Abstract
Proteins in food and personal care products can pose a risk for an immediate immunoglobulin E (IgE)-mediated allergic response. Bioinformatic tools can assist to predict and investigate the allergenic potential of proteins. Here we present AllerCatPro 2.0, a web server that can be used to predict protein allergenicity potential with better accuracy than other computational methods and new features that help assessors making informed decisions. AllerCatPro 2.0 predicts the similarity between input proteins using both their amino acid sequences and predicted 3D structures towards the most comprehensive datasets of reliable proteins associated with allergenicity. These datasets currently include 4979 protein allergens, 162 low allergenic proteins, and 165 autoimmune allergens with manual expert curation from the databases of WHO/International Union of Immunological Societies (IUIS), Comprehensive Protein Allergen Resource (COMPARE), Food Allergy Research and Resource Program (FARRP), UniProtKB and Allergome. Various examples of profilins, autoimmune allergens, low allergenic proteins, very large proteins, and nucleotide input sequences showcase the utility of AllerCatPro 2.0 for predicting protein allergenicity potential. The AllerCatPro 2.0 web server is freely accessible at https://allercatpro.bii.a-star.edu.sg.
Graphical Abstract
Graphical Abstract.

AllerCatPro 2.0 predicts protein allergenicity potential with new features for identifying potential cross-reactivity, protein information, functionality, allergen information, sequence similarities to autoimmune allergens and low allergenic proteins, and clinical relevance.
INTRODUCTION
Evaluating the allergenic potential of proteins with the help of bioinformatic tools has gained increasing interest for foods, consumer and personal care products (1). However, the accurate prediction and evaluation of the allergenic potential of novel proteins is still a challenge. The computational approaches using the FAO/WHO guidelines of linear sequence window identity thresholds and short peptide hits for predicting protein allergenicity potential misclassify a large number of protein sequences as allergens (2). In addition, the FAO/WHO linear-window rule (3) and the current popular methods of PREAL (4), AllerHunter (5), AllergenFP (6), AllerTOPv2 (7) and AlgPred 2.0 (8) only achieved the overall accuracies from 51% (FAO/WHO rules (3)) to respectable 73% (7) on our benchmark datasets (2).
Many different factors can contribute to the overall allergenicity of proteins, such as: cleavage sites, protein stability, physico-chemical properties, and post-translational modifications (9). Nevertheless, a protein allergen still needs to be recognized by the immune system and thus, in order to sensitize an individual, induce a T cell and B cell response to produce allergen-specific Immunoglobulin (IgE) and/or, in order to elicit an IgE-mediated allergic response, interact with IgE on mast cells and basophils. These interactions and recognitions of allergenic proteins can be captured by their amino acid sequences and 3D structures. Here, we have developed AllerCatPro 2.0 web server using the computational method AllerCatPro established in our previous study (AllerCatPro 1.7) (2) to predict the allergenic potential of protein sequences based on similarity of both their amino acid sequences and 3D structures compared with the most comprehensive datasets of reliable proteins associated with allergenicity. Our AllerCatPro method was extensively benchmarked and compared with other popular methods for predicting protein allergenicity potential. Evaluation of the benchmark datasets used, AllerCatPro achieved an overall accuracy of 84%, significantly better than other methods (2).
Moreover, in this study we have improved AllerCatPro by enabling nucleotide alongside protein sequences as input and extending the dataset of 4180 (2) to 4979 proteins associated with allergenicity to improve the accuracy of AllerCatPro 2.0 for predicting protein allergenicity potential. We have also identified and added 165 human proteins associated with autoimmune diseases (in the following called ‘autoimmune allergens’) and 162 proteins of low allergenic potential (‘low allergenic proteins’). The latter were identified from a dataset derived from six abundant and commonly consumed protein sources, resulting in a list of 178 characterized proteins with no evidence for allergenicity, despite opportunities for human exposure (10). The autoimmune allergens were extracted and curated from the databases of WHO/International Union of Immunological Societies (IUIS), Comprehensive Protein Allergen Resource (COMPARE), Food Allergy Research and Resource Program (FARRP), UniProtKB and Allergome. For our allergen database, we carefully selected annotated allergens from available sources that provide further evidence and/or have established procedures for manual expert curation, and hence the number of false annotations should be marginal and not having significant influence on performance estimates.
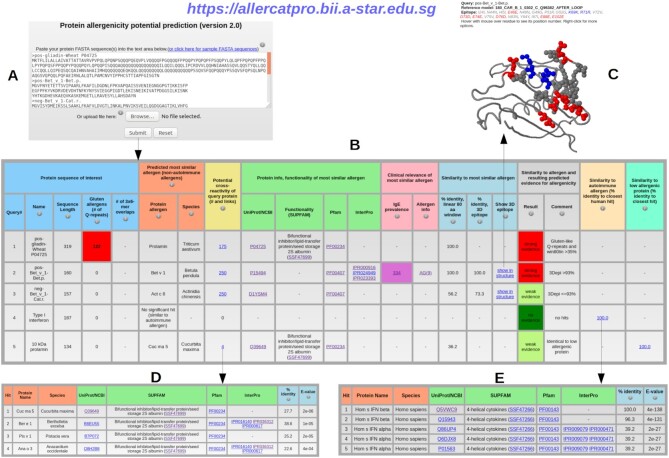
The prediction for the most similar protein allergen, similarity scores in protein sequence (‘% identity, linear 80 aa window’) and 3D structure (‘% identity, 3D epitope’), Gluten-like repeats of Glutamine (‘Gluten allergens (# of Q-repeats)’), and hexamer hits (‘# of 3 × 6-mer overlaps’) are based on the output of AllerCatPro 1.7 as presented earlier (2). For AllerCatPro 2.0, we now developed a more comprehensive output and extended the result to potential cross-reactivity, protein information (UniProt/NCBI) and functionality (Pfam, InterPro, SUPFAM), as well as the clinical relevance which refers to IgE prevalence data from Allergome and allergen information related to the most similar allergen (Figure 1). The allergen information refers to the allergenicity scoring used by Allergome and represents the number of positive evidence types annotated in the database of Allergome. Furthermore, we extended the applicability domain of AllerCatPro 2.0 to perform well with very long input sequences (>1000 amino acids), which was a limitation in the AllerCatPro 1.7. Various examples showcase the utility of the AllerCatPro 2.0 web server for predicting protein allergenicity potential from protein sequences as well as present the improvement of AllerCatPro 2.0 compared to AllerCatPro 1.7.
Figure 1.
Submitting one or more protein/nucleotide sequences in FASTA format (A) leads to the AllerCatPro 2.0 output table with the result for strong, weak or no evidence for allergenicity per protein based on corresponding workflow decisions and, in case of a hit, the possibility to view the most similar allergens with the detailed results for cross-reactivity, protein information (UniProt/NCBI), functionality (Pfam, InterPro, SUPFAM), as well as clinical relevance of IgE prevalence (Allergome) and allergen information (B), the most similar 3D surface epitope via links with the structural view showing identical epitope residues as beads colored as blue for positive charges, red for negative charges and gray for all other amino acid types (C). AllerCatPro 2.0 also identifies all similar allergens that have significant sequence similarity to the query protein and refers to the number with the link in potential cross-reactivity of the output table (D), as well as all possible similar autoimmune allergens displayed in the link (E) and all possible similar low allergens in the link of output table.
IMPLEMENTATION
Program overview
Our web server is developed using the AllerCatPro method established in our previous study (AllerCatPro 1.7) (2). The input of AllerCatPro 2.0 is the query protein/nucleotide sequences and the output of those queried sequences is the similarity to protein allergens and the resulting predicted strong, weak, or no evidence for allergenicity accompanied with a comment section to clarify the rule triggering the result. The allergenicity potential for the query protein is predicted using different measures of similarity of amino acid sequence and 3D structure compared with our dataset of known allergens. Since Gluten-like repeats of Glutamine (Gluten-like Q-repeats) are independent of any other similarity scores, the presence of Gluten-like Q-repeats is first evaluated in our workflow (Figure 2). The Gluten-like prediction of AllerCatPro 2.0 does not lead to ‘strong evidence’ unless there is similarity to a known protein allergens. Subsequently, AllerCatPro 2.0 checks the similarity of the queried sequence with representatives in our 3D model/structure database of known allergens. Currently, there are 714 representative protein allergens in our 3D model/structure database (2). If there is significant sequence similarity to the BLASTP search (11) for the input sequence against our 3D model/structure database of known allergens (E-value < 0.001), then the similarity of the 3D surface epitope is applied to predict as ‘strong evidence’ as the sequence identity of 3D surface epitope is >93% (this cutoff is 92% if Gluten-like Q-repeats are found in the query sequence) or as ‘weak evidence’ otherwise (Figure 2). If AllerCatPro 2.0 cannot find any structure hit, the linear-window approach is then applied to predict the queried sequence as protein allergen with strong evidence if the rule of 35% identity over 80 residues is found. After using the linear-window rule, and no hit is found, the hexamer hit approach with at least three short hexamers with known allergens is applied to evaluate the queried sequence (Figure 2). Finally, if AllerCatPro 2.0 still cannot find any hit, the prediction of ‘no evidence’ for allergenicity is assigned (Figure 2).
Figure 2.
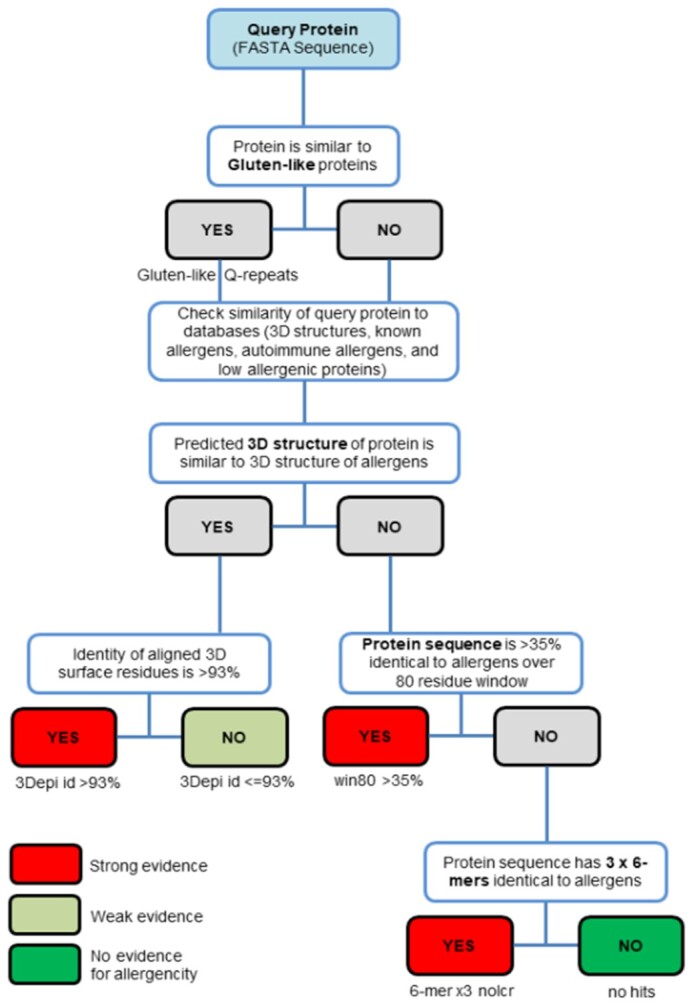
Decision workflow of AllerCatPro 2.0 from the query protein to the results of either strong, weak or no evidence for allergenic potential. AllerCatPro 2.0 checks the similarity of the query protein with 714 representatives in our 3D model/structure database of known allergens as well as the most comprehensive dataset of reliable proteins associated with allergenicity (4979 protein allergens). In addition to only comparing the similarity of the query protein with the dataset of known allergens in AllerCatPro 1.7, AllerCatPro 2.0 now predicts the similarity of the query sequence to datasets of 165 autoimmune allergens and 162 low allergenic proteins separately. If a significant sequence similarity is found, then AllerCatPro 2.0 identifies hits of similar proteins associated with autoimmune diseases and/or similar proteins of low allergenic potential and presents the sequence identity to the closest hit.
In addition to comparing the similarity of the query protein with the dataset of known allergens, our new web server AllerCatPro 2.0 now predicts the similarity of the query sequence to datasets of autoimmune allergens and low allergenic proteins separately. If a significant sequence similarity is found, then AllerCatPro 2.0 identifies hits of similar proteins associated with autoimmune diseases and/or similar proteins of low allergenic potential, and presents the sequence identity to the closest hit. As shown in Supplementary Figures S3A and S4, AllerCatPro 2.0 correctly predicts the query protein sequences as autoimmune allergens and low allergenic proteins while AllerCatPro misclassified these input sequences as allergenic proteins with ‘strong’ or ‘no evidence’ for allergenicity, respectively.
Recently, Sharma et al. have developed AlgPred 2.0 for predicting allergenic proteins based on a combination of different scores generated from machine learning (random forest) method, motif-emerging and with classes-identification, and BLAST search (8). Benchmark datasets are crucial to evaluate the performance of prediction models and they should include a set of allergens and a set of non-allergens with the same structure fold (2). Thus, AllerCatPro 2.0 was compared to AlgPred 2.0 on our benchmark datasets of 218 positive (known allergen and selected to be structurally non-redundant using our CLICK method (12–15)) and 212 negative (likely non-allergen) sequences. These benchmark datasets are available on the help page (https://allercatpro.bii.a-star.edu.sg/help.html) or via the help button at the AllerCatPro 2.0 website and are extracted from the benchmark datasets of 221 positive and 221 negative sequences (2) with the following adaptations: We removed one low allergenic protein (‘Spi o RuBisCo’ of Spinacia oleracea) and two proteins associated with autoimmune diseases in the positive set, and nine protein sequences related to the protein allergen ‘Ory s 14’ (‘Ory s 14’ of Oryza sativa) in the negative set due to its allergenic potential (16).
On our datasets, AllerCatPro 2.0 achieves an accuracy of 84.7% at 100% sensitivity and 68.9% specificity and Matthews correlation coefficient (MCC) of 0.727 (Supplementary Table S3) while AlgPred 2.0 only obtains an accuracy of 52.3% at 97.2% sensitivity and 6.1% specificity and MCC of 0.08. Also, we compare AllerCatPro 2.0 with other popular methods of PREAL (4), AllerHunter (5), AllergenFP (6), and AllerTOPv2 (7) on these benchmark datasets. We emphasize that these datasets are small but well representative of protein allergens with structures (2). The accuracy and MCC of AllerCatPro 2.0 are significantly better than those of the other methods with accuracies from 52.3% (AlgPred 2.0) to 75.0% (AllerTOPv2) and MCC from 0.08 (AlgPred 2.0) to 0.504 (AllerTOPv2) (Supplementary Figures S1A and B).
In addition, we perform AllerCatPro 2.0 on the larger validation datasets of 2003 positive (allergen) and 2015 negative (non-allergen) sequences from AlgPred 2.0 (8). These datasets are extracted from the validation datasets of 2015 positive and 2015 negative sequences of AlgPred 2.0 (https://webs.iiitd.edu.in/raghava/algpred2/stand.html) with the following adaptations: We remove one protein (P_10228) that is identical to low allergenic protein (Phoenix dactylifera) and eleven proteins (P_7089, P_7095, P_7102, P_7098, P_7101, P_7108, P_7125, P_7117, P_7114, P_7092, P_7111) associated with autoimmune diseases in the positive set. On these datasets, AllerCatPro 2.0 achieves high accuracy of 96.0% at 93.2% sensitivity and 98.8% specificity and MCC of 0.921 that are better than those of AllerCatPro 1.7 with accuracy of 93.0% at 91.1% sensitivity and 94.8% specificity and MCC of 0.860 (Supplementary Table S2). Since both AllerCatPro 2.0 and AllerCatPro 1.7 use the same algorithm and measures of similarity of amino acid sequence and 3D structure compared with the database of known allergens, the improvements of accuracy and MCC of AllerCatPro 2.0 compared to those of AllerCatPro 1.7 are due to the 1132 sequences of protein allergens, low allergenic proteins, and autoimmune allergens newly included into the database of AllerCatPro 2.0.
We also test AllerCatPro 2.0 on all recent IUIS allergens (twenty proteins) that has been created/modified from December 2021 to April 2022. For these new IUIS allergens, AllerCatPro 2.0 predicts their allergenicity potential correctly for fourteen proteins with ‘strong evidence’, five proteins with ‘weak evidence’ and incorrectly for only one protein with 'no evidence’.
Furthermore, we perform AllerCatPro 2.0 with and without using 3D structure similarity on the benchmark datasets of 218 allergens and 212 non-allergens with the same structure fold. On these benchmark datasets, AllerCatPro 2.0 achieves accuracy of 84.7% and MCC of 0.727 while AllerCatPro 2.0 without using 3D structure similarity only obtains accuracy of 52.8% and MCC of 0.148 (Supplementary Table S3). These results indicate that 3D structure similarity contributes significantly to the performance of AllerCatPro 2.0.
Our all datasets are provided on the help page (https://allercatpro.bii.a-star.edu.sg/help.html).
Server description
Input
Query sequences can be submitted by specifying their sequences in FASTA format, or by uploading a FASTA file of sequences (Figure 1A). The FASTA format has a description line starting with ‘>’ for each entry followed by one or more lines of sequence. In our AllerCatPro 2.0 web server, we have improved the AllerCatPro method to allow users to input nucleotide sequences (Supplementary Figures S6A and B). In addition, AllerCatPro 2.0 has been developed to perform well with very long input sequences (>1000 amino acids) and allow users to submit more than 50 protein/nucleotide sequences at a time, which were limitations of AllerCatPro 1.7.
A detailed explanation of input sequences for predicting the allergenicity potential of AllerCatPro 2.0 is provided on the help page (https://allercatpro.bii.a-star.edu.sg/help.html).
Output
The output of AllerCatPro 2.0 for input protein/nucleotide sequences is a result table following the workflow in the ‘Program Overview’ section and allergenicity prediction for one protein per line (Figure 1). The output table presents the detailed results, i.e. Gluten-like Q-repeats, hexamer hits, the most similar allergen (in case there is significant sequence similarity), potential hits for cross-reactivity, protein information (UniProt/NCBI), functionality (Pfam, InterPro, SUPFAM), as well as the clinical relevance of IgE prevalence (derived from Allergome) and allergen information related to the most similar allergen (Figure 1B). The IgE prevalence result shows the total number of studied individuals used for IgE tests and provide also the link to the ‘Epidemiology from Literature’ page on the Allergome website. The allergen information for the most similar allergen refers to the allergenicity scoring used by Allergome and represents the number of positive evidence types annotated in the database of Allergome (e.g. Functional Test, Non-Functional Test, Skin Test, Conjunctival Provocation Test, Nasal Provocation Test, Bronchial Provocation Test, Oral Challenge, Epidemiology from Literature, ReTiME). In addition, this allergen information is accompanied with an annotation, if the most similar allergen is a known food or insect allergen and related to anaphylaxis.
AllerCatPro 2.0 also checks the similarity score of the query protein with the most similar allergen based on the linear-window approach and identifies the similarity on a 3D structure level, if there is significant sequence similarity based on the BLASTP search (11) for the query protein against the 3D model/structure database of known allergens. These results are shown in the output table in case of a 3D similarity hit (Figure 1B). If the 3D surface epitope similarity is found, AllerCatPro 2.0 presents a link to view the potential 3D epitope with the highest percent amino acid identity relative to the query and visualizes on the closest known 3D structure (Figure 1C). Since charge similarity can be important, positions of identical positively charged residues of 3D epitope are shown as blue, negatively charged as red and all other identical residues as grey beads. The 3D rendering of the epitope and the closest known structure is displayed using JSMol (http://www.jmol.org/). Finally, AllerCatPro 2.0 classifies the query protein based on the weight of evidence coming from the different levels of similarity to known allergens (Gluten-like Q-repeats, 3D epitopes, linear-window rule, and hexamer hits). A red label of ‘strong evidence’ presents that there is a clear similarity to known allergens. In contrast, a light green label of ‘weak evidence’ shows some similarity to known allergens but with clear divergence in relevant sequence and structure features (Figure 1B). The dark green label of ‘no evidence’ means that no significant similarity of sequence and structure features has been found against our current most comprehensive dataset of 4979 known allergens (Figure 1B).
Moreover, instead of providing only the most similar allergen for the query protein in AllerCatPro 1.7, AllerCatPro 2.0 now identifies all similar allergens within the AllerCatPro 2.0 dataset that have significant sequence similarity to the query protein (E-value of BLASTP search < 0.001) in the column ‘Potential cross-reactivity of query protein (# and links)’ as a number and a link to the according list of allergens (Figure 1B). The list, which opens in a separate browser tab, shows protein information (UniProt/NCBI), functionality (Pfam, InterPro, SUPFAM), sequence identity and E-value of all similar allergens (Figure 1D). In order to differentiate between typical environmental allergens versus autoimmune allergens as well as increasing the confidence in predicting the absence of allergenicity, AllerCatPro 2.0 now provides the similarity of the query proteins with the datasets of autoimmune allergens as well as low allergenic proteins. If there is significant sequence similarity, then AllerCatPro 2.0 finds hits of similar proteins associated with autoimmune diseases and/or similar proteins of low allergenic potential and presents the sequence identity to the closest hit with the link to a separate browser tab showing protein information, functionality, sequence identity, and E-value of all hits in the output table (Figure 1E).
The output table also includes a download link for the results of the query protein in comma-separated CSV format which can be opened by Excel or other popular spreadsheet programs. A detailed explanation of the output for predicting allergenicity potential of AllerCatPro 2.0 is provided on the help page (https://allercatpro.bii.a-star.edu.sg/help.html).
RESULTS
Case study 1: differentiation of allergenic from low allergenic profilin proteins
The profilin protein family is one of the biggest families associated with food and pollen allergies, profilins are ubiquitous and highly conserved with sequence identities of 70% to 85% among allergenic plant profilins (17). Profilin is a clinically relevant aeroallergen and given its ubiquity, sensitized individuals might react to multiple profilin-containing sources with potentially even more severe symptoms, which makes this protein family highly relevant for both diagnosis and treatment of patients with plant food and pollen allergy (18).
In this example, we analyse and predict the allergenicity potential of different profilin proteins using AllerCatPro 2.0. Figure S2A in Supplementary shows all four pollen profilins from European white birch (Betula pendula), olive (Olea europaea), Timothy (Phleum pratense), and rice (Oryza sativa) are classified correctly as allergenic proteins with ‘strong evidence’ for allergenicity. Among these four proteins, three pollen profilins of Betula pendula, Olea europaea, Phleum pratense demonstrate high numbers of individuals tested for allergen-specific IgE (Supplementary Figure S2A), which indicates clinical relevance for these allergens. Figure S2B in Supplementary displays the 3D model of pollen profilin from Betula pendula based on the similarity of the 3D surface epitope. In this 3D model, blue, red and grey beads present positions of identical positively, negatively charged and the other identical residues of the 3D epitope, respectively (Supplementary Figure S2B).
As shown in Figure S2C in Supplementary, AllerCatPro 2.0 predicts correctly allergenic plant profilins from peach, apple, hazelnut, potato, spinach, and Para rubber tree with ‘strong evidence’ for allergenicity. All except the potato allergen Sola t 8, show high numbers of individuals tested for allergen-specific IgE.
For a set of five low allergenic profilins from yeast, human, cow, chicken, and fungus, AllerCatPro 2.0 predicts their allergenicity potential correctly with ‘weak evidence’ (Supplementary Figure S2D). On this set, AllerCatPro 1.7 overpredicted two profilins from human and cow with ‘strong evidence’ for allergenicity.
Case study 2: identification of autoimmune allergens
Many human proteins such as HLA (human leukocyte antigen)-DR and type I interferons importantly contribute to the pathogenesis of autoimmune diseases for type-1 diabetes, rheumatoid arthritis, and systemic lupus erythematosus (19,20). In this example, AllerCatPro 2.0 is used to identify the similarity to human proteins of HLA-DR alpha, alpha-galactosidase, nascent polypeptide-associated complex subunit alpha, granulocyte colony-stimulating factor, B-cell CLL/lymphoma 7 protein family member B, and type I interferon as autoimmune allergens. Since there are significant sequence similarity in BLASTP search for these input sequences against our dataset of autoimmune allergens, AllerCatPro 2.0 identified these proteins with 100% similarity to proteins associated with autoimmune diseases and shows in the output table that no significant hit (‘no evidence’) for environmental (non-human) allergens have been found (Supplementary Figure S3A). Compared to AllerCatPro 2.0, AllerCatPro 1.7 did not distinguish between environmental and autoimmune allergens, but assigned query proteins similar or identical to human proteins as allergens with ‘strong evidence’. AllerCatPro 2.0 now can identify if a protein is similar to environmental allergens and/or similar to human proteins, which provides additional information for users.
In addition, AllerCatPro 2.0 identifies all proteins from the dataset of autoimmune allergens with significant sequence similarity to the query protein sequences. Figure S3B in Supplementary shows the list of related human protein sequences (all related to interferon alpha and beta) that are associated with autoimmune diseases and have significant sequence similarity to the input sequence of type I interferon.
Case study 3: identification of proteins with low allergenic potential
Since different levels of similarity are applied to identify evidence for allergenic potential (Gluten-like Q-repeats, 3D epitopes, linear-window rule, and hexamer hits) in AllerCatPro and the dataset of known allergenic proteins grows with each new version, the likelihood to predict similarity with weak evidence (especially due to the linear-window rule) to a query protein increases steadily. However, not all proteins predicted with weak evidence for allergenicity are necessarily of allergenic concern. In order to improve the output and prediction of low allergenic potential in AllerCatPro 2.0, we implemented a dataset of proteins derived from our previous publication (10) for which we have confidence that these proteins have only a low allergenic potential. In this example, we use AllerCatPro 2.0 to predict the similarity of 10 kDa prolamin from rice (Oryza sativa), a flower-specific gamma-thionin-like protein from tomato (Solanum lycopersicum), ribulose bisphosphate carboxylase from spinach (Spinacia oleracea), late embryogenesis abundant proteins 14, 17, 19 and group 3 from wheat (Triticum aestivum) to low allergenic proteins. The results show that AllerCatPro 2.0 provides the comment ‘Identical to low allergenic protein’ in addition to the ‘weak evidence’ for allergenic potential (Supplementary Figure S4), which is triggered if AllerCatPro 2.0 identifies that they have very high sequence similarity and identity to other proteins in our dataset of low allergenic proteins.
Nevertheless, this is the very first time that a prediction tool provides the feature of predicting low allergenic potential of proteins. Science is still at the early beginning to better understand what makes a protein not an allergen and thus, every user needs to be aware that such result needs to be interpreted cautiously. Here, there feature ‘potential cross-reactivity of query protein’, which provides additional results on sequence similarity to other known allergens, may be important to evaluate in addition to the result of similarity to low allergenic proteins.
Compared to AllerCatPro 2.0, the previous version AllerCatPro 1.7 inappropriately assigned e.g. the input protein of ribulose bisphosphate carboxylase, which can be considered as low allergenic protein (10), as protein with ‘strong evidence’ for allergenicity. Additionally, in case of ‘no evidence’ or ‘weak evidence’ for allergenicity, the version 1.7 did not provide any additional evidence to substantiate low allergenic potential. AllerCatPro 2.0 now provides the similarity to low allergenic proteins, which are found to be appropriate to be used as negative controls to aid the development and validation of alternative methods for more robust testing strategies (10).
Case study 4: Prediction of similarity to allergens for very large proteins
One of the limitations of AllerCatPro 1.7 was the low performance and high running time for very large proteins (>1000 amino acids). AllerCatPro 2.0 is now able to predict protein allergenicity potential for very long input sequences in shorter time. As shown in Figure S5A in Supplementary, AllerCatPro 2.0 predicts allergenic potential for a set of very large proteins with their lengths varying from 834 to 3967 residues. The average running time for each protein was approximately 8.5 seconds. Similarity towards allergenic proteins for these large proteins is predicted using the usual levels based on Gluten-like Q-repeats, 3D epitopes, linear-window rule, but excluding the hexamer rule as it is overpredicting similarity for large proteins and takes more time to finalize a run.
Among the input sequences shown in Figure S5A, eleven proteins (all except Query# 9, 11, 13) are predicted to be similar to an allergen. For ten proteins (Query# 1 to 8, 12, and 14) allergen information is shown for the most similar protein allergen with the number of positive evidence types indicating allergenicity annotated in the database of Allergome. Two proteins (Query# 2 and 6) are similar to allergens for which more than 100 individuals have been tested for allergen-specific IgE (Supplementary Figure S5A).
Figure S5B in Supplementary displays the 3D structure for the input protein with the UniProtID A0A5F4DEY2 (Query# 2) and its 3D similarity to the most similar protein allergen ‘Cuc p AscO’ with blue and grey beads showing positions of identical positively charged and the other identical residues, respectively.
Case study 5: extended input capability to nucleotide sequences
In the new version of AllerCatPro 2.0, we have improved AllerCatPro 1.7 by enabling the input of nucleotide sequences in addition to protein sequences. For this case study, we apply AllerCatPro 2.0 to predict the allergenic potential of three known allergens (ABD51778, CAB02154, AAA34278) from honey bee (Apis mellifera), European white birch (Betula pendula), and wheat (Triticum aestivum) using their nucleotide sequences as input (Supplementary Figure S6A). As shown in Figure S6B in Supplementary, AllerCatPro 2.0 classifies correctly as allergens with ‘strong evidence’ for all these nucleotide input sequences. The predicted most similar allergen for the nucleotide sequence CAB02154 from Betula pendula is annotated with a high number of individuals tested for IgE prevalence. AllerCatPro 2.0 predicts the nucleotide input sequence (AAA34278) from Triticum aestivum with strong evidence for allergenicity due to the presence of 122 Gluten-like Q-repeats and 100% sequence identity over the window of 80 amino acid residues are found towards the most similar protein allergen Prolamin (Supplementary Figure S6B).
CONCLUSION
Here, we present AllerCatPro 2.0, a web server that can be used to predict protein allergenicity potential for protein or nucleotide sequences with outperforming accuracies (84.7% at 100% sensitivity and 68.9% specificity on the difficult benchmark datasets) and a user-friendly interface with new features that provide detailed results for potential cross-reactivity, protein information (UniProt/NCBI), functionality (Pfam, InterPro, SUPFAM), as well as the clinical relevance with regards to IgE prevalence and allergen information related to the most similar allergen. To the best of our knowledge, this is the first web server for predicting protein allergenicity potential equipped with the capability for identifying the clinical relevance, allergen information, sequence similarities to autoimmune allergens and low allergenic proteins that help assessors making informed decisions.
AllerCatPro 2.0 is developed based on our previous established AllerCatPro method (AllerCatPro 1.7) (2). AllerCatPro 2.0 predicts protein allergenicity potential using similarity of both their amino acid sequences and 3D structures towards the most comprehensive datasets (4979 protein allergens, 162 low allergenic proteins, and 165 autoimmune allergens) of reliable proteins associated with allergenicity from the databases of WHO/IUIS, COMPARE, FARRP, UniProtKB and Allergome.
Various examples of profilins, autoimmune diseases, low allergenic proteins, very large proteins, and nucleotide input sequences showcase the utility of AllerCatPro 2.0 for predicting protein allergenicity potential from protein sequences as well as demonstrate the improvement of AllerCatPro 2.0 compared to our previous version AllerCatPro 1.7.
Supplementary Material
ACKNOWLEDGEMENTS
This study has been supported by the Agency of Science, Technology and Research (A*STAR) Singapore and the Procter & Gamble (P&G) company. We would like to acknowledge the Biomedical Research Council (BMRC)-Economic Development Board (EDB), Industry Alignment Fund, A*STAR, Singapore for support. Minh N. Nguyen would like to thank HBMS Domain Industry Alignment Fund Pre-Positioning (IAF-PP), A*STAR (H2001a0P14) for funding. Vachiranee Limviphuvadh would like to thank Industry Alignment Fund BMRC, A*STAR/P&G (APG2013/096) for funding.
Contributor Information
Minh N Nguyen, Bioinformatics Institute, 30 Biopolis Street, #07-01, Matrix 138671, Singapore; IFCS Programme, Singapore Institute for Food and Biotechnology Innovation, Agency for Science, Technology and Research, Singapore.
Nora L Krutz, NV Procter & Gamble Services Company SA, Strombeek-Bever, Belgium.
Vachiranee Limviphuvadh, Bioinformatics Institute, 30 Biopolis Street, #07-01, Matrix 138671, Singapore; IFCS Programme, Singapore Institute for Food and Biotechnology Innovation, Agency for Science, Technology and Research, Singapore.
Andreas L Lopata, Bioinformatics Institute, 30 Biopolis Street, #07-01, Matrix 138671, Singapore; Molecular Allergy Research Laboratory, Australian Institute of Tropical Health and Medicine, James Cook University, Townsville, QLD, Australia; Tropical Futures Institute, James Cook University, Singapore.
G Frank Gerberick, GF3 Consultancy LLC, West Chester, OH, USA.
Sebastian Maurer-Stroh, Bioinformatics Institute, 30 Biopolis Street, #07-01, Matrix 138671, Singapore; IFCS Programme, Singapore Institute for Food and Biotechnology Innovation, Agency for Science, Technology and Research, Singapore; Department of Biological Sciences, National University of Singapore, 14 Science Drive 4, 117543, Singapore.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agency for Science, Technology and Research (A*STAR) HBMS Domain Industry Alignment Fund Pre-Positioning (IAF-PP) [H2001a0P14]; Industry Alignment Fund BMRC/P&G [APG2013/096]. Funding for open access charge: OC/EFSA/GMO/2021/04.
Conflict of interest statement. None declared.
REFERENCES
- 1. Krutz N.L., Kimber I., Maurer-Stroh S., Gerberick G.F.. Determination of the relative allergenic potency of proteins: hurdles and opportunities. Crit. Rev. Toxicol. 2020; 50:521–530. [DOI] [PubMed] [Google Scholar]
- 2. Maurer-Stroh S., Krutz N.L., Kern P.S., Gunalan V., Nguyen M.N., Limviphuvadh V., Eisenhaber F., Gerberick G.F.. AllerCatPro-prediction of protein allergenicity potential from the protein sequence. Bioinformatics. 2019; 35:3020–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Organization W.H., Nations F.A.O. of the U. and Joint FAO/WHO Expert Consultation on Allergenicity of Foods Derived from Biotechnology 2001: Rome, I . Evaluation of allergenicity of genetically modified foods: report of a Joint FAO/WHO Expert Consultation on Allergenicity of Foods Derived from Biotechnology. 2001; Rome: 22–25 January 2001 Food and Agriculture Organization of the United Nations. [Google Scholar]
- 4. Wang J., Zhang D., Li J.. PREAL: prediction of allergenic protein by maximum relevance minimum redundancy (mRMR) feature selection. BMC Syst. Biol. 2013; 7:S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Muh H.C., Tong J.C., Tammi M.T.. AllerHunter: a SVM-Pairwise system for assessment of allergenicity and allergic cross-reactivity in proteins. PLoS One. 2009; 4:e5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dimitrov I., Naneva L., Doytchinova I., Bangov I.. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014; 30:846–851. [DOI] [PubMed] [Google Scholar]
- 7. Dimitrov I., Bangov I., Flower D.R., Doytchinova I.. AllerTOP v.2–a server for in silico prediction of allergens. J. Mol. Model. 2014; 20:2278. [DOI] [PubMed] [Google Scholar]
- 8. Sharma N., Patiyal S., Dhall A., Pande A., Arora C., Raghava G.P.S.. AlgPred 2.0: an. improved method for predicting allergenic proteins and mapping of IgE epitopes. Brief. Bioinform. 2021; 22:bbaa294. [DOI] [PubMed] [Google Scholar]
- 9. Huby R.D., Dearman R.J., Kimber I.. Why are some proteins allergens. Toxicol. Sci. 2000; 55:235–246. [DOI] [PubMed] [Google Scholar]
- 10. Krutz N.L., Winget J., Ryan C.A., Wimalasena R., Maurer-Stroh S., Dearman R.J., Kimber I., Gerberick G.F.. Proteomic and bioinformatic analyses for the identification of proteins with low allergenic potential for hazard assessment. Toxicol. Sci. 2019; 170:210–222. [DOI] [PubMed] [Google Scholar]
- 11. Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nguyen M.N., Madhusudhan M.S.. Biological insights from topology independent comparison of protein 3D structures. Nucleic Acids Res. 2011; 39:e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nguyen M.N., Tan K.P., Madhusudhan M.S.. CLICK–topology-independent comparison of biomolecular 3D structures. Nucleic Acids Res. 2011; 39:W24–W8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nguyen M.N., Sim A.Y.L., Wan Y., Madhusudhan M.S., Verma C.. Topology independent comparison of RNA 3D structures using the CLICK algorithm. Nucleic Acids Res. 2017; 45:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nguyen M.N., Verma C.S., Zhong P.. AppA: a web server for analysis, comparison, and visualization of contact residues and interfacial waters of antibody-antigen structures and models. Nucleic Acids Res. 2019; 47:W482–W489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Asero R., Amato S., Alfieri B., Folloni S., Mistrello G.. Rice: another potential cause of food allergy in patients sensitized to lipid transfer protein. Int. Arch. Allergy Immunol. 2007; 143:69–74. [DOI] [PubMed] [Google Scholar]
- 17. Radauer C., Breiteneder H.. Pollen allergens are restricted to few protein families and show distinct patterns of species distribution. J. Allergy Clin. Immunol. 2006; 117:141–147. [DOI] [PubMed] [Google Scholar]
- 18. Rodríguez Del Río P., Díaz-Perales A., Sánchez-García S., Escudero C., Ibáñez M.D., Méndez-Brea P., Barber D.. Profilin, a change in the paradigm. J. Investig. Allergol. Clin. Immunol. 2018; 28:1–12. [DOI] [PubMed] [Google Scholar]
- 19. Gough S.C.L., Simmonds M.J.. The HLA region and autoimmune disease: associations and mechanisms of action. Curr. Genomics. 2007; 8:453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Crow M.K. Autoimmunity: interferon α or β: which is the culprit in autoimmune disease. Nat. Rev. Rheumatol. 2016; 12:439–440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.