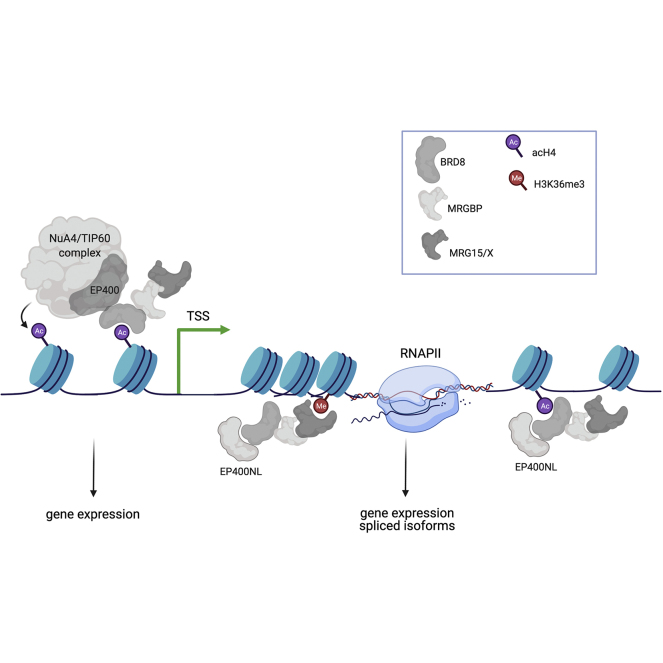
Abstract
MRG15/MORF4L1 is a highly conserved protein in eukaryotes that contains a chromodomain (CHD) recognizing methylation of lysine 36 on histone H3 (H3K36me3) in chromatin. Intriguingly, it has been reported in the literature to interact with several different factors involved in chromatin modifications, gene regulation, alternative mRNA splicing, and DNA repair by homologous recombination. To get a complete and reliable picture of associations in physiological conditions, we used genome editing and tandem affinity purification to analyze the stable native interactome of human MRG15, its paralog MRGX/MORF4L2 that lacks the CHD, and MRGBP (MRG-binding protein) in isogenic K562 cells. We found stable interchangeable association of MRG15 and MRGX with the NuA4/TIP60 histone acetyltransferase/chromatin remodeler, Sin3B histone deacetylase/demethylase, ASH1L histone methyltransferase, and PALB2–BRCA2 DNA repair protein complexes. These associations were further confirmed and analyzed by CRISPR tagging of endogenous proteins and comparison of expressed isoforms. Importantly, based on structural information, point mutations could be introduced that specifically disrupt MRG15 association with some complexes but not others. Most interestingly, we also identified a new abundant native complex formed by MRG15/X-MRGBP-BRD8-EP400NL (EP400 N-terminal like) that is functionally similar to the yeast TINTIN (Trimer Independent of NuA4 for Transcription Interactions with Nucleosomes) complex. Our results show that EP400NL, being homologous to the N-terminal region of NuA4/TIP60 subunit EP400, creates TINTIN by competing for BRD8 association. Functional genomics indicate that human TINTIN plays a role in transcription of specific genes. This is most likely linked to the H4ac-binding bromodomain of BRD8 along the H3K36me3–binding CHD of MRG15 on the coding region of transcribed genes. Taken together, our data provide a complete detailed picture of human MRG proteins–associated protein complexes, which are essential to understand and correlate their diverse biological functions in chromatin-based nuclear processes.
Keywords: MRG domain, chromodomain, histone methylation, histone acetylation, transcription
Abbreviations: ASH1L, absent, small, or homeotic discs 1-like; BRD, bromodomain; BSA, bovine serum albumin; Cas9, CRISPR-associated protein 9; cDNA, complementary DNA; CHD, chromodomain; ChIP, chromatin immunoprecipitation; EP400NL, EP400 N-terminal like; FDR, false discovery rate; gRNA, guide RNA; GST, glutathione-S-transferase; H3K36me, methylation of lysine 36 on histone H3; HA, hemagglutinin; KAT, lysine acetyltransferase; KD, knockdown; MRG15, MORF-related gene on chromosome 15; MS, mass spectrometry; NuA4, nucleosome acetyltransferase of H4; qPCR, quantitative PCR; RNAPII, RNA polymerase II; SETD2, SET domain containing 2; siLuc, siLuciferase; TINTIN, Trimer Independent of NuA4 for Transcription Interactions with Nucleosomes; TIP60, Tat interactive protein 60 kDa; U2OS, human osteosarcoma epithelial cell
Graphical Abstract
Highlights
-
•
MRG proteins are found in different complexes linked to transcription and DNA repair.
-
•
Several MRG-containing complexes are chromatin modifiers.
-
•
Point mutations in the MRG domain differentially affect associated complexes.
-
•
A major human complex homologous to the yeast TINTIN complex is identified
-
•
EP400NL competes with EP400 to separate TINTIN from the NuA4/TIP60 complex.
In Brief
MRG proteins have been reported to interact with several different factors involved in chromatin modifications, gene regulation, alternative mRNA splicing, and DNA repair. Using genome editing, we tagged endogenous protein to clearly identify stable protein complexes containing these factors. Doing so, we uncovered a new conserved tetrameric complex, named TINTIN, that regulates transcription with reader domains for epigenetic marks.
The eukaryote genome is organized into a structure called chromatin, with the nucleosomes being its basic units, playing central roles in the regulation of all DNA-based processes for genome expression and maintenance (1). One major element governing chromatin structure and functions is the diverse post-translational modifications deposited on the external tails of the nucleosomal histones (2). Post-translational modifications on specific residues of specific histones include acetylation, methylation, phosphorylation, and ubiquitination, which are recognized by dedicated reader domains in factors to regulate biological processes (3, 4). Disruption of the enzymes that “write” or “erase” these histone marks, or their “readers,” occurs in several human diseases, including cancer, leading to intense research efforts during the past several years to understand these regulatory interactions during cellular life (5).
One mark that has been studied extensively is the methylation of lysine 36 on histone H3 (H3K36me), which can occur as mono- (me1), di- (me2), or tri- (me3) methylation, levels being linked to distinct functions. While all methylation levels are generated by Set2 in Saccharomyces cerevisiae (6), mammalian methyltransferases like ASH1L (absent, small, or homeotic discs 1-like) and NSD1/2/3 only do H3K36me1/me2. Mammalian SET domain containing 2 (SETD2) on the other hand is responsible for H3K36me3 (7). Set2 associates with the elongating RNA polymerase II (RNAPII) phosphorylated on Ser2 of its C-terminal repeats (C-terminal domain). This leads to enrichment of H3K36me3 on the coding region of transcribed genes, which recruits the RPD3S histone deacetylase complex to stabilize chromatin in the wake of RNAPII in order to avoid cryptic initiation and spurious transcription (8, 9). Mammalian SETD2 does not seem to affect global chromatin acetylation levels (7), even if H3K36me3 plays a central role in transcriptional activation and is distributed throughout the coding regions of active genes (10, 11). More precisely, H3K36me3 has been observed to be specifically enriched on exons, compared with introns, independent of nucleosome occupancy, in both yeast and mammals (12, 13, 14). These data were used to define a function of this histone mark in alternative mRNA splicing (15). It has been proposed that H3K36me3 is used as a platform recognized by the factor MORF-related gene on chromosome 15 (MRG15, a.k.a. MORF4L1) through its N-terminal chromodomain (CHD), an interaction that recruits the splicing machinery (16, 17). The yeast homolog of MRG15, Eaf3, is also known to affect splicing through its binding with the splicing factor Prp45/SKIP, thereby leading to the recruitment of spliceosome on intron-containing genes (18).
Mammalian MRG15 has a paralog expressed from the X-chromosome, MRGX (a.k.a. MORF4L2), which lacks the CHD. MRG15/X seem to regulate cell proliferation and senescence in specific contexts, whereas only MRG15 is required for mouse development (19, 20). They have been found associated with both lysine acetyltransferase (KAT) and deacetylase complexes, namely nucleosome acetyltransferase of H4 (NuA4)/Tat interactive protein 60 kDa (TIP60) and Sin3B, also conserved in yeast (NuA4 and RPD3S) and containing the homologous protein Eaf3 (21, 22, 23, 24, 25). While MRGX does not possess an N-terminal CHD, it shares with MRG15 a highly conserved C-terminal MRG domain, crucial for their nonhistone protein–protein interactions (20). Interestingly, it has been suggested that MRG15 can form a homodimer, which is dissociated by its acetylation, regulating MRG15 association with other factors (26). MRG15 is also involved in DNA repair through its interaction with DNA repair factor PALB2, thereby favoring repair of double-strand breaks by homologous recombination (27, 28, 29). It has been recently described that the MRG15–PALB2 complex is associated with undamaged chromatin on active genes to protect them from genotoxic/replicative stress (30). Finally, MRG15 was very recently implicated in diurnal rhythm of epigenomic remodeling through chromatin acetylation as well as lipid metabolism through gene activation with LRH-1 (31).
Although current knowledge on MRG15 points toward quite distinct important roles in the cell and during development, its described functions and associated cofactors are often studied in very artificial nonphysiological conditions. Here, we aimed at defining the complete detailed interactomes of MRG15/X in the most native conditions possible. Tandem affinity purification from isogenic cell lines identified several stable complexes containing MRG15 or MRGX. Endogenous protein tagging by CRISPR allowed validation and characterization of a new tetrameric protein complex with striking resemblance to the yeast TINTIN (Trimer Independent of NuA4 for Transcription Interactions with Nucleosomes) complex that we have previously identified (25). The presence of a bromodomain (BRD) and a CHD in this complex and its effect on transcription support the claim that we have identified a human TINTIN complex that associates with the body of active genes.
Experimental Procedures
Recombinant Proteins and Pull-Down Assay
Recombinant proteins (all human) 6×His-BRD8, glutathione-S-transferase (GST)-MRG15 CHD long and short, GST-EP400 (amino acids 194–446), GST-EP400NL (EP400 N-terminal like) (amino acids 51–297), and GST-ASH1L (amino acids 2040–2634) were expressed in Escherichia coli BL21 cells grown in LB media (from pET15b, pGEX4T3, or pGEX6P2 vectors). After induction with IPTG (30 μM) overnight at 16 °C, bacteria were harvested, and GST or 6×His-tagged proteins were purified as previously described with minor modifications (32).
Protein and chromatin pull-down experiments were performed as previously described (33). The GST pull-down assays were performed using 300 to 600 ng of GST-fused protein and an equivalent amount of His-tagged protein or 5 μg of native H1-depleted chromatin from HeLa cells (prepared as described (34)), which was precleared using glutathione-Sepharose beads. Equivalent protein levels were estimated through Coomassie blue–stained SDS-PAGE by comparison with known amounts of bovine serum albumin (BSA) standards. After preclearing, the His-tagged proteins were incubated with GST-immobilized protein in binding buffer (25 mM Hepes [pH 7.9], 10% glycerol, 100 μg/ml BSA, 1 mM PMSF, 0.5 mM DTT, 0.1% Tween-20, and protease inhibitors, 250 mM NaCl was used for chromatin pull down) at 4 °C for 1 h to 4 h followed by washing the beads three times. To visualize the proteins, the beads were loaded on SDS-PAGE gels, followed by Western blotting with anti-GST (Sigma; catalog no.: G1160), anti-His (Clontech; catalog no.: 631212), anti-H3K36me2 (Upstate; catalog no.: 07-369), anti-H3K36me3 (Abcam; catalog no.: ab9050), anti-H3K79me3 (Abcam; catalog no.: ab2621), and anti-H4 (Abcam; catalog no.: ab7311). A GST-only protein and beads were used as a control.
Experimental Model and Subject Details
K562 and human osteosarcoma epithelial (U2OS) cells were obtained from the American Type Culture Collection and maintained at 37 °C under 5% CO2. K562 were cultured in RPMI medium supplemented with 10% newborn calf serum and GlutaMAX. U2OS cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
Establishment of Isogenic Cell Lines Using the AAVS1 Safe Harbor or CRISPR–CRISPR-Associated Protein 9–Mediated Genome Editing
K562 cells stably expressing 3xFLAG-2xStrep-tagged MRG15 spliced variants (long CHD or short CHD), MRGBP (MRG-binding protein), MRGX, BRD8 spliced variants (one or two BRDs), EPC1 or EP400NL spliced variants (isoforms Q6ZTU2-5 and Q6ZTU2-6) were established as described before (35). BRD8, ASH1L, MRGX, and EP400NL were also endogenously 3xFLAG-2xStrep tagged using CRISPR/CRISPR-associated protein 9 (Cas9) with ouabain selection as previously described (36). The following guide RNAs (gRNAs) were used to generate these cell lines:
| Gene | Part | Sequence for | Sequence rev |
|---|---|---|---|
| BRD8 | N-terminal | CCTCAGGGTTGGAGACTTCG | CGAAGTCTCCAACCCTGAGG |
| BRD8 Q9H0E9-1 | C-terminal | AAAGGCTCAAGTAGTCTGGA | TCCAGACTACTTGAGCCTTT |
| MRGX | C-terminal | TGTGAGCGTCTACAGACAGC | GCTGTCTGTAGACGCTCACA |
| EP400NL | C-terminal | TTTGAGGACTCGATGTGCTG | CAGCACATCGAGTCCTCAAA |
| ASH1L | C-terminal | CTCAAAGAATGAGAACCTCAA | TTGAGGTTCTCATTCTTTGAG |
Affinity Purification of Complexes
Native complexes were purified essentially as previously described (35). Briefly, nuclear extracts were prepared from 2.5.109 cells and precleared with CL6B sepharose beads. FLAG immunoprecipitations with anti-FLAG agarose affinity resin (Sigma; clone M2) were performed followed by two elutions with 3xFLAG peptide in elution buffer (20 mM Hepes–KOH [pH 7.9], 10% glycerol, 150 mM KCl, 0.1% Tween-20, 1 mM DTT, 1 mM PMSF, 2 mg/ml leupeptin, 5 mg/ml, 2 mg/ml pepstatin, 10 mM sodium butyrate, and 10 mM β-glycerophosphate) with 200 μg/ml 3xFLAG peptide (Sigma). Then, STREP immunoprecipitations with Strep-tactin sepharose beads (Cedarlane) were performed followed by two elutions with elution buffer supplemented with 4 mM biotin. Typically, 20 μl of the first Strep elution was loaded on SDS-Nu-PAGE 4 to 12% Bis–Tris gels (Invitrogen) and analyzed via silver staining. Fractions were then analyzed by mass spectrometry (MS) and Western blotting. For purification after inducing DNA damage, cells were treated with 50 ng/ml of neocarzinostatin for 3 h before being collected to prepare the nuclear extract.
MS Analysis
To produce the peptides for MS analysis, purified fractions were loaded on a 10% SDS-PAGE, ran until the dye migrated about 1 cm, stained with Sypro Ruby Red, and a gel slice containing the entire protein signal was cut and processed for in-gel digestion with trypsin. MS analyses were performed by the Proteomics Platform of the CHU de Québec-Université Laval Research Center. Samples were analyzed by nano-LC/MS–MS either with a 5600 triple TOF or an Orbitrap Fusion.
5600 Triple TOF
Peptides were analyzed using an Ekspert NanoLC425 coupled to a 5600+ triple TOF mass spectrometer (Sciex). Peptides were trapped at 4 μl/min in loading solvent (0.1% formic acid) on a 5 mm × 300 μm C18 PepMap cartridge precolumn (Thermo Fisher Scientific/Dionex Softron GmbH) for 10 min. Then, the precolumn was switched online with a self-packed PicoFrit column (New Objective) packed with reprosil 3u, 120A C18, 15 cm × 0.075 mm internal diameter (Dr Maisch). Peptides were eluted with a linear gradient from 5 to 35% solvent B (A: 0.1% formic acid, B: acetonitrile and 0.1% formic acid) in 35 min, at 300 nl/min for a total run time of 60 min. Mass spectra were acquired using a data-dependent acquisition mode using Analyst software, version 1.7 (Sciex). Each full scan mass spectrum (m/z 400–1250) was followed by collision-induced dissociation of the 20 most intense ions. Dynamic exclusion was set for 12 s and tolerance of 100 ppm.
Orbitrap Fusion
Peptides were analyzed using a Dionex UltiMate 3000 nanoRSLC chromatography system (Thermo Fisher Scientific) connected to an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific). Peptides were trapped at 20 μl/min in loading solvent (2% acetonitrile and 0.05% TFA) on a 5 mm × 300 μm C18 PepMap cartridge precolumn (Thermo Fisher Scientific/Dionex Softron GmbH) for 5 min. Then, the precolumn was switched online with a PepMap Acclaim column (Thermo Fisher Scientific) 50 cm × 75 μm internal diameter separation column, and the peptides were eluted with a linear gradient from 5 to 40% solvent B (A: 0.1% formic acid, B: 80% acetonitrile and 0.1% formic acid) in 60 min, at 300 nl/min for a total runtime of 90 min. Mass spectra were acquired using a data-dependent acquisition mode using Thermo XCalibur software, version 4.3.73.11 (Thermo Fisher Scientific). Full scan mass spectra (m/z 350–1800) were acquired in the Orbitrap using an automatic gain control target of 4e5, a maximum injection time of 50 ms, and a resolution of 120,000. Internal calibration using lock mass on the m/z 445.12003 siloxane ion was used. Each MS scan was followed by MS–MS fragmentation of the most intense ions for a total cycle time of 3 s (top speed mode). The selected ions were isolated using the quadrupole analyzer in a window of m/z 1.6 and fragmented by higher energy collision-induced dissociation with 35% of collision energy. The resulting fragments were detected by the linear ion trap at a rapid scan rate with an automatic gain control target of 1e4 and a maximum injection time of 50 ms. Dynamic exclusion of previously fragmented peptides was set for 20 s and a tolerance of 10 ppm.
Database Searching
Mascot generic format peak list files were created using Protein Pilot, version 4.5 software (Sciex) for the data obtained with the 5600+ triple TOF and with Proteome Discoverer 2.3 software (Thermo) for the Orbitrap data. The Mascot generic format sample files were then analyzed using Mascot (Matrix Science; version 2.5.1). Mascot was set up to search a contaminant database and UniProtKB Homo sapiens database assuming the digestion enzyme trypsin. Mascot was searched with a fragment ion mass tolerance of 0.60 Da (Orbitrap) or 0.1 Da (5600+) and a parent ion tolerance of 10.0 ppm (Orbitrap) and 0.1 Da (5600+). Carbamidomethyl of cysteine was specified in Mascot as a fixed modification. Deamidation of asparagine and glutamine and oxidation of methionine were specified in Mascot as variable modifications. Two missed cleavages were allowed.
Criteria for Protein Identification
Scaffold (version Scaffold_4.8.7; Proteome Software, Inc) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 8.0% probability to achieve a false discovery rate (FDR) less than 1.0% by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 99.0% probability to achieve an FDR less than 1.0% and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (37). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Data were compared to mock purified fractions obtained from cells expressing an empty TAP tag from the AAVS1 site. Data were further analyzed using the CRAPome online tool and filtered against similar experiments (www.crapome.org). For all samples analyzed, protein identifications, corresponding accession numbers, numbers of spectral counts/distinct peptides, and percentage of protein coverage are listed in Supplemental Excel Tables included in one workbook file.
Antibodies and siRNAs
The following antibodies were used for Western blotting at the indicated dilution: anti-FLAG-horseradish peroxidase conjugate (Sigma; clone M2; 1:10,000 dilution); anti-Brd8 (Bethyl; catalog no.: A300-219A; 1:10,000 dilution); anti-DMAP1 (DNA methyltransferase 1–associated protein 1; Thermo Fisher Scientific; catalog no.: PA1-886; 1:1000 dilution); anti-P400 (Abcam; catalog no.: ab5201; 1:1000 dilution); anti-MRG15 (Active Motif; catalog no.: 39361; 1:1000 dilution); anti-MRGBP (Abnova; catalog no.: H00055257-B01; 1:2000 dilution); anti-MRGX (Abnova; catalog no.: PAB6152; 1:1000 dilution); anti-KAT5–TIP60 (Abcam; catalog no.: ab137518; 1:1000 dilution); anti-ASH1L (Bethyl; catalog no.: A301-749; 1:1000 dilution); anti-KDM5A (Cell signaling; catalog no.: 3876; 1:1000 dilution); anti-Pf1 (Bethyl; catalog no.: A301-647; 1:1000 dilution); anti-PALB2 (Bethyl; catalog no.: A301-246; 1:3000 dilution); anti–A Kinase Anchoring protein 8 (AKAP8) (Abcam; catalog no.: ab72196; 1:2500 dilution); anti-GAPDH (Thermo Fisher Scientific; catalog no.: 39-8600; 1:10,000 dilution); and anti-KAP1 (catalog no.: MAB3662; 1/4000 dilution).
The following siRNAs were used against the indicated proteins: KAT5/Tip60 SMARTpool (Dharmacon; CCACAGAUCACCAUCAAUG, GAACAAGAGUUAUUCCAG, CACAGGAACUCACCACAUU, and GGA CAGCUCUGAUGGAAUA), BRD8 from Sigma (SASI_Hs01_00131635; target sequences start at nucleotide 2223); MRG15 SMARTpool (Dharmacon; GAAGAGCCUUGCUUUAUUA, UAAAGUACCUGGCAA AGAA, GUACCCAGCUACUCUAUAA, and GAUGACUGGGACUUAA UUA); MRGX SMARTpool (Dharmacon; GCACUCAGCUGCUCUAC AA, GAGGAGGCGUUUAAGAAUA, GCAGGGAAAUGUUGAUAAU, and GCUCCAAUGUCCCAGGUUU), MRGBP SMARTpool (Dharmacon; GAAGAACUCCUCAGACUUG, GAACUUCGUCCUUCCAGAA, G AACCGACACUUCCACAUG, and ACAAAGUCCUGACCGCAAA), and siLuciferase (siLuc) as control is used (DOYYA; AACUGACGCGG AAUACUUCGA). Knockdown (KD) efficiency was validated by RT–quantitative PCR (qPCR) with the following primers:
| Targets | Primers forward | Primers reverse |
|---|---|---|
| BRD8 | ATGGTGGGGAGATACAGCAA | AGTATGTGGATCCCCCACAG |
| KAT5/TIP60 | CATCCTCCAGGCAATGAGAT | ACTTGGCCAAAAGACACAGG |
| MRG15 | CACCCATGTCCCAGGTGTAT | GCAAGGCTCTTCTCATCCAG |
| MRGX | GTGTGCTTGCTTGGAGATCA | TCAGGGAAGGTTCTGCAATC |
| MRGBP | GGAGGAGACAGTGGTGTGG | CATGTGGAAGTGTCGGTTCA |
In Vitro Histone Methyltransferase Assay
Purified endogenous ASH1L (or truncated recombinant GST-ASH1L [amino acids 2040–2634]) was first preincubated with recombinant MRG15 purified from baculovirus (or the same amount of BSA) and 0.5 μg of native short oligonucleosomes purified from HeLa cells for 4 h at 30 °C in a 20 μl reaction containing 100 mM Tris–HCl (pH 8.0), 25% glycerol, 0.5 mM EDTA, 5 mM DTT, 5 mM PMSF, and 10 mM MgCl2 for 30 min at 4 °C. Then, 2 μl of 3H S-adenosyl methionine (0.55 μCi/μl) was added, and the mixture was incubated for 3 h at 30 °C. The reaction was then spotted onto p81 filter paper, washed in carbonate buffer, and scintillation counting was used to determine the incorporation.
RNA-Sequencing Analysis
Two days post-KD of MRG proteins, BRD8 or KAT5 in U2OS cells, RNA was extracted with the Monarch Total RNA Miniprep Kit (NEB; catalog no.: T2010), following the manufacturer’s indications. Then, at least 1.5 μg of RNA were depleted of rRNAs using the NEB rRNA-depletion kit (HMR) followed by Illumina library preparation. Finally, complementary DNAs (cDNAs) were sequenced using the NovaSeq6000 S2 for 50M of PE100 reads. Two replicates of each KD were sequenced. Raw sequences and processed data of short read RNA sequencing from U2OS cells were deposited in the Gene Expression Omnibus database under accession number GSE181533.
Gene Expression Analysis
Raw fast5 files were base called using Guppy, v.4.2.2 with the guppy_basecaller command and the dna_r9.4.1_450bps_fast.cfg configuration file and default settings. Barcodes were detected, and reads were separated with the guppy_barcoder using the default settings. The resulting FASTQ reads were aligned to the hg19 human reference with Minimap2 with the following parameters: -aLx splice --cs=long. Raw read counts were obtained with the featureCounts tool from the Subread package, v. 2.0.0, using the exon counting mode (38, 39). EdgeR R-package (version 3.12.1) was then used to normalize the data, to calculate RNA abundance at the gene and transcript levels (as counts per million reads), and perform statistical analysis (40). Briefly, a common biological coefficient of variation and dispersion (variance) were estimated based on a negative binomial distribution model. This estimated dispersion value was incorporated into the final EdgeR analysis for differential gene expression, and the generalized linear model likelihood ratio test was used for statistics, as described in EdgeR user guide. Analysis of variations in gene expression was performed by comparing KDs BRD8, MRGBP, MRG15, MRGX, and KAT5/Tip60 to siControl (siLuc). From all genes showing differential expression, a cutoff of twofold difference was applied to the Log2(fold change) values between the selected sample and control.
mRNA Splice Variant Analysis
Reads were trimmed using fastp, version 0.20.0 (41). Quality check was performed on raw and trimmed data to ensure the quality of the reads using FastQC, version 0.11.8 and MultiQC, version 1.8 (42, 43). The quantification was performed with Kallisto, version 0.46.2 (44). Differential expression analysis was performed in R, version 4.0.0 (GNU) using the DESeq2, version 1.28.1 (Bioconductor) (45, 46). Analysis of variations in splice isoform expression was performed by comparing differential expression at the gene and transcript levels. From all genes showing no differential expression with the selected control (padj ≥ 0.1), those with significant differences in transcript expression (padj ≤ 0.1) were selected. To highlight the genes with high changes in transcript expression compared with gene expression, a cutoff of twofold difference was applied to the Log2(fold change) values between the selected sample and control.
RT–qPCR
siRNAs for KD of BRD8 or MRG15 or MRGBP or MRGX or KAT5/Tip60 were transfected in U2OS cells. siLuc was used as control. About 48 h later, total RNA was extracted with the RNeasy Plus Mini kit (QIAGEN). About 500 ng of RNA was reverse transcribed by oligo-dT and random priming into cDNA with a qScript cDNA SuperMix kit (QuantaBio-VWR), according to the manufacturer’s instructions. Quantification of the amount of cDNA was done with SYBR Green I (Roche) on a LightCycler 480 (Roche) for real-time PCR. The RPLP0 gene was used as a housekeeping internal control for normalization. The error bars represent the range based on independent experiments. The oligonucleotide sequences used for expression analysis by RT–qPCR are listed below:
| Targets | Primers forward | Primers reverse |
|---|---|---|
| RFX8 | GTCATGCTGATGTCATTGCCT | CCAAGAAGTCATGAAGGAGAACA |
| HHIP | GGGCGCCTGGAGAATAAGATA | GGCTTTGAGAATGTGGAGAGC |
| SYTL5 | TATGCAGGGAGTGTCGAGTTG | GTGCCGAGAACATTGACTTGC |
| CD180 | TGAAGGGTCTGAACTTGCTCA | TAGAGGTGTGTAAGGGAGGGG |
| AKT2 | CCGCTACTACGCCATGAAGAT | AACGGGTGCCTGGTGTTC |
| PRRG4 | GCATTTTGGCAGGAATATTCAGC | CCAGCAGCAATTAATCCAGTCAG |
| PCDHB9 | AGGATCTGGGACTAGCAGAGG | CAGCTTCTCTCGGTCCAGTTT |
| RPLP0 | CGACCTGGAAGTCCAACTAC | ATCTGCTGCATCTGCTTG |
Experimental Design and Statistical Rationale
A mock K562 cell line expressing an empty 3xFLAG-2xStrep tag from the AAVS1 locus is used as control. Most purification of tagged proteins shown are from single representative replicate experiments, whereas protein IDs (and loss in mutants) were carefully validated in purified fractions by gels and Western analyses. Wildtype MRG15S purified fractions included are from three biological replicates, whereas the W172A/Y235A double mutants are from two biological replicates. Each KD in U2OS cells was performed in biological duplicates. Control with an siLuc was generated concomitantly to experimental samples. RT–qPCRs to validate KD and RNA sequencing were performed in biological duplicates. Error bars represent the range from two biological replicates. Statistical analyses were performed via two-way ANOVA using Prism, version 7 (GraphPad Software, Inc) followed by Tukey’s test. p Values <0.05 were considered significant.
Recruitment-Activator Assay
This assay is described (47). The reporter human embryonic kidney 293T cell line with a TRE3G-enhanced GFP reporter was a kind gift from Stanley Qi (Stanford University). The reporter cell line was transduced with a lentivirus expressing ABI-dCas9 followed by two rounds of selection under 6 μg/ml blasticidin. These cells were then transduced with a lentivirus expressing EBFP2 and a gRNA targeting seven tetO repeats in the TRE3G promoter (gRNA sequence: GTACGTTCTCTATCACTGATA). Single cell–derived clonal cell lines were generated, and a clone showing robust enhanced GFP induction by a strong transcriptional activator VPR was selected for downstream assays. About 96-well plates were seeded with 3 × 104 cells per well 1 day prior to transfection. About 150 ng of each construct was transfected using polyethyleneimine. Transfected cells were induced 24 h after transfection by treatment with 100 μM abscisic acid. About 48 h after induction, cells were dissociated and resuspended in flow buffer using a liquid handling robot and analyzed by LSRFortessa (BD). Flow cytometry data were analyzed using FlowJo (BD Biosciences) by gating for positive gRNA (EBFP2), then further for construct (TagRFP) expression. At least 25,000 cells were analyzed for each replicate.
Results
Impact of the CHD on the Binding of MRG Proteins to Chromatin
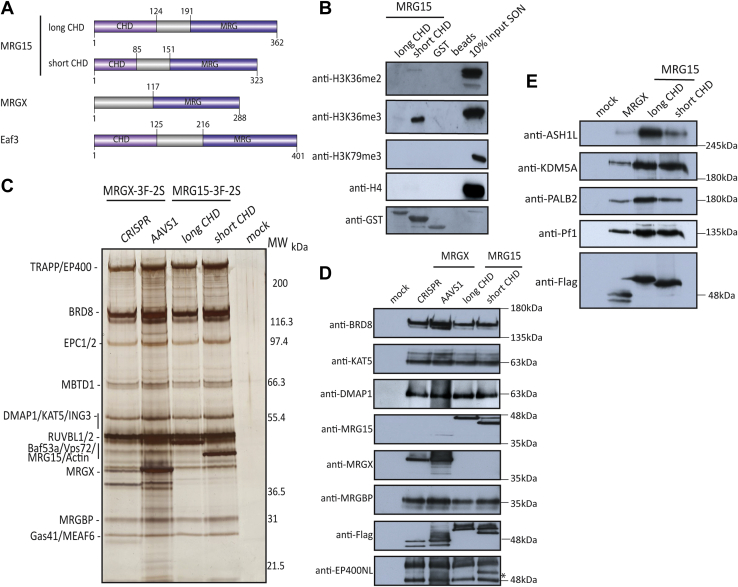
Although MRG15 is highly conserved from yeast to human, it is interesting to observe that two isoforms are produced in mammals through alternative mRNA splicing and that a paralogous protein, MRGX, is produced by another gene. Interestingly, the major difference between these three proteins resides in the CHD. MRG15 isoforms contain either a “long CHD” (Q9UBU8-1 in UniProtKB) with an extra 39 amino acids insert or a “short CHD” (Q9UBU8-2 in UniProtKB), which is considered canonical. In contrast, MRGX lacks the CHD region at its N terminus (Fig. 1A). Yeast Eaf3 in comparison also has a “long CHD,” but the extra insert is located on the opposite side of the hydrophobic cage that recognizes H3K36me3 compared with MRG15 (supplemental Fig. S1A). The proteins are characterized by a ∼175 amino acid MRG domain at their C terminus, important for their nonhistone protein–protein interactions (20). Eaf3 CHD has been shown to bind H3K36me2/me3, which is required for suppression of spurious intragenic transcription by the RPD3S histone deacetylase complex (24, 48, 49). While Eaf3 CHD has been argued to also bind H3K4me3 in vitro, MRG15 short CHD does not show any interaction with this mark (50). We then wondered if the insert in MRG15 long CHD isoform could change its binding properties. To test that, we performed GST pull-down experiments with purified native chromatin and recombinant proteins containing the short or long CHD of MRG15. The short CHD showed great specificity for H3K36me3, compared with H3K36me2, whereas the long CHD seem to have weaker affinity (Fig. 1B).
Fig. 1.
The interactome of MRG proteins highlights the association with several distinct complexes.A, schematic representation of MRG15 spliced variants, MRGX, and the yeast Eaf3 functional domains. B, short chromodomain MRG15 binds preferentially H3K36me3 in vitro. Recombinant GST-tagged MRG15 isoforms or GST or beads were incubated with LON followed by washes and Western blotting. Anti-H4 and anti-H3K79me3 are used as control. C, 3xFLAG-2xStrep elutions of indicated purifications from K562 cells were migrated on 4 to 12% SDS-PAGE and analyzed by silver staining. MRGX and MRG15 spliced variants bind the acetyltransferase NuA4/TIP60 complex. Subunits were identified by mass spectrometry (supplemental Table S1). Mock is a purification from cells expressing an empty tag. D and E, purified complexes from (C) were analyzed by Western blot with the indicated antibodies to confirm the equivalent presence of known subunits of the NuA4/TIP60 complex (D) and the other known interactors of MRG proteins (E) (∗nonspecific band). GST, glutathione-S-transferase; H3K36me, methylation of lysine 36 on histone H3; MRG15, MORF-related gene on chromosome 15; NuA4, nucleosome acetyltransferase of H4; TIP60, Tat interactive protein 60 kDa.
To further examine the potential impact of the CHD size and or its absence on chromatin binding in vivo, we generated isogenic K562 cell lines with a single copy of the epitope-tagged (3xFLAG-2xStrep) MRG15 isoforms or MRGX cDNA inserted at the AAVS1 safe-harbor locus, as described previously (supplemental Fig. S1B) (35). We then analyzed their genome-wide locations using chromatin immunoprecipitation (ChIP)-Seq experiments with an anti-FLAG antibody. MRG15-S, MRG15-L, and MRGX seem they share the vast majority (>70%) of their genomic targets, independently of the CHD status (supplemental Fig. S1C). On the other hand, MRG15-L had a relatively much smaller number of bound regions that could be mapped, possibly reflecting a looser interaction with chromatin. It remains to be seen if the small proportion of genomic regions uniquely bound by MRGX or MRG15 are functionally significant.
MRG Proteins Are Associated With Several Distinct Protein Complexes But Are Interchangeable
As mentioned previously, several associated factors have been identified for mammalian canonical MRG15 (short CHD/MRG15-S), often using methods like transient or stable overexpression and simple immunoprecipitations. Thus, we wanted to get a full picture of the associated complexes in the most native conditions using our validated approaches. At the same time, we investigated if MRG15-L, MRG15-S, and MRGX have different interactomes. Even though the AAVS1 system usually allows near physiological level of expression (within 2–2.5-fold (35)), it does not recapitulate native regulation by endogenous promoters and 5’/3′ UTRs. We were able to tag the endogenous MRGX gene using an improved CRISPR–Cas9 genome editing method (36). We used a gRNA targeting the stop codon of MRGX to introduce the 3xFLAG-2xStrep tag (supplemental Fig. S1D). Comparing endogenous versus AAVS1-mediated expression levels indicates significantly lower endogenous expression but similar to MRG15 expression from AAVS1 (supplemental Fig. S1B).
We then performed tandem affinity purifications with native elution at both steps, as previously described (35, 51). The final elutions with biotin were then analyzed by silver staining, MS, and Western blotting (Fig. 1, C–E and supplemental Table S1). In agreement with the highly conserved MRG domain between MRG15 and MRGX, we observed very similar interactomes for all MRG proteins. We could detect the full set of subunits for the NuA4/TIP60 acetyltransferase complex as well as the Sin3B deacetylase complex (homologous to yeast RPD3S) that includes the KDM5A H3K4 demethylase. It is interesting to point out that MRG15 and MRGX proteins seem largely mutually exclusive in their associations since we could detect only one or two spectral counts specific for MRGX in MRG15 preps and vice versa (Fig. 1D and supplemental Table S1). The data are also arguing that the reported ability of the MRG domain to homodimerize in an acetylation-regulated manner is not a major event (26). MRG15-S and MRGX have also been previously linked to PALB2–BRCA2, playing a crucial function in DNA repair by homologous recombination (27, 29, 30), and we confirmed that (Fig. 1E and supplemental Table S1). This interaction seems stable and not modulated by a cellular DNA damage response since we obtained very similar results purifying MRG15-S from cells treated with DNA double-strand break–inducing agent neocarzinostatin, for PALB2–BRCA2 or any other interacting partners (supplemental Fig. S2A and supplemental Table S1), in agreement with the proposed role of MRG15–PALB2–BRCA2 constitutive association in protecting undamaged chromatin on active genes from replication-associated stress (30). MRFAP1 is also an interesting interactor of both MRG15 spliced isoforms and MRGX as it was known to form a complex with MRG15-S, which is regulated by NEDDylation (52). Finally, a particularly interesting shared interaction is with the H3K36 methyltransferase ASH1L (Fig. 1E and supplemental Table S1).
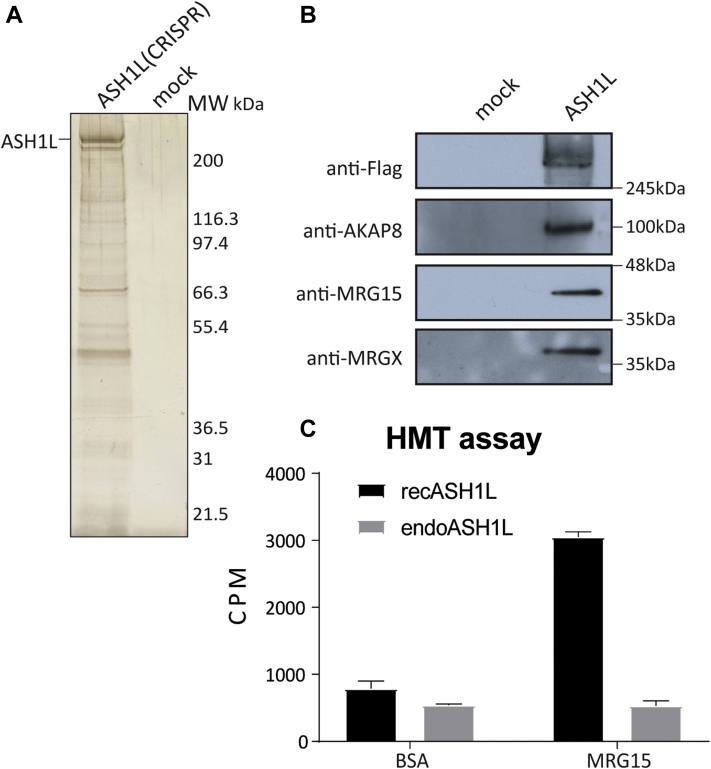
Endogenous ASH1L Is Associated With MRG Proteins
ASH1L is a member of the trithorax group proteins involved in activation of Hox genes in Drosophila. Its methyltransferase activity is crucial for H3K36me1/me2 maintenance, thereby leading to mixed lineage leukemia recruitment on its specific promoter targets (53, 54). The catalytic activity of ASH1L seems also important for DNA repair via nucleotide excision (55). Moreover, previous studies have shown that MRG15 and MRGX enhance the methyltransferase activity of a truncated version of ASH1L (56, 57). Indeed, this binding can affect the autoinhibitory loop of ASH1L (58, 59). Importantly, this stimulatory effect is therefore independent of the H3K36me-binding CHD of MRG15. To confirm this association and biochemical studies in the physiological context, we were able to tag and purify endogenous ASH1L using the CRISPR–Cas9 system in K562 cells (Fig. 2A and supplemental Fig. S1E). We then analyzed by Western blots and MS the ASH1L interactome. We confirmed association of MRG15 and MRGX as well as Nurf55/RBBP4, an already known factor link to ASH1L (Fig. 2B and supplemental Table S2). Interestingly, the AKAP8 and multiple RNA-processing factors were also found associated with ASH1L, suggesting a potential function in alternative splicing, as it was already described for AKAP8 (60).
Fig. 2.
Endogenous ASH1L is stably associated with MRG proteins.A, purification of endogenous ASH1L from K562 cells modified by CRISPR–Cas9 system to add 3xFLAG 2xStrep. The elution was analyzed on 4 to 12% SDS-PAGE followed by silver staining. B, purified ASH1L from (A) was analyzed by Western blot with the indicated antibodies to confirm the interaction with ASH1L and its interactors such as MRG proteins and AKAP8. C, in vitro histone methyltransferase assay performed with recombinant ASH1L (amino acids 2040–2634)-pGEX6P2 (recAHS1L) and strep elution from endogenous ASH1L (endoASH1L). The graph shows the scintillation counts of the liquid assays with 3H SAM with native short oligonucleosome (SON). An empty vector pGEX6P2 and a mock cell line were used as control and subtracted from final counts. Error bars represent the range of two technical replicates. ASH1L, absent, small, or homeotic discs 1-like; Cas9, CRISPR-associated protein 9; MRG, MORF-related gene.
To confirm the role of MRG15 in stimulating ASH1L methyltransferase activity, we performed an in vitro histone methyltransferase assay to measure its activity. We used a recombinant truncated version of ASH1L (amino acids 2040–2684), preincubated with recombinant MRG15 or BSA. As expected, MRG15 greatly enhanced the methyltransferase activity of ASH1L (Fig. 2C). In parallel, the methyltransferase activity of the purified endogenous ASH1L was not affected by the preincubation with recombinant MRG15. This strongly suggests that the vast majority of endogenous ASH1L is stably bound to an MRG protein in vivo, being fully active and nonresponsive to addition of exogenous MRG15.
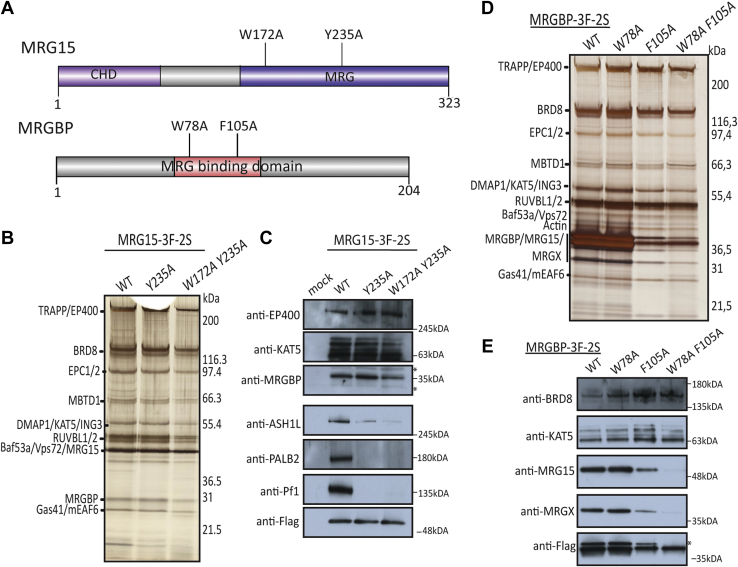
Genetic Mutations to Modulate MRG15 Association With Distinct Complexes
Previous work has been done to characterize the structure of the MRG domain as an interface for protein–protein interactions (20, 58, 59, 61, 62). Based on this information, we investigated if we could manipulate the association of MRG proteins with distinct complexes in native conditions. We generated K562 cell lines expressing tagged MRG15 carrying the W172A and Y235A substitutions in its MRG domain, which have been shown to affect some interactions (Pf1, MRGBP, and ASH1L) (20) in binary recombinant experiments (Fig. 3A) (20, 59, 61). After purification and analysis by silver-stained gel, Western blot, and MS, we can conclude that these mutations specifically disrupt MRG15 association with the Sin3B deacetylase complex, BRCA2-PALB2, MRFAP1, and ASH1L (Fig. 3, B and C, supplemental Fig. S2B and supplemental Table S3). Strikingly, MRG15 association with the NuA4/TIP60 complex is still very stable based on Western signals, maybe a slight decrease in recovery based on spectral counts. This clearly indicates that distinct molecular features of the MRG domain allow the multispecificity of protein interactions and therefore associations to complexes. These mutants could help address the functional implication of MRG protein association with these partners versus its presence in NuA4/TIP60. To see if it would be possible to address the opposite question, we again used published structural information (20) and introduced point mutations in MRGBP, a subunit of NuA4/TIP60 directly associated with MRG15/X in the complex. We generated K562 cell lines expressing tagged wildtype and mutant MRGBP carrying the W78A F105A substitutions, mutations known to affect MRG15 binding in binary recombinant experiments (Fig. 3A) (20). Analysis of the purified fractions showed that the double mutant disrupted the association of MRG15 and MRGX with the native NuA4/TIP60 complex without affecting other components (Fig. 3, D and E). Together, these MRG mutants have the potential to help us understand the specific function of MRG protein in distinct complexes. For example, as mentioned before, MRG15 is known to have a function in the repair of DNA double-strand breaks by homologous recombination, certainly in part through its association with PALB2–BRCA2 (27, 29, 30, 63). Furthermore, the NuA4–TIP60 complex also plays a role in DNA repair, promoting homologous recombination (64, 65, 66). Thus, taking advantage of its mutants, it will be possible to characterize the function of MRG15 in DNA repair, distinguishing its role from within the NuA4/TIP60 complex versus its association with PALB2–BRCA2.
Fig. 3.
Structure-based mutations can differentially affect specific MRG15-containing complexes.A, schematic representation of the W172A Y235A mutations in MRG15 predicted to affect MRGBP/ASH1L/Pf1 interactions and the W78A F105A mutations in MRGBP predicted to affect MRG15 interaction. B, 3xFLAG-2xStrep elutions of indicated MRG15 purifications from K562 cells were migrated on 4 to 12% SDS-PAGE and analyzed by silver staining. Strep elutions were analyzed by mass spectrometry in total spectrum count (supplemental Table S3). See also biological replicate in supplemental Fig. S2B. C, purified complexes from (B) were analyzed by Western blot with the indicated antibodies (∗nonspecific band). D, 3xFLAG-2xStrep elutions of indicated MRG15 purifications from K562 cells were migrated on 4 to 12% SDS-PAGE and analyzed by silver staining. E, purified complexes from (D) were analyzed by Western blot with the indicated antibodies (∗nonspecific band). ASH1L, absent, small, or homeotic discs 1-like; MRG15, MORF-related gene on chromosome 15; MRGBP, MRG-binding protein.
The MRG Interactomes Identify a New Tetrameric Protein Complex
When we analyzed the interactomes of MRG15-S/L and MRGX, we noticed by gel, Western, and MS the over-representation of specific subunits of NuA4–TIP60 (67). This was the case for MRGBP and the BRD-containing protein BRD8. This was confirmed by exponentially modified protein abundance index (67), similar to RUVBL1/2 subunits, which are known to form a hexameric ring in protein complexes (supplemental Fig. S3, A–C). That drew a parallel with a similar observation we made with yeast NuA4 components a few years ago (25). At that time, we noticed that MRG15 and MRGBP homologs, Eaf3 and Eaf7, were preferentially enriched when used as bait, along the protein Eaf5, which has no clear homolog in higher eukaryotes. We went on to characterize a yeast trimeric complex of Eaf5–Eaf7–Eaf3 that exists independently of NuA4, binds the coding region of active genes, and interacts with the elongating RNAPII (25). TINTIN is involved in transcription elongation presumably by facilitating disruption and recycling of nucleosomes from the front to the back of the elongating polymerase, also helping to suppress spurious transcription (68).
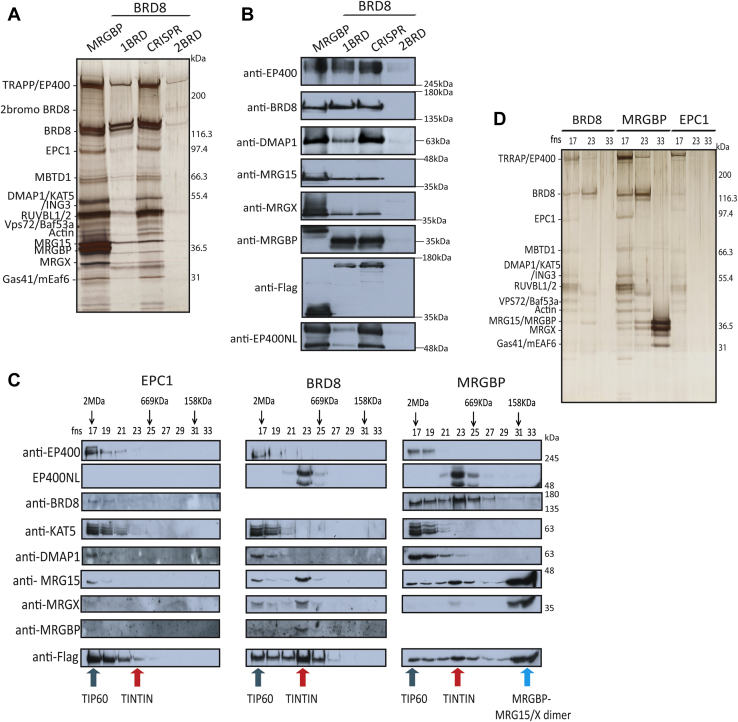
Since our purification of MRGBP, the human homolog of Eaf7, also yielded over over-representation of MRG15/X and BRD8 compared with the rest of NuA4/TIP60 subunits (Figs. 3D and 4A), we wondered if MRG15 in higher eukaryotes could also form an independent functional complex like yeast TINTIN, in which BRD8 would be the functional homolog of yeast Eaf5. To test this hypothesis, we first used Sf9 cells and coinfected them with baculovirus vectors coding for BRD8 (1BRD), MRG15 (S), and hemagglutinin (HA)-tagged MRGBP. Fractionation of extracts with anti-HA beads and HA peptide elution clearly showed copurification of BRD8 and MRG15 with MRGBP (supplemental Fig. S2C). We then generated K562 cell lines using the AAVS1 system to purify BRD8 in comparison to MRGBP (supplemental Fig. S4A). Interestingly, BRD8 possesses two isoforms produced by alternative mRNA splicing, a shorter version with one H4ac-binding BRD (Q9H0E9-2 to -4) and a longer version with two BRDs (Q9H0E9-1). Using the CRISPR–Cas9 system, we were able to tag both endogenous isoforms. We designed gRNAs to target the C-terminal region of the 2BRD isoform or the N-terminal of the BRD8 gene, tagging both isoforms (supplemental Fig. S4B). Although clonal selection and sequencing confirm the efficient introduction of our tag in the C terminus of the long isoform, we were not able to clearly detect expression of this specific variant compared with the N-terminal tagging (supplemental Fig. S4C, maybe a very faint signal). Strangely, even expressing the 2BRD isoform from the AAVS1 locus yielded very low signal, arguing that this isoform is unstable in K562 cells. Data in public databases confirm that expression of the 2BRD isoform is not really detected in most tissues except testis, whereas the 1BRD isoform is ubiquitously expressed (e.g., GTExportal.org).
Fig. 4.
Identification of the TINTIN complex in human cells, composed of BRD8–MRGBP–MRG15/X and new subunit EP400NL.A, purification of native NuA4/TIP60 complex from K562 cells. Strep elutions of indicated purifications were analyzed on 4 to 12% SDS-PAGE followed by silver staining. MRGBP and spliced variants of BRD8 (1BRD, one bromodomain, 2BRD, two bromodomains, and CRISPR for N-terminal-tagged BRD8) are linked to NuA4/TIP60 complex. B, Western blots of purified complexes in (A) confirmed the association with the NuA4/TIP60 complex and the new subunit EP400NL. C, Western blots of gel filtration fractions 17 to 33 (calibrated Superose 6, FPLC) with FLAG elutions from EPC1, BRD8, and MRGBP AAVS1 K562 cells (EPC1 purified fraction is used as a control reflecting the native NuA4–TIP60 complex (64)). Arrows point to specific annotated complexes with different elution profiles based on size. D, 4 to 12% SDS-PAGE followed by silver staining of annotated fractions from (C) showing the NuA4/TIP60 complex (17), the TINTIN complex (23), and a dimer MRGBP–MRG15 or MRGX (33). See also supplemental Table S5 for mass spectrometry results. EP400NL, EP400 N-terminal like; MRGBP, MRG-binding protein; NuA4, nucleosome acetyltransferase of H4; TINTIN, Trimer Independent of NuA4 for Transcription Interactions with Nucleosomes; TIP60, Tat interactive protein 60 kDa.
We tandem affinity purified MRGBP-3xFLAG-2xStrep and BRD8-3xFLAG-2xStrep expressed from AAVS1 or endogenously tagged and analyzed the fractions by silver staining, MS, and Western blots with the annotated antibodies (Fig. 4, A and B and supplemental Table S4). Surprisingly, in not only those purifications, but also in our previous MRG15/X purifications, we detected an uncharacterized factor, called EP400NL (Figs. 1D and 4B, supplemental Tables S1, S3 and S4). EP400NL was first described as a pseudogene and is homologous to a short region at the 5′ of the EP400 gene (supplemental Fig. S4D). It can produce multiple spliced variants, but only two of them (Q6ZTU2-5 and Q6ZTU2-6) with a specific C-terminal sequence were found associated with MRG proteins and BRD8 by MS results (supplemental Tables S1, S3 and S4). Thus, EP400NL proteins share a similar N-terminal sequence with the very large EP400 protein, an essential ATP-dependent remodeler and scaffolding subunit of the NuA4/TIP60 complex (supplemental Fig. S4D) (35, 69).
EP400NL Creates a Human TINTIN Complex by Competing With EP400 for Association to Trimeric BRD8–MRGBP–MRG15/X
To prove the existence of this functional module independent of the NuA4/TIP60 complex, we purified BRD8 and MRGBP as aforementioned, but in parallel to the NuA4/TIP60-specific subunit EPC1 (64). We then loaded the tandem affinity–purified fractions on a Superose 6 size exclusion column to visualize distinct assemblies. Fractions from the gel filtration were analyzed by Western blot, silver-stained gel, and MS (Fig. 4, C and D and supplemental Table S5). While the purified EPC1 fraction eluted early as expected with the large size of the NuA4/TIP60 (∼2 MDa), BRD8 eluted as two populations with the strongest signal associated with a smaller protein complex (∼700–800 KDa) (Fig. 4, C and D). This smaller population also coeluted with the strongest signal of MRGBP and MRG15/X proteins. Importantly, in contrast to these proteins, EP400NL was only detected in the smaller size fraction, with no signal eluting with NuA4/TIP60. These results demonstrate the existence of a human TINTIN complex formed by EP400NL–BRD8–MRGBP–MRG15/X. Gel filtration of the purified MRGBP fraction gave similar results but with an important difference. In this case, a third much smaller population was detected below 150 kDa and containing likely only MRGBP and MRG15/X (Fig. 4, C and D and supplemental Table S5). Altogether, these data support the idea that the human TINTIN exists independently of the NuA4/TIP60 complex with potential functions in transcription as it was observed in yeast.
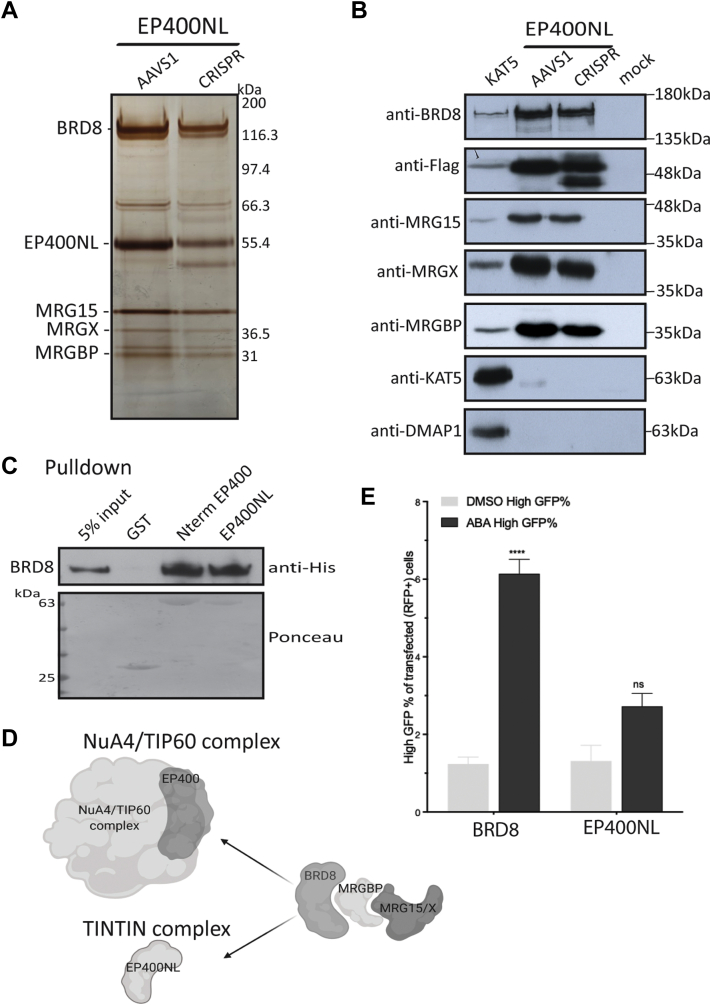
Since EP400NL was previously described as a pseudogene, we generated K562 cell lines using both AAVS1 and CRISPR–Cas9 to, on one hand, tag the specific isoform Q6ZTU2-5 and, on the other hand, the endogenous gene at the C terminus to tag the two specific isoforms found by MS (supplemental Fig. S4, E and F). We then used those cell lines to demonstrate physiological expression of the endogenous EP400NL gene and characterize the encoded protein interactome. Analysis of the tandem affinity–purified fractions confirmed expression of EP400NL by the endogenous gene and showed strong, apparently stoichiometric, association with BRD8, MRGBP, and MRG15/X (Fig. 5, A and B and supplemental Table S6) (note that MRGBP does not stain efficiently with silver, whereas showing strong signal by MS) (see also exponentially modified protein abundance index analysis in supplemental Fig. S3D). Importantly, EP400NL did not purify with any other known components of NuA4/TIP60. Interestingly, an unusually high number of peptides from mRNA processing factors were found, suggesting a potential function of human TINTIN in mRNA processing or at least a close proximity during transcription elongation (supplemental Table S6).
Fig. 5.
Endogenous EP400NL isolates TINTIN from NuA4–TIP60 through an interaction with BRD8.A, 3xFLAG-2xStrep elutions of Q6ZTU2-5 (AAVS1) or endogenous (CRISPR) EP400NL purified from K562 cells and analyzed on gel by silver staining. B, purified complexes from (A) were analyzed by Western blot with the indicated antibodies to confirm the association of EP400NL with BRD8, MRGBP, and MRG15/X representing the human TINTIN complex. A KAT5/Tip60 purified fraction is used as a comparison for the NuA4/TIP60 complex (64). C, BRD8 directly interacts with EP400NL and the homologous N-terminal region of EP400 in vitro. Recombinant His-tagged BRD8 was incubated with the indicated GST fusion proteins on beads followed by washes and Western blotting. D, schematic representation of the distinct associations of BRD8–MRGBP–MRG15/X with EP400 to form NuA4/TIP60 complex or EP400NL to form an independent TINTIN complex. E, EP400NL is a weak transcriptional activator when artificially targeted to a promoter, in contrast to BRD8, which can be linked to the known coactivator function of NuA4/TIP60. Recruitment–activator reporter assay was quantified by flow cytometry analysis (47). It shows the percent of cells transfected with EP400NL and BRD8 that express high level of GFP upon ABA-mediated targeting of the proteins to the reporter. At least 25,000 cells were analyzed for each replicate (mean ± SEM, n = 3). DMSO is used as a negative control. Statistical analyses were performed by two-way ANOVA test followed by Tukey’s test. ∗∗∗∗p < 0.0001. BRD8, bromodomain 8; DMSO, dimethyl sulfoxide; EP400NL, EP400-terminal like; GST, glutathione-S-transferase; MRGBP, MRG-binding protein; NuA4, nucleosome acetyltransferase of H4; TINTIN, Trimer Independent of NuA4 for Transcription Interactions with Nucleosomes; TIP60, Tat interactive protein 60 kDa.
If EP400NL competes with EP400 to keep human TINTIN separated from the NuA4/TIP60 complex, it is important to identify its binding target. Direct physical interactions between BRD8 and MRGBP as well as MRGBP and MRG15 have been reported (20, 70). We have shown that Eaf5 is responsible for anchoring TINTIN to the yeast NuA4 complex, where it binds the first 85 amino acids of Eaf1, the scaffolding subunit homologous to EP400 (25, 71, 72). Thus, if BRD8 is really the human functional homolog of yeast Eaf5, it should be responsible to bridge MRGBP–MRG15/X to EP400 in NuA4/TIP60 or EP400NL in TINTIN. To answer this question, we performed a pull-down assay with full-length recombinant BRD8 and GST fusions with homologous N-terminal regions of EP400 (amino acids 194–446) and EP400NL (amino acids 51–297) (supplemental Fig. S4D). BRD8 was very efficiently pulled down by both fusions but not GST control, thereby confirming our hypothesis (Fig. 5C). Thus, BRD8 acts like the bridge that can anchor the MRG proteins to EP400 in the NuA4/TIP60 complex or to EP400NL to form the independent TINTIN complex. Thus, since the binding levels of BRD8 to EP400 N terminus or EP400NL seem similar in vitro (Fig. 5C), relative protein abundance of EP400/EP400NL and BRD8 may be the determinants for the ratio of independent TINTIN complex in the cell (Fig. 5D). Analysis of public databases indicates that EP400NL mRNA is ubiquitously expressed, like EP400, but threefold less in average (e.g., GTExportal.org). To characterize a specific role of EP400NL/TINTIN in transcription, we used a recently described dCas9-based recruitment–activator reporter assay (47). This system measures transcription activation upon ABA-induced recruitment of factors at a promoter controlling the expression of GFP. Interestingly, EP400NL showed weak activator activity in this assay (Fig. 5E). In contrast, BRD8 showed significant transcription activation, likely in part because of the known coactivator function of NuA4/TIP60. This result suggests distinct functions of TINTIN and NuA4/TIP60 in the transcription process. As NuA4/TIP60 is known to function at gene promoters/start sites to favor transcription initiation (readout of the assay in Fig. 5E), the weak effect of EP400NL may reflect a more important role on the body of genes to favor elongation, like we suggested for yeast TINTIN (25).
The Human TINTIN Complex Regulates Both Transcription and Transcript Isoforms of Specific Genes
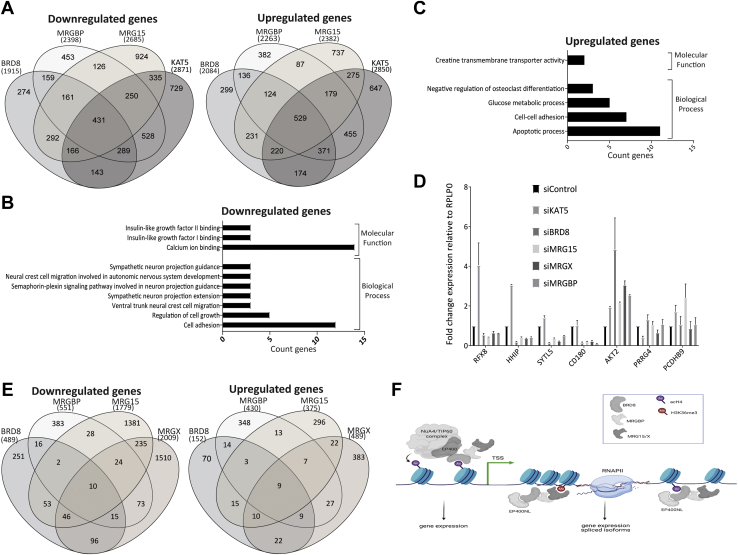
The yeast TINTIN was shown to interact with the elongating RNAPII and to be involved in recycling disrupted nucleosomes in its wake to suppress spurious transcription (25). To test if human TINTIN also has a role in transcription, we performed siRNA-mediated KDs of TINTIN components in comparison to KAT5/Tip60 KD, in an attempt to distinguish functions associated with NuA4/TIP60 versus independent TINTIN (supplemental Fig. S5, A and B). RNAs were extracted 48 h post-KDs, and mRNA sequencing was performed from samples depleted either for BRD8, MRGBP, MRG15, or KAT5/Tip60. About 431 common genes were found downregulated in all KDs compared with siControl, whereas 529 common genes were upregulated (Fig. 6A). These likely reflect the function of the NuA4–TIP60 complex. Because of the mutual exclusion of MRG15 and MRGX, we also compared KD of MRGX with the other KDs and found over 500 common genes downregulated or upregulated (supplemental Fig. S5C). Thus, some specific genes seem differently affected by the MRGX-containing NuA4/TIP60 complex versus the MRG15-containing one (1263 genes are commonly downregulated by MRG15/X KDs and 869 are upregulated, leaving two sets of nearly 2900 genes specifically affected by each one independently). Furthermore, when we excluded specific targets affected by the KD of KAT5/Tip60, we observed 161 downregulated genes and 124 upregulated genes commonly affected by KDs of TINTIN members BRD8, MRGBP, and MRG15 (Fig. 6A). These numbers change to 253 downregulated and 142 upregulated when MRG15 KD is replaced by MRGX KD (supplemental Fig. S5C). Gene Ontology term enrichment analyses were performed on those gene sets in order to identify specific biological processes and molecular function pathways in which TINTIN could be implicated (Fig. 6, B and C and supplemental Fig. S5D). We also used RT–qPCR to validate some targets specifically regulated by the TINTIN complex but not the NuA4/TIP60 complex, such as RFX8, HHIP, SYTL5, CD180, which are downregulated by the KDs, and AKT2, which is upregulated. PRRG4 and PCDHB9 are used as control, where all KDs were similar to siControl (Fig. 6D). Interestingly, our ChIP-Seq data (supplemental Fig. S1C), while in a different cell line, showed binding of MRG15/X to the AKT2 gene.
Fig. 6.
Human TINTIN is implicated in the expression of specific genes.A, Venn diagrams showing that 161 genes are commonly downregulated (left) and 124 are upregulated in U2OS cells depleted (siRNA-mediated KDs) of BRD8, MRGBP, and MRG15 but not in cells depleted of Tip60/KAT5. Genes showing high changes in expression, using a cutoff of twofold difference (Log2 [fold change values]), were tabulated between the selected KDs and control (siLuc). B and C, Gene Ontology analysis for downregulated (B) and upregulated genes (C) showing a significant enrichment (p < 0.01) using DAVID 6.8. D, RT–qPCR to validate target genes of TINTIN. RFX8, HHIP, SYTL5, and CD180 are found downregulated, and AKT2 is upregulated when TINTIN subunits are depleted. PRRG4 and PCDHB9 are used as control. Values represent means relative to control (siLuc). Error bars represent the range from two biological replicates. E, Venn diagrams of downregulated (left) and upregulated (right) transcripts (versus genes) specifically regulated by BRD8, MRGBP, MRG15, or MRGX after exclusion of transcripts regulated by KAT5/Tip60, using a cutoff of twofold difference. siLuc was used as control. Two replicates of each KD were performed in U2OS cells. F, model of BRD8–MRGBP–MRG15/X function during transcription. BRD8 brings MRG proteins into the acetyltransferase NuA4/TIP60 complex because of its direct interaction with EP400 N-terminal region. This association is important for expression of specific gene targets. In addition, these subunits form a new independent functional module, called TINTIN, when BRD8 interacts instead with a new factor, EP400NL. TINTIN is also important for regulation of specific genes, maybe during the transcription elongation stage. BRD8, bromodomain 8; EP400NL, EP400 N-terminal like; KAT5, lysine acetyltransferase 5; KD, knockdown; MRG15, MORF-related gene on chromosome 15; MRGBP, MRG-binding protein; NuA4, nucleosome acetyltransferase of H4; qPCR, quantitative PCR; siLuc, siLuciferase; TINTIN, Trimer Independent of NuA4 for Transcription Interactions with Nucleosomes; TIP60, Tat interactive protein 60 kDa; U2OS, human osteosarcoma epithelial cell.
MRG15, its yeast homolog Eaf3, and MRGBP having already been linked to mRNA splicing processes (16, 17, 18, 73), we determined if the ratio of alternative transcripts per gene were changed by KDs of TINTIN components. We first compared differential transcript expressions in KDs of BRD8, MRGBP, MRG15/X, and KAT5/Tip60 to siControl. Then, we again excluded the transcripts that were significantly changed in KD KAT5–TIP60 (padj ≤0.1). The components of the TINTIN complex seem to commonly affect the regulation of only a few alternative transcripts (12 downregulated and 12 upregulated for TINTIN-MRG15 versus 25 down and 18 up for TINTIN-MRGX), even if each KD by itself shows a defect in the expression of much higher numbers of transcripts (Fig. 6E). Overall, these data demonstrate a significant function of the human TINTIN complex, independently of the NuA4/TIP60, in transcription regulation by modulating the expression of specific genes (Fig. 6F). We also showed that BRD8 and MRG proteins play also likely important roles in the regulation of some genes from within the NuA4/TIP60 acetyltransferase/remodeling complex or through colocalization on the same target of NuA4/TIP60 at the promoter and TINTIN on the coding region, as we have described in yeast (Fig. 6F) (25).
Discussion
In this work, we characterized the interactomes of human MRG domain–containing proteins MRG15 and MRGX, factors that have been implicated in several distinct nuclear processes through association with diverse proteins. These experiments were done as to reflect the most native interactions, avoiding overexpression and when possible, using endogenous proteins as bait. We observed that MRG15 isoforms with long or short CHD and MRGX share the same interactome, such as stable association with the NuA4/TIP60 histone acetyltransferase complex and the Sin3B histone deacetylase/demethylase complex. Interestingly, MRG15 and MRGX are mutually exclusive in their associated complexes. Since MRGX does not possess the H3K36me3-binding CHD, it will be important to characterize the impact of this difference on the function of their partners. MRG15 knockout is embryonic lethal in mice, whereas MRGX knockout does not lead to any obvious phenotype in development or cell proliferation, which may be in part because of some unidirectional compensation mechanism (19, 74, 75).
Interestingly, our data show that MRGX and MRG15 are stably associated with the endogenous H3K36me1/me2 methyltransferase ASH1L, in an apparent stoichiometric ratio since its activity cannot be stimulated in vitro by addition of exogenous MRG15 to relieve an autoinhibitory loop (Fig. 2) (56, 57, 58, 59). It will be interesting to study the functional impact of the MRG15 mutations that disrupt its association with endogenous ASH1L in vivo, through genome-wide location analysis of ASH1L and H3K36me2. The same mutants also disrupt association with the Sin3B complex. It was shown in yeast that Eaf3 plays a crucial role in the function of the homologous RPD3S complex to deacetylate nucleosomes in the wake of the elongating polymerase to suppress spurious transcription (24). A related function was proposed for the mammalian Sin3B complex (22, 76). Thus, it will be interesting to do transcriptomics to analyze if spurious transcription, that is, cryptic initiation, antisense transcription, is detected with the MRG15 mutants. Since Sin3B also contains the KDM5 H3K4me3 demethylase that blocks the spreading of this mark from the transcription start site region to the body of the genes, it will also be important to verify the impact of the MRG15 mutant on the localization of this histone mark (76).
Most importantly, this study allowed the identification and characterization of a new functional MRG-containing complex, human TINTIN. We discovered a new partner of MRG15, EP400NL, previously dismissed as a probable pseudogene. EP400NL is key to create a BRD8–MRGBP–MRG15/X-containing TINTIN complex physically independent of NuA4/TIP60. EP400NL is homologous to the N-terminal region of NuA4/TIP60 scaffold subunit EP400, the interface on which the BRD8–MRGBP–MRG15/X trimer is normally anchored to the complex through BRD8. EP400NL can bind BRD8 presumably as efficiently as EP400 N terminus and by doing so blocks the trimer from associating with NuA4/TIP60, creating an independent TINTIN complex. Interestingly, the mouse and human EP400 genes are also known to produce a short splice variant encoding a truncated N-terminal protein corresponding to the same region where BRD8 binds, homologous to EP400NL (National Center for Biotechnology Information). This short splice variant of EP400 could have the same function as EP400NL to produce an NuA4/TIP60-independent TINTIN complex. Importantly, these proteins lack the HSA and SANT domains of EP400 that are required to assemble the NuA4/TIP60 complex (71, 72).
Since the yeast TINTIN complex is found on the coding region of active genes and plays a role during transcription elongation, we investigated if human TINTIN also affects gene expression. Indeed, several genes are deregulated in the absence of TINTIN components, independently of NuA4/TIP60 (Fig. 6). Further work will be necessary to better understand the function of TINTIN in mammals. EP400NL is likely an important target to use but, as mentioned previously, EP400 short isoforms produced by alternative splicing may play a redundant role. Mapping the precise region of interaction between BRD8 and EP400NL/EP400 may be useful to design better tools to study TINTIN-specific function.
Although MRG15 and MRGBP have been proposed to regulate mRNA splicing (16, 17, 73), we could not find a significant number of alternate transcripts specifically affected by the human TINTIN complex. Our MS data also did not detect previously identified partners of MRG15 (PTB) and MRGBP (VEZF) for such function. It is possible that these interactions mostly occur on chromatin, making them difficult to detect in extracts. Independent KDs of MRG15, MRGX, and MRGBP do show a larger number of transcripts being misregulated (Fig. 6E). This may be linked instead to the function of the small but abundant MRGBP–MRG15/X dimer that our gel filtration experiment identified (Fig. 4).
In conclusion, this study presented a highly confident native stable interactome of MRG proteins and uncovered a new mammalian complex implicated in gene regulation. It highlights the multispecificity of the MRG domain for protein–protein interactions and associations with distinct protein complexes implicated in several chromatin-based nuclear processes. The presence of H4ac-binding BRD and H3K36me3-binding CHD in TINTIN also underlines again the crosstalk between transcription and chromatin modifications.
Data Availability
All MS files generated as part of this study were deposited at MassIVE (http://massive.ucsd.edu). The MassIVE ID is MSV000089533, and the MassIVE link for download is: http://massive.ucsd.edu/ProteoSAFe/status.jsp?task=69398380312a47a79e171cd742ddb03e.
ChIP-Seq data from K562 cells and RNA sequencing from U2OS cells were deposited in the Gene Expression Omnibus database under accession number GSE181533.
Supplemental data
This article contains supplemental data (77, 78).
Conflict of interest
The authors declare no competing interests.
Acknowledgments
We thank Suk Min Jang for work linked to this study and Tatiana Kutateladze for the recombinant ASH1L construct.
Funding and additional information
This work was supported by grants from the Quebec Breast Cancer Foundation, Canada and the Canadian Institutes of Health Research, Canada (grant no.: FDN-143314) to J. C., the Natural Sciences and Engineering Research Council of Canada, Canada (grant no.: 1304616-2017), the Cancer Research Society, Canada (grant no.: 25123), and Genome Quebec to J.-P. L., University of Toronto, Canada startup funds to M. T., J.-P. L., and S. M. I. H. were supported by Junior 2 salary awards from the Fonds de Recherche du Québec-Santé, Canada, and J. C. held the Canada Research Chair in Chromatin Biology and Molecular Epigenetics.
Author contributions
M. D. and J. C. methodology; A. L., J.-P. L., M. D., J. C., C. J.-B., and S. M. I. H. formal analysis; M. D., C. R., C. L., K. J., and A. P. investigation; M. D. and J. C. writing–original draft; A. D., J.-P. L., and J. C. supervision; A. D., J.-P. L., and J. C. funding acquisition.
Supplemental data
References
- 1.Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.Torres I.O., Fujimori D.G. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr. Opin. Struct. Biol. 2015;35:68–75. doi: 10.1016/j.sbi.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothbart S.B., Strahl B.D. Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta. 2014;1839:627–643. doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson M.A., Kouzarides T., Huntly B.J. Targeting epigenetic readers in cancer. N. Engl. J. Med. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- 6.Venkatesh S., Workman J.L. Set2 mediated H3 lysine 36 methylation: regulation of transcription elongation and implications in organismal development. Wiley Interdiscip. Rev. Dev. Biol. 2013;2:685–700. doi: 10.1002/wdev.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edmunds J.W., Mahadevan L.C., Clayton A.L. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kizer K.O., Phatnani H.P., Shibata Y., Hall H., Greenleaf A.L., Strahl B.D. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J.S., Shilatifard A. A site to remember: h3K36 methylation a mark for histone deacetylation. Mutat. Res. 2007;618:130–134. doi: 10.1016/j.mrfmmm.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Bannister A.J., Schneider R., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 11.Vakoc C.R., Sachdeva M.M., Wang H., Blobel G.A. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell. Biol. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolasinska-Zwierz P., Down T., Latorre I., Liu T., Liu X.S., Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilhelm B.T., Marguerat S., Aligianni S., Codlin S., Watt S., Bahler J. Differential patterns of intronic and exonic DNA regions with respect to RNA polymerase II occupancy, nucleosome density and H3K36me3 marking in fission yeast. Genome Biol. 2011;12 doi: 10.1186/gb-2011-12-8-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spies N., Nielsen C.B., Padgett R.A., Burge C.B. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Almeida S.F., Grosso A.R., Koch F., Fenouil R., Carvalho S., Andrade J., et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat. Struct. Mol. Biol. 2011;18:977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 16.Luco R.F., Pan Q., Tominaga K., Blencowe B.J., Pereira-Smith O.M., Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwamori N., Tominaga K., Sato T., Riehle K., Iwamori T., Ohkawa Y., et al. MRG15 is required for pre-mRNA splicing and spermatogenesis. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E5408–E5415. doi: 10.1073/pnas.1611995113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung C.S., Douglass S.M., Morselli M., Obusan M.B., Pavlyukov M.S., Pellegrini M., et al. H3K36 methylation and the chromodomain protein Eaf3 are required for proper cotranscriptional spliceosome assembly. Cell Rep. 2019;27:3760–3769.e4. doi: 10.1016/j.celrep.2019.05.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen M., Tominaga K., Pereira-Smith O.M. Emerging role of the MORF/MRG gene family in various biological processes, including aging. Ann. N. Y. Acad. Sci. 2010;1197:134–141. doi: 10.1111/j.1749-6632.2010.05197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie T., Zmyslowski A.M., Zhang Y., Radhakrishnan I. Structural basis for multi-specificity of MRG domains. Structure. 2015;23:1049–1057. doi: 10.1016/j.str.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyon Y., Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Jelinic P., Pellegrino J., David G. A novel mammalian complex containing Sin3B mitigates histone acetylation and RNA polymerase II progression within transcribed loci. Mol. Cell. Biol. 2011;31:54–62. doi: 10.1128/MCB.00840-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayakawa T., Ohtani Y., Hayakawa N., Shinmyozu K., Saito M., Ishikawa F., et al. RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells. 2007;12:811–826. doi: 10.1111/j.1365-2443.2007.01089.x. [DOI] [PubMed] [Google Scholar]
- 24.Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 25.Rossetto D., Cramet M., Wang A.Y., Steunou A.L., Lacoste N., Schulze J.M., et al. Eaf5/7/3 form a functionally independent NuA4 submodule linked to RNA polymerase II-coupled nucleosome recycling. EMBO J. 2014;33:1397–1415. doi: 10.15252/embj.201386433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y., Li J., Dunn S., Xiong S., Chen W., Zhao Y., et al. Histone deacetylase 2 (HDAC2) protein-dependent deacetylation of mortality factor 4-like 1 (MORF4L1) protein enhances its homodimerization. J. Biol. Chem. 2014;289:7092–7098. doi: 10.1074/jbc.M113.527507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sy S.M., Huen M.S., Chen J. MRG15 is a novel PALB2-interacting factor involved in homologous recombination. J. Biol. Chem. 2009;284:21127–21131. doi: 10.1074/jbc.C109.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ducy M., Sesma-Sanz L., Guitton-Sert L., Lashgari A., Gao Y., Brahiti N., et al. The tumor suppressor PALB2: inside out. Trends Biochem. Sci. 2019;44:226–240. doi: 10.1016/j.tibs.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 29.Hayakawa T., Zhang F., Hayakawa N., Ohtani Y., Shinmyozu K., Nakayama J., et al. MRG15 binds directly to PALB2 and stimulates homology-directed repair of chromosomal breaks. J. Cell Sci. 2010;123:1124–1130. doi: 10.1242/jcs.060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bleuyard J.Y., Fournier M., Nakato R., Couturier A.M., Katou Y., Ralf C., et al. MRG15-mediated tethering of PALB2 to unperturbed chromatin protects active genes from genotoxic stress. Proc. Natl. Acad. Sci. U. S. A. 2017;114:7671–7676. doi: 10.1073/pnas.1620208114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei Y., Tian C., Zhao Y., Liu X., Liu F., Li S., et al. MRG15 orchestrates rhythmic epigenomic remodelling and controls hepatic lipid metabolism. Nat. Metab. 2020;2:447–460. doi: 10.1038/s42255-020-0203-z. [DOI] [PubMed] [Google Scholar]
- 32.Lalonde M.E., Avvakumov N., Glass K.C., Joncas F.H., Saksouk N., Holliday M., et al. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 2013;27:2009–2024. doi: 10.1101/gad.223396.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saksouk N., Avvakumov N., Champagne K.S., Hung T., Doyon Y., Cayrou C., et al. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol. Cell. 2009;33:257–265. doi: 10.1016/j.molcel.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Côté J., Utley R.T., Workman J.L. In: Methods in Molecular Genetics. Adolph K.W., editor. Vol. 6. Academic Press; Cambridge, MA: 1995. [6] Basic analysis of transcription factor binding to nucleosomes; pp. 108–128. [Google Scholar]
- 35.Dalvai M., Loehr J., Jacquet K., Huard C.C., Roques C., Herst P., et al. A scalable genome-editing-based approach for mapping multiprotein complexes in human cells. Cell Rep. 2015;13:621–633. doi: 10.1016/j.celrep.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Agudelo D., Duringer A., Bozoyan L., Huard C.C., Carter S., Loehr J., et al. Marker-free coselection for CRISPR-driven genome editing in human cells. Nat. Methods. 2017;14:615–620. doi: 10.1038/nmeth.4265. [DOI] [PubMed] [Google Scholar]
- 37.Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 38.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 40.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Sena Brandine G., Smith A.,D. Falco: high-speed FastQC emulation for quality control of sequencing data. F1000Res. 2021;8:1874. doi: 10.12688/f1000research.21142.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ewels P., Magnusson M., Lundin S., Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 45.Kirby K.N., Gerlanc D. BootES: an R package for bootstrap confidence intervals on effect sizes. Behav. Res. Methods. 2013;45:905–927. doi: 10.3758/s13428-013-0330-5. [DOI] [PubMed] [Google Scholar]
- 46.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alerasool N., Leng H., Lin Z.Y., Gingras A.C., Taipale M. Identification and functional characterization of transcriptional activators in human cells. Mol. Cell. 2022;82:677–695.e7. doi: 10.1016/j.molcel.2021.12.008. [DOI] [PubMed] [Google Scholar]
- 48.Ruan C., Lee C.H., Cui H., Li S., Li B. Nucleosome contact triggers conformational changes of Rpd3S driving high-affinity H3K36me nucleosome engagement. Cell Rep. 2015;10:204–215. doi: 10.1016/j.celrep.2014.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venkatesh S., Smolle M., Li H., Gogol M.M., Saint M., Kumar S., et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 50.Zhang P., Du J., Sun B., Dong X., Xu G., Zhou J., et al. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 2006;34:6621–6628. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doyon Y., Cote J. Preparation and analysis of native chromatin-modifying complexes. Methods Enzymol. 2016;573:303–318. doi: 10.1016/bs.mie.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 52.Larance M., Kirkwood K.J., Xirodimas D.P., Lundberg E., Uhlen M., Lamond A.I. Characterization of MRFAP1 turnover and interactions downstream of the NEDD8 pathway. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu L., Li Q., Wong S.H., Huang M., Klein B.J., Shen J., et al. ASH1L links histone H3 lysine 36 dimethylation to MLL leukemia. Cancer Discov. 2016;6:770–783. doi: 10.1158/2159-8290.CD-16-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.An S., Yeo K.J., Jeon Y.H., Song J.J. Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism. J. Biol. Chem. 2011;286:8369–8374. doi: 10.1074/jbc.M110.203380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balbo Pogliano C., Gatti M., Ruthemann P., Garajova Z., Penengo L., Naegeli H. ASH1L histone methyltransferase regulates the handoff between damage recognition factors in global-genome nucleotide excision repair. Nat. Commun. 2017;8:1333. doi: 10.1038/s41467-017-01080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang C., Yang F., Zhang Z., Zhang J., Cai G., Li L., et al. Mrg15 stimulates Ash1 H3K36 methyltransferase activity and facilitates Ash1 Trithorax group protein function in Drosophila. Nat. Commun. 2017;8:1649. doi: 10.1038/s41467-017-01897-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmahling S., Meiler A., Lee Y., Mohammed A., Finkl K., Tauscher K., et al. Regulation and function of H3K36 di-methylation by the trithorax-group protein complex AMC. Development. 2018;145 doi: 10.1242/dev.163808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee Y., Yoon E., Cho S., Schmahling S., Muller J., Song J.J. Structural basis of MRG15-mediated activation of the ASH1L histone methyltransferase by releasing an autoinhibitory loop. Structure. 2019;27:846–852.e3. doi: 10.1016/j.str.2019.01.016. [DOI] [PubMed] [Google Scholar]
- 59.Hou P., Huang C., Liu C.P., Yang N., Yu T., Yin Y., et al. Structural insights into stimulation of Ash1L's H3K36 methyltransferase activity through Mrg15 binding. Structure. 2019;27:837–845.e3. doi: 10.1016/j.str.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 60.Hu X., Harvey S.E., Zheng R., Lyu J., Grzeskowiak C.L., Powell E., et al. The RNA-binding protein AKAP8 suppresses tumor metastasis by antagonizing EMT-associated alternative splicing. Nat. Commun. 2020;11:486. doi: 10.1038/s41467-020-14304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie T., Graveline R., Kumar G.S., Zhang Y., Krishnan A., David G., et al. Structural basis for molecular interactions involving MRG domains: implications in chromatin biology. Structure. 2012;20:151–160. doi: 10.1016/j.str.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Redington J., Deveryshetty J., Kanikkannan L., Miller I., Korolev S. Structural insight into the mechanism of PALB2 interaction with MRG15. Genes (Basel) 2021;12:2002. doi: 10.3390/genes12122002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu J., Chen Y., Lu L.Y., Wu Y., Paulsen M.T., Ljungman M., et al. Chfr and RNF8 synergistically regulate ATM activation. Nat. Struct. Mol. Biol. 2011;18:761–768. doi: 10.1038/nsmb.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jacquet K., Fradet-Turcotte A., Avvakumov N., Lambert J.P., Roques C., Pandita R.K., et al. The TIP60 complex regulates bivalent chromatin recognition by 53BP1 through direct H4K20me binding and H2AK15 acetylation. Mol. Cell. 2016;62:409–421. doi: 10.1016/j.molcel.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bassi C., Li Y.T., Khu K., Mateo F., Baniasadi P.S., Elia A., et al. The acetyltransferase Tip60 contributes to mammary tumorigenesis by modulating DNA repair. Cell Death Differ. 2016;23:1198–1208. doi: 10.1038/cdd.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li M.L., Jiang Q., Bhanu N.V., Wu J., Li W., Garcia B.A., et al. Phosphorylation of TIP60 suppresses 53BP1 localization at DNA damage sites. Mol. Cell. Biol. 2019;39 doi: 10.1128/MCB.00209-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 68.Bhat W., Ahmad S., Cote J. TINTIN, at the interface of chromatin, transcription elongation, and mRNA processing. RNA Biol. 2015;12:486–489. doi: 10.1080/15476286.2015.1026032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pradhan S.K., Su T., Yen L., Jacquet K., Huang C., Cote J., et al. EP400 deposits H3.3 into promoters and enhancers during gene activation. Mol. Cell. 2016;61:27–38. doi: 10.1016/j.molcel.2015.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamaguchi K., Sakai M., Shimokawa T., Yamada Y., Nakamura Y., Furukawa Y. C20orf20 (MRG-binding protein) as a potential therapeutic target for colorectal cancer. Br. J. Cancer. 2010;102:325–331. doi: 10.1038/sj.bjc.6605500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Setiaputra D., Ahmad S., Dalwadi U., Steunou A.L., Lu S., Ross J.D., et al. Molecular architecture of the essential yeast histone acetyltransferase complex NuA4 redefines its multimodularity. Mol. Cell. Biol. 2018;38 doi: 10.1128/MCB.00570-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X., Ahmad S., Zhang Z., Cote J., Cai G. Architecture of the Saccharomyces cerevisiae NuA4/TIP60 complex. Nat. Commun. 2018;9:1147. doi: 10.1038/s41467-018-03504-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gowher H., Brick K., Camerini-Otero D.R., Felsenfeld G. Vezf1 protein binding sites genome-wide are associated with pausing of elongating RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2370–2375. doi: 10.1073/pnas.1121538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tominaga K., Matzuk M.M., Pereira-Smith O.M. MrgX is not essential for cell growth and development in the mouse. Mol. Cell. Biol. 2005;25:4873–4880. doi: 10.1128/MCB.25.12.4873-4880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tominaga K., Kirtane B., Jackson J.G., Ikeno Y., Ikeda T., Hawks C., et al. MRG15 regulates embryonic development and cell proliferation. Mol. Cell. Biol. 2005;25:2924–2937. doi: 10.1128/MCB.25.8.2924-2937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xie L., Pelz C., Wang W., Bashar A., Varlamova O., Shadle S., et al. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011;30:1473–1484. doi: 10.1038/emboj.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feng J., Liu T., Qin B., Zhang Y., Liu X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All MS files generated as part of this study were deposited at MassIVE (http://massive.ucsd.edu). The MassIVE ID is MSV000089533, and the MassIVE link for download is: http://massive.ucsd.edu/ProteoSAFe/status.jsp?task=69398380312a47a79e171cd742ddb03e.
ChIP-Seq data from K562 cells and RNA sequencing from U2OS cells were deposited in the Gene Expression Omnibus database under accession number GSE181533.