Abstract
Pathway substrates and some structural analogues directly activate the regulatory protein DmpR to promote transcription of the dmp operon genes encoding the (methyl)phenol degradative pathway of Pseudomonas sp. strain CF600. While a wide range of phenols can activate DmpR, the location and nature of substituents on the basic phenolic ring can limit the level of activation and thus utilization of some compounds as assessed by growth on plates. Here we address the role of the aromatic effector response of DmpR in determining degradative properties in two soil matrices that provide different nutritional conditions. Using the wild-type system and an isogenic counterpart containing a DmpR mutant with enhanced ability to respond to para-substituted phenols, we demonstrate (i) that the enhanced in vitro biodegradative capacity of the regulator mutant strain is manifested in the two different soil types and (ii) that exposure of the wild-type strain to 4-methylphenol-contaminated soil led to rapid selection of a subpopulation exhibiting enhanced capacities to degrade the compound. Genetic and functional analyses of 10 of these derivatives demonstrated that all harbored a single mutation in the sensory domain of DmpR that mediated the phenotype in each case. These findings establish a dominating role for the aromatic effector response of DmpR in determining degradation properties. Moreover, the results indicate that the ability to rapidly adapt regulator properties to different profiles of polluting compounds may underlie the evolutionary success of DmpR-like regulators in controlling aromatic catabolic pathways.
Naturally occurring soil and water microorganism have an enormous metabolic versatility towards aromatic compounds (2, 14, 45). However, many factors, including the distribution of desired catabolic traits, incomplete degradation or production of toxic dead-end products, biovailability, and inhibition of microbial activity by toxicity of a polluting compound, can lead to low efficiency of degradation by indigenous microflora. When degradation by indigenous microbial populations is unsatisfactory, bioaugmentation with more efficient exogenous organisms is often considered for bioremediation purposes (51). Generation of new or improved microbial catabolic activities can be achieved by many different strategies, including modulation of (i) components of the specific degradation pathway, (ii) properties that enhance bioavailability of the substrate, or (iii) survival and distribution traits of the bacteria in soils (9, 48, 49). The regulatory circuits that control the expression of degradative pathways lie at the top of the hierarchy of events that lead to efficient biodegradation of aromatic compounds, since they determine under what conditions and in response to what compound the catabolic enzymes are expressed. The regulatory circuits therefore provide a prime target for generating strains with enhanced degradative potential. Here we examine the role of the DmpR-mediated regulatory circuit in bacterial biodegradation in soil matrices using the (methyl)phenol-degrading strain Pseudomonas sp. strain CF600.
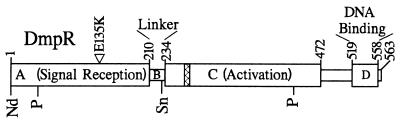
Pseudomonas sp. strain CF600 is a natural isolate harboring the self-transmissible IncP-2 catabolic megaplasmid pVI150 (40). The 15 genes encoding enzymes for the conversion of (methyl)phenols to pyruvate and acetyl coenzyme A are organized in the dmp operon of pVI150 (43). Expression of the dmp operon is tightly controlled by the divergently transcribed dmpR gene product (39, 41). The DmpR regulatory protein belongs to the ς54-dependent family of transcriptional activators, which have discrete domains involved in signal reception, transcriptional activation, and DNA binding (Fig. 1) (24). DmpR and XylR, a regulator of toluene and xylene catabolism (1, 18), typify a subgroup of this family that respond directly to the presence of aromatic effector molecules (reviewed in reference 38). Proteins homologous to DmpR and XylR are frequently found associated with catabolism of aromatic compounds and include AphR (4), BphR (32), HbpR (19), MopR (36), PheR (M. Takeo, unpublished data [GenBank accession no. D63814]), PhhR (27), PhlR Pseudomonas putida H (5), PhlR from Ralstonia eutropha JMP134 (25), PhnR (23), PoxR (17), TbuT (7), TmbR (12), and TouR (6).
FIG. 1.
Schematic representation of the domain structure of DmpR. The hatched box represents the extent of the NTP binding motif found in this class of regulators (consensus, G--G-GKE---A---H--S [24]). The position of the E135K mutation that confers an enhanced response to para-substituted methylphenols is indicated. The locations of restriction sites used in dmpR manipulations are shown relative to the domain structure (Nd, NdeI; P, PstI; Sn, SnaBI).
Substrates of the dmp pathway and some structural analogues serve as effector molecules that directly control the activity of DmpR (41, 42). The aromatic effectors are bound through a single common binding site on the signal reception A domain of DmpR (28, 29). Binding of the aromatic compound releases interdomain repression, resulting in depression of the transcriptional activation property of the protein with consequent initiation of transcription of dmp catabolic genes (26, 28, 29, 42). DmpR can be activated by a wide range of phenolics, however, the location and nature of substituents on the basic phenol molecule determine the level of activation, which can be limiting for utilization of some compounds (29, 31). Pseudomonas strain CF375 is a derivative of Pseudomonas strain CF600 harboring a single point mutation in the signal reception-encoding region of dmpR, resulting in a Glu-to-Lys replacement of residue 135 (DmpR-E135K). Due to modulation of the regulatory properties of DmpR, the resulting strain has a greatly enhanced ability to sense and degrade para-substituted methylphenols such as 4-methylphenol and 3,4-dimethylphenol under laboratory conditions while retaining wild-type ability to sense and degrade phenol and 2-methylphenol (31). These properties of CF375 suggest that regulatory mutants may serve as efficient bioaugmentation reagents. However, soils provide heterogeneous nutritional environments, and microbial degradation performance is dependent on varying physicochemical properties of a given soil (50, 51). Here we address whether the enhanced in vitro biodegradative efficiency of the CF375 regulatory mutant translates to increased biodegradative efficiency in two different soil matrices that provide different nutritional conditions. These experiments led to the isolation and analysis of spontaneous mutants that arose from 4-methylphenol-amended soils. The results from these studies provide evidence that CF600 naturally rapidly adapts its suboptimal ability to degrade 4-methylphenol in soil via enhancement of the ability of DmpR to respond to 4-methylphenol.
MATERIALS AND METHODS
General procedures.
Escherichia coli strains were grown at 37°C, and Pseudomonas strains were grown at 30°C. E. coli DH5 (16) was used for RSF1010-based plasmids, while the replication-permissive host S17-1λpir (11) was used for propagation of R6K-based suicide donor plasmids. Unless otherwise stated, Luria-Bertani medium was used as rich medium and M9 salts supplemented with the indicated carbon source was used as minimal medium (34). Kanamycin (50 μg ml−1), rifampin (100 μg ml−1), streptomycin (1 mg ml−1), and carbenicillin (100 μg ml−1 for E. coli strains and 1 mg ml−1 for Pseudomonas strains) were added to the media as required.
Strain construction.
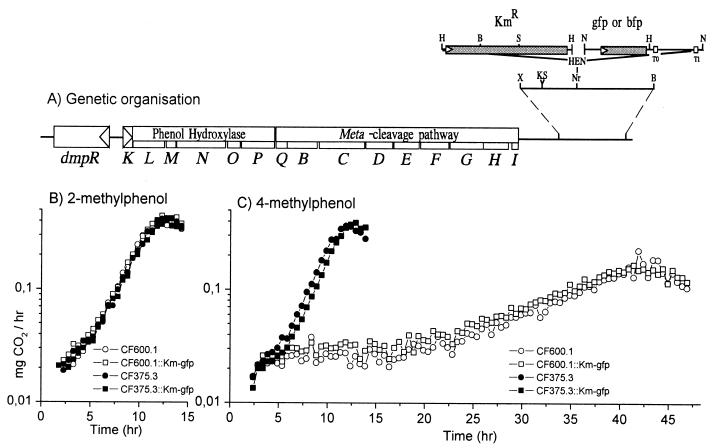
Spontaneous antibiotic-resistant derivatives of the (methyl)phenol-catabolizing prototrophic Pseudomonas sp. strain CF600 (40) and its isogenic counterpart CF375 (carrying the DmpR-E135K mutation) (31) were selected on rich medium supplemented with rifampin or streptomycin. The resident pVI150 plasmids of the resulting Rifr derivative CF600.1 and Smr derivative CF375.3 were then tagged by insertion of a kanamycin resistance gene and a green fluorescent protein (GFP) variant or blue fluorescent protein (BFP) gene. Insertion was made in each case at the NruI site located 2.2 kb downstream of the last gene of the dmp operon. This was achieved in a sequential process schematically illustrated in Fig. 2. First, a 2.125-kb XhoI-to-BglII fragment from downstream of the dmp operon was cloned between the XhoI and BamHI sites of pBluescript SK (Stategene) to generate pVI650. Second, a blunt-ended HindIII-EcoRV-NotI linker was inserted into the unique NruI site of this fragment, simultaneously destroying the NruI site, to give plasmid pVI651. A 2-kb HindIII fragment carrying a kanamycin resistance gene from mini-Tn5Km (10) was then inserted into the HindIII site of the linker to generate pVI652. A XhoI-to-EcoRI fragment, spanning the entire XhoI-to-BglII fragment with the Kmr insert, was then cloned between the SalI and EcoRI sites of the Cbr R6K-based suicide plasmid pGP704L (31), resulting in plasmid pVI653. NotI fragments encoding the GFP mutant 3 derivative from pJBA27 (3) or BFP (Novo Nordisk, Bagsvæed, Denmark) from an analogous plasmid (pJBA68) (J. B. Anderson, unpublished data) were then cloned into the NotI site of the linker, resulting in plasmids pVI654 (carrying a Km-GFP insert) and pVI655 (carrying a Km-BFP insert). These insertions were introduced into the resident pVI150 plasmid of Pseudomonas strains by recombination from the suicide plasmids following conjugation from E. coli S17-1λpir. Kmr was used to select for first-site recombination, and second-site recombinants were detected by screening for Cbs. The resulting strains were designated CF600.1::Km-gfp, CF600.1::Km-bfp, CF375.3::Km-gfp, and CF375.3::Km-bfp.
FIG. 2.
(A) Schematic representation of the genetic organization of dmp genes (open boxes) relative to the kanamycin resistance and fluorescent protein genes (closed boxes) in tagged pVI150 plasmids. Arrowheads indicate the direction of transcription. The different illustrated DNA manipulation steps leading to the final conformation of the DNA in the tagged strains are described in Materials and Methods. Restriction sites: B, BglII; E, EcoRV; H, HindIII; K, KpnI; N, NotI; Nr, NruI; S, SauI; X, XhoI. T0 and T1 indicate the locations of strong transcriptional terminators within the cloned NotI fragments (3). (B and C) Rates of CO2 production of the indicated strains upon growth on minimal medium supplemented with 2.5 mM 2-methylphenol (B) or 4-methylphenol (C) as the sole carbon and energy source. Values are the averages from duplicate experiments. Data for CF600.1::Km-bfp and CF375.3::Km-bfp were indistinguishable from those for their Km-gfp counterparts on both aromatic test compounds (data not shown).
DNA sequencing and plasmid generation.
Mutations in the A-domain-encoding DNA were initially identified by direct sequencing of PCR-generated fragments from target strains (from 178 bp upstream of the ATG start codon to codon 246 of dmpR) using custom-designed oligonucleotides. Cloning of the mutant A domains was performed using the plasmid pVI546, which is a Cbr broad-host-range RSF1010-based plasmid that expresses a DmpR derivative with an 8-amino-acid Flag epitope tag fused to the carboxy-terminal of the protein. In this plasmid, dmpR is expressed from its native promoter and has been manipulated to contain a silent NdeI site overlapping the ATG initiation codon and a silent SnaBI site overlapping codons 221 and 222 of dmpR to allow replacement of the entire A domain and regeneration of full-length DmpR regulators (Fig. 1) (44). Introduction of PCR fragments generated using primers that include these site and template DNAs from the different mutants resulted in plasmids pVI656 (DmpR-F42Y), pVI657 (DmpR-R109C), pVI658 (DmpR-L113V), pVI659 (DmpR-D116N), pVI660 (DmpR-F122L), pVI661 (DmpR-E135K), pVI662 (DmpR-C137Y), and pVI663 (DmpR-179(CG)180). The nucleotide sequences of all PCR-derived DNAs in these plasmids were confirmed. These plasmids thus encode mutants of DmpR expressed from the same cis elements as wild-type DmpR on plasmid pVI455 (42). To introduce the mutations into pVI150 of Pseudomonas strain, CF600, NotI fragments spanning the entire inserts of the above-described plasmids were cloned into the R6K-based Cbr suicide vector pGP704L-NotI (31). The resulting suicide plasmids, pVI664 (DmpR-F42Y), pVI665 (DmpR-R109C), pVI666 (DmpR-L113V), pVI667 (DmpR-D116N), pVI668 (DmpR-F122L), pVI669 (DmpR-C137Y), and pVI670 [DmpR-179(CG)180], were introduced from the permissive host E. coli S17-1λpir into a CF600 derivative (CF427 [31]) in which the internal PstI fragment had been replaced by a Kmr cassette (Fig. 1). The PstI fragment spans codons 40 to 422 of dmpR and thus all of the mutant codons present in the donor suicide plasmids. Recombinants were identified on the basis of the ability to restore growth on 2-methylphenol and sensitivity to kanamycin and carbenicillin
CO2 production measurements.
For determination of CO2 production upon growth with aromatic carbon sources as the sole carbon and energy sources, cultures were grown overnight in M9 medium supplemented with 2.5 mM test compound and 0.1% casein amino acids. Cells were then washed in the same medium lacking casein amino acids and diluted to an A650 of 0.2. Approximately 106 cells were plated on 50-mm-diameter M9 minimal plates containing the different aromatic compounds at 2.5 mM. The plates were then incubated in a Respicon III respirometer (Norgren Innovation AB, Umeå, Sweden), and CO2 production was monitored every 30 min for up to 48 h.
Soil properties and sterilization.
Physical and chemical properties of the soils were determined using standard procedures (Soil Analysis Service Ltd., Helsinki, Finland). The sandy soil water content was 9.1%, the organic carbon content was 1.6%, the total nitrogen content was 0.13%, and the pH was 6.3. The pine forest humus had a water content of 61.5, an organic carbon content of 22.4%, and a total nitrogen content of 0.75%. Its original pH was 3.9; however, since CF600 derivatives could not be established at this pH (data not shown), the pH was increased to 6.4 by liming with CaCO3 (12 mg g [fresh weight] of soil−1). The forest humus (before liming) and the sandy soil contained, respectively, 269 and 1,560 mg of calcium (Ca) liter−1, 4.1 and 31 mg of phosphorus (P) liter−1, 71.5 and 150 mg of potassium (K) liter−1, 38 and 107 mg of magnesium (Mg) liter−1, 11 and 50 mg of nitrate (NO3−) liter−1 and 21 and <2 mg of ammonia (NH4+) liter−1. Quartz sand (Sigma) had a pH of 7.0 when measured in sand-water suspension. Soil samples were sterilized in 0.3- to 1-liter aliquots by autoclaving (121°C, 20 min).
Soil inoculation and sampling.
Sterilized limed humus (1.5 g [fresh weight]) and sandy soil (2 g [fresh weight]) were dispensed in 14-ml polypropylene tubes and 20-ml glass scintillation vials. Quartz sand was dispensed in 2-g aliquots, and its water content was adjusted to 5.6%. The soils were amended with 595 μg of 2- or 4-methylphenol (Aldrich) made up as aqueous solutions prepared from 1 M stock solutions in dimethyl sulfoxide (DMSO) (Merck). Thus, test samples and controls received 5 μl of DMSO per 1.5 g (sandy soil) or 2 g (limed humus) (fresh weight) of soil. This level of DMSO did not affect the population numbers of controls (see Results). Supplements were added to soils 1 to 1.5 h prior to initiation of the experiment. Inverting and shaking before bacterial inoculation vigorously mixed soils and the supplements. Bacteria were pregrown overnight in semirich medium (1/4KSN-MYE [35]) in the presence of 2 mM 2-methylphenol. Cell pellets were resuspended in phosphate-buffered saline, and approximately 106 or 108 bacterial cells in 50 μl were inoculated into soils as indicated. The amended soil samples were subsequently kept at room temperature (20 to 22°C). After the additions, the final water contents were 63% for humic soil, 15% for sandy soil, and 11% for quartz sand.
The bacterial cell numbers were determined from three replicates for each time point. Nine milliliters of phosphate-buffered saline solution was added to each soil-containing tube and vortexed for 2 min. Appropriate dilutions were plated on 1/4KSN-MYE containing kanamycin or on minimal medium (KSN [53]) containing 2 mM 2- or 4-methylphenol as the sole carbon and energy source. In competition experiments, Luria-Bertani medium LB in the presence or absence of antibiotic selection was used to distinguish the two strains (Kmr and Rifr for CF600.1::Km-gfp and Kmr and Smr for CF375.3::Km-gfp). After growth on the selection plates, the presence of the appropriate GFP or BFP fluorescence was confirmed by visual inspection under UV light.
To determine the levels of methylphenols remaining in the soil samples at the different time points, extractions of triplicate samples were performed in glass scintillation vials. As internal standards, soil samples were amended 1 h prior to the extraction with an appropriate aromatic compound (2-methylphenol when determining 4-methylphenol levels and vice versa). Three milliliters of acetone amended with nitric acid was mixed with the samples for 1 h with rotation. Water was removed by adding dehydrated Na2SO4, and the samples were then filtered through nylon Acrodisc filters (0.45-μm pore size; Pall Gelman Sciences). Analysis was performed with a gas chromatograph (5890A; Hewlett-Packard) equipped with an HP-5MS column (30 m by 0.25 mm by 0.25 μm) using a flame ionization detector. The oven was programmed so that after 4 min the initial temperature of 45°C was increased to 180°C at 10°C/min and then to 250°C at 35°C/min.
Luciferase assays.
Plate test screening was performed as previously described (44). In brief, colonies of P. putida KT2440::Po-luxAB (31) harboring various plasmids were replica plated and grown overnight on Luria agar plates with antibiotic selection for the resident plasmid and supplemented with the indicated quantities of the different aromatics. Inverted plates were exposed to decanal vapor and the light emission was recorded by placing a film over the plates. For quantitative measurements, cells were grown and treated as previously described (46), and values were determined using a PhL microtiter plate luminometer (Mediators Diagnostika, Vienna, Austria).
Protein analysis.
Crude extracts of cytosolic proteins, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transfer to nitrocellulose filters, and Western blot analysis with polyclonal rabbit anti-DmpR sera were as previously described (42). Antibody-decorated bands were revealed using chemiluminescence reagents as directed by the supplier (Amersham Pharmacia Biotech). Differences in expression levels were assessed by comparison of dilution series of the test samples with those of the wild-type DmpR extract.
RESULTS
Growth of genetically tagged derivatives of Pseudomonas strain CF600.
To enable identification of Pseudomonas sp. strain CF600 and the dmpR-regulatory mutant strain CF375 from soil samples, differentially tagged derivatives were generated as described in Materials and Methods. The resulting antibiotic-resistant derivatives CF600.1 (Rifr) and CF375.3 (Smr) harbor pVI150 plasmids carrying either Km-GFP or Km-BFP insertions downstream of the dmp operon (Fig. 2A). To test that these derivatives retained their respective parental biodegradative properties, their ability to utilize 2-methylphenol and 4-methylphenol as sole carbon and energy sources was tested. 2-Methylphenol was chosen as the control aromatic compound since this is the best effector of DmpR and strains harboring either wild-type DmpR or DmpR-E135K respond and degrade this compound with equal efficiency. As shown in Fig. 2B, the genetically tagged strains also grow on 2-methylphenol equally well, with generation times like those found previously for the untagged derivatives (generation time of approximately 1.5 h [31]). Importantly, the tagged CF375.3 (DmpR-E135K mutant) strains retain the previously observed phenotype of enhanced degradation efficiency with 4-methylphenol, resulting in rates equivalent to those of the wild-type strain on 2-methylphenol. Hence, these results demonstrate that the genetic markers do not confer any detectable detrimental genetic load under these minimal growth conditions.
Propagation and biodegradation by CF600 derivatives in contaminated soils.
Since the ability of microbes to degrade aromatic compounds can be greatly influenced by abiotic factors determined by the soil composition, we chose to analyze bacterial degradation of target substrates in two different soil types. Limed forest humus (hereafter referred to as humic soil) represents a comparatively nutrient-rich soil, while the sandy soil is a nutrient-poor environment (see Materials and Methods). Preliminary studies showed that monomethylated phenols were transformed and/or degraded by indigenous microflora in these soils (data not shown). Therefore, in order to monitor removal of the two test compounds, 2- and 4-methylphenol, by the added bacteria, sterilized autoclaved soils were used.
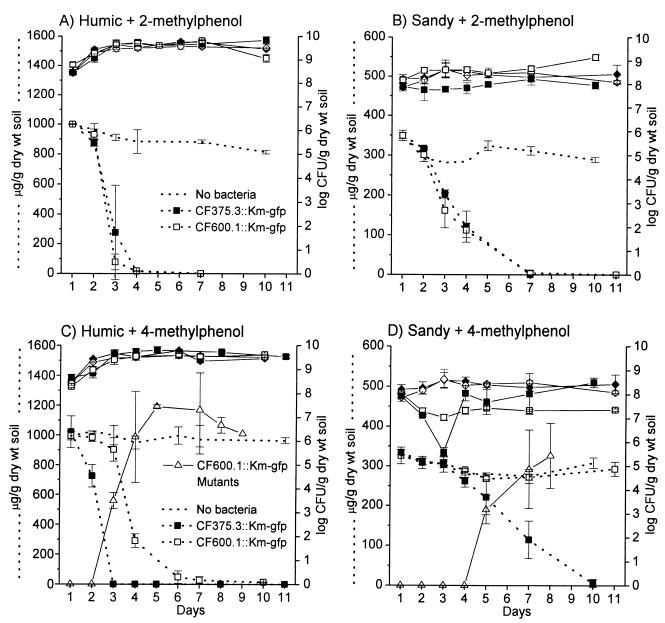
The strains CF600.1::Km-gfp and CF375.3::Km-gfp were added to both soil matrices with or without preamendment with the test aromatic compounds. The same quantity of aromatic compound was added to each soil type, which resulted in final concentrations of 1.0625 mg/g (dry weight) for humic soil and 0.3324 mg/g (dry weight) for sandy soil. In humic soil, the number of viable bacteria increased approximately 10-fold over the first 2 days of the experiment. This increase in biomass was not dependent on the added phenols (Fig. 3A and C) and, since no significant increase in viable cell numbers was observed in unamended quartz sand (data not shown), is attributable to the degradation of organic soil compounds. The same reason probably underlies the slight increase in biomass observed in the unamended sandy soil (Fig. 3B and D). In amended sandy soils, after an initial drop the viable counts of the introduced bacteria increased to approximately the initial inoculation density. In addition, CF375.3::Km-gfp was observed to be more sensitive than CF600.1::Km-gfp to the toxic effects of phenolics, notably 4-methylphenol (Fig. 3D), in the sandy soil. The difference in sensitivity to aromatic compounds in the two different soil types is probably due to the quenching effect of components of the higher organic content of humic soil (21, 30, 37) and/or to the higher concentration of the added aromatic compounds in the water phase of the sandy soil.
FIG. 3.
Degradation and survival in two soil types. Logarithms of viable colony-forming units of CF600.1::Km-gfp (open symbols) and CF375.3::Km-gfp (closed symbols) are shown with continuous lines. Counts were determined from both amended soils (squares) and non-amended controls (diamonds). The number of CFU of CF600.1::Km-gfp mutants detected by virtue of comparatively large colony formation on 4-methylphenol is shown with open triangles. Concentrations of 2-methylphenol (A and B) and 4-methylphenol (C and D) in the limed pine forest humic (A and C) and sandy (B and D) soils are shown with dotted lines. The concentrations of methylphenols in control soils without bacterial inoculation (dotted line, no symbols) are also shown. Values are the averages of triplicate determinations. Error bars indicate standard deviations.
The observed decreases in concentrations of the methylated phenols in soils are shown in Fig. 3 and are clearly attributed to the activity of inoculated bacteria rather than to some abiotic factors related to chemical or photochemical reactions (compare noninoculated and inoculated soil samples). As expected, CF600.1::Km-gfp and CF375.3 degraded 2-methylphenol with comparable efficiencies in both soils (Fig. 3A and B). Degradation of 2- and 4-methylphenol was more efficient in the humic soil than in sandy soil, consistent with the high bacterial population achieved in humic soil. Most importantly, the DmpR-E135K regulatory mutant CF375.3::Km-gfp showed a clearly enhanced ability to degrade 4-methylphenol compared with its wild-type counterpart CF600.1::Km-gfp in both soil types (Fig. 3C and D). These results show that the enhanced ability of the regulatory mutant to degrade 4-methylphenol on plates (Fig. 2) is also observed under the two different nutritional conditions provided by the two soil matrices.
Propagation of CF600.1::Km-gfp into soils amended with 4-methylphenol resulted in accumulation of a subpopulation which formed large colonies on minimal 4-methylphenol plates (Fig. 3C and D) and that also showed improved growth on 3,4-dimethylphenol. The rapid accumulation of this population in humic soil accompanied 4-methylphenol degradation. In sandy soil inoculated with CF600.1::Km-gfp, the maximal number of this subpopulation was 2 orders of magnitude less than that detected in humus (compare Fig. 3C and D), and no change in 4-methylphenol concentration was observed over the time course of the experiment. No CF600.1::Km-gfp derivatives exhibiting the phenotype of enhanced growth on 4-methylphenol or 3,4-dimethylphenol were observed in unamended soils or in soils amended with 2-methylphenol (Fig. 3A and B and data not shown). These results suggest that this subpopulation was directly selected in response to the presence of 4-methylphenol.
Competition between CF600.1::Km-gfp and CF375.3::Km-gfp strains in soils.
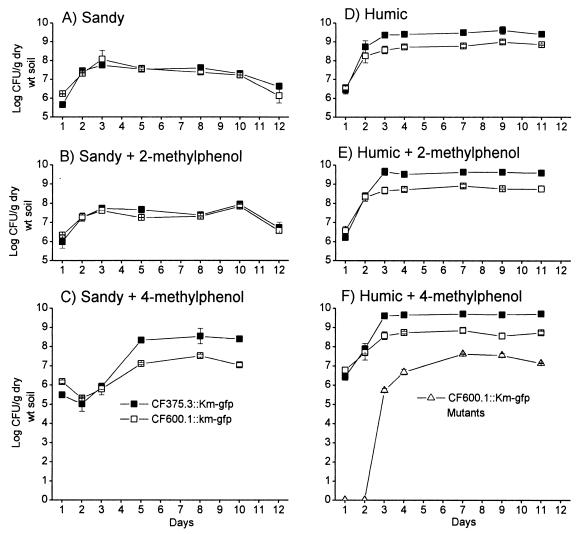
The specific DmpR-mediated regulatory circuit is subservient to global regulation in response to the availability of alternative carbon sources in the medium (46, 47), leading to various degrees of silencing depending on the nutrients supplied. Therefore, to test if the conditional growth advantage of the regulatory mutant strain leads to enhanced competitive growth in the two soil types, 106 cells of strains CF600.1::Km-gfp and CF375.3::Km-gfp were coinoculated into humic and sandy soils with or without methylphenol amendment. In the experiment shown in Fig. 4, the two strains were directly distinguished on the basis of their differential antibiotic resistance markers (Rifr and Smr). Growth of CF375.3::Km-gfp and CF600.1::Km-gfp in sandy soil with or without 2-methylphenol was indistinguishable (Fig. 4A and B). However, the DmpR-E135K regulatory mutant strain CF375.3::Km-gfp had a clear advantage over CF600.1::Km-gfp in the presence of 4-methylphenol. After an initial decrease, the cell numbers of CF375.3::Km-gfp rose to 10 times higher than those of CF600.1::Km-gfp (Fig. 4C). In humic soil, the DmpR-E135K mutant strain showed a growth advantage over the wild-type strain under all conditions studied, i.e., in the absence and presence of either 2-methylphenol or 4-methylphenol, and there was no significant difference in bacterial cell numbers between the treatments (Fig. 4D to F). This result may be attributable to the natural phenolic content of humic soil, which can contain up to 556 μg of water-soluble phenolic compounds per g, i.e., approximately 50% of the specific phenolic amendments used (15, 22). As seen previously, accumulation of CF600.1::Km-gfp derivatives with improved growth on para-substituted phenols was observed in humic soil amended with 4-methylphenol (Fig. 4F), again suggesting that the presence of 4-methylphenol selects for variants with an enhanced capacity to grow and/or survive in the presence of 4-methylphenol. Similar results were obtained in competition experiments between CF600.1::Km-bfp and CF375.3::Km-gfp in which bacteria were plated in the absence of selection or selected on the basis on their common antibiotic resistance marker (Kmr) and subsequently distinguished on the basis of their differential fluorescent protein tags (data not shown). The fact that the results obtained with the various strategies were similar demonstrates that the different antibiotic selections for the two strains in this and the preceding experiment has no significant effect on bacterial count determinations.
FIG. 4.
Competition between CF600.1::Km-gfp (open symbols) and CF375.3::Km-gfp (closed symbols) in sandy soil (left panels) and limed pine forest humus (humic soil) (right panels) with or without 2-methylphenol or 4-methylphenol as indicated. The number of CF600.1::Km-bfp mutants with enhanced growth on 4-methylphenol is shown with open triangles. Values are the averages of triplicate determinations.
CF600.1::Km-gfp derivatives with improved growth on para-substituted phenols.
Ten mutant CF600.1::Km-gfp derivatives isolated from 4-methylphenol-amended soils that exhibited improved growth on 4-methylphenol and 3,4-dimethylphenol were selected for analysis. The colonies were derived from both soil types and represent different sampling days. Since the present study demonstrates that the regulatory mutant DmpR-E135K enhanced degradation by CF600 in these soil matrices, we reasoned that the derivatives might likewise harbor mutations in dmpR. Indeed, DNA sequence analysis of the 10 derivatives of CF600.1::Km-gfp revealed that all harbored mutations within the A domain of dmpR (Table 1). Seven different mutations, representing transitions and transversions that result in single amino acid substitutions and a 6-bp insertion (TGCGGC, a duplication of the preceding sequence) that results in a 2-amino-acid insertion between residues 179 and 180 of DmpR, were found. Furthermore, in two cases the same mutation was found in derivatives from both soil types (i.e., L113V and F122L [Table 1]).
TABLE 1.
Spectrum of mutations detected in DmpR
| Mutationa | Type of genetic rearrangement | No. of cases detected | Sampling day and sourceb |
|---|---|---|---|
| F42Y | T→A | 1 | S2 |
| R109C | C→T | 1 | S4 |
| L113V | C→G | 2 | S6, H7 |
| D116N | G→A | 1 | S7 |
| F122L | T→C | 3 | S8, H5, H6 |
| C137Y | G→A | 1 | H2 |
| 179(CG)180 | 6-bp insertion | 1 | H4 |
Mutation designations use the one-letter amino acid code to indicate the wild-type residue and position followed by the resulting substitution. In the case of the 6-bp insertion, two additional amino acid residues (C and G) are present between residues 179 and residues 180 of the wild-type DmpR sequence.
S, sandy soil; H, humic soil; numbers, days of sampling.
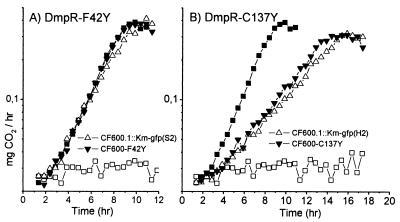
To confirm that the identified changes in the dmpR gene were responsible for the phenotype of improved catabolism of para-substituted phenols, one representative of each of the seven mutants was introduced by recombination into the pVI150 plasmid of CF600 as described in Materials and Methods. The strategy used ensures that the identified mutation is the sole mutation introduced into the wild-type system. Plate tests showed that all the recombinants grew better than wild-type CF600 on both minimal 4-methylphenol and 3,4-dimethylphenol plates. To directly compare the enhanced 4-methylphenol-catabolizing abilities of the mutant derivatives, the growth profiles of the original isolate and its cognate regenerated strain were compared. Examples of two of these comparisons are shown in Fig. 5. The growth profiles of the strain pairs on 4-methylphenol were indistinguishable in each case, demonstrating that the identified mutations conferred the phenotype. Six of the mutations [F42Y, R109C, L113V, D116N, F122L, and the insertion 179(CG)180], exemplified by F42Y in Fig. 5A, conferred growth profiles similar to that conferred by the E135K mutation, while the C137Y mutant had a lower growth rate that was nevertheless much higher than that of the wild type (Fig. 5B). Experiments with 2-methylphenol as the sole carbon and energy source showed that all mutants grew at the expense of this substrate with rates similar to that of the wild type (data not shown).
FIG. 5.
Effect of DmpR mutations on growth rates with 4-methylphenol. The rates of CO2 roduction by the indicated strains were measured upon growth on minimal medium supplemented with 2.5 mM 4-methylphenol. The rate achieved for each original mutant isolate of CF600.1::Km-gfp is compared with that of a regenerated CF600 derivative harboring the detected DmpR mutation. The profiles for CF375.3::Km-gfp (closed squares) and CF600.1::Km-gfp (open squares) are also shown to aid comparison. Values are the averages from duplicate experiments. Results similar to those shown in panel A were obtained with strains harboring DmpR-R109C,-L113V, -D116N, -F122L, and -179(CG) 180 (data not shown).
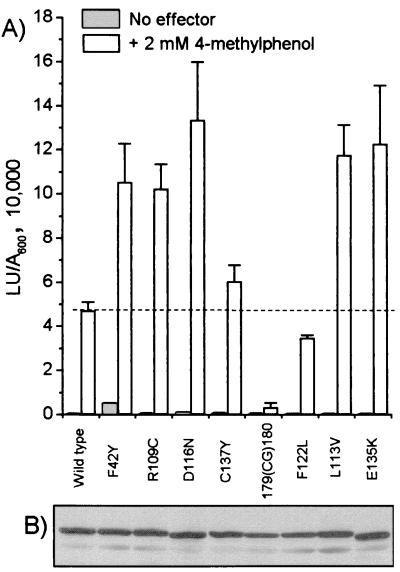
The above results indicate that the mutations identified in this study, like DmpR-E135K, modulate the response to 4-methylphenol. To directly test this idea, the previously constructed luciferase reporter strain P. putida KT2440::Po-luxAB, which carries a chromosomal copy of the luxAB genes under the control of the dmp operon promoter Po, was used. Plasmids expressing the different DmpR derivatives from its native promoter complete the system. The results shown in Fig. 6 demonstrate that five of the mutations (F42Y, R109C, L113V, D116N, and C137Y), like E135K, are expressed at levels comparable to that of wild-type DmpR and show an expected enhanced ability to respond to 4-methylphenol. DmpR-C137Y exhibits the lowest enhancement above the wild-type level in response to 4-methylphenol (Fig. 6A) and is the derivative which exhibits the lowest enhancement of growth rates at the expense of 4-methylphenol (Fig. 5B).
FIG. 6.
In vivo transcriptional response mediated by DmpR derivatives in the presence of 4-methylphenol. (A) The luciferase transcriptional response of P. putida KT2440::Po-luxAB harboring plasmids expressing the indicated derivatives was measured in the presence or absence of effector. Figures are the averages and standard deviations of triplicate determinations in each of two independent experiments. LU, luciferase units. (B) Western analysis of DmpR levels. Crude extracts (30 μg of cellular protein) derived from the cells exposed to 4-methylphenol used for panel A were separated by Sodium dodecyl sulfate–11% polyacrylamide gel electrophoresis and probed with anti-DmpR serum as described in Materials and Methods.
Unexpectedly, DmpR-F122L, which represents the mutation in 3 of the 10 strains isolated, did not show an enhanced response to 4-methylphenol in the luciferase reporter system, and the response with DmpR-179(CG)180 was comparatively extremely low. All of the DmpR derivatives are expressed from the same cis-acting regulatory elements. However, we have previously observed that single amino acid substitutions of the A domain can result in different protein stabilities (26) and that disruption of the integrity of the A domain can result in nonfunctional unfolded proteins (44), which may be differentially targeted for proteolysis under different growth regimes. From Western blot analysis (Fig. 6B), it is clear that these two derivatives are present at lower levels, and quantitative Western analysis showed that they are present at 30 to 50% of the levels of wild-type DmpR (data not shown). Therefore, it appears that the protein level of the regulator can account for the lower response observed with DmpR-F122L but not DmpR-197(CG)180 in the reporter system.
Effector specificity profiles of DmpR mutants.
The effector specificity mutant DmpR-E135K was independently isolated during a genetic selection for mutants that respond to either 2,4-dimethylphenol or 4-ethylphenol. Upon screening, the E135K mutation was found to confer an enhanced response to other mono- or disubstituted phenols with substituents in the para position (31, 42). Therefore, the mutants identified in this study were screened using a simple semiquantitative plate test assay for the ability to respond to a range of para-substituted phenols. The results summarized in Table 2 show that all but DmpR-C137Y and DmpR-179(CG)180 had gained a detectable ability to respond to one or more of the compounds that are noneffectors of wild-type DmpR. The mutations DmpR-R109C, DmpR-D116N, and DmpR-E135K show the broadest spectrum of response to para-substituted compounds, which includes the ability to respond to the priority-list pollutants 2,4-dimethylphenol, 2,4-dichlorophenol, and 4-nitrophenol. Thus, this class of mutants have potential utility in whole-cell biosensor applications for pollution monitoring (44, 52).
TABLE 2.
Effector response profiles of DmpR derivatives in KT2440::Po-luxAB
| Derivative | Luciferase activity witha:
|
|||||
|---|---|---|---|---|---|---|
| 4-MPb | 3, 4-DMPb | 2, 4-DMP | 4-EP | 4-NP | 2, 4-DCP | |
| Wild type | ++ | ++ | − | − | − | − |
| F42Y | +++ | +++ | ++ | − | − | +++ |
| R109C | +++ | ++ | ++ | + | ++ | +++ |
| L113V | +++ | +++ | + | − | − | +++ |
| D116N | +++ | +++ | ++ | + | ++ | +++ |
| F122L | ++ | ++ | ++ | ++ | − | +++ |
| E135K | +++ | +++ | ++ | ++ | +++ | +++ |
| C137Y | ++ | ++ | − | − | − | − |
| 179(CG)180 | ++ | ++ | − | − | − | − |
All compounds were present at a final concentration of 0.5 mM. Abbreviations: MP, methylphenol; DMP, dimethylphenol; EP, ethylphenol; NP, nitrophenol; DCP, dichlorophenol. Plus symbols represent the intensity of the luciferase activity as observed on film, a minus symbol indicates no activity above background in the absence of any aromatic effector.
Compounds can serve as a growth substrate.
DISCUSSION
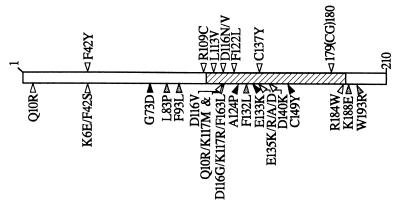
Here, using the (methyl)phenol degradative system of Pseudomonas strain CF600, we examined the potential to improve bacterial biodegradative efficiencies in the soil via modulation of the ability of a regulator to respond to the target compound. Utilizing strains that differ only by a defined point mutation in the dmpR regulatory gene, we could rigorously test the assumption that enhanced biodegradative capacity under laboratory conditions translates to improved performance and competitiveness in structurally and nutritionally heterogeneous soil matrix conditions. The DmpR-E135K regulatory mutant, which has an enhanced ability to respond to para-substituted phenols, exhibited both enhanced biodegradative and enhanced competitive properties compared to the wild type in two very different soil types amended with 4-methylphenol (Fig. 3 and 4). Interestingly, the regulatory mutant strain CF375.3::Km-gfp has a competitive growth advantage over the wild-type strain in the pine forest-derived humic soil matrix irrespective of specific amendments (Fig. 4). In soil from under pine trees, the humus layer is enriched in phenolics as a result of decomposition and humification, and monomeric para-substituted phenols are predominant in the water-soluble phenolic fraction (22). Thus, the universal competitive advantage of CF375.3::Km-gfp in the pine forest humus may be attributed to the more efficient degradation of a range of para-substituted phenolic compounds already present in the soil. The above findings indicate that manipulation of the regulatory circuit can indeed have significant beneficial effects on both biodegradative efficiencies and competitiveness under soil matrix conditions. Consistent with this interpretation, incubation of CF600.1::Km-gfp in soils amended with 4-methylphenol was accompanied by accumulation of spontaneous mutants with an enhanced ability to degrade 4-methylphenol (Fig. 3 to 5). DNA sequence analysis of 10 derivatives revealed seven different individual mutations in the signal receptor domain of DmpR, and the enhanced growth phenotypes in each case could be attributed solely to the single amino acid substitutions or insertion within DmpR (Fig. 5). A number of different molecular genetic strategies have previously been used to isolate mutations within the A domain of DmpR that positively or negatively modulate the response to aromatic effectors. These include positive genetic selection systems and random PCR mutagenesis to generate mutants with enhanced sensitivity and/or novel sensory capacity (31, 42, 52), genetic selection of second-site suppressors of constitutively active mutations of DmpR (26), and DNA shuffling between the A domain of DmpR and that of the toluene-xylene response regulator XylR (44). The locations of the mutations identified in this study relative to previously identified mutations are shown in Fig. 7. The mutations identified in this study are from spontaneous mutants that were isolated from soils contaminated, albeit deliberately, with a toxic aromatic compound, and thus more closely reflect selection pressures found in the soil environment. Of the seven different mutations identified, five target residues that had not previously been implicated in effector recognition. However, two mutations (F42Y and D116N) target residues previously implicated by molecular genetic manipulations.
FIG. 7.
Locations of the A-domain dmpR mutations identified in the present study (upper arrowheads) in relation to previously identified A-domain mutations (lower arrowheads). Open arrowheads indicate mutations that positively affect the response to aromatic compounds as identified by enhanced responses to an effector(s) of wild-type DmpR and/or expanded specificity range (31, 42, 52). Grey and black arrowheads indicate mutations that negatively affect the aromatic response of DmpR. Grey arrowheads represent second-site suppressors that restore aromatic control to semiconstitutive mutants of DmpR by tightening the A-C domain interaction (26) or mutations that constrict the aromatic response profile (F132L [44]). Black arrowheads indicate mutations that result in the inability to respond to aromatic compounds (26; L.C.Ng and V. Shingler, unpublished data). The hatched box indicates the region (residues 110 to 186) that distinguishes the effector specificity profiles of DmpR and XylR (44).
During aromatic effector activation of DmpR, productive binding of aromatic effectors alleviates inhibitory interactions between the A and C domains, thus releasing the transcriptional activating property of DmpR (26, 28, 42). Not all effectors are equally efficient in activating DmpR, and modulation of the A-C domain inhibitory interaction through mutations in either the A or C domain can alter the magnitude of the response to a given effector (26, 31). In addition, mutations within the signal reception A domain can expand the range of compounds that can activate DmpR to promote transcription (31, 42, 52). As illustrated in Fig. 7, residues involved in modulating effector specificity and maintaining the A-C domain interaction are interdispersed on the linear sequence, and replacement of some residues results in modulation of both the effector specificity and A-C domain interaction (e.g., E135R/A/D and D140K [26, 42]). Thus, the phenotype of individual A-domain mutations identified here could have arisen by influencing (i) the aromatic effector binding properties, (ii) the repressive regulatory function of the A domain and thus the functional consequences of binding, or (iii) a combination of both, as was found for DmpR-E135K (29). Given the interdependence of the A-domain-mediated properties, it is difficult to infer the precise mechanism by which each of the A-domain mutations may be mediating its effect. Nevertheless, it is interesting (i) that five of the mutations [L113V, D116N, F122L, C137Y, and 179(CG)180] lie within residues 110 to 186, which were identified by DNA shuffling to distinguish the effector activation profiles of (methyl)phenol-responsive DmpR and toluene-xylene-responsive XylR (44), (ii) that the R109C mutation lies directly adjacent to this region in a residue conserved between DmpR and XylR, and (iii) that the F42Y mutation confers the ability to promote a low level of transcriptional activation in the absence of effectors (Fig. 6), which is indicative of loosening of the A-C domain interaction (26).
When the individual DmpR sensory mutations were introduced into a luciferase reporter system, five of the mutations (F42Y, R109C, L113V, D116N, and C137Y) mediated enhanced responses to 4-methylphenol, consistent with their ability to promote enhanced rate of degradation by their CF600 counterpart (Fig. 5 and 6). Unexpectedly, however, the DmpR-F122L and DmpR-179(CG)180 mutants, while promoting enhanced growth on 4-methylphenol (Fig. 5), did not give an enhanced response to 4-methylphenol in the luciferase reporter system (Fig. 6). These apparently conflicting results have been rigorously checked with independent isolates in both systems (data not shown). While we can only speculate on the reason for the disparity of the results in the two systems, a possible explanation may lie in the different physiological states of the cells under the two experimental conditions. The DmpR-mediated regulatory circuit is subservient to global regulation in response to the nutritional and physiological status of the cell, resulting in optimal expression under conditions likely to prevail in the environment (46). One major mechanism that links the physiological status of the cell to DmpR-mediated transcriptional control has been identified and involves the metabolic alarmone ppGpp (47). Since ppGpp regulates the levels of transcription of many genes, including those of some global regulatory proteins (8), it is plausible that stability, folding, and/or action of a given mutant may differ depending on the medium composition and growth phase of the cell.
The physiological status of the cell also appears to have profound effects on mutational rates (33). The appearance of mutants with different degrees of fitness as a result of the increased mutational rate in starved or stationary-phase bacterial cultures has been shown (13, 20). Furthermore, a higher mutation rate creating desired catabolic phenotype was found with nongrowing bacterial cultures in the presence of potentially useful substrates (13). Therefore, the rapid appearance of the fitter mutants able to efficiently utilize a polluting substrate might be expected in natural soil environments, since these provide suboptimal nutrient levels and growth conditions. As shown in Fig. 3C, incubation of CF600.1::Km-gfp in humic soil contaminated with 4-methylphenol for as little as 4 days results in a fitter mutant subpopulation comprising about 0.1 to 1% of the total population. Although we analyzed only 10 mutants, all were found to harbor mutations in the sensory A domain of DmpR and mediate the enhanced ability to degrade the pollutant. Thus, it follows that the majority of the observed fitter population have gained their phenotype by virtue of modulating the specific regulatory circuit. Hence, it is likely that mutation to optimize the effector response to a given pollutant(s) is a major adaptation mechanism for aromatic catabolism of compounds that are controlled by DmpR-like regulators. Our results show that selection of DmpR-regulatory mutants with an optimized response to the contaminant present might be a natural ongoing process in polluted environments that can occur in a short time frame. As outlined in the introduction, DmpR- and XylR-like regulators are frequently associated with aromatic catabolic pathways. Our findings suggest that genetic variation in the A domains may be feature of this class of regulator and that the concomitant ability to rapidly adapt transcriptional levels via the regulator response to aromatics may account for the predominance of the DmpR-XylR family of regulators in controlling aromatic catabolic pathways.
ACKNOWLEDGMENTS
This work was supported by grants from the Swedish Foundation for Strategic Research, the Swedish Research Councils for Natural and Engineering Sciences, and the Academy of Finland.
We thank Mirja Salkinoja-Salonen for advice and access to equipment for the chemical analysis, Rainer Peltola and Irina Tsitko for advice and help in performing the chemical analysis, Chun-Mei Li for technical assistance, Jens B. Andersen for plasmids, and Martin Gullberg for critical reading of the manuscript.
REFERENCES
- 1.Abril M-A, Michan C, Timmis K N, Ramos J L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for the degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn Y, Sanseverino J, Sayler G S. Analyses of polycyclic aromatic hydrocarbon-degrading bacteria isolated from contaminated soils. Biodegradation. 1999;10:149–157. doi: 10.1023/a:1008369905161. [DOI] [PubMed] [Google Scholar]
- 3.Andersen J B, Sternberg C, Poulsen L K, Bjorn S P, Givskov M, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai H, Akahira S, Ohishi T, Maeda M, Kudo T. Adaptation of Comamonas testosteroni TA441 to utilize phenol: organization and regulation of the genes involved in phenol degradation. Microbiology. 1998;144:2895–2903. doi: 10.1099/00221287-144-10-2895. [DOI] [PubMed] [Google Scholar]
- 5.Ayoubi P J, Harker A R. Whole-cell kinetics of trichloroethylene degradation by phenol hydroxylase in a Ralstonia eutropha JMP134 derivative. Appl Environ Microbiol. 1998;64:4353–4356. doi: 10.1128/aem.64.11.4353-4356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertoni G, Martino M, Galli E, Barbieri P. Analysis of the gene cluster encoding toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl Environ Microbiol. 1998;64:3626–3632. doi: 10.1128/aem.64.10.3626-3632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrne A M, Olsen R H. Cascade regulation of the toluene-3-monooxygenase operon (tbuA1UBVA2C) of Burkholderia pickettii PKO1: role of the tbuA1 promoter (PtbuA1) in the expression of its cognate activator, TbuT. J Bacteriol. 1996;178:6327–6337. doi: 10.1128/jb.178.21.6327-6337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhart F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1458–1496. [Google Scholar]
- 9.Chen W, Brühlmann F, Richins R D, Mulchandani A. Engineering of improved microbes and enzymes for bioremediation. Curr Opin Biotechnol. 1999;10:137–141. doi: 10.1016/s0958-1669(99)80023-8. [DOI] [PubMed] [Google Scholar]
- 10.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertional mutagenesis, promoter probing, and chromosomal insertion of cloned DNA into gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5 and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 12.Favaro R, Bernasconi C, Passini N, Bertoni G, Bestetti G, Galli E, Deho G. Organisation of the tmb catabolic operons of Pseudomonas putida TMB and evolutionary relationship with the xyl operons of the TOL plasmid pWW0. Gene. 1996;182:189–193. doi: 10.1016/s0378-1119(96)00552-5. [DOI] [PubMed] [Google Scholar]
- 13.Finkel S E, Kolter R. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci USA. 1999;96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fulthorpe R R, Rhodes A N, Tiedje J M. Pristine soils mineralize 3-chlorobenzoate and 2,4-dichlorophenoxyacetate via different microbial populations. Appl Environ Microbiol. 1996;62:1159–1166. doi: 10.1128/aem.62.4.1159-1166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallet C, Lebreton P. Evolution of phenolic patterns in plants and associated litters and humus of a mountain forest ecosystem. Soil Biol Biochem. 1995;27:157–165. [Google Scholar]
- 16.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning, a practical approach. Vol. 1. Oxford, United Kingdom: IRL Press Ltd.; 1985. pp. 109–136. [Google Scholar]
- 17.Hino S, Watanabe K, Takahashi N. Phenol hydroxylase cloned from Ralstonia eutropha strain E2 exhibits novel kinetic properties. Microbiology. 1998;144:1765–1772. doi: 10.1099/00221287-144-7-1765. [DOI] [PubMed] [Google Scholar]
- 18.Inouye S, Nakazawa A, Nakazawa T. Determination of the transcription initiation site and identification of the protein product of the gene xylR for xyl operons on the TOL plasmid. J Bacteriol. 1985;163:863–869. doi: 10.1128/jb.163.3.863-869.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaspers M C, Suske W A, Schmid A, Goslings D A, Kohler H P, van der Meer J R. HbpR, a new member of the XylR/DmpR subclass within the NtrC family of bacterial transcriptional activators, regulates expression of 2-hydroxybiphenyl metabolism in Pseudomonas azelaica HBP1. J Bacteriol. 2000;182:405–417. doi: 10.1128/jb.182.2.405-417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasak L, Hõrak R, Kivisaar M. Promoter-creating mutations in Pseudomonas putida: a model system for the study of mutation in starving bacteria. Proc Natl Acad Sci USA. 1997;94:3134–3139. doi: 10.1073/pnas.94.7.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kästner M, Mahro B. Microbial degradation of polycyclic aromatic hydrocarbons in soils affected by the organic matrix of compost. Appl Microbiol Biotechnol. 1996;44:668–675. doi: 10.1007/BF00172501. [DOI] [PubMed] [Google Scholar]
- 22.Kuiters A T, Denneman C A J. Water-soluble phenolic substances in soils under several coniferous and deciduous tree species. Soil Biol Biochem. 1987;19:765–769. [Google Scholar]
- 23.Laurie A D, Lloyd-Jones G. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J Bacteriol. 1999;181:531–540. doi: 10.1128/jb.181.2.531-540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;178:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller C, Petruschka L, Cuypers H, Burchhardt G, Herrmann H. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J Bacteriol. 1996;178:2030–2036. doi: 10.1128/jb.178.7.2030-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng L C, O'Neill E, Shingler V. Genetic evidence for inter-domain regulation of the phenol responsive ς54-dependent activator DmpR. J Biol Chem. 1996;271:17281–17286. doi: 10.1074/jbc.271.29.17281. [DOI] [PubMed] [Google Scholar]
- 27.Ng L C, Poh C L, Shingler V. Aromatic effector activation of the NtrC-like transcriptional regulator PhhR limits the catabolic potential of the (methyl)phenol degradative pathway it controls. J Bacteriol. 1995;177:1485–1490. doi: 10.1128/jb.177.6.1485-1490.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Neill E, Ng L C, Sze C C, Shingler V. Aromatic ligand binding and intramolecular signalling of the phenol-responsive ς54-dependent regulator DmpR. Mol Microbiol. 1998;28:131–141. doi: 10.1046/j.1365-2958.1998.00780.x. [DOI] [PubMed] [Google Scholar]
- 29.O'Neill E, Sze C C, Shingler V. Novel effector control through modulation of a preexisting binding site of the aromatic-responsive ς54-dependent regulator DmpR. J Biol Chem. 1999;274:32425–32432. doi: 10.1074/jbc.274.45.32425. [DOI] [PubMed] [Google Scholar]
- 30.Ortega-Calvo J-J, Saiz-Jimenez C. Effect of humic fractions and clay on biodegradation of phenanthrene by a Pseudomonas fluorescens strain isolated from soil. Appl Environ Microbiol. 1998;64:3123–3126. doi: 10.1128/aem.64.8.3123-3126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavel H, Forsman M, Shingler V. An aromatic effector specificity mutant of the transcriptional regulator DmpR overcomes the growth constraints of Pseudomonas sp. strain CF600 on para-substituted methylphenols. J Bacteriol. 1994;176:7550–7557. doi: 10.1128/jb.176.24.7550-7557.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romine M F, Stillwell L C, Wong K K, Thurston S J, Sisk E C, Sensen C, Gaasterland T, Fredrickson J K, Saffer J D. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J Bacteriol. 1999;181:1585–1602. doi: 10.1128/jb.181.5.1585-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg S M. Mutation for survival. Curr Opin Genet Dev. 1997;6:829–834. doi: 10.1016/s0959-437x(97)80047-0. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual, 2nded. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Sarand I, Haario H, Jrgensen K S, Romantschuk M. Effect of inoculation of a TOL plasmid containing mycorrhizosphere bacterium on development of Scots pine seedlings, their mycorrhizosphere and the microbial flora in m-toluate-amended soil. FEMS Microbiol Ecol. 2000;31:127–141. doi: 10.1111/j.1574-6941.2000.tb00678.x. [DOI] [PubMed] [Google Scholar]
- 36.Schirmer F, Ehrt S, Hillen W. Expression, inducer spectrum, domain structure, and function of MopR, the regulator of phenol degradation in Acinetobacter calcoaceticus NCIB8250. J Bacteriol. 1997;179:1329–1336. doi: 10.1128/jb.179.4.1329-1336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarzenbach R P, Gschwend P M, Imboden D M. Environmental organic chemistry. New York, N.Y: John Wiley & Sons, Inc.; 1993. [Google Scholar]
- 38.Shingler V. Signal sensing by ς54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–416. doi: 10.1046/j.1365-2958.1996.388920.x. [DOI] [PubMed] [Google Scholar]
- 39.Shingler V, Bartilson M, Moore T. Cloning and nucleotide sequence of the gene encoding the positive regulator (DmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J Bacteriol. 1993;175:1596–1604. doi: 10.1128/jb.175.6.1596-1604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shingler V, Franklin F C H, Tsuda M, Holroyd D, Bagdasarian M. Molecular analysis of a plasmid encoded phenol hydroxylase from Pseudomonas CF600. J Gen Microbiol. 1989;135:1083–1092. doi: 10.1099/00221287-135-5-1083. [DOI] [PubMed] [Google Scholar]
- 41.Shingler V, Moore T. Sensing of aromatic compounds by the DmpR transcriptional activator of phenol-catabolizing Pseudomonas sp. strain CF600. J Bacteriol. 1994;176:1555–1560. doi: 10.1128/jb.176.6.1555-1560.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shingler V, Pavel H. Direct regulation of the ATPase activity of the transcriptional activator DmpR by aromatic compounds. Mol Microbiol. 1995;17:505–513. doi: 10.1111/j.1365-2958.1995.mmi_17030505.x. [DOI] [PubMed] [Google Scholar]
- 43.Shingler V, Powlowski J, Marklund U. Nucleotide sequence and functional analysis of the complete phenol/3,4-dimethylphenol catabolic pathway of Pseudomonas sp. strain CF600. J Bacteriol. 1992;174:711–724. doi: 10.1128/jb.174.3.711-724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skärfstad E, O'Neill E, Garmendia J, Shingler V. Identification of an effector specificity subregion within the aromatic-responsive regulators DmpR and XylR by DNA shuffling. J Bacteriol. 2000;182:3008–3016. doi: 10.1128/jb.182.11.3008-3016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solano-Serena F, Marchal R, Blanchet D, Vandecasteele J-P. Intrinsic capacities of soil microflorae for gasoline degradation. Biodegradation. 1998;9:319–326. doi: 10.1023/a:1008305906032. [DOI] [PubMed] [Google Scholar]
- 46.Sze C C, Moore T, Shingler V. Growth phase-dependent transcription of the ς54-dependent Po promoter controlling the Pseudomonas-derived (methyl)phenol dmp operon of pVI150. J Bacteriol. 1996;178:3727–3735. doi: 10.1128/jb.178.13.3727-3735.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sze C C, Shingler V. The alarmone (p)ppGpp mediates physiological control at the ς54-dependent Po promoter. Mol Microbiol. 1999;31:1217–1228. doi: 10.1046/j.1365-2958.1999.01264.x. [DOI] [PubMed] [Google Scholar]
- 48.Timmis K N, Steffan R J, Unterman R. Designing microorganisms for the treatment of toxic wastes. Annu Rev Microbiol. 1994;48:525–557. doi: 10.1146/annurev.mi.48.100194.002521. [DOI] [PubMed] [Google Scholar]
- 49.Timmis K N, Pieper D H. Bacteria designed for bioremediation. Trends Biotechnol. 1999;17:201–204. doi: 10.1016/s0167-7799(98)01295-5. [DOI] [PubMed] [Google Scholar]
- 50.van Veen J A, van Overbeek L S, van Elsas J D. Fate and activity of microorganisms introduced into soil. Microbiol Mol Biol Rev. 1997;61:121–135. doi: 10.1128/mmbr.61.2.121-135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogel T M. Bioaugmentation as a soil bioremediation approach. Curr Opin Biotechnol. 1996;7:311–316. doi: 10.1016/s0958-1669(96)80036-x. [DOI] [PubMed] [Google Scholar]
- 52.Wise A A, Kuske C R. Generation of novel bacterial regulatory proteins that detect priority pollutant phenols. Appl Environ Microbiol. 2000;66:163–169. doi: 10.1128/aem.66.1.163-169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaitsev G M, Uotila J S, Tsitko I V, Lobanok A G, Salkinoja-Salonen M S. Utilization of halogenated benzenes, phenols, and benzoates by Rhodococcus opacus GM-14. Appl Environ Microbiol. 1995;61:4191–4201. doi: 10.1128/aem.61.12.4191-4201.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]