Abstract
Chronic administration of methamphetamine (METH) leads to physical and psychological dependence. It is generally accepted that METH exerts rewarding effects via competitive inhibition of the dopamine transporter (DAT), but the molecular mechanism of METH addiction remains largely unknown. Accumulating evidence shows that mitochondrial function is important in regulation of drug addiction. In this study, we investigated the role of Clk1, an essential mitochondrial hydroxylase for ubiquinone (UQ), in METH reward effects. We showed that Clk1+/− mutation significantly suppressed METH-induced conditioned place preference (CPP), accompanied by increased expression of DAT in plasma membrane of striatum and hippocampus due to Clk1 deficiency-induced inhibition of DAT degradation without influencing de novo synthesis of DAT. Notably, significantly decreased iron content in striatum and hippocampus was evident in both Clk1+/− mutant mice and PC12 cells with Clk1 knockdown. The decreased iron content was attributed to increased expression of iron exporter ferroportin 1 (FPN1) that was associated with elevated expression of hypoxia-inducible factor-1α (HIF-1α) in response to Clk1 deficiency both in vivo and in vitro. Furthermore, we showed that iron played a critical role in mediating Clk1 deficiency-induced alteration in DAT expression, presumably via upstream HIF-1α. Taken together, these data demonstrated that HIF-1α-mediated changes in iron homostasis are involved in the Clk1 deficiency-altered METH reward behaviors.
Keywords: methamphetamine, drug addiction, conditioned place preference, Clk1, dopamine transporter, iron, ferroportin 1, hypoxia-inducible factor-1α, striatum, hippocampus
Introduction
Drug addiction is considered to be a disorder of reward and cognitive systems characterized by compulsive drug use [1, 2]. Methamphetamine (METH), one of the most commonly used psychostimulants worldwide, competitively inhibits the dopamine transporter (DAT), which results in elevated dopamine concentrations and activation of reward circuitry [3, 4]. Furthermore, METH is also transported into dopaminergic neurons through the DAT to enhance dopamine efflux and prolong neurotransmission [3, 5]. Thus, the DAT is regarded as a major target in METH-induced drug dependence and a potential target for addiction treatments [6, 7].
Mitochondria are known as cellular energy generators that are essential for cellular functions. Mitochondrial function is particularly important for brain tissue because it requires considerable energy for maintaining the ion gradient, neurotransmission and neuroplasticity. Recent studies indicate that energy metabolism may play a critical role in drug dependence; alterations in enzymes related to energy metabolism have been confirmed in morphine addiction [8, 9] and substances of abuse, including alcohol, are reported to cause mitochondrial stress that is believed to contribute to reward processes [10, 11]. In relation to drug abuse, alterations in mitochondrial DNA copy numbers have been reported in both cocaine and METH abuse [12, 13]. METH has been shown to injure dopaminergic neurons by stimulating production of reactive oxygen species (ROS), which may contribute to METH-induced inhibition of respiratory complexes [14]. Moreover, METH may increase striatal expression and DNA binding activity of p53, an important transcriptional factor that mediates signaling between mitochondria and nuclei [15, 16]. However, the detailed molecular pathways that regulate drug dependence via mitochondrial energy metabolism and function remain largely unknown.
Clk1, also known as Coq7, is a critical mitochondrial hydroxylase that catalyzes the hydroxylation of demethoxyubiquinone for ubiquinone (UQ) synthesis, which is involved in the mitochondrial electron transport chain as well as in generating mitochondrial ATP and ROS [17, 18]. Previous work has demonstrated that elevated mitochondrial ROS and expression of hypoxia-inducible factor-1α (HIF-1α) contributes to extended lifespans in C. elegans clk-1 null mutants and Clk1+/− (heterozygous) mutant mice [19–21]. In addition, Clk1+/− mutant mice display altered mitochondrial metabolism, such as reduced mitochondrial oxygen consumption, reduced electron transport, reduced mitochondrial ATP synthesis and increased generation of mitochondrial ROS (mtROS). Moreover, Clk1-deficient macrophages display increased pro-inflammatory cytokine and inflammatory responses to lipopolysaccharide (LPS) through stabilization of HIF-1α [20]. We have recently reported that Clk1 modulates microglial activation via regulation of metabolic reprogramming, and additionally demonstrated that mTOR/HIF-1α signaling pathways are involved in Clk1 deficiency-induced aerobic glycolysis [22]. We further found that Clk1 regulates the autophagy-lysosome pathway and TFEB nuclear translocation via the AMPK/mTORC1 pathway [23]. Taken together, these studies indicate that alteration in Clk1 produces significant changes in mitochondrial functions and in multiple biological responses.
Several lines of evidence suggest dysregulation of iron homeostasis in stimulant drug addiction. Both cocaine and METH have been demonstrated to alter permeability to iron in the blood–brain barrier [24] and chronic drug abuse is known to induce accumulation of iron in the basal ganglia [25, 26]. While several clinical studies have found alterations in peripheral blood iron concentration in drug abusers, these changes are dependent on the type and duration of the drugs abused [27, 28]. A recent report in patients with cocaine addiction showed excess iron accumulation in the globus pallidus, which correlated strongly with duration of cocaine use, and mild iron deficiency in the periphery, which was associated with low iron levels in the red nucleus and may reflect reduced iron intake from food sources in chronic drug abuse [25]. Interestingly, iron plays important roles in the mitochondrial respiration chain and as a central component of electronic iron-sulfur cluster carriers [29, 30]. As mitochondria were found to be the central regulator of biogenesis of Fe-S clusters [31], iron appears crucial to the respiration of mitochondria and iron over-loading causes oxidative stress of mitochondria [32].
Given the importance of mitochondrial function in drug addiction, with both Clk1 and iron appearing essential in mitochondrial electron transport, alteration in Clk1 may provide a novel approach to investigating how mitochondrial function modulates drug addiction. In this context, it would be important to clarify whether and how changes in iron homeostasis may contribute to addictive drug–mediated neuroplasticity in response to alterations in mitochondrial function induced by mutant Clk1.
Materials and methods
Animals and cells
The Clk1 mutant mice (Clk1+/−) were donated by Rugen Therapeutics Co., Ltd (Suzhou, China) and housed in groups of 4–6 per cage in a humidity and temperature-controlled vivarium on a 12:12 h light/dark cycle with free access to food and water. WT mice were bred from Clk1+/− mutant mice and all the animals used in this study are 6–8 week male mice. All animal protocols were proved by the Animal Care and Use Committee of Soochow University and were in compliance with Guidelines for the Care and Use of Laboratory Animals (Chinese National Research Council, 2006) and the ‘ARRIVE’ (Animals in Research: Reporting In Vivo Experiments) guidelines. Every effort was made to minimize animal suffering and to reduce the number of animals used in the experiments. PC12 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured with Dulbecco’s modified Eagle’s medium (GIBCO, Grand Island, NY, USA) containing 10% FBS and 1% penicillin/streptomycin.
Conditioned place preference
Apparatus
The apparatus for conditioned place preference (CPP) conditioning and testing (Jiliang Ltd., Shanghai, China) consisted of two distinct visual and textural cue compartments. These two compartments had different wall colors (white or black) and floors (fine wire mesh floor or wide grid floor) and a removable wall; an arched gateway between the two compartments granted access to each compartment during the test session. As described previously [33], time spent in each compartment was measured and recorded via infrared beams and an automated analysis system.
Procedure
The CPP procedure was performed as described previously with minor modifications, as shown in the timeline of Fig. 1a [33]. The 6-day CPP procedure included three main phases of pre-conditioning, conditioning and post-conditioning. In the pre-conditioning phase (day 1), also known as the habituation session, wild-type (WT) and Clk1+/− mice were placed into the apparatus and allowed to freely explore the two compartments for 15 min. Exploration time spent in each compartment was recorded and mice showing strong unconditioned preference (≥70% of time in one compartment) were excluded. During the conditioning phase (day 2–5), sessions were performed using an unbiased, balanced protocol. Briefly, mice in each group received intraperitoneal (i.p.) injections of either METH or saline and were confined in the drug-paired compartment for 1 h in the morning for four consecutive days. In the evening, mice were injected with saline and confined in the saline-paired compartment (the opposite compartment) for 1 h. In post-conditioning phase (day 6), the arched gateway was open and mice were free to explore the two compartments again. During exploration, time spent in each compartment was recorded in a 15 min preference test with results presented as a preference score.
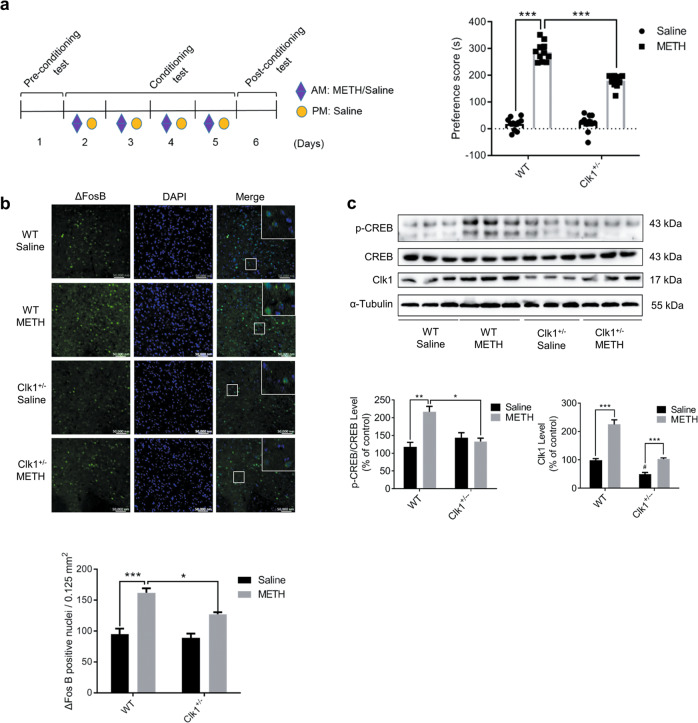
Fig. 1. Clk1 deficiency inhibits methamphetamine (METH)-induced conditioned place preference (CPP) and suppresses the expression of ΔFosB and phosphorylation of CREB in mice.
a CPP schematic diagram. WT and Clk1+/− mutant mice were each divided into saline and METH groups. During the conditioning test, mice in the drug group received repeated i.p. injections of METH (2 mg/kg) in the drug-paired compartment and saline in the other compartment to develop CPP over 4 consecutive days, while controls received saline on each occasion. In the post-conditioning test, time spent in the two compartments was recorded and a preference score was calculated as the difference in time spent in the drug-paired compartment between post- and pre-conditioning tests (n = 11 animals per group). b Immunofluorescence images (scale bar, 50,000 nm) of ΔFosB in NAc from WT or Clk1+/− mutant mice that had received the above METH/saline treatments, with tissue collected 24 h after CPP testing (at least 3 animals per group). c Whole striatal tissue from each group was dissected 24 h after CPP testing and processed for immunoblot assay as described in Methods. Representative images for immunofluorescence and immunoblots are shown in the upper panel and quantitative data are shown in the lower panels. Bar graphs are presented as means ± SEM with data obtained from at least 3 animals. Statistical analyses were performed using two-way ANOVA followed by Bonferroni-corrected tests (*P < 0.05, **P < 0.01, ***P < 0.001; #P < 0.05 vs WT).
Drugs and reagents
Methamphetamine was obtained from Shanghai Standard Biotech Co., Ltd. and diluted with saline for i.p. injection (2 or 4 mg/kg). Deferoxamine (DFO, 100 μM treatment for 0–10 h [34]), cycloheximide (CHX, 10 μM treatment for 0–10 h [35, 36]), lificiguat (YC-1, 100 μM treatment for 24 h [37, 38]) and MG132 (10 μM treatment for 12 h [39]) were purchased from Sigma-Aldrich, St. Louis, MO, USA. Phenylmethanesulfonyl fluoride (PMSF) was purchased from Cell Signaling Technology, Danvers, MA, USA. All other reagents were purchased from standard suppliers at highest quality available.
Western blotting
Tissues or PC12 cells were lysed in RIPA buffer (Cell Signaling Technology, Danvers, MA, USA) for 30 min on ice. Following 5 min incubation at 95 °C before centrifuge, the supernatants were used to detect protein concentrations using Bradford Protein Assay Kit (Bio-Rad, Hercules, CA, USA). Proteins were separated by sodium dodecyl sulfate polyacrylamide gel and then transferred to polyvinylidene difluoride membranes. After 1 h blocking in 5% milk, the membranes were incubated with primary antibodies. Blots were probed with anti-p-CREB antibody (1:1000, Cell Signaling Technology, Danvers, MA, USA), anti-CREB antibody (1:1000, Cell Signaling Technology, Danvers, MA, USA), anti-Clk1 antibody (1:1000, Proteintech, Wuhan, China), anti-DAT antibody (1:1000, Abcam, Cambridge, MA, USA), anti-D1R antibody (1:1000, Abcam, Cambridge, MA, USA), anti-HIF-1α antibody (1:1000, Proteintech, Wuhan, China), anti-E-cadherin antibody (1:500, Genscript, Nanjing China), anti-Ub antibody (1:1000, Santa Cruz, CA, USA), anti-EEA1 antibody (1:1000, Santa Cruz, CA, USA), anti-LAMP 1 antibody (1:1000, Santa Cruz, CA, USA), anti-Rab 5 antibody (1:1000, Santa Cruz, CA, USA), anti-Ferroportin1 antibody (1:500, Abcam, Cambridge, MA, USA), or anti-Ferritin-L antibody (1:1000, Proteintech, Wuhan, China), respectively. Loading controls were probed with monoclonal anti-α-tubulin antibody (1:10000, Sigma, St. Louis, MO, USA). After incubation with respective secondary antibodies, results were analyzed using ChemiScope 3300 Mini (CLINX, Shanghai, China). ImageJ software (version 1.52 a) was used for densitometry of Western blot gels and values were normalized to the control group (% of control).
Enzyme-linked immunosorbent assay (ELISA)
WT and Clk1+/− mutant mouse brain tissues and PC12 cells were collected for hepcidin protein detection. Samples were homogenized ultrasonically in PBS with PMSF and supernatants were collected after centrifugation (5000 × g for 10 min at 4 °C). All ELISA assays were performed according to manufacturer’s instructions (Elabscience, TX, USA). Briefly, 100 μL of sample or standard was added to each well and incubated for 90 min at 37 °C. Then, the liquid was removed and 100 μL biotinylated detection antibody was added in each well for 60 min at 37 °C. Thereafter, 100 μL HRP conjugate was added and incubated for 30 min at 37 °C, followed by addition of 90 μL substrate regent and incubation for 15 min at 37 °C. Finally, 50 μL of stop solution was added and OD values were assessed at 450 nm using Microplate Reader (Infinite M200 PRO, Tecan, Switzerland).
RNA extraction and quantitative real-time PCR
RNAiso Plus reagent (TaKaRa, Dalian, China) was used to isolate total RNA from tissues and PC12 cells. RNA (1000 ng) was reverse-transcribed into cDNA Oligod(T) primers (TaKaRa, Dalian, China) as reported previously [40]. cDNAs were amplified using specific primers in Table 1. Quantitative real-time PCR procedures were 30 s at 95 °C, followed by 5 s at 95 °C and 30 s at 60 °C for 40 cycles. Thereafter, melt curve analysis was performed by heating from 60 °C to 95 °C. GAPDH or β-actin was used as reference gene and the 2−ΔΔCT formula was used to assess relative expression of target genes.
Table 1.
Sequences of the primers used for RT-PCR.
| Species | Gene | Primer | Sequence |
|---|---|---|---|
| Mouse | DAT | Forward | 5’-CTTCTCCTCTGGCTTCGTTGT-3' |
| DAT | Reverse | 5’-CAGGGTAGATGATGAAGATCAACC-3' | |
| Mouse | hepcidin | Forward | 5’-TTGCGATACCAATGCAGAAGAG-3' |
| hepcidin | Reverse | 5’-AATTGTTACAGCATTTACAGCAGAAGA-3' | |
| Mouse | TfR1 | Forward | 5’-GTTTCTGCCAGCCCCTTATTAT-3' |
| TfR1 | Reverse | 5’-GCAAGGAAAGGATATGCAGCA-3' | |
| Mouse | DMT1 | Forward | 5’-TCTATGTGGTGGCTGCAGTG-3' |
| DMT1 | Reverse | 5’-GCTGGTATCTTCGCTCAGCA-3' | |
| Mouse | FTL | Forward | 5’-CCATCTGACCAACCTCCGC-3' |
| FTL | Reverse | 5’-CGCTCAAAGAGATACTCGCC-3' | |
| Mouse | GAPDH | Forward | 5’-TGTGTCCGTCGTGGATCTGA-3' |
| GAPDH | Reverse | 5’-TTGCTGTTGAAGTCGCAGGAG-3’ | |
| Rat | hepcidin | Forward | 5’-GAAGGCAAGATGGCACTAAGCA-3' |
| hepcidin | Reverse | 5’-TCTCGTCTGTTGCCGGAGATAG-3' | |
| Rat | TfR1 | Forward | 5’-CGAAGTCCAGTGTGGGAACA-3' |
| TfR1 | Reverse | 5’-TGGCACCAACAGCTCCATAG-3' | |
| Rat | DMT1 | Forward | 5’-GGCAGTGTTTGATTGCGTTG-3' |
| DMT1 | Reverse | 5’-TAGTATTGCCACCGCTGGTATC-3' | |
| Rat | FPN1 | Forward | 5’-ATGTATCATGGATGGGTCAAAAC-3' |
| FPN1 | Reverse | 5’-TACGACAACAATCCAGTCC-3' | |
| Rat | β-actin | Forward | 5’-CCCTAAGGCCAACCGTGAAAAG-3' |
| β-actin | Reverse | 5’-TACGTACATGGCTGGGGTGT-3' | |
| Rat | FTL | Forward | 5’-GATTCGTCAGAATTATTCCA-3' |
| FTL | Reverse | 5’-AGAGAGGTAGGTGTAAGA-3' | |
| Rat | Clk1 | Forward | 5’-CGGTTTTGATGCCCTTGTGG-3' |
| Clk1 | Reverse | 5’-AGGCCATTGCTCCTTCCTTC-3' |
Membrane protein extraction
Membrane protein was extracted using a Mem-PER Plus Membrane Protein Extraction Kit (Thermo Scientific, Waltham, MA, USA) according to manufacturer’s instructions. Briefly, 5 × 106 cells were collected and washed with cell wash solution. The samples were then incubated with permeabilization buffer including PMSF and the homogenate was transferred to a new tube and incubated for 10 min at 4 °C with constant mixing. After centrifugation (16,000 × g for 15 min at 4 °C), the supernatant containing cytosolic proteins was transferred to another tube and the pellet was re-suspended in buffer containing PMSF followed by 30 min incubation at 4 °C with constant mixing. After centrifugation (16,000 × g for 15 min at 4 °C), the supernatant containing solubilized membrane and membrane-associated proteins was collected for further experiments. Thermo Scientific™ BCA Protein Assay (Waltham, MA, USA) was used to quantify proteins in the cytosolic and membrane fractions.
Lentivirus transfection
Lentivirus (LV) gene transfer vectors encoding shClk1 (LV-shClk1, 5’-CAGCCATATACTTATCAGAAA-3’) and nonspecific control (LV-shNC, 5’-TTCTCCGAACGTGTCACGT-3’) were prepared by GeneChem (Shanghai, China). Cells were transfected with LV-shRNA according to manufacturer’s instructions. In the process of LV-shRNA transduction, the titer of lentivirus used was 5 × 108 units and polybrene (5 μg/mL) was applied to promote transduction efficiency. Successfully infected cells were selected using puromycin 72 h later. Total RNA and protein were extracted to assess knockdown efficiency by RT-PCR and Western blot.
In vitro protein ubiquitination assay
LV-shNC or LV-shClk1 PC12 cells were treated with the proteasome inhibitor MG132 (10 μM) for 12 h before lysis. In order to purify DAT proteins, cell lysates were incubated with DAT antibody-conjugating agarose beads overnight at 4 °C with constant mixing, followed by four washes with lysis buffer. Then nonspecific binding proteins were removed. Ubiquitination levels of DAT and DAT expression in the purified samples were detected with anti-ubiquitin and anti-DAT immunoblotting, respectively. Anti-DAT, anti-Clk1 or anti-α-Tubulin antibody was used to detect total expression of DAT, Clk1 and α-Tubulin, respectively, in the whole cell lysates.
Immunofluorescence staining
Immunofluorescence staining was performed as described previously [33, 41]. Briefly, for brain samples, under anesthesia with 4% chloral hydrate, mice were perfused with PBS and 4% paraformaldehyde. Then, the brain was collected and kept in paraformaldehyde overnight at 4 °C, followed by 30% sucrose overnight at 4 °C. Thereafter, the brain was serially cut into 20 µm coronal sections with a freezing microtome. PC12 cells were washed 3 times with 0.01 M PBST and fixed with iced methanol for 20 min. Both brain samples and PC12 cells were blocked by blocking solution (PBS containing 3% albumin from bovine serum and 0.3% Triton-X100) for at least 2 h at 37 °C and incubated overnight at 4 °C with primary antibodies anti-ΔFosB (1:1000, Santa Cruz, CA, USA), anti-EEA1 (1:500, Santa Cruz, CA, USA), anti-LAMP 1 (1:500, Santa Cruz, CA, USA), anti-Rab 5 (1:500, Santa Cruz, CA, USA) and anti-DAT (1:300, Abcam, Cambridge, MA, USA). After 3 times washes with 0.01 M PBST, sections and cells were incubated with secondary antibodies conjugated to Alex Fluor 488 or 594 (1:1000, Invitrogen, Carlsbad, CA, USA). After dyeing with DAPI and a further 3 times washes, images were taken using a confocal microscope (Zeiss LSM710 META, Germany). Quantification of ΔFosB positive nuclei was performed using ImageJ software (version 1.52 a) [42]. Colocalization analysis was performed using ImageJ software (version 1.52 a) with the plugin JACoP. Quantification of colocalization was performed by calculating the Manders overlap coefficient, as described previously [43].
Measurement of iron content
Total iron content of mouse tissues was detected by inductively coupled plasma mass spectrometry (ICP-MS) (Thermo Fisher Scientific, Waltham, MA, USA). Before measurement, all tissues were weighed and BCA Protein Assay was used to quantify proteins for PC12 cells. Then, samples were put in conical flasks and treated with 5 mL mixed acid solution consisting of nitric acid, hydrochloric acid and perchloric acid, followed by heating to 260 °C. Samples diluted in 5% nitric acid (5 mL) were filtered with 0.22 μm microfiltration membranes. The standard curve was prepared and ranged from 0 to 500 ppb using iron standard (1 mg iron/mL) and triplicates were performed for each sample by ICP-MS [44, 45].
Statistical analysis
Data were analyzed by Graph Pad Prism® (Version 6.0) software and presented as means ± SEM. All molecular biological and biochemical assays were repeated at least three times on different days and the animal numbers used in behavioral experiments are noted in figure legends. One-way and two-way ANOVAs followed by Bonferroni-corrected tests were utilized for multiple-group comparisons, with Student’s t test used for comparisons between two groups; P < 0.05 was considered statistically significant.
Results
Clk1 deficiency inhibits METH-induced CPP
We employed Clk1+/− mutant mice to study the effect of Clk1 deficiency on METH-induced CPP. The CPP procedure was as depicted in Fig. 1a. After 4 days of successive intraperitoneal (i.p.) injection of 2 mg/kg METH, time spent in the drug-paired compartment was significantly increased in WT mice. However, marked inhibition of METH-induced CPP was found in Clk1+/− mutant mice (Fig. 1a). In association with impaired CPP formation, we detected significant decreases in expression of ΔFosB and phosphorylation of CREB (S133), endogenous biomarkers for CPP formation, in nucleus accumbens (NAc) and striatum of Clk1+/− mutant mice relative to WT mice (Fig. 1b, c). In addition, expression of Clk1 was elevated in response to METH treatment in both WT and Clk1+/− mutant mice (Fig. 1c), also revealing the importance of Clk1 in METH-induced reward.
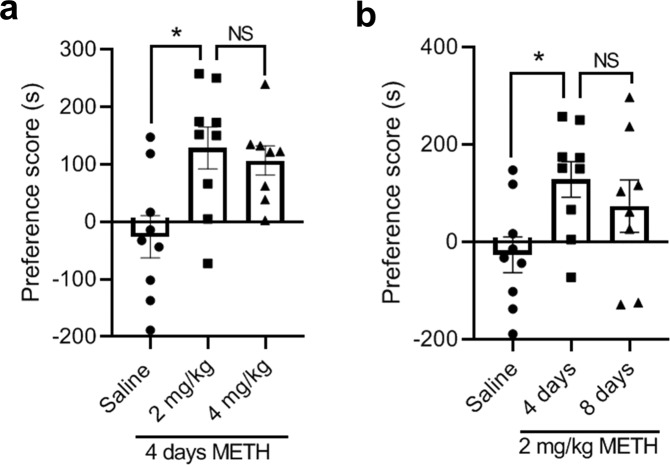
In order to rule out the possibility that impaired CPP formation in the Clk1+/− mice might be attributed to time or dose of METH, we conducted additional experiments using either a higher dose of METH or a longer treatment time. Neither a high dose (4 mg/kg) nor a long treatment time (8 days) improved CPP formation in Clk1+/− mice (Fig. 2a, b). Preference scores for METH-treated mice at 8 days were somewhat lower relative to 4 days; while they did not differ significantly from METH-treated mice at 4 days, they did not differ significantly from saline-treated mice. Whether this reflects some effect of an excessive treatment period or the reduced number of mice studied at this time point remains to be determined. Regardless, these results indicate that Clk1 deficiency markedly inhibits acquisition of METH-induced CPP.
Fig. 2. Attenuation of METH-induced CPP is not dependent on dose or time of METH treatment in Clk1-deficient mice.
The CPP procedure was performed as described in Fig. 1a and Methods. a Clk1+/− mutant mice were divided into three groups: saline, METH 2 mg/kg, and METH 4 mg/kg. Each group received 4 consecutive days of injections in the conditioning phase. Then, following a post-conditioning test, CPP scores were calculated (n = 8–9 animals per group). b After this post-conditioning test, the group receiving METH 2 mg/kg received an additional 4 consecutive days of injections in the conditioning phase (i.e., a total of 8 days). Then, following another post-conditioning test, CPP scores were again calculated (n = 8–9 animals per group). Bar graphs are presented as means ± SEM. Statistical analysis was performed using one-way ANOVA followed by Bonferroni-corrected tests (*P < 0.05 vs saline; NS not significant).
Clk1 deficiency increases plasma membrane DAT expression in vivo and in vitro
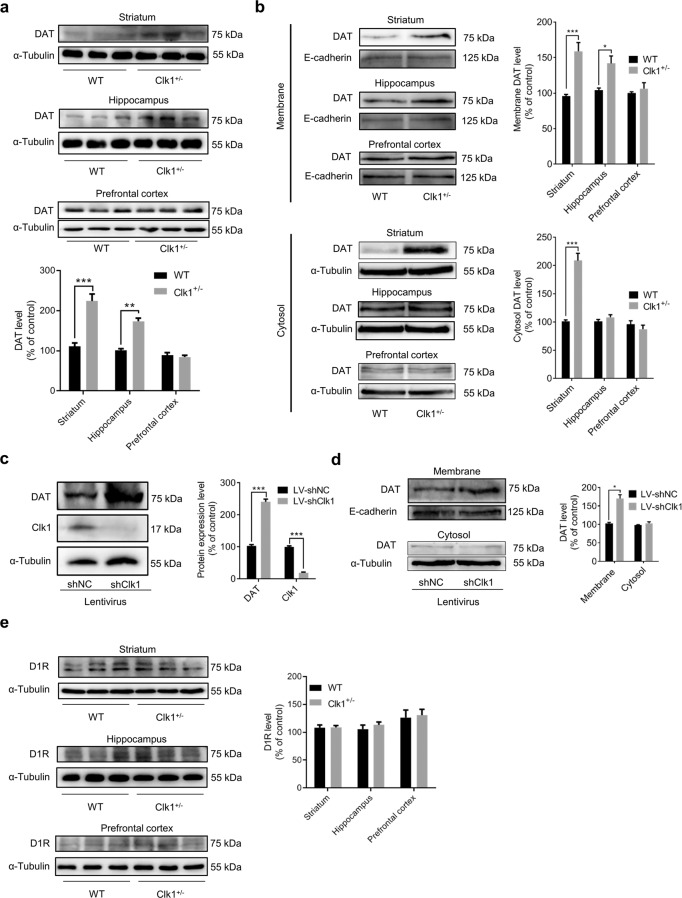
METH binds competitively to DAT to inhibit dopamine re-uptake and therefore increase dopamine concentrations in the synaptic cleft, which is believed to be key mechanism of METH reward. Surprisingly, we detected a marked increase in the expression of DAT in striatum and hippocampus, but not in prefrontal cortex (Fig. 3a), in Clk1+/− mutant mice relative to WT mice. Furthermore, while increase in DAT expression in the striatum was found in both the plasma membrane fraction and the cytosol preparation, increase in DAT expression in the hippocampus was found in the plasma membrane fraction but not in the cytosol preparation (Fig. 3b); the reason for these distinct patterns of DAT expression across brain regions in Clk1+/− mutant mice remains to be determined. To further confirm this observation, we employed PC12 cells to determine if knockdown of Clk1 could alter DAT expression. As expected, Clk1 knockdown in PC12 cells also increased expression of DAT in the plasma membrane fraction but not in the cytosol preparation (Fig. 3c, d). In contrast, the expression of the dopamine D1 receptor (D1R) was not altered in any of these three brain regions in Clk1+/− mutant mice (Fig. 3e).
Fig. 3. Clk1 deficiency increases plasma dopamine transporter (DAT) expression without alteration of dopamine D1 receptors.
a, b WT or Clk1+/− mutant mice were sacrificed and striatum, hippocampal and prefrontal cortex tissues were dissected and processed for immunoblot assays of DAT. c, d At 72 h after LV-shNC or LV-shClk1 transduction, successfully infected PC12 cells were selected and cultured as described in Methods. Then, PC12 cells with stable expression of LV-shNC or LV-shClk1 was collected for immunoblot assays. e WT or Clk1+/− mutant mice were sacrificed and brain tissue collected for immunoblot assays of D1R in striatum, hippocampus and prefrontal cortex. Membrane proteins were isolated as described in Methods. For a, representative images for immunoblots are shown in the upper panels and quantitative data are shown in the lower panel. For each of b–e, representative images for immunoblots are shown in the left panels and quantitative data are shown in the right panels. Bar graphs are presented as means ± SEM, with 3 independent experiments conducted for each immunoblot assay. Statistical analyses were performed using two-way ANOVA followed by Bonferroni-corrected tests (*P < 0.05, ***P < 0.001).
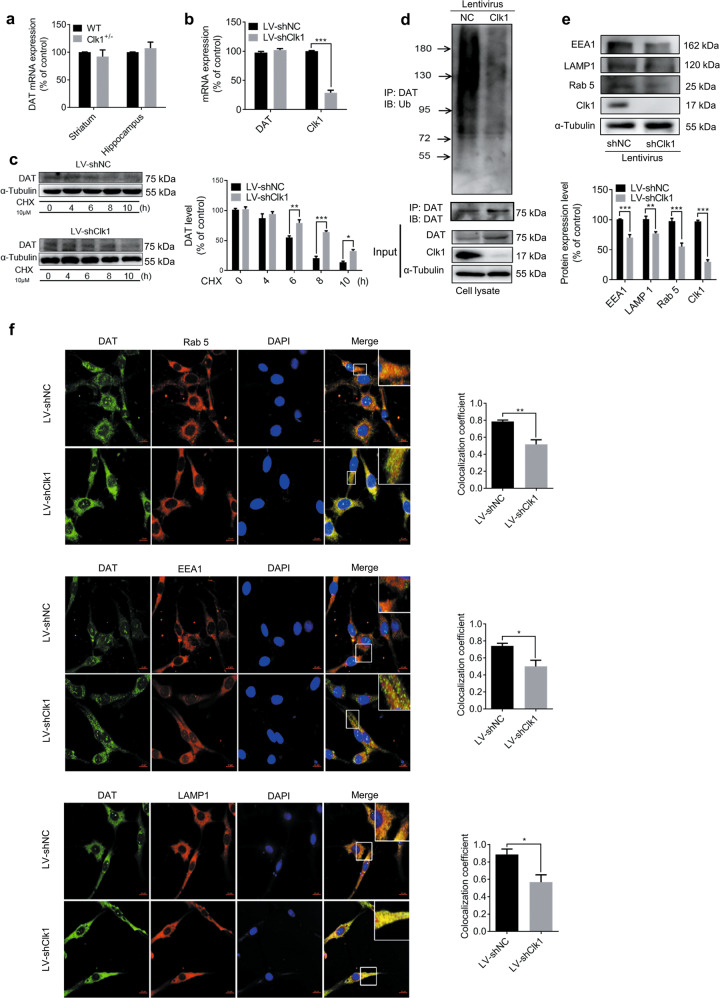
In order to elucidate the mechanism by which Clk1 deficiency upregulates plasma membrane expression of DAT, we first determined the mRNA expression of DAT. As shown in Fig. 4a, b, transcriptional levels of DAT in striatum and hippocampus were not altered in Clk1+/− mutant mice or in PC12 cells with Clk1 knockdown. However, the degradation rate of DAT was significantly inhibited in LV-shClk1 PC12 cells when the protein synthesis inhibitor cycloheximide (CHX) was added (Fig. 4c). Taken together, these findings indicate that Clk1 deficiency-induced elevation in expression of plasma membrane DAT can be attributed to impairment in DAT degradation rather than increase in de novo synthesis of DAT.
Fig. 4. Clk1 deficiency suppresses the DAT lysosomal degradation pathway.
a WT or Clk1+/− mutant mice were sacrificed and striatum and hippocampal tissues were collected for assay of DAT mRNA expression. b LV-shNC and LV-shClk1 PC12 cells were collected for assay of DAT mRNA expression. c LV-shNC and LV-shClk1 PC12 cells were treated with 10 μM cycloheximide (CHX) for indicated periods and cell lysates were prepared for immunoblotting against specific antibodies. d PC12 cell lysates were precipitated with anti-ubiquitin beads followed by immunoblotting with anti-DAT antibody. Total cell lysate was used as control with specific antibodies as indicated. e Expression of Rab 5, EEA1 and LAMP 1 and f immunofluorescence images (scale bar, 10 μm) of co-localization (Rab 5, EEA1 and LAMP 1) with DAT-positive intracellular punctums in PC12 cells (3 fields of view per sample). For c and f, representative images for immunoblots and immunofluorescence are shown in the left panel and quantitative data are shown in the right panel. For e, representative images for immunoblots are shown in the upper panel and quantitative data are shown in the lower panel. Bar graphs are presented as means ± SEM for at least 3 independent experiments. Statistical analyses for a–e were performed using two-way ANOVA followed by Bonferroni-corrected tests and statistical analyses for f was performed using Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001).
Clk1 deficiency suppresses the DAT lysosomal degradation pathway
We next investigated the underlying mechanism responsible for impairment in DAT degradation associated with Clk1 deficiency. It is known that membrane DAT is subjected to dynamic recycling, and that ubiquitination is required for DAT internalization [46, 47]. Internalized DAT is either degraded via a lysosomal pathway mediated by ubiquitination or recycled to the plasma membrane [6, 46]. We first examined changes in DAT ubiquitination with ubiquitin antibody in PC12 cells transfected with either LV-shNC or LV-shClk1 to knockdown Clk1. As shown in Fig. 4d, e, a marked decrease of DAT-ubiquitin conjugates was found in PC12 cells with Clk1 knockdown; the expression of early endosome markers Rab 5 and early endosome antigen 1 (EEA1) and the late endosome marker lysosomal-associated membrane protein 1 (LAMP 1) were also significantly decreased in response to Clk1 deficiency. Moreover, co-distribution of DAT-positive intracellular punctum with Rab 5, EEA1 or LAMP 1 was also markedly reduced in LV-shClk1 PC12 cells (Fig. 4f). This further supported that Clk1 deficiency-mediated inhibition of lysosomal degradation of DAT may underlie the observed upregulation of plasma membrane DAT in Clk1 deficiency.
Clk1 deficiency alters iron homeostasis
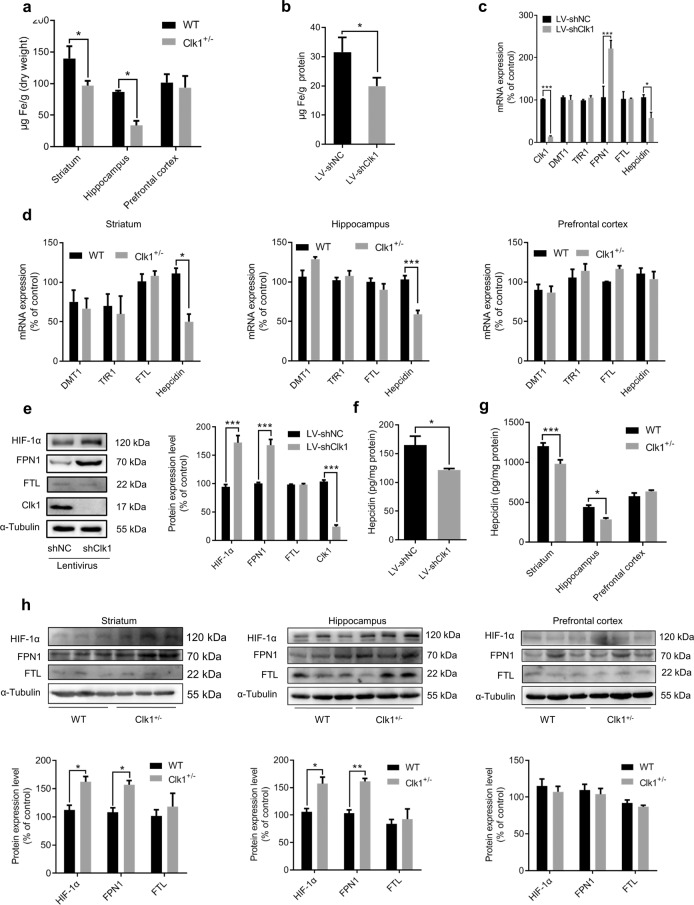
Given that both iron and Clk1 play critical roles in mitochondrial function, we examined changes in iron metabolism in response to Clk1 deficiency. Clk1+/− mutant mice exhibited brain region-selective changes in iron content: significant decreases in total iron content were found in hippocampus and striatum but not prefrontal cortex (Fig. 5a); interestingly, we also detected a differential expression of Clk1 across these brain regions, with the lowest level in frontal cortex relative to hippocampus and striatum (Supplementary Fig. S1). To confirm this observation, we employed PC12 cells transfected with LV-shNC or LV-shClk1 and found that knockdown of Clk1 in PC12 cells also induced a significant decrease in intracellular total iron content (Fig. 5b). It is known that iron homeostasis is under the control of iron transporters and uptake regulators. We first analyzed mRNA expression of the iron transporter and found that in PC12 cells with LV-shClk1 transfection, FPN1, the only known iron exporter, was upregulated while expression of hepcidin, an iron negative regulatory factor that binds to FPN1 to result in its internalization and degradation, was significantly decreased; expression of ferritin-L (FTL), which is involved in the storage of overloaded iron, as well as divalent metal transporter (DMT1) and transferrin receptor (TfR1), two vital iron uptake-related proteins was unaltered (Fig. 5c). In Clk1+/− mutant mice, expression of hepcidin was significantly decreased in striatum and hippocampus, but not in prefrontal cortex, while expression levels of DMT1, TfR1 and FTL were unaltered in each of these brain regions relative to WT (Fig. 5d). Enhancement of FPN1 and reduction in hepcidin was further confirmed by measuring protein expression in Clk1 knockdown PC12 cells and in the striatum and hippocampus of Clk1+/− mutant mice (Fig. 5e–h). These data indicate that enhanced iron exporter activity, as evidenced by increase in FPN1 expression and downregulation in hepcidin level, may be responsible for the brain region-selective decreases in iron content.
Fig. 5. Clk1 deficiency alters iron homeostasis and upregulates HIF-1α expression.
WT or Clk1+/− mutant mice were sacrificed and striatum, hippocampus and prefrontal cortex tissues, together with LV-shNC and LV-shClk1 PC12 cells, were used as follows: a, b ICP-MS analysis of total iron levels; c, d mRNA expression of DMT1, TfR1, FTL and Hepcidin; e Western Blotting for HIF-1α, FPN1, FTL and Clk1; f, g ELISA assay of hepcidin; h Western Blotting for HIF-1α, FPN1 and FTL. For e, representative images for immunoblots are shown in the left panel and quantitative data are shown in the right panel. For h, representative images are shown in the upper panels and quantitative data are shown in the lower panels. Bar graphs are presented as means ± SEM for at least 3 independent experiments. Statistical analyses for a, c, d, e, g and h were performed using two-way ANOVA followed by Bonferroni-corrected tests and statistical analyses for b and f were performed using Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001).
HIF-1α appears to be an upstream modulator for iron deficiency-induced plasma membrane DAT expression
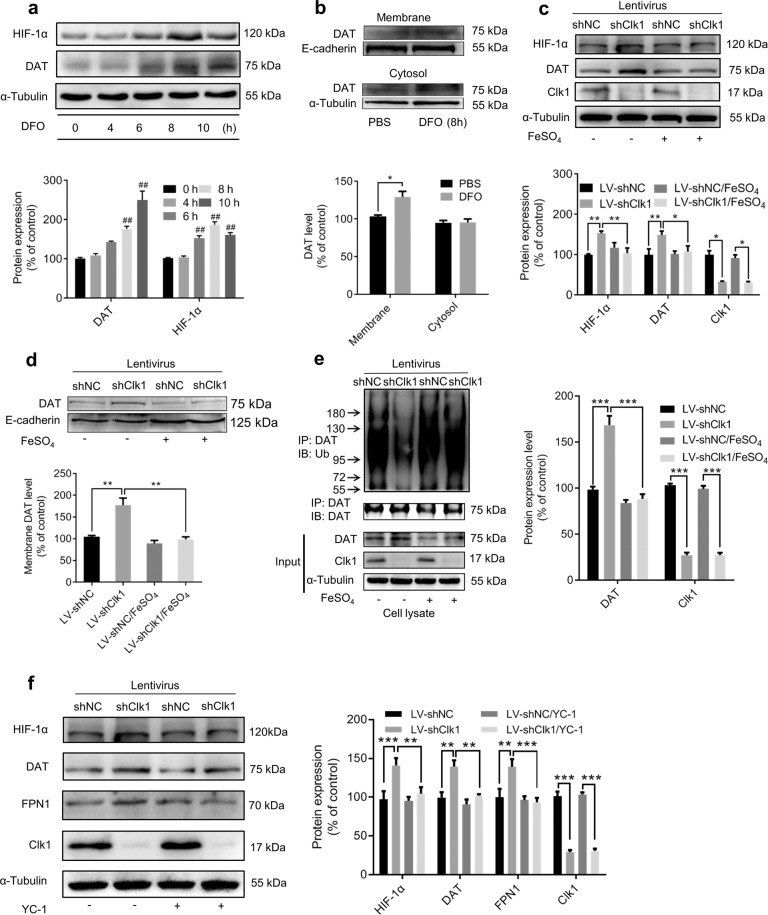
HIF-1α is an important transcription factor in response to hypoxia. It is known that knockdown of Clk1 induces enhanced expression of HIF-1α in association with production of ROS [19, 20] and we confirmed that expression of HIF-1α was upregulated in both Clk1+/− mutant mice and Clk1 knockdown PC12 cells (Fig. 5e, h). Downregulation of iron content by deferoxamine (DFO), a high affinity iron chelator for treatment of iron overload, was reported to induce accumulation of HIF-1α in vivo and in vitro [48–51]. We confirmed elevated expression of HIF-1α in PC12 cells in response to DFO treatment (Fig. 6a) and additionally detected significantly enhanced plasma membrane expression of DAT similar to that observed in Clk1 deficiency (Fig. 6b). This indicated that iron deficiency mimicked Clk1 deficiency in elevating expression of both HIF-1α and plasma membrane DAT. To further elucidate the role of iron in Clk1 deficiency-mediated enhancement of DAT expression, we treated LV-shClk1 PC12 cells with 100 μM FeSO4 to restore the iron content of PC12 cells and found that Clk1 deficiency-induced elevations in plasma membrane DAT and HIF-1α expression were no longer present (Fig. 6c, d). Moreover, increased DAT ubiquitin in response to Clk1 deficiency was also reversed (Fig. 6e). We then employed YC-1, an inhibitor of HIF-1α, to inhibit HIF-1α expression. In PC12 cells with Clk1 knockdown YC-1 treatment decreased expression of HIF-1α, as expected, and also inhibited the elevated expression of DAT and FPN1 (Fig. 6f). These data clearly demonstrate that iron plays a critical role in mediating Clk1 deficiency-induced alteration in DAT expression, in which upregulation of HIF-1α in response to Clk1 deficiency is a critical signaling mechanism in altered iron homeostasis.
Fig. 6. HIF-1α modulates iron deficiency-induced plasma membrane DAT expression.
a PC12 cells were treated with 100 μM DFO for indicated periods prior to immunoblot assay and b membrane protein of PC12 cells was isolated as described in Methods. Cell lysates were prepared for immunoblotting against specific antibodies. c, d LV-shNC and LV-shClk1 PC12 cells were treated with 100 μM FeSO4 for 8 h prior to immunoblot assays and e DAT-ubiquitin assay. f YC-1 (100 μM) was added to LV-shNC and LV-shClk1 PC12 cells for 24 h prior to immunoblot assays. Bar graphs are presented as means ± SEM for at least 3 independent experiments. For each of a–d, representative images for immunoblots are shown in the upper panels and quantitative data are shown in the lower panels. For e and f, representative images for immunoblots are shown in the left panels and quantitative data are shown in the right panels. Statistical analyses for a–c, e and f were performed using two-way ANOVA and statistical analysis for d was performed using one-way ANOVA, each followed by Bonferroni-corrected tests (*P < 0.05, **P < 0.01, ***P < 0.001; ##P < 0.01 vs 0 h).
Discussion
In the present study, we utilized Clk1+/− mutant mice to study how mitochondrial function alters METH reward behaviors and found that Clk1+/− mutant mice exhibited marked impairment in METH-induced CPP. Clk1 deficiency resulted in increased expression of plasma membrane DAT, which was attributed to Clk1 deficiency-induced inhibition of DAT lysosomal degradation but not in de novo synthesis of DAT. Furthermore, we identified brain region-selective changes in iron content in Clk1+/− mutant mice, i.e., decrease in total iron content in hippocampus and striatum but not in prefrontal cortex. Such change in iron content was also confirmed in vitro using PC12 cells transfected with LV-shClk1.
To further elucidate the underlying molecular mechanisms, we found that FPN1 was upregulated while expression of hepcidin was decreased in both Clk1+/− mutant mice and in PC12 cells with Clk1 knockdown. We further demonstrated that depletion of iron content in PC12 cells by deferoxamine mimicked Clk1 deficiency-elevated plasma membrane DAT and HIF-1α expression, whereas supplement with FeSO4 to restore iron content in PC12 cells blocked the changes induced by Clk1 deficiency. Furthermore, application of an inhibitor of HIF-1α, YC-1, blocked the enhanced expression of HIF-1α, DAT and FPN1 in PC12 cells by Clk1 knockdown. The present data thus provide the first clear experimental evidence that alteration of iron homeostasis is associated with METH reward effects.
It is well known that METH-mediated reward circuit activation, via competitive inhibition of DAT to increase transmission via dopaminergic neurons projecting from the ventral tegmental area, plays a key role in METH addiction [52]. On investigating the effect of Clk1 on DAT expression, Clk1 deficiency-induced elevation in plasma membrane expression of DAT, both in vitro and in Clk1+/− mutant mice, which was associated with inhibition of DAT lysosomal degradation but not of de novo synthesis. DAT is subject to dynamic recycling, hence internalized DAT could be either degraded via the ubiquitination-mediated lysosomal pathway or recycled to the plasma membrane [6, 46]. Thus, disturbed DAT ubiquitination could impair lysosomal degradation and lead to increased DAT accumulation in the plasma membrane, contributing to DA re-uptake by the presynaptic membrane and elimination of DA from the synaptic cleft. In agreement with previous reports, our data indicate that inhibition of lysosomal degradation of DAT may be responsible for the upregulation of plasma membrane DAT in response to Clk1 deficiency, which results in decreased dopaminergic activity and ultimately contributes to attenuation of METH-induced CPP.
Iron is an essential element in regulating a large range of biochemical processes in the brain. Changes in iron homeostasis are associated with deficits in memory/learning capacity and motor skills, together with other psychological disturbances [53–55]. Disturbance of iron metabolism has been shown in substance abuse [25, 26, 56–58] and a recent study has reported altered iron levels in METH abusers [28]. Notably, application of the iron chelator DFO also attenuated METH-induced hyperthermia and hyperactivity in rats [59]. Interestingly, we found that Clk1 deficiency selectively decreases iron levels in striatum and hippocampus but not in prefrontal cortex. FPN1 is the only known iron export protein negatively regulated by hepcidin. The FPN1-hepcidin system plays an important role in the regulation of iron homeostasis [45, 60–63] and we found that FPN1 was upregulated while expression of hepcidin was decreased both in Clk1+/− mutant mice and in PC12 cells with Clk1 knockdown, indicating that altered FPN1 and hepcidin may be responsible for the observed changes in iron homeostasis. We further demonstrated that altered iron homeostasis is associated with Clk1 deficiency-induced suppression of DAT lysosomal degradation via ubiquitin. This is in line with previous reports showing that iron homeostasis plays an important role in PKC-mediated ubiquitination-dependent degradation of DAT [46, 64–66].
We next investigated the key signaling molecules mediating iron/DAT changes in response to Clk1 deficiency. Previous reports from our group and others have shown that Clk1 deficiency enhances expression of HIF-1α in brain [20, 22], with changes in HIF-1α known to alter iron metabolism [67, 68]. Also, alteration of iron content altered the expression of HIF-1α, revealing that iron is also an oxygen sensor [69]. The present data depict a mechanism for HIF-1α-modulated iron homeostasis via enhanced iron exporter activity, which consequently alters plasma DAT expression by affecting DAT ubiquitin in response to Clk1 deficiency. Since it is known that DAT ubiquitin is a key step for DAT internalization and degradation [46, 66, 70], impaired DAT ubiquitin due to iron homeostasis would disturb the internalization of DAT in the plasma membrane and consequently result in elevation in plasma membrane DAT. Given that DAT is a key target of METH, elevated DAT expression in the plasma membrane will lead to enhanced reuptake of DA from the synaptic cleft and result in decreased DA activity, which may underlie impairment in CPP development in Clk1-deficient animals [71, 72].
Clk1, also known as Coq7, is an important hydroxylase for the biosynthesis of ubiquinone, a critical electron carrier from complex I and complex III to complex II in the mitochondrial respiratory chain [73, 74]. Thus, Clk1 is a critical regulator of mitochondrial functions. Notably, Clk1 deficiency results in selective elevation in plasma membrane DAT expression in hippocampus and striatum but not prefrontal cortex, which parallels alteration in iron content. These differential changes across brain regions may be related to distinct processes identified in these same brain regions [75]. They may also be related to distinct responses of individual brain regions to METH administration [76–80]. In addition, we detected significant differences in basal Clk1 expression across brain regions in naïve animals. As prefrontal expression of Clk1 is the lowest among these three brain regions, whereas prefrontal iron content was not altered by Clk1 deficiency. This may imply that prefrontal Clk1 may play a less important role in regulation of DAT and iron homeostasis.
We also found that METH induces elevated expression of brain Clk1 in both WT and Clk1+/- mutant mice. It is well known that systemic administration of METH results in increased central respiratory activity, heart rate and basal metabolism [81], which would result in increased energy consumption. As Clk1-catalyzed ubiquinone biosynthesis is critical for mitochondrial energy generation [82, 83], elevated Clk1 expression in response to METH treatment may be an adaptive response to such enhanced requirement for energy.
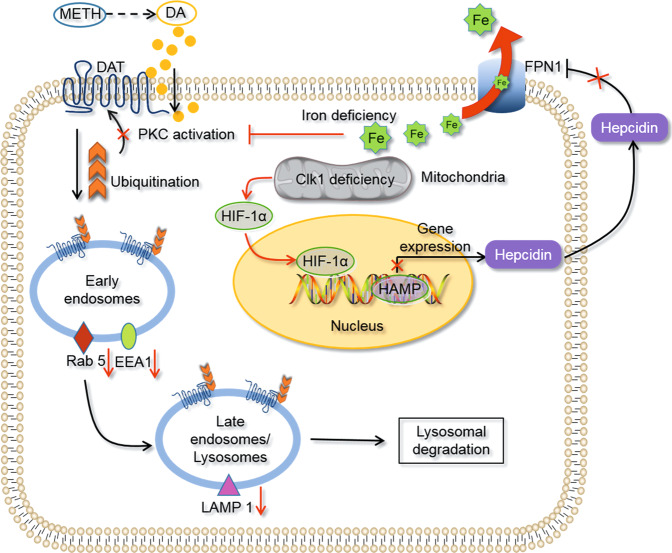
In summary, Clk1 deficiency inhibited the development of METH-induced CPP. Mechanistically, altered mitochondrial function induced by Clk1 deficiency results in elevated HIF-1α expression, which promotes iron exporter activity by enhancing FPN1 expression to decrease iron content in selective brain regions involved in reward circuitry (Fig. 7). Furthermore, disturbed iron homeostasis stimulates HIF-1α expression and inhibits ubiquitination of DAT and impairs its lysosomal degradation; this ultimately leads to elevation of plasma membrane DAT that may be responsible for decreased DA activity and suppression of METH-induced CPP in Clk1+/− mutant mice. The current study provides robust experimental evidence and a potentially novel mechanism for how iron is involved in METH-induced reward behaviors, which may in turn serve as a novel approach for the treatment of METH addiction. However, several issues remain regarding such mechanisms: How does Clk1 regulate HIF-1α expression? Why does Clk1 deficiency result in selective alteration in DAT and iron content? Further studies are required to clarify these processes.
Fig. 7. Schematic diagram to indicate possible mechanisms underlying Clk1 deficiency-mediated inhibition in acquisition of METH-induced CPP.
Clk1 deficiency inhibits transcription of hepcidin by HIF-1α and then efflux of iron is promoted by upregulation of FPN1. In addition, expression of each of Rab 5, EEA1 and LAMP 1 is reduced. Finally, iron deficiency down-regulates PKC activity and suppresses DAT lysosomal degradation, thus decreasing DA concentration to inhibit acquisition of METH-induced CPP.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81973334, 81773702) and the Priority Academic Program Development of the Jiangsu Higher Education Institutes (PAPD). We thank Guo-qiang Xu from Soochow University for his patient guidance on DAT-ubiquitination assays.
Author contributions
XCZ designed the research. PJY and ZXR performed the molecular biological and biochemical experiments, analyzed the data and drafted the manuscript. ZXR performed the CPP experiments. ZFS, CLW, CJH, LSZ and NNL performed experiments and collected data. JLW and XCZ elaborated the manuscript. All authors reviewed the manuscript and approved the submission for publication.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Peng-ju Yan, Zhao-xiang Ren
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-021-00806-1.
References
- 1.Bali P, Kenny PJ. MicroRNAs and drug addiction. Front Genet. 2013;4:43. doi: 10.3389/fgene.2013.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J, Ham S, Hong H, Moon C, Im HI. Brain reward circuits in morphine addiction. Mol Cells. 2016;39:645–53. doi: 10.14348/molcells.2016.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sulzer D, Chen TK, Lau YY, Kristensen H, Rayport S, Ewing A. Amphetamine redistributes dopamine from synaptic vesicles to the cytosol and promotes reverse transport. J Neurosci. 1995;15:4102–8. doi: 10.1523/JNEUROSCI.15-05-04102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prakash MD, Tangalakis K, Antonipillai J, Stojanovska L, Nurgali K, Apostolopoulos V. Methamphetamine: effects on the brain, gut and immune system. Pharmacol Res. 2017;120:60–67. doi: 10.1016/j.phrs.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Rifkin RA, Moss SJ, Slesinger PA. G protein-gated potassium channels: a link to drug addiction. Trends Pharmacol Sci. 2017;38:378–92. doi: 10.1016/j.tips.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu S, Zhao C, Wu Y, Yang Q, Shao A, Wang T, et al. Identification of a Vav2-dependent mechanism for GDNF/Ret control of mesolimbic DAT trafficking. Nat Neurosci. 2015;18:1084–93. doi: 10.1038/nn.4060. [DOI] [PubMed] [Google Scholar]
- 7.Dubol M, Trichard C, Leroy C, Sandu AL, Rahim M, Granger B, et al. Dopamine transporter and reward anticipation in a dimensional perspective: a multimodal brain imaging study. Neuropsychopharmacology. 2018;43:820–7. doi: 10.1038/npp.2017.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen XL, Lu G, Gong YX, Zhao LC, Chen J, Chi ZQ, et al. Expression changes of hippocampal energy metabolism enzymes contribute to behavioural abnormalities during chronic morphine treatment. Cell Res. 2007;17:689–700. doi: 10.1038/cr.2007.63. [DOI] [PubMed] [Google Scholar]
- 9.Antolak A, Bodzoń-Kułakowska A, Cetnarska E, Pietruszka M, Marszałek-Grabska M, Kotlińska J, et al. Proteomic data in morphine addiction versus real protein activity: Metabolic enzymes. J Cell Biochem. 2017;118:4323–30. doi: 10.1002/jcb.26085. [DOI] [PubMed] [Google Scholar]
- 10.Tapia-Rojas C, Carvajal FJ, Mira RG, Arce C, Lerma-Cabrera JM, Orellana JA, et al. Adolescent binge alcohol exposure affects the brain function through mitochondrial impairment. Mol Neurobiol. 2018;55:4473–91. doi: 10.1007/s12035-018-1268-5. [DOI] [PubMed] [Google Scholar]
- 11.Valenzuela CF. Alcohol and neurotransmitter interactions. Alcohol Health Res World. 1997;21:144–8. [PMC free article] [PubMed] [Google Scholar]
- 12.Var SR, Day TR, Vitomirov A, Smith DM, Soontornniyomkij V, Moore DJ, et al. Mitochondrial injury and cognitive function in HIV infection and methamphetamine use. AIDS. 2016;30:839–48. doi: 10.1097/QAD.0000000000001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadakierska-Chudy A, Frankowska M, Filip M. Mitoepigenetics and drug addiction. Pharmacol Ther. 2014;144:226–33. doi: 10.1016/j.pharmthera.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Riddle EL, Fleckenstein AE, Hanson GR. Mechanisms of methamphetamine-induced dopaminergic neurotoxicity. AAPS J. 2006;8:E413–8. doi: 10.1007/BF02854914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asanuma M, Miyazaki I, Higashi Y, Cadet JL, Ogawa N. Methamphetamine-induced increase in striatal p53 DNA-binding activity is attenuated in Cu,Zn-superoxide dismutase transgenic mice. Neurosci Lett. 2002;325:191–4. doi: 10.1016/S0304-3940(02)00291-4. [DOI] [PubMed] [Google Scholar]
- 16.Imam SZ, Itzhak Y, Cadet JL, Islam F, Slikker W, Jr, Ali SF. Methamphetamine-induced alteration in striatal p53 and bcl-2 expressions in mice. Brain Res Mol Brain Res. 2001;91:174–8. doi: 10.1016/S0169-328X(01)00139-5. [DOI] [PubMed] [Google Scholar]
- 17.Zheng H, Lapointe J, Hekimi S. Lifelong protection from global cerebral ischemia and reperfusion in long-lived Mclk1(+/)(-) mutants. Exp Neurol. 2010;223:557–65. doi: 10.1016/j.expneurol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lapointe J, Stepanyan Z, Bigras E, Hekimi S. Reversal of the mitochondrial phenotype and slow development of oxidative biomarkers of aging in long-lived Mclk1+/- mice. J Biol Chem. 2009;284:20364–74. doi: 10.1074/jbc.M109.006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol. 2010;20:2131–6. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D, Malo D, Hekimi S. Elevated mitochondrial reactive oxygen species generation affects the immune response via hypoxia-inducible factor-1alpha in long-lived Mclk1+/− mouse mutants. J Immunol. 2010;184:582–90. doi: 10.4049/jimmunol.0902352. [DOI] [PubMed] [Google Scholar]
- 21.Wang D, Wang Y, Argyriou C, Carrière A, Malo D, Hekimi S. An enhanced immune response of Mclk1+/− mutant mice is associated with partial protection from fibrosis, cancer and the development of biomarkers of aging. PLoS ONE. 2012;7:e49606. doi: 10.1371/journal.pone.0049606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu R, Zhang F, Chen G, Han C, Liu J, Ren Z, et al. Clk1 deficiency promotes neuroinflammation and subsequent dopaminergic cell death through regulation of microglial metabolic reprogramming. Brain Behav Immun. 2017;60:206–19. doi: 10.1016/j.bbi.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Yan Q, Han C, Wang G, Waddington JL, Zheng L, Zhen X. Activation of AMPK/mTORC1-mediated autophagy by metformin reverses Clk1 deficiency-sensitized dopaminergic neuronal death. Mol Pharmacol. 2017;92:640–52. doi: 10.1124/mol.117.109512. [DOI] [PubMed] [Google Scholar]
- 24.Dietrich JB. Alteration of blood-brain barrier function by methamphetamine and cocaine. Cell Tissue Res. 2009;336:385–92. doi: 10.1007/s00441-009-0777-y. [DOI] [PubMed] [Google Scholar]
- 25.Ersche KD, Acosta-Cabronero J, Jones PS, Ziauddeen H, van Swelm RP, Laarakkers CM, et al. Disrupted iron regulation in the brain and periphery in cocaine addiction. Transl Psychiatry. 2017;7:e1040. doi: 10.1038/tp.2016.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melega WP, Lacan G, Harvey DC, Way BM. Methamphetamine increases basal ganglia iron to levels observed in aging. Neuroreport. 2007;18:1741–5. doi: 10.1097/WNR.0b013e3282f0d4f4. [DOI] [PubMed] [Google Scholar]
- 27.Elnimr T, Hashem A, Assar R. Heroin dependence effects on some major and trace elements. Biol Trace Elem Res. 1996;54:153–62. doi: 10.1007/BF02786262. [DOI] [PubMed] [Google Scholar]
- 28.Akbari A, Mosayebi G, Solhi H, Rafiei M, Asadi A, Ghazavi A. Serum zinc, copper, iron, and magnesium levels in Iranian drug users: a case control study. J Addict Med. 2015;9:304–7. doi: 10.1097/ADM.0000000000000134. [DOI] [PubMed] [Google Scholar]
- 29.Paul BT, Manz DH, Torti FM, Torti SV. Mitochondria and iron: current questions. Expert Rev Hematol. 2017;10:65–79. doi: 10.1080/17474086.2016.1268047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stiban J, So M, Kaguni LS. Iron-sulfur clusters in mitochondrial metabolism: multifaceted roles of a simple cofactor. Biochemistry. 2016;81:1066–80. doi: 10.1134/S0006297916100059. [DOI] [PubMed] [Google Scholar]
- 31.Wachnowsky C, Fidai I, Cowan JA. Iron-sulfur cluster biosynthesis and trafficking - impact on human disease conditions. Metallomics. 2018;10:9–29. doi: 10.1039/C7MT00180K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volani C, Doerrier C, Demetz E, Haschka D, Paglia G, Lavdas AA, et al. Dietary iron loading negatively affects liver mitochondrial function. Metallomics. 2017;9:1634–44. doi: 10.1039/C7MT00177K. [DOI] [PubMed] [Google Scholar]
- 33.Mu Y, Ren Z, Jia J, Gao B, Zheng L, Wang G, et al. Inhibition of phosphodiesterase10A attenuates morphine-induced conditioned place preference. Mol Brain. 2014;7:70. doi: 10.1186/s13041-014-0070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coutts AS, Pires IM, Weston L, Buffa FM, Milani M, Li JL, et al. Hypoxia-driven cell motility reflects the interplay between JMY and HIF-1α. Oncogene. 2011;30:4835–42. doi: 10.1038/onc.2011.188. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Chen J, Ning D, Liu Q, Wang C, Zhang Z, et al. FBXO22 promotes the development of hepatocellular carcinoma by regulating the ubiquitination and degradation of p21. J Exp Clin Cancer Res. 2019;38:101. doi: 10.1186/s13046-019-1058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.An L, Peng LY, Sun NY, Yang YL, Zhang XW, Li B, et al. Tanshinone IIA activates nuclear factor-erythroid 2-related factor 2 to restrain pulmonary fibrosis via regulation of redox homeostasis and glutaminolysis. Antioxid Redox Signal. 2019;30:1831–48. doi: 10.1089/ars.2018.7569. [DOI] [PubMed] [Google Scholar]
- 37.Li SH, Shin DH, Chun YS, Lee MK, Kim MS, Park JW. A novel mode of action of YC-1 in HIF inhibition: stimulation of FIH-dependent p300 dissociation from HIF-1{alpha} Mol Cancer Ther. 2008;7:3729–38. doi: 10.1158/1535-7163.MCT-08-0074. [DOI] [PubMed] [Google Scholar]
- 38.Mao X, Nanzhang, Xiao J, Wu H, Ding K. Hypoxia-induced autophagy enhances cisplatin resistance in human bladder cancer cells by targeting hypoxia-inducible factor-1α. J Immunol Res. 2021;2021:8887437. doi: 10.1155/2021/8887437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singha PK, Pandeswara S, Geng H, Lan R, Venkatachalam MA, Saikumar P. TGF-βinduced TMEPAI/PMEPA1 inhibits canonical Smad signaling through R-Smad sequestration and promotes non-canonical PI3K/Akt signaling by reducing PTEN in triple negative breast cancer. Genes Cancer. 2014;5:320–36. doi: 10.18632/genesandcancer.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ni J, Wang X, Chen S, Liu H, Wang Y, Xu X, et al. MicroRNA let-7c-5p protects against cerebral ischemia injury via mechanisms involving the inhibition of microglia activation. Brain Behav Immun. 2015;49:75–85. doi: 10.1016/j.bbi.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 41.Ren ZX, Zhao YF, Cao T, Zhen XC. Dihydromyricetin protects neurons in an MPTP-induced model of Parkinson’s disease by suppressing glycogen synthase kinase-3 beta activity. Acta Pharmacol Sin. 2016;37:1315–24. doi: 10.1038/aps.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weishaupt N, Riccio P, Dobbs T, Hachinski VC, Whitehead SN. Characterization of behaviour and remote degeneration following thalamic stroke in the rat. Int J Mol Sci. 2015;16:13921–36. doi: 10.3390/ijms160613921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franco IS, Shohdy N, Shuman HA. The legionella pneumophila effector VipA is an actin nucleator that alters host cell organelle trafficking. PLoS Pathog. 2012;8:e1002546. doi: 10.1371/journal.ppat.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu X, Du T, Song N, He Q, Shen Y, Jiang H, et al. Decreased iron levels in the temporal cortex in postmortem human brains with Parkinson disease. Neurology. 2013;80:492–5. doi: 10.1212/WNL.0b013e31827f0ebb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.You LH, Yan CZ, Zheng BJ, Ci YZ, Chang SY, Yu P, et al. Astrocyte hepcidin is a key factor in LPS-induced neuronal apoptosis. Cell Death Dis. 2017;8:e2676. doi: 10.1038/cddis.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong WC, Amara SG. Differential targeting of the dopamine transporter to recycling or degradative pathways during amphetamine- or PKC-regulated endocytosis in dopamine neurons. FASEB J. 2013;27:2995–3007. doi: 10.1096/fj.12-218727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sorkina T, Richards TL, Rao A, Zahniser NR, Sorkin A. Negative regulation of dopamine transporter endocytosis by membrane-proximal N-terminal residues. J Neurosci. 2009;29:1361–74. doi: 10.1523/JNEUROSCI.3250-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim J, Kim HI, Bang Y, Seol W, Choi HS, Choi HJ. Hypoxia-inducible factor-1alpha upregulates tyrosine hydroxylase and dopamine transporter by nuclear receptor ERRgamma in SH-SY5Y cells. Neuroreport. 2015;26:380–6. doi: 10.1097/WNR.0000000000000356. [DOI] [PubMed] [Google Scholar]
- 49.Tai TC, Wong-Faull DC, Claycomb R, Wong DL. Hypoxic stress-induced changes in adrenergic function: role of HIF1 alpha. J Neurochem. 2009;109:513–24. doi: 10.1111/j.1471-4159.2009.05978.x. [DOI] [PubMed] [Google Scholar]
- 50.Guo C, Hao LJ, Yang ZH, Chai R, Zhang S, Gu Y, et al. Deferoxamine-mediated up-regulation of HIF-1α prevents dopaminergic neuronal death via the activation of MAPK family proteins in MPTP-treated mice. Exp Neurol. 2016;280:13–23. doi: 10.1016/j.expneurol.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 51.Guo C, Wang T, Zheng W, Shan ZY, Teng WP, Wang ZY. Intranasal deferoxamine reverses iron-induced memory deficits and inhibits amyloidogenic APP processing in a transgenic mouse model of Alzheimer’s disease. Neurobiol Aging. 2013;34:562–75. doi: 10.1016/j.neurobiolaging.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 52.Wang KH, Penmatsa A, Gouaux E. Neurotransmitter and psychostimulant recognition by the dopamine transporter. Nature. 2015;521:322–7. doi: 10.1038/nature14431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J, Wessling-Resnick M. Iron and mechanisms of emotional behavior. J Nutr Biochem. 2014;25:1101–7. doi: 10.1016/j.jnutbio.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z, Zeng YN, Yang P, Jin LQ, Xiong WC, Zhu MZ, et al. Axonal iron transport in the brain modulates anxiety-related behaviors. Nat Chem Biol. 2019;15:1214–22. doi: 10.1038/s41589-019-0371-x. [DOI] [PubMed] [Google Scholar]
- 55.Thirupathi A, Chang YZ. Brain iron metabolism and CNS diseases. Adv Exp Med Biol. 2019;1173:1–19. doi: 10.1007/978-981-13-9589-5_1. [DOI] [PubMed] [Google Scholar]
- 56.Jones BC, Wheeler DS, Beard JL, Grigson PS. Iron deficiency in rats decreases acquisition of and suppresses responding for cocaine. Pharmacol Biochem Behav. 2002;73:813–9. doi: 10.1016/S0091-3057(02)00906-1. [DOI] [PubMed] [Google Scholar]
- 57.Adisetiyo V, McGill CE, DeVries WH, Jensen JH, Hanlon CA, Helpern JA. Elevated brain iron in cocaine use disorder as indexed by magnetic field correlation imaging. Biol Psychiatry Cogn Neurosci Neuroimaging. 2019;4:579–88. doi: 10.1016/j.bpsc.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 58.Jenney CB, Alexander DN, Jones BC, Unger EL, Grigson PS. Preweaning iron deficiency increases non-contingent responding during cocaine self-administration in rats. Physiol Behav. 2016;167:282–8. doi: 10.1016/j.physbeh.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park MJ, Lee SK, Lim MA, Chung HS, Cho SI, Jang CG, et al. Effect of alpha-tocopherol and deferoxamine on methamphetamine-induced neurotoxicity. Brain Res. 2006;1109:176–82. doi: 10.1016/j.brainres.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 60.Singh B, Arora S, Agrawal P, Gupta SK. Hepcidin: a novel peptide hormone regulating iron metabolism. Clin Chim Acta. 2011;412:823–30. doi: 10.1016/j.cca.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 61.Vela D. Hepcidin, an emerging and important player in brain iron homeostasis. J Transl Med. 2018;16:25. doi: 10.1186/s12967-018-1399-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Varghese J, James JV, Sagi S, Chakraborty S, Sukumaran A, Ramakrishna B, et al. Decreased hepatic iron in response to alcohol may contribute to alcohol-induced suppression of hepcidin. Br J Nutr. 2016;115:1978–86. doi: 10.1017/S0007114516001197. [DOI] [PubMed] [Google Scholar]
- 63.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–3. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 64.Kuvibidila S, Baliga BS, Murthy KK. Impaired protein kinase C activation as one of the possible mechanisms of reduced lymphocyte proliferation in iron deficiency in mice. Am J Clin Nutr. 1991;54:944–50. doi: 10.1093/ajcn/54.5.944. [DOI] [PubMed] [Google Scholar]
- 65.Kuvibidila SR, Kitchens D, Baliga BS. In vivo and in vitro iron deficiency reduces protein kinase C activity and translocation in murine splenic and purified T cells. J Cell Biochem. 1999;74:468–78. doi: 10.1002/(SICI)1097-4644(19990901)74:3<468::AID-JCB14>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 66.Miranda M, Wu CC, Sorkina T, Korstjens DR, Sorkin A. Enhanced ubiquitylation and accelerated degradation of the dopamine transporter mediated by protein kinase C. J Biol Chem. 2005;280:35617–24. doi: 10.1074/jbc.M506618200. [DOI] [PubMed] [Google Scholar]
- 67.Voit RA, Sankaran VG. Stabilizing HIF to ameliorate anemia. Cell. 2020;180:6. doi: 10.1016/j.cell.2019.12.010. [DOI] [PubMed] [Google Scholar]
- 68.Xu MM, Wang J, Xie JX. Regulation of iron metabolism by hypoxia-inducible factors. Sheng Li Xue Bao. 2017;69:598–610. [PubMed] [Google Scholar]
- 69.Huang LE, Willmore WG, Gu J, Goldberg MA, Bunn HF. Inhibition of hypoxia-inducible factor 1 activation by carbon monoxide and nitric oxide. Implications for oxygen sensing and signaling. J Biol Chem. 1999;274:9038–44. doi: 10.1074/jbc.274.13.9038. [DOI] [PubMed] [Google Scholar]
- 70.Sorkina T, Miranda M, Dionne KR, Hoover BR, Zahniser NR, Sorkin A. RNA interference screen reveals an essential role of Nedd4-2 in dopamine transporter ubiquitination and endocytosis. J Neurosci. 2006;26:8195–205. doi: 10.1523/JNEUROSCI.1301-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Madras BK, Miller GM, Fischman AJ. The dopamine transporter and attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57:1397–409. doi: 10.1016/j.biopsych.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 72.Fumagalli F, Gainetdinov RR, Valenzano KJ, Caron MG. Role of dopamine transporter in methamphetamine-induced neurotoxicity: evidence from mice lacking the transporter. J Neurosci. 1998;18:4861–9. doi: 10.1523/JNEUROSCI.18-13-04861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, Hekimi S. Mitochondrial respiration without ubiquinone biosynthesis. Hum Mol Genet. 2013;22:4768–83. doi: 10.1093/hmg/ddt330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, Oxer D, Hekimi S. Mitochondrial function and lifespan of mice with controlled ubiquinone biosynthesis. Nat Commun. 2015;6:6393. doi: 10.1038/ncomms7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heysieattalab S, Naghdi N, Hosseinmardi N, Zarrindast MR, Haghparast A, Khoshbouei H. Methamphetamine-induced enhancement of hippocampal long-term potentiation is modulated by NMDA and GABA receptors in the shell-accumbens. Synapse. 2016;70:325–35. doi: 10.1002/syn.21905. [DOI] [PubMed] [Google Scholar]
- 76.Broom SL, Yamamoto BK. Effects of subchronic methamphetamine exposure on basal dopamine and stress-induced dopamine release in the nucleus accumbens shell of rats. Psychopharmacology. 2005;181:467–76. doi: 10.1007/s00213-005-0007-6. [DOI] [PubMed] [Google Scholar]
- 77.Sabrini S, Russell B, Wang G, Lin J, Kirk I, Curley L. Methamphetamine induces neuronal death: evidence from rodent studies. Neurotoxicology. 2020;77:20–8. doi: 10.1016/j.neuro.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 78.Krasnova IN, Hodges AB, Ladenheim B, Rhoades R, Phillip CG, Cesena A, et al. Methamphetamine treatment causes delayed decrease in novelty-induced locomotor activity in mice. Neurosci Res. 2009;65:160–5. doi: 10.1016/j.neures.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tokunaga I, Ishigami A, Kubo S, Gotohda T, Kitamura O. The peroxidative DNA damage and apoptosis in methamphetamine-treated rat brain. J Med Invest. 2008;55:241–5. doi: 10.2152/jmi.55.241. [DOI] [PubMed] [Google Scholar]
- 80.Kerdsan W, Thanoi S, Nudmamud-Thanoi S. Changes in glutamate/NMDA receptor subunit 1 expression in rat brain after acute and subacute exposure to methamphetamine. J Biomed Biotechnol. 2009;2009:329631. doi: 10.1155/2009/329631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hassan SF, Wearne TA, Cornish JL, Goodchild AK. Effects of acute and chronic systemic methamphetamine on respiratory, cardiovascular and metabolic function, and cardiorespiratory reflexes. J Physiol. 2016;594:763–80. doi: 10.1113/JP271257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kirby CS, Patel MR. Elevated mitochondrial DNA copy number found in ubiquinone-deficient clk-1 mutants is not rescued by ubiquinone precursor 2-4-dihydroxybenzoate. Mitochondrion. 2021;58:38–48. doi: 10.1016/j.mito.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cristina D, Cary M, Lunceford A, Clarke C, Kenyon C. A regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000450. doi: 10.1371/journal.pgen.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.