Abstract
During transmission of malaria‐causing parasites from mosquitoes to mammals, Plasmodium sporozoites migrate rapidly in the skin to search for a blood vessel. The high migratory speed and narrow passages taken by the parasites suggest considerable strain on the sporozoites to maintain their shape. Here, we show that the membrane‐associated protein, concavin, is important for the maintenance of the Plasmodium sporozoite shape inside salivary glands of mosquitoes and during migration in the skin. Concavin‐GFP localizes at the cytoplasmic periphery and concavin(−) sporozoites progressively round up upon entry of salivary glands. Rounded concavin(−) sporozoites fail to pass through the narrow salivary ducts and are rarely ejected by mosquitoes, while normally shaped concavin(−) sporozoites are transmitted. Strikingly, motile concavin(−) sporozoites disintegrate while migrating through the skin leading to parasite arrest or death and decreased transmission efficiency. Collectively, we suggest that concavin contributes to cell shape maintenance by riveting the plasma membrane to the subtending inner membrane complex. Interfering with cell shape maintenance pathways might hence provide a new strategy to prevent a malaria infection.
Keywords: cell migration, cell shape maintenance, gliding motility, malaria transmission, pellicle
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Microbiology, Virology & Host Pathogen Interaction
Concavin, a membrane‐associated Plasmodium protein, is essential for sporozoite shape maintenance. Parasites lacking concavin deform in the mosquito salivary gland and disintegrate while migrating in the skin, which limits transmission.

Introduction
Malaria is still prevalent in tropical countries where it infects over 200 million people every year killing over 400,000, mostly young African children (WHO, 2020). While the symptoms of the disease are caused by the parasite stages infecting red blood cells, the only licenced malaria vaccine has been derived from the surface circumsporozoite protein (CSP) of the mosquito‐transmitted parasite stage, the Plasmodium sporozoite (Clemens & Moorthy, 2016; Cowman et al, 2016). CSP is essential for sporozoite formation and functions at different steps of the sporozoite journey from the mosquito gut to the mammalian liver (Ménard et al, 1997; Thathy et al, 2002; Coppi et al, 2011; Aliprandini et al, 2018; Singer & Frischknecht, 2021). Unfortunately, the CSP‐based vaccine has failed to deliver the long‐sought efficient protection from malaria infections and new vaccine candidates are urgently needed for exploration (Matuschewski, 2017). Plasmodium sporozoites are deposited into the dermis during a mosquito bite and migrate at high speed to enter both blood or lymph vessels (Amino et al, 2006; Hopp et al, 2021). Those entering the blood can ultimately exit the circulation in the liver and infect hepatocytes to further develop into red blood cell infecting merozoites (Prudêncio et al, 2006; Tavares et al, 2013). Anti‐CSP antibodies target sporozoites in the skin prior to entering blood vessels (Aliprandini et al, 2018; Flores‐Garcia et al, 2018; Hopp et al, 2021). Secreted and/or plasma membrane associated sporozoite proteins might therefore constitute additional good candidates for next generation vaccines.
Similar to other extracellular zoites of Plasmodium and related apicomplexan parasites, sporozoites are highly polarized and slender cells with a chiral sub‐pellicular cytoskeleton that defines parasite length and curvature linked to the inner membrane complex (IMC) that subtends the plasma membrane (Khater et al, 2004; Gould et al, 2008; Kudryashev et al, 2012; Volkmann et al, 2012; Tremp et al, 2013; Spreng et al, 2019; Harding & Frischknecht, 2020). The IMC is found at a constant distance to and hence likely linked to the plasma membrane as shown for Toxoplasma gondii tachyzoites (Frénal et al, 2010). Disruption of the IMC‐proteins IMC1a and IMC1h leads to parasite swelling around the nucleus, which impacts motility and infectivity (Khater et al, 2004; Volkmann et al, 2012). However, many proteins of the pellicle of apicomplexan zoites remain to be described and we are still awaiting a complete understanding of how these complex interactions form and maintain the cellular shape (Harding & Frischknecht, 2020).
Sporozoite surface proteins, similar to CSP, can be essential for the escape of sporozoites from oocysts at the Anopheles midgut wall into the mosquito haemolymph, to enter salivary glands, for migration within the skin, to enter blood vessels and ultimately hepatocytes (Silvie et al, 2004; Wang et al, 2005; Carey et al, 2014; Risco‐Castillo et al, 2015; Klug & Frischknecht, 2017; Ishino et al, 2019). Sporozoite migration within the skin provides the first possible target for intervention against an infection with Plasmodium (Aliprandini et al, 2018; Douglas et al, 2018; Murugan et al, 2020; Vaughan, 2021). Antibodies targeting CSP can inhibit sporozoite motility (Vanderberg & Frevert, 2004; Aliprandini et al, 2018) and induce self‐killing of sporozoites via secreted pore‐forming proteins (e.g., SPECT2), which are essential for sporozoite migration through cells (Bhanot et al, 2003; Amino et al, 2008; Risco‐Castillo et al, 2015; Aliprandini et al, 2018). Other secreted proteins include TRP1, LIMP and CelTOS, as well as members of the transmembrane TRAP (thrombospondin related anonymous protein) family. TRP1 (thrombospondin related protein 1) is essential for the release of sporozoites from oocysts and invasion of salivary glands (Klug & Frischknecht, 2017). LIMP (named after a limping phenotype) and TRAP are essential for sporozoite invasion of salivary glands and liver cells (Sultan et al, 1997; Santos et al, 2017) and CelTOS (cell traversal protein for ookinetes and sporozoites) is important for sporozoite motility and migration through cells (Kariu et al, 2006; Jimah et al, 2016; Steel et al, 2018). CelTOS and TRAP are investigated as possible vaccine candidates (Tiono et al, 2018; Pirahmadi et al, 2019).
Proteomic analysis of sporozoites revealed a number of yet uncharacterized proteins including proteins on the parasite surface (Lindner et al, 2013b, 2019; Swearingen et al, 2016). However, the identification of surface proteins has been a challenge due to contamination from cytoplasmic proteins. Here, we selected the PbANKA_1422900 protein that was recently shown to be important for Plasmodium transmission (Ukegbu et al, 2021) for further analysis by gene deletion, GFP‐tagging and functional analyses. We found that this protein is localized at the cytosolic side of the plasma membrane. PbANKA_1422900 is important for the maintenance of the sporozoite shape during their salivary gland residency and is essential for efficient transmission to the vertebrate host. While deformed sporozoites could still move, they were not ejected through the narrow salivary ducts and failed to penetrate through skin and skin‐like environments showing the importance of the slender shape for parasite transmission. Due to its impact on the convex‐concave polarity of sporozoites, we named the protein concavin. Strikingly, during sporozoite migration through narrow constrictions in the skin, we observed concavin(−) parasites to disintegrate by the apparent shedding of large membrane‐delimited parts of the parasite.
Results
Concavin is a conserved Apicomplexan protein important for sporozoite shape maintenance
PBANKA_1422900 is expressed with high abundance in ookinetes and sporozoites according to RNA seq data obtained from Plasmodb.org (Appendix Fig S1A). It is a 393 amino acid long protein in Plasmodium berghei and conserved among Apicomplexa (Appendix Fig S1B). Concavin shares 96% amino acid residue identity with P. yoelii, 80% identity with P. vivax, 76% with Plasmodium falciparum (Appendix Fig S2A) and 36% identity with the orthologue from T. gondii (Appendix Fig S2B). The only recognizable feature in this protein was a potential palmitoylation site at the N‐terminus, which suggests that it could be associated to the IMC or plasma membrane (Appendix Fig S2B). To test for a function of concavin, we disrupted the gene through double homologous recombination in P. berghei (Appendix Fig S3A–C) and T. gondii (Appendix Fig S4A–D). The deletion of concavin readily yielded clonal P. berghei parasite lines, which grew at similar multiplication rates per 24 h as wild‐type parasites in the blood stage (Fig 1A). Similarly, we could not detect a phenotypic difference in a plaque assay between wild‐type and transgenic T. gondii (Appendix Fig S4E and F). Gametocyte to ookinete conversion as well as ookinete motility and cell shape of P. berghei concavin(−) ookinetes did not show any difference to wild type (Fig 1B–D) as was also shown in a recent study (Ukegbu et al, 2021). Yet, the transmission of the parasites to Anopheles stephensi mosquitoes showed slightly reduced numbers of oocysts in infected mosquitoes (Fig 1E). We regularly found large numbers of concavin(−) sporozoites within the salivary glands. However, a large proportion of sporozoites showed an abnormal shape. While wild‐type sporozoites usually keep the typical curved and slender shape at any time post salivary gland entry, concavin(−) sporozoites rounded up over time. The rounding up of concavin(−) sporozoites was initiated at the posterior end of the cell (Fig 1F) and hence appeared different to the rounding observed after liver cell entry (Jayabalasingham et al, 2010). This loss of curvature led us to name PbANKA_1422900 concavin. Curiously, over 90% of oocyst‐derived concavin(−) sporozoites and almost 80% of sporozoites in the haemolymph, the circulatory fluid of the mosquito, were normally formed. Yet with prolonged residency in salivary glands more sporozoites became deformed or rounded up completely (Fig 1G). The apparent asynchrony of deformation hints towards an effect of sporozoite maturation on concavin function that affects individual sporozoites to different degrees. Similar to a recent study (Ukegbu et al, 2021), infection of A. gambiae mosquitoes with concavin(−) parasites revealed a more drastic reduction in oocyst numbers compared to A. stephensi infections (Fig EV1A). The few sporozoites able to reach the A. gambiae salivary gland also showed the same deformation phenotype (Fig EV1A and B). In contrast, we never observed deformed wild‐type sporozoites, neither in the midgut nor in the salivary gland. Curiously, both deformed and normally shaped sporozoites were still able to move (Fig 1H). While normally shaped concavin(−) sporozoites displayed circular movement in a wild‐type manner with nearly wild‐type speed, deformed sporozoites progressed with significantly slower speed (Fig 1H) and completely round sporozoites did not move at all. Deformed sporozoites also moved on less curved paths as did wild‐type or normally formed concavin(−) parasites (Fig 1I).
Figure 1. Deletion of concavin leads to rounding of sporozoites.

- Blood‐stage growth rate of concavin(−) parasites in comparison to wild‐type. Data points represent parasites growing in individual mice, and n indicates total number of mice. P‐value calculated using the Mann–Whitney test. Shown is the mean ± SEM.
- Gametocyte to ookinete conversion of three biological replicates. P‐value calculated using the Mann–Whitney test. Shown is the mean ± SEM.
- Average speed of moving ookinetes. Data points represent individual ookinetes, and n indicates total number of cells from three biological replicates. P‐value calculated using the Kruskal–Wallis test followed by Dunns multiple comparison.
- Ookinete images of wild‐type and concavin(−) parasites revealing similar cell shapes. Scale bar 5 µm.
- Oocyst development of concavin(−) parasites compared to wild‐type. Data points represent individual midguts observed between d12‐17 post infection from 3 independent cage feeds. Shown is the mean ± SEM. P‐value calculated using the Kruskal–Wallis test followed by Dunns multiple comparison.
- Mosquito infections resulted in deformed sporozoites in the salivary gland. Shown are example images of different sporozoites arranged to illustrate their rounding up over time.
- Quantification of sporozoite rounding up over time in the midgut (MG) the haemolymph (HL) and salivary gland (SG) at the indicated days post infection from three biological replicates. Sporozoites were classified as either normal, deformed or round as illustrated in F.
- Average speed of salivary gland sporozoites. Data points represent individual sporozoites, and n indicates total number from three biological replicates. P‐values calculated using the Kruskal–Wallis test followed by the Dunns multiple comparison test.
- Selected trajectories of manually tracked sporozoites.
Source data are available online for this figure.
Figure EV1. Infection of A. gambiae mosquitoes compared to A. stephensi .

- Oocyst development of wild‐type and concavin(−) parasites in A. gambiae mosquitoes. Data points represent individual midguts observed between d12‐17 post infection (left). Shown is the mean ± SEM. P‐value calculated using the Mann–Whitney test. Quantification of sporozoite forms in the midgut and the salivary gland at indicated time points (right).
- Quantification of sporozoite forms in the midgut, the haemolymph and the salivary gland at indicated time points in A. stephensi infections.
Concavin localizes at the cytosolic side of the periphery
To localize concavin, we generated two parasite lines expressing a GFP‐tagged version of the protein. To this end, we recycled the resistance cassette in concavin(−) parasites using negative selection (Appendix Fig S3) (Braks et al, 2006) and reintroduced a P. berghei concavin‐gfp sequence (Appendix Fig S5A and B) as well as a gene encoding for P. falciparum concavin‐gfp (Appendix Fig S5A and C). The resulting parasite lines were named concavin(−)Pbconcavin‐gfp and concavin(−)Pf3D7concavin‐gfp . Furthermore, we tagged the ortholog from Toxoplasma gondii in that parasite (Appendix Fig S4A–D). Both Plasmodium lines were able to establish mosquito infections comparable to wild‐type levels including the colonization of salivary glands by highly motile sporozoites suggesting that both proteins are fully functional (Fig 2A–C). Concavin‐GFP could be detected in gametocytes, ookinetes, sporozoites and liver stages and was absent in blood stage parasites (Fig 2D and E). In non‐activated as well as activated male and female gametocytes the protein localized diffusely (Appendix Fig S6), while in ookinetes and salivary gland derived sporozoites, concavin‐GFP localized at the periphery suggesting an association with the plasma membrane. In T. gondii tachyzoites, we also found a peripheral signal (Appendix Fig S4B and C). A peripheral localization could derive from a protein resident in the sub‐pellicular network, IMC or supra‐alveolar space, the narrow space between IMC and the plasma membrane (Khater et al, 2004; Bane et al, 2016). To specify concavin‐GFP localization, we next fixed concavin(−)Pbconcavin‐gfp expressing sporozoites and labelled them with anti‐GFP antibodies with or without membrane permeabilization. Antibodies only detected concavin‐GFP after permeabilization, suggesting an internal localization of the protein (Fig 2F).
Figure 2. Concavin‐GFP localizes to the periphery of ookinetes and sporozoites.

-
AOocyst development of concavin(−) parasites complemented with either the Plasmodium berghei gene or the P. falciparum orthologue fused to GFP. Data points represent individual midguts observed between d12‐17 post infection of three independent cage feeds. Shown is the mean ± SEM. P‐values are calculated using the Kruskal–Wallis test followed by the Dunns multiple comparison test.
-
BAverage speed of salivary gland sporozoites. Data points represent individual sporozoites of three independent biological replicates. Shown is the mean ± SEM. P‐values are calculated using the Kruskal–Wallis test followed by the Dunns multiple comparison test.
-
CSum table of mosquito infections of wild‐type, concavin(−) and complemented lines expressing either P. berghei (Pb) or P. falciparum (Pf) concavin‐GFP. Numbers determined on d17 post mosquito infection. Infection rate contains three different countings of at least 20 mosquitoes from different mosquito infections. Note the difference of normally shaped and total sporozoites for concavin(−) parasites. Images show infection of salivary glands. Scale bars: 100 µm.
-
DPb concavin‐GFP and Pf concavin‐GFP localization in blood and mosquito stages. Nuclei (blue) stained with Hoechst. Scale bar: 5 µm.
-
ELocalization of P. berghei concavin‐GFP in liver stages. Nuclei (blue) stained with Hoechst. Scale bar: 5 µm.
-
FImmunofluorescence images of permeabilized and unpermeabilized concavin‐GFP expressing salivary gland sporozoites stained with an anti‐GFP antibody. Note that a GFP signal could only be detected after permeabilization, excluding concavin localization on the parasite surface as illustrated in the model. Scale bar: 5 µm.
Source data are available online for this figure.
Concavin‐GFP is highly mobile and does not localize to the cytoskeleton
To test if concavin‐GFP is associated with a cytoskeletal structure, we employed fluorescence bleaching and monitored the recovery of the signal (FRAP). As a control, we generated a PHIL1‐GFP line (Appendix Fig S7A and B) as PHIL1 is a constituent of the sub‐pellicular network. This line recapitulated the published localization of the protein at the periphery of merozoites, ookinetes and sporozoites (Appendix Fig S7B, Saini et al, 2017). Concavin‐GFP expressing sporozoites showed a rapid recovery of the fluorescence signal after a bleached spot was introduced by a high energy laser, suggesting high lateral diffusion of concavin‐GFP (Fig 3A, Movie EV1). In contrast, in bleached PHIL1‐GFP sporozoites, there was no detectable recovery as expected from a protein anchored in the sub‐pellicular network (Fig 3B, Movie EV2). These data suggest that concavin‐GFP is not associated to the sub‐pellicular network or another stable cytoskeletal structure.
Figure 3. FRAP reveals dynamics of concavin‐GFP and stasis of PhiLl1‐GFP.

-
A, BTime series of concavin‐GFP (A) PhiL1‐GFP (B) sporozoites before and after bleaching and quantification of the fluorescence signal over time of three technical replicates. * indicates time of bleaching. Shown is the mean ± SD.
-
CSuper resolution (STED) imaging of PhiL1‐GFP and CSP as well as concavin‐GFP and CSP. Cells were stained with an anti‐GFP antibody in combination with Atto‐594 (green) in addition to an anti‐CSP staining in combination with Atto‐647 (red). Images were deconvolved using the Richardson‐Lucy algorithm. The distance between the 2 signal peaks was measured using the plot profile of the respective channels in Fiji. Data points represent distance in individual sporozoites at the center of the cell from two independent biological replicates. P‐values are calculated using the Kruskal Wallis test followed by the Dunns multiple comparison test. Scale bar: 1 µm.
-
DLocalization of PhiL1‐GFP (green) in normal and deformed concavin(−) parasites. Nuclei (blue) stained with Hoechst. Scale bar 5 µm.
-
ELocalization of SiR‐Tubulin (red) in concavin(−) parasites. Nuclei (blue) stained with Hoechst. Scale bar 5 µm.
-
FLocalization of CSP (red) in concavin(−) parasites. Nuclei (blue) stained with Hoechst. Scale bar 5 µm.
Data information: Asterisks in D‐F indicate apical part of the sporozoite.
Source data are available online for this figure.
Next, we performed super‐resolution (STED) co‐localization experiments with antibodies against concavin‐GFP, CSP and PHIL1‐GFP. STED imaging showed that the signals of anti‐GFP antibodies detecting PHIL1‐GFP and antibodies against CSP were spatially separated (Fig 3C; Appendix Fig S8). In contrast, anti‐GFP antibodies detecting concavin‐GFP co‐localized with anti‐CSP antibodies. Similarly, antibodies recognizing CSP but stained with two different colours also co‐localized (Fig 3C). This suggests that concavin‐GFP is localized closer to the plasma membrane than PHIL1, probably within the alveolar space between IMC and plasma membrane or at the inner leaflet of the plasma membrane.
We next generated a non‐clonal parasite line via single homologous recombination that expresses PHIL1‐GFP in the concavin(−) parasite to investigate the sub‐pellicular network in these parasites (Appendix Fig S7C). PHIL1‐GFP localization of this parasite was similar to that of PHIL1‐GFP in wild‐type parasites in blood stages (Appendix Fig S7B). However, in sporozoites, PHIL1‐GFP appeared in an aberrantly localized manner, as would be expected from the changed shape of the sporozoite (Figs 3D and EV2). We compared this staining to concavin(−) parasites, where the microtubules were labelled with SiR tubulin or with anti‐CSP antibodies. This showed that microtubules appeared largely intact (Figs 3E and EV2) and that CSP was found on the surface of the deformed sporozoites (Figs 3F and EV2). Curiously, however, close inspection of the PHIL1‐GFP labelling showed unexpected accumulations of signal. To investigate the deformed sporozoites at higher resolution, sporozoite containing salivary glands were examined by transmission and scanning electron microscopy. This showed the IMC subtending the plasma membrane in both wild‐type (Fig 4A) and concavin(−) sporozoites (Fig 4B and C). In rounded concavin(−) sporozoites, the IMC was in addition partially not associated with the plasma membrane anymore, but extended deep into the sporozoite cytoplasm (Figs 4B and C, and EV3A and B). Using array tomography (Horstmann et al, 2012; Araki et al, 2020), we next reconstructed serial sections of a complete rounded concavin(−) sporozoite. This clearly revealed an intact tube‐like “rolled up” structure separated by the IMC from the remaining plasma membrane (Fig 4C). This suggests that the IMC is still intact but detached from the subtending plasma membrane.
Figure EV2. Image gallery of concavin(−) parasites.

Concavin(−) parasites expressing phil1‐gfp (left) or stained with SiR‐tubulin (middle) or anti‐CSP antibody (right). Scale bars: 5 µm.
Figure 4. Cytoplasmic IMC extensions in concavin(−) sporozoites.

- Transmission electron micrographs of wild‐type sporozoites with highlighted secretory organelles in magenta, IMC in yellow, microtubules in green and plasma membrane in blue. Scale bar 500 nm.
- Transmission electron microscopy of concavin(−) sporozoites with highlighted secretory organelles as in A; rhoptries in red and nucleus in turquoise. Scale bars: 500 nm.
- Reconstruction of serial sections of a complete rounded up concavin(−) sporozoite; plasma membrane in blue and IMC in yellow. Scale bar 1 µm. In total, 6 parasites were reconstructed from two different preparations.
Figure EV3. Electron microscopy Image gallery of concavin(−) parasites.
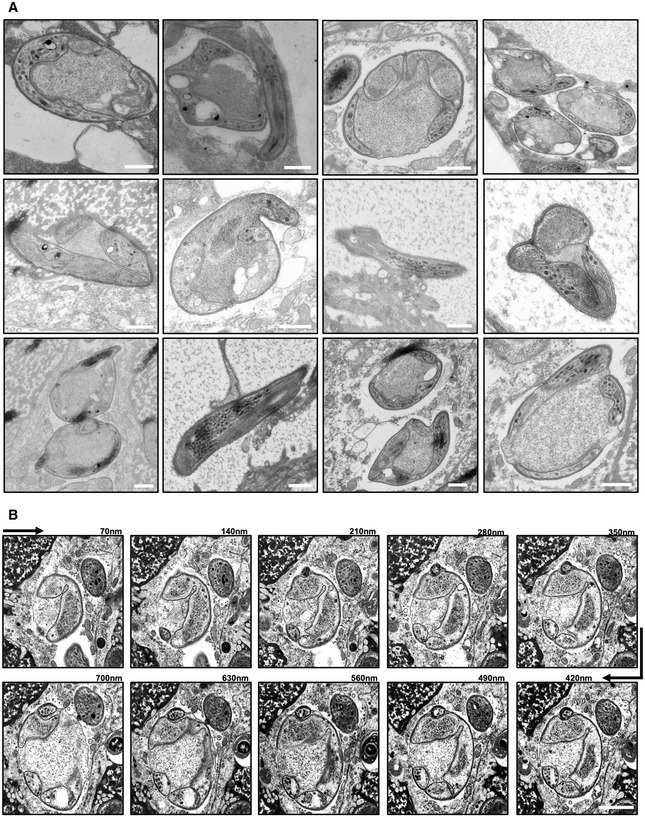
- Transmission electron microscopy images of different concavin(−) sporozoites. Scale bars: 500 nm.
- Scanning electron microscopy images from serial sections of a concavin(−) sporozoites. Scale bar: 500 nm.
Complementation with a palmitoylation site mutant partially restores sporozoite form
We next complemented concavin(−) parasites with a concavin version that expresses an alanine instead of the cysteine of the likely palmitoylation site fused to GFP (Fig 5A, Appendix Fig S5A and D). In blood stage parasites, concavinC7A‐GFP displayed a cytoplasmic signal in gametocytes but was absent in asexual stages, identical to what we observed in wild‐type concavin‐GFP parasites (Fig 5B). This concavinC7A‐GFP parasite readily yielded sporozoites in oocysts (Fig 5C) and salivary glands. Quantitation of the sporozoite shape from both locations showed that at any time point assessed, over 80% of sporozoites showed a normal shape, even as late as 22 days after infection, when the vast majority of concavin(−) sporozoites showed aberrant shapes (Fig 5D, compare to Fig 1D). Only very few sporozoites were completely rounded, while 5–20% of sporozoites showed a rounded proximal end or rounded off around the nucleus. Similar to concavin(−), normal shaped concavinC7A‐GFP sporozoites were able to move in a wild‐type manner, while deformed sporozoites were only able to move at reduced speed (Fig 5E). This suggests that palmitoylation alone is not essential, but does contribute to concavin function. Furthermore, concavinC7A‐GFP showed a peripheral localization in both normally shaped and also slightly rounded sporozoites, suggesting that localization was also not impaired by the mutation (Fig 5F). The GFP signal in this line appeared similar to the signal obtained by anti‐CSP antibodies in concavin(−) sporozoites. This suggests that concavinC7A‐GFP localizes to the plasma membrane and not the IMC.
Figure 5. Limited impact of potential palmitoylation for shape maintenance.

- Cysteine 7 is predicted to be palmitoylated and was changed into alanine in the concavinC7A‐GFP mutant.
- Expression and localization of concavinC7A‐GFP in blood stage parasites. Scale bar 5 µm.
- Oocysts number in infected mosquitoes. Data points represent individual midguts observed between d12‐17 post infection of three independent cage feeds. Shown is the mean ± SEM. P‐value calculated using the Mann Whitney test.
- Quantification of concavinC7A‐GFP cell shapes at the indicated days from midgut (MG) or salivary gland (SG) derived sporozoites. Numbers above bars indicate investigated sporozoites.
- Average speed of salivary gland sporozoites. Data points represent individual sporozoites from three independent biological replicates. Shown is the mean ± SEM. P‐values are calculated using the Kruskal Wallis test followed by the Dunns multiple comparison test.
- Localization of concavinC7A‐GFP in normal and deformed sporozoites (arrowheads point to deformations). Scale bar: 5 µm.
Source data are available online for this figure.
Concavin is essential for efficient transmission
To test if the rounded parasites could still transmit to mice, sets of ten infected A. stephensi mosquitoes were allowed to bite C57BL/6 mice, which are highly sensitive to P. berghei infections. All three mice that were bitten by mosquitoes infected with wild‐type parasites showed the typical blood stage infection in these experiments starting 3 days after the bites (Fig 6A and B). In contrast, only 8 of 12 mice that were bitten by concavin(−) infected mosquitoes ever became infected. In these 8 mice, the development of the blood stage infection was delayed by over 1 day compared to the wild‐type controls, in itself a loss of infectivity by 90% (Fig 6A and B). In contrast, mice infected with 10,000 concavin(−) sporozoites by intravenous injection did not show any difference in time to blood stage appearance compared to wild‐type (Fig 6A and B). To test if deformed sporozoites could enter into cells, we performed an infection experiment, where sporozoites were added to cultured HeLa cells, which are as susceptible to P. berghei infection as hepatocytes (Kaiser et al, 2016). After incubation of 1 h, cells were fixed and labelled with anti‐CSP antibodies, without permeabilization, to distinguish sporozoites within and outside of cells. While the first three biological replicates showed a higher percentage of cell invasion for wild‐type than concavin(−) parasites, a fourth experiment lowered the level of statistical confidence. Importantly, however, the deformed parasites could enter into host cells, albeit concavin(−) parasites probably entered at an overall lower rate as wild‐type parasites (Fig 6C). Liver‐stage development of concavin(−) parasites did not show any difference to wild‐type parasites (Fig 6D).
Figure 6. Concavin is essential for efficient transmission by mosquitoes.

-
AGrowth curve of blood stage parasites in C57BL/6 mice infected by the bite of 10 mosquitoes (left) or 10,000 sporozoites intra venously (right) at two different times post mosquito infection as indicated. Shown is the mean ± SEM. Mice infected with concavin(−) parasites by bite represent two independent biological replicates with three mice each. Wild‐type bite back and concavin(−) intra venous injections represent one biological replicate using 3 or 4 mice respectively.
-
BSum table of infected mice from A with pre‐patency period (time to detect a blood stage infection) and parasitemia on day 6. Note that in concavin(−) by bite infected mice not all develop a blood stage infection.
-
CLiver cell invasion assay of concavin‐gfp (WT) and concavin(−) parasites. Parasites are positively stained for CSP in case they remain extracellular. Both, normal and deformed parasites were detected intracellularly. Graph shows quantification of CSP positive (red, hence extracellular) and negative (grey, hence intracellular) sporozoites. Small graph shows the percentage of deformed and normally shaped intracellular concavin(−) sporozoites. Data points represent the 4 individual biological replicates with n indicating the numbers of sporozoites observed. Shown is the mean ± SEM. P‐values are calculated using the Mann–Whitney test. Scale bar: 5 µm.
-
DLiver‐stage development of concavin(−) parasites compared to wild‐type, both expressing cytoplasmic GFP (white). Parasite size was measured 24 and 48 h post infection. Data points represent individual parasites from four independent biological replicates. Shown is the mean ± SEM. P‐values calculated using the Mann Whitney test. Scale bar: 5 µm.
-
ESporozoite ejection of immobilized concavin(−) infected mosquitoes on glass slides and quantification of ejected sporozoites from 20 mosquitoes. * indicates individual sporozoites in the ejected saliva. Scale bar: 10 µm.
-
F, GOnly normally shaped concavin(−) sporozoites released by salivary glands move on helical paths (arrows) through polyacrylamide gels that mimic the skin (F). Scale bar: 10 µm. (G): Quantification of two individual experiments; only normal shaped sporozoites were able to migrate through the gel.
Source data are available online for this figure.
But could deformed concavin(−) parasites at all reach the liver in vivo? Two obstacles might block the progression of the deformed or rounded parasites from the salivary gland to the liver: the narrow salivary ducts through which the parasites are ejected and the dermis in the skin, through which the parasites need to pass prior to entering the blood stream. To test transmission efficiency, we first immobilized concavin(−) infected mosquitoes on glass slides and observed the ejection of sporozoites (Fig 6E). As expected (Frischknecht et al, 2004; Aleshnick et al, 2020), the ejection of sporozoites was highly irregular (Fig 6E). Yet, at days 17 and 21, in four different experimental sessions, we always observed at least one mosquito salivating many dozens of sporozoites while most ejected just a few or none. While not all ejected sporozoites could be readily classified according to their shape, the vast majority of sporozoites showed a normal crescent shaped (Movie EV3). Some appeared rounded but often revealed themselves as being normally shaped once they reoriented in the focal plane after ejection into the droplet of saliva (Movie EV3). We could not confidently see ejected sporozoites that were deformed or rounded suggesting that these cannot enter the salivary canal, albeit we cannot exclude that about 10–20% of ejected sporozoites were deformed. Yet, when investigating the sporozoites resident in the salivary gland of the mosquitoes that ejected many sporozoites, over 80% of concavin(−) sporozoites were deformed. This suggested that rounded sporozoites could not efficiently enter into and pass through the salivary canals. To test if sporozoites fail to migrate through confined spaces, we squeezed a salivary gland between a glass slide and a polyacrylamide gel such that sporozoites were liberated and able to enter into the gel (Fig 6F) (Ripp et al, 2021). On the surface of the gel, both deformed and normally shaped parasites are readily visible, while at the bottom end of the gel, only normally shaped sporozoites were found (Fig 6F and G), indicating that deformed sporozoites could not cross the dense matrix of the gel. These data suggest that deformed sporozoites cannot enter and move through confined spaces such as the salivary canals and probably also not in the skin.
Disintegration of sporozoites migrating in the skin
To investigate if concavin(−) sporozoites could progress through the skin, we let mosquitoes infected with wild‐type or concavin(−) sporozoites expressing a cytosolic fluorescent protein bite the ear pinnae of mice and imaged with a spinning disk confocal microscope. This showed that mosquitoes were less successful in transmitting concavin(−) than wild‐type sporozoites. Fewer mosquitoes transmitted concavin(−) sporozoites and at lower numbers (Fig 7A), although the lower level of fluorescence in the concavin(−) sporozoites might have impeded precise quantification. Intriguingly, of 90 concavin(−) sporozoites observed over 13 different bite sites, 72 (80%) appeared to have a normal morphology, suggesting that indeed fewer abnormal sporozoites pass through the mosquito proboscis during a bite (Appendix Fig S9A and B). The remaining 18 (20%) concavin(−) sporozoites were abnormally shaped from the beginning of the recording, some being only half as long as normal sporozoites, appearing completely roundish or exhibiting a rounded posterior end (Appendix Fig S9B). Analysis of time‐lapse series showed fewer migrating mutant sporozoites, which however progressed with the same speed as wild‐type parasites (Fig 7B; Movie EV4). In addition, fewer blood vessel invasion events were observed in motile micro‐syringe inoculated concavin(−) sporozoites (Fig 7C). Strikingly, some sporozoites formed a bulky dot at their rear (deformation) during migration, mostly associated with passing through a narrow opening as indicated by their body constriction and decrease in speed (Fig 7D; Movie EV5). This round posterior structure sometimes detached as the parasites kept moving forward and this loss of body integrity was defined as disintegration (Fig 7E; Movie EV5). Of 54 wild‐type sporozoites, only 2 (4%) showed disintegration, while 32% of 57 migrating concavin(−) sporozoites disintegrated and 7% exhibited posterior deformation only (Fig 7F). This suggests that in their effort to migrate in the skin, around 30–40% of concavin(−) sporozoites lose their shape or cellular integrity. Most of those sporozoites stop migrating while some appear to suffer no impairment for the duration of imaging, and around 10% die as evidenced by the loss of GFP fluorescence (Fig 7G). Disintegration most frequently occurred when sporozoites were gliding inside hair follicles, in the upper part of the dermis or after sustained circular gliding, which is usually associated with cell traversal (Formaglio et al, 2014). This suggests that sporozoite disintegration is a consequence of tight interactions between migrating parasites and host cells either because the sporozoites squeeze themselves between or through cells.
Figure 7. Migrating concavin(−) sporozoites lose their cellular integrity.

-
APercentage of mosquitoes depositing WT or concavin(−) sporozoites in the skin of a mouse during a bite (left) and sporozoites deposited during a mosquito bite (right). P‐values are calculated using the Fishers exact test (left) and the Mann Whitney test (right). Bite transmission experiments were repeated over three independent mosquito infections for the KO and two independent mosquito infections for the WT. For each mosquito infection 2–3 mice were imaged, for a total of 3–5 ears.
-
BMaximum fluorescence intensity projections encoded by color for time from movies showing migrating sporozoites after mosquito‐bite transmission. Graphs and camembert diagrams below show fraction of motile and immotile sporozoites after 10, 20 and 30 min of recording, numbers analysed as well as sporozoite speed. Pooled data from 4–5 WT or 5–8 concavin(−) bite sites per time point. Scale bar: 50 µm. P‐values are calculated using the Mann Whitney test.
-
CPercentage of motile WT or concavin(−) sporozoites entering blood vessels during the first hour following micro‐injection. Number above bars shows numbers of sporozoites observed. Pooled data from four WT or five concavin(−) experiments. P‐values are calculated using the Fishers exact test.
-
D, E(D) Deformation and (E) disintegration of concavin(−) sporozoites migrating in the skin. Individual images corresponding to the frames shown on the right are indicated in distinct colours in the maximum projection (left) of 13‐min movies. Arrowheads indicate constrictions of the parasites. Graphs below time‐lapse show that deformation and disintegration are preceded by a decrease in speed. Color of dots correspond to the time‐points displayed in the time‐lapse images. Scale bars: 10 µm.
-
FPercentage of deformation and disintegration events observed in 54 WT and 57 concavin(−) sporozoites that were tracked for at least 10 min. Pooled data from 5 WT or 10 concavin(−) bite sites.
-
GConsequence of deformation and disintegration from the 22 sporozoites in F include immobilization and parasite death.
Source data are available online for this figure.
Together these data strongly suggest that concavin(−) sporozoites fail to maintain their shape, which blocks migration into the salivary ducts and leads to lower levels of transmission. Of the transmitted sporozoites, a large proportion loses their cellular integrity and cannot migrate efficiently in the skin explaining the reduced transmission to the mammalian host.
Discussion
Here, we show that a protein named concavin contributes to cell shape maintenance in Plasmodium berghei sporozoites and is essential for efficient transmission of malaria parasites from mosquitoes to the mammalian host. Although already affecting colonization of the midgut and being expressed before sporozoites enter salivary glands, the major phenotypic difference of concavin(−) parasites only becomes apparent upon prolonged residency within this organ (Figs 1 and 2). The parasites appear to rely on concavin to keep the IMC closely subtending the plasma membrane as loss of the protein leads to blebbing of plasma membrane and invaginations of the IMC (Figs 3C and 4). The rounding of the sporozoite inhibits ejection from the narrow salivary canals and leads to less efficient deposition of parasites in the skin (Figs 6E and 7A). Transmitted concavin(−) parasites are not yet rounded and can move in the skin as wild‐type parasites do, but upon squeezing through narrow spaces they lose their cellular integrity much more frequently than wild‐type parasites (Fig 7B–G). It appears that restraining forces from the environment act upon the motile parasites, which necessitated the evolution of a machinery that keeps the parasites from disintegration. Likely the skin provides a formidable challenge for the parasite as they move for tens of minutes past collagen fibres as well as through and between cells. We only observed a portion of concavin(−) sporozoites disintegrating, which might be due to the limited observation time and volume during in vivo imaging or reflect a heterologous population of sporozoites, some of which are not impacted as strongly by the lack of concavin. Indeed, sporozoites exhibit a distribution of shapes with curvatures ranging from 0.13 to 0.35/µm (Muthinja et al, 2017), which might indicate different strengths in the shape maintenance of the individual parasites and hence a different impact of the gene deletion. Based on the number of newly transcribed genes upon salivary gland entry (Matuschewski et al, 2002), the parasite clearly prepares actively for its journey through the skin to the liver. Hence, it might be possible that other proteins also contribute to one or multiple complexes that involve concavin and gliding associated proteins for riveting the plasma membrane and IMC.
A strikingly similar loss of cellular integrity during migration of sporozoites is observed in the presence of antibodies targeting CSP (Aliprandini et al, 2018). This surprising similarity shows that the skin is a harmful environment for the moving sporozoite and indicates that the lack of concavin and the crosslinking of CSP by antibodies can make the parasite fragile. Could these processes be linked? Antibodies crosslink CSP into large aggregates, which are shed at the rear of the parasite and can literally strip a migrating sporozoite. The force acting upon the parasite by these large aggregates being stuck within the tissue as the parasite pushes forward might well lead to a rupture of the plasma membrane‐IMC link and hence blebbing. Detailed electron microscopy images of disintegrating parasites would be needed to address if the processes are similar. We postulate that concavin is anchored in the plasma membrane by palmitoylation at its N‐terminus, possibly by the palmitoyl‐S‐acyl transferase DHHC3 (Hopp et al, 2016) or DHHC2, which is located at the sporozoite periphery with a bias towards the rear end (Santos et al, 2015). It might hence interact within the membrane with the GPI‐anchor of CSP, which could influence the mobility within the membrane of both proteins.
Interestingly, disrupting a predicted palmitoylation site in concavin led to only a very mild phenotypic difference from the wild‐type with less than 20% of the salivary gland derived concavinC7A‐GFP parasites showing an aberrant shape (Fig 5). Importantly, the localization of the protein was not changed suggesting that palmitoylation alone is not responsible for localization and hence also only partially contributes to concavin function. This hints towards the presence of other protein‐protein interactions that are essential for concavin localization and functionality as well as sporozoite shape maintenance. This finding of partial impact of palmitoylation is reminiscent of the recent work on palmitoylation of the myosin light chain in Toxoplasma gondii (Rompikuntal et al, 2021), where disruption of palmitoylation of myosin light chain 1 led to the partial disassembly of the gliding motor complex but had little impact on motility itself. Considering the EM images showing large invaginations of the IMC away from the plasma membrane in concavin(−) sporozoites, we postulate that concavin might contribute towards riveting the plasma membrane to the subtending IMC, which are a constant distance of 25 nm apart. In T. gondii, the gliding associated protein 45 (GAP45) was shown to bridge the gap between IMC and plasma membrane (Frénal et al, 2010) and fulfil a similar function. GAP45 is associated with the PM through N‐terminal palmitoylation and myristoylation and interacts with the IMC at the C‐ terminus. Intriguingly, gap45(−) T. gondii parasites develop normally but show aberrant IMC invaginations upon host cell invasion, reminiscent of the deformed concavin(−) Plasmodium sporozoites (Frénal et al, 2010; Egarter et al, 2014). Strikingly, concavin is the first protein essential for sporozoite shape maintenance that is not fixed to or appears as part of a cytoskeletal structure. Compared to Plasmodium berghei GAP45, which consists of 184 aa, concavin is about twice as large (393 aa) and hence might well span the distance between plasma membrane and IMC.
In ookinetes, the deletion of the phosphodiesterase PDEδ, which breaks down cyclic nucleotides such as cGMP, revealed a remarkably similar phenotype to deletion of concavin in sporozoites. The pdeδ(−) parasites formed normally shaped ookinetes, which then progressively rounded up, where not capable of migrating across the midgut epithelium and largely failed to establish a mosquito infection (Moon et al, 2009). Single section transmission electron microscopy showed the partial absence of the IMC. In contrast, concavin(−) ookinetes showed no morphological difference. To investigate why concavin(−) ookinetes establish fewer oocysts (see also: Ukegbu et al, 2021), and in analogy to the disruption of normally shaped migrating concavin(−) sporozoites, it would be interesting to image migrating ookinetes as they pass from the blood meal to the epithelium. How either PDEδ or concavin contribute to keeping the IMC in place remains elusive. Understanding how concavin remains mobile while contributing to cellular shape maintenance clearly needs further work. The presence of a potential palmitoylation site at the N‐terminus of concavin, in combination with the ability of the protein to recover after FRAP suggest an incorporation into the PM rather than the IMC. The ability of normal and deformed sporozoites to remain motile excludes most likely a function of concavin in glideosome formation. We speculate that concavin is either directly involved in PM and IMC organisation or through transient interactions with GAP45. Open questions include: does concavin interact with other proteins and if yes, what are the binding partners? Does it interact with proteins linked to the IMC or the actin‐myosin motor machinery that drives the parasite? Likely more proteins are important in keeping the plasma membrane at a constant distance from the IMC and maintaining the shape of these parasites. Finding concavin interacting proteins could also reveal why the change in shape becomes more dramatic with prolonged salivary gland residency.
Sporozoites are not born with a final shape, but mature from long and slender ones in the oocysts to crescent‐shaped slightly thicker ones in the salivary glands (Kudryashev et al, 2012; Muthinja et al, 2017). Investigations by cryogenic electron tomography revealed that the sub‐pellicular network (SPN) is only robustly detectable in sporozoites isolated from the salivary glands (Kudryashev et al, 2012) and the tagging of SPN proteins revealed their peripheral localization only in salivary gland derived sporozoites (Khater et al, 2004). This suggests that the SPN and possibly its linkage to microtubules plays a key role in the generation of the crescent shape observed for transmission ready sporozoites. In turn, this suggests that the shape has a key function for sporozoite infectivity. Indeed, mutants where the shape of sporozoites is altered have been shown to transmit less efficiently or not at all to rodent hosts (Montagna et al, 2012; Volkmann et al, 2012; Tremp et al, 2013; Spreng et al, 2019). Yet, concavin(−) sporozoites reveal for the first time a loss of cellular integrity as a phenotypic consequence of deleting a Plasmodium gene. The movies of sporozoites migrating in the mouse skin in vivo suggest that plasma membrane, containing considerable amounts of cytosol, is lost as the parasites migrate through tight obstacles.
Upon liver cell invasion, the sporozoite naturally changes its shape, a likely active process depending on newly translated proteins from stored transcripts (Gomes‐Santos et al, 2011). The deletion of the RNA binding protein pumilio‐2 led to a progressive rounding of sporozoite already in the salivary glands and premature expression of liver stage specific genes (Gomes‐Santos et al, 2011; Lindner et al, 2013a). In contrast to pumilio‐2 mutants, which only round up after several days of salivary gland residence, the lack of concavin led to early rounding after salivary gland invasion (Fig 1C–E). This, and the different localization of the two proteins, suggests a completely different function of the two proteins. Also, pumilio‐2(−) sporozoites “round up” in a way reminiscent of the shape changes after liver cell invasion: the sporozoites bleb in the center with their ends initially keeping their sporozoite shape (Gomes‐Santos et al, 2011). This is similar to the rounding of mutants lacking the IMC‐1 and IMC‐1h proteins (Khater et al, 2004; Volkmann et al, 2012). In contrast, concavin(−) sporozoites round up from their proximal ends (Figs 1B, 5C–E and EV2).
We found that, in T. gondii, disrupting the concavin orthologue has no visible impact on in vitro life in cultured fibroblasts. This however does not rule out that during other parts of the life cycle of this parasite more constraining barriers encountered by the parasite might impact on cellular integrity too. We observed a slight reduction in oocyst numbers, which hints at a possible function of concavin also for ookinetes. Ookinetes need to pass the peritrophic matrix that forms around the ingested blood meal and through one layer of epithelial cells. Ookinetes move much slower than sporozoites and hence concavin might not be as important during their short life as it is for the much longer living and faster migrating sporozoites facing many more constrictions on their journey from oocyst to liver.
Lastly, as malaria parasites can be targeted within mosquitoes, e.g., through mosquito uptake of antiparasitic drugs (Paton et al, 2019), it might be feasible to target the parasite cell shape maintenance pathways with small molecules within the insect to prevent the transmission of the parasite. While concavin might not directly be targetable, enzymes involved in overall cell shape maintenance such as palmitoyl transferases (Wang et al, 2005; Santos et al, 2015; Hopp et al, 2021; Schlott et al, 2021) could be good candidate targets.
In conclusion, we showed here the disintegration of migrating Plasmodium sporozoites due to the lack of a novel protein, concavin. Functional analyses through GFP‐tagging, FRAP, gene deletion and in vivo imaging showed the importance of concavin for the maintenance and integrity of Plasmodium sporozoite shape and hence efficient transmission from mosquitoes to mammals.
Materials and Methods
Generation of parasite lines
Concavin(−)
The 3′UTR (779 bp) of PbANKA_1422900 was amplified from wild‐type gDNA using primers JK57 and JK58 and inserted into a plasmid (pL22) containing the recyclable yFCU/ hDHFR selection cassette and gfp expressed under the hsp70 promoter digested with NotI and SacII. The 5′UTR (554 bp) was amplified using primers JK55 and JK56 and inserted into the plasmid using KpnI and HindIII. The resulting plasmid pL24 was linearized with KpnI and SacII prior transfection for double crossover integration (Appendix Fig S3; Appendix Table S1).
Marker free concavin(−) NS
The drinking water of mice infected with concavin(−) parasites was supplemented with 2 mg/ml 5‐FC (5‐fluorocytosine). Clonal parasites which looped out the selection cassette were obtained by limiting dilution. (Appendix Fig S3; Appendix Table S1).
Plasmodium berghei complementation
The 5′UTR together with the entire ORF of PbANKA_1422900 was amplified from wild‐type gDNA using primers JK55 and JK176 and inserted into a plasmid (pL59) containing gfp and the TgDHFR selection cassette using KpnI and NdeI. Resulting in plasmid pL79. For transfection of concavin(−)NS parasites via double crossover, the plasmid was digested with KpnI and SacII. (Appendix Fig S5; Appendix Table S1).
PF3D7 complementation
The P. berghei 5′UTR was amplified using primers JK55 and JK179 and inserted into pL59 using KpnI and BstbI, followed by the insertion of PF3D7_0814600 amplified with primers JK177 and JK178 from P. falciparum gDNA and digested with BstbI and NdeI. For the transfection of concavin(−)NS parasites via double crossover, the plasmid was digested with KpnI and SacII (Appendix Fig S5; Appendix Table S1).
ConcavinC7A complementation
ConcavinC7A together with gfp was amplified from pl79 using primers JK236 and JK237. The resulting PCR product was digested with BamHI and ligated into pL82 digested with BamHI and BstBI with filled in overhangs, leading to the final plasmid pL120. For transfection of concavin(−)NS parasites via double crossover, the plasmid was digested with KpnI and SacII (Appendix Fig S5; Appendix Table S1).
Phil1‐GFP
PhIL1‐GFP (PBANKA_020460) parasites with GFP at the C‐terminus were generated via single homologous recombination. A region of the PhIL1 gene 54 bp downstream of the ATG start codon and lacking the stop codon was amplified using primers P969 and P970 (Appendix Fig S7; Appendix Table S1). The PCR product was digested using EcoR1 and BamH1 enzymes and ligated into a vector containing the TgDHFR selection cassette as a positive selection marker. For transfection, the final vector was linearized using BsaB1.
Transfection of linearized plasmids was performed as previously described (Janse et al, 2006). After electroporation of schizonts, positive selection was performed using pyrimethamine. All resulting monoclonal lines were obtained through limiting dilution using NMRI or CD1 mice. Briefly, 0.9 parasites were injected into 10 naïve mice followed by genotyping of positive animals on day 9 post infection.
Mosquito infections
All experiments were performed with Anopheles stephensi mosquitoes, fed with 1% salt/water and 10% sucrose/water solution containing 0.05% para‐aminobenzoic acid (PABA) and pads containing alcohol free Paulaner. Naïve mosquitoes were kept at 28°C and 70% humidity and subsequently transferred to 21°C after infection. Infection of mosquitoes was done with mice infected with 20 million blood stage parasites, 4 days post infection. Gametocyte formation was monitored by counting exflagellation events in peripheral blood. Mice were anesthetized with a combination of ketamin/ xylazin and mosquitoes were allowed to bite for 20 min.
Gametocyte activation and Ter‐119 staining
Six million blood stage parasites were injected i.v. into a naïve Swiss CD1 mouse. 4 days post infection, the blood was harvested by cardiac puncture: 100 µl of blood together with 500 µl of ookinete medium (see below) was incubated for 15 min. After fixation of the parasites with 4% PFA at 4°C for at least overnight, staining of the red blood cell membrane was performed with the anti‐Ter119::Alexa647 antibody (Biolegend, 0.5 mg/ml; 1/1,000) for 1 h. Cells were washed twice with PBS and resuspended in PBS containing Hoechst for observation under the microscope.
In vitro ookinete cultures
6 million blood stage parasites were injected i.v. into a naïve Swiss CD1 mouse. 4 days post infection the blood was harvested by cardiac puncture to set up ookinete cultures. The blood of one mouse was added to 10 ml of ookinete medium (RPMI containing 25 mM Hepes and 300 mg/l, l‐glutamine, 10 mg/l hypoxanthine, 50,000 units/l penicillin, 50 mg/l streptomycin, 2 g/l NaHCO3, 20.48 mg/l xanthurenic acid, 20% FCS; pH 7.8) at 21°C and incubated for 21 h. Fully developed ookinetes were purified on a 63% Nycodenz cushion and resuspended with ookinete medium.
Sporozoite motility assay
Gliding assays of isolated sporozoites were performed in 96‐well optical bottom plates (Nunc) using 3% BSA/ RPMI with a frame rate of 3 s for 3 min on a Zeiss CellObserver widefield microscope with 25× magnification. Speed was determined with the manual tracking tool in ImageJ.
Immunofluorescence staining of sporozoites
Sporozoites were seeded with 3% BSA/ RPMI into an 8‐well Labtek chambered cover glass. After fixation with 4% PFA for 20 min, cells were permeabilized with 0.5% TritonX for 15 min. Primary antibodies were incubated for 1 h and washed twice with PBS. Secondary antibodies together with Hoechst were incubated for 1 h. Cells were washed twice with PBS and observed under the microscope. Images were either taken on a Zeiss CellObserver widefield (63×) or Nikon/PerkinElmer spinning disc (100×) microscope. Image processing was performed with ImageJ. Antibodies: rabbit anti‐GFP for IFA 1/40 (abfinity 0.4 µg/µl), mouse anti CSP (mAb 3D11, 1/100, Yoshida et al, 1980), goat anti‐mouse Alexa 594 1/1,000 (Invitrogen 2 mg/ml), goat anti‐rabbit Alexa 594 1/1,000 (Invitrogen 2 mg/ml). For STED imaging, anti‐mouse Atto 594 1/300 (Sigma) and anti‐rabbit Atto 647N 1/300 (Sigma) was used. The staining of sporozoites with antibodies or SiR‐tubulin was performed as described previously (Spreng et al, 2019).
FRAP
FRAP experiments were performed using a PerkinElmer Nikon spinning disc microscope with FRAP module. Images were taken with a 100× objective (NA 1.4) every 0.5 s. Selected regions were bleached using a 405 nm laser at 100% laser power.
STED imaging
Super‐resolution imaging was performed on a STED microscope from Abberior Instruments GmbH (Göttingen) using a 100× objective. STED‐illumination was done using a 594 or 640 nm excitation laser in combination with 775 nm depletion. Images were taken with 15 nm pixel size and a line accumulation of 2. Deconvolution was done using the Imspector software provided by Abberior using the Richardson‐Lucy algorithm with 30 iterations. Further image processing was done using Fiji.
Electron microscopy
Isolated salivary glands from infected Anopheles stephensi mosquitoes were directly dissected into 2% glutaraldehyde and 2% paraformaldehyde in 100 mM Cacodylate buffer and fixed at 4°C overnight. After rinsing in buffer, the samples were further fixed in 1% osmium in 100 mM Cacodylate buffer for 1 h, washed in water and incubated in 1% uranylacetate in water overnight at 4°C. Dehydration was done in 10 min steps in an acetone gradient followed by stepwise Spurr resin infiltration at room temperature and polymerization at 60°C.
The blocks were trimmed to get cross‐sections of the salivary glands and sectioned using a Leica UC6 ultramicrotome (Leica Microsystems Vienna) in 70 nm thin sections. The sections were placed on formvar coated slot grids, post‐stained in uranyl acetate and Reynold’s lead citrate and imaged on a JEOL JEM‐1400 electron microscope (JEOL, Tokyo) operating at 80 kV and equipped with a 4K TemCam F416 camera (Tietz Video and Image Processing Systems GmBH, Gautig).
Array tomography
3D reconstruction of the round sporozoites was done using array tomography. Ultrathin serial sections (70 nm) were cut on an UC7 ultramicrotome (Leica Microsystems, Vienna, Austria) equipped with a section receiver (cd‐fh, Heidelberg) enabling a smooth pick up of serial sections on wafers (Si‐Mat SiliconMaterials) treated by glow discharge. The sections were post‐stained by placing the wafer pieces in uranyl acetate for 20 min and lead citrate for 10 min, each in a closed tube. The wafers with sections were washed repeatedly in water after each step. Serial sections through one sporozoite (around 40 sections) were imaged by scanning electron microscopy with a LEO Gemini 1530 equipped with a field emission gun and an ATLAS scanning generator (Zeiss). Images of 10,000 × 10,000 pixels and 2 nm resolution were taken at the same area in consecutive sections using the in‐lens detector. Imaging parameters used: 3 mm working distance, 30 µm aperture and 2 kV acceleration voltage. The images through each parasite were aligned and the membranes segmented manually using the IMOD software package (Kremer et al, 1996). In total, 6 sporozoites were reconstructed.
Sporozoite invasion into cells and liver‐stage development
Hela cells were seeded into 8‐well Labtek chamber slides with glass bottom. Once the cells reached about 95% confluency, sporozoites were added on top of the cells. The entire slide was centrifuged for 5 min at 800 rpm to make the sporozoites adhere to the cells and incubated at 37°C. After 1 h, the cells were fixed with 4% PFA/ PBS for 30 min, followed by staining of the cells against CSP (mouse anti CSP mAb 3D11, 1/ 100, Yoshida et al, 1980). The primary antibody was incubated for 1 h and washed twice with PBS. The secondary antibody (goat anti‐mouse Alexa 594 1/1,000 (Invitrogen 2 mg/ml)) together with Hoechst were incubated for 1 h. Cells were washed twice with PBS and observed under the microscope. Images were either taken on a Zeiss CellObserver widefield (63×) or Nikon/PerkinElmer spinning disc (100×) microscope. Image processing was performed with Fiji.
For liver‐stage development, cells were split into 2 wells 1 h post infection and fixed with 4% PFA/ PBS after 24 and 48 h. Nuclei were stained with Hoechst and images taken on a Zeiss Axio‐Observer widefield (63×) microscope. Image processing and quantification of parasite size was done with Fiji. Cell lines were authenticated and tested for mycoplasma contamination.
Live imaging of Mosquito sporozoite ejection
For imaging of ejected sporozoites, mosquitoes were immobilized with small drops of superglue on the wings and thorax on a 24 × 60 mm cover glass. The labrum was carefully removed using 2 needles to liberate the stylets of the mosquito which were ideally pressed flat onto the cover glass. Imaging was done on a Zeiss Axio‐Observer with 25× objective at a frame rate of 3 frames per second.
3D imaging in polyacrylamide gels
Polyacrylamide hydrogels were prepared as described before (Pelham & Wang, 1997). Briefly, APS and TEMED were added to a prepolymer solution containing 3% acrylamide and 0.03% bis‐acrylamide in PBS. The solution was pipetted onto a silanized glass coverslip immediately and covered with a second glass coverslip. After polymerization, the top coverslip was removed in PBS. Whole salivary glands were dissected into 30 µl 3% BSA/RPMI and covered with the hydrogel. Images were recorded on top of the hydrogel as well as at the hydrogel‐glass interface using a 25× objective and a frame rate of 3s.
In vivo imaging of sporozoites in the mouse skin
In vivo imaging in the skin was performed on a spinning‐disk confocal system (UltraView ERS, Perkin Elmer) controlled by Volocity (Perkin Elmer) and composed of 4 Diode Pumped Solid State Lasers (excitation wavelengths: 405, 488, 561 and 640 nm), a Yokogawa Confocal Scanner Unit CSU22, a Z‐axis piezoelectric actuator and a Hamamatsu Orca‐Flash 4.0 camera mounted on a Axiovert 200 microscope (Zeiss). Z‐stacks of 6 plans covering 25–30 µm were acquired using a LCI “Plan‐ Neofluar” 25×/0.8 Imm Korr DIC objective (Zeiss) at a rate of 2.7 frames per second for up to 80 min following sporozoite transmission.
For bite transmission experiments, infected Anopheles stephensi were selected under an epifluorescence stereomicroscope 14–15 days after the infectious blood meal and used between day 18 to 21 post‐infection. To enhance the bite rate, mosquitoes were deprived of sucrose one day before the experiment. For intravital imaging of syringe‐inoculated sporozoites, infected salivary glands were harvested in 1× DPBS 13–21 days following the infectious blood meal and were kept intact on ice. Shortly before the experiment, they were crushed, filtered on a 35‐µm strainer and further diluted in 1× DPBS. All experiments were performed using either concavin KO sporozoites or a control P. berghei ANKA strain expressing GFP under the control of the hsp70 promoter (Ishino et al, 2006).
Prior to imaging, 4‐ to 6‐week old female C57BL/6JRj mice (Janvier Labs) were injected intravenously into the tail vein with 10–15 µg of Alexa Fluor™ 647‐coupled anti‐CD31 antibody (clone 390, Biolegend) to label blood vessels. Animals were then anesthetized with a mixture of ketamine (125 mg/kg body weight, Imalgene 1000, Merial) and xylazine (12.5 mg/kg body weight, Rompun 2%, Bayer), and their ear pinnae were gently epilated with a piece of tape. The dorsal side of their ears was then either exposed to mosquito bites for 2 min or injected with 0.2 µl of sporozoite suspension using a micro syringe (NanoFil 10 μl syringe mounted with a 35G beveled needle, World Precision Instruments), yielding fields of view containing in average 100 parasites. Animals were then immediately transferred onto the microscope stage to localize and image the inoculation sites. During acquisition, mice were kept warm with a heating blanket (Harvard Apparatus) and their anaesthesia status was regularly monitored (Amino et al, 2007).
Image files were processed and quantified using Fiji (Schindelin et al, 2012). Parasite morphology in the skin was determined on the first images obtained immediately after the localization of the bite site. Sporozoite movements were manually tracked over 2‐min movies recorded at the indicated time points after inoculation and mean velocity was determined using the MTrackJ plug‐in (Meijering et al, 2012). Sporozoites whose speed was inferior to 0.25 µm/s were considered immotile. Blood vessel invasion events were quantified over the first hour following sporozoite micro‐injection and normalized to the number of motile sporozoites in the field at 10 min post‐infection. To quantify the frequency of parasite disintegration after bite transmission, only sporozoites which could be tracked for at least 10 min were taken into consideration. Following loss of parasite integrity, gradual disappearance of fluorescence was considered a hallmark of sporozoite death.
Toxoplasma gondii parasite culture
Parasites were cultured in human foreskin fibroblasts (HFFs) using Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum, 2 mM l‐glutamine and 10 mg/ml gentamycin. Cell culture was maintained at 37°C and 5% CO2.
Toxoplasma gondii parasite strain generation
Parasites with TGGT1_216650 endogenously tagged and floxed
Endogenous tagging was done as described in (Stortz et al, 2019). Briefly, RHΔku80DiCre tachyzoites (created by Dr Moritz Treeck (Hunt et al, 2019) were used for endogenous tagging. Guide RNAs to target both the C‐terminal region, and upstream of the predicted 5′UTR of TGGT1_216650 were designed. These guides were cloned into a plasmid which expressed Cas9‐YFP. The 5′loxP was introduced into the parasites as an oligo flanked by 33 bp of sequences homologous to the region that the Cas9 targeted. The repair templates used for the introduction of the C‐terminal tags (YFP and SNAP) were amplified by PCR (Q5 polymerase, New England Biolabs) from template plasmids. The primers used for the amplification of these tags were designed in such a way that an LIC sequence was used as a linker between the protein and the tags, whereas a loxP sequence was introduced downstream of the tags. Homology sequences 50 bp long were added on either side of the tags to facilitate homologous recombination. The 5′loxP and 3′ tags were introduced into the parasites in two separate successive transfections. 10 µg of vector (encoding the guide RNA and Cas9‐YFP) and the respective repair templates were transfected into 1 × 107 newly egressed parasites, using 4D AMAXA electroporation. Following transfection, the parasites were allowed to invade and replicate for approximately 40 h, at which point they were mechanically egressed, filtered and sorted for transient Cas9‐YFP expression using a cell sorter (FACSAria IIIu, BD Biosciences) into 96‐well plates. Successfully genetically modified parasite clones were identified by IFA, PCR and sequencing (using primers designed to bind as indicated by the blue and red arrows in Appendix Fig S4.
Induction of TGGT1_216650 knock‐out
RHΔku80DiCre tachyzoites with a floxed TGGT1_216650 C‐terminally tagged with a SNAP‐tag were cultured in the presence of 50 nM rapamycin for 1 week and then cloned out by serial dilution in a 96‐well plate. Successful knockouts were identified by PCR and sequencing using primers designed to bind as indicated by the black arrows in Appendix Fig S4.
Toxoplasma gondii Plaque assay
5 × 102 parasites were used to inoculate 6‐well plates in the presence of dimethyl sulfoxide (DMSO) as vehicle control or 50 nM rapamycin. After 7 days of undisturbed incubation, the cells were fixed with 100% ice cold methanol for 20 min at room temperature, and washed with phosphate buffered saline. The cells were then left in eosin for 1 min, followed by 2 min in methylene blue and were finally washed thoroughly with water.
Toxoplasma gondii Immunofluorescence assays and microscopy
HFFs were inoculated with parasites for 24 h, after which they were either imaged live or fixed with 4% paraformaldehyde (PFA) for 20 min at room temperature. For live imaging, the dyes (SNAP‐Cell 647‐SiR, and HALO‐tag Oregon Green) were used as per the manufacturer’s instructions. For fixed samples, the cells were washed three times with phosphate buffered saline (PBS) following fixation, and then permeabilized and blocked for 45 min at room temperature with 3% bovine serum albumin (BSA), 0.2% Triton X‐100 in PBS. The cells were then labelled with the following primary antibodies for 1 h at room temperature; mouse anti‐GFP (1:500, monoclonal Anti‐GFP, Roche), rabbit anti‐GFP (1:1,000, polyclonal Anti‐GFP, abcam), rabbit anti‐GAP45 (1:1,000, a generous gift from Dominique Soldati Favre; (Plattner et al, 2008)), mouse anti‐IMC1 (1:2,000, a generous gift from Gary Ward; (Tilley et al, 2014)). After incubating with primary antibodies, the cells were washed three times with PBS and then labelled with the following secondary antibodies for 1 h at room temperature in the dark; STAR 635P anti‐rabbit (Abberior), STAR 635P anti‐mouse (Abberior), Alexa Fluor488 anti‐mouse (Life Technologies), and Alexa Fluor Plus 488 anti‐rabbit (Invitrogen). Following the incubation with the secondary antibodies, the cells were incubated with 0.4 µM Hoechst for 5 min, and were finally washed three times with PBS and mounted with ProLongTM Gold antifade mounting medium (ThermoFisher Scientific). Widefield images were taken using a 100× objective on a Leica DMi8 widefield microscope attached to a Leica DFC9000 GTC camera. Z‐stacks were taken and then processed with ImageJ.
Ethics statement
All animal experiments were performed according to European guidelines and regulations and the German Animal Welfare Act (Tierschutzgesetz) and executed following the guidelines of the Society of Laboratory Animal Science (GV‐SOLAS) and of the Federation of European Laboratory Animal Science Associations (FELASA). All experiments were approved by the responsible German authorities (Regierungspräsidium Karlsruhe) or Animal Care and Use ommittee of Institut Pasteur (CETEA 2013‐0093) and the French Ministry of Higher Education and Research (MESR 01324).
Animal work
For all experiments, female 4–6‐week‐old Naval Medical Research Institute (NMRI) mice, Swiss or C57BL/6 mice obtained from Charles River or Janvier laboratories were used. Transgenic parasites were generated in the Plasmodium berghei ANKA background (Vincke & Bafort, 1968) either directly in wild‐type or from wild‐type derived. Parasites were cultivated in NMRI or Swiss CD1 mice while transmission experiments with sporozoites were performed in C57Bl/6 mice only. Animal experiments conducted at the Institut Pasteur were approved by the Animal Care and Use Committee of Institut Pasteur (CETEA 2013‐0093) and the French Ministry of Higher Education and Research (MESR 01324) and were performed in accordance with European guidelines and regulations.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8.0 (GraphPad, San Diego, CA, USA). Data sets were either tested with a Mann–Whitney, Fischers Exact or Kruskal–Wallis test. A value of P < 0.05 was considered significant. No blinding of experiments has been done.
Author contributions
Jessica Kehrer: Conceptualization; Data curation; Formal analysis; Investigation; Visualization; Methodology; Writing—original draft; Writing—review & editing. Pauline Formaglio: Formal analysis; Validation; Investigation; Visualization; Methodology; Writing—review & editing. Julianne Mendi Muthinja: Investigation; Visualization. Sebastian Weber: Investigation. Danny Baltissen: Investigation. Christopher Lance: Software; Investigation. Johanna Ripp: Investigation; Methodology. Janessa Grech: Investigation; Visualization. Markus Meissner: Conceptualization; Supervision; Funding acquisition; Project administration; Writing—review & editing. Charlotta Funaya: Conceptualization; Supervision; Funding acquisition; Investigation; Methodology; Project administration. Rogerio Amino: Conceptualization; Supervision; Funding acquisition; Visualization; Project administration; Writing—review & editing. Friedrich Frischknecht: Conceptualization; Supervision; Funding acquisition; Investigation; Writing—original draft; Project administration; Writing—review & editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
JK and FF designed the project; JK, PF. CF, SW, JMM, DB, CL, JR, JG and FF performed research; all authors analysed data; MM and RA supervised the Toxoplasma and in vivo imaging parts, respectively. JK and FF wrote the paper with input from all authors.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
We thank members of the Frischknecht and Ingham Lab and the team of the Centre for the Production and Infection of Anopheles (CEPIA, Institut Pasteur) for rearing Anopheles stephensi/ gambiae mosquitoes as well as Markus Ganter for helpful discussions and comments on the manuscript. We acknowledge the microscopy support from the Infectious Diseases Imaging Platform (IDIP) at the Center for Integrative Infectious Disease Research.
This work was funded by grants from the Human Frontier Science Program (RGY0071/2011), the Institut Pasteur, the Agence National de la Recherche (ANR, French National Research Agency) / Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—project number SporoSTOP ANR‐19‐CE15‐0027, the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases”—project number ANR‐10‐LABX‐62‐IBEID, the DFG project number 240245660—SFB 1129—and the European Research Council (ERC StG 281719). FF is a member of CellNetworks Cluster of excellence at Heidelberg University and SFB 1129. Open Access funding enabled and organized by Projekt DEAL.
EMBO reports (2022) 23: e54719.
Data availability
No large primary datasets have been generated and deposited. All new vectors and parasite lines are available from the corresponding author upon request.
References
- Aleshnick M, Ganusov VV, Nasir G, Yenokyan G, Sinnis P (2020) Experimental determination of the force of malaria infection reveals a non‐linear relationship to mosquito sporozoite loads. PLoS Pathog 16: e1008181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliprandini E, Tavares J, Panatieri RH, Thiberge S, Yamamoto MM, Silvie O, Ishino T, Yuda M, Dartevelle S, Traincard F et al (2018) Cytotoxic anti‐circumsporozoite antibodies target malaria sporozoites in the host skin. Nat Microbiol 3: 1224–1233 [DOI] [PubMed] [Google Scholar]
- Amino R, Giovannini D, Thiberge S, Gueirard P, Boisson B, Dubremetz JF, Prévost MC, Ishino T, Yuda M, Ménard R (2008) Host cell traversal is important for progression of the malaria parasite through the dermis to the liver. Cell Host Microbe 3: 88–96 [DOI] [PubMed] [Google Scholar]
- Amino R, Thiberge S, Martin B, Celli S, Shorte S, Frischknecht F, Ménard R (2006) Quantitative imaging of Plasmodium transmission from mosquito to mammal. Nat Med 12: 220–224 [DOI] [PubMed] [Google Scholar]
- Amino R, Thiberge S, Blazquez S, Baldacci P, Renaud O, Shorte S, Ménard R (2007) Imaging malaria sporozoites in the dermis of the mammalian host. Nat Protoc 2: 1705–1712 [DOI] [PubMed] [Google Scholar]
- Araki T, Kawai S, Kakuta S, Kobayashi H, Umeki Y, Saito‐Nakano Y, Sasaki T, Nagamune K, Yasutomi Y, Nozaki T et al (2020) Three‐dimensional electron microscopy analysis reveals endopolygeny‐like nuclear architecture segregation in Plasmodium oocyst development. Parasitol Int 76: 102034 [DOI] [PubMed] [Google Scholar]
- Bane KS, Lepper S, Kehrer J, Sattler JM, Singer M, Reinig M, Klug D, Heiss K, Baum J, Mueller A‐K et al (2016) The actin filament‐binding protein coronin regulates motility in Plasmodium sporozoites. PLoS Pathog 12: e1005710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhanot P, Frevert U, Nussenzweig V, Persson C (2003) Defective sorting of the thrombospondin‐related anonymous protein (TRAP) inhibits Plasmodium infectivity. Mol Biochem Parasitol 126: 263–273 [DOI] [PubMed] [Google Scholar]
- Braks JAM, Franke‐Fayard B, Kroeze H, Janse CJ, Waters AP (2006) Development and application of a positive ‐ gative selectable marker system for use in reverse genetics in Plasmodium . Nucleic Acids Res 34: e39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey AF, Singer M, Bargieri D, Thiberge S, Frischknecht F, Ménard R, Amino R (2014) Calcium dynamics of Plasmodium berghei sporozoite motility. Cell Microbiol 16: 768–783 [DOI] [PubMed] [Google Scholar]
- Clemens J, Moorthy V (2016) Implementation of RTS, S/AS01 malaria vaccine — the need for further evidence. N Engl J Med 374: 2596–2597 [DOI] [PubMed] [Google Scholar]
- Coppi A, Natarajan R, Pradel G, Bennett BL, James ER, Roggero MA, Corradin G, Persson C, Tewari R, Sinnis P (2011) The malaria circumsporozoite protein has two functional domains, each with distinct roles as sporozoites journey from mosquito to mammalian host. J Exp Med 208: 341–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman AF, Healer J, Marapana D, Marsh K (2016) Malaria: biology and disease. Cell 167: 610–624 [DOI] [PubMed] [Google Scholar]
- Douglas RG, Reinig M, Neale M, Frischknecht F (2018) Screening for potential prophylactics targeting sporozoite motility through the skin. Malar J 17: 319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egarter S, Andenmatten N, Jackson AJ, Whitelaw JA, Pall G, Black JA, Ferguson DJP, Tardieux I, Mogilner A, Meissner M (2014) The toxoplasma acto‐MyoA motor complex is important but not essential for gliding motility and host cell invasion. PLoS One 9: e91819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores‐Garcia Y, Nasir G, Hopp CS, Munoz C, Balaban AE, Zavala F, Sinnis P (2018) Antibody‐mediated protection against Plasmodium sporozoites begins at the dermal inoculation site. mBio 9: e02194–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formaglio P, Tavares J, Ménard R, Amino R (2014) Loss of host cell plasma membrane integrity following cell traversal by Plasmodium sporozoites in the skin. Parasitol Int 63: 237–244 [DOI] [PubMed] [Google Scholar]
- Frénal K, Polonais V, Marq JB, Stratmann R, Limenitakis J, Soldati‐Favre D (2010) Functional dissection of the apicomplexan glideosome molecular architecture. Cell Host Microbe 8: 343–357 [DOI] [PubMed] [Google Scholar]
- Frischknecht F, Baldacci P, Martin B, Zimmer C, Thiberge S, Olivo‐Marin JC, Shorte SL, Ménard R (2004) Imaging movement of malaria parasites during transmission by Anopheles mosquitoes. Cell Microbiol 6: 687–694 [DOI] [PubMed] [Google Scholar]
- Gomes‐Santos CSS, Braks J, Prudêncio M, Carret C, Gomes AR, Pain A, Feltwell T, Khan S, Waters A, Janse C et al (2011) Transition of Plasmodium sporozoites into liver stage‐like forms is regulated by the RNA binding protein Pumilio. PLoS Pathog 7: e1002046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Tham WH, Cowman AF, McFadden GI, Waller RF (2008) Alveolins, a new family of cortical proteins that define the protist infrakingdom Alveolata. Mol Biol Evol 25: 1219–1230 [DOI] [PubMed] [Google Scholar]
- Harding CR, Frischknecht F (2020) The riveting cellular structures of apicomplexan parasites. Trends Parasitol 36: 979–991 [DOI] [PubMed] [Google Scholar]
- Hopp CS, Balaban AE, Bushell ES, Billker O, Rayner JC, Sinnis P (2016) Palmitoyl transferases have critical roles in the development of mosquito and liver stages of Plasmodium. Cell Microbiol 18: 1625–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopp CS, Kanatani S, Archer NK, Miller RJ, Liu H, Chiou KK, Miller LS, Sinnis P (2021) Comparative intravital imaging of human and rodent malaria sporozoites reveals the skin is not a species‐specific barrier. EMBO Mol Med 13: e11796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstmann H, Körber C, Sätzler K, Aydin D, Kuner T (2012) Serial section scanning electron microscopy (S3EM) on silicon wafers for ultra‐structural volume imaging of cells and tissues. PLoS One 7: e35172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A, Russell MRG, Wagener J, Kent R, Carmeille R, Peddie CJ, Collinson L, Heaslip A, Ward GE, Treeck M (2019) Differential requirements for cyclase‐associated protein (CAP) in actin‐dependent processes of Toxoplasma gondii . Elife 8: e50598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino T, Murata E, Tokunaga N, Baba M, Tachibana M, Thongkukiatkul A, Tsuboi T, Torii M (2019) Rhoptry neck protein 2 expressed in Plasmodium sporozoites plays a crucial role during invasion of mosquito salivary glands. Cell Microbiol 21: e12964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino T, Orito Y, Chinzei Y, Yuda M (2006) A calcium‐dependent protein kinase regulates Plasmodium ookinete access to the midgut epithelial cell. Mol Microbiol 59: 1175–1184 [DOI] [PubMed] [Google Scholar]
- Janse CJ, Franke‐Fayard B, Mair GR, Ramesar J, Thiel C, Engelmann S, Matuschewski K, Van GGJ, Sauerwein RW, Waters AP (2006) High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol Biochem Parasitol 145: 60–70 [DOI] [PubMed] [Google Scholar]
- Jayabalasingham B, Bano N, Coppens I (2010) Metamorphosis of the malaria parasite in the liver is associated with organelle clearance. Cell Res 20: 1043–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimah JR, Salinas ND, Sala‐Rabanal M, Jones NG, David Sibley L, Nichols CG, Schlesinger PH, Tolia NH (2016) Malaria parasite CelTOS targets the inner leaflet of cell membranes for pore‐ dependent disruption. Elife 5: e20621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser G, De Niz M, Zuber B, Burda PC, Kornmann B, Heussler VT, Stanway RR (2016) High resolution microscopy reveals an unusual architecture of the Plasmodium berghei endoplasmic reticulum. Mol Microbiol 102: 775–791 [DOI] [PubMed] [Google Scholar]
- Kariu T, Ishino T, Yano K, Chinzei Y, Yuda M (2006) CelTOS, a novel malarial protein that mediates transmission to mosquito and vertebrate hosts. Mol Microbiol 59: 1369–1379 [DOI] [PubMed] [Google Scholar]
- Khater EI, Sinden RE, Dessens JT (2004) A malaria membrane skeletal protein is essential for normal morphogenesis, motility, and infectivity of sporozoites. J Cell Biol 167: 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug D, Frischknecht F (2017) Motility precedes egress of malaria parasites from oocysts. Elife 6: e19157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three‐dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Kudryashev M, Münter S, Lemgruber L, Montagna G, Stahlberg H, Matuschewski K, Meissner M, Cyrklaff M, Frischknecht F (2012) Structural basis for chirality and directional motility of Plasmodium sporozoites. Cell Microbiol 14: 1757–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner SE, Mikolajczak SA, Vaughan AM, Moon W, Joyce BR, Sullivan WJ, Kappe SHI (2013a) Perturbations of Plasmodium Puf2 expression and RNA‐seq of Puf2‐deficient sporozoites reveal a critical role in maintaining RNA homeostasis and parasite transmissibility. Cell Microbiol 15: 1266–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner SE, Swearingen KE, Harupa A, Vaughan AM, Sinnis P, Moritz RL, Kappe SHI (2013b) Total and putative surface proteomics of malaria parasite salivary gland sporozoites. Mol Cell Proteomics 12: 1127–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner SE, Swearingen KE, Shears MJ, Walker MP, Vrana EN, Hart KJ, Minns AM, Sinnis P, Moritz RL, Kappe SHI (2019) Transcriptomics and proteomics reveal two waves of translational repression during the maturation of malaria parasite sporozoites. Nat Commun 10: 4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuschewski K (2017) Vaccines against malaria—still a long way to go. FEBS J 284: 2560–2568 [DOI] [PubMed] [Google Scholar]
- Matuschewski K, Ross J, Brown SM, Kaiser K, Nussenzweig V, Kappe SHI (2002) Infectivity‐associated changes in the transcriptional repertoire of the malaria parasite sporozoite stage. J Biol Chem 277: 41948–41955 [DOI] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I (2012) Methods for cell and particle tracking. Methods Enzymol 504: 183–200 [DOI] [PubMed] [Google Scholar]
- Ménard R, Sultan AA, Cortes C, Altszuler R, Van Dijk MR, Janse CJ, Waters AP, Nussenzweig RS, Nussenzweig V (1997) Circumsporozoite protein is required for development of malaria sporozoites in mosquitoes. Nature 385: 336–340 [DOI] [PubMed] [Google Scholar]
- Montagna GN, Buscaglia CA, Münter S, Goosmann C, Frischknecht F, Brinkmann V, Matuschewski K (2012) Critical role for heat shock protein 20 (HSP20) in migration of malarial sporozoites. J Biol Chem 287: 2410–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon RW, Taylor CJ, Bex C, Schepers R, Goulding D, Janse CJ, Waters AP, Baker DA, Billker O (2009) A cyclic GMP signalling module that regulates gliding motility in a malaria parasite. PLoS Pathog 5: e1000599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan R, Scally SW, Costa G, Mustafa G, Thai E, Decker T, Bosch A, Prieto K, Levashina EA, Julien J‐P et al (2020) Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs. Nat Med 26: 1135–1145 [DOI] [PubMed] [Google Scholar]
- Muthinja MJ, Ripp J, Hellmann JK, Haraszti T, Dahan N, Lemgruber L, Battista A, Schütz L, Fackler OT, Schwarz US et al (2017) Microstructured blood vessel surrogates reveal structural tropism of motile malaria parasites. Adv Healthc Mater 6: e1601178 [DOI] [PubMed] [Google Scholar]
- Paton DG, Childs LM, Itoe MA, Holmdahl IE, Buckee CO, Catteruccia F (2019) Exposing Anopheles mosquitoes to antimalarial blocks Plasmodium parasite transmission. Nature 567: 2239–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham RJ, Wang YL (1997) Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA 94: 13661–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirahmadi S, Zakeri S, Mehrizi AA, Karimi L, Djadid ND (2019) Heterogeneity in the acquisition of naturally acquired antibodies to cell‐traversal protein for ookinetes and sporozoites (CelTOS) and thrombospondin‐related adhesion protein (TRAP) of Plasmodium falciparum in naturally infected patients from unstable mal. Acta Trop 190: 365–374 [DOI] [PubMed] [Google Scholar]
- Plattner F, Yarovinsky F, Romero S, Didry D, Carlier M‐F, Sher A, Soldati‐Favre D (2008) Toxoplasma profilin is essential for host cell invasion and TLR11‐dependent induction of an interleukin‐12 response. Cell Host Microbe 3: 77–87 [DOI] [PubMed] [Google Scholar]
- Prudêncio M, Rodriguez A, Mota MM (2006) The silent path to thousands of merozoites: the Plasmodium liver stage. Nat Rev Microbiol 4: 849–856 [DOI] [PubMed] [Google Scholar]
- Ripp J, Kehrer J, Smyrnakou X, Tisch N, Tavares J, Amino R, Ruiz de Almodovar C, Frischknecht F (2021) Malaria parasites differentially sense environmental elasticity during transmission. EMBO Mol Med 13: e13933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco‐Castillo V, Topçu S, Marinach C, Manzoni G, Bigorgne AE, Briquet S, Baudin X, Lebrun M, Dubremetz JF, Silvie O (2015) Malaria sporozoites traverse host cells within transient vacuoles. Cell Host Microbe 18: 593–603 [DOI] [PubMed] [Google Scholar]
- Rompikuntal PK, Kent RS, Foe IT, Deng B, Bogyo M, Ward GE (2021) Blocking palmitoylation of Toxoplasma gondii myosin light chain 1 disrupts glideosome composition but has little impact on parasite motility. mSphere 6: 300823–300920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini E, Zeeshan M, Brady D, Pandey R, Kaiser G, Koreny L, Kumar P, Thakur V, Tatiya S, Katris NJ et al (2017) Photosensitized INA‐Labelled protein 1 (PhIL1) is novel component of the inner membrane complex and is required for Plasmodium parasite development. Sci Rep 7: 15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Egarter S, Zuzarte‐Luís V, Kumar H, Moreau CA, Kehrer J, Pinto A, da Costa M, Franke‐Fayard B, Janse CJ et al (2017) Malaria parasite LIMP protein regulates sporozoite gliding motility and infectivity in mosquito and mammalian hosts. Elife 6: e24109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Kehrer J, Franke‐Fayard B, Frischknecht F, Janse CJ, Mair GR (2015) The Plasmodium palmitoyl‐S‐acyl‐transferase DHHC2 is essential for ookinete morphogenesis and malaria transmission. Sci Rep 5: 16034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlott AC, Knuepfer E, Green JL, Hobson P, Borg AJ, Morales‐Sanfrutos J, Perrin AJ, Maclachlan C, Collinson LM, Snijders AP et al (2021) Inhibition of protein N‐myristoylation blocks Plasmodium falciparum intraerythrocytic development, egress and invasion. PLoS Biol 19: e3001408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvie O, Franetich J‐F, Charrin S, Mueller MS, Siau A, Bodescot M, Rubinstein E, Hannoun L, Charoenvit Y, Kocken CH et al (2004) A role for apical membrane antigen 1 during invasion of hepatocytes by Plasmodium falciparum sporozoites. J Biol Chem 279: 9490–9496 [DOI] [PubMed] [Google Scholar]
- Singer M, Frischknecht F (2021) Fluorescent tagging of Plasmodium circumsporozoite protein allows imaging of sporozoite formation but blocks egress from oocysts. Cell Microbiol 23: e13321 [DOI] [PubMed] [Google Scholar]
- Spreng B, Fleckenstein H, Kübler P, Di Biagio C, Benz M, Patra P, Schwarz US, Cyrklaff M, Frischknecht F (2019) Microtubule number and length determine cellular shape and function in Plasmodium . EMBO J 38: e100984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel RWJ, Pei Y, Camargo N, Kaushansky A, Dankwa DA, Martinson T, Nguyen T, Betz W, Cardamone H, Vigdorovich V et al (2018) Plasmodium yoelii S4/CelTOS is important for sporozoite gliding motility and cell traversal. Cell Microbiol 20: e12817 [DOI] [PubMed] [Google Scholar]
- Stortz JF, Del Rosario M, Singer M, Wilkes JM, Meissner M, Das S (2019) Formin‐2 drives polymerisation of actin filaments enabling segregation of apicoplasts and cytokinesis in Plasmodium falciparum . Elife 8: e49030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan AA, Thathy V, Frevert U, Robson KJH, Crisanti A, Nussenzweig V, Nussenzweig RS, Ménard R (1997) TRAP is necessary for gliding motility and infectivity of Plasmodium sporozoites. Cell 90: 511–522 [DOI] [PubMed] [Google Scholar]
- Swearingen KE, Lindner SE, Shi L, Shears MJ, Harupa A, Hopp CS, Vaughan AM, Springer TA, Moritz RL, Kappe SHI et al (2016) Interrogating the Plasmodium sporozoite surface: identification of surface‐exposed proteins and demonstration of glycosylation on CSP and TRAP by mass spectrometry‐based proteomics. PLoS Pathog 12: e1005606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares J, Formaglio P, Thiberge S, Mordelet E, Van Rooijen N, Medvinsky A, Ménard R, Amino R (2013) Role of host cell traversal by the malaria sporozoite during liver infection. J Exp Med 210: 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thathy V, Fujioka H, Gantt S, Nussenzweig R, Nussenzweig V, Ménard R (2002) Levels of circumsporozoite protein in the Plasmodium oocyst determine sporozoite morphology. EMBO J 21: 1586–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley LD, Krishnamurthy S, Westwood NJ, Ward GE (2014) Identification of TgCBAP, a novel cytoskeletal protein that localizes to three distinct subcompartments of the Toxoplasma gondii pellicle. PLoS One 9: e98492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiono AB, Nébié I, Anagnostou N, Coulibaly AS, Bowyer G, Lam E, Bougouma EC, Ouedraogo A, Yaro JBB, Barry A et al (2018) First field efficacy trial of the ChAd63 MVA ME‐TRAP vectored malaria vaccine candidate in 5–17 months old infants and children. PLoS One 13: e0208328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremp AZ, Carter V, Saeed S, Dessens JT (2013) Morphogenesis of Plasmodium zoites is uncoupled from tensile strength. Mol Microbiol 89: 552–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukegbu CV, Christophides GK, Vlachou D (2021) Identification of three novel Plasmodium factors involved in ookinete to oocyst developmental transition. Front Cell Infect Microbiol 11: 141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderberg JP, Frevert U (2004) Intravital microscopy demonstrating antibody‐mediated immobilisation of Plasmodium berghei sporozoites injected into skin by mosquitoes. Int J Parasitol 34: 991–996 [DOI] [PubMed] [Google Scholar]
- Vaughan A (2021) Motile mosquito stage malaria parasites: ready for their close‐up. EMBO Mol Med 13: e13975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincke L, Bafort J (1968) Results of 2 years of observation of the cyclical transmission of Plasmodium berghei . Ann Soc Belges Med Trop Parasitol Mycol 48: 439–454 [PubMed] [Google Scholar]
- Volkmann K, Pfander C, Burstroem C, Ahras M, Goulding D, Rayner JC, Frischknecht F, Billker O, Brochet M (2012) The alveolin IMC1h is required for normal ookinete and sporozoite motility behaviour and host colonisation in Plasmodium berghei . PLoS One 7: e41409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Fujioka H, Nussenzweig V (2005) Exit of plasmodium sporozoites from oocysts is an active process that involves the circumsporozoite protein. PLoS Pathog 1: e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2020) WHO World Malaria Report 2020
- Yoshida N, Nussenzweig RS, Potocnjak P, Nussenzweig V, Aikawa M (1980) Hybridoma produces protective antibodies directed against the sporozoite stage of malaria parasite. Science 207: 71–73 [DOI] [PubMed] [Google Scholar]


