Abstract
Background
Genes implicated in the Golgi and endosomal trafficking machinery are crucial for brain development, and mutations in them are particularly associated with postnatal microcephaly (POM).
Methods
Exome sequencing was performed in three affected individuals from two unrelated consanguineous families presenting with delayed neurodevelopment, intellectual disability of variable degree, POM and failure to thrive. Patient-derived fibroblasts were tested for functional effects of the variants.
Results
We detected homozygous truncating variants in ATP9A. While the variant in family A is predicted to result in an early premature termination codon, the variant in family B affects a canonical splice site. Both variants lead to a substantial reduction of ATP9A mRNA expression. It has been shown previously that ATP9A localises to early and recycling endosomes, whereas its depletion leads to altered gene expression of components from this compartment. Consistent with previous findings, we also observed overexpression of ARPC3 and SNX3, genes strongly interacting with ATP9A.
Conclusion
In aggregate, our findings show that pathogenic variants in ATP9A cause a novel autosomal recessive neurodevelopmental disorder with POM. While the physiological function of endogenous ATP9A is still largely elusive, our results underline a crucial role of this gene in endosomal transport in brain tissue.
Keywords: congenital, hereditary, and neonatal diseases and abnormalities, genetics, medical, sequence analysis, DNA, sequence analysis, RNA
Introduction
Intellectual disability (ID) has a prevalence of approximately 1% in the general population. It is often diagnosed in childhood due to neurodevelopmental delay (NDD) and represents one of the most common paediatric conditions.1 Most ID/NDD disorders occur in combination with additional malformations or other clinical features, such as behavioural abnormalities, epilepsy and microcephaly. Using exome sequencing (ES), a causative genetic variant can be detected in about 40%–60% of cases.2 In particular, the discovery of new disease genes has markedly increased the diagnostic yield in recent years.3 Currently, more than 1300 genes are listed as primary ID-associated genes (ID-AGs, SysID database) and approximately 25% of these genes are associated with microcephaly.4 Proteins encoded by microcephaly-associated genes play a role in diverse biological processes and molecular pathways such as transcriptional regulation, DNA repair, microtubule organisation and endosome regulation.5 Genes encoding proteins of the Golgi and endosomal trafficking machinery are of particular interest because they are crucial for brain development, and mutations in these genes are associated with postnatal microcephaly (POM).6
The endosomal genes include the family of P4-ATPases, which regulates asymmetric membrane lipid distribution in eukaryotic cells by ATP-dependent translocation of phospholipids from the exofacial to the cytosolic leaflet.7 8 Active lipid remodelling of the plasma membrane and intracellular membranes is a critical organisational prerequisite for various cellular processes such as vesicle trafficking, cell signalling and neuronal cell survival.9 The P4-ATPase-mediated flipping of phospholipids initiates vesicle biogenesis and facilitates the formation of various transport carriers.10 The human genome encodes 14 members of the P4-ATPase family that differ in substrate specificity and tissue expression and are divided into five subclasses.11 12 So far, three P4-ATPases (ATP8A2, ATP8B1 and ATP11C) have been linked to genetic diseases. Pathogenic variants in ATP8A2 cause autosomal recessive ‘cerebellar ataxia, mental retardation and disequilibrium syndrome 4’ (MIM: 615268); ATP8B1 is associated with autosomal recessive and dominant cholestasis (MIM: 243300, MIM: 147480 and MIM: 211600), and mutations in ATP11C have been shown to be causal for X-linked haemolytic anaemia (MIM: 301015).13–15
Using ES, we identified biallelic truncating variants in ATP9A (ATPase phospholipid transporting 9A, HGNC:13540) in three children affected with POM and ID in two unrelated families. ATP9A and ATP9B in humans and its orthologues TAT-5 (c. elegans) and Neo1 (s. cerevisiae) form the phylogenetic distinct subclass 2 of the P4-ATPase family.11 16 17 Phospholipid translocase activity has not yet been directly proven for this distinct subclass.18
ATP9A is located on chromosome 20q13.2 (GRCh37/hg19). The canonical transcript (ENST00000338821.5, NM_006045.3) consists of 28 exons, encodes a 1047 residue protein and is mainly expressed in the brain. Detailed knowledge about the function and subcellular distribution of endogenous ATP9A in physiologically relevant tissue is lacking. In HeLa cells, overexpressed ATP9A localises to early/recycling endosomes and the trans-Golgi network (TGN) and is required for endocytic recycling of the transferrin receptor to the plasma membrane.19 In contrast to ATP9A, ATP9B contains a specific N-terminal region and localises exclusively to the TGN.17 ATP9A as its orthologues TAT-5 and Neo1 form a highly conserved complex with DOP1B (DOP1 leucine zipper like protein B, HGNC:21194) and MON2 (MON2 homologue, regulator of endosome-to-Golgi trafficking, HGNC:29177) and are functionally intertwined with the SNX3-retromer (SNX3, sorting nexin 3, HGNC:11174).20–22 Gene expression analysis in an ATP9A knockdown cell line showed a significant upregulation and downregulation of 75 and 50 genes, respectively, where ARPC3 (actin-related protein 2/3 complex subunit 3, HGNC:706) was one of the most strongly upregulated genes (3.5-fold overexpression) and CORO1A (coronin 1A, HGNC:2252) was the most significantly downregulated (2.0-fold). In addition, as a result of its role in endosome trafficking pathways, ATP9A appears to inhibit the secretion of exosomes at the plasma membrane, possibly due to structural alteration of the actin cytoskeleton.23
Here we show that individuals with biallelic truncating ATP9A variants display NDD, POM and failure to thrive. To the best of our knowledge, this is the first time that mutations in ATP9A are described as causative for a human phenotype.
Material and methods
ES and segregation analysis
Individual A-II-1 was included in TRANSLATE-NAMSE, an innovation project of the German Federal Joint Committee to improve the care of patients with rare diseases. Genomic DNA was isolated from peripheral blood and enriched with SureSelect Human All Exon V6 (Agilent) for ES on an Illumina system. Reads were aligned to human genome build GRCh37/hg19. Sequence reads were called and analysed according to an in-house standard operating procedure using the VarFish platform.24 In brief, variants were filtered by minor allele frequency, mode of inheritance, predicted functional impact (i.e., missense, nonsense and splicing) and ranked by their gene-phenotype similarity score (exomiser: HiPhive)25 and pathogenicity score (CADD, MutationTaster).26 27
In family B, genomic DNA from individual B-II-1 was used for library preparation (kit: Nextera DNA Flex). Subsequently, ES was performed on a NextSeq 550 (Illumina). Sequence alignment of raw fastq files to the human reference sequence (GRCh37/hg19 assembly) and variant calling was performed using the DRAGEN Germline Pipeline V.3.2.8 on Illumina BaseSpace (https://basespace.illumina.com/). Variant annotation and filtering was performed using VarSeq V.2.2 (Golden Helix, Bozeman, Montana, USA; www.goldenhelix.com).
Segregation analysis was performed via Sanger sequencing in family members of both families.
The GeneMatcher tool allowed the identification of these two families.28
Homozygosity mapping
Due to the known parental consanguinity, homozygosity mapping was performed using AutozygosityMapper, an updated version of HomozygosityMapper.29 For homozygosity mapping, a joint variant calling was performed on the BAM files of the two individuals (A-II-1 and B-II-1). The resulting VCF file was analysed by AutozygosityMapper using the default settings. Different shared homozygous regions could be detected. Genome-wide runs of homozygosity and the genotypes surrounding the ATP9A gene are shown in online supplemental figure S1.
jmedgenet-2021-107843supp001.pdf (279.5KB, pdf)
Dermal fibroblasts, splice site analysis and mRNA expression
For RNA extraction, cells from the affected individuals and controls were seeded in a confluent status of 2×105 cells per well. After 3 days, cells were lysed in TRI Reagent, and total RNA was prepared using Direct-zol RNA Miniprep Kit (Zymo Research). Total cDNA was reverse transcribed by RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas). Quantitative PCR (qPCR) was performed using EVAgreen (Solis BioDyne) on a QuantStudio V.3 Real-Time-PCR System (Thermo Fisher Scientific). ATP9A, ARPC3 and SNX3 expression levels were measured using the comparative C T method in comparison to GAPDH. Primer sequences for qPCR and reverse transcription PCR (RT-PCR) are provided in online supplemental tables S1 and S2.
To analyse the consequence of the splice site variant in B-II-1, RT-PCR amplicons were Sanger sequenced.
Results
Clinical report
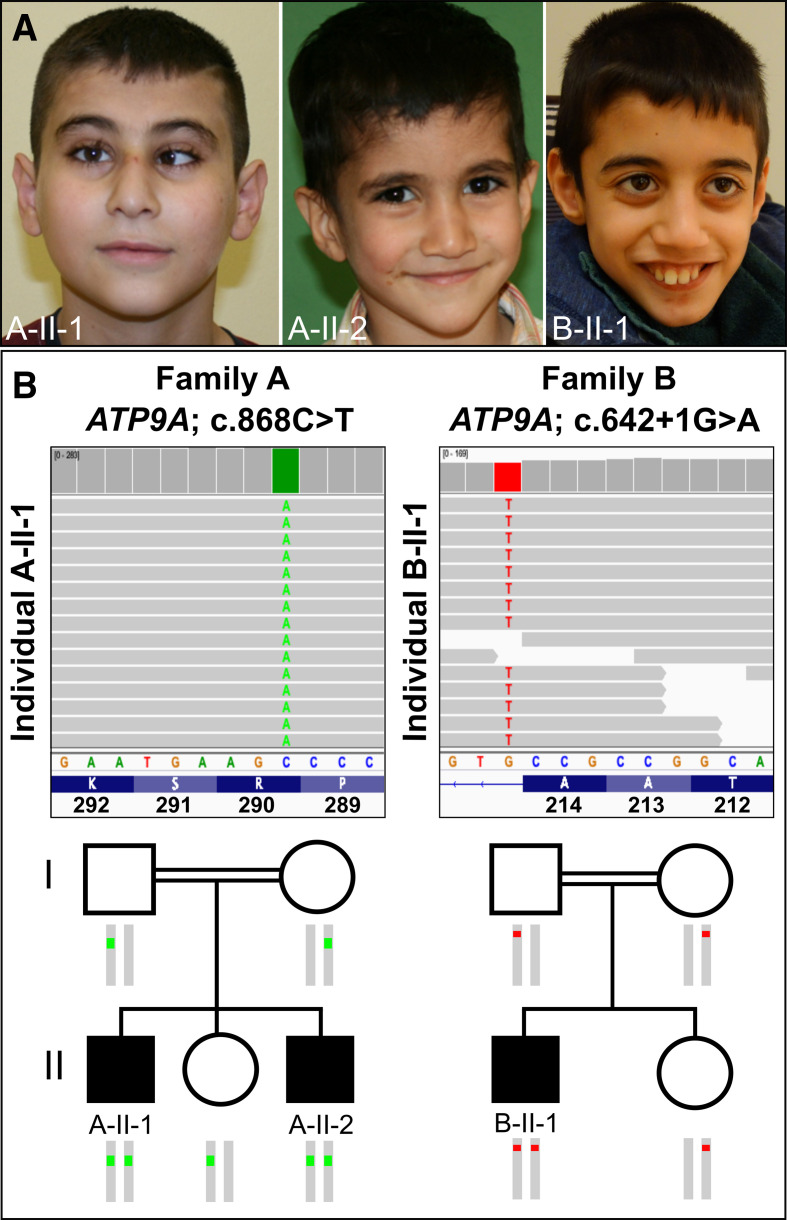
In this study, we report on three affected individuals from two unrelated, consanguineous families. The clinical features of the affected individuals are summarised in table 1 and illustrated in figure 1A.
Table 1.
Clinical features of affected individuals with biallelic ATP9A pathogenic variants
| Affected individual | A-II-1 | A-II-2 | B-II-1 |
| Sex | Male | Male | Male |
| Ethnicity | Syrian | Turkish | |
| Consanguinous | Yes | Yes | |
| Age at last evaluation | 12 years, 3 months | 4 years, 5 months | 9 years, 7 months |
| Clinical phenotype | |||
| Pregnancy | |||
| Gestation | Unknown | 38+1 | 41+2 |
| Birth weight (g) | Unknown | 3492 g (0.36 SD) | 3570 g (−0.41 SD) |
| Birth length (cm) | Unknown | 56 cm (2.08 SD) | 50 cm (−1.37 SD) |
| Birth OFC (cm) | Unknown | 34.5 cm (−0.33 SD) | 34 cm (−1.47 SD) |
| Growth (last assessment) | |||
| OFC (cm) | 49.5 cm (−3.10 SD) | 48 cm (−2.33 SD) | 48 cm (−3.58 SD) |
| Height (cm) | 125 cm (−3.71 SD) | 101 cm (−1.66 SD) | 121 cm (−3.10 SD) |
| Weight (kg) | 26.3 kg (−2.95 SD) | 13.8 kg (−2.50 SD) | 19.6 kg (−4.01 SD) |
| Neurodevelopment | |||
| Motor delay HP:0001270 | + | + | + |
| Speech delay HP:0000750 | + | + | + |
| Fine motor impairment HP:0007010 | + | + | + |
| ID, mild HP:0001256 | + | +/− | – |
| ID, severe HP:0010864 | – | – | + |
| Hyperactivity HP:0000752 | + | – | – |
| Short attention span HP:0000736 | + | + | + |
| Sleep disturbance HP:0002360 | + | – | + |
| Dysmorphism | |||
| Postnatal microcephaly HP:0005484 | + | + | + |
| Smooth philtrum HP:0000319 | + | + | + |
| Thin upper lip vermilion HP:0000219 | + | + | + |
| Strabismus HP:0000486 | + | – | – |
| 2–3 toe cutaneous syndactyly HP:0005709 | + | – | + |
| High palate HP:0000218 | – | – | + |
| Brain MRI | |||
| Hypoplasia of the cerebellar vermis HP:0006817 | – | n.d. | + |
| Hypoplasia of the corpus callosum HP:0002079 | – | n.d. | + |
| Delayed myelination HP:0012448 | – | n.d. | + |
| Gastrointestinal features | |||
| Nausea and vomiting HP:0002017 | + | + | +/– |
| Gastro-oesophageal reflux HP:0002020 | + | + | + |
| Failure to thrive HP:0001508 | + | + | + |
| ATP9A (NC_000020.1, NM_006045.3, NP_006036.1) genotypes and variant description | |||
| gDNA | g.50292679G>A | g.50310546C>T | |
| cDNA | c.868C>T | c.642+1G>A | |
| RNA | n.a. | r.547_642del (exon7) | |
| Protein | p.(Arg290*) | p.(Ser184Profs*16) | |
| Genotype | Homozygous | Homozygous | |
| gnomAD frequency | Absent | Absent | |
+ indicates presence; − indicates absence; +/− means features might be present.
HP, Helicobacter pylori; ID, intellectual disability; n.a., not applicable; n.d., not determined; OFC, head circumference.
Figure 1.
(A) Clinical appearance of the three affected individuals. A-II-1: at the age of 12 years, our proband showed microcephaly, esotropia, a smooth philtrum and a thin upper lip vermilion. A-II-2: his brother, aged 4 years, with smooth philtrum and thin lips. B-II-1: at the age of 10 years, the individual from family B displayed microcephaly, a thin upper lip and a smooth philtrum. (B) Integrative Genomics Viewer screenshot of exome sequencing showing the variants in ATP9A (NM_006045.3): homozygous nonsense variant c.868C>T, p.(Arg290*) in individual A-II-1 and homozygous intronic variant c.642+1G>A in individual B-II-1. Pedigree showing the segregation of the variants within the two families.
Proband A-II-1 was delivered at term by C-section after an uneventful pregnancy as the first child of healthy consanguineous Syrian parents. Birth parameters were not available. The boy showed psychomotor developmental delay and growth retardation. He was able to speak his first words at the age of 18 months and walked unaided at 24 months. Microcephaly was first documented at the age of 7 years when the family moved to Germany. The ophthalmic evaluation showed visual impairment with hyperopia and astigmatism. X-ray examination of the hand showed a delayed bone age. A developmental test (Snijders-Oomen non verbal-R2-7) at 8 years of age revealed a score in the range of 50–60, representing mild ID. He attends a special needs school. Parents reported frequent emesis and epigastric pain. Esophagogastroduodenoscopy (EGD) at the age of 11 years and 5 months revealed antrum gastritis. Although Helicobacter pylori (HP)-eradication therapy was initiated, recurrent vomiting continued. At the last physical examination (12 years and 3 months), he showed deficits in coordination and fine motor skills. Furthermore, he had attention deficit, motor restlessness and sleep disturbance. Anthropometric measurements were as follows: weight, 26.3 kg (−2.95 SD); length, 125 cm (−3.71 SD); and head circumference (OFC), 49.5 cm (−3.10 SD). An MRI of the brain, performed at 11 years, was normal. Karyotype, array CGH and transferrin electrophoresis were unremarkable, thus excluding congenital disorders of glycosylation (CDG syndromes).
Proband A-II-2 is the third child of family A. He was born at term by C-section. Apgar scores were 9/10/10. Birth measurements were normal: weight, 3492 g (0.36 SD); length, 56 cm (2.08 SD); and OFC, 34.5 cm (−0.33 SD). Microcephaly was first documented at the age of 6 weeks. Echocardiography revealed an atrial septal defect (ASD) II and a left ventricular septum hypertrophy with sufficient function. Cardiological follow-up at the age of 3 years and 10 months confirmed the known ASD II with a left–right shunt and mild right ventricular enlargement. So far, an interventional occlusion was not carried out because his overall heart function was sufficient. He required recurrent hospital admissions due to respiratory infections, feeding difficulties as well as daily emesis. At the age of 3 years and 7 months, EGD was performed due to abdominal pain and melena, which diagnosed a chronic gastritis without evidence of HP colonisation. He was able to walk unaided at the age of 21 months. He had a speech delay, attention deficit and impaired fine motor skills. At the last examination (4 years and 5 months), he could speak single words but no sentences. Anthropometric measurements were weight of 13.8 kg (−2.50 SD), height of 101 cm (−1.66 SD) and OFC of 48 cm (−2.33 SD). Weekly emesis and feeding difficulties due to gastritis despite therapy with proton-pump inhibitors persisted.
Proband B-II-1 is the first child of consanguineous parents of Turkish descent. Polyhydramnios was diagnosed in pregnancy. The boy was born at 41+2 weeks of gestation by C-section with a weight of 3570 g (−0.41 SD), a length of 50 cm (−1.37 SD) and an OFC of 34 cm (−1.47 SD). Apgar scores were 9/10/10. In the first weeks of life, the boy suffered from oral thrush. In the first year, insufficient weight gain was noted because of severe oesophageal reflux and a fundoplication was performed with 10 months and repeated with 15 months due to a paraoesophageal hernia. Tube feeding was necessary for the first years of life and in the further course, a gastrostomy was placed. MRI of the brain at the age of 9 months showed a mild delay of myelination, mild widening of the temporofrontal ventricular system, and mild hypoplasia of the corpus callosum and the vermis. Hypotonia and motor delay were diagnosed in infancy. Sitting was achieved at 10 months, independent walking at about 2.5 years. He was noted to have severe cognitive impairment. At the age of 1 year, OFC was 43.3 cm (−3.01 SD). At the age of 9 years and 7 months, his body height was 120.8 cm (−3.10 SD); his weight was 19.6 kg (−4.01 SD); and his OFC was 48 cm (−3.58 SD). He used only single words. Excessive salivation was treated with scopolamine. He showed mild scoliosis, foot malposition, a high palate, a long face with a smooth philtrum, and thin upper lip vermillion. Chromosomal analysis, array CGH and transferrin electrophoresis revealed normal results. His younger sister showed normal development.
ES and cosegregation
ES in individual A-II-1 revealed a homozygous variant in exon 10 of ATP9A (NM_006045.3:c.868C>T, NC_000020.10:g.50292679G>A), predicted to result in a premature stop of translation NP_006036.1:p.(Arg290*) (figure 1B). Sanger sequencing confirmed homozygosity for the variant in the affected brother (A-II-2) and showed that the unaffected sister and the parents are heterozygous carriers (figure 1B).
ES in individual B-II-1 detected a different homozygous variant in intron 7 of ATP9A (NM_006045.3:c.642+1G>A, NC_000020.10:g.50310546C>T) (figure 1B). Bioinformatic in silico prediction using the Ensembl VEP tool indicated a loss of function of the authentic donor splice site (SpliceAI, MaxEntScan and dbscSNV scores are shown in online supplemental table S3).30 The variant was found in a heterozygous state in both parents and the unaffected sister (figure 1B). Both variants have not been described previously and are not listed in gnomAD v2.1.1.31
Homozygosity mapping
These rare ATP9A variants map within a homozygous block of 4.42 Mb (chr20[GRCh37]:46279866–50705211) in the affected individual A-II-1 and 7.46 Mb (chr20[GRCh37]:47686723–55151240) in the affected individual B-II-1 (online supplemental figure S1), likely resulting from identity by descent due to parental consanguinity.
Splicing analysis
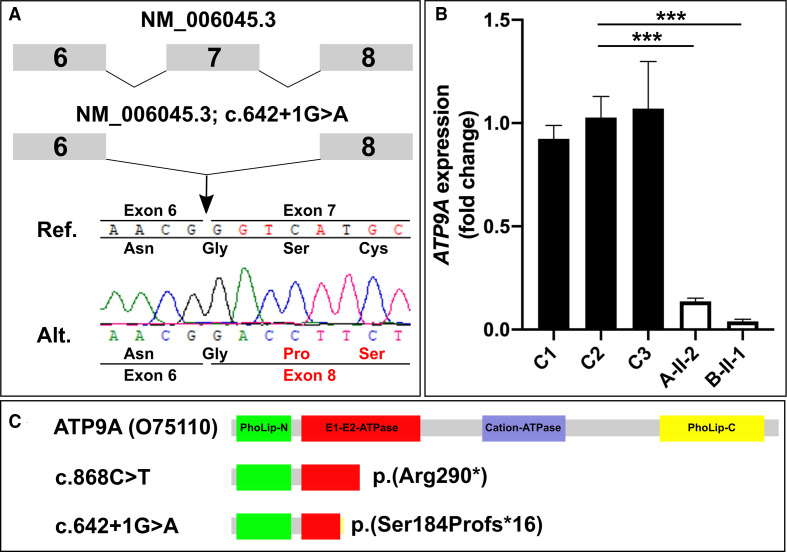
Direct sequencing of RT-PCR amplicons confirmed the loss of this splice site affected by the variant c.642+1G>A, resulting in skipping of exon 7 (figure 2A). This is predicted to result in a frameshift and premature stop of translation p.(Ser184Profs*16).
Figure 2.
(A) Direct sequencing of cDNA of individual B-II-1 confirmed that the splice variant results in skipping of exon 7 (r.547_642del). This deletion is predicted to result in a frameshift and premature stop of translation p.(Ser184Profs*16). (B) Quantitative PCR revealed strongly reduced ATP9A mRNA expression in skin fibroblasts of the affected individuals A-II-1 and B-II-1 compared with controls C1, C2 and C3. ***P<0.0005 (quantified by Student’s t-test). (C) Protein domains of ATP9A (O75110) and schematic representation of the synthesised truncated protein in case of nonsense-mediated mRNA decay escape.
mRNA expression of ATP9A and interacting genes
We measured the ATP9A expression levels in dermal fibroblasts of individual A-II-2 and individual B-II-1 by qPCR, which revealed the downregulation of ATP9A mRNA expression down to 14% in cells of A-II-2 and to 4% in B-II-1 (figure 2B). In conclusion, both variants resulted in almost absent ATP9A mRNA expression in both affected individuals.
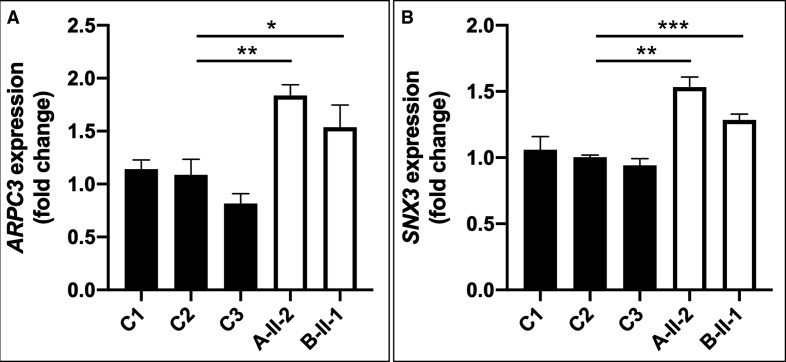
Previously, ATP9A knockdown experiments in HepG2 cells resulted in an increased gene expression of ARPC3 and downregulation of CORO1A.23 Furthermore, in HEK293T cells, ATP9A, MON2 and DOP1B have been shown to form a complex binding to SNX3, a regulator of endosomal trafficking.20 We checked RNAseq data available from multiple control fibroblast lines for expression levels of the aforementioned putative interaction partners of ATP9A and verified them using qPCR. Unfortunately, CORO1A, DOP1B and MON2 are only poorly expressed in human fibroblasts (data not shown). Hence, we could only measure relative ARPC3 RNA expression, which was increased by 1.8-fold in individual A-II-2 and 1.5-fold in cells of individual B-II-1 (figure 3A), and SNX3 expression, which was increased by 1.5-fold in individual A-II-2 and 1.3-fold in individual B-II-1 (figure 3B).
Figure 3.
(A) qPCR detected overexpression of ARPC3 [HGNC:706; NM_001278556] in patient-derived fibroblasts compared with controls. (B) Upregulation of SNX3 mRNA expression levels [HGNC:11174; NM_003795] in patients fibroblast compared with controls. *P<0.05, *P<0.005, ***P<0.0005 (quantified by Student’s t-test).
Discussion
Our study identified homozygous loss-of-function variants in ATP9A as the cause of a novel neurodevelopmental disorder associated with NDD/ID, POM, failure to thrive and gastrointestinal symptoms. Complex neurodevelopmental and behavioural disabilities, including poor fine motor skills, restriction of expressive language, attention deficit, hyperactivity and abnormal sleep patterns are common features in children with POM.32 33 The affected individuals in this study displayed NDD/ID and POM of variable degrees. Individuals A-II-1 and A-II-2 had mild ID, whereas individual B-II-1 was severely affected. Individuals A-II-1 and A-II-2 showed impaired fine motor skills, an attention deficit, hyperactivity and sleep disturbances. All three patients had feeding difficulties in infancy, showed gastrointestinal symptoms (i.e., unclear emesis, gastritis and gastro-oesophageal reflux) and suffered from failure to thrive. Furthermore, individuals A-II-1 and B-II-1 displayed short stature.
Microcephaly is a common clinical feature in NDD/ID disorders and can either be primary (i.e., congenital) or secondary (i.e., POM).4 While primary microcephaly is often caused by early disturbance of neurogenesis, POM probably results from impairment of later neuronal processes, for example, synapse formation or myelination.5 34 Endosome regulation and vesicular routing are essential for neuronal differentiation and synaptic plasticity.35 Mutations in different genes with a role in the regulation of the endocytic network have been described as causal for monogenetic disorders including ID, POM of variable degrees and brain abnormalities.33 One of the genes, TRAPPC9, is highly expressed in the postmitotic neurons of the cerebral cortex and encodes a subunit of the transport protein particle II, which is implicated in endoplasmic reticulum-to-Golgi and intra-Golgi vesicle trafficking.36 In line with the clinical features of the individuals reported herein, biallelic truncating variants of TRAPPC9 have been described in individuals with an ID of variable severity, POM, normal motor development, non-specific cranial MRI signs and hyperactivity (MIM: 613192).37 Recently, biallelic pathogenic variants in the gene VPS51, which encodes subunits of the endosome-associated recycling protein (EARP) and the Golgi-associated retrograde protein, have been described as causal for a neurodevelopmental disorder, characterised by POM, failure to thrive, cortical visual impairment with strabismus, epilepsy and pontocerebellar abnormalities (MIM: 618606).38 EARP facilitates tethering and fusion of early endosome-derived cargos with recycling endosomes.39 Additionally, biallelic mutations in VPS11, another crucial gene of the endosomal pathway, cause a neurodevelopmental syndrome with POM and hypomyelination (MIM: 616683).40 A consistent feature of POM syndromes are white matter defects, likely due to insufficient cargo supply in neurons or oligodendrocytes at the postmitotic stage, which results in delayed myelination and impaired synaptic function.33 In patient B-II-1, there was evidence of delayed myelination in a brain MRI, whereas we noticed no structural changes in individual A-II-1. In contrast to other syndromes associated with POM, we observed no impairment of ambulation, no movement disorder (ataxic gait and stereotypies) or epilepsy in our patients.
Notably, in the recently published Mouse Organogenesis Cell Atlas, sci-RNA-seq3 measured particularly strong expression of Atp9a in excitatory, inhibitory, granule and postmitotic premature neurons (online supplemental figure S2).41 Atp9a−/ − mice show hyperactivity and attention deficit and thus resemble the neurobehavioural disabilities of the individuals reported here (www.mousephenotype.org).42 In this context, we demonstrate that the variants identified in the probands are also loss-of-function variants: the variant c.868C>T, detected in family A, causes a premature stop codon in exon 10. mRNA expression determined by qPCR was reduced by >85% in the affected individual A-II-2 compared with controls. The splice variant c.642+1G>A in family B leads to skipping of exon 7, resulting in a frameshift and the creation of a premature stop codon. Nearly absent mRNA expression in cells harbouring one of the described homozygous variants is likely due to nonsense-mediated mRNA decay. No antibody against endogenous ATP9A is available, a problem that has already been addressed by other researchers.19 Therefore, we cannot rule out the possibility that some mutant protein is still produced. However, both premature stop codons would presumably result in a truncated protein lacking many functional domains (figure 2C).
ATP9A is mainly expressed in neuronal cells (online supplemental figure S3), but there is still limited knowledge about the subcellular localisation and physiological function of endogenous ATP9A in humans. In HeLa cells, overexpressed ATP9A localises to the early/recycling endosomes and is crucial for the endocytic recycling of the transferrin receptor and glucose transporter 1 from the endosome to the plasma membrane. Recently, Jyoti Naik and colleagues demonstrated that shRNA-mediated ATP9A knockdown in HepG2cells resulted in enhanced exosome release and structural changes of actin fibres. It is speculated that this is secondary to induced ARPC3 and reduced coronin1A activity as well as to altered actin nucleation.23 We could only study the expression of ARPC3 because CORO1A is not sufficiently expressed in fibroblasts. Our qPCR data also show a significant upregulation of ARPC3 in fibroblasts of individuals A-II-2 and B-II-1 (figure 3A), supporting the hypothesis that ATP9A impacts the endocytic recycling of proteins that regulate cytoskeletal structures such as the docking sites of multivesicular late endosomes.
ATP9A, MON2 and DOP1B form an evolutionary conserved endosome associated complex and interact with the SNX3-retromer, which orchestrates vesicular trafficking within the endosomal network.43 Because MON2 and DOP1B are not adequately expressed in fibroblasts, we studied SNX3 expression, which was also shown to be upregulated in the fibroblasts of the subjects of this study (figure 3B). Thus, our data provide further evidence that ATP9A is functionally involved in the regulation of the endocytic system. We hypothesise that loss of ATP9A impairs diversifying secretory routes, which would be in line with previous reports. It remains unclear whether ATP9A is localised to a specialised microdomain of early and recycling endosomes and whether ATP9A actively participates in translocation of phospholipids. Thus, the role of ATP9A in membrane composition and its involvement in vesicular transport carrier formation and exosome release needs to be further elucidated. For further studies, the examination of neuronal tissue (mice or induced pluripotent stem cellderived) would be beneficial.
In conclusion, this work describes a novel neurodevelopmental disorder caused by biallelic truncating variants in ATP9A. Identification of further individuals affected by pathogenic variants in this gene will help to expand the molecular and phenotypical spectrum of this disorder.
Acknowledgments
We are grateful to the affected individuals of this report and their parents for their participation in this study. We thank Susanne Rothe, Gabriele Hildebrand and Naji El Choubassi for excellent technical assistance.
Footnotes
Twitter: @BZirnsak
Contributors: Conceptualisation: BF-Z and FB; methodology: MH, NE and DS; formal analysis and investigation: FB, GV, SV, SS, JB, BP, AS-N and CP; writing, original draft preparation: FP; writing, review and editing: BF-Z, GV, SV, MRS, DS, DH and BP; resources: SM and UK.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information. For further information, contact the corresponding author (felix.boschann@charite.de).
Ethics statements
Patient consent for publication
Not required.
Ethics approval
All studies and investigations were performed according to the declaration of Helsinki principles of medical research involving human subjects, and the study was approved by institutional Ethics Committees of Charité -Universitätsmedizin (EA2/177/18).
References
- 1. Maulik PK, Mascarenhas MN, Mathers CD, Dua T, Saxena S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res Dev Disabil 2011;32:419–36. 10.1016/j.ridd.2010.12.018 [DOI] [PubMed] [Google Scholar]
- 2. Vissers LELM, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet 2016;17:9–18. 10.1038/nrg3999 [DOI] [PubMed] [Google Scholar]
- 3. Liu P, Meng L, Normand EA, Xia F, Song X, Ghazi A, Rosenfeld J, Magoulas PL, Braxton A, Ward P, Dai H, Yuan B, Bi W, Xiao R, Wang X, Chiang T, Vetrini F, He W, Cheng H, Dong J, Gijavanekar C, Benke PJ, Bernstein JA, Eble T, Eroglu Y, Erwin D, Escobar L, Gibson JB, Gripp K, Kleppe S, Koenig MK, Lewis AM, Natowicz M, Mancias P, Minor L, Scaglia F, Schaaf CP, Streff H, Vernon H, Uhles CL, Zackai EH, Wu N, Sutton VR, Beaudet AL, Muzny D, Gibbs RA, Posey JE, Lalani S, Shaw C, Eng CM, Lupski JR, Yang Y. Reanalysis of clinical exome sequencing data. N Engl J Med 2019;380:2478–80. 10.1056/NEJMc1812033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kochinke K, Zweier C, Nijhof B, Fenckova M, Cizek P, Honti F, Keerthikumar S, Oortveld MAW, Kleefstra T, Kramer JM, Webber C, Huynen MA, Schenck A. Systematic phenomics analysis Deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am J Hum Genet 2016;98:149–64. 10.1016/j.ajhg.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boonsawat P, Joset P, Steindl K, Oneda B, Gogoll L, Azzarello-Burri S, Sheth F, Datar C, Verma IC, Puri RD, Zollino M, Bachmann-Gagescu R, Niedrist D, Papik M, Figueiro-Silva J, Masood R, Zweier M, Kraemer D, Lincoln S, Rodan L, Passemard S, Drunat S, Verloes A, Horn AHC, Sticht H, Steinfeld R, Plecko B, Latal B, Jenni O, Asadollahi R, Rauch A, Undiagnosed Diseases Network (UDN) . Elucidation of the phenotypic spectrum and genetic landscape in primary and secondary microcephaly. Genet Med 2019;21:2043–58. 10.1038/s41436-019-0464-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rasika S, Passemard S, Verloes A, Gressens P, El Ghouzzi V. Golgipathies in neurodevelopment: a new view of old defects. Dev Neurosci 2018;40:396–416. 10.1159/000497035 [DOI] [PubMed] [Google Scholar]
- 7. Holthuis JCM, Levine TP. Lipid traffic: floppy drives and a superhighway. Nat Rev Mol Cell Biol 2005;6:209–20. 10.1038/nrm1591 [DOI] [PubMed] [Google Scholar]
- 8. Sebastian TT, Baldridge RD, Xu P, Graham TR. Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochim Biophys Acta 2012;1821:1068–77. 10.1016/j.bbalip.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Best JT, Xu P, Graham TR. Phospholipid flippases in membrane remodeling and transport carrier biogenesis. Curr Opin Cell Biol 2019;59:8–15. 10.1016/j.ceb.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muthusamy B-P, Natarajan P, Zhou X, Graham TR. Linking phospholipid flippases to vesicle-mediated protein transport. Biochim Biophys Acta 2009;1791:612–9. 10.1016/j.bbalip.2009.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Andersen JP, Vestergaard AL, Mikkelsen SA, Mogensen LS, Chalat M, Molday RS. P4-Atpases as phospholipid Flippases-Structure, function, and enigmas. Front Physiol 2016;7:275. 10.3389/fphys.2016.00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roland BP, Graham TR. Decoding P4-ATPase substrate interactions. Crit Rev Biochem Mol Biol 2016;51:513–27. 10.1080/10409238.2016.1237934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Onat OE, Gulsuner S, Bilguvar K, Nazli Basak A, Topaloglu H, Tan M, Tan U, Gunel M, Ozcelik T. Missense mutation in the ATPase, aminophospholipid transporter protein ATP8A2 is associated with cerebellar atrophy and quadrupedal locomotion. Eur J Hum Genet 2013;21:281–5. 10.1038/ejhg.2012.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Klomp LWJ, Vargas JC, van Mil SWC, Pawlikowska L, Strautnieks SS, van Eijk MJT, Juijn JA, Pabón-Peña C, Smith LB, DeYoung JA, Byrne JA, Gombert J, van der Brugge G, Berger R, Jankowska I, Pawlowska J, Villa E, Knisely AS, Thompson RJ, Freimer NB, Houwen RHJ, Bull LN. Characterization of mutations in ATP8B1 associated with hereditary cholestasis. Hepatology 2004;40:27–38. 10.1002/hep.20285 [DOI] [PubMed] [Google Scholar]
- 15. Arashiki N, Takakuwa Y, Mohandas N, Hale J, Yoshida K, Ogura H, Utsugisawa T, Ohga S, Miyano S, Ogawa S, Kojima S, Kanno H. Atp11C is a major flippase in human erythrocytes and its defect causes congenital hemolytic anemia. Haematologica 2016;101:559–65. 10.3324/haematol.2016.142273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bublitz M, Morth JP, Nissen P. P-Type ATPases at a glance. J Cell Sci 2011;124:2515–9. 10.1242/jcs.088716 [DOI] [PubMed] [Google Scholar]
- 17. Takatsu H, Baba K, Shima T, Umino H, Kato U, Umeda M, Nakayama K, Shin H-W. ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50 protein-independent manner. J Biol Chem 2011;286:38159–67. 10.1074/jbc.M111.281006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Segawa K, Kurata S, Nagata S. Human type IV P-type ATPases that work as plasma membrane phospholipid flippases and their regulation by caspase and calcium. J Biol Chem 2016;291:762–72. 10.1074/jbc.M115.690727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tanaka Y, Ono N, Shima T, Tanaka G, Katoh Y, Nakayama K, Takatsu H, Shin H-W. The phospholipid flippase ATP9A is required for the recycling pathway from the endosomes to the plasma membrane. Mol Biol Cell 2016;27:3883–93. 10.1091/mbc.E16-08-0586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McGough IJ, de Groot REA, Jellett AP, Betist MC, Varandas KC, Danson CM, Heesom KJ, Korswagen HC, Cullen PJ. SNX3-retromer requires an evolutionary conserved MON2:DOPEY2:ATP9A complex to mediate Wntless sorting and Wnt secretion. Nat Commun 2018;9:3737. 10.1038/s41467-018-06114-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dalton LE, Bean BDM, Davey M, Conibear E. Quantitative high-content imaging identifies novel regulators of Neo1 trafficking at endosomes. Mol Biol Cell 2017;28:1539–50. 10.1091/mbc.e16-11-0772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beer KB, Rivas-Castillo J, Kuhn K, Fazeli G, Karmann B, Nance JF, Stigloher C, Wehman AM. Extracellular vesicle budding is inhibited by redundant regulators of TAT-5 flippase localization and phospholipid asymmetry. Proc Natl Acad Sci U S A 2018;115:E1127–36. 10.1073/pnas.1714085115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Naik J, Hau CM, Ten Bloemendaal L, Mok KS, Hajji N, Wehman AM, Meisner S, Muncan V, Paauw NJ, de Vries HE, Nieuwland R, Paulusma CC, Bosma PJ. The P4-ATPase ATP9A is a novel determinant of exosome release. PLoS One 2019;14:e0213069. 10.1371/journal.pone.0213069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Holtgrewe M, Stolpe O, Nieminen M, Mundlos S, Knaus A, Kornak U, Seelow D, Segebrecht L, Spielmann M, Fischer-Zirnsak B, Boschann F, Scholl U, Ehmke N, Beule D. VarFish: comprehensive DNA variant analysis for diagnostics and research. Nucleic Acids Res 2020;48:W162–9. 10.1093/nar/gkaa241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robinson PN, Köhler S, Oellrich A, Wang K, Mungall CJ, Lewis SE, Washington N, Bauer S, Seelow D, Krawitz P, Gilissen C, Haendel M, Smedley D, Sanger Mouse Genetics Project . Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res 2014;24:340–8. 10.1101/gr.160325.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 2019;47:D886–94. 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014;11:361–2. 10.1038/nmeth.2890 [DOI] [PubMed] [Google Scholar]
- 28. Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting Investigators with an interest in the same gene. Hum Mutat 2015;36:928–30. 10.1002/humu.22844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seelow D, Schuelke M. HomozygosityMapper2012--bridging the gap between homozygosity mapping and deep sequencing. Nucleic Acids Res 2012;40:W516–20. 10.1093/nar/gks487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F. The Ensembl variant effect predictor. Genome Biol 2016;17:122. 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O'Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Neale BM, Daly MJ, MacArthur DG, Genome Aggregation Database Consortium . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–43. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seltzer LE, Paciorkowski AR. Genetic disorders associated with postnatal microcephaly. Am J Med Genet C Semin Med Genet 2014;166C:140–55. 10.1002/ajmg.c.31400 [DOI] [PubMed] [Google Scholar]
- 33. Passemard S, Perez F, Colin-Lemesre E, Rasika S, Gressens P, El Ghouzzi V. Golgi trafficking defects in postnatal microcephaly: The evidence for "Golgipathies". Prog Neurobiol 2017;153:46–63. 10.1016/j.pneurobio.2017.03.007 [DOI] [PubMed] [Google Scholar]
- 34. Alcantara D, O'Driscoll M. Congenital microcephaly. Am J Med Genet C Semin Med Genet 2014;166C:124–39. 10.1002/ajmg.c.31397 [DOI] [PubMed] [Google Scholar]
- 35. Anggono V, Huganir RL. Regulation of AMPA receptor trafficking and synaptic plasticity. Curr Opin Neurobiol 2012;22:461–9. 10.1016/j.conb.2011.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barrowman J, Bhandari D, Reinisch K, Ferro-Novick S. TRAPP complexes in membrane traffic: convergence through a common Rab. Nat Rev Mol Cell Biol 2010;11:759–63. 10.1038/nrm2999 [DOI] [PubMed] [Google Scholar]
- 37. Mochida GH, Mahajnah M, Hill AD, Basel-Vanagaite L, Gleason D, Hill RS, Bodell A, Crosier M, Straussberg R, Walsh CA. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am J Hum Genet 2009;85:897–902. 10.1016/j.ajhg.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gershlick DC, Ishida M, Jones JR, Bellomo A, Bonifacino JS, Everman DB. A neurodevelopmental disorder caused by mutations in the VPS51 subunit of the GARP and EARP complexes. Hum Mol Genet 2019;28:1548–60. 10.1093/hmg/ddy423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schindler C, Chen Y, Pu J, Guo X, Bonifacino JS. EARP is a multisubunit tethering complex involved in endocytic recycling. Nat Cell Biol 2015;17:639–50. 10.1038/ncb3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Edvardson S, Gerhard F, Jalas C, Lachmann J, Golan D, Saada A, Shaag A, Ungermann C, Elpeleg O. Hypomyelination and developmental delay associated with VPS11 mutation in Ashkenazi-Jewish patients. J Med Genet 2015;52:749–53. 10.1136/jmedgenet-2015-103239 [DOI] [PubMed] [Google Scholar]
- 41. Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, Trapnell C, Shendure J. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019;566:496–502. 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, Baker CN, Bower L, Brown JM, Caddle LB, Chiani F, Clary D, Cleak J, Daly MJ, Denegre JM, Doe B, Dolan ME, Edie SM, Fuchs H, Gailus-Durner V, Galli A, Gambadoro A, Gallegos J, Guo S, Horner NR, Hsu C-W, Johnson SJ, Kalaga S, Keith LC, Lanoue L, Lawson TN, Lek M, Mark M, Marschall S, Mason J, McElwee ML, Newbigging S, Nutter LMJ, Peterson KA, Ramirez-Solis R, Rowland DJ, Ryder E, Samocha KE, Seavitt JR, Selloum M, Szoke-Kovacs Z, Tamura M, Trainor AG, Tudose I, Wakana S, Warren J, Wendling O, West DB, Wong L, Yoshiki A, MacArthur DG, Tocchini-Valentini GP, Gao X, Flicek P, Bradley A, Skarnes WC, Justice MJ, Parkinson HE, Moore M, Wells S, Braun RE, Svenson KL, de Angelis MH, Herault Y, Mohun T, Mallon A-M, Henkelman RM, Brown SDM, Adams DJ, Lloyd KCK, McKerlie C, Beaudet AL, Bućan M, Murray SA, International Mouse Phenotyping Consortium, Jackson Laboratory, Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS), Charles River Laboratories, MRC Harwell, Toronto Centre for Phenogenomics, Wellcome Trust Sanger Institute, RIKEN BioResource Center . High-Throughput discovery of novel developmental phenotypes. Nature 2016;537:508–14. 10.1038/nature19356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burd C, Cullen PJ. Retromer: a master conductor of endosome sorting. Cold Spring Harb Perspect Biol 2014;6. 10.1101/cshperspect.a016774. [Epub ahead of print: 01 Feb 2014]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jmedgenet-2021-107843supp001.pdf (279.5KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information. For further information, contact the corresponding author (felix.boschann@charite.de).