Abstract
Since January 1, 2001, the only acceptable nomenclatural type for species under the International Code of Nomenclature of Prokaryotes (ICNP) has been pure cultures. Here, we argue that this requirement is discordant with the more inclusive nature of nomenclatural types accepted under other codes of nomenclature and posit that the unique rigidity of the ICNP has failed to serve the broad research community and has stifled progress. This case is based on the axiom that many archaea and bacteria are interdependent in nature and therefore difficult, if not impossible, to grow, preserve, and distribute as pure cultures. As such, a large proportion of Earth's biodiversity cannot be named under the current system, which limits our ability to communicate about microbial diversity within and beyond the microbiology research community. Genome sequence data are now encouraged for valid publication of new taxa in microbial systematics journals, and metagenome-assembled genomes and single cell-amplified genomes are being generated rapidly from every biome on Earth. Thus, genome sequences are available for both cultivated and uncultivated microorganisms and can readily serve as a new category of nomenclatural type, allowing for a unified nomenclature for all archaea and bacteria, whether or not they are available as pure cultures. Ideally this would be under a single code of nomenclature but, as we review here, the newly established SeqCode will operate in parallel with the ICNP as a first step toward this goal.
Keywords: Systematics, taxonomy, genomics, ICNP, metagenomics, single-cell genomics, SeqCode, nomenclatural type, type
What are nomenclatural types and why are they important?
A nomenclatural type, or type, can be defined as an “element to which the name of a taxon is permanently attached” [1]. Nomenclatural types are critical for most, but not all, modern codes of nomenclature because they anchor names to something concrete, rather than impermanent concepts, such as monophyletic groups, which may change through the normal course of scientific inquiry. The type is thus the reference against which taxonomic comparisons can be made, which, along with rules of priority, ensures that a unique name should be applied to one and only one taxon, that which includes the nomenclatural type [2,3].
The ICNP requirement for viable and distributable pure cultures as types is discordant with other codes
The International Code of Nomenclature for Prokaryotes (ICNP) is unique among the codes of nomenclature by now accepting only one category of nomenclatural type for species and subspecies: viable pure cultures that have been successfully submitted, stored, and made available from at least two culture collections in different countries [3]. Because lower taxonomic ranks serve as nomenclatural types of higher ranks under the ICNP, formal proposals of names for higher taxa also require pure cultures. For comparison, the International Code of Nomenclature for algae, fungi and plants (ICN) [1] and the International Code of Zoological Nomenclature (ICZN) [4] each accept several categories of types, such as museum or herbarium specimens of animals or plants, fossils of extinct taxa, viable cultures of algae or fungi, and slides of micro-eukaryotes (Table 1; Fig. 1). By doing so, these other codes serve broad communities of botanists and zoologists by empowering them to use resources that are appropriate for the organisms they study. Organisms named under the ICN and ICZN span >21 orders of magnitude in mass and range from single cells, in the case of micro-eukaryotes, to about 1018 cells for blue whales (and many more for the largest trees). The inclusion of fossils under the ICN and ICZN as a category of type enables formal taxonomic names to extend to taxa that have long been extinct, even for a billion years or more [5]. Critically, they also include slides of micro-eukaryotes, which allows abundant and diverse, but notoriously under-sampled [6], micro-eukaryotes to be formally named.
Table 1.
Some codes of nomenclature and acceptable types for species
| Code | First/current version | Governing body | Relevant text | Acceptable types for species under current versiona |
|---|---|---|---|---|
| International Code of Nomenclature for algae, fungi and plants (ICN) [1] | First: 1867 Current: 2008 |
Nomenclature Section of the International Botanical Congress provided with logistical and financial support by the International Association for Plant Taxonomy (IAPT) | Article 8 | Single specimen in an herbarium or other collection, or a published or unpublished illustration. Metabolically inactive but viable cultures of algae and fungi are also eligible as types.b,c |
| International Code of Zoological Nomenclature (ICZN) [4] (for fungi [50]) | First: 1905 Current: 2018 |
“Section of Zoological Nomenclature” International Union of Biological Sciences | 72.5.1–72.5.6.73.1.4 | An animal, or any part of an animal (including DNA), or an example of the fossilized work of an animal; a colony or part of a colony of animals that exists in nature as a single entity; in the case of fossils, a natural replacement, natural impression, natural mold, or natural cast of an animal or colony, or part of either; in extant species of protistans, one or more preparations of directly related individuals representing differing stages in the life cycle; a preparation for microscope examination (e.g., a “type slide”) containing one or more individual organisms, in which the name-bearing types are clearly indicated and identifiable; moreover an illustration or photograph can serve as a proxy for a type specimen (https://www.iczn.org/outreach/faqs/). |
| International Code of Nomenclature of Prokaryotes (ICNP) [3] | First: 1958 Current: 2008 |
International Committee on Systematics of Prokaryotes (ICSP) | Rule 18a | Since 1 January 2001: Whenever possible, the type of a species or subspecies is a designated straind. |
| International Code of Virus Classification and Nomenclature ICVCN) [51] | First: 1971 Current: 2021 |
International Committee on Taxonomy of Viruses (ICTV) | N/A | Nomenclatural types are not discussed in the ICVCN and rules of priority are not observed. |
| International Code of Phylogenetic Nomenclature (PhyloCode) [52] | First: 2000 Current: 2008 |
Committee on Phylogenetic Nomenclature (CPN) | Articles 9.5, 9.6 | Types are not used; instead, specifiers are used to delimit members of a clade to which the name is attached. Specifiers are different from types because there are multiple specifiers per taxon, and each can define the taxon either positively or negatively by using appropriate operators. |
| Code of Nomenclature of Prokaryotes Described from Sequence Data | 2022 | To be determined | To be determined | Genome sequences derived from pure cultures, single cells, or communities. |
Text is taken directly from the codes with minor edits for simplification. The table focuses on forward-looking rules and excludes retroactive rules.
Illustrations are not acceptable nomenclatural types for fossils under Article 8.5 of the ICN.
Illustrations are “encouraged” in cases where microscopic algae or fungi cannot be effectively preserved under Article 40.5 of the ICN.
Prior to January 2001, a “description, preserved specimen, or illustration” could serve as type. The phrase “whenever possible” is not explicitly addressed in the code and the meaning is uncertain. The Note to Rule 30.3. b. States that in “exceptional cases” (considered on an individual basis) exceptions may be made to the rule requiring type strain deposition” but this provision has not been widely used and has mainly been used to allow naming of strains that have not been deposited into two culture collections.
Fig. 1.
Different types of types. Examples of categories of types acceptable under different codes of nomenclature. (A) Tyrannosaurus rex holotype specimen CM 9380/AMNH 973 at the Carnegie Museum of Natural History, originally collected from the Hell Creek Formation, Montana, USA. Although the skeleton was <25% complete, it is sufficient to identify the species. (Photo: Wikimedia Commons.) (B) Finch types collected in the Galapagos Islands by Darwin during the second voyage of the HMS Beagle in the 1830s, used with permission from the Natural History Museum in London. (C) Paratype slide of the ciliate Euplotes rariseta, compliments of Emma Sherlock of the Natural History Museum in London. (D) Herbarium type sheet for Acer saccharum, commonly known as sugar maple. Photo: Wikimedia Commons. (E) Lyophilized culture of Kallotenue papyrolyticumDSM 26889T [48] from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). Currently, viable axenic cultures are the only category of nomenclatural type acceptable under the ICNP. (F) Representation of the whole genome of Thermus oshimai JL-2 in comparison with two other T. oshimai genomes; high-quality genome sequence data would be eligible under the SeqCode, which is under development. (Image reproduced from [49]).
Many archaea and bacteria are also notoriously under-sampled from a taxonomic perspective. For example, both phylogenomic studies [7,8] and 16S rRNA gene meta-analyses have estimated that yet-uncultivated taxa account for ≥80% of the diversity of archaea and bacteria [9,10]. RS202 of the Genome Taxonomy Database (RS202) lists 20 phyla of archaea and 149 phyla of bacteria, of which only four (20%) and 46 (31%) include one or more type strains that are available as pure cultures [11]. Recent estimates suggest that <0.5% of species of archaea and bacteria have been formally named under the ICNP and, of those that have, ∼70% belong to just four phyla [12]. Thus, the uncultured majority is not limited to extreme environments, “weird” taxa, or the rare biosphere. Rather, it pervades the tree of life and includes abundant and active microorganisms in nearly every biome [9,10].
As a somewhat frivolous but potentially illuminating exercise, we could imagine the consequences if the ICN and ICZN required purified and viable reproductive material to designate types under their codes (e.g., cryopreserved fertile mating pairs, frozen embryos, or viable seeds or spores). Few could dispute that such materials would be valuable, which is why plant seed banks [13] and yeast culture collections [14] exist; however, the deposition of axenic, viable, and readily distributable material into these collections is not mandatory to name taxa. If it were, then the costs of eukaryotic systematics research would greatly increase, both for primary researchers and for the collections themselves, and the rate and value of progress of eukaryotic taxonomy research would suffer significantly. Of course, the scope and scale of taxa that could be named would also be greatly limited. In such a world, we could not formally name Tyrannosaurus rex, Carcharodon megalodon, or Homo habilis, myriads of orchids and other plants that are difficult to cultivate, many obligate mycorrhizal fungi, or most micro-eukaryotes. By limiting the only dedicated code of nomenclature for archaea and bacteria to pure cultures, microbiologists are unique in limiting the scope, scale, and value of our systems of nomenclature and taxonomy and, arguably, the entire scientific enterprise of studying microorganisms.
Not allowing the uncultured majority to be named hinders the microbiology research community
The requirements for deposition of viable type strains, specialized approaches, and relatively high costs associated with characterizing and naming new taxa under the ICNP (compared, for example, to systematics of plants and metazoans), along with challenges in funding such research, have limited progress in formal microbial systematics. For example, in 2015, we reported that the U.S. National Science Foundation's Systematics and Biodiversity Science Cluster was only allocating ∼2% of their projects and ∼2% of the program's funding to projects studying archaea and/or bacteria [15], and this problem persists today (see: https://www.nsf.gov/awardsearch/advancedSearchResult?ProgEleCode=7374, 1171, 1198, 7375,%207,376&BooleanElement = ANY&BooleanRef = ANY&). Simply stated, microbial systematics is poorly supported, and grant proposals focused on isolating, naming, and taxonomically describing new species of archaea and/or bacteria are non-competitive in the U.S. and, anecdotally, also in many other territories.
In addition to funding challenges, some countries are also challenged by barriers to sharing biological material, including pure cultures, under their implementations of the Nagoya Protocol. Despite the overreaching goal of the Nagoya Protocol to ensure access and equitable sharing of benefits derived from biological resources and preventing biopiracy, the dangers posed to biodiversity research have been extensively argued [[16], [17], [18]]. More than 100 countries are signatory parties to the Nagoya protocol (https://www.cbd.int/abs/nagoya-protocol/signatories/); however, the implementation of domestic legislation and regulatory criteria differ greatly among signatories. For example, submitting pure cultures of archaea and bacteria to international culture collections may be challenging for scientists in India, Brazil, Costa Rica, South Africa, and elsewhere, or for international researchers working in those countries. These countries require that permission should first be obtained from authorities before a culture collection can distribute the type culture to a third party, even if only used for taxonomic studies. According to Rule 30 (4) of the ICNP, any restrictions on access to a strain disqualify it as a nomenclature type [19], and this may further extend to issues with overly restrictive Material Transfer Agreements. This problem will be greatly exacerbated if restrictions are extended to digital sequence information [16,20]. These countries are home to several of the world's biodiversity hotspots [21], and the research community cannot afford to estrange communities of scientists exploring biodiversity in these countries.
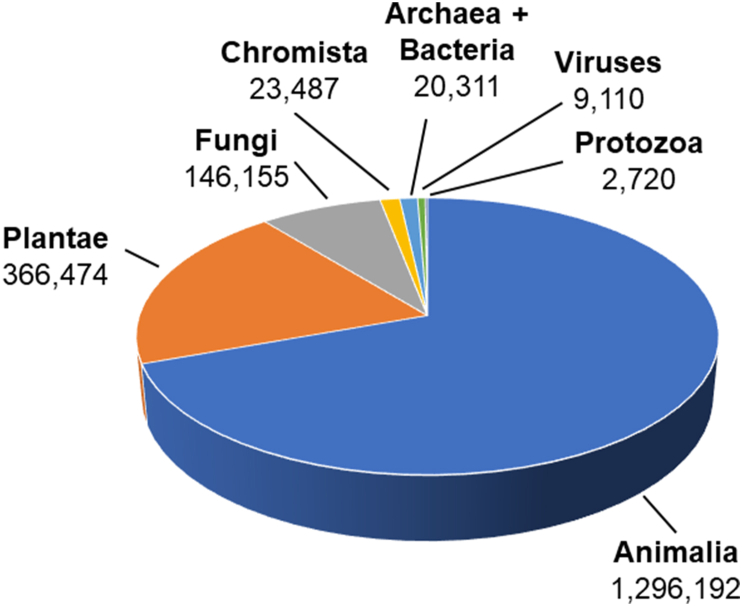
These challenges, in addition to scientific and technological challenges, are responsible for the slow pace of microbial systematics. To date, very few species of archaea and bacteria have been formally named, and those that have are highly skewed toward lineages that are easy to grow and maintain in the laboratory [8,12,22]. Thus, the 20,311 validly named species of archaea and bacteria [ [23]; https://lpsn.dsmz.de/text/numbers] account for only ∼1% of total formally named species across all domains of life (Fig. 2) [24]. As argued above, these are not only obscure or rare archaeal and bacterial species, and the problem extends from species all the way up to the rank of phylum. Even if strategies to increase the rate of naming cultivated taxa can be adopted quickly (reviewed in [12]), as could be enabled by high-throughput culturomics approaches (reviewed in [25]), the current rules of the ICNP will still exclude difficult-to-cultivate microorganisms from being formally named. Indeed, despite General Consideration 5 of the ICNP stating the code “applies to all prokaryotes,” since January 2001 only species with type strains available in pure culture can be validly named [3], meaning that the code itself would be more accurately named the “International Code of Nomenclature of Cultivated Prokaryotes.”
Fig. 2.
Validly published species names. Names per “kingdom” according to the Catalogue of Life 2019 Annual Checklist. Data for Archaea + Bacteria and viruses were updated per the LPSN count of validly published species names without synonyms plus 2020 cyanobacterial species names listed in the Botanical Code section, which will be validated under the ICNP (updated 4/6/2022) and per the ICTV Master Species Lists 2020. v1 (9110).
A system of nomenclature that accepts DNA sequence data as the nomenclatural type will decrease chaos and improve communication
Whitman [26,27] proposed modification of the ICNP to recognize DNA sequence data as a category of nomenclatural type for species and subspecies proposals, and others have additionally suggested that a parallel code of nomenclature should be developed if Whitman's proposal did not gain approval [28,29]. Oren and Garrity [30] argued that a parallel system of nomenclature would cause chaos. However, the current state of prokaryotic taxonomy is already chaotic because most organisms cannot be named, as argued previously [31], and the restriction of the ICNP to pure cultures effectively prohibits the ICNP from resolving the situation. Microbiologists studying biodiversity within the context of ecology or human health, both of which are much better funded than systematics, have unwittingly come face to face with the multitude of microbial taxa in nature that have never been isolated and therefore cannot be named under the ICNP. In the face of the enormous scale of microbial diversity, and in the absence of a system to formally name, organize, and communicate that diversity, the research community has turned to poorly ordered, ambiguous, and partially or fully synonymous names and alphanumeric codes. As pointed out elsewhere [31], many of those names and codes are rankless, which can lead to misleading interpretations of the biology of member organisms, particularly when taxa of higher ranks are interpreted as species and vice versa. Additionally, those terms typically refer to clades with no designated type [2], and therefore lack precision and permanence. Moreover, none are protected by the rules of priority, which also undermines the use of the compromise Candidatus status for uncultured taxa [32,33]. The informal names and alphanumeric codes applied to archaea and bacteria are further limited because many refer to paraphyletic groups. Finally, linguistics experts have long known that alphanumeric codes have lowmnemonic value because they are remembered as strings of words, letters, and numbers, each contributing to a limited memory or digit span, which limits their effectiveness as vehicles for communication [34]. In contrast, a single word in the case of a taxonomic name is easier to remember especially if the elements of the name are familiar and/or descriptive.
A more inclusive code of nomenclature for archaea and bacteria that recognizes DNA sequence data as a category of nomenclatural type would decrease the current chaos [35] and better serve the microbiology research community by enabling the community to use the resources that are available and appropriate to explore, organize, name, and communicate the entirety of archaeal and bacterial diversity. DNA that is extracted from a specimen and sequenced is already eligible to serve as the type under the ICZN because the ICZN allows parts of animals to serve as types. Under that code, DNA that has not been amplified before sequencing is considered part of the animal, and therefore is able to serve as the type in the same way that a part of a complete fossil can serve as a type [4]. Genomic data derived from archaea and bacteria is even more information-rich and permanent compared to a physical sample of eukaryotic DNA, regardless of whether the DNA was purified from axenic cultures, amplified from single cells, or isolated from a microbial community, provided that standards for use of such sequences as types are high. Table 2 outlines the utility of genomes and strains as nomenclatural types.
Table 2.
Utility of strains versus genome sequences as nomenclatural types for species in microbiology
| Criterion | Strains | Genomes | Explanation |
|---|---|---|---|
| Unambiguous means as a reference to identify the species | High | High | Genomes of sufficient quality are considered the gold standard for species circumscription for both strains and environmental genomes. |
| Experimental value | High | Moderate | Strains are unquestionably more valuable because they are sources for laboratory experiments, although valuable insights can still be obtained from genomic analysis. |
| Ability to scale to most or all taxa evident in nature | Very low | High | Due to problems with cultivability, funding limitations, and specialized workflows to name taxa under the ICNP, scalability is currently very low. The ability to obtain an isolate genome, SAG, or MAG is much more scalable (as evidenced by large numbers already being reported) but still may be limited based on accessibility of difficult-to-culture members of the rare biosphere or members of highly even, complex communities from which it may be difficult to obtain high-quality MAGs. |
| Archival permanence through time | Moderate | Very high | Submission of type strains to two international culture collections generates archival material; however, high costs and unstable funding structures may undermine the long-term future of culture collections. Moreover, the technology for cultivation of some fastidious microorganisms is not available in most culture collections. Also, viability of cultures over decadal time scales across many taxa is a significant challenge. Genomic data in International Sequence Database Collaboration (INSDC) databases are stored on multiple backup systems in at least three continents and are expected to be future-proofed. |
| Findabilitya | High | High | Online culture collection catalogs, LPSN, and INSDC data are all easily findable via internet. |
| Accessibilitya | Moderate | High | Strains are considered moderately accessible here because of costs, time, administrative (including MTAs), and technical requirements to obtain and maintain them. Genomic data only require a stable internet connection. |
| Cost of access to the type | $150–$500 | $0 | Costs were estimated from the DSMZ pricing. The cost of accessing genome sequences is negligible given internet access. |
| Time to access the type | Days to months | Seconds to minutes | Timelines to obtain type strains was estimated based on experience of the authors. Genome download/upload speeds depend on internet speed and genome size. |
| Interoperabilitya | Low | High | DNA sequence data can be used to compare types and other sequences to a query in seconds to hours using well-defined pipelines. The comparison of strains vis-à-vis polyphasic taxonomy is typically focused on properties perceived to be important for the comparison group, lowering interoperability across the diversity of archaea and bacteria. |
| Globally appropriate | Moderate | High | As described in the text, some implementations of the Nagoya Protocol conflict with the ability to designate type strains under the ICNP. Genomes are considered high here, but some implementations of the Nagoya Protocol may limit genomic data sharing in the future. |
These elements are used here to reflect FAIR practices in data science [52], although these principles do not translate seamlessly to strains.
Regrettably the ICSP rejected Whitman's proposals to allow DNA sequence data to serve as nomenclatural types [26,27] and a related proposal to grant priority to Candidatus names [36,37], thus triggering the development of a parallel code to accommodate prokaryotes that cannot be accommodated by the ICNP (“Plan B″ of [28])—the SeqCode (https://www.isme-microbes.org/seqcode-initiative). Although a single, inclusive nomenclatural code for prokaryotes is most desirable [34], the SeqCode represents the first step toward providing a formal system of nomenclature that grants priority to uncultivated taxa and can therefore fully accommodate the known diversity of archaea and bacteria. As outlined in the consensus statement by Murray et al. [29], the establishment of the SeqCode could potentially lead to a merger of the two codes into a single unified code, which would allow genome sequences or cultures as nomenclatural types. Importantly, to promote harmony with the ICNP, the SeqCode will recognize the priority of names that are validly published under the ICNP; reciprocity from the ICNP will be highly desirable for a functional coexistence of the two codes.
Will pure cultures have value in a code of nomenclature that accepts genome sequences as types?
Absolutely! It has been argued that a code of nomenclature accepting DNA sequence data as nomenclatural types would negatively impact the motivation to study and archive pure cultures [38]. We acknowledge this concern and take it seriously. However, we agree with a previous counterargument that metagenomics and other systems approaches focused on uncultivated microorganisms can provide invaluable support to guide cultivation efforts [31,35], not least by helping researchers compile “most wanted” lists of taxa to bring into culture and providing insights into their metabolism to inform culture strategies. We also suggest that the motivation to isolate and study taxa without a strong ecological, biotechnological, or health-related framework is already low, because in many parts of the world this kind of work is neither fundable nor glorious [39]. In contrast, we argue that compelling research on the properties of novel, abundant, highly active, or otherwise noteworthy microorganisms should be attractive to funding agencies if they proceed using the best technical approaches possible, which includes, but is not limited to, pure culture studies.
The value inherent in studying pure cultures was developed some ∼138 years ago by Koch and Loeffler through development of Koch's Postulates [40,41] and parallels the merits of studying purified systems in chemistry, geology, materials science, and other fields of science. Those merits and the collective advantages of sharing pure cultures across international borders will remain strong into the future. However, we urge the community to embrace previous arguments [31] that technology has come a long way since the days of Koch, and we are increasingly able to probe the properties of microorganisms living in their natural habitat. We also point out here that Koch's postulates did not end with pure cultures but circled around to test hypotheses about the activities of the potential pathogens in appropriate natural settings. We believe that many modern pure culture studies have lost touch with this critical aspect of microbiology and urge the systematics community not to underestimate the value of high-quality, data-rich science that modern microbial ecology enables. Thus, we believe that the state of the art for any taxon is to combine studies of the organism in pure culture, where leveraging the ease, control, and precision offered by a purified system is possible, with studies of how that taxon behaves in its natural habitat. Thus, we strongly advocate for pure culture studies and the use of culture collections whenever feasible, in combination with the study of the organisms in situ, and we will continue to prioritize both in our own research. We also note that most journals already mandate sharing of materials, including strains, which will continue to promote resource sharing.
But, of critical importance, we believe that studies of uncultivated microorganisms have reached a point where they can provide more than adequate data to guide both taxonomy and nomenclature decisions [42]. Those decisions leverage the power of average nucleotide identity to unambiguously identify species [43] and marker gene phylogenetic approaches to guide decisions on taxonomic placement and rank [11]. These are the same basic approaches that drive modern systematics of pure cultures [44].
How will genome sequences be implemented as nomenclatural types under the SeqCode?
Under the SeqCode, as under the ICNP, the evidence for a taxon, including the quality of type genomes and the uniqueness, rank, and position, must be published in the scientific literature (i.e., the effective publication); however, a distinction is that under the SeqCode, the effective publication must be published in a peer-reviewed journal or book. In parallel, scientists must register the proposed taxonomic names in the SeqCode Registry (https://seqco.de/), which captures the name, etymology, nomenclatural type, and associated metadata. The SeqCode Registry also employs curators to guide the user community and implements automated checks, including public availability of type genomes and effective publications, uniqueness of proposed names, and Latinization. Ultimately, the validation of names proposed under the SeqCode requires both an effective publication and completion of the registration process in the SeqCode Registry.
Under all codes of nomenclature that employ nomenclatural types, assessment of the quality of types is critical for the operability and stability of names. Under the ICN and ICZN, most nomenclatural types are non-viable, preserved material (Fig. 1), and the quality of material that can serve as types differs according to the nature of the resources available and norms and standards developed by the scientific community. Key to these standards is that nomenclatural types must be of sufficient quality to identify (i.e., diagnose) the taxon. For example, in hominid paleontology where the fossil record is sparce, the consensus of the research community is that skull structure and dentition alone can be sufficient for species and subspecies diagnosis. The result of this implementation of the ICZN is that the hominid paleontology community is enabled to progress despite what some might consider to be low-quality nomenclatural types. For example, in the case of H. habilis, the type specimen OH7 consists of around two dozen bone fragments and 14 teeth, including two parietals, and a mostly complete lower mandible [45].
In prokaryotic microbiology, the decision that types validated under the ICNP must be a pure and viable culture is in itself a value judgement concerning the “quality” of type material. Under the SeqCode, the quality of genomes available to serve as types varies according to the source of the genome. There is currently very little technical challenge to assemble complete or high-quality draft genomes from pure cultures; however, it can be difficult to assemble high-quality genomes from a taxon of interest from a complex microbial community. Thus, it may ultimately be reasonable to consider different standards for different taxa, yet such a decision would have to be made by the scientific community. As an initial implementation of the SeqCode, high-quality genomes, essentially per the Genomic Standards Consortium's Minimum Information About a Single Amplified Genome (MISAG) and Minimum Information About a Metagenome-Assembled Genome (MIMAG) standards [46], are required regardless of the source of the genome. These standards are very high relative to many implementations of the ICN and ICZN (e.g., see Fig. 1 and discussion of H. habilis above) and sufficient for diagnosis of the species. For example, ANI experiments conducted with isolate genomes deliberately contaminated with other genomic data (i.e., modeling a contaminated MAG) demonstrated genomes with completeness of only 60% and contamination of up to 50% are sufficient to reproducibly place genomes into 95% ANI clusters [43], thus enabling species diagnosis. By comparison, the MISAG/MIMAG criteria for high-quality genomes are >90% estimated completeness and <5% estimated contamination, with additional requirements for tRNAs and rRNAs. Similarly, concatenation of 70 conserved marker genes can reliably overcome noise due to inter-species recombination and discordance of individual gene trees, resulting in stable and robust species trees, regardless of which conserved marker genes are used [47]. For reference, the GTDB employs conserved positions from 120 marker genes for bacteria and conserved positions from 122 marker genes for archaea [11]. At 90% completeness, genomes should have ∼108 of those conserved marker genes, which is more than sufficient for the construction of reliable species trees. In the near future, we will furthermore code automated data quality checks (i.e., estimated completeness and contamination) into the SeqCode Registry, which would produce warnings that would prevent validation unless manually overridden by curators based on rational justification (e.g., confirmation of missing or duplicated marker genes used to estimate genome completeness and contamination.)
Outlook
The use of a “new type of type” is for the moment embodied through the development of the “SeqCode,” but ideally in future, the ICNP and “SeqCode” may be rationalized into a single unified code that encompasses all archaeal and bacterial diversity. Thus, by broadening our current restrictive criteria for what passes muster as nomenclatural type and ensuring a more inclusive system of nomenclature, we believe the broad microbiology community will strongly benefit in the long run.
Credit author statement
Brian P. Hedlund: Writing – original draft, Writing – review & editing, Project administration, Funding acquisition. Marike Palmer: Writing – original draft, Writing – review & editing; Iain Sutcliffe: Writing – review & editing. Stephanus N. Venter: Writing – review & editing.
Transparency declaration
The authors declare no conflicts of interest.
Acknowledgements
Funding was provided by the US National Science Foundation (DEB 1841658) and the US National Institute of General Medical Sciences (GM103440) from the National Institutes of Health.
References
- 1.Turland N.J., Wiersema J.H., Barrie F.R., Greuter W., Hawksworth D.L., Herendeen P.S., Knapp S., Kusber W.-H., Li D.-Z., Marhold K., May T.W., McNeill J., Monro A.M., Prado J., Price M.J., Smith G.F., editors. Regnum vegetabile 159. Koeltz Botanical Books; Glashütten: 2018. International code of nomenclature for algae, fungi, and plants (shenzhen code) adopted by the nineteenth international botanical congress shenzhen, China, july 2017. [DOI] [Google Scholar]
- 2.Chuvochina M., Rinke C., Parks D.H., Rappé M.S., Tyson G.W., Yilmaz P., Whitman W.B., Hugenholtz P. The importance of designating type material for uncultured taxa. Syst Appl Microbiol. 2019;42:15–21. doi: 10.1016/j.syapm.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Parker C.T., Tindall B.J., Garrity G.M. International code of nomenclature of prokaryotes. Prokaryotic Code (2008 Revision) Int J Syst Evol Microbiol. 2019;69:S1–S111. doi: 10.1099/ijsem.0.000778. [DOI] [PubMed] [Google Scholar]
- 4.Ride W.D.L., Cogger H.G., Dupuis C., Kraus O., Minelli A., Thompson F.C., Tubbs P.K. 4th ed. International Trust for Zoological Nomenclature, c/o Natural History Museum; London: 1999. International code of zoological nomenclature.https://www.iczn.org/the-code/the-code-online/ [Google Scholar]
- 5.Strother P.K., Brasier M.D., Wacey D., Timpe L., Saunders M., Wellman C.H. A possible billion-year-old holozoan with differentiated multicellularity. Curr Biol. 2021;31:2658–2665. doi: 10.1016/j.cub.2021.03.051. [DOI] [PubMed] [Google Scholar]
- 6.Leray M., Knowlton N. Censusing marine eukaryotic diversity in the twenty-first century. Philos Trans R Soc Lond B Biol Sci. 2016;371 doi: 10.1098/rstb.2015.0331. 20150331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nayfach S., Roux S., Seshadri R., Udwary D., Varghese N., Schulz F., et al. A genomic catalog of Earth's microbiomes. Nat Biotechnol. 2021;39:499–509. doi: 10.1038/s41587-020-0718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinke C., Schwientek P., Sczyrba A., Ivanova N.N., Anderson I.J., et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 9.Lloyd K.G., Steen A.D., Ladau J., Yin J., Crosby L. Phylogenetically novel uncultured Microbial cells dominate Earth microbiomes. mBio. 2018;3 doi: 10.1128/mSystems.00055-18. e00055-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steen A.D., Crits-Christoph A., Carini P., DeAngelis K.M., Fierer N., Lloyd K.G., Thrash J.C. High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 2019;13:3126–3130. doi: 10.1038/s41396-019-0484-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parks D.H., Chuvochina M., Chaumeil P.-A., Rinke C., Mussig A.J., Hugenholtz P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat Biotechnol. 2020:1079–1086. doi: 10.1038/s41587-020-0501-8. [DOI] [PubMed] [Google Scholar]
- 12.Sutcliffe I.C., Rosselló-Mora R., Trujillo M. Addressing the sublime scale of the microbial world: reconciling an appreciation of microbial diversity with the need to describe species. New Microbe. New Infect. 2021;43:100931. doi: 10.1016/j.nmni.2021.100931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruttimann J. Doomsday food store takes pole position. Nature. 2006;441:912–913. doi: 10.1038/441912b. [DOI] [PubMed] [Google Scholar]
- 14.Boundy-Mills K.L., Glantschnig E., Roberts I.N., Yukov A., Casaregola S., Daniel H.-M., Groenewald M., Turchetti B. Yeast culture collections in the twenty-first century: new opportunities and challenges. Yeast. 2016;33:243–260. doi: 10.1002/yea.3171. [DOI] [PubMed] [Google Scholar]
- 15.Hedlund B.P., Dodsworth J.A., Staley J.T. The changing landscape of microbial biodiversity exploration and its implications for systematics. Syst Appl Microbiol. 2015;38:231–236. doi: 10.1016/j.syapm.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Karger E.J., Scholz A.H. DSI, the Nagoya protocol, and stakeholders’ concerns. Trend Biotechnol. 2021;39:110–112. doi: 10.1016/j.tibtech.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Overmann J., Scholz A.M. Microbiological research under the Nagoya Protocol: facts and fiction. Trends Microbiol. 2017;25:85–88. doi: 10.1016/j.tim.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Prathapan K.D., et al. When the cure kills - CBD limits biodiversity research. Science. 2018;360(6396):1405–1406. doi: 10.1126/science.aat9844. [DOI] [PubMed] [Google Scholar]
- 19.Tindall B.J., de Vos P., Trüper H.G. Judicial commission of the international committee on systematics of prokaryotes XIth international (IUMS) congress of bacteriology and applied microbiology. Int J Syst Evol Microbiol. 2008;58:1737–1745. doi: 10.1099/ijs.0.2008/005074-0. [DOI] [Google Scholar]
- 20.Bond M.R., Scott D. Digital biopiracy and the (dis)assembling of the Nagoya Protocol. Geoforum. 2020;117:24–32. doi: 10.1016/j.geoforum.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers N., Mittermeier R.A., Mittermeier C.G., da Fonseca G.A.B., Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 22.Reimer L.C., Vetcininova A., Sardà Carbasse J., Söhngen C., Gleim D., Ebeling C., Overmann J. BacDive in 2019: bacterial phenotypic data for High-throughput biodiversity analysis. Nucleic Acids Res. 2019;47:D631. doi: 10.1093/nar/gky879. –6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parte A.C., Sardà Carbasse J., Meier-Kolthoff J.P., Reimer L.C., Göker M. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. 2020;70(11):5607–5612. doi: 10.1099/ijsem.0.004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roskov Y., Ower G., Orrell T., Nicolson D., Bailly N., Kirk P.M., et al. 2021. Species 2000 & ITIS Catalogue of life, 25th march 2019. Digital resource at.www.catalogueoflife.org/col Species 2000: Naturalis, Leiden, the Netherlands. ISSN 2405-8858. www.catalogueoflife.org/col. [Google Scholar]
- 25.Lewis W.H., Tahon G., Geesink P., Sousa D.Z., Ettema T.J.G. Innovations to culturing the uncultured microbial majority. Nat Rev Microbiol. 2021;21:225–240. doi: 10.1038/s41579-020-00458-8. [DOI] [PubMed] [Google Scholar]
- 26.Whitman W.B. Genome sequences as the type material for taxonomic descriptions of prokaryotes. Syst Appl Microbiol. 2015;38:217–222. doi: 10.1016/j.syapm.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Whitman W.B. Modest proposals to expand the type material for naming of prokaryotes. Int J Syst Evol Microbiol. 2016;66:2018–2112. doi: 10.1099/ijsem.0.000980. [DOI] [PubMed] [Google Scholar]
- 28.Konstantinidis K.T., Rossello-Mora R., Amann R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017;11:2399–2406. doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray A.E., Freudenstein J., Gribaldo S., Hatzenpichler R., Hugenholtz P., Kämpfer P., et al. Roadmap for naming uncultivated archaea and bacteria. Nat Microbiol. 2020;5:987–994. doi: 10.1038/s41564-020-0733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oren A., Garrity G. Uncultivated microbes—in need of their own nomenclature? ISME J. 2018;12:309–311. doi: 10.1038/ismej.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konstantinidis K.T., Rossello-Mora R., Amann R. Advantages outweigh concerns about using genome sequence as type material for prokaryotic taxonomy. Env Microbiol. 2020;22:819–822. doi: 10.1111/1462-2920.14934. [DOI] [PubMed] [Google Scholar]
- 32.Oren A. Nomenclature of prokaryotic ‘Candidatus’ taxa: establishing order in the current chaos. New Microbe New Infections. 2021;44:200932. doi: 10.1016/j.nmni.2021.100932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pallen M. The status Candidatus for uncultured taxa of Bacteria and Archaea: SWOT analysis. Int J Syst Evol Microbiol. 2021;71:5000. doi: 10.1099/ijsem.0.005000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller G. The magical number seven, plus or minus two–the magical number seven, plus or minus two: some limits on our capacity for processing information. Psychol Rev. 1956;63:81–97. [PubMed] [Google Scholar]
- 35.Rossello-Mora R., Konstantinidis K.T., Sutcliffe I.C., Opinion Whitman WB. 2020. Response to concerns about the use of DNA sequences as types in the nomenclature of prokaryotes. 43: 126070. [DOI] [PubMed] [Google Scholar]
- 36.Whitman W.B., Sutcliffe I.C., Rossello-Mora R. Proposal for changes in the international code of nomenclature of prokaryotes: granting priority to Candidatus names. Int J Syst Evol Microbiol. 2019;69(7):2174–2175. doi: 10.1099/ijsem.0.003419. [DOI] [PubMed] [Google Scholar]
- 37.Sutcliffe I.C., Dijkshoorn L., Whitman W.B., Executive Board I.C.S.B. Minutes of the International Committee on Systematics of Prokaryotes online discussion on the proposed use of gene sequences as type for naming of prokaryotes, and outcome of vote. Int J Syst Evol Microbiol. 2020;70:4416–4417. doi: 10.1099/ijsem.0.004303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bisgaard M., Christensen H., Clermont D., Dijkshoorn L., Janda J.M., Moore E.R.B., et al. The use of genomic DNA sequences as type material for valid publication of bacterial species names will have severe implications for clinical microbiology and related disciplines. Diagn Microbiol Infect Dis. 2019;95:102–103. doi: 10.1016/j.diagmicrobio.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 39.Sutcliffe I.C., Trujillo M.E., Goodfellow M. A call to arms for systematists: revitalising the purpose and practises underpinning the description of novel microbial taxa. Antonie Van Leeuwenhoek. 2012;101(1):13–20. doi: 10.1007/s10482-011-9664-0. [DOI] [PubMed] [Google Scholar]
- 40.Koch R. Die aetiologie der Tuberkulose. Mittheilungen aus dem Kaiserlichen Gesundheitsamte. 1884;2:1–88. [Google Scholar]
- 41.Loeffler F. Untersuchungen über die Bedeutung der Mikroorganismen für die Entstehung der Diphtherie beim Menschen, bei der Taube und beim Kalbe. Mittheilungen aus dem Kaiserlichen Gesundheitsamte. 1883;11:421–499. [Google Scholar]
- 42.Hugenholtz P., Chuvochina M., Oren A., Parks D.H., Soo R.M. Prokaryotic taxonomy and nomenclature in the age of big sequence data. ISME J. 2021;15:1879–1892. doi: 10.1038/s41396-021-00941-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jain C., Rodriguez -R.L.M., Phillippy A.M., Konstantinidis K.T., Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9(1):5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chun J., Oren A., Ventosa A., Christensen H., Arahal D.R., da Costa M.S., Rooney A.P., Yi H., Xu X.-W., De Meyer S., Trujillo M.E. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 45.Leakey L.S.B., Tobias P.V., Napier J.R. A new species of the genus Homo from Olduvai Gorge. Nature. 1964;202:8. doi: 10.1038/202007a0. [DOI] [PubMed] [Google Scholar]
- 46.Bowers R.M., Kyrpides N.C., Stepanauskas R., Harmon-Smith M., Doud D., Reddy T.B.K. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol. 2017;35:725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmer M., Venter S.N., McTaggart A.R., Coetzee M.P.A., Van Wyk S., Avontuur J.R., Beukes C.W., Fourie G., Santana Q.C., Van Der Nest M.A., Blom J., Steenkamp E.T. The synergistic effect of concatenation in phylogenomics: the case in Pantoea. PeerJ. 2019;7:e6698. doi: 10.7717/peerj.6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cole J.K., Gieler B.A., Heisler D.L., Palisoc M.M., Williams A.J., Dohnalkova A.C., Ming H., Yu T.T., Dodsworth J.A., Li W.J., Hedlund B.P. Kallotenue papyrolyticum gen. nov., sp. nov., a cellulolytic and filamentous thermophile that represents a novel lineage (Kallotenuales ord. nov., Kallotenuaceae fam. nov.) within the class Chloroflexia. Int J Syst Evol Microbiol. 2013;63:4675–4682. doi: 10.1099/ijs.0.053348-0. [DOI] [PubMed] [Google Scholar]
- 49.Murugapiran S.K., Huntemann M., Wei C.L., Han J., Detter J.C., Han C.S., et al. Thermus oshimai JL-2 and T. thermophilus JL-18 genome analysis illuminates pathways for carbon, nitrogen, and sulfur cycling. Stand Genomic Sci. 2013;7:449–468. doi: 10.4056/sigs.3667269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.May T.W., Redhead S.A., Bensch K., Hawksworth D.L., Lendemer J., Lombard L., Turland N.J. Chapter F of the international code of nomenclature for algae, fungi, and plants as approved by the 11th international mycological congress, san juan, Puerto Rico, july 2018. IMA Fungus. 2019;10:21. doi: 10.1186/s43008-019-0019-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuhn J.H., Radoshitzky S.R., Jarhling P.B. The International Code of Virus Classification and Nomenclature (ICVCN): proposal for text changes for improved differentiation of viral taxa and viruses. Arch Virol. 2013;158:1621–1629. doi: 10.1007/s00705-012-1582-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Queiroz The PhyloCode and the distinction between taxonomy and nomenclature. Syst Biol. 2006;55:160–162. doi: 10.1080/10635150500431221. [DOI] [PubMed] [Google Scholar]