Abstract
Malaria is the most serious mosquito‐borne parasitic disease, caused mainly by the intracellular parasite Plasmodium falciparum. The parasite invades human red blood cells and releases extracellular vesicles (EVs) to alter its host responses. It becomes clear that EVs are generally composed of sub‐populations. Seeking to identify EV subpopulations, we subject malaria‐derived EVs to size‐separation analysis, using asymmetric flow field‐flow fractionation. Multi‐technique analysis reveals surprising characteristics: we identify two distinct EV subpopulations differing in size and protein content. Small EVs are enriched in complement‐system proteins and large EVs in proteasome subunits. We then measure the membrane fusion abilities of each subpopulation with three types of host cellular membranes: plasma, late and early endosome. Remarkably, small EVs fuse to early endosome liposomes at significantly greater levels than large EVs. Atomic force microscope imaging combined with machine‐learning methods further emphasizes the difference in biophysical properties between the two subpopulations. These results shed light on the sophisticated mechanism by which malaria parasites utilize EV subpopulations as a communication tool to target different cellular destinations or host systems.
Keywords: AFM, asymmetric flow field‐flow fractionation, extracellular vesicles, malaria, membrane fusion
Subject Categories: Membranes & Trafficking; Microbiology, Virology & Host Pathogen Interaction; Signal Transduction
Plasmodium falciparum invades human red blood cells and releases two extracellular vesicle subsets secreted by infected cells. These EV subpopulations harbor different protein cargo and have specific mechanical membrane properties, suggesting distinct host cell targets.
Introduction
Extracellular vesicles (EVs) are membranous particles released by cells of a wide range of living organisms, including pathogens, into their microenvironment carrying bioactive cargo such as RNA, DNA, proteins and lipids (Coakley et al, 2015; Schorey et al, 2015; Malda et al, 2016; Osteikoetxea et al, 2016; Théry et al, 2018; Ofir‐Birin & Regev‐Rudzki, 2019; Pegtel & Gould, 2019; Wortzel et al, 2019; Torrecilhas et al, 2020). These particles are often classified according to their size, biogenesis and biochemical features into three main groups: exosomes, microvesicles (MVs) (Chaavez et al, 2019) and apoptotic bodies (Willms et al, 2018). EVs are isolated using various methods, including differential ultracentrifugation, size exclusion chromatography (SEC), density gradient flotation and gel filtration (Gardiner et al, 2016; Théry et al, 2018). Until recently, three main groups of secreted vesicles were classified; exosomes, derived from multivesicular bodies originating from late endosomes with typical diameters in the range of 30–150 nm, microvesicles resulting from the budding of the cell membrane which measures from 100 to 1,000 nm (Doyle & Wang, 2019), and apoptotic bodies released from the cell surface during apoptosis which reach sizes of 5,000 nm (Osteikoetxea et al, 2016; Willms et al, 2018; Doyle & Wang, 2019). Previously, cancer‐derived exosomes have been classified into three subpopulations, large exosome vesicles (Exo‐L) 90–120 nm, small exosome vesicles (Exo‐S) 60–80 nm and non‐membranous nanoparticles termed (exomeres) ~35 nm (Zhang et al, 2018, 2019). Given this evolving notion that the ensemble of secreted vesicles consists of several sub‐groups (Mathieu et al, 2021), numerous technologies have been established to identify the distinct EV sub‐groups including flow cytometry and asymmetric flow field‐flow fractionation (AF4) (Gardiner et al, 2016; Willms et al, 2018).
AF4 separates particles based on their radius of gyration (i.e., size) in a channel consisting of a permeable and a semi‐permeable membrane, with two different flows across the chamber: a transverse cross‐flow that pushes the molecules toward the semi‐permeable membrane, and a longitudinal flow that moves the molecules forward to the detector (Willms et al, 2018). In this way, small particles with high diffusion coefficients are eluted earlier than larger particles that are accumulated in the membrane. Therefore, the equilibrium between the two orthogonal flows will dictate the resolution of a complex sample into distinct particle subpopulations (Zhang & Lyden, 2019). In addition, AF4 devices are coupled with UV and multi‐angle light scattering (MALS) detectors, which provide further information about the protein content and estimated radius of the particles. Indeed, AF4 has been used for the isolation of EVs from neurons (Kang et al, 2008), lymphoblastoid B cells (Sitar et al, 2015), urine (Oeyen et al, 2018) and for the analysis and identification of EV sub‐populations from various cancer cell lines (Zhang et al, 2018).
The Apicomplexa parasite Plasmodium falciparum (Pf) is one of the most virulent malaria species and is responsible for most clinical cases, mainly among children and pregnant women (Cowman et al, 2016). Malaria‐infected RBCs secrete EVs containing parasite‐derived DNA, RNA and protein cargo (Sisquella et al, 2017; Toda et al, 2020; Avalos‐Padilla et al, 2021; Dekel et al, 2021; Ofir‐Birin et al, 2021). These secreted EVs have been implicated in host pathogen crosstalk with cells of the immune system (Mantel et al, 2013; Sisquella et al, 2017; Sampaio et al, 2018; Ye et al, 2018; Demarta‐Gatsi et al, 2019; Ofir‐Birin et al, 2021), in parasite–parasite communication (Mantel et al, 2013; Regev‐Rudzki et al, 2013), in host RBC priming (Dekel et al, 2021) and in erythropoiesis (Neveu et al, 2020). Mature RBCs lack internal organelles, endocytosis and exocytosis machinery (Moras et al, 2017), indeed it has been demonstrated that the Plasmodium parasite utilizes its own protein network to release EVs (Regev‐Rudzki et al, 2013; Avalos‐Padilla et al, 2021). It was demonstrated that the parasitic protein PfPTP2 is involved in the budding of the EVs from Pf‐iRBCs (Regev‐Rudzki et al, 2013). Additionally, Pf possesses functional ESCRT‐III machinery to produce EVs, involving the action of the parasitic proteins PfBro1 and PfVps32/PfVps60 (Avalos‐Padilla et al, 2021).
In this study, using the AF4 apparatus combined with both atomic force microscopy (AFM) and cryo‐transmission electron microscopy (cryo‐TEM), we resolved the EV subpopulations released by Pf‐iRBCs. Interestingly, we found two distinct subpopulations of EVs that differed in their average size, one being 26 nm (EV fraction 3) and the other 69 nm (EV fraction 4). We next performed proteomic analysis on these EV subpopulations and showed that the enriched cellular pathways are different for each subpopulation with complement system proteins in the small EVs and proteasome proteins in the largest EVs. Moreover, we employed Laurdan staining assay and AFM puncture analysis combined with machine learning methods to show that fraction 3 (F3‐EVs) exhibit a more tightly packed membrane as compared to fraction 4 (F4‐EVs), expressed by the larger force required to pierce the membrane, with two distinct sub‐populations comprising the different samples. Lastly, using Förster resonance energy transfer (FRET)‐based fusion assay we demonstrated that the EV subpopulations are able to fuse to liposomes, mimicking the plasma membrane and early and late endosomes. Surprisingly, F3‐EVs fuse in early endosomal conditions at significantly greater levels as compared to F4‐EVs, suggesting cargo release at different subcellular localizations for each of these EV subsets. Our data demonstrate that Pf‐iRBCs release two distinct subpopulations of EVs which differ in their protein content, biophysical properties and fusion capacity. Therefore, this study may shed light on how malaria parasites utilize the powerful EV pathway to secrete particles responsible for discrete functions in the parasite–host interaction.
Results
EVs derived from Pf‐iRBCs are divided into two distinct subpopulations
To identify EV subpopulations released by Pf‐iRBCs we set up the AF4 system (schematic and workflow illustration Fig 1A), in which the entire EV population harvested from Pf‐iRBCs (Fig EV1A) was injected into the apparatus. The analysis revealed a separation profile of four main particle fractions (Fig 1B) based on the separation of the Pf‐derived EVs via hydrodynamic radius (Fig 1C and D using the AF4 cross and longitudinal flows). The distribution of fraction sizes ranged from 10 nm up to 350 nm (Fig 1C). The particles from the two initial fractions, 1 and 2 were collected from 9 to 12.5 min and from 13 to 17.5 min (Fig 1B), respectively. The average radius of particles from fraction 1 was 18 ± 8 nm and for fraction 2 it was 10 ± 10 nm (Fig 1D). Fraction 3 ranged from 25 to 45 min and particles were 31 ± 9 nm. Lastly, fraction 4 appeared between 50 and 75 min and the average particle size was 168 ± 22 nm (Fig 1D). Samples from each fraction were collected and concentrated for further analysis. The estimated radii of gyration obtained from the AF4 apparatus were calculated by applying an intensity distribution function fitted to a random coil model to monitor the size throughout the separation process. The random coil model cannot, however, reliably deduce sizes for dispersed proteins in solution and is suboptimal for measurement of vesicle radii, therefore it was only employed for monitoring and not to obtain a true measurement of the EV fractions, which were more accurately quantified with AFM and cryo‐TEM analysis.
Figure 1. AF4 fractionation profile of Pf‐derived EVs.
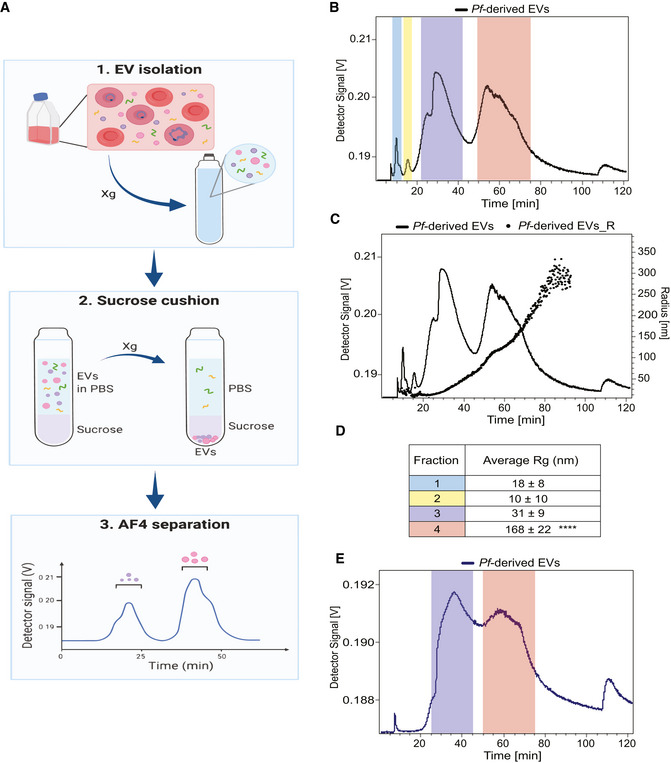
- Illustration demonstrating the workflow of an AF4 separation. The xg refers to centrifugation at 150,000 g or 100,000 g.
- Graph shows the four peaks collected from Pf‐derived EVs (continuous black line) post AF4 separation. Fractions 1 and 2 were collected from 9 to 12.5 and from 13 to 17.5 min respectively, fraction 3 from 25 to 45 and fraction 4 from 50 to 75 min.
- Representative overlay profile from (B) with the radius of gyration (Rg) of the Pf‐derived EVs, latter displayed as dots.
- Table summarizing the average size distribution and standard deviation of the four peaks based on their Rg for three separate biological replicates. The F4‐EV Rg is significantly larger than the others (2‐way ANOVA, accounting for batch effect followed by Tukey's post hoc test; p (4‐3) = 0.0000923, p (4‐2) = 0.0000405 and p (4‐1) = 0.0000558), ****P < 0.0001.
- Continuous blue trace shows the two peaks collected from Pf‐derived EVs post sucrose cushion for further analysis. Color‐coding as in (B) and (D) above. These results are a representative sample taken from one of three independent biological repeats.
Figure EV1. AF4 analysis separation of Pf‐derived EVs.
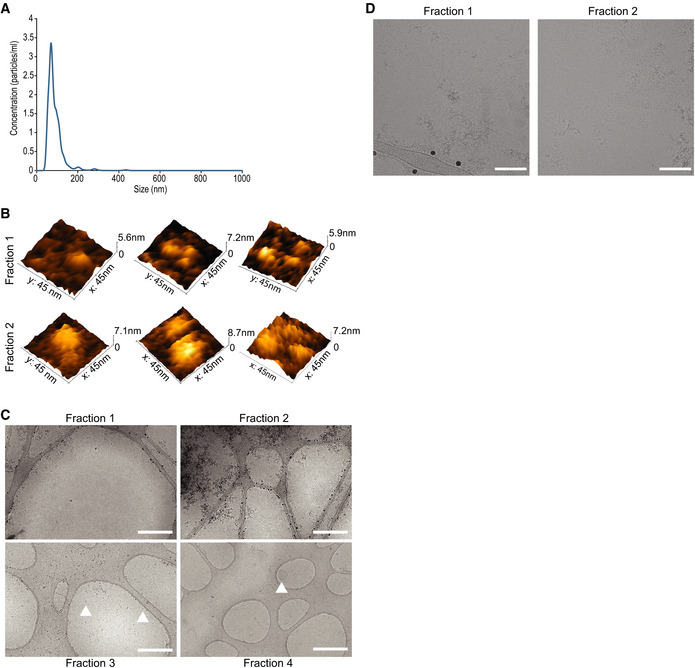
- Nanotrack particle analysis (NTA) of the input Pf‐derived EVs. Assessment of total Pf‐derived EVs using NanoSight® particle analyzer detecting particles in a diameter mode of 70 nm. The concentration is approximately 1012 particles/ml. This experiment is representative of three biological replicates.
- Representative 3D AFM images of particles from fractions 1 and 2.
- Representative cryo‐TEM images at low magnification of fractions 1–4 to illustrate sample purity and isolation of extracellular proteins (fractions 1 and 2) from extracellular vesicles (fractions 3 and 4). Scale bar—500 nm. Arrowheads indicate EVs.
- Cryo‐TEM images of fractions 1 and 2 highlighting bio‐molecular aggregates. Scale bar—100 nm.
Based on the size analysis we hypothesized that the first two fractions are protein aggregates while the last two fractions contain two types of EV subpopulations. Thus, we set the system to further purify the particle subpopulations by subjecting the fractions to a sucrose cushion separation (Alexander et al, 2016). Isolating EVs by ultracentrifugation is known to lead to co‐sedimentation of protein aggregates with the EVs (Langevin et al, 2019). Thus, to further separate vesicle structures prior to AF4 analysis, EVs were subjected to a 20% sucrose cushion (Alexander et al, 2016; Langevin et al, 2019), a method that reduces protein aggregate sediments. Indeed, AF4 separation post‐sucrose cushion revealed only two major peaks (Fig 1E).
Atomic force microscopy imaging analysis showed a clear difference in the particle size distribution of the four different fractions prior to the sucrose cushion (Figs 2A and B, and EV1B). The average radii of particles from fractions 1 to 4 were 14 ± 5, 19 ± 8, 26 ± 14 and 69 ± 46 nm, respectively.
Figure 2. AFM analysis of Pf‐derived EV subpopulations.
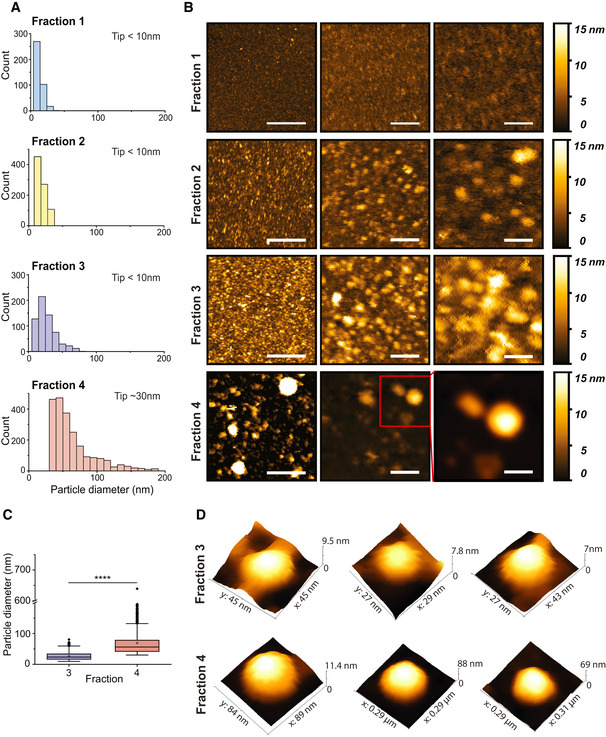
- Distributions of EV sizes in fractions 1–4 measured from AFM images. Size distributions were quantified combining three separate biological replicates, for a total of fraction 1 n = 391, fraction 2 n = 853, fraction 3 n = 623 and fraction 4 n = 2251.
- Representative AFM images for each fraction at different magnifications. Scale bars are (left to right)—400, 100 and 50 nm.
- EV size analysis results for fractions 3 and 4 presented as box plots. The black dots represent outliers and the black horizontal line represents the median, the black diamonds represent outliers, boxes represent 25–75% of the data and the lines inside the boxes are the medians. Minimum and maximum values are indicated by the whiskers. F4‐EV values were larger than F3‐EVs (t‐test on log‐transformed values; t 978 = 42.3, P = 2.2e−16; effect size: Cohen's D = 1.94). ****P < 0.0001.
- Representative AFM 3D images of single EVs for each fraction.
Biophysical characterizations of Pf‐derived EV subpopulations
Atomic force microscopy 3D imaging analysis confirmed that EVs are present in both fractions 3 and 4 but not in fractions 1 and 2 (Figs 2C and D, and EV1B); the results presented in the box plot (Fig 2C) clearly show that a wider distribution of sizes is present in fraction 4 compared to fraction 3. Particle size distributions of 10–70 and 30–300 nm were imaged in fractions 3 and 4, respectively. Additionally, 3D images of EV subpopulations obtained by the AFM revealed particles with a morphology (diameter and height) typical for EV structures (Fig 2D).
Cryo‐TEM imaging of the resulting AF4 fractions was employed to further validate the AFM measurements and indeed demonstrated that both fractions 3 and 4 contained EVs with distinct size distributions. No vesicular structures were detected in fractions 1 and 2 (Fig EV1C and D). Vesicles isolated in fraction 3 (F3‐EVs) displayed a smaller size (Fig 3A top panel), and a narrow size distribution, with an average diameter of 55 ± 11 nm (Fig 3B, median: 53 nm), compared to EVs from fraction 4 (F4‐EVs), which were larger (Fig 3A bottom panel) and presented a broader size distribution (108 ± 53 nm; median: 81 nm, Fig 3B).
Figure 3. Cryo‐TEM imaging of Pf‐derived EV fractions reveals two distinct size populations.
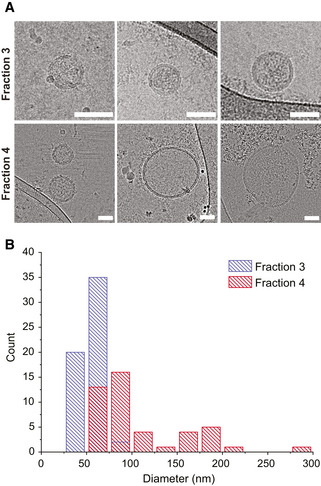
- Representative cryo‐TEM micrographs for fraction 3 (top row) and fraction 4 (bottom row) for Pf‐derived EVs post AF4 fractionation. Both subpopulations display a clear vesicular structure with a visible 4 nm bilayer. Fraction 3 shows a typical smaller diameter in the 30–70 nm range, whereas fraction 4 displays a much broader size, ranging from 70 to 300 nm. Scale bar—50 nm.
- Diameter distribution of both fractions measured from cryo‐TEM images of the two EV subpopulations. Fraction 3 (red bars), consistent with representative images, has a narrow distribution centered at 50 nm. Fraction 4 (blue bars), presents a broader size distribution with two putative peaks, centered at 70 and 170 nm, respectively. The two subpopulations display a size overlap in the 75 nm range. Size distributions were quantified from three separate biological replicates of the two subpopulations, for a total of n = 57 vesicles for fraction 3 and n = 45 vesicles for fraction 4, with at least 7 vesicles for each biological repeat. Data presented as count distributions with a 25 nm bin size.
Measurements of the size distribution from Cryo‐TEM images for the two fractions show a partial overlap between F3‐EVs and F4‐EVs in the 60–70 nm size range (Fig 3B). These results are in agreement with those of AFM (although the size ranges are slightly different here due to statistical fluctuations for the cryo‐TEM where fewer images were obtained), indicating that Pf‐derived EVs are composed of two subpopulations with different typical sizes.
Distinct cellular protein enrichments of Pf‐derived EV subpopulations
Having established that Pf‐iRBCs secrete two distinct EV subpopulations we directed efforts to investigate their protein cargo, by performing quantitative label‐free proteomics analysis (Fig 4A–C). Importantly, one of the major limitations of AF4 technology is that each EV subpopulation is collected at a relatively low concentration, which leads to insufficient protein quantities for western blot analysis. We thus pursued other means for the proteomic analysis for the two EV fractions (F3‐EVs and F4‐EVs). The EV proteins were extracted, trypsinized, separated by capillary liquid chromatography and analyzed by tandem mass spectrometry (LC/MS/MS). The abundance profile was compared: 132 proteins were identified, of which 23 were Pf‐derived proteins and the rest (109) were human (Fig 4A, Dataset EV1). Pf proteins included ribosomal proteins, histones, chaperones, metabolic enzymes, transmembrane proteins and several coiled‐coil proteins with unknown functions (N = 23) (Dataset EV1). Only proteins that were quantified in at least two repeats from the same group (F3‐EVs or F4‐EVs) were taken for further analysis. Sixty‐six proteins were significantly different comparing the F3 and F4 fractions at q‐value <0.05 and 20 were unique to one of the groups (Fig 4B). Two of the Pf proteins were unique to the F4‐EVs: PFNF54_02665 which is a Ring‐exported protein 1, and PFNF54_04990, a 60S ribosomal protein L27. Two other highly differential proteins are PFNF54_01656 which is a small exported membrane protein 1 and PFNF54_05498, a chaperonin (HSP60). On the other hand, PFNF54_03023, Histone H4 and PFNF54_04974, glyceraldehyde‐3‐phosphate dehydrogenase were unique to F3‐EVs (Dataset EV2).
Figure 4. Proteomic analysis of Pf‐derived EV subpopulations.
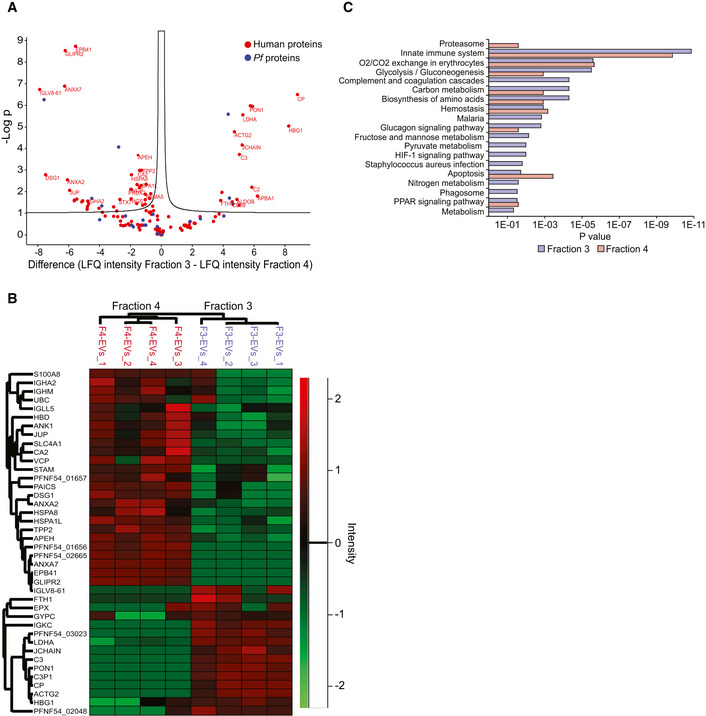
- Volcano Plot of differentially expressed EV proteins between F3‐EVs and F4‐EVs. The Volcano Plot presents significant results of a t‐test with permutation‐based FDR calculation by Log2 fold change (difference) and significance (−Log P) using the Perseus software.
- Euclidean heat map diagram presenting an unsupervised hierarchical clustering of the different EV groups. The clustering was based on the intensities of the identified proteins and was generated by the Perseus software—a zoom on the differential proteins is presented.
- Pathway analysis of the two EV fractions based on STRING. Quantification and averaging of four independent biological replicates. Reference list: Homo sapiens or Plasmodium falciparum (all genes in the database).
Among the human host proteins, many enzymes involved in cell signaling, endocytosis and cell motility were identified (Fig EV2A and B). Notably, proteolysis related proteins were more prevalent in the F4‐EVs (Fig 4C) including proteasome subunits (PSMB6, PSMA5, VCP and PSMA1) and these results are in line with a previous study which found that the 20S proteasome complexes are encapsulated within Pf‐derived EVs (Dekel et al, 2021; Sharon & Regev‐Rudzki, 2021). We also identified within the EVs, ESCRT proteins (HGS, STAM) and proteins involved in metabolic processes (APEH, VCP, TPP2, UBC). In addition, cell redox homeostasis (PRDX2, PRDX6 and TXN) was identified mainly in F4‐EVs (Dataset EV3). On the other hand, proteins that regulate the complement system activation (C3, C2 and CD59) were mainly identified in the F3‐EV sample (Fig 4C, Dataset EV3). General examination of the subcellular localization annotation of proteins revealed comparable localizations of the two fractions (Fig EV2C).
Figure EV2. Proteomic analysis of Pf‐derived EV subpopulations.
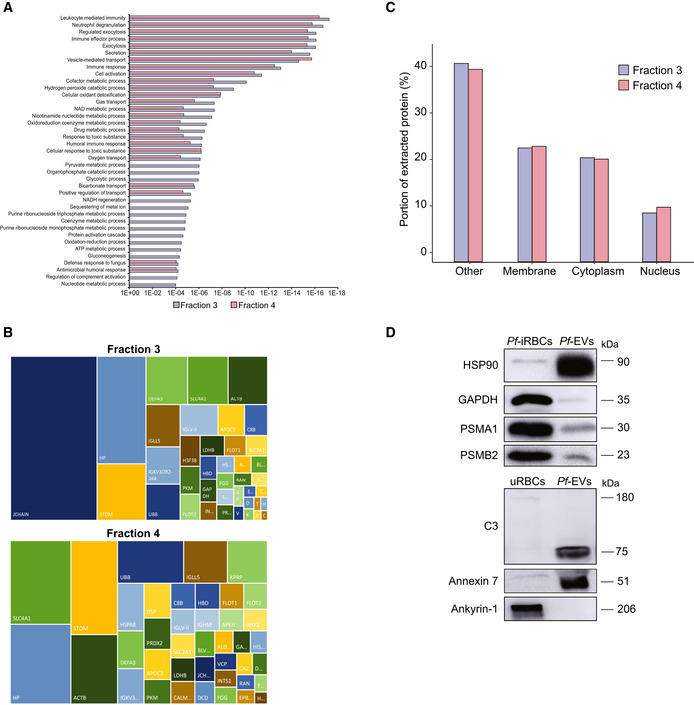
- GO annotation analysis of the two EV fractions.
- The figure presents the most intense proteins in the EV repertoire (not including the HBB and HBA dominant proteins). The rectangular areas are proportional to the median intensity of the protein in either F3 or F4 EVs.
- Subcellular localization of identified proteins by GO annotation. Other—mitochondria, early and late endosome, lysosome, cytoskeleton, secreted, and so on. Quantification and averaging of four independent biological replicates. Reference list: Homo sapiens or Plasmodium falciparum (all genes in the database).
- Western blot analysis of uRBCs or iRBCs and Pf‐derived EVs. Anti‐PSMA1, anti‐PSMB2 (proteasome subunits) and anti‐C3 (complement system protein) antibodies were used. As a positive control, antibodies against the EV markers HSP90 and Annexin 7 were used. C3b subunit is detected within Pf‐derived EVs. As a negative control, antibodies against the RBC membrane marker Ankyrin‐1 were used.
Although EV levels in each fraction of the post AF4 separation process were relatively low, we further confirmed the presence of EV markers (HSP90, Annexin 7 and GAPDH), RBC membrane marker (Ankyrin‐1), complement (C3) and proteasome (PSMA1 and PSMB2) proteins using WB analysis for the EV pool (Fig EV2D). These data indicate that the malaria parasite releases two EV subpopulations that differ in size and protein content, suggesting a distinct role for each subpopulation.
Pf‐derived EV subpopulations have distinct fusion capabilities toward different subcellular membranes
The existence of two separate vesicle subpopulations, with different size distributions and distinct protein cargo enrichment, suggests that each individual subset may be dedicated to a specific function, and therefore might display specificity or preference toward certain cellular targets. Secreted EVs interacting with recipient cells will encounter different environments across their internalization pathway. The initial contact for EVs will be the cell plasma membrane, and subsequently uptake will occur leading to endocytosis, characterized by progressive acidification of the pH and a change of the endosomal lipid composition as maturation from early to late endosome takes place (Huotari & Helenius, 2011).
We therefore aimed to examine whether the EV subpopulations might possess different preferences in fusing with membranes of such cellular compartments. We used a FRET‐based membrane mixing assay with the membrane dye pair DiI and DiD. Membrane mixing is probed by monitoring the increase of DiI intensity upon fusion or hemifusion between membranes, where both the pH and apposing membrane composition can be tailored to match different cellular destinations (Holt et al, 2008; Marsden et al, 2011; Kiessling et al, 2015; Lira et al, 2019; Dekel et al, 2021; preprint: Morandi et al, 2022) (illustrated in Fig 5A).
Figure 5. Pf‐derived EV subpopulations fuse with liposomes (LUVs) with a composition resembling plasma membrane, early endosome and late endosome.
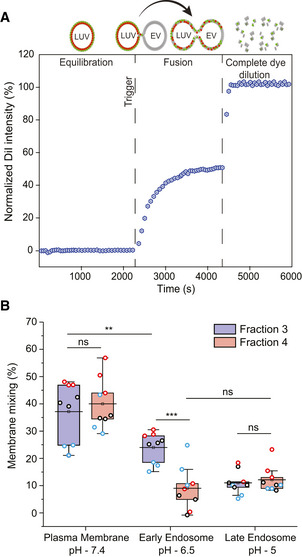
- A schematic diagram illustrating the principle of the FRET‐based two‐color dye for detection of fusion between EVs and LUVs.
- Box plot of membrane mixing assay between LUVs mimicking the plasma membrane, early endosome and late endosome lipid composition and between F3‐EVs and F4‐EVs. Three independent biological experiments were performed. Each dot represents one data point, with the whole dataset obtained from n = 3 biological repeats (colored symbols) with three technical repeats each. Box layouts represent 25–75% of the distribution, whiskers highlight outlier's data points, and horizontal black lines represent the mean of the distribution. For all three conditions, a two‐sample t‐test was employed, with a significance value of 0.05. Plasma membrane and late endosomal conditions show no difference between fractions 3 and 4 (P = 0.55 and P = 0.60, respectively). Plasma membrane and early endosome show a significant difference for F3‐EVs (P = 0.0049). Early endosomal conditions show fraction 3 to fuse significantly more than F4‐EVs (P = 0.00026). For F4‐EVs, early and late endosomes show no difference in fusion (P = 0.32). **P ≤ 0.01, ***P < 0.001.
We sought to investigate the fusion capabilities of each EV subpopulation with three different cellular membranes, following the internalization pathway of EVs interacting with recipient cells. We therefore produced two different types of liposomes mimicking the (i) plasma membrane (Virtanen et al, 1998) and early endosomal membrane (which possess the same lipid composition), and (ii) late endosomal membrane (Erazo‐Oliveras et al, 2016) (Fig 5B). The lipid composition of the plasma membrane's outer leaflet (and the luminal leaflet of early endosomes as well) is composed primarily (~80% of lipid species) of phosphatidylcholine (PC), sphingomyelin (SM), and cholesterol (chol) (Van Meer et al, 2008). Although phosphatidylethanolamine (PE) and the negatively charged phosphatidylserine (PS) are predominantly confined to the plasma membranes' inner leaflet, several studies have highlighted exposure of PS in uninfected RBCs neighboring malaria‐infected RBCs (Totino et al, 2010; Fernandez‐Arias et al, 2016). Similarly, PS in the membrane outer leaflet was observed in immune cells during an immune response (Fischer et al, 2006; Shin & Takatsu, 2020). Thus, to factor in PS exposure during malaria infection, we composed plasma membrane/early endosome large unilamellar vesicles (LUVs) with PC, PE, PS, SM and cholesterol. To mimic progressive acidification of the endosomes, for the early and late endosomal conditions the liposome‐EV mixture was acidified to a pH of 6.5 and pH of 5, respectively (Huotari & Helenius, 2011), and compared to non‐acidified EV interacting with LUVs.
Using FRET analysis, we demonstrated that F3‐EVs and F4‐EVs display fusion and membrane mixing toward all three milieus, albeit at different efficiency (Fig 5B), whereas no mixing is observed for liposomes only (Fig EV3A). We found that maximal fusion probability for both fractions occurs at plasma membrane conditions, with F3‐EVs and F4‐EVs displaying similar values (37 ± 10% for F3‐EVs, 40 ± 9% for F4‐EVs, P = 0.55) (Fig 5B). Upon acidification to both early and late endosomal conditions, we surprisingly detected different trends for the two EV subsets (Fig 5B). F4‐EVs displayed a decrease in fusion both at early and late endosomal conditions (9 ± 8% and 12 ± 5%, respectively, P = 0.32) compared to the plasma membrane, suggesting that acidification reduces the fusion capability of this specific EV subpopulation. Contrarily, we observed that fusion of F3‐EVs at early endosomal conditions was significantly higher than F4‐EVs (24 ± 6% for F3‐EVs, 9 ± 8% for F4‐EVs, P = 0.00026) despite showing a significant reduction in membrane mixing compared to plasma membrane conditions (37 ± 10% for plasma membrane and 24 ± 6% for early endosome, P = 0.0049) (Fig 5B). Further acidification to late endosomal conditions resulted in an additional decrease of F3‐EVs fusion, comparable to F4‐EVs (11 ± 4% for F3‐EVs, 12 ± 5% for F4‐EVs, P = 0.6).
Figure EV3. FRET assay controls.
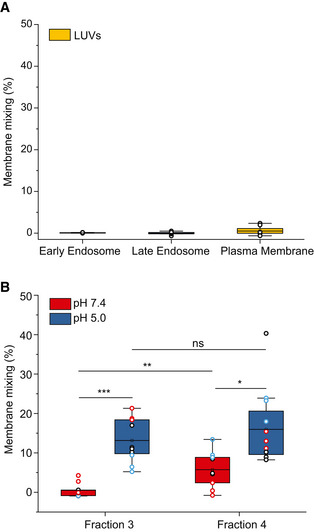
- FRET‐based membrane mixing for LUVs–LUVs interaction (negative control) under the three tested conditions (plasma membrane, early endosome and late endosomes). No significant fusion is observed. Each dot represents one data point, with the whole dataset obtained from n = 3 biological repeats (colored symbols) with three technical repeats each. Box layouts represent 25–75% of the distribution, whiskers highlight outlier' data points, and horizontal black lines represent the mean of the distribution.
- Comparison of membrane mixing between F3‐EVs and F4‐EVs at late endosomal conditions (pH 5.0, blue) and interaction with the same liposomes at neutral pH (red). Significant membrane mixing is observed post acidification for F3‐EVs (P = 0.0009) and F4‐EVs (P = 0.017). A significant difference in membrane mixing between the two fractions is observed at pH 7.4 (P = 0.006). No significant difference in membrane mixing between the two fractions is observed at pH 5 (P = 0.375). Three independent experiments were performed and a two‐sample t‐test was applied. Each dot represents one data point, with the whole dataset obtained from n = 3 biological repeats (colored symbols) with three technical repeats each. Box layouts represent 25–75 percentiles of the distribution, whiskers highlight outlier' data points and horizontal black lines represent the mean of the distribution. *P ≤ 0.05, *P ≤ 0.01, ***P < 0.001.
Furthermore, we showed that F3‐EVs displayed a higher variation in fusion between pH 7.4 and pH 5.0 upon interaction with late endosomal LUVs, as compared to F4‐EVs, suggesting that F3‐EVs are more resistant to pH changes (Fig EV3B).
These different fusion values indicate that while these EV subpopulations exhibit similarly good fusion to the plasma membrane and poor fusion to the late endosome, F3‐EVs retain more capacity to fuse at early endosome conditions at pH 6.5 than F4‐EVs, which hardly fuse in such conditions. Overall, these results suggest the existence of distinct protein machinery mediating membrane fusion for the two EV subsets.
Pf‐derived EV subpopulations display different lipid membrane order
In addition to the demonstration of different fusion efficiencies, we investigated whether the two EV subpopulations displayed distinct membrane mechanical properties which could in turn reflect on their lipid composition. We therefore stained EVs with the fluorescent dye Laurdan and measured each fraction's fluorescent spectra as in previous studies (Laulagnier et al, 2004; Parolini et al, 2009; Szempruch et al, 2016). Laurdan has been extensively applied to determine the gel‐to‐liquid crystalline transition of lipid bilayers due to its sensitivity to the polarization state of its environment (Parasassi et al, 1993), and more generally to measure differences in lipid packing in various membranes as a function of composition (Stott et al, 2008; Amaro et al, 2017). Indeed, the spectra revealed a higher generalized polarization (GP) in F3‐EVs at 0.35 ± 0.03, as compared to F4‐EVs, which displayed a GP value of 0.03 ± 0.12 (Fig 6A). This difference is also clearly visible in the Laurdan spectra (Figs 6B and EV4), where we observe a shift from a fluorescence peak near 440 nm peak (indicative of high membrane lipid packing), to 490 nm (characteristic of a more fluid and less packed membrane).
Figure 6. Analysis of EV fractions membrane fluidity by Laurdan fluorescence measurement.
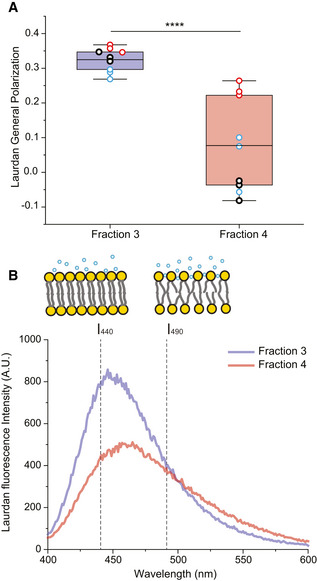
- Laurdan general polarization (GP) values obtained for F3‐EVs and F4‐EVs at physiological temperature showing higher membrane order (higher GP values) for F3‐EVs than F4‐EVs (P = 0.00007). Each dot represents one data point, with the whole dataset obtained from n = 3 biological repeats (colored symbols) with three technical repeats each. Box layouts represent 25–75 percentiles of the distribution, whiskers highlight outlier's data points and horizontal black lines represent the mean of the distribution. ****P < 0.0001.
- Characteristic Laurdan spectra for F3‐EVs and F4‐EVs, showing a predominant peak at 440 nm for F3‐EVs, indicating a high lipid packing, and a shift toward 490 nm emission for F4‐EVs. The illustration shows differences in membrane packing measured by Laurdan according to the two emission peaks.
Data information: Three independent experiments were performed. Statistical significance was quantified with a two‐sample t‐test, with a significance value of 0.05.
Figure EV4. Laurdan spectra for additional biological repeats.
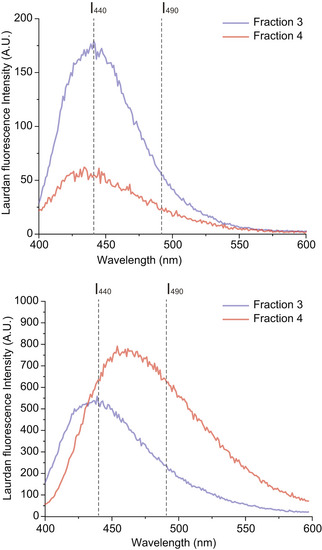
Characteristic Laurdan spectra for F3‐EVs and F4‐EVs for the second and third biological repeat, as shown in Fig 6, display a predominant peak at 440 nm for F3‐EVs, indicating a high lipid packing, and a shift towards 490 nm emission for F4‐EVs.
Overall, these results suggest that the malaria parasite secretes distinct EVs harboring different lipid compositions and thus distinct membrane organization, resulting in specific biophysical membrane properties.
Puncture force determination and quantification: feature engineering and unsupervised machine learning
To further corroborate the differences in membrane biophysical properties of the two EV subpopulations we have performed puncture tests on supported lipid bilayers created from F3‐EVs and F4‐EVs using AFM (Fig 7).
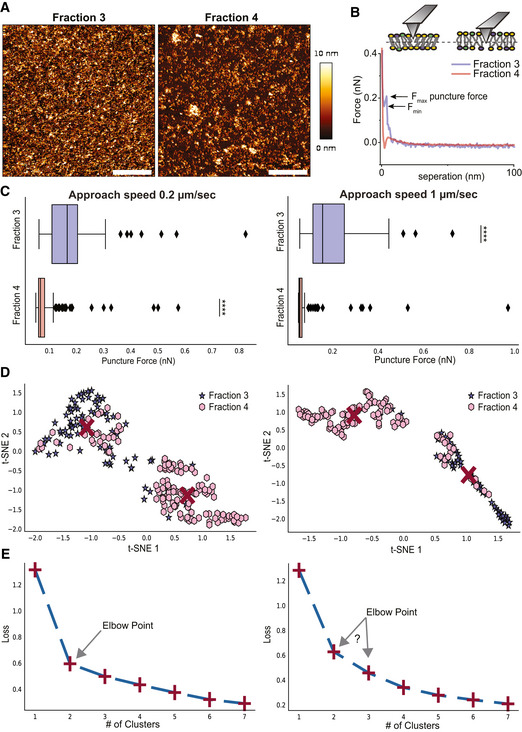
Figure 7. AFM puncture measurements and unsupervised machine learning for further insights on membrane nanomechanical behavior.
-
A, B(A) Representative images (scale bar—1 µm) and (B) force‐separation curves for supported lipid bilayer of F3‐EVs and F4‐EVs. F max and F min are defined on the curve of F3‐EVs.
-
CStatistics of puncture forces for the two fractions at two different approach speeds presented as box plots, and tested with Mann–Whitney statistical analysis (Mann & Whitney, 1947) accounting for skewed distribution, P‐values are < 10−19 for the 0.2 μm/s approach speed (n = 81 for F3‐EVs and 131 for F4‐EVs) and < 10−14 for 1 μm/s (n = 49 events for fraction 3 and 105 for fraction 4). The black diamonds represent outliers, boxes represent 25–75% of the data and the lines inside the boxes are the medians. Minimum and maximum values are indicated by the whiskers. ****P < 0.0001.
-
D, EUnsupervised machine learning grouping of features engineered from the puncture data features by t‐distributed stochastic neighbor embedding analysis (t‐SNE) and K‐means. (D) In the t‐SNE analysis, two clusters are found at each speed which is predominantly F3‐EVs (purple) or F4‐EVs (pink). Red crosses are the centers of the clusters determined by K‐means. (E) K‐means loss function for the analysis shown in (D), displaying loss function as a function of a number of clusters, shown as the red + symbols, with a dashed line added to guide the eye. The Elbow point indicates the most probable number of clusters that best represent the data. The results presented are a representative set taken from three independent biological repetitions. All independent repetitions gave similar results.
Supported lipid bilayers are known to undergo a local collapse when pressed by the AFM tip. Typical puncture events are observed in the force indentation curve by a dip when the tip breaks through the lipid layer. The force at which this occurs represents the highest force the membrane can withstand. Puncture events were detected for many supported lipid bilayers of different lipid compositions (Dufrêne et al, 1999; Mueller et al, 2000; Schneider et al, 2000; Garcia‐Manyes & Sanz, 2010; Sorkin et al, 2014; Unsay et al, 2015; Saavedra et al, 2020; Vorselen et al, 2020) and provide direct insight into the mechanical stability of supported lipid membranes (Dufrêne et al, 1999; Schneider et al, 2000; Garcia‐Manyes & Sanz, 2010) which is governed by the molecular interactions between neighboring lipids in the membrane. The breakthrough force is related to the membrane composition (Garcia‐Manyes et al, 2010; Unsay et al, 2013) and state, that is, phase (Garcia‐Manyes et al, 2005b; Chiantia et al, 2006; Sullan et al, 2009). For example, ionic strength (Garcia‐Manyes et al, 2005a) and temperature (Garcia‐Manyes et al, 2005b; Alessandrini et al, 2012) were shown to have a direct effect on the value of the breakthrough force. Moreover, a solid‐like phase (higher membrane packing) is indicated by higher puncture force values than a liquid‐like phase (lower membrane packing) (Garcia‐Manyes et al, 2005b; Alessandrini et al, 2012; Unsay et al, 2013). Studies on mixed lipid systems revealed that the puncture force differs significantly in the different phase‐separated domains. Overall, ordered phases require a higher puncture force than disordered phases (Chiantia et al, 2006; Alessandrini et al, 2012). Puncture event assay could also provide insights into more complex natural membranes, which are composed of different distinct lipids, proteins and sugars (Watson, 2015). Puncture events were observed in force versus separation curves measured on such natural membranes, that is, on unilamellar vesicles made from PR8 influenza lipids (Li et al, 2011), RBC‐EVs (Sorkin et al, 2018; Vorselen et al, 2018), supported lipid bilayers (SLBs) obtained from prostate cancer derived‐EVs (Montis et al, 2018) and from Escherichia coli lipid extract (Garcia‐Manyes et al, 2005a).
The SLBs from F3‐EVs and F4‐EVs were prepared by adsorbing each EV fraction on Mg2+ modified mica and washing them with pure water. AFM images of the surface topography are shown in Fig 7A. The F3‐EV sample has a nearly uniform coating whereas SLBs prepared from F4‐EVs display an irregular morphology with scattered holes reminiscent of those seen for prostate cancer derived‐EVs SLB (Montis et al, 2018). We have then applied puncture tests on SLBs obtained from F3‐EVs and F4‐EVs to discern differences in the membrane lipid packing and relative rigidity. Representative force separation curves recorded on each sample are shown in Fig 7B.
Only a fraction of the force curves exhibited a significant inflection (Fig EV5), indicating a valid puncture event, as determined through the procedure defined in the methods section. Each such valid event was then associated with a force value. The membrane puncture event is a dynamic process and is related to the response of the lipid molecules to dissipate the applied force. It has been shown that approach speed can affect the probability of a puncture event (Butt & Franz, 2002) and this is seen clearly in our results—at lower speeds more puncture events are observed (Fig EV5). In our procedure, force curves are performed on the entire sample, including both empty regions (no puncture, therefore, does not contribute to the statistics) and regions of higher topography, which could be stacked bilayers. The latter is seen to comprise only a small fraction of the total area (Fig 7A).
Figure EV5. Percentage of detected events as a function of the AFM tip approach speed for fractions 3 and 4.
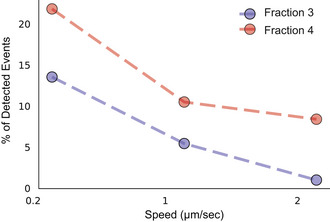
Under all conditions measured the characteristic yield force for a puncture event differs significantly between the different fractions, with higher forces required to puncture F3‐EV SLBs (Fig 7C). This is direct evidence of differences in membrane compactness arising from compositional differences between the two populations.
To objectively verify the presence of two distinct populations, the data were analyzed by unsupervised machine learning, t‐SNE and K‐means, which autonomously search for similarities in force behavior between the curves without the use of prior knowledge of the curve assignment to a specific fraction. The data clusters into two groups which to a large extent correspond to the two fractions, F3‐EV and F4‐EV SLBs (Fig 7D). The t‐SNE embedding and K‐means loss functions (Fig 7D and E) indicate that the best description of the data is the division into two unique clusters (that can be further divided into sub‐clusters) and this can also be visualized by the elbow point (Fig 7E).
In summary, these data demonstrate that the two EV fractions are different in their resistance to puncture, indicating a higher level of compactness in F3‐EV SLBs than in F4‐EV SLBs.
Discussion
There is increasing evidence, primarily from mammalian systems, that EVs are a heterogeneous pool (Zhang et al, 2018; Mathieu et al, 2021) and current advanced technologies, such as AF4 analysis, enable the distinction between different EV subpopulations. In the case of cancer‐derived EVs this technique revealed two exosome subpopulations (Exo‐L and Exo‐S) and a population of non‐membranous nanoparticles termed “exomeres” (Zhang et al, 2018). Those EV subpopulations have distinct biophysical properties and protein, lipid, N‐glycosylation, DNA and RNA profiles. Furthermore, these three subsets demonstrated diverse organ distribution patterns, suggesting distinct biological functions (Zhang et al, 2018). Another study distinguished between exosomes and small ectosomes (plasma membrane‐derived EVs) by following the intracellular trafficking of the EV markers CD9 and CD63, further identifying additional protein surface markers for the two EV populations by comparative proteomic analysis (Mathieu et al, 2021).
Here, we demonstrate for the first time the existence of two distinct subpopulations in the Pf‐derived EV pool, which differ in size, protein content, biophysical properties and membrane fusion capabilities. Using the AF4 technology we were able not only to efficiently separate the EVs based on size (Fig 1) but also collect the subpopulations for further analyses. The subpopulation of fraction 3, F3‐EVs, displayed a size distribution between 30 and 70 nm while the second EV type of fraction 4, F4‐EVs, has a size distribution of 70–300 nm. The wider size distribution of F4‐EVs compared to F3‐EVs was confirmed by both AFM and cryo‐TEM analysis (Figs 2A and C, and 3B). The high‐resolution cryo‐TEM characterization revealed that the two EV subpopulations indeed have typical EV morphology (Fig 3A).
The low concentrations of EV fractions obtained from the AF4 system challenged the proteomic analysis. Nonetheless, sensitive chromatographic and mass spectrometric analyses were achieved. While we identified a relatively short list of proteins, we distinguished between the two EV subpopulations and identified unique human or parasitic proteins in each fraction (Fig 4A). F3‐EVs were enriched with immune system proteins and more specifically the complement system proteins (e.g., C2 and C3), which suggests the RBCs are not as inert as originally considered and might have a response against invading pathogens (Darbonne et al, 1991; Neote et al, 1993; Anderson et al, 2018; Hotz et al, 2018; Lam et al, 2021). On the other hand, F4‐EVs were enriched with proteasome subunits (e.g., PSMA1) (Fig 4C) in agreement with previous work which demonstrated that Pf‐derived EVs carry active 20S proteasome complexes (Dekel et al, 2021).
Our results indicate that the two EV subpopulations display different biophysical properties, both at the functional (fusion) and the structural level (membrane organization). We observed significantly different trends between the two subpopulations towards endosomal conditions, with F3‐EVs exhibiting less reduction in fusion at early endosomal conditions than F4‐EVs (Fig 5). Late endosomal membrane conditions resulted in comparably reduced fusion for both subsets.
Although the FRET‐based assay does not fully distinguish between full fusion and hemifusion, we demonstrated that measurements obtained for membrane mixing clearly reflect the merging of membranes and pore formation between the two vesicles (preprint: Morandi et al, 2022).
We found that the smaller size EVs (F3‐EVs) are capable of mediating significant fusion under early endosomal pH, as opposed to F4‐EVs, suggesting that the F3‐EV subset would deliver its cargo more efficiently than F4‐EVs in cells with high endocytosis or internalization rate. Interestingly, previous studies reported a fusion‐enhancing effect of pH for human‐derived EVs, both in vitro using isolated membranes (Bonsergent & Lavieu, 2019), or in cells with internal compartments (Bonsergent et al, 2021). Consistent with a previous report (Dekel et al, 2021), we show that the plasma membrane also serves as a target for Pf derived EV fusion. While further work is needed to understand the molecular nature of such differences, we suggest that they might be associated with the studied organism, as the malaria parasite interacts with a plethora of target cells (including naïve RBCs) with different degrees of endocytosis or no endocytic pathways.
Differences in the fusion behavior between F3‐EVs and F4‐EVs could be partially ascribed to a specific preference toward certain lipid compositions. Both plasma membrane and early endosome contain a significant amount of cholesterol (Gagescu et al, 2000) and sphingomyelin (Gagescu et al, 2000; Yang et al, 2015), which are known to create liquid‐ordered domains in lipid membranes, whereas late endosome liposomes are enriched with the anionic lipid LBPA and contain no cholesterol. However, neither the pH nor the membrane composition is sufficient to fully elucidate the underlying molecular mechanism. Moreover, protein‐protein interactions are likely involved, which are not observed in the FRET assay which probes only mixing between EVs and synthetic liposomes. Further studies would therefore be required to fully elucidate all the factors contributing to the fusion.
Overall, our results do suggest that the two EV populations may possess distinct protein machinery to mediate vesicle‐fusion, with smaller EVs possessing proteins capable of withstanding acidification and maintaining their fusogenic capability, a feature that has been observed for viruses displaying pH‐independent fusion (Miyauchi et al, 2009).
The difference in Laurdan GP values between F3‐EVs and F4‐EVs indicated a tighter lipid packing and membrane order in F3‐EVs. This is consistent with the AFM puncture results and highlights a different membrane composition between the two EV subpopulations. Moreover, the unsupervised machine learning analysis independently determined the existence of two different populations with distinct membrane properties. The data gained from the combination of the Laurdan spectra (performed on intact vesicles) with the AFM results (probed on supported bilayers from the EVs) confirm that these differences are not due to size differences, but rather reflect different biophysical characteristics and distinct lipid compositions for F3‐EVs and F4‐EVs. These findings are particularly interesting as they not only suggest that there might be dedicated biogenesis pathways for the production of the two EV subpopulations, but they also provide a mechanistic rationale for the differences in protein content between the two EV groups. In this context, our results suggest separate preferences on cellular delivery for the two EV subsets, with a specific response to the subcellular surroundings, further reinforcing the concept that these subpopulations contain distinct proteomes.
Indeed, the specific membrane configurations not only contribute to the bilayer physical properties like rigidity (Gracià et al, 2010; Takechi‐Haraya et al, 2016) but also lead to preferential partitioning and activity of membrane proteins, due to differences in membrane thickness and lipid species. This phenomenon has been well characterized in lipid model systems (Schlebach et al, 2016), for example, on the adenosine A2A membrane receptors (Gutierrez et al, 2019), ion channels (Seeger et al, 2010; Kimchi et al, 2018) and has also been shown extensively for viral fusion proteins where biogenesis of virions and clustering of viral proteins occurs only in specific lipid domains (Bajimaya et al, 2017; Sengupta et al, 2019). It is therefore consistent that two EV subpopulations with significantly different lipid compositions would result in distinct membrane structures leading to preferential protein content and particularly dedicated membrane fusion proteins.
Overall, our results shed light on how malaria parasites utilize the different EV subpopulations as a communication tool to alter several host systems, by employing different subsets of its own EV pool with specific membrane and protein compositions to target different cells or pathways.
Materials and Methods
Pf parasite line
The malaria parasite line used in the experiments is NF54, which was generously provided by the Malaria Research Reference Reagent Resource Center, MR4, obtained through BEI Resources, NIAID, NIH: P. falciparum, Strain NF54 (Patient Line E), MRA‐1000, contributed by Megan G. Dowler.
Pf parasite culture
Plasmodium falciparum parasites were cultured in human red blood cells (RBCs) using a standard method (Trager & Jensen, 1976). Briefly, parasites were grown at 4% hematocrit in pooled healthy uninfected RBCs, provided by the Israeli blood bank (Magen David Adom blood donations, Israel, IRB ‐ 1634‐1), and incubated at 37°C in a gas mixture of 1% O2 and 5% CO2 in N2. Parasites were maintained in RPMI medium pH 7.4, supplemented with 25 mg/ml 4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid, 50 μg/ml hypoxanthine, 2 mg/ml sodium bicarbonate, 20 μg/ml gentamycin and 0.5% AlbumaxII (Gibco, Cat#: 11021045). Parasite growth was monitored using methanol‐fixed Giemsa stained blood smears. Pf‐iRBC cultures were tested for Mycoplasma infections twice a month using the commercial kit MycoAlert Plus (Lonza, Cat#: LT07‐318).
Isolation of extracellular vesicles
Growth media of high parasitemia (≥ 5%) Pf‐iRBC cultures was collected. EV extraction was performed as reported previously (Sisquella et al, 2017). Briefly, cellular debris was removed by differential centrifugation at 413 g for 5 min, 1,900 g for 10 min and 15,180 g for 1 h, supernatants were filtered through a 0.45 µm pore filter and later concentrated using Vivacell® units with molecular weight cut‐off (MWCO) of 100 kDa (Sartorius AG, Germany), according to the instructions of the manufacturer. Then, the EVs contained in Pf‐iRBC (Pf‐derived EVs) suspension were pelleted by ultracentrifugation at 150,000 g for 16 h at 4°C. Finally, the pellet was carefully re‐suspended in sterile PBS (Ca2+‐/Mg2+‐) (Biological Industries) for further analysis.
Extracellular vesicle isolation onto a sucrose cushion
Re‐suspended EVs were slowly loaded over a 2 ml sterile 20% sucrose solution (prepared in PBS (Ca2+‐/Mg2+‐)), forming a layer. EVs were pelleted by ultracentrifugation using a swinging bucket rotor (Beckman Coulter) at 100,000 g for 4 h at 4°C. After this, the supernatant was discarded and EVs were re‐suspended in sterile PBS (Ca2+‐/Mg2+‐) for further analysis.
Nanoparticle tracking analysis
Vesicle size distribution and concentration were calculated using nanoparticle tracking analysis (NTA) (Filipe et al, 2010) with the NanoSight NS300 device (Malvern Panalytical Ltd., Instruments, UK) using a 405 nm filter. Sample size distributions were calibrated in a liquid suspension by the analysis of Brownian motion via light scattering and the size of the particles was estimated based on their hydrodynamic radius (Filipe et al, 2010).
AF4 analysis
AF4 experiments and data collection were performed in an AF2000 MT instrument (Postnova Analytics, Landsberg, Germany). Separations were performed at 25°C in a channel containing a 350 μm spacer. A regenerated cellulose membrane with an MWCO of 100 kDa was used and the channel flow was monitored by UV absorption at 280 nm. The temperature in the auto‐sampler and fraction collector modules was maintained stable at 4°C. Channel flow was kept constant at 1.5 ml/min and sterile PBS (Ca2+‐/Mg2+‐) was used as the mobile phase in the AF4. Then, 50 µl of Pf‐derived EVs adjusted to 4*1011 particles/ml with PBS were injected into the system using the partial‐loop mode for 6 min at a flow of 0.2 ml/min. After injection, a particle focusing step was held for 6 min with 1.5 ml/min flow. Then, the sample was eluted in a separation window as follows: an initial power decay (exponent = 9.3) for 0.5 min with a cross flow of 1.5 ml/min, followed by a linear decay of 40 min with a cross flow of 1.15 ml/min, a power decay (exponent = 0.3) for 0.5 min with a cross flow of 1.0 ml/min and a final step of constant cross flow at 0.075 ml/min for 40 min to allow all particles to elute. Between runs, the channel was rinsed with the mobile phase for 15 min without cross flow. Fractions were collected at the following time ranges: 9–12.5, 13–17.5, 25–45 and 50–75 min.
Multi‐angle light scattering data were collected using a Vout of 0.5 ml/min for elution. The radius of gyration (Rg) was calculated from the measured MALS signal intensities by applying an intensity distribution function P(θ) (random coil model) of the region of interest from 9 to 93 min of the run.
The collected fractions were kept at 4°C and concentrated to a volume of approximately 100 µl with Amicon filter units of 10 kDa MWCO (Sartorius AG, Göttingen, Germany). Aliquots of these concentrated fractions were subjected to AFM, cryo‐TEM, FRET assay, and Laurdan staining or proteomic analysis.
Atomic force microscopy
A freshly cleaved mica surface was incubated with 10 mM MgCl2 solution for 2 min then rinsed with 200 µl PBS (Ca2+‐/Mg2+‐). Fifty microliters of F3 or F4 were placed on the Mg modified mica for 15 min. Prior to scanning, 3 ml of PBS (Ca2+‐/Mg2+‐) was added to the sample, then 2 ml were removed and fresh 2 ml were added. This washing procedure was done carefully to avoid a passing interface. AFM imaging was performed on a JPK Nanowizard III AFM microscope (Bruker Nano GmbH, Berlin, Germany) in QI mode. Measurements were conducted with two different probes, a probe with a rounded tip apex and rated radius of 30 nm, spring constant ≈ 0.06 N/m (qp‐BioAC‐CI, Nanosensors, Neuchâtel, Switzerland) and a second probe with a tip radius of curvature smaller than 10 nm (qpBioAC, Nanosensors, Neuchâtel, Switzerland), and nominal spring constant same as the first probe type. Detector sensitivity and spring constant were determined for each probe using the JPK software.
Image analysis was performed using Gwyddion (Nečas & Klapetek, 2012) or JPK‐SPM data processing software. Particle size analysis was conducted using particle analysis in ImageJ software (Rueden et al, 2017). Plots were constructed with OriginPro 2018 and images were assembled in Adobe Illustrator 2019. For all the images acquired with the 30 nm tip, the tip convolution artifact was removed by surface reconstruction from the Gwyddion software, using a modeled tip shape (conical 22deg tip with 30 nm tip apex) prior to the size analysis. Particles below 30 nm were excluded from the analysis. For the sharp tip < 10 nm, particles below 9 nm were excluded from the analysis.
AFM force spectroscopy and machine learning
For the puncture assay test, the Mg2+ modified mica (freshly cleaved mica incubated for 2 min with 10 mM MgCl2 solution and then rinsed with 200 µl PBS (Ca2+‐/Mg2+‐) was incubated with F3‐EVs or F4‐EVs for 30 min). The adsorbed EVs were ruptured by washing the sample with DDW by infinite dilution. The puncture measurements were performed using the same AFM and qpBioAC probes as described above, working in force spectroscopy mode. Specifically, thousands of force versus distance curves were recorded in different locations on each sample in batches of 100, each made over a 10 × 10 grid in a 4 × 4 µm2 area. To distinguish attractive “pop‐in” events the same analysis was performed on a control sample—Mg2+ modified mica, and the force threshold for a puncture event was defined above the force of the highest event captured for the control.
Three different extension speeds were used (0.2, 1 and 2 µm/s). The force curves were analyzed using functions in the package SciPy.signal (Virtanen et al, 2020) in Python by first searching the curves for “dips” (F max–F min, see Fig 7B) which exceeded 35 pN, with the second condition that they occurred above the force threshold described above (50 pN). In cases where there were two puncture events in one approach, the puncture occurring at the higher force was considered in the analysis. Statistical analysis of these results was performed using the package SciPy.stats (Virtanen et al, 2020). Subsequently, the data were subjected to unsupervised machine‐learning analysis with t‐distributed stochastic neighbor embedding (t‐SNE) (van der Maaten & Hinton, 2008), as implemented in Scikit‐learn (Pedregosa et al, 2011) and K‐means algorithm as implemented in SciPy.cluster in Python. Ten different features were generated from the curve data related to the puncture events. These features were standardized and fed into the t‐SNE algorithm. Initiation for this algorithm was the PCA matrix derived from the ten features. The resulting t‐SNE components were divided by their respective standard deviations and subjected to clustering with the K‐means algorithm to determine the optimal number of clusters representing the data.
Cryo‐transmission electron microscopy
Samples were vitrified using a Vitrobot plunger system Mark IV (FEI, USA; Humidity 80%, temperature 24°C, blotting time 4.5 s; blot force – 10). Four µl of EVs suspension (~5*1010 particles/ml) were dispensed onto a lacey carbon grid (LACEY copper 200 mesh; Electron Microscopy Sciences, Hatfield, PA, USA) and incubated for 5 m. Grids were first rendered hydrophilic using a glow discharge system (PELCO easiGlow™, Redding, CA, USA; 30 s, 25 mA). Sample imaging was conducted on a Talos Arctica G3 TEM/STEM (FEI, USA) cryo‐electron microscope, equipped with a OneView camera (GATAN) at an accelerating voltage of 200 kV. Images were acquired in low‐dose mode (total dose 70 e−/Å2) using the SerialEM software (FEI, USA) (Mastronarde, 2003) to avoid radiation damage to the samples, at 73,000× magnification with a defocus value of −4 µm.
Size quantification of recorded EV micrographs was performed with Fiji (ImageJ) (Schindelin et al, 2012) by measuring the area covered by each vesicle and extrapolating the corresponding diameter. For each fraction, three separate biological replicates were plunge‐frozen and visualized via cryo‐TEM on separate days.
MS analysis and bioinformatics analysis
Purified EVs were dissolved in 10 mM dithiothreitol (DTT), 100 mM Tris and 5% sodium dodecyl sulfate (SDS), sonicated and boiled at 95°C for 5 min and precipitated in 80% acetone. Protein pellets were dissolved in 9 M Urea and 400 mM ammonium bicarbonate, and their concentrations were determined using the Bradford assay. Five microgram protein of each sample was reduced with 3 mM DTT (60°C for 30 min), modified with 10 mM iodoacetamide in 100 mM ammonium bicarbonate for 30 min at room temperature in the dark and digested in 2 M urea, 25 mM ammonium bicarbonate with modified trypsin (Promega), overnight at 37°C in a 1:50 (M/M) enzyme‐to‐substrate ratio. Tryptic peptides were desalted using C18 tips (Top tip, Glygen) dried and re‐suspended in 0.1% formic acid.
Lug peptides were resolved by reverse‐phase chromatography on 0.075 × 180‐mm fused silica capillaries (J&W) packed with Reprosil reversed‐phase material (Dr Maisch GmbH, Germany). Peptides were eluted with a linear 60‐min gradient of 5 to 28%, followed by a 15‐min gradient of 28–95% and then 25 min at 95% acetonitrile with 0.1% formic acid in water at flow rates of 0.15 μl/min. MS was performed by Q Exactive HF mass spectrometer (Thermo) in a positive mode (m/z 300–1,800, resolution 120,000 for MS1 and 15,000 for MS2) using a repetitively full MS scan followed by collision‐induced dissociation (HCD, at 27 normalized collision energy) of the 18 most dominant ions (> 1 charges) selected from the first MS scan. The AGC settings were 3 × 106 for the full MS and 1 × 105 for the MS/MS scans. The intensity threshold for triggering MS/MS analysis was 1 × 104. A dynamic exclusion list was enabled with an exclusion duration of 20 s.
MS data were analyzed using the MaxQuant software 1.5.2.8 (1) for peak picking and identification using the Andromeda search engine, searching against the human and Pf sections from the Uniprot database with a mass tolerance of 6 ppm for the precursor masses and 20 ppm for the fragment ions. Methionine oxidation and protein N‐terminus acetylation were accepted as variable modifications and carbamidomethyl on cysteine was accepted as a static modification. Minimal peptide length was set to six amino acids and a maximum of two miscleavages was allowed. Data were quantified by label‐free analysis using the same software (Cox et al, 2014). Peptide‐ and protein‐level false discovery rates (FDRs) were filtered to 1% using the target‐decoy strategy. Protein tables were filtered to eliminate the identifications from the reverse database, common contaminants and single peptide identifications. Statistical analysis of the identification and quantitation results was done using Perseus 1.6.10.43 software (Tyanova et al, 2016). Additional annotation enrichment was done using the String software (https://string‐db.org/).
Protein extraction
Pf‐iRBCs were washed with iso‐osmotic buffer (103 mM Na2HPO4, 155 mM NaH2PO4, Sigma–Aldrich), followed by lysis with RIPA buffer (150 mM NaCl, 1% Triton X‐100, 0.1% SDS, 50 mM Tris, 0.5% sodium deoxycholate, pH 8.0) for 15 min on ice, followed by 10‐min centrifugation at the maximum speed at 4°C. The supernatant was collected and hemoglobin was depleted using TALON metal affinity beads (635502 Takara‐Clontech). For ghost purification, the RBC pellet was re‐washed with 1 ml iso‐osmotic buffer until the membrane pellet became clear.
Proteins derived from total EVs were obtained by incubating the EVs with RIPA buffer 5× for 15 min on ice, followed by 10‐min centrifugation at the maximum speed at 4°C. Protein concentrations for Pf‐iRBCs and EVs were calculated with the BCA method with BSA as standard.
Western blot analysis
Protein extracts in the sample buffer were loaded into 10% polyacrylamide gels. Proteins in the gel were transferred to a nitrocellulose membrane and blocked for 1 h at 25°C with 5% skimmed milk in PBS with 0.05% Tween (PBS‐T). The membranes were then washed five times with PBS‐T for 5 min each in constant agitation and probed overnight at 4°C against different protein targets using the following primary antibodies: mouse anti‐HSP90 (ab13492 Abcam, dilution 1:1,000), mouse anti‐GAPDH (ab8245, dilution 1:1,000), rabbit anti‐Annexin 7 (QC5531, Antibody Verify, dilution 1:1,000), mouse anti‐C3 (sc‐28294, Santa Cruz, dilution 1:1,000), rabbit anti‐PSMA1 (ab140499, Abcam, dilution 1:1,000), rabbit anti‐PSMB2 (ab137108, Abcam, dilution 1:1,000) and mouse anti‐ankyrin‐1 (ab212053, Abcam, dilution 1:1,000). After this, membranes were washed in the same conditions and re‐probed for 1 h at 25°C with secondary antibodies Goat anti‐Rabbit IgG‐HPR (111‐035‐003, Jackson, dilution 1:7,500), and Goat anti‐mouse IgG‐HRP (115‐035‐003, Jackson, dilution 1:10,000). Image collection was performed using the ThermoScientific MyECL Imager V. 2.2.0.1250 and Amersham Imager 680 (GE Healthcare Life Sciences).
Preparation of LUVs
Large unilamellar vesicles were prepared as previously described (Virtanen et al, 1998). In brief, lipid solutions in chloroform of the different phospholipid species were mixed to the desired molar ratios in a glass vial, and the organic solvent was evaporated by 12 h of vacuum pumping. For labeled LUVs, the lipids were stained with 2% mol of DiI and DiD in chloroform before evaporation. The lipid film was then hydrated with PBS (Ca2+‐/Mg2+‐) at 40°C to reach the desired concentration and gently vortexed. The resulting multilamellar vesicle suspension was then sonicated for 10 min to disperse larger aggregates and the liposomal suspension was extruded 21 times through polycarbonate filters (100 nm pore size, Avanti Polar Lipids) using a mini‐extruder (Avanti Polar Lipids). Size and concentration were verified using NTA and the liposomal suspension was used within 2 weeks from the extrusion. DiI (1,1′‐dioctadecyl‐3,3,3′,3′‐tetramethylindocarbocyanine perchlorate) and DiD (1,1′‐dioctadecyl‐3,3,3′,3′‐tetramethylindodicarbocyanine 4‐chlorobenzenesulfonate salt) membrane dyes (Thermo Fisher) were dissolved in chloroform at 1 mM final concentration. All chemicals had high purity (>99%) and were used without further purification. The plasma membrane and early endosomal membrane lipid composition was DOPC:DOPE:DOPS:SM:cholesterol at 20:5:15:25:35 molar ratio. For late endosomal membrane composition LBPA:DOPE:DOPC 70:5:25 molar ratio LUVs were prepared, mimicking the cholesterol sequestration and enrichment of late endosomal lipid LBPA.
Membrane mixing assay
All experiments were performed using an Infinite M Plex multimode plate reader (TECAN) with a 96‐well plate. DiI‐DiD labeled liposomes were diluted in 200 µl PBS (Ca2+‐/Mg2+‐) per well to reach a final concentration of 10 µM, and fluorescence intensity of the donor (DiI) was recorded every 60 s for 30 min, with an excitation wavelength of 530 nm and emission wavelengths of 570 nm. Subsequently, unlabeled AF4 fractions (F3‐EVs and F4‐EVs) were added to each well to reach a labeled:unlabeled ratio of 1:9 particles and DiI fluorescence intensity was recorded for 1 h every 60 s. Finally, Triton X‐100 was added to each well to reach 0.1% final concentration and fluorescence intensity was recorded for 15 min every 60 s.
For early or late endosomal conditions, labeled liposomes were mixed with either of the unlabeled AF4 fractions (F3‐EVs and F4‐EVs) to reach a LUV:EV ratio of 1:9, and incubated at 4°C for 60 min. Subsequently, mixed LUV and EV samples were placed in wells of a 96‐well plate. The fluorescence intensity of the donor (DiI) was recorded every 60 s for 30 min, with an excitation wavelength of 530 nm and emission wavelengths of 570 nm. Subsequently, a volume fraction of HCl 100 mM was added to reach the desired pH (6.5 for early endosomal conditions, and pH 5.0 for late endosomal conditions) and DiI fluorescence intensity was recorded for 1 h every 60 s. Finally, Triton X‐100 was added to each well to reach 0.1% final concentration and fluorescence intensity was recorded for 15 min every 60 s. The emission fluorescence for each time point was measured as In. The emission fluorescence of the untreated liposomes was measured as I0, and that of the liposomes solubilized with 0.1% TRITON X‐100 was defined as I100. The percentage of membrane mixing at each time point was defined as donor relative intensity (% of TRITON X‐100) = (In − I0) × 100/(I100 − I0). The membrane mixing recorded for EV – LUV interaction was subsequently subtracted by an EV‐only blank measurement to obtain the effective fusion efficiency. All measurements were performed at 37°C and performed on three separate biological replicates (n = 3) with three technical repeats each.
Laurdan staining
6‐dodecanoyl‐2‐dimethylaminonaphthalene (Laurdan) was dissolved in DMSO at a stock solution concentration of 0.5 mM, and staining was performed on F3‐EVs and F4‐EVs by incubating Laurdan stock solution (final concentration 2 µM) at 37°C for 30 min (Molecular Probes, Eugene, OR). The fluorescent spectra of labeled EVs were subsequently measured using a Cary Eclipse Fluorescence Spectrophotometer (Agilent) at 37°C, using an excitation wavelength of 340 nm (slit width 5 nm) and emission recorded between 400 nm and 600 nm (slit width 10 nm). The resulting Laurdan General Polarization (GP) was quantified using the previously established equation (Parasassi et al, 1998): GP = (I440 – I490)/(I440 + I490), with I440 and I490 representing the fluorescent emission intensity at 440 and 490 nm, respectively. All measurements were performed on three separate biological replicates (n = 3) with three technical repeats each.
Statistical analysis
When comparing several groups, we used ANOVA followed by Tukey's post hoc test. When comparing two groups, we used an independent‐samples t‐test. Puncture forces were compared between the two fractions using a Mann–Whitney test with Python SciPy.stats package v. 1.5.0. All other statistics were done in R, v. 4.1.2.
Author contributions
Paula Abou Karam: Conceptualization; Validation; Investigation; Visualization; Methodology; Writing—original draft; Writing—review & editing. Irit Rosenhek‐Goldian: Conceptualization; Investigation; Visualization; Methodology; Formal Analysis; Writing—original draft; Writing—review & editing. Tamar Ziv: Data curation; Formal analysis; Visualization; Methodology; Writing—original draft. Hila Ben Ami Pilo: Investigation; Visualization. Ido Azuri: Software; Formal analysis; Validation; Visualization; Writing—original draft. Anna Rivkin: Investigation; Methodology. Edo Kiper: Visualization. Ron Rotkopf: Formal analysis; Writing—review & editing. Sidney R Cohen: Formal analysis; Methodology; Writing—original draft; Writing—review & editing. Ana Claudia Torrecilhas: Writing—original draft. Ori Avinoam: Resources; Funding acquisition; Writing—original draft. Alicia Rojas: Conceptualization; Validation; Investigation; Methodology; Writing—original draft. Mattia I Morandi: Conceptualization; Investigation; Visualization; Methodology; Writing—original draft; Writing—review & editing. Neta Regev‐Rudzki: Conceptualization; Resources; Supervision; Funding acquisition; Writing—original draft; Project administration; Writing—review & editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
PAK, MIM and NR‐R designed the experiments and analyzed the data. IR‐G and SRC designed the AFM experiments, analyzed the force curves and size distributions, and assisted in writing. IRG performed the AFM experiments. PAK performed AF4 setup. MIM performed and analyzed the cryo‐TEM imaging, FRET fusion assay and the Laurdan staining assay. TZ performed the proteomic experiments and data analysis. RR assisted in the statistical analysis, IA analyzed the force curves and established the machine learning approach and HBAP and EK assisted in protein subcellular localization analysis. HBAP, AnR and AlR assisted in Pf parasite culturing, EV isolation and biochemical assays. OA and ACT assisted in writing. PAK, MIM, IRG, TZ and NRR analyzed the data and wrote the manuscript.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Source Data for Figure EV2
Acknowledgements
We thank the Malaria Research Reference Reagent Resource Center (MR4) for their generous supply of parasite strains. We thank Yael Fridmann Sirkis and the protein analysis unit for their assistance with the AF4 system. We thank Smoler Proteomics center at the Technion—Israel Institute of Technology for their work on EV proteomic analysis. The research of NR‐R is supported by the Benoziyo Endowment Fund for the Advancement of Science, the Jeanne and Joseph Nissim Foundation for Life Sciences Research and the Samuel M. Soref and Helene K. Soref Foundation. NR‐R is the incumbent of the Enid Barden and Aaron J. Jade President's Development Chair for New Scientists in Memory of Cantor John Y. Jade. NRR is grateful for the support from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement No. 757743), the Minerva Program support (grant number 714142), the Weizmann—Sao Paulo Research Foundation (FAPESP) Brazil; supported by a research grant from the Instituto Serrapilheira and the Israel Science Foundation (ISF) (Grant Application no. 570/21). This research was supported by the Israel Science Foundation (ISF) (grant No. 1637/20), within the Israel Precision Medicine Partnership (IPMP) program to O.A. and N.R‐R. O.A. is an incumbent on the Miriam Berman Presidential Development Chair.
EMBO reports (2022) 23: e54755.
See also: F Blow & AH Buck (July 2022)
Contributor Information
Mattia I Morandi, Email: mattia.morandi@weizmann.ac.il.
Neta Regev‐Rudzki, Email: neta.regev-rudzki@weizmann.ac.il.
Data availability
The datasets generated during this study are available from the corresponding author on reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD032012. http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD032012.
References
- Alessandrini A, Seeger HM, Caramaschi T, Facci P (2012) Dynamic force spectroscopy on supported lipid bilayers: effect of temperature and sample preparation. Biophys J 103: 38–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RP, Chiou N‐T, Ansel KM (2016) Improved exosome isolation by sucrose gradient fractionation of ultracentrifuged crude exosome pellets. Protocol Exchange 1–4 10.1038/protex.2016.057 [DOI] [Google Scholar]
- Amaro M, Reina F, Hof M, Eggeling C, Sezgin E (2017) Laurdan and Di‐4‐ANEPPDHQ probe different properties of the membrane. J Phys D Appl Phys 50: 134004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson HL, Brodsky IE, Mangalmurti NS (2018) The evolving erythrocyte: RBCs as modulators of innate immunity. J Immunol 201: 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalos‐Padilla Y, Georgiev VN, Lantero E, Pujals S, Verhoef R, Borgheti‐Cardoso LN, Albertazzi L, Dimova R, Fernàndez‐Busquets X (2021) The ESCRT‐III machinery participates in the production of extracellular vesicles and protein export during Plasmodium falciparum infection. PLoS Pathog 17: 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajimaya S, Frankl T, Hayashi T, Takimoto T (2017) Cholesterol is required for stability and infectivity of influenza A and respiratory syncytial viruses. Virol J 510: 234–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsergent E, Grisard E, Buchrieser J, Schwartz O, Théry C, Lavieu G (2021) Quantitative characterization of extracellular vesicle uptake and content delivery within mammalian cells. Nat Commun 12: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsergent E, Lavieu G (2019) Content release of extracellular vesicles in a cell‐free extract. FEBS Lett 593: 1983–1992 [DOI] [PubMed] [Google Scholar]
- Butt HJ, Franz V (2002) Rupture of molecular thin films observed in atomic force microscopy. I. Theory. Phys Rev E Stat Nonlin Soft Matter Phys 66: 1–9 [DOI] [PubMed] [Google Scholar]
- Chávez ASO, O'Neal AJ, Santambrogio L, Kotsyfakis M, Pedra JHF (2019) Message in a vesicle‐trans‐kingdom intercommunication at the vector‐host interface. J Cell Sci 132: jcs224212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiantia S, Ries J, Kahya N, Schwille P (2006) Combined AFM and two‐focus SFCS study of raft‐exhibiting model membranes. ChemPhysChem 7: 2409–2418 [DOI] [PubMed] [Google Scholar]
- Coakley G, Maizels RM, Buck AH (2015) Exosomes and other extracellular vesicles: the new communicators in parasite infections. Trends Parasitol 31: 477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman AF, Healer J, Marapana D, Marsh K (2016) Malaria: biology and disease. Cell 167: 610–624 [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome‐wide label‐free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13: 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbonne WC, Rice GC, Mohler MA, Apple T, Hébert CA, Valente AJ, Baker JB (1991) Red blood cells are a sink for interleukin 8, a leukocyte chemotaxin. J Clin Invest 88: 1362–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel E, Yaffe D, Rosenhek‐Goldian I, Ben‐Nissan G, Ofir‐Birin Y, Morandi MI, Ziv T, Sisquella X, Pimentel MA, Nebl T et al (2021) 20S proteasomes secreted by the malaria parasite promote its growth. Nat Commun 12: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarta‐Gatsi C, Rivkin A, Di Bartolo V, Peronet R, Ding S, Commere P‐H, Guillonneau F, Bellalou J, Brûlé S, Abou Karam P et al (2019) Histamine releasing factor and elongation factor 1 alpha secreted via malaria parasites extracellular vesicles promote immune evasion by inhibiting specific T cell responses. Cell Microbiol 21: 1–18 [DOI] [PubMed] [Google Scholar]
- Doyle LM, Wang MZ (2019) Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 8: 41–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne YF, Boland T, Schneider JW, Barger WR, Lee GU (1999) Characterization of the physical properties of model biomembranes at the nanometer scale with the atomic force microscope. Faraday Discuss 111: 79–94 [DOI] [PubMed] [Google Scholar]
- Erazo‐Oliveras A, Najjar K, Trouong D, Wang T‐Y, Brock DJ, Prater AR, Pellois J‐P (2016) The late endosome and its lipid BMP act as gateways for efficient cytosolic access of the delivery agent dfTAT and its macromolecular cargos. Cell Chem Biol 23: 598–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Arias C, Rivera‐Correa J, Gallego‐Delgado J, Rudlaff R, Fernandez C, Roussel C, Götz A, Gonzalez S, Mohanty A, Mohanty S et al (2016) Anti‐self phosphatidylserine antibodies recognize uninfected erythrocytes promoting malarial anemia. Cell Host Microbe 19: 194–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipe V, Hawe A, Jiskoot W (2010) Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res 27: 796–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K, Voelkl S, Berger J, Andreesen R, Pomorski T, Mackensen A (2006) Antigen recognition induces phosphatidylserine exposure on the cell surface of human CD8+ T cells. Blood 108: 4094–4101 [DOI] [PubMed] [Google Scholar]
- Gagescu R, Demaurex N, Parton RG, Hunziker W, Huber LA, Gruenberg J (2000) The recycling endosome of Madin‐Darby canine kidney cells is a mildly acidic compartment rich in raft components. Mol Biol Cell 11: 2775–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Manyes S, Oncins G, Sanz F (2005a) Effect of ion‐binding and chemical phospholipid structure on the nanomechanics of lipid bilayers studied by force spectroscopy. Biophys J 89: 1812–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Manyes S, Oncins G, Sanz F (2005b) Effect of temperature on the nanomechanics of lipid bilayers studied by force spectroscopy. Biophys J 89: 4261–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Manyes S, Redondo‐Morata L, Oncins G, Sanz F (2010) Nanomechanics of lipid bilayers: heads or tails? J Am Chem Soc 132: 12874–12886 [DOI] [PubMed] [Google Scholar]
- Garcia‐Manyes S, Sanz F (2010) Nanomechanics of lipid bilayers by force spectroscopy with AFM: a perspective. Biochim Biophys Acta 1798: 741–749 [DOI] [PubMed] [Google Scholar]
- Gardiner C, Vizio DD, Sahoo S, Théry C, Witwer KW, Wauben M, Hill AF (2016) Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles 5: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracià RS, Bezlyepkina N, Knorr RL, Lipowsky R, Dimova R (2010) Effect of cholesterol on the rigidity of saturated and unsaturated membranes: fluctuation and electrodeformation analysis of giant vesicles. Soft Matter 6: 1472–1482 [Google Scholar]
- Gutierrez MG, Deyell J, White KL, Dalle Ore LC, Cherezov V, Stevens RC, Malmstadt N (2019) The lipid phase preference of the adenosine A2A receptor depends on its ligand binding state. Chem Commun 55: 5724–5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt M, Riedel D, Stein A, Schuette C, Jahn R (2008) Synaptic vesicles are constitutively active fusion machines, which function independently of Ca2+ . Curr Biol 18: 715–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz MJ, Qing D, Shashaty MGS, Zhang P, Faust H, Sondheimer N, Rivella S, Worthen GS, Mangalmurti NS (2018) Red blood cells homeostatically bind mitochondrial DNA through TLR9 to maintain quiescence and to prevent lung injury. Am J Respir Crit Care Med 197: 470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari J, Helenius A (2011) Endosome maturation. EMBO J 30: 3481–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Oh S, Ahn SM, Lee BH, Moon MH (2008) Proteomic analysis of exosomes from human neural stem cells by flow field‐flow fractionation and nanoflow liquid chromatography‐tandem mass spectrometry. J Proteome Res 7: 3475–3480 [DOI] [PubMed] [Google Scholar]
- Kiessling V, Liang B, Tamm LK (2015) Reconstituting SNARE‐mediated membrane fusion at the single liposome level. Methods Cell Biol 128: 339–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi O, Veatch SL, Machta BB (2018) Ion channels can be allosterically regulated by membrane domains near a de‐mixing critical point. J Gen Physiol 150: 1769–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LKM, Murphy S, Kokkinaki D, Venosa A, Sherrill‐Mix S, Casu C, Rivella S, Weiner A, Park J, Shin S et al (2021) DNA binding to TLR9 expressed by red blood cells promotes innate immune activation and anemia. Sci Transl Med 13: eabj1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin SM, Kuhnell D, Orr‐Asman MA, Biesiada J, Zhang X, Medvedovic M, Thomas HE (2019) Balancing yield, purity and practicality: a modified differential ultracentrifugation protocol for efficient isolation of small extracellular vesicles from human serum. RNA Biol 16: 5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulagnier K, Motta C, Hamdi S, Roy S, Fauvelle F, Pageaux JF, Kobayashi T, Salles JP, Perret B, Bonnerot C et al (2004) Mast cell‐ and dendritic cell‐derived display a specific lipid composition and an unusual membrane organization. Biochemical Journal 380: 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Eghiaian F, Sieben C, Herrmann A, Schaap IAT (2011) Bending and puncturing the influenza lipid envelope. Biophys J 100: 637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira RB, Robinson T, Dimova R, Riske KA (2019) Highly efficient protein‐free membrane fusion: a giant vesicle study. Biophys J 116: 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, Hinton G (2008) Visualizing data using t‐SNE. J Mach Learn Res 2579–2605 [Google Scholar]
- Malda J, Boere J, Van De Lest CHA, Van Weeren PR, Wauben MHM (2016) Extracellular vesicles — new tool for joint repair and regeneration. Nat Rev Rheumatol 12: 243–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann HB, Whitney DR (1947) On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat 18: 50–60 [Google Scholar]
- Mantel P‐Y, Hoang A, Goldowitz I, Potashnikova D, Hamza B, Vorobjev I, Ghiran I, Toner M, Irimia D, Ivanov A et al (2013) Malaria‐infected erythrocyte‐derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe 13: 521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden HR, Tomatsu I, Kros A (2011) Model systems for membrane fusion. Chem Soc Rev 40: 1572–1585 [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2003) SerialEM: a program for automated tilt series acquisition on Tecnai microscopes using prediction of specimen position. Microsc Microanal 9(Suppl. 2): 1182–1183 [Google Scholar]
- Mathieu M, Névo N, Jouve M, Valenzuela JI, Maurin M, Verweij FJ, Palmulli R, Lankar D, Dingli F, Loew D et al (2021) Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat Commun 12: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB (2009) HIV enters cells via endocytosis and dynamin‐dependent fusion with endosomes. Cell 137: 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montis C, Busatto S, Valle F, Zendrini A, Salvatore A, Gerelli Y, Berti D, Bergese P (2018) Biogenic supported lipid bilayers from nanosized extracellular vesicles. Advanced Biosystems 2: 1–7 [Google Scholar]
- Morandi MI, Busko P, Ozer‐partuk E, Khan S, Zarfati G, Elbaz‐ Y (2022) Extracellular vesicle fusion visualized by cryo‐EM. bioRxiv 10.1101/2022.03.28.486013 [PREPRINT] [DOI] [PMC free article] [PubMed]
- Moras M, Lefevre SD, Ostuni MA (2017) From erythroblasts to mature red blood cells: organelle clearance in mammals. Front Physiol 8: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller H, Butt HJ, Bamberg E (2000) Adsorption of membrane‐associated proteins to lipid bilayers studied with an atomic force microscope: myelin basic protein and cytochrome c. J Phys Chem B 104: 4552–4559 [Google Scholar]
- Nečas D, Klapetek P (2012) Gwyddion: an open‐source software for SPM data analysis. Cent Eur J Phys 10: 181–188 [Google Scholar]
- Neote K, Darbonne W, Ogez J, Horuk R, Schall TJ (1993) Identification of a promiscuous inflammatory peptide receptor on the surface of red blood cells. J Biol Chem 268: 12247–12249 [PubMed] [Google Scholar]
- Neveu G, Richard C, Dupuy F, Behera P, Volpe F, Subramani PA, Marcel‐Zerrougui B, Vallin P, Andrieu M, Minz AM et al (2020) Plasmodium falciparum sexual parasites develop in human erythroblasts and affect erythropoiesis. Blood 136: 1381–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeyen E, Van Mol K, Baggerman G, Willems H, Boonen K, Rolfo C, Pauwels P, Jacobs AN, Schildermans K, Cho WC et al (2018) Ultrafiltration and size exclusion chromatography combined with asymmetrical‐flow field‐flow fractionation for the isolation and characterisation of extracellular vesicles from urine. J Extracell Vesicles 7: 1490143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir‐Birin Y, Ben Ami Pilo H, Cruz Camacho A, Rudik A, Rivkin A, Revach O‐Y, Nir N, Block Tamin T, Abou Karam P, Kiper E et al (2021) Malaria parasites both repress host CXCL10 and use it as a cue for growth acceleration. Nat Commun 12: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir‐Birin Y, Regev‐Rudzki N (2019) Extracellular vesicles in parasite survival. Science 363: 817–818 [DOI] [PubMed] [Google Scholar]
- Osteikoetxea X, Németh A, Sódar BW, Vukman KV, Buzás EI (2016) Extracellular vesicles in cardiovascular disease: are they Jedi or Sith? J Physiol 594: 2881–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasassi T, Krasnowska EK, Bagatolli L, Gratton E (1998) Laurdan and prodan as polarity‐sensitive fluorescent membrane probes. J Fluoresc 8: 365–373 [Google Scholar]
- Parasassi T, Ravagnan G, Rusch RM, Gratton E (1993) Modulation and dynamics of phase properties in phospholipid mixtures detected by Laurdan fluorescence. Photochem Photobiol 57: 403–410 [DOI] [PubMed] [Google Scholar]
- Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, Coscia C, Iessi E, Logozzi M, Molinari A et al (2009) Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem 284: 34211–34222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Grisel O, Weiss R, Passos A, Brucher M, Varoquax G, Gramfort A, Michel V, Thirion B, Grisel O et al (2011) Scikit‐learn: machine learning in Python. J Mach Learn Res 12: 2825–2830 [Google Scholar]
- Pegtel DM, Gould SJ (2019) Exosomes. Annu Rev Biochem 88: 487–514 [DOI] [PubMed] [Google Scholar]
- Regev‐Rudzki N, Wilson D, Carvalho T, Sisquella X, Coleman B, Rug M, Bursac D, Angrisano F, Gee M, Hill A et al (2013) Cell‐cell communication between malaria‐infected red blood cells via exosome‐like vesicles. Cell 153: 1120–1133 [DOI] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW (2017) Image J2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18: 1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra V. O, Fernandes TFD, Milhiet P‐E, Costa L (2020) Compression, rupture, and puncture of model membranes at the molecular scale. Langmuir 36: 5709–5716 [DOI] [PubMed] [Google Scholar]
- Sampaio NG, Emery SJ, Garnham AL, Tan QY, Sisquella X, Pimentel MA, Jex AR, Regev‐Rudzki N, Schofield L, Eriksson EM (2018) Extracellular vesicles from early stage Plasmodium falciparum‐infected red blood cells contain PfEMP1 and induce transcriptional changes in human monocytes. Cell Microbiol 20: 1–18 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlebach JP, Barrett PJ, Day CA, Kim JH, Kenworthy AK, Sanders CR (2016) Topologically diverse human membrane proteins partition to liquid‐disordered domains in phase‐separated lipid vesicles. Biochemistry 55: 985–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Dufrêne YF, Barger WR, Lee GU (2000) Atomic force microscope image contrast mechanisms on supported lipid bilayers. Biophys J 79: 1107–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorey JS, Cheng Y, Singh PP, Smith VL (2015) Exosomes and other extracellular vesicles in host‐pathogen interactions. EMBO Rep 16: 24–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger HM, Aldrovandi L, Alessandrini A, Facci P (2010) Changes in single K+ channel behavior induced by a lipid phase transition. Biophys J 99: 3675–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P, Seo AY, Pasolli HA, Song YE, Johnson MC, Lippincott‐Schwartz J (2019) A lipid‐based partitioning mechanism for selective incorporation of proteins into membranes of HIV particles. Nat Cell Biol 21: 452–461 [DOI] [PubMed] [Google Scholar]
- Sharon M, Regev‐Rudzki N (2021) Cell communication and protein degradation: all in one parasitic package. J Extracell Vesicles 10: e12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HW, Takatsu H (2020) Phosphatidylserine exposure in living cells. Crit Rev Biochem Mol Biol 55: 166–178 [DOI] [PubMed] [Google Scholar]
- Sisquella X, Ofir‐Birin Y, Pimentel MA, Cheng L, Abou Karam P, Sampaio NG, Penington JS, Connolly D, Giladi T, Scicluna BJ et al (2017) Malaria parasite DNA‐harbouring vesicles activate cytosolic immune sensors. Nat Commun 8: 1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitar S, Kejžar A, Pahovnik D, Kogej K, Tušek‐Žnidarič M, Lenassi M, Žagar E (2015) Size characterization and quantification of exosomes by asymmetrical‐flow field‐flow fractionation. Anal Chem 87: 9225–9233 [DOI] [PubMed] [Google Scholar]
- Sorkin R, Dror Y, Kampf N, Klein J (2014) Mechanical stability and lubrication by phosphatidylcholine boundary layers in the vesicular and in the extended lamellar phases. Langmuir 30: 5005–5014 [DOI] [PubMed] [Google Scholar]
- Sorkin R, Huisjes R, Bošković F, Vorselen D, Pignatelli S, Ofir‐Birin Y, Freitas Leal JK, Schiller J, Mullick D, Roos WH et al (2018) Nanomechanics of extracellular vesicles reveals vesiculation pathways. Small 14: 1–8 [DOI] [PubMed] [Google Scholar]
- Stott BM, Vu MP, McLemore CO, Lund MS, Gibbons E, Brueseke TJ, Wilson‐Ashworth HA, Bell JD (2008) Use of fluorescence to determine the effects of cholesterol on lipid behavior in sphingomyelin liposomes and erythrocyte membranes. J Lipid Res 49: 1202–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullan RMA, Li JK, Zou S (2009) Direct correlation of structures and nanomechanical properties of multicomponent lipid bilayers. Langmuir 25: 7471–7477 [DOI] [PubMed] [Google Scholar]
- Szempruch A, Sykes S, Kieft R, Dennison L, Becker A, Gartrell A, Martin W, Nakayasu E, Almeida I, Hajduk S et al (2016) Extracellular vesicles from Trypanosoma brucei mediate virulence factor transfer and cause host anemia. Cell 164: 246–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takechi‐Haraya Y, Sakai‐Kato K, Abe Y, Kawanishi T, Okuda H, Goda Y (2016) Atomic force microscopic analysis of the effect of lipid composition on liposome membrane rigidity. Langmuir 32: 6074–6082 [DOI] [PubMed] [Google Scholar]
- Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin‐Smith GK et al (2018) Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7: 1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda H, Diaz‐Varela M, Segui‐Barber J, Roobsoong W, Baro B, Garcia‐Silva S, Galiano A, Gualdrón‐López M, Almeida ACG, Brito MAM et al (2020) Plasma‐derived extracellular vesicles from Plasmodium vivax patients signal spleen fibroblasts via NF‐kB facilitating parasite cytoadherence. Nat Commun 11: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrecilhas AC, Soares RP, Schenkman S, Fernández‐Prada C, Olivier M (2020) Extracellular vesicles in trypanosomatids: host cell communication. Front Cell Infect Microbiol 10: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totino PRR, Magalhães AD, Silva LA, Banic DM, Daniel‐Ribeiro CT, Ferreira‐Da‐Cruz MDF (2010) Apoptosis of non‐parasitized red blood cells in malaria: a putative mechanism involved in the pathogenesis of anaemia. Malar J 9: 8–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager W, Jensen J (1976) Human malaria parasites in continuous culture. Science 193: 673–675 [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740 [DOI] [PubMed] [Google Scholar]
- Unsay JD, Cosentino K, Subburaj Y, García‐Sáez AJ (2013) Cardiolipin effects on membrane structure and dynamics. Langmuir 29: 15878–15887 [DOI] [PubMed] [Google Scholar]
- Unsay JD, Cosentino K, García‐Sáez AJ (2015) Atomic force microscopy imaging and force spectroscopy of supported lipid bilayers. J vis Exp 2015: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meer G, Voelker DR, Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9: 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen JA, Cheng KH, Somerharju P (1998) Phospholipid composition of the mammalian red cell membrane can be rationalized by a superlattice model. Proc Natl Acad Sci USA 95: 4964–4969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J et al (2020) SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17: 261–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorselen D, Piontek MC, Roos WH, Wuite GJL (2020) Mechanical characterization of liposomes and extracellular vesicles, a protocol. Front Mol Biosci 7: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorselen D, van Dommelen SM, Sorkin R, Piontek MC, Schiller J, Döpp ST, Kooijmans SAA, van Oirschot BA, Versluijs BA, Bierings MB et al (2018) The fluid membrane determines mechanics of erythrocyte extracellular vesicles and is softened in hereditary spherocytosis. Nat Commun 9: 4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson H (2015) Biological membranes. Essays Biochem 59: 43–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willms E, Cabañas C, Mäger I, Wood MJA, Vader P (2018) Extracellular vesicle heterogeneity: subpopulations, isolation techniques, and diverse functions in cancer progression. Front Immunol 9: 738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortzel I, Dror S, Kenific CM, Lyden D (2019) Exosome‐mediated metastasis: communication from a distance. Dev Cell 49: 347–360 [DOI] [PubMed] [Google Scholar]
- Yang S‐T, Kiessling V, Simmons JA, White JM, Tamm LK (2015) HIV gp41‐Mediated membrane fusion occurs at edges of cholesterol‐rich lipid domains. Nat Chem Biol 11: 424–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Chew M, Hou J, Lai F, Leopold SJ, Loo HL, Ghose A, Dutta AK, Chen Q, Ooi EE et al (2018) Microvesicles from malaria‐infected red blood cells activate natural killer cells via MDA5 pathway. PLoS Pathog 14: 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L et al (2018) Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field‐flow fractionation. Nat Cell Biol 20: 332–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lyden D (2019) Asymmetric‐flow field‐flow fractionation technology for exomere and small extracellular vesicle separation and characterization. Nat Protoc 14: 1027–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Higginbotham JN, Jeppesen DK, Yang Y‐P, Li W, McKinley ET, Graves‐Deal R, Ping J, Britain CM, Dorsett KA et al (2019) Transfer of functional cargo in exomeres. Cell Rep 27: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]


