Abstract
Parasite cryptic species are morphologically indistinguishable but genetically distinct organisms, leading to taxa with unclear species boundaries. Speciation mechanisms such as cospeciation, host colonization, taxon pulse, and oscillation may lead to the emergence of cryptic species, influencing host-parasite interactions, parasite ecology, distribution, and biodiversity. The study of cryptic species diversity in helminth parasites of human and veterinary importance has gained relevance, since their distribution may affect clinical and epidemiological features such as pathogenicity, virulence, drug resistance and susceptibility, mortality, and morbidity, ultimately affecting patient management, course, and outcome of treatment. At the same time, the need for recognition of cryptic species diversity has implied a transition from morphological to molecular diagnostic methods, which are becoming more available and accessible in parasitology. Here, we discuss the general approaches for cryptic species delineation and summarize some examples found in nematodes, trematodes and cestodes of medical and veterinary importance, along with the clinical implications of their taxonomic status. Lastly, we highlight the need for the correct interpretation of molecular information, and the correct use of definitions when reporting or describing new cryptic species in parasitology, since molecular and morphological data should be integrated whenever possible.
Keywords: Speciation, Cryptic species, Taxonomy, Phylogeny, Species complex, Parasitology
Graphical abstract
Highlights
-
•
Cryptic diversity has been described in helminths of human and animal importance.
-
•
Cryptic species are morphologically indistinguishable but genetically distinct organisms.
-
•
These entities emerge by different evolutionary and speciation mechanisms.
-
•
Analysis of molecular and morphological data is needed for cryptic species delimitation.
-
•
Cryptic diversity may affect pathogenicity, virulence and drug resistance of helminths.
1. Introduction
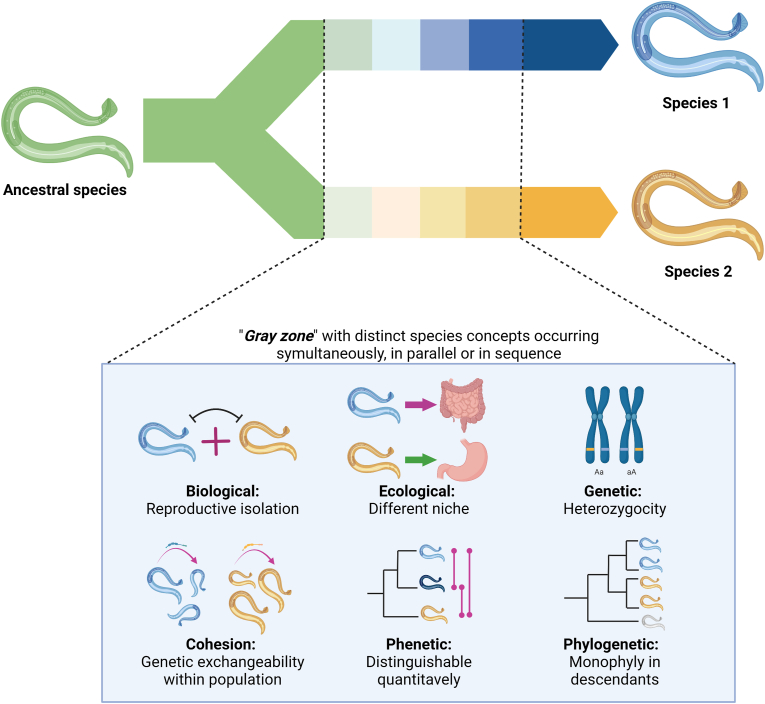
Parasites represent a large percentage of global biodiversity, distributed virtually in all vertebrate species and geographical areas of the world (Nadler & Pérez-Ponce de León, 2011). Parasites are a burden for approximately a billion humans suffering from neglected tropical diseases and billions of companion and production animals, as well as wildlife (Fenwick, 2012; Ondrejicka et al., 2014). Classification of parasites into nominal species is an essential task for biodiversity assessment, restoration ecology, conservation biology and for improving the understanding of host biogeography and parasite evolution. Nominal species are distinct genetic and morphological clusters separated from other taxa by speciation or separately evolving metapopulation lineages (De Queiroz, 2007; Shapiro et al., 2016) (Fig. 1). This evolutionary process in parasites is driven by multiple factors, including host colonization, taxon pulse, ecological fitting, host speciation, host population genetics, human intervention, landscape changes or parasite spillover (Hoberg & Brooks, 2008, 2010, 2013; Thompson, 2013).
Fig. 1.
Graphical representation of species concepts adapted from De Queiroz (2007), depicting an ancestor and its descendants sharing derived character states (Rosen, 1979; Donoghue, 1985; Mishler, 1985). One parasitic lineage (ancestral species indicated in green) originates two separate lineages (Species 1 and Species 2), where all three are reciprocally exclusive. The color gradient illustrates different species concepts (non-exhaustive: biological, ecological, genetic, cohesion, phenetic or phylogenetic) occurring simultaneously, in parallel or sequentially that lead to speciation. This figure was created using BioRender.com.
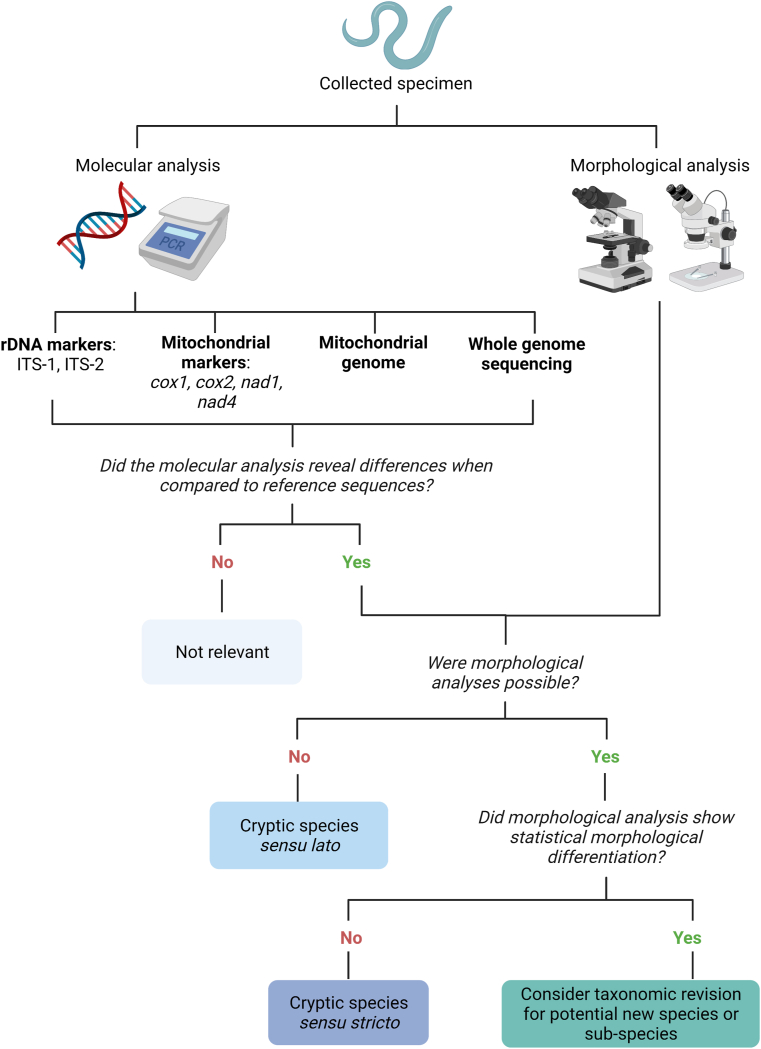
Historically, taxonomists largely relied on morphological, ecological, evolutionary, phenetic, and phylogenetic features to classify organisms (i.e. taxa), or a combination of two or more of these features (Aldhebiani, 2018) (Fig. 1). Scientific advances, especially the use of molecular biology tools, allowed the reassessment of many of these nominal species, revealing that many in fact contained two or more distinct taxa. These separate entities are referred to as cryptic species (Knowlton, 1993), which are morphologically identical but genetically divergent (Daly et al., 2021). Cryptic species may be the result of host-parasite coevolutionary events, as well as host-independent events (Xavier et al., 2015). However, cryptic parasite diversity is much more complex, and several definitions have derived from this concept. For instance, cryptic species sensu stricto (s.s.) are delimited when there is no morphological differentiation of the cryptic organism when compared to reference specimens, but molecular analysis reveals significant divergence (Fig. 2). Cryptic species sensu lato (s.l.) are reported when molecular analyses reveal unexpectedly high genetic divergence when compared to other genetic sequences, while morphological differences have not yet been verified. Furthermore, putative cryptic species sensu stricto/sensu lato may apply when genetic speciation is suggested by data, but there is lack of convincing evidence (Fig. 2). Additionally, cryptic genetically isolated units (CGIs) represent a category of cryptic species which are reproductively isolated but may potentially interbreed due to host range expansion, as well as the disappearance of a geographical barrier (Chenuil et al., 2019). However, definition of CGIs involves experimental data to test fertilization between the nominal and cryptic species, and fitness of their offspring, therefore, is a term less used for parasitic helminths due to their complex biology.
Fig. 2.
Recommended general flowchart for cryptic species definition for a collected specimen. This figure was created using BioRender.com.
Delimitation of cryptic species of parasites has changed in the last decade to an almost all-molecular approach (Fišer et al., 2018) with implications in health sciences and clinical practice, since this knowledge may lead to a better understanding of parasite epidemiology, and implementation of diagnostics, control and prevention (Nadler & Pérez-Ponce de León, 2011). In addition, cryptic species can vary in pathogenicity, resulting in a different course of infection and outcome, as suggested for the protozoan Tetratrichomonas gallinarum (Cepicka et al., 2005; Nadler & Pérez-Ponce de León, 2011), but less understood for helminths. Furthermore, cryptic species may also be geographically segregated, leading to epidemiological implications for parasite control and prevention, such as the case of the foodborne trematode Opisthorchis viverrini (Saijuntha et al., 2007; Nadler & Pérez-Ponce de León, 2011). Another relevant feature of cryptic species is how they are distributed among higher-level parasite taxa. In this sense, considering helminths, cryptic species are predominantly found in trematodes, followed by cestodes, and to a lesser extent in nematodes (Pérez-Ponce de León & Poulin, 2018). Lastly, other related features yet to be understood for helminth cryptic species are as follows: differences in virulence; applicability of molecular diagnostics for cryptic entities; drug susceptibility and resistance of the cryptic species; and their associated mortality and morbidity on host populations (Sithithaworn et al., 2015). Due to all these unsolved questions, parasitologists have advised the recognition of cryptic diversity as a medical priority (Sithithaworn et al., 2015; Pérez-Ponce de León & Nadler, 2016).
In this review, we focus on speciation mechanisms previously described for helminth parasites, molecular approaches towards delimitation of cryptic helminth species, and the usefulness of various molecular markers and approaches for such a purpose (see Table 1). Finally, we discuss some of the most fascinating examples found in parasitic nematodes, trematodes and cestodes of human and veterinary importance that have implications for both public health and animal production systems.
Table 1.
Summary of cryptic diversity reported in the literature for nematodes, trematodes and cestodes of human and veterinary importance
| Species | Host | Origin | Studied stage | Molecular markers employeda | Proposed status | Reference |
|---|---|---|---|---|---|---|
| Phylum Nematoda | ||||||
| Ascaris lumbricoides, Ascaris suum | Humans | Kenia | Adults | Mitogenome (4 × ratio < 4), cox1 (1–4 nucleotide differences), nad4, nuclear genome (1% polymorphism in major nuclear alleles) | CGIs | Easton et al. (2020) |
| Toxocara cati, Toxocara malaysiensisb | Cats | Malaysia | Adults | Mitogenome | Cryptic species | Jex et al. (2008) |
| Toxascaris leonina species complex (3 undescribed species) | Dogs, wolves, wild felids, red foxes | Poland | Adults | ITS1, cox1 (1.7–6.58% K2P), nad1 | Cryptic species complex | Fogt-Wyrwas et al. (2019) |
| Strongyloides spp. | Philippine slow loris/humans and dogs | Malaysian Borneo/Australia, Cambodia, Japan, and Myanmar | Third-stage larvae | cox1/18S and cox1 | Cryptic species sensu lato | Frias et al. (2018)/Beknazarova et al. (2019); Jaleta et al. (2017); Nagayasu et al. (2017) |
| Trichinella chanchalensis | Wolverine | Canada | Larval stage not specified | cytb, mitogenome, 15 SCNs | Cryptic species sensu stricto | Sharma et al. (2020) |
| Dirofilaria sp. “Thailand II”/Dirofilaria sp. (D. immitis-like) (2 undescribed species) | Carnivores/humans | Thailand | Adults | ITS1 (17%)/12S (5%), cox1 (6%) | Cryptic species sensu lato | Yilmaz et al. (2016) |
| Onchocerca sp. (1 undescribed species) | Cervids | North America | Microfilariae and adult females | 12S, 16S, cox1 | Cryptic species sensu lato | McFrederick et al. (2013); Verocai et al. (2018) |
| Oesophagostomum sp. (undescribed species) | Human and non-human primates | Uganda | Eggs | ITS2: 2.9% Clade I vs O. bifurcum; 0.6% Clade II vs O stephanostomum; Clade III 7.0–7.6% vs O. bifurcum and 6.4–7.0% vs O. stephanostomum | Cryptic species sensu stricto | Ghai et al. (2014) |
| Teladorsagia boreoarticusb | Sheep and goats | Holarctic | Adults | nad4 (13%) | Cryptic species | Hoberg et al. (1999) |
| Class Trematoda | ||||||
| Opisthorchis viverrini | Cyprinid fish (2nd intermediate hosts) and rodents | Thailand, Laos PDR | Adults | 38 enzyme loci (MEE) (60%) | Cryptic species sensu lato | Saijuntha et al. (2007) |
| Echinostoma “revolutum” species complex (7 described species and 10 cryptic species-level lineages) | Gastropods (1st intermediate hosts), mammals and birds | Europe, North America, South America, Africa, Australia, New Zealand | Cercariae and adults | nad1 (intraspecific divergence: 0–3.6%; interspecific divergence: 4.2–21.5%) | Cryptic species complex | Georgieva et al. (2014) |
| Class Cestoda | ||||||
| Echinococcus granulosus (s.l.) species complex (E. granulosus (s.s.), Echinococcus equinus, Echinococcus ortleppi, Echinococcus canadensis, Echinococcus felidis) | Sheep, dogs, dogs, reindeer/moose, and lion, respectively | Germany, UK, South Africa, Canada and South Africa, respectively | Eggs | E. felidis vs E. granulosus (s.s.), E. equinus, E. ortleppi and E. canadensis G6, G7 and G8, respectively: cox1 (8.4%, 8.1%, 10.6%, 10.6%. 10.5%, 11.1%); nad1 (17.1%, 16.7%, 18.4%, 17.9%, 17.9%, 19.3%); cytb (11.6%, 12.4%, 14.8%, 14.8%, 15.0%, 15.0%); mitochondrial rrn (6.6%, 7.6%, 8.9%, 9.2%, 9.2%, 8.6%); elf-α (1.4%, NA, 0.9%, 1.0%, 1.0%, 0.9%) | Species complex | Hüttner et al. (2008); Romig et al. (2015) |
| Moniezia benedeni, Moniezia expansa | Sheep and cattle | Australia | Adults | 15 enzyme loci (MEE) (92% within M. benedeni, 33% within M. expansa) | Cryptic lineages within groups | Chilton et al. (2007) |
Abbreviation: NA, not available.
Genetic divergence in parentheses.
Despite described as cryptic in the literature, the species possesses a valid morphological diagnosis.
2. General speciation mechanisms in parasites
2.1. Coevolution, cophylogeny, cophylogenetic patterns and host-parasite interactions
Speciation refers to the process in which a species accumulates changes until it is considered a new taxonomic entity. This process is usually led by geographical, temporal, phenotypic, or phylogenetic changes and can be affected by climate and environmental alterations, biotic and geographical expansion, as well as host habitat invasion that leads to faunal mosaics (Hua & Wiens, 2013) (Fig. 1).
Cospeciation occurs when the speciation of a host or parasite takes place simultaneously to its counterpart, keeping their ecological relationship (Hayward et al., 2021). Nomenclature has been ever changing on this subject, and concepts can vary from one research group to another, but generally, main definitions on the subject can be separated as follows: (i) coevolution; (ii) cophylogeny; (iii) cophylogenetic events; and (iv) cospeciation processes.
Coevolution can be defined as the selective pressure of two species exerted on one another, resulting in a mutual evolutionary influence. In principle, the two species evolving are influenced by beneficial or unfavorable associations such as mutualism and parasitism, respectively (Hugot, 2006). Coevolutionary events can be studied using multiple tools, cophylogeny being the most widely used, which is described as the study of the phylogenetic congruence between two different organisms due to their long-standing interactions (Avino et al., 2019), through cophylogenetic signal. Phylogenetic congruence, explained by Fahrenholzʼs rule as “Parasite phylogeny mirrors that of its host” (Fahrenholz, 1913), could be defined as the extent to which each node and branch length in the parasite tree mirrors an associated taxon in the host tree (Blasco-Costa et al., 2021).
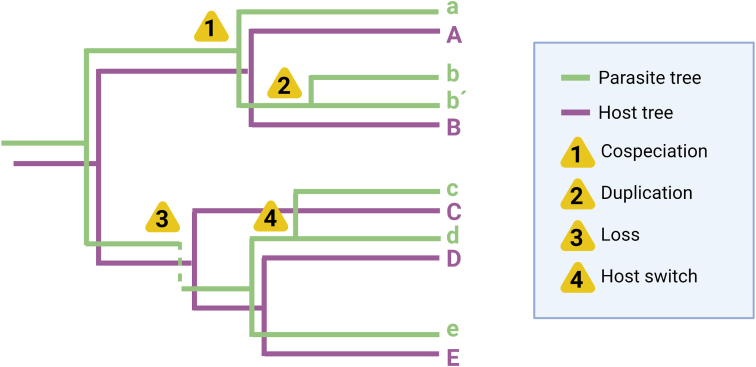
Coevolutionary events are predicted based on the branching of the parasite and host trees. These events based on phylogenetic relationships are mainly cospeciation, host switch, duplication, and loss (Fig. 3). Cospeciation or co-divergence, is the concomitant speciation of parasites and hosts (Brooks et al., 2015). Host switch, also known as host shift, horizontal transfer or host colonization, occurs when parasites speciate by occupying new hosts in a different ancestral lineage of its original host as a result of the parasiteʼs low host specificity (De Vienne et al., 2013; Brooks et al., 2015). Two main conditions and stages are necessary for host switch speciation to occur, namely opportunity and compatibility (Araujo et al., 2015). Contrary to host switching, duplication happens when parasites speciate in the absence of a host change, meaning that parasite lineages duplicate within the same host (Johnson et al., 2003; Garamszegi, 2009). Finally, if the evolutionary fate of parasites and their hosts fail to compile with the processes mentioned above, they might suffer a loss, which can also be referred to as sorting or failure. If this happens repeatedly, a given parasite species may become extinct (Garamszegi, 2009). Interestingly, all these events are promoted by changes in the host-parasite interactions, also stated in the Red Queen Hypothesis. In biology, this hypothesis explains a dynamic set of antagonistic interactions of defence and invasion where the species that fails to catch up with its partner may become extinct (Rabajante et al., 2016).
Fig. 3.
Graphical representation of host-parasite coevolutionary events. Overlapped parasite (in green) and host (in purple) phylogenetic trees depicting the following coevolutionary events: (1) cospeciation; (2) duplication; (3) loss; and (4) host switch. This figure was created using BioRender.com.
The cophylospace is a tool for comparing different host-parasite systems and provides mechanistic explanations of cospeciation patterns (Russo et al., 2018; Blasco-Costa et al., 2021). As portrayed in these analyses, cospeciation may be exerted by one of the following events: (i) coevolution facilitated by mutual changes in host-parasite interactions; (ii) phylogenetic tracking in which speciation of either host or parasite is followed by speciation of its counterpart in an asymmetrical way; or (iii) vicariance where the detected phylogenetic congruence is due to geographical isolation, given ecological barriers to gene flow and dispersal as the result of host-parasite sympatry, as a counterpart to cospeciation (Althoff et al., 2014; Hoberg et al., 2017). Importantly, these mechanisms are not exclusionary, and a combination of each with different intensities is expected (Blasco-Costa et al., 2021).
2.2. Main drivers of parasite diversification: the Stockholm Paradigm
Even though cospeciation has been linked to parasite diversification, as mentioned previously, it is not the main generator of diversity. In turn, the Stockholm Paradigm has been proposed, which integrates macro- and microevolutionary dynamics, along with ecological and biogeographical information into faunal assembly and diversification. This paradigm is constituted by four main diversification drivers: (i) taxon pulse (TP); (ii) ecological fitting (EF); (iii) oscillation and (iv) geographic mosaics of coevolution (GMC) (Hoberg et al., 2017). It has been proposed that both EF and TP are primary mediators of macroevolutionary structure (i.e. evolution above the species level), followed by oscillation and GMC (Hoberg & Brooks, 2010).
Taxon pulse proposes that organism lineages suffer adaptive shifts through geographical or ecological spaces from their monophyletic beginning to their more derived end (Erwin, 1985). For this reason, TP describes the context for biotic mixing, or ecological collisions that increase the opportunity of host-parasite contact that are further explained by EF. In addition, TP explains the dynamics of episodic niche perturbation, stability and recurring host invasion that occur as a consequence of geographical isolation and geographical colonization (Halas et al., 2005; Lim, 2008; Hoberg & Brooks, 2010; Hoberg et al., 2017).
Ecological fitting (EF) initiates host switching or host colonization, either by resource tracking when the new host is similar to the original one, or by “sloppy fitness space”, meaning that the new host offers novel resources (Agosta et al., 2010). Therefore, EF depends on the conditions explained above of opportunity, compatibility and conflict resolution, where opportunity is defined by expansion in TP (Janzen, 1985; Brooks & McLennan, 2002; Agosta et al., 2010; Hoberg & Brooks, 2010; Hoberg et al., 2017).
Oscillation explains that specialist parasites will eventually become generalists, and then these will produce new specialists (Araujo et al., 2015). This alternation of host range will depend on the use of resources with the ultimate narrowing of host range or ecological associations (Janz et al., 2006; Janz & Nylin, 2008; Hoberg & Brooks, 2010). Since oscillation largely defines host range, it may interact with EF, thereby determining host exploitation and colonization (Brooks & McLennan, 2002; Hoberg & Brooks, 2008; Hoberg et al., 2017).
The fourth and last driver is GMC, a framework describing coevolution between hosts and parasites in real populations and species (Brooks, 1979; Thompson, 2005; Hoberg et al., 2017). The GMC has been defined as a tripartite hypothesis, i.e. (i) different environmental conditions may lead to genetic variations of populations under those conditions, thus, selection may favor distinct evolutionary trajectories; (ii) there are coevolutionary hot spots within communities where reciprocal coevolutionary selection occurs embedded in cold spots where selection is non-reciprocal; and (iii) trait remixing in populations facilitated by constant genomic alterations, gene flow and genetic drift, change the distribution of coevolving characters within and among populations over time. Finally, this hypothesis suggests that trait coevolution in hosts and parasites will be influenced by the favorable or unfavorable environmental conditions to which populations are exposed to, with few traits spreading to all populations of interacting species (Thompson, 1999).
Other authors have suggested specific evolutionary processes to explain how cryptic species may have originated. Herein, it must be considered that cryptic species are the opposite of species resulting from adaptive radiation, given that in face of different ecological niches or barriers, these exhibit a reduced variation or disparity in phenotypic characteristics (e.g. morphology) (Struck & Cerca, 2019). These mechanisms may be referred to as recent divergence, convergence, parallelism, and stasis. Accordingly, recent divergence explains that speciation has occurred so recently that visible phenotypic changes have not yet taken place, excluding physiological, immunological, reproductive or behavioral changes (Knowlton, 1993; Struck & Cerca, 2019). Next, convergence and parallelism, while being antagonists in operation, result in the formation of cryptic species. The former implies the evolution of a derived character from different ancestral sets of traits given that these converge, while the latter implies the independent evolution of a character in different taxa from similar and shared ancestral set of traits (arising sets of parallel traits). Finally, stasis occurs when a specific phenotype is conserved during long time scales, even when environmental conditions fluctuate and highlight the potential deceleration of phenotypic evolution. These mechanisms demonstrate that while phenotypic variation may be subtle, it is not absent in cryptic species (Struck & Cerca, 2019).
2.3. General classification of helminths according to phylogenetic analyses and cryptic species distribution
Taxonomy, systematics, and phylogeny in helminthology have transitioned from classic morphological approaches to the era of molecular data. As stated by Brooks (1985), clinical diagnosis in parasitology has focused on the study of unique traits found in organisms for separating taxa. This highlights the importance of unifying classification approaches in medical and veterinary helminthology, in order to determine zoonotic potential, implement control strategies, and study the spread of anthelminthic resistance (Betson & Stothard, 2016).
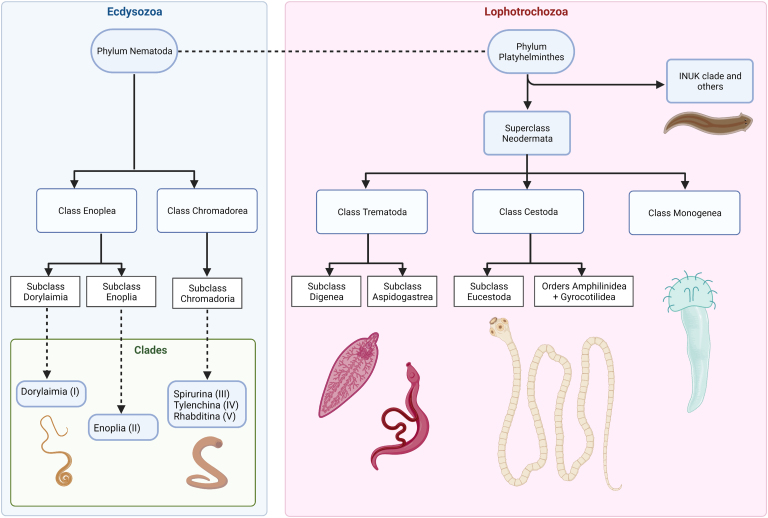
Helminths are classified into two main phyla: Nematoda and Platyhelminthes (Fig. 4). The phylum Nematoda represents free-living or parasitic pseudocoelomate organisms of animals and plants which are, in the vast majority, sexually dimorphic (Basyoni & Rizk, 2016). While nematodes were originally classified into classes Adenophorea and Chromadorea, phylogenetic approaches using the 18S rRNA gene (Blaxter et al., 1998; Kampfer et al., 1998; De Ley & Blaxter, 2002, 2004; Meldal et al., 2007; Holterman et al., 2009; Van Megen et al., 2009; Bik et al., 2010; Blaxter & Koutsovoulos, 2015) have resulted in a new reclassification (Fig. 4). Moreover, five clades have been proposed according to the 18S rDNA phylogeny of nematodes: (i) Dorylaimia; (ii) Enoplia; (iii) Spirurina; (iv) Tylenchina; and (v) Rhabditina. The greatest number of parasitic nematodes are members of the clade Rhabditina, followed by Dorylaimia and lastly, Enoplia, with only one known parasitic species of animals, namely Ironus macrocephalum (Blaxter & Koutsovoulos, 2015). In addition, phylogenomic and transcriptomic analyses have rendered the same major nematode clades as obtained with the 18S-derived phylogeny by De Ley & Blaxter (2002, 2004), with the advantage of including a larger number of taxa leading to a better resolution of branching distances in the major Nematoda clades (Blaxter & Koutsovoulos, 2015; Smythe et al., 2019).
Fig. 4.
Schematical phylogeny of helminths. The phylum Nematoda is composed of classes Enoplea and Chromadorea, which are subdivided into subclasses Dorylaimia, Enoplia and Chromadoria and their respective clades, as illustrated. Parasitic organisms in the phylum Platyhelminthes are represented by the superclass Neodermata, which is divided into classes Trematoda, Cestoda and Monogenea. Note that Cestodaria is not used to refer to Amphilinidea + Gyrocotilidea since it is no longer a valid taxon. This figure was created using BioRender.com.
On the other hand, platyhelminths are an immensely diverse group of parasitic and free-living acoelomate and monoecious organisms (except for schistosomes) (Collins, 2017). This morphology-based classification has been confirmed with genomic data from 44 neodermatan platyhelminths and one free-living turbellarian worm, which has shown the same subdivision (Coghlan et al., 2019). According to phylogenetic analyses based on 18S and 28S rRNA genes, parasitic platyhelminths are classified in two major groups: Superclass Neodermata and the clade INUK (Carranza et al., 1997; Mollaret et al., 1997; Littlewood & Bray, 2001; Lockyer et al., 2003; Egger et al., 2015) (Fig. 4) that includes parasitic rhabditophoran platyhelminths of the genera Ichthyophaga, Notentera, Urastoma and Kronborgia (Littlewood et al., 1999a, b; Baguñà & Riutort, 2004). Superclass Neodermata is subdivided into three main classes: Trematoda, Cestoda and Monogenea. The first two contain parasites of human and veterinary importance, while the latter comprises mainly ectoparasites of fish (Lockyer et al., 2003; Baguñà & Riutort, 2004).
Parasites in the class Trematoda (flukes) and are subdivided in subclasses Digenea (involved in trematodiases in humans and other vertebrate hosts) and Aspidogastrea (parasites of fish, reptiles, and bivalves) (Alves et al., 2015; Mehlhorn, 2016) (Fig. 4). Organisms belonging to the class Cestoda (tapeworms) are divided into the subclass Eucestoda, a monophyletic group composed of 17 orders, of which Cyclophyllidea is the most important in human and veterinary medicine; and a group composed of the orders Amphilinidea and Gyrocotylidea, formerly known as the subclass Cestodaria, for which there is no support of monophyly to date and is henceforth no longer considered a valid taxon (Caira et al., 2017).
As mentioned before in this review, the distribution of cryptic species in helminth species is uneven, given that the class Trematoda encompasses the greatest number of cryptic species reported, followed by the class Cestoda, and finally the phylum Nematoda (Pérez-Ponce de León & Poulin, 2018; Chan et al., 2022). This can be explained by a greater probability of somatic mutations occurring during the asexual reproduction of trematodes, to the subtler morphological differences exhibited in trematode species than in other taxa, and less specialized morphological studies (i.e. scanning electron micrographs) employed to describe such species, possibly leading to their initial misdiagnosis as cryptic species (Pérez-Ponce de León & Poulin, 2018).
3. Cryptic diversity in helminth parasites
3.1. Elucidation of cryptic species through morphology, behavior or ecology
For many years, parasitologists have described, classified, and diagnosed species and higher taxa using solely morphological traits. However, this strategy has been proven problematic when cryptic entities have been recognized for a species (Pérez-Ponce de León & Nadler, 2010; Nadler & Pérez-Ponce de León, 2011). In the past, several non-morphological and non-molecular methodologies such as host ethology, parasite mating behavior, resource use, geographical distribution, biochemistry or natural history were widely employed for the detection of cryptic species (Nadler & Pérez-Ponce de León, 2011). For instance, Ascaris suum was originally described by Goeze (1782) as the roundworm of pigs (Sus scrofa), since it was found in this host for the first time, even though it did not exhibit morphological differences from A. lumbricoides (Goeze, 1782). The term cryptic species was used for the first time in helminthology in 1978 when differentiating Parascaris equorum from P. univalens using isoenzyme electrophoresis (Bullini et al., 1978). To date, PCR-based assays employing both mitochondrial and ribosomal genes are the most employed protocols for deciphering cryptic entities in different fields, including helminthology (Nadler & Pérez-Ponce de León, 2011).
3.2. Detection of cryptic species with gene sequencing: mitochondrial vs ribosomal genes
Prospecting cryptic species using DNA markers must follow two basic criteria. First, the locus needs to evolve fast enough among individuals so genetic divergence can be quickly spotted even within populations with a low number of individuals. Secondly, the marker must have low intra-specific and intra-individual variability, in order to differentiate true events of speciation from normal intra-individual variability (Vilas et al., 2005).
Mitochondrial and ribosomal loci have been used for phylogeographic studies to identify cryptic species (Nadler & Pérez-Ponce de León, 2011). Mitochondrial DNA (mtDNA) genes such as the NADH dehydrogenase subunit 4 (nad4) and the cytochrome c oxidase subunit 1 (cox1) evolve rapidly and rarely recombine. These genes have proven to be excellent candidates to search for cryptic diversity because of their resolution power that reveals monophyly within very closely related species (Morgan & Blair, 1998; Nadler & Hudspeth, 2000; Blouin, 2002). Other mitochondrial genes, namely the 12S and 16S rRNA genes show an interspecific genetic difference comparable to that of cox1 for the molecular identification of helminths (i.e. trematodes), and can discriminate closely related species (Chan et al., 2022). Additionally, these markers are suitable for discriminating possible cryptic species at initial molecular prospecting, given that their reported genetic divergence between different parasitic nematode genera ranges from approximately 10% to 20% (Blouin et al., 1998), and between 6.9% and 16.0% in congeneric species (Blouin, 2002). Furthermore, it has been observed that the maximum percentage of genetic divergence in an interbreeding population of the cattle parasitic nematode Ostertagia ostertagi is 6% when using nad4 (Anderson et al., 1998; Blouin, 2002). However, intra-specific and intra-individual variation can range from 1% to 7% in members of the family Trichostrongylidae when using nad4 (Blouin et al., 1998), which equals the degree of variation often found between closely related taxa. This situation may pose a difficulty when using mtDNA to identify cryptic species, since the discrimination of the intra-specific variability from the emergence of monophyly cannot be achieved, even when using partial cox1 and nad4 sequences (Blouin, 2002). For this reason, additional markers and larger DNA fragments are recommended to provide higher resolution regarding a species’ taxonomic position. As for cestodes, mitochondrial markers such as cox1 and nad1 have been used to study cryptic diversity in the family Taeniidae, while multilocus enzyme electrophoresis (MEE) has been employed in the family Anoplocephalidae (Chilton et al., 2007; Lavikainen et al., 2010; Jia et al., 2012).
Another powerful mitochondria-associated tool is whole mitogenome sequencing, which has increased the resolution of unclear taxonomic relationships, even though only just over 200 nematode species have been sequenced to date (Kern et al., 2020; Roe et al., 2021). For instance, mitogenome sequences and morphological differences in reproductive structures were useful to distinguish the monogenean parasite Benedenia seriolae from B. humboldti (Baeza et al., 2019). In addition, suborders Spirurina and Tylenchina have been proposed as monophyletic according to 18S rDNA phylogenies, whereas mitogenome information has separated them into several independent clusters (Kern et al., 2020). Still, discordances between mitogenome and nuclear gene phylogenies are observed, and the combination of several markers is recommended for a more robust interpretation of phylogenetic relationships.
Ribosomal loci like the internal transcribed spacers (ITS1 and ITS2) have been extensively used to distinguish between helminth species due to the high variability they hold, and the availability of universal primers targeting these regions (Blouin, 2002). For these reasons, ITS2 sequences have been useful for characterizing gastrointestinal Clade V nematode communities using deep amplicon sequencing in cattle, sheep and bison (Avramenko et al., 2015, 2017, 2018). Additionally, the presence of fixed insertions and deletions (indels) in ITS sequences have made them appealing for diagnostics (Blouin, 2002). Nevertheless, exceptions have been detected for the carcinogenic nematode of canids Spirocerca lupi, where ITS1 intra-individual variation ranges from 0.37 to 2.85%, with each specimen showing two or more different copies of ITS1 (Rojas et al., 2018), an event that has also been reported in ITS2 of the nematodes Varestrongylus alces, Varestrongylus cf. capreoli and Varestrongylus sagittatus (Verocai et al., 2014) and Trichuris spp. (Callejón et al., 2012), the cestode Echinococcus granulosus (Blair & McManus, 1995), and the trematode Paragonimus westermani (Van Herwerden et al., 1999) as a result of incomplete rDNA homogenization.
Genetic divergence obtained using mtDNA markers has been compared to that obtained using the ITS loci. For instance, the nad4 gene offers a better resolution when prospecting for cryptic species (Morgan & Blair, 1998; Blouin, 2002; Vilas et al., 2005) in nematodes (such as Ancylostoma spp. and Haemonchus spp.) than in trematodes (such as Echinostoma spp., Fasciola spp., Schistosoma spp.), and taeniid cestodes (Echinococcus and Taenia). Nonetheless, examples are found where neither ribosomal nor mitochondrial markers have been able to solve the cryptic species status. In these cases, whole genome sequencing (WGS) has offered a better resolution.
3.3. Advances of the whole genome sequencing
Whole genome sequencing (WGS) has improved the study of single copy nuclear genes (SCN). SCNs have not been routinely used for cryptic species prospecting, since their amplification from genomic DNA is challenging for some parasitic taxa, as described by Nadler & Pérez-Ponce de León (2011). Since 2007, when the genome of Brugia malayi was obtained for the first time in any helminth species, 81 parasitic nematodes and 31 flatworms have been sequenced to date (Ghedin et al., 2007; McVeigh, 2020). Furthermore, WGS has been applied to better understand the phylogenomics and phylogenetic relationship between the major parasitic nematodes of humans and pigs, Ascaris lumbricoides and Ascaris suum, respectively, which are morphologically indistinguishable from each other. In this case, WGS was necessary to elucidate the evolutionary history of both taxa (Easton et al., 2020). This illustrates that a wider look into a parasiteʼs genetic composition can improve species resolution and reduce the uncertainty of using a limited number of genes. Overall, WGS techniques have expanded the available information for helminth phylogenetic reconstructions, simultaneously increasing the complexity for analysing large datasets (Maldonado et al., 2019; Easton et al., 2020).
3.4. Limitations of gene-based analyses: alternative methods for delimiting cryptic species
Cryptic species are usually delimited with the support of DNA differences. However, it is not advisable to delimit cryptic entities on a threshold of genetic divergence or a genetic yardstick alone. This is suggested based on the heterogenous relative rates of evolution in each species, since increased relative rates of gene change in a particular lineage may result in falsely elevated pairwise nucleotide distances (Nadler & Pérez-Ponce de León, 2011). Nevertheless, genetic thresholds may be useful for prospecting cryptic diversity in the same species, since high intraspecific genetic divergence is not expected (Blouin, 2002; Vilas et al., 2005).
Interpretation of molecular data alone, without analysing morphological characters when available (as opposed to cryptic species sensu lato) has occasionally led to the mislabelling of entities as cryptic. For instance, Anisakis simplex, Anisakis pegreffi and Anisakis berlandi, were resolved as new species based on allozyme electrophoresis and ITS, 12S and cox1 sequence analyses, after being considered cryptic (Nascetti et al., 1986; Mattiucci et al., 2014). Another example is the report of a cryptic species within Eucoleus, based on a female adult collected from the bronchoalveolar lavage of a 12-week-old kitten (Calvani et al., 2021). Herein, researchers found up to 15–19% divergence in the cox1 sequence of this Eucoleus sp. isolate when compared to other available Eucoleus sequences; and a 14–23% divergence in mitochondrial protein coding gene sequences between the kitten isolate and E. aerophilus. However, differences in egg morphology found between the kitten isolate and E. aerophilus imply that the species is not strictly cryptic, even if adult morphology was not examined (Calvani et al., 2021). Moreover, the tapeworm of brown bears (Ursus arctos), Taenia arctos, was described as a nominal species with its own diagnosis after being originally reported as cryptic (Lavikainen et al., 2010; Haukisalmi et al., 2011). Similar evidence was reported for Hydatigera taeniaeformis (s.s.) and H. kamiyai, wherein both had been regarded as cryptic species of H. taeniaeformis (s.l.); however, rostellar hook analysis, deciphered their real identity (Jia et al., 2012; Lavikainen et al., 2016). These examples support the concept that classifying cryptic entities by their genetic divergence alone may not always serve as a yardstick in assessing cryptic diversity (Nadler & Pérez-Ponce de León, 2011).
There are alternatives to gene sequencing for detecting cryptic diversity, such as MEE, allozyme electrophoresis, restriction fragment length polymorphism (RFLPs), single strand conformational polymorphism (SSCPs), amplified fragment length polymorphism (AFLPs), randomly amplified polymorphic DNA (RAPDs) and microsatellites, some of which may be coupled with PCR (RFLPs, RAPDs) (Nadler & Pérez-Ponce de León, 2011). Each strategy has its advantages and disadvantages. For example, MEE may not be suitable in studies detecting genetic variation, since electrophoretic patterns depend only on factors altering electrophoretic mobility, such as charge and protein conformation (Nadler, 1990; Andrews & Chilton, 1999). In addition, computational resources for species delimitation, like the Bayesian Phylogenetics and Phylogeography (BPP) program have been used (Yang, 2015). This software, previously applied to the study of Baylisascaris phylogenetics and systematics, integrates independent evolutionary histories of different loci through the multispecies coalescent model (MSC), considering the population size of ancestral and modern species, and species divergence times (Camp et al., 2018). Other studies dealing with cryptic diversity of trematodes, such as Stegodexamene anguillae and Phyllodistomum have also employed a BPP approach to delimit cryptic diversity (Herrmann et al., 2014; Pinacho-Pinacho et al., 2021).
4. Examples of cryptic diversity in parasites of human and veterinary importance
In this section we discuss some of the most relevant examples of cryptic diversity found among helminths of human and veterinary importance, due to their impact in public health or production systems. These are listed in higher-level taxonomic order below.
4.1. Phylum Nematoda: Ascaris spp.
Ascariasis is a widespread geohelminthiasis caused by the roundworms A. lumbricoides and A. suum in humans and pigs, respectively. These two ascarids are morphologically indistinguishable from one another, and molecular markers have failed to resolve their phylogenetic relationship. The lack of resolution between these two species of Ascaris has been a cause of debate for decades (Da Silva Alves et al., 2016).
Four hypotheses have been proposed to correctly assess the taxonomic status of A. suum and A. lumbricoides and explain their evolutionary history: (i) A. lumbricoides and A. suum are not monophyletic and are thus two separate and valid species, where speciation occurred before the domestication of pigs by humans from a common ancestor (Leles et al., 2012); (ii) following pig domestication, A. suum infected humans and derived to A. lumbricoides by host-switch or host colonization and the latter persisted as a species (Leles et al., 2012; Zhou et al., 2020); (iii) after pig domestication, A. lumbricoides from humans switched to pigs and derived to A. suum (Leles et al., 2012); and (iv) these two nominal species of ascarids are in fact conspecific. Previously, it has been suggested that human and pig ascarids share a common recent ancestor with Parascaris equorum, an ascarid of equids (Nadler & Hudspeth, 2000) based on morphology, mitochondrial and nuclear genes, indicating that one did not originate from the other (Leles et al., 2012), but rather that they comprise a sister group, deriving from their most common recent ancestor, along with P. equorum. However, further research is needed to determine the support associated to each of the mentioned hypotheses.
The low genetic divergence in A. lumbricoides and A. suum mitogenomes and paleoparasitological evidence (i.e. the close contact between humans and pigs, before and during their domestication, facilitating host switch or colonization, cross-infections and hybridization events) (Leles et al., 2012) has provided further evidence to the hypothesis that both ascarids are conspecific. However, other studies have pointed towards derivation of one ascarid following infection with the other ascarid species. In this regard, hybrid genotypes of A. lumbricoides and A. suum have been recognized circulating among human and pig populations in the USA, Thailand, Lao PDR, Myanmar, Guatemala and China suggesting anthropozoonotic transmission to pigs has occurred in the past (Criscione et al., 2007; Jesudoss Chelladurai et al., 2017; Sadaow et al., 2018).
A high-quality Ascaris reference genome was constructed in 2020 from a single representative female worm collected from a human in Kenya, assumed to be infected by A. lumbricoides due to the lack of pig husbandry in their village (Easton et al., 2020). This study revealed that A. suum and A. lumbricoides are an inter-breeding and cross-infecting genetic complex after analysing 68 mitogenomes from Ascaris specimens collected from humans and the resemblance of the cox1 and nad4 haplotypes to other A. suum and A. lumbricoides sequences. Remarkably, the same study confirmed the presence of three genetic clades based on the genetic diversity at the whole genome, nuclear and mitochondrial level: A (mainly pig-derived specimens); B (mainly human-derived specimens); and C (pig-derived specimens from Europe and Asia) (Cavallero et al., 2013; Easton et al., 2020).
Ascaris hybrid populations may behave as CGIs, since these groups have been reproductively isolated, and are able to interbreed, as a consequence of host range expansion (Chenuil et al., 2019). In this sense, A. suum was originally described as a separate species from A. lumbricoides based on host specificity, and it was believed that A. lumbricoides solely parasitized humans (Goeze, 1782). Interestingly, experimental cross-infections of pig and human Ascaris have been demonstrated in both hosts, indicating the ability of this parasite to infect several types of hosts. Furthermore, ascariasis in humans is considered a zoonotic disease in Denmark, where prevalence of this parasite is low in humans and thus, pigs may be the main source of infection to humans (Anderson, 1995; Nejsum et al., 2005). This confirms the findings of Easton et al. (2020) that Ascaris can cross-infect pigs and humans and interbreed, and highlights the possible transfer of anthelminthic resistance genes from one Ascaris population in humans or pigs to another.
4.2. Phylum Nematoda: Toxocara spp.
Species of the genus Toxocara are intestinal parasites of mammals, with some species holding zoonotic importance. Canids serve as definitive hosts of T. canis, whereas felids are the definitive hosts of Toxocara cati and Toxocara malaysiensis. Definitive hosts harbour adults in the small intestine with the consequent excretion of their eggs in feces (Fialho & Corrêa, 2016; Chen et al., 2018). On the other hand, adult development is arrested in humans and a strong inflammatory response is released against migrating larval stages (Fialho & Corrêa, 2016) leading to covert toxocariasis or visceral, neural, ocular larva migrans (Chen et al., 2018).
Cryptic diversity in the genus has been regarded since the description of the Toxocara sp. variant found in cats in Malaysia (Rohde, 1962; Lee et al., 1993) which could not be morphologically discriminated from T. canis (Zhu et al., 1998). ITS1, ITS2 and 5.8S ribosomal markers and restriction analysis (PCR-RFLP) were used for comparative analysis of this Malaysian Toxocara isolate from T. canis and T. cati. The results of Zhu et al. (1998) initially indicated that the Malaysian Toxocara sp. isolates were closer to T. cati than to T. canis when all molecular markers were considered, contrary to the initial morphological diagnosis. This led to the report of Toxocara cf. canis for the Malaysian specimens, which were later formally described as Toxocara malaysiensis based on morphological traits of lips, cervical alae, spicule length and female reproductive system (Gibbons et al., 2001). Furthermore, research on the T. canis mitogenome suggested that T. malaysiensis and T. cati represented cryptic species (Jex et al., 2008). Nonetheless, a closer look might aid in evaluating cryptic species in the genus since T. malaysiensis has unique morphological characters that distinguish it from the other species of Toxocara (Gibbons et al., 2001).
4.3. Phylum Nematoda: Toxascaris leonina
Toxascaris is a monotypic nematode genus, with Ta. leonina as its only representative. This parasite commonly infects a large variety of definitive canid and felid hosts, such as dogs, cats, red foxes, tigers, lions and wolves worldwide, where they develop into adults in the small intestine potentially causing disease in young animals (Okulewicz et al., 2002, 2012). Additionally, humans can potentially become infected with larval stages of Ta. leonina and develop visceral, ocular or cerebral larva migrans (Robertson & Thompson, 2002; Okulewicz et al., 2012).
Several molecular studies have demonstrated the presence of cryptic species among Ta. leonina isolates, with apparent species delimitation according to host (Song et al., 2015; Li et al., 2018; Fogt-Wyrwas et al., 2019; Jin et al., 2019; Xie et al., 2020). In a preliminary study, cox1 was employed to determine the genetic relationship between Ta. leonina from dogs and cheetahs, revealing a difference of 6.8% between collected specimens from different hosts (Li et al., 2018). Another study found a difference of 9.0% and 10.8% in the nad1 and nad4 genes, respectively, between worms obtained from canid and felid hosts from South China (Song et al., 2015). A subsequent study obtained a 7.2% divergence in the coding regions of Ta. leonina mitogenomes isolated from cheetahs and dogs, suggesting the presence of cryptic species (Jin et al., 2019). Importantly, those specimens were morphologically identified as Ta. leonina, and it was suggested that further detailed morphological analyses of this species from different hosts should be undertaken. Additionally, ITS1, cox1 and nad1 sequence analysis of 35 Ta. leonina collected from red foxes (Vulpes vulpes), domestic dogs (Canis lupus familiaris), grey wolves (C. lupus), tigers (Panthera tigris amoyensis, P. t. altaica, P. t. corbetti), lions (P. leo spelaea), and the Eurasian lynx (Lynx lynx) found three Ta. leonina subclades clustered according to the host phylogenetic groups: (i) dogs and wolves; (ii) wild felids; and (iii) red foxes (Fogt-Wyrwas et al., 2019). Therefore, Ta. leonina from dogs and wolves clustered together (Kimura 2-parameter (K2P) barcode gap distance > 1.7%), while those from wild felids (K2P barcode gap distance of 3%) and red foxes (K2P barcode gap distance of 6.58%) formed two separate groups, suggesting the existence of three cryptic species within the complex. Interestingly, this research also pointed out that gene flow among wild felid isolates could be low (Fogt-Wyrwas et al., 2019) as supported by complete ITS sequences and partial cox2 and nad1 analyses (Xie et al., 2020).
Is it currently unknown which Ta. leonina cryptic species may infect humans, as well as their differences in pathogenicity, clinical course of infection and diagnosis. This is an issue that should be addressed in the future, since epidemiological research suggests that Ta. leonina is a potentially emerging zoonotic pathogen due to its close contact with humans, dogs and cats (Xie et al., 2019). Monitoring for cryptic species in human and animal cases is especially important considering the greater chance of morphology-based misdiagnosis associated with the genus, which is unlikely to be monotypic (Gasser, 2006; Chen et al., 2012; Fogt-Wyrwas et al., 2019; Xie et al., 2019).
4.4. Phylum Nematoda: Strongyloides spp.
Strongyloides is a speciose genus composed of over 50 valid species, that dwell between free-living and parasitic generations. The infective L3 larvae penetrate the host skin, migrate, and establish themselves in the small intestine mucosa, with only parthenogenetic females being parasitic. Most species of this genus are host-specific and have been reported in humans, non-human primates (NHP), various mammals, birds, amphibians and reptiles (Thamsborg et al., 2017; Jaleta & Lok, 2019). A few exceptions are S. stercoralis and S. fuelleborni, which have a wide host range (Thamsborg et al., 2017).
There is evidence of cryptic diversity in the genus, in isolates from NHP, humans and dogs. One study depicting the diversity of Strongyloides spp. in various NHP from Malaysian Borneo detected cryptic diversity in L3 obtained from fresh fecal samples of one Philippine slow loris (Nycticebus menagensis) (Frias et al., 2018). Through cox1 analysis, the sequences obtained from these animals clustered into two haplotypes apart from S. stercoralis or any other Strongyloides spp. sequences, possibly constituting a cryptic subpopulation of the former, with pairwise distances suggesting that the loris-associated clade may be distinct from those found in free and captive NHP, dogs, and humans. In addition, S. fuelleborni was also found in slow loris from Borneo, separated from isolates of other geographical regions.
These findings have epidemiological implications, since the host range of Strongyloides isolates belonging to the loris clade remains unknown and there is no information regarding whether this host may promote persistent parasite populations through autoinfection or if these parasites are better adapted to the free-living cycle (Frias et al., 2018). Favorable conditions for zoonotic transmission of this clade could be an emerging issue given the close phylogenetic relationship of the loris cluster to S. stercoralis, the potential parasite spillover facilitated by human activity, and that lorises are a popular target for wildlife trade, leading to an increased strongyloidiasis burden (Thompson, 2013). Additionally, evidence of cryptic and potentially zoonotic Strongyloides strains have been reported from dogs in Australia, and from humans and dogs in rural Cambodia, Japan, and Myanmar, on account of 18S rDNA and cox1 phylogenies (Jaleta et al., 2017; Nagayasu et al., 2017; Barratt et al., 2019; Beknazarova et al., 2019). These studies detected existing zoonotic and canine-specific cryptic Strongyloides lineages, emphasizing the importance of studying cryptic diversity among different hosts to further comprehend the extent of species distribution (Jaleta & Lok, 2019). Lastly, variation in pathogenicity and virulence in cryptic strains should be assessed, being of the utmost importance in parasites such as S. stercoralis and S. fuelleborni, considering their wide host range (Hasegawa et al., 2010; Frias et al., 2018; Wulcan et al., 2019).
4.5. Phylum Nematoda: Trichinella spp.
Species within the genus Trichinella are divided into two clades, namely the encapsulated (T1, T2, T3, T5, T6, T7, T8, T9, T12 and T13) and non-encapsulated (T4, T10, T11) clades; depending on whether they possess a collagen capsule arranging the nurse cell in the hostʼs muscle. Out of these 13 genotypes, 10 have been recognized and named as species, and three remain formally undescribed (i.e. T6, T8 and T9) (Pozio & Zarlenga, 2013; Korhonen et al., 2016; Zarlenga et al., 2020). Most species in the genus Trichinella are zoonotic nematodes of mammals, while T. pseudospiralis (T4) may parasitize avian hosts, and T. papuae (T10) and T. zwimbabwensis (T11) can parasitize reptiles as well as homeothermic hosts (Sharma et al., 2020; Zarlenga et al., 2020). Trichinellosis occurs after ingestion of raw or undercooked meat, containing the infective first-stage larvae (L1) (Gottstein et al., 2009). The larval stages develop into adults and sexually reproduce in the small intestine of the definitive host, with the new-born larvae reaching the muscle later on, finally becoming encapsulated depending on their genotype (Zarlenga et al., 2020).
A novel PCR-RFLP-based assay targeting the cytb gene detected the presence of a new cryptic species, T. chanchalensis (T13), in the frozen tissue of wolverines (Gulo gulo) from Canada, in single infections and coinfections with T. nativa (T2) and T6. Additionally, morphometric analysis of 25 specimens was unable to differentiate T13 when compared to other species, implying that T. chanchalensis is a cryptic species sensu stricto; supported by phylogenetic inference based on whole mitogenome and 15 SCNs (Sharma et al., 2020).
Trichinella chanchalensis has proven to be a freeze-resistant species, as well as T2 and T6. This may pose a relevant epidemiological implication, since freezing is an acceptable post-harvest treatment for T. spiralis inactivation in domestic pig meat (Noeckler et al., 2019; Johne et al., 2020). Moreover, naturally occurring wild hybrids of T2 and T6, and those of T. spiralis and T. britovi (T3) have already been reported (Dunams-Morel et al., 2012; Franssen et al., 2015). The existence of such hybrids supports the possibility of transferring genes conferring freeze-resistance (i.e. from T2, T6, T13) into freeze-susceptible genotypes, which may result in parasites that are better adapted to meat preservation conditions and may successfully infect their hosts (Zarlenga et al., 2020). These findings highlight the importance of accurately detecting cryptic species, since their biological features may change the infective potential of parasites.
4.6. Phylum Nematoda: Dirofilaria spp.
Species of Dirofilaria are vector-borne pathogens of carnivores, with many species proven to be zoonotic. Dirofilaria immitis is distributed worldwide and leads to the heartworm disease in canids, whereas Dirofilaria repens produces subcutaneous dirofilariasis and is mostly restricted to the Old World, with few cases reported in the Americas (Dantas-Torres & Otranto, 2013). Dirofilaria immitis and D. repens can infect humans and produce nodular lesions in lungs, subcutaneous tissues or eyes (Antolová et al., 2015).
A high genetic diversity has been observed in D. repens isolates from Asia, as opposed to specimens collected from Europe (Yilmaz et al., 2016). Four complete mitochondrial genomes of specimens identified as D. repens from Europe (n = 3) and India (n = 1), and 46 D. repens mitochondrial genome fragments from adults and microfilariae of blood samples from Italy, Vietnam, Thailand, and India were analyzed. The three European mitogenomes were identified as D. repens, while the Indian mitogenome was tentatively referred to as “Dirofilaria hongkongensis”, an agent of subcutaneous and subconjunctival human and animal dirofilariasis based on cox1 analysis. Moreover, the authors demonstrated the presence of a possible cryptic species detected in blood samples from cats, namely Dirofilaria sp. “Thailand II”, which could also belong to very divergent isolates referred to as “D. hongkongensis”. Dirofilaria repens-like parasites have been identified as D. hongkongensis based on ITS1 sequences (To et al., 2012). In addition, D. hongkongensis specimens collected from humans and canids have shown ITS2 sequences identical to D. repens, adding further evidence for the existence of cryptic diversity (Suzuki et al., 2015; Liesner et al., 2016; Yilmaz et al., 2016).
In addition, the existence of cryptic diversity was suggested for a Dirofilaria specimen recovered from a 16-year-old Brazilian male suffering ocular dirofilariasis. In this case, a 5% and 6% genetic difference was found in 12S rDNA and cox1 genes, respectively, in comparison with publicly available sequences of other species within the genus (Otranto et al., 2011). Interestingly, the specimen was morphologically similar to D. immitis and D. spectans, therefore, this could represent an undescribed cryptic species sensu lato. These findings underline the rich species diversity in the genus with no molecular data matching morphologically identical specimens and increase the interest for molecular prospecting of cryptic species in dirofilarial vectors such as Culex spp. in the Americas and their possible roles in contributing to hybridization.
4.7. Phylum Nematoda: Onchocerca spp.
Onchocerca volvulus, the agent of river blindness, produces ocular and subcutaneous infections in humans, endemic in several African countries, Yemen and the Amazonian focus straddling Venezuela and Brazil (Milton et al., 2020). The congeneric Onchocerca lupi induces ocular lesions in dogs, cats and humans. Onchocerca volvulus is transmitted by arthropod vectors of the family Simuliidae (Basáñez et al., 2009), while the vector intermediate host of O. lupi is still unknown (Rojas et al., 2021). It has been suggested that domestication of Onchocerca vertebrate hosts may have guided the parasiteʼs speciation into different taxa, since host switch events may have occurred between the host families Bovidae, Canidae and humans with recent speciation into O. lupi, Onchocerca gutturosa, Onchocerca lienalis, Onchocerca ochengi and O. volvulus (Lefoulon et al., 2015).
Cryptic diversity has been described in Onchocerca spp. associated with wild animals, especially hosts of the family Cervidae in the Nearctic (McFrederick et al., 2013; Verocai et al., 2018; Kulpa et al., 2021). Adult worms and microfilariae collected from the moose Alces alces have been molecularly identified as O. cervipedis, which was for decades assumed to represent the only species infecting North American cervids (Verocai et al., 2012; McFrederick et al., 2013). However, microfilariae obtained from the white-tailed deer Odocoileus virginianus analyzed using 12S, 16S and cox1 mitochondrial genes, did not cluster with O. cervipedis obtained from moose. This has suggested that the Onchocerca isolates collected from deer could represent a different or cryptic species of O. cervipedis, a matter that will remain unsolved until morphological analysis is completed (McFrederick et al., 2013). Additionally, a molecular surveillance of Onchocerca spp. in 1434 blackflies from California using the nad5 gene revealed an uncharacterized Onchocerca sp. infecting Simulium tescorum and S. vittatum (s.l.) (presumably S. tribulatum) (Verocai et al., 2018). Onchocerca sp. sequences from simuliids grouped together with the Onchocerca sp. previously found in the white-tailed deer discussed above. Moreover, partial cox1 sequences exhibited a difference greater than the intraspecific variation for this marker. Although no morphological analyses have been performed, both organisms may represent the same cryptic species sensu lato.
Host switch leading to parasite speciation has been suggested before for the genus Onchocerca (Lefoulon et al., 2015). Thus, it is imperative that cryptic diversity is assessed throughout the genus and its hosts, since this information may provide a better understanding of onchocercid genetic distribution, a feature that may aid in further comprehending onchocerciasis epidemiology and pathophysiology.
4.8. Phylum Nematoda: Oesophagostomum spp.
Species of the genus Oesophagostomum are parasitic strongylids of cattle, ruminants, pigs (Stewart & Gasbarre, 1989) and non-human primates (Polderman et al., 1991). Infection with these nematodes results in large economic losses in the production animal industry (Rashid et al., 2019). Oesophagostomum spp. induce the formation of nodules in the digestive subserosa of their hosts and can parasitize humans from endemic regions of West Africa, such as Togo and Ghana (Polderman et al., 1991; Polderman & Blotkamp, 1995).
Oesophagostomum diversity in Ugandan human and non-human primates is large, due to the existence of three cryptic clades according to ITS2 loci, that are morphologically indistinguishable. Clade 1 was 97.1–100% similar to Oesophagostomum bifurcum, while Clade 2 sequences from other primate species were 99.4% identical to Oesophagostomum stephanostomum and Clade 3 contained sequences from humans and other primates 92.4–93.0% and 93.0–93.6% similar to O. bifurcum and O. stephanostomum, respectively, with no similarity to any other Oesophagostomum spp. reference sequence (Ghai et al., 2014). In this study, the need for morphological analysis of L3-larvae and adult was highlighted to accurately confirm that sequences in Clade 3 belong to a cryptic lineage. Importantly, analysis of mitochondrial markers might render a greater resolution on Clade 3ʼs phylogenetic relationships to other Oesophagostomum spp. since the study of Ghai et al. (2014) was based on ITS2 analysis alone.
Implications of cryptic diversity within the genus Oesophagostomum are yet to be assessed. It has been reported that Oesophagostomum spp. induce a weak immune response with a late Th2 response in hosts and a different cytokine profile when compared to the conventional Th2 profile expected in helminthiases, resulting in immune system modulation and evasion (Roepstorff et al., 2011). It is not known if cryptic Oesophagostomum lineages present antigenic differences and whether these could derive in isolates with different immunogenicity, ultimately contributing to determine the fate of the clinical outcome.
4.9. Phylum Nematoda: Teladorsagia spp.
Teladorsagia circumcincta is a trimorphic nematode species (i.e. a morphospecies containing three morphotypes) that infects caprines and cervids in the Holartic, with cryptic diversity in the genus proposed in the past (Hoberg et al., 2001). Moreover, T. circumcincta has been suggested as a complex of cryptic species, composed of two strains irrespective of their morphology, one occurring in goats and the other in both sheep and goats (Wyrobisz et al., 2016; Wyrobisz-Papiewska et al., 2018). In a morphometric and molecular analysis conducted by Hoberg et al. (1999), Teladorsagia boreoarticus was described as a dimorphic species which was later found to be trimorphic (Wyrobisz et al., 2016). Its description was based on integrated morphological characters, including the synlophe, esophageal valve, spicules, gubernaculum and bursa, and molecular characterization of the nad4 gene, showing a 13% divergence when compared to sequences of other Teladorsagia spp. Therefore, T. boreoarcticus represented a separate nominal species on the grounds of a valid morphological diagnosis, in a genus permeated of cryptic diversity, instead of a cryptic species.
4.10. Class Trematoda: Opisthorchis viverrini
Opisthorchis viverrini is a parasitic trematode of humans, endemic in Southeast Asia and a type 1 carcinogen. This parasite infects the liver and is responsible for opisthorchiasis-associated cholangiocarcinoma in chronic infections (Saijuntha et al., 2007; Khuntikeo et al., 2018).
Cryptic diversity has been suggested in O. viverrini isolates from cyprinid fish in Thailand and Lao PDR using MEE (Saijuntha et al., 2007). Analysis of 32 enzyme loci showed that 19 loci were different between O. viverrini isolates, with separation into two major groups and at least one species with differences in more than 30% of the loci. Additionally, MEE analysis of the gastropod first intermediate host, Bithynia siamensis goniomphalos, isolates revealed 17% fixed genetic variation, separating them into two major genetic groups, consistent with the division seen in O. viverrini fish isolates, which suggests host-parasite cospeciation (Saijuntha et al., 2007). These results indicate that different O. viverrini (s.l.) populations or species exist in Thailand and Lao PDR which may influence epidemiological changes in prevalence and morbidity, as well as in opistorchiid control, prevention and treatment effectiveness (Saijuntha et al., 2007).
Overall, the identification of cryptic diversity and cryptic species within trematodes is key to understand the risks associated with the spread and extent of foodborne trematodiases such as opisthorchiasis (Pérez-Ponce de León & Nadler, 2016). Aquacultural activities and fish commerce may be facilitators for parasite spillover and may enhance human contact with these pathogens. This may ultimately lead to the spread of cryptic species (or origin thereof) outside areas of endemicity (Thompson, 2013; Pérez-Ponce de León & Nadler, 2016).
4.11. Class Trematoda: Echinostoma revolutum
Echinostomiasis is an intestinal food-borne parasitic infection caused by Echinostoma spp. in humans and animals associated with ingestion of raw molluscs and fish (Sah et al., 2018). Seventeen species and species-level genetic lineages have been delineated using nad1 mitochondrial gene within the Echinostoma “revolutum” species complex with cryptic lineages radiating in North America and in central and northern Europe (Georgieva et al., 2014). Morphological and molecular analysis of the nad1 gene of Echinostoma spp. cercariae collected from snail intermediate hosts in Germany and Iceland revealed the occurrence of a novel cryptic species sensu stricto in the “revolutum” group, namely Echinostoma sp. IG (p-distance range 17.2–21.6%) (Georgieva et al., 2013).
4.12. Class Cestoda: Echinococcus granulosus
Echinococcus spp. belong to the family Taeniidae and use carnivores of the families Canidae, Felidae and Hyaenidae as their definitive hosts (Romig et al., 2017). These cestodes can also parasitize humans due to their wide intermediate host range (Romig et al., 2017). For instance, Echinococcus granulosus (s.l.) induces cystic echinococcosis, Echinococcus multilocularis leads to alveolar echinococcosis and E. oligarthra and E. vogeli produce polycystic echinococcosis (Wen et al., 2019).
In the past, it was believed that E. granulosus consisted of a single species with an enormous genotypic and phenotypic variation. However, mitochondrial gene and whole mitogenome analyses have confirmed that E. granulosus is not a single species comprising strains or variants, but rather a cryptic species complex, with species exhibiting fascinating differences in their life-cycle and epidemiological settings (Romig et al., 2017). Altogether, the species complex is comprised by E. granulosus (s.s.) (strains G1, G2 and G3), E. equinus (G4, horse strain), E. ortleppi (G5, cattle strain), E. canadensis (G6 or camel strain, G7 or pig strain, G8 or cervid strain along with G10, and G9 now considered as a G7 microvariant) and E. felidis (formerly considered the lion strain) (Romig et al., 2015). Importantly, E. granulosus (s.s.) is the main species parasitizing humans in cystic echinococcosis but it has been shown that E. canadensis can also infect humans and cause disease to a lesser extent than the former species (Romig et al., 2015). To date, whole genome sequences from E. granulosus, E. multilocularis and E. oligarthra have been published, allowing further genomic comparisons and analyses of the species complex (Maldonado et al., 2019).
The correct identification of cryptic Echinococcus diversity might aid in determining which epidemiological factors are key for disease control and prevention, which becomes highly relevant due to the diverse range of intermediate hosts that may change transmission dynamics (Sithithaworn et al., 2015). This species complex also exhibits differences in factors such as antigenicity, chemotherapeutic sensitivity, and pathology, further illustrating the relevance of a correct diagnosis and research regarding cryptic diversity (McManus, 2013).
4.13. Class Cestoda: Moniezia spp.
Moniezia is a member of the family Anoplocephalidae that utilizes ruminants, pigs, rodents and birds as definitive hosts (Beveridge, 1994; Chilton et al., 2007). While rarely associated with clinical disease, it has been implicated as a cause of unthriftiness and emaciation in ungulates (Kutz et al., 2012). A strong support for cryptic species within two groups of Moniezia benedeni and two groups of Moniezia expansa from sheep and cattle in Southern Australia was demonstrated in a study employing MEE (Chilton et al., 2007). In this study, MEE analysis showed a 92% and 33% genetic divergence within two M. benedeni and two M. expansa groups, respectively, while divergence between the two species reached 77%. Those results support the existence of true cryptic lineages in the groups; however, mitochondrial analyses are needed to further validate the cryptic species status (Chilton et al., 2007).
5. Concluding remarks
Cryptic diversity in parasitic helminths commonly results from several evolutionary events. Speciation occurs in large parasite and host communities, driven by climate, space and time, and aided by the influence of human activities which facilitates parasite spillover. Currently, the wide availability and relative low cost of molecular techniques and bioinformatic tools has facilitated the sequencing of mitochondrial and whole parasite genomes leading to the identification of cryptic diversity within various helminth species of human and veterinary importance, as outlined throughout this review. In this regard, mitogenomes and mitochondrial markers have proven extremely valuable in cryptic species discrimination, as they evolve rapidly and do not recombine. Moreover, these sequences are useful in identifying the appearance of monophyly before being detected by ribosomal markers. In spite of this, when delimiting cryptic species, molecular divergence data should not be used as yardsticks, due to the absence of such universal reference range, considering the existing variation in evolutionary rates for markers in specific parasite lineages.
Identification of cryptic diversity and cryptic species should be assessed as a general medical priority, since these taxa may vary in clinical and epidemiological features such as distribution, pathogenicity, virulence, drug resistance or susceptibility, and may weigh in clinical decision making. The recognition of this diversity may influence patient management, course of treatment (drug and dose of choice), anthelmintic resistance emergence, disease control and prevention, and the development of educational programmes to mitigate parasite transmission and infection. Moreover, cryptic diversity monitoring is necessary to detect emergent zoonotic species and further study their implications in human medicine. Furthermore, the burden of cryptic diversity in the veterinary setting could reflect the expansion of infections in a wider host range by novel cryptic species, most likely due to host colonization. The repercussions of these phenomena could manifest as a heavy economic loss in animal productive systems, or a decline in wildlife populations.
Lastly, we highlight the need for the correct use of definitions and concepts when describing new cryptic species in parasitology, as molecular and morphological data should be integrated whenever possible. A standardized nomenclature will ease the description of the status of potential cryptic entities to the scientific community.
Funding
This work was funded by the Helminthology Collection of Costa Rica project B7733-21, University of Costa Rica.
CRediT author statement
LEC-G and AR: conceptualization, writing - original draft, writing - review & editing. FM-C: writing - original draft. JM, AS-B, GGV: conceptualization, writing - review & editing. All authors read and approved the final manuscript.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr Steven A. Nadler, University of California, Davis, USA, for his critical review of the manuscript and his guidance about the use of molecular markers for elucidating cryptic species, and Dr Mark Blaxter, Wellcome Sanger Institute, UK, for his comments about the origin of parasitism in vertebrate nematodes.
References
- Agosta S.J., Janz N., Brooks D.R. How specialists can be generalists: Resolving the “parasite paradox” and implications for emerging infectious disease. Zoologia. 2010;27:151–162. doi: 10.1590/S1984-46702010000200001. [DOI] [Google Scholar]
- Aldhebiani A.Y. Species concept and speciation. Saudi J. Biol. Sci. 2018;25:437–440. doi: 10.1016/j.sjbs.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althoff D.M., Segraves K.A., Johnson M.T.J. Testing for coevolutionary diversification: Linking pattern with process. Trends Ecol. Evol. 2014;29:82–89. doi: 10.1016/j.tree.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Alves P.V., Vieira F.M., Santos C.P., Scholz T., Luque J.L. A checklist of the Aspidogastrea (Platyhelminthes: Trematoda) of the world. Zootaxa. 2015;3918:339–396. doi: 10.11646/zootaxa.3918.3.2. [DOI] [PubMed] [Google Scholar]
- Anderson T.J.C. Ascaris infections in humans from North America: Molecular evidence for cross-infection. Parasitology. 1995;110:215–219. doi: 10.1017/S0031182000063988. [DOI] [PubMed] [Google Scholar]
- Anderson T.J.C., Blouin M.S., Beech R.N. Population biology of parasitic nematodes: Applications of genetic markers. Adv. Parasitol. 1998;41:219–283. doi: 10.1016/s0065-308x(08)60425-x. [DOI] [PubMed] [Google Scholar]
- Andrews R.H., Chilton N.B. Multilocus enzyme electrophoresis: A valuable technique for providing answers to problems in parasite systematics. Int. J. Parasitol. 1999;29:213–253. doi: 10.1016/S0020-7519(98)00168-4. [DOI] [PubMed] [Google Scholar]
- Antolová D., Miterpáková M., Paraličová Z. Case of human Dirofilaria repens infection manifested by cutaneous larva migrans syndrome. Parasitol. Res. 2015;114:2969–2973. doi: 10.1007/s00436-015-4499-7. [DOI] [PubMed] [Google Scholar]
- Araujo S.B.L., Braga M.P., Brooks D.R., Agosta S.J., Hoberg E.P., Von Hartenthal F.W., Boeger W.A. Undestanding host-switching by ecological fitting. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avino M., Ng G.T., He Y., Renaud M.S., Jones B.R., Poon A.F.Y. Tree shape-based approaches for the comparative study of cophylogeny. Ecol. Evol. 2019;9:6756–6771. doi: 10.1002/ece3.5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramenko R.W., Bras A., Redman E.M., Woodbury M.R., Wagner B., Shury T., et al. High species diversity of trichostrongyle parasite communities within and between Western Canadian commercial and conservation bison herds revealed by nemabiome metabarcoding. Parasit. Vectors. 2018;11 doi: 10.1186/s13071-018-2880-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramenko R.W., Redman E.M., Lewis R., Bichuette M.A., Palmeira B.M., Yazwinski T.A., Gilleard J.S. The use of nemabiome metabarcoding to explore gastro-intestinal nematode species diversity and anthelmintic treatment effectiveness in beef calves. Int. J. Parasitol. 2017;47:893–902. doi: 10.1016/j.ijpara.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Avramenko R.W., Redman E.M., Lewis R., Yazwinski T.A., Wasmuth J.D., Gilleard J.S. Exploring the gastrointestinal “nemabiome”: deep amplicon sequencing to quantify the species composition of parasitic nematode communities. PLoS One. 2015;10 doi: 10.1371/journal.pone.0143559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J.A., Sepúlveda F.A., González M.T. The complete mitochondrial genome and description of a new cryptic species of Benedenia Diesing, 1858 (Monogenea: Capsalidae), a major pathogen infecting the yellowtail kingfish Seriola lalandi Valenciennes in the South-East Pacific. Parasit. Vectors. 2019;12 doi: 10.1186/s13071-019-3711-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguñà J., Riutort M. Molecular phylogeny of the Platyhelminthes. Can. J. Zool. 2004;82:168–193. doi: 10.1139/z03-214. [DOI] [Google Scholar]
- Barratt J.L.N., Lane M., Talundzic E., Richins T., Robertson G., Formenti F., et al. A global genotyping survey of Strongyloides stercoralis and Strongyloides fuelleborni using deep amplicon sequencing. PLoS Negl. Trop. Dis. 2019;13 doi: 10.1371/JOURNAL.PNTD.0007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basáñez M.G., Churcher T.S., Grillet M.E. Onchocerca-Simulium interactions and the population and evolutionary biology of Onchocerca volvulus. Adv. Parasitol. 2009;68:263–313. doi: 10.1016/S0065-308X(08)00611-8. [DOI] [PubMed] [Google Scholar]
- Basyoni M.M.A., Rizk E.M.A. Nematodes ultrastructure: Complex systems and processes. J. Parasit. Dis. 2016;40:1130–1140. doi: 10.1007/s12639-015-0707-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beknazarova M., Barratt J.L.N., Bradbury R.S., Lane M., Whiley H., Ross K. Detection of classic and cryptic Strongyloides genotypes by deep amplicon sequencing: A preliminary survey of dog and human specimens collected from remote Australian communities. PLoS Negl. Trop. Dis. 2019;13 doi: 10.1371/JOURNAL.PNTD.0007241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betson M., Stothard J.R. Ascaris lumbricoides or Ascaris suum: Whatʼs in a name? J. Infect. Dis. 2016;213:1355–1356. doi: 10.1093/infdis/jiw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge I. In: Keys to the cestode parasites of vertebrates. Khalil L., Jones A., Bray R., editors. CAB International; Wallingford: 1994. 17. Family Anoplocephalidae Cholodkovsky, 1902; pp. 315–366. [Google Scholar]
- Bik H.M., Lambshead P.J.D., Thomas W.K., Lunt D.H. Moving towards a complete molecular framework of the Nematoda: A focus on the Enoplida and early-branching clades. BMC Evol. Biol. 2010;10 doi: 10.1186/1471-2148-10-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair D., McManus D.P. A molecular phylogeny of the genus Echinococcus. Parasitology. 1995;3:317–328. doi: 10.1017/S0031182000080902. [DOI] [PubMed] [Google Scholar]
- Blasco-Costa I., Hayward A., Poulin R., Balbuena J.A. Next-generation cophylogeny: Unravelling eco-evolutionary processes. Trends Ecol. Evol. 2021;36:907–918. doi: 10.1016/J.TREE.2021.06.006. [DOI] [PubMed] [Google Scholar]
- Blaxter M., Koutsovoulos G. The evolution of parasitism in Nematoda. Parasitology. 2015;142:S26–S39. doi: 10.1017/S0031182014000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M.L., De Ley P., Garey J.R., Llu L.X., Scheldeman P., Vierstraete A., et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- Blouin M.S. Molecular prospecting for cryptic species of nematodes: Mitochondrial DNA versus internal transcribed spacer. Int. J. Parasitol. 2002;32:527–531. doi: 10.1016/S0020-7519(01)00357-5. [DOI] [PubMed] [Google Scholar]
- Blouin M.S., Yowell C.A., Courtney C.H., Dame J.B. Substitution bias, rapid saturation, and the use of mtDNA for nematode systematics. Mol. Biol. Evol. 1998;15:1719–1727. doi: 10.1093/oxfordjournals.molbev.a025898. [DOI] [PubMed] [Google Scholar]
- Brooks D.R. Testing the context and extent of host-parasite coevolution. Syst. Zool. 1979;28:299–307. doi: 10.2307/2412584. [DOI] [Google Scholar]
- Brooks D.R. Phylogenetics and the future of helminth systematics. J. Parasitol. 1985;71:719–727. doi: 10.2307/3281702. [DOI] [PubMed] [Google Scholar]
- Brooks D.R., Hoberg E.P., Boeger W.A. In the eye of the cyclops: The classic case of cospeciation and why paradigms are important. Comp. Parasitol. 2015;82:1–8. doi: 10.1654/4724C.1. [DOI] [Google Scholar]
- Brooks D.R., McLennan D.A. University of Chicago Press; Chicago: 2002. The nature of diversity. An evolutionary voyage of diversity. [Google Scholar]
- Bullini L., Nascetti G., Carrè S., Rumore F., Biocca E. Matematiche e Naturali; Rendiconti: 1978. Ricerche cariologiche ed elettroforetiche su Parasacaris univalens e Parascaris equorum. Atti Della Accademia Nazionale Dei Lincei. Classe Di Scienze Fisiche. [Google Scholar]
- Caira J.N., Jensen K., Georgiev B.B., Kuchta R., Littlewood T., Mariaux J., Scholz T., Tkach V.V., Waeschenbach A. In: Planetary Biodiversity Inventory (2008–2017): Tapeworms from vertebrate bowels of the earth. Caira J.N., Jensen K., editors. University of Kansas, Natural History Museum, Special Publication No. 25; Lawrence, KS, USA: 2017. An overview of the tapeworms of vertebrate bowels of the earth; pp. 1–20.https://nhm.openrepository.com/handle/10141/622379 [Google Scholar]
- Callejón R., Halajian A., de Rojas M., Marrugal A., Guevara D., Cutillas C. 16S partial gene mitochondrial DNA and internal transcribed spacers ribosomal DNA as differential markers of Trichuris discolor populations. Vet. Parasitol. 2012;186:350–363. doi: 10.1016/j.vetpar.2011.11.033. [DOI] [PubMed] [Google Scholar]
- Calvani N.E.D., Wright M., White J., Stepkovitch B., Francis E., Rivory P., et al. What the fox? Cryptic Eucoleus [Capillaria] sp. in the respiratory tract of a cat from Australia. Curr. Res. Parasitol. Vector Borne Dis. 2021;1 doi: 10.1016/j.crpvbd.2021.100028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp L.E., Radke M.R., Shihabi D.M., Pagan C., Yang G., Nadler S.A. Molecular phylogenetics and species-level systematics of Baylisascaris. Int. J. Parasitol. Parasites Wildl. 2018;7:450–462. doi: 10.1016/J.IJPPAW.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carranza S., Baguñà J., Riutort M. Are the platyhelminthes a monophyletic primitive group? An assessment using 18S rDNA sequences. Mol. Biol. Evol. 1997;14:485–497. doi: 10.1093/oxfordjournals.molbev.a025785. [DOI] [PubMed] [Google Scholar]
- Cavallero S., Snabel V., Pacella F., Perrone V., D’Amelio S. Phylogeographical studies of Ascaris spp. based on ribosomal and mitochondrial DNA sequences. PLoS Negl. Trop. Dis. 2013;7 doi: 10.1371/journal.pntd.0002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepicka I., Kutišová K., Tachezy J., Kulda J., Flegr J. Cryptic species within the Tetratrichomonas gallinarum species complex revealed by molecular polymorphism. Vet. Parasitol. 2005;128:11–21. doi: 10.1016/j.vetpar.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Chan A.H.E., Saralamba N., Saralamba S., Ruangsittichai J., Thaenkham U. The potential use of mitochondrial ribosomal genes (12S and 16S) in DNA barcoding and phylogenetic analysis of trematodes. BMC Genomics. 2022;23 doi: 10.1186/S12864-022-08302-4/TABLES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Liu Q., Liu G.H., Zheng W.B., Hong S.J., et al. Toxocariasis: A silent threat with a progressive public health impact. Inf. Dis. Poverty. 2018;7:59. doi: 10.1186/s40249-018-0437-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zhou D.H., Nisbet A.J., Xu M.J., Huang S.Y., Li M.W., et al. Advances in molecular identification, taxonomy, genetic variation and diagnosis of Toxocara spp. Infect. Genet. Evol. 2012;12:1344–1348. doi: 10.1016/J.MEEGID.2012.04.019. [DOI] [PubMed] [Google Scholar]
- Chenuil A., Cahill A.E., Délémontey N., Du Salliant du Luc E., Fanton H. In: Casetta E., Marques da Silva J., Vecchi D., editors. vol. 24. Springer; Cham: 2019. Problems and questions posed by cryptic species. A framework to guide future studies. (From assessing to conserving biodiversity. History, philosophy and theory of the life sciences). [Google Scholar]
- Chilton N.B., O'Callaghan M.G., Beveridge I., Andrews R.H. Genetic markers to distinguish Moniezia expansa from M. benedeni (Cestoda: Anoplocephalidae) and evidence of the existence of cryptic species in Australia. Parasitol. Res. 2007;100:1187–1192. doi: 10.1007/s00436-006-0388-4. [DOI] [PubMed] [Google Scholar]
- Coghlan A., Tyagi R., Cotton J.A., Holroyd N., Rosa B.A., Tsai I.J., et al. Comparative genomics of the major parasitic worms. Nat. Genet. 2019;51:163–174. doi: 10.1038/s41588-018-0262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J.J. Platyhelminthes. Curr. Biol. 2017;27:R252–R256. doi: 10.1016/j.cub.2017.02.016. [DOI] [PubMed] [Google Scholar]
- Criscione C.D., Anderson J.D., Sudimack D., Peng W., Jha B., Williams-Blangero S., Anderson T.J.C. Disentangling hybridization and host colonization in parasitic roundworms of humans and pigs. Proc. R. Soc. B. Biol. Sci. 2007;274:2669–2677. doi: 10.1098/rspb.2007.0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva Alves E.B., Conceição M.J., Leles D. Ascaris lumbricoides, Ascaris suum, or “Ascaris lumbrisuum”? J. Infect. Dis. 2016;213 doi: 10.1093/infdis/jiw027. [DOI] [PubMed] [Google Scholar]
- Daly A.J., De Meester N., Baetens J.M., Moens T., De Baets B. Untangling the mechanisms of cryptic species coexistence in a nematode community through individual-based modelling. Oikos. 2021;130:587–600. doi: 10.1111/oik.07989. [DOI] [Google Scholar]
- Dantas-Torres F., Otranto D. Dirofilariosis in the Americas: a more virulent Dirofilaria immitis? Parasit. Vectors. 2013;6 doi: 10.1186/1756-3305-6-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ley P., Blaxter M.L. In: The biology of nematodes. Lee D., editor. Taylor & Francis; London: 2002. Systematic position and phylogeny; pp. 1–30. [Google Scholar]
- De Ley P., Blaxter M.L. In: Nematology monographs and perspectives. Cook R., Hunt D.J., editors. E.J. Brill; Leiden: 2004. A new system for Nematoda: combining morphological characters with molecular trees, and translating clades into ranks and taxa. [Google Scholar]
- De Queiroz K. Species concepts and species delimitation. Syst. Biol. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- De Vienne D.M., Refrégier G., López-Villavicencio M., Tellier A., Hood M.E., Giraud T. Cospeciation vs host-shift speciation: Methods for testing, evidence from natural associations and relation to coevolution. New Phytol. 2013;198:347–385. doi: 10.1111/nph.12150. [DOI] [PubMed] [Google Scholar]
- Donoghue M.J. A critique of the biological species concept and recommendations for a phylogenetic alternative. Bryologist. 1985;88:172. doi: 10.2307/3243026. [DOI] [Google Scholar]
- Dunams-Morel D.B., Reichard M.V., Torretti L., Zarlenga D.S., Rosenthal B.M. Discernible but limited introgression has occurred where Trichinella nativa and the T6 genotype occur in sympatry. Infect. Genet. Evol. 2012;12:530–538. doi: 10.1016/J.MEEGID.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Easton A., Gao S., Lawton S.P., Bennuru S., Khan A., Dahlstrom E., et al. Molecular evidence of hybridization between pig and human Ascaris indicates an interbred species complex infecting humans. eLife. 2020;9 doi: 10.7554/eLife.61562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B., Lapraz F., Tomiczek B., Müller S., Dessimoz C., Girstmair J., et al. A transcriptomic-phylogenomic analysis of the evolutionary relationships of flatworms. Curr. Biol. 2015;25:1347–1353. doi: 10.1016/j.cub.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin T.L. In: Taxonomy, phylogeny, and zoogeography of beetles and ants: A Volume Dedicated to the Memory of Philip Jackson Darlington Jr. 1904–1983. Ball G.E., editor. Dr. W. Junk b.v. Publishers; The Hague: 1985. The taxon pulse: A general pattern of lineage radiation and extinction among carabid beetles; pp. 437–472.http://repository.si.edu/xmlui/handle/10088/3381 [Google Scholar]
- Fahrenholz H. Ectoparasiten und Abstammungslehre. Zool. Anz. 1913;41:371–374. https://www.biodiversitylibrary.org/part/68763 [Google Scholar]
- Fenwick A. The global burden of neglected tropical diseases. Publ. Health. 2012;126:233–236. doi: 10.1016/J.PUHE.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Fialho P.M.M., Corrêa C.R.S. A systematic review of toxocariasis: A neglected but high-prevalence disease in Brazil. Am. J. Trop. Med. Hyg. 2016;94:1193–1199. doi: 10.4269/ajtmh.15-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fišer C., Robinson C.T., Malard F. Cryptic species as a window into the paradigm shift of the species concept. Mol. Ecol. 2018;27:613–635. doi: 10.1111/mec.14486. [DOI] [PubMed] [Google Scholar]
- Fogt-Wyrwas R., Dabert M., Jarosz W., Rząd I., Pilarczyk B., Mizgajska-Wiktor H. Molecular data reveal cryptic speciation and host specificity in Toxascaris leonina (Nematoda: Ascarididae) Vet. Parasitol. 2019;266:80–83. doi: 10.1016/j.vetpar.2019.01.002. [DOI] [PubMed] [Google Scholar]
- Franssen F., Bilska-Zajac E., Deksne G., Sprong H., Pozio E., Rosenthal B., et al. Genetic evidence of interspecies introgression of mitochondrial genomes between Trichinella spiralis and Trichinella britovi under natural conditions. Infect. Genet. Evol. 2015;36:323–332. doi: 10.1016/J.MEEGID.2015.10.005. [DOI] [PubMed] [Google Scholar]
- Frias L., Stark D.J., Lynn M.S., Nathan S.K., Goossens B., Okamoto M., MacIntosh A.J.J. Lurking in the dark: Cryptic Strongyloides in a bornean slow loris. Int. J. Parasitol. Parasites Wildl. 2018;7:141–146. doi: 10.1016/J.IJPPAW.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garamszegi L.Z. Patterns of co-speciation and host switching in primate malaria parasites. Malar. J. 2009;8:110. doi: 10.1186/1475-2875-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser R.B. Molecular tools - advances, opportunities and prospects. Vet. Parasitol. 2006;136:69–89. doi: 10.1016/J.VETPAR.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Georgieva S., Selbach C., Faltýnková A., Soldánová M., Sures B., Skírnisson K., Kostadinova A. New cryptic species of the “revolutum” group of Echinostoma (Digenea: Echinostomatidae) revealed by molecular and morphological data. Parasit. Vectors. 2013;6 doi: 10.1186/1756-3305-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva S., Faltýnková A., Brown R., Blasco-Costa I., Soldánová M., Sitko J., et al. Echinostoma ‘revolutum’ (Digenea: Echinostomatidae) species complex revisited: species delimitation based on novel molecular and morphological data gathered in Europe. Parasit. Vectors. 2014;7 doi: 10.1186/s13071-014-0520-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghai R.R., Chapman C.A., Omeja P.A., Davies T.J., Goldberg T.L. Nodule worm infection in humans and wild primates in Uganda: Cryptic species in a newly identified region of human transmission. PLoS Negl. Trop. Dis. 2014;8:39. doi: 10.1371/journal.pntd.0002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E., Wang S., Spiro D., Caler E., Zhao Q., Crabtree J., et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756. doi: 10.1126/SCIENCE.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons L.M., Jacobs D.E., Sani R.A. Toxocara malaysiensis n. sp. (Nematoda: Ascaridoidea) from the domestic cat (Felis catus Linnaeus, 1758) J. Parasitol. 2001;87:660–665. doi: 10.1645/0022-3395(2001)087[0660:TMNSNA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Goeze J. Versuch einer Naturgeschichte der Eingeweidewürmer thierischer Körper. P.A. Pape. 1782 [Google Scholar]
- Gottstein B., Pozio E., Nöckler K. Epidemiology, diagnosis, treatment, and control of trichinellosis. Clin. Microbiol. Rev. 2009;22:127–145. doi: 10.1128/CMR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halas D., Zamparo D., Brooks D.R. A historical biogeographical protocol for studying biotic diversification by taxon pulses. J. Biogeogr. 2005;32:249–260. doi: 10.1111/J.1365-2699.2004.01147.X. [DOI] [Google Scholar]
- Hasegawa H., Sato H., Fujita S., Nguema P.P.M., Nobusue K., Miyagi K., et al. Molecular identification of the causative agent of human strongyloidiasis acquired in Tanzania: Dispersal and diversity of Strongyloides spp. and their hosts. Parasitol. Int. 2010;59:407–413. doi: 10.1016/J.PARINT.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Haukisalmi V., Lavikainen A., Laaksonen S., Meri S. Taenia arctos n. sp. (Cestoda: Cyclophyllidea: Taeniidae) from its definitive (brown bear Ursus arctos Linnaeus) and intermediate (moose/elk Alces spp.) hosts. Syst. Parasitol. 2011;80:217–230. doi: 10.1007/S11230-011-9324-9. [DOI] [PubMed] [Google Scholar]
- Hayward A., Poulin R., Nakagawa S. A broadscale analysis of host-symbiont cophylogeny reveals the drivers of phylogenetic congruence. Ecol. Lett. 2021;24:1681–1696. doi: 10.1111/ele.13757. [DOI] [PubMed] [Google Scholar]
- Herrmann K.K., Poulin R., Keeney D.B., Blasco-Costa I. Genetic structure in a progenetic trematode: Signs of cryptic species with contrasting reproductive strategies. Int. J. Parasitol. 2014;44:811–818. doi: 10.1016/J.IJPARA.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Hoberg E., Monsen K., Kutz S., Blouin M. Structure, biodiversity, and historical biogeography of nematode faunas in Holarctic ruminants: Morphological and molecular diagnoses for Teladorsagia boreoarticus n. sp. (Nematoda: Ostertagiinae), dimorphic cryptic species in muskoxen (Ovibos moschatus) J. Parasitol. 1999;85:910–934. https://digitalcommons.unl.edu/parasitologyfacpubs/658 [PubMed] [Google Scholar]
- Hoberg E.P., Kocan A.A., Rickard L.G. In: Parasitic diseases of wild mammals. Samuel W.M., Pybus M.J., Kocan A.A., editors. Iowa State University Press; Ames: 2001. Gastrointestinal strongyles in wild ruminants; pp. 193–227.https://digitalcommons.unl.edu/parasitologyfacpubs/623 [Google Scholar]
- Hoberg E.P., Brooks D.R. A macroevolutionary mosaic: Episodic host-switching, geographical colonization and diversification in complex host-parasite systems. J. Biogeogr. 2008;35:1533–1550. doi: 10.1111/J.1365-2699.2008.01951.X. [DOI] [Google Scholar]
- Hoberg E.P., Brooks D. In: The geography of host-parasite interactions. Morand S., Krasnov B., editors. Oxford University Press; Oxford, UK: 2010. Beyond vicariance: Integrating taxon pulses, ecological fitting and oscillation in evolution and historical biogeography; pp. 7–20. [Google Scholar]
- Hoberg E.P., Brooks D.R. In: The balance of nature and human impact. Rohde K., editor. Cambridge University Press; Cambridge: 2013. Episodic processes, invasion and faunal mosaics in evolutionary and ecological time; pp. 199–214. [Google Scholar]
- Hoberg E.P., Cook J.A., Agosta S.J., Boeger W., Galbreath K.E., Laaksonen S., et al. Arctic systems in the Quaternary: Ecological collision, faunal mosaics and the consequences of a wobbling climate. J. Helminthol. 2017;91:409–421. doi: 10.1017/S0022149X17000347. [DOI] [PubMed] [Google Scholar]
- Holterman M., Karssen G., Van Den Elsen S., Van Megen H., Bakker J., Helder J. Small subunit rDNA-based phylogeny of the Tylenchida sheds light on relationships among some high-impact plant-parasitic nematodes and the evolution of plant feeding. Phytopathology. 2009;99:227–235. doi: 10.1094/PHYTO-99-3-0227. [DOI] [PubMed] [Google Scholar]
- Hua X., Wiens J.J. How does climate influence speciation? Am. Nat. 2013;182:1–12. doi: 10.1086/670690. [DOI] [PubMed] [Google Scholar]
- Hugot J.P. In: Micromammals and macroparasites: From evolutionary ecology to management. Morand S., Krasnov B.R., Poulin R., editors. Springer Tokyo; Tokyo: 2006. Coevolution of macroparasites and their small mammalian hosts: Cophylogeny and coadaptation; pp. 257–276. [Google Scholar]
- Hüttner M., Nakao M., Wassermann T., Siefert L., Boomker J.D.F., Dinkel A., et al. Genetic characterization and phylogenetic position of Echinococcus felidis (Cestoda: Taeniidae) from the African lion. Int. J. Parasitol. 2008;38:861–868. doi: 10.1016/j.ijpara.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Jaleta T.G., Lok J.B. Advances in the molecular and cellular biology of Strongyloides spp. Curr. Trop. Med. Rep. 2019;6:161–178. doi: 10.1007/S40475-019-00186-X/FIGURES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleta T.G., Zhou S., Bemm F.M., Schär F., Khieu V., Muth S., et al. Different but overlapping populations of Strongyloides stercoralis in dogs and humans - dogs as a possible source for zoonotic strongyloidiasis. PLoS Negl. Trop. Dis. 2017;11 doi: 10.1371/JOURNAL.PNTD.0005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz N., Nylin S. In: Specialization, speciation, and radiation: The evolutionary biology of herbivorous insects. Tilmon K., editor. University of California Press; California: 2008. The oscillation hypothesis of host-plant range and speciation; pp. 203–215. [Google Scholar]
- Janz N., Nylin S., Wahlberg N. Diversity begets diversity: Host expansions and the diversification of plant-feeding insects. BMC Evol. Biol. 2006;6:4. doi: 10.1186/1471-2148-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen D.H. On ecological fitting. Oikos. 1985;45:308. doi: 10.2307/3565565. [DOI] [Google Scholar]
- Jesudoss Chelladurai J., Murphy K., Snobl T., Bader C., West C., Thompson K., Brewer M.T. Molecular epidemiology of Ascaris infection among pigs in Iowa. J. Infect. Dis. 2017;215:131–138. doi: 10.1093/infdis/jiw507. [DOI] [PubMed] [Google Scholar]
- Jex A.R., Waeschenbach A., Littlewood D.T.J., Hu M., Gasser R.B. The mitochondrial genome of Toxocara canis. PLoS Negl. Trop. Dis. 2008;2:273. doi: 10.1371/journal.pntd.0000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W., Yan H., Lou Z., Ni X., Dyachenko V., Li H., Littlewood D.T.J. Mitochondrial genes and genomes support a cryptic species of tapeworm within Taenia taeniaeformis. Acta Trop. 2012;123:154–163. doi: 10.1016/j.actatropica.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Jin Y.C., Li X.Y., Liu J.H., Zhu X.Q., Liu G.H. Comparative analysis of mitochondrial DNA datasets indicates that Toxascaris leonina represents a species complex. Parasit. Vectors. 2019;12:194. doi: 10.1186/s13071-019-3447-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne A., Filter M., Gayda J., Buschulte A., Bandick N., Nöckler K., Mayer-Scholl A. Survival of Trichinella spiralis in cured meat products. Vet. Parasitol. 2020;287 doi: 10.1016/J.VETPAR.2020.109260. [DOI] [PubMed] [Google Scholar]
- Johnson K.P., Adams R.J., Page R.D.M., Clayton D.H. When do parasites fail to speciate in response to host speciation? Syst. Biol. 2003;52:37–47. doi: 10.1080/10635150390132704. [DOI] [PubMed] [Google Scholar]
- Kampfer S., Sturmbauer C., Ott J. Phylogenetic analysis of rDNA sequences from adenophorean nematodes and implications for the Adenophorea-Secernentea controversy. Invertebr. Biol. 1998;117:29–36. doi: 10.2307/3226849. [DOI] [Google Scholar]
- Kern E.M.A., Kim T., Park J.-K. The mitochondrial genome in nematode phylogenetics. Front. Ecol. Evol. 2020:250. doi: 10.3389/FEVO.2020.00250. [DOI] [Google Scholar]
- Khuntikeo N., Titapun A., Loilome W., Yongvanit P., Thinkhamrop B., Chamadol N., et al. Current perspectives on opisthorchiasis control and cholangiocarcinoma detection in Southeast Asia. Front. Med. 2018;5:117. doi: 10.3389/fmed.2018.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton N. Sibling species in the sea. Annu. Rev. Ecol. Systemat. 1993;24:189–216. doi: 10.1146/annurev.es.24.110193.001201. [DOI] [Google Scholar]
- Korhonen P.K., Pozio E., La Rosa G., Chang B.C.H., Koehler A.V., Hoberg E.P., et al. Phylogenomic and biogeographic reconstruction of the Trichinella complex. Nat. Commun. 2016;7 doi: 10.1038/NCOMMS10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulpa M., Nelson K.J., Morales A.M., Ryan B.M., Koschik M.L., Scott J.J., Verocai G.G. Presence of a cryptic Onchocerca species in black flies of northern California, USA. Parasit. Vectors. 2021;14:478. doi: 10.1186/s13071-021-04990-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutz S.J., Ducrocq J., Verocai G.G., Hoar B.M., Colwell D.D., Beckmen K.B., et al. Parasites in ungulates of Arctic North America and Greenland. A view of contemporary diversity, ecology, and impact in a world under change. Adv. Parasitol. 2012;79:99–252. doi: 10.1016/B978-0-12-398457-9.00002-0. [DOI] [PubMed] [Google Scholar]
- Lavikainen A., Haukisalmi V., Lehtinen M.J., Laaksonen S., Holmström S., Isomursu M., et al. Mitochondrial DNA data reveal cryptic species within Taenia krabbei. Parasitol. Int. 2010;59:290–293. doi: 10.1016/j.parint.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Lavikainen A., Iwaki T., Haukisalmi V., Konyaev S.V., Casiraghi M., Dokuchaev N.E., et al. Reappraisal of Hydatigera taeniaeformis (Batsch, 1786) (Cestoda: Taeniidae) sensu lato with description of Hydatigera kamiyai n. sp. Int. J. Parasitol. 2016;46:361–374. doi: 10.1016/J.IJPARA.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Lee C.C., Cheng N.A.B.Y., Bohari Y. Toxocara canis in domestic cats in Kuala Lumpur. Trop. Biomed. 1993;10:79–80. [Google Scholar]
- Lefoulon E., Bain O., Bourret J., Junker K., Guerrero R., Cañizales I., et al. Shaking the tree: Multi-locus sequence typing usurps current onchocercid (filarial nematode) phylogeny. PLoS Negl. Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0004233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leles D., Gardner S.L., Reinhard K., Ĩiguez A., Araujo A. Are Ascaris lumbricoides and Ascaris suum a single species? Parasit. Vectors. 2012;5:42. doi: 10.1186/1756-3305-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.Y., Jin Y.C., Nie Y., Su Y.G., Tang C.H., Li F. Sequence comparison and phylogenetic analysis of mithocondrial cox1 gene of Toxascaris leonina in Cheetah. China Anim. Health Insp. 2018;35:97–100. [Google Scholar]
- Liesner J.M., Krücken J., Schaper R., Pachnicke S., Kohn B., Müller E., et al. Vector-borne pathogens in dogs and red foxes from the federal state of Brandenburg, Germany. Vet. Parasitol. 2016;224:44–51. doi: 10.1016/j.vetpar.2016.05.012. [DOI] [PubMed] [Google Scholar]
- Lim B.K. Historical biogeography of New World emballonurid bats (tribe Diclidurini): Taxon pulse diversification. J. Biogeogr. 2008;35:1385–1401. doi: 10.1111/J.1365-2699.2008.01888.X. [DOI] [Google Scholar]
- Littlewood D.T.J., Bray R.A., editors. Interrelationships of the Platyhelminthes. CRC Press; Boca Raton, FL, USA: 2001. [Google Scholar]
- Littlewood D.T.J., Rohde K., Bray R.A., Herniou E.A. Phylogeny of the Platyhelminthes and the evolution of parasitism. Biol. J. Linn. Soc. 1999;68:257–287. doi: 10.1006/bijl.1999.0341. [DOI] [Google Scholar]
- Littlewood D.T.J., Rohde K., Clough K.A. The interrelationships of all major groups of Platyhelminthes: phylogenetic evidence from morphology and molecules. Biol. J. Linn. Soc. 1999;66:75–114. doi: 10.1006/bijl.1998.0276. [DOI] [Google Scholar]
- Lockyer A.E., Olson P.D., Littlewood D.T.J. Utility of complete large and small subunit rRNA genes in resolving the phylogeny of the Neodermata (Platyhelminthes): Implications and a review of the cercomer theory. Biol. J. Linn. Soc. 2003;78:155–171. doi: 10.1046/j.1095-8312.2003.00141.x. [DOI] [Google Scholar]
- Maldonado L.L., Arrabal J.P., Rosenzvit M.C., Oliveira G.C. De, Kamenetzky L. Revisiting the phylogenetic history of helminths through genomics, the case of the new Echinococcus oligarthrus genome. Front. Genet. 2019;10:708. doi: 10.3389/fgene.2019.00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiucci S., Cipriani P., Webb S.C., Paoletti M., Marcer F., Bellisario B., et al. Genetic and morphological approaches distinguish the three sibling species of the Anisakis simplex species complex, with a species designation as Anisakis berlandi n. sp. for A. simplex sp. C (Nematoda: Anisakidae) J. Parasitol. 2014;100:199–214. doi: 10.1645/12-120.1. [DOI] [PubMed] [Google Scholar]
- McFrederick Q.S., Haselkorn T.S., Verocai G.G., Jaenike J. Cryptic Onchocerca species infecting North American cervids, with implications for the evolutionary history of host associations in Onchocerca. Parasitology. 2013;140:1201–1210. doi: 10.1017/S0031182012001758. [DOI] [PubMed] [Google Scholar]
- McManus D.P. Current status of the genetics and molecular taxonomy of Echinococcus species. Parasitology. 2013;140:1617–1623. doi: 10.1017/S0031182013000802. [DOI] [PubMed] [Google Scholar]
- McVeigh P. Post-genomic progress in helminth parasitology. Parasitology. 2020;147:835. doi: 10.1017/S0031182020000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlhorn H. Encyclopedia of parasitology. 2016. [DOI]
- Meldal B.H.M., Debenham N.J., De Ley P., De Ley I.T., Vanfleteren J.R., Vierstraete A.R., et al. An improved molecular phylogeny of the Nematoda with special emphasis on marine taxa. Mol. Phylogenet. Evol. 2007;42:622–636. doi: 10.1016/j.ympev.2006.08.025. [DOI] [PubMed] [Google Scholar]
- Milton P., Hamley J.I.D., Walker M., Basáñez M.G. Moxidectin: An oral treatment for human onchocerciasis. Expert Rev. Anti Infect. Ther. 2020;18:1067–1081. doi: 10.1080/14787210.2020.1792772. [DOI] [PubMed] [Google Scholar]
- Mishler B.D. The morphological, developmental, and phylogenetic basis of species concepts in bryophytes. Bryologist. 1985;88:207. doi: 10.2307/3243030. [DOI] [Google Scholar]
- Mollaret I., Jamieson B.G.M., Adlard R.D., Hugall A., Lecointre G., Chombard C., Justine J.-L. Phylogenetic analysis of the Monogenea and their relationships with Digenea and Eucestoda inferred from 28S rDNA sequences. Mol. Biochem. Parasitol. 1997;90:433–438. doi: 10.1016/S0166-6851(97)00176-X. [DOI] [PubMed] [Google Scholar]
- Morgan J.A.T., Blair D. Relative merits of nuclear ribosomal internal transcribed spacers and mitochondrial CO1 and ND1 genes for distinguishing among Echinostoma species (Trematoda) Parasitology. 1998;116:289–297. doi: 10.1017/S0031182097002217. [DOI] [PubMed] [Google Scholar]
- Nadler S.A. Molecular approaches to studying helminth population genetics and phylogeny. Int. J. Parasitol. 1990;20:11–29. doi: 10.1016/0020-7519(90)90168-M. [DOI] [PubMed] [Google Scholar]
- Nadler S.A., Hudspeth D.S.S. Phylogeny of the Ascaridoidea (Nematoda: Ascaridida) based on three genes and morphology: Hypotheses of structural and sequence evolution. J. Parasitol. 2000;86:380–393. doi: 10.1645/0022-3395(2000)086[0380:POTANA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Nadler S.A., Pérez-Ponce de León G. Integrating molecular and morphological approaches for characterizing parasite cryptic species: Implications for parasitology. Parasitology. 2011;138:1688–1709. doi: 10.1017/S003118201000168X. [DOI] [PubMed] [Google Scholar]
- Nagayasu E., Aung M.P.P.T.H.H., Hortiwakul T., Hino A., Tanaka T., Higashiarakawa M., et al. A possible origin population of pathogenic intestinal nematodes, Strongyloides stercoralis, unveiled by molecular phylogeny. Sci. Rep. 2017;7:4844. doi: 10.1038/S41598-017-05049-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascetti G., Paggi L., Orecchia P., Smith J.W., Mattiucci S., Bullini L. Electrophoretic studies on the Anisakis simplex complex (Ascaridida: Anisakidae) from the Mediterranean and North-East Atlantic. Int. J. Parasitol. 1986;16:633–640. doi: 10.1016/0020-7519(86)90032-9. [DOI] [PubMed] [Google Scholar]
- Nejsum P., Parker E.D., Frydenberg J., Roepstorff A., Boes J., Haque R., et al. Ascariasis is a zoonosis in Denmark. J. Clin. Microbiol. 2005;43:1142–1148. doi: 10.1128/JCM.43.3.1142-1148.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeckler K., Pozio E., van der Giessen J., Hill D.E., Gamble H.R. International Commission on Trichinellosis: Recommendations on post-harvest control of Trichinella in food animals. Food Waterborne Parasitol. 2019;14 doi: 10.1016/J.FAWPAR.2019.E00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okulewicz A., Lonc E., Borgsteede F.H. Ascarid nematodes in domestic and wild terrestrial mammals. Polish J. Vet. Sci. 2002;5:277–281. [PubMed] [Google Scholar]
- Okulewicz A., Perec-Matysiak A., Buńkowska K., Hildebrand J. Toxocara canis, Toxocara cati and Toxascaris leonina in wild and domestic carnivores. Helminthologia. 2012;49:3–10. doi: 10.2478/s11687-012-0001-6. [DOI] [Google Scholar]
- Ondrejicka D.A., Locke S.A., Morey K., Borisenko A.V., Hanner R.H. Status and prospects of DNA barcoding in medically important parasites and vectors. Trends Parasitol. 2014;30:582–591. doi: 10.1016/J.PT.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Otranto D., Diniz D.G., Dantas-Torres F., Casiraghi M., de Almeida I.N.F., de Almeida L.N.F., et al. Human intraocular filariasis caused by Dirofilaria sp. nematode, Brazil. Emerg. Infect. Dis. 2011;17:863–865. doi: 10.3201/eid1705.100916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ponce de León G., Nadler S.A. What we don’t recognize can hurt us: A plea for awareness about cryptic species. J. Parasitol. 2010;96:453–464. doi: 10.1645/GE-2260.1. [DOI] [PubMed] [Google Scholar]
- Pérez-Ponce de León G., Nadler S.A. The importance of recognising parasite cryptic diversity for research programmes on foodborne trematodiases. Trans. R. Soc. Trop. Med. Hyg. 2016;110:4–5. doi: 10.1093/trstmh/trv090. [DOI] [PubMed] [Google Scholar]
- Pérez-Ponce de León G., Poulin R. An updated look at the uneven distribution of cryptic diversity among parasitic helminths. J. Helminthol. 2018;92:197–202. doi: 10.1017/S0022149X17000189. [DOI] [PubMed] [Google Scholar]
- Pinacho-Pinacho C.D., Sereno-Uribe A.L., Hernández-Orts J.S., García-Varela M., Pérez-Ponce de León G. Integrative taxonomy reveals an even greater diversity within the speciose genus Phyllodistomum (Platyhelminthes: Trematoda: Gorgoderidae), parasitic in the urinary bladder of Middle American freshwater fishes, with descriptions of five new species. Invertebr. Systemat. 2021;35:754–775. doi: 10.1071/IS21007. [DOI] [Google Scholar]
- Polderman A.M., Blotkamp J. Oesophagostomum infections in humans. Parasitol. Today. 1995;11:451–456. doi: 10.1016/0169-4758(95)80058-1. [DOI] [PubMed] [Google Scholar]
- Polderman A.M., Krepel H.P., Baeta S., Blotkamp J., Gigase P. Oesophagostomiasis, a common infection of man in Northern Togo and Ghana. Am. J. Trop. Med. Hyg. 1991;44:336–344. doi: 10.4269/ajtmh.1991.44.336. [DOI] [PubMed] [Google Scholar]
- Pozio E., Zarlenga D.S. New pieces of the Trichinella puzzle. Int. J. Parasitol. 2013;43:983–997. doi: 10.1016/J.IJPARA.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Rabajante J.F., Tubay J.M., Ito H., Uehara T., Kakishima S., Morita S., et al. Host-parasite Red Queen dynamics with phase-locked rare genotypes. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1501548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M., Rashid M.I., Akbar H., Ahmad L., Hassan M.A., Ashraf K., et al. A systematic review on modelling approaches for economic losses studies caused by parasites and their associated diseases in cattle. Parasitology. 2019;146:129–141. doi: 10.1017/S0031182018001282. [DOI] [PubMed] [Google Scholar]
- Robertson I.D., Thompson R.C. Enteric parasitic zoonoses of domesticated dogs and cats. Microb. Infect. 2002;4:867–873. doi: 10.1016/S1286-4579(02)01607-6. [DOI] [PubMed] [Google Scholar]
- Roe C.C., Urbanz J., Andrews L., Verocai G.G., Engelthaler D.M., Hepp C.M., Sahl J.W. Complete mitochondrial genome of Onchocerca lupi (Nematoda, Onchocercidae) Mitochondrial DNA B Resour. 2021;6:2572–2574. doi: 10.1080/23802359.2021.1960211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepstorff A., Mejer H., Nejsum P., Thamsborg S.M. Helminth parasites in pigs: New challenges in pig production and current research highlights. Vet. Parasitol. 2011;180:72–81. doi: 10.1016/J.VETPAR.2011.05.029. [DOI] [PubMed] [Google Scholar]
- Rohde K. Helminthen aus Katzen und Hunden in Malaya; Bemerkungen zu ihrer epidemiologischen Bedeutung für den Menschen. Z. Parasitenk. 1962;22:237–244. doi: 10.1007/BF00260009. [DOI] [Google Scholar]
- Rojas A., Dvir E., Farkas R., Sarma K., Borthakur S., Jabbar A., et al. Phylogenetic analysis of Spirocerca lupi and Spirocerca vulpis reveal high genetic diversity and intra-individual variation. Parasit. Vectors. 2018;11:639. doi: 10.1186/s13071-018-3202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A., Morales-Calvo F., Salant H., Otranto D., Baneth G. Focus: Zoonotic disease: Zoonotic ocular onchocercosis by Onchocerca lupi. Yale J. Biol. Med. 2021;94:331. [PMC free article] [PubMed] [Google Scholar]
- Romig T., Deplazes P., Jenkins D., Giraudoux P., Massolo A., Craig P.S., et al. Ecology and life cycle patterns of Echinococcus species. Adv. Parasitol. 2017;95:213–314. doi: 10.1016/bs.apar.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Romig T., Ebi D., Wassermann M. Taxonomy and molecular epidemiology of Echinococcus granulosus sensu lato. Vet. Parasitol. 2015;213:76–84. doi: 10.1016/j.vetpar.2015.07.035. [DOI] [PubMed] [Google Scholar]
- Rosen D.E. Fishes from the uplands and intermontane basins of Guatemala: Revisionary studies and comparative geography. Bull. Am. Mus. Nat. Hist. 1979;162:267–376. https://digitallibrary.amnh.org/handle/2246/1281 [Google Scholar]
- Russo L., Miller A.D., Tooker J., Bjornstad O.N., Shea K. Quantitative evolutionary patterns in bipartite networks: Vicariance, phylogenetic tracking or diffuse co-evolution? Methods Ecol. Evol. 2018;9:761–772. doi: 10.1111/2041-210X.12914. [DOI] [Google Scholar]
- Sadaow L., Sanpool O., Phosuk I., Rodpai R., Thanchomnang T., Wijit A., et al. Molecular identification of Ascaris lumbricoides and Ascaris suum recovered from humans and pigs in Thailand, Lao PDR, and Myanmar. Parasitol. Res. 2018;117:2427–2436. doi: 10.1007/s00436-018-5931-6. [DOI] [PubMed] [Google Scholar]
- Sah R., Khadka S., Hamal R., Poudyal S. Human echinostomiasis: A case report. BMC Res. Notes. 2018;11:17. doi: 10.1186/s13104-018-3133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijuntha W., Sithithaworn P., Wongkham S., Laha T., Pipitgool V., Tesana S., et al. Evidence of a species complex within the food-borne trematode Opisthorchis viverrini and possible co-evolution with their first intermediate hosts. Int. J. Parasitol. 2007;37:695–703. doi: 10.1016/j.ijpara.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B.J., Leducq J.-B., Mallet J. What is speciation? PLoS Genet. 2016;12 doi: 10.1371/JOURNAL.PGEN.1005860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R., Thompson P.C., Hoberg E.P., Brad Scandrett W., Konecsni K., Harms N.J., et al. Hiding in plain sight: Discovery and phylogeography of a cryptic species of Trichinella (Nematoda: Trichinellidae) in wolverine (Gulo gulo) Int. J. Parasitol. 2020;50:277–287. doi: 10.1016/j.ijpara.2020.01.003. [DOI] [PubMed] [Google Scholar]
- Sithithaworn P., Petney T.N., Andrews R.H. What significance do helminths species-complexes have for the prevention, diagnosis and treatment of human infections? Trans. R. Soc. Trop. Med. Hyg. 2015;109:289–290. doi: 10.1093/TRSTMH/TRV029. [DOI] [PubMed] [Google Scholar]
- Smythe A.B., Holovachov O., Kocot K.M. Improved phylogenomic sampling of free-living nematodes enhances resolution of higher-level nematode phylogeny. BMC Evol. Biol. 2019;19:121. doi: 10.1186/s12862-019-1444-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.R., Li K.X., Shi X., Hu W., Tan L.P., Luo Q. Phylogenetic analysis of three mithocondrial genes of Toxascaris leonina from South China tiger. Chin. J. Anim. Infect. Dis. 2015;23:68–75. [Google Scholar]
- Stewart T.B., Gasbarre L.C. The veterinary importance of nodular worms (Oesophagostomum spp.) Parasitol. Today. 1989;5:209–213. doi: 10.1016/0169-4758(89)90269-X. [DOI] [PubMed] [Google Scholar]
- Struck T.H., Cerca J. Cryptic species and their evolutionary significance. eLS. 2019:A0028292. doi: 10.1002/9780470015902.A0028292. [DOI] [Google Scholar]
- Suzuki J., Kobayashi S., Okata U., Matsuzaki H., Mori M., Chen K.R., Iwata S. Molecular analysis of Dirofilaria repens removed from a subcutaneous nodule in a Japanese woman after a tour to Europe. Parasite. 2015;22:2. doi: 10.1051/parasite/2015002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamsborg S.M., Ketzis J., Horii Y., Matthews J.B. Strongyloides spp. infections of veterinary importance. Parasitology. 2017;144:274–284. doi: 10.1017/S0031182016001116. [DOI] [PubMed] [Google Scholar]
- Thompson J.N. Specific hypotheses on the geographic mosaic of coevolution. Am. Nat. 1999;153:S1–S14. doi: 10.1086/303208. [DOI] [Google Scholar]
- Thompson J.N. University of Chicago Press; Chicago: 2005. The geographic mosaic of coevolution. [Google Scholar]
- Thompson R. Parasite zoonoses and wildlife: One health, spillover and human activity. Int. J. Parasitol. 2013;43:1079–1088. doi: 10.1016/J.IJPARA.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.W., Wong S.S.Y., Poon R.W.S., Trendell-Smith N.J., Ngan A.H.Y., Lam J.W.K., et al. A novel Dirofilaria species causing human and canine infections in Hong Kong. J. Clin. Microbiol. 2012;50:3534–3541. doi: 10.1128/JCM.01590-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Herwerden L., Blair D., Agatsuma T. Intra- and interindividual variation in ITS1 of Paragonimus westermani (Trematoda: Digenea) and related species: iImplications for phylogenetic studies. Mol. Phylogenet. Evol. 1999;12:67–73. doi: 10.1006/mpev.1998.0572. [DOI] [PubMed] [Google Scholar]
- Van Megen H., Van Den Elsen S., Holterman M., Karssen G., Mooyman P., Bongers T., et al. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology. 2009;11:927–950. doi: 10.1163/156854109X456862. [DOI] [Google Scholar]
- Verocai G.G., Kutz S.J., Simard M., Hoberg E.P. Varestrongylus eleguneniensis sp. n. (Nematoda: Protostrongylidae): A widespread, multi-host lungworm of wild North American ungulates, with an emended diagnosis for the genus and explorations of biogeography. Parasit. Vectors. 2014;7:556. doi: 10.1186/S13071-014-0556-9/TABLES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verocai G.G., Lejeune M., Beckmen K.B., Kashivakura C.K., Veitch A.M., Popko R.A., et al. Defining parasite biodiversity at high latitudes of North America: New host and geographic records for Onchocerca cervipedis (Nematoda: Onchocercidae) in moose and caribou. Parasit. Vectors. 2012;5:242. doi: 10.1186/1756-3305-5-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verocai G.G., Nelson K.J., Callahan R.T., Wekesa J.W., Hassan H.K., Hoberg E.P. A cryptic species of Onchocerca (Nematoda: Onchocercidae) in blackflies (Simulium spp.) from southern California, USA. Parasit. Vectors. 2018;11:547. doi: 10.1186/s13071-018-3133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilas R., Criscione C.D., Blouin M.S. A comparison between mitochondrial DNA and the ribosomal internal transcribed regions in prospecting for cryptic species of platyhelminth parasites. Parasitology. 2005;131:839–846. doi: 10.1017/S0031182005008437. [DOI] [PubMed] [Google Scholar]
- Wen H., Vuitton L., Tuxun T., Li J., Vuitton D.A., Zhang W., McManus D.P. Echinococcosis: Advances in the 21st century. Clin. Microbiol. Rev. 2019;32:e00075. doi: 10.1128/CMR.00075-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulcan J.M., Dennis M.M., Ketzis J.K., Bevelock T.J., Verocai G.G. Strongyloides spp. in cats: A review of the literature and the first report of zoonotic Strongyloides stercoralis in colonic epithelial nodular hyperplasia in cats. Parasit. Vectors. 2019;12:349. doi: 10.1186/S13071-019-3592-7/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrobisz A., Kowal J., Nosal P. Insight into species diversity of the Trichostrongylidae Leiper, 1912 (Nematoda: Strongylida) in ruminants. J. Helminthol. 2016;90:639–646. doi: 10.1017/S0022149X15001017. [DOI] [PubMed] [Google Scholar]
- Wyrobisz-Papiewska A., Kowal J., Nosal P., Chovancová G., Rehbein S. Host specificity and species diversity of the Ostertagiinae Lopez-Neyra, 1947 in ruminants: A European perspective. Parasit. Vectors. 2018;11:369. doi: 10.1186/s13071-018-2958-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier R., Faria P.J., Paladini G., Van Oosterhout C., Johnson M., Cable J. Evidence for cryptic speciation in directly transmitted gyrodactylid parasites of Trinidadian guppies. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Li H., Wang C., Li Y., Liu Y., Meng X., et al. Characterization of the complete mitochondrial genome sequence of the dog roundworm Toxascaris leonina (Nematoda, Ascarididae) from China. Mitochondrial DNA Part B. 2019;4:3517–3519. doi: 10.1080/23802359.2019.1675545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Li Y., Gu X., Liu Y., Zhou X., Wang L., et al. Molecular characterization of ascaridoid parasites from captive wild carnivores in China using ribosomal and mitochondrial sequences. Parasit. Vectors. 2020;13:382. doi: 10.1186/s13071-020-04254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. The BPP program for species tree estimation and species delimitation. Curr. Zool. 2015;61:854–865. doi: 10.1093/CZOOLO/61.5.854. [DOI] [Google Scholar]
- Yilmaz E., Fritzenwanker M., Pantchev N., Lendner M., Wongkamchai S., Otranto D., et al. The mitochondrial genomes of the zoonotic canine filarial parasites Dirofilaria (Nochtiella) repens and Candidatus Dirofilaria (Nochtiella) hongkongensis provide evidence for presence of cryptic species. PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/JOURNAL.PNTD.0005028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarlenga D., Thompson P., Pozio E. Trichinella species and genotypes. Res. Vet. Sci. 2020;133:289–296. doi: 10.1016/J.RVSC.2020.08.012. [DOI] [PubMed] [Google Scholar]
- Zhou C., Chen J., Niu H., Ouyang S., Wu X. Study on the population evolution of Ascaris lumbricoides and Ascaris suum based on whole genome resequencing. Vet. Parasitol. 2020;279 doi: 10.1016/j.vetpar.2020.109062. [DOI] [PubMed] [Google Scholar]
- Zhu X.Q., Jacobs D.E., Chilton N.B., Sani R.A., Cheng N.A.B.Y., Gasser R.B. Molecular characterization of a Toxocara variant from cats in Kuala Lumpur, Malaysia. Parasitology. 1998;117:155–164. doi: 10.1017/S0031182098002856. [DOI] [PubMed] [Google Scholar]