Abstract
Escherichia coli NZN111 is blocked in the ability to grow fermentatively on glucose but gave rise spontaneously to a mutant that had this ability. The mutant carries out a balanced fermentation of glucose to give approximately 1 mol of succinate, 0.5 mol of acetate, and 0.5 mol of ethanol per mol of glucose. The causative mutation was mapped to the ptsG gene, which encodes the membrane-bound, glucose-specific permease of the phosphotransferase system, protein EIICBglc. Replacement of the chromosomal ptsG gene with an insertionally inactivated form also restored growth on glucose and resulted in the same distribution of fermentation products. The physiological characteristics of the spontaneous and null mutants were consistent with loss of function of the ptsG gene product; the mutants possessed greatly reduced glucose phosphotransferase activity and lacked normal glucose repression. Introduction of the null mutant into strains not blocked in the ability to ferment glucose also increased succinate production in those strains. This phenomenon was widespread, occurring in different lineages of E. coli, including E. coli B.
Under anaerobic conditions and in the absence of exogenous electron acceptors, Escherichia coli ferments glucose to a mixture of products consisting primarily of acetate, formate, and ethanol, as well as smaller amounts of lactate and succinate (2). In this process, NADH generated during glycolysis is reoxidized through the reduction of organic intermediates derived from glucose. The relative proportions of the end products vary with the strain and growth conditions, but in all cases the distribution of products is balanced so that the reducing equivalents generated are fully consumed (7). Normally, no more than 0.2 mol of succinate is formed per mol of glucose consumed by E. coli (2, 3, 7).
Succinate is of commercial interest as a potential precursor of industrial chemicals. Purified, biologically produced succinic acid can be converted chemically to numerous commercial chemicals ranging from low-volume, high-value products such as malic acid to commodity chemicals such as 1,4-butanediol (18, 34). Two bacteria that naturally produce succinate as their major fermentation product, Anaerobiospirillum succiniciproducens and Actinobacillus succinogenes, generate up to 1 mol of succinate per mol of glucose and have been developed for commercial production of succinic acid (11, 14). The possibility of increasing succinate production in E. coli through genetic engineering has been investigated recently. Expression of plasmid-encoded phosphoenolpyruvate (PEP) carboxylase (12, 20) or pyruvate carboxylase (12) resulted in increased succinate formation, but the highest yield of succinate was less than 0.5 mol per mol of glucose (20). Expression of the malic enzyme in a nonfermenting mutant of E. coli, NZN111, resulted in very slow formation of succinate and a higher yield (29).
Recently, we described a mutant strain of E. coli, designated AFP111, that ferments glucose to an unusual mixture of products consisting of succinate, acetate, and ethanol (9). AFP111 arose by spontaneous chromosomal mutation in strain NZN111, which is unable to ferment glucose due to inactivation of the genes encoding pyruvate:formate lyase and the fermentative lactate dehydrogenase (5). In this study we identified the mutation that restored the ability of NZN111 to grow fermentatively on glucose as a lesion in the ptsG gene. When a null mutation of the ptsG gene was introduced into various strains of E. coli not blocked in the ability to ferment glucose, the resulting strains also produced more succinate.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All E. coli strains used in this study (Table 1) were routinely cultured in Luria-Bertani (LB) medium (27) at 37°C. Antibiotics were included as necessary at the following concentrations: carbenicillin, 100 μg per ml; kanamycin, 30 μg per ml; tetracycline, 10 μg per ml; and chloramphenicol, 30 μg per ml. Rich broth contained (per liter) 10 g of tryptone, 5 g of NaCl, and 1 g of yeast extract. Solid media for plates contained 1.5% (wt/vol) Difco Bacto Agar. Minimal medium E was prepared as described by Vogel and Bonner (31).
TABLE 1.
E. coli strains used in this study
| Strain(s) | Genotype | Source and/or reference(s) |
|---|---|---|
| W1485 | F+ λ−rpoS396(Am) rph-1 | CGSC 5024 |
| FMJ123 | ΔpflB::Cam of W1485 | 5 |
| NZN111 | ldhA::Kan of FMJ123 | 5 |
| AFP111 | Spontaneous ptsG mutant of NZN111 | 9; this study |
| SE1752 | ldhA::Tn10 | L. O. Ingram |
| DC1327 | ldhA::Tn10 of FMJ123 | This study |
| BW6156 | λ−relA1 spoT1 metB1 zje-2005::Tn10 | CGSC 6754 |
| BW7623 | purK79::Tn10 λ−relA spoT1 | CGSC 6815 |
| CAG12078 | λ−zce-726::Tn10 rph-1 | CGSC 7361 |
| CAG18463 | λ−zcf-117::Tn10 rph-1 | CGSC 7363 |
| CAG18468 | λ−nupC510::Tn10 rph-1 | CGSC 7410 |
| CAG18470 | λ−purC80::Tn10 rph-1 | CGSC 7413 |
| LA-12G | λ−ptsG21 relA1 spoT1 thi-1 | CGSC 5085 |
| AFP301–AFP306 | zce-726::Tn10 ptsG+ of AFP111 | P1(CAG12078) × AFP111; this study |
| AFP308 | zce-726::Tn10 (ptsG21) of LA-12G | P1(CAG12078) × LA-12G; this study |
| AFP310–AFP313 | ptsG21 zce-726::Tn10 of NZN111 | P1(AFP308) × NZN111; this study |
| AFP400 | ptsG::Kan of DC1327 | This study |
| AFP414 | ptsG::Kan of W1485 | This study |
| AFP402 | ptsG::Kan of FMJ123 | This study |
| LCB320 | F−thr-1 leu-6 thi-1 lacY tonA22 strA | M. Pascal |
| AFP412 | ptsG::Kan of LCB320 | This study |
| E. coli B | Wild type | ATCC 11303 |
| AFP411 | ptsG::Kan of E. coli B | This study |
Fermentations were carried out in sealed serum tubes containing 10 ml of LB medium supplemented with 0.5 g of MgCO3 (added in order to maintain the pH of the medium during fermentation), the appropriate antibiotic(s), and approximately 10 g of glucose per liter. Other sugars were tested at concentrations of 5 to 6 g/liter. The headspace in the sealed tubes was CO2, established by means of a gassing manifold (1). Inocula for the anaerobic liquid cultures were prepared by growing the strains aerobically overnight in LB medium supplemented with the appropriate antibiotics. A sample of each overnight culture was diluted 100-fold in fresh medium and allowed to grow aerobically to an A600 of 1, and an anaerobic serum tube was inoculated with 1 ml of this culture. Samples for analysis were removed at intervals anoxically with a syringe. For anaerobic growth studies, cultures were grown in serum tubes lacking MgCO3 in a 1:1 mixture of LB medium and M9 medium (to provide buffering) supplemented with glucose. For anaerobic growth on solid media, agar plates were incubated at 37°C in an anaerobic jar under an H2-CO2 atmosphere generated by use of a Gas-Pak (Becton Dickinson).
A modification of the plate assay for β-galactosidase activity (27) was employed as a test for the presence of normal catabolite repression in strains. LB agar or medium E agar was supplemented with 4 g of glucose per liter, 4 g of lactose per liter, 30 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) per liter, and the appropriate antibiotic(s). The resulting plates are referred to below as glucose/lactose/X-Gal plates. Formation of blue colonies indicated that expression of β-galactosidase occurred in the presence of glucose due to the absence of catabolite repression, and formation of white colonies indicated that normal catabolite repression occurred.
Genetic methods.
Hfr conjugations for mapping of the mutation were performed by using the set of Hfr donor strains constructed in the laboratory of B. Wanner (32) with modifications of methods described previously (21, 33). AFP111 was used as the recipient, and 15- to 20-min mating periods were typically allowed. Exconjugants were selected for resistance to tetracycline (to select for transfer of Tn10 into the recipient) and kanamycin (to select against the donor) and were screened for formation of white colonies on LB medium–glucose/lactose/X-Gal plates, which indicated that transfer of a locus that restored normal catabolite repression had occurred. Exconjugant colonies were purified by restreaking on the same selective medium; well-isolated white colonies were analyzed for fermentation of glucose.
For more detailed mapping of the mutation, P1 phage transductions were carried out by using the method of Miller (21). Transductants were selected by the procedure described above for exconjugants, except that medium E agar plates were used in place of LB agar plates to prevent further lysis of the host by residual P1 phage. Transductants were purified prior to analysis by fermentation as described above for the exconjugants.
To create a strain with Tn10 located near a defective ptsG gene, zce::Tn10 was transduced from CAG12078 into LA-12G (ptsG21) (Table 1). LA-12G formed blue colonies on glucose/lactose/X-Gal plates. Transductants were screened for resistance to tetracycline and the formation of blue colonies (indicating transfer of Tn10 into LA-12G without replacement of the ptsG21 allele). One such transductant, designated AFP308, was subsequently used to transduce ptsG21 into various strains. Transductants were screened for resistance to tetracycline and for the formation of blue colonies on glucose/lactose/X-Gal agar plates and purified as described above.
A wild-type allele of ptsG was introduced in trans by transforming AFP111 with plasmid pCB10 (4, 27). Plasmid pCB10 contains a 3.3-kb segment of E. coli genomic DNA that includes the ptsG gene and approximately 1 kb of flanking DNA on either side of the gene. The plasmid was maintained by repeated additions of carbenicillin to replenish that lost by hydrolysis over the course of fermentation studies.
Construction and introduction of an insertionally inactivated ptsG gene.
The native ptsG gene of E. coli was cloned by PCR from genomic DNA prepared from W1485 by using primers targeting the N and C termini of the protein, and no additional genomic sequences were amplified. The gene was cloned into the vector pFJ118EH (10) to give pJFptsG. The gene was disrupted by insertion of the kanamycin resistance cassette of pUC-4K (Pharmacia), excised with EcoRI, into the MfeI site of the ptsG gene in pJFptsG to give plasmid pPTSGK. Because NZN111 already included a kanamycin resistance marker, an equivalent strain was constructed by transducing a Tn10-inactivated ldhA gene from strain SE1752 (Table 1) into FMJ123. The physiology of the resulting strain, DC1327, was indistinguishable from that of NZN111. The disrupted ptsG gene was transferred into DC1327 by transforming the cells with pPTSGK, growing the cells for approximately 30 generations in the presence of kanamycin and absence of ampicillin, and then plating the culture on LB agar plates containing glucose and incubating the plates anaerobically. Colonies that were able to grow fermentatively were purified and screened for sensitivity to the two antibiotics. Strain AFP400 was isolated as a stable kanamycin-resistant, ampicillin-sensitive strain that fermented glucose to succinate, acetate, and ethanol. Proper integration of the disrupted ptsG gene was confirmed by PCR. The disrupted gene was amplified from AFP400 DNA by using primers that matched flanking sequences approximately 110 bp outside the coding region of the gene. These sequences were not present in the integration vector. The resulting product was 3.0 kb long, as predicted from the known sequences of ptsG, its flanking regions, and the kanamycin insert. The product was digested with ClaI (site in the kanamycin cassette) and AgeI (site in ptsG), and this generated the fragments expected for insertion of the cassette into the MfeI site of ptsG (1.95 and 1.05 kb for ClaI and 2.3 and 0.7 kb for AgeI).
Analytical methods and enzyme assays.
Substrate consumption and product formation during fermentation were quantified by high-performance liquid chromatography using a Bio-Rad Aminex HPX-87H ion-exchange column (7.8 by 300 mm) and a Shimadzu LC-10A chromatography system equipped with UV absorbance and refractive index detectors. Samples of the anaerobic culture were removed anoxically and centrifuged at 7,000 × g for 1 min. Each supernatant was diluted with 2 volumes of 5 mM H2SO4, and 200 μl of the diluted sample was injected. The column was eluted isocratically at a rate of 0.5 ml/min with 5 mM H2SO4, and data were analyzed with an EZChrom data system (Scientific Software, Inc.). The approximate retention times were as follows: glucose, 11.8 min; mannose, 12.6 min; succinic acid, 15.0 min; lactic acid, 16.5 min; formic acid, 17.9 min; acetic acid, 19.3 min; and ethanol, 27 min. Glucose levels were also monitored enzymatically by using a commercial kit obtained from Stanbio, Inc. Pyruvate levels were determined enzymatically by the lactate dehydrogenase reaction (17).
Phosphotransferase system (PTS) activities were measured by a modification of the assay of Kornberg and Reeves (19). This assay monitors the consumption of NADH by lactate dehydrogenase acting on pyruvate formed from PEP, the metabolite that donates the phosphate group to the phosphoprotein cascade of the PTS. Cells were grown in 20 ml of LB medium containing 20 mM glucose or 20 mM fructose in 125-ml notched flasks shaken at 250 rpm. Exponentially growing cultures were harvested by centrifugation, washed with cold 0.1 M phosphate–MgCl2 buffer (pH 7.5) (28 ml of 1 M NaH2PO4 per liter, 72 ml of 1 M K2HPO4 per liter, 20 ml of 1 M MgCl2 per liter), and then resuspended in 2 ml of the same buffer. The suspension was then diluted with buffer to give an optical density at 600 nm (OD600) of 10. The cells were permeabilized by adding 10 μl of a toluene-acetone (1:9) mixture per ml of suspension while the suspension was vortexed vigorously. This concentration was previously determined empirically to result in permeabilization of the cells without disruption. The cells were maintained on ice. The rate of glucose-dependent formation of pyruvate by partially permeabilized cells reflects the activity of the PTS. Control assay mixtures lacking glucose, PEP, or lactate dehydrogenase were included to validate the protocol and account for background NADH-oxidizing activities. Control cultures grown on fructose were also assayed for both fructose- and glucose-dependent PTS activities. The values reported below are the sugar-dependent rates of NADH oxidation minus the background rate observed in the absence of sugar (typically about 1 nmol/min/OD600 unit of cells).
RESULTS
Physiological characteristics of AFP111.
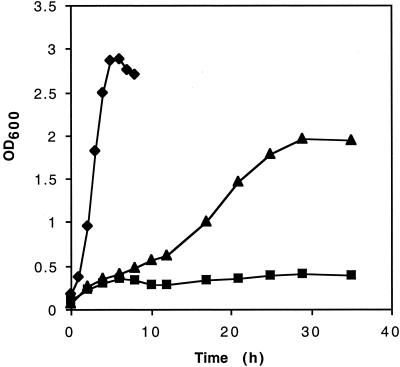
Strain AFP111 arose by spontaneous chromosomal mutation of strain NZN111 that restored the ability to grow fermentatively on glucose (9). The fermentation carried out by AFP111 was well balanced in terms of accounting for both carbon atoms and electrons and generated an unusual mixture of products consisting of approximately 1 mol of succinate, 0.5 mol of acetate, and 0.5 mol of ethanol per mol of glucose consumed (Table 2). AFP111, however, did not grow as well on glucose as its ancestral strain, W1485, did (Fig. 1). Whereas W1485 grew with a generation time of less than 1 h, AFP111 grew with a generation time of approximately 7 h and reached a lower final cell density. After a small amount of growth after inoculation, NZN111 grew with a generation time of more than 25 h (in the interval between 12 and 25 h) (Fig. 1).
TABLE 2.
Fermentation balance in the fermentation of glucose by AFP111
| Producta | Amt
|
Redox stateb | |
|---|---|---|---|
| mol/mol of glucose | mol of C atoms | ||
| Succinate | 0.92 | 3.66 | 0.92 |
| Lactate | 0.02 | 0.05 | 0 |
| Formate | 0.02 | 0.02 | 0.02 |
| Acetate | 0.50 | 1.00 | 0 |
| Ethanol | 0.45 | 0.91 | −0.91 |
| Total | 1.91 | 5.64 | 0.03 |
Fermentative growth of strains is described in Materials and Methods. The medium contained approximately 10 g of glucose per liter. The average values for four strains are shown. The standard deviations for succinate, acetate, and ethanol were 0.09, 0.08, and 0.08 mol/mol of glucose, respectively. The values were corrected for the small amounts of products formed in the absence of glucose.
The redox state was calculated by multiplying the amount of product (in moles formed per mole of glucose) by the oxidation state.
FIG. 1.
Anaerobic growth of strain AFP111 (▴), its immediate parent, NZN111 (■), and wild-type ancestor W1485 (⧫) on LB medium supplemented with 10 g of glucose per liter.
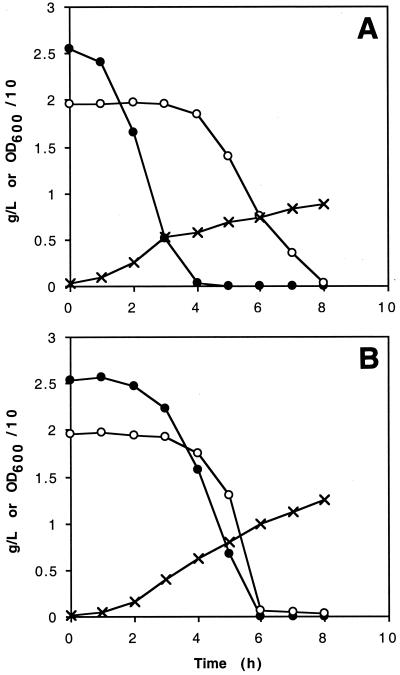
AFP111 was also altered in terms of aerobic metabolism of sugars. When grown on a mixture of glucose and mannose, AFP111 metabolized both sugars simultaneously, in contrast to NZN111, which exhibited the expected sequential consumption of glucose and then mannose (Fig. 2). This difference was corroborated by plate tests designed to detect expression of chromosomal β-galactosidase. When grown on glucose/lactose/X-Gal plates, AFP111 produced blue-green colonies due to expression of β-galactosidase, whereas NZN111 and other precursor strains produced pale yellow colonies, as expected due to the normal repression of the lac operon in the presence of glucose.
FIG. 2.
Aerobic growth and consumption of a mixture of glucose and mannose by NZN111 (A) and mutant AFP111 (B). LB medium was supplemented with approximately 2 g of glucose per liter (●) or 2 g of mannose per liter (○). The cell density values (×) are culture OD600 values divided by 10.
Mapping the causative mutation.
The causative mutation was mapped by first screening for restoration of normal glucose repression of β-galactosidase (see above) and then screening for fermentation of glucose. Exconjugants and transductants were evaluated based on the color of colonies produced on aerobic LB medium–glucose/lactose/X-Gal plates and subsequently were evaluated based on growth on glucose under anaerobic conditions. In every case throughout the mapping experiments, these two traits were found to be linked; blue-green colonies were able to grow fermentatively on glucose, but yellow colonies were not.
Conjugation of AFP111 with various Hfr strains localized the mutation to the region between 7 and 32 min on the chromosome. Conjugation with BW7623 (origin of transfer, 32 min; direction of transfer, counterclockwise) resulted in restoration of repression of β-galactosidase in 14% of the exconjugants, whereas conjugation with BW6156 (origin of transfer, 7 min; direction of transfer, counterclockwise) gave none. Transduction of AFP111 with selection for various Tn10 markers located in that region resulted in restoration of repression in two cases. Phage prepared from CA12078 (Tn10 at 24.6 min) restored repression in 86% of the transductants, and phage prepared from CAG18468 (Tn10 at 25.5 min) restored repression in 13% of the transductants. All of the white colonies initially detected (white colonies with normal repression of β-galactosidase as determined by this criterion) failed to grow fermentatively on glucose. Several representative white transductants, AFP301 through AFP306, were analyzed by high-performance liquid chromatography to determine whether they fermented glucose in liquid medium; like parental strain NZN111, all failed to convert significant amounts of glucose in 20 h (Table 3) and all excreted pyruvate into the medium at a concentration of approximately 1 mM.
TABLE 3.
Effects of ptsG gene alleles on strains lacking functional pflB and ldhA genes
| Strain | ptsG allele | Amt of glucose remaining and amt of products formed (mol/mol of glucose)a
|
|||||
|---|---|---|---|---|---|---|---|
| Glucose | Succinate | Lactate | Formate | Acetate | Ethanol | ||
| AFP111 | Mutant of NZN111 | 0 | 0.88 | 0.01 | 0.07 | 0.53 | 0.38 |
| AFP301–AFP306b | ptsG+ of CAG12078 | 0.94 | 0.03 | 0.02 | 0 | 0.01 | 0.01 |
| NZN111 | ptsG+ of W1485 | 0.87 | 0.04 | 0 | 0 | 0 | 0 |
| AFP310–AFP313c | ptsG21 of LA-12A | 0 | 0.91 | 0.01 | 0.11 | 0.61 | 0.39 |
| AFP111(pCB10)d | Plasmid-borne ptsG+ | 0.89 | 0.11 | 0.02 | 0.02 | 0.10 | 0.05 |
| DC1327 | ptsG+ of W1485 | 0.84 | 0.10 | 0.03 | 0.03 | 0.11 | 0.05 |
| AFP400e | ptsG::Kan | 0 | 0.93 | 0.01 | 0 | 0.51 | 0.41 |
Fermentation products were formed in 20 h from approximately 9 g of glucose per liter.
Functional ptsG of CAG12078 was introduced by P1 transduction. Values are averages for all six strains.
Mutant ptsG21 of LA-12A was introduced by P1 transduction. Values are averages for all four strains.
Carbenicillin was included in the medium to maintain the plasmid. Values are averages of the results for two transformants.
Insertionally inactivated ptsG was introduced into DC1237 as described in Materials and Methods.
Effects of various alleles of ptsG.
The mapping results indicated that the causative gene very likely lay between 24.6 and 25.5 min on the chromosome. The ptsG gene lies squarely in this region at 25.0 min and could contribute to the observed phenotypic changes. Its product, protein EIICBglc, is the glucose-specific permease of the PTS. Mutation of ptsG could affect both the growth rate and the repression of other operons in the presence of glucose (25). To evaluate this possibility, we transferred various mutant and functional forms of ptsG into appropriate strains and observed the physiological effects of their presence (Table 3). Introduction of a mutant allele, ptsG21, into NZN111 eliminated repression of β-galactosidase and restored the ability to grow fermentatively on glucose. Analysis of the fermentation products of representative transductants showed that all of them converted glucose to the succinate-acetate-ethanol mixture observed in AFP111. Introduction of a wild-type, plasmid-borne ptsG gene under control of its own promoter (4) into AFP111 eliminated the ability of the organism to ferment glucose (Table 3). These results strongly suggest that mutation of ptsG was responsible for phenotypic changes observed in AFP111.
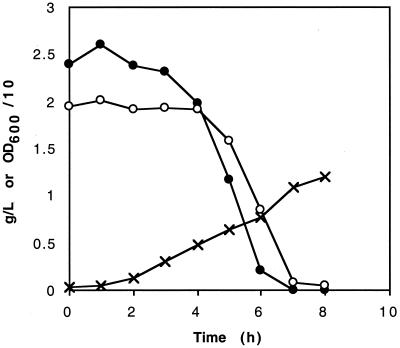
Because EIICBglc is a complex protein with multiple functions, it was essential to determine the effect of a null mutation. We constructed an insertionally disrupted form of the gene by using the kanamycin resistance cassette from plasmid pUC4K and introduced it into strain DC1327 (Table 1). DC1327 is identical to NZN111 except that its ldhA gene is inactivated by a Tn10 transposon rather than by a kanamycin cassette. Its physiological characteristics were indistinguishable from those of NZN111. Introduction of the disrupted ptsG gene into DC1327 restored the ability to ferment glucose, and the resulting strain, AFP400, produced succinate, acetate, and ethanol in molar yields equivalent to those observed for AFP111 (Table 3). The ptsG null mutant showed the same lack of glucose repression of the use of other sugars that AFP111 showed. When grown aerobically on a mixture of glucose and mannose, AFP400 also consumed both sugars simultaneously (Fig. 3). Proper integration of the disrupted ptsG gene was confirmed by PCR amplification of the integrated gene from AFP400 (see Materials and Methods).
FIG. 3.
Aerobic growth and consumption of a mixture of glucose and mannose by AFP400. LB medium was supplemented with approximately 2 g of glucose per liter (●) or 2 g of mannose per liter (○). The cell density values (×) are culture OD600 values divided by 10.
Enzymatic analysis of the EIICBglc permease activities of strains.
The glucose-specific phosphotransferase activities of various strains were assayed by using permeabilized cells grown in the presence of glucose (see Materials and Methods). In this assay, cells are mildly permeabilized to allow exchange of small molecules while retaining intracellular enzymes and are washed to remove endogenous small molecules. In these cells, glucose-dependent conversion of PEP to pyruvate, adjusted for appropriate controls, measures the glucose-dependent phosphotransferase activity and depends on the activity of the EIICBglc permease, the product of the ptsG gene. Controls lacking PEP or glucose or controls in which cells grown on fructose were used were included to ensure that the activity reported reflected EIICBglc activity. A comparison of exponentially growing cells of representative strains showed that all of the succinate-producing mutants had approximately 10-fold less EIICBglc activity than their parental strains (Table 4). The reductions in EIICBglc activity were consistently associated with the lack of repression of the β-galactosidase by glucose. The fructose-dependent phosphotransferase activities of fructose-grown NZN111 and AFP111 were 5.4 and 5.9 nmol/min/OD600 unit, respectively, indicating that the mutation in AFP111 did not affect the PTS in general but was specific for the glucose PTS.
TABLE 4.
Glucose phosphotransferase activities of strainsa
| Strain | Relevant genotype | Repression of β-galactosidaseb | PTS activity (nmol/min/OD600 unit) |
|---|---|---|---|
| W1485 | Wild type | + | 23.8 |
| NZN111 | W1485 ΔpflB::Cam ldhA::Kan | + | 18.3 |
| AFP111 | NZN111 ptsG | − | 1.7 |
| AFP301 | AFP111 ptsG+ | + | 24.3 |
| AFP310 | NZN111 ptsG21 | − | 2.8 |
| AFP111(pCB10) | Plasmid-borne ptsG+ | + | 19.5 |
| DC1327 | W1485 ΔpflB::Cam ldhA::Tet | + | 23.6 |
| AFP400 | DC1327 ptsG::Kan | − | 1.6 |
Phosphotransferase activity was measured in permeabilized cells as described in Materials and Methods.
The presence (+) or absence (−) of catabolite repression was determined by the color of individual colonies on X-Gal-containing agar as described in Materials and Methods.
Effect of the null ptsG allele in other strains.
Strains NZN111 and DC1327 are blocked in normal fermentation of glucose. To evaluate possible effects of ptsG in nonblocked strains, we transduced insertionally inactivated ptsG into ancestral strains of NZN111 and into unrelated E. coli lineages (Table 5). In all cases, inactivation of ptsG shifted the fermentative metabolism strongly toward succinate. Even in the presence of a functional pyruvate:formate lyase, succinate production increased from about 0.2 to about 0.6 to 0.8 mol per mol of glucose in all strains tested. This result occurred not only in K-12 lineages but also in E. coli B. In the absence of a functional pyruvate:formate lyase, an even larger shift occurred. The immediate precursor of NZN111, FMJ123, produces lactic acid almost exclusively. Upon introduction of the ptsG null mutation, the resulting strain, AFP402, produced a mixture of products very similar to that produced by AFP111 but containing more lactate and less ethanol (Tables 2 and 5).
TABLE 5.
Effect of the ptsG::Kan insertion in E. coli strains
| Strain | Genotype | Amt of products formed (mol/mol of glucose)a
|
||||
|---|---|---|---|---|---|---|
| Succinate | Lactate | Formate | Acetate | Ethanol | ||
| W1485 | Wild type | 0.20 | 0.06 | 0.48 | 0.74 | 0.93 |
| AFP414 | W1485 ptsG::Kan | 0.74 | 0.03 | 0.18 | 0.72 | 0.36 |
| FMJ123 | W1485 pflB::Cam | 0.11 | 1.79 | 0.02 | 0.09 | 0.06 |
| AFP402 | FMJ123 ptsG::Kan | 0.94 | 0.14 | 0.01 | 0.52 | 0.31 |
| LCB320 | —b | 0.29 | 0.01 | 0.40 | 0.93 | 0.88 |
| AFP412 | LCB320 ptsG::Kan | 0.53 | 0.00 | 0.23 | 0.67 | 0.76 |
| E. coli B | Wild type | 0.18 | 0.00 | 0.77 | 0.74 | 0.85 |
| AFP411 | E. coli B ptsG::Kan | 0.73 | 0.01 | 0.11 | 0.58 | 0.64 |
Fermentative growth of strains is described in Materials and Methods. Approximately 12 g of glucose per liter was included in the media; the amounts of the products formed were measured after all of the glucose was consumed.
See Table 1.
Metabolism of other sugars by NZN111.
Ancestral strain W1485 can ferment a number of sugars and sugar derivatives. Anaerobic plate tests revealed that NZN111 was also able to grow fermentatively on sugars other than glucose. The only sugar that failed to support comparable growth of both AFP111 and NZN111 was glucose. Sugars that supported growth of both strains were mannose, lactose, fructose, and trehalose. When tested in liquid medium, NZN111 converted these sugars primarily to succinate, acetate, and ethanol with approximately the same yields as those obtained with glucose (data not shown). The sugar derivatives glucuronate and sorbitol also supported growth of NZN111, giving alternative distributions of products that were consistent with the redox balance required because of the different oxidation states of the substrates. Glucuronate was converted primarily to the more oxidized product acetate (and presumably carbon dioxide) and to smaller amounts of succinate and ethanol, whereas sorbitol generated higher yields of the reduced products succinate and ethanol. Substrates at the same oxidation state as glucose (mannose, lactose, fructose, and trehalose) yielded approximately 1 mol of succinate per mol of glucose.
DISCUSSION
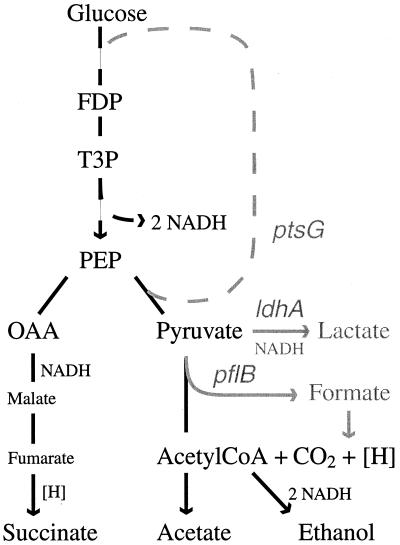
E. coli normally carries out mixed-acid fermentation of glucose to give primarily acetate, formate, and ethanol, all of which are derived from pyruvate in a series of reactions initiated by pyruvate:formate lyase (Fig. 4). Succinate, derived via carboxylation and subsequent reduction of PEP, is a minor product. The ancestral precursor of the mutants investigated here produces approximately 0.2 mol of succinate per mol of glucose. The naturally occurring succinate-producing bacterium A. succiniciproducens produces only succinate and acetate from glucose and theoretically can generate 1.3 mol of succinate per mol of glucose (28). In this case, two-thirds of the PEP derived from glucose is converted via the reductive arm of the tricarboxylic acid cycle to succinate, and one-third is oxidized to acetate via pyruvate:formate lyase to establish the necessary redox balance. The results described here indicate that E. coli possesses a latent ability to carry out a similar fermentation when the genes encoding pyruvate:formate lyase (pflB), the fermentative lactate dehydrogenase (ldhA), and EIICBglc (ptsG) are inactivated. Under these conditions, 1 mol of glucose is converted to approximately 1 mol of succinate, 0.5 mol of acetate, and 0.5 mol of ethanol. The difference between these two fermentations may result from the established coupling of acetate formation and ethanol formation in E. coli (15); mutation of either pta or adhE, the two genes whose products partition acetyl coenzyme A to acetate and ethanol, eliminates the ability to grow fermentatively in wild-type strains. Mutation of adhE in AFP111 similarly eliminated the ability to ferment glucose (Champion, unpublished observation), indicating that the same restraint applies to the partitioning of acetyl coenzyme A in the new succinate fermentation pathway. If ethanol must be formed in equimolar yields with acetate, reducing equivalents that could be used in succinate production are consumed, lowering the theoretical maximum yield of succinate to 1 mol of succinate per mol of glucose.
FIG. 4.
Key intermediates, genes, and products relevant to the fermentation of glucose by AFP111. In parental strain NZN111, inactivation of pflB and ldhA eliminated the ability to grow fermentatively on glucose. Mutation of ptsG restored growth and resulted in the formation of succinate, acetate, and ethanol. See text for details. FDP, fructose diphosphate; T3P, triose phosphates; OAA, oxaloacetic acid; Acetyl CoA, acetyl coenzyme A.
The potential of E. coli to generate this alternative distribution of fermentation products is widespread, occurring in different K-12 lineages, in strains that are not blocked in the fermentation of glucose, and in E. coli B (Table 5). In all of these examples, inactivation of the ptsG gene shifts metabolism toward succinate production, resulting in approximately 0.5 to 1 mol of succinate formed per mol of glucose. The differences in succinate yield in part reflect the presence of competing pathways; strains containing pyruvate:formate lyase make less succinate because they continue to produce large amounts of acetate and ethanol, but strain-dependent variation in the relative amounts of the products was observed. The most dramatic shift occurs in strain FMJ123. This strain lacks a functional pflB gene and carries out nearly homolactic fermentation of glucose with a yield approaching the theoretical maximum of 2 mol of lactate per mol of glucose. Inactivation of ptsG almost completely replaces this fermentation with the succinate-acetate-ethanol fermentation first observed in AFP111. Only a small amount of lactate is still produced. In strains which still possess a functional pflB gene, disruption of ptsG results in superimposition of the original pathway (which generates primarily acetate, formate, and ethanol) and the succinate-acetate-ethanol pathway (Table 5).
The evidence presented here establishes that a null mutation of ptsG alters the fermentative metabolism of E. coli and results in increased succinate formation, but the mechanism of this effect is not obvious. The product of ptsG, the glucose-specific PTS permease EIICBglc, catalyzes efficient transport and phosphorylation of glucose using phosphate derived originally from PEP. Inactivation of EIICBglc in principle could cause an increase in the PEP pool, favoring formation of succinate (Fig. 4). However, under aerobic conditions, a null mutation of ptsG in a different strain of E. coli had exactly the opposite consequence; the sizes of the PEP pools decreased, and the amounts of the products derived from PEP decreased (6). Alternatively, rapid glucose uptake by a functional glucose PTS in the absence of efficient conversion to products could lead to detrimental accumulation of intermediates. Fructose diphosphate, PEP, and NADH have regulatory roles at both the enzyme level and the gene level (13, 26), and the triose phosphates can give rise to toxic methylglyoxal (23). Perhaps the slower metabolism of the ptsG mutants prevents such metabolic imbalances.
Inactivation of EIICBglc could also modify metabolism indirectly by influencing the phosphorylation state of the immediate phosphate donor in the PTS, EIIAglc (25, 26). During normal glucose uptake, EIIAglc is largely dephosphorylated and represses uptake of other sugars by a process known as inducer exclusion (16). In addition, under these conditions the phosphorylated form of EIIAglc is depleted, which interrupts a regulatory cascade that activates many operons (26). This loss of activation is known as catabolite repression. Both mechanisms act to reduce expression of the lac operon in the presence of glucose and contribute to the preferential use of glucose in mixtures of sugars. The expression of β-galactosidase in the presence of glucose by the ptsG mutants described here reflects the loss of these regulatory controls, as does the simultaneous consumption of glucose and mannose by AFP111 and AFP400 (Fig. 2 and 3). With respect to succinate production, loss of a functional EIICBglc might similarly allow expression of enzymes normally repressed in the presence of glucose that could be crucial to the fermentation. EIICBglc has recently been proposed to have regulatory functions itself as well, including a direct effect on the expression of several genes, including its own regulatory gene, and possibly a role as a global sensor of glucose (8, 24, 30).
Regardless of the mechanism of the shift in metabolism, the results reveal that many strains of E. coli have a latent ability to produce good yields of succinate, which provides a foundation for developing E. coli as an organism for producing succinic acid. AFP111 can reproducibly generate up to 50 g of succinate per liter at a scale of 500 liters by using inexpensive agricultural feedstocks (22). Given the widespread distribution of the capacity to make succinate and the numerous tools available for manipulating E. coli genetically, it should be possible to improve succinate production significantly. Identification of a mutation of ptsG as the cause that unmasks this fermentation constitutes the first step in understanding how the novel distribution of products is generated. The homolactic fermenting strain FMJ123 and its derivative with the ptsG null mutation, AFP402, both ferment glucose well, but these organisms make drastically different fermentation products. Thus, these strains provide an ideal isogenic pair for further study.
ACKNOWLEDGMENTS
This work was supported by the Alternative Feedstocks Program, Office of Industrial Technology, U.S. Department of Energy Assistant Secretary for Energy Efficiency and Renewable Energy, under contract W-31-109-Eng-38.
We thank Carolyn Bouma for providing plasmid pCB10, Hend Samaha for constructing plasmid pJFptsG, and Ed St.Martin for helpful discussions.
REFERENCES
- 1.Balch W, Wolfe R S. New approach to the cultivation of methanogenic bacteria. Appl Environ Microbiol. 1976;32:781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackwood A C, Neish A C, Ledingham G A. Dissimilation of glucose at controlled pH values by pigmented and non-pigmented strains of Escherichia coli. J Bacteriol. 1956;72:497–499. doi: 10.1128/jb.72.4.497-499.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bock A, Sawers G. Fermentation. In: Neidhardt F C, editor. Escherichia coli and Salmonella. Washington, D.C.: American Society for Microbiology; 1996. pp. 262–282. [Google Scholar]
- 4.Bouma C L, Meadow N D, Stover E N, Roseman S. II-Bglc, a glucose receptor of the bacterial phosphotransferase system: molecular cloning of ptsG and purification of the receptor from an overproducing strain of E. coli. Proc Natl Acad Sci USA. 1987;84:930–934. doi: 10.1073/pnas.84.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunch P K, Mat-Jan F, Lee N, Clark D P. The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology. 1997;143:187–195. doi: 10.1099/00221287-143-1-187. [DOI] [PubMed] [Google Scholar]
- 6.Chen R, Hatzimanikatis V, Yap W M, Postma P W, Bailey J E. Metabolic consequences of phosphotransferase (PTS) mutation in phenylalanine-producing recombinant Escherichia coli. Biotechnol Prog. 1997;13:768–775. doi: 10.1021/bp970060h. [DOI] [PubMed] [Google Scholar]
- 7.Clark D P. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev. 1989;63:223–234. doi: 10.1016/0168-6445(89)90033-8. [DOI] [PubMed] [Google Scholar]
- 8.DeReuse H, Danchin A. Positive regulation of the pts operon of Escherichia coli: genetic evidence of a signal transduction mechanism. J Bacteriol. 1991;173:727–733. doi: 10.1128/jb.173.2.727-733.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnelly M I, Millard C S, Clark D P, Chen M J, Rathke J W. A novel fermentation pathway in an Escherichia coli mutant producing succinic acid, acetic acid and ethanol. Appl Biochem Biotechnol. 1998;70–72:187–198. doi: 10.1007/BF02920135. [DOI] [PubMed] [Google Scholar]
- 10.Furste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 11.Glassner, D. A., and R. Datta. September 1992. U.S. patent 5,143,834.
- 12.Gokarn R R, Eiteman M A, Altman E. Metabolic analysis of Escherichia coli in the presence and absence of carboxylating enzymes phosphoenolpyruvate carboxylase and pyruvate carboxylase. Appl Environ Microbiol. 2000;66:1844–1850. doi: 10.1128/aem.66.5.1844-1850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottschalk G. Bacterial metabolism. 2nd ed. New York, N.Y: Springer-Verlag; 1985. [Google Scholar]
- 14.Guettler, M. V., M. K. Jain, and D. Rumler. November 1996. U.S. patent 5,573,931.
- 15.Gupta S, Clark D P. Escherichia coli derivatives lacking both alcohol dehydrogenase and phosphotransacetylase grow anaerobically by lactate fermentation. J Bacteriol. 1989;171:3650–3655. doi: 10.1128/jb.171.7.3650-3655.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoischen C, Levin J, Pitaknarongphorn S, Reizer J, Saier M H. Involvement of the central loop of the lactose permease of Escherichia coli in its allosteric regulation by the glucose-specific enzyme IIA of the phosphoenolpyruvate-dependent phosphotransferase system. J Bacteriol. 1996;178:6082–6086. doi: 10.1128/jb.178.20.6082-6086.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holbrook J J, Liljas A, Steindel S J, Roseman M G. Lactate dehydrogenase. In: Boyer P D, editor. The enzymes. 3rd ed. New York, N.Y: Academic Press; 1975. pp. 191–292. [Google Scholar]
- 18.Jain M K, Datta R, Zeikus J G. High-value organic acids fermentation—emerging processes and products. In: Ghose T K, editor. Bioprocess engineering: the first generation. United Kingdom: Norwood, Chichester; 1989. pp. 366–389. [Google Scholar]
- 19.Kornberg H L, Reeves R E. Inducible phosphenol pyruvate-dependent hexose phosphotransferase activities in Escherichia coli. Biochem J. 1972;128:1339–1344. doi: 10.1042/bj1281339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millard C S, Chao Y-P, Liao J C, Donnelly M I. Enhanced production of succinic acid by overexpression of phosphoenolpyruvate carboxylase in Escherichia coli. Appl Environ Microbiol. 1996;62:1808–1810. doi: 10.1128/aem.62.5.1808-1810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 22.Nghiem, N., M. Donnelly, C. S. Millard, and L. Stols. February 1999. U.S. patent 5,869,301.
- 23.Phillips S A, Thornalley P S. The formation of methylglyoxal from triose phosphates. Investigation using a specific assay for methylglyoxal. Eur J Biochem. 1993;212:101–105. doi: 10.1111/j.1432-1033.1993.tb17638.x. [DOI] [PubMed] [Google Scholar]
- 24.Plumbridge J. Expression of the phosphotransferase system both mediates and is mediated by Mlc regulation in Escherichia coli. Mol Microbiol. 1999;33:260–273. doi: 10.1046/j.1365-2958.1999.01462.x. [DOI] [PubMed] [Google Scholar]
- 25.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems. In: Neidhardt F C, editor. Escherichia coli and Salmonella. Washington, D.C.: American Society for Microbiology; 1996. pp. 1149–1174. [Google Scholar]
- 26.Saier M H, Ramseier T M, Reizer J. Regulation of carbon utilization. In: Neidhardt F C, editor. Escherichia coli and Salmonella. Washington, D.C.: American Society for Microbiology; 1996. pp. 1325–1343. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 28.Samuelov N S, Lamed R, Lowe S, Zeikus J G. Influence of CO2-HCO3− levels and pH on growth, succinate production, and enzyme activities of Anaerobiospirillum succiniciproducens. Appl Environ Microbiol. 1991;57:3013–3019. doi: 10.1128/aem.57.10.3013-3019.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stols L, Donnelly M I. Production of succinic acid through overexpression of NAD+-dependent malic enzyme in an Escherichia coli mutant. Appl Environ Microbiol. 1997;63:2695–2701. doi: 10.1128/aem.63.7.2695-2701.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeda S, Matsushika A, Mizuno T. Repression of the gene encoding succinate dehydrogenase in response to glucose is mediated by the EIICBglc protein in Escherichia coli. J Biochem (Tokyo) 1999;126:354–360. doi: 10.1093/oxfordjournals.jbchem.a022457. [DOI] [PubMed] [Google Scholar]
- 31.Vogel H J, Bonner D M. Acetylornithinase in E. coli. J Biol Chem. 1956;218:97–103. [PubMed] [Google Scholar]
- 32.Wanner B L. Novel regulatory mutants of the phosphate regulon in E. coli. J Mol Biol. 1986;191:39–59. doi: 10.1016/0022-2836(86)90421-3. [DOI] [PubMed] [Google Scholar]
- 33.Winkelman J W, Clark D C. Anaerobically induced genes of Escherichia coli. J Bacteriol. 1986;167:362–367. doi: 10.1128/jb.167.1.362-367.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeikus J G, Jain M K, Elankovan P. Biotechnology of succinic acid production and markets for derived industrial products. Appl Microbiol Biotechnol. 1999;51:545–552. [Google Scholar]