Abstract
Growing evidence shows that Baihu-Guizhi decoction (BHGZD), a traditional Chinese medicine (TCM)-originated disease-modifying anti-rheumatic prescription, may exert a satisfying clinical efficacy for rheumatoid arthritis (RA) therapy. In our previous studies, we verified its immunomodulatory and anti-inflammatory activities. However, bioactive compounds (BACs) of BHGZD and the underlying mechanisms remain unclear. Herein, an integrative research strategy combining UFLC-Q-TOF-MS/MS, gene expression profiling, network calculation, pharmacokinetic profiling, surface plasmon resonance, microscale thermophoresis, and pharmacological experiments was carried out to identify the putative targets of BHGZD and underlying BACs. After that, both in vitro and in vivo experiments were performed to determine the drug effects and pharmacological mechanisms. As a result, the calculation and functional modularization based on the interaction network of the “RA-related gene–BHGZD effective gene” screened the TLR4/PI3K/AKT/NFκB/NLRP3 signaling-mediated pyroptosis to be one of the candidate effective targets of BHGZD for reversing the imbalance network of “immune-inflammation” during RA progression. In addition, both mangiferin (MG) and cinnamic acid (CA) were identified as representative BACs acting on that target, for the strong binding affinities between compounds and target proteins, good pharmacokinetic features, and similar pharmacological effects to BHGZD. Notably, both BHGZD and the two-BAC combination of MG and CA effectively alleviated the disease severity of the adjuvant-induced arthritis-modified rat model, including elevating pain thresholds, relieving joint inflammation and bone erosion via inhibiting NF-κB via TLR4/PI3K/AKT signaling to suppress the activation of the NLRP3 inflammasome, leading to the downregulation of downstream caspase-1, the reduced release of IL-1β and IL-18, and the modulation of GSDMD-mediated pyroptosis. Consistent data were obtained based on the in vitro pyroptosis cellular models of RAW264.7 and MH7A cells induced by LPS/ATP. In conclusion, these findings offer an evidence that the MG and CA combination identified from BHGZD may interact with TLR4/PI3K/AKT/NFκB signaling to inhibit NLRP3 inflammasome activation and modulate pyroptosis, which provides the novel representative BACs and pharmacological mechanisms of BHGZD against active RA. Our data may shed new light on the mechanisms of the TCM formulas and promote the modernization development of TCM and drug discovery.
Keywords: rheumatoid arthritis (RA), traditional Chinese medicine-originated disease-modifying anti-rheumatic prescription, pyroptosis, bioactive compound, NLRP3 inflammasome
Introduction
Rheumatoid arthritis (RA), characterized by hyperplasia and inflammation of the synovial joints, is an intractable and highly prevalent autoimmune disease with unknown pathogenic triggers (1, 2). Currently, the key therapeutic approaches to RA are mainly based on drugs and surgeries (3), including disease-modifying anti-rheumatic drugs (DMARDs), non-steroidal anti-inflammatory drugs (NSAIDs), glucocorticoids, and biological response modifiers, which may reduce synovitis and systemic inflammation (4). However, there are no specific drugs for RA therapy due to its various comorbidities, such as cardiovascular diseases (5–8), osteoporosis (9, 10), interstitial lung disease (9, 11), malignancies (11, 12), and hypertension (13), as well as the heterogeneous and complex clinical manifestations with wide individual differences. Traditional Chinese medicine (TCM), as a complementary and alternative medicine, has its unique advantages in the clinical treatment of RA, especially at the active stage with moist heat arthralgia spasm syndrome (active RA), which is characterized by the hyperactive immune response and excessive inflammatory cytokines, subsequently leading to osteoclast differentiation and active joint inflammation (14, 15). Among TCM-originated DMARDs, Baihu-Guizhi decoction (BHGZD) has been indicated to efficiently reverse the aggressive progression of active RA with moist heat arthralgia spasm syndrome and to reach a remarkable response rate (90%) (16–19). Our previous studies identified the chemical profiling of BHGZD in vitro and verified its immunomodulatory and anti-inflammatory activities (20–22). However, the bioactive compounds (BACs) and the underlying mechanisms of BHGZD against active RA have not been fully elucidated.
In the current study, the candidate BACs of BHGZD were identified by the ultra-fast liquid chromatography-quadrupole-time-of-flight tandem mass spectrometry (UFLC-Q-TOF-MS/MS) system in vivo. Then, we carried out transcriptomic profiling, target prediction, and network calculation to screen the candidate targets of BHGZD for reversing the imbalance network of “immune-inflammation” during the progression of active RA. After that, the binding affinities between candidate BACs and candidate targets of BHGZD were calculated using the molecular docking simulation, the results of which were subsequently verified by surface plasmon resonance (SPR) assay and microscale thermophoresis (MST) analysis. Moreover, the in vivo biodistribution and pharmacokinetic characteristics of candidate BACs were also determined. Furthermore, the pharmacological effects and the molecular mechanisms of BHGZD, and its representative BACs against active RA were further validated by a series of experiments based on the adjuvant-induced arthritis-modified rat model (AIA-M) as well as LPS/ATP-induced pyroptosis cellular models on both RAW264.7 macrophage and MH7A cells.
Materials and Methods
Preparation of BHGZD
Crude drugs of BHGZD formula were purchased from Beijing Tongrentang Co., Ltd. (Beijing, China). BHGZD was prepared according to the original composition of the formula recorded in Chinese pharmacopoeia 2020 edition and our previous study (22) ( Supplementary Materials Section 1 , and Table S1 ).
RA Modeling, Grouping, and Treatment
A total of 49 male Lewis rats (200 ± 20 g in weight, 6–8 weeks old) were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. (SCXK 2019-0003, Beijing, China) and randomly divided into two independent clusters: a discovery cluster (n = 24) and a validation cluster (n = 25). All animals were maintained in specific pathogen-free conditions with a constant temperature of 24 ± 1°C with a 12-h light/dark cycle, and had free access to standard rodent diet and water. The discovery and validation clusters were respectively used for gene expression profiling and experimental validation. In the discovery cluster, 24 rats were randomly divided into three groups [3 per group for microarray and 5 per group for mRNA sequencing (mRNA-Seq)] (1): the normal control group, (2) the AIA-M model group, and (3) the BHGZD treatment group. In the validation cluster, 25 rats were randomly divided into five groups (5 per group): (1) the normal control group, (2) the AIA-M model group, (3) the BHGZD treatment group, (4) the MG+CA treatment group, and (5) the methotrexate (MTX) treatment group.
The AIA-M rat model was established simulating the pathological changes and characteristics of active RA with moist heat arthralgia spasm syndrome based on our previous studies (20–23). Please see the detailed information on the AIA-M rat model establishment in Supplementary Materials Section 2 . The dosage of BHGZD for the corresponding treatment group was 21.4 g/kg, equivalent to two times the clinical dose, which has been proved to exert the most prominent therapeutic efficacy in our previous studies (21, 22). The dosages for the two-BAC combination of MG and CA for the corresponding treatment group were respectively 600.912 mg/kg and 46.652 mg/kg, equivalent to their content in 21.4 g/kg BHGZD. The dosage for MTX was 0.2 mg/kg. All treatments were performed for 30 days via oral administration from the day of the primary immunization (20–23).
On the 31st day, all rats were anesthetized by intraperitoneal injection of pentobarbital sodium (50 mg/kg). The blood samples were collected using one-time anticoagulant negative pressure blood collection tubes, and then placed at room temperature for 20 min and centrifuged at 3,000 rpm for 15 min. The sera were collected, vortexed, and centrifuged again at high speed at low temperature (4°C) for 15 min, and stored at −20°C for subsequent assays. Whole blood cells were freshly isolated from blood, then frozen in liquid nitrogen overnight, and stored at −80°C for mRNA-Seq. The synovium tissues were isolated and immediately frozen in liquid nitrogen for microarray analysis. The right joints of rats were removed and preserved in 4% paraformaldehyde (PFA) combined with phosphate-buffered saline (PBS) for 72 h for subsequent assays. The left joints and organs of rats were kept at −80°C for subsequent assays.
Assessment of Arthritis Severity
The severity of arthritis was evaluated by arthritis incidence, hind paw thickness (the diameter of limb), arthritis score, and arthritis surface temperature as in our previous studies (20–24), and is provided in Supplementary Materials Section 3 . Pain thresholds were evaluated by mechanical-, acetone-, and thermal-induced hyperalgesia as previously described (20–25), and are provided in Supplementary Materials Section 4 .
Viscera Index and Histology
The viscera indexes were calculated by the weight of the thymus, spleen, liver, and kidney relative to total brain weight. Histological changes were examined using hematoxylin and eosin (H&E), safranin O-fast green (Solarbio, Beijing, China), and Masson trichrome staining (Baso, Wuhan, China) according to routine protocols. Please see detailed information on the protocol in Supplementary Materials Section 5 .
Micro-Computed Tomography Analysis
To quantitatively evaluate bone formation within the defects, the specimens were scanned using a micro-CT instrument (GE healthcare, USA) at a resolution of 45 μm. The x-ray settings and cylinder region of interest (ROI) are provided in Supplementary Materials Section 6 .
Gene Expression Profiling
Whole blood cells and the synovium tissues representing the pathological characteristics of systemic disease and target organs during active RA progression were respectively used for whole rat genome microarray analysis (Agilent Technologies, design ID: 014879, Santa Clara, CA, USA) and mRNA-Seq Illumina NovaSeq 6000 (Illumina, CA, USA) to screen RA-related genes [the significant differentially expressed genes (DEGs) between the AIA-M model group and the normal control group] and BHGZD therapeutic effect-related genes (DEGs between the BHGZD treatment group and the AIA-M model group). The gene expression data have been uploaded and are publicly available in the NCBI GEO database (GEO No. GSE189942, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE189942, December 3, 2021, and GEO No. GSE190523, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE190523, December 11, 2021), and the DEGs were identified by referring to the criteria of t-test p-value < 0.05 and fold change (FC)>2/<0.5. Please see the detailed information in Supplementary Materials Section 7 .
Prediction of Putative BHGZD Targets
Drugs with a high similarity score (>0.80) to structures of composite compounds of each ingredient contained in BHGZD were selected based on the TCMIP v2.0 database (Integrative Pharmacology-based Research Platform of Traditional Chinese Medicine, last update in 2021-09-10, http://www.tcmip.cn/TCMIP/index.php/Home) and the Encyclopedia of Traditional Chinese Medicine (ETCM, http://www.tcmip.cn/ETCM/index.php/Home/) (26). Therapeutic targets of the similar drugs approved by the Food and Drug Administration (FDA) according to DrugBank Online (Version 5.1.8, released 2021-01-03, https://go.drugbank.com/) were identified as putative targets of BHGZD.
Network Construction and Analysis
The RA-related gene interaction network was constructed using the links among the DEGs between the AIA-M model and normal control groups to identify the key RA-related genes. Then, the interaction network of the “key RA-related gene–BHGZD-effective gene” was constructed using the links between the key RA-related genes and BHGZD effective genes. The gene–gene interaction data were collected from the String database (version 10.0, http://string-db.org/) (27), and the interaction networks were all visualized by the Cytoscape platform (version 3.9.0, https://cytoscape.org/) (28). The network topological properties of nodes, including “Degree”, “Betweenness”, and “Closeness”, were calculated for screening the key network targets according to our previous descriptions (20–25, 29) and Supplementary Materials Section 8 .
Determining the Chemical Compounds of BHGZD and Absorption Distribution Metabolism Excretion Evaluation
A rapid, sensitive, and reliable method by the UFLC-Q-TOF-MS/MS system was used to identify the chemical profiling of BHGZD as described in Supplementary Materials Section 9 (30). The reference standards of these compounds ( Supplementary Table S2 ) were purchased from Chengdu Must Bio-technology Co., Ltd. (Chengdu, China), the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China), and Chengdu DeSiTe Biological Technology Co., Ltd. (Chengdu, China). Then, the ADME information on 19 BACs included in BHGZD was collected from the ETCM database (The Encyclopedia of Traditional Chinese Medicine, http://www.tcmip.cn/ETCM/) (26). In addition, in silico ADME models were used to identify the candidate BACs of BHGZD by calculating the passive intestinal permeability of the Caco-2 module, the oral bioavailability, and the apparent permeability coefficient (Papp) of each BACs as described in Supplementary Materials Section 10 .
Molecular Docking Simulation
The molecular docking and virtual screening program was carried out to investigate the direct binding efficiencies of the two BACs contained in BHGZD and the corresponding putative targets. The structures of human TLR4 (PDB ID: 2z65), AKT1 (PDB ID: 3o96), and NFκB (PDB ID: 1nfi) were obtained from the PDB website (The Protein Data Bank, https://www.rcsb.org/). For docking analysis, the ligand mangiferin (MG) and cinnamic acid (CA) were downloaded from the ZINC database (last update in 2018-02-14, http://zinc.docking.org/) (31) in mol2 format and converted to pdb.files using OpenBabel GUI (last update in 2016-09-21, version 2.4.1, http://openbabel.org/wiki/Main_Page). AutoDock Tools (Version 1.5.6, https://ccsb.scripps.edu/mgltools/) was used to convert pdb to pdbqt format. Docking calculations were performed using AutoDock Vina (The Scripps Research Institute, version 1.1.2) and AutoDock (The Scripps Research Institute, version 4.2.6). The visualization and analysis of the results were obtained from AutoDock Vina by PyMOL (Version 2.5, https://pymol.org/2/).
SPR Assay
To confirm the binding affinity of recombinant AKT1 protein and MG, SPR assay was performed with Biacore 8K (Biacore, Cytiva), and the K D value was calculated using the Biacore 8K evaluation software 2.0 (GE Healthcare). Detailed information on SPR assay is provided in Supplementary Materials Section 11 .
MST Assay
The binding affinity between recombinant TLR4 protein and CA was measured by a NanoTemper Monolith NT.115 instrument (NanoTemper Technologies, Germany). The K D value was fitted by NanoTemper Monolith affinity software (NanoTemper Technologies, Germany) using 1:1 binding mode. Detailed information on the MST assay is provided in Supplementary Materials Section 12 .
In Vivo Pharmacokinetic Analysis
To detect the pharmacokinetic parameters of MG and CA in vivo, quantification analysis was performed by an ultra-high-performance liquid chromatography (Shimadzu Corp., Japan) tandem ion trap quadrupole QTRAP 6500 plus mass spectrometry (AB Sciex, USA). The quantitative analyses for MG and CA were approved by the Guidance for Bioanalytical Method Validation issued by the Chinese Pharmacopoeia Commission in 2020. The pharmacokinetic parameters were calculated by Drug and Statistic software (Shanghai, China). Detailed information on the collection of blood samples and UHPLC-QTRAP-MS/MS conditions is provided in Supplementary Materials Section 13 .
Cell Culture, Induction, and Treatment
Mouse macrophage cell line (RAW264.7 cells, Bio-Swamp, Wuhan, China) and fibroblast-like synoviocytes in patients with RA (MH7A cells, Riken cell bank, Ibaraki, Japan) at passage numbers four to eight were used in the current experiment validations. The culture conditions are provided in Supplementary Materials Section 14 .
The conventional NLRP3 inflammasome activation cellular model was induced by lipopolysaccharide [LPS, Escherichia coli (O111:B4), Sigma-Aldrich, St Louis, MO, USA] and adenosine triphosphate (ATP disodium salt hydrate, Sigma-Aldrich, St Louis, MO, USA) as in our previous study (22) ( Supplementary Materials Section 15 ).
For the drug treatment, both RAW264.7 and MH7A cells were divided into the following groups: (1) Normal control group: with no stimulation and treatment; (2) LPS/ATP-induced cellular model group: cells were stimulated with the corresponding concentrations of LPS and ATP; (3) BHGZD treatment group: after stimulation, cells were treated with 28.54 μg/ml BHGZD for 24 h, which was determined to be the most effective as in our previous study (22); (4) MG+CA treatment group: after stimulation, cells were treated with the two-BAC combination (1.69 ng/ml MG and 0.13 ng/ml CA, with the same content as that in the 5 μg/ml BHGZD formula) for 24 h; (5) MG treatment group: after stimulation, cells were treated with 1.69 ng/ml MG for 24 h; and (6) CA treatment group: after stimulation, cells were treated with 0.13 ng/ml CA for 24 h. The concentration of DMSO was less than 1‰ of the solution for the in vitro experiment.
Cell Viability Assay
Cell viability was analyzed with the cell counting kit-8 (CCK-8) assay kit (Beyotime Biotechnology, Shanghai, China) as described in Supplementary Materials Section 16 .
Flow Cytometry
To identify and quantify the pyroptosis population of RAW264.7 cells, FITC Apoptosis Detection kit I staining with both the Annexin V-fluorescein isothiocyanate and propidium iodide (PI, BD Biosciences, San Jose, CA, USA) was used according to the manufacturer’s guidelines. Flow cytometry was performed using NovoCyte 2040R (ACEA Bioscience, San Diego, CA, USA), and analyzed using the NovoExpress 1.4.1 software (ACEA Bioscience, San Diego, CA, USA). Detailed information of this experiment is provided in Supplementary Materials Section 17 .
Terminal Deoxynucleotidyl Transferase-Mediated dUTP Biotin Nick End Labeling Staining
The occurrence of pyroptosis in both MH7A cells and synovium tissues in AIA-M rats was determined by in situ TUNEL staining (TUNEL Andy Fluor™ 594 Apoptosis Detection Kit, ABP Biosciences, Wuhan, China) in accordance with the manufacturer’s protocol. Fluorescence images were visualized and photographed with inverted fluorescence microscopes (MF53, MSHOT, Guangzhou, China; Olympus, BX51, Tokyo, Japan). Detailed information on TUNEL assay is provided in Supplementary Materials Section 18 .
FAM-FLICA Caspase-1 Assay
Active caspase-1 was visualized by a FAM-FLICA caspase-1 assay kit using a FAM-YVAD-FMK inhibitor probe (ImmunoChemistry Technologies, Bloomington, MN, USA), according to the manufacturer’s guidelines. Fluorescence images were visualized and photographed with a confocal microscope (Zeiss LSM 880, Carl Zeiss, Jena, Germany). Detailed information on FAM-FLICA capase-1 assay is provided in Supplementary Materials Section 19 .
Immunofluorescence Staining
Double fluorescence staining was performed as described previously (22) ( Supplementary Materials Section 20 ). The images were visualized and photographed with a confocal microscope (Zeiss LSM 880, Carl Zeiss, Jena, Germany) or an inverted fluorescence microscope (MF53, MSHOT, Guangzhou, China).
Immunohistochemical Staining
To detect the expression levels of NLPR3/ASC of the knee joints in AIA-M rats and corresponding treatment groups, immunohistochemical staining was carried out using a DAB kit (Cat No. AR1027, Boster Biological Technology Co., Ltd., Wuhan, China) and a rabbit/mouse two-step detection kit (Cat No. SV0002/SV0001, Boster Biological Technology Co., Ltd., Wuhan, China) according to the routine protocols. Immunohistochemistry quantification was performed using ImageJ (Image Progressing and Analysis in Java, version 1.42q, https://imagej.nih.gov/ij/), following the ImageJ User Guide.
Western Blotting Analysis
To evaluate the regulation of BHGZD on the candidate targets in arthritic tissue samples, as well as RAW264.7 and MH7A cells, Western blotting analysis was carried out following the protocol as in our previous studies (21, 24, 29). TLR4, phospho-PI3K (p-PI3K), PI3K, p-AKT, p-NFκB-p65, NFκB-p65, NLRP3, ASC, caspase-1, the N-terminal domain of GSDMD (GSDMD-NT), and IL-1β antibodies were used as shown in Supplementary Table S3 . GAPDH (GAPDH Mouse monoclonal antibody, Abcam, Cambridge, UK) and β-actin (Anti-beta-Actin/β-Actin Antibody, Boster Biological Technology, California, USA) were used as loading controls for arthritic tissue samples and cultured cells, respectively.
Cytokine Analysis and Lactate Dehydrogenase Measurement
To evaluate the therapeutic effects of two BACs from BHGZD on the regulation of the “immune-inflammation” system, the levels of cytokines, proteins, and LDH release in RAW264.7 and MH7A cellular supernatant, as well as the sera of AIA-M rats were determined using enzyme-linked immunosorbent assay (ELISA) to assess the integrity of membranes in accordance with the manufacturer’s guidelines.
Statistical Analyses
Statistical analyses were performed using GraphPad Prism 8.0 Software (San Diego, CA, USA). Data are expressed as the mean ± SD and analyzed by one-way ANOVA with Bonferroni’s or Dunnett’s post-hoc test for comparison of multiple columns. Differences were considered statistically significant when the p-value was less than 0.05.
Results
BHGZD Reverses the Imbalance of the “Immune-Inflammation” System in Active RA Rats
The AIA-M rat model was successfully established (the achievement ratio of this model was 100%) and verified according to its reliable and rapid-onset characteristics, and severe disease progression by external stimulus of wind, dampness, and heat, which may be the major pathological changes and features of active RA with moist heat arthralgia spasm syndrome (22). In contrast, the administration of BHGZD effectively alleviated the disease severity in arthritis of active RA rats. Detailed information on the characterizations of the active RA rat model and the therapeutic effects of BHGZD in active RA severity based on this model was reported in our previous study (22).
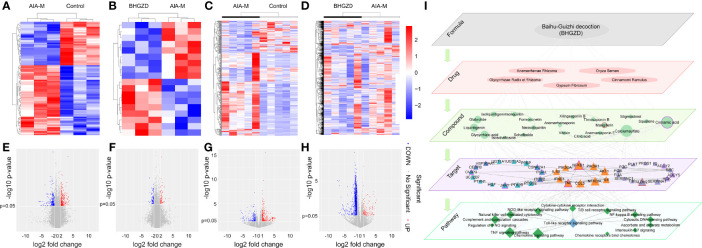
To determine the candidate targets of BHGZD against active RA, the gene expression profiles of whole blood cells and synovium tissues were carried out. A total of 473 RA-related genes (275 upregulated and 178 downregulated genes) and 1,802 BHGZD therapeutic effect-related genes (456 upregulated and 1,346 downregulated genes) were identified. The heatmap and volcano plot of DEGs are shown in Figures 1A–H , suggesting a distinct separation between the AIA-M model and normal control groups, as well as the BHGZD treatment and AIA-M model groups. Among them, there were 135 RA-related genes reversely regulated by the treatment of BHGZD ( Table 1 ). Detailed information on the RA-related genes and BHGZD therapeutic effect-related genes is respectively provided in Supplementary Tables S4 and S5 .
Figure 1.
Transcriptomic profiling-based differential data analysis and biomolecular network-based investigation of AIA-M candidate markers and candidate targets of Baihu-Guizhi decoction (BHGZD) against AIA-M (Control, AIA-M, BHGZD). (A) The heatmap of the significant differentially expressed genes (DEGs) between the AIA-M model group and the normal control group in synovium using microarray analysis (n = 3 per group). (B) The heatmap of DEGs between the BHGZD treatment group and the AIA-M model group in synovium using microarray analysis (n = 3 per group). (C) The heatmap of the DEGs between the AIA-M model group and the normal control group in whole blood cells using RNA sequencing (RNA-Seq) analysis (n = 5 per group). (D) The heatmap of DEGs between the BHGZD treatment group and the AIA-M model group in whole blood cells using RNA-Seq analysis (n = 5 per group). (E) The volcano plot of the DEGs between the AIA-M model group and the normal control group in synovium using microarray analysis (n = 3 per group). (F) The volcano plot of the DEGs between the BHGZD treatment group and the AIA-M model group in synovium using microarray analysis (n = 3 per group). (G) The volcano plot of the DEGs between the AIA-M model group and the normal control group in whole blood cells using RNA sequencing (RNA-Seq) analysis (n = 5 per group). (H) The volcano plot of the DEGs between the BHGZD treatment group and the AIA-M model group in whole blood cells using RNA-Seq analysis (n = 5 per group). (I) The subnetwork of the “formula-drug-compound-target-pathway” network is constructed using the links between key RA-related genes and BHGZD effective genes. The ellipse refers to BHGZD. Light orange hexagons refer to drugs contained in BHGZD. Green circles refer to compounds contained in BHGZD. Triangles refer to targets. Green diamonds refer to pathways. Triangles with red circles refer to the targets of mangiferin (MG). Triangles with purple circles refer to the targets cinnamic acid (CA).
Table 1.
The information on 135 RA-related genes reversely regulated by the treatment of Baihu-Guizhi decoction (BHGZD).
| Samples | Gene Symbol | AIA-M vs. Con | p-value | log2 Fold Change | BHGZD vs. AIA-M | p-value | log2 Fold Change |
|---|---|---|---|---|---|---|---|
| Synovium | ACKR4 | Down | 0.0389 | −1.4327 | Up | 0.0011 | 1.1346 |
| Synovium | AQP3 | Down | 0.0419 | −4.6637 | Up | 0.0003 | 4.6114 |
| Synovium | ARSI | Up | 0.0157 | 1.1593 | Down | 0.0043 | −1.9063 |
| Synovium | C7 | Up | 0.0046 | 1.0014 | Down | 0.0142 | −1.2088 |
| Synovium | CAR13 | Up | 0.0411 | 1.3595 | Down | 0.0136 | −1.9177 |
| Synovium | CFB | Up | 0.0087 | 2.3269 | Down | 0.0100 | −1.9143 |
| Synovium | CHL1 | Up | 0.0266 | 1.3184 | Down | 0.0082 | −1.7127 |
| Synovium | CUBN | Down | 0.0440 | −1.5011 | Up | 0.0272 | 2.0115 |
| Synovium | CYP26A1 | Up | 0.0410 | 1.2507 | Down | 0.0032 | −1.5831 |
| Synovium | CYP2W1 | Up | 0.0302 | 1.6286 | Down | 0.0320 | −2.3456 |
| Synovium | EGLN3 | Down | 0.0224 | −1.5984 | Up | 0.0216 | 1.4110 |
| Synovium | EXPH5 | Down | 0.0254 | −1.2262 | Up | 0.0204 | 1.6917 |
| Synovium | GAS2 | Up | 0.0374 | 3.1797 | Down | 0.0478 | −1.2633 |
| Synovium | MARCKS | Up | 0.0360 | 1.1328 | Down | 0.0142 | −1.3571 |
| Synovium | HHIP | Down | 0.0135 | −1.6433 | Up | 0.0307 | 1.4088 |
| Synovium | ID4 | Down | 0.0244 | −1.6145 | Up | 0.0089 | 1.3183 |
| Synovium | PKHD1L1 | Down | 0.0199 | −1.6321 | Up | 0.0298 | 1.6867 |
| Synovium | TNMD | Up | 0.0418 | 3.3639 | Down | 0.0245 | −5.1964 |
| Synovium | TUBB3 | Up | 0.0089 | 1.5756 | Down | 0.0031 | −2.1800 |
| Synovium | UNC5CL | Down | 0.0172 | −1.8548 | Up | 0.0164 | 1.3891 |
| Synovium | UTS2R | Up | 0.0234 | 3.6773 | Down | 0.0391 | −3.3398 |
| Synovium | VSNL1 | Down | 0.0251 | −1.1707 | Up | 0.0161 | 1.0142 |
| Whole Blood Cells | ADCY5 | Up | 1.5517 | 0.0011 | Down | −1.4196 | 0.0001 |
| Whole Blood Cells | AGRP | Up | 1.4030 | 0.0234 | Down | −2.2359 | 0.0001 |
| Whole Blood Cells | ALDOC | Up | 2.4691 | 0.0010 | Down | −1.3832 | 0.0036 |
| Whole Blood Cells | ANGPTL2 | Up | 3.7609 | 0.0417 | Down | −2.6154 | 0.0384 |
| Whole Blood Cells | ASTN1 | Up | 1.1875 | 0.0034 | Down | −1.4422 | 0.0000 |
| Whole Blood Cells | ATF5 | Up | 1.7382 | 0.0006 | Down | −1.7147 | 0.0144 |
| Whole Blood Cells | BFSP2 | Up | 1.3618 | 0.0039 | Down | −1.5460 | 0.0004 |
| Whole Blood Cells | BGLAP | Up | 1.1948 | 0.0072 | Down | −1.3003 | 0.0003 |
| Whole Blood Cells | BRICD5 | Up | 4.9630 | 0.0180 | Down | −4.5770 | 0.0112 |
| Whole Blood Cells | BTLA | Down | −1.0068 | 0.0352 | Up | 1.0449 | 0.0059 |
| Whole Blood Cells | CASP12 | Down | −3.1532 | 0.0011 | Up | 2.2173 | 0.0236 |
| Whole Blood Cells | CBLN2 | Up | 1.1423 | 0.0021 | Down | −1.5381 | 0.0000 |
| Whole Blood Cells | CCDC92 | Up | 1.7220 | 0.0000 | Down | −1.7041 | 0.0001 |
| Whole Blood Cells | CD177 | Down | −1.9882 | 0.0031 | Up | 2.0804 | 0.0098 |
| Whole Blood Cells | CD79AL | Down | −1.1535 | 0.0081 | Up | 1.1442 | 0.0045 |
| Whole Blood Cells | CDCA2 | Down | −3.2077 | 0.0117 | Up | 2.8039 | 0.0013 |
| Whole Blood Cells | CELA1 | Up | 1.5977 | 0.0012 | Down | −1.6140 | 0.0007 |
| Whole Blood Cells | CLU | Up | 1.0155 | 0.0039 | Down | −1.4820 | 0.0000 |
| Whole Blood Cells | CRABP1 | Up | 1.9562 | 0.0243 | Down | −1.9772 | 0.0081 |
| Whole Blood Cells | CRABP2 | Down | −4.8634 | 0.0057 | Up | 3.8683 | 0.0101 |
| Whole Blood Cells | CTDSPL | Up | 1.0889 | 0.0348 | Down | −1.0297 | 0.0089 |
| Whole Blood Cells | CTTN | Up | 1.0143 | 0.0137 | Down | −1.9100 | 0.0000 |
| Whole Blood Cells | CXCL2 | Up | 1.9025 | 0.0012 | Down | −1.2825 | 0.0001 |
| Whole Blood Cells | CXXC4 | Up | 1.1896 | 0.0126 | Down | −1.5581 | 0.0000 |
| Whole Blood Cells | DACT1 | Up | 3.5048 | 0.0091 | Down | −2.7562 | 0.0059 |
| Whole Blood Cells | DDX25 | Down | −3.1180 | 0.0396 | Up | 2.9352 | 0.0077 |
| Whole Blood Cells | DENND2C | Up | 1.1059 | 0.0242 | Down | −1.3200 | 0.0006 |
| Whole Blood Cells | DNAH1 | Up | 1.1723 | 0.0186 | Down | −1.6980 | 0.0002 |
| Whole Blood Cells | DOK4 | Up | 1.2929 | 0.0109 | Down | −1.2531 | 0.0079 |
| Whole Blood Cells | ECE2 | Up | 3.5704 | 0.0297 | Down | −7.0144 | 0.0000 |
| Whole Blood Cells | EHD2 | Up | 1.0293 | 0.0323 | Down | −1.7746 | 0.0001 |
| Whole Blood Cells | ELN | Up | 4.4083 | 0.0335 | Down | −6.1763 | 0.0005 |
| Whole Blood Cells | ERFE | Up | 3.3489 | 0.0315 | Down | −2.7202 | 0.0101 |
| Whole Blood Cells | F11R | Up | 1.1213 | 0.0037 | Down | −1.5369 | 0.0000 |
| Whole Blood Cells | F2RL3 | Up | 1.1166 | 0.0056 | Down | −1.5841 | 0.0000 |
| Whole Blood Cells | FAM184A | Up | 1.6124 | 0.0416 | Down | −1.5781 | 0.0007 |
| Whole Blood Cells | GADD45G | Up | 1.1221 | 0.0463 | Down | −1.1012 | 0.0309 |
| Whole Blood Cells | GAS2L1 | Up | 1.1011 | 0.0021 | Down | −1.4515 | 0.0001 |
| Whole Blood Cells | GNAZ | Up | 1.0121 | 0.0086 | Down | −1.4201 | 0.0000 |
| Whole Blood Cells | GP1BB | Up | 1.9090 | 0.0008 | Down | −1.5758 | 0.0431 |
| Whole Blood Cells | GP9 | Up | 1.0026 | 0.0038 | Down | −1.4034 | 0.0002 |
| Whole Blood Cells | GRB10 | Up | 2.6721 | 0.0005 | Down | −1.3209 | 0.0028 |
| Whole Blood Cells | GSTA1 | Up | 1.0067 | 0.0053 | Down | −1.3929 | 0.0008 |
| Whole Blood Cells | HIVEP3 | Down | −1.7890 | 0.0017 | Up | 1.0399 | 0.0267 |
| Whole Blood Cells | KATNAL1 | Up | 1.5441 | 0.0248 | Down | −1.2866 | 0.0069 |
| Whole Blood Cells | KCNS3 | Up | 1.0348 | 0.0105 | Down | −1.4733 | 0.0003 |
| Whole Blood Cells | KLHL13 | Up | 2.3729 | 0.0216 | Down | −2.3771 | 0.0009 |
| Whole Blood Cells | KLRA22 | Down | −2.1066 | 0.0132 | Up | 1.6770 | 0.0175 |
| Whole Blood Cells | LARGE1 | Up | 1.0049 | 0.0112 | Down | −1.4699 | 0.0001 |
| Whole Blood Cells | LHFPL6 | Up | 2.1809 | 0.0305 | Down | −1.2276 | 0.0290 |
| Whole Blood Cells | LIPG | Down | −1.2935 | 0.0374 | Up | 1.3060 | 0.0134 |
| Whole Blood Cells | LOC100912228 | Up | 1.0055 | 0.0074 | Down | −1.4317 | 0.0003 |
| Whole Blood Cells | LOC102556092 | Up | 1.1719 | 0.0026 | Down | −1.5513 | 0.0000 |
| Whole Blood Cells | LOXL3 | Up | 1.3740 | 0.0022 | Down | −1.4840 | 0.0002 |
| Whole Blood Cells | LRRC71 | Up | 1.4124 | 0.0178 | Down | −1.5799 | 0.0003 |
| Whole Blood Cells | LY6G6F | Up | 1.0222 | 0.0043 | Down | −1.4740 | 0.0000 |
| Whole Blood Cells | LYVE1 | Up | 1.0159 | 0.0023 | Down | −1.3934 | 0.0000 |
| Whole Blood Cells | MAPK8IP1 | Up | 2.5304 | 0.0317 | Down | −2.4659 | 0.0016 |
| Whole Blood Cells | MCPT1L1 | Up | 1.8206 | 0.0003 | Down | −1.7125 | 0.0000 |
| Whole Blood Cells | MEST | Up | 1.5656 | 0.0296 | Down | −1.7263 | 0.0007 |
| Whole Blood Cells | MMRN1 | Up | 1.0609 | 0.0026 | Down | −1.5474 | 0.0000 |
| Whole Blood Cells | MMRN2 | Down | −2.8201 | 0.0275 | Up | 3.0209 | 0.0015 |
| Whole Blood Cells | MRVI1 | Up | 1.0361 | 0.0113 | Down | −1.4740 | 0.0000 |
| Whole Blood Cells | MSRA | Up | 1.8396 | 0.0266 | Down | −1.1511 | 0.0301 |
| Whole Blood Cells | MYCT1 | Up | 1.1718 | 0.0058 | Down | −1.5488 | 0.0000 |
| Whole Blood Cells | MYL9 | Up | 1.2004 | 0.0009 | Down | −1.5114 | 0.0000 |
| Whole Blood Cells | NRGN | Up | 1.1404 | 0.0008 | Down | −1.4248 | 0.0001 |
| Whole Blood Cells | NRIP3 | Up | 1.1768 | 0.0188 | Down | −1.0978 | 0.0016 |
| Whole Blood Cells | OLFM4 | Down | −1.0349 | 0.0065 | Up | 1.1084 | 0.0336 |
| Whole Blood Cells | PCDHGB5 | Down | −3.9559 | 0.0262 | Up | 3.5018 | 0.0274 |
| Whole Blood Cells | PDE3A | Up | 1.0241 | 0.0116 | Down | −1.4264 | 0.0001 |
| Whole Blood Cells | PF4 | Up | 1.2675 | 0.0007 | Down | −1.4629 | 0.0004 |
| Whole Blood Cells | PKIA | Up | 1.2370 | 0.0016 | Down | −1.3519 | 0.0002 |
| Whole Blood Cells | PLA2G2A | Up | 1.3067 | 0.0003 | Down | −1.5044 | 0.0001 |
| Whole Blood Cells | PLOD2 | Up | 1.3673 | 0.0338 | Down | −1.8337 | 0.0000 |
| Whole Blood Cells | PLTP | Up | 1.4100 | 0.0003 | Down | −1.3760 | 0.0012 |
| Whole Blood Cells | PPIF | Up | 1.0631 | 0.0030 | Down | −1.3334 | 0.0001 |
| Whole Blood Cells | PROSER2 | Up | 2.4378 | 0.0106 | Down | −1.4237 | 0.0073 |
| Whole Blood Cells | PTPN13 | Up | 4.1086 | 0.0158 | Down | −2.6389 | 0.0331 |
| Whole Blood Cells | RASL10A | Up | 1.1205 | 0.0028 | Down | −1.3945 | 0.0001 |
| Whole Blood Cells | REEP2 | Up | 1.0181 | 0.0048 | Down | −1.4944 | 0.0000 |
| Whole Blood Cells | RPAP1 | Up | 1.0304 | 0.0027 | Down | −1.4851 | 0.0000 |
| Whole Blood Cells | RPP25 | Up | 1.8057 | 0.0048 | Down | −1.5779 | 0.0320 |
| Whole Blood Cells | RT1-HA | Down | −1.3847 | 0.0248 | Up | 1.1581 | 0.0230 |
| Whole Blood Cells | RTP4 | Down | −1.1421 | 0.0350 | Up | 1.1822 | 0.0025 |
| Whole Blood Cells | SCAI | Down | −1.2765 | 0.0173 | Up | 1.1195 | 0.0207 |
| Whole Blood Cells | SCARF1 | Up | 1.7774 | 0.0006 | Down | −1.5817 | 0.0020 |
| Whole Blood Cells | SEC14L5 | Up | 1.3750 | 0.0134 | Down | −1.8066 | 0.0013 |
| Whole Blood Cells | SEMA5A | Up | 2.8501 | 0.0110 | Down | −4.0576 | 0.0003 |
| Whole Blood Cells | SEPT10 | Down | −1.7620 | 0.0426 | Up | 1.5958 | 0.0271 |
| Whole Blood Cells | SERPINE2 | Up | 2.2082 | 0.0383 | Down | −1.4497 | 0.0397 |
| Whole Blood Cells | Sh3bgr | Up | 2.4272 | 0.0004 | Down | −1.3007 | 0.0077 |
| Whole Blood Cells | SHPK | Up | 1.0305 | 0.0005 | Down | −1.1246 | 0.0000 |
| Whole Blood Cells | SLC6A4 | Up | 1.0946 | 0.0021 | Down | −1.5201 | 0.0000 |
| Whole Blood Cells | SMPDL3B | Up | 1.0133 | 0.0071 | Down | −1.0129 | 0.0042 |
| Whole Blood Cells | STON2 | Up | 1.8772 | 0.0063 | Down | −1.6397 | 0.0004 |
| Whole Blood Cells | SYT5 | Up | 1.1579 | 0.0003 | Down | −1.3477 | 0.0002 |
| Whole Blood Cells | SYTL4 | Up | 1.6712 | 0.0402 | Down | −1.6984 | 0.0001 |
| Whole Blood Cells | TAL1 | Up | 1.0319 | 0.0073 | Down | −1.5858 | 0.0000 |
| Whole Blood Cells | THBS1 | Up | 1.0434 | 0.0086 | Down | −1.5613 | 0.0000 |
| Whole Blood Cells | TMEM56 | Up | 1.7935 | 0.0015 | Down | −2.3856 | 0.0015 |
| Whole Blood Cells | TP53TG5 | Up | 4.6235 | 0.0109 | Down | −3.3568 | 0.0168 |
| Whole Blood Cells | TREML1 | Up | 1.0076 | 0.0069 | Down | −1.4372 | 0.0002 |
| Whole Blood Cells | TRIM17 | Up | 1.6098 | 0.0278 | Down | −1.1120 | 0.0370 |
| Whole Blood Cells | TYRO3 | Up | 1.0424 | 0.0372 | Down | −1.7972 | 0.0000 |
| Whole Blood Cells | UNC13B | Up | 1.2061 | 0.0047 | Down | −1.4375 | 0.0000 |
| Whole Blood Cells | VSIG2 | Up | 1.4849 | 0.0204 | Down | −1.2263 | 0.0039 |
| Whole Blood Cells | VWF | Up | 1.0792 | 0.0005 | Down | −1.4591 | 0.0000 |
| Whole Blood Cells | WNT2 | Up | 2.4945 | 0.0406 | Down | −2.3451 | 0.0055 |
| Whole Blood Cells | XKR8 | Up | 1.0216 | 0.0050 | Down | −1.4189 | 0.0002 |
| Whole Blood Cells | YAP1 | Up | 2.6466 | 0.0060 | Down | −1.9990 | 0.0002 |
| Whole Blood Cells | ZCCHC12 | Up | 3.7146 | 0.0010 | Down | −2.1065 | 0.0024 |
| Whole Blood Cells | ZFP780B-PS1 | Down | −1.6098 | 0.0081 | Up | 1.3243 | 0.0131 |
AIA-M vs. Con: the significant differentially expressed genes (DEGs) between the AIA-M model group and the normal control group. BHGZD vs. BHGZD: the DEGs between the BHGZD treatment group and the AIA-M model group.
In addition, 238 putative targets of Anemarrhena asphodeloides Bge., Cinnamomum cassia Presl, and Glycyrrhiza uralensis Fisch., and 197 putative targets of Gypsum, contained in BHGZD with a high similarity score (>0.80), were predicted based on the TCMIP v2.0 platform, such as NAHD, HSD17B1, CFTR, TTGR, MIF, OXYR, CYP1A2, ESRRA, JAK1, ESRRB, CSNK2A1, ANXA5, ARSA, ATSK, C3L, CACNA1C, CACNA1S, CACNA2D1, CACNB1, CACNB2, CACNG1, CCKAR, CCL5, CD1D, CGIA, CRCA, CSLA, CSLB, and EGF. On the basis of the ETCM database, 184 putative targets of 31 bioactive chemical compounds contained in BHGZD, and 53 putative targets of Oryza sativa L. were collected, including ACO2, AKR1B1, ANG, APRT, BHMT, C8G, CPB1, CS, CTDSP1, GNMT, HGS, HS3ST3A1, VDR, CYP27B1, KCND1, KCNA3, PRKAB1, ADH1A, KCNA10, and GAMT. There were a total of 635 genes, including RA-related genes reversely regulated by BHGZD, putative targets of bioactive chemical compounds, and the known therapeutic targets of drug contained in BHGZD, considered as BHGZD effective genes.
Then, the interaction network of the “RA-related gene–BHGZD effective gene” was constructed using the links among RA-related genes and BHGZD effective genes. Following the calculation of the nodes’ topological features in the network (the median values of “Degree”, “Betweenness”, and “Closeness” were 7.0000, 0.0009, and 0.2860, respectively), four BHGZD candidate targets (PI3K, AKT1, NFκB, and IL-1β) against active RA ( Figure 1I , and Supplementary Table S6 ) were identified due to their topological importance. In addition, previous data obtained from our research group and from other research groups indicated that BHGZD may restore the imbalance of the “immune-inflammation” system via inhibiting TLR4-induced NLRP3 inflammasome signaling (21, 22), and TLR4/NFκB signaling may play a vital role in the regulation of the inflammatory response (32, 33). Moreover, PI3K/AKT signaling is involved in the process and release of pro-inflammatory cytokines (34, 35), and the activation of this signaling leads to autoimmunity, showing the increased activity in some autoimmune diseases, including RA (36). On this basis, we hypothesize that BHGZD might reverse the main pathological changes of active RA, including synovial inflammation, cartilage damage, and bone erosion, via regulating TLR4/PI3K/AKT/NFκB/NLRP3 signaling.
MG and CA May Be the Representative BACs Contained in BHGZD Against Active RA
To screen the potential BACs contained in BHGZD against active RA, herein we identified the chemical profiling of this herbal formula in sera using the UFLC-Q-TOF-MS/MS system. As a result, a total of 31 chemical compounds were identified in the serum samples 2 h after the treatment of BHGZD. Among them, 14, 16, and 1 chemical compound were from Anemarrhenae Rhizoma, Glycyrrhizae Radix et Rhizoma, and Cinnamomi Ramulus, respectively, and belong to glycosides, flavonoids, organic acid, triterpenoids, and saponins. Quantitatively, timosaponin B II, mangiferin, glycyrrhizic acid, neomangiferin, 7-O-methyl mangiferin, anemarrhensaponin I, vitexin, formononetin, liquiritigenin, isoliquiritin, isoliquiritin apioside, isoschaftoside, and isovitexin were all detected at 30 min after the treatment of BHGZD. Detailed information on chemical profiling is provided in Supplementary Table S7 , and the quantitative detection data are provided in Supplementary Table S8 . In addition, the drug-like properties of the chemical compounds contained in BHGZD, including the intestinal absorption rate and oral bioavailability, were evaluated using ADME models in silico, and a total of 8 candidate BACs with good drug-likeness were identified ( Supplementary Table S9 ).
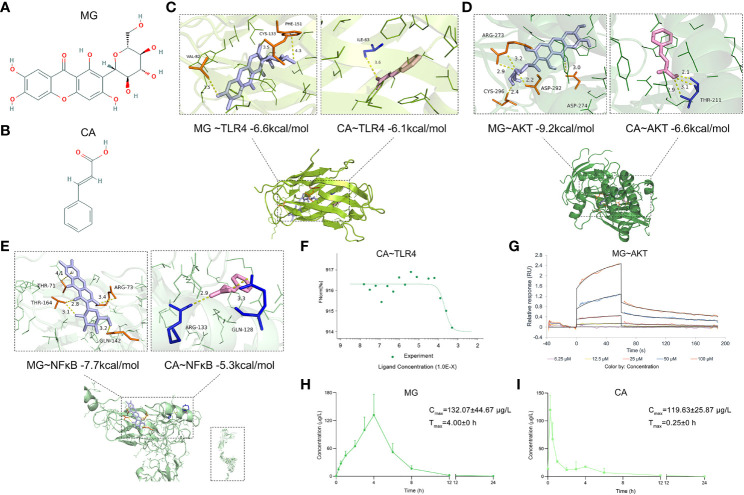
After that, molecular docking was performed to determine the binding affinities of candidate BACs contained in BHGZD to corresponding proteins of TLR4/PI3K/AKT/NFκB/NLRP3 signaling. As a result, a total of 9 chemical compounds were found to bind these proteins with binding affinities of more than −4.0 kcal/mol ( Supplementary Table S10 ). In particular, MG ( Figure 2A ) from Anemarrhenae Rhizoma and CA ( Figure 2B ) from Cinnamomi Ramulus docked well into the TLR4, AKT, and NFκB binding cavity with more robust interactions and stronger binding affinity (MG–TLR4 −6.6, CA–TLR4 −6.1, MG–AKT −9.2, CA–AKT −6.6, MG–NFκB −7.7, and CA–NFκB −5.3 kcal/mol, Figures 2C–E ). Experimentally, MST and SPR assays were carried out to verify the direct binding efficiency of CA to TLR4 and MG to AKT, respectively. As shown in Figures 2F and G , the mean K D values of 2.11e−4 and 3.83e−5 M for CA–TLR4 and MG–AKT, respectively, indicated strong binding affinities.
Figure 2.
Chemical structures of MG and CA, molecular docking simulation of the binding patterns of them with corresponding proteins, as well as the mean plasma concentration–time curves of MG and CA after the treatment of BHGZD. (A) Chemical structure of MG (obtained from PubChem databases, CID 5281647). (B) Chemical structure of CA (obtained from PubChem databases, CID 444539). (C) Molecular docking simulation of the binding patterns of TLR4 with MG and CA, respectively. (D) Molecular docking simulation of the binding pattern of AKT with MG and CA, respectively. (E) Molecular docking simulation of the binding pattern of NFκB with MG and CA, respectively. (F) Microscale thermophoresis (MST)-determined binding affinity between CA and TLR4 protein. (G) Surface plasmon resonance (SPR) assay of the interaction between MG and AKT1 protein. (H) The mean plasma concentration–time curves of MG after the treatment of BHGZD for 12 days at the dosage of 21.4 g/kg each day (n = 3 per group). (I) The mean plasma concentration–time curves of CA after the treatment of BHGZD for 12 days at the dosage of 21.4 g/kg each day (n = 3 per group).
Moreover, the content of MG and CA was determined to be 336.96 ng and 26.14 ng in 1 mg of BHGZD lyophilized powder, respectively, with a conversion ratio of 8.33% (calculated by the weight of BHGZD lyophilized powder divided by the weight of crude herbs contained in BHGZD; 1 g of BHGZD lyophilized powder is equivalent to the amount of 12 g of BHGZD).
The UHPLC-QTRAP-MS/MS system was further used to detect the pharmacokinetic properties of MG and CA in sera after the treatment of BHGZD ( Supplementary Figure S1 ). A summary of MG and CA pharmacokinetic parameters using a non-compartment model analysis is presented in Table 2 , and the mean plasma concentration–time profiles from time 0 to 24 h after the treatment of BHGZD at a dosage of 21.4 g/kg each day are shown in Figures 2H and I . Briefly, the maximum plasma concentration (Cmax) of MG and CA was 132.01 μg/L and 119.63 μg/L, respectively. The time to reach the maximum plasma concentration (Tmax) for MG and CA in rats receiving BHGZD was 4.00 h and 0.25 h, respectively. The plasma concentration–time curve AUC (0-24h) of MG and CA was 578.42 μg/L·h and 102.48 μg/L·h, respectively. The apparent elimination half-life (T1/2z) and mean retention time (MRT) of MG (1.26 h and 4.23 h, respectively) and CA (1.65 h and 1.93 h, respectively) in rats after the treatment of BHGZD were measured.
Table 2.
Pharmacokinetic parameters analyzed by the UHPLC-QTRAP-MS/MS system.
| Pharmacokinetic Parameter | Mangiferin (MG) | Cinnamic acid (CA) | ||
|---|---|---|---|---|
| Mean ± SD | RSD/% | Mean ± SD | RSD/% | |
| Cmax (μg/L) | 132.07 ± 44.67 | 33.80 | 119.63 ± 25.87 | 21.60 |
| Tmax (h) | 4.00 ± 0.00 | 0.00 | 0.25 ± 0.00 | 0.00 |
| AUC(0-t) (μg/L*h) | 578.42 ± 182.51 | 31.60 | 102.48 ± 67.77 | 66.10 |
| AUC(0-∞) (μg/L*h) | 579.07 ± 181.70 | 31.40 | 144.23 ± 30.36 | 21.00 |
| t1/2z (h) | 1.26 ± 0.23 | 17.90 | 1.65 ± 1.34 | 81.10 |
| MRT(0-t) (h) | 4.23 ± 0.30 | 7.10 | 1.93 ± 0.35 | 18.30 |
| MRT(0-∞) (h) | 4.25 ± 0.28 | 6.50 | 2.26 ± 0.80 | 35.20 |
Cmax, the maximum plasma concentration. Tmax, the time to reach the maximum plasma concentration. AUC (0-t), area under the concentration–time curve from zero to the last sampling time. AUC (0-∞), area under the concentration–time curve from zero to infinity. T1/2z, apparent elimination half-life. MRT (0-t), mean residence time from zero to the last sampling time. MRT (0-∞), mean residence time from zero to infinity. Data are expressed as the mean ± SD.
Both BHGZD and the Two-BAC Combination of MG and CA Improve Arthritis Severity in Active RA Rats
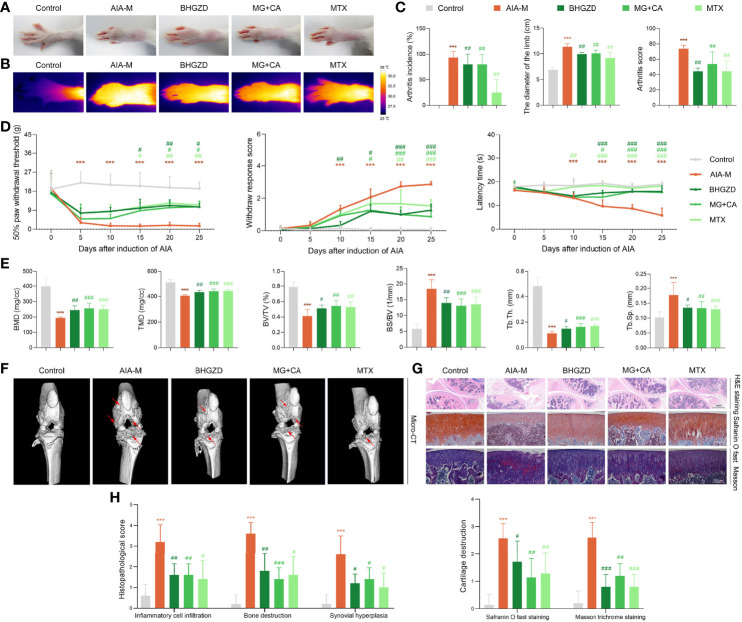
To verify the pharmacological effects of the two-BAC combination of MG and CA against active RA, the in vivo experiments were performed based on the AIA-M rat model (22, 24). Similar pathological characteristics and changes in AIA-M rats were observed from the validation cluster to the discovery cluster ( Figure 3 ). As a result, the ankles and knuckle joints of AIA-M rats with severe redness and swelling were remarkably improved by the treatment of the two-BAC combination (equivalent to BHGZD at a dose of 21.4 g/kg) and BHGZD (dose of 21.4 g/kg, Figures 3A and B ). Importantly, both the treatment of BHGZD and the two-BAC combination significantly alleviated disease severity, including reducing arthritis incidence, the diameter of the limb, and arthritis score (all p < 0.05, on the 27th day after immunization), and simultaneously elevated pain thresholds (mechanical-, acetone-, and thermal-induced hyperalgesia), all of which were similar to the pharmacological effects of MTX (positive drug, dose of 0.2 mg/kg, Figures 3C and D ).
Figure 3.
Therapeutic effects of BHGZD, two-BAC combination, and MTX on the severity of arthritis in AIA-M rats (Control, AIA-M, BHGZD, MG+CA, and MTX). (A) Representative images of arthritis. (B) Infrared thermography. (C) Arthritis incidence (n = 3 per group), the diameter of the limb (n = 5 per group), and arthritis score on the 27th day after immunization (n ≥ 4 per group, all experiments were performed in triplicate). (D) The pain thresholds (mechanical-, acetone-, and thermal-induced hyperalgesia, n ≥ 4 per group, all experiments were performed in triplicate). (E) Quantitative micro-computed tomography (micro-CT) analysis of bone mineral density (BMD), tissue mineral density (TMD), bone volume/tissue volume (BV/TV), bone surface/bone volume (BS/BV), trabecular separation (Tb.Sp), and trabecular thickness (Tb.Th) on the 31st day after immunization (n ≥ 5 per group, all experiments were performed in triplicate). (F) Representative micro-CT images of knee joints showing bone erosion levels (the red arrow indicates the position of the bone destruction). (G) Pathological changes in the knee joints using hematoxylin and eosin (H&E, scale bar represents 1 mm), safranin O fast green (scale bar represents 200 μm), and Masson trichrome staining (scale bar represents 200 μm) in different groups (n = 5 per group). (H) Quantitative analysis of H&E staining in inflammatory cell infiltration, bone destruction, and synovial hyperplasia. Cartilage destruction of safranin O fast, and Masson trichrome staining (n = 5 per group). Data are expressed as the mean ± SD. ***, p < 0.001, comparison with the normal control group; #, ##, ###, p < 0.05, p < 0.01, p < 0.001, respectively, comparison with the AIA-M model group.
In order to investigate the joint destruction and synovial inflammation, both the ankle and knee joints of AIA-M rats in different groups were scanned using Micro-CT analysis on the 31st day after immunization. The quantified data revealed that BMD, TMD, BV/TV ratio, and Tb.Th were dramatically decreased, and BS/BV ratio and Tb.Sp was observably increased in the AIA-M model group (all p < 0.05, Figure 3E and Supplementary Figure S2B ) with rough bone surfaces and severe bone erosion ( Figure 3F , and Supplementary Figure S2A ). As shown in Figure 3F , both BHGZD and the two-BAC combination treatment groups showed smooth bone surface with a significantly increased BMD, TMD, BV/TV ratio, and Tb.Th, as well as a significantly decreased BS/BV ratio and Tb.Sp (all p < 0.05, Figure 3E and Supplementary Figure S2B ), suggesting that the two-BAC combination may efficiently reverse bone erosion, which was similar to the pharmacological effects of MTX.
Moreover, H&E, safranin O fast green, and Masson trichrome staining were carried out to evaluate the degree of joint lesions in AIA-M rats. As shown in Figures 3G and H , the treatment of the two-BAC combination, BHGZD, and MTX all apparently reversed the histopathological changes of knee joints in AIA-M rats, including the decreased inflammatory cell infiltration, the prevention of cartilage and bone destruction, and synovial hyperplasia (all p < 0.05). Consistently, a significant reduction of the safranin O fast green-positive area and an elevation of the Masson-positive area were observed on the cartilage surface of AIA-M rats, reflecting a loss of articular cartilage, but reversed by the two-BAC combination, similar to the pharmacological effects of BHGZD and MTX (all p < 0.05, Figures 3G and H ). These findings revealed that BHGZD and the two-BAC combination of MG and CA may effectively alleviate the progression of bone damages, repair bone erosion, and simultaneously improve pathological changes of articular cartilage and synovial inflammation in the arthritic joints of AIA-M rats.
In terms of the response to inflammation, pathological changes of the thymus and spleen and viscera indexes in different groups were examined. As shown in Figure S2C , the lower proportion of white pulp in the spleen with decreased cell density lymphatic sheath, lymphoid follicular hyperplasia, marginal zone, red pulp, and germinal center was observed in AIA-M rats, which were significantly improved by the treatment of BHGZD and the two-BAC combination (all p < 0.05, Supplementary Figure S2D ). A thinner thymic cortex, less thymic lobule, an unclear boundary, and more vacuoles in the cytoplasm of epithelial reticular cells were also observed in AIA-M rats, which were remarkably improved by the treatment of BHGZD and the two-BAC combination ( Supplementary Figure S2C). The treatment with BHGZD and the two-BAC combination protected the morphological structure of the spleen and thymus, and simultaneously decreased spleen and thymus indexes significantly (all p < 0.05, Supplementary Figure S2E ). Furthermore, H&E-stained tissues of liver and kidney, and the liver and kidney indexes showed no toxic damages ( Supplementary Figures S2C and F ).
Both BHGZD and the Two-BAC Combination of MG and CA Suppress NLRP3 Inflammasome-Induced Pyroptosis Via Regulating TLR4/PI3K/AKT/NFκB Signaling
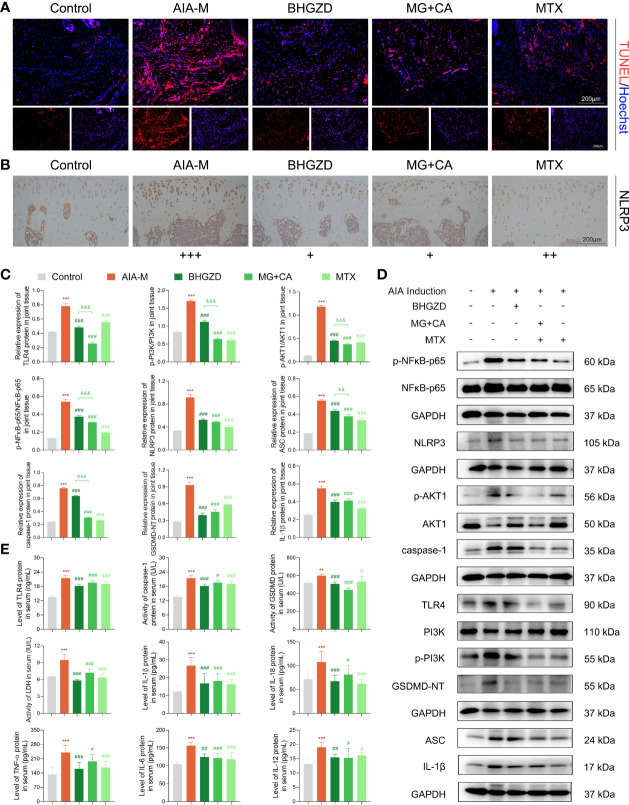
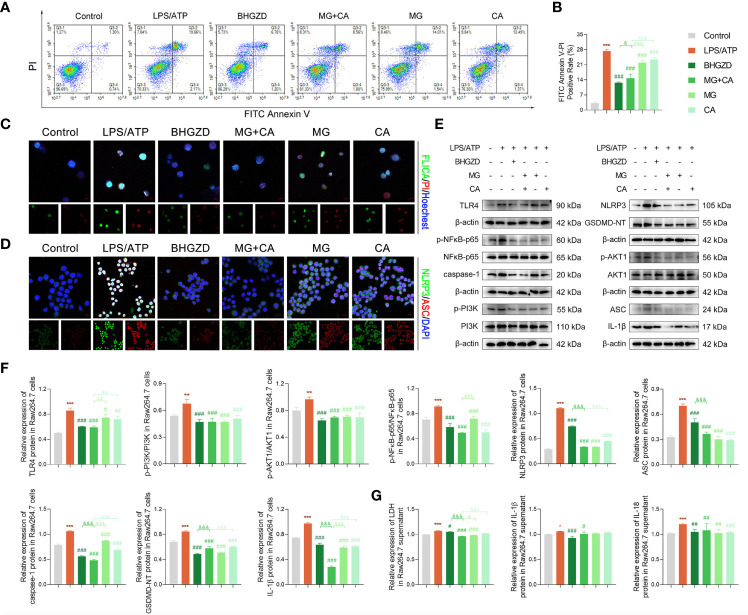
The above network-based data imply that BHGZD might reverse the imbalance of the “immune-inflammation” system during active RA progression via regulating TLR4/PI3K/AKT/NFκB/NLRP3 signaling. To the best of our knowledge, pyroptosis is a kind of NLRP3 inflammasome-induced inflammatory cell death, characterized by cell swelling and release of pro-inflammatory cytokines depending on the activation of caspase-1, contributing to inflammation in arthritis (37). The formation of the NLRP3 inflammasome complex requires the interaction of NLRP3 with ASC and caspase-1, which are vital for the assembly and activation of the NLRP3 inflammasome. In our previous study, the immunomodulatory and anti-inflammatory activities of BHGZD were verified, especially in inhibiting pyroptotic death, which may be attributed to the activation of TLR4-NLRP3 inflammasome signaling (22). Currently, our TUNEL data showed a significant proportion of pyroptosis cells in the arthritic joints of AIA-M rats, which was improved by the treatment of BHGZD and the two-BAC combination ( Figure 4A ). Similar to the pharmacological effects of MTX, high expression levels of NLRP3 and ASC, high expression and enhanced activity of caspase-1, and increased expression of IL-1β and IL-18 (the signature inflammatory cytokines in pyroptosis) in the AIA-M model group (all p < 0.05) were all significantly decreased by the treatment of BHGZD and the two-BAC combination (all p < 0.05, Figures 4B–E ). Pyroptosis was assessed by LDH activity, which may be used to verify cell membrane integrity and the release of intracellular soluble component. Consistently, the enhancing activity of LDH in the sera of AIA-M rats was observed, which was significantly decreased by the treatment of BHGZD, the two-BAC combination, and MTX (all p < 0.05, Figure 4E ), suggesting the alleviation of downstream membrane damage.
Figure 4.
Regulatory effects of BHGZD, the two-BAC combination, and MTX on the expression of TLR4, p-PI3K/PI3K, p-AKT1/AKT1, p-NFκB-p65/NFκB-p65, NLRP3, ASC, caspase-1, GSDMD-NT, IL-1β, IL-18, IL-6, TNF-α, and IL-12 in AIA-M rats of different groups (Control, AIA-M, BHGZD, MG+CA, and MTX). (A) Representative images of pyroptosis in the knee joints of AIA-M rats detected by deoxynucleotidyltransferase-mediated UTP end labeling (TUNEL) assay (scale bar represents 200 μm, TUNEL red, Hoechst blue). (B) The expression of NLRP3 protein in the knee joints of AIA-M rats (scale bar represents 200 μm). (C, D) The protein expression of TLR4, p-PI3K/PI3K, p-AKT1/AKT1, p-NFκB-p65/NFκB-p65, NLRP3, ASC, caspase-1, GSDMD-NT, and IL-1β in the knee joints of AIA-M rats using Western botting analysis (n = 3 per group). (E) The expression levels of TLR4, TNF-α, IL-6, IL-1β, IL-18, and IL-12 in the sera of AIA-M rats using enzyme-linked immunosorbent assay (ELISA) analysis, and the activities of caspse-1, GSDMD-NT, and LDH in the sera of AIA-M rats using ELISA analysis (n ≥ 3 per group, all experiments were performed in triplicate). Data are expressed as the mean ± SD. **, ***, p < 0.01, p < 0.001, respectively, comparison with the normal control group; #, ##, ###, p < 0.05, p < 0.01, p < 0.001, respectively, comparison with the AIA-M model group; &&, &&&, p < 0.01, p < 0.001, comparison with the treatment of BHGZD.
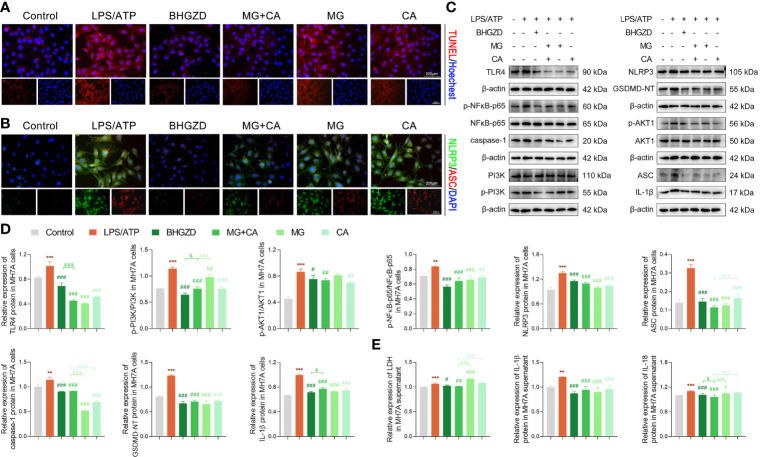
To verify the in vivo findings based on AIA-M rats, an established method (LPS plus ATP) was applied to induce NLRP3 inflammasome activation in both RAW264.7 and MH7A cells. The cell viability on RAW264.7 cells with the treatment of MG and CA was initially examined using the CCK8 assay, and the results exhibited no cell toxicity under 0.21–6.74 ng/ml MG or 0.02–0.52 ng/ml CA treatment alone, as shown in Supplementary Figure S2G . Thus, 1.69 ng/ml MG and 0.13 ng/ml CA treatment were chosen in the following assays (the same content as that in the 5 μg/ml BHGZD formula). Flow cytometry analysis revealed that MG, CA, and the two-BAC combination treatment prominently reduced the PI [a marker of cells that stains necrotic, dead, and membrane-compromised cells (38)] positive cell rate of RAW264.7 cells induced by LPS/ATP, which was inferior to that in the BHGZD treatment group (all p < 0.05, Figures 5A and B ). As shown in Figures 5C and 6A , LPS/ATP induced an increase in the number of both FLICA-positive RAW264.7 cells and TUNEL-positive MH7A cells, which was reduced by the treatment of BHGZD, MG, CA, and the two-BAC combination. Intriguingly, the inhibition of MG or CA alone on cell pyroptosis induced by LPS/ATP was weaker than that in the two-BAC combination treatment group (all p < 0.05, Figures 5A–C and 6A ). In addition, the expression levels of NLRP3, ASC, and caspase-1 were significantly increased in both RAW264.7 and MH7A cells induced by LPS/ATP, the same as the levels of IL-1β, IL-18, and LDH release in the supernatant of RAW264.7 and MH7A cells, which were all reduced by the treatment of BHGZD, MG, CA, and the two-BAC combination (all p < 0.05, Figures 5D–G and 6B–E ). GSDMD, a crucial mediator of pyroptosis downstream of canonical and non-canonical inflammasomes (39–41), is cleaved by caspase-1 at a specific site (GSDMD-NT) and subsequently causes cell lysis and IL-1β release (42, 43). In the current study, the activities of GSDMD-NT in sera and its levels in knee joints were both significantly elevated in AIA-M rats, which was reduced by the treatment of BHGZD, the two-BAC combination, and MTX (all p < 0.05, Figures 4C–E ). Notably, GSDMD-NT occurred in both RAW264.7 and MH7A cells induced by LPS/ATP, which was decreased by the treatment of BHGZD, MG, CA, and the two-BAC combination ( Figures 5E, F and 6C, D ).
Figure 5.
BHGZD inhibits LPS/ATP-induced pyroptosis in RAW264.7 macrophage by downregulating TLR4, p-PI3K/PI3K, p-AKT1/AKT1, p-NFκB-p65/NFκB-p65, NLRP3, ASC, caspase-1, GSDMD-NT, IL-1β, and IL-18 (Control, AIA-M, BHGZD, MG+CA, MG, and CA). (A, B) Flow cytometry analysis for Annexin V/PI staining in RAW264.7 cells (n ≥ 3 per group, all experiments were performed in triplicate). (C) Representative images of FAM-FLICA Casapse-1 that binds only to activated caspase-1 (scale bar represents 50 μm, FAM-FLICA green, PI red, Hoechst blue). (D) Expression of NLRP3 and ASC protein measured by immunofluorescent staining and confocal microscopy in RAW264.7 cells (scale bar represents 50 μm, NLRP3 FITC green, ASC CY3 red, DAPI blue). (E, F) Expression levels of TLR4, p-PI3K/PI3K, p-AKT1/AKT1, p-NFκB-p65/NFκB-p65, NLRP3, ASC, caspase-1, GSDMD-NT, and IL-1β in RAW264.7 cells measured by Western blotting (n = 3 per group). (G) Levels of IL-1β, IL-18, and LDH release in RAW264.7 cells detected by ELISA (n ≥ 3 per group, all experiments were performed in triplicate). *, **, ***, p < 0.05, p < 0.01, p < 0.001, respectively, comparison with the normal control group; #, ##, ###, p < 0.05, p < 0.01, p < 0.001, respectively, comparison with LPS/ATP-induced model; &, &&&, p < 0.05, p < 0.001, comparison with the treatment of BHGZD; △, △△, △△△, p < 0.005, p < 0.01, p < 0.001, comparison with the treatment of the two-BAC combination.
Figure 6.
BHGZD inhibits LPS/ATP-induced pyroptosis in MH7A cells by downregulating TLR4, p-PI3K/PI3K, p-AKT1/AKT1, p-NFκB-p65/NFκB-p65, NLRP3, ASC, caspase-1, GSDMD-NT, IL-1β, and IL-18 (Control, AIA-M, BHGZD, MG+CA, MG, and CA). (A) TUNEL staining of MH7A cells (scale bar represents 200 μm, TUNEL red, Hoechst blue). (B) Expression of NLRP3 and ASC protein measured by immunofluorescent staining and confocal microscopy in MH7A cells (scale bar represents 200 μm, NLRP3 FITC green, ASC CY3 red, DAPI blue). (C, D) Expression levels of TLR4, p-PI3K/PI3K, p-AKT1/AKT1, p-NFκB-p65/NFκB-p65, NLRP3, ASC, caspase-1, GSDMD-NT, and IL-1β in MH7A cells measured by Western blotting (n = 3 per group). (E) Levels of IL-1β, IL-18, and LDH release in MH7A cells evaluated by ELISA (n ≥ 3 per group, all experiments were performed in triplicate). **, ***, p < 0.01, p < 0.001, respectively, comparison with the normal control group; #, ##, ###, p < 0.05, p < 0.01, p < 0.001, respectively, comparison with LPS/ATP-induced model; &, &&&, p < 0.05, p < 0.001, comparison with the treatment of BHGZD; △△△, p < 0.001, comparison with the treatment of two-BAC combination.
After determining the inhibitory effects of BHGZD and the two-BAC combination on NLRP3 inflammasome-induced pyroptosis, we further investigated the changes of its upstream determinant TLR4/PI3K/AKT/NFκB signaling in different groups. Importantly, the expression levels of TLR4 in both sera and knee joints of AIA-M rats were increased, and subsequently were effectively reversed by the treatment of the two-BAC combination, with similar trends to BHGZD and MTX (all p < 0.05, Figures 4C–E ). The protein expression ratios of p-PI3K/PI3K, p-NFκB-p65/NFκB-p65, and p-AKT1/AKT1 were all significantly increased in AIA-M rats, which were distinctively reduced by the treatment of BHGZD and the two-BAC combination (all p < 0.05, Figures 4C, D ), indicating their inhibitory effects on TLR4/PI3K/AKT/NFκB signaling activation. In addition to the increased accumulation of inflammatory cells, the expression levels of inflammatory cytokines in AIA-M rats sera, including TNF-α, IL-6, and IL-12, were abnormally elevated in AIA-M rats, and reduced by the treatment of the two-BAC combination, in accordance with the effects of BHGZD and MTX (all p < 0.05, Figure 4E ). Interestingly, we also achieved the same findings in the in vitro experiment validations based on both RAW264.7 and MH7A cells induced by LPS/ATP (all p < 0.05, Figures 5E–G and 6C–E ).
These findings demonstrated that both BHGZD and the two-BAC combination of MG and CA may suppress TLR4/PI3K/AKT/NFκB signaling-related protein activation, and subsequently inhibit NLRP3 inflammasome-induced pyroptosis.
Discussion
Growing clinical evidence shows that various immune cells sustainably influx and migrate into the joints via secreting different types of immunomodulatory cytokines, leading to pyroptosis-induced persistent synovitis and cartilage degradation (44, 45). Thus, reversing the imbalance of the “immune-inflammation” system, especially alleviating pyroptosis, may be a promising therapeutic strategy for active RA. The current study performed an integrative research combining UFLC-Q-TOF-MS/MS, gene expression profiling, network calculation, pharmacokinetic profiling, SPR/MST assay, and pharmacological experiment validations, and identified TLR4/PI3K/AKT/NFκB/NLRP3 signaling-induced pyroptosis as one of the candidate effective targets of BHGZD for reversing the imbalance network of “immune-inflammation” during active RA progression. In addition, both MG and CA were identified as representative BACs acting on that target, for the strong binding affinities between compounds and target proteins, good pharmacokinetic features, and similar pharmacological effects to BHGZD. Notably, both BHGZD and the two-BAC combination of MG and CA effectively improved disease severity of active RA rats including elevating pain thresholds, relieving joint inflammation and bone erosion via inhibiting TLR4/PI3K/AKT/NFκB signaling to suppress the activation of the NLRP3 inflammasome, leading to the downregulation of downstream caspase-1, the reduced release of IL-1β, and the modulation of GSDMD-mediated pyroptosis. Consistent data were obtained based on the in vitro pyroptosis models of RAW264.7 and MH7A cells induced by LPS/ATP. Thus, RAW264.7 macrophages might be the probable immune cells targeted by both BHGZD and the two-BAC combination of MG and CA.
Currently, the AIA-M rat model was established simulating the pathogenetic characteristics of active RA with moist heat arthralgia spasm syndrome on the basis of male Lewis rats according to our previous studies (20–23). The abnormal changes in indicators reflecting the imbalance of the “immune-inflammation” system, including distinct redness and swelling, an increase in arthritis surface temperature, arthritis incidence, the diameter of the limb, arthritis score, pathologic changes of the thymus and spleen, as well as severe inflammatory cell infiltration, cartilage and bone destruction, synovial hyperplasia, and the high levels of inflammatory mediators, such as TLR4, IL-6, IL-12, IL-1β, IL-18, and TNF-α, were observed as the distinctive characteristics and biological basis of AIA-M rats, which may be in line with the clinical manifestations in active RA patients. Following the transcriptomic profiling and biomolecular network analyses, a series of active RA-related genes were also identified, and functionally involved in the regulation of the “immune-inflammation” system accordingly.
Considering that TCM contributed to the multi-target interactions of the complex ingredients of its herbal drugs (46–48), pharmaceutical development has consistently been an urgent challenge. Therefore, a new method that combines BACs contained in herbal formulas has been strongly indicated for new drug discovery. Herein, the two BACs, namely, MG from Anemarrhenae Rhizoma and CA from Cinnamomi Ramulus, were identified as the representative BACs of BHGZD for the strong binding affinities between compounds and target proteins, good pharmacokinetic properties, the high content and the importance of the formula, and similar pharmacological effects to BHGZD. Interestingly, the treatment of MG or CA alone did not exert prominently therapeutic effects compared to that of the two-BAC combination. Similar to the BHGZD formula, the two-BAC combination treatment of MG and CA may exert satisfying therapeutic efficacy on both in vivo and in vitro experiments via regulating TLR4/PI3K/AKT/NFκB signaling, which plays a vital role in synovial inflammation, cartilage degradation, and bone erosion by regulating inflammation response, immune disorder cells, and pyroptosis (37, 49–52). More importantly, the two-BAC combination of MG and CA offers key potential advantages over BHGZD for the following points. Firstly, the two-BAC combination may exert similar pharmacological effects in treating AIA-M rats with a definite material basis. Secondly, the two-BAC combination may be flexible in design, easily synthesized on a large scale, easily absorbed, and relatively stable, not dependent on the cultivations of crude herbs. Thirdly, it is better to understand the underlying molecular mechanisms of TCM-based RA therapeutics.
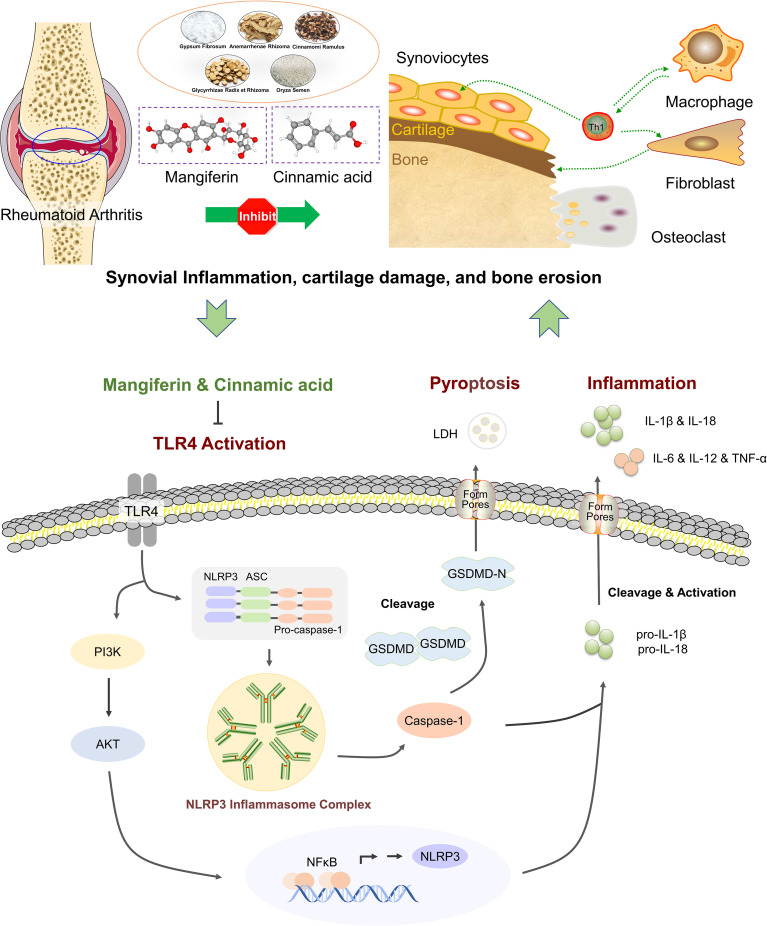
In conclusion, our data offer an evidence that the MG and CA combination from BHGZD may interact with TLR4/PI3K/AKT/NFκB signaling to inhibit NLRP3 inflammasome activation and modulate pyroptosis, which provides the novel representative BACs and pharmacological mechanisms of BHGZD against active RA ( Figure 7 ). The findings may shed new light on the mechanisms of the TCM formula, and promote the modernization development of TCM and drug discovery.
Figure 7.
The schematic diagram of the underlying mechanisms of the two-BAC combination of mangiferin (MG) and (CA) against active RA via reversing the imbalance of the “immune-inflammation” system by inhibiting TLR4/NFκB/NLRP3 activation-induced pyroptosis. “⟂“: Inhibition. “→“: Activation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number(s) can be found below: NCBI Gene Expression Omnibus; GSE190523, GSE189942.
Ethics Statement
The study was approved by the Research Ethics Committee of the Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China [Ethics Approval Number: 2019-026 and IBTCMCACMS21-2105-01, certificate number of the facility: SYXK (Beijing) 2021-0017]. All animal studies were treated in accordance with the guidelines and regulations for the use and care of animals of the Center for Laboratory Animal Care, China Academy of Chinese Medical Sciences. All animal-handling procedures were performed according to the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and followed the guidelines of the Animal Welfare Act.
Author Contributions
NL and YZ engaged in study design and coordination, material support for obtained funding, and supervised study. YZ designed the experimental validation and revised the manuscript. WL performed most of the experiments and statistical analysis, as well as wrote the manuscript. SY and WS performed parts of the experiments, reviewed and approved the final manuscript. The other authors performed parts of the experiments. All authors contributed to the article and approved the final manuscript.
Funding
This study is funded by the National Natural Science Foundation of China (81630107), the Scientific and Technological Innovation Project of China Academy of Chinese Medical Sciences (CI2021A03808 and CI2021A01508), and the National Key Research and Development Program of China (2018YFC1705201). No study sponsors are involved in the research process of this project.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
- ADME
absorption distribution metabolism excretion
- AIA
adjuvant-induced arthritis
- AKT1: RAC
alpha serine/threonine-protein kinase
- ANOVA
analysis of variance
- ASC
apoptosis-associated speck-like protein containing a CARD
- ATP
adenosine triphosphate
- BAC
bovine serum albumin
- BACs
bioactive compounds
- BHGZD
Baihu-Guizhi decoction
- BMD
bone mineral density
- BS/BV
bone surface/bone volume
- BV/TV
bone volume/tissue volume
- CA
cinnamic acid
- CASP1
caspase-1
- CCK-8
cell counting kit-8
- Cmax
the maximum plasma concentration
- DAPI
4’, 6-diamidino-2-phenylindole
- DMARDs
disease-modifying anti-rheumatic drugs
- ELISA
enzyme-linked immunosorbent assay
- FBS
fetal bovine serum
- FDA
Food and Drug Administration
- GSDMD
gasdermin
- H&E staining
hematoxylin and eosin staining
- IL-12
interleukin-12
- IL-18
interleukin-18
- IL-1β
interleukin-1 beta
- IL-6
interleukin-6
- LDH
lactate dehydrogenase
- LPS
lipopolysaccharide
- MG
mangiferin
- Micro-CT
micro-computed tomography
- MST
Microscale thermophoresis
- MTX
methotrexate
- NFκB-P65
NF-kappa B transcription factor p65 subunit
- NLRP3
NOD-like receptor pyrin domain containing 3
- NSAIDs
non-steroidal anti-inflammatory drugs
- PBS
phosphate-buffered saline
- PFA
paraformaldehyde
- PI3K
phosphatidylinositol 3 kinase
- PIK3CG
phosphatidylinositol 4, 5-bisphosphate 3-kinase catalytic subunit gamma isoform
- PK
pharmacokinetic
- RA
rheumatoid arthritis
- RNA-Seq
RNA sequencing
- ROI
region of interest
- SPR
surface plasmon resonance
- Tb.Sp
trabecular separation
- Tb.Th
trabecular thickness
- TCM
traditional Chinese medicine
- TLR4
toll-like receptor 4
- Tmax
the time to reach the maximum plasma concentration
- TMD
tissue mineral density
- TNF
tumor necrosis factor
- TUNEL assay
deoxynucleotidyltransferase-mediated UTP end labeling
- TwHF
Tripterygium wilfordii Hook F
- UFLC-Q-TOF-MS/MS
the ultra-fast liquid chromatography-quadrupole-time-of-flight tandem mass spectrometry
- WTD
Wu-Tou decoction
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.912933/full#supplementary-material
References
- 1. Firestein GS, McInnes IB. Immunopathogenesis of Rheumatoid Arthritis. Immunity (2017) 46(2):183–96. doi: 10.1016/j.immuni.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Catrina AI, Svensson CI, Malmstrom V, Schett G, Klareskog L. Mechanisms Leading From Systemic Autoimmunity to Joint-Specific Disease in Rheumatoid Arthritis. Nat Rev Rheumatol (2017) 13(2):79–86. doi: 10.1038/nrrheum.2016.200 [DOI] [PubMed] [Google Scholar]
- 3. Tak PP, Kalden JR. Advances in Rheumatology: New Targeted Therapeutics. Arthritis Res Ther (2011) 13 Suppl 1:S5. doi: 10.1186/1478-6354-13-S1-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burmester GR, Pope JE. Novel Treatment Strategies in Rheumatoid Arthritis. Lancet (2017) 389(10086):2338–48. doi: 10.1016/S0140-6736(17)31491-5 [DOI] [PubMed] [Google Scholar]
- 5. England BR, Thiele GM, Anderson DR, Mikuls TR. Increased Cardiovascular Risk in Rheumatoid Arthritis: Mechanisms and Implications. BMJ (2018) 361:k1036. doi: 10.1136/bmj.k1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Semb AG, Kvien TK, Aastveit AH, Jungner I, Pedersen TR, Walldius G, et al. Lipids, Myocardial Infarction and Ischaemic Stroke in Patients With Rheumatoid Arthritis in the Apolipoprotein-Related Mortality Risk (Amoris) Study. Ann Rheum Dis (2010) 69(11):1996–2001. doi: 10.1136/ard.2009.126128 [DOI] [PubMed] [Google Scholar]
- 7. Solomon DH, Karlson EW, Rimm EB, Cannuscio CC, Mandl LA, Manson JE, et al. Cardiovascular Morbidity and Mortality in Women Diagnosed With Rheumatoid Arthritis. Circulation (2003) 107(9):1303–7. doi: 10.1161/01.cir.0000054612.26458.b2 [DOI] [PubMed] [Google Scholar]
- 8. Bordy R, Totoson P, Prati C, Marie C, Wendling D, Demougeot C. Microvascular Endothelial Dysfunction in Rheumatoid Arthritis. Nat Rev Rheumatol (2018) 14(7):404–20. doi: 10.1038/s41584-018-0022-8 [DOI] [PubMed] [Google Scholar]
- 9. Sapir-Koren R, Livshits G. Postmenopausal Osteoporosis in Rheumatoid Arthritis: The Estrogen Deficiency-Immune Mechanisms Link. Bone (2017) 103:102–15. doi: 10.1016/j.bone.2017.06.020 [DOI] [PubMed] [Google Scholar]
- 10. Adami G, Saag KG. Osteoporosis Pathophysiology, Epidemiology, and Screening in Rheumatoid Arthritis. Curr Rheumatol Rep (2019) 21(7):34. doi: 10.1007/s11926-019-0836-7 [DOI] [PubMed] [Google Scholar]
- 11. Simon TA, Thompson A, Gandhi KK, Hochberg MC, Suissa S. Incidence of Malignancy in Adult Patients With Rheumatoid Arthritis: A Meta-Analysis. Arthritis Res Ther (2015) 17:212. doi: 10.1186/s13075-015-0728-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. England BR, Hershberger D. Management Issues in Rheumatoid Arthritis-Associated Interstitial Lung Disease. Curr Opin Rheumatol (2020) 32(3):255–63. doi: 10.1097/bor.0000000000000703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qindeel M, Ullah MH, Fakhar Ud D, Ahmed N, Rehman AU. Recent Trends, Challenges and Future Outlook of Transdermal Drug Delivery Systems for Rheumatoid Arthritis Therapy. J Control Release (2020) 327:595–615. doi: 10.1016/j.jconrel.2020.09.016 [DOI] [PubMed] [Google Scholar]
- 14. Guo D, Lv J, Chen X, Yan X, Ma F, Liu Y, et al. Study of Mirna Interactome in Active Rheumatoid Arthritis Patients Reveals Key Pathogenic Roles of Dysbiosis in the Infection-Immune Network. Rheumatol (Oxford) (2021) 60(3):1512–22. doi: 10.1093/rheumatology/keaa369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang P, Li J, Han Y, Yu XW, Qin L. Traditional Chinese Medicine in the Treatment of Rheumatoid Arthritis: A General Review. Rheumatol Int (2010) 30(6):713–8. doi: 10.1007/s00296-010-1370-0 [DOI] [PubMed] [Google Scholar]
- 16. Fang L. The Clinical Effect of Baihu Plus Guizhi Decoction in Treating Rheumatoid Arthritis With Damp-Heat Arthralgia Syndrome and its Influence on the Expression of RF, IL-22, IL-35 and GPI. Global Tradit Chin Med (2018) 11(06):964–7. doi: 10.14164/j.cnki.cn11-5581/r.2022.02.078 [DOI] [Google Scholar]
- 17. Yuan L, Wu J, Tang J, Chen Y, Zhang Z. Clinical Observation on Baihu Plus Guizhi Decoction Combined With Western Medicine in Treating Rheumatoid Arthritis of Rheumatic Fever Arthralgia Sydrome. J Liaoning Univ Tradit Chin Med (2019) 21(12):168–71. doi: 10.13194/j.issn.1673-842x.2019.12.044 [DOI] [Google Scholar]
- 18. Yuan L. Clinical Observation on Baihu Plus Guizhi Decoction Combined With Western Medicine in Treating Rheumatoid Arthritis of Rheumatic Fever Arthralgia Sydrome. Guangzhou Chin Med (2020). doi: 10.27879/d.cnki.ggxzy.2020.000306 [DOI] [Google Scholar]
- 19. Wu D. Efficacy of Baihu-Guizhi Decoction Combined With Western Medicine in Treating Rheumatoid Arthritis. Chin J Urban Rural Enterp Hyg (2021) 36(01):158–60. doi: 10.16286/j.1003-5052.2021.01.067 [DOI] [Google Scholar]
- 20. Li W, Lu J, Mao X, Guo Q, Wang X, Guo M, et al. A Comparative Study on the Mechanisms of Two Classical Herbal Formulae for Rheumatoid Arthritis Applying Cold and Heat Patterns Based on Target Network. Acta Pharm Sin A (2018) 53(09):1387–97. doi: 10.16438/j.0513-4870.2018-0519 [DOI] [Google Scholar]
- 21. Li W, Mao X, Wu H, Guo M, Su X, Lu J, et al. Deciphering the Chemical Profile and Pharmacological Mechanisms of Baihu-Guizhi Decoction Using Ultra-Fast Liquid Chromatography-Quadrupole-Time-Of-Flight Tandem Mass Spectrometry Coupled With Network Pharmacology-Based Investigation. Phytomed Int J phytother phytopharmacol (2020) 67:153156. doi: 10.1016/j.phymed.2019.153156 [DOI] [PubMed] [Google Scholar]
- 22. Li W, Mao X, Wang X, Liu Y, Wang K, Li C, et al. Disease-Modifying Anti-Rheumatic Drug Prescription Baihu-Guizhi Decoction Attenuates Rheumatoid Arthritis Via Suppressing Toll-Like Receptor 4-Mediated Nlrp3 Inflammasome Activation. Front Pharmacol (2021) 12:743086. doi: 10.3389/fphar.2021.743086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mao X. The Key Material Basis Identification and Pharmacological Mechanism Investigation of Chinese Herbal Formula Wutou Decoction Against Rheumatoid Arthritis With Cold Syndrome. Chin Acad Chin Med Sci (2020). doi: 10.27658/d.cnki.gzzyy.2020.000124 [DOI] [Google Scholar]
- 24. Mao X, Li W, Chen W, Li Y, Wang Q, Wang X, et al. Exploring and Characterizing a Novel Combination of Paeoniflorin and Talatizidine for the Treatment of Rheumatoid Arthritis. Pharmacol Res (2020) 153:104658. doi: 10.1016/j.phrs.2020.104658 [DOI] [PubMed] [Google Scholar]
- 25. Guo Q, Mao X, Zhang Y, Meng S, Xi Y, Ding Y, et al. Guizhi-Shaoyao-Zhimu Decoction Attenuates Rheumatoid Arthritis Partially by Reversing Inflammation-Immune System Imbalance. J Trans Med (2016) 14(1):165. doi: 10.1186/s12967-016-0921-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu HY, Zhang YQ, Liu ZM, Chen T, Lv CY, Tang SH, et al. Etcm: An Encyclopaedia of Traditional Chinese Medicine. Nucleic Acids Res (2019) 47(D1):D976–D82. doi: 10.1093/nar/gky987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. String V10: Protein-Protein Interaction Networks, Integrated Over the Tree of Life. Nucleic Acids Res (2015) 43(Database issue):D447–52. doi: 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res (2003) 13(11):2498–504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y, Mao X, Guo Q, Bai M, Zhang B, Liu C, et al. Pathway of Ppar-Gamma Coactivators in Thermogenesis: A Pivotal Traditional Chinese Medicine-Associated Target for Individualized Treatment of Rheumatoid Arthritis. Oncotarget (2016) 7(13):15885–900. doi: 10.18632/oncotarget.7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. He Y, Su W, Chen T, Zeng X, Yan Z, Guo J, et al. Identification of Prototype Compounds and Derived Metabolites of Naoxintong Capsule in Beagle Dog Urine and Feces by Uflc-Q-Tof-Ms/Ms. J Pharm Biomed Anal (2019) 176:112806. doi: 10.1016/j.jpba.2019.112806 [DOI] [PubMed] [Google Scholar]
- 31. Sterling T, Irwin JJ. Zinc 15-Ligand Discovery for Everyone. J Chem Inf Model (2015) 55(11):2324–37. doi: 10.1021/acs.jcim.5b00559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pandey MK, Sung B, Ahn KS, Kunnumakkara AB, Chaturvedi MM, Aggarwal BB. Gambogic Acid, a Novel Ligand for Transferrin Receptor, Potentiates Tnf-Induced Apoptosis Through Modulation of the Nuclear Factor-Kappab Signaling Pathway. Blood (2007) 110(10):3517–25. doi: 10.1182/blood-2013-03-494385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y, Cui Y, Cao F, Qin Y, Li W, Zhang J. Ganglioside Gd1a Suppresses Lps-Induced Pro-Inflammatory Cytokines in Raw264.7 Macrophages by Reducing Mapks and Nf-Kappab Signaling Pathways Through Tlr4. Int Immunopharmacol (2015) 28(1):136–45. doi: 10.1016/j.intimp.2015.05.044 [DOI] [PubMed] [Google Scholar]
- 34. Villegas SN, Gombos R, Garcia-Lopez L, Gutierrez-Perez I, Garcia-Castillo J, Vallejo DM, et al. Pi3k/Akt Cooperates With Oncogenic Notch by Inducing Nitric Oxide-Dependent Inflammation. Cell Rep (2018) 22(10):2541–9. doi: 10.1016/j.celrep.2018.02.049 [DOI] [PubMed] [Google Scholar]
- 35. Khan H, Sureda A, Belwal T, Cetinkaya S, Suntar I, Tejada S, et al. Polyphenols in the Treatment of Autoimmune Diseases. Autoimmun Rev (2019) 18(7):647–57. doi: 10.1016/j.autrev.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ma Z, Yu R, Zhu Q, Sun L, Jian L, Wang X, et al. Cxcl16/Cxcr6 Axis Promotes Bleomycin-Induced Fibrotic Process in Mrc-5 Cells Via the Pi3k/Akt/Foxo3a Pathway. Int Immunopharmacol (2020) 81:106035. doi: 10.1016/j.intimp.2019.106035 [DOI] [PubMed] [Google Scholar]
- 37. Spel L, Martinon F. Inflammasomes Contributing to Inflammation in Arthritis. Immunol Rev (2020) 294(1):48–62. doi: 10.1111/imr.12839 [DOI] [PubMed] [Google Scholar]
- 38. Yang J, Zhao Y, Zhang P, Li Y, Yang Y, Yang Y, et al. Hemorrhagic Shock Primes for Lung Vascular Endothelial Cell Pyroptosis: Role in Pulmonary Inflammation Following Lps. Cell Death Dis (2016) 7(9):e2363. doi: 10.1038/cddis.2016.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, et al. Cleavage of Gsdmd by Inflammatory Caspases Determines Pyroptotic Cell Death. Nature (2015) 526(7575):660–5. doi: 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- 40. Micaroni M, Stanley AC, Khromykh T, Venturato J, Wong CX, Lim JP, et al. Rab6a/a' Are Important Golgi Regulators of Pro-Inflammatory Tnf Secretion in Macrophages. PloS One (2013) 8(2):e57034. doi: 10.1371/journal.pone.0057034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kayagaki N, Stowe IB, Lee BL, O'Rourke K, Anderson K, Warming S, et al. Caspase-11 Cleaves Gasdermin D for Non-Canonical Inflammasome Signalling. Nature (2015) 526(7575):666–71. doi: 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- 42. Evavold CL, Ruan J, Tan Y, Xia S, Wu H, Kagan JC. The Pore-Forming Protein Gasdermin D Regulates Interleukin-1 Secretion From Living Macrophages. Immunity (2018) 48(1):35–44.e6. doi: 10.1016/j.immuni.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. DiPeso L, Ji DX, Vance RE, Price JV. Cell Death and Cell Lysis Are Separable Events During Pyroptosis. Cell Death Discovery (2017) 3:17070. doi: 10.1038/cddiscovery.2017.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yap HY, Tee SZ, Wong MM, Chow SK, Peh SC, Teow SY. Pathogenic Role of Immune Cells in Rheumatoid Arthritis: Implications in Clinical Treatment and Biomarker Development. Cells (2018) 7(10):161. doi: 10.3390/cells7100161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alenzi FQ. The Significance and Occurrence of Tnf Receptor Polymorphisms in the Saudi Population. Saudi J Biol Sci (2016) 23(6):767–72. doi: 10.1016/j.sjbs.2016.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang X, Morris-Natschke SL, Lee KH. New Developments in the Chemistry and Biology of the Bioactive Constituents of Tanshen. Med Res Rev (2007) 27(1):133–48. doi: 10.1002/med.20077 [DOI] [PubMed] [Google Scholar]
- 47. Jiang M, Lu C, Chen G, Xiao C, Zha Q, Niu X, et al. Understanding the Molecular Mechanism of Interventions in Treating Rheumatoid Arthritis Patients With Corresponding Traditional Chinese Medicine Patterns Based on Bioinformatics Approach. Evid Based Complement Alternat Med (2012) 2012:129452. doi: 10.1155/2012/129452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fan W, Fan L, Peng C, Zhang Q, Wang L, Li L, et al. Traditional Uses, Botany, Phytochemistry, Pharmacology, Pharmacokinetics and Toxicology of Xanthium Strumarium L.: A Review. Molecules (2019) 24(2):359. doi: 10.3390/molecules24020359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hennessy EJ, Parker AE, O'Neill LA. Targeting Toll-Like Receptors: Emerging Therapeutics? Nat Rev Drug Discovery (2010) 9(4):293–307. doi: 10.1038/nrd3203 [DOI] [PubMed] [Google Scholar]
- 50. Feng FB, Qiu HY. Effects of Artesunate on Chondrocyte Proliferation, Apoptosis and Autophagy Through the Pi3k/Akt/Mtor Signaling Pathway in Rat Models With Rheumatoid Arthritis. Biomed pharmacother = Biomed pharmacotherapie (2018) 102:1209–20. doi: 10.1016/j.biopha.2018.03.142 [DOI] [PubMed] [Google Scholar]
- 51. Antica M, Kusic B, Hranilovic D, Dietz AB, Vuk-Pavlovic S. Cloning the Cdna for Murine U2 Snrnp-A' Gene and Its Differential Expression in Lymphocyte Development. Immunol Lett (2002) 82(3):217–23. doi: 10.1016/s0165-2478(02)00064-0 [DOI] [PubMed] [Google Scholar]
- 52. Behl T, Chadha S, Sachdeva M, Kumar A, Hafeez A, Mehta V, et al. Ubiquitination in Rheumatoid Arthritis. Life Sci (2020) 261:118459. doi: 10.1016/j.lfs.2020.118459 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number(s) can be found below: NCBI Gene Expression Omnibus; GSE190523, GSE189942.