Introduction
NPM1-mutated acute myeloid leukemia (AML) represents about one-third of all adult AML [1] and, due its unique clinicopathological and genetic features [2], is recognized as a leukemia entity of the World Health Organization (WHO) Classification of myeloid neoplasms. Nucleophosmin (NPM1) is a multifunctional protein physiologically located in the nucleolus [2]. NPM1 mutations, the most common genetic lesion in AML, abrogate the ability of the protein to localize in the nucleolus and create a new nuclear export signal (NES) at the C-terminus, leading to enhanced nuclear export of mutant NPM1 and its aberrant accumulation in the cytoplasm of leukemic cells [1, 3, 4].
We demonstrated that the interaction between mutant NPM1 and the nuclear exporter Exportin-1 (XPO1) causes the aberrant cytoplasmic delocalization of mutant NPM1 and is responsible for the high expression of HOX genes in NPM1-mutated AML, since relocalization of the NPM1 mutant by XPO1 inhibitors causes early downregulation of HOX genes that is followed by cell differentiation and growth arrest [2, 5]. The rapid loss of HOX expression, despite XPO1 inhibition does not restore the physiologic localization of NPM1 to the nucleolus [5], strongly suggest that the interaction between XPO1 and mutant NPM1 (rather than its localization) is responsible for maintaining high HOX levels.
Another characteristic feature of NPM1-mutated AML is the absent/low expression of CD34 [6, 7] that yet remains poorly investigated. This feature, combined with the low HLA-DR [8], strong CD33 expression [9] and presence of abnormal PML bodies [10], is reminiscent of acute promyelocytic leukemia (APL) and have inspired APL-like treatment strategies (i.e., all-trans retinoic acid and arsenic trioxide) also in NPM1-mutated AML both preclinically [10] and in patients (NCT04689815, NCT03031249). Unlike HOX genes, CD34 expression seems to be independent from XPO1-mediated cytoplasmic dislocation of mutant NPM1. This is supported by the finding that in most NPM1-mutated AML patients, a small subset of early CD34+ hematopoietic precursors carrying NPM1 mutations/cytoplasmic NPM1 is usually present [11], suggesting a derivation from CD34+ hemopoietic stem cells, with the potential of multilineage differentiation. On the other hand, the observation that at least a percentage of NPM1-mutated AML may derive from CD34-negative hematopoietic stem cells, raises the question of a possible relationship between absent/low expression of CD34 and cytoplasmic dislocation of mutant NPM1 [12]. Clinically, CD34 expression in NPM1-mutated AML has been mostly associated with an adverse outcome [13].
To address this issue, we performed functional studies to assess whether the nuclear relocalization of the mutant NPM1 could result in the re-expression of CD34. Moreover, we searched for CD34+/NPM1 cytoplasmic precursors in the bone marrow (BM) biopsies of NPM1-AML patients at diagnosis and relapse, using a highly specific monoclonal antibody against mutant NPM1. The results of these studies are presented below.
Results and discussion
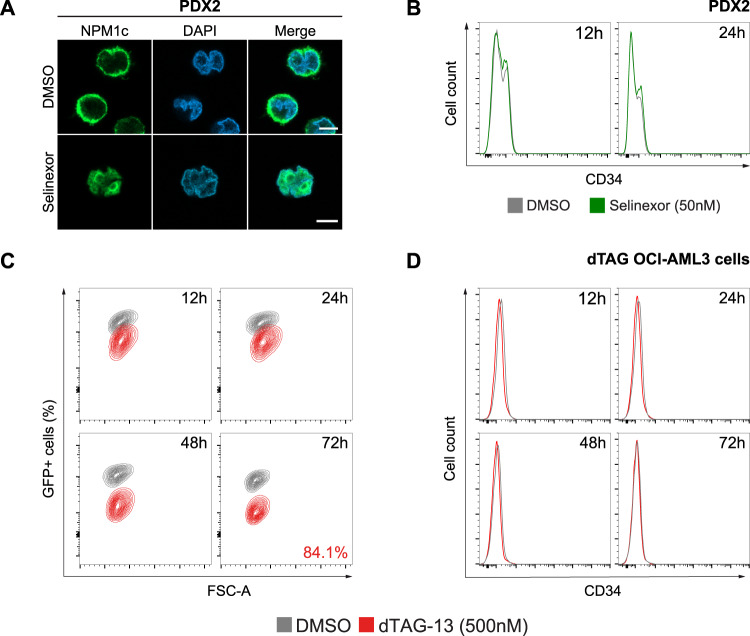
We first investigated whether cytoplasmic delocalization of mutant NPM1 could explain the absent/low expression of CD34 in NPM1-mutated AML. To this end, we assessed the effect of nuclear relocalization of mutant NPM1 on CD34 expression, using the selective XPO1 inhibitor selinexor, which blocks the nuclear export of XPO1 cargo proteins, including NPM1. We treated the NPM1-mutated OCI-AML3 cell line that does not express CD34 and two NPM1-mutated patient-derived xenograft (PDX) cells (PDX2 and PDX3) [5] that partially express CD34 with selinexor for 24 h. Immunofluorescence with a monoclonal antibody specific for mutant NPM1 showed that after 12 h of XPO1 inhibition, mutant NPM1 was completely relocated to the nucleus of AML cells (Fig. 1A, PDX2). Flow cytometry analysis performed at 12 and 24 h of treatment revealed that CD34 expression levels remained unchanged (Fig. 1B). CD34 expression was also studied by real-time PCR, confirming its low/absent expression in NPM1-mutated AML cells and showing that levels did not change following XPO1 inhibition (not shown).
Fig. 1. Nuclear relocalization or selective degradation of mutant NPM1 does not alter CD34 expression.
A. Representative fluorescence microscopy images of PDX2 cells treated for 12 h with DMSO (control) or selinexor 50 nM. Cells were labeled with anti-NPM1 mutant antibody (NPM1c, green). Cell nuclei were stained with DAPI (blue). 63x magnification. Scale bar, 5 μm. B Representative histogram plots for CD34 expression analyzed by flow cytometry at 12 and 24 h in PDX2 cells treated with either DMSO (control, grey) or selinexor 50 nM (green). C Flow cytometry contour plots showing the percentage of GFP+ cells at 12, 24, 48 and 72 h in dTAG OCI-AML3 cells treated with DMSO (control, grey) or dTAG-13 500 nM (red). The degradation rate of mutant NPM1 at 72 h is shown. D Overlaid histogram plots representing CD34 expression analyzed by flow cytometry at 12, 24, 48 and 72 h in dTAG OCI-AML3 cells treated with either DMSO (control, grey) or dTAG-13 500 nM (red).
Selinexor does not specifically block the nucleo-cytoplasmic shuttling of mutant NPM1, abrogating also the nuclear export of all XPO1 cargo proteins. Therefore, we tested the effect of selective mutant NPM1 degradation on CD34 expression. For this purpose, we exploited two NPM1-mutated CRISPR-engineered cell lines (OCI-AML3 and IMS-M2) in which endogenous mutant NPM1 is fused to the FKBPF36V degron tag and GFP (NPM1c-FKBPF36V-GFP, Supplementary Fig. S1A) [5]. This system enables fast and specific degradation of mutant NPM1 by addition of the small compound dTAG-13. We treated cells with either DMSO (control) or dTAG-13 for 72 h. As expected, dTAG-13 induced efficient degradation of mutant NPM1 in both CRISPR-engineered cell lines (Fig. 1C and Supplementary Fig. S1B). Immunoblotting confirmed the loss of fusion protein without changes in wild-type NPM1 (Supplementary Figs. S1D, E). Nonetheless, flow cytometry and overlaid histograms of CD34 expression showed that CD34 levels were not changed at all time points in both cell lines (Fig. 1D and Supplementary Fig. S1C). Altogether, these findings strongly suggest that CD34 expression is not dependent upon mutant NPM1 expression and/or localization.
We then investigated the expression of mutant NPM1 in CD34+ cells of BM biopsies from NPM1-mutated AML patients by double immunostaining. In a previous study [11], we had addressed this issue using a monoclonal antibody recognizing both the wild-type and mutant NPM1 protein. In this study, double immunostaining for CD34/NPM1 was carried out using the highly specific anti-NPM1 mutant antibody used for immunofluorescence, suitable for automated immunoperoxidase staining of paraffin-embedded BM biopsies fixed in formalin and decalcified in Osteodec (full details are given in the Supplementary Materials).
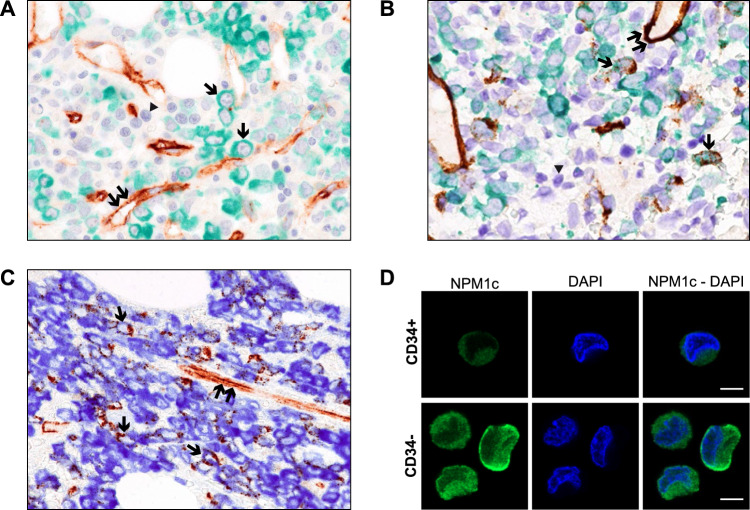
BM paraffin sections from 15 NPM1-mutated AML patients (n = 10 at first diagnosis and n = 5 at relapse) were double stained for CD34 and mutant NPM1. Fourteen out of the 15 cases showed the presence of no or rare CD34/mutant NPM1 double-positive cells (Fig. 2A, B). One case of NPM1-mutated AML at relapse (FLT3-ITD positive) showed a higher percentage (up to 20%) of leukemic cells double stained for CD34 and cytoplasmic mutant NPM1 (Fig. 2C). Moreover, we performed immunofluorescence using the same anti-NPM1 mutant antibody in FACS-sorted CD34+ and CD34− leukemic cells from a 32-year-old female patient with high count NPM1-mutated AML (Supplementary Fig. S2A). Purified CD34+ and CD34− cells were spotted onto poly-L-lysine coated glass slides and immunostained with the specific anti-NPM1 mutant monoclonal antibody followed by anti-rabbit Alexa Fluor 488-conjugated secondary antibody (full details are provided in Supplementary Materials). Both CD34+ and CD34− leukemic cells showed cytoplasm-restricted positivity for the NPM1 mutant protein without nuclear staining (Fig. 2D).
Fig. 2. Co-expression of NPM1 mutant and CD34+ in NPM1-mutated AML.
A. Formalin-fixed BM paraffin section from NPM1-mutated AML at diagnosis double stained for CD34 (brown, peroxidase with DAB chromogen) and mutant NPM1 protein (green, peroxidase with green chromogen, DC9913, Leica) plus BOND Polymer Refine HRP PLEX Detection and counterstaining in hematoxylin (blue). Single arrows point to leukemic cells expressing cytoplasmic NPM1 (green) but not CD34. Cells stained only in blue (hematoxylin) represent normal residual hematopoietic cells that do not express CD34 and the NPM1 mutant protein (arrowhead). Double arrows indicate CD34+ vessel endothelial cells (brown) that serve as positive control (magnification, x400). B Formalin-fixed BM paraffin section from another NPM1-mutated AML sample at diagnosis double stained with CD34 (brown) and mutant NPM1 (green) and counterstained in hematoxylin (blue) as indicated in panel A. Most leukemic cells show cytoplasmic NPM1 (green) in the absence of CD34 (brown) whilst a few blast cells (single arrows) are double stained (brown/green) for CD34 and cytoplasmic NPM1 mutant protein. Cells stained only in blue (hematoxylin) represent normal residual hematopoietic cells that do not express CD34 and the NPM1 mutant protein (arrowhead). Double arrows indicate CD34+ vessel endothelial cells (brown) that serve as positive control (magnification, x400). C Formalin-fixed BM paraffin section from a NPM1-mutated AML patient at relapse double stained for CD34 (brown, peroxidase with DAB chromogen) and mutant NPM1 protein (blue, peroxidase with blue chromogen, DC9896, Leica) plus BOND Polymer Refine Detection, without hematoxylin counterstaining. A significant percentage of leukemic cells (single arrows) are double stained (brown/blue) for CD34 and NPM1 mutant protein. Double arrows indicate CD34+ vessel endothelial cells (brown) that serve as positive control (magnification, x400). D Immunofluorescence with anti-NPM1 mutant antibody (green) on the CD34+ and CD34− sorted cells. Nuclei were stained with DAPI (blue). 63x magnification. Scale bar, 5 μm.
Collectively, the above results strongly suggest that, unlike HOX genes [5], CD34 expression is not dependent on the expression or the delocalization of NPM1 mutant protein from the nucleus to the cytoplasm. In fact, neither nuclear relocalization nor selective degradation of mutant NPM1 induced changes in CD34 expression in our parental and CRISPR-engineered AML cell lines.
In conclusion, based on our functional and immunohistochemical studies, we hypothesize that most NPM1-mutated cells originate from an immature CD34+ hematopoietic cell [10], but the leukemic bulk population is detected at an abnormal differentiation stage in which HOX expression is still high whereas CD34 is already silenced. In this context, a minor “stem-like” population, which contains leukemia-initiating cells, and which may expand at relapse [11, 14], maintains both CD34 and high HOX levels. Indeed, as in the patient with high percentage of CD34+ cells described here, an increased CD34 expression at relapse has been reported in NPM1-mutated AML, particularly in association with FLT3-ITD [11, 15].
Supplementary information
Author contributions
GP, FR and BF conceived the study. GP and FR performed in vitro experiments. BB performed IHC analysis. DS performed cell sorting. MDI provided patient samples and performed NGS analysis. SP, MC, VA, PS, MPM and BTG provided reagents and critical inputs. GP, FR, LB and BF analyzed the results and wrote the manuscript with the input from all the authors.
Funding
This work has been supported by the European Research Council (ERC-2016 ADG n. 740230 to BF), the Italian Association for Cancer research (AIRC; IG n. 23604 to BF and Start-Up 2019 n. 22895 to LB), the Norwegian Cancer Society (grants n. 303445 and 190175 to BTG) and the Trond Mohn Foundation recruitment program grant (to VA).
Competing interests
BF licensed a patent on NPM1 mutants (n. 102004901256449). BF and MPM declare honoraria from Rasna Therapeutics, Inc for scientific advisor activities. PS declares honoraria/consultancy at scientific advisory board for Abbvie, Janssen, Novartis, AstraZeneca, Incyte. MPM also declares honoraria/consultancy at scientific advisory board for Abbvie, Amgen, Celgene, Janssen, Novartis, Pfizer, Jazz Pharmaceuticals. LB declares consultancy at scientific advisory boards for Abbvie and Amgen.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Giulia Pianigiani, Francesca Rocchio.
These authors jointly supervised this work: Lorenzo Brunetti, Brunangelo Falini.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-022-01593-2.
References
- 1.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl J Med. 2005;352:254–66. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 2.Falini B, Brunetti L, Sportoletti P, Martelli MP. NPM1-mutated acute myeloid leukemia: from bench to bedside. Blood. 2020;136:1707–21. doi: 10.1182/blood.2019004226. [DOI] [PubMed] [Google Scholar]
- 3.Falini B, Martelli MP, Bolli N, Bonasso R, Ghia E, Pallotta MT, et al. Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood. 2006;108:1999–2005. doi: 10.1182/blood-2006-03-007013. [DOI] [PubMed] [Google Scholar]
- 4.Falini B, Bolli N, Liso A, Martelli MP, Mannucci R, Pileri S, et al. Altered nucleophosmin transport in acute myeloid leukaemia with mutated NPM1: molecular basis and clinical implications. Leukemia. 2009;23:1731–43. doi: 10.1038/leu.2009.124. [DOI] [PubMed] [Google Scholar]
- 5.Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang YH, Ramabadran R, et al. Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell. 2018;34:499–512. doi: 10.1016/j.ccell.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcalay M, Tiacci E, Bergomas R, Bigerna B, Venturini E, Minardi SP, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;106:899–902. doi: 10.1182/blood-2005-02-0560. [DOI] [PubMed] [Google Scholar]
- 7.Verhaak RG, Goudswaard CS, van Putten W, Bijl MA, Sanders MA, Hugens W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106:3747–54. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 8.Oelschlaegel U, Mohr B, Schaich M, Schakel U, Kroschinsky F, Illmer T, et al. HLA-DRneg patients without acute promyelocytic leukemia show distinct immunophenotypic, genetic, molecular, and cytomorphologic characteristics compared to acute promyelocytic leukemia. Cytom B Clin Cytom. 2009;76:321–7. doi: 10.1002/cyto.b.20475. [DOI] [PubMed] [Google Scholar]
- 9.De Propris MS, Raponi S, Diverio D, Milani ML, Meloni G, Falini B, et al. High CD33 expression levels in acute myeloid leukemia cells carrying the nucleophosmin (NPM1) mutation. Haematologica. 2011;96:1548–51. doi: 10.3324/haematol.2011.043786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martelli MP, Gionfriddo I, Mezzasoma F, Milano F, Pierangeli S, Mulas F, et al. Arsenic trioxide and all-trans retinoic acid target NPM1 mutant oncoprotein levels and induce apoptosis in NPM1-mutated AML cells. Blood. 2015;125:3455–65. doi: 10.1182/blood-2014-11-611459. [DOI] [PubMed] [Google Scholar]
- 11.Martelli MP, Pettirossi V, Thiede C, Bonifacio E, Mezzasoma F, Cecchini D, et al. CD34+ cells from AML with mutated NPM1 harbor cytoplasmic mutated nucleophosmin and generate leukemia in immunocompromised mice. Blood. 2010;116:3907–22. doi: 10.1182/blood-2009-08-238899. [DOI] [PubMed] [Google Scholar]
- 12.Taussig DC, Vargaftig J, Miraki-Moud F, Griessinger E, Sharrock K, Luke T, et al. Leukemia-initiating cells from some acute myeloid leukemia patients with mutated nucleophosmin reside in the CD34(−) fraction. Blood. 2010;115:1976–84. doi: 10.1182/blood-2009-02-206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang H, Chen Y, Kamel-Reid S, Brandwein J, Chang H. CD34 expression predicts an adverse outcome in patients with NPM1-positive acute myeloid leukemia. Hum Pathol. 2013;44:2038–46. doi: 10.1016/j.humpath.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Perriello VM, Gionfriddo I, Rossi R, Milano F, Mezzasoma F, Marra A, et al. CD123 is consistently expressed on NPM1-mutated AML cells. Cancers (Basel). 2021;13:496. [DOI] [PMC free article] [PubMed]
- 15.Angelini DF, Ottone T, Guerrera G, Lavorgna S, Cittadini M, Buccisano F, et al. A leukemia-associated CD34/CD123/CD25/CD99+ immunophenotype identifies FLT3-mutated clones in acute myeloid leukemia. Clin Cancer Res. 2015;21:3977–85. doi: 10.1158/1078-0432.CCR-14-3186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.