Abstract
In the context of global pandemic Coronavirus disease 2019 (COVID-19) that threatens life of all human beings, it is of vital importance to achieve early detection of COVID-19 among symptomatic patients. In this paper, a computer aided diagnosis (CAD) model Cov-Net is proposed for accurate recognition of COVID-19 from chest X-ray images via machine vision techniques, which mainly concentrates on powerful and robust feature learning ability. In particular, a modified residual network with asymmetric convolution and attention mechanism embedded is selected as the backbone of feature extractor, after which skip-connected dilated convolution with varying dilation rates is applied to achieve sufficient feature fusion among high-level semantic and low-level detailed information. Experimental results on two public COVID-19 radiography databases have demonstrated the practicality of proposed Cov-Net in accurate COVID-19 recognition with accuracy of 0.9966 and 0.9901, respectively. Furthermore, within same experimental conditions, proposed Cov-Net outperforms other six state-of-the-art computer vision algorithms, which validates the superiority and competitiveness of Cov-Net in building highly discriminative features from the perspective of methodology. Hence, it is deemed that proposed Cov-Net has a good generalization ability so that it can be applied to other CAD scenarios. Consequently, one can conclude that this work has both practical value in providing reliable reference to the radiologist and theoretical significance in developing methods to build robust features with strong presentation ability.
Keywords: COVID-19, Computer aided diagnosis (CAD), Feature learning, Image recognition, Machine vision
1. Introduction
In the past few years, the coronavirus disease 2019 (COVID-19) has posed a world-wide threat to the life of all human beings which leads to millions of deaths. According to report of the World Health Organization (WHO), up to May 2022, there have been over 513 millions confirmed cases covering hundreds of countries (real-time data can be traced at https://covid19.who.int). Consequently, there has been constant and increasing focus on mechanisms for coping with this disaster, for example through development and application of effective diagnostic tools, treatments, and vaccination programmes. As part of this, the role of early-stage detection of COVID-19 has been fundamental in facilitating timely treatment and reducing transmission through isolation (Shaban et al., 2021, Vianello et al., 2021, Yu et al., 2021).
Currently in practice, reverse transcription-polymerase chain reaction (RT-PCR) is the most popular screening method of COVID-19 detection since the beginning of the epidemic, which is regarded as a reliable gold-standard. While in addition, imaging can serve as a complementary method to assist radiologist in diagnosing COVID-19 so as to achieve greater diagnostic certainty (Ai et al., 2020, Ismael and Şengür, 2021, Kassani et al., 2021), where the most remarkable merits include convenient acquisition and time-saving detection (Keidar et al., 2021). Consequently, the application of computer aided diagnosis (CAD) framework incorporating chest X-ray (CXR) or computed tomography (CT) images can circumvent issues associated with existing diagnostic procedures and enable enhanced recognition of COVID-19.
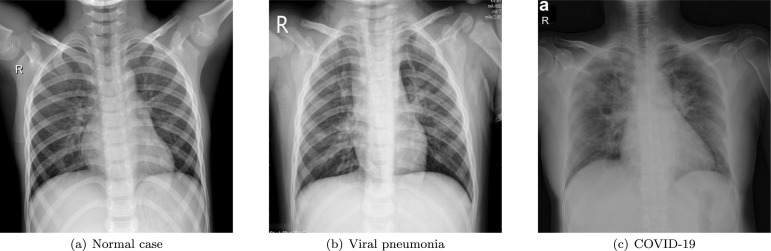
Deep learning methods in modeling and classification have been successfully applied in a variety of fields especially medical imaging domain (Albu et al., 2019, Borlea et al., 2021, Hsiu-Sen et al., 2014, Leong et al., 2013, Upadhyay and Nagpal, 2020, Zeng et al., 2021b). In the context of early COVID-19 diagnosis, CXR imaging methods are generally formulated as a classification problem, as shown in Fig. 1 where three categories are included. It should be pointed out that so far plenty of researches have been carried out on identifying COVID-19 from CXR images (Bouchareb et al., 2021, Chandra et al., 2021, Fan et al., 2021, Fusco et al., 2021, Roberts et al., 2021, Signoroni et al., 2021, Xu et al., 2021).
Fig. 1.
Three categories of chest X-ray images, which from left to right are normal case, viral pneumonia and COVID-19, respectively.
In Hu et al. (2021), the authors establish a deep convolutional neural network (CNN) structure. Specifically, LeNet-5 framework is employed as the backbone to learn features from X-ray images, and a single-layer extreme learning machine serves as the classifier to differentiate normal case, viral pneumonia, and COVID-19. A deep-learning based multi-modal system is proposed in Sait et al. (2021), which makes use of not only CXR images but also the breathing sounds for COVID-19 diagnosis, whereas the feature extractor therein is the CNN model as well.
In Minaee et al. (2020), by means of transfer learning, an end-to-end framework for COVID-19 prediction from chest X-ray images without additional feature extraction steps is proposed, where performances of several classical CNN variants have been evaluated, including CNN with residual, dense block and “fire” module (i.e., the ResNet, DenseNet and SqueezeNet), respectively. Furthermore, in Signoroni et al. (2021), another end-to-end deep learning architecture is developed, which realizes a multi-regional scoring system so that degree of lung injury in COVID-19 patient could be estimated in a semi-quantitative manner. According to the real-world application results, it is proven a reliable and practical method with significant prognostic value.
Another notable thing is that transfer learning paradigm has been adopted in Minaee et al. (2020) to overcome the data-imbalanced problem. In Sheykhivand et al. (2021), generative adversarial network (GAN) is employed to generate CXR images of COVID-19 category, which has enlarged the sample capacity to nearly 7 times and facilitated more robust feature learning. Notice that the relatively less COVID-19 training samples can lead to model bias, and aiming at this phenomenon, work (Garcia Santa Cruz et al., 2021) has presented a systematic inspection on public COVID-19 X-ray imaging datasets and provided effective guidance accordingly.
The role that attention mechanism plays in images classification is highlighted in literature (Fan et al., 2021), where spatial and channel attention mechanism with multi-kernel-size is adopted to obtain abundant semantic information over multiple scopes so as to promote accurate detection of COVID-19. In Xu et al. (2021), the authors propose a mask attention mechanism, which is used to achieve segment-based classification of COVID-19 on CXR images. Generated masks are regarded as spatial attention maps and subsequently the CNN is able to concentrate more on specific regions (like lesions) for classification. A multi-receptive field attention module is proposed in Ma et al. (2021). Although the data used in Ma et al. (2021) are CT images of COVID-19, the common idea is applying spatial and channel attention mechanism to make extracted features focus more on lesion areas so as to improve the final classification performance. Moreover, comprehensive reviews on applications of artificial intelligence method for COVID-19 recognition from radiological images are presented in Fusco et al. (2021) and Bouchareb et al. (2021) where one can find more related works.
From above discussions, it is found that a feature extractor together with a classifier constitutes a universal structure for COVID-19 recognition from CXR images. Consequently, the most challenging issue is how to build strong and robust features with abundant information and high discrimination from the relatively limited COVID-19 samples, so as to facilitate a highly-efficient feature learning and achieve accurate classification results. As is shown in Fig. 1, there is high similarity in CXR images between COVID-19 and viral pneumonia (Lascu, 2021, Nabizadeh-Shahre-Babak et al., 2021), thus discriminative features with strong presentation ability no doubts can benefit an accurate classification.
To handle above challenge in COVID-19 recognition from CXR images, a computer vision (CV) based model Cov-Net is proposed in this study, of which the primary motivation is to make full use of CV techniques to build robust and highly-distinctive features, so that eventual classification results can provide more accurate and reliable reference for the radiologists. It should be pointed out that some of our previous works have already laid solid foundation and accumulated valuable experiences for this study (Wu et al., 2022, Zeng et al., 2021a, Zeng et al., 2021b, Zeng et al., 2022).
Particularly, in our previous work (Wu et al., 2022), an effective self-attention mechanism is developed in proposed model FMD-Yolo to detect face masks and is proven efficient; in Zeng et al. (2022) a practical feature fusion method is put forward to realize precise localization and recognition of tiny objects, which also turns out a successful implementation. Mainly inspired by above two instances, in proposed Cov-Net, a feature learning and a feature fusion module are meticulously designed, and it is the delicate implementation manner of the two modules that makes Cov-Net distinct and superior.
To be specific, residual network (ResNet) structure, a well-known CNN variant, is adopted as the backbone of Cov-Net to extract features, where asymmetric convolution substitutes the vanilla configurations, which can adapt to the disturb noises introduced by various shooting angles. Attention mechanism is realized by squeeze-and-excitation operation which enables the network to focus more on the lesion areas so that strengthen the discrimination ability. Moreover, in order to fully excavate representative features in COVID-19 samples like bilateral pulmonary parenchyma ground-glass and consolidative pulmonary opacities (Yu et al., 2021), a feature fusion module is deployed after above backbone, which applies skip connection among a set of dilated convolution kernels with varying dilation rates.
It should be pointed out that it is the powerful feature extraction ability that is concentrated on while establishing the Cov-Net. Compared with other existing works, innovations of Cov-net are reflected on the subtly designed components therein, which are eventually proven effective, superior and practical. More importantly, this study mainly aims at providing a complementary method to commonly used diagnosis tool of COVID-19 like RT-PCR, so that greater diagnostic certainty can be achieved. For a better view of this paper, major contributions of proposed Cov-Net have been summarized as follows:
(1) Building highly discriminative features from chest X-ray images to boost classification of COVID-19 is emphasized in this work, and proposed Cov-Net is proven reliable and effective on two radiography databases.
(2) Feature learning ability is enhanced by a meticulously designed attention mechanism, which applies asymmetric convolution operations in a residual structure.
(3) A set of skip-connected dilated convolution kernels with different dilation rates is deployed to sufficiently fuse multi-scale feature maps including both high-level textural and low-level morphological information.
The rest of this paper is organized as follows. Preliminaries of this work are outlined in Section 2. The complete Cov-Net architecture is elaborately presented in Section 3. Experimental results have been reported in Section 4, and comprehensive discussions on both advantages and disadvantages of proposed model are offered. Finally, conclusions are drawn in Section 5.
2. Preliminaries
In this section, some brief preliminaries of proposed Cov-Net are provided for a better understanding, including background information about the residual building block, dilated and asymmetric convolution.
2.1. Residual Network (ResNet)
Convolutional neural network and its variants have been acknowledged making great progresses in computer vision, where one of the famous structures is known as residual network (ResNet) which is firstly designed to handle the degradation problem in network training, i.e., accuracy will decline with the depth of a network increases (He et al., 2016). Formally in a residual structure, a mapping relationship will be recast into via a shortcut connection, where is the so-called residual. It is assumed that fitting the mapping is much easier than approximating during the same training process, while effects of two learning are equivalent essentially. Furthermore, even under an extreme case where , both input and output of the structure are , which at least will not lead to worse results.
2.2. Dilated convolution
Dilated convolution (Yu & Koltun, 2016) extends the local receptive field by adding a hyper-parameter on standard kernel, of which the physical meaning refers to distance between adjacent parameters, hence in a standard convolution kernel, is default as 1. The most advantage of dilated convolution is that it can provide a larger receptive field under same computational condition, given a dilation rate , then an kernel will generate a receptive field with size of , whereas the number of kernel parameters remains because all added “holes” are padded with 0. However, it should be pointed out that a large dilation rate is not always beneficial, which may lead to severe loss of information.
2.3. Asymmetric convolution
Asymmetric convolution network (Ding et al., 2019) adopts 1-dimensional kernels to strengthen central skeleton parts of traditional convolution kernels, which usually occur in the form of squares with size . There is evidence showing that convolution can be factorized into two 1-rank filters in order of a vertical filter and a horizontal one as follows (Jaderberg et al., 2014):
| (1) |
where is input, symbol denotes convolution operator, and stand for kernels with scale marked in subscript. Albeit computational complexity becomes less with parameters shrunk from to , performance also declines to some extend (Jin et al., 2014), hence a better alternative is to combine the factorized components with initial square kernels as shown in Fig. 2. Mathematically, the dark synthetic skeleton kernel is obtained as:
| (2) |
where is the hadamard product operator, is input and denotes the two 1-rank filters.
Fig. 2.
Reinforced square-convolution-kernel with two 1-rank filters.
3. Detailed implementations of proposed Cov-Net
In this section, proposed Cov-Net is elaborated with details, and specifically, major innovations of Cov-Net have been highlighted. As aforementioned, the core components in Cov-Net contain one feature learning module used to extract abundant and valuable information from input chest X-ray images, and another feature fusion module set for further generating and merging multi-level feature maps. It is noteworthy that both high-level architectures and low-level units in proposed Cov-Net have been comprehensively presented, and an illustration of the overall framework is shown in Fig. 3, where boxes in the top right part reveal the implementation details of applied blocks with corresponding color. Following subsections will provide more explicit explanations around Fig. 3 combining with the specific work procedure.
Fig. 3.
Framework of the proposed Cov-Net, via matching color with boxes one can find implementation details of corresponding applied blocks.
3.1. Modified residual structure for feature extraction
Feature learning module in Cov-Net consists of stacked bottlenecks where residual structure is adopted to eliminate degradation as the network deepens. Unlike traditional ResNet deployment, asymmetric convolution has been employed for enhanced feature extraction ( block in Fig. 3). To be specific, after incorporating two 1-dimensional kernels, a square kernel with reinforced skeleton part is obtained which enables greater focus on center areas in corresponding receptive fields. During convolution, a kernel will slide over the input feature map with a fixed stride. Pixels can therefore serve as both center and corner during an entire convolution procedure. It is believed that in each individual receptive field, center areas take more important information while the corners can be relatively ignored. A reinforced skeleton part makes it possible to strengthen partial connections between a convolution kernel and input feature map, resulting in more concentrated and efficient learning without laying extra computational burdens.
Given that COVID-19 samples are relatively limited, it is desirable to build highly-distinctive features by making full use of finite images. It is a challenging task and is further compounded by the high visual similarity between COVID-19 and viral pneumonia classes. That is, some features observed in most COVID-19 cases can also be seen in other pneumonia, which no doubts makes the accurate diagnosis a dilemma. Over-focusing on common features may lead to misclassification but paying less attention to them can result in information loss. It is desirable to focus more on the unique characteristics of COVID-19 such as the ground-glass opacity and pleural effusion.
To circumvent these issues, attention mechanism is introduced to the Cov-Net feature learning module after asymmetric convolution ( block in Fig. 3). Essentially, the squeeze and excitation are two cascaded fully connected layers (denoted as and ). Firstly a global average pooling is performed to shrink input in the spatial, then refined feature map will be forwarded to the layers for element-wise weight generation. Following equations describe above squeeze and excitation process (Hu et al., 2017):
| (3) |
where and are number of channels, and are two matrices, and denotes activation functions. is initial input feature maps and stands for the weights. Notice that symbols written in boldface refer to vectors, and others are scalars.
Applying attention mechanism mainly aims at suppressing unimportant information and simultaneously highlighting important ones, which reassigns weights of different feature maps according to their importance so that selective integration of abundant information is possible and finally feature maps with significant specificity of COVID-19 can be acquired. In Algorithm 1, complete workflow of designed feature learning module is provided.
3.2. Skip-connected dilated convolutions for feature fusion
In proposed Cov-Net, a feature fusion module is concatenated after the feature learning module to further merge extracted information of different levels. A set of dilated convolution kernels with varying dilation rates is applied to acquire higher-level features based on extracted maps ( block in Fig. 3). Multiple dilation rates enable focus on lesions of different size and overcome the drawback of local information loss in large receptive field. Moreover, this setting enables generation of multi-scale feature maps, which is helpful for building distinctive features and beneficial when tackling the similarity between COVID-19 and viral pneumonia images.
As shown in Fig. 3, let denote operation of five dilated convolution blocks () from left to right, respectively. and are input and output of , and so on. Obviously is , which is the final output of feature learning module mentioned in Algorithm 1. Based on above annotations, for each block there is:
| (4) |
According to Fig. 3, dilation rates increase as the structure deepens; and consequently, output of each dilated convolution block will be gradually shrunk in size. Hence a larger receptive field is helpful for capturing more global information in later stage, which to some extent avoids losing information so that a through coarse to fine feature extraction is realized. In addition, dropout technique is introduced in each dilated convolution block with a probability of 0.1 to alleviate the over-fitting problem. Simultaneously, generated feature maps are in different sizes and include both high-level semantic information and low-level detail features. Furthermore, skip connection has been applied in designed feature fusion module, which can be reflected by:
| (5) |
where is element-wise addition.
The application of skip connection among multi-scale feature maps facilitates the sufficient integration between high-resolution textural and low-resolution morphological features, which also contributes to recovering the lost information during the down-sampling procedure. Afterwards, the final output of the feature fusion module can be obtained as:
| (6) |
So far, an input image has been successively fed into the feature learning and fusion modules, resulting in . After normalization, is fed into a fully connected layer for final classification. Algorithm 2 presents the work principles of designed feature fusion module.
3.3. Supplementary explanations of computational details
In addition to the information outlined above, this subsection constitutes additional details which supplement Fig. 3 for a comprehensive understanding of the proposed Cov-Net.
-
•
Before entering the first bottleneck, the input has been performed a down-sampling operation (7 × 7 convolution and max-pooling), which not only preserves initial information but also decreases the computational complexity via dimension reduction.
-
•
Considering the problem of gradient vanishing in training process, Relu is adopted as the activation function, of which the derivation always equals to 1 in the positive real number domain.
-
•
1 × 1 convolution is applied for the purpose of adjusting channels so as to guarantee different feature maps can be added in element wise.
-
•
In order to accelerate the training and facilitate the convergence, normalization techniques are also employed in proposed Cov-Net, i.e., and in Fig. 3, which stand for batch and group normalization, respectively.
-
•
Since the COVID-19 classification task has three categories, the output layer of the classifier correspondingly has three neurons ( in Fig. 3), and function is used for calculating the category probability.
Notice that specific configurations of entire Cov-Net are determined empirically, including structures of modules and the blocks therein. Whereas partial hyper-parameters are finally selected via several attempts with the best performance, like dilation rate, dropout probability, etc.
3.4. Loss function and optimizer of Cov-Net
Proposed Cov-Net belongs to the supervised learning model, thereby training samples should be equipped with corresponding labels, and each training sample can be denoted as where is the ground-truth label. During model training, the loss function adopts following cross-entropy form:
| (7) |
where is the predicted result of sample , and denotes all training parameters in Cov-Net. It is noteworthy that applying cross-entropy loss function can effectively alleviate the vanishing gradient problem.
Essentially, training the Cov-Net is equivalent to find the optimal so that reaches its minimal value. To this end, the stochastic gradient descent algorithm with momentum is employed as optimizer of Cov-Net to update in following manner:
| (8) |
where Hamilton operator refers to the gradient, is velocity vector for update; and stand for momentum and learning rate, respectively.
Moreover, it should be pointed out that learning rate has played an important role in the optimizer. To promote convergence and maintain a stable performance simultaneously, in this study, is set to decay in cosine manner, one can refer to Bello et al. (2017) for more information.
4. Experiments and discussions
In this section, proposed Cov-Net is comprehensively evaluated on two public COVID-19 radiography datasets (Chowdhury et al., 2020, Rahman et al., 2021) and the experimental results have been reported in detail. To begin with, a brief introduction of the experiment environment is provided, including database information, evaluation metrics and experimental settings. Afterwards, in-depth discussions around reported results are presented, and comparisons with other six advanced machine vision models are carried out. At last, limitations existing in this study are discussed and an outlook of potential improvements in future works has been provided.
4.1. Experiment environment
4.1.1. Databases
Two public COVID-19 radiography datasets available on website https://www.kaggle.com/tawsifurrahman/covid19-radiography-database have been adopted to evaluate performance of proposed Cov-Net, which are denoted as D1 and D2, respectively. It should be pointed out that both D1 and D2 are created by integrating multiple accessible data sources including publicly available datasets, online repository, and published articles, etc. Moreover, severity of the COVID-19 disease is not marked in neither database, hence regarding to clinical practice, applied datasets may not be authoritative enough to make an objective evaluation. However, it is remarkable that benchmark evaluations in this work mainly orient to examine the classification accuracy of proposed Cov-Net, which is closely related to its feature extraction ability, while to build highly discriminative features with strong presentation ability is right the main contribution of Cov-Net.
In particular, classification task on D1 contains three categories, including COVID-19, normal and viral pneumonia. The latter two classes are collected from Kaggle chest X-ray database which has totally 5247 CXR images with different resolutions, whereas the former one is established by collecting data from other five COVID-19 sub-databases, and more information can refer to Chowdhury et al. (2020). Database D2 provides a four-category classification problem, which has an extra lung opacity class in comparison to D1 and hence is more challenging. To be specific, samples of normal and lung opacity classes are selected from RSNA pneumonia detection challenge dataset, while the left two classes are built by integrating different data sources, and more details can be found in Rahman et al. (2021).
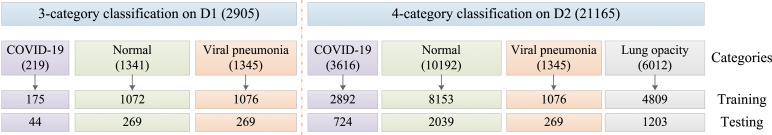
In addition, specific sample capacity of both D1 and D2 is shown in Fig. 4. Details of data partition for training and testing are also presented in Fig. 4, which is based on the ratio . Notice that regular hold-out validation is performed during the model training, where 10% of training samples are selected for independent validation. It is also noteworthy that basic data augmentation operations including flipping and cropping have been performed on the training images, which enlarge the sample capacity to some extend and hence benefit a more robust feature learning.
Fig. 4.
Data partition for training and testing.
4.1.2. Evaluation metrics
COVID-19 detection is essentially a classification problem, hence confusion matrix has been selected as the fundamental metric, which is illustrated in Fig. 5 where “0” and “1” are two labels.
Fig. 5.
Illustration of confusion matrix, where 1 and 0 stand for positive and negative, respectively.
Furthermore, based on the confusion matrix, following indicators have been adopted for evaluation and comparison, which are , , , and , respectively. The definitions of mentioned metrics are provided as follows:
| (9) |
| (10) |
| (11) |
| (12) |
| (13) |
For all the five indices above, the larger their values are, the better performance a model will have. In particular, false positive is a vital concept in clinic, which is also known as the misdiagnosis. Based on Eq. (10), the false positive rate can be obtained as . Accordingly, false negative rate (i.e., the missed diagnosis) is calculated as .
4.1.3. Experimental settings
Table 1 presents the specific experimental settings of training the proposed Cov-Net, other implementation details including structural configuration and model training have already been elaborated in Section 3, which will hence not be repeated here.
Table 1.
Experimental settings of proposed Cov-Net.
| Parameters | Settings |
|---|---|
| 0.0375, and decays in cosine manner | |
| 4 in training and 1 in testing stage | |
| 20 | |
| regularization and | |
| Stochastic gradient descent (SGD) with momentum |
Additionally, since it is the feature learning ability of Cov-Net that has been emphasized in this work, other six state-of-the-art computer vision models have been employed for comparison to estimate performance of Cov-Net from the perspective of methodology, which are denoted as ResNet50_vd (He et al.), MobileNetV2 (Sandler et al., 2018), DarkNet53 (Redmon & Farhadi, 2018), GhostNet (Han et al., 2020), InceptionV4 (Szegedy et al., 2017) and DenseNet161 (Huang et al., 2017), respectively. For the sake of fairness, above models are trained and tested on same data shown in Fig. 4, and all of them are parameterized according to corresponding literature. Moreover, all experiments are run under the paddlepaddle 1.8.0 framework with a NVIDIA Tesla V100 GPU (32 GB).
4.2. Experimental results and discussions
4.2.1. Classification results
At first, confusion matrix obtained by proposed Cov-Net on database D1 is provided in Fig. 6, where Fig. 6, Fig. 6 show the ratio and specific number respectively. Corresponding data have also been provided in Table 2 for a better inspection.
Fig. 6.
Confusion matrix of proposed Cov-Net on database D1, where class 0, 1, 2 stand for COVID-19, normal, viral pneumonia, respectively.
Table 2.
Confusion matrix of Cov-Net on database D1.
| Ground-truth | Prediction results |
||
|---|---|---|---|
| COVID-19 | Normal | Viral pneumonia | |
| COVID-19 | 43 | 0 | 1 |
| Normal | 0 | 269 | 0 |
| Viral pneumonia | 1 | 1 | 267 |
As shown in Fig. 6, the misclassification rates for samples of normal and viral pneumonia classes are 0% and 0.74% respectively, which implies that extracted features from these two classes of images already possess high discrimination. When it comes to COVID-19 samples, this value raises to 2.27%, however, only 1 sample is wrongly recognized as the viral pneumonia class, which further validates that it is the similarity between COVID-19 and viral pneumonia that makes the differentiation a tough work. In general, proposed Cov-Net has obtained satisfactory results on database D1.
In the same way, confusion matrix on D2 and corresponding data are shown in Fig. 7 and Table 3. Notice that classification task on database D2 has four categories, where the extra lung opacity category brings more difficulties in accurate recognition. As is shown, most misclassifications occur between lung opacity and normal category, which may call for a more targeted preprocessing, but the overall performance is still satisfactory. In particular, only 2 COVID-19 cases are wrongly regarded as viral pneumonia, and none of the latter is misclassified. As a result, one can still conclude that Cov-Net performs well on database D2 with total 20 misdiagnosis and 22 missed diagnosis in terms of COVID-19, which demonstrates that Cov-Net does have a remarkable feature learning ability.
Fig. 7.
Confusion matrix of proposed Cov-Net on database D2, where class 0, 1, 2, 3 stand for COVID-19, lung opacity, normal, viral pneumonia, respectively.
Table 3.
Confusion matrix of Cov-Net on database D2.
| Ground-truth | Prediction results |
|||
|---|---|---|---|---|
| COVID-19 | Lung opacity | Normal | Viral pneumonia | |
| COVID-19 | 702 | 12 | 8 | 2 |
| Lung opacity | 10 | 1045 | 148 | 0 |
| Normal | 10 | 96 | 1930 | 3 |
| Viral pneumonia | 0 | 0 | 8 | 261 |
In addition, loss function curve of proposed Cov-Net during the training on database D2 is presented in Fig. 8, from which one can see that the model has a well convergence trend.
Fig. 8.
Loss function curve of Cov-Net.
4.2.2. Comprehensive and in-depth discussions
On the basis of fusion matrix, evaluation metrics in Eqs. (9)–(13) are calculated, and notice that all of the metrics are presented in class-wise. Obtained results on database D1 are reported in Table 4, Table 5, respectively, and Fig. 9 has presented an intuitive illustration.
Table 4.
Class-wise performance of proposed Cov-Net on database D1.
| Metrics | COVID-19 |
Normal |
Viral pneumonia |
All |
||||
|---|---|---|---|---|---|---|---|---|
| Cov-Net | Cov-Net* | Cov-Net | Cov-Net* | Cov-Net | Cov-Net* | Cov-Net | Cov-Net* | |
| Sensitivity | 0.9772 | 1 | 1 | 0.9814 | 0.9926 | 0.9814 | 0.9948 | 0.9828 |
| Specificity | 0.9981 | 0.9926 | 0.9968 | 0.9872 | 0.9968 | 0.9936 | 0.9974 | 0.9914 |
| Precision | 0.9772 | 0.9167 | 0.9963 | 0.9851 | 0.9963 | 0.9925 | – | – |
| Accuracy | 0.9966 | 0.9931 | 0.9983 | 0.9845 | 0.9948 | 0.9880 | 0.9966 | 0.9885 |
| F1 Score | 0.9772 | 0.9565 | 0.9981 | 0.9832 | 0.9944 | 0.9869 | – | – |
Cov-Net* means there is no data augmentation applied.
Table 5.
Class-wise performance of proposed Cov-Net on database D2.
| Metrics | COVID-19 | Lung opacity | Normal | Viral pneumonia | All |
|---|---|---|---|---|---|
| Sensitivity | 0.9696 | 0.8687 | 0.9465 | 0.9703 | 0.9299 |
| Specificity | 0.9943 | 0.9644 | 0.9253 | 0.9987 | 0.9766 |
| Precision | 0.9723 | 0.9063 | 0.9217 | 0.9812 | – |
| Accuracy | 0.9901 | 0.9372 | 0.9355 | 0.9969 | 0.9649 |
| F1 score | 0.971 | 0.8871 | 0.9339 | 0.9757 | – |
Fig. 9.
Class-wise evaluations of proposed Cov-Net on two datasets.
It should be pointed out that the last column “All” in Table 4, Table 5 represents the global classification results without considering specific class, which can be regarded as an evaluation of the model performance. In this way, while calculating based on the confusion matrix, and will always be equal; consequently values of , and will be same. Hence for simplicity, when referring to overall performance of a model, only , and three indicators are reported in this work.
As can be seen, proposed Cov-Net provides an outstanding overall performance on both datasets, especially on the viral pneumonia and COVID-19 classes. To be specific, five metrics all exceed 0.99 in recognizing viral pneumonia on D1, and the worst score occurs in recognizing COVID-19 on D2 is the sensitivity of 0.9696, which implies only 3.04% false negative rate, hence it is still a remarkable achievement. Another noteworthy phenomenon is that sensitivity of normal class on D1 reaches 100%, which indicates that proposed Cov-Net generates no misclassification of normal cases on this database. However, as for database D2, it is found that almost all indicators have degradation to different extent, which implies that the extra category does bring more challenges in accurate diagnosis, and in future work, more robust preprocessing like Gamma correction (Rahman et al., 2021) and subtler feature fusion deserve more attentions. From the overall performance, Cov-Net still achieves accuracy of 96.49% on the much harder dataset D2, which demonstrates that Cov-Net is able to construct robust features for classification.
4.2.3. Influences of data augmentation
Another issue should be pointed out is that fundamental data augmentation process has been applied to Cov-Net during training, which is regarded especially helpful on database D1 due to the relatively limited samples (see Fig. 4) since deep learning model works mainly in data driven way. To find out whether this operation is helpful, comparison results are reported in Table 4 as well, where Cov-Net* stands for training the model on database D1 without further data augmentation.
Apparently, Cov-Net outperforms Cov-Net* regarding to all metrics of almost each category, which implies that diversifying training images can benefit learning more robust features so as to promote the final classification accuracy. In addition, it is found that after introducing data augmentation, the sensitivity value of COVID-19 class even declines from 1 to 0.9772. This phenomenon may be explained as data augmentation operations enlarge the number of training samples but not limited to the COVID-19 category, which is highly possible to exacerbate the data-imbalance problem and leads to a even worse performance.
Given that the calculation of sensitivity is , hence there is none of samples in Cov-Net* because sensitivity equals to 1. Therefore, the only way to reduce sensitivity value is to generate new samples. As aforementioned, it is the similarity between viral pneumonia and COVID-19 that makes them tough to differentiate, and as a result, one can infer that data augmentation has generated more viral pneumonia samples, which leads to over-fitting problem in Cov-Net and eventually, partial COVID-19 instances are recognized as viral pneumonia ones. This analysis indicates that above inference of intensifying data-imbalance is reasonable; and moreover, it also implies that handling the mentioned imbalanced data distribution in COVID-19 recognition will also be a promising work.
To sum up, according to Table 4, one can conclude that applying data augmentation is helpful, which increases sensitivity, specificity and accuracy by 1.2%, 0.6% and 0.81% on database D1, respectively. Although it only brings in a slight improvement, it suggests another beneficial work direction. To be specific, in addition to simply enlarging data capacity, one can also generate multi-modal samples to encourage building more robust and discriminative features, which will also be useful to deal with challenges in database D2, where multi-modal information can be considered to integrate characteristics of lung opacity and normal samples so as to facilitate more accurate recognition.
4.2.4. Comparisons with other state-of-the-art models in machine vision areas
In this part, six state-of-the-art models in machine vision areas have been trained and tested on the same database. It is remarkable that the major purpose is to validate the effectiveness and superiority of proposed Cov-Net from the perspective of methodology via comparing the experiment results under the same condition. For fairness, data partition and relevant experimental settings both maintain unchanged, and each comparison model is parameterized according to corresponding literature. In particular, comparison results on overall performance are presented in Fig. 10 and Table 6, whereas Fig. 11 and Table 7 have presented the comparison results in terms of the classification performance on COVID-19 category.
Fig. 10.
Comparisons on overall performance between proposed Cov-Net and other state-of-the-art models.
Table 6.
Comparison on overall performance among different computer vision models.
| Databases | Metrics | Models |
||||||
|---|---|---|---|---|---|---|---|---|
| ResNet50_vd | MobileNetV2 | DarkNet53 | GhostNet | InceptionV4 | DenseNet161 | Cov-Net | ||
| D1 | Specificity | 0.9872 | 0.9880 | 0.9828 | 0.9725 | 0.9725 | 0.9759 | 0.9974 |
| Sensitivity | 0.9656 | 0.9759 | 0.9656 | 0.9450 | 0.9450 | 0.9519 | 0.9948 | |
| Accuracy | 0.9771 | 0.9840 | 0.9771 | 0.9633 | 0.9633 | 0.9679 | 0.9966 | |
| D2 | Specificity | 0.9690 | 0.9601 | 0.9609 | 0.9485 | 0.9592 | 0.9665 | 0.9707 |
| Sensitivity | 0.9318 | 0.9109 | 0.9140 | 0.8734 | 0.8982 | 0.9215 | 0.9388 | |
| Accuracy | 0.9628 | 0.9529 | 0.9530 | 0.9380 | 0.9497 | 0.9594 | 0.9649 | |
Fig. 11.
Comparisons on classification performance of COVID-19 category between proposed Cov-Net and other state-of-the-art models.
Table 7.
Performance comparison on COVID-19 recognition among different computer vision models.
| Databases | Metrics | Models |
||||||
|---|---|---|---|---|---|---|---|---|
| ResNet50_vd | MobileNetV2 | DarkNet53 | GhostNet | InceptionV4 | DenseNet161 | Cov-Net | ||
| D1 | Sensitivity | 0.9318 | 0.9545 | 0.9318 | 0.9318 | 1 | 0.9545 | 0.9772 |
| Specificity | 0.9926 | 0.9888 | 0.9963 | 0.9888 | 0.9628 | 0.9926 | 0.9981 | |
| Precision | 0.9111 | 0.8750 | 0.9535 | 0.8723 | 0.6875 | 0.9130 | 0.9772 | |
| Accuracy | 0.9880 | 0.9863 | 0.9914 | 0.9845 | 0.9656 | 0.9897 | 0.9966 | |
| F1 score | 0.9213 | 0.9130 | 0.9425 | 0.9011 | 0.8148 | 0.9333 | 0.9772 | |
| D2 | Sensitivity | 0.9572 | 0.9448 | 0.9517 | 0.9213 | 0.8950 | 0.9613 | 0.9696 |
| Specificity | 0.9920 | 0.9886 | 0.9912 | 0.9826 | 0.9866 | 0.9912 | 0.9943 | |
| Precision | 0.9612 | 0.9448 | 0.9569 | 0.9162 | 0.9324 | 0.9574 | 0.9723 | |
| Accuracy | 0.9861 | 0.9811 | 0.9844 | 0.9721 | 0.9710 | 0.9861 | 0.9901 | |
| F1 score | 0.9592 | 0.9448 | 0.9543 | 0.9187 | 0.9133 | 0.9593 | 0.9710 | |
As illustrated in Fig. 10, proposed Cov-Net has overwhelmed the other six popular methods in terms of three metrics on both datasets. To be specific, proposed Cov-Net achieves the highest accuracy of 99.66% on D1, which exceeds the second one 98.40% by 1.26%; and moreover, sensitivity and specificity of Cov-Net also rank first on D1 as shown in Table 6, which surpasses the second one by 1.89% and 0.94% respectively. Whereas on database D2 where the task is more tough, above three metrics have all degraded to different extent. However, proposed Cov-Net still performs best out of the seven models. These results imply that the feature extractor along with the feature fusion module in Cov-Net does accomplish a comprehensive and sufficient feature learning so that constructed features are highly discriminative for accurate classification.
Furthermore, considering that the overall performance is regardless of specific class whereas the accurate COVID-19 recognition is of vital significance in practice, especially the discrimination between COVID-19 and viral pneumonia patients, it is necessary to figure out how proposed model performs on COVID-19 recognition.
According to Table 7, Cov-Net has obtained four out of five and all best results on database D1 and D2, respectively, which verifies the superiority and competitiveness of Cov-Net in feature extraction work. Additionally, it should be pointed out that it is generally hard to make the trade-off between precision and sensitivity so as to form a comprehensive evaluation, thus a harmonic average in Eq. (13) is usually adopted in practice, which attaches equivalent importance on both precision and sensitivity. On D1, in spite of that InceptionV4 has ranked first in terms of sensitivity with 1, its precision value is however only 0.6875. Consequently, InceptionV4 has obtained the of 0.8148, which is inferior to Cov-Net by 16.62%. While on the more tough four-category classification work, Cov-Net also achieves the best of 0.9710 and exceeds the second one (0.9593 by DenseNet161) by 0.0117, which validates the well generalization ability of proposed Cov-Net.
In addition, notice that Cov-Net possesses a specificity of 0.9981 on D1 and 0.9943 on D2, which are 0.18% and 0.23% over the corresponding second-ranked models (0.9963 by DarkNet53 on D1 and 0.9920 by ResNet50_vd on D2), respectively. Due to higher specificity corresponds to lower FPR, it is implied that proposed Cov-Net is competent in accurate diagnosing COVID-19. Moreover, FNR provided by Cov-Net are only 2.28% and 3.04% on D1 and D2, respectively. As a conclusion, being able to generate less missed diagnosis and misdiagnosis instances further demonstrates the practicality of proposed Cov-Net, which can provide accurate and reliable reference to the radiologists as a helpful CAD framework.
4.3. Limitations and future outlooks
Albeit proposed Cov-Net has presented satisfactory performance, there are spaces for further improvement. Consequently, this subsection has discussed existing limitations and provided outlooks for future enhancement.
4.3.1. Internal deficiencies
From the perspective of methodology, notice that the whole structure configuration of Cov-Net mainly relies on our previous studies, which is empirically determined and only a few parameters have been explored via trial and errors. Therefore in future work, applying swarm intelligence based heuristic algorithm to realize structure or parameter optimization of Cov-Net is a promising work, which can effectively alleviate adverse effect caused by subjectivity.
Furthermore, it seems that inadequate data processing procedure is implemented in the pipeline of Cov-Net, where only simple data augmentation is performed. However, whether there is redundant information has not been thoroughly inspected in both the feature leaning and fusion module. Consequently, in future work, pipeline of Cov-Net can be further enhanced by concatenating other data preprocessing or postprocessing module to facilitate generating more class-dependent robust features.
4.3.2. External limitations
In addition, it is remarkable that benchmark evaluations in this study mainly concentrate on validating whether proposed Cov-Net has the ability to build highly discriminative features for classification, and the major challenges in public datasets lie in the similarity among different classes. Therefore, obtained results can verify an outstanding classification capability but may not be sufficient to prove the practicality of Cov-Net in real-world application scenarios, and some related important caveats can refer to (Maguolo and Nanni, 2021, Tabik et al., 2020, Tartaglione et al., 2020).
Therefore, it is of vital importance to inspect and enhance the clinical validity of proposed Cov-Net in future work, where validation on some private databases of hospital may be helpful. Furthermore, it is better to attach hierarchical scores on applied data according to severity of COVID-19, and the database is also supposed to be representative enough to cover a variety of patient information such as gender and age, etc. In this way, one can further examine and accordingly refine Cov-Net so as to make it a competent, reliable and practical computer-aided diagnosis method.
5. Conclusion
In this paper, a novel computer aided diagnosis method Cov-Net is proposed for accurate recognition of COVID-19 from chest X-ray images, which can serve as a complementary diagnosis method to current advanced techniques such as RT-PCR so that higher diagnostic certainty can be achieved. In particular, proposed Cov-Net has taken full advantages of machine vision methods, which implements a modified residual network with asymmetric convolution and attention mechanism for feature extraction, and employs skip-connected dilated convolution with various dilation rates for feature fusion.
Experiments on two public COVID-19 radiography datasets have demonstrated the practicality and reliability of proposed Cov-Net in accurate COVID-19 recognition, which achieves overall accuracy of 0.9966 and 0.9649 on the three-category and four-category classification problems, respectively. Furthermore, Cov-Net has presented an overwhelming superiority in comparison to other six state-of-the-art models in machine vision areas, which further verifies that Cov-Net is competitive in building robust and highly discriminative features from the perspective of methodology. Therefore, one can infer that proposed Cov-Net can adapt to other task-specific scenarios via constructing high-level semantic features with strong presentation ability.
In future works, optimizing the architecture and integrating other data preprocessing process so as to improve performance of Cov-Net are both promising. More importantly, it is quite essential to apply Cov-Net into real-world applications, like training it on the private database of a hospital, so that one can thoroughly inspect and accordingly enhance its clinical validity. From the aspect of practicality, how to make Cov-Net a competent CAD framework deserves further in-depth considerations.
CRediT authorship contribution statement
Han Li: Conceptualization, Resources, Writing – original draft. Nianyin Zeng: Conceptualization, Methodology, Software, Validation, Investigation. Peishu Wu: Methodology, Software, Investigation, Writing – review & editing. Kathy Clawson: Validation, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
This work was supported in part by the Engineering and Physical Sciences Research Council (EPSRC) of the UK, in part by the Natural Science Foundation of China under Grant 62073271, in part by the UK-China Industry Academia Partnership Programme under Grant UK-CIAPP-276, in part by the Fundamental Research Funds for the Central Universities under Grant 20720190009.
Data availability
This study adopts publicly available databases, and their links are provided in our manuscript.
References
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albu A., Precup R., Teban T. Results and challenges of artificial neural networks used for decision-making and control in medical applications. Facta Universitatis-Series Mechanical Engineering. 2019;17(3):285–308. doi: 10.22190/FUME190327035A. [DOI] [Google Scholar]
- Bello, I., Zoph, B., Vasudevan, V., & Le, Q. (2017). Neural Optimizer Search with Reinforcement Learning. In D. Precup, & Y. Teh (Eds.), Proceedings of machine learning research: Vol. 70, International conference on machine learning. Sydney, Australia.
- Borlea I., Precup R., Borlea A., Iercan D. A unified form of fuzzy C-means and K-means algorithms and its partitional implementation. Knowledge-Based Systems. 2021;214 doi: 10.1016/j.knosys.2020.106731. [DOI] [Google Scholar]
- Bouchareb Y., Moradi Khaniabadi P., Al Kindi F., Al Dhuhli H., Shiri I., Zaidi H., Rahmim A. Artificial intelligence-driven assessment of radiological images for COVID-19. Computers in Biology and Medicine. 2021;136 doi: 10.1016/j.compbiomed.2021.104665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra T.B., Verma K., Singh B.K., Jain D., Netam S.S. Coronavirus disease (COVID-19) detection in chest X-Ray images using majority voting based classifier ensemble. Expert Systems with Applications. 2021;165 doi: 10.1016/j.eswa.2020.113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury M., Rahman T., Khandakar A., Mazhar R., Kadir M., Bin Mahbub Z., Islam K., Khan M., Iqbal A., Al Emadi N., Reaz M., Islam M. Can AI help in screening viral and COVID-19 pneumonia. IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- Ding, X., Guo, Y., Ding, G., & Han, J. (2019). ACNet: Strengthening the Kernel Skeletons for Powerful CNN via Asymmetric Convolution Blocks. In IEEE/CVF international conference on computer vision (ICCV), (pp. 1911–1920). Seoul, South Korea: 10.1109/ICCV.2019.00200. [DOI]
- Fan Y., Liu J., Yao R., Yuan X. Covid-19 detection from X-ray images using multi-kernel-size spatial-channel attention network. Pattern Recognition. 2021;119 doi: 10.1016/j.patcog.2021.108055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco R., Grassi R., Granata V., Setola S.V., Grassi F., Cozzi D., Pecori B., Izzo F., Petrillo A. Artificial intelligence and COVID-19 using chest CT scan and chest X-ray images: Machine learning and deep learning approaches for diagnosis and treatment. Journal of Personalized Medicine. 2021;11(10) doi: 10.3390/jpm11100993. URL https://www.mdpi.com/2075-4426/11/10/993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Santa Cruz B., Bossa M.N., Sölter J., Husch A.D. Public Covid-19 X-ray datasets and their impact on model bias – a systematic review of a significant problem. Medical Image Analysis. 2021;74 doi: 10.1016/j.media.2021.102225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, K., Wang, Y., Tian, Q., Guo, J., Xu, C., & Xu, C. (2020). GhostNet: More Features from Cheap Operations. In IEEE/CVF conference on computer vision and pattern recognition (CVPR), (pp. 1577–1586). Electr Network: 10.1109/CVPR42600.2020.00165. [DOI]
- He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep Residual Learning for Image Recognition. In IEEE conference on computer vision and pattern recognition (CVPR), (pp. 770–778). Las Vegas, USA: 10.1109/CVPR.2016.90. [DOI]
- He, T., Zhang, Z., Zhang, H., Zhang, Z., Xie, J., & Li, M. Bag of Tricks for Image Classification with Convolutional Neural Networks. In 2019 IEEE/CVF conference on computer vision and pattern recognition (CVPR) (pp. 558–567). 10.1109/CVPR.2019.00065. [DOI]
- Hsiu-Sen C., Dong-Her S., Binshan L., Ming-Hung S. An APN model for arrhythmic beat classification. Bioinformatics. 2014;30:1739–1746. doi: 10.1093/bioinformatics/btu101. [DOI] [PubMed] [Google Scholar]
- Hu T., Khishe M., Mohammadi M., Parvizi G.-R., Taher Karim S.H., Rashid T.A. Real-time COVID-19 diagnosis from X-Ray images using deep CNN and extreme learning machines stabilized by chimp optimization algorithm. Biomedical Signal Processing and Control. 2021;68 doi: 10.1016/j.bspc.2021.102764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Shen L., Albanie S., Sun G., Wu E. 2017. Squeeze-and-excitation networks. arXiv:1709.01507v4. [DOI] [PubMed] [Google Scholar]
- Huang, G., Liu, Z., Maaten, L., & Weinberger, K. (2017). Densely Connected Convolutional Networks. In IEEE conference on computer vision and pattern recognition (CVPR), (pp. 2261–2269). Honolulu, USA: 10.1109/CVPR.2017.243. [DOI]
- Ismael A.M., Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Systems with Applications. 2021;164 doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaderberg M., Vedaldi A., Zisserman A. 2014. Speeding up convolutional neural networks with low rank expansions. arXiv:1405.3866v1. [Google Scholar]
- Jin J., Dundar A., Culurciello E. 2014. Flattened convolutional neural networks for feedforward acceleration. arXiv:1412.5474v1. [Google Scholar]
- Kassani S., Kassani P., Wesolowski M., Schneider K., Deters R. Automatic detection of coronavirus disease (COVID-19) in X-ray and CT images: a machine learning based approach. Biocybernetics and Biomedical Engineering. 2021;41:867–879. doi: 10.1016/j.bbe.2021.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keidar D., Yaron D., Goldstein E., Shachar Y., Blass A., Charbinsky L., Aharony I., Lifshitz L., Lumelsky D., Neeman Z., Mizrachi M., Hajouj M., Eizenbach N., Sela E., Weiss C., Levin P., Benjaminov O., Bachar G., Tamir S.…Eldar Y. COVID-19 classification of X-ray images using deep neural networks. European Radiology. 2021;31:9654–9663. doi: 10.1007/s00330-021-08050-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascu M. Deep learning in classification of Covid-19 coronavirus, pneumonia and healthy lungs on CXR and CT images. Journal of Medical and Biological Engineering. 2021;41:514–522. doi: 10.1007/s40846-021-00630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong L.-Y., Ooi K.-B., Chong A.Y.-L., Lin B. Modeling the stimulators of the behavioral intention to use mobile entertainment: Does gender really matter. Computers in Human Behavior. 2013;29(5):2109–2121. doi: 10.1016/j.chb.2013.04.004. [DOI] [Google Scholar]
- Ma X., Zheng B., Zhu Y., Yu F., Zhang R., Chen B. Covid-19 lesion discrimination and localization network based on multi-receptive field attention module on CT images. Optik. 2021;241 doi: 10.1016/j.ijleo.2021.167100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguolo G., Nanni L. A critic evaluation of methods for COVID-19 automatic detection from X-ray images. Information Fusion. 2021;76:1–7. doi: 10.1016/j.inffus.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minaee S., Kafieh R., Sonka M., Yazdani S., Jamalipour Soufi G. Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Medical Image Analysis. 2020;65 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabizadeh-Shahre-Babak Z., Karimi N., Khadivi P., Roshandel R., Emami A., Samavi S. Detection of COVID-19 in X-ray images by classification of bag of visual words using neural networks. Biomedical Signal Processing and Control. 2021;68 doi: 10.1016/j.bspc.2021.102750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman T., Khandakar A., Qiblawey Y., Tahir A., Kiranyaz S., Abul Kashem S.B., Islam M.T., Al Maadeed S., Zughaier S.M., Khan M.S., Chowdhury M.E. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Computers in Biology and Medicine. 2021;132 doi: 10.1016/j.compbiomed.2021.104319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmon J., Farhadi A. 2018. YOLOV3: an incremental improvement. arXiv:1804.02767v1. [Google Scholar]
- Roberts M., Driggs D., Thorpe M. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nature Machine Intelligence. 2021;3:199–217. doi: 10.1038/s42256-021-00307-0. [DOI] [Google Scholar]
- Sait U., K.V. G.L., Shivakumar S., Kumar T., Bhaumik R., Prajapati S., Bhalla K., Chakrapani A. A deep-learning based multimodal system for Covid-19 diagnosis using breathing sounds and chest X-ray images. Applied Soft Computing. 2021;109 doi: 10.1016/j.asoc.2021.107522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler, M., Howard, A., Zhu, M., Zhmoginov, A. Chen, L. (2018). MobileNetV2: inverted residuals and linear bottlenecks. In IEEE/CVF conference on computer vision and pattern recognition (CVPR), (pp. 4510–4520). Salt Lake City, USA: 10.1109/CVPR.2018.00474. [DOI]
- Shaban W.M., Rabie A.H., Saleh A.I., Abo-Elsoud M. Accurate detection of COVID-19 patients based on distance biased naïve Bayes (DBNB) classification strategy. Pattern Recognition. 2021;119 doi: 10.1016/j.patcog.2021.108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheykhivand S., Mousavi Z., Mojtahedi S., Yousefi Rezaii T., Farzamnia A., Meshgini S., Saad I. Developing an efficient deep neural network for automatic detection of COVID-19 using chest X-ray images. Alexandria Engineering Journal. 2021;60(3):2885–2903. doi: 10.1016/j.aej.2021.01.011. [DOI] [Google Scholar]
- Signoroni A., Savardi M., Benini S., Adami N., Leonardi R., Gibellini P., Vaccher F., Ravanelli M., Borghesi A., Maroldi R., Farina D. Bs-net: Learning COVID-19 pneumonia severity on a large chest X-ray dataset. Medical Image Analysis. 2021;71 doi: 10.1016/j.media.2021.102046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegedy, C., Ioffe, S., ; Vanhoucke, V., & Alemi, A. (2017). Inception-v4 inception-resnet and the impact of residual connections on learning. In 31st AAAI conference on artificial intelligence, (pp. 4278–4284). San Francisco, CA.
- Tabik S., Gomez-Rios A., Martin-Rodriguez J., Sevillano-Garcia I., Rey-Area M., Charte D., Guirado E., Suarez J., Luengo J., Valero-Gonzalez M., Garcia-Villanova P., Olmedo-Sanchez E., Herrera F. COVIDGR dataset and COVID-sdnet methodology for predicting COVID-19 based on chest X-ray images. COVIDGR Dataset and COVID-SDNet Methodology for Predicting COVID-19 Based on Chest X-Ray Images. 2020;24:3595–3605. doi: 10.1109/JBHI.2020.3037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglione E., Barbano C.A., Berzovini C., Calandri M., Grangetto M. Unveiling COVID-19 from CHEST X-Ray with deep learning: A hurdles race with small data. International Journal of Environmental Research and Public Health. 2020;17(18) doi: 10.3390/ijerph17186933. URL https://www.mdpi.com/1660-4601/17/18/6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay P., Nagpal C. Wavelet based performance analysis of SVM and RBF kernel for classifying stress conditions of sleep EEG. Science and Technology. 2020;23:292–310. [Google Scholar]
- Vianello C., Strozzi F., Mocellin P., Cimetta E., Fabiano B., Manenti F., Pozzi R., Maschio G. A perspective on early detection systems models for COVID-19 spreading. Biochemical and Biophysical Research Communications. 2021;538:244–252. doi: 10.1016/j.bbrc.2020.12.010. COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Li H., Zeng N., Li F. FMD-Yolo: An efficient face mask detection method for COVID-19 prevention and control in public. Image and Vision Computing. 2022;117 doi: 10.1016/j.imavis.2021.104341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Lam H.-K., Jia G. MANet: A two-stage deep learning method for classification of COVID-19 from chest X-ray images. Neurocomputing. 2021;443:96–105. doi: 10.1016/j.neucom.2021.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Koltun V. 2016. Multi-scale context aggregation by dilated convolutions. arXiv:1511.07122v3. [Google Scholar]
- Yu X., Lu S., Guo L., Wang S.-H., Zhang Y.-D. ResGNet-C: A graph convolutional neural network for detection of COVID-19. Neurocomputing. 2021;452:592–605. doi: 10.1016/j.neucom.2020.07.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng N., Li H., Peng Y. A new deep belief network-based multi-task learning for diagnosis of Alzheimer’s disease. Neural Computing and Applications. 2021 doi: 10.1007/s00521-021-06149-6. [DOI] [Google Scholar]
- Zeng N., Li H., Wang Z., Liu W., Liu S., Alsaadi F.E., Liu X. Deep-reinforcement-learning-based images segmentation for quantitative analysis of gold immunochromatographic strip. Neurocomputing. 2021;425:173–180. doi: 10.1016/j.neucom.2020.04.001. [DOI] [Google Scholar]
- Zeng N., Wu P., Wang Z., Li H., Liu W., Liu X. A small-sized object detection oriented multi-scale feature fusion approach with application to defect detection. IEEE Transactions on Instrumentation and Measurement. 2022;71:1–14. doi: 10.1109/TIM.2022.3153997. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study adopts publicly available databases, and their links are provided in our manuscript.