Abstract
Diet can impact on gut health and disease by modulating intestinal stem cells (ISCs). However, it is largely unknown if and how the ISC niche responds to diet and influences ISC function. Here, we demonstrate that Lepr+ mesenchymal cells (MCs) surrounding intestinal crypts sense diet change and provide a novel niche signal to maintain ISC and progenitor cell proliferation. The abundance of these MCs increases upon administration of a high-fat diet (HFD) but dramatically decreases upon fasting. Depletion of Lepr+ MCs resulted in fewer intestinal stem/progenitor cells, compromised the architecture of crypt–villus axis and impaired intestinal regeneration. Furthermore, we showed that IGF1 secreted by Lepr+ MCs is an important effector that promotes proliferation of ISCs and progenitor cells in the intestinal crypt. We conclude that Lepr+ MCs sense diet alterations and, in turn, modulate intestinal stem/progenitor cell function via a stromal IGF1–epithelial IGF1R axis. These findings reveal that Lepr+ MCs are important mediators linking systemic diet changes to local ISC function and might serve as a novel therapeutic target for gut diseases.
Subject terms: Intestinal stem cells, Stem-cell niche
Introduction
Diet is a significant lifestyle factor that impacts on gut homeostasis, intestinal stem cells (ISCs) and disease.1,2 High-fat diet (HFD) is a high risk factor that is linked to many gut diseases, such as colorectal cancer (CRC) and inflammatory bowel disease (IBD).3–5 HFD activates ISCs and increases the tumorigenicity of intestinal progenitors.6 In contrast, fasting attenuates cancer development and is considered an effective strategy in cancer prevention.7,8 Diet exerts its effects on gut health and disease at least in part by modulating stem cell function. ISCs serve as a central mediator that integrates dietary and energy cues to maintain homeostatic turnover and responds to stress. However, the cellular and molecular mechanisms that connect diet to ISCs are not fully understood. A deeper understanding of these mechanisms will be instrumental to prevent diet-induced gut diseases and to develop valid intervention strategies for these diseases.
The fate of ISCs relies on the interplay of multiple signaling pathways that are regulated by niche signals secreted by surrounding cells. Increasing evidence indicates that the underlying mesenchyme plays critical roles in modulating ISCs.9–14 Several mesenchymal cell (MC) types have been identified to provide the signals required for the proliferation and differentiation of ISCs in intestinal crypts. Among them, Pdgfra+, Foxl1+ and Gli1+ MCs express Wnt ligands and R-spondins, providing the source of WNT that promotes ISC self-renewal.9,10,12 Cd34+Gp38+ MCs function as a niche by producing both R-spondin 1 and the BMP signaling antagonist Grem1.11 Most recently, a single-cell RNA-sequencing (scRNA-seq) approach led to the identification of three functionally distinct groups of Pdgfra+ MCs. Pdgfrahigh telocytes provide a reservoir of BMP ligands at the villus base, whereas Pdgfralow MCs beneath crypts express high mRNA levels of the BMP antagonist Grem1.15 Thus, the mesenchymal sources of WNT agonists and BMP inhibitors have been predominantly defined for ISC function at homeostasis. Furthermore, a recent study demonstrated that MC-derived Nrg1 enhanced intestinal regeneration,16 suggesting the importance of growth factors in regulating ISC function during tissue repair. However, despite these efforts, we have a limited understanding of the regulatory effect of dietary and energy cues on mesenchymal niche signals that govern ISC function, particularly during tissue repair.
Leptin is a metabolic regulator that senses nutrients to regulate energy homeostasis and promote tissue growth.17 Leptin exerts its effects by binding to its receptor Lepr that localizes both in the hypothalamus as well as in peripheral tissues, including intestine. Leptin modulates food intake when it binds to Lepr in the hypothalamus, whereas it promotes cell proliferation in other tissues. However, it is not known whether Lepr+ cells link diet to intestinal homeostasis. Here we identified Lepr+ MCs as a novel niche source for ISC and enterocyte (EC) precursor cells. Lepr+ MCs are able to sense diet alterations to modulate ISC status via a stromal Igf1–epithelial Igf1r axis during homeostasis and regeneration.
Results
Lepr+ MCs surround the intestinal crypts
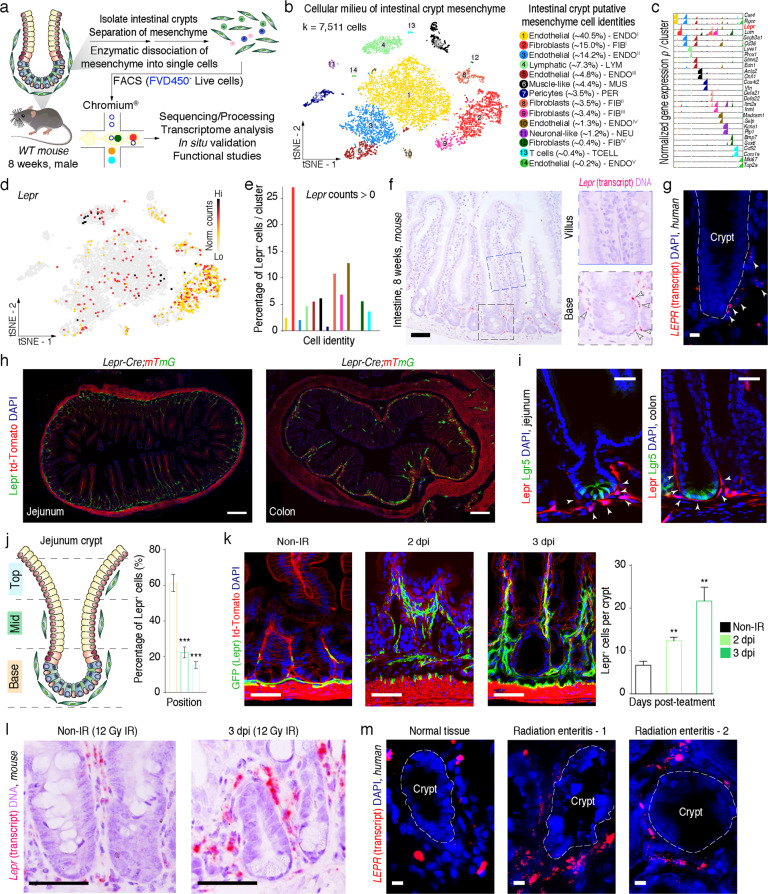
To dissect the cellular features of the mesenchymal niche of ISCs, we first scraped the villi off and isolated MCs from the vicinity of intestinal crypts and subjected them to droplet-based 3ʹ-scRNA-seq (Fig. 1a). After quality control and filtering, 7511 ‘viable’ cells remained and were used for downstream query analyses (Supplementary information, Fig. S1a). Our analysis identified fourteen cell types using t-distributed stochastic neighbor embedding (t-SNE) assay. Based on expression of known marker genes, we defined their putative cell identities, which include four types of fibroblasts, five types of endothelial cells, a small proportion of lymphatic cells, pericytes, T cells, muscle-like cells and neuronal-like cells (Fig. 1b; Supplementary information, Fig. S1b). Two representative signature genes were shown for each cluster (Fig. 1c). Of great interest, Lepr, which encodes a protein for Leptin receptor functioning on energy metabolism, is highly enriched in fibroblasts, and the majority of Lepr+ cells belong to cluster 2 fibroblast I (Fig. 1c–e). Notably, Lepr+ stromal cells are a source of niche signals for hematopoietic stem cells in the bone marrow.18 These observations prompted us to investigate whether Lepr+ fibroblasts may also serve as a niche source for ISCs. To test this idea, in situ hybridization assay was performed. Results demonstrate that Lepr is primarily expressed in the MCs surrounding intestinal crypts but rarely present at the upper region of the intestinal villi in mice (Fig. 1f), and that it is primarily distributed in the lower region of human colonic crypts (Fig. 1g). To label Lepr+ cells in vivo, we generated Lepr-Cre;mTmG mice (Supplementary information, Fig. S1c), in which Cre activity is restricted to cells transcribing the LeprB isoform,19 allowing the labeling of Lepr+ cells by GFP. In agreement with the in situ expression patterns of Lepr transcript, GFP+ cells resided primarily at the base of the intestinal crypts, intermediately at the transit-amplifying (TA) zone, and rarely in the upper region of the villi (Fig. 1h–j; Supplementary information, Fig. S1d). We referred to GFP+ cells as Lepr+ cells in the following study. Consistently, Lepr+ cells are primarily located in the lower region of colonic crypts (Fig. 1h, i; Supplementary information, Fig. S1d). Almost all Lepr+ cells are overlapped with Vimentin+ (Vim+) cells, but completely not with epithelial cells (Supplementary information, Fig. S1e, f), suggesting that they are MCs. Interestingly, only 8.9% of Lepr+ cells are Acta2 (encoding α-SMA)-positive, indicating that the majority of Lepr+ cells are not muscle-like MCs (Supplementary information, Fig. S1e, g). Furthermore, Lepr+ cells were primarily adjacent to Lgr5+ ISCs in both intestine and colon from Lepr-Cre;td-Tomato;Lgr5-eGFP-CreERT2 mice (Fig. 1i). Thus, based on their spatial distribution around the base of the crypt, Lepr+ cells appear to be a potential mesenchymal niche for ISCs.
Fig. 1. Lepr+ MCs surround intestinal crypts and expand upon irradiation.
a Schematic diagram of MC isolation from WT intestinal crypts and capture of single cells by droplet-based device, 3ʹ-scRNA-seq, and downstream query analyses. b t-SNE plot reveals cellular heterogeneity of intestinal crypt MCs. Fourteen distinct clusters are identified and annotated (k = 7511 viable cells). The putative identity of each cell cluster is defined on the right. c Relative expression of selected cluster-specific genes shown as high-density bar charts (n = 2 genes/cluster). d Feature plots of expression distribution for Lepr. Expression levels are color-coded as in t-SNE. e Quantification of the percentage of Lepr+ cells per cluster. f, g In situ hybridization for Lepr in intestinal crypts and villi from 8-week-old mice (f, n = 3) and human (g, n = 2). Arrowheads point to Lepr+ signal in single cells. The dashed line indicates the border between intestinal epithelium and mesenchyme in crypts. Scale bar, 50 μm (f); 10 μm (g). h Fluorescence images for GFP (refers to Lepr+ cells) and td-Tomato in cross sections of jejunum and colon from Lepr-Cre;mTmG mice (n = 3). Scale bar, 200 μm. i Fluorescence images for td-Tomato (refers to Lepr+ cells) and GFP (refers to Lgr5+ cells) in intestine and colon from Lepr-Cre;td-Tomato;Lgr5-eGFP-CreERT2 mice (n = 3). Arrowheads point to Lepr+ cells. Scale bar, 25 μm. j Schematic diagram showing defined base, middle and top regions in jejunum. Graph showing quantification for Lepr+ cells that are spatially distributed at base, middle and top regions of jejunum. Lepr+ cells were counted in defined areas along the crypt–villus axis (n = 120 intestinal architectures, n = 3 mice). k Fluorescence images for GFP (refers to Lepr+ cells) and td-Tomato in cross sections of jejunum from Lepr-Cre;mTmG mice after 12 Gy of irradiation at the indicated time points. Graph represents quantification of Lepr+ cells per crypt at indicated time points. n = 3 at each time point. Scale bar, 50 μm. l In situ hybridization for Lepr in 8-week-old mice before and 3 days post-irradiation (3 dpi, n = 3). Scale bar, 50 μm. m In situ hybridization for LEPR in intestinal crypts from normal and enteritis human tissues (n = 2). Scale bar, 10 μm. Values in the graphs represent means ± SD. Unpaired Student’s t-test was used for calculating P values in j, k. **P < 0.01; ***P < 0.001.
Foxl1+, Gli1+ and Pdgfra+ cells have been identified as mesenchymal niche sources for ISCs;9,10,12 thus, we asked whether Lepr+ cells overlap with these previously identified mesenchymal niche cells. Foxl1 was specifically enriched in cluster 6 muscle-like MCs characterized by expression of Acta2 (Supplementary information, Fig. S1g, h), which is consistent with the previous report.10 By comparison, no Foxl1+ cells are overlapped with Lepr+ cells among intestinal MCs (Supplementary information, Fig. S1h). To validate this result, we performed in situ hybridization for Lepr and Foxl1 in serial sections, corroborating the notion that Lepr+ cells do not overlap with Foxl1+ cells (Supplementary information, Fig. S1k). Gli1 was mainly enriched in cluster 6 muscle-like MCs, cluster 9 fibroblast III and cluster 7 pericytes, while only sporadically distributed in cluster 2 fibroblast I cells in which Lepr is primarily presented. Quantification analysis showed that 8.5% of Lepr+ cells are overlapped with Gli1+ cells, while 91.5% of Lepr+ cells and 73.9% of Gli1+ cells are unique (Supplementary information, Fig. S1i). Pdgfra is extensively and broadly expressed in MCs through the crypt–villus axis, and Pdgfra+ cells in intestinal crypts are considered as a niche for ISCs.12,15 In line with the previous reports, we found that Pdgfra is broadly presented in clusters 2, 8, 9 and 12 that are all the four clusters of fibroblasts (Supplementary information, Fig. S1j). Quantification analysis showed that 54.9% of Lepr+ cells are overlapped with Pdgfra+ cells, while 45.1% of Lepr+ cells and 71.5% of Pdgfra+ cells are unique (Supplementary information, Fig. S1j). Taken together, these findings indicate that Lepr+ cells are generally distinct from Foxl1+ and Gli1+ MCs, while around half of Lepr+ cells are overlapped with Pdgfra+ cells.
Niche cells are also important for intestinal regeneration under stress and injury conditions, thus we sought to test whether Lepr+ cells are responsive to irradiation. Strikingly, we observed that the abundance of Lepr+ cells remarkably increased three days post-irradiation when epithelial cells rapidly divide in regenerative foci (Fig. 1k). The increase in Lepr+ cells three days post-irradiation was further validated by flow cytometry (Supplementary information, Fig. S1l). Consistently, the levels of Lepr transcript were markedly upregulated in these cells (Fig. 1l). Importantly, the abundance of LEPR+ cells also increased in colonic tissues from patients with radiation enteritis compared with that in normal human colonic tissues (Fig. 1m). Collectively, these data suggest that Lepr+ cells might be important for intestinal regeneration.
Ablation of Lepr+ cells leads to impaired homeostasis and regeneration of the intestinal epithelium
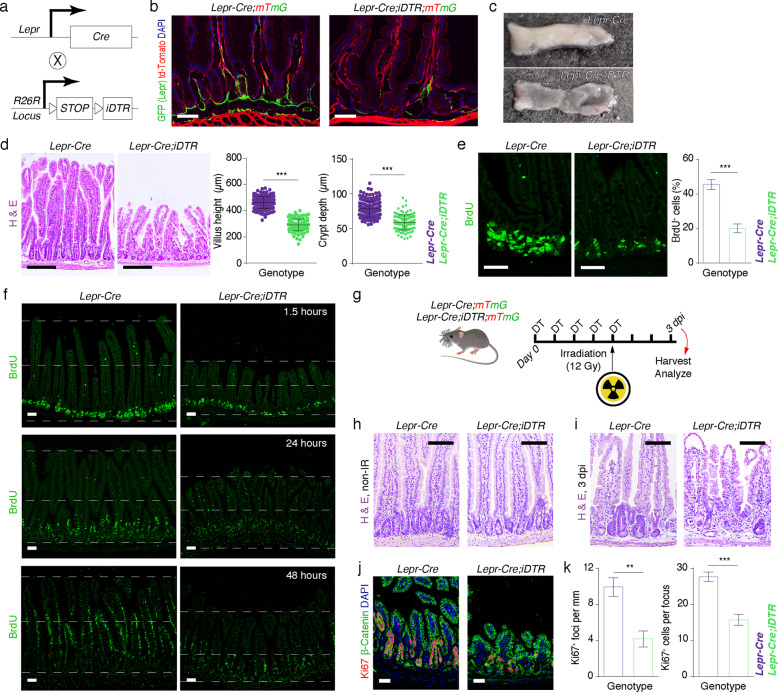
To investigate the function of Lepr+ cells, we generated Lepr-Cre;Rosa26-iDTR (Lepr-Cre;iDTR) mice in which Lepr+ cells are susceptible to diphtheria toxin (DT) treatment (Fig. 2a). After five days of DT administration, almost all Lepr+ cells were ablated in Lepr-Cre;iDTR;mTmG mice, while no other obvious gross phenotypes emerged (Fig. 2b). After ten days of DT administration, ablation of Lepr+ cells resulted in a thinned intestinal mucosa (Fig. 2c) and shortened architecture of the crypt–villus axis in the intestine (Fig. 2d). The number of proliferative cells in intestinal crypts remarkably reduced upon ablation of Lepr+ cells (Fig. 2e). Upward movement of BrdU+ cells along the crypt–villus axis was impaired upon ablation of Lepr+ cells (Fig. 2f), suggesting a defect in tissue turnover. Despite the necessity of Lepr+ cells for promoting cell proliferation in intestinal epithelium, Lepr+ cells are dispensable for cell differentiation, as evidenced by a lack of alteration in the ratio of goblet cells, enteroendocrine cells (EECs) and Tuft cells in Lepr+ cell-ablated intestine (Supplementary information, Fig. S2a–c). The number of Paneth cells slightly reduced in the Lepr+ cell-ablated intestine (Supplementary information, Fig. S2d), which is most likely due to the compromised intestinal architecture. Similar phenotypes were also found in the colons of Lepr-Cre;iDTR mice upon DT treatment (Supplementary information, Fig. S2e–g). Together, these data demonstrate that Lepr+ cells are essential for cell proliferation in intestinal epithelium to maintain its rapid turnover.
Fig. 2. Ablation of Lepr+ cells led to disrupted intestinal homeostasis and regeneration.
a Schematic diagram of the generation of Lepr-Cre;iDTR mice. b Fluorescence images showing ablation of Lepr+ cells (green) in the jejunum from Lepr-Cre;iDTR;mTmG mice (n = 4). Mice were injected intraperitoneally with diphteria toxin (DT) for 5 consecutive days. Scale bar, 50 μm. c Gross images of intestinal mucosa from Lepr-Cre and Lepr-Cre;iDTR mice (n = 4). Mice were injected intraperitoneally with DT for 10 consecutive days. d Histology of intestine from Lepr-Cre and Lepr-Cre;iDTR mice after 10 days of DT treatment. Villus length and crypt depth were quantified. Lepr-Cre, n = 160 villi, n = 176 crypts, n = 5 mice; Lepr-Cre;iDTR, n = 105 villi, n = 122 crypts, n = 5 mice. Scale bar, 200 μm. e Immunofluorescence for BrdU in the jejunum from Lepr-Cre and Lepr-Cre;iDTR mice after a 90-min pulse of BrdU. The percentages of BrdU+ epithelial cells per crypt were quantified. Lepr-Cre, n = 120 crypts, n = 3 mice; Lepr-Cre;iDTR, n = 120 crypts, n = 3 mice. Scale bar, 50 μm. f Immunofluorescence for BrdU in intestines from Lepr-Cre and Lepr-Cre;iDTR mice at the indicated time points after BrdU pulse. The dashed lines mark the top of the villi, middle line of the intestine, and the base of the crypt, respectively. Scale bar, 25 μm. n = 3 for each time point. g Schematic representation of experimental design for irradiation-mediated injury in Lepr-Cre and Lepr-Cre;iDTR mice. h Histology of intestines from Lepr-Cre and Lepr-Cre;iDTR mice before irradiation (n = 3). Scale bar, 100 μm. i Histology of intestines from Lepr-Cre and Lepr-Cre;iDTR mice 3 days after exposure of 12 Gy γ-irradiation (γ-IR). Mice were pre-treated with DT for 5 consecutive days before irradiation (n = 5). Scale bar, 100 μm. j Immunofluorescence of Ki67 and β-Catenin in intestines of Lepr-Cre and Lepr-Cre;iDTR mice 3 days after 12 Gy γ-IR (n = 3). Scale bar, 50 μm. k Quantification of Ki67+ regenerative foci and the number of Ki67+ cells per focus in j. Values in the graphs represent means ± SD. Unpaired Student’s t-test was used for calculating P values in d, e, and k. **P < 0.01; ***P < 0.001.
The increase in Lepr+ cells upon irradiation prompted us to investigate its function during regeneration. Following 5 days of DT treatment, the intestinal crypts were generally identical between control and Lepr-Cre;iDTR mice before irradiation (Fig. 2g, h). However, the regenerative capacity of intestinal epithelium was remarkably impaired after ablation of Lepr+ cells (Fig. 2i). The numbers of regenerative foci and proliferative cells per regenerative focus were markedly reduced three days post-irradiation (Fig. 2j, k). Taken together, these findings demonstrate that Lepr+ cells are critical for intestinal regeneration.
Lepr+ MCs sustain proliferation of intestinal stem/progenitor cells
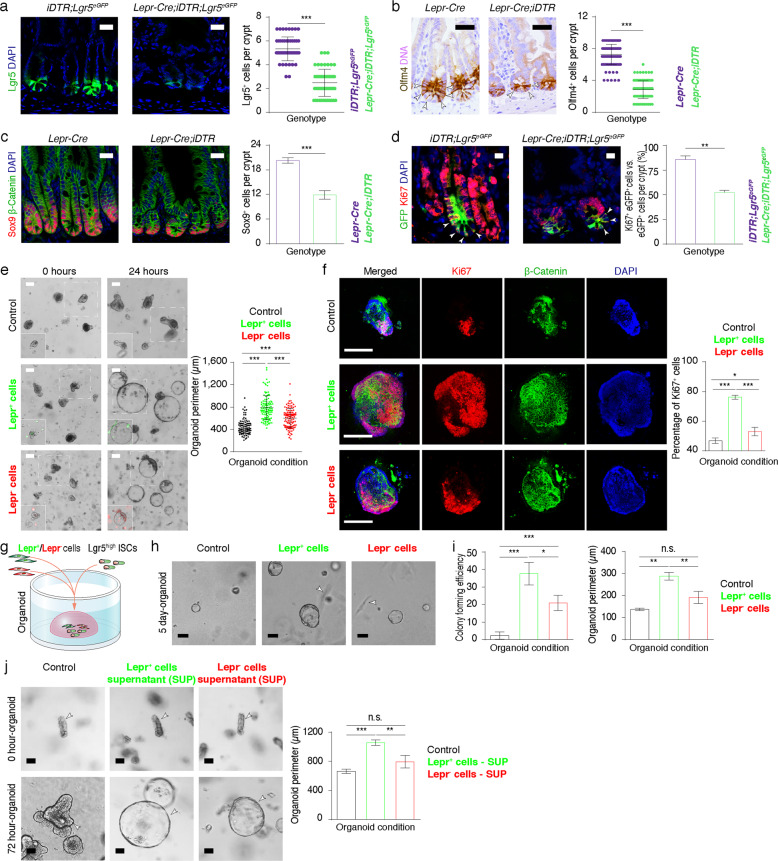
The rapid proliferation of ISCs is critical for intestinal homeostasis and efficient regeneration; hence, we sought to investigate the effect of Lepr+ cells on ISC proliferation. We observed that the number of Lgr5+ ISCs dramatically decreased in both small intestine and colon from DT-treated Lepr-Cre;iDTR mice (Fig. 3a; Supplementary information, Fig. S3a). Olfm4 and Sox9 are known markers for intestinal stem/progenitor cells.20,21 In agreement with this observation, reductions in Olfm4+ and Sox9+ cells were also found both in small intestine and colon upon ablation of Lepr+ cells (Fig. 3b, c; Supplementary information, Fig. S3b). Furthermore, the proportion of proliferative Lgr5+ ISCs was markedly reduced upon ablation of Lepr+ cells (Fig. 3d), which could account for the decrease of intestinal stem/progenitor cells and the shortening of both crypt depth and villus length. In line with the in vivo findings, crypts from Lepr-Cre;iDTR mice appear to be smaller than those from controls; and the intestinal organoids from Lepr-Cre;iDTR mice grew slower and gave rise to fewer buds than those from control mice (Supplementary information, Fig. S3c, d). However, after passaging, the proliferative capacity of organoids from Lepr-Cre;iDTR mice was recovered (Supplementary information, Fig. S3c). These data suggest that the proliferative capacity of intestinal stem/progenitor cells rather than stemness is modulated by Lepr+ cells.
Fig. 3. Lepr+ MCs sustain the proliferation of ISCs.
a Immunofluorescence of GFP (Lgr5-GFP) in jejunum from iDTR;Lgr5-eGFP-CreERT2 (iDTR;Lgr5eGFP) and Lepr-Cre;iDTR;Lgr5-eGFP-CreERT2 (Lepr-Cre;iDTR;Lgr5eGFP) mice after 10 days of consecutive DT treatment. Quantification of Lgr5+ cells per crypt under both conditions (n = 3). Scale bar, 25 μm. b, c Immunostaining for Olfm4 and Sox9/β-Catenin in the jejunum from Lepr-Cre and Lepr-Cre;iDTR mice after 10 days of consecutive DT treatment. Quantification of Olfm4+ cells and Sox9+ cells per crypt under both conditions (n = 3). Scale bar, 25 μm. d Double immunofluorescence for GFP and Ki67 in jejunum from iDTR;Lgr5eGFP and Lepr-Cre;iDTR;Lgr5eGFP mice after 10 days of consecutive DT treatment. Quantification of percentage of Ki67+Lgr5+ cells versus Lgr5+ cells under both conditions (n = 3). Scale bar, 10 μm. e Representative images and quantification of intestinal organoids co-cultured with vehicle control, Lepr+ cells or Lepr– cells at indicated time points after seeding. Crypts were seeded at the same initial density. High-resolution images are shown in the left bottom corner as insets, in which Lepr– or Lepr+ cells can be observed. n = 108 organoids in each group. Scale bar, 100 μm. f Immunofluorescence for Ki67 and β-Catenin for organoids 24 h after seeding in e. Quantification of percentage of Ki67+ cells under the indicated conditions. n = 30 organoids in control group, n = 40 organoids in experimental groups co-cultured with Lepr– or Lepr+ cells. Scale bar, 100 μm. g Schematic diagram showing the co-culture system of Lgr5high single cells with Lepr+ or Lepr– cells. h Representative images of spheroids derived from Lgr5high cells under distinct culture conditions for control, Lepr+ or Lepr– cells after 5 days of co-culture. n = 2 biologically independent experiments, 3 technical replicates each. Scale bar, 20 μm. i Quantification of the colony-forming efficiency and perimeter of Lgr5high cell-derived spheroids in h. j Representative images of intestinal organoids cultured with the supernatants from Lepr+ or Lepr– cells at the indicated time points after seeding. Crypts were seeded at the same initial density. Quantification of the perimeter of organoids at 72 h. n = 2 biologically independent experiments, 3 technical replicates each. Scale bar, 50 μm. Values in the graphs represent means ± SD. Unpaired Student’s t-test was used for calculating P values in a–f, i and j. *P < 0.05; **P < 0.01; ***P < 0.001.
Next, to test the direct effect of Lepr+ cells on intestinal stem/progenitor cells, we co-cultured WT crypts with Lepr+ or Lepr– MCs. The organoids grew faster under the Lepr+ cell co-culture condition than under Lepr– cell and vehicle control conditions (Fig. 3e). In agreement, the percentage of proliferative cells was higher under the Lepr+ cell co-culture condition compared to the Lepr− cell and vehicle control conditions (Fig. 3f). Furthermore, we co-cultured Lgr5high ISCs with Lepr+ or Lepr– MCs (Supplementary information, Fig. S3e, f). Lgr5high cells had higher colony-forming efficiency and gave rise to larger spheroids under the Lepr+ MC co-culture condition than under the Lepr− MC or vehicle control conditions (Fig. 3g–i). It suggests that Lepr+ MCs enhanced the clonogenic and proliferative capacity of ISCs. Moreover, we sought to examine whether secreted factors from Lepr+ cells function in this enhancement. We treated the cultured WT organoids with supernatant from the culture medium of Lepr+ or Lepr– cells. Compared to the supernatant from Lepr– cells, the combination of supernatant from Lepr+ cells and organoid growth medium (OGM) gave rise to larger spheroids (Fig. 3j). These findings suggested that some putative secreted factors derived from Lepr+ cells retain intestinal stem/progenitor cells in a proliferative state.
scRNA-seq analysis identified Lepr+ cell-derived Igf1 as an important niche signal
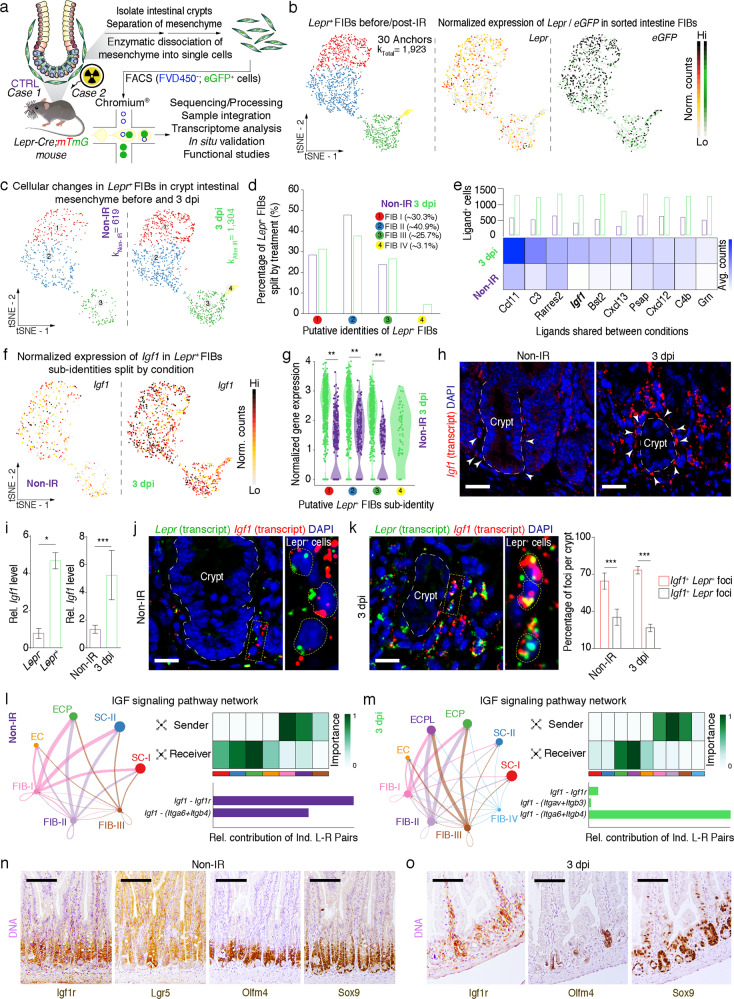
Next, we sought to identify the key secreted factors from Lepr+ cells that have pro-proliferative function. GFP+ MCs from Lepr-cre;mTmG mice before and three days after irradiation were isolated, and subjected to droplet-based 3ʹ-scRNA-seq (Fig. 4a). After quality control filtering, 2232 cells before and 2167 cells after irradiation were used for downstream comparative analysis (Supplementary information, Fig. S4a, b). We first processed the datasets with SCTransform22 and then used Seurat for anchoring and integration (Supplementary information, Fig. S4c). We identified and removed contaminating immune, muscle, endothelial and epithelial cells based on known biomarkers (Supplementary information, Fig. S4d). The remaining fibroblasts displayed high eGFP transcript positivity and most cells were Lepr positive (Fig. 4b), confirming Lepr+ MC identity. The unsupervised clustering analysis identified four distinct eGFP+Lepr+ fibroblast clusters (Fig. 4c, d; Supplementary information, Fig. S4e, f). The majority of these cells were equivalently projected to clusters 1–3 when normalized by number of cells per library post-filtering. These results suggest that the major types of eGFP+Lepr+ fibroblasts were not altered in response to irradiation, with the exception of cluster 4 which appeared to be unique in the irradiation condition. Gene ontology analysis revealed that cluster 4 cells function in cellular response to cytokine stimulus, regulation of inflammatory response, regulation of angiogenesis, response to stress and cellular response to chemokine (Supplementary information, Fig. S4g), supporting the notion that it represents a unique population upon irradiation.
Fig. 4. scRNA-seq analysis identified Lepr+ cell-derived Igf1 as an important niche signal.
a Schematic diagram of Lepr+ MCs sorted from intestinal crypts of Lepr-Cre;mTmG mice before or 3 days post-irradiation, capture of single cells by droplet-based device, 3ʹ-scRNA-seq, and downstream query and comparative analyses. b Sub-cluster of anchored and integrated (n = 30 anchors each) Lepr+ fibroblasts (k = 1923 cells) visualized in t-SNE embedding. Feature plots of Lepr and eGFP expression based on normalized counts and visualized on t-SNE embedding. c Lepr+ fibroblasts split by condition (before IR, k = 619 viable cells; 3 dpi, k = 1304 viable cells). d Quantification of the percentage of Lepr+ fibroblasts per cluster split by condition. e Heatmap showing top 10 ligands expressed in Lepr+ fibroblasts before and 3 days post-irradiation and percentage of cells in each condition expressing such ligand at gene/features counts > 0. f Feature plots of Igf1 expression based on normalized counts and visualized on t-SNE embedding and split by condition. g Violin plot of Igf1 expression based on normalized counts in Lepr+ fibroblast clusters across conditions. **P < 0.01 (two-sided Wilcoxon rank-sum test). h In situ hybridization for Igf1 in jejunum of 8-week-old mice before and 3 days post-irradiation (n = 3). Scale bar, 25 μm. i qRT-PCR analysis for Igf1 in Lepr+ and Lepr– cells from Lepr-Cre;mTmG mice before or 3 days post-irradiation (n = 3). j, k In situ hybridization for Igf1 and Lepr in jejunum of 8-week-old mice before (j) or 3 days post-irradiation (k). The white dashed line represents the border between intestinal epithelium and mesenchyme in crypts. Insets on the right represent large magnification of yellow selected areas. Yellow dash lines in the insets indicate the border of individual cells. Graph showing quantification of the percentage of Lepr+Igf1+ and Lepr–Igf1+ cells in each crypt from WT mice (n = 3). Scale bar, 15 μm. l, m Circle plots showing inferred IGF signaling between Lepr+ fibroblasts (FIB1-IV) and Msi1+ intestinal epithelial cells before irradiation (l) and 3 days post irradiation (m) (cluster stem cells-I (SC-I)), cluster stem cells-II (SC-II), cluster EC precursors (ECP), cluster EC and cluster EC precursor-like cells (ECPL). Data of epithelial cells were adapted from our previous study.29 The size of the circle is proportional to the number of cells in each cell cluster and the edge connecting the circles represent the communication probability between any two cell groups. The color of the edge denotes directionality (i.e., senders vs receivers). The heatmap shows the relative importance of each cell group based on the computed network centrality for sender, receiver, mediator and influencer for IGF signaling pathway network. Graphs represent the quantification of the relative contributions of individual ligand–receptor pairs to the overall IGF communication network. n Immunohistochemistry for Igf1r, Lgr5-GFP, Olfm4 and Sox9 in serial sections of jejunum from Lgr5-eGFP-CreERT2 mice (n = 3). Scale bar, 100 μm. o Immunohistochemistry for Igf1r, Olfm4 and Sox9 in serial sections of jejunum from WT mice 3 days post-irradiation (n = 3). Scale bar, 100 μm. Values in the graphs represent means ± SD. Unpaired Student’s t-test was used for calculating P values in i and k. *P < 0.05; ***P < 0.001.
Next, we identified differentially secreted ligands from Lepr+ cells in either condition. These identified ligands were ranked by their gene expression levels and the proportion of ligand-positive cells against total cells in Lepr+ fibroblasts. This approach led to the identification of top10 ligands, including Ccl11, C3, Rarres2, Igf1, Bst2, Cxcl13, Psap, Cxcl12, C4b and Grm (Fig. 4e). From this list, Psap, Bst2 and Grn are not specifically expressed in Lepr+ fibroblasts (Supplementary information, Fig. S4h). C3 and C4b are complement components,23 and Ccl11, Cxcl13, Cxcl12 and Rarres2 are chemokines,24–26 which are important for immune function, while Igf1 has been implicated to be important in regulating ISC function.27 Thus, Igf1 was selected for downstream query. Igf1 was broadly expressed in Lepr+ cells under both conditions as compared to the other ligands (Fig. 4f), and it was markedly upregulated three days post-irradiation compared to pre-irradiation condition (Fig. 4g–i). Notably, the level of Igf1 was approximately four times higher in Lepr+ MCs than in Lepr– MCs (Fig. 4i). Consistently, the level of Igf1 in the medium from cultured Lepr+ MCs is significantly higher than that from Lepr– MCs (Supplementary information, Fig. S4i). Furthermore, Igf1 transcript co-localized with Lepr in intestinal MCs during both homeostasis and regeneration (Fig. 4j, k), as well as in the colonic tissues of patients with radiation enteritis (Supplementary information, Fig. S4m). These data suggest that Igf1 might be an important secretory effector of Lepr+ cells and may exert its effect on ISCs.
To investigate whether Lepr+ cell-derived Igf1 is an important niche signal for ISCs, we implemented CellChat28 to model potential cell–cell interactions between Lepr+ cells and Msi1+ cells containing their progeny both at homeostasis and three days after irradiation, respectively. Msi1+ cells are a heterogeneous population in the intestinal crypts with the majority being ISCs (Supplementary information, Fig. S4j, k, adapted from our previously published manuscript).29 Using this approach, the network of IGF signaling pathway was found to exert a strong effect on stem cells and EC precursor cells at homeostasis, particularly through the ligand–receptor pairs Igf1–Igf1r and Igf1–(Itga6 + Itgb4) (Fig. 4l; Supplementary information, Fig. S4j). During regeneration, the IGF1 signaling pathway has a stronger effect on EC precursors and EC precursor-like cells than on ISCs (Fig. 4m; Supplementary information, Fig. S4k). In line with this idea, Igf1 receptor Igf1r is strongly expressed in intestinal crypts and is co-localized with the stem/progenitor cell markers Lgr5, Sox9 and Olfm4 both at homeostasis (Fig. 4n) and during regeneration (Fig. 4o; Supplementary information, Fig. S4n). The expression pattern of Igf1r is further supported by scRNA-seq (Supplementary information, Fig. S4l). Taken together, these findings suggest that Lepr+ cell-induced epithelial Igf1 signaling activation plays a role in the proliferation of ISCs and progenitor cells during both homeostasis and regeneration.
Deletion of Igf1 in Lepr+ cells resembles Lepr+ cell-ablation phenotypes in vivo
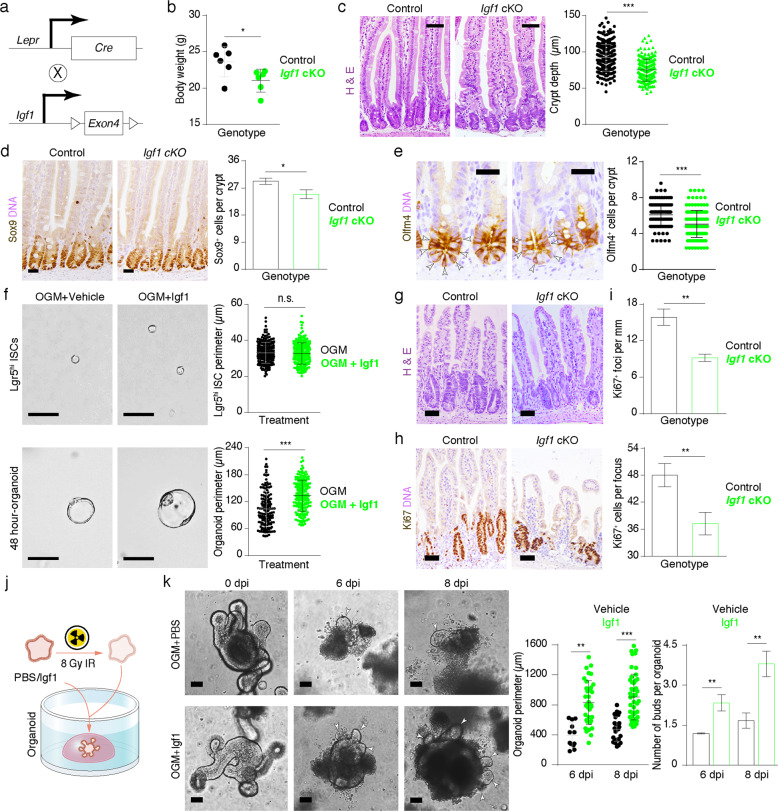
scRNA-seq analyses and cell–cell interaction modeling suggest that the stromal Igf1–epithelial Igf1r axis is potentially important for maintaining ISC function. To verify the in vivo function of Lepr+ cell-derived Igf1, we generated Igf1 conditional knockout (cKO) Lepr-Cre;Igf1fl/fl mice (Fig. 5a; Supplementary information, Fig. S5a), in which Igf1 is specifically deleted in Lepr+ cells. Igf1 cKO mice are viable and fertile with no apparent gross phenotypes. The body weight of cKO mice is slightly lighter than that of their littermate counterparts (Fig. 5b). Deletion of Igf1 in Lepr+ cells led to a mild but significant reduction in crypt depth (Fig. 5c). The number of Sox9+ and Olfm4+ stem/progenitor cells was also significantly reduced in cKO intestine (Fig. 5d, e), and mRNA levels of ISC marker genes Lgr5, Ascl2 and Sox9 were markedly downregulated in Igf1 cKO mice (Supplementary information, Fig. S5b). In line with this, there were fewer proliferative cells in intestinal crypts from cKO mice (Supplementary information, Fig. S5c). In contrast, the addition of recombinant Igf1 protein enhanced the growth of Lgr5high ISC-derived spheroids (Fig. 5f). Collectively, these intestinal phenotypes of Igf1 cKO mice partially resemble the compromised architecture of intestine caused by ablation of Lepr+ cells.
Fig. 5. Deletion of Igf1 in Lepr+ cells partially recapitulates Lepr+ cell-ablated gross phenotypes during homeostasis and regeneration.
a Schematic diagram for the generation of Igf1 cKO mice. b Quantification of body weight in control and Igf1 cKO mice at 8 weeks of age (n = 6). c H&E staining showing the histology of the jejunum from control and Igf1 cKO mice. Crypt depth was quantified (n = 3). Scale bar, 40 μm. d, e Immunohistochemistry for Sox9 (d) and Olfm4 (e) in the jejunum of control and Igf1 cKO mice. Quantification of Sox9+ cells and Olfm4+ cells per crypt was shown on the right (n = 3). Scale bar, 25 μm. f Representative images of spheroids derived from Lgr5high ISCs under culture conditions of OGM + PBS and OGM + Igf1. Top images show individual Lgr5high cells. Bottom images show spheroids 48 h after seeding. The perimeter of Lgr5high ISC-derived spheroids was quantified. n = 2 biologically independent experiments, 3 technical replicates each. Scale bar, 50 μm. g Histology of the jejunum from control and Igf1 cKO mice three days after irradiation. Scale bar, 50 μm. h Immunohistochemistry for Ki67 in jejunum from control and Igf1 cKO mice 3 days post-irradiation. Scale bar, 50 μm. i Quantification of Ki67+ regenerative foci per mm and number of Ki67+ cells per regenerative focus in h (n = 5 each). j Schematic representation of Igf1-treated irradiated organoids. k The intestinal organoids were pre-cultured for three days, and then irradiated with 8 Gy γ-IR and eventually cultured with addition of recombinant Igf1. The perimeter of organoid and the number of organoid buds were quantified 6 and 8 days after recombinant Igf1 protein treatment. Scale bar, 50 μm. n = 2 biologically independent experiments, 3 technical replicates each. Values in the graphs represent means ± SD. Unpaired Student’s t-test was used for calculating P values in b–f, i and k. n.s., not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
Upon exposure to irradiation, deletion of Igf1 in Lepr+ cells resulted in a remarkable reduction in regenerative foci and proliferative cells in each focus (Fig. 5g–i), accompanied by suppression of p-AKT, p-ERK and mTORC1 activity (Supplementary information, Fig. S5d–f). This impaired regenerative capacity resembles the Lepr+ cell-ablated phenotypes more robustly. Furthermore, we irradiated the cultured organoids and then treated them with exogeneous Igf1 recombinant protein (Fig. 5j). Igf1 treatment significantly enhanced organoid growth and budding (Fig. 5k). These findings demonstrated that Lepr+ MC-derived Igf1 plays an important role in promoting intestinal regeneration.
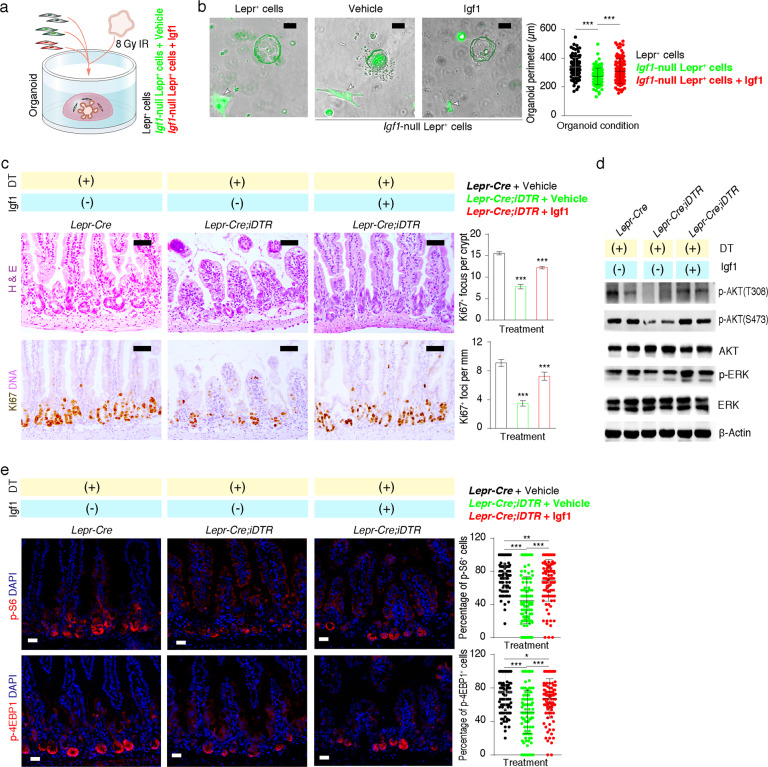
To test whether Igf1 functionally mediates the regenerative role of Lepr+ cells, we irradiated cultured organoids and then co-cultured them with Lepr+ cells, Igf1-null Lepr+ cells, or Igf1-null Lepr+ cells supplemented with Igf1 recombinant protein. Compared to normal Lepr+ cells, Igf1-null Lepr+ cells significantly lost the ability to promote organoid growth, while the addition of Igf1 rescued the organoid-promoting effect (Fig. 6a, b). Furthermore, Igf1 administration rescued the impaired regeneration caused by ablation of Lepr+ cells in vivo (Fig. 6c), accompanied by enhancement of p-AKT, p-ERK levels and mTORC1 signaling activity (Fig. 6d, e). Taken together, these data demonstrate that Igf1 functions as a niche signal that promotes the proliferation of stem/progenitor cells both at homeostasis and regeneration.
Fig. 6. Exogenous Igf1 rescued the impaired regeneration of intestine caused by ablation of Lepr+ cells.
a Schematic representation of irradiated organoids under distinct co-culture conditions. b Images of irradiated organoids co-cultured with Lepr+ cells, Igf1-null Lepr+ cells, or Igf1-null Lepr+ cells + Igf1 captured 3 days post-irradiation with 8 Gy γ-IR. Quantification of the perimeter of regenerative organoids. n = 2 biologically independent experiments, n = 3 each. Scale bar, 50 μm. c Representative images of H&E staining and Ki67 immunohistochemistry of jejunum from Lepr-Cre and Lepr-Cre;iDTR mice 3 days post-irradiation with 12 Gy γ-IR under the indicated conditions. Mice were administered with DT intraperitonially for 5 consecutive days, followed by irradiation and recombinant mouse Igf1 protein treatment. Scale bar, 50 μm. Ki67+ regenerative foci per mm and number of Ki67+ cells per regenerative focus were quantified (n = 3). d Western blotting for p-AKT (T308), p-AKT (S473), AKT, p-ERK and ERK in intestine from Lepr-Cre and Lepr-Cre;iDTR mice. β-Actin was used as loading control. e Immunofluorescence for p-S6 and p-4EBP1, and quantification of the percentage of p-S6+ and p-4EBP1+ cells in each focus from Lepr-Cre and Lepr-Cre;iDTR mice (n = 3). Scale bar, 25 μm. Values in the graphs represent means ± SD. Unpaired Student’s t-test was used for calculating P values in b, c and e. *P < 0.05; **P < 0.01; ***P < 0.001.
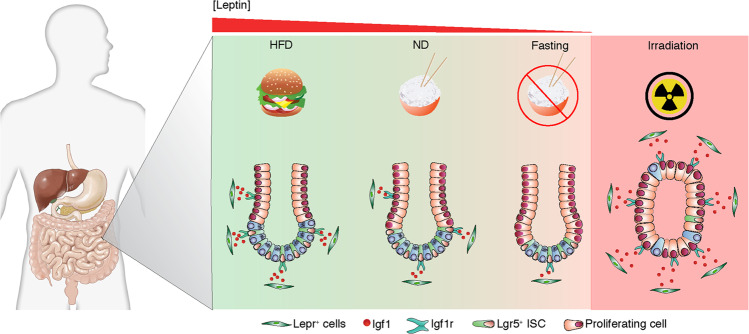
Lepr+ niche cells are modulated by diet
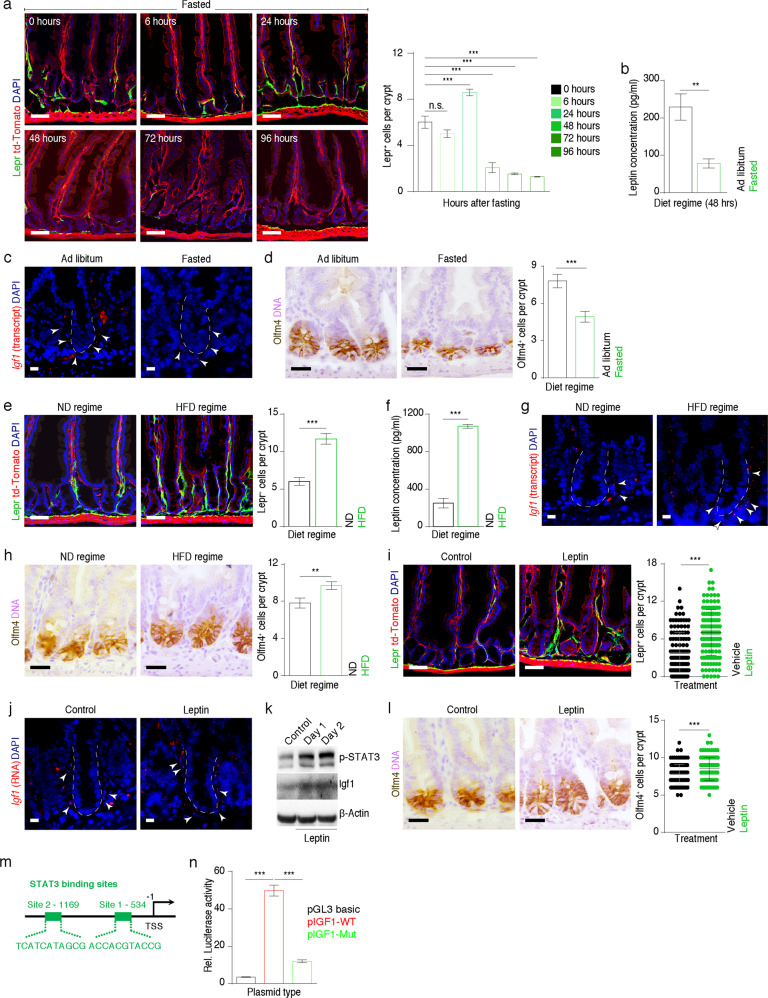
Considering that Leptin, a ligand of Lepr, is a sensor for macronutrition, we thus asked whether Lepr+ cells are responsive to alterations in diet. We subjected Lepr-Cre;mTmG mice to distinct diet regimes. The number of Lepr+ cells decreased 6 h after initiation of fasting, returned to higher counts in abundance at the 24th hour, then sharply declined after the 48th hour, and ultimately only a small number of Lepr+ cells remained (Fig. 7a). The decrease in serum Leptin validates this fasting process (Fig. 7b). In line with the reduction in Lepr+ cells, the level of Igf1 was markedly downregulated in the mesenchyme of the intestinal crypts from fasted mice (Fig. 7c; Supplementary information, Fig. S6k), consequently resulting in a significant reduction in the numbers of Olfm4+ ISCs, Sox9+ stem/progenitor cells and Ki67+ proliferative cells (Fig. 7d; Supplementary information, Fig. S6a, b). Next, we examined cell apoptosis at successive time points after fasting. However, no apoptotic cells were detected in the MCs of intestinal crypts upon fasting (Supplementary information, Fig. S6c), suggesting that Lepr+ cells reduce in an apoptosis-independent manner.
Fig. 7. The abundance of Lepr+ cells was reduced upon fasting and increased upon HFD administration.
a Fluorescent images of the jejunum from Lepr-Cre;mTmG mice under fasting condition at indicated time points (n = 3 mice per time point). Green signal indicates Lepr+ cells. Scale bar, 50 μm. Quantification of Lepr+ cells per crypt. n = 105 crypts for each time point. b Serum levels of Leptin from ad libitum-fed and fasted mice at 48 h (n = 3). c In situ hybridization for Igf1 in jejunum from ad libitum-fed and fasted mice at 48 h (n = 3). Scale bar, 10 μm. d Immunochemistry for Olfm4 in the jejunum from ad libitum-fed and fasted mice at 48 h (n = 3). Number of Olfm4+ cells per crypt was quantified. Scale bar, 25 μm. e Fluorescent images of jejunum from Lepr-Cre;mTmG mice placed under conditions of normal diet (ND) and 90-day HFD regimes. Green signal indicates Lepr+ cells. Number of Lepr+ cells per crypt was quantified (n = 3). Ctrl, n = 105 crypts; HFD, n = 105 crypts. Scale bar, 50 μm. f Serum levels of Leptin from mice placed under ND and HFD regimes (n = 3). g In situ hybridization for Igf1 in jejunum from mice placed under ND and HFD regimes (n = 3). Scale bar, 10 μm. h Immunochemistry for Olfm4 in the jejunum of mice placed under ND and HFD regimes. Number of Olfm4+ cells per crypt was quantified (n = 3). Scale bar, 25 μm. i Fluorescent images of the jejunum of Lepr-Cre;mTmG mice treated with recombinant mouse Leptin protein. Green signal indicates Lepr+ cells. Scale bar, 50 μm. Quantification of Lepr+ cells per crypt (n = 3). Ctrl, n = 129 crypts; Leptin, n = 148 crypts. j In situ hybridization for Igf1 in the jejunum of mice treated with recombinant mouse Leptin protein. Scale bar, 10 μm. k Western blotting for Igf1 and p-STAT3 from the intestines of mice treated with recombinant mouse Leptin protein. β-Actin was used as a loading control. l Immunochemistry for Olfm4 in the jejunum of mice treated with recombinant mouse Leptin protein. Number of Olfm4+ cells per crypt was quantified (n = 3). Scale bar, 25 μm. m The schematic diagram showing two potential STAT3-binding sites in the IGF1 promoter. n Luciferase activity in lysates of 293T cells transfected with luciferase reporter plasmids of pGL3-basic empty vector (basic), wild-type IGF1 promoter, or promoter with mutations of STAT3-binding sites; n = 3. Values in the graphs represent means ± SD. Unpaired Student’s t-test was used for calculating P values in a, b, d–f, h, i, l and n. *P < 0.05; **P < 0.01; ***P < 0.001.
In contrast, when Lepr-Cre;mTmG mice were treated with a HFD for 90 days, Lepr+ cells markedly expanded in the vicinity of the intestinal crypts (Fig. 7e), accompanied by an increase in serum Leptin (Fig. 7f). Consistently, there are more LEPR+ cells in colonic crypts from obese human individuals than lean individuals (Supplementary information, Fig. S6d). In agreement with these findings, the level of Igf1 was upregulated in the mesenchyme surrounding the intestinal crypts from HFD-fed mice (Fig. 7g; Supplementary information, Fig. S6l), leading to a significant increase in stem/progenitor cells and proliferative cells (Fig. 7h; Supplementary information, Fig. S6e, f). To test whether diet-induced Leptin alterations regulate the abundance of Lepr+ cells, we administered recombinant Leptin protein directly into Lepr-Cre;mTmG mice via intraperitoneal injection. Administration of recombinant Leptin protein led to a significant increase in the abundance of Lepr+ cells (Fig. 7i) and Igf1 levels in the mesenchyme of intestinal crypts (Fig. 7j, k; Supplementary information, Fig. S6m). Consistently, it also resulted in the increase of stem/progenitor cells and proliferative cells (Fig. 7l; Supplementary information, Fig. S6g, h). Furthermore, recombinant Leptin treatment induced an enhancement of Igf1 at both RNA and protein levels in cultured primary MCs (Supplementary information, Fig. S6i, j).
To further understand whether Leptin directly regulates Igf1 expression, we first analyzed the potential binding sites of transcription factors in the 2 kb region upstream of the transcription start site (TSS) of the IGF1 gene locus using the JASPAR database and identified two STAT3-binding sites (Fig. 7m). Leptin administration upregulated p-STAT3 levels both in vivo and in vitro (Fig. 7k; Supplementary information, Fig. S6j). Mutating two STAT3-binding sites markedly downregulated IGF1 promoter luciferase activity (Fig. 7n). These findings suggest that Leptin directly upregulates IGF1 by activating STAT3 signaling pathway. Furthermore, we showed that addition of recombinant Igf1 protein significantly promotes the growth of primary MCs (Supplementary information, Fig. S6n). Considering that HFD can enhance circulating Leptin levels, it is likely that HFD can at least partially increase the abundance of Lepr+ cells via a Leptin–STAT3–Igf1 axis.
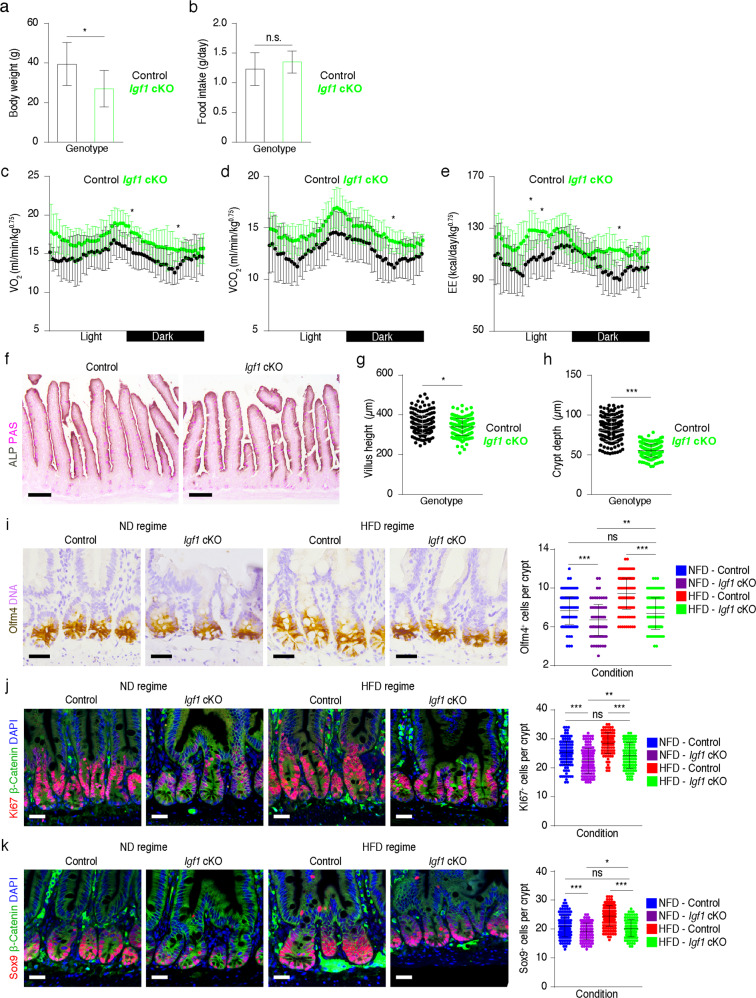
In order to evaluate the physiological significance of Igf1 in Lepr+ MCs in response to diet, we examined physiological features of control and Igf1 cKO mice under the HFD regime. Compared to control, deletion of Igf1 in Lepr+ MCs led to significant reduction in body weight, while food intake was not significantly influenced (Fig. 8a, b). In line with the reduction in body weight, a significant increase in VO2, VCO2 and energy expenditure (EE) was found in Igf1 cKO mice compared to controls (Fig. 8c–e), suggesting an enhanced systematic energy metabolism. Furthermore, we examined differentiated ECs and found that the distribution pattern of differentiated ECs is not changed in cKO mice (Fig. 8f). However, both crypt depth and villus height became significantly shorter upon deletion of Igf1 in Lepr+ MCs (Fig. 8g, h), implying a reduction in absorptive area. Taken together, these findings suggest that HFD-induced Igf1 increase in Lepr+ MCs enhances the absorptive capacity for nutrients, and concomitantly restricts systemic energy expenditure.
Fig. 8. The physiological significance of Igf1 in Lepr+ MCs upon HFD.
a, b Body weight (a) and food intake (b) of control (n = 8) and Igf1 cKO (n = 8) mice under HFD regimes. c–e Indirect calorimetry analysis of oxygen consumption (VO2) (c), exhaled carbon dioxide (VCO2) (d) and energy expenditure (EE) (e) in control and Igf1 cKO mice under HFD regime (n = 3). f ALP-PAS staining in the jejunum from control and Igf1 cKO mice under HFD regime (n = 3). Scale bar, 100 μm. g Villus height of control and Igf1 cKO mice under HFD regime (control, n = 217 villi; Igf1 cKO mic, n = 196 villi). h Crypt depth of control and Igf1 cKO mice under HFD regime (control, n = 180 crypts; Igf1 cKO mice, n = 180 crypts). i Immunohistochemistry for Olfm4 in the jejunum from control and cKO mice treated with ND or HFD. Number of Olfm4+ cells per crypt was quantified (ND regime-Control, n = 137 crypts; ND regime-Igf1 cKO, n = 137 crypts; HFD regime-Control, n = 137 crypts; HFD regime-Igf1 cKO, n = 103 crypts). Scale bar, 25 μm. j Immunofluorescence for Ki67/β-Catenin in the jejunum from control and Igf1 cKO mice treated with ND or HFD. Number of Ki67+ cells per crypt was quantified (ND regime-Control, n = 156 crypts; ND regime-Igf1 cKO, n = 150 crypts; HFD regime-Control, n = 105 crypts; HFD regime-Igf1 cKO, n = 105 crypts). Scale bar, 25 μm. k Immunofluorescence for Sox9/β-Catenin in the jejunum from control and Igf1 cKO mice treated with or without HFD. Number of Sox9+ cells per crypt was quantified (ND regime-Control, n = 151 crypts; ND regime-Igf1 cKO, n = 105 crypts; HFD regime-Control, n = 188 crypts; HFD regime-Igf1 cKO, n = 105 crypts). Scale bar, 25 μm. Values in the graphs represent means ± SD. Unpaired Student’s t-test was used for calculating P values in a–e, g–k. *P < 0.05; **P < 0.01; ***P < 0.001.
We further investigate whether Igf1 mediates the effect of HFD on intestinal stem/progenitor cells. Deletion of Igf1 abrogates the HFD-induced increase in Olfm4+ cells, Sox9+ cells and Ki67+ cells (Fig. 8i–k), suggesting that Lepr+ MC-derived Igf1 is an important effector of HFD to modulate intestinal stem/progenitor cells. Taken together, these data indicate that Lepr+ MCs can be modulated by diet-induced Leptin alteration and function as a niche for ISCs and progenitor cells to govern intestinal homeostasis and regeneration through secretion of Igf1.
Discussion
Here, we demonstrated that Lepr+ MCs function as a novel niche source for ISCs that promotes the proliferation of stem/progenitor cells. Recently, several types of mesenchymal niche cells, including Pdgfra+, Foxl1+, Gli1+ and Cd90+ cells, have been identified.9–12 Our work adds Lepr+ MCs as a novel niche for ISCs to this list. Ablation of Lepr+ cells resulted in shortened crypt depth/villus length and fewer ISCs due to suppression of cell proliferation. In support of its pro-proliferative function, Igf1 was identified as an important effector of Lepr+ MCs that promotes the proliferation of ISCs and EC precursor cells. Deletion of Igf1 in Lepr+ cells, at least partially, recapitulated Lepr+ cell-ablated phenotypes during homeostasis and regeneration. In agreement with our findings, Igf1 has been proposed to enhance ISC activation by modulating the mTORC1 pathway.27,30,31 This work proposes that Lepr+ MC-derived Igf1 is an important niche signal that activates epithelial IGF signaling to maintain the proliferative capacity of ISCs and EC precursors. Previous reports have demonstrated that Wnt and R-spondin, which are generated by Pdgfrα+, Foxl1+ and Gli1+ MCs, function as niche signals for ISCs.9–12 A recent study has proposed that Lepr+ cell-derived Wnt2b promotes regeneration during colitis.32 In addition to Wnt agonists, the present study indicates that the growth factor Igf1 is also an important component of niche signals for ISCs.
It is striking that Lepr+ MCs are capable of sensing diet alterations, as their abundance dramatically decreased upon fasting, while, conversely, increased upon HFD feeding. The alteration of Lepr+ cells is possibly caused by the level change of Leptin, which is regulated by diet. It suggests that diet could modulate ISCs via niche cells. Interestingly, the fasting-induced decrease of Lepr+ MCs is independent of apoptosis. Another interesting finding is that the abundance of Lepr+ cells dramatically increase upon irradiation. Surprisingly, the proliferative capacity of the cells is not enhanced after irradiation. This suggests that the increase of Lepr+ cells may result from a conversion from Lepr– MCs to Lepr+ cells under certain conditions. Another possibility is that Lepr+ cells could be recruited from the vinicity or other organs. It turns out that the mechanism underlying the alteration of Lepr+ cells is complicated and merits future investigation.
It has been reported that fasting results in reduced intestinal wet weight and shortened villi,33 which could be explained by fasting-induced reduction in Lepr+ cells. However, Mihaylova et al. have reported that fasting has pro-regenerative effects on ISC function by activating fatty acid oxidation,34 which is contrary to our findings. This difference can be explained by the different fasting regimes, as they used a short-term fasting (24 h) regime, a time window (6–24 h) when the Lepr+ cells increased in our study. In contrast, it has been reported that a HFD enhances ISC activity and organoid formation capacity by regulating PPAR-δ transcriptional program in ISCs.6,35 Consistently, the HFD-induced enhancement of ISC activity and regenerative capacity could also be explained by the increase in Lepr+ cells. It appears that, in addition to direct regulation of ISCs, diet could also modulate ISC activity via indirect regulation of their niche. Furthermore, it is well-known that HFD is a high-risk factor for many gut diseases.3 The present study proposes a model in which Lepr+ cells are the key mediator that links diet to ISCs (Fig. 9). Considering that IGF1 is a high risk factor for colorectal cancer,36,37 this finding might partially explain the increased incidence of CRC in obese individuals. We also notice that Lepr+ cells can secret chemokines that recruit immune cells, pointing to the potential importance of Lepr+ cells in inflammation-related diseases such as IBD. Thus, Lepr+ cells represent a potential therapeutic target for life style-related gut diseases.
Fig. 9. The working model of the mechanism by which Lepr+ cells modulate ISCs under conditions of HFD regime, fasting regime and post-irradiation.
Lepr+ cells are identified as a mesenchymal niche source for ISCs, and they can sense diet alterations to modulate ISCs via a stromal IGF1–epithelial IGF1R axis. Upon fasting, the abundance of Lepr+ cells dramatically decreases due to the reduced Leptin, leading to the reduction of Lepr+ cell-secreted Igf1. Consequently, the proliferation of ISCs and progenitor cells declines under fasting regime. Conversely, the abundance of Lepr+ cells increases upon HFD regime and post-irradiation, and the increased Lepr+ cells promote the proliferation of ISCs and progenitor cells by enhancing the secretion of Igf1.
Importantly, the finding that HFD-induced Igf1 increase in Lepr+ MCs enhances the absorptive capacity for nutrients and concomitantly restricts systemic EE reveals a novel signal network underlying how animals expedite energy store under the condition of sufficient nutrients. Supporting this notion, oral administration of IGF-I enhances nutrient and electrolyte absorption in neonatal piglet intestine.38 Thus, this work provides significant insights into designing ideal nutritional formula in pig feeding industry.
Materials and methods
Ethics
All mouse experimental procedures and protocols were authorized by the Beijing Laboratory Animal Management and were performed strictly in accordance with the guidelines of the Institutional Animal Care and Use Committee of China Agricultural University (approval number: SKLAB-2019-04-03).
Mice
Lepr-Cre (JAX stock number: 008320), Rosa26-mTmG (JAX stock number: 007676), Rosa26-tdTomato (JAX stock number: 007914), Lgr5-eGFP-CreERT2 (JAX stock number: 008875) and Igf1fl/fl (JAX stock number: 016831) mice were obtained from Jackson Laboratories. Rosa26-iDTR mice were obtained from Hua Zhang’s laboratory at China Agricultural University.
Subjects
Colonoscopic biopsies were obtained from two female patients with radiation enteritis as well as from the normal colonic mucosa in healthy individuals. All patients were from Department of Gastroenterology, XiangYa Hospital of Central South University (Changsha, China). The colonoscopic biopsies of obese and lean individuals were from the Shanghai Tenth People’s Hospital of Tongji University (Shanghai, China). Each subject has provided written informed consent. The diagnosis of radiation enteritis was based on clinical, radiological, endoscopic, and pathological examinations.
Diet treatment
For diet experiments, fasting was achieved by withholding food only from mice, while water remained available. HFD administration was achieved by feeding the mice with a dietary chow consisting of 60% kcal fat for 90 days. Ad libitum-fed controls were gender- and age-matched.
Irradiation injury
Two-month-old mice were subjected to 12 Gy γ-IR and sacrificed at the indicated time points. The cultured organoids were subjected to 8 Gy γ-IR and used for downstream experiments.
Histology, immunohistochemistry and immunofluorescence assays
For histological analysis, paraffin-embedded tissues were sectioned at 5-μm. Tissue sections were stained with hematoxylin and eosin (H&E) using standard methods. Periodic Acid-Schiff (PAS)-alkaline phosphatase (ALP) staining was performed using standard methods. For immunohistochemistry, the sections were deparaffinized with xylene followed by treatment with serial dilutions of ethanol. Antigen retrieval was performed by heating slides in 0.01 M citrate buffer (pH 6.0) for 10 min in a microwave oven. After cooling to room temperature (RT), the sections were incubated with blocking solution for 1 h after administration of 3% H2O2 to eliminate endogenous peroxidase activity. The sections were incubated with primary antibodies overnight at 4 °C. The sections were then immunostained by the ABC peroxidase method (Vector Laboratories) with diaminobenzidine as the enzyme substrate and hematoxylin as counterstain. For immunofluorescence staining, paraffin sections were heated as before, pretreated in 0.01 M citrate buffer (pH 6.0), and then incubated with primary antibodies, followed by incubation with secondary antibodies (Invitrogen) and counterstained with DAPI in mounting media. The primary antibodies used in this study include: Ki67 (Thermo Fisher, RM-9106-S1, 1:1000), Lysozyme C (Santa Cruz, sc-27958, 1:500),
ChgA (Abcam, ab254322, 1:500), Dclk1 (Cell Signaling Technology, 62257, 1:500), GFP (Abcam, ab13970, 1:800), RFP (ROCKLAND, 600-401-379, 1:200), β-Catenin (Sigma, C7082, 1:1000), Olfm4 (Cell Signaling Technology, 39141, 1:800), α-SMA (Santa Cruz, sc-53142, 1:1000), Vim (Santa Cruz, sc-6260, 1:1000), Sox9 (Cell Signaling Technology, 82630, 1:800), Igf1r (Abcam, ab182408, 1:500), p-S6 (Cell Signaling Technology, 4858, 1:800), Caspase 3 (Cell Signaling Technology, 14220, 1:1000) and p-4EBP1 (Cell Signaling Technology, 2855, 1:800).
For BrdU staining, BrdU solution was intraperitoneally injected into mice at 50 μg/g body weight. Paraffin-embedded tissues were sectioned at 5-μm sections and treated with 2× SSC: formamide (Amresco, Solon, OH) at 1:1 ratio at 65 °C for 2 h. Sections were then immersed in 2 M HCl at 37 °C for 30 min, rinsed with 0.1 M boracic acid and washed in 1% PBST. To block nonspecific epitopes, tissues were treated with 5% normal goat serum in 1% PBST with 0.1 M glycine at RT for 1 h. Anti-BrdU antibody (Abcam, ab6326, 1:100) was incubated overnight at RT. The sections were washed three times and incubated with 488-conjugated secondary antibodies (Thermo Fisher, A11006, 1:400) for 1 h at RT, stained with DAPI for 5 min, and finally mounted with anti-fluorescence quenching sealing medium.
Confocal imaging
Samples were embedded in OCT (Tissue-Tek; Sakura Finetek), and frozen in dry-ice for cryosectioning. Tissues were cryosectioned at 10-μm, air-dried for 30 min, washed with PBS three times, and stained with DAPI. Images were captured using a Leica laser scanning confocal microscope (Leica TCS SP8).
Isolation and culture of MCs
Intestinal MCs were isolated as described previously with minor modifications.39 Briefly, intestines were placed in cold HBSS to remove debris. They were opened longitudinally and cut into 0.5-cm pieces, which were then incubated in PBS containing 10 mM EDTA for 30 min at 4 °C to remove primary intestinal epithelial cells. After removing the epithelial cells by vigorous shaking, the remaining tissue was rinsed several times in cold modified DMEM and incubated in digestion solution containing 0.63 mg/mL collagenase II (Thermo Fisher, 17101015) and 0.05 U/mL dispase II for 35 min at 37 °C on a rotating platform. After vigorous shaking, single MCs were pelleted at 350× g for 5 min. The supernatant was discarded and each pellet was resuspended with modified DMEM containing 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin (Sigma), 1× l-glutamine, and 1× nonessential amino acids (NEAAs). The isolated MCs were then subjected to droplet-based 3ʹ-scRNA-seq (10× Genomics), or seeded in a 6-cm dish and cultured in a 5% CO2 incubator at 37 °C. After 2 h, the floating debris and nonadherent cells were washed off with PBS. The adherent cells were intestinal MCs consisting of fibroblasts and myofibroblasts. The cells were passaged 1-week post-seeding at a ratio of 1:3. After passaging three times, MCs or supernatant was collected from each dish for co-culture experiments.
Processing of raw 3ʹ-scRNA-seq data
A single-cell suspension of intestinal MCs was prepared as described above. GFP+ cells were sorted into EP tubes in single-cell mode by FACS. The collected cells were held on ice before loaded for GemCode single cell platform (10×). Chromium Single Cell 3ʹ v2 libraries were sequenced with a Novaseq 6000 sequencer (Illumina), with the following sequencing parameters: read 1, 150 cycles; i7 index, 8 cycles and read 2, 150 cycles.
Whole intestinal MCs from WT mice
Raw FASTQ reads were mapped to the mouse mm10 reference genome (GRCm38.93.dna/GRCm38.93.gtf) using Cell Ranger Version 3.1.0. The alignment metrics based on web summary were as follows. Total number of reads: 339,183,860; reads mapped to genome: 90.3%; reads mapped confidently to the genome: 88.5%; reads confidently mapped to transcriptome: 62.0%; estimated number of cells: 7968; fraction of reads in cells: 94.3%; mean reads per cells: 42,568; median genes per cell: 1222; total genes detected: 18,151; median UMI counts per cell: 2274.
GFP+ cells from Lepr-Cre;mTmG mice before irradiation
Raw FASTQ reads were mapped to a modified version of the mouse mm10 reference genome (GRCm38.93.dna/GRCm38.93.gtf) using Cell Ranger Version 2.1.1. Briefly, the mm10 genome and GTF annotations were modified to include the GFP nucleotide sequence as exon (1–5556 nts) with the following common gene, transcript ID and name: “eGFP”. The alignment metrics based on web summary were as follows. Total number of reads: 399,597,554; reads mapped to genome: 95.1%; reads mapped confidently to the genome: 92.3%; reads confidently mapped to transcriptome: 75.1%; estimated number of cells: 2395; fraction of reads in cells: 75.3%; mean reads per cells: 166,846; median genes per cell: 1586; total genes detected: 17,718; median UMI counts per cell: 4505.
GFP+ cells from Lepr-Cre;mTmG mice 3 days post-irradiation
Raw FASTQ reads were mapped to a modified version of the mouse mm10 reference genome (GRCm38.93.dna/GRCm38.93.gtf) using Cell Ranger Version 2.1.1. Briefly, the mm10 genome and GTF annotations were modified to include the GFP nucleotide sequence as exon (1–5556 nts) with the following common gene, transcript ID and name: “eGFP”. The alignment metrics based on web summary were as follows. Total number of reads: 380,801,379; reads mapped to genome: 92.6%; reads mapped confidently to the genome: 88.9%; reads confidently mapped to transcriptome: 70.5%; estimated number of cells: 2363; fraction of reads in cells: 73.4%; mean reads per cells: 161,151; median genes per cell: 2381; total genes detected: 18,631; median UMI counts per cell: 7687.
Doublet/multiplet simulation
Digitized count matrices were pre-processed and doublets/multiplets were simulated using Scrublet40 (version 0.2.1) with default parameters. The doublet/multiplet score threshold was adjusted manually. Both whole unsorted and GFP+ cells were subjected to the doublet/multiplet simulation and filtering.
Low-quality cell filtering
Whole unsorted and GFP+ singlet cells were kept and used for downstream query analyses if and only if they met the following collective quality control metrics: (a) 350 < genes/cell < 5000; (b) cells contained no more than 10% of mitochondrial gene expression; or (c) cells were not identified as outliers falling outside a prediction confidence interval of features and counts (P value 1e–3).
Bioinformatic analysis
Whole intestinal MCs from WT mice
A Seurat object was created using the filtered count matrix (Version 3.2.2).41 Data were Log-normalized. Variable genes/features were identified using the FindVariableFeatures function with the following functions enabled: selection.method = “vst” and using a total of 2000 features. The data were then scaled using the ScaleData function using all feature genes. Significant principal components (PCs) were used for clustering and neighbors. PCs were identified using a combination of statistical and heuristic methods. Neighbors and clusters were identified using the FindNeighbors and FindClusters functions, respectively, with dimensions specified by the user and visualized using two-dimensional t-SNE. Cluster features were identified using the FindAllMarkers function with the following parameters enabled: test.use = “wilcox”, logfc.threshold = 0.25, only.pos = T. The minimum required LogF.C. in gene expression and minimum percentage of cells expressing a feature were 2× and 0.25%, respectively. Gene biomarkers were Log-normalized and visualized as normalized counts in high-density and feature plots.42 Co-expression of features in individual cells were calculated based on expression of each feature (normalized counts > 0) and visualized in feature plots. Putative cell identities were assigned based on bona fide biomarker feature expression.
Anchoring, integration and downstream analysis
We anchored and integrated sorted GFP+ single cells from non-irradiation and three days post-irradiation conditions using Seurat (Version 3.2.2). Briefly, Seurat objects were created using individual, digitized count matrices post doublet removal and cell pruning. To minimize cell–cell variation across data sets owed to non-biological and technical factors, we performed Single Cell Transform (SCTransform).22 We then performed anchoring (n = 30 anchors) and integration (n = 30 anchors) using the FindIntegrationAnchors and IntegrateData functions, respectively, using SCT as a normalization method. PCs were identified using a combination of statistical and heuristic methods as before. Neighbors and clusters were identified using the FindNeighbors and FindClusters functions, respectively, with dimensions specified by the user and visualized using two-dimensional t-SNE. Cluster features were identified on individual data sets using the FindAllMarkers function with parameters described before. Gene biomarkers were Log-normalized and visualized as normalized counts in feature plots. Putative cell community identities were assigned based on bona fide biomarker feature expression.
Modeling cell–cell communication networks
Cell–cell communication networks were modeled based on abundance of known ligand–receptor pair transcripts using CellChat (version 0.0.1) as previously described with minor modifications. To identify potential cell–cell communication networks between Lepr+ MCs and Msi1+ cells before irradiation or Msi1+ cell progeny three days post-irradiation, we sub-clustered Lepr+ fibroblasts from our data sets and selected epithelial cells from our previous study.29 Seurat objects for either condition (i.e., before or three days post-irradiation) were merged and normalized. Overexpressed ligands and receptors and signaling pathways were identified using CellChat. The mouse CellChat database provided by CellChat was used for identification of ligands and receptors. Pre-processing and downstream inference of cell–cell communication was performed with default parameters on each independent data set. Modeled cell–cell communications were visualized as circle plots. The contribution of each ligand–receptor pair to the overall signaling pathway of interest were identified and graphed. Signaling roles for cells based on network centrality analysis were identified using heatmaps.
Organoid and cell co-culture
Intestinal crypts were isolated from small intestine samples by incubating the tissues in PBS containing 10 mM EDTA for 30 min at 4 °C as described previously.43 The crypt fractions were collected by pipetting and filtered through a 70-μm cell strainer (BD Biosciences). The gathered crypts were washed twice with PBS and collected by centrifugation at 700 rpm for 5 min. The supernatant was removed and the crypts were resuspended into Matrigel (Corning, 356231) and IntestiCult OGM (STEMCELL Technologies, 06005) (ratio of 1:1), and plated into 48-well plates. After Matrigel polymerization, 200 μL of OGM was added to each well. The medium was changed every 2 days, and cultured organoids were passaged every 4 days. For co-culture with MCs, organoids were seeded at the same initial density, and mixed with 2 × 104 Lepr+ cells or Lepr– cells in each well.
To treat organoids with supernatant, 200 μL standard OGM medium was replaced with a mixed medium containing 150 μL OGM and 50 μL supernatant from Lepr+ cells or Lepr– cells, and the mixed medium was changed daily.
For Lgr5high cell culture or co-culture with MCs, intestinal crypts were isolated from Lgr5-eGFP-CreERT2 mice. The isolated crypts were pipetted with prepared 1.8 U/mL dispase for 5 min to generate single-cell suspensions, dissociated cells were filtrated through 40 μm cell strainer (BD Biosciences). Then cells were resuspended in 500 μL of PBS with 2% FBS on ice and sorted on a BD FACS Arial 3.0 machine. According to the co-culture strategy described previously,44 a total of 2000 sorted Lgr5high cells were collected into 1.5-mL tubes containing 2% FBS, mixed with 2000 Lepr+ cells or Lepr– cells at a ratio of 1:1, pelleted by centrifugation, and embedded in Matrigel. After Matrigel polymerization, the cells were seeded on 48-well plates. For the first three days, the sorted cells were incubated in OGM containing Y-27632 (10 μM). To test the effect of Igf1 on Lgr5high ISCs or irradiated organoids, 1 μg/mL of Igf1 (Absin, abs04634) was added to the OGM medium, and then the mixed medium was changed daily. The perimeters of all organoids are quantified by software ImageJ.
Flow cytometry and cell sorting
Single-cell suspensions were stained using Fixable Viability Dye (eBioscience, 65-0863-14) for 15 min to remove dead cells. Flow cytometry analysis was performed on a BD FACS Arial 3.0. Lgr5high cells were sorted by flow cytometry from Lgr5-eGFP-CreERT2 mice.
qRT-PCR analysis
Total RNA was extracted using TRIzol reagent (Life Technologies) according to the manufacturer’s instructions. To detect mRNA levels, reverse transcription was carried out using oligo (dT) primers. qRT-PCR was performed using LightCycler 480 SYBR Green I Master Mix on a LightCycler 480 Real-Time PCR System (Roche, Mannheim, Germany). Relative expression was calculated based on the 2–ΔΔCT method, and Gapdh was used as internal control. The primers used for gene assessment were as follows:
Igf1-forward, 5ʹ-AGTCCCCGTCCCTATCGACAAA-3ʹ;
Igf1-reverse, 5ʹ-AAATGTACTTCCTTTCCTTCT-3ʹ;
Sox9-forward, 5ʹ-GAGCCGGATCTGAAGAGGGA-3ʹ;
Sox9-reverse, 5ʹ-GCTTGACGTGTGGCTTGTTC-3ʹ;
Ascl2-forward, 5ʹ-AAGCACACCTTGACTGGTACG-3ʹ;
Ascl2-reverse, 5ʹ-AAGTGGACGTTTGCACCTTCA-3ʹ;
Lgr5-forward, 5ʹ-CAGCCTCAAAGTGCTTATGCT-3ʹ;
Lgr5-reverse, 5ʹ-GTGGCACGTAACTGATGTGG-3ʹ;
Gapdh-forward, 5ʹ-AGGTCGGTGTGAACGGATTTG-3ʹ;
Gapdh-reverse, 5ʹ-TGTAGACCATGTAGTTGAGGTCA-3ʹ.
In situ hybridization
Small intestines were harvested from 8-week-old mice and fixed in neutral buffered formalin at RT for 24 h before paraffin embedding. The tissues were paraffin-embedded and sectioned at a thickness of 5 μm. In situ hybridization was performed using RNAscope as per the manufacturer’s instructions (322360-USM). An RNAscope 2.5 HD detection Reagents-RED kit and an RNAscope Multiplex Fluorescent v2 kit (ACD) with Mm-Lepr probe (ACD), 402731-C1, Mm-Foxl1 probe (ACD, 407401-C1), Hs-LEPR (ACD, 406371-C1), Hs-IGF1 (ACD, 313031-C3) and Mm-Igf1 probe (ACD, 443901-C2) were used in this experiment.
Western blotting
Western blotting assays were performed according to standard procedures. Briefly, fresh tissues were homogenized using RIPA buffer (Beyotime Biotechnology, Shanghai, China) in the presence of protease and phosphatase inhibitor cocktails (Roche), followed by treatment with homogenizer work center (T10 basic, IKA). Protein concentration was measured with the BCA protein assay kit (Beyotime) as per the manufacturer’s directions. Thirty micrograms of total denatured protein were loaded into an 8% SDS–PAGE gel and transferred to PVDF membranes (GE Healthcare). The PVDF membranes were blocked with 5% nonfat dry milk, followed by incubation with primary and secondary antibodies. The antibodies used were: anti-β-Actin (YEASEN, 30101, 1:1000), anti-AKT (pan) (Cell Signaling Technology, 4821, 1:1000), anti-p-AKT (S473) (Cell Signaling Technology, 4060, 1:1000), anti-p-AKT (T308) (Cell Signaling Technology, 13038, 1:1000), anti-ERK (Cell Signaling Technology, 4695, 1:1000), anti-p-ERK (Cell Signaling Technology, 4370, 1:1000), anti-p-STAT3 (Cell Signaling Technology, 9145, 1:1000) and anti-Igf1 (GeneTex, GTX100521, 1:500).
Luciferase assay for IGF1 promoter activity
The sequence for IGF1 is located on Chromosome 12 (NC_000012.12, base pairs 102395860..102481839, complement) in the human genome. A ~2-kb region upstream of TSS was identified as the IGF1 promoter in this study, which is located at chromosome 12 (NC_000012.12: base pairs 102482380…102480379) and cloned into the pGL3-Basic reporter constructs. Binding sites 1 and 2 of STAT3 are located at base pairs 102481743…102481732 and base pairs 102481109…102481098, respectively. The construct was verified by DNA sequencing. Nucleotides at STAT3-binding sites were mutated by carrying out site-directed mutagenesis (BGI, Shenzhen, China). The plasmids with wild-type IGF1 promoter and mutants in STAT3-binding sites were transfected with phRL-TK control plasmid to 293 T cells which were purchased from AmericanType Culture Collection (ATCC) (Manassas, VA), respectively. The luciferase activities of Firefly and Renilla were measured after 24 h of transfection by using Dual-Glo luciferase assay kit (Promega) according to the manufacturer’s instructions.
Indirect calorimetry
Indirect calorimetry was performed using the oxylet System (Panlab, Barcelona, Spain) and the METABOLISM software suite. Adult (8–12 months old) male Igf1 cKO and their littermate controls were used. All comparisons are based on animals studied simultaneously in different chambers connected to the same O2 and CO2 sensors in an effort to minimize the effect of environmental variations and calibration on data. After 24-h acclimation period, mice were monitored in the metabolic chambers for 24 h with ad-libitum access to standard chow and water. Output parameters include oxygen consumption (VO2) and carbon dioxide production (VCO2) and EE.
Cell growth curve assay
Primary MCs were seeded into a 96-well plate in triplicate, then treated with the CCK8 reagent and further cultured for 1 h. The optical density was measured at 450 nm with a spectrophotometer at indicated time points after seeding.
Igf1 and Leptin treatments
For Igf1 treatment, Lepr-Cre;iDTR mice and controls were intraperitoneally injected with 3 mg/kg recombinant mouse Igf1 protein (Absin, abs04634) dissolved in 0.9% sodium chloride daily for three consecutive days. For Leptin treatment, Lepr-Cre;mTmG mice were intraperitoneally injected with recombinant mouse Leptin protein (R&D Systems, 498-OB-01M) at a concentration of 2 mg/kg body weight twice daily for two consecutive days.
Enzyme-linked immunosorbent assay
Serum levels of Leptin were measured from mouse blood using enzyme-linked immunosorbent assay (ELISA) kit as per the manufacturer’s instructions (Abcam, ab100718). Supernatent levels of Igf1 were measured from primary intestinal MCs using ELISA kit as per the manufacturer’s instructions (Abcam, ab100695).
Statistical analysis
Unless noted, unpaired Student’s t-test was used for calculating P value using Graphpad Prism. All data are reported as the means ± standard deviation (SD). Asterisks denote statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001). Wilcox test was used for calculating P value in Fig. 4g.
Supplementary information
Acknowledgements
This work is funded by grants from the National Key R&D Program of China (2021YFF1000603), the National Natural Science Foundation of China (82025006, 82000498, 82103115), SKLAB Open Grant (2022SKLAB6-03), the fellowship of China National Postdoctoral Program for Innovative Talents (BX20200369) and Plan111 (B12008). C.F.G.-J. is supported by UC Irvine Chancellor’s ADVANCE Postdoctoral Fellowship Program, NSF-Simons Postdoctoral Fellowship, and by a kind gift from the Howard Hughes Medical Institute Hanna H. Gray Postdoctoral Fellowship Program.
Author contributions
Z.Y. and C.L. designed the experiments. M.D., C.L., X.S., J. Xu, X.Y., X.W., K.Y., M.L., G.L., J. Xiao performed research. C.F.G.-J. conceptualized and performed computational experiments and analysis. C.F.G.-J., X.L., K.W., F.R., Q.N., M.V.P., C.L. and Z.Y. analyzed the data. M.D., C.F.G.-J., C.L. and Z.Y. wrote the manuscript.
Data availability
The authors declare that all supporting data are available within the Article and its Supplementary Information files. 3ʹ-scRNA-seq data sets have been deposited in the Gene Expression Omnibus (GEO) database under the accession code: GSE165806. All generic and custom R, Python and MATLAB scripts are available on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Min Deng, Christian F. Guerrero-Juarez.
Contributor Information
Zhengquan Yu, Email: zyu@cau.edu.cn.
Cong Lv, Email: lvc@cau.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-022-00643-9.
References
- 1.Francescangeli F, De Angelis ML, Zeuner A. Dietary factors in the control of gut homeostasis, intestinal stem cells, and colorectal cancer. Nutrients. 2019;11:2936. doi: 10.3390/nu11122936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso S, Yilmaz OH. Nutritional regulation of intestinal stem cells. Annu. Rev. Nutr. 2018;38:273–301. doi: 10.1146/annurev-nutr-082117-051644. [DOI] [PubMed] [Google Scholar]
- 3.Ocvirk S, Wilson AS, Appolonia CN, Thomas TK, O’Keefe SJD. Fiber, fat, and colorectal cancer: new insight into modifiable dietary risk factors. Curr. Gastroenterol Rep. 2019;21:62. doi: 10.1007/s11894-019-0725-2. [DOI] [PubMed] [Google Scholar]
- 4.Ruemmele FM. Role of diet in inflammatory bowel disease. Ann. Nutr. Metab. 2016;68(Suppl 1):33–41. doi: 10.1159/000445392. [DOI] [PubMed] [Google Scholar]
- 5.Shivashankar R, Lewis JD. The role of diet in inflammatory bowel disease. Curr. Gastroenterol. Rep. 2017;19:22. doi: 10.1007/s11894-017-0563-z. [DOI] [PubMed] [Google Scholar]
- 6.Beyaz S, et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 2016;531:53–58. doi: 10.1038/nature17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weng ML, et al. Fasting inhibits aerobic glycolysis and proliferation in colorectal cancer via the Fdft1-mediated AKT/mTOR/HIF1alpha pathway suppression. Nat. Commun. 2020;11:1869. doi: 10.1038/s41467-020-15795-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Flanagan CH, Smith LA, McDonell SB, Hursting SD. When less may be more: calorie restriction and response to cancer therapy. BMC Med. 2017;15:106. doi: 10.1186/s12916-017-0873-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shoshkes-Carmel M, et al. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature. 2018;557:242–246. doi: 10.1038/s41586-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Degirmenci B, Valenta T, Dimitrieva S, Hausmann G, Basler K. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature. 2018;558:449–453. doi: 10.1038/s41586-018-0190-3. [DOI] [PubMed] [Google Scholar]
- 11.Stzepourginski I, et al. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc. Natl. Acad. Sci. USA. 2017;114:E506–E513. doi: 10.1073/pnas.1620059114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greicius G, et al. PDGFRalpha(+) pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc. Natl. Acad. Sci. USA. 2018;115:E3173–E3181. doi: 10.1073/pnas.1713510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu N, et al. MAP3K2-regulated intestinal stromal cells define a distinct stem cell niche. Nature. 2021;592:606–610. doi: 10.1038/s41586-021-03283-y. [DOI] [PubMed] [Google Scholar]
- 14.Holloway EM, et al. Mapping development of the human intestinal niche at single-cell resolution. Cell Stem Cell. 2021;28:568–580.e4. doi: 10.1016/j.stem.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarthy N, et al. Distinct mesenchymal cell populations generate the essential intestinal BMP signaling gradient. Cell Stem Cell. 2020;26:391–402.e5. doi: 10.1016/j.stem.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarde T, et al. Mesenchymal niche-derived neuregulin-1 drives intestinal stem cell proliferation and regeneration of damaged epithelium. Cell Stem Cell. 2020;27:646–662.e7. doi: 10.1016/j.stem.2020.06.021. [DOI] [PubMed] [Google Scholar]
- 17.Park HK, Ahima RS. Physiology of leptin: energy homeostasis, neuroendocrine function and metabolism. Metabolism. 2015;64:24–34. doi: 10.1016/j.metabol.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asada N, et al. Differential cytokine contributions of perivascular haematopoietic stem cell niches. Nat. Cell Biol. 2017;19:214–223. doi: 10.1038/ncb3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeFalco J, et al. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608–2613. doi: 10.1126/science.1056602. [DOI] [PubMed] [Google Scholar]
- 20.van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 21.Furuyama K, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat. Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- 22.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noris M, Remuzzi G. Overview of complement activation and regulation. Semin. Nephrol. 2013;33:479–492. doi: 10.1016/j.semnephrol.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teixeira AL, Gama CS, Rocha NP, Teixeira MM. Revisiting the role of eotaxin-1/CCL11 in psychiatric disorders. Front. Psychiatry. 2018;9:241. doi: 10.3389/fpsyt.2018.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lisignoli G, et al. CXCL12 (SDF-1) and CXCL13 (BCA-1) chemokines significantly induce proliferation and collagen type I expression in osteoblasts from osteoarthritis patients. J. Cell Physiol. 2006;206:78–85. doi: 10.1002/jcp.20435. [DOI] [PubMed] [Google Scholar]
- 26.Helfer G, Wu QF. Chemerin: a multifaceted adipokine involved in metabolic disorders. J. Endocrinol. 2018;238:R79–R94. doi: 10.1530/JOE-18-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bohin N, et al. Insulin-like growth factor-1 and mTORC1 signaling promote the intestinal regenerative response after irradiation injury. Cell. Mol. Gastroenterol. Hepatol. 2020;10:797–810. doi: 10.1016/j.jcmgh.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin S, et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheng X, et al. Cycling stem cells are radioresistant and regenerate the intestine. Cell Rep. 2020;32:107952. doi: 10.1016/j.celrep.2020.107952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou W, Rowitz BM, Dailey MJ. Insulin/IGF-1 enhances intestinal epithelial crypt proliferation through PI3K/Akt, and not ERK signaling in obese humans. Exp. Biol. Med. (Maywood) 2018;243:911–916. doi: 10.1177/1535370218785152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Landeghem L, et al. IGF1 stimulates crypt expansion via differential activation of 2 intestinal stem cell populations. FASEB J. 2015;29:2828–2842. doi: 10.1096/fj.14-264010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsumura S, et al. Stratified layer analysis reveals intrinsic leptin stimulates cryptal mesenchymal cells for controlling mucosal inflammation. Sci. Rep. 2020;10:18351. doi: 10.1038/s41598-020-75186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokolovic M, et al. Fasting induces a biphasic adaptive metabolic response in murine small intestine. BMC Genomics. 2007;8:361. doi: 10.1186/1471-2164-8-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mihaylova MM, et al. Fasting activates fatty acid oxidation to enhance intestinal stem cell function during homeostasis and aging. Cell Stem Cell. 2018;22:769–778.e4. doi: 10.1016/j.stem.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie Y, et al. Impact of a highfat diet on intestinal stem cells and epithelial barrier function in middleaged female mice. Mol. Med. Rep. 2020;21:1133–1144. doi: 10.3892/mmr.2020.10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiao C, et al. IGF1-mediated HOXA13 overexpression promotes colorectal cancer metastasis through upregulating ACLY and IGF1R. Cell Death Dis. 2021;12:564. doi: 10.1038/s41419-021-03833-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy N, et al. Circulating levels of insulin-like growth factor 1 and insulin-like growth factor binding protein 3 associate with risk of colorectal cancer based on serologic and mendelian randomization analyses. Gastroenterology. 2020;158:1300–1312.e20. doi: 10.1053/j.gastro.2019.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexander AN, Carey HV. Oral IGF-I enhances nutrient and electrolyte absorption in neonatal piglet intestine. Am. J. Physiol. 1999;277:G619–G625. doi: 10.1152/ajpgi.1999.277.3.G619. [DOI] [PubMed] [Google Scholar]
- 39.Xu J, et al. Secreted stromal protein ISLR promotes intestinal regeneration by suppressing epithelial Hippo signaling. EMBO J. 2020;39:e103255. doi: 10.15252/embj.2019103255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8:281–291.e9. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guerrero-Juarez CF, et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat. Commun. 2019;10:650. doi: 10.1038/s41467-018-08247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 44.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all supporting data are available within the Article and its Supplementary Information files. 3ʹ-scRNA-seq data sets have been deposited in the Gene Expression Omnibus (GEO) database under the accession code: GSE165806. All generic and custom R, Python and MATLAB scripts are available on reasonable request.