Abstract
Snail is implicated in tumour growth and metastasis and is up-regulated in various human tumours. Although the role of Snails in epithelial-mesenchymal transition, which is particularly important in cancer metastasis, is well known, how they regulate tumour growth is poorly described. In this study, the possible molecular mechanisms of Snail in tumour growth were explored. Baculoviral inhibitor of apoptosis protein (IAP) repeat-containing protein 3 (BIRC3), a co-activator of cell proliferation during tumourigenesis, was identified as a Snail-binding protein via a yeast two-hybrid system. Since BIRC3 is important for cell survival, the effect of BIRC3 binding partner Snail on cell survival was investigated in ovarian cancer cell lines. Results revealed that Bax expression was activated, while the expression levels of anti-apoptotic proteins were markedly decreased by small interfering RNA (siRNA) specific for Snail (siSnail). siSnail, the binding partner of siBIRC3, activated the tumour suppressor function of p53 by promoting p53 protein stability. Conversely, BIRC3 could interact with Snail, for this reason, the possibility of BIRC3 involvement in EMT was investigated. BIRC3 overexpression resulted in a decreased expression of the epithelial marker and an increased expression of the mesenchymal markers. siSnail or siBIRC3 reduced the mRNA levels of matrix metalloproteinase (MMP)-2 and MMP-9. These results provide evidence that Snail promotes cell proliferation by interacting with BIRC3 and that BIRC3 might be involved in EMT via binding to Snail in ovarian cancer cells. Therefore, our results suggested the novel relevance of BIRC3, the binding partner of Snail, in ovarian cancer development.
Keywords: Snail, BIRC3, Yeast two-hybrid, P53, Proliferation, Ovarian cancer
INTRODUCTION
Ovarian tumours are the leading cause of gynecologic cancer deaths in Western countries. Their risk factors include fertility, obesity, hormone replacement therapy after menopause, and smoking (Clarfield et al., 2022). In 2020, ovarian cancer was diagnosed in 239,000 females and resulted in 207,252 deaths (Elshami et al., 2022).
Epithelial-mesenchymal transition (EMT) is a pivotal mechanism in the initial steps of ovarian cancer metastasis and invasion (Lee, 2018, 2019; Cai et al., 2021). Kurrey et al. (2005) reported that over-expression of Snail or Slug results in EMT, which promoted cell motility, invasiveness and tumorigenicity in SKOV3 cells. Yuan et al. (2013) also reported that ALX1, the homeobox transcription factor ALX1 provokes Snail expression to promote EMT of ovarian cancer cells. In addition, Blechschmidt et al. (2008) demonstrated that Snail, an E-cadherin repressor, is related to the reduced overall survival of patients with ovarian tumours. Snail also plays an important role in ovarian tumour growth and metastasis via the regulation of MMP activity (Jin et al., 2010).
Snail is a transcription factor that participates in EMT (Nam et al., 2022). The Snail family of zinc-finger transcription factors includes Snail1 (Snail), Snail2 (Slug), and Snail3 (Smuc) in vertebrates (Debnath et al., 2022). Snail1 (Snail) and Snail2 (Slug) are necessary to trigger EMT during embryonic development and cancer progression (Nieto, 2002; Peiro et al., 2006; Wu and Zhou, 2010; Debnath et al., 2022). The C-terminal region of Snail1 binds to DNA and the N-terminal region represses transcription (Peinado et al., 2004). In the central domain, a serine-rich sequence (SRS) and a nuclear export sequence (NES) control Snail subcellular localization and protein stability, respectively (Domínguez et al., 2003). Although the biological functions of Snail in malignancies have been characterized extensively, its molecular mechanisms are unclear.
In this study, we used a yeast two-hybrid system to screen a human cDNA library for novel Snail-interacting proteins and characterize the biological functions of snails in ovarian cancer. We also reported novel insights into the roles of the Snails and its new binding partner in modulating cancer cell proliferation.
MATERIALS AND METHODS
Cell lines, culture, and antibodies
Human carcinoma cell lines (SKOV-3, HeLa, HCT116) were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). Cells were seeded in Roswell Park Memorial Institute (RPMI) 1640 medium, supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin (100 units/mL and 100 µg/mL, respectively) in 5% CO2 at 37°C. Other chemicals were obtained from Sigma (St. Louis, MO, USA). The following primary antibodies were used in this study: anti-Snail, anti-BIRC3, anti-Bax, anti-Bcl-xL, anti-Mcl-1, anti-Bcl-2, anti-p53, anti-α-tubulin, N-cadherin, anti-activated Ras-related C3 botulinum toxin substrate 1 (Rac1), anti-Rac1 (Cell Signaling, Beverly, MA, USA), anti-green fluorescent protein (GFP), anti-p53 up-regulated modulator of apoptosis (PUMA), anti-E-cadherin, anti-β-catenin, anti-vimentin, anti-fibronectin, anti-activated-Rho, anti-Rho, anti-activated cell division control protein 42 homolog (Cdc42), anti-Cdc42, and anti-β-actin (Sigma).
Cell viability analysis
Cell viability was evaluated using 3-(4,5-dimethylthiazol-2-yl)-2.5-diphenyl-2H-tetrazolium bromide (MTT) assays (Rho et al., 2021a). Cells were grown at a density of 4.3×103 per well in 96-well plates. Three days after transfection, a fresh medium including 10% FBS and 20-µL MTT solution (5 µg/mL; Sigma) were added to each well, followed by incubation for an additional 4 h at 37°C. After centrifugation at 500 g for 10 min, the supernatant was decanted and the formazan was dissolved with dimethyl sulfoxide (DMSO). The amounts of MTT-formazan generated were determined by measuring the absorbance at 540 nm using a microplate reader.
RNA isolation and reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated from cells using TRIzol® (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols (Kim et al., 2021). Total RNA (1 µg) was reverse transcribed to cDNA using a First-Strand cDNA Synthesis Kit with oligo-dT primers (SuperScript™ RT, Invitrogen). To detect human Snail, BIRC3, MMP-2, MMP-9, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), mRNA transcripts were isolated by PCR as described above. The primers were as follows: Snail, 5’-TTC TTC GCT ACT GCT GCG-3’ (forward) and 5’-GGG CAG GTA TGG AGA GGA AGA-3’ (reverse); BIRC3, 5’-AGG TGT TGG GAA TCT GGA GAT-3’ (forward) and 5’- GCA GCA TTA ATC ACA GGA GTA-3’ (reverse); MMP-2, 5’-ATG GGG AAT CGG TTG AAG G-3’ (forward) and 5’-AAT TGC ATT TCC TGA CAG AAG G-3’ (reverse); MMP-9, 5’-ACG CAG ACA TCG TCA TCC AGT-3’ (forward) and 5’-GGA CCA CAA CTC GTC ATC GTC-3’ (reverse); and GAPDH, 5’-GAG TCA ACG GAT TTG GTC GTA-3’ (forward) and 5’-CTT CTC CAT GGT TGG TGA AGA-3’ (reverse). The specificity of the amplified PCR fragments was confirmed by sequencing.
Silencing of gene expression by small interfering RNA (siRNA)
Oligonucleotides including an RNA interference (RNAi) sequence [Snail: 5’- GCC UUC AAC UGC AAA UAC U-3’ (sense) and 5’-AGU AUU UGC AGU UGA AGG C-3’ (antisense); BIRC3, 5’-UAA GGG AAG AGG AGA GAG AA-3’ (sense) and 5’-UUC UCU CUC CUC UUC CCU UA-3’ (antisense)] were synthesized using an RNAi construction kit (Ambion, Austin, TX, USA), and were then transfected with Lipofectamine® 2000 reagent (Invitrogen) according to the manufacturer’s protocols (Rho et al., 2021b). The following unrelated non-specific scrambled oligonucleotide was used as a control: [Snail, 5’-TAT TGC CTA GCA TTA CGT T-3’ (sense) and 5’-GTA TTG CCT AGC ATT ACG T-3’ (antisense); BIRC3, 5’-CCT GGT AGC AGC GAG TGA G-3’ (sense) and 5’-CTC ACT CGC TGC TAC CAG G-3’ (antisense)]. Cells grown for 24 h were transfected with 150 nM siSnail or siBIRC3. At 24 h after transfection, cells were prepared for MTT assay and immunoblotting analysis.
Western blot analysis
Western blot assay was performed as previously described (Yu et al., 2021a). Cells were collected, rinsed twice with cold phosphate-buffered saline (PBS), centrifuged, and lysed in cold lysis buffer containing a protease inhibitor cocktail (Sigma) at 4°C for 50 min. The cell lysates were subsequently separated with 8%-12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto Hybond enhanced chemiluminescence (ECL) nitrocellulose membrane (GE Healthcare, Little Chalfont, Buckinghamshire, UK). After blocking, the membranes were incubated with primary antibodies at 4°C overnight. The membranes were rinsed thrice with Tris-buffered saline and Tween 20 (TBST) wash buffer and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling). Immunoreactive bands were developed using an ECL detection system (GE Healthcare).
Yeast two-hybrid assay system
The full-length cDNA of the human Snail gene was subcloned into the EcoRI and XhoI restriction enzyme sites of the pGilda vector. A full-length human BIRC3 cDNA was introduced into a pJG4-5 activation vector at the EcoRI and XhoI sites to generate B42 fusion proteins. Positive interactions were indicated by the formation of blue colonies on an X-gal-containing medium, as described previously (Kim et al., 2021). Interaction binding activity was compared by calculating the relative expression level of o-nitrophenyl β-D-galactopyranoside (ONPG) β-galactosidase (Rho et al., 1999; Dong et al., 2012).
Promoter-reporter gene analysis
In vitro promoter luciferase activity was assayed as described previously (Rho et al., 2020). In brief, cells were transfected with vector DNA containing Bcl-2-, p21-, or p53-luciferase, in which luciferase is expressed under the control of the respective promoters. After reaching 85% confluency, cells were transfected with the indicated reporter constructs. As an internal control to correct for variations in transfection efficiency, 20 ng of pRL-TK (Promega, Madison, WI, USA) were co-transfected. The transfections were accomplished using Oligofectamine™ (Invitrogen). After lysis with Reporter Lysis Buffer (Promega), the cell extracts were incubated with the luciferase substrate at room temperature for 30 min. Then, a 5-µL aliquot of each sample was transferred into a MicroLumat Plus LB96 V luminometer (Berthold Technologies, Bad Wildbad, Germany). Reporter activity was measured using a Dual-Luciferase® Reporter (DLR™) Assay System (Promega) according to the manufacturer’s directions, and reporter activity was normalized to that of Renilla luciferase to correct for variations in the transfection efficiency.
Statistical analysis
Results are presented as means ± standard deviation (SD) of four independent experiments. Student’s t-test was used for comparisons between the two groups. Statistically significant differences (p<0.05) are indicated by asterisks (*).
RESULTS
Snail directly binds with BIRC3
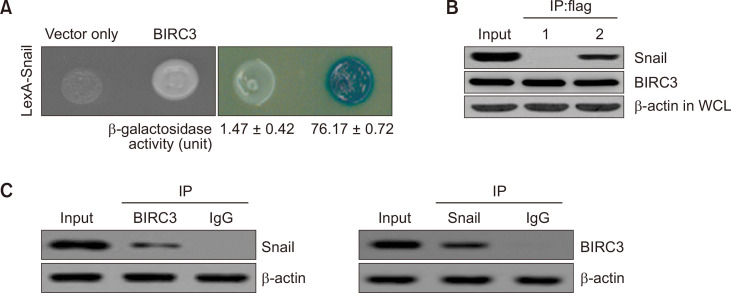
A yeast two-hybrid method was used to determine whether BIRC3 is a binding partner of Snail. In Fig. 1A, a positive binding activity was identified in the Snail/BIRC3-binding assay system (76.17 ± 0.72), whereas a low β-galactosidase binding activity was detected in the Snail/empty vector expression system (1.47 ± 0.42). Then, Snail and BIRC3 were co-immunoprecipitated (Co-IP) to confirm the direct interaction between Snail and BIRC3 observed in cells. The cDNA constructs of BIRC3 (pcDNA3.1/Flag-BIRC3) and Snail (pcDNA3.1-Snail), or pcDNA3.1/Flag-BIRC3 and empty vector only (pcDNA3.1), were co-transfected into cells. Snail precipitated with BIRC3, whereas Co-IP failed to detect any protein-protein interaction between pcDNA3.1 (empty vector-only) or pcDNA3.1 with Flag-BIRC3 (Fig. 1B). Subsequently, the cellular interactions of the two proteins were validated by the Co-IP of endogenous Snail and BIRC3. Subsequently, endogenously expressed Snail protein directly co-immunoprecipitated with BIRC3 (Fig. 1C).
Fig. 1.
Physical interaction between Snail and the Baculoviral inhibitor of apoptosis protein (IAP) repeat-containing protein 3 (BIRC3). (A) Positive interactions were observed by monitoring cell growth over 3 days at 30°C on a medium lacking leucine. The values of β-galactosidase activity (unit) calculated by adding O-nitrophenyl β-D-galactopyranoside (ONPG) are indicated below the corresponding lanes. (B) Co-immunoprecipitation (Co-IP) of Snail with BIRC3. Immunoprecipitation was carried out using an anti-Flag antibody with lysates from transfected breast carcinoma cells. After immunoprecipitation, precipitated proteins were immunoblotted with anti-Snail and anti-BIRC3 antibodies: lane 1, pcDNA3.1 (vector only) and pcDNA3.1/Flag-BIRC3 transfectant; lane 2, pcDNA3.1-Snail and pcDNA3.1/Flag-BIRC3 transfectant. (C) Co-IP of endogenous BIRC3 and Snail shows the interaction between the two proteins in HEK 293T cells. β-actin was used as a loading control.
siSnail and siBIRC3 induce apoptotic death of carcinoma cells
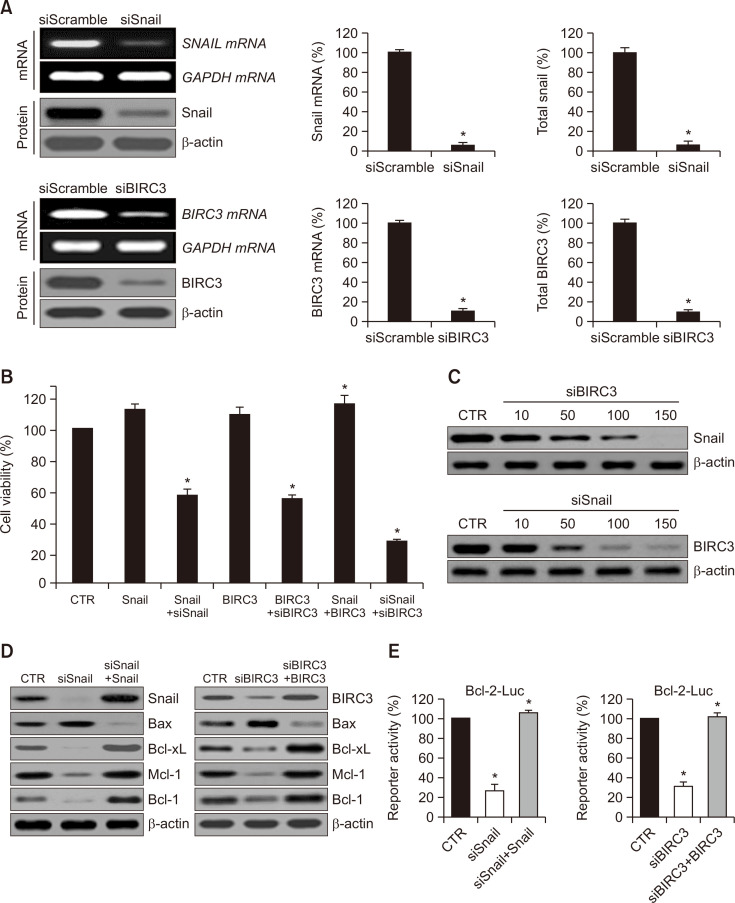
The effect of Snail and BIRC3 on ovarian tumour cell proliferation was examined. Snail and BIRC3 were highly expressed at mRNA and protein levels. Snail expression was inhibited dramatically after Snail siRNA transfection (Fig. 2A), and cellular proliferation was reduced significantly (Fig. 2B). BIRC3 overexpression promoted cell growth, whereas cell growth was suppressed by BIRC3 siRNA. The proliferation of cells transfected with siSnail were inhibited to ~40% compared with that of the cells transfected with the control (expression vector only). siBIRC3 transfection reduced cell viability by 42%, and the transfection of siSnail plus siBIRC3 additively inhibited cellular viability. Therefore, Snail directly interacts with BIRC3 under biological cellular conditions (Fig. 2B).
Fig. 2.
Effects of snail siRNA and BIRC3 siRNA on the proliferation of cancer cells and effects of BIRC3 siRNA on snail expression. (A) Reverse transcription-polymerase chain reaction (RT-PCR) showing decreased Snail and BIRC3 mRNA levels following transfection with plasmid constructs targeting Snail and BIRC3. Representative Western blot of Snail and BIRC3 proteins in control (siScramble) and siSnail and siBIRC3. Quantification of RT-PCR and Western blot data (right panel). Results represent the means ± SD of at least three independent experiments performed in triplicate. *p<0.05 compared to the siScramble control group. (B) Cells were transfected with control, Snail, Snail plus siSnail, BIRC3, BIRC3 plus siBIRC3, Snail plus BIRC3, and siSnail plus siBIRC3 plasmid, respectively. Relative cell proliferation rates were calculated by MTT assay. MTT-formazan amounts were calculated by measuring the absorbance at 540 nm in a microplate reader, and absorbance values were converted to relative proliferation rates. Each data point represents triplicate samples, and the bars indicate the means ± standard deviation (SD). *p<0.05. (C) Cells were transfected with various concentrations of siBIRC3 or siSnail, and then total cell lysates were prepared. Snail and BIRC3 proteins in cells were visualized using immunoblotting. (D) Protein levels of B-cell lymphoma (Bcl-2) family genes were analyzed by immunoblot analysis of the SKOV-3 cells transfected with siSnail and siSnail plus Snail or siBIRC3 and siBIRC3 plus BIRC3. β-actin was used as a loading control and was immunoblotted with antibodies to Snail, BIRC3, Bax, Bcl-xL, Mcl-1, and Bcl-2. (E) Promoter activities of Bcl-2 were measured through a luciferase reporter gene assay system with Bcl-2 (Bcl-2-Luc) promoter reporters, respectively. For instance, the Bcl-2-luciferase reporter gene constructs or promoter-less plasmid vectors only were transiently transfected into SKOV-3 cells. Cells were incubated for 24 h and then resuspended in lysis buffer. After collection by centrifugation, cells were lysed, mixed with the luciferase reaction substrate, and then assayed for luciferase activity. Results represent the means ± SD of at least three independent experiments performed in triplicate. *p<0.05. The experiments were repeated three times with similar results. IP, immunoprecipitation; WB, Western blot; WCL, whole-cell lysates.
To explore the biological function of siBIRC3 during Snail-induced cell proliferation, we investigated Snail protein expression through transient siBIRC3 transfection. As presented in Fig. 2C, siBIRC3 transfection reduced Snail protein levels in a concentration-dependent manner (upper panel). In the case of siSnail transfection, BIRC3 protein level patterns were similar to those of the Snail protein (lower panel). Subsequently, we assessed Snail- and BIRC3-induced cell proliferation by determining the activation levels of apoptosis-associated proteins, such as Bax, B-cell lymphoma-extra large (Bcl-xL), myeloid cell leukaemia 1 (Mcl-1), and B-cell lymphoma 2 (Bcl-2), which are well-known pivotal modulators of cell death and proliferation. Fig. 2D shows that the Bax expression in the transfections was reduced compared with that in the CTR (control), whereas Bcl-xL, Mcl-1, and Bcl-2 were activated by Snail and BIRC3. We transfected the cells with specific RNAi (siSnail and siBIRC3) to verify these results. Bax expression was activated, while the expression levels of Bcl-xL, Mcl-1, and Bcl-2 were decreased markedly by siSnail and siBIRC3. To assess the biological effects of siSnail on Snail-stimulated proliferation, we transfected the cells with siSnail and Snail, or with siBIRC3 and BIRC3 expression plasmids. We then measured the promoter activity by using a dual-luciferase reporter gene assay system. Fig. 2E illustrates that the Bcl-2 activity was noticeably reduced by siSnail and siBIRC3. Therefore, Snail and BIRC3 knockdown effectively inhibited the established tumour growth in vitro by inducing apoptosis.
Effects of Snail and BIRC3 knockdown by siRNA on p53
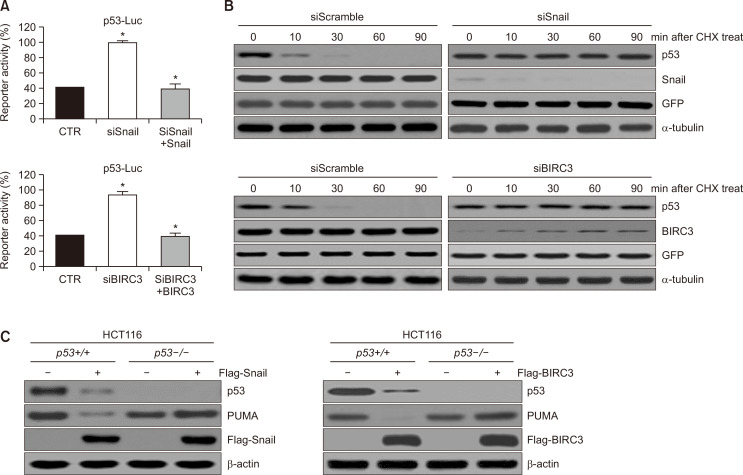
We additional conformed that p53 promoter activity was inhibited by siSnail and siBIRC3 by dual-luciferase reporter gene assay (Fig. 3A). To better understand the effects of Snail and BIRC3 on p53, cells were transfected with siSnail and Snail or with siBIRC3 and BIRC3 for increasing periods after cycloheximide (CHX) treatment. Interestingly, endogenous p53 levels were significantly reduced by CHX treatment in a time-dependent manner (Fig. 3B, left panel). Conversely, Snail and BIRC3 knockdown stabilized the p53 protein level (Fig. 3B, right panel). These results suggested that siSnail and siBIRC3 activated the p53 tumour-suppressor function by promoting p53 protein stability. A Flag-tagged Snail and BIRC3 plasmid were introduced into HCT116 p53+/+ and p53–/– cells to assess whether the increase in PUMA was attributed to the direct regulation of p53 by Snail and BIRC3. PUMA expression was reduced in p53+/+ cells but not in p53–/– cells (Fig. 3C), indicating that PUMA regulation by Snail and BIRC3 was dependent on p53.
Fig. 3.
Snail and BIRC3 reduce the promoter activity of p53 and decrease the half-life of the p53 protein. (A) Promoter activities of p53 were measured through a luciferase reporter gene assay system with a p53 (p53-Luc) promoter-reporter. For instance, the p53 luciferase reporter gene constructs or promoter-less plasmid vectors only were transiently transfected into SKOV-3 cells. Cells were incubated for 24 h and then resuspended in lysis buffer. After collection by centrifugation, cells were lysed, mixed with the luciferase reaction substrate, and then assayed for luciferase activity. Results represent the means ± SD of at least three independent experiments performed in triplicate. *p<0.05. (B) p53 protein stability was evaluated in HeLa cells transfected with siScramble (control), siSnail, or siBIRC3. Twenty-four hours after transfection, cells were treated with cycloheximide (50 mg/mL) for the indicated periods. Cell lysates were developed by immunoblotting with an anti-p53 antibody. α-tubulin served as a loading control. (C) One microgram of Flag-Snail and Flag-control (plasmid vector-only) or Flag-BIRC3 and Flag-control was transfected into p53+/+ or p53–/– HCT116 cells. At 48 h post-transfection, cell lysates were visualized by immunoblotting with the indicated specific antibodies.
Involvement of BIRC3 in the EMT
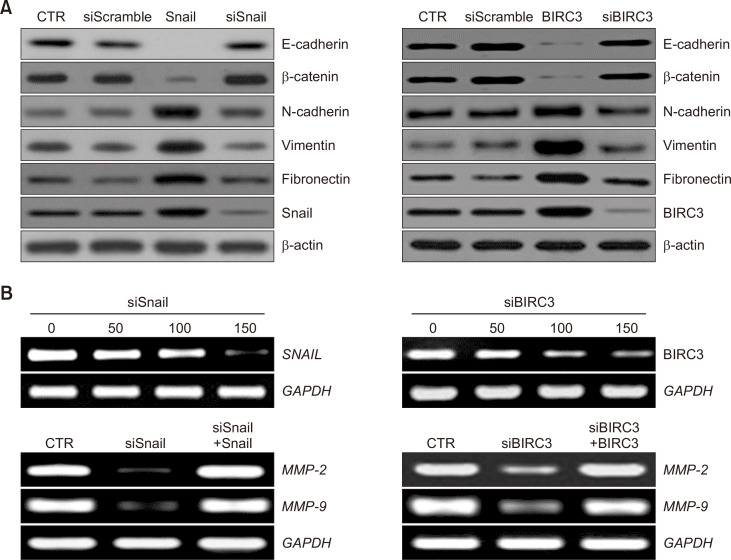
Since Snail is a transcription factor involved in EMT, the possibility of the EMT involvement of BIRC3 binding to Snail was investigated. E-cadherin expression was decreased when BIRC3 was overexpressed (Fig. 4A, right panel). This reduction was similar to that when Snail was overexpressed (Fig. 4A, left panel). Conversely, the expression of mesenchymal proteins, particularly N-cadherin, vimentin, and fibronectin, was significantly increased in Snail- and BIRC3-expressing cells compared with that in control-expressing cells (Fig. 4A). Conversely, when gene silencing was performed with siBRC3, the E-cadherin expression was restored, and the N-cadherin and vimentin expression levels were decreased (Fig. 4A, right panel). This recovery was similar to that of Snail gene silencing (Fig. 4A, left panel).
Fig. 4.
Changes in epithelial–mesenchymal transition (EMT) markers, regulatory factors and MMP2 and MMP9 following ovarian tumor cell transfection with Snail and BIRC3. (A) Expression of epithelial markers (E-cadherin and β-catenin) and mesenchymal markers (N-cadherin, vimentin, and fibronectin) in tumour cells with or without Snail and BIRC3 transfection were investigated by immunoblotting. β-actin was used as a loading control. (B) Snail and BIRC3 mRNA expression following transfection with various concentrations of siSnail and siBIRC3, respectively (upper panel). MMP-2 and MMP-9 mRNA expression was evaluated using RT-PCR (lower panel).
MMPs play a pivotal role in invasion (Nagase and Woessner, 1999) and are frequently up-regulated in EMT (Scheau et al., 2019). SKOV-3 cells were transfected with siSnail or siBIRC3 to demonstrate the effect of the siRNA-mediated down-regulation of Snail and BIRC3 on MMP-2 and MMP-9, and target gene expression was evaluated through RT-PCR and Western blot. As shown in Fig. 4B, the knockdown of Snail or BIRC3 by siRNA markedly reduced the mRNA levels of MMP-2 and MMP-9 (Fig. 4B).
DISCUSSION
EMT plays a critical role in ovarian tumour progression, including enhancing cell migration and invasive ability (Vergara et al., 2010; Lili et al., 2013; Lambert and Weinberg, 2021). The relationship between Snail and BIRC3 in cellular biological function was previously unknown. Thus, in the present study, we revealed that the BIRC3 protein, an inhibitor of apoptotic proteins, was a novel Snail-binding protein.
In this study, the yeast two-hybrid method revealed that Snail interacted with BIRC3 (Fig. 1). BIRC3 is an IAP family member that suppresses apoptotic cell death by interfering with caspase activation (Frazzi, 2021). In tumours, BIRC3 is often up-regulated in various cancers, including cervical cancer (Imoto et al., 2002; Wu et al., 2021b), hepatocellular carcinoma (Zender et al., 2006; Jiang et al., 2021), non-small and small cell lung cancer (Ferreira et al., 2001; Dai et al., 2003; Liu et al., 2022), oesophageal squamous cell cancer (Imoto et al., 2001; Wu et al., 2021a), oral squamous cell carcinoma (Snijders et al., 2005; Bi et al., 2021), and pancreatic cancer (Esposito et al., 2007; Yu et al., 2021b). BIRC3 interacts with molecules other than Snail. For example, BIRC3 interacts with various intracellular proteins, such as receptor-interacting protein 1 (RIP1) (Bertrand et al., 2008), tumour necrosis factor receptor-associated factor 1 (TRAF1) (Li et al., 2002), TRAF2 (Yoneda et al., 2000; Zheng et al., 2010), and ubiquitin-conjugating enzyme E2 D2 (UBE2D2) (Mace et al., 2008).
Snail binds to various intracellular proteins, including the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex (Peinado et al., 2004), polycomb repressive complex 2 (PRC2) (Herranz et al., 2008), 14-3-3 and protein arginine methyltransferase 5 (PRMT5)/AJUBA complex (Hou et al., 2008, 2010), lysine-specific demethylase 1 (LSD1) (Lin et al., 2010), and small C-terminal domain phosphatase (SCP) (Wu et al., 2009). This binding causes the inhibition of Snail dephosphorylation and stabilization following the loss of the Snail C-terminal domain (Wu et al., 2009). Dong et al. (2013) reported that Snail interacts with the suppressor of variegation 3-9 homolog 1 (Suv39H1) in vivo, and its expression is correlated with breast cancer metastasis. Therefore, BIRC3 is also a new member that combines with Snail.
Interestingly, the decrease in the BIRC3 expression induced a decrease in the Snail expression, and the decrease in the Snail expression inversely caused the decrease in the BIRC3 expression (Fig. 2C). However, studies have yet to clarify how such a relationship can be established.
BIRC3 affects cell viability, as mentioned earlier (Frazzi, 2021). In the present study, the effect of the interaction of Snail with BIRC3 on the survival of ovarian cancer cell lines was investigated, and the results indicated that siSnail treatment significantly reduced cell survival. The simultaneous siBIRC3 treatment further increased this reduced survival (Fig. 2B). The treatment with siSnail also induced a decrease in the expression of cell survival-related molecules, such as Bax, Bcl-XL, Mcl-1 and Bcl-2, and Snail re-expression reversed this observation and restored the expression of these cell survival molecules (Fig. 2D, left panel). Likewise, the treatment with siBIRC3 resulted in a decrease in cell survival-related molecules similar to siSnail, and BIRC3 re-expression similarly reversed this result (Fig. 2D, right panel). In our investigation into how BIRC3 and Snail expressed Bcl-2, treatment with siBIRC3 and siSnail induced a decrease in the Bcl-2 luciferase activity. Thus, siBIRC3 and siSnail appear to inhibit the Bcl-2 expression at the transcriptional level (Fig. 2E). Studies have yet to determine how snail and BIRC3 regulate the Bcl-2 expression at the transcriptional level. However, MnSOD overexpression confers cisplatin resistance in lung adenocarcinoma via the NF-κB/Snail/Bcl-2 pathway (Chen et al., 2015). Therefore, Snail is a member of an NF-κB/Snail/Bcl-2 axis that increases Bcl-2 expression.
Herbert et al. (2010) demonstrated that mammary epithelial cells harbouring the TP53 M133T mutation displayed a significant increase (20-40 fold) in the expression of the anti-apoptotic gene BIRC3. These results suggested an association between p53 and BIRC3. The siBIRC3 expression increased the p53 expression, similarly, the knockdown of Snail, a binding partner of BIRC3, increased the p53 expression (Fig. 3A). Snail and p53 were inversely correlated, and BIRC3 was likely involved in this relationship. Snail and BIRC3 possibly affected p53 protein stabilization (Fig. 3B). Nevertheless, a study on the correlation between BIRC3, p53, and Snail is currently underway.
As Snail plays an important role in EMT, the role of its binding protein BIRC3 in EMT was investigated. Similar to Snail expression, BIRC3 overexpression decreased the expression of E-cadherin and increased the expression of N-cadherin, vimentin, and fibronectin (Fig. 4A). IL-1β induces the expression of BIRC3, which is involved in doxorubicin resistance in breast cancer cells. This resistance is a phenomenon related to EMT (Mendoza-Rodríguez et al., 2017). BIRC3 induces EMT in hepatocellular carcinoma (Fu et al., 2019). Although this study did not present a direct mechanism of how to induce EMT, our results provided some relevant insights.
The knockdown of Snail expression by siSnail reduced the expressions of MMP-2 and MMP-9 (Fig. 4B). In an animal model, knockdown of Snail expression inhibited the growth and metastasis of ovarian tumours (Hojo et al., 2018). Overall, Snail plays a critical role in ovarian tumour growth and metastasis via the regulation of MMP expression (Yokoyama et al., 2003; Jin et al., 2010). Snail-binding BIRC3 knockdown also reduced the expression of MMP2 and MMP9. (Fig. 4B). These results suggested that BIRC3 was also involved in MMP expression and likely implicated in cancer invasion.
In summary, our study provided new insights into the critical role and regulatory function of Snail in cancer cell proliferation via BIRC3 and the relevant mechanism of BIRC3 in EMT via Snail. Studies on the triangle relationship between Snail, BIRC3, and p53 could yield interesting results.
ACKNOWLEDGMENTS
This work was supported by grants from the National Cancer Center (NCC-2112500-1) and the Basic Science Research Program NRF (NRF-2020R1A2C3004973), NRF (NRF-2018R1A5A2023127), and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Korea (HP20C0131).
REFERENCES
- Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., Barker P. A. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Bi L., Zhang C., Yao Y., He Z. Circ-HIPK3 regulates YAP1 expression by sponging miR-381-3p to promote oral squamous cell carcinoma development. J. Biosci. 2021;46:20. doi: 10.1007/s12038-021-00142-w. [DOI] [PubMed] [Google Scholar]
- Blechschmidt K., Sassen S., Schmalfeldt B., Schuster T., Hofler H., Becker K. F. The E-cadherin repressor Snail is associated with lower overall survival of ovarian cancer patients. Br. J. Cancer. 2008;98:489–495. doi: 10.1038/sj.bjc.6604115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Gong L., Li G., Guo J., Yi X., Wang Z. Exosomes in ovarian cancer ascites promote epithelial-mesenchymal transition of ovarian cancer cells by delivery of miR-6780b-5p. Cell Death Dis. 2021;12:210. doi: 10.1038/s41419-021-03490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. M., Cheng Y. W., Wu T. C., Chen C. Y., Lee H. MnSOD overexpression confers cisplatin resistance in lung adenocarcinoma via the NF-κB/Snail/Bcl-2 pathway. Free Radic. Biol. Med. 2015;79:127–137. doi: 10.1016/j.freeradbiomed.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Clarfield L., Diamond L., Jacobson M. Risk-reducing options for high-grade serous gynecologic malignancy in BRCA1/2. Curr. Oncol. 2022;29:2132–2140. doi: 10.3390/curroncol29030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., Zhu W. G., Morrison C. D., Brena R. M., Smiraglia D. J., Raval A., Wu Y. Z., Rush L. J., Ross P., Molina J. R., Otterson G. A., Plass C. A comprehensive search for DNA amplification in lung cancer identifies inhibitors of apoptosis cIAP1 and cIAP2 as candidate oncogenes. Hum. Mol. Genet. 2003;12:791–801. doi: 10.1093/hmg/ddg083. [DOI] [PubMed] [Google Scholar]
- Debnath P., Huirem R. S., Dutta P., Palchaudhuri S. Epithelial-mesenchymal transition and its transcription factors. Biosci. Rep. 2022;42:BSR20211754. doi: 10.1042/BSR20211754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez D., Montserrat-Sentís B., Virgós-Soler A., Guaita S., Grueso J., Porta M., Puig I., Baulida J., Francí C., Garci, a de Herreros A. Phosphorylation regulates the subcellular location and activity of the snail transcriptional repressor. Mol. Cell. Biol. 2003;23:5078–5089. doi: 10.1128/MCB.23.14.5078-5089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Wu Y., Wang Y., Wang C., Kang T., Rychahou P. G., Chi Y. I., Evers B. M., Zhou B. P. Interaction with Suv39H1 is critical for Snail-mediated E-cadherin repression in breast cancer. Oncogene. 2013;32:1351–1362. doi: 10.1038/onc.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S. M., Byun H. J., Kim B. R., Lee S. H., Trink B., Rho S. B. Tumor suppressor BLU enhances pro-apoptotic activity of sMEK1 through physical interaction. Cell. Signal. 2012;24:1208–1214. doi: 10.1016/j.cellsig.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Elshami M., Tuffaha A., Yaseen A., Alser M., Al-Slaibi I., Jabr H., Ubaiat S., Khader S., Khraishi R., Jaber I., Abu Arafeh Z., Al-Madhoun S., Alqattaa A., Abd El Hadi A., Barhoush O., Hijazy M., Eleyan T., Alser A., Abu Hziema A., Shatat A., Almakhtoob F., Mohamad B., Farhat W., Abuamra Y., Mousa H., Adawi R., Musallam A., Abu-El-Noor N., Bottcher B. Awareness of ovarian cancer risk and protective factors: a national cross-sectional study from Palestine. PLoS ONE. 2022;17:e0265452. doi: 10.1371/journal.pone.0265452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito I., Kleeff J., Abiatari I., Shi X., Giese N., Bergmann F., Roth W., Friess H., Schirmacher P. Overexpression of cellular inhibitor of apoptosis protein 2 is an early event in the progression of pancreatic cancer. J. Clin. Pathol. 2007;60:885–895. doi: 10.1136/jcp.2006.038257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira C., Van Der Valk P., Span S., Jonker J., Postmus P., Kruyt F., Giaccone G. Assessment of IAP (inhibitor of apoptosis) proteins as predictors of response to chemotherapy in advanced non-small-cell lung cancer patients. Ann. Oncol. 2001;12:799–805. doi: 10.1023/A:1011167113067. [DOI] [PubMed] [Google Scholar]
- Frazzi R. BIRC3 and BIRC5: multi-faceted inhibitors in cancer. Cell Biosci. 2021;11:8. doi: 10.1186/s13578-020-00521-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu P. Y., Hu B., Ma X. L., Yang Z. F., Yu M. C., Sun H. X., Huang A., Zhang X., Wang J., Hu Z. Q., Zhou C. H., Tang W. G., Ning R., Xu Y., Zhou J. New insight into BIRC3: a novel prognostic indicator and a potential therapeutic target for liver cancer. J. Cell. Biochem. 2019;120:6035–6045. doi: 10.1002/jcb.27890. [DOI] [PubMed] [Google Scholar]
- Herbert B. S., Chanoux R. A., Liu Y., Baenziger P. H., Goswami C. P., McClintick J. N., Edenberg H. J., Pennington R. E., Lipkin S. M., Kopelovich L. A molecular signature of normal breast epithelial and stromal cells from Li-Fraumeni syndrome mutation carriers. Oncotarget. 2010;1:405–422. doi: 10.18632/oncotarget.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz N., Pasini D., Díaz V. M., Francí C., Gutierrez A., Dave N., Escrivà M., Hernandez-Muñoz I., Di Croce L., Helin K., García, de Herreros A., Peiró S. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol. Cell. Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojo N., Huisken A. L., Wang H., Chirshev E., Kim N. S., Nguyen S. M., Campos H., Glackin C. A., Ioffe Y. J., Unternaehrer J. J. Snail knockdown reverses stemness and inhibits tumour growth in ovarian cancer. Sci. Rep. 2018;8:8704. doi: 10.1038/s41598-018-27021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Peng H., Ayyanathan K., Yan K. P., Langer E. M., Longmore G. D., Rauscher F. J., III The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol. Cell. Biol. 2008;28:3198–3207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Peng H., White D. E., Wang P., Lieberman P. M., Halazonetis T., Rauscher F. J. 14-3-3 binding sites in the snail protein are essential for snail-mediated transcriptional repression and epithelial-mesenchymal differentiation. Cancer Res. 2010;70:4385–4393. doi: 10.1158/0008-5472.CAN-10-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto I., Tsuda H., Hirasawa A., Miura M., Sakamoto M., Hirohashi S., Inazawa J. Expression of cIAP1, a target for 11q22 amplification, correlates with resistance of cervical cancers to radiotherapy. Cancer Res. 2002;62:4860–4866. [PubMed] [Google Scholar]
- Imoto I., Yang Z. Q., Pimkhaokham A., Tsuda H., Shimada Y., Imamura M., Ohki M., Inazawa J. Identification of cIAP1 as a candidate target gene within an amplicon at 11q22 in esophageal squamous cell carcinomas. Cancer Res. 2001;61:6629–6634. [PubMed] [Google Scholar]
- Jiang Y., Nan H., Shi N., Hao W., Dong J., Chen H. Chlorogenic acid inhibits proliferation in human hepatoma cells by suppressing noncanonical NF-κB signaling pathway and triggering mitochondrial apoptosis. Mol. Biol. Rep. 2021;48:2351–2364. doi: 10.1007/s11033-021-06267-3. [DOI] [PubMed] [Google Scholar]
- Jin H., Yu Y., Zhang T., Zhou X., Zhou J., Jia L., Wu Y., Zhou B. P., Feng Y. Snail is critical for tumor growth and metastasis of ovarian carcinoma. Int. J. Cancer. 2010;126:2102–2111. doi: 10.1002/ijc.24901. [DOI] [PubMed] [Google Scholar]
- Kim H. J., Kim B., Byun H. J., Yu L., Nguyen T. M., Nguyen T. H., Do P. A., Kim E. J., Cheong K. A., Kim K. S., Huy Phùng H., Rahman M., Jang J. Y., Rho S. B., Kang G. J., Park M. K., Lee H., Lee K., Cho J., Han H. K., Kim S. G., Lee A. Y., Lee C. H. Resolvin D1 suppresses H2O2-induced senescence in fibroblasts by inducing autophagy through the miR-1299/ARG2/ARL1 axis. Antioxidants. 2021;10:1924. doi: 10.3390/antiox10121924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurrey N. K., Bapat S. A. Snail and Slug are major determinants of ovarian cancer invasiveness at the transcription level. Gynecol. Oncol. 2005;97:155–165. doi: 10.1016/j.ygyno.2004.12.043. [DOI] [PubMed] [Google Scholar]
- Lambert A. W., Weinberg R. A. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat. Rev. Cancer. 2021;21:325–338. doi: 10.1038/s41568-021-00332-6. [DOI] [PubMed] [Google Scholar]
- Lee C. H. Epithelial-mesenchymal transition: initiation by cues from chronic inflammatory tumor microenvironment and termination by anti-inflammatory compounds and specialized pro-resolving lipids. Biochem. Pharmacol. 2018;158:261–273. doi: 10.1016/j.bcp.2018.10.031. [DOI] [PubMed] [Google Scholar]
- Lee C. H. Reversal of epithelial-mesenchymal transition by natural anti-inflammatory and pro-resolving lipids. Cancers (Basel) 2019;11:1841. doi: 10.3390/cancers11121841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Yang Y., Ashwell J. D. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature. 2002;416:345–347. doi: 10.1038/416345a. [DOI] [PubMed] [Google Scholar]
- Lili L. N., Matyunina L. V., Walker L., Wells S. L., Benigno B. B., McDonald J. F. Molecular profiling supports the role of epithelial-to-mesenchymal transition (EMT) in ovarian cancer metastasis. J. Ovarian Res. 2013;6:49. doi: 10.1186/1757-2215-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Wu Y., Li J., Dong C., Ye X., Chi Y. I., Evers B. M., Zhou B. P. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Chen Z., Ding X., Qiao Y., Li B. Ubiquitin-specific protease 35 (USP35) mediates cisplatin-induced apoptosis by stabilizing BIRC3 in non-small cell lung cancer. Lab. Invest. 2022;102:524–533. doi: 10.1038/s41374-021-00725-z. [DOI] [PubMed] [Google Scholar]
- Mace P. D., Linke K., Feltham R., Schumacher F.-R., Smith C. A., Vaux D. L., Silke J., Day C. L. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J. Biol. Chem. 2008;283:31633–31640. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- Mendoza-Rodríguez M., Romero H. A., Fuentes-Panana E. M., Ayala-Sumuano J.-T., Meza I. IL-1β induces up-regulation of BIRC3, a gene involved in chemoresistance to doxorubicin in breast cancer cells. Cancer Lett. 2017;390:39–44. doi: 10.1016/j.canlet.2017.01.005. [DOI] [PubMed] [Google Scholar]
- Nagase H., Woessner J. F., Jr. Matrix metalloproteinases. J. Biol. Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- Nam M. W., Kim C. W., Choi K. C. Epithelial-mesenchymal transition-inducing factors involved in the progression of lung cancers. Biomol. Ther. (Seoul) 2022;30:213–220. doi: 10.4062/biomolther.2021.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M. A. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Peinado H., Ballestar E., Esteller M., Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell. Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiro S., Escriva M., Puig I., Barbera M. J., Dave N., Herranz N., Larriba M. J., Takkunen M., Francí C., Munoz A., Virtanen I., Baulida J., García, de Herreros A. Snail1 transcriptional repressor binds to its own promoter and controls its expression. Nucleic Acids Res. 2006;34:2077–2084. doi: 10.1093/nar/gkl141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho S. B., Byun H. J., Kim B. R., Lee C. H. Knockdown of LKB1 sensitizes endometrial cancer cells via AMPK activation. Biomol. Ther. (Seoul) 2021a;29:650–657. doi: 10.4062/biomolther.2021.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho S. B., Kim M. J., Lee J. S., Seol W., Motegi H., Kim S., Shiba K. Genetic dissection of protein-protein interactions in multi-tRNA synthetase complex. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4488–4493. doi: 10.1073/pnas.96.8.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho S. B., Lee K. W., Lee S. H., Byun H. J., Kim B. R., Lee C. H. Novel anti-angiogenic and anti-tumour activities of the N-terminal domain of NOEY2 via binding to VEGFR-2 in ovarian cancer. Biomol. Ther. (Seoul) 2021b;29:506–518. doi: 10.4062/biomolther.2021.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho S. B., Lee S. H., Byun H. J., Kim B. R., Lee C. H. IRF-1 inhibits angiogenic activity of HPV16 E6 oncoprotein in cervical cancer. Int. J. Mol. Sci. 2020;21:7622. doi: 10.3390/ijms21207622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheau C., Badarau I. A., Costache R., Caruntu C., Mihai G. L., Didilescu A. C., Constantin C., Neagu M. The role of matrix metalloproteinases in the epithelial-mesenchymal transition of hepatocellular carcinoma. Anal. Cell. Pathol. 2019;2019:9423907. doi: 10.1155/2019/9423907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders A. M., Schmidt B. L., Fridlyand J., Dekker N., Pinkel D., Jordan R. C., Albertson D. G. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–4242. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- Vergara D., Merlot B., Lucot J. P., Collinet P., Vinatier D., Fournier I., Salzet M. Epithelial-mesenchymal transition in ovarian cancer. Cancer Lett. 2010;291:59–66. doi: 10.1016/j.canlet.2009.09.017. [DOI] [PubMed] [Google Scholar]
- Wu Q., Zhang Y., An H., Sun W., Wang R., Liu M., Zhang K. The landscape and biological relevance of aberrant alternative splicing events in esophageal squamous cell carcinoma. Oncogene. 2021a;40:4184–4197. doi: 10.1038/s41388-021-01849-8. [DOI] [PubMed] [Google Scholar]
- Wu S., Zang Q., Xing Z., Li X., Leng J., Liu Y., Wang X., Yang J. A pan-cancer analysis of the BIRC gene family and its association with prognosis, tumor microenvironment, and therapeutic targets. Crit. Rev. Eukaryot. Gene Expr. 2021b;31:35–48. doi: 10.1615/CritRevEukaryotGeneExpr.2021038714. [DOI] [PubMed] [Google Scholar]
- Wu Y., Evers B. M., Zhou B. P. Small C-terminal domain phosphatase enhances snail activity through dephosphorylation. J. Biol. Chem. 2009;284:640–648. doi: 10.1074/jbc.M806916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhou B. P. Snail: more than EMT. Cell Adh. Migr. 2010;4:199–203. doi: 10.4161/cam.4.2.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K., Kamata N., Fujimoto R., Tsutsumi S., Tomonari M., Taki M., Hosokawa H., Nagayama M. Increased invasion and matrix metalloproteinase-2 expression by Snail-induced mesenchymal transition in squamous cell carcinomas. Int. J. Oncol. 2003;22:891–898. doi: 10.3892/ijo.22.4.891. [DOI] [PubMed] [Google Scholar]
- Yoneda T., Imaizumi K., Maeda M., Yui D., Manabe T., Katayama T., Sato N., Gomi F., Morihara T., Mori Y., Miyoshi K., Hitomi J., Ugawa S., Yamada S., Okabe M., Tohyama M. Regulatory mechanisms of TRAF2-mediated signal transduction by Bcl10, a MALT lymphoma-associated protein. J. Biol. Chem. 2000;275:11114–11120. doi: 10.1074/jbc.275.15.11114. [DOI] [PubMed] [Google Scholar]
- Yu L., Kim H. J., Park M. K., Byun H. J., Kim E. J., Kim B., Nguyen M. T., Kim J. H., Kang G. J., Lee H., Kim S. Y., Rho S. B., Lee C. H. Ethacrynic acid, a loop diuretic, suppresses epithelial-mesenchymal transition of A549 lung cancer cells via blocking of NDP-induced WNT signaling. Biochem. Pharmacol. 2021a;183:114339. doi: 10.1016/j.bcp.2020.114339. [DOI] [PubMed] [Google Scholar]
- Yu Q., Jobin C., Thomas R. M. Implications of the microbiome in the development and treatment of pancreatic cancer: thinking outside of the box by looking inside the gut. Neoplasia. 2021b;23:246–256. doi: 10.1016/j.neo.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Kajiyama H., Ito S., Yoshikawa N., Hyodo T., Asano E., Hasegawa H., Maeda M., Shibata K., Hamaguchi M., Kikkawa F., Senga T. ALX1 induces snail expression to promote epithelial-to-mesenchymal transition and invasion of ovarian cancer cells. Cancer Res. 2013;73:1581–1590. doi: 10.1158/0008-5472.CAN-12-2377. [DOI] [PubMed] [Google Scholar]
- Zender L., Spector M. S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S.-T., Luk J. M., Wigler M., Hannon G. J., Mu D., Lucito R., Powers S., Lowe S. W. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Kabaleeswaran V., Wang Y., Cheng G., Wu H. Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes: affinity, specificity, and regulation. Mol. Cell. 2010;38:101–113. doi: 10.1016/j.molcel.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]