Abstract
The tumor suppressor gene BAP1 encodes a widely expressed deubiquitinase for histone H2A. Both hereditary and acquired mutations are associated with multiple cancer types, including cutaneous melanoma (CM), uveal melanoma (UM), and clear cell renal cell carcinoma (ccRCC). However, there is no personalized therapy for BAP1-mutant cancers. Here, we describe an epigenetic drug library screening to identify small molecules that exert selective cytotoxicity against BAP1 knockout CM cells over their isogenic parental cells. Hit characterization reveals that BAP1 loss renders cells more vulnerable to bromodomain and extraterminal (BET) inhibitor-induced transcriptional alterations, G1/G0 cell cycle arrest and apoptosis. The association of BAP1 loss with sensitivity to BET inhibitors is observed in multiple BAP1-deficient cancer cell lines generated by gene editing or derived from patient tumors as well as immunodeficient xenograft and immunocompetent allograft murine models. We demonstrate that BAP1 deubiquitinase activity reduces sensitivity to BET inhibitors. Concordantly, ectopic expression of RING1A or RING1B (H2AK119 E3 ubiquitin ligases) enhances sensitivity to BET inhibitors. The mechanistic study shows that the BET inhibitor OTX015 exerts a more potent suppressive effect on the transcription of various proliferation-related genes, especially MYC, in BAP1 knockout cells than in their isogenic parental cells, primarily by targeting BRD4. Furthermore, ectopic expression of Myc rescues the BET inhibitor-sensitizing effect induced by BAP1 loss. Our study reveals new approaches to specifically suppress BAP1-deficient cancers, including CM, UM, and ccRCC.
Keywords: BRCA1-associated protein 1, bromodomain extraterminal inhibitors, clear cell renal cell carcinoma, cutaneous melanoma, individualized therapy, uveal melanoma
Introduction
Heterozygous germline mutations in the deubiquitinase BAP1 predispose carriers to the development of uveal melanoma (UM), cutaneous melanoma (CM), clear cell renal cell carcinoma (ccRCC), mesothelioma, and potentially other cancers, a condition known as BAP1 cancer syndrome [1]. In line with BAP1 being a classic two-hit tumor suppressor gene, patient tumors often show inactivating somatic mutation of the remaining functional BAP1 allele. In parallel, somatic BAP1 mutations and loss of BAP1 expression have also been reported in sporadic cancers of various origins, including the abovementioned cancer types [1]. Thus, BAP1 mediates tumor suppression in both hereditary and sporadic cases [2]. BAP1 loss may combine with other genetic aberrations to promote tumorigenesis given that BAP1 is often comutated with BRAF in CM [3], GNAQ or GNA11 in UM [4, 5], and VHL in ccRCC [6]. Moreover, accumulating evidence has shown that tumors with BAP1 mutations have a worse prognosis than those without BAP1 mutations [1]. For example, mutations in BAP1 are strongly correlated with a higher metastatic risk and reduced survival rate, with up to 84% of metastatic UM cases harboring BAP1 inactivating mutations [7].
BAP1 loss-of-function mutations limit the development of therapies directly targeting BAP1, and no personalized treatments are currently in use for cancers carrying BAP1 mutations. BAP1 is a deubiquitinating hydrolase associated with multiprotein complexes that are involved in various key cellular functions, including protein deubiquitination, cell cycle progression, DNA damage repair, and transcriptional regulation [1, 8]. Of note, increasing evidence suggests a critical role of BAP1 in epigenetic regulation. In this respect, BAP1 and ASXL1/2/3 (additional sexcombs like 1/2/3) form the polycomb repressive deubiquitinase (PR-DUB) complex, which removes monoubiquitin from ubiquitinated histone 2A at lysine 119 (H2AK119ub1); thus, BAP1 loss leads to hyperubiquitination of H2AK119 [9].
We hypothesized that an altered epigenetic status, such as a histone hyperubiquitination status, could serve as the Achilles’ heel of cancer cells with BAP1 mutations. In this scenario, further epigenetic disturbance is likely to induce synthetic lethality with BAP1 loss. Therefore, we conducted epigenetic compound library screening using BAP1 knockout (KO) isogenic cells generated with CRISPR/Cas9. One advantage of screening a compound library with defined targets is that the compounds, which are structurally diverse, can function as probes to cross-validate each other. This strategy led to the discovery that BAP1 KO CM cells display heightened sensitivity to bromodomain and extraterminal (BET) inhibitors. Similar results were observed in BAP1-deficient UM and ccRCC cell lines. We used in vitro and in vivo models to explore the potential of BET inhibitors as an individualized therapy for cancers with BAP1 mutations.
Materials and methods
Epigenetic compound library
The TargetMol library of epigenetic compounds (Catalog No. L1200) contains 303 compounds related to epigenetic regulation. Each compound was supplied as 250 μL at 10 mM in DMSO. The compounds were screened at a final concentration of 10 μM in this study.
Cell lines
KMRC1, VMRC-RCW, and VMRC-RCZ cells were purchased from JCRB Cell Bank (Japan). Caki-1 and 786O cells were from the Cell Resource Center of the Chinese Academy of Sciences (Shanghai, China). B16-F10, A498 and OS-RC2 cells were from Cellcook Biotech Co., Ltd. (Guangzhou, China). M14 and OMM2.3 cells were kindly provided by Dr. Kun-Liang Guan (University of California, San Diego). B16-F10 and B16-F10 BAP1 KO clones were maintained as previously described [10]. A498, VMRC-RCW, and VMRC-RCZ cells were cultured in MEM (Corning). KMRC1 cells were cultured in DMEM (Thermo Fisher Scientific). Caki-1 cells were cultured in McCoys 5A (Thermo Fisher Scientific). Other cells were cultured in RPMI-1640 (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Gemini) and 1% penicillin/streptomycin (Invitrogen). All cell lines and cultures were tested for mycoplasma contamination and confirmed to be negative.
Cell cytotoxicity assay
We established isogenic BAP1 KO clones directly from BAP1 wild-type (wt) cell lines by targeting the BAP1 gene utilizing CRISPR/Cas9. M14 and its isogenic BAP1 KO#1 clone were paired to screen for compounds that exhibited selective cytotoxicity toward BAP1 KO#1 clone. Cell cytotoxicity was measured with MTT assays. In brief, cells were seeded in 96-well plates and allowed to attach overnight before being incubated for 6 d with the test compounds. MTT at a final concentration of 0.5 mg/mL was then added, and the cells were further incubated for 4 h. The optical density was measured spectrophotometrically at 570 nm in a multiwell plate reader (Bio-Rad Laboratories). Each condition consisted of three replicate wells, and data are expressed as the percentage of survival compared to vehicle control (DMSO) cells. The fold change was used to compare compound selectivity for M14 BAP1 KO#1 clone over parental M14 cells. The fold change was calculated as the ratio of the survival percentage of treated M14 cells to that of treated M14 BAP1 KO#1 cells. Positive hits (BET inhibitors) were further validated in BAP1 wt and KO isogenic cancer cell lines (B16-F10, OMM2.3, and 786O). Detailed information on the tested compounds and the results are shown in Supplementary Table 1.
Colony formation assay
M14 (1500 cells per well), OMM2.3 (2000 cells per well), and VMRC-RCW (1500 cells per well) cells were seeded in six-well plates. Colonies were stained using 0.1% crystal violet and counted 10–14 days after plating.
Immunoblot
Western blotting was performed following standard methods. Antibodies against BAP1 (sc-28383, 1:100), CDK4 (sc-601, 1:1000), CDK6 (sc-7961, 1:200), RING1B (sc-101109, 1:100), BRD2 (sc-393720, 1:100), BRD3 (sc-81202, 1:100), and GAPDH (sc-25778, 1:10000) were purchased from Santa Cruz Biotechnology. Antibodies against H2AK119ub1 (8240S, 1:200000), Myc (13987S, 1:1000), Bcl-2 (15071S, 1:1000), CDK2 (2546S, 1:1000), RING1A (2820S, 1:1000), BRD4 (13440S, 1:1000), HA (2367S, 1:1000), p21 (2947T, 1:1000), and p-Rb (ser780) (9307T, 1:1000) were purchased from Cell Signaling Technology. The antibody against vinculin (V9131, 1:100,000) was purchased from Sigma-Aldrich.
Immunofluorescence
Cells on coverslips were fixed with 4% paraformaldehyde in PBS for 15 min followed by permeabilization with 0.1% Triton-X for 5 min. After blocking, the cells were stained with anti-BAP1 (SC-28383, 1:50, Santa Cruz Biotechnology), Alexa Fluor 555-conjugated secondary antibody (A31570, 1:500, Thermo Fisher Scientific) and Alexa Fluor 488 phalloidin (A12379, 1:400, Thermo Fisher Scientific). Slides were mounted with ProLong Gold antifade reagent with DAPI (P36931, Thermo Fisher Scientific) for observation. Images were captured with a Zeiss LSM880 with Airyscan confocal microscope. Images were exported from ZEN software and processed in ImageJ.
Gene deletion with the CRISPR/Cas9 system
The CRISPR/Cas9 system was used to delete genes in M14, OMM2.3, and 786O cells. The single guide RNA (sgRNA) sequences were cloned into the plasmids px459 v2 (Addgene #62988) or lentiCRISPR v2 (Addgene #52961). Sequences of the sgRNAs targeting human BAP1, BRD2, BRD3, BRD4, RING1A, and RING1B from 5′ to 3′ are provided as follows: ACCCACCCTGAGTCGCATGA (sgBAP1), GCTGCAGTAACAGCAAGGAG (sgBRD2), TCGTGGCGGTGGACATCCTC (sgBRD3), TTCAGCTTGACGGCATCCAC (sgBRD4), TCTCCTATTACCCGCTCCGT (sgRING1A), and CCCTTGACTAGACTGCAGCG (sgRING1B).
Retroviral infection and mutagenesis
Complementary DNAs (cDNAs) encoding BAP1 (BC001596, Transomic), Myc, Bcl-2, RING1A, RING1B and H2A (G152835, G157085, F110731, F105989, and F11945, respectively, Youbio Technologies) were subcloned into the pQCXIH retroviral plasmid (Clontech). Retroviral packaging and infection were performed as described previously [11]. BAP1 (C91S and K116K) and H2A (K118R and K119R) point mutation constructs were generated by site-directed mutagenesis using Q5 Hot Start High Fidelity DNA Polymerase (New England Biolabs) and the following primers: C91S-BAP1-forward, 5′-ACCCAACTCTAGCGCAACTCATGC-3′; C91S-BAP1-reverse, 5′-ATCAGCTGGTGGGCAAAG-3′; K116K-BAP1-forward, 5′-GTCGCATGAATGACTTCACCAAGGGTTTTAG-3′, K116K-BAP1-reverse, 5′-TCAGGGTGGGTCCCAGGT-3′; K118R-H2A-forward, 5′-GCTACTGCCCAGGAAGACCGAGA-3′, K118R-H2A-reverse, 5′-ACAGCCTGGATGTTGGGC-3′; and K119R-H2A-forward, 5′-ACTGCCCAAGAGGACCGAGAGTC-3′, K119R-H2A-reverse, 5′-AGCACAGCCTGGATGTTG-3′. The BAP1 K116K mutation impairs the PAM recognition site, which can avoid CRISPR/Cas9-mediated gene editing. This synonymous mutation was introduced by utilizing degeneracy of codons and used in cells stably expressing BAP1 sgRNA and Cas9 in the rescue experiments.
RNA extraction and quantitative real-time PCR with reverse transcription
Total RNA was extracted using an RNAsimple Total RNA kit (DP419, Qiagen), and reverse transcription was performed using PrimeScript RT Master Mix (RR036A, Takara). Primer sequences were obtained from PrimerBank (http://pga.mgh.harvard.edu/primerbank/). Real-time PCR was performed using GoTaq qPCR Master Mix (A6002, Promega) and an Applied LightCycler480 real-time PCR system. RT-PCR experiments were carried out with technical and biological triplicates. Relative mRNA levels were determined by normalization to endogenous GAPDH mRNA.
Cell cycle analysis
Cells were treated with OTX015 at the indicated concentrations for 48 h. Cells were then fixed with ice-cold 75% ethanol in PBS followed by incubation with 50 μg/mL propidium iodide (Sigma-Aldrich) and 10 μg/mL RNase A (Sigma-Aldrich) at 37 °C for 1 h. The distribution of cells in different phases of the cell cycle was analyzed by flow cytometry (BD FACSCanto II).
Annexin V staining for apoptosis analysis
Cells were treated with OTX015 at the indicated concentrations for 96 h. The cells were harvested, rinsed with precooled PBS, and then stained with Annexin V-FITC and PI according to the manufacturer’s protocol (Annexin V-FITC Apoptosis Detection Kit, AD10, Dojindo). Flow cytometry was used to analyze the stained cells within 1 h (BD LSRFortessa X-20).
RNA sequencing and differential expression analysis
Total RNA was extracted by an RNAsimple Total RNA kit (DP419, Qiagen) from M14 and BAP1 KO#1 cells treated with vehicle control (DMSO) or 5 μM OTX015 for 24 h. Three replicates for each sample were generated and analyzed. RNA was sent to a sequencing company (Novogene) for sequencing. The quality of sequencing reads was evaluated using FastQC. Read pseudoalignment to the human hg38 reference transcriptome and read counting were performed using Kallisto [12]. The abundance values were then imported into the R platform using the tximport package [13]. DEGs (differentially expressed genes) were identified by the DESeq2 package [14]. Genes with adjusted P < 0.05 and absolute log2-fold-change > 1 were considered significant DEGs. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the DEGs was performed using the online database KOBAS 3.0 [15].
Animal work
All animal procedures were carried out according to protocols approved by the Institutional Animal Care and Use Committee of Southern Medical University (2015-0056). Male nude mice (6 weeks old) and female C57BL/6 mice (6–8 weeks old) purchased from Guangdong Medical Laboratory Animal Center (Guangzhou, China) were used to evaluate the in vivo anticancer efficacy of OTX015 against M14, OS-RC2, VMRC-RCW and B16-F10 cells. For subcutaneous xenograft experiments, M14 (3 × 106), M14 BAP1 KO#1 (3 × 106), OS-RC2 (3 × 106), and VMRC-RCW (5 × 106) cells were grafted subcutaneously into the right back flank of nude mice. For the orthotopic allograft model, B16-F10 (5 × 105) and B16-F10 BAP1 KO#1 (5 × 105) cells were intradermally injected into the abdominal walls of C57BL/6 mice. When tumors were just palpable, mice were randomly assigned to the vehicle control (200 μL water) or OTX015 (n = 10 or 6 per group) group. OTX015 was administered by oral gavage at 50 mg/kg body weight twice daily. Tumor height and width were measured with a caliper to calculate tumor volume (=width2 × height × 0.5). Mice were sacrificed after OTX015 treatment (22 days for M14 cells; 14 days for OS-RC2 and VMRC-RCW cells; 10 days for B16-F10 cells). Tumor xenografts were excised, recorded, and fixed.
Statistical analysis
All data analysis was performed using GraphPad Prism version 6.0c for Mac (GraphPad Software, CA).
Results
Epigenetic compound library screening identifies BET inhibitors exhibiting selective cytotoxicity toward BAP1-deficient cancer cells
We performed cytotoxicity screening with an epigenetic compound library using wt and BAP1 KO M14 CM cells. We deleted BAP1 in M14 cells using CRISPR/Cas9 [16]. The epigenetic drug library has 303 compounds, including the majority of currently available inhibitors of epigenetic regulatory enzymes (Fig. 1a and Supplementary Table 1).
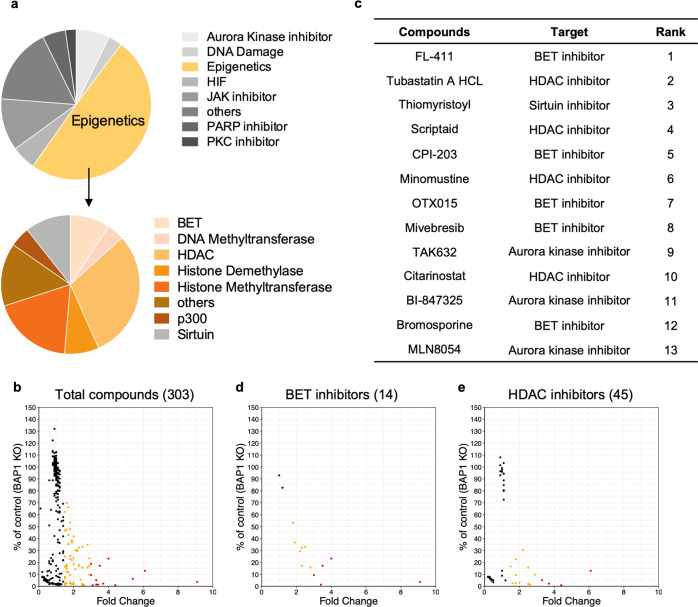
Fig. 1. Epigenetic compound library screening to identify compounds selectively suppressing the viability of BAP1-deficient cancer cells.
a Summary of compound classification in the epigenetic compound library. The library comprises 303 compounds directly targeting epigenetic enzymes and other related factors. b Scatter plot for 303 compounds grouped by fold change in selectivity for BAP1 KO#1 clone over parental M14 cells. The fold change was calculated by dividing the percentage survival of parental cells by that of M14 BAP1 KO#1 cells exposed to the individual compound at 10 μM for 6 d. Each dot represents one compound from the library, and the colors represent different fold changes (black dots: <1.5, orange dots: ≥1.5 and <3 and red dots: ≥3). c Rank of the 13 compounds displaying strong selectivity with fold change ≥3. See also Supplementary Table 1. Scatter plot for 14 BET inhibitors (d) and 45 HDAC inhibitors (e) grouped by fold change in selectivity for BAP1 KO#1 clone over parental M14 cells.
The output of the screen was the percentage survival of cells treated with individual compounds (10 μM) compared to that of cells treated with the vehicle control (DMSO). We ranked the compounds by fold change in selectivity for BAP1 KO#1 clone over wt cells. Applying a selection threshold of fold change ≥1.5, we identified 72 candidates, of which 13 exhibited strong selectivity with fold change ≥3 (Fig. 1b and Supplementary Table 1). These 13 compounds included five BET inhibitors, four HDAC inhibitors, three Aurora kinase inhibitors and one Sirtuin inhibitor (Fig. 1c). We focused our study on BET inhibitors based on the percentage hit rates for the compounds in each class. In this regard, 12 of 14 BET inhibitors showed selective cytotoxicity against BAP1 KO#1 clone. Two BET inhibitors (apabetalone and CeMMEC1) that failed to show selectivity displayed noncytotoxicity at the tested dose (Fig. 1d). In contrast, 16 of 45 HDAC inhibitors (Fig. 1e), 10 of 22 Aurora kinase inhibitors (Supplementary Fig. S1a) and one of 16 Sirtuin inhibitors (Supplementary Fig. S1b) displayed selectivity (Supplementary Table 1).
We also deleted BAP1 in OMM2.3 UM cells and 786O ccRCC cells and then determined the cellular selectivity of OTX015, an oral BET inhibitor that has entered phase I/II clinical trials (http://clinicaltrials.gov). The obtained BAP1 KO clones were verified to have both a lack of protein expression of BAP1 and elevated H2AK119ub1 (Fig. 2a). We generated BAP1 KO clones from the murine CM cell line B16-F10 in our previous study [10]. OTX015 selectively suppressed the viability of BAP1 KO cells compared with their parental M14, B16-F10, OMM2.3, and 786O cells (Fig. 2b). In addition, JQ1, a widely used BET inhibitor in laboratory applications, similarly exhibited selective cytotoxicity against the tested BAP1 KO clones (Supplementary Fig. S2). These findings demonstrate that deletion of BAP1 in CM, UM and ccRCC cells confers sensitivity to BET inhibitors in vitro.
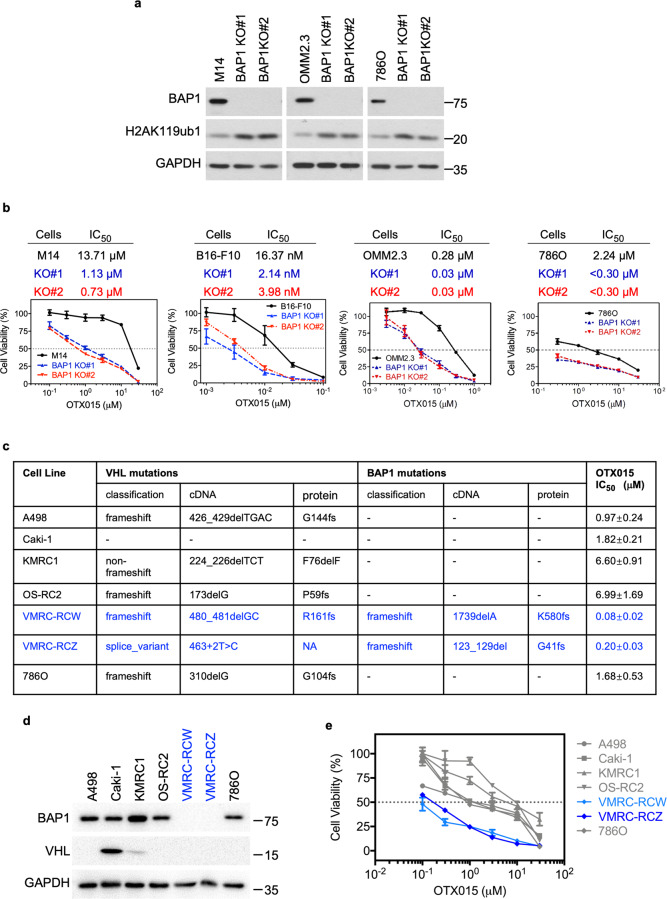
Fig. 2. BAP1 loss augments sensitivity to BET inhibitors.
a Immunoblot analysis showing the deletion of BAP1 and increased H2AK119ub1 in M14, OMM2.3, and 786O cells. Two clones for each cell line (#1 and #2) are shown. b The BET inhibitor OTX015 exhibited potent and selective cytotoxicity against BAP1-deficient M14, B16-F10, OMM2.3, and 786O cells. Cells were treated with OTX015 at the indicated concentrations for 6 days. The data are presented as the mean ± SD (n = 3) from one representative experiment out of three. c The mutation status of VHL and BAP1 as well as OTX015 IC50 values are shown for each ccRCC cell line. The mutation data are from Cell Model Passport Databases (http://cellmodelpassports.sanger.ac.uk). The IC50 values for each cell line are shown as the mean ± SD (n = 3) from three independent experiments. d Immunoblot analysis showing VHL and BAP1 expression in multiple ccRCC cell lines. e Cell viability of multiple ccRCC cell lines with wt or mutant BAP1 in response to OTX015 treatment at the indicated concentrations for 6 days. The data are presented as the mean ± SD (n = 3) from one representative experiment out of three.
We next evaluated the impact of OTX015 on the viability of a panel of ccRCC cell lines harboring wt or mutant BAP1. In this regard, BAP1-deficient ccRCC cell lines (VMRC-RCW and VMRC-RCZ) were more sensitive to OTX015 than the tested BAP1-competent ccRCC cell lines (786O, A498, Caki-1, KMRC1, and OS-RC2) (Fig. 2c–e).
On the basis of the above data, we speculate that BAP1 loss generated by gene editing or occurring in patient tumors renders cells more sensitive to BET inhibitors.
Deubiquitinating activity of BAP1 is required to reverse the sensitivity of BAP1-deficient cells to BET inhibitors
We performed rescue experiments in BAP1 KO cells. Reexpression of wt BAP1 protein (synonymous mutation K116K) decreased H2AK119ub1 and completely reversed the sensitivity of BAP1 KO M14 cells to OTX015 (Fig. 3a, b). We next assessed whether BAP1 function requires its deubiquitinase activity. wt BAP1 and a catalytically dead version (BAP1 C91S) were re-expressed at a similar level in OMM2.3 BAP1 KO clones. Consistent with previous studies [9], both versions of BAP1 were predominantly localized in the nucleus, while BAP1 C91S failed to deubiquitinate H2AK119 (Fig. 3c, d). Reexpression of wt BAP1 reversed the sensitivity of BAP1 KO OMM2.3 cells to OTX015 and JQ1, while BAP1 C91S did not exhibit any effect (Fig. 3e and Supplementary Fig. S3a), implying that BAP1 catalytic activity is required. Similarly, exogenous expression of wt BAP1 conferred resistance to OTX015 and JQ1 in BAP1-mutant VMRC-RCW cells (Fig. 3f, g and Supplementary Fig. S3b). In contrast, exogenous expression of BAP1 C91S did not alter H2AK119ub1 levels or the cellular sensitivity to OTX015 of VMRC-RCW cells (Fig. 3f, g). These data suggest that BAP1 modulates the cellular response to BET inhibitors via its deubiquitinase activity.
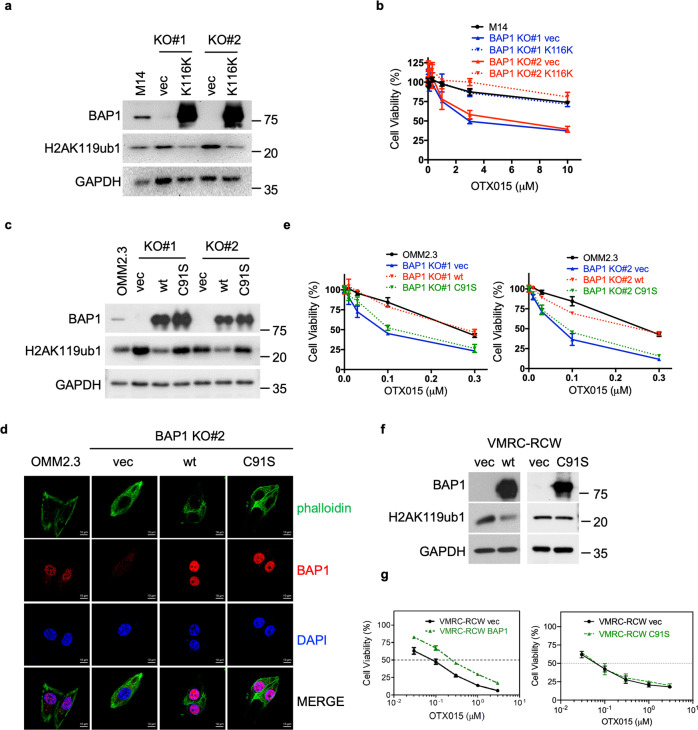
Fig. 3. BAP1 confers resistance to BET inhibitors in a manner dependent on functional deubiquitinases.
a Immunoblot analysis showing reexpression of BAP1 (with the synonymous mutation K116K) in M14 BAP1 KO cells. b Reintroduction of BAP1 completely reversed the OTX015 sensitivity of M14 BAP1 KO cells. Cells were treated with OTX015 at the indicated concentrations for 6 days. The data are presented as the mean ± SD (n = 3) from one representative experiment out of three. c Immunoblot analysis showing reexpression of wild-type (wt) or catalytically dead (C91S) BAP1 in OMM2.3 BAP1 KO cells. d Representative immunofluorescence images showing nuclear localization of BAP1 wt and BAP1 C91S in OMM2.3 cells. e Reintroduction of wt but not BAP1 C91S reversed the OTX015 sensitivity of OMM2.3 BAP1 KO cells. Cells were treated with OTX015 at the indicated concentrations for 6 days. The data are presented as the mean ± SD (n = 3) from one representative experiment out of three. f, g Exogenous expression of wild-type BAP1 but not mutant BAP1 (C91S) induced OTX015 resistance in VMRC-RCW cells carrying BAP1 nonsense mutation. Immunoblot analysis showing the expression of BAP1 in VMRC-RCW cells. Cells were treated with OTX015 at the indicated concentrations for 6 days. The data are presented as the mean ± SD (n = 3) from one representative experiment out of three.
OTX015 exhibits selective cytotoxicity in BAP1-deficient cells via transcriptional repression of genes critical to cell survival and proliferation
To explore the molecular mechanisms by which BAP1 deletion confers sensitivity to BET inhibitors, we performed RNA-sequencing analysis of parental M14 and BAP1 KO#1 clone with or without OTX015 (5 μM) treatment for 24 h. BAP1 deletion resulted in preferential downregulation of genes (1070 downregulated genes of 1474 total DEGs; Supplementary Fig. S4a and Supplementary Table 2), consistent with the reported gene activation function of BAP1 [9]. We noticed that OTX015 similarly caused many genes to be suppressed or upregulated in BAP1 KO#1 and parental M14 cells, yielding a Pearson correlation coefficient of 0.64 (Supplementary Fig. S4b). We identified 2388 DEGs between OTX015-treated and OTX015-untreated M14 cells and 3549 DEGs between OTX015-treated and OTX015-untreated M14 BAP1 KO#1 clone. Strikingly, M14 BAP1 KO#1 clone showed more dramatic transcriptome alterations in response to OTX015 treatment (781 uniquely downregulated DEGs in BAP1 KO#1 clone versus 303 in M14 cells; 1021 uniquely upregulated DEGs in BAP1 KO#1 clone versus 338 in M14 cells) (Fig. 4a and Supplementary Table 2).
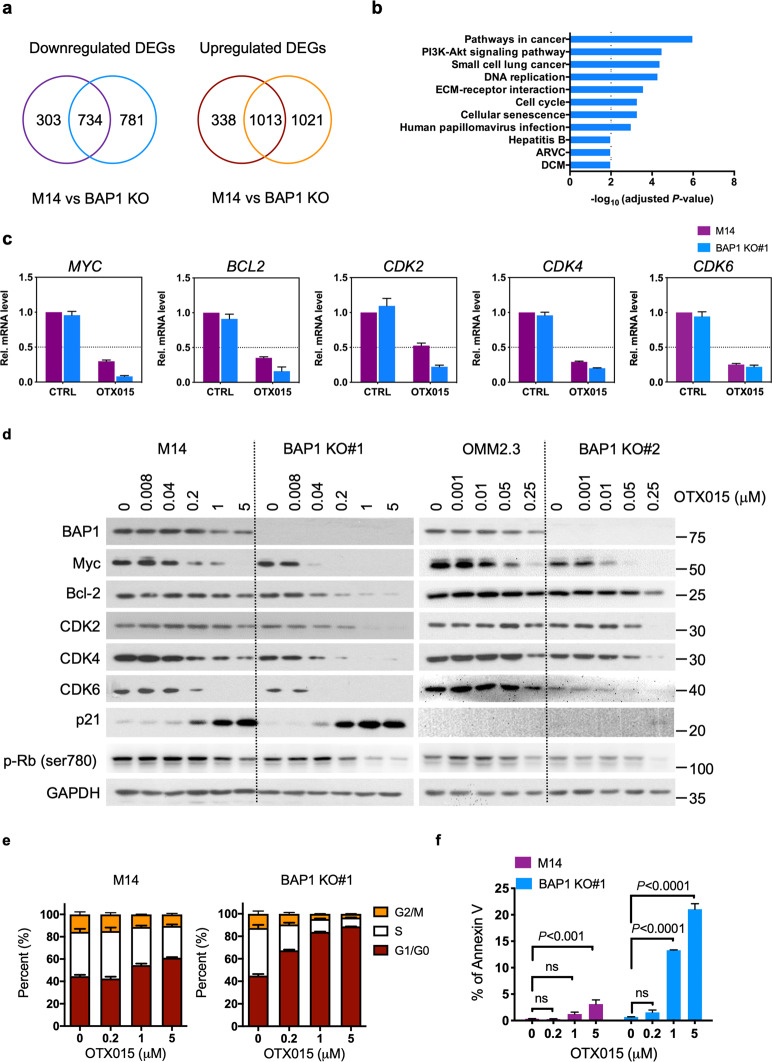
Fig. 4. Functional consequences of BAP1 deletion on the transcriptome, cell cycle progression and apoptosis in response to OTX015 treatment.
a Venn diagram showing the overlap between differentially expressed genes (DEGs) downregulated or upregulated in parental M14 and BAP1 KO#1 cells upon 24 h OTX015 treatment (5 μM). b KEGG pathway enrichment analysis of the downregulated DEGs in M14 BAP1 KO#1 cells upon 24 h OTX015 treatment (5 μM). The bars represent the enrichment scores as −log10 (adjusted P values). Pathways with adjusted P value <0.01 are shown. c The expression levels of the indicated genes (downregulated DEGs in M14 BAP1 KO#1 cells upon OTX015 treatment) were determined by qRT–PCR. Expression was normalized to GAPDH expression. The data are presented as the mean ± SD (n = 3) from one representative experiment out of three. d Immunoblot analysis was performed to measure the indicated protein expression upon 48 h OTX015 treatment in BAP1 KO clones and their isogenic parental cells. OTX015 concentrations are indicated in the figure. e Parental M14 or BAP1 KO#1 cells were treated with OTX015 at the indicated concentrations for 48 h, and their DNA content was determined by flow cytometry analysis. The data are presented as the mean ± SD (n = 3) from one representative experiment out of three. f Parental M14 or BAP1 KO#1 cells were treated with OTX015 at the indicated concentrations for 96 h. Cell apoptosis was assessed by phosphatidylserine externalization and binding of Annexin V-FITC. The data are presented as the mean ± SD (n = 3) from one representative experiment out of three. One-way ANOVA test was used.
To understand the cellular processes involving the identified DEGs, we performed enrichment analysis of KEGG pathways. Downregulated DEGs in M14 BAP1 KO#1 clone upon OTX015 treatment were mainly enriched in terms related to cell survival and proliferation, including the PI3K-Akt signaling pathway, DNA replication, ECM–receptor interactions, the cell cycle, and cellular senescence (Fig. 4b and Supplementary Table 3). In contrast, no significantly enriched terms were identified for the downregulated DEGs identified in M14 cells treated with OTX015 (Supplementary Table 3). The upregulated DEGs in M14 cells and BAP1 KO#1 clone upon OTX015 exposure were associated with similar terms (Supplementary Fig. S4c, d; Supplementary Table 3). Quantitative PCR with reverse transcription analysis confirmed that OTX015 decreased the expression of most selected genes (MYC, BCL-2, CKD2 and CKD4) in M14 BAP1 KO#1 clone to a greater extent than in M14 cells (Fig. 4c). In agreement with their mRNA downregulation, the reduction in the protein levels of Myc, Bcl-2, CKD2, CDK4, and CDK6 by OTX015 was much greater in BAP1 KO#1 clone than in parental M14 cells at the indicated concentrations. Similarly, a more dramatic reduction in these proteins was observed in BAP1 KO#2 clone than in parental OMM2.3 cells (Fig. 4d). We also determined the expression of p21 and p-Rb (Ser780), cell cycle regulatory proteins mediating the G1-to-S-phase transition [17]. The protein levels of p21 and p-Rb (Ser780) were more profoundly affected by OTX015 in BAP1 KO#1 clone than in parental M14 cells. The expression of p-Rb (Ser780) was also decreased more by OTX015 in BAP1 KO#2 clone than in parental OMM2.3 cells. The expression of p21 was weak or undetectable in OMM2.3 cells with or without OTX015 treatment, suggesting that p21 might not contribute to cell cycle arrest in this cell line (Fig. 4d).
Since the enriched pathways were closely associated with cell proliferation and cell survival, we examined whether OTX015 affects cell cycle progression and apoptosis. OTX015 dramatically increased the percentage of cells in G1/G0 phase in a dose-dependent manner in M14 BAP1 KO#1 clone, indicating that OTX015 induced G1/G0 phase arrest. In contrast, OTX015 induced much milder G1/G0 phase arrest in parental M14 cells (Fig. 4e and Supplementary Fig. S5a). Furthermore, BAP1 deletion rendered cells more susceptible to OTX015-induced apoptosis (Fig. 4f and Supplementary Fig. S5b). We also employed BAP1-proficient cell lines (KMRC1 and OS-RC2) and BAP1-deficient cell lines (VMRC-RCW and VMRC-RCZ). OTX015-induced G1/G0 phase arrest and apoptosis were observed in VMRC-RCW and VMRC-RCZ cells but not in KMRC1 and OS-RC2 cells (Supplementary Figs. S6, S7). Taken together, these data indicate that BAP1-deficient cells are more susceptible to OTX015-induced transcriptional modulation, G1/G0 phase arrest and apoptosis.
Exogenous expression of Myc rescues BAP1-deficient cells from OTX015-induced cytotoxicity
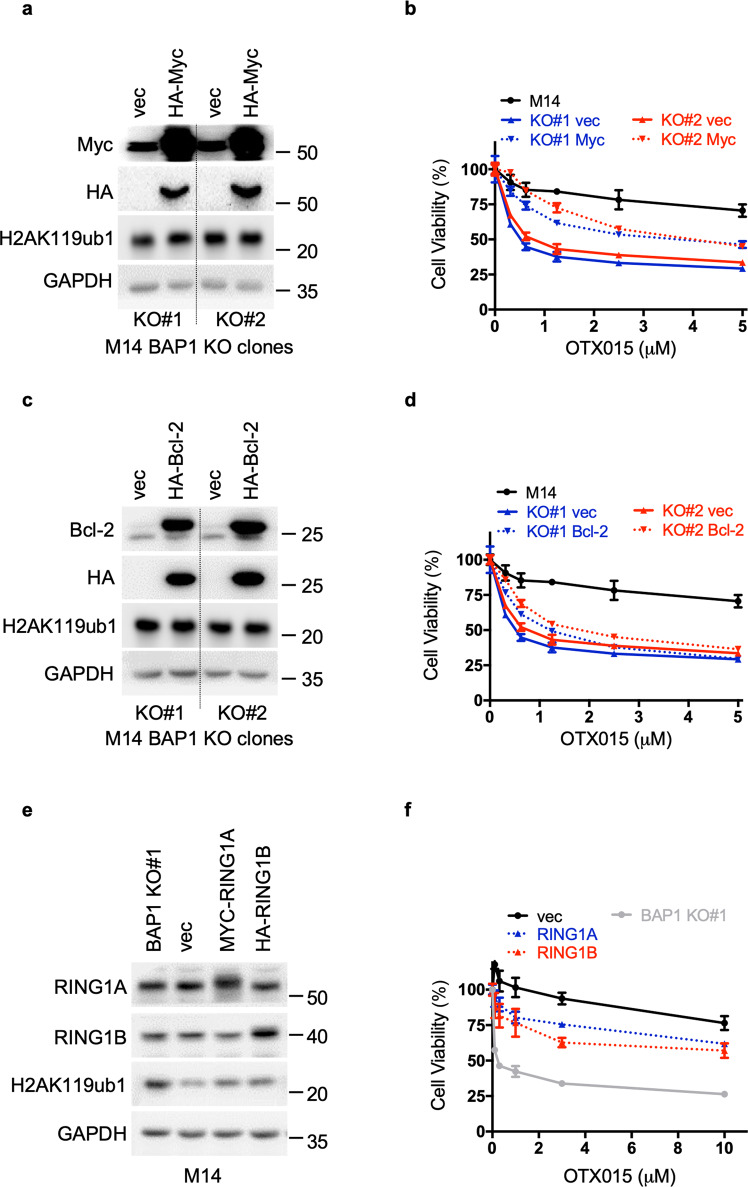
Since Myc is a well-known target of BET inhibitors [18], we tested whether exogenous expression of Myc could rescue BAP1-deficient cells from the cytotoxic effects of OTX015. Two BAP1 KO clones from M14 cells were stably transduced with retroviral vectors that express Myc cDNA, and the expression was confirmed by Western blotting (Fig. 5a). Ectopic expression of Myc substantially but not completely rescued BAP1 KO clones from the deleterious effects of OTX015 (Fig. 5b). In contrast, ectopic Bcl-2 expression displayed a minor effect (Fig. 5c, d). Consistently, ectopic expression of Myc significantly attenuated the G1/G0 phase arrest and apoptosis induced by OTX015 treatment (Supplementary Fig. S8). In addition, ectopic expression of Myc or Bcl-2 rendered VMRC-RCW cells less sensitive to OTX015 (Supplementary Fig. S9). These data suggest that downregulation of Myc and Bcl-2 contributes to the cytotoxic effect of BET inhibitors on BAP1-deficient cells.
Fig. 5. Exogenous expression of Myc protects BAP1-deficient M14 cells from OTX015 inhibition, while ectopic expression of RING1A/1B enhances the sensitivity of M14 cells to OTX015.
M14 BAP1 KO clones (#1 and #2) were stably transduced with control (pQCXIH-vec), Myc (pQCXIH-HA-Myc) (a) or Bcl-2 (pQCXIH-HA-Bcl-2) (c) retroviruses. Myc and Bcl-2 protein levels were determined by immunoblot analysis. Ectopic expression of Myc (b) or Bcl-2 (d) rescued M14 BAP1 KO clones (#1 and #2) from the cytotoxic effects of OTX015 to different extents. Cells were treated with OTX015 at the indicated concentrations for 6 days. The data are presented as the mean ± SD (n = 3) from one representative experiment out of three. e Immunoblot analysis showing ectopic expression of RING1A and RING1B in M14 cells. Cells were stably transduced with control (pQCXIH-vec), RING1A (pQCXIH-MYC-RING1A), or RING1B (pQCXIH-HA-RING1B) retroviruses. f Ectopic expression of RING1A or RING1B in M14 cells augmented sensitivity to OTX015. Cells were treated with OTX015 at the indicated concentrations for 6 days. The data are presented as the mean ± SD (n = 3) from one representative experiment out of three.
Ectopic expression of RING1A or RING1B renders cells more sensitive to OTX015
PRC1 contains the RING finger domain proteins RING1A (encoded by RING1) and RING1B (encoded by RNF2), which mediate the monoubiquitination of H2AK119 via E3 ubiquitin ligase activity [9]. As expected, deletion of either RING1A or RING1B by CRISPR/Cas9 decreased H2AK119ub1 levels (Supplementary Fig. S10a), while ectopic expression of RING1A or RING1B increased H2AK119ub1 levels (Fig. 5e). Ectopic expression of RING1A or RING1B comparable to endogenous levels rendered M14 cells more sensitive to OTX015 (Fig. 5e, f). Of note, the enhancing effects of ectopic expression of RING1A or RING1B on cellular sensitivity to OTX015 were weaker than those of BAP1 deletion, which were proportional to their different potencies in affecting H2AK119 ubiquitination levels (Fig. 5e, f).
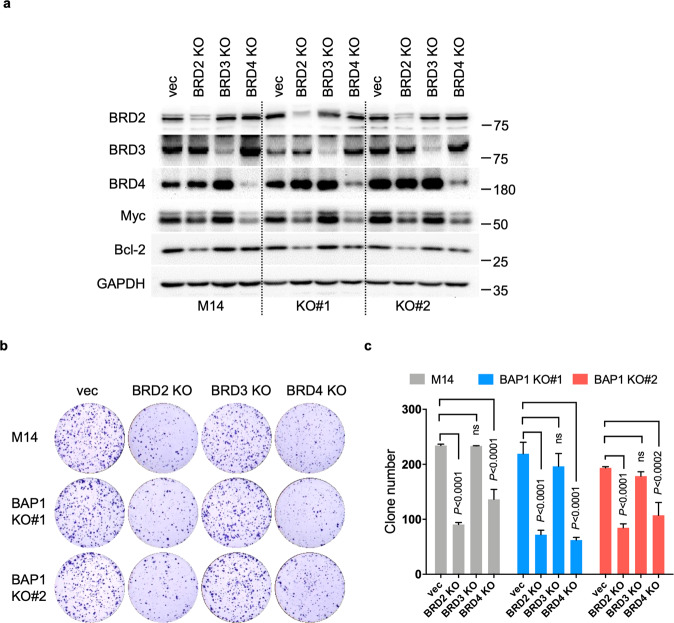
Deletion of BRD2 or BRD4 but not BRD3 suppresses colony formation
We investigated inhibition of which BRD member or members could phenocopy the cytotoxic action of BET inhibitors. Deletion of BRD2 or BRD4 decreased Myc protein expression, while BRD4 deletion exhibited a stronger effect. BRD2 deletion exerted a stronger effect on reducing Bcl-2 than BRD4 deletion. In contrast, BRD3 deletion did not alter the protein levels of Myc and Bcl-2 (Fig. 6a). In agreement with these observations, BRD2 or BRD4 deletion obviously decreased colony formation in BAP1 KO clones and parental M14 cells; however, BRD3 deletion exhibited no effect (Fig. 6b, c). The downregulation of Myc and Bcl-2 as well as the reduction in colony formation by BRD2 or BRD4 deletion were similarly observed in VMRC-RCW cells (Supplementary Fig. S11).
Fig. 6. Deletion of BRD2 or BRD4 reduces colony formation.
a Immunoblot analysis showing the specific deletion of BRD2, BRD3, or BRD4 by CRISPR/Cas9 in M14 cells. Images (b) and corresponding quantification of colony formation assays (c) assessing the effects of BRD2/3/4 deletion on the colony formation of M14 cells. The data are presented as the mean ± SD (n = 3) from one representative experiment out of three. One-way ANOVA test was used.
We noticed that deletion of BRD2 or BRD4 induced no selective cytotoxicity toward BAP1 KO cells but reduced colony-forming capacity in both parental M14 and BAP1 KO cells to a similar extent. It has been reported that cells that are resistant to BET inhibitors are still sensitive to genetic silencing of BRD4, suggesting that BRD4 has both bromodomain-dependent and bromodomain-independent roles [19]. Thus, a bromodomain-independent mechanism may also contribute to the cytotoxic effects induced by BRD2 or BRD4 deletion in parental M14 and BAP1 KO cells.
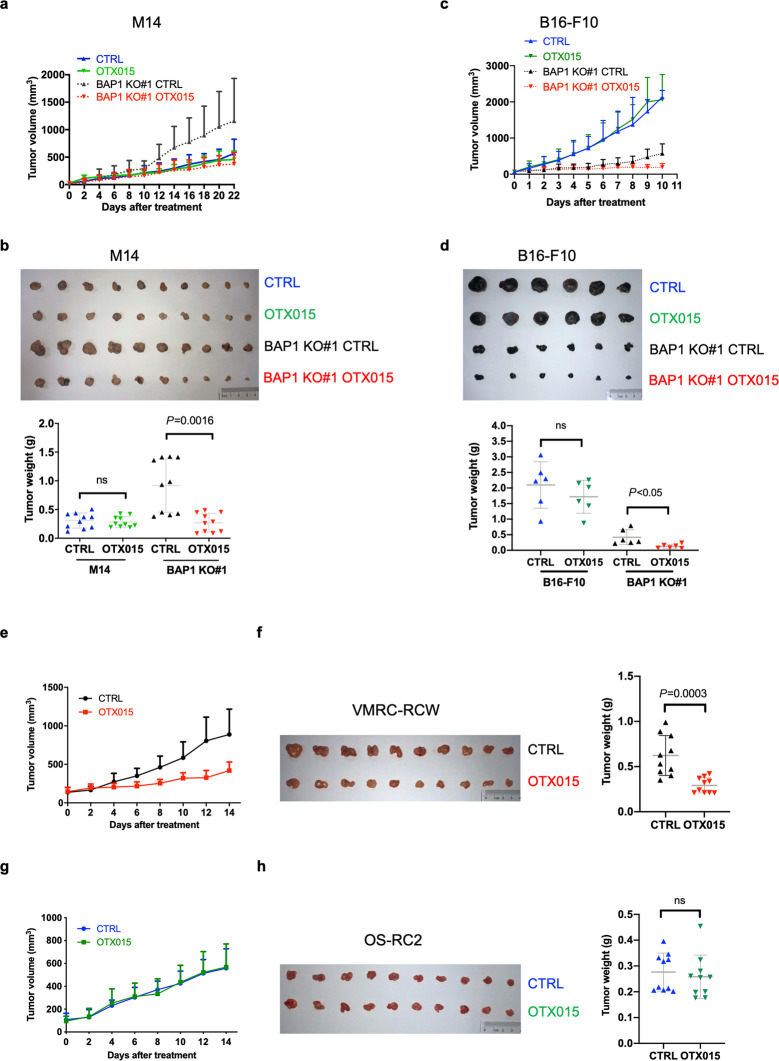
BAP1-deficient xenografts and allografts are sensitive to OTX015 treatment
To assess the effects of OTX015 on BAP1-deficient tumor growth, we first performed xenograft experiments in nude mice. When injected into athymic nude mice, M14 BAP1 KO cells formed tumors that were much larger than those obtained with parental M14 cells (Fig. 7a, b). Notably, OTX015 reduced both the size and weight of BAP1-deficient tumors compared to those of tumors treated with the vehicle control, whereas their wt counterparts were not responsive to OTX015 treatment (Fig. 7a, b). We also tested the antitumor effect of OTX015 by using a syngeneic allograft model. In contrast to the result from M14 cells, BAP1 deletion in B16-F10 cells strongly inhibited tumor growth in C57BL/6 mice. Remarkably, we still observed a suppressive effect of OTX015 on BAP1 KO#1 B16-F10 tumor growth but not on the tumor growth of wt cells (Fig. 7c, d). We next assessed the effects of OTX015 in nude mice xenografted with BAP1-proficient OS-RC2 cells and BAP1-deficient VMRC-RCW cells. Similarly, OTX015 treatment significantly suppressed the growth of VMRC-RCW xenografts but not of OS-RC2 xenografts in nude mice (Fig. 7e–h). These data indicate that BET inhibitors represent potential therapeutic drugs for BAP1-deficient tumors.
Fig. 7. OTX015 selectively suppresses tumor growth of BAP1-deficient cells in vivo.
a Tumor size of tumors formed from parental and BAP1 KO#1 M14 cells subcutaneously implanted into nude mice and treated with either vehicle or OTX015. The data are presented as the mean ± SD (n = 10). b Tumor weight of parental and BAP1 KO#1 M14 xenografts on day 22 posttreatment with vehicle or OTX015. The data are presented as the mean ± SD (n = 10). Student’s t test was used. c Tumor size of tumors formed from parental and BAP1 KO#1 B16-F10 cells intradermally injected into the abdominal walls of C57BL/6 mice and treated with either vehicle or OTX015. The data are presented as the mean ± SD (n = 6). d Tumor weight of parental and BAP1 KO#1 B16-F10 allografts on day 10 posttreatment with vehicle or OTX015. The data are presented as the mean ± SD (n = 6). Student’s t test was used. e Tumor size of tumors formed from BAP1-deficient VMRC-RCW cells subcutaneously implanted into nude mice and treated with either vehicle or OTX015. The data are presented as the mean ± SD (n = 10). f Tumor weight of VMRC-RCW xenografts on day 14 posttreatment with vehicle or OTX015. The data are presented as the mean ± SD (n = 10). Student’s t test was used. g Tumor size of tumors formed from BAP1-proficient OS-RC2 cells subcutaneously implanted into nude mice and treated with either vehicle or OTX015. The data are presented as the mean ± SD (n = 10). h Tumor weight of OS-RC2 xenografts on day 14 posttreatment with vehicle or OTX015. The data are presented as the mean ± SD (n = 10). Student’s t test was used.
Discussion
Individualized therapies have impressively improved the prognosis for selected patient populations carrying certain genetic or epigenetic aberrations, although this strategy has yet to impact cancers with BAP1 mutations. Our data suggest that BAP1-mutated cancers such as CM, UM, or ccRCC could benefit from BET inhibitors. We propose that the underlying mechanism involves transcriptional downregulation of genes critical for proliferation and survival upon the combination of BAP1 loss and BRD2/4 inhibition.
BAP1 has multifaceted functions. Thus, it is not unexpected that BAP1 deletion would sensitize cancer cells to some screened compounds with different mechanisms of action, including BET inhibitors, PARP inhibitors (Supplementary Fig. S1c), HDAC inhibitors and Aurora kinase inhibitors. A previous study demonstrated a link between BAP1 status and sensitivity to PARP inhibitors [20] and HDAC inhibitors or shRNAs [21, 22]. According to our screening data, PARP inhibitors either had no selectivity or showed only moderately selective inhibitory activity in BAP1 KO M14 cells compared to parental cells. We also found that more than half of the tested HDAC inhibitors (29 out of 45) failed to display selectivity toward BAP1 KO cells. Considering the tested HDAC inhibitors, including nonselective broad-spectrum and selective inhibitors, inhibition of different HDAC subtypes or classes with varying efficiency might contribute to the observed heterogeneity. It is important to identify which HDAC subtype/subtypes when inhibited could preferentially suppress BAP1-deficient cancer cells. Of note, our screening data suggest that BET inhibitors are a promising individualized therapy for BAP1-deficient cancers given that all the tested BET inhibitors exhibited moderate to high selectivity toward BAP1 KO M14 cells, in which they showed cytotoxicity.
The bromodomain and extraterminal (BET) domain family proteins BRD2, BRD3, BRD4, and BRDT bind via their tandem bromodomains to acetylated lysine residues, particularly in histones, and thereby have important roles in transcription [23]. Unlike the other three BET proteins that are ubiquitously expressed, BRDT expression is normally testis-specific [24]. BET inhibitors occupy the acetyl-lysine binding pocket of BET proteins, preventing BET bromodomains from binding to acetylated histone tails and thus from localizing to chromatin. As a result, BET inhibitors cause extensive transcriptional changes [25]. Although our data show a positive correlation between transcriptome alterations of M14 BAP1 KO cells and those of parental cells upon OTX015 treatment, the number of upregulated and downregulated DEGs in BAP1 KO cells is much higher than that in parental cells, indicating that the loss of BAP1 renders cells more vulnerable to OTX015-induced transcriptional alteration. The upregulated DEGs in BAP1 KO and parental cells were found to be enriched in similar pathways; however, enrichment of the downregulated DEGs in pathways related to cell proliferation and survival was exclusively found in BAP1 KO cells. This finding suggests that repressed DEGs account for the heightened sensitivity of BAP1-deficient cells to BET inhibitors. In agreement with its different abilities to repress transcription, OTX015 treatment resulted in reduced levels of multiple proteins, including Myc and Bcl-2, as well as cell cycle arrest and apoptosis in BAP1 KO cells at concentrations at which OTX015 failed to exert effects on parental cells. BET inhibitors have previously been shown to exhibit broad antiproliferative effects in various cancer types through either Myc-dependent or Myc-independent mechanisms [18, 26, 27]. Our data show that ectopic expression of Myc significantly, although not completely, rescued the cytotoxic effect of OTX015 on BAP1 KO cells, while overexpression of Bcl-2 exerted a marginal rescue effect. We therefore conclude that BET inhibitors exert cytotoxicity against BAP1-deficient cancer cells primarily via transcriptional repression of Myc and possibly other genes.
Despite their similarity, BRD2, BRD3, and BRD4 have been shown to have some overlap in their chromatin binding sites by genome-wide analysis, but significant differences in these sites could contribute to their differing functions [28, 29]. In line with this, our data show that BRD4 deletion preferentially decreased Myc over Bcl-2, while BRD2 deletion showed the opposite effect. BRD3 deletion exhibited no effect. Concordantly, deletion of either BRD2 or BRD4 but not BRD3 inhibited clonogenic growth. Although our data indicate that BRD3 deletion exerted no effect on cell proliferation, a recent study suggests a growth-repressive function of BRD3, raising the concern that pan-BET inhibitors may have unintended consequences of inhibiting BRD3 in parallel with inhibiting the desired BRD4 functions [30]. Thus, we propose that inhibitors that specifically target BRD2/4 may have an advantage in this situation.
Some targeted therapies are only effective for a limited number of tumor types or even a certain subtype within a tumor type due to genetic and epigenetic heterogeneity seen in tumors. For instance, BAP1-deficient mesothelioma cells were found to be more sensitive to EZH2 inhibition [31], whereas BAP1-deficient UM cells were not [32]. Moreover, GNAQ/GNA11-mutant UM cells were more sensitive to BET inhibitors than UM cells with wt GNAQ and GNA11 [26]. Our data demonstrate that BET inhibitors preferentially suppress BAP1-deficient CM, UM, and ccRCC cells. These findings suggest that the association between BAP1 deficiency and sensitivity to BET inhibitors might be highly penetrant across different backgrounds. It would be interesting to determine whether other tumor types with BAP1 mutations are more sensitive to BET inhibitors. Notably, the specific phenotype that results from BAP1 loss has been demonstrated to be context- and cell-dependent [2, 33]. We also observed that BAP1 deletion exerted different effects on the tumor growth of M14 and B16-F10 cells. However, more importantly, OTX015 treatment suppressed the in vivo growth of BAP1-deficient M14 and B16-F10 cells regardless of the effect of BAP1 deletion on tumor growth.
It should be noted that our hypothesis that the hyperubiquitination status of H2AK119 might contribute to BAP1 loss-induced sensitivity to OTX015 mainly relies on indirect evidence. Indeed, it is challenging to definitively establish the causal link between BAP1 deletion-induced high H2AK119ub1 levels and OTX015 sensitivity. A typical approach to address this issue would investigate whether BAP1 loss-induced OTX015 sensitivity is attenuated in H2AK119R (nonubiquitinated) mutation-expressing cells compared to H2AK118R mutation-expressing cells or H2A wt cells. However, this approach is rarely used in the epigenetic field to study the functional relevance of a histone modification (including H2AK119ub1), mainly because of the abundance of histone proteins and the potential of introducing histone mutants to induce undesired global effects [34]. In this respect, we found it extremely challenging to overexpress H2AK119R to sufficient levels to efficiently suppress H2AK119ub1 expression (Supplementary Fig. S12), which has also been reported by a previous study [34]. More importantly, genome-wide analyses have shown that BAP1 and RING1B occupy similar regions (i.e., the Bcl-2 promoter) across the genome [2]. As expected, ectopic expression of H2AK119R globally diminished H2AK119ub1 but did not specifically diminish H2AK119ub1 induced by BAP1 loss or RING1A/1B overexpression. Interestingly, the H2AK119ub1 level was decreased upon OTX015 treatment, possibly due to the downregulation of RING1A and RING1B (Supplementary Fig. S10b, c), implying a complex association between H2AK119ub1 and the cellular response to OTX015. Future studies to compare dynamic H2AK119ub1 occupancy on the promoters of critically regulated genes (i.e., Myc and Bcl-2) by chromatin immunoprecipitation (ChIP) sequencing upon OTX015 treatment in BAP1-deficient and wt cells will shed new light on our hypothesis.
BAP1 alterations (including genetic and mRNA alterations) were mutually exclusive with RING1A mRNA upregulation (P < 0.001) and RING1B mRNA upregulation (P = 0.0031) in the TCGA UM cohort (Supplementary Fig. S13a). Although the mechanism underlying this mutual exclusivity is unclear, the findings suggest that a considerable number of UM patients with RING1A or RING1B upregulation in the absence of BAP1 alterations may also benefit from treatment with BET inhibitors. Strikingly, a pancancer analysis of genetic alterations of BAP1, RING1A, and RING1B revealed that the major genetic aberration of RING1A or RING1B is amplification, which seems to occur primarily in tumors without BAP1 alterations among all available TCGA cohorts (10950 cases in 32 studies) (Supplementary Fig. S13b). Accumulating evidence demonstrates that elevated RING1A or RING1B expression contributes to cancer initiation and progression [35, 36]. However, to date, there are no therapeutic drugs that target RING1A or RING1B aberrations. It would be interesting to explore the potential application of BET inhibitors for cancers with RING1A or RING1B upregulation.
More than 20 clinical trials have been initiated to investigate the efficacy of BET inhibitors in a wide range of cancer types (http://clinicaltrials.gov), yielding mixed results [37]. BAP1 immunohistochemistry provides a rapid, reliable and economical way to screen for biallelic BAP1 inactivating mutations in patients [38]. Our findings suggest that BAP1 mutation status stratification may improve drug response rates in ongoing clinical trials exploring the use of BET inhibitor therapy in cancers, especially CM, UM, and ccRCC, in which BAP1 status is not considered.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81973357), Guangdong Basic and Applied Basic Research Foundation (No. 2020A1515010027), Wu Jieping Medical Foundation (No. 320.6750.2020-10-120) and Bethune Charitable Foundation Research Program (B-19-H-20200622). We thank Dr. Kun-Liang Guan (University of California, San Diego) for critically reading and revising the paper.
Author contributions
YYX and ZLR substantially contributed to designing the study, performing experiments, and analyzing the data. XLL, GMZ, SSH, WHS, LXY, and XL performed the experiments and analyzed the data. SWL provided experimental resources. YLL and LY conceived and designed the study and experiments, assembled and interpreted the data, wrote and revised the paper, and approved the paper. All authors read and approved the final paper.
Data availability
The RNA-seq data have been deposited in the National Center for Biotechnology Information (NCBI) via the Sequence Read Archive (SRA) database repository under the accession code PRJNA629640. All other relevant data supporting the findings of this study are available upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yu-yan Xu, Zhong-lu Ren
Contributor Information
Yi-lei Li, Email: lei@smu.edu.cn.
Le Yu, Email: yulezy@smu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-021-00783-5.
References
- 1.Carbone M, Harbour JW, Brugarolas J, Bononi A, Pagano I. Biological mechanisms and clinical significance of BAP1 mutations in human cancer. Cancer Discov. 2020;10:1103–20.. doi: 10.1158/2159-8290.CD-19-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He M, Chaurushiya MS, Webster JD, Kummerfeld S, Reja R, Chaudhuri S, et al. Intrinsic apoptosis shapes the tumor spectrum linked to inactivation of the deubiquitinase BAP1. Science. 2019;364:283–5. doi: 10.1126/science.aav4902. [DOI] [PubMed] [Google Scholar]
- 3.Rabbie R, Ferguson P, Molina-Aguilar C, Adams D, Robles-Espinoza C. Melanoma subtypes: genomic profiles, prognostic molecular markers and therapeutic possibilities. J Pathol. 2019;247:539–51. doi: 10.1002/path.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jager MJ, Shields CL, Cebulla CM, Abdel-Rahman MH, Grossniklaus HE, Stern MH, et al. Uveal melanoma. Nat Rev Dis Prim. 2020;6:24. doi: 10.1038/s41572-020-0158-0. [DOI] [PubMed] [Google Scholar]
- 5.Robertson A, Shih J, Yau C, Gibb E, Oba J, Mungall K, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32:204–20.e15. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dizman N, Philip E, Pal S. Genomic profiling in renal cell carcinoma. Nat Rev Nephrol. 2020;16:435–51. doi: 10.1038/s41581-020-0301-x. [DOI] [PubMed] [Google Scholar]
- 7.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. BAP1 and cancer. Nat Rev Cancer. 2013;13:153–9. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campagne A, Lee MK, Zielinski D, Michaud A, Le Corre S, Dingli F, et al. BAP1 complex promotes transcription by opposing PRC1-mediated H2A ubiquitylation. Nat Commun. 2019;10:348. doi: 10.1038/s41467-018-08255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo X, Xu Y, Li Y, Zhang G, Huang S, Liu X, et al. BAP1 deletion abrogates growth and metastasis of murine cutaneous melanoma. Melanoma Res. 2021;31:119–29.. doi: 10.1097/CMR.0000000000000714. [DOI] [PubMed] [Google Scholar]
- 11.Yu FX, Luo J, Mo JS, Liu G, Kim YC, Meng Z, et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell. 2014;25:822–30. doi: 10.1016/j.ccr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bray NL, Pimentel H, Melsted P, Pachter L. Erratum: near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:888. doi: 10.1038/nbt0816-888d. [DOI] [PubMed] [Google Scholar]
- 13.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–22. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308.. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–7. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- 18.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108:16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529:413–7. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, Pak H, Hammond-Martel I, Ghram M, Rodrigue A, Daou S, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci USA. 2014;111:285–90. doi: 10.1073/pnas.1309085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuznetsov JN, Aguero TH. BAP1 regulates epigenetic switch from pluripotency to differentiation in developmental lineages giving rise to BAP1-mutant cancers. Sci Adv. 2019;5:eaax1738. doi: 10.1126/sciadv.aax1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landreville S, Agapova OA, Matatall KA, Kneass ZT, Onken MD, Lee RS, et al. Histone deacetylase inhibitors induce growth arrest and differentiation in uveal melanoma. Clin Cancer Res. 2012;18:408–16. doi: 10.1158/1078-0432.CCR-11-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujisawa T, Filippakopoulos P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat Rev Mol Cell Biol. 2017;18:246–62.. doi: 10.1038/nrm.2016.143. [DOI] [PubMed] [Google Scholar]
- 24.Jones MH, Numata M, Shimane M. Identification and characterization of BRDT: a testis-specific gene related to the bromodomain genes RING3 and Drosophila fsh. Genomics. 1997;45:529–34. doi: 10.1006/geno.1997.5000. [DOI] [PubMed] [Google Scholar]
- 25.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13:337–56. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 26.Ambrosini G, Sawle AD, Musi E, Schwartz GK. BRD4-targeted therapy induces Myc-independent cytotoxicity in Gnaq/11-mutatant uveal melanoma cells. Oncotarget. 2015;6:33397–409. doi: 10.18632/oncotarget.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci USA. 2012;109:19408–13. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB, et al. Genome-wide localization of small molecules. Nat Biotechnol. 2014;32:92–6. doi: 10.1038/nbt.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung M, Gelato KA, Fernández-Montalván A, Siegel S, Haendler B. Targeting BET bromodomains for cancer treatment. Epigenomics. 2015;7:487–501. doi: 10.2217/epi.14.91. [DOI] [PubMed] [Google Scholar]
- 30.Lambert JP, Picaud S, Fujisawa T, Hou H, Savitsky P, Uusküla-Reimand L, et al. Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains. Mol Cell. 2019;73:621–38.e17. doi: 10.1016/j.molcel.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lafave LM, Béguelin W, Koche R, Teater M, Spitzer B, Chramiec A, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med. 2015;21:1344–9. doi: 10.1038/nm.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoumacher M, Le Corre S, Houy A, Mulugeta E, Stern MH, Roman-Roman S, et al. Uveal melanoma cells are resistant to EZH2 inhibition regardless of BAP1 status. Nat Med. 2016;22:577–8. doi: 10.1038/nm.4098. [DOI] [PubMed] [Google Scholar]
- 33.Kumar R, Taylor M, Miao B, Ji Z, Njauw JC, Jönsson G, et al. BAP1 has a survival role in cutaneous melanoma. J Invest Dermatol. 2015;135:1089–97.. doi: 10.1038/jid.2014.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Koppula P, Gan B. Regulation of H2A ubiquitination and SLC7A11 expression by BAP1 and PRC1. Cell Cycle. 2019;18:773–83.. doi: 10.1080/15384101.2019.1597506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen J, Li P, Shao X, Yang Y, Liu X, Feng M, et al. The E3 ligase RING1 targets p53 for degradation and promotes cancer cell proliferation and survival. Cancer Res. 2018;78:359–71. doi: 10.1158/0008-5472.CAN-17-1805. [DOI] [PubMed] [Google Scholar]
- 36.Vidal M. Role of polycomb proteins Ring1A and Ring1B in the epigenetic regulation of gene expression. Int J Dev Biol. 2009;53:355–70. doi: 10.1387/ijdb.082690mv. [DOI] [PubMed] [Google Scholar]
- 37.Alqahtani A, Choucair K, Ashraf M, Hammouda DM, Alloghbi A, Khan T, et al. Bromodomain and extra-terminal motif inhibitors: a review of preclinical and clinical advances in cancer therapy. Future Sci OA. 2019;5:Fso372. doi: 10.4155/fsoa-2018-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalirai H, Dodson A, Faqir S, Damato BE, Coupland SE. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. Br J Cancer. 2014;111:1373–80. doi: 10.1038/bjc.2014.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in the National Center for Biotechnology Information (NCBI) via the Sequence Read Archive (SRA) database repository under the accession code PRJNA629640. All other relevant data supporting the findings of this study are available upon reasonable request.