Abstract
Siglec-15, a member of sialic-acid binding immunoglobulin type lectins, is normally expressed by myeloid cells and upregulated in some human cancers and represents a promising new target for immunotherapy. While PD-L1 blockade is an important strategy for immunotherapy, its effectiveness is limited. The expression of Siglec-15 has been demonstrated to be predominantly mutually exclusive to PD-L1 in certain cancer histologies. Thus, there is significant opportunity for Siglec-15 as an immunotherapeutic target for patients that do not respond to PD-1/PD-L1 inhibition. The aim of this study was to prospectively develop an immunohistochemical (IHC) assay for Siglec-15 to be used as a companion diagnostic for future clinical trials. Here, we create and validate an IHC assay with a novel recombinant antibody to the cytoplasmic domain of Siglec-15. To find an enriched target, this antibody was first used in a quantitative fluorescence (QIF) assay to screen a broad range of tumor histologies to determine tumor types where Siglec-15 demonstrated high expression. Based on this and previous data, we focused on development of a chromogenic IHC assay for lung cancer. Then we developed a scoring system for this assay that has high concordance amongst pathologist readers. We then use this chromogenic IHC assay to test the expression of Siglec-15 in two cohorts of NSCLC. We found that this assay shows a higher level of staining in both tumor and immune cells compared to previous QIF assays utilizing a polyclonal antibody. However, similar to that study, only a small percentage of positive Siglec-15 cases showed high expression for PD-L1. This validated assay for Siglec-15 expression may support development of a companion diagnostic assay to enrich for patients expressing the Siglec-15 target for therapy.
Keywords: Siglec-15, NSCLC, breast cancer, companion diagnostic test, immunohistochemistry
Introduction
Siglec-15 is a member of the sialic acid-binding immunoglobulin type lectins, an important cell surface glycan recognition protein. It has been well described for its role in osteoclast differentiation and as a potential target for the treatment of osteoporosis. Structurally, it consists of an extracellular domain, a lysine-residue containing transmembrane domain and a cytoplasmic domain 1–3. Normally expressed by cells of the myeloid lineage, it has been shown to be upregulated in some human cancers 4.
Among the various human cancers in which Siglec-15 upregulation has been observed, of note are cancers of colon, thyroid and endometrioid cancers. It is also significantly upregulated in liver, lung, bladder, and kidney cancers 5. A recently published study by Wang et al showed that over-expression of Siglec-15 by macrophages played an important role in the inhibition of T-cells, and mice deficient of Siglec-15, or treated with a Siglec-15 blocking antibody, can overcome this inhibition limiting tumor growth. Thus, like programmed death ligand (PD-L1), which is a major immune evasion mechanism for many cancers, over-expression of Siglec-15 in the tumor microenvironment (TME) has been proposed to be a suppressor of the immune response. IFN-gamma, an important cytokine required for PD-L1 induction, was seen to inhibit Siglec-15 expression, thereby indicating that PD-L1 regulation pathways are distinct from those of Siglec-15. Wang et al also observed that Siglec-15 and PD-L1 expression was seen to be somewhat mutually exclusive, which could be explained by its inhibition by IFN-gamma and activation by macrophage colony stimulating factor (CSF-1) 4.
Blocking PD-1/PD-L1 is widely regarded as an important strategy in normalization immunotherapy 6. However, the anti-tumor effect of a single immune checkpoint inhibitor is more effective in some tumors than others, for example in lung cancer where the effective response rate is less than 30% 7–9. Hence, targeting Siglec-15 may be an effective alternative therapy for patients that do not respond to PD-1/PD-L1 inhibition 10. A first-in-human phase I clinical trial evaluating an anti-Siglec-15 mAb, NC318, in solid tumors showed clinical benefit, and a phase II clinical trial is ongoing. While it is too early to assess activity of the anti-Siglec-15 antibody, it seems likely that a companion diagnostic test will be helpful to enrich for responders.
It is required that clinical trials are analyzed using a prescribed statistical plan. We believe companion diagnostic tests should be similarly prescribed and tested, rather than developed post hoc in a manner that may result in non-reproducibility as occurred with the PD-L1 IHC tests. The aim of this study was to construct a robust and reproducible, pathologist-read IHC assay to evaluate Siglec-15 expression on tumor cells and in the tumor microenvironment to inform future clinical development, and potentially be used as a future companion diagnostic test for Siglec-15 therapeutics. Furthermore, we illustrate prospective use of our previously described method of determination of the number of Observers Needed to Evaluate a Subjective Test (ONEST) for this assay11, 12.
Materials and Methods
Patient cohort, tissue procurement and immunohistochemistry
A multi-tumor tissue microarray (YTMA 395) with 210 patient samples from 13 different tumor types was assessed to determine the frequency of Siglec-15 protein expression in a range of tumor types. Two lung cancer cohorts were also tested. The first represented a serial collection of two hundred and thirty non-small cell lung cancer (NSCLC) cases collected from 2011 to 2016 (YTMA 423) with long term follow up and has been previously described13, 14. A second cohort with known mutation status comprised of 120 NSCLC cases (YTMA 310, retrospectively NSCLC collected from 2011 to 2013) also previously described 13 was evaluated. Tissue cores of 0.6 mm were used to prepare the tissue microarrays (TMAs), using standard procedures 14, 15. Core samples were obtained from the representative areas of tumor selected by reviewing the hematoxylin and eosin-stained whole tissue slides.
After antibody validation as described below, serial sections were stained for Siglec-15 and PD-L1 (Siglec-15, clone 1F7, rabbit monoclonal, NextCure Inc., Maryland, USA, and PD-L1, clone E1L3N, rabbit monoclonal, Cell Signaling Technology Inc., MA, USA). Clone 1F7 was raised in rabbits against a peptide (ENLSQMNPRSPPATMCSP) in the intracellular domain of Siglec-15. For chromogenic immunohistochemistry, the optimal concentration used was 0.1μg/ml for Siglec-15 and 3.37μg/ml for PD-L1. All TMA slides were digitized using the high-resolution slide scanner Aperio AT2 (Leica Biosystems Inc., IL, USA) and reviewed by three pathologists (DLR, HW, SS) using the software Aperio ImageScope12.4, Leica Biosystems Inc., IL, USA. Multiple scoring systems were tested as described below. All tissue samples were collected with the approval from the Yale Human Investigation Committee protocol #9505008219. Written informed consent, or waiver of consent, was obtained from all patients with the approval of the Yale Human Investigation Committee.
Quantitative immunofluorescence
For immunostaining, we used a protocol for PD-L1 described previously by our group 16. Briefly, slides were heated at 60°C in an oven, de-waxed in xylenes twice (20 minutes each time), then rehydrated with graded ethanol (100% 1 min; 100% 1 min, 70% 1 min) and washed in tap water for 5 min. Antigen retrieval was done using EDTA (0.74 mg in 2 liters of dist. water, pH=8) at 97°C for 20 min in a Lab Vision PT Module (Thermo Scientific, Waltham, MA, USA). Endogenous peroxidases were blocked with 2.5% hydrogen peroxide in methanol for 30 min, followed by additional 30 min of incubation with 0.3% bovine serum albumin with 0.05% Tween-20 blocking solution. For fluorescence staining, optimal signal was seen at 0.1μg/ml for Siglec-15 and 1.1μg/ml for PD-L1. Image analysis was performed using AQUA method (NavigateBP), which generates a QIF score by dividing the sum of target pixel intensities by the area of the molecularly designated compartment. Scores are automatically normalized to lamp hours, bit depth and CC intensity of the microscope 17.
Validation of Siglec-15 in cell lines
For validation of the antibodies, we used the guidelines for pillars of validation from Uhlen et al and specifically for IHC by MacNeil et al 18, 19. Twelve different antibodies, consisting of both commercial and custom monoclonal clones, were tested to find an antibody with high affinity and low cross reactivity, and good performance in a chromogenic assay system that could be further developed as a potential companion diagnostic test. Initial characterization of Siglec-15 expression was previously performed with a polyclonal antibody which is no longer commercially available 4. For each antibody, first we assessed membrane localization (Figure 1), then optimized the titration, and then tested on human embryonic kidney derived cell lines. HEK/293T cells, both non-transduced and transduced with Siglec-15 (HEK/293T.S15) were used as negative and positive controls respectively. A Siglec-15 test array (YTMA 403) was used, which in addition to transfected and non-transfected cell lines, contained non-small cell lung cancers (NSCLC) showing a wide range of dynamic expression of Siglec-15 (Figure 1). This process identified 1F7, a rabbit monoclonal, as the best candidate with the highest signal to noise, but similar in signal localization to a second monoclonal NP411 (Figure 1E, F).
Figure 1. Validation of Siglec-15 antibody.
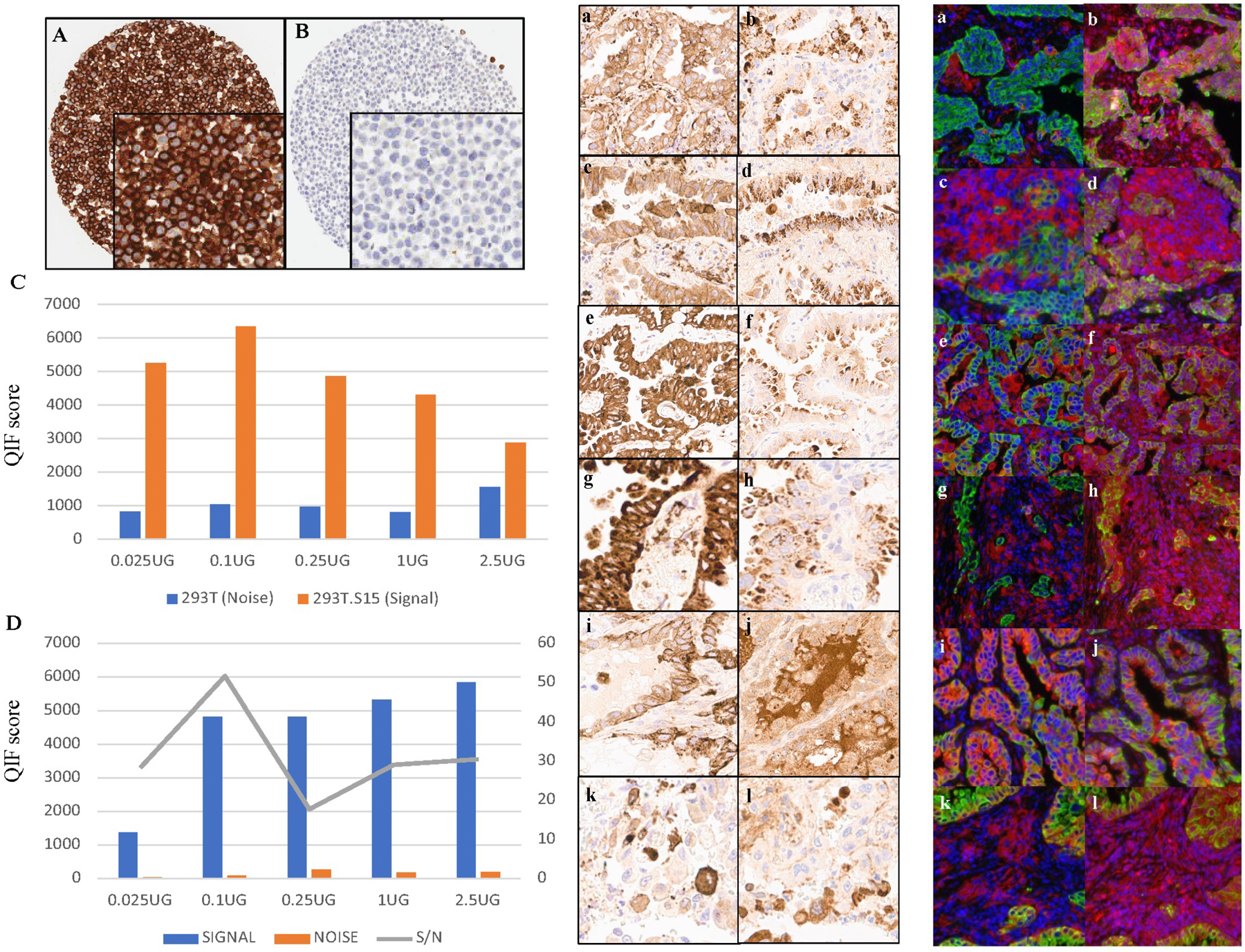
A Membrane staining seen in the transfected cell line 293T.S15 (positive control), confirming the architectural localization of Siglec-15. B No Siglec-15 is seen in non-transfected cell line 293T (negative control). C Signal-to-noise plot for Siglec-15 in cell lines 293T (noise) and 293T.S15 (signal). D Signal-to-noise plot in tumor cores (YTMA 403) showing an optimal concentration at 0.1μg/ml. E, F Comparison of two clones of Siglec-15 in NSCLC by IHC (E) and QIF (F). The images on the left panel are stained with 1F7 (a, c, e, g, i, k). The images on the right are serial sections of the same spots stained with NP411 (b, d, f, h, j, l); red-Siglec-15, blue-DAPI (4′,6-diamidino-2-phenylindole), green-Cytokeratin.
Pathologist Scoring
PD-L1 scoring has been conducted in many ways by IHC depending on the assay and vendor 20. It can be distilled into a score for tumor cells and immune cells using a three-category system with each value representing the range of PD-L1 expression as percentage of tumor cells (TCs) showing positive membrane staining as follows: 0, ≥1% and ≥50% of tumor cells. The scoring does not consider the intensity of staining. Immune cell scoring has been done in many ways as well but can be distilled into the following categories: 0, ≥1% and ≥10%. This system is used by our clinical service here at Yale even though the immune cell scoring system has been shown to be poorly reproducible 12.
We began using the PD-L1 scoring system for Siglec-15, but concordance between pathologist readers was poor. To maximize agreement amongst pathologists, a new categorical system for scoring of Siglec-15 was derived based on the different patterns of staining observed in the tumor and immune cells and loosely based on the currently accepted system for estrogen receptor scoring in breast cancer 21. Our system included 3 categories and were defined as follows: Category 1-tumor cell positive (granular, membranous, or cytoplasmic expression in at least 10% of tumor cells at any intensity); Category 2-immune cell positive (granular expression in at least 10% of immune cells at any intensity) and Category 3-no specific staining (negative for Siglec-15). When there is staining in both the tumor and the immune cells, the predominant pattern is chosen. Cases which were not scoreable (missed spot, inadequate sample, i.e., less than 100 viable tumor cells, distorted morphology, artifactual staining etc.) were excluded from further analyses. Faint cytoplasmic staining was ignored, any granular staining in less than 10% of cells as well as normal pulmonary macrophages entrapped within the tumor cells were excluded. In cases where two different staining patterns were noticed, the predominant pattern was selected. The diaminobenzidine (DAB) slides were reviewed independently by five pathologists, after a brief training session, and then the ONEST analysis was performed. Figure 2 illustrates this scoring system.
Figure 2.
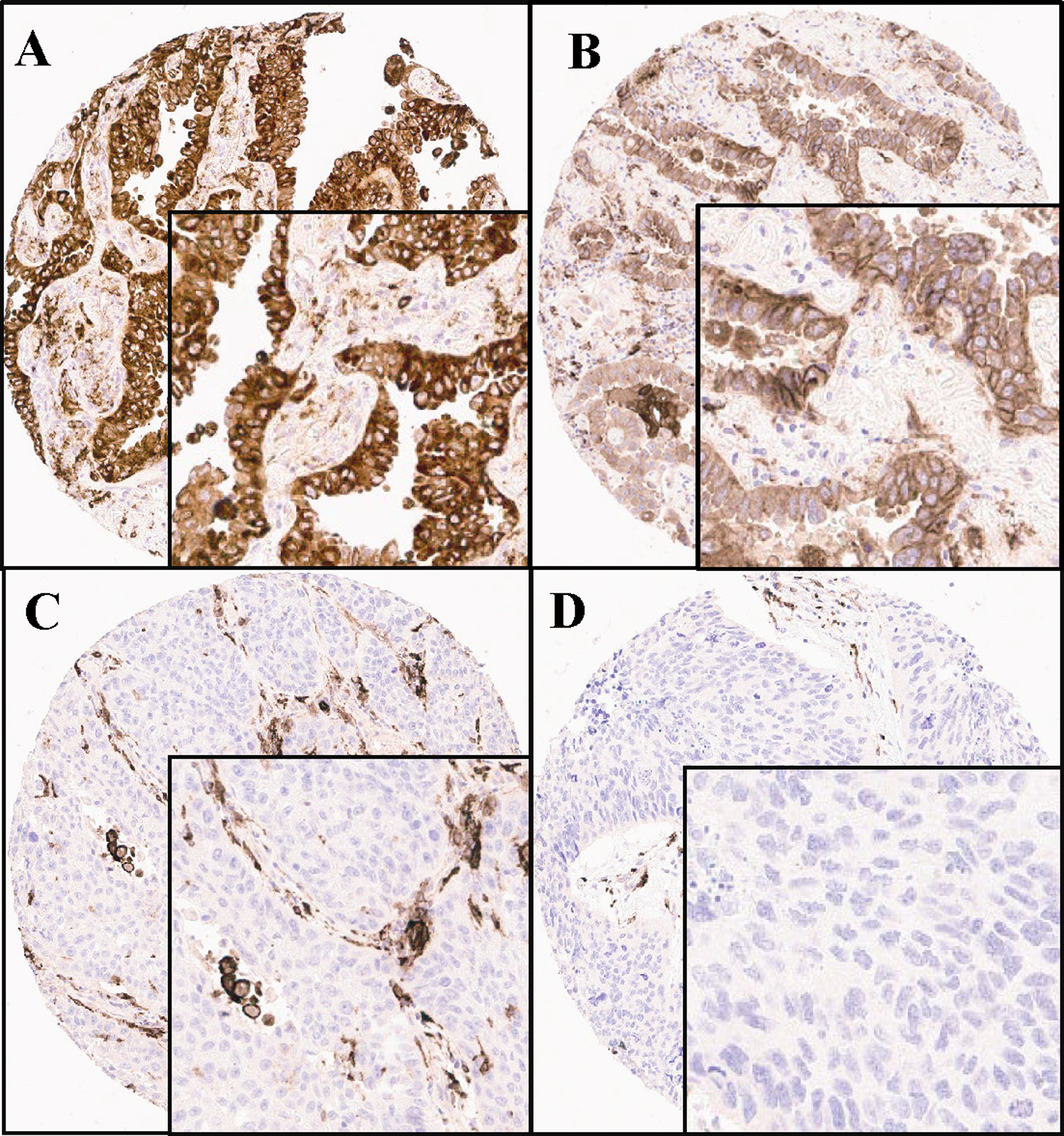
Ordinal (binary) Scoring System for Siglec-15 with Category 1, which shows staining in at least 10% of tumor cells regardless of intensity (A, B), Category 2 with staining in at least 10% of immune cells (C) and Category 3 with no evidence of any staining (D).
Statistical analyses
The inter-reader concordance rates were estimated as overall percent agreement (OPA) calculated as the total number of times in which the raters agree divided by the total number of readings/classifications made. Ordinal scoring, essentially binary scoring, we compared between pathologist readers by the Fleiss Kappa statistic 22. All the graphs, including the Kaplan-Meier survival plots, were generated using GraphPad Prism v8.4.1(460) (GraphPad Software, Inc., CA, USA). A p-value <0.05 was considered statistically significant.
Results
Siglec-15 protein expression in a range of cancer histologies.
Increased levels Siglec-15 expression, as assessed by QIF, was noted in subpopulations of a range of cancer types, notably, lung, breast, bladder, colon, ovarian and head and neck cancers (Figure 3A). In lung cancer, Siglec-15 expression was observed in both tumor cells and immune cells, most likely of myeloid lineage4 (Figure 3B). Similarly, in bladder cancer, tumors were positive for Siglec-15 expression in tumor cells (Figure 3C) or immune cells (Figure 3D). Furthermore, breast tumors were primarily positive for expression of Siglec-15 in immune cells (Figure 3E). Given that Siglec-15 expression was seen to be the highest in lung cancers, we further investigated expression in this cancer type in larger cohorts.
Figure 3. Expression of Siglec-15 in different cancers by immunofluorescence.
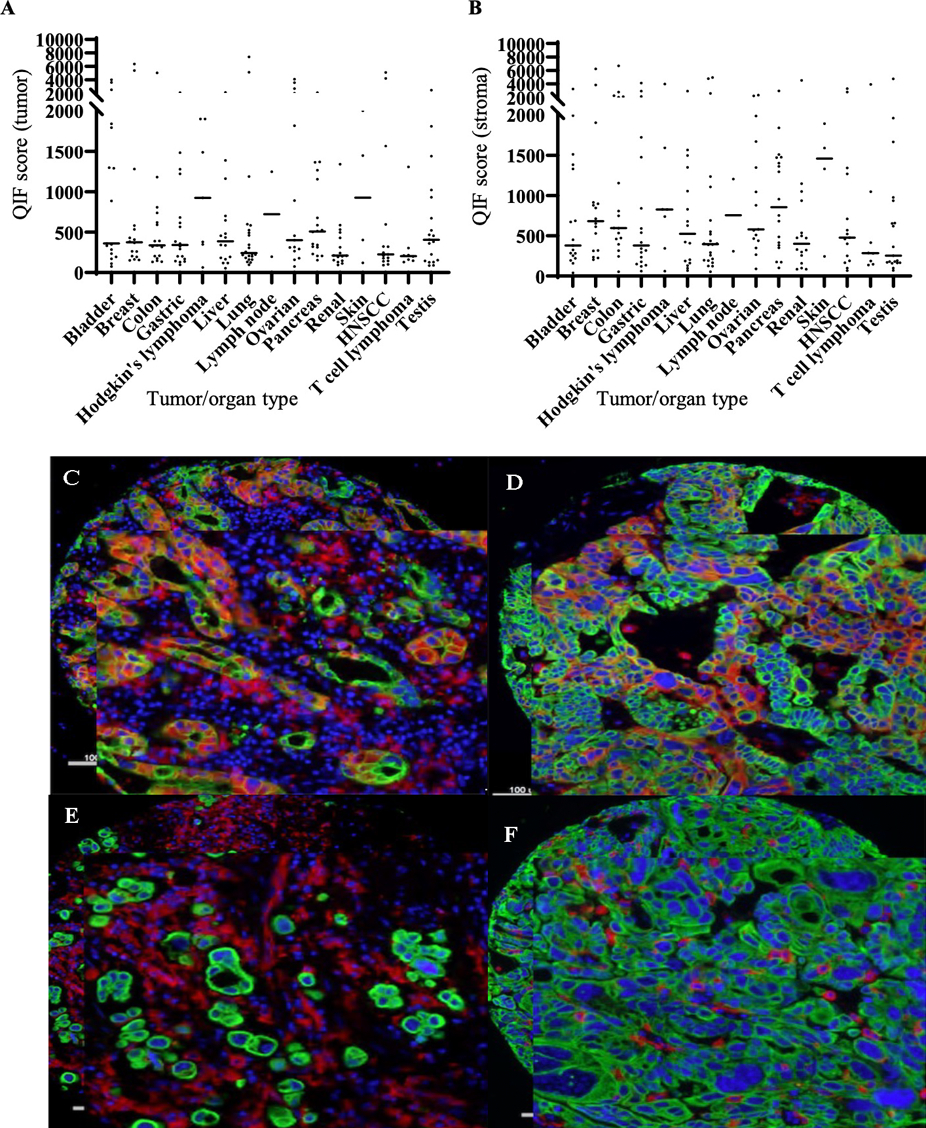
Expression of Siglec-15 in different cancers by immunofluorescence. A, B Lung, bladder, and breast cancers are seen to express high Siglec-15 seen as high QIF scores by AQUA in tumor (A) and stroma (B). Representative images of Siglec-15 expression (red) in tumor and stroma in lung (C), predominantly in tumor cells in bladder cancer (D), mainly in stromal immune cells in bladder cancer (E) and predominantly in stromal immune cells in breast cancer (F); red-Siglec-15, blue-DAPI (4′,6-diamidino-2-phenylindole), green-Cytokeratin.
Expression of Siglec-15 in NSCLC by IHC.
To further support development of a Siglec-15 companion diagnostic test, traditional IHC was performed on an NSCLC cohort. After testing multiple systems, including that used for PD-L1, we found that binary scoring by pathologists as described above in either tumor cells or immune cells with >10% of cells showing expression appeared most promising. Using this system, Siglec-15 expression was common in both tumor cells (42.8% of cases) and stromal immune cells (46.7% of cases) (Figure 4A). The inter-reader concordance using this scoring system was very good with an overall Fleiss Kappa value of 0.676 between three readers. The inter-reader concordance was highest for Category1 (tumor cell positive) and lowest for Category 3 (Fleiss Kappa for Category 1: 0.773, Category 2: 0.652, Category 3: 0.395). Table 1 shows the category-wise percent agreement between 3 pathologists.
Figure 4.
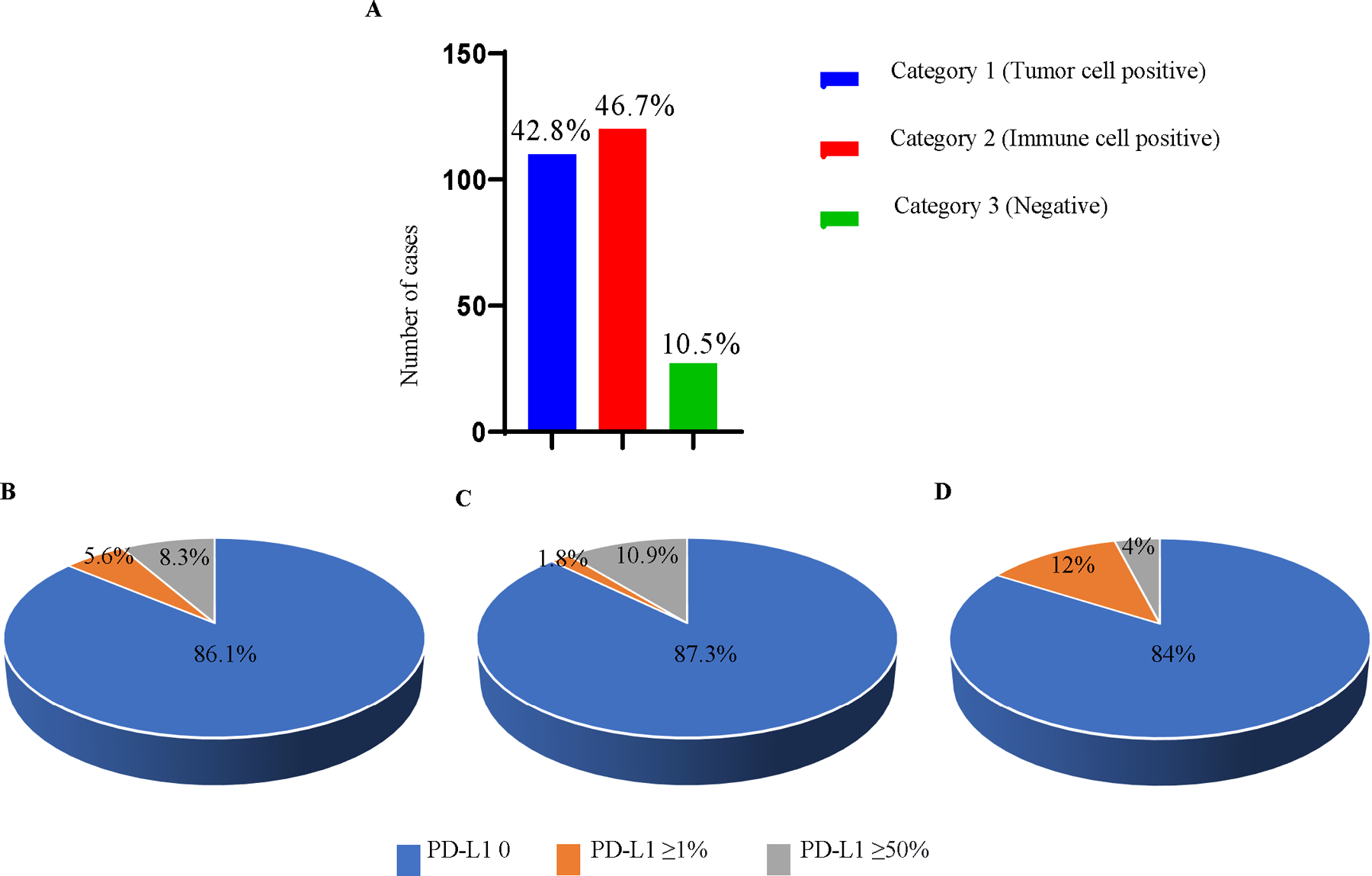
Comparison of Siglec-15 and PD-L1 in tumor showed some-what mutually exclusive expression. A Overall expression of Siglec-15 in NSCLC (YTMA 423). B PD-L1 expression in Category 1 (tumor cell positive for Siglec-15). C PD-L1 in Category 2 (immune cell positive for Siglec-15). D PD-L1 in Category 3 (no staining for Siglec-15).
Table 1.
Category-wise agreement between three pathologists for scoring Siglec-15 in NSCLC (YTMA 423)
| Agreement on Individual Categoriesa | |||||||
|---|---|---|---|---|---|---|---|
| Rating Category | Conditional Probability | Kappa | Asymptotic | Asymptotic 95% Confidence Interval | |||
| Standard Error | z | Sig. | Lower Bound | Upper Bound | |||
| 1.00 | .515 | .773 | .038 | 20.253 | .000 | .770 | .775 |
|
| |||||||
| 2.00 | .415 | .652 | .038 | 17.095 | .000 | .650 | .655 |
|
| |||||||
| 3.00 | .070 | .395 | .038 | 10.360 | .000 | .393 | .398 |
Sample data contains 229 effective subjects and 3 raters.
The addition of two more pathologists reading the same cases allowed us to analyze the assay performance using the ONEST method. Figure 5A shows an ONEST curve and figure 5B shows the 95% confidence interval around the mean of the overall percent agreement (OPA) of the 5 observers. Tumor vs not tumor appears to plateau around 73% OPA. Stroma vs not stroma performs somewhat worse at 61% and may not be valid and may be re-evaluated after clinical trial response data is collected. The highest agreement (OPA = 80%) is seen for cases with no expression. The ONEST method 11 allows a statistical model to be built based on five observers that shows that 5 observers are sufficient since the change in the 95% confidence interval is <2%.
Figure 5. ONEST plots for assessment of tumor staining of Siglec 15.
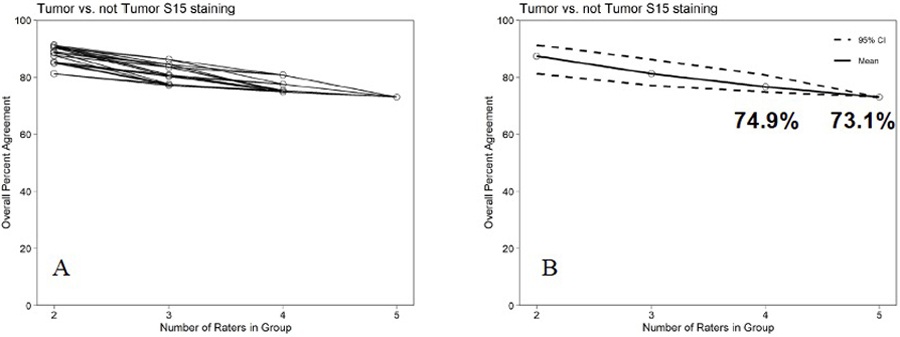
A. The OPA for set of 2 to 5 raters for tumor staining by Siglec 15. B. The mean and 95% lower bound for the OPAs for 2 to 5 observers.
Using this system to assess expression, no associations are seen between clinical variables or patient survival in the cohort treated with standard of care therapy pre-dating immunotherapy (see supplemental figure S1). Also, assessment of two older cohorts (>10 years old) showed loss of detection of expression by this assay with time (Supplemental figure S2).
Comparison of Siglec-15 and PD-L1 in NSCLC.
Previous evaluation of expression of Siglec-15 and PD-L1 has been described to be predominantly mutually exclusive 4. To further evaluate this relationship, we assessed expression of both proteins in our NSCLC cohort (YTMA 423) using both IHC assessment methods. Distribution of Siglec-15 expression observed in the cohort was split between category 1 (tumor cell positive) and category 2 (immune cell positive) with ~43% positive for tumor expression and 47% positive for immune expression. The remaining 10% were negative for both (Figure 4A). Note that this scoring system finds more “positive” cases than previously described in the original work by Wang et al most likely due to a broader scoring system and an antibody with enhanced sensitivity 4. Evaluation of PD-L1 and Siglec-15 expression revealed that for each category, most positive samples were negative for PD-L1 (Figure 4B, C, D) with similar levels of PD-L1 expression observed for all categories. This observation is similar to the initial finding4 in where a polyclonal antibody was used (see supplemental figure 3).
Siglec-15 in different mutation subtypes of NSCLC.
To determine the association between Siglec-15 expression and mutation type in NSCLC, we used a smaller cohort where each case had been sequenced for mutation type (YTMA 310). Siglec-15 expression was detected across mutation types as well as in non-mutated cases (Figure 6), suggesting no association between Siglec-15 expression and the common KRAS and EGFR mutations, in NSCLC. Notably, the presence of samples with mutations were observed at a higher frequency in category 2 (immune cell) positive samples, but frequencies were similar for KRAS and EGFR.
Figure 6. Siglec-15 in different mutation types of NSCLC (YTMA 310).
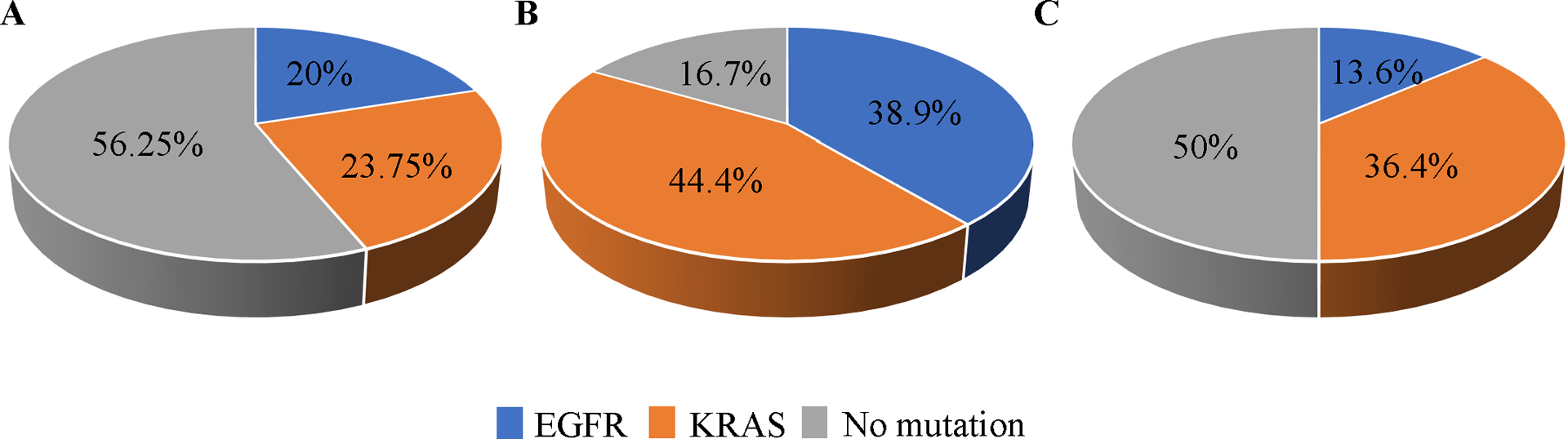
A Siglec-15 expression in Category 1 (tumor cell positive for Siglec-15). B Siglec-15 expression in Category 2 (immune cell positive for Siglec-15). C Siglec-15 expression in Category 3 (no positive
Discussion
Targeted therapies are becoming a cornerstone of treatment modalities utilized in oncology. However, with targeted therapies comes the requirement of selecting the appropriate patient population. For immunotherapy, PD-L1 immunohistochemistry is critical for triaging treatment for patients with NSCLC, as those with PD-L1 ≥ 50% tumoral staining are often treated with the PD-1 inhibitor pembrolizumab alone as a first-line drug treatment 23–25. However, there are several challenges with developing an IHC assay that have been highlighted with the assays developed for PD-L1, and low pathologists’ concordance rates for immune cell assessment have been extensively described26, 27. In designing a system for assessment of Siglec-15 expression, we aimed to avoid some of the key issues suffered by the PD-L1 test. As a result, the expression of Siglec-15 in lung cancers was categorized similar to the estrogen receptor in breast cancer 21 where categories are binary (with a 10% cut-point rather than a 1% cut-point). While the clinical trial (NC318) to determine the predictive power of expression in these categories is ongoing, we show here that, like estrogen receptor, there is high concordance amongst pathologist readers for assessment of Siglec-15 expression and that using the ONEST method, 5 observers are sufficient to validate this scoring system for Siglec 15 expression in tumor cells.
The current study describes the expression of Siglec-15 in NSCLC using a new, high affinity rabbit monoclonal antibody against the intracellular domain of Siglec-15. Siglec-15 expression in lung cancer has been previously described in a non-small cell lung cancer (NSCLC) cohort with QIF using a polyclonal anti-Siglec-15 antibody PA5–48221 28. Utilizing this antibody, a wide range of expression of Siglec-15 both in tumor cells as well as immune cells was observed. This agrees with the study conducted by Wang et al showing both tumor and immune cell expression of the antibody, put the percent patients expressing Siglec-15, as read by pathologists is over twice that seen by QIF 28. This is most likely due to a rabbit monoclonal with higher affinity and lower noise than seen in previous studies with a polyclonal antibody. Nevertheless, the predictive value of expression or the need to stratify expression levels to associate with drug response is not yet established.
Previous studies of Siglec 15 expression 6, 28, 29 showed that patients with high Siglec-15 expression generally have low PD-L1 expression in a pattern of incomplete, but predominant mutual exclusivity between Siglec-15 and PD-L1. The higher sensitivity and ordinal classification seen with this new assay maintains but diminishes the mutual exclusivity. Specifically, while most cases with either high tumor or stromal expression of Siglec-15 show no PD-L1 expression, the cases that express high levels of both markers is increased from the very low levels described when assessment was done on a continuous scale by QIF with a polyclonal Siglec-15 antibody and PD-L1 assessed quantitatively rather than by pathologist readers. A future paper will address this issue in a more quantitative manner
This study has several limitations to consider. Perhaps the most significant is that the trial is only now being run with sufficient tissue to assess response to Siglec-15 therapy. Thus, the categorical scoring system used in this work has not been shown to correlate with response to therapy. However, we believe prospective assay design in as important as prospective design of the statistical analysis, and thus have developed and validated an assay prior to collection of tissue from clinical trial subjects. Second, as is common in developmental studies, we used TMA rather than whole tissue slides for the evaluation of Siglec-15 expression. The TMAs provide a small sample of tumor to be evaluated and the intra-tumoral heterogeneity would be more accurately captured in a larger tumor sample. Heterogeneity of expression of Siglec-15 appears to be similar to PD-L1 and other checkpoint inhibitor molecules in other studies in our group using continuous assessment of expression levels 30. Heterogeneity is harder to assess with an ordinal or binary scoring system, and thus, future work will further characterize the heterogeneity observed in more detail. Finally, the tissues analyzed here are not all recently collected. The age of the tissue is one of hundreds of pre-analytic variables that are uncontrolled in all IHC studies. This is another reason why prospective studies are important for a complete characterization for this and all IHC assays.
In conclusion, our study describes a new IHC companion diagnostic assay for Siglec-15 with a good pathologist concordance. We show somewhat mutual exclusivity of expression between Siglec-15 and PD-L1 expression in NSCLC as seen previously. However, the number of cases expressing both targets have increased, which may suggest more heterogeneity in expression than previously reported. Future efforts are underway to quantify this heterogeneity of expression observed in NSCLC and other tumor types and assess the prognostic and predictive value of this new Siglec-15 assay.
Supplementary Material
Acknowledgements
We would like to thank Lori Charette, Deirdre H. Salemme, Amos Brooks, Patricia Gaule and the team at the Yale Pathology Tissue Service and Developmental Histology Facility for production of the high-quality tissue sections and IHC staining.
Funding
This study was supported by NextCure (DLR) and the Yale SPORE in Lung Cancer.
Footnotes
Ethics Approval and Consent to Participate All tissue samples were collected with the approval from the Yale Human Investigation Committee protocol #9505008219. Written informed consent, or waiver of consent, was obtained from all patients with the approval of the Yale Human Investigation Committee.
Competing Interests DLR has served as an advisor for AstraZeneca, Agendia, Amgen, BMS, Cell Signaling Technology, Cepheid, Daiichi Sankyo, Novartis, GSK, Konica Minolta, Merck, NanoString, PAIGE.AI, Perkin Elmer, Roche, Sanofi, Ventana and Ultivue. Amgen, Cepheid, Konica Minolta, NavigateBP, NextCure, and Lilly fund research in his lab. Sue Niu, Linda, Liu, Zac Cusumano and Sol Langermann are employees of NextCure. J. Zugazagoitia has served as a consultant for Astra Zeneca, BMS, Roche, Pfizer, Novartis, and Guardant Health. Reports speakers’ honoraria from BMS, Pfizer, Roche, Astra Zeneca, NanoString and Guardant Health. Reports travel honoraria from BMS, Pfizer, Roche, Astra Zeneca, and NanoString. Receives research support/funds from BMS, Astra Zeneca, and Roche.
Contributor Information
Saba Shafi, Department of Pathology, Yale University School of Medicine, New Haven, CT, USA.
Thazin Nwe Aung, Department of Pathology, Yale University School of Medicine, New Haven, CT, USA.
Charles Robbins, Department of Pathology, Yale University School of Medicine, New Haven, CT, USA.
Jon Zugazagoitia, Department of Medical Oncology, Hospital Universitario 12 de Octubre Hospital, Madrid, Spain.
Ioannis Vathiotis, Department of Medicine, School of Medicine, National and Kapodistrian University of Athens, 15772 Athens Greece.
Niki Gavrielatou, Department of Pathology, Yale University School of Medicine, New Haven, CT, USA.
Vesal Yaghoobi, Department of Pathology, Yale University School of Medicine, New Haven, CT, USA.
Aileen Fernandez, Department of Pathology, Yale University School of Medicine, New Haven, CT, USA.
Shuqiong Niu, NextCure Inc., Beltsville, MD, USA.
Linda N. Liu, NextCure Inc., Beltsville, MD, USA
Zachary T. Cusumano, NextCure Inc., Beltsville, MD, USA
Nalin Leelatian, Department of Pathology, Yale University School of Medicine, New Haven, CT, USA.
Kimberley Cole, Department of Pathology, Yale University School of Medicine, New Haven, CT, USA.
He Wang, Department of Pathology, Yale University School of Medicine, New Haven, CT, USA.
Robert Homer, Department of Pathology, Yale University School of Medicine, New Haven, CT, USA.
Roy S. Herbst, Department of Medicine, Yale University School of Medicine, New Haven, CT, USA
Sol Langermann, NextCure Inc., Beltsville, MD, USA.
David L. Rimm, Department of Pathology, Yale University School of Medicine, New Haven, CT, USA
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request after publication.
References
- 1.Crocker PR &Varki, A. Siglecs, sialic acids and innate immunity. Trends Immunol 22, 337–342 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Adams OJ, Stanczak MA, von Gunten S & Laubli H Targeting sialic acid-Siglec interactions to reverse immune suppression in cancer. Glycobiology 28, 640–647 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Varki A & Angata T Siglecs--the major subfamily of I-type lectins. Glycobiology 16, 1R–27R (2006). [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Sun J, Liu LN, Flies DB, Nie X, Toki M et al. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat Med 25, 656–666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angata T, Tabuchi Y, Nakamura K &Nakamura, M. Siglec-15: an immune system Siglec conserved throughout vertebrate evolution. Glycobiology 17, 838–846 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Li B, Zhang B, Wang X, Zeng Z, Huang Z, Zhang L et al. Expression signature, prognosis value, and immune characteristics of Siglec-15 identified by pan-cancer analysis. Oncoimmunology 9, 1807291 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gettinger S, Horn L, Jackman D, Spigel D, Antonia S, Hellmann M et al. Five-Year Follow-Up of Nivolumab in Previously Treated Advanced Non-Small-Cell Lung Cancer: Results From the CA209–003 Study. J Clin Oncol 36, 1675–1684 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Pan C, Liu H, Robins E, Song W, Liu D, Li Z et al. Next-generation immuno-oncology agents: current momentum shifts in cancer immunotherapy. J Hematol Oncol 13, 29 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R et al. Five-year survival outcomes for patients with advanced melanoma treated with pembrolizumab in KEYNOTE-001. Ann Oncol 30, 582–588 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellone S, Buza N, Choi J, Zammataro L, Gay L, Elvin J et al. Exceptional Response to Pembrolizumab in a Metastatic, Chemotherapy/Radiation-Resistant Ovarian Cancer Patient Harboring a PD-L1-Genetic Rearrangement. Clin Cancer Res 24, 3282–3291 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han G, Schell MJ, Reisenbichler ES, Guo B & Rimm DL Determination of the number of observers needed to evaluate a subjective test and its application in two PD-L1 studies. Stat Med (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reisenbichler ES, Han G, Bellizzi A, Bossuyt V, Brock J, Cole K et al. Prospective multi-institutional evaluation of pathologist assessment of PD-L1 assays for patient selection in triple negative breast cancer. Mod Pathol 33, 1746–1752 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vathiotis IA, MacNeil T, Zugazagoitia J, Syrigos KN, Aung TN, Gruver AM et al. Quantitative Assessment of CD200 and CD200R Expression in Lung Cancer. Cancers (Basel) 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camp RL, Charette LA & Rimm DL Validation of tissue microarray technology in breast carcinoma. Lab Invest 80, 1943–1949 (2000). [DOI] [PubMed] [Google Scholar]
- 15.McCabe A, Dolled-Filhart M, Camp RL & Rimm DL Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. J Natl Cancer Inst 97, 1808–1815 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Schalper KA, Velcheti V, Carvajal D, Wimberly H, Brown J, Pusztai L et al. In Situ Tumor PD-L1 mRNA Expression Is Associated with Increased TILs and Better Outcome in Breast Carcinomas. Clin Cancer Res 20, 2773–2782 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Camp RL, Chung GG & Rimm DL Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med 8, 1323–1327 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Uhlen M, Bandrowski A, Carr S, Edwards A, Ellenberg J, Lundberg E et al. A proposal for validation of antibodies. Nat Methods 13, 823–827 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacNeil T, Vathiotis IA, Martinez-Morilla S, Yaghoobi V, Zugazagoitia J, Liu Y et al. Antibody validation for protein expression on tissue slides: a protocol for immunohistochemistry. Biotechniques 69, 460–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol 18, 345–362 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Allison KH, Hammond MEH, Dowsett M, McKernin SE, Carey LA, Fitzgibbons PL et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J Clin Oncol 38, 1346–1366 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Williams GH, Nicholson AG, Snead DRJ, Thunnissen E, Lantuejoul S, Cane P et al. Interobserver Reliability of Programmed Cell Death Ligand-1 Scoring Using the VENTANA PD-L1 (SP263) Assay in NSCLC. J Thorac Oncol 15, 550–555 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 375, 1823–1833 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 372, 2018–2028 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman J, Chirieac LR et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 15, 504–535 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Tsao MS, Kerr KM, Kockx M, Beasley M-B, Borczuk AC, Botling J et al. PD-L1 immunohistochemistry comparability study in real-life clinical samples: results of blueprint phase 2 project. Journal of Thoracic Oncology 13, 1302–1311 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rimm DL, Han G, Taube JM, Eunhee SY, Bridge JA, Flieder DB et al. A prospective, multi-institutional, pathologist-based assessment of 4 immunohistochemistry assays for PD-L1 expression in non–small cell lung cancer. JAMA oncology 3, 1051–1058 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corredor G, Wang X, Zhou Y, Lu C, Fu P, Syrigos K et al. Spatial Architecture and Arrangement of Tumor-Infiltrating Lymphocytes for Predicting Likelihood of Recurrence in Early-Stage Non-Small Cell Lung Cancer. Clin Cancer Res 25, 1526–1534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao JQ, Nong JY, Zhao D, Li HY, Su D, Zhou LJ et al. The significance of Siglec-15 expression in resectable non-small cell lung cancer. Neoplasma 67, 1214–1222 (2020). [DOI] [PubMed] [Google Scholar]
- 30.McLaughlin J, Han G, Schalper KA, Carvajal-Hausdorf D, Pelekanou V, Rehman J et al. Quantitative Assessment of the Heterogeneity of PD-L1 Expression in Non-Small-Cell Lung Cancer. JAMA Oncol 2, 46–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request after publication.


