotypes of ammonia-oxidizing bacteria in sewage plants by flow cytometry. Appl Environ Microbiol. 1990;140:153–158. doi: 10.1128/aem.56.8.2430-2435.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ward B B, Carlucci A F. Marine ammonia- and nitrite-oxidizing bacteria: serological diversity determined by immunofluorescence in sewage plants by flow cytometry. Appl Environ Microbiol. 1985;56:2430–2435. doi: 10.1128/aem.50.2.194-201.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watson S W, Waterbury J B. Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Arch Microbiol. 1971;77:203–230. [Google Scholar]
- 58.Whittenbury R, Phillips K C, Wilkinson J F. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 1970;61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 59.Wilhelm R, Abeliovich A, Nejidat A. Effect of long-term ammonia starvation on the oxidation of ammonia and hydroxylamine by Nitrosomonas europaea. J Biochem. 1998;124:811–815. doi: 10.1093/oxfordjournals.jbchem.a022184. [DOI] [PubMed] [Google Scholar]
- 60.Wood P M. Nitrification as a bacterial energy source. In: Prosser J I, editor. Nitrification. Oxford, England: IRL Press; 1986. pp. 39–62. [Google Scholar]