Abstract
Bacteria, viruses, and parasites are harmful microorganisms that cause infectious diseases. Early detection of diseases is critical to prevent disease transmission and provide epidemic preparedness, as these can cause widespread deaths and public health crises, particularly in resource-limited countries. Lateral flow assay (LFA) systems are simple-to-use, disposable, inexpensive diagnostic devices to test biomarkers in blood and urine samples. Thus, LFA has recently received significant attention, especially during the pandemic. Here, first of all, the design principles and working mechanisms of existing LFA methods are examined. Then, current LFA implementation strategies are presented for communicable disease diagnoses, including COVID-19, zika and dengue, HIV, hepatitis, influenza, malaria, and other pathogens. Furthermore, this review focuses on an overview of current problems and accessible solutions in detecting infectious agents and diseases by LFA, focusing on increasing sensitivity with various detection methods. In addition, future trends in LFA-based diagnostics are envisioned.
Keywords: Lateral flow assay, Viruses detection, SARS-CoV-2, Biosensor, Immunoassay
Graphical abstract
1. Introduction
Microorganisms such as bacteria, viruses, fungi, algae, and protozoa affect human life in beneficial and harmful ways, depending on the environment and natural conditions [1]. Infectious diseases constitute a large part of global medical problems, and developing countries are most affected by this situation. Humans have been exposed to several novel viral infectious agents and re-emerging infectious agents during the last two decades. Many factors cause the emergence of an infectious disease. The factors contributing to its emergence can be divided into three. These are (i) virus evolution and adaptation, (ii) human factors, and (iii) ecological changes [2]. Most of the viral agents are RNA viruses. As a result of this fast evolution and environmental flexibility, RNA viruses can quickly reach adaptive equilibrium within their host species. Surprisingly, it is not believed that the genetic evolution of viruses is the primary reason for creating the virus in question. Viruses appear to remain stable within the boundaries of their ecological niche. Human factors are widely considered the most crucial element in the emergence of viral infectious diseases. A disease's pathogen first is introduced to humans, then spread and maintained in the population. After the 19th century, with the development of medicine and increased welfare, there has a significant increase in the human population. The size of the human population negatively favors the spread and persistence of diseases. Increasing population density and urbanization bring about housing, sanitation, pollution, drinking water, and health facilities. Moreover, the enormous expansion in the human population necessitates additional living space. As a result, human expansion into virgin forests may disrupt virus reservoirs and increase the probability of viral transmission from animals to people. Also, unplanned migration resulting from war or natural catastrophes has been a significant factor in spreading infectious illnesses among people. Airplanes, which are the product of developing technology, can easily transport infected animals, humans, or reptiles from country to country [3]. With such a decrease in the distance between the continents, the transmission can cause worldwide pandemics [4]. It is the most obvious example of how quickly the emerging SARS-CoV-2 virus has spread from China to the world. The COVID-19 pandemic is considered the most crucial global health calamity of the century and the greatest challenge humankind has faced since the 2nd World War. In addition to its sad effects on human life, COVID-19 has also seriously damaged its economy [5]. Education, trade, and sports have been halted in almost all countries to limit further disease transmission. People stayed at home for months and suffered material/moral damage. Most importantly, many people faced the threat of unemployment. For this reason, health personnel, scientists, and governments have shown great interest in rapid and accurate detection methods for the prevention and control of the pandemic [6].
Consequently, cost-effective diagnoses are significant to monitor and detect diseases. Traditional detection methods for detecting specific pathogens include pathogen culture, gram stain, enzyme immunoassays, enzyme-linked immunosorbent assays, biochemical methods, other nucleic acid-based amplification methods, and real-time polymerase chain reaction (RT-PCR) [7,8]. These techniques have contributed significantly to diagnosing, preventing, and treating several infectious diseases. Nevertheless, it also has several drawbacks, including time-consuming, costly, needing advanced analytical hardware, and requiring expert professionals. For example, although the RT-PCR is the “gold standard method” in the clinical diagnosis of SARS-CoV-2, it has its disadvantages and limitations. Sensitivity may be reduced due to sampling errors or low viral load (false negatives). Inactive virus and viral fragments may also test positive (false positives) [9]. As a result, a novel and reliable point-of-care diagnostic method is required. Rapid diagnostic tests are of great importance to prevent the spread of the virus to the public and allow timely intervention. In addition to rapid diagnostic decision-making, this system reduces potential analytical interference by promptly minimizing complex sample processing and transporting the test sample to the diagnostic facility. More importantly, continuous PCR testing to large audiences costs the healthcare system large sums. LFA tests have the potential to reduce the financial burden as they are relatively inexpensive, practical, and do not require qualified personnel. Also known as immunochromatographic or lane tests, LFA is an immunoassay designed to operate along a single axis. Immunoassay is defined as a bioanalytical method. LFA strips have been widely used to test various analytes, including nucleic acids [[10], [11], [12]], proteins [[13], [14], [15], [16]], and infectious viruses [[17], [18], [19]]. LFA platform can be seen as the best alternative for the rapid and accurate diagnosis of several viral infectious diseases [[20], [21], [22]]. Since it does not require any knowledge, the LFA method is apparent as a convenient point-of-care (POC) diagnostic technique [[23], [24], [25], [26], [27], [28]].
The first example of an immunoassay for diagnosing infectious disease was reported in a study by Dochez and Avery in 1917. According to the study, pneumococcal polysaccharides can be detected by LFA of both urine and serum taken from patients with lobar pneumonia [29]. The authors suggested that antibody/antigen detection can quickly detect the presence/absence of infection. Disadvantages and inadequacy of immunoassays such as enzyme-linked immunosorbent assay (ELISA) and RT-PCR for disease diagnosis have prompted many researchers and biotech companies to find ways to perform rapid testing in POC. Subsequently, various companies developed a new method that led to the novel LFA platform [[30], [31], [32]]. Home pregnancy testing using the lateral flow platform provided the first clear evidence for home use of antigen testing. Subsequently, rapid tests developed for diagnosing streptococcal pharyngitis popularized the lateral flow immunoassay (LFIA) test for diagnosing infectious diseases. Emerging new diagnostics systems promise to solve these drawbacks by providing low-cost, quick, sensitive, specific, and straightforward diagnoses. After a brief overview of LFA structures and working principles in this review, we reviewed LFA studies on infectious diseases in the literature.
2. Structure of the lateral flow immunoassay platform
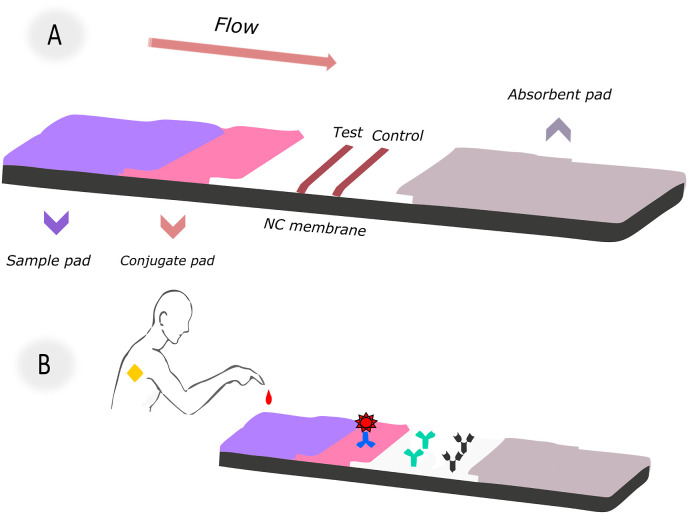
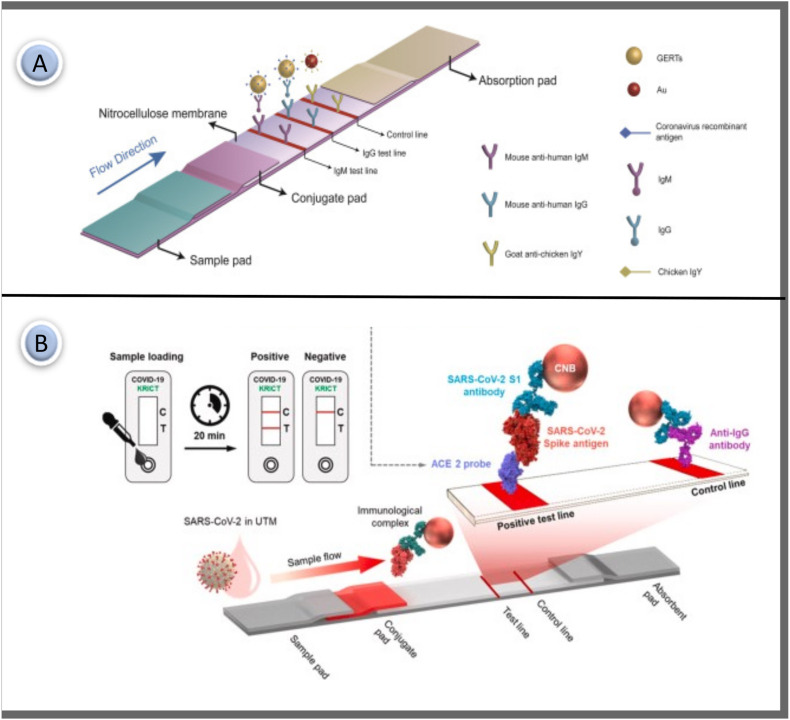
LFA's standard structure consists of functional zones constructed of different materials. All LFA systems work using the same basic principle. A commercially available LFA strip comprises four components: a sample pad, conjugation pad, nitrocellulose (NC) membrane, and an absorption pad. They usually overlap and install on a backing card in sequence [33]. The main steps in preparing the LFA system are; (a) antibody/antigen preparation against target analyte (b) selection and preparation of colored label (c) process of labeling biorecognition molecules (d) immobilization of detector reagents to the appropriate pad (e) immobilization of test and control lines to the NC membrane (f) mounting all components on a backing card (g) sample sending and receiving results in minutes. The schematic diagram of the essential components of the lateral flow system is shown in Fig. 1 . A-B. Each component serves one or more purposes.
Fig. 1.
(A) Components of a conventional LFA; (B) Schematic illustration for test design and application method.
A sample pad is a pad that is used to apply the samples (serum/urine/blood). The sample pad has two essential functions. First, it enables the sample's controlled-release towards the conjugated analyte contained in the conjugate pad. So the primary purpose is to process the sample to match the rest of the test. Some sample contents, such as urine and saliva, might differ dramatically based on things like time of day, food, and age. These changes may hamper the detection of the target analyte in composition and pH. The second purpose of the sample pad, the first component of the LFA strip that comes into contact with the sample, is to control these inconsistencies [34].
The tagged biosensing molecules are dispersed on the conjugate pad. The conjugate pad's role in LFA is to take the conjugate, keep it stable for the duration of the shelf life, and release it quickly and consistently once the assay is run [35]. It should have a high release ability and little protein binding. The conjugate pad should show low non-specific binding so that the detection reagent does not remain trapped in the pad. The conjugate pad material immediately releases the labeled conjugate from the surface when the mobile liquid comes into contact with the sample. Some conjugate pads support slow-release, while others support fast release. However, the release speed should always be internally consistent. Inefficient mixing and separation of the conjugate from the pad can result in negative results in test sensitivity and reproducibility. Due to the materials' nature, it is needed to pretreat the conjugate pads to provide optimal release, reduce non-specific interactions, and control pH. Conjugate pretreatment buffer components move faster in the lane than the conjugate. They may also help to block the binding sites of the capture reagent present on the membrane prior to conjugate interaction. Pretreatment of the conjugate pad with blocking agents such as proteins, surfactants, or polymers may also help restore signal intensity [36]. The conjugate pad pretreatment can be carried out by dipping the buffer or spraying it homogeneously with an automatic dispenser. A dispensing platform ensures that a controlled and consistent amount of conjugate is loaded into each pad. The immersion technique results in an uncontrolled amount of conjugate loaded into a place. This technique is only recommended when a distribution platform is not available. The labeled conjugate should remain stable for the entire life of the lateral flow strip. Under controlled humidity, the dried conjugate pad can remain stable for up to 3 years. Conjugated liquid reagent can remain stable for a maximum of 1 month at 4 °C.
Nitrocellulose is a highly flammable material. It consists of cellulose nitrated with strong nitrating agents such as nitric acid. Three nitrate groups are esterified on each glucose unit in the cellulose polymer. The negative charge of nitrocellulose at neutral pH and the excellent flammability of dry nitrocellulose are both due to these nitrate groups in the structure [36].
If the NC membrane is treated harshly, it can become fragile. Thus membranes cast on a commercially available inert synthetic support are generally preferred. Each NC membrane has unique capillary flow properties depending on the manufacturing process, which affects its physical properties, specificity, flow mechanics, sensitivity, and stability of the LFA [37,38]. The capillary flow rate is influenced by the pore size and porosity of the membrane. As the pore size increases, the flow rate of the sample increases accordingly. The size of the analyte and the sample type are taken into account when deciding the pore size of the NC membrane. Each producer treats membranes with a specialized mixture of surfactants and chemicals to hydrophilic nitrocellulose. These processes also affect the achievement, and performance of the LFA, depending on the biomolecule used. As a result, membranes with the same physical attributes (e.g., pore size, flow rate) acquired from different suppliers may behave differently [35,38]. There are three essential parameters for striping NC membranes. These are the reagent amount, the dispersion amount, and the dispense rate. All parameters vary depending on the physical properties of the membrane used and the reagent being tested. The electrostatic interaction between the strong dipoles of the nitrate ester and the peptide bonds during lane formation can immobilize various proteins [39]. While non-ionic detergents like Tween-20 and Triton X-100 remove protein from the membrane, raising the salt content improves protein binding to the NC membrane by enhancing hydrophobic binding and electrostatic interaction.
The absorbent pad is the last part of the lateral flow strips. Generally, highly polar cellulose pads are used. The absorbent pad acts as a fluid trap at the end of the LFA. It absorbs the excess liquid and prevents the backflow of the sample solution. It is important not to return the liquid to the test; otherwise, false-positive results may occur.
3. LFA principles and assay formats
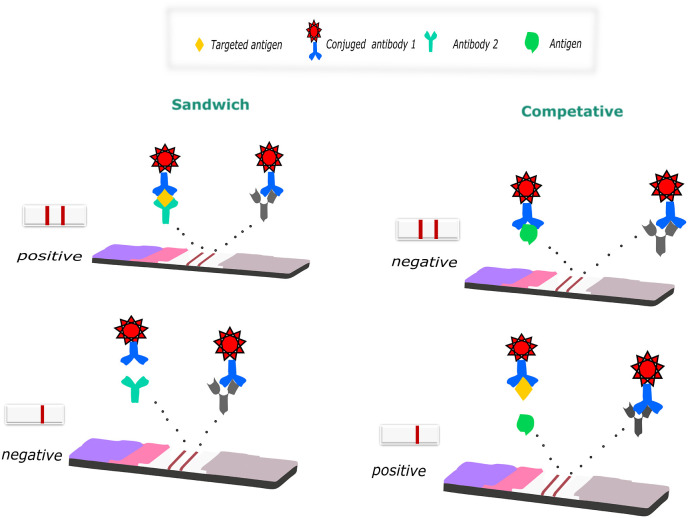
There are several possible stream formats on the LFA platform, depending on the type of target analyte. The two most common forms are competitive assay and sandwich assay. The working principle of the two different LFA formats is simple, as shown in Fig. 2 . In a sandwich assay, when a liquid sample containing the target analyte is loaded onto the LFA strip, the mixture flows towards the absorption pad via capillary force. The first pad neutralizes the sample and filters out unwanted particles such as red blood cells. In short, it acts as a pre-filter. Then, the sample flows into the conjugate pad with conjugated golds on its surface. When the liquid reaches the conjugate pad, the dried gold nanoparticles begin to be released from the surface. Mix the sample with the conjugated prob. During this mixing, if there is no target analyte in the liquid sample with which the conjugated gold will interact, the flow continues towards the NC membrane without any interaction. If the target analyte is present, the conjugated golds bind to the antibody and continue to flow in the NC membrane, which is the region where the sample and capture reagent will interact. The second biological component of the test, the capture reagent, is previously immobilized there. The capture reagent consists of a test region and a control reagent strip. The test line is the primary readout of the diagnosis. The target analyte interacting with the conjugated prob on the conjugation pad interacts with the antibody pair in the test line, resulting in a red zone on the strip. The control biomolecule is the control region that indicates the test is run successfully. NC membrane also provides a qualitative analysis by creating color formation due to the reaction between antigen and antibody in the test and control region. After the sample passes through the test and control lines, it continues to be carried until it reaches the absorbent pad at the other end of the strip, and the test is completed. So, the user can read the results. Consequently, when the target analyte is present, two red lines appear at the end of the test, but only one red line appears on the control line when it is absent. A sandwich test is often used for analyte antigens with two epitopes that bind to two different types of antibodies. Since the targeted antigen is captured between the two antibodies at the test region, the signal strength is proportional to the analyte concentration in the sample.
Fig. 2.
The sandwich assay and competitive assay models.
On the other hand, the competitive format is generally used when testing analytes with low molecular weight or presenting a single antigenic determinant. Also, unlike the sandwich format, the result is positive when the signal intensity in the test line decreases or disappears in the presence of the target biomolecule.
4. LFIA application strategies for infectious disease diagnosis
4.1. COVID-19
Following its first appearance in China, the pandemic of COVID-19 has spread worldwide at an unprecedented rate. Coronaviruses (CoVs) are RNA viruses enclosed and positive-strained and may infect animals. CoVs usually causes moderate upper respiratory symptoms in humans [[40], [41], [42], [43]]. CoVs have caused several epidemics across the world to date. In 2020, SARS-CoV-2 caused a pandemic all over the globe [44]. Although SARS-CoV-2 is very similar to other coronaviruses, its precise reservoir has not been determined [[45], [46], [47]]. Also, the constant mutation of these viruses makes it difficult for researchers to identify a precise reservoir.
SARS-CoV-2 contains four structural proteins: point surface (S) glycoprotein, nucleocapsid (N) protein, matrix (M) protein, and envelope (E) protein [48]. The immune system produces immunoglobulins to fight pathogens. First of all, an antigen test is preferred to determine whether the person has a disease. Antigens are far more stable than RNA, making them less prone to degradation during transport and storage. Therefore, it may be a useful biomolecule for the early detection of an asymptomatic population. After a while, antibody tests follow the body's battle against diseased cells. Typically, antibody levels rise significantly in the second and third weeks after initial onset. This feature prevents antibody testing from being used to diagnose COVID-19 promptly. However, detecting IgG and IgM is useful for studying the course of the disease post-illness or after vaccination [49]. Considering the advantages and disadvantages of antibody and antigen tests, there are many different studies for antibody detection and antigen detection.
Many LFA studies have been carried out to diagnose SARS-CoV-2 in the last three years. This review provides an overview of the latest developments in COVID-19 LFA testing. This section discussed multiple COVID-19 antibody tests, new nanoparticle applications, and antigen tests respectively. Reporter type, target analytes, limits of detection (LOD), sensitivities, and test times of COVID-19 tests are compiled in a table.
IgM is identified in serum shortly after infection and can be used as a biomarker in the early stages of infection detection. IgG indicates the previous or current state of the infection. So, IgG can show whether a person has developed immunity to a disease. Most LFA studies detect IgM and IgG antibodies independently of each other. The IgM-IgG combined assay has better utility and sensitivity compared with a single IgM or IgG test. The first LFA-based study for early detection of SARS-CoV-2 was reported in 2020 by Li et al. It was pioneering work to simultaneously detect two different SARS-CoV-2 antibodies. The researchers described an LFA immunoassay to detect IgM and IgG antibodies against CoVs in human blood for 15 min. Commercial AuNP of 40 nm diameter was conjugated with SARS-CoV-2 recombinant protein and rabbit IgG. The prepared AuNP-COVID-19 recombinant antigen conjugate and AuNP-rabbit-IgG mixture were activated by spraying onto the conjugate pad. Anti-human-IgM, anti-human-IgG, and anti-rabbit-IgG were immobilized in the test M, G, and control lines. The working principle of the test is based on the presence of SARS-CoV-2 IgG and IgM antibodies, the red/purple line appearing in the test areas on the device. The assay's specificity and sensitivity were determined using blood samples from approximately 400 COVID-19 positive patients and approximately 130 negative patients. The testing sensitivity was % 88.7 and specificity was % 90.6. It was seen as a pioneering and successful study as it was the first study to detect two important COVID-19 antibodies in whole blood samples. Li and coworkers recommended the development of IgG-IgM combined antibody test kits instead of separate IgG or IgM antibody test kits if a reliable technical system is available [50].
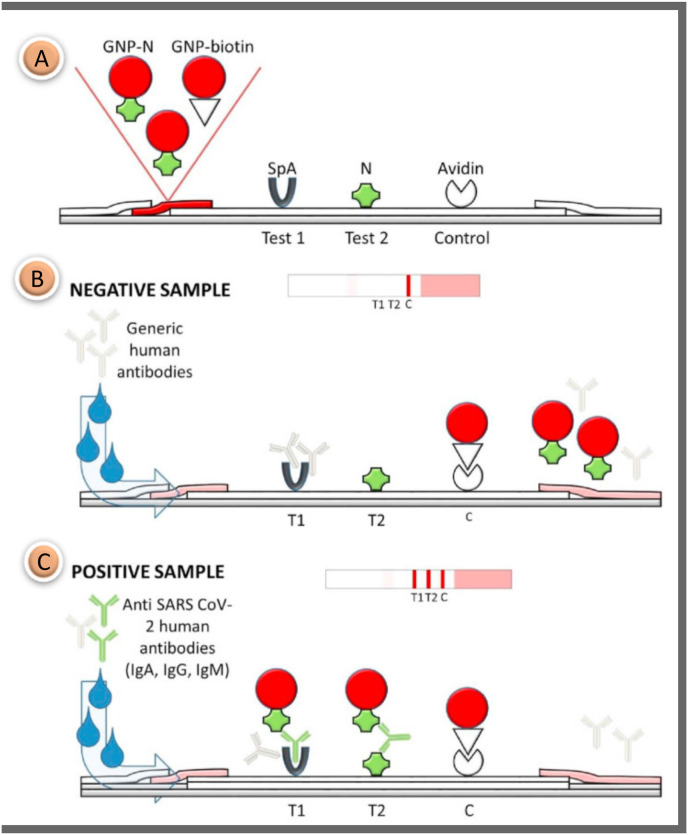
SARS CoV-2, like other viruses that impact respiratory features, causes the development of another class of particular immunoglobulins A (IgA) in respiratory specimens and the presence of specific anti-SARS CoV-2 IgA in respiratory samples in the blood [51,52]. A dual test-line LFA capable of indiscriminate binding to human IgG, IgM, and IgA was developed to improve the specificity of the SARS CoV-2 assay. The study differs from its counterparts in that it is the first lateral flow test designed to determine total immunoglobulins for the N protein of SARS CoV-2 (Fig. 3 ). The protein A was applied to the nitrocellulose membrane to form the first test line and the protein N was applied to form a second test line. Both test lines are linked to several immunoglobulin classes (G, M, and A). Sensitivity enhancement was achieved by using broad specific agents to capture total immunoglobulins. Avidin was used as the capturing reagent for the AuNP-biotin conjugate at the control line. It was a different study from other COVID-19 tests in terms of using the avidin-biotin interaction to form the control line. Measurement with an accuracy of % 93.33 was achieved in as little as 20 min. Although the sensitivity is quite good, the test time is relatively long [53].
Fig. 3.
(A) Schematic representation of the LFIA test for the diagnosis of SARS CoV-2. Antibodies; (B) For the negative sample, only the control line color is expected; (C) 2 test regions and 1 control regions appear in the asset of anti-N antibodies (IgG, IgM and IgA) [53]. Reproduced with permission from ref. 53, Elsevier Ltd.
Another study developed for the simultaneous detection of two different antibodies has good analytical performance. Black et al. successfully developed a lateral flow assay to detect SARS-CoV-2 IgM and IgG antibodies in whole blood simultaneously. According to the results obtained, the specificity for IgM and IgG was % 92 and % 100, respectively. As a result, red's intensity discoloration in the IgM and IgG test zones alone or together was considered positive and read after 10 min. Since the results were obtained from whole blood samples, they can consider quite successful [54].
AuNPs are the most widely used labeling system for lateral flow tests. In addition to their unique color properties, AuNPs have long been indispensable in LFA studies. Moreover, it can be easily conjugated with biomolecules via simple adsorption processes or gold-thiol surface chemistry to prepare for the preparation of bio labels [55,56]. Traditional colloidal gold-based LFAs do not have optimal performance or quantification capability. In recent years, many fluorescence-based LFAs have been developed for quantitative or semiquantitative detection. Fluorescent microspheres [57], quantum dots (QD) [58,59], europium nanoparticles [60,61], carbon nanoparticles [62,63] and plasmonic nanoparticles [64] as alternatives to AuNP is used in LFA. Also, several promising nanoparticles have been introduced and successfully applied in LFA studies.
Lanthanide doped nanoparticles (LNPs) exhibit unique optical properties such as a long luminescence lifetime, sharp emission peaks, and upconversion luminescence in the wavelength range from near-infrared to visible. In addition, these nanoparticles have the advantage of a high signal-to-noise ratio for highly sensitive and selective diagnostic detection. A group of researchers presented the development of a rapid and sensitive LFIA using lanthanide-doped polystyrene nanoparticles to determine SARS-CoV-2 IgG antibodies in serum. The LNPs were synthesized by miniemulsion polymerization. LNPs (615 nm) were conjugated with Rabbit-IgG and mouse anti-IgG and then sprayed onto the conjugate pad. Recombinant nucleocapsid phosphoprotein was stripped in the test region, and goat anti-rabbit IgG (2 mg/mL) was stripped in the control region. The use of high concentrations of antibodies in the control line can be a disadvantage of the study as it is reflected in the cost of assay production. The assay results were obtained in less than 10 min, indicating that diagnostics might be used in clinical practice [65].
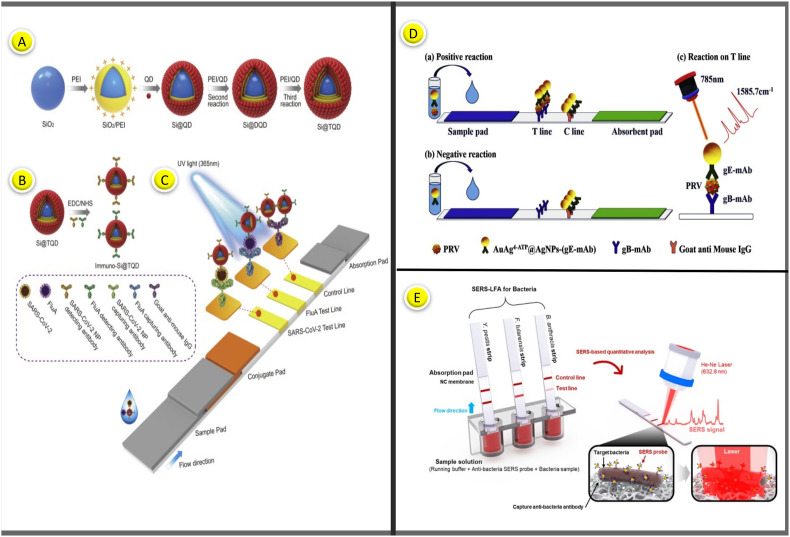
Quantum dots (QD) have become popular as fluorescent tags in the LFA system because of their excellent optical features such as wide excitation, quantifiable fluorescence intensity, and great stability. Many QD-based LFAs have been developed to accomplish high-level detection [[66], [67], [68]]. According to a recent study, some blood samples from COVID-19 patients cross-reacted to the SARS-CoV NP protein. There was no cross-reactivity between antibodies specific to SARS-CoV-2 (IgG and IgM) and the S antigen of SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) [69,70]. Moreover, the anti-SARS-CoV-2 IgM ELISA based on S proteins has proven to be more sensitive than the N protein-based test [71,72]. These findings revealed that the S protein is the best antigen for serodiagnosis of COVID-19. In 2020, a study proposed an LFA assay that enables simultaneous detection of IgM and IgG antibodies in human serum using S protein conjugated SiO2@Au@QD nanobeads as tags. The proposed SiO2@Au@QD NBs consisted of three parts. 200 nm monodisperse SiO nanoparticles were used as the hydrophilic core, while AuNP (4 nm) was used to provide a strong colorimetric signal. QD was used to present a large surface area. Only 1 μL of serum sample is required for the test, and the result can be read within 15 min. As a result, using SiO2@Au@QD nanobeads is almost 100 times more sensitive than AuNPs-based LFA [73].
In another similar study, a new LFA test method was proposed to simultaneously detect SARS-CoV-2-specific IgM and IgG by conjugating S protein with a QD nanotag. PEI-mediated electrostatic adsorption was used to create the SiO2@DQD, which has a monodisperse SiO2 core and a dual QD-formed shell. The carbodiimide chemistry used in the SiO2@DQD label was functionalized with the SARS-CoV-2 S protein. The test result was obtained using a portable fluorescent reader instrument at excitation and emission wavelengths of 365 and 615 nm, respectively. The overall detection sensitivity and specificity were reported as % 97.37 and % 95.54, respectively [74].
In 2010, Baker and colleagues discovered that N-acetyl neuraminic acid has a high binding affinity for the SARS-CoV-2 S glycoprotein. Based on this interaction, they developed an LFA test. Unlike traditional LFAs, which utilize antibodies, the glycan is bound in gold nanoparticles (as a BSA-glycoconjugate) on the test strip and in the mobile phase. After optimization, the ready test can present the result to the reader within 30 min. The LOD of the proposed glyco-LFA, S protein is approximately 5 μg/mL. This study is proof that glycan-binding can be used to generate rapid point-of-care diagnostics in a format that requires no infrastructure and limited training. The researchers claim it to be the first reported glycan lateral flow system. In this regard, it seems to be an innovative study [75].
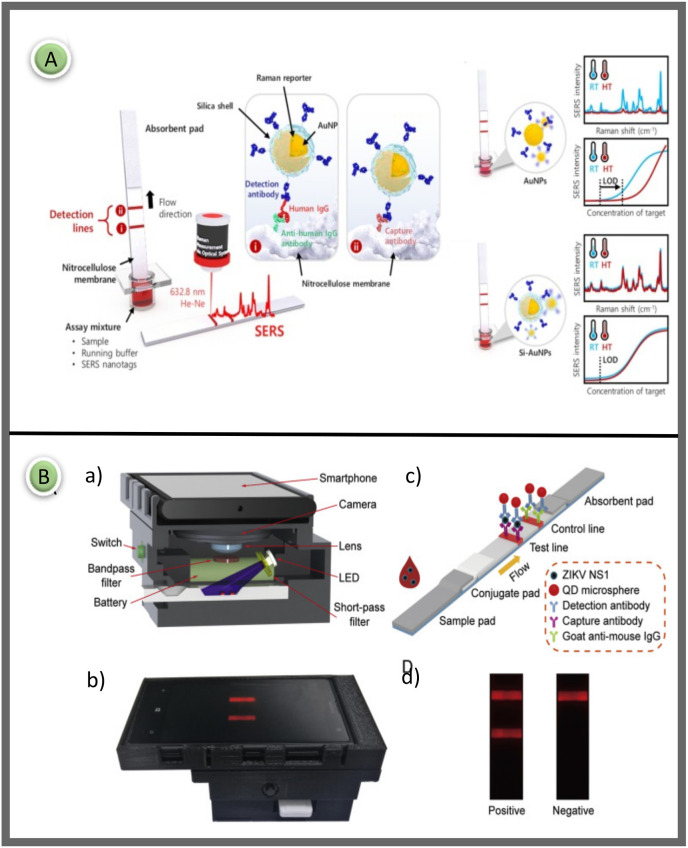
In 2021, Gap-enhancement nanotags were used to improve the performance of SERS-based LFA. Ultra-low LODs of 1 ng/mL and 0.1 ng/mL for IgM and IgG were obtained, respectively. Obviously, the study is important because it reports the interference effect of IgM on IgG. A particularly high IgM concentration makes it difficult to measure IgG in the system. Therefore, using dual-plex LFA with nano-tags modified by the same antigen to analyze antibodies quantitatively is not rigorous and reliable in multiplex detection. Therefore, it was concluded that the dual model quantitative analysis of SERS-based LFA for simultaneous detection of IgM and IgG should be reconsidered (Fig. 4 A). [76].
Fig. 4.
(A) Schematic diagram of the SERS based LFA strip prepared using gap enhancement nano-tags for IgM and IgG detection [76]; (B) Working principle of LFA that can test SARS-CoV-2 S protein using ACE2 receptor in test line [78]. Reproduced with permission from ref. 76–78, Elsevier Ltd.
Antigens can be defined as diseased cells found in the body. Antibodies are proteins that fight these diseased cells in the body. The task of antibodies is to settle on the antigens that appear in the body and neutralize them. The antigen test is performed in the first place in order to determine whether the person has the disease. Then, antibody tests follow the body's fight against diseased cells. Antibody tests give information about the course of the disease and the state of the immune system; antigen tests are performed to detect the presence of the disease instantly. When comparing antibody and antigen tests, antigen tests are preferred for early diagnosis. Benjamin et al. suggested that antigen-based COVID-19 LFA may have considerably better clinical sensitivity than traditional tests. Until now, only a few numbers of antigen-based LFAs have been proposed. Furthermore, the design of the half-tape LFA has been seen as a necessary precursor to the development of any LFA format. The researchers demonstrated a half-tape LFA using commercially available antibodies to identify the SARS-CoV-2 antigen in this study. The sample pad and conjugate pad were not mounted in the system in the assay. Only the NC membrane and absorbent pad were used in the test. Therefore, it has the potential to be a pioneering work for many researchers who want to develop LFIA for the first time [77].
While antigen-based assays are consistent diagnostics for detecting the previous infection, infection progression, and contagion dynamics, no matching antibody pair has been reported for immunoassay of SARS-CoV-2 antigens. In a study by Lee and colleagues, a new rapid detection technic for the SARS-CoV-2 S1 protein was designed using the SARS-CoV-2 receptor ACE2, which can couple with commercially available antibodies to produce suitable pairings. As indicated in Fig. 4B, the test line was immobilized ACE2, and the antibodies were tagged with red cellulose nanobeads as detection probes. S1-mAb and ACE2 were used to capture and identify S1 protein in LFA without cross-reaction with other CoV S1 proteins. The proposed LFA platform did not have cross-reaction between SARS-CoV S1 or MERS-CoV S1 protein. This is significant since it is the first research to use an LFA with a matched combination of ACE2 and antibody to identify the SARS-CoV-2 S1 antigen [78].
Many SARS-CoV-2 antibody detection tests were quickly commercialized due to the urgent need with minimal validation requirements. Most detect IgM, IgA, and IgG against the nucleocapsid protein or different areas of the spike glycoprotein (S1, S2, and RBD) [79]. The excellent specificity of the IgG test indicates that we can use it as a confirmatory test for the SARS-CoV-2 antibody test. However, using these tests in the field, which does not give % 100 reliable results on real samples, may cause some concern. Therefore, it is better to use these tests as part of our daily workflow along with ELISA until their analytical performance improves, reaching acceptable specificity and sensitivity. Efforts to increase analytical performance continue rapidly. Looking at Table 1 , many studies used the same labeling agent (AuNPs, LNPs, etc.). Although studies using AuNP as color labels appear similar, studies that generally result in the use of different target analytes (IgG or IgM) and better sensitivity are presented. Researchers also developed new AuNP production methods to improve analytical performance in existing COVID-19 LFA assays and reported improvements in test sensitivity. Thus, similar studies have gained superiority in analytical performance. New nanoparticles and reporters (Carboxylic red latex beads, SiO2@Au@QD nanobeads, LNPs, Gap-enhancement nanotags, Eu(III) fluorescent microsphere etc.) have contributed significantly to the sensitivity of the LFA test and offered new solutions. Continued development will lead to more sensitive, rapid, and cost-effective SARS-CoV-2 detection and more effective treatments and vaccines against viral infection (see Table 2).
Table 1.
LFA-based studies for COVID-19 detection.
| Disease | Reporter | Detection element | Assay type | Analyte/Real Sample | Specificity or LOD | Assay time | References |
|---|---|---|---|---|---|---|---|
| COVID-19 | AuNPs | IgM/IgG Antibody | LFA | Serum | Sensitivity IgM 88% Sensitivity IgG 90% |
15 min | [50] |
| COVID-19 | AuNPs | IgM/IgG Antibody | LFA | Whole Blood | Sensitivity IgM 92% Sensitivity IgG100% |
10 min | [54] |
| COVID-19 | Carboxylic red latex beads | Nucleocapsid proteins | Half-LFA | – | 3.03 ng/mL | 20 min | [77] |
| COVID-19 | IgM/IgG Antibody | Fluorescent LFA | Serum | IgM 100% IgG 100% |
15 min | [73] | |
| COVID-19 | AuNPs | Spike glycoprotein | LFA | – | 5 μg/mL | 30 min | [75] |
| COVID-19 | SiO2@DQD fluorescent probe | IgM/IgG Antibody | Fluorescent LFA | Serum | Sensitivity 97.37% Specificity 95.54% |
15 min | [74] |
| COVID-19 | LNPs | IgG Antibody | LFA | Serum | – | 10 min | [52] |
| COVID-19 | AuNPs | Total antibody | LFA | Serum | Sensitivity 94.6% Specificity 100% |
20 min | [53] |
| COVID-19 | Gap-enhancement nanotags | IgM/IgG Antibody | SERS-based LFIA | – | IgM 1 ng/mL IgG 0.1 ng/mL |
15 min | [76] |
| COVID-19 | Red cellulose nanobeads | S protein | LFA | Nasal swabs | 1.86 × 105 copies/mL | 20 min | [78] |
| COVID-19 | AuNPs | IgM Antibody | LFA | Whole blood | Sensitivity 100% Specificity 93.3% |
15 min | [188] |
| COVID-19 | AuNPs | IgG Antibody | LFA | Serum | Sensitivity 69.1% Specificity 100% |
15–20 min | [189] |
| COVID-19 | Eu(III) fluorescent microsphere | IgM/IgG Antibody | Fluorescent LFA | Serum and plasma | IgM 98.68% IgG 98.72% |
10 min | [190] |
| COVID-19 | AuNPs | IgM/IgG Antibody | LFA | Whole blood | Sensitivity 85.29% Specificity 100% |
15 min | [191] |
| COVID-19 | AuNPs | IgM/IgG Antibody | LFA | Serum | Specificity 97,47% | 15 min | [192] |
| COVID-19 | Selenium nanoparticle | IgM/IgG Antibody | LFA | Whole blood and serum | Sensitivity 93.33% Specificity 97.34% |
5 min | [193] |
| COVID-19 | Europium (III) chelate microparticles | Nucleocapsid proteins | LFA | Oropharyngeal swabs | Sensitivity 100% Specificity 100% |
10 min | [194] |
| COVID-19 | Co–Fe@hemin-peroxidase nanoenzyme | S protein | LFA | Saliva and nasal swab | 0.1 ng/mL | 16 min | [195] |
| COVID-19 | Gold nanostars | IgM Antibody | SERS-based LFA | – | 100 fg/mL | – | [196] |
| COVID-19 | AuNPs | S protein | LFA | – | 61 pg/mL | 8 min | [197] |
| COVID-19 | AuNPs | IgM/IgG Antibody | LFA | Serum | Sensitivity 90% Specificity 96.6% | 10–15 min | [198] |
| COVID-19 | Fluorescence | SARS-CoV-2 viral RNA | Catalytic hairpin assembly-LFA | Nasal swabs | 2000 copies/mL | 90 min | [199] |
| COVID-19 | Simple split luciferase | S protein | LFA | Serum | Sensitivity 98% Specificity 100% | 30 min | [200] |
| COVID-19 | Carbon dot-based silica spheres | Nucleocapsid proteins | LFA | Serum | 10 pg mL | 20 min | [201] |
Table 2.
LFA-based studies for the detection of viral infectious diseases.
| Disease | Reporter | Detection element | Assay type | Analyte/Real Sample | Specificity or LOD | Assay time | References |
|---|---|---|---|---|---|---|---|
| Dengue | Dextrin-capped AuNPs | Dengue-1 RNA | LFA | Serum | cut-off value of 1.2 × 104 pfu/mL | 20 min | [94] |
| Zika and Dengue | Gold nanostars | Zika NS1, Dengue NS1 |
SERS-based LFA | Serum | Zika NS1 0,72 ng/mL Dengue NS1 7,67 ng/mL |
– | [99] |
| Zika | Si-AuNPs | Zika NS1 | SERS-based LFA | – | 1.906 μg/mL | [100] | |
| Dengue | Carboxyl-Adembeads | DENV-1, DENV-2, DENV-3, DENV-4 |
Magneto enzyme LFA | – | 0.1 ng/mL 1 ng/mL ng/mL 1 ng/mL |
25 min | [101] |
| Zika and Dengue | Quantum dot microspheres | ZIKV NS1, DENV NS1 |
Fluorescent-LFA | – | 0.045 ng/mL and 0.15 ng/mL | 20 min | [102] |
| Zika | Silanized carbon dots | Zika NS1 | LFA | – | 10 pg/mL | 20 min | [202] |
| Dengue | Gold decorated graphene oxide sheets | Dengue NS1 | LFA | – | 4.9 ng/mL | 10 min | [203] |
| AIDS | AuNPs | HIV-1 DNA | Surface modified AuNP by MGITC as Raman reporter | – | 0.24 pg/mL | 15 min | [105] |
| AIDS | AuNP | HIV-1 p24 antigen | LFA | Serum | 30 pg/mL | 40 min | [108] |
| AIDS | AuNPs | HIV-1 RNA | LFA | – | 0.5–13 log10 copies/mL (50 copies of gag RNA) | 20 min | [106] |
| AIDS | QDs | HIV-DNA | LFA | Serum | 0.76 pM | 15 min | [107] |
| AIDS,Treponema pallidum virus, Hepatitis B and Hepatitis C |
QDs | HIVTreponema pallidum antibody, hepatitis C virus antibody, hepatitis B virus surface antigen |
Fluorescent LFA | Serum | 0.11 NCU/mL, 0.62 IU/L, 0.14 NCU/mL, 0.22 IU/mL |
20 min | [110] |
| AIDS | GNS | HIV-1 p24 antigen | TCA- LFA | Serum | 8 pg/mL | – | [109] |
| AIDS | Carbon nanoparticles | HIV-1 p24 antigen | LFA | – | 50 pg/mL Sensitivity 90% Specificity 100% |
40 min | [204] |
| AIDS | PtNCs | HIV-1 p24 antigen | LFA | Serum | 8 pg/mL | 20 min | [205] |
| AIDS | AuNPs | HIV-1 DNA | LFA | – | 0.1 nM | 15 min | [206] |
| AIDS | AuNPs | HIV gp41 antigen | LFA | – | Sensitivity 100% Specificity 100% |
15 min | [207] |
| AIDS | UCNPs | anti-HIV-1/2 antibodies | UCNPs LFS | Serum | Sensitivity 96.6% Specificity 98.7% |
20–40 min | [208] |
| Hepatitis | Carboxyl modified CdSe/ZnS QDs | HbsAg | LFA | – | 75 pg/mL | 15 min | [117] |
| Hepatitis B | Fluoro-Max fluorescent nanoparticles | HBV A HBV B HBV C HBV D |
Fluorescent -LFA | Serum | 2.5 IU/mL, 5 IU/mL, 5 IU/mL, 10 IU/mL |
20 min | [118] |
| Hepatitis B | Fluoro-Max fluorescent nanoparticles | HBV DNA | Fluorescent -LFA | Serum | 2.5–10.0 IU/mL | 20 min | [119] |
| AIDS Hepatitis C and Hepatitis A |
AuNPs | HIV, HCV, HAV | LFA | Serum | Sensitivity 100% | 30 min | [125] |
| Hepatitis | Up-converting phosphor | Hepatitis B surface antigen (HbsAg) | LFA | – | 60 mIU/mL Sensitivity 99.19% |
10 min | [209] |
| Hepatitis | ZnSe/CdSe/CdS/Cd xZn1−xS/ZnS QDs | Hepatitis B surface antigen | QD-LFIA | – | 0.05 ng/mL | 10 min | [210] |
| Influenza A | AuNPs | Hemagglutinin | LFA | – | 0.25 HA units | 10 min | [134] |
| Influenza A | Cy5 doped silica nanoparticles | Hemagglutinin | LFA | – | 250 ng/mL | 30 min | [138] |
| Influenza A and Human adenovirus | Fe3O4@Ag magnetic tags | H1N1 and HadV | SERS- based LFA | – | 50 PFU/mL, 10 PFU/mL |
30 min | [141] |
| COVID-19 and Influenza A | QDs | Sars-CoV-2 IgG Antibody and FluA H1N1 antigens |
Fluorescent -LFA | Serum | IgG 5 pg/mL FluA 50 pfu/mL |
15 min | [142] |
| Influenza B | Cy5-loaded silica nanoparticles | Influenza B virus protein | Fluorescent -LFA | – | 0.55 μg | 30 min | [211] |
| Influenza A and Influenza B | Streptavidin-coated gold colloid | H1N1 and H3N2 | Lateral flow dipsticks | – | H1N1 78.57% H3N2 87.50% |
20 min | [212] |
| Malaria | AuNPs | pLDH | Dipstick LFA | Serum | 1 ng/μL | 20 min | [148] |
| Malaria | AuNPs | pLDH | LFA | – | 10 ng/mL | – | [213] |
| PRV | AuAg4−ATP@AgNPs | wild-type PRV | SERS-based LFA | – | 5 ng/mL | 15 min | [153] |
| Y. pestis, F. Tularensis, and B. Anthracis | SERS nanotags | Y. pestis, F. Tularensis, and B. Anthracis | SERS- based LFA | – | 43.4 CFU/mL, 45.8 CFU/mL, 357 CFU/mL |
15 min | [157] |
| Ebola | AuNPs | EBOV Glycoprotein | LFA | Serum | Sensitivity 100% Selectivity 98% | 15 min | [214] |
| Tuberculosis | AuNPs | Tuberculosis antigen | LFA | Serum | Specificity 95% Sensitivity 40% |
25 min | [215] |
Antigen testing is a method of confirming SARS-CoV-2 infection by examining viral proteins in various sample types. As the COVID-19 pandemic continues, multiple variants of the SARS-CoV-2 virus have been found and identified that are more infectious than the original virus. The emergence of these variations puts the reliability and accuracy of antigen rapid recognition tests to the test in different situations. However, there is a lack of information regarding antigen performance in clinics. The antigen accuracy and performance of these LFA-based antigen tests reported in clinical trials should be further evaluated.
With the quick spread of COVID-19, the FDA has begun to issue Emergency Use Authorizations (EUA) for several COVID-19 diagnostic tests [80]. In fact, tests with FDA-EUA status have not received full FDA approval. Instead, permissions for these tests are only valid for the duration of the pandemic [193]. Cellex developed the first rapid antibody blood test (SARS-CoV-2 IgG/IgM Rapid Test) for SARS-CoV-2 approved by the FDA [81]. The test was developed to detect patient IgG and IgM antibodies against SARS-CoV-2 results within 15–20 min. The test can be used on serum, plasma, or whole blood samples. 128 COVID-19 positive patients and 250 negative control patients were tested to evaluate the test's clinical performance. The clinical sensitivity of the test was % 93.8, and the clinical specificity of the test was % 96. The Cellex assay has pioneered other lateral flow immunoassay tests to detect IgG and IgM, such as the Autobio Diagnostics Anti-SARS-CoV2 Rapid Test [82]. Autobio Diagnostics Anti-SARS-CoV2 Rapid test is advantageous as it does not rely on visual IgG/IgM detection. Instead, it uses the micro reader, which provides a qualitative readout to eliminate the possibility of user bias or misinterpretation. The test's clinical specificity was reported to be % 97.6 for IgM, % 96.8 for IgG, and % 94.4 for the combination of IgM and IgG. In 2020, Ortho Clinical Diagnostics developed the VITROS Immunodiagnostic Products Anti-SARS-CoV-2 Total Reagent Pack/Total Calibrator, one of the first CLIA tests approved by the FDA for EUA. In the test that detects total IgG/IgM, clinical sensitivity was reported as % 83 and clinical specificity as % 100. This test takes approximately 50 min [83]. Roche developed another test similar to the CLIA test. The Elecsys® _Anti-SARS-CoV-2 Test detects total antibody to N protein, and the test time takes only 18 min. After PCR validation, clinical sensitivity was reported as % 100 and clinical specificity as % 99.81 [84]. If a detailed comparison of FDA-EUA-approved major serological tests is desired, all tests can be used with serum or plasma samples. Tests developed by both Cellex and Bio-Rad have relatively lower sensitivity than ELISA tests [85]. The Bio-Rad test also appeared to have the longest turnaround time, as ELISA tests have longer incubation times than CLIA tests. Premkumar et al. showed that the S protein is highly sensitive to antibody detection in patients [86]. Therefore, tests that detect total antibodies against the RBD region of the S protein are particularly important [83,[87], [88], [89]]. There was pressure to develop rapid diagnostic tests, as the healthcare system was under severe pressure during the pandemic. Consequently, a holistic review and evaluation of all available diagnostic tests for COVID-19 are required.
4.2. Zika and dengue
Zika virus is a mosquito-borne flavivirus and was first described in monkeys in Uganda in 1947. It was later identified in humans in 1952 in Uganda and the United Republic of Tanzania [90]. It is silent in most infected people, usually remains benign, and can last for up to a week. The disease presents 3–12 days after the bite of the insect vector, with various symptoms such as fever, headache, skin rash, fatigue, and muscle and joint pain. When the zika virus infects pregnant women, it causes their babies to be born with smaller than normal heads [91].
Dengue fever, transmitted by the same mosquito species, is much more common and the most dangerous. It causes high fever, headache, muscle aches, and severe joint pain. Symptoms usually appear 4–10 days after being bitten. Most people recover within a week, but sometimes the symptoms are life-threatening because the disease can damage blood vessels [92].
The important point here is the close relationship between zika and dengue fever. If someone who has had dengue fever is infected with zika, the disease is severe. This is because the body's antibodies against dengue respond to the patient with zika [93]. Scientists have yet to develop a specific drug or vaccine for zika and dengue virus. Consequently, it is crucial to detect these two infections simultaneously using a fast, reliable, and inexpensive technology. LFA tests are significant in monitoring disease progression by assessing antibody abundance and short- and long-term antibody responses. It allows the use of different targets in a single device for reliable and specific pathogen detection. LFA is seen as the most suitable platform to meet these expectations. Therefore, many LFA studies have been conducted by researchers in this area.
In 2019, a lateral flow test was developed to detect Dengue-1 RNA using dextrin-capped AuNPs in conjugation. This study first demonstrated the feasibility of dextrin-capped AuNP as a label for lateral flow testing. The detection principle is based on the nucleic acid sandwich reaction. A simple one-step signal amplification strategy using oligonucleotide-linked gold nanoparticle clusters has been developed to increase the sensitivity of the nucleic acid LFA assay. Three DNA probes were used in the biosensor: an AuNPs-labeled reporter probe (10 nm), a DENV-1-specific capture probe (dsDNA), and a control probe (cDNA). The probes detect negative-sense target RNA amplicons from nucleic acid sequence-based amplification (NASBA). Performing RNA amplification with isothermal reactions at 41°C without the need for heat cycling with NASBA makes it advantageous compared to RT-PCR. Moreover, it can be applied directly to LFA for detection. As a result of detailed optimization studies, target Dengue-1 RNA can be successfully detected in LFA within 20 min [94].
Recently, a new concept of LFA based on surface-enhanced Raman scattering (SERS) has been developed to address the shortcomings of conventional AuNP-based LFA [95,96]. SERS is a method of detecting molecules adsorbed on a rough metal surface by amplifying Raman scattering. SERS nanotags modified with AuNPs as signal reporters are an important aspect of the SERS-based LFA system. Representative SERS nanotags consists of three components: (i) charged Raman dye molecules to provide a strong and specific SERS signal, (ii) core based on gold or silver material as the enhanced substrate, and (iii) surface modified antibodies [97,98]. Additionally, the SERS-based LFA strip offers remarkable multi-channel detection capability in one test.
Dengue and zika are mosquito-borne infections with similar symptoms but quite different outcomes. Sanchez and his team reported an LFA-based system that takes advantage of the high sensitivity of SERS in a multiplex assay capable of distinguishing nonstructural protein 1 (NS1) biomarkers of dengue and zika viruses. The gold nanostar was used as a marker probe. Monoclonal antibodies against zika or dengue NS1 protein were immobilized at the test site. The control area was labeled with a goat antibody capable of binding to the Fc portion of mouse IgG antibodies on the gold nanostar (GNS). The biomarker NS1 for acute dengue is present at elevated ∼15 μg/mL levels in patient serum two days after infection. This high concentration is a great advantage for reading results in LFA tests. However, NS1 levels for zika are still largely unknown but are expected to be much lower than Dengue NS1 from the data available to date. Therefore, increased test sensitivity is required for zika detection. Thus, it can help detect both viruses early. This study successfully reported a new SERS-based LFA assay that can distinguish between dengue fever and zika fever with a lower LOD than that obtained by colorimetric reading. The compared lanes showed a 15-fold increase in LODs for zika and a 7.2-fold increase in dengue LODs [99].
Jeon and colleagues used silica-encapsulated AuNPs (Si-AuNPs) in LFA to increase thermal stability and maintain reproducibility in temperature domains (Fig. 5 A). Thermally stable SERS nanotags are required to identify dengue and zika viruses common in tropical regions. The researchers performed SERS-LFA using Si-coated AuNPs as a marker probe on an LFA strip, and the results were compared with those obtained using AuNPs alone to overcome this problem. Si-AuNPs were synthesized by a layer-by-layer method. AuNPs were prepared according to the kinetically controlled seed growth method. First, the surface of AuNPs was labeled with Raman reporter molecules (MGITC) to create SERS-active nanoprobes. The SH-PEG-COOH solution was added dropwise to the MGITC-labeled AuNPs with vigorous mixing and made ready. Here, the SH-PEG-COOH solution was used as a coupling agent, in which silica could be grown directly on particle surfaces. In conventional LFA strips using AuNPs, sensitivity decreased when the temperature exceeded 45°C, while strips prepared using Si-coated AuNPs retained similar LOD values regardless of temperature increase. In the light of the obtained data, it was concluded that this SERS-LFA strip using Si coated AuNPs can be used as an ideal platform for zika, dengue, HIV, and malaria endemic to high-temperature regions [100].
Fig. 5.
(A) Schematic illustration of the SERS-based LFA for human IgG using thermally stable Si-AuNPs. The Si-AuNPs are trapped by anti-human IgG antibodies on the test region, whereas the excess SERS nanoprobes are captured by capture antibodies on the control region [100]; (B) Reading the proposed fluorescent LFIA test result with a smartphone and the general principle of the design. a) 3D schematic of the imaging device showing the internal structure and components, b) İmage of the designed LFA reader device, c) Detailed schematic representation of fluorescent based LFA for determination of Zika NS1, d) Strip images of positive and negative results [102]. Reproduced with permission from ref. 100–102, Elsevier Ltd.
In 2019, researchers developed a magneto-enzyme LFA that combines superparamagnetic nanoparticles as tags and the streptavidin-biotin signal amplification method to identify Dengue NS1. This study uses magnetic nanoparticles attached to monoclonal antibodies as the detection complex in the magneto-enzyme LFA developed. When the TMB substrate is introduced, the biotin biomolecules allow further interaction with Streptavidin and polyhistidine conjugates, resulting in an enhanced signal. Unlike previous similar studies, this newly developed assay can detect Dengue NS1. LOD was detected as low as 0.25 ng/mL for DENV-1 and DENV-3, as low as 0.1 ng/mL for DENV-2 and 1.0 ng/mL for DENV-4. The LOD obtained for DENV-2 exhibited a 50-fold over the previously reported best values [101].
Rong et al. developed a smartphone-based fluorescent LFA platform for high-sensitivity detection of Zika NS1 and Dengue NS1 proteins (Fig. 5B). A plug-in was designed, and 3D printed to connect the smartphone with external optical and electrical components. The smartphone attachment was designed using SolidWorks software. Emitting a high-power UV LED at 365 nm, this device contains light to excite the QD microspheres deposited in the LFA strips. This approach allowed the device to be miniaturized and reduced cost and complexity. Fluorescence-based LFA uses QD microspheres as tags as they produced extremely bright fluorescent signals. This assay determined the detection limits of Zika NS1 and Dengue NS1 at 0.045 ng/mL and 0.15 ng/mL within 20 min [102].
4.3. HIV
The human immune deficiency virus type 1 (HIV-1) is a retrovirus that affects the body's immune system, rendering it incapable of resisting infection. HIV is primarily a sexually transmitted infection. However, it may also be transmitted by blood contact or transfer. The HIV-1 virus can develop into acquired immune deficiency syndrome (AIDS), which causes illnesses and infections and generally results in death [103]. The morbidity, mortality, and social disruption caused by the HIV pandemic continue to be exacerbated by resource shortages in poor areas [104].
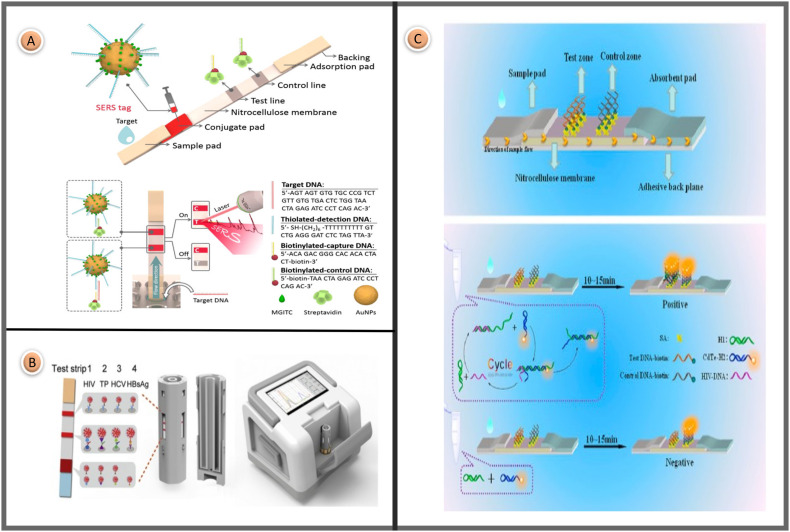
Fu et al. reported a SERS-LFA platform for the sensitive determination of HIV-1 DNA. The researchers suggested that this study is the first report of a SERS-based HIV-1 DNA LFA biosensor. Oligonucleotide-linked gold nanoparticle clusters are used to increase the sensitivity of the LFA assay. When reporter molecules are adsorbed on the AuNP surface, SERS signals are greatly enhanced due to electromagnetic and chemical amplification effects. AuNPs were prepared according to the citrate reduction method. A Raman reporter (MGITC molecules) was adsorbed on the surface of AuNPs to prepare a Raman active probe. Next, thiolated DNAs were conjugated on the surface of MGITC-functionalized AuNPs. The measurement principle of the SERS-based lateral flow test for the quantification of HIV-1 DNA is shown in Fig. 6 A. Under optimum conditions, SERS-LFA with a LOD of 0.24 pg/mL was approximately 1000 times more sensitive than colorimetric or fluorescent detection methods [105].
Fig. 6.
(A) Installation image and measuring principle of the SERS-LFA for quantification of HIV-1 DNA biomarker [105]; (B) Demonstration of design of integrated fluorescence lateral flow assay platform with 4-sided cassette for multiplex detection of HIV, TP, HCV markers [110]; (C) The elaboration of a LFA strip and detail schematic of strand displacement amplification and positive or negative expression on strips [107]. Reproduced with permission from ref. 105-110-107, Elsevier Ltd.
It was desired to improve the sensitivity of nucleic acid detection by signal amplification to avoid the difficulties associated with enzymatic amplification of target RNA. Rohrman et al. developed a quantitative LFA that detects amplified HIV RNA using AuNPs probes and gold enhancement. The technique uses an AuNP-labeled nucleotide sequence to detect an amplified 142 bp RNA sequence. When an RNA sequence is present in a sample, the strand binds to the probe labeled AuNPs and the nucleotide sequence found on the NC membrane. The test uses a 20 μL sample and only needs three steps over 20 min. The LFA test achieved a resolution of 0.5 log10 copies/mL [106].
The use of QDs in LFA strips has become very common due to the potential to increase the sensitivity and accuracy of the system and its biocompatibility. In 2018, Deng and colleagues designed highly reproducible fluorescent LFA strips to detect HIV 1 DNA in serum (Fig. 6C). Transmission electron microscopy (TEM) images showed that the QD nano-composite particles had an average diameter of 4 ± 1.5 nm with good homogeneity and dispersion. Quantum dots were combined with DNA based on a one-step hydrothermal process. The complex nanocomposite was gradually cooled to room temperature and purified by centrifugal ultrafiltration. Quantum dots were combined with DNA based on a one-step hydrothermal process. The complex nanocomposite was gradually cooled to room temperature and purified by centrifugal ultrafiltration. Streptavidin (5 mg/mL) was added into biotinylated t-DNA (50 μM) and c-DNA (50 μM) and stripped to the test and control lines. The detection limit of this assay is between 1 pM and 10 nM. The researchers also predicted that this method could be applied prospectively in clinical applications [107].
LFA was designed to detect HIV-1 p24 antigen using superparamagnetic particles as the identifying marker. The lower detection limit for HIV-1 p24 and % 50 serum added to the test sample buffer was 30 pg/mL. The test can be completed in 40 min. This testing process is considered quite lengthy for a traditional LFA. However, compared to typical LFA readers, the equipment used to read these tests is economical. It has already received CE certification in the Southeast Asian and European markets. A battery-powered, lower-cost handheld variant is currently in development [108].
In another study targeting the detection of HIV p24 protein, Zhan et al. aimed to develop a thermal contrast amplification (TCA) LFA to supply ultrasensitive detection of HIV p24 protein. As its operating principle, the TCA reader device interprets the thermal signal of the AuNPs in the NC membrane when a laser irradiates the particle at the plasmon resonance wavelength. Various optimizations were made, such as the effect of laser power, particle size, shape, and appropriate laser wavelength selection, and 100 nM spheres with a 100 mW 532 nM laser were reported to provide the best performance. As a result, a successful TCA-LFA with a LOD of 8 pg/mL was designed [109].
Rong and colleagues presented an integrated fluorescent LFA strip technology for the multiplex detection of four infectious (hepatitis B, hepatitis C, Treponema pallidum antibody, and AIDS) disease markers at point-of-care in 2021. Fluorescent probes containing hundreds of QDs in a single polymer microsphere with a diameter of 150 nm and exhibiting an intense sharp red fluorescence spectrum were used as colored markers. As seen in Fig. 6B, the cassette has four sides. Different biomolecules were stripped onto the test line to detect other analytes on each surface. The cartridge, which included four test strips, was inserted into a rotating holder driven by a stepper motor. The centrifugal force may guide the sample solution fed from a single point into each reaction chamber with a maximum flow rate of 1 mm/s in the channel center at 420 revolutions per minute. A specially designed Si photodiode-based optical signal acquisition and amplification module consists of a rotating stage and a vertically oriented slide. The developed method allowed highly sensitive optical scanning of many strips in 5 s with a wide dynamic range. The disposable multichannel LFA strip cartridge was devised and manufactured at a low cost for automated fluid supply. A miniaturized tape reader device with electrical, mechanical, and optical components and software was custom-made for as little as $200. In the proposed assay, the LODs were 0.11 NCU/mL, 0.62 IU/L, 0.14 NCU/mL, and 0.22 IU/mL, respectively, over a 20-min assay time [110].
Detection of HIV-1 RNA, capsid antigen (p24), and anti-HIV antibodies are used to diagnose HIV-1 infection, which is the cause of AIDS. Molecular diagnostic analyses of genomic sequences have provided a highly sensitive and quantitative method for detecting infectious diseases, pathogens, and genetic variation. For decades, the simple, hypersensitive detection of sequence-specific DNA has been the target of many studies and technologies [111]. For this reason, when LFA-based HIV tests were examined, it was seen that HIV DNA/RNA was preferred as the target molecule in many studies. During the early, acute phase of infection and the final stage of AIDS, the HIV-1 p24 antigen level is significantly higher. It is also an important marker of choice for predicting CD4+ [112]. However, it is also known that high concentrations of biotin prevent the detection of HIV-1 p24 antigen in serum [113]. This significantly reduces the specificity of the tests. Since the possible inhibitory effect of biotin in HIV-1 p24 antigen tests has not been investigated, the reliability of the tests is doubtful. For this reason, necessary selectivity tests for biotin selectivity should be performed to eliminate cross-interactions in developed LFA tests.
4.4. Hepatitis
Hepatitis B is an infectious disease that damages the liver caused by the hepatitis B virus. Many Hepatitis B patients report no symptoms during the first infection, but the disease can progress over time, leading to cirrhosis and liver cancer. Hepatitis A virus produces acute hepatitis via a fecal-oral infection pathway and can live in an abiotic environment for a few months. It can withstand rapid changes in the environment, such as low pH or cold [[114], [115], [116]]. Even though developing an effective vaccine in the early 1990s, around 1.5 million individuals become sick each year. As a result, rapid and reliable diagnostic methods are critical for reducing the risk of infectious diseases.
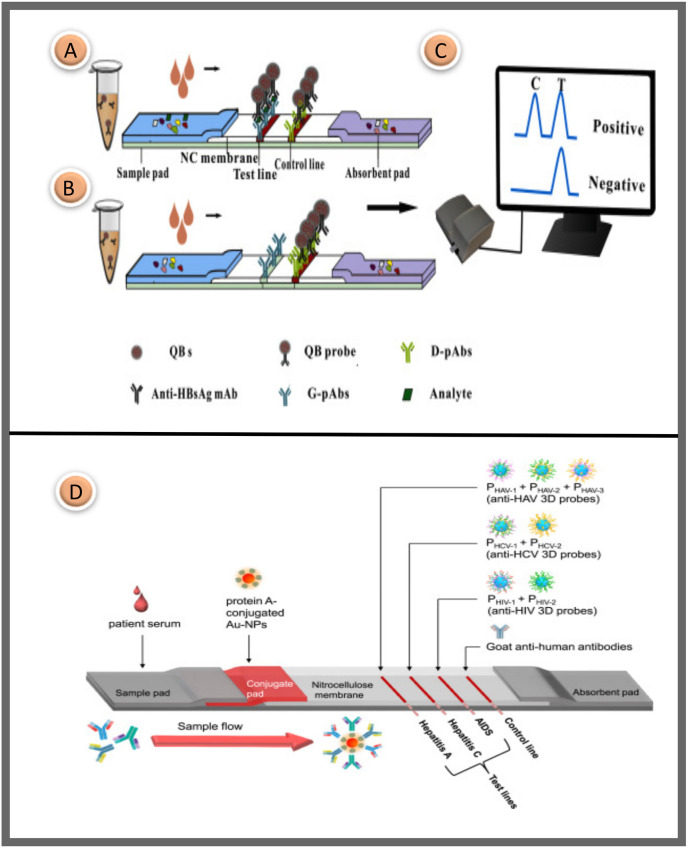
In a 2015 study, highly luminescent quantum dot beads (QBs) were used as marker probes in an immunochromatographic sandwich format for the high-sensitivity quantitative detection of hepatitis B virus surface antigen (HBsAg) in serum. The QBs were prepared by applying a microemulsion technique. Goat anti-HBsAg polyclonal antibodies (G-pAbs) and Donkey anti-mouse polyclonal antibodies (D-pAbs) on the NC membrane were striped to the test and control line. When the target analyte HBsAg was included in the human blood sample, the QB probe on the conjugation pad binds with HBsAg to form a QB-mAb-HBsAg complex. Subsequently, the combination was captured by G-pAbs and D-pAbs on the NC membrane to generate fluorescence. It can be seen in Fig. 7 A. In contrast, when the serum target analyte was not present, the QB probes were not fixed by the G-pAbs to form a sandwich formation, resulting in no fluorescent signal in the test region. A commercial fluorescent strip reader was used to capture fluorescence density signals on both lines for quantitative assessment of HBsAg levels in the blood (Fig. 1C). Also, a step motor controller was used to transport the strip into the reader device, and then a high power 450 nm light-emitting diode was used to activate the QBs on both lines. The absorption fluorescence of the QBs was focused using a cylindrical lens and signaled by a photosensitive cell after passing through an optical slit. As a result, the suggested QB-LFA sensor provided outstanding performance for the quick determination amount of serum HBsAg within 15 min. Its high sensitivity of 75 pg/mL outstripped the standard approach [117].
Fig. 7.
The sandwich approach for detecting HBsAg utilizing a QB based LFA platform is shown schematically; (A) The five-component assembly of a traditional LFA, with a test region and a control region due to positive testing; (B) An immunochromatographic test with a control line yields a negative result; (C) The reader for fluorescence strips [117]; (D) Schematic representation of the developed LFA system. The NC membrane contains three test zones and a control zone. The test regions include the protein probe to detect anti-HIV, anti-HCV, and anti-HAV antibodies, respectively, while the control region contains goat anti-human secondary antibodies [125]. Reproduced with permission from ref. 117–125, Elsevier Ltd.
Jiang et al. published a Hepatitis B virus (HBV) diagnostic and genotyping test based on the LFA platform. They used four genotype-specific monoclonal antibodies (mAbs 2B2, 16D12, 6H3, 3E6) to distinguish between the HBV kinds (A, B, C, and D). Using an artificial intelligence system and the multiplex detection module, they used a smartphone camera to measure fluorescence signals and calculate the findings [118]. In a previous study, Song and colleagues similarly reported that multiplex LFA provides multiple simultaneous diagnoses. They developed a new DNA-independent HBV genotyping tool based on a one-step fluorescent LFA. In addition, there are efforts to increase the sensitivity of the HBV LFA platform. The novel assay can provide HBV genotyping test results in 20 min [119]. The same team also undertook work to improve the sensitivity of the HBV LFA platform. To this end, they made AuNPs with a well-controlled, restricted size distribution. AuNPs were conjugated with antibodies, and it was reported that more sensitive results were obtained in the LFA biosensor at the end of this modification [120].
HIV and HCV have similar transmission routes [121]. HIV infection often causes the progression of liver fibrosis, while HCV infection is linked to faster onset of AIDS [122]. Co-infected patients need to be treated with two different anti-viral therapies. Multiplex diagnosis requires simultaneous detection of HIV and HCV to detect co-infected individuals easily [123]. In addition, Hepatitis A virus reasons acute hepatitis by fecal-oral infection. It may thrive in an abiotic environment for many months while resisting rapid environmental changes such as low pH or freezing [124]. For the LFA-based multiplex diagnosis of three viral intractable diseases (HIV, hepatitis C, and A), 7 different protein-based 3D probes that display various viral antigens on their surface were developed. In Fig. 7D, the arrangement of biomolecules on the strip is visualized in detail. These probes were much more sensitive than peptide probes, and the peptide probe-based LFA correctly identified all 20 patient samples per illness without a false negative signal. In contrast, peptide probe-based LFA had % 65–90 diagnostic sensitivity. The researchers suggested that protein-based 3D probes could solve the problems of probes that generally lower the diagnostic accuracy of LFA, such as clustering, irregular flow, inactivation, uncontrolled orientation, and test surface instability [125].
4.5. Influenza
Another well-known example of cross-species transmission is the influenza virus [126,127]. Different seasonal influenza viruses are classified as A, B, and C types [128]. Type A influenza is next separated into some subtypes based on the surface proteins, including the neuraminidase (N protein) and the hemagglutinin (H protein), for example, A(H1N1), A(H3N2), A(H5N1), and so on. Influenza A is a highly contagious virus that causes severe respiratory infections and has re-emerged in 1918, 1957, and 1968 [129]. It's critical to detect the virus early, treat patients appropriately, and vaccinate high-risk people to avoid a pandemic [130]. Therefore, the virology, clinical manifestations, laboratory diagnosis, and treatment of these viral infections have been well examined and studied extensively [[131], [132], [133]].
Peng et al. developed an immunochromatographic strip for detecting the H9 subtype of avian influenza viruses in poultry using two mAbs (4C4 and 4D4) for H9AIV hemagglutinin. Conjugation was prepared with the classical AuNP protein conjugation procedure. 4D4 was stripped to the test line and a goat anti-mouse antibody was used in the control region of the nitrocellulose membrane. Within 10 min, the results can be read with the naked eye. The tests yielded no false positive or negative results for any LFA strips. In conclusion, the H9AIV immunochromatographic strip has been developed with high specificity, sensitivity, and stability [134].
A few researchers reported on the LFA test using the fluorescent (Cy5)-doped silica nanoparticles as a marker for influenza A antigen detection. Cy5 is known to be one of the most fundamental dyes for labeling in biological research disciplines such as fluorescent biosensors, flow cytometry, and microarray analysis [135]. The absorption and emission wavelengths of Cy5 dyes are in the red spectral region and cause low background noise. Cy5 dyes are ideal marker candidates because they eliminate background contamination in LFA tests. It was revealed by Hee et al. that Cy5-doped silica nanoparticles indicated excellent photostability. Also, it has been proven that fluorescence-doped silica nanoparticles are more sensitive and photostable than organic fluorophores as fluorescent probes for bioanalysis [136]. Thousands of fluorescent molecules are present in the silica matrix during production and protect them from the oxygenated environment. As a result, both dye photostability and probe sensitivity are significantly improved [137]. Within 30 min, LFIA detected recombinant nucleoprotein as low as 250 ng/mL using a 100 μL sample. Compared to a commercial influenza A test using gold nanoparticles as the reporter, this technique showed 8 times greater sensitivity. Based on their unique optical properties and versatility in synthesis and surface modification, this study demonstrated that fluorescent doped silica nanoparticles are good candidates for an LFIA system as fluorescent probes [138].
In 2019, the first study to insert magnetic SERS tags into a lateral flow immunoassay strip was published in the Journal Nature Communications. A sensitive and quantitative SERS-LFIA strip was designed to rapidly analyze human adenovirus (HAdV) and influenza A H1N1 virus using Fe3O4@AgNPs (150 nm) as magnetic SERS nanotags. These magnetic SERS substrates can be used to separate the target analyte from a complex solution with an external magnetic field. Owing to the sample enrichment property of Fe3O4 MNPs, excellent SERS activity, and the stability of magnetic NPs, it has the potential to serve as stable SERS substrates to enhance Raman signaling [139,140]. The magnetic SERS biosensor is based on antibody-antigen interaction and consists of three lanes (two test regions and one control region). In the developed lateral flow test, the conjugate pad found in traditional LFA platforms was not used. After the flow was completed, the virus was captured by virus detection antibodies immobilized on the test regions (test region 1 for H1N1 and test region 2 for HAdV). Wang et al. show that the visualization limit of the magnetic SERS strip is 10 times and the SERS detection limit is 2000 times lower than that of conventional AuNP biosensors [141].
Wang et al. proposed a two-channel fluorescent immunochromatographic assay for ultrasensitive and parallel qualification of SARS-CoV-2 antigen and influenza A virus in biological samples (Fig. 8 A-B-C). A highly luminescent nanobead was used as a colored label to ensure the high sensitivity and stability of the LFA test. High-performing QDs nanobeads were developed through the adsorption of multilayer dense QDs onto the SiO2 surface (180 nm). Under optimal conditions, LOD of SARS-CoV-2 antigen and FluA H1N1 in throat swab samples were 5 pg/mL and 50 pfu/M. According to the research results, the developed LFA test results are approximately 100 times more susceptible than AuNP-based tests [142].
Fig. 8.
(A) Synthesis of silica-QD nanobead probes for detection of SARS-CoV-2 and FluA at the same time, (B) conjugation of silica-QD nanobeads with SARS-CoV-2 NP and FluA, (C) application of developed beads to LFA-based biosensor and test working principle [142]; (D) Detail illustration of the SERS-LFA. (a) Positive result, the AuAg4−ATP@AgNPs-(gE-mAb) is captured at both the detector region (test and the control). (b) Negative result, the AuAg4−ATP@AgNPs-(gE-mAb) is captured at the control region. (c) Basic drawing and measurement of the positive reaction on the T line [153]; (E) Operating concept of the SERS-LFA biosensing platform. Y. pestis, F. tularensis, and B. anthracis LFA strips were dipped into wells of a 96-well ELISA plate containing mixes of SERS nanotags and varying quantities of bacteria in buffer solution. The generated immunocomplexes moved to the test line through capillary action, where their Raman signals were recorded and evaluated [157]. Reproduced with permission from ref. 142-153-157, Elsevier Ltd.
4.6. Malaria
A parasitic human disease spread by female Anopheles mosquitoes, malaria is still a major worldwide problem. The disease is more prevalent in the tropical and subtropical parts of the globe [143,144]. The causative agent is a parasitic protozoan of the genus Plasmodium. Five species (P. falciparum, P. vivax, P. malariae, P. Ovale, and P. knowlesi) are known to affect humans. There were 228 million cases reported globally, with 405,000 fatalities [145]. Anemia and jaundice may develop in cases where diagnosis and treatment are delayed. In some types of parasites that cause malaria, if treatment is not started within 24 h, it can progress and lead to death. The World Health Organization aims to eradicate malaria by 2030. However, the goal can be achieved when all cases are correctly diagnosed and treated appropriately [146]. Although many tests are available today to diagnose malaria, the tests suffer from many limitations, such as variability in results, lack of quantification, and poor storage stability in the tropics [147]. Therefore, LFA is a good alternative device.
Pereira et al. used Triton X-114 micelle aqueous biphasic system (ATPS) to determine the malaria protein biomarker, Plasmodium lactate dehydrogenase (pLDH). The group has previously introduced using a micellar ATPS of nonionic Triton X-114 detergent to concentrate biomarkers in a sample and facilitate their recognition by LFA. Nevertheless, complete phase separation and achieving the target intensity using the Triton X-114 system required long hours. Manual extraction of the high concentrate sample and application to LFA needed approximately 8 h. Through the proposed process, they showed for the first time that a micelle ATPS could be directly applied to a fiberglass paper membrane to outstandingly accelerate its macroscopic separation from 8 h to about 3 h. They created a 3D paper-based diagnostic strip that provides a 10-fold increase in detection limit over LFA. It efficiently processes undiluted human serum, supsuppliesfast test results with minimum user involvement, and does not require electricity or huge or complex laboratory equipment. According to the researchers, integrating a mycelial ATPS and LFA could improve the use of paper-based diagnostic assays for malaria and similar illness in countries where resources are scarce [148].
Microscopy and LFA tests are the main diagnostic devices for malaria but fail to detect low-intensity parasitemias important for maintaining malaria transmission. Albert and colleagues integrated LFA with an isothermal reverse transcription-recombinase polymerase amplification to complement existing diagnostic methods. Recombinase polymerase amplification (RPA) is a versatile isothermal amplification technology that uses minimally prepared materials and takes less than 20 min [149]. This method uses recombinase, single-stranded DNA-binding protein, and DNA polymerase enzyme with chain displacement activity for amplification. As in PCR, at least one pair of primers specific to the target site is needed, and reactions can be carried out at low constant temperatures (37–42 °C). The design of fluorescent-marked probes particular to the region to be examined can be designed, and analysis with lateral current tests can be provided to detect the products formed after RPA. These characteristics may make RPA the ideal alternative for use as a PCR replacement at the point of need, particularly in low-resource situations. Reliable detection of asymptomatic low-intensity infections is possible with this technique, based on the combination of RPA with LFA tests. Assay time is 10 min, and the lower detection limit was 64 parasites/mL [150].
Multiplex loop-mediated isothermal amplification (mLAMP) method, like the RPA method, is one of the nucleic acid amplification methods that can be fast and economical in the diagnosis of infectious diseases. In another similar study, researchers identified the two most common malaria strains by integrating mLAMP and LFA. Results were visualized on LFA within 60 min for both species. Compared with conventional PCR, % 100 sensitivity and specificity have been reported. A LOD of 0.15 pg/μL was obtained. mLAMP-LFA assays can be a potential tool for early detection in non-laboratory conditions, with reduced assay time and ease of simple interpretation of results. Also, the specificity of the results is higher as many oligonucleotides are used in the reaction compared to RPA-LFA [151]. Another study using the LAMP-LFA technique for rapid malaria diagnosis reported a very low LOD value of 0.01 pg/mL, with assay time reduced to 42 min [152].
4.7. Other viruses
Pseudorabies virus (PRV) is a pathogen that causes infectious diseases in pigs and can cause huge financial losses to the farming industry. Recently, researchers developed a SERS-based LFA strip to detect PRV, which causes a serious infectious disease among wild animals. As shown in Fig. 8D, anti-PRV glycoprotein B (gB) mAbs (0.78 mg/mL) and goat anti-mouse IgG (0.5 mg/mL) were recruited into the test and control lines, respectively. Unlike conventional LFA platforms, a colored marker probe and a conjugation pad containing detector antibodies were not used in the designed system. Instead, the sample is added to the antibody's liquid labeled with the marker probe and mixed. After sufficient interaction time, the mixture is loaded onto the sample pad; then the flow is allowed. The result can be read in the test line after the stream. AuAg@Ag SERS nanotags were captured in both test and control sites in the presence of a sample containing wild-type PRV. In contrast, SERS nanotags were captured only in the control region for a sample without the target PRV virus. The linear detection range was 41–650 ng/mL, while the LOD was reported to be 5 ng/mL [153].
PRV contains at least 11 glycoproteins, gB, gC, gD, gE, gG, gH, gI, gK, gL, gM, and gN, each having various physiological activities [154]. Pigs infected with PRV are differentiated from those vaccinated using tests that detect antibodies to gE [155]. Hui et al. developed a new lateral flow immunoassay by conjugating PRV gE monoclonal antibodies (PRV gE-mAb) with EuNPs. EuNPs-PRV gE probe in the conjugation pad, PRV gE-mAb in the test line, and chicken IgY in the control line were immobilized. In the case of a positive serum administration, specific epitopes of the gE protein on the EuNPs-PRV probe in the conjugation pad were blocked. In this case, binding to PRV gE-mAb on the T line was inhibited, resulting in a low or negligible fluorescent signal. When a negative sample was applied, EuNPs-PRV probes could bind the target antibody on the T line, resulting in a high fluorescent signal. At 15 min, images were captured using a fluorescent camera, and fluorescence intensity data were collected using a fluorescence reader. This approach showed % 96.1 concordance with the most popular gE-ELISA kit [156].
The Ebola virus causes a febrile illness that causes life-threatening bleeding and results in death in % 50 to % 90 of cases. This virus is an RNA virus from the filoviruses family [157]. Hu et al. developed a new LFA test for the detection of the Ebola virus glycoprotein. A novel multifunctional nanosphere (RNs@Au) containing hundreds of quantum dots and dozens of Au nanoparticles was used as a reporter. The antibody and streptavidin were simultaneously modified on RNs@Au to label the target biomolecule (Ebola virus glycoprotein) and act as signal enhancers. The test can detect 2 ng/mL glycoprotein visually in 20 min, while it can quantitatively detect 0.18 ng/mL. RNs@Au is considered a successful study as it can provide both a colorimetric signal and a fluorescent signal to meet the requirements of different clinical conditions [158].
5. Discussion
Infectious diseases such as malaria, HIV, COVID-19, zika, dengue, hepatitis, and influenza account for more than a quarter of all fatalities globally every year (about 57 million) [159,160]. Many organisms that cause infectious diseases, especially RNA viruses, mutate quickly. Therefore, it is almost impossible to develop effective treatment and vaccine strategies. Due to the lack of a vaccine and limited treatment options for COVID-19, fast diagnostics must be developed and validated as soon as possible. It is essential to test target analytes as quickly as possible in a format that can predict them in the early diagnosis of diseases. Test methods such as RT-PCR, ELISA, or microfluidic system-based assays are not widely available in developing countries. Since these tests require equipment/devices and skilled personnel, they are only available in private clinical facilities [161,162]. As a result, these technologies are not suitable for POC use. Therefore, it is essential to decrease recognition time and develop a POC detection system outside of the clinical research laboratory. Compared to other commonly used diagnostic methods, LFA is pretty simple and cost-effective, with the added benefit of quick diagnosis at home. Moreover, the platform provides detection in a short time (10–15 min). Because of these characteristics, LFAs have become commonplace in various settings, including public and private clinics, hospitals, laboratories, and homes. The most important advantage is that it eliminates the obligation of patients in the risk group to go to the hospital during the pandemic process and reduces the burden on healthcare workers.
The sensitivity and specificity of LFAs have been significantly improved. Most LFAs are developed by targeting only one biomarker at a time. On the other hand, the most desired iteration of LFA is being able to detect many biomarkers simultaneously [163]. Given the complex nature of human diseases, coinfection states, and overlapping symptoms, the demand for multiplex systems that can identify multiple markers simultaneously has been increasing in recent years. When literature data is evaluated statistically, simultaneous diagnosis of infectious diseases with multiple biomarkers is limited due to inherent limitations in the design of conventional membrane-based LFAs. The optimum test condition for one disease may not be optimal for another or may have the risk of cross-reactivity. We propose that combining alternative biomolecules such as recombinant antibodies, aptamers, and engineered protein scaffolds in a single LFA assay may be suitable for designing high-specification multiplex LFAs while avoiding any potential cross-reactivity in the assays.
The immune response usually takes a long time to mature in vivo, followed by dynamic variations of the antibody response [164]. During this duration, a few specific immunoglobulins, including IgG, IgA, and IgM, emerge and reach peak values at different times [165,166]. Thus, theoretically, one or all immunoglobulins could be labeled to diagnose infectious diseases. Studies on the variability of serological response to infection have found that personal immunoglobulin testing is useless for monitoring the process of infectious disease. However, when the literature data is examined statistically, studies on the detection of IgM and IgG show intensity. IgM antibodies can indicate imminent exposure to infection, while IgG antibodies point out prolonged exposure and are often a precursor to immunity [167]. Additionally, the IgA response is relatively limited. Almost all of the COVID-19 rapid diagnostic tests in the market and literature offer simultaneously determined amounts of IgM and IgG antibodies. In these studies, the use of a blocking agent during conjugation and blocking the NC membrane was generally recommended to prevent cross-reactions.
Appropriate antibody/antigen labeling methods on the LFA platform are required to obtain sensitive and specific signal readings. In LFA devices, nanoparticles are commonly utilized as label or probe molecules. Although AuNPs are the most widely used nanoparticle for labeling, other nanoparticles have also been used. Colored or fluorescent nanoparticles with a diameter of 20–500 nm are appropriate because their modest size does not obstruct the flow or the flow direction. Recently, progress in fluorescent labeling has provided a better way to replace the traditional colloidal gold assay. The fluorescent label is quantitative and can be prepared in multiplex with different dyes. Some of the novel technologies for enhanced sensitivity and quantitative analysis include quantum dots [168], upconverting phosphorus technology [169], carbon nanoparticles, selenium nanoparticles [170], lanthanide-doped nanoparticles [171], and SERS reporters [172,173]. Magnetic NPs are highly attractive probes for the enrichment of target analytes. These desirable agents provide highly stable signals to sensitive strips, but expensive readout devices are required to read the results. For this reason, it may be considered in future studies to integrate more affordable devices into LFA systems. As described above, LFA tests provide an easily accessible and user-friendly instrument. Although many methods have been developed that provide technical progress, SERS-based detection methods provide a highly sensitive detection tool because they can provide analytical results beyond qualitative measurement. Because it monitors the characteristic Raman peak density of SERS nanotags collected in the test line, SERS-LFA outperforms colloidal gold. Thus, the combination of LFA with SERS detection tools makes them a prevalent diagnostic method for the accurate and rapid diagnosis of viral infectious diseases. Nevertheless, it is limited in providing diagnoses at home. From this perspective, it is essential to learn more about SERS-LFA platforms' current situation and make measuring instruments portable. Also, more research needs to be done on more consistent and reproducible SERS nanotags. As a result of research, it has been seen that conventional LFA biosensors using AuNPs are thermally unsteady in temperature regions and can cause false effects [174,175]. As a result, the use of AuNPs in diagnosing diseases commonly seen in tropical areas (dengue and zika) has been restricted. Various silica or polymer-coated SERS nano-tags have recently been developed to address this issue [176,177]. However, the reproducibility in the test region of SERS nano-tags is challenging to obtain, thus hampering the application of SERS-LFA in quantitative analysis. Therefore, more reproducible and stable performing SERS nanotags must be continually improved to make SERS-LFA durable for in vitro diagnostic. Because of its popularity and benefits, the SERS approach is now progressing toward downsizing and cost reduction. The effective creation and deployment of portable Raman spectrometers for the LFA of different biomarkers were recently proven [178,179]. Due to their miniaturization and cost reduction, SERS-based readers are a viable and possible tool for producing quick and quantitative on-site LFA testing due to their miniaturization and cost reduction.
LFA analysis consists of four main components; the sample pad, conjugate pad, NC membrane, and absorbent pad. In their LFA studies to date, researchers have generally not sought to improve these four main components to increase the specificity of the test line and obtain strong signals. He refrained from making changes to these four main elements. In particular, studies should be carried out to improve the structure of the NC membrane, the nature of which has not yet been fully elucidated. Control line is actively used in lateral flow tests to understand whether many diseases are passed, whether there is a virus infection and the disease's course. Research on alternative methods and biomolecules should be focused on, aiming to eliminate the high cost of the high concentration of antibodies used in this region.
Due to the complexity of antigen-antibody interactions that occur in a complex matrix, immunoassays are susceptible to interference [180]. Such intervention leads to misinterpretation of immunoassay results, leading to unnecessary investigations or administration of inappropriate therapy. The use of high concentrations of biotin as a nutritional supplement to improve hair, skin, and nail quality has increased in recent years. However, high concentrations of biotin have been found to interfere with detecting HIV-1 p24 antigen in serum and plasma. However, the examination of biotin interference in any LFA test developed so far has been seen as a major shortcoming. Where additional biotin supplementation cannot be ruled out, it is important to consider these potential sources of false test results [181]. Similarly, Dengue NS1 can be cited as an example. Because false-positive results for this dengue biomarker have been reported in patients with acute zika infection, caution should be exercised when interpreting dengue NS1 results [182]. A group of researchers has shown that the combined use of Dengue NS1 and antibodies (IgM and IgG) improves diagnosis rates for dengue fever [183,184]. Even if these and similar solution suggestions are applied, more optimizations should be made for cross-reaction in immunoassay tests. Laboratory professionals should not ignore or underestimate immunoassay weaknesses. They should engage in a two-way, open, and persistent dialogue with clinicians to quickly identify questionable outcomes before making further diagnoses or treatment decisions [185].
CRISPR and CRISPR-related (Cas) genes have been utilized in various practices, including diagnostics biosensing, and were awarded the 2020 Nobel Prize in Chemistry. CRISPR-based diagnostics tool can identify the attomolar levels of viral nucleic acids. They are more sensitive than RT-PCR-based approaches that only detect femtomolar concentrations [186]. LF CRISPR-based biosensing allows fast and sensitive identification of nucleic acids extracted from these diverse samples. In a study reported by researchers in 2020, they designed the LFA strip and combined this technique with the CRISPR-Cas system to make it cost-effective for on-site diagnosis [187]. According to the researchers, Cas-LFA can accomplish quite a low detection limit with % 100 specificity in less than an hour, indicating a promising option for future POC testing. Nevertheless, more research is needed to improve the CRISPR-Cas identification step and assay time.
As a result of the literature review, many kit studies have been carried out to diagnose infectious diseases, and most of them have been introduced to the market. This detection method does not need any reading instrument as it relies solely on visual detection. However, it suffers from “limits of quantitative analysis” and “low sensitivity” limitations. Many researchers have tried to solve these problems. Diverse optical strip readers and associated detection tags have advanced the analytes' quantification capability and sensitivity. Smartphones have become a necessary personal gadget in today's world. Smartphones may help with contact tracing, patient isolation, disease control, and surveillance monitoring during a pandemic. Smartphones have been utilized as viral infectious disease surveillance tools to identify people who have come into contact with a patient.
6. Conclusion and future prospects
This review article has collected and discussed the history, mechanism, and recent developments of various LFA techniques to diagnose infectious diseases (HIV, COVID-19, hepatitis, zika, dengue, malaria, and influenza). There have been a lot of pandemics and epidemics caused by various infectious diseases worldwide. Not all of them could produce epidemics on a scale that would affect several continents of the world at the same time. However, a new SARS-CoV-2 has recently resulted in a continuing global epidemic. Given the very high person-to-person transmission rate of the virus, many countries have taken extraordinary measures (isolation, quarantine, and social distancing) to reduce the spread of the disease. However, these efforts fall short. LFA-based diagnostic devices provide a very suitable platform in this regard. While the LFA platform holds great promise for infectious disease diagnosis, several challenges need to be addressed to facilitate LFA's performance in controlling epidemics. Although gold nanoparticles and latex microbeads are the primary tags used in LFA platforms, they sometimes fail to provide the required sensitivity. Technologies such as catalytic enhancement and SERS-based recognition are being used to achieve higher sensitivity and detect more pathogens to overcome this obstacle. However, stable and efficient new labels should be developed, and more interdisciplinary studies on detection techniques are needed. In addition, the use of portable, electronic reader instruments gives quantitative results in a much shorter time than traditional methods. However, these electronic devices should be reduced in size and made portable by the user. In this way, the detection method to be presented will promise the diagnosis of many diseases, especially cancer diagnosis. Finally, under the guidance of the authorities and regulatory agencies, it can be ensured that the studies in the laboratories meet the industry, allowing the LFA diagnostic tests to be used to diagnose infectious diseases in resource-constrained environments to become more accessible. In summary, we emphasize the need to continue to enhance quick diagnostic tests.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Anderson R.M., May R.M. Population biology of infectious diseases: Part I. Nature. 1979;280:361–367. doi: 10.1038/280361a0. [DOI] [PubMed] [Google Scholar]
- 2.Hui E.K.W. Reasons for the increase in emerging and re-emerging viral infectious diseases. Microb. Infect. 2006;8:905–916. doi: 10.1016/j.micinf.2005.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russell R.C. Survival of insects in the wheel bays of a Boeing 747B aircraft on flights between tropical and temperate airports. Bull. World Health Organ. 1987;65:659. [PMC free article] [PubMed] [Google Scholar]
- 4.May R.M., Gupta S., McLean A.R. Infectious disease dynamics: what characterizes a successful invader? Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2001;356:901–910. doi: 10.1098/rstb.2001.0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakraborty I., Maity P. COVID-19 pandemic: migration, its effects on society, the global environment and protection. Total Environ. Sci. 2020;728 doi: 10.1016/j.scitotenv.2020.138882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Candela R.A., Geloso V. Economic freedom, epidemics, and sound political economy. Southern Econ. Mag. 2021;87:1250–1266. doi: 10.1002/soej.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ritzi-Lehnert M. Development of chip-compatible sample preparation to diagnose infectious diseases. Expert Rev. Mol. Diagn. 2012;12:189–206. doi: 10.1586/erm.11.98. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y., Yu L., Kong X., Sun L. Application of nanodiagnostics in point-of-care tests for infectious diseases. Int. J. Nanomed. 2017;12:4789. doi: 10.2147/IJN.S137338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravi N., Cortade D.L., Ng E., X Wang S. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukama O., Wu J., Li Z., Liang Q., Yi Z., Lu X., Zeng L. An ultrasensitive and specific point-of-care CRISPR/Cas12 based lateral flow biosensor for the rapid detection of nucleic acids. Biosens. Bioelectron. 2020;159 doi: 10.1016/j.bios.2020.112143. [DOI] [PubMed] [Google Scholar]
- 11.He X., Liu Z., Yang Y., Li L., Wang L., Li A., Xu F. Sensitivity enhancement of nucleic acid lateral flow assays through a physical–chemical coupling method: dissoluble saline barriers. ACS Sens. 2019;4:1691–1700. doi: 10.1021/acssensors.9b00594. [DOI] [PubMed] [Google Scholar]
- 12.Wang X., Xiong E., Tian T., Cheng M., Lin W., Wang H., Zhou X. Clustered regularly interspaced short palindromic repeats/Cas9-mediated lateral flow nucleic acid assay. ACS Nano. 2020;14:2497–2508. doi: 10.1021/acsnano.0c00022. [DOI] [PubMed] [Google Scholar]
- 13.Shen M., Li N., Lu Y., Cheng J., Xu Y. An enhanced centrifugation-assisted lateral flow immunoassay for the point-of-care detection of protein biomarkers. Lab Chip. 2020;20:2626–2634. doi: 10.1039/d0lc00518e. [DOI] [PubMed] [Google Scholar]
- 14.Lee K.W., Kim K.R., Chun H.J., Jeong K.Y., Hong D.K., Lee K.N., Yoon H.C. Time-resolved fluorescence resonance energy transfer-based lateral flow immunoassay using a raspberry-type europium particle and a single membrane for the detection of cardiac troponin I. Biosens. Bioelectron. 2020;163 doi: 10.1016/j.bios.2020.112284. [DOI] [PubMed] [Google Scholar]
- 15.Grant B.D., Anderson C.E., Williford J.R., Alonzo L.F., Glukhova V.A., Boyle D.S., P Nichols K. SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal. Chem. 2020;92:11305–11309. doi: 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- 16.Brazaca L.C., Moreto J.R., Martín A., Tehrani F., Wang J., Zucolotto V. Colorimetric paper-based immunosensor for simultaneous determination of fetuin B and clusterin toward early Alzheimer's diagnosis. ACS Nano. 2019;13:13325–13332. doi: 10.1021/acsnano.9b06571. [DOI] [PubMed] [Google Scholar]
- 17.Ivanov A.V., Safenkova I.V., Zherdev A.V., Dzantiev B.B. Nucleic acid lateral flow assay with recombinase polymerase amplification: solutions for highly sensitive detection of RNA virus. Talanta. 2020;210 doi: 10.1016/j.talanta.2019.120616. [DOI] [PubMed] [Google Scholar]
- 18.Le T.T., Chang P., Benton D.J., McCauley J.W., Iqbal M., Cass A.E. Dual recognition element lateral flow assay toward multiplex strain specific influenza virus detection. Anal. Chem. 2017;89:6781–6786. doi: 10.1021/acs.analchem.7b01149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phan J.C., Pettitt J., George J.S., Fakoli L.S., III, Taweh F.M., Bateman S.L., Schoepp R.J. Lateral flow immunoassays for Ebola virus disease detection in Liberia. J. Infect. Dis. 2016;214:222–228. doi: 10.1093/infdis/jiw251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi D.H., Lee S.K., Oh Y.K., Bae B.W., Lee S.D., Kim S., Kim M.G. A dual gold nanoparticle conjugate-based lateral flow assay (LFA) method for the analysis of troponin I. Biosens. Bioelectron. 2010;25:1999–2002. doi: 10.1016/j.bios.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 21.Kim J.H., Cho J.H., Cha G.S., Lee C.W., Kim H.B., Paek S.H. Conductimetric membrane strip immunosensor with polyaniline-bound gold colloids as signal generator. Biosens. Bioelectron. 2000;14:907–915. doi: 10.1016/s0956-5663(99)00063-9. [DOI] [PubMed] [Google Scholar]
- 22.Li Z., Wang Y., Wang J., Tang Z., Pounds J.G., Lin Y. Rapid and sensitive detection of protein biomarker using a portable fluorescence biosensor based on quantum dots and a lateral flow test strip. Anal. Chem. 2010;82:7008–7014. doi: 10.1021/ac101405a. [DOI] [PubMed] [Google Scholar]
- 23.Li F., You M., Li S., Hu J., Liu C., Gong Y., Xu F. Based point-of-care immunoassays: recent advances and emerging trends. Biotechnol. Adv. 2020;39 doi: 10.1016/j.biotechadv.2019.107442. [DOI] [PubMed] [Google Scholar]
- 24.Mahmoudi T., de la Guardia M., Shirdel B., Mokhtarzadeh A., Baradaran B. Recent advancements in structural improvements of lateral flow assays towards point-of-care testing. TrAC, Trends Anal. Chem. 2019;116:13–30. [Google Scholar]
- 25.Nguyen V.T., Song S., Park S., Joo C. Recent advances in high-sensitivity detection methods for paper-based lateral-flow assay. Biosens. Bioelectron. 2020;152 doi: 10.1016/j.bios.2020.112015. [DOI] [PubMed] [Google Scholar]
- 26.Mahmoudi T., de la Guardia M., Baradaran B. Lateral flow assays towards point-of-care cancer detection: a review of current progress and future trends. TrAC, Trends Anal. Chem. 2020;125 [Google Scholar]
- 27.Huang X., Aguilar Z.P., Xu H., Lai W., Xiong Y. Membrane-based lateral flow immunochromatographic strip with nanoparticles as reporters for detection: a review. Biosens. Bioelectron. 2016;75:166–180. doi: 10.1016/j.bios.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 28.Bahadır E.B., Sezgintürk M.K. Lateral flow assays: principles, designs and labels. TrAC, Trends Anal. Chem. 2016;82:286–306. [Google Scholar]
- 29.Dochez A.R., Avery O.T. The elaboration of specific soluble substance by pneumococcus during growth. J. Exp. Med. 1917;26:477. doi: 10.1084/jem.26.4.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R.L. Campbell, D.B. Wagner, J.P. O'connell, U.S. Washington, DC: U.S. Patent and Trademark Office, (1987), Patent No. 4,703,017.
- 31.R.W Rosenstein, T.G. Bloomster, U.S. Washington, DC: U.S. Patent and Trademark Office., (1989), Patent No. 4, 855, 240.
- 32.K. May, M.E. Prior, I. Richards, U.S. Patent No. 5,622,871. Washington, DC: U.S. Patent and Trademark Office, (1997), Patent No. 5, 622, 871.
- 33.Jing J., Wang Z. Research advance of lateral flow assay labels. Sheng wu yi xue gong cheng xue za zhi= Journal of biomedical engineering= Shengwu yixue gongchengxue zazh. 2018;35:661–664. doi: 10.7507/1001-5515.201703005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bissonnette L., Bergeron M.G. Diagnosing infections—current and anticipated technologies for point-of-care diagnostics and home-based testing. Clin. Microbiol. Infect. 2010;16:1044–1053. doi: 10.1111/j.1469-0691.2010.03282.x. [DOI] [PubMed] [Google Scholar]
- 35.O'Farrell B. Lateral Flow Immunoassay. Humana Press; 2009. B. Evolution in lateral flow–based immunoassay systems; pp. 1–33. [Google Scholar]
- 36.Koczula K.M., Gallotta A. Lateral flow assays. Essays Biochem. 2016;60:111–120. doi: 10.1042/EBC20150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong R.C., Tse H.Y. Vol. 393. Analytical Bioanalytical Chemistry; 2009. pp. 569–582. (Lateral Flow Immunoassay). [Google Scholar]
- 38.NanoComposix Nitrocellulose membrane selection striping for lateral flow assays. 2020. https://nanocomposix.com/pages/nitrocellulose-membrane-selection--striping-for-lateral-flow-assays Access: 5 Mart 2020.
- 39.Linares E.M., Kubota L.T., Michaelis J., Thalhammer S. Enhancement of the detection limit for lateral flow immunoassays: evaluation and comparison of bioconjugates. J. Immunol. Methods. 2012;375:264–270. doi: 10.1016/j.jim.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W. SARS Study Group. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuen K.Y., Chan P.K.S., Peiris M., Tsang D.N.C., Que T.L., Shortridge K.F. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 42.Ahmad T., Khan M., Haroon T.H.M., Nasir S., Hui J., Bonilla-Aldana D.K., Rodriguez-Morales A.J. COVID-19: zoonotic aspects. Trav. Med. Infect. Dis. 2020;36 doi: 10.1016/j.tmaid.2020.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tiwari R., Dhama K., Sharun K., Iqbal Yatoo M., Malik Y.S., Singh R., Rodriguez-Morales A.J. COVID-19: animals, veterinary and zoonotic links. Vet. Q. 2020;40:169–182. doi: 10.1080/01652176.2020.1766725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Morales A.J., Cardona-Ospina J.A., Gutiérrez-Ocampo E., Villamizar-Peña R., Holguin-Rivera Y., Escalera-Antezana J.P., Sah R. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Trav. Med. Infect. Dis. 2020;34 doi: 10.1016/j.tmaid.2020.101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Latinne A., Hu B., Olival K.J., Zhu G., Zhang L., Li H., Daszak P. Origin and cross-species transmission of bat coronaviruses in China. Nat. Commun. 2020;11:1–15. doi: 10.1038/s41467-020-17687-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Lau S.K., Luk H.K., Wong A.C., Li K.S., Zhu L., He Z., Woo P.C. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26:1542. doi: 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morrison J.H., Miller C., Bankers L., Crameri G., Wang L.F., Poeschla E.M. A potent postentry restriction to primate lentiviruses in a yinpterochiropteran bat. mBio. 2020;11 doi: 10.1128/mBio.01854-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Espejo A.P., Akgun Y., Al Mana A.F., Tjendra Y., Millan N.C., Gomez-Fernandez C., Cray C. Review of current advances in serologic testing for COVID-19. Am. J. Clin. Pathol. 2020;154:293–304. doi: 10.1093/ajcp/aqaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghazal K., Brabant S., Prie D., Piketty M.L. Hormone immunoassay interference: a 2021 Update. Ann. Lab. Med. 2022;42(1):3–23. doi: 10.3343/alm.2022.42.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Z. Li, Y. Yi, X. Luo, N. Xiong,Y. Liu, S. Li, F. Ye, Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol., 92(2020), p.1518-1524. [DOI] [PMC free article] [PubMed]
- 51.Z. Li, Y. Yi, X. Luo, N. Xiong, Y. Liu, S. Li, F. Ye, Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol., 92(2020), p.1518-1524. [DOI] [PMC free article] [PubMed]
- 52.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Huang A.L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 53.Cavalera S., Colitti B., Rosati S., Ferrara G., Bertolotti L., Nogarol C., Anfossi L. A multi-target lateral flow immunoassay enabling the specific and sensitive detection of total antibodies to SARS COV-2. Talanta. 2021;223 doi: 10.1016/j.talanta.2020.121737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Black M.A., Shen G., Feng X., Beltran W.F.G., Feng Y., Vasudevaraja V., Snuderl M. Analytical performance of lateral flow immunoassay for SARS-CoV-2 exposure screening on venous and capillary blood samples. J. Immunol. Methods. 2021;489 doi: 10.1016/j.jim.2020.112909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeon J., Lee S.H., Joung Y., Kim K., Choi N., Choo J. Improvement of reproducibility and thermal stability of surface-enhanced Raman scattering-based lateral flow assay strips using silica-encapsulated gold nanoparticles. Sensor. Actuator. B Chem. 2020;321 [Google Scholar]
- 56.Borzenkov M., Chirico G., d'Alfonso L., Sironi L., Collini M., Cabrini E., Denat F. Thermal and chemical stability of thiol bonding on gold nanostars. Langmuir. 2015;31:8081–8091. doi: 10.1021/acs.langmuir.5b01473. [DOI] [PubMed] [Google Scholar]
- 57.Xie Q.Y., Wu Y.H., Xiong Q.R., Xu H.Y., Xiong Y.H., Liu K., Lai W.H. Advantages of fluorescent microspheres compared with colloidal gold as a label in immunochromatographic lateral flow assays. Biosens. Bioelectron. 2014;54:262–265. doi: 10.1016/j.bios.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 58.Taranova N.A., Berlina A.N., Zherdev A.V., Dzantiev B.B. Traffic light’immunochromatographic test based on multicolor quantum dots for the simultaneous detection of several antibiotics in milk. Biosens. Bioelectron. 2015;63:255–261. doi: 10.1016/j.bios.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 59.Berlina A.N., Taranova N.A., Zherdev A.V., Vengerov Y.Y., Dzantiev B.B. Quantum dot-based lateral flow immunoassay for detection of chloramphenicol in milk. Anal. Bioanal. Chem. 2013;405:4997–5000. doi: 10.1007/s00216-013-6876-3. [DOI] [PubMed] [Google Scholar]
- 60.Juntunen E., Myyryläinen T., Salminen T., Soukka T., Pettersson K. Performance of fluorescent europium (III) nanoparticles and colloidal gold reporters in lateral flow bioaffinity assay. Anal. Biochem. 2012;428:31–38. doi: 10.1016/j.ab.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 61.Rundstrom G., Jonsson A., Martensson O., Mendel-Hartvig I., Venge P. Lateral flow immunoassay using europium (III) chelate microparticles and time-resolved fluorescence for eosinophils and neutrophils in whole blood. Clin. Chem. 2007;53:342–348. doi: 10.1373/clinchem.2006.074021. [DOI] [PubMed] [Google Scholar]
- 62.Blažková M., Mičková-Holubová B., Rauch P., Fukal L. Immunochromatographic colloidal carbon-based assay for detection of methiocarb in surface water. Biosens. Bioelectron. 2009;25:753–758. doi: 10.1016/j.bios.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 63.Noguera P., Posthuma-Trumpie G.A., Van Tuil M., Van der Wal F.J., De Boer A., H Moers A.P., Van Amerongen A.A. Carbon nanoparticles in lateral flow methods to detect genes encoding virulence factors of Shiga toxin-producing Escherichia coli. Anal. Bioanal. Chem. 2011;399:831–838. doi: 10.1007/s00216-010-4334-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwartzberg A.M., Oshiro T.Y., Zhang J.Z., Huser T., Talley C.E. Improving nanoprobes using surface-enhanced Raman scattering from 30-nm hollow gold particles. Anal. Chem. 2006;78:4732–4736. doi: 10.1021/ac060220g. [DOI] [PubMed] [Google Scholar]
- 65.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Lin G. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 66.Qie Z., Liu Q., Yan W., Gao Z., Meng W., Xiao R., Wang S. Universal and ultrasensitive Immunochromatographic assay by using an antigen as a Bifunctional element and Antialbumin antibody on a test line. Anal. Chem. 2019;91:9530–9537. doi: 10.1021/acs.analchem.9b00673. [DOI] [PubMed] [Google Scholar]
- 67.You P.Y., Li F.C., Liu M.H., Chan Y.H. Colorimetric and fluorescent dual-mode immunoassay based on plasmon-enhanced fluorescence of polymer dots for detection of PSA in whole blood. ACS Appl. Mater. Interfaces. 2019;11:9841–9849. doi: 10.1021/acsami.9b00204. [DOI] [PubMed] [Google Scholar]
- 68.Hu J., Zhang Z.L., Wen C.Y., Tang M., Wu L.L., Liu C., Pang D.W. Sensitive and quantitative detection of C-reaction protein based on immunofluorescent nanospheres coupled with lateral flow test strip. Anal. Chem. 2016;88:6577–6584. doi: 10.1021/acs.analchem.6b01427. [DOI] [PubMed] [Google Scholar]
- 69.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 70.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Zheng S. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00461-20. e00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Infantino M., Damiani A., Gobbi F.L., Grossi V., Lari B., Macchia D., Manfredi M. Serological assays for SARS-CoV-2 infectious disease: benefits, limitations and perspectives. Isr. Med. Assoc. J. 2020;22:203–210. [PubMed] [Google Scholar]
- 72.Meyer B., Drosten C., Müller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang C., Yang X., Gu B., Liu H., Zhou Z., Shi L., Wang S. Sensitive and simultaneous detection of SARS-CoV-2-specific IgM/IgG using lateral flow immunoassay based on dual-mode quantum dot nanobeads. Anal. Chem. 2020;92:15542–15549. doi: 10.1021/acs.analchem.0c03484. [DOI] [PubMed] [Google Scholar]
- 74.Wang C., Shi D., Wan N., Yang X., Liu H., Gao H., Wang S. Development of spike protein-based fluorescence lateral flow assay for the simultaneous detection of SARS-CoV-2 specific IgM and IgG. Analyst. 2021;146:3908–3917. doi: 10.1039/d1an00304f. [DOI] [PubMed] [Google Scholar]
- 75.Baker A.N., Richards S.J., Guy C.S., Congdon T.R., Hasan M., Zwetsloot A.J., Gibson M.I. The SARS-COV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent. Sci. 2020;6:2046–2052. doi: 10.1021/acscentsci.0c00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen S., Meng L., Wang L., Huang X., Ali S., Chen X., Huang G. SERS-based lateral flow immunoassay for sensitive and simultaneous detection of anti-SARS-CoV-2 IgM and IgG antibodies by using gap-enhanced Raman nanotags. Sensor. Actuator. B Chem. 2021;348 doi: 10.1016/j.snb.2021.130706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grant B.D., Anderson C.E., Williford J.R., Alonzo L.F., Glukhova V.A., Boyle D.S., Nichols K.P. SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal. Chem. 2020;92:11305–11309. doi: 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- 78.Lee J.H., Choi M., Jung Y., Lee S.K., Lee C.S., Kim J., Kim H.G. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin-converting enzyme 2 (ACE2) Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020;71:778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.FDA . 2020. Emergency Use Authorizations for Medical Devices. [Google Scholar]
- 81.Cellex Inc qSARS-CoV-2 IgG/IgM rapid test. 2020. https://www.fda.gov/media/136625/download
- 82.Autobio Diagnostics Co, Ltd, https://www.fda.gov/media/137364/download, (2020).
- 83.Ortho Clinical Diagnostics, Inc . 2020. Instructions for Use: VITROS Immunodiagnostic Products Anti SARS-CoV-2 Total 619 9922 Reagent Pack. [Google Scholar]
- 84.Roche Diagnostics Elecsys® anti-SARS-Cov 2. https://diagnostics.roche.com/us/en/products/params/elecsys-anti-sars-cov-2.html#productInfo
- 85.Ravi N., Cortade D.L., Ng E., Wang S.X. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A.J., Silva A.M. The receptor-binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ong D.S., Fragkou P.C., Schweitzer V.A., Chemaly R.F., Moschopoulos C.D., Skevaki C. How to interpret and use COVID-19 serology and immunology tests. Clin. Microbiol. Infect. 2021;27:981–986. doi: 10.1016/j.cmi.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.DiaSorin L.L.C. 2020. Molecular Simplexa TM COVID-19 Direct.https://www.fda.gov/media/136286/download [Google Scholar]
- 89.Siemens Medical Solutions Inc . 2020. SARS-CoV-2 Total Assay.https://www.siemens-healthineers.com/en-us/laboratory diagnostics/assays-by-diseases-conditions/infectious-disease assays/cov2t-assay#The_Scale [Google Scholar]
- 90.Fritzell C., Rousset D., Adde A., Kazanji M., Van Kerkhove M.D., Flamand C. Current challenges and implications for dengue, chikungunya and Zika seroprevalence studies worldwide: a scoping review. PLoS Neglected Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jones R., Kulkarni M.A., Davidson T.M., RADAM-LAC Research Team, Talbot B. Arbovirus vectors of epidemiological concern in the Americas: a scoping review of entomological studies on Zika, dengue and chikungunya virus vectors. PLoS One. 2020;15 doi: 10.1371/journal.pone.0220753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Masel J., McCracken M.K., Gleeson T., Morrison B., Rutherford G., Imrie A., Pollett S. Does prior dengue virus exposure worsen clinical outcomes of Zika virus infection? A systematic review pooled analysis and lessons learned. PLoS Neglected Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oliver G.F., Carr J.M., Smith J.R. Emerging infectious uveitis: chikungunya, dengue, Zika and Ebola: a review. Clin. Exp. Ophthalmol. 2019;47:372–380. doi: 10.1111/ceo.13450. [DOI] [PubMed] [Google Scholar]
- 94.Yrad F.M., Castañares J.M., Alocilja E.C. Visual detection of dengue-1 RNA using gold nanoparticle-based lateral flow biosensor. Diagnostics. 2019;9:74. doi: 10.3390/diagnostics9030074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hwang J., Lee S., Choo J. Application of a SERS-based lateral flow immunoassay strip for the rapid and sensitive detection of staphylococcal enterotoxin B. Nanoscale. 2016;8:11418–11425. doi: 10.1039/c5nr07243c. [DOI] [PubMed] [Google Scholar]
- 96.Fu X., Cheng Z., Yu J., Choo P., Chen L., Choo J. A SERS-based lateral flow assay biosensor for highly sensitive detection of HIV-1 DNA. Biosens. Bioelectron. 2016;78:530–537. doi: 10.1016/j.bios.2015.11.099. [DOI] [PubMed] [Google Scholar]
- 97.Khlebtsov B.N., Bratashov D.N., Byzova N.A., Dzantiev B.B., Khlebtsov N.G. SERS-based lateral flow immunoassay of troponin I by using gap-enhanced Raman tags. Nano Res. 2019;12:413–420. [Google Scholar]
- 98.V. Tran, B. Walkenfort, M. König, M. Salehi, S. Schlücker, Rapid, quantitative, and ultrasensitive point-of-care testing: a portable SERS reader for lateral flow assays in clinical chemistry. Angew. Chem. Int. Ed., 58(2019), p.442-446. [DOI] [PMC free article] [PubMed]
- 99.Sánchez-Purrà M., Carré-Camps M., de Puig H., Bosch I., Gehrke L., Hamad-Schifferli K. Surface-enhanced Raman spectroscopy-based sandwich immunoassays for multiplexed detection of Zika and Dengue viral biomarkers. ACS Infect. Dis. 2017;3:767–776. doi: 10.1021/acsinfecdis.7b00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jeon J., Lee S.H., Joung Y., Kim K., Choi N., Choo J. Improvement of reproducibility and thermal stability of surface-enhanced Raman scattering-based lateral flow assay strips using silica-encapsulated gold nanoparticles. Sensor. Actuator. B Chem. 2020;321 [Google Scholar]
- 101.Tran T.V., Nguyen B.V., Nguyen T.T., Tran T.T., Pham K.G., Le Q.B., Le H.Q. Development of a highly sensitive magneto-enzyme lateral flow immunoassay for dengue NS1 detection. PeerJ. 2019;7:7779. doi: 10.7717/peerj.7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rong Z., Wang Q., Sun N., Jia X., Wang K., Xiao R., Wang S. Smartphone-based fluorescent lateral flow immunoassay platform for highly sensitive point-of-care detection of Zika virus nonstructural protein 1. Anal. Chim. Acta. 2019;1055:140–147. doi: 10.1016/j.aca.2018.12.043. [DOI] [PubMed] [Google Scholar]
- 103.Douek D.C., Roederer M., Koup R.A. Emerging concepts in the immunopathogenesis of AIDS. Annu. Rev. Med. 2009;60:471–484. doi: 10.1146/annurev.med.60.041807.123549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mayer K.H., Karp C.L., Auwaerter P.G., Mayer K.H. Coinfection with HIV and tropical infectious diseases. II. Helminthic, fungal, bacterial, and viral pathogens. Clin. Infect. Dis. 2007;45:1214–1220. doi: 10.1086/522180. [DOI] [PubMed] [Google Scholar]
- 105.Fu X., Cheng Z., Yu J., Choo P., Chen L., Choo J. A SERS-based lateral flow assay biosensor for highly sensitive detection of HIV-1 DNA. Biosens. Bioelectron. 2016;78:530–537. doi: 10.1016/j.bios.2015.11.099. [DOI] [PubMed] [Google Scholar]
- 106.Rohrman B.A., Leautaud V., Molyneux E., Richards-Kortum R.R. 2012. A Lateral Flow Assay for Quantitative Detection of Amplified HIV-1 RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Deng X., Wang C., Gao Y., Li J., Wen W., Zhang X., Wang S. Applying strand displacement amplification to quantum dots-based fluorescent lateral flow assay strips for HIV-DNA detection. Biosens. Bioelectron. 2018;105:211–217. doi: 10.1016/j.bios.2018.01.039. [DOI] [PubMed] [Google Scholar]
- 108.Workman S., Wells S.K., Pau C.P., Owen S.M., Dong X.F., LaBorde R., Granade T.C. Rapid detection of HIV-1 p24 antigen using magnetic immuno-chromatography (MICT) J. Virol Methods. 2009;160:14–21. doi: 10.1016/j.jviromet.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 109.Zhan L., Granade T., Liu Y., Wei X., Youngpairoj A., Sullivan V., Bischof J. Development and optimization of thermal contrast amplification lateral flow immunoassays for ultrasensitive HIV p24 protein detection. Microsyst. Nanoeng. 2020;6:1–11. doi: 10.1038/s41378-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rong Z., Xiao R., Peng Y., Zhang A., Wei H., Ma Q., Wang S. Integrated fluorescent lateral flow assay platform for point-of-care diagnosis of infectious diseases by using a multichannel test cartridge. Sensor. Actuator. B Chem. 2021;329 [Google Scholar]
- 111.Abu-Salah K.M., Zourob M.M., Mouffouk F., Alrokayan S.A., Alaamery M.A., Ansari A.A. DNA-based nanobiosensors as an emerging platform for detection of disease. Sensors. 2015;15:14539–14568. doi: 10.3390/s150614539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tang S., Hewlett I. Nanoparticle-based immunoassays for sensitive and early detection of HIV-1 capsid (p24) antigen. JID (J. Infect. Dis.) 2010;201:59–64. doi: 10.1086/650386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Haleyur Giri Setty M.K., Lee S., Lathrop J., Hewlett I.K. Biotin interference in point of care HIV immunoassay. BioResearch Open Access. 2010;9:243–246. doi: 10.1089/biores.2020.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jeong S.H., Lee H.S. Hepatitis A: clinical manifestations and management. Intervirology. 2010;53:15–19. doi: 10.1159/000252779. [DOI] [PubMed] [Google Scholar]
- 115.Mahboobi N., Porter S.R., Karayiannis P., Alavian S.M. Oral fluid and hepatitis A, B and C: a literature review. J. Oral Pathol. Med. 2012;41:505–516. doi: 10.1111/j.1600-0714.2011.01123.x. [DOI] [PubMed] [Google Scholar]
- 116.Koslap-Petraco M.B., Shub M., Judelsohn R. Hepatitis A: disease burden and current childhood vaccination strategies in the United States. J. Pediatr. Health Care. 2008;22:3–11. doi: 10.1016/j.pedhc.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 117.Shen J., Zhou Y., Fu F., Xu H., Lv J., Xiong, Wang A. Immunochromatographic assay for quantitative and sensitive detection of hepatitis B virus surface antigen using highly luminescent quantum dot-beads. Talanta. 2015;142:145–149. doi: 10.1016/j.talanta.2015.04.058. [DOI] [PubMed] [Google Scholar]
- 118.Jiang H., Wu D., Song L., Yuan Q., Ge S., Min X., Qiu X. A smartphone-based genotyping method for hepatitis B virus at point-of-care settings. SLAS Technol.: Transl. Life Sci. Innovat. 2017;22:122–129. doi: 10.1177/2211068216680163. [DOI] [PubMed] [Google Scholar]
- 119.Song L.W., Wang Y.B., Fang L.L., Wu Y., Yang L., Chen J.Y., Xia N.S. Rapid fluorescent lateral-flow immunoassay for hepatitis B virus genotypes. Anal. Chem. 2015;87:5173–5180. doi: 10.1021/ac504832c. [DOI] [PubMed] [Google Scholar]
- 120.Kim D.S., Kim Y.T., Hong S.B., Kim J., Heo N.S., Lee M.K., Choi B.G. Development of lateral flow assay based on size-controlled gold nanoparticles for detection of hepatitis B surface antigen. Sensors. 2016;16:2154. doi: 10.3390/s16122154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Operskalski E.A., Kovacs A. HIV/HCV co-infection: pathogenesis, clinical complications, treatment, and new therapeutic technologies. Curr. HIV AIDS Rep. 2011;8:12–22. doi: 10.1007/s11904-010-0071-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Macías J., Berenguer J., Japón M.A., Girón J.A., Rivero A., López-Cortés L.F., Pineda J.A. Fast fibrosis progression between repeated liver biopsies in patients coinfected with human immunodeficiency virus/hepatitis C virus. Hepatology. 2009;50:1056–1063. doi: 10.1002/hep.23136. [DOI] [PubMed] [Google Scholar]
- 123.Kuehlkamp V.M., Schuelter-Trevisol F. Prevalence of human immunodeficiency virus/hepatitis C virus co-infection in Brazil and associated factors: a review. Braz. J. Infect. Dis. 2013;17:455–463. doi: 10.1016/j.bjid.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mahboobi N., Porter S.R., Karayiannis P., Alavian S.M. Oral fluid and hepatitis A, B and C: a literature review. J. Oral Pathol. Med. 2012;41:505–516. doi: 10.1111/j.1600-0714.2011.01123.x. [DOI] [PubMed] [Google Scholar]
- 125.Lee J.H., Seo H.S., Kwon J.H., Kim H.T., Kwon K.C., Sim S.J., Lee J. Multiplex diagnosis of viral infectious diseases (AIDS, hepatitis C, and hepatitis A) based on point of care lateral flow assay using engineered proteinticles. Biosens. Bioelectron. 2015;69:213–225. doi: 10.1016/j.bios.2015.02.033. [DOI] [PubMed] [Google Scholar]
- 126.Gensheimer K.F., Meltzer M.I., Postema A.S., Strikas R.A. Influenza pandemic preparedness. Emerg. Infect. Dis. 2003;9:1645. doi: 10.3201/eid0912.030289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Snacken R., Kendal A.P., Haaheim L.R., Wood J.M. The next influenza pandemic: lessons from Hong Kong. Emerg. Infect. Dis. 1997;5:195. doi: 10.3201/eid0502.990202. 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Qiu X., Zhang S., Xiang F., Wu D., Guo M., Ge S., Qian S. Instrument-free point-of-care molecular diagnosis of H1N1 based on microfluidic convective PCR. Sensor. Actuator. B Chem. 2017;243:738–744. [Google Scholar]
- 129.Hui E.K.W. Reasons for the increase in emerging and re-emerging viral infectious diseases. Microb. Infect. 2006;8:905–916. doi: 10.1016/j.micinf.2005.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mounts A.W., Kwong H., Izurieta H.S., Ho Y.Y., Au T.K., Lee M., Fukuda K. Case-control study of risk factors for avian influenza A (H5N1) disease, Hong Kong. J. Infect. Dis. 1999;180:505–508. doi: 10.1086/314903. 1997. [DOI] [PubMed] [Google Scholar]
- 131.Avian A. Influenza, infection in humans. The writing committee of the WHO consultation on human influenza A/H5. N. Engl. J. Med. 2005;353:1374–1385. [Google Scholar]
- 132.Dandekar A.A., Perlman S. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005;5:917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Horimoto T., Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 2005;3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 134.Peng F., Wang Z., Zhang S., Wu R., Hu S., Li Z., Bi D. Development of an immunochromatographic strip for rapid detection of H9 subtype avian influenza viruses. Clin. Vaccine Immunol. 2008;15:569–574. doi: 10.1128/CVI.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bae S.W., Tan W., Hong J.I. Fluorescent dye-doped silica nanoparticles: new tools for bioapplications. Chem. Commun. 2012;48:2270–2282. doi: 10.1039/c2cc16306c. [DOI] [PubMed] [Google Scholar]
- 136.Mahmoudi T., de la Guardia M., Baradaran B. Lateral flow assays towards point-of-care cancer detection: a review of current progress and future trends. TrAC, Trends Anal. Chem. 2020;125 [Google Scholar]
- 137.Tan W., Wang K., He X., Zhao X.J., Drake T., Wang L., Bagwe R.P. Bionanotechnology based on silica nanoparticles. Med. Res. Rev. 2004;24:621–638. doi: 10.1002/med.20003. [DOI] [PubMed] [Google Scholar]
- 138.Bamrungsap S., Apiwat C., Chantima W., Dharakul T., Wiriyachaiporn N. Rapid and sensitive lateral flow immunoassay for influenza antigen using fluorescently-doped silica nanoparticles. Microchim. Acta. 2014;181:223–230. [Google Scholar]
- 139.Wang Y., Wang K., Zou B., Gao T., Zhang X., Du Z., Zhou S. Magnetic-based silver composite microspheres with nanosheet-assembled shell for effective SERS substrate. J. Mater. Chem. C. 2013:1. [Google Scholar]
- 140.2441-2447.
- 141.Wang C., Wang, Li P., Rong Z., Jia X., Ma Q., Wang S. Sonochemical synthesis of highly branched flower-like Fe 3 O 4@ SiO 2@ Ag microcomposites and their application as versatile SERS substrates. Nanoscale. 2016;8:19816–19828. doi: 10.1039/c6nr07295j. [DOI] [PubMed] [Google Scholar]
- 142.Wang C., Wang C., Wang X., Wang K., Zhu Y., Rong Z., Wang S. Magnetic SERS strip for sensitive and simultaneous detection of respiratory viruses. ACS Appl. Mater. Interfaces. 2019;11:19495–19505. doi: 10.1021/acsami.9b03920. [DOI] [PubMed] [Google Scholar]
- 143.Wang C., Yang X., Zheng S., Cheng X., Xiao R., Li Q., Wang S. Development of an ultrasensitive fluorescent immunochromatographic assay based on multilayer quantum dot nanobead for simultaneous detection of SARS-CoV-2 antigen and influenza A virus. Sensor. Actuator. B Chem. 2021;345 doi: 10.1016/j.snb.2021.130372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jain P., Chakma B., Patra S., Goswami P. BioMed research international; 2014. Potential Biomarkers and Their Applications for Rapid and Reliable Detection of Malaria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.WHO, UNICEF . World Health Organization and UNICEF; Geneva: 2005. World Malaria Report 2005. [Google Scholar]
- 146.WHO, UNICEF . World Health Organization and UNICEF; Geneva: 2005. World Malaria Report 2005. [Google Scholar]
- 147.Feachem R.G., Chen I., Akbari O., Bertozzi-Villa A., Bhatt S., Binka F., Mpanju-Shumbusho W. Malaria eradication within a generation: ambitious, achievable, and necessary. Lancet. 2019;394:1056–1112. doi: 10.1016/S0140-6736(19)31139-0. [DOI] [PubMed] [Google Scholar]
- 148.Jain P., Chakma B., Patra S., Goswami P. Potential biomarkers and their applications for rapid and reliable detection of malaria. BioMed Res. Int. 2014 doi: 10.1155/2014/852645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pereira D.Y., Chiu R.Y., Zhang S.C., Wu B.M., Kamei D.T. Single-step, paper-based concentration and detection of a malaria biomarker. Anal. Chim. Acta. 2015;882:83–89. doi: 10.1016/j.aca.2015.04.040. [DOI] [PubMed] [Google Scholar]
- 150.Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lalremruata A., Nguyen T.T., McCall M.B., Mombo-Ngoma G., Agnandji S.T., Adegnika A.A., Mordmüller B. Recombinase polymerase amplification and lateral flow assay for ultrasensitive detection of low-density Plasmodium falciparum infection from controlled human malaria infection studies and naturally acquired infections. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01879-19. e01879-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sharma S., Kumar S., Ahmed M.Z., Bhardwaj N., Singh J., Kumari S., Das J. Advanced multiplex loop mediated isothermal amplification (mLAMP) combined with lateral flow detection (LFD) for rapid detection of two prevalent malaria species in India and melting curve analysis. Diagnostics. 2021;12:32. doi: 10.3390/diagnostics12010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mallepaddi P.C., Lai M.Y., Podha S., Ooi C.H., Liew J.W.K., Polavarapu R., Lau Y.L. Development of loop-mediated isothermal amplification–based lateral flow device method for the detection of malaria. Am. J. Trop. Med. Hyg. 2018;99:704. doi: 10.4269/ajtmh.18-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hampl H., Ben-Porat T., Ehrlicher L., Habermehl K.O., Kaplan A.S. Characterization of the envelope proteins of pseudorabies virus. J. Virol. 1984;52:583–590. doi: 10.1128/jvi.52.2.583-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sun Y., Liang W., Liu Q., Zhao T., Zhu H., Hua L., Wu B. Epidemiological and genetic characteristics of swine pseudorabies virus in mainland China between 2012 and 2017. PeerJ. 2018;6:5785. doi: 10.7717/peerj.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen H., Zhang X., Jin Z., Huang L., Dan H., Xiao W., Tang Y. Differential diagnosis of PRV-infected versus vaccinated pigs using a novel EuNPs-virus antigen probe-based blocking fluorescent lateral flow immunoassay. Biosens. Bioelectron. 2020;155 doi: 10.1016/j.bios.2020.112101. [DOI] [PubMed] [Google Scholar]
- 157.Hoenen T., Groseth A., Falzarano D., Feldmann H. Ebola virus: unravelling pathogenesis to combat a deadly disease. Trends Mol. Med. 2016;12:206–215. doi: 10.1016/j.molmed.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 158.Hu J., Jiang Y.Z., Wu L.L., Wu Z., Bi Y., Wong G., Zhang Z.L. Dual-signal readout nanospheres for rapid. Anal. Chem. 2017;89:13105–13111. doi: 10.1021/acs.analchem.7b02222. [DOI] [PubMed] [Google Scholar]
- 159.Hong W., Huang L., Wang H., Qu J., Guo Z., Xie C., Zhou L. Yersinia pestis'e karşı antikorların profilini çıkarmak için yukarı dönüştürücü fosfor teknolojisi tabanlı 10 kanallı yanal akış testinin geliştirilmesi. Mikrobiyolojik yöntemler dergisi. 2010;83:133–140. [Google Scholar]
- 160.Bissonnette L., Bergeron M.G. Diagnosing infections-Current and anticipated technologies for point-of-care diagnostics and home-based testing. Clin. Microbiol. Infect. 2010;16:1044–1053. doi: 10.1111/j.1469-0691.2010.03282.x. [DOI] [PubMed] [Google Scholar]
- 161.Morens D.M., Folkers G.K., Fauci A.S. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.MacArthur G. Global Health Diagnostics: Research, Development, and Regulation. Workshop Report. Academy of Medical Sciences; 2009. Global health diagnostics: research, development, and regulation. Workshop report. [Google Scholar]
- 163.Mabey D., Peeling R.W., Ustianowski A., Perkins M.D. Diagnostics for the developing world. Nat. Rev. Microbiol. 2004;2:231–240. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- 164.St John A., Price C.P. Existing and emerging technologies for point-of-care testing. Clin. Biochem. Rev. 2014;35:155. [PMC free article] [PubMed] [Google Scholar]
- 165.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.M.A., Esswein S.R., Gristick H.B., Bjorkman P.J. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gaebler C., Wang Z., Lorenzi J.C., Muecksch F., Finkin S., Tokuyama M., Nussenzweig M.C. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Kyratsous C.A. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2021;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cristina S., Concetta R., Francesco R., Annalisa C. SARS-Cov-2 infection: response of human immune system and possible implications for the rapid test and treatment. Int. Immunopharm. 2020;84 doi: 10.1016/j.intimp.2020.106519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Goldman E.R., Clapp A.R., Anderson G.P., Uyeda H.T., Mauro J.M., Medintz I.L., Mattoussi H. Anal. Chem. 2004;76:684–688. doi: 10.1021/ac035083r. [DOI] [PubMed] [Google Scholar]
- 170.Corstjens P., Zuiderwijk M., Brink A., Li S., Feindt H., Niedbala R.S., Tanke H. Use of up-converting phosphor reporters in lateral-flow assays to detect specific nucleic acid sequences: a rapid, sensitive DNA test to identify human papillomavirus type 16 infection. Clin. Chem. 2001;47:1885–1893. [PubMed] [Google Scholar]
- 171.Wang Z.Z., Zheng Z., Wang X.C., Zheng P.M., Cui F.C., Zhou Q.W., Ma Y.F. MedRxiv; 2020. Rapid Detection of Anti-SARS-CoV-2 IgM and IgG Using a Selenium Nanoparticle-Based Lateral Flow Immunoassay. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Lin G. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 173.Ahi E.E., Torul H., Zengin A., Sucularlı F., Yıldırım E., Selbes Y., Tamer U. A capillary driven microfluidic chip for SERS based hCG detection. Biosens. Bioelectron. 2022;195 doi: 10.1016/j.bios.2021.113660. [DOI] [PubMed] [Google Scholar]
- 174.Clarke O.J., Goodall B.L., Hui H.P., Vats N., Brosseau C.L. Development of a SERS-based rapid vertical flow assay for point-of-care diagnostics. Anal. Chem. 2017;89:1405–1410. doi: 10.1021/acs.analchem.6b04710. [DOI] [PubMed] [Google Scholar]
- 175.Jeon J., Lee S.H., Joung Y., Kim K., Choi N., Choo J. Improvement of reproducibility and thermal stability of surface-enhanced Raman scattering-based lateral flow assay strips using silica-encapsulated gold nanoparticles. Sensor. Actuator. B Chem. 2020;321 [Google Scholar]
- 176.Borzenkov M., Chirico G., d'Alfonso L., Sironi L., Collini M., Cabrini E., Denat F. Thermal and chemical stability of thiol bonding on gold nanostars. Langmuir. 2015;31:8081–8091. doi: 10.1021/acs.langmuir.5b01473. [DOI] [PubMed] [Google Scholar]
- 177.Huang J., Kim K.H., Choi N., Chon H., Lee S., Choo J. Preparation of silica-encapsulated hollow gold nanosphere tags using layer-by-layer method for multiplex surface-enhanced Raman scattering detection. Langmuir. 2011;27:10228–10233. doi: 10.1021/la201739n. [DOI] [PubMed] [Google Scholar]
- 178.Fernández-López C., Mateo-Mateo C., Alvarez-Puebla R.A., Pérez-Juste J., Pastoriza-Santos I., Liz-Marzán L.M. Highly controlled silica coating of PEG-capped metal nanoparticles and preparation of SERS-encoded particles. Langmuir. 2009;25:13894–13899. doi: 10.1021/la9016454. [DOI] [PubMed] [Google Scholar]
- 179.Li Y., Liu X., Guo J., Zhang Y., Guo J., Wu X., Ma X. Simultaneous detection of inflammatory biomarkers by SERS nanotag-based lateral flow assay with portable cloud Raman spectrometer. Nanomaterials. 2021;11:1496. doi: 10.3390/nano11061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Liu X., Yang X., Li K., Liu H., Xiao R., Wang W., Wang S. Fe3O4@ Au SERS tags-based lateral flow assay for simultaneous detection of serum amyloid A and C-reactive protein in unprocessed blood sample. Sensor. Actuator. B Chem. 2020;320 [Google Scholar]
- 181.Sturgeon C.M., Viljoen A. Analytical error and interference in immunoassay: minimizing risk. Ann. Clin. Biochem. 2011;48:418–432. doi: 10.1258/acb.2011.011073. [DOI] [PubMed] [Google Scholar]
- 182.Haleyur Giri Setty M.K., Lee S., Lathrop J., Hewlett I.K. Biotin interference in point of care HIV immunoassay. BioResearch Open Access. 2020;9:243–246. doi: 10.1089/biores.2020.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Khlebtsov B.N., Bratashov D.N., Byzova N.A., Dzantiev B.B., Khlebtsov N.G. SERS-based lateral flow immunoassay of troponin I by using gap-enhanced Raman tags. Nano Res. 2019;12:413–420. [Google Scholar]
- 184.Dussart P., Labeau B., Lagathu G., Louis P., Nunes M.R., Rodrigues S.G., Baril L. Evaluation of an enzyme immunoassay for detection of dengue virus NS1 antigen in human serum. Clin. Vaccine Immunol. 2006;13:1185–1189. doi: 10.1128/CVI.00229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang S.M., Sekaran S.D. Evaluation of a commercial SD dengue virus NS1 antigen capture enzyme-linked immunosorbent assay kit for early diagnosis of dengue virus infection. J. Clin. Microbiol. 2010;48:2793–2797. doi: 10.1128/JCM.02142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ghazal K., Brabant S., Prie D., Piketty M.L. Hormone immunoassay interference: a 2021 Update. Ann. Lab. Med. 2022;42:3–23. doi: 10.3343/alm.2022.42.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Aman R., Mahas A., Mahfouz M. Nucleic acid detection using CRISPR/Cas biosensing technologies. ACS Synth. Biol. 2020;9:1226–1233. doi: 10.1021/acssynbio.9b00507. [DOI] [PubMed] [Google Scholar]
- 189.Huang C., Wen T., Shi F.J., Zeng X.Y., Jiao Y.J. Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega. 2020;5:12550–12556. doi: 10.1021/acsomega.0c01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.T. Wen, C. Huang, F.J. Shi, X.Y. Zeng, T. Lu, S.N. Ding, Y.L. Jiao, Y. J. Development of a lateral flow immunoassay strip for rapid detection of IgG antibody against SARS-CoV-2 virus. Analyst, 145(2020), p.5345-5352. [DOI] [PubMed]
- 191.Feng M., Chen J., Xun J., Dai R., Zhao W., Lu H., Cheng X. Development of a sensitive immunochromatographic method using lanthanide fluorescent microsphere for rapid serodiagnosis of COVID-19. ACS Sens. 2020;5:2331–2337. doi: 10.1021/acssensors.0c00927. [DOI] [PubMed] [Google Scholar]
- 192.Zeng L., Li Y., Liu J., Guo L., Wang Z., Xu X., Xu C. Rapid, ultrasensitive and highly specific biosensor for the diagnosis of SARS-CoV-2 in clinical blood samples. Mater. Chem. Front. 2020;4:2000–2005. [Google Scholar]
- 193.Liu C., Mao B., Martinez V., Chen X., Li Y., He L., Li C. A facile assay for rapid detection of COVID-19 antibodies. RSC Adv. 2020;10:28041–28048. doi: 10.1039/d0ra04107f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang Z., Zheng Z., Hu H., Zhou Q., Liu W., Li X., Ma Y. A point-of-care selenium nanoparticle-based test for the combined detection of anti-SARS-CoV-2 IgM and IgG in human serum and blood. Lab Chip. 2020;20:4255–4261. doi: 10.1039/d0lc00828a. [DOI] [PubMed] [Google Scholar]
- 195.Zhang C., Zhou L., Du K., Zhang Y., Wang J., Chen L., He K. Foundation and clinical evaluation of a new method for detecting SARS-CoV-2 antigen by fluorescent microsphere immunochromatography. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.553837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Liu D., Ju C., Han C., Shi R., Chen X., Duan D., Yan X. bioRxiv; 2020. Ultra-sensitive Nanozyme-Based Chemiluminescence Paper Test for Rapid Diagnosis of SARS-CoV-2 Infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Srivastav S., Dankov A., Adanalic M., Grzeschik R., Tran V., Pagel-Wieder S., Schlücker S. Rapid and sensitive SERS-based lateral flow test for SARS-CoV2-specific IgM/IgG antibodies. Anal. Chem. 2021;93:12391–12399. doi: 10.1021/acs.analchem.1c02305. [DOI] [PubMed] [Google Scholar]
- 198.Panferov V.G., Byzova N.A., Biketov S.F., Zherdev A.V., Dzantiev B.B. Comparative study of in situ techniques to enlarge gold nanoparticles for highly sensitive lateral flow immunoassay of SARS-CoV-2. Biosensors. 2021;11:229. doi: 10.3390/bios11070229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ahmadi A., Mirzaeizadeh Z., Omidfar K. Simultaneous detection of SARS-CoV-2 IgG/IgM antibodies, using gold nanoparticles-based lateral flow immunoassay. Monoclon. Antibodies Immunodiagn. Immunother. 2021;40:210–218. doi: 10.1089/mab.2021.0027. [DOI] [PubMed] [Google Scholar]
- 200.Zou M., Su F., Zhang R., Jiang X., Xiao H., Yan X., Wu G. Rapid point-of-care testing for SARS-CoV-2 virus nucleic acid detection by an isothermal and nonenzymatic Signal amplification system coupled with a lateral flow immunoassay strip. Sensor. Actuator. B Chem. 2021;342 doi: 10.1016/j.snb.2021.129899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Elledge S.K., Zhou X.X., Byrnes J.R., Martinko A.J., Lui I., Pance K., Wells J.A. Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection. Nat. Biotechnol. 2021:1–8. doi: 10.1038/s41587-021-00878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Xu L.D., Zhu J., Ding S.N. Immunoassay of SARS-CoV-2 nucleocapsid proteins using novel red emission-enhanced carbon dot-based silica spheres. Analyst. 2021;146:5055–5060. doi: 10.1039/d1an01010g. [DOI] [PubMed] [Google Scholar]
- 203.Xu L.D., Du F.L., Zhu J., Ding S.N. Luminous silica colloids with carbon dot incorporation for sensitive immunochromatographic assay of Zika virus. Analyst. 2021;146:706–713. doi: 10.1039/d0an02017f. [DOI] [PubMed] [Google Scholar]
- 204.Kumar S., Bhushan P., Krishna V., Bhattacharya S. Tapered lateral flow immunoassay based point-of-care diagnostic device for ultrasensitive colorimetric detection of dengue NS1. Biomicrofluidics. 2018;12 doi: 10.1063/1.5035113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Parpia Z.A., Elghanian R., Nabatiyan A., Hardie D.R., Kelso D.M. p24 antigen rapid test for diagnosis of acute pediatric HIV infection. JAIDS J. Acquir. Immune Defic. Syndr. 2010;55:413–419. doi: 10.1097/QAI.0b013e3181f1afbc. [DOI] [PubMed] [Google Scholar]
- 206.Loynachan C.N., Thomas M.R., Gray E.R., Richards D.A., Kim J., Miller B.S., Stevens M.M. Platinum nanocatalyst amplification: redefining the gold standard for lateral flow immunoassays with ultrabroad dynamic range. ACS Nano. 2018;12:279–288. doi: 10.1021/acsnano.7b06229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hu J., Wang L., Li F., Han Y.L., Lin M., Lu T.J., Xu F. Oligonucleotide-linked gold nanoparticle aggregates for enhanced sensitivity in lateral flow assays. Lab Chip. 2013;13:4352–4357. doi: 10.1039/c3lc50672j. [DOI] [PubMed] [Google Scholar]
- 208.Kwon J.H., Kim H.T., Sim S.J., Cha Y.J., Lee J. Performance of point-of-care diagnosis of AIDS: label-free one-step-immunoassay vs. lateral flow assay. Analyst. 2018;143:936–942. doi: 10.1039/c7an01748k. [DOI] [PubMed] [Google Scholar]
- 209.Martikainen I., Juntunen E., Salminen T., Vuorenpa K., Bayoumy S., Vuorinen T., Talha S.M. Double-antigen lateral flow immunoassay for the detection of anti-HIV-1 and-2 antibodies using upconverting nanoparticle reporters. Sensors. 2021;21:330. doi: 10.3390/s21020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Li L., Zhou L., Yu Y., Zhu Z., Lin C., Lu C., Yang R. Development of up-converting phosphor technology-based lateral-flow assay for rapidly quantitative detection of hepatitis B surface antibody. Diagn. Microbiol. Infect. Dis. 2009;63:165–172. doi: 10.1016/j.diagmicrobio.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 211.Shen H., Yuan H., Niu J.Z., Xu S., Zhou C., Ma L., Li L.S. Phosphine-free synthesis of high-quality reverse type-I ZnSe/CdSe core with CdS/CdxZn1− xS/ZnS multishell nanocrystals and their application for detection of human hepatitis B surface antigen. Nanotechnology. 2011;22 doi: 10.1088/0957-4484/22/37/375602. [DOI] [PubMed] [Google Scholar]
- 212.Wiriyachaiporn N., Sirikaew S., Bamrungsap S., Limcharoen T., Polkankosit P., Roeksrungruang P., Ponlamuangdee K. A simple fluorescence-based lateral flow test platform for rapid influenza B virus screening. Anal. Methods. 2021;13:1687–1694. doi: 10.1039/d0ay01988g. [DOI] [PubMed] [Google Scholar]
- 213.Sun N., Wang Y., Yao X., Chen F., Gao D., Wang W., Li X. Visual signal generation for the detection of influenza viruses by duplex recombinase polymerase amplification with lateral flow dipsticks. Anal. Bioanal. Chem. 2019;411:3591–3602. doi: 10.1007/s00216-019-01840-z. [DOI] [PubMed] [Google Scholar]
- 214.Mthembu C.L., Sabela M.I., Mlambo M., Madikizela L.M., Kanchi S., Gumede H., Mdluli P.S. Google Analytics and quick response for advancement of gold nanoparticle-based dual lateral flow immunoassay for malaria–Plasmodium lactate dehydrogenase (pLDH) Anal. Methods. 2017;9:5943–5951. [Google Scholar]
- 215.Brangel P., Sobarzo A., Parolo C., Miller B.S., Howes P.D., Gelkop S., Stevens M.M. A serological point-of-care test for the detection of IgG antibodies against Ebola virus in human survivors. ACS Nano. 2018;12:63–73. doi: 10.1021/acsnano.7b07021. [DOI] [PubMed] [Google Scholar]