Abstract
The effect of developing chrysanthemum roots on the presence and activity of bacterial populations in the rhizosphere was examined by using culture-independent methods. Nucleic acids were extracted from rhizosphere soil samples associated with the bases of roots or root tips of plants harvested at different stages of development. PCR and reverse transcriptase (RT) PCR were used to amplify 16S ribosomal DNA (rDNA) and 16S rRNA, respectively, and the products were subjected to denaturing gradient gel electrophoresis (DGGE). Prominent DGGE bands were excised and sequenced to gain insight into the identities of predominantly present (PCR) and predominantly active (RT-PCR) bacterial populations. The majority of DGGE band sequences were related to bacterial genera previously associated with the rhizosphere, such as Pseudomonas, Comamonas, Variovorax, and Acetobacter, or typical of root-free soil environments, such as Bacillus and Arthrobacter. The PCR-DGGE patterns observed for bulk soil were somewhat more complex than those obtained from rhizosphere samples, and the latter contained a subset of the bands present in bulk soil. DGGE analysis of RT-PCR products detected a subset of bands visible in the rDNA-based analysis, indicating that some dominantly detected bacterial populations did not have high levels of metabolic activity. The sequences detected by the RT-PCR approach were, however, derived from a wide taxonomic range, suggesting that activity in the rhizosphere was not determined at broad taxonomic levels but rather was a strain- or species-specific phenomenon. Comparative analysis of DGGE profiles grouped all DNA-derived root tip samples together in a cluster, and within this cluster the root tip samples from young plants formed a separate subcluster. Comparison of rRNA-derived bacterial profiles showed no grouping of root tip samples versus root base samples. Rather, all profiles derived from 2-week-old plant rhizosphere soils grouped together regardless of location along the root.
Plant roots influence soilborne microbial communities via several mechanisms, including excretion of specific organic compounds, competition for nutrients, and providing a solid surface for attachment. The nature of this influence is highly variable and depends upon both the amount and the composition of organic materials released by the plants (17). Since such root-released products can be highly specific for a given plant species or even a particular cultivar, plants are thought to selectively enrich their rhizospheres for microorganisms that are well adapted to utilization of specific released organic compounds (4, 18, 24). Since production of root-released materials can also vary during plant and root development (36), one might also expect microbial communities in the rhizosphere to be influenced by the developmental stage and age of a plant, as well as the location in particular parts of the root system.
Thus, specific bacterial populations, including those that antagonize pathogen development, may be stimulated in the rhizosphere and be different in different plant species, genotypes (28, 29), plant developmental stages, or root parts (base versus tip) (23). Despite the longstanding realization that plant roots affect microbial communities in the rhizosphere, the study of this interaction has proven to be difficult, due chiefly to the complexity of soil ecosystems and the limitations of the traditional pure-culture techniques (37, 38). Studies based upon characterization of culturable rhizosphere bacteria have suggested that plants can have specific effects on microbial communities (16). Unfortunately, such approaches only address the culturable bacteria, which are thought to represent only a small proportion (0.1 to 10%) of the total bacteria present in the rhizosphere (34, 38). Similarly, microscopic techniques can be used to obtain information about bacterial numbers and, potentially, spatial distribution, but these approaches lack the discriminating ability to assess diversity and distinguish between multiple bacterial populations. However, the introduction of nucleic acid-based techniques into microbial ecology has allowed characterization of microbial communities without a pure-culture isolation step. The use of PCR to specifically amplify 16S ribosomal DNA (rDNA) molecules from DNA extracted directly from the environment has allowed assessment of microbial diversity in a wide range of habitats, including microbial lineages for which there are no known pure cultures (15, 21). Furthermore, the use of denaturing gradient gel electrophoresis (DGGE) to separate mixed PCR products recovered from the environment by specific amplification of bacterial 16S rDNA sequences offers a culture-independent method for tracking dominant bacterial populations in space and time (27).
The dynamics of the dominant bacterial communities inhabiting the rhizosphere of developing chrysanthemum plants were examined in previous studies by using both a community profiling approach and culture methods (8, 9). These studies revealed some differences in the physiological characteristics and numbers of culturable bacteria at different times and locations within the chrysanthemum root system. However, the bacterial community fingerprints based upon PCR-amplified DNA isolated from the rhizosphere were similar to those observed for bulk soil, and differences among rhizosphere samples were small, suggesting that root effects (3, 6) were minor with respect to determining the dominant bacterial populations. However, detection and identification of active bacterial populations are of greatest interest for understanding the microbial ecology of the rhizosphere and for discovering potentially useful microbial antagonists of pathogens.
The aim of this study was, therefore, to increase our understanding of the distribution, diversity, and activity of dominant bacterial groups associated with the roots of developing chrysanthemum plants. As rRNA content represents a first approximation of bacterial activity, it has been proposed that this target is appropriate for assessing changes in active bacterial populations (41). We, therefore, chose to target 16S rRNA and 16S rDNA extracted directly from rhizosphere soil in reverse transcriptase PCR (RT-PCR) and PCR analyses, respectively. The community fingerprints produced by DGGE analysis of the mixed RT-PCR and PCR products were then compared in order to gain insight into changes in the active and total bacterial communities in rhizosphere soil samples taken at different stages of plant development and at different locations along the root (root tip and root base). Bulk soil samples were also included in the analysis for comparison. Prominent DGGE bands were excised and used for nucleotide sequence determination (13) in order to confirm band identity between samples and provide preliminary identification. Our results are discussed with respect to the effects of plants on bacterial rhizosphere communities and the search for bacterial groups that show good rhizosphere colonization, a prerequisite for effective biocontrol.
MATERIALS AND METHODS
Soil conditions.
Loamy sand soil, referred to as Beekeerd soil, was collected near Ede, The Netherlands, and used in all of the experiments described below. This soil had a pH (-KCl) of 5.5 and contained 3.5% organic matter. Additional soil characteristics and soil preparation details have been described by Duineveld et al. (8). During plant growth, the moisture content of the soil was maintained at approximately 12% (wt/wt). The soil was sieved prior to use by using a sieve with a 6- by 3-mm mesh. To enhance aeration, the soil was mixed with perlite (20%, vol/vol). After mixing, the soil was put into 1-liter pots at a density of approximately 1 g/cm3 and incubated under plant growth conditions for 2 weeks before seedlings were planted.
Plant growth conditions.
The chrysanthemum (Dendranthema grandiflora Tzvelev) cultivar used was cultivar Majoor Bosshardt and was obtained from Fides Inc. (De Lier, The Netherlands). Treatment of the chrysanthemum seedlings and the plant growth conditions were as described by Duineveld et al. (8). Briefly, cuttings of the cultivar were treated with insecticides and hormones, placed in peat blocks, and allowed to develop in a growth room. After 2 weeks each peat block containing one seedling was placed on top of the Ede loamy sand soil in a 1-liter pot. The plants were grown at 20°C with 70% humidity and a photoperiod consisting of 16 h of light and 8 h of darkness. The plants were watered once a day. The plants also received 75 ml of a nutrient solution twice a week and a trace element mixture (containing iron, manganese, boron, zinc, copper, and molybdenum) once a week. The day length was changed from 16 to 8 h 3 weeks after planting in order to induce flowering. Flowering occurred approximately 9 weeks after planting.
Sampling.
Samples of chrysanthemum root tip and base parts were collected 2, 4, 6, and 10 weeks after planting. The rhizosphere soil from a total of 10 individually grown plants was harvested on each sampling date. From these samples two pooled samples, each containing the rhizosphere soil of five plants, were prepared in order to ensure that there was sufficient material for DNA and RNA isolation. Plants and soil were removed from the pots. Subsequently, excess bulk soil was removed from the roots by shaking, leaving roots and firmly adhering soil, which was defined as the rhizosphere soil. The young parts of the roots (i.e., the 1 to 2 cm at the end of each root) were dissected with a double cutting knife with a cutting space of 1 cm. These parts were separated from the older root parts, which were dissected with a double cutting knife with a cutting space of 4 cm. The root parts with adhering soil were weighed. Samples were stored overnight at 7°C, and DNA and RNA extractions were initiated the following day.
Extraction of DNA and RNA.
Nucleic acid extraction from the rhizosphere soil was performed by the method of Moran et al. (26), as modified by Duarte et al. (7). For purification of DNA from crude extracts we used CsCl with potassium acetate precipitation, followed by further purification with the Wizard DNA Clean-up system (Promega, Madison, Wis.). DNA was removed from RNA by treatment with DNase I (10 U/μl; RNase free; Boehringer), and for RNA purification we used an RNeasy mini kit (Qiagen).
PCR amplification.
PCR amplification targeting bacterial 16S rDNA was performed with the 968f-GC and 1401r primers (20). A touchdown thermocycling program was used for PCR as described by Rosado et al. (33). Reverse transcription of 16S rRNA and subsequent PCR amplification were performed by using a two-step reaction scheme, as follows. One microliter of a total-RNA sample (containing approximately 5 ng of RNA) was added to a 49-μl RT-PCR mixture, which consisted of 19 μl of RNase-free H2O, 10 μl of 5 × GeneAmp EZ buffer (Perkin-Elmer), 10 μl of a deoxynucleoside triphosphate (dNTP) mixture, 1.5 μl of 10 μM primer 1401r, 2.5 μl of dimethyl sulfoxide, 2 μl of rTth DNA polymerase (Perkin-Elmer), and 5 μl of 25 mM manganese diacetate. Reverse transcription was carried out at 60°C for 10 min and at 62°C for an additional 20 min. Five microliters of the RT-PCR mixture was then added to a 45-μl PCR mixture containing 21.33 μl of MilliQ H2O, 5 μl of 10× Stoffel buffer (Perkin-Elmer), 10 μl of a dNTP mixture containing each dNTP at a concentration of 1 mM, 7.5 μl of 25 mM MgCl2, 0.12 μl of 10 μM primer 968f-GC, 0.5 μl of formamide, 0.05 μl of T4 gene 32 protein, and 0.5 μl (1 u) of Stoffel fragment (Perkin-Elmer). For PCR amplification we used the thermocycling program described above (33), except that 0.87 μl of 10 μM primer 1401r and 0.87 μl of 10 μM primer 968f-GC were added to each sample after three cycles. PCR and RT-PCR products were examined by agarose electrophoresis (1.5% agarose; 0.5× TBE gel [1× TBE is 90 mM Tris-Borate plus 2 mM EDTA, pH 8.3]) with standard ethidium bromide staining to check for recovery of products of the expected size (approximately 450 bp) and to estimate product concentrations relative to those of known standards.
DGGE analysis.
DGGE was performed by using 6% acrylamide gels (ratio of acrylamide to bisacrylamide, 37:1) with a 45 to 65% denaturant gradient (20), where 100% denaturant was defined as 7 M urea plus 40% formamide (27). Approximately 2 μg of PCR or RT-PCR product was loaded per sample in a final volume of 20 μl. The gels were electrophoresed at 60°C at 80 V for 16 h by using the D-Gene system (Bio-Rad Laboratories, Hercules, Calif.). The gels were stained for 20 min with ethidium bromide and washed twice for 10 min with MilliQ H2O prior to UV transillumination. The DGGE gels were digitized by using the Imager system (Ampligene, Illkirch, France). Gel images were examined with the ImageMaster Elite software package (version 3.1; Pharmacia). The background was first subtracted by using a rolling circle algorithm (circle diameter, 30), and the lanes were normalized so that all lanes contained the same amount of total signal. Bands were called automatically and controlled visually. Band positions were then converted to Rf values between 0 and 1 by using the uppermost and lowermost bands in the marker lanes as boundaries. Profile similarity was calculated by determining a Pearson's coefficient for the total lane pattern after background subtraction with the ImageMaster Elite Database program (version 2.0), and a dendrogram was constructed by using the unweighted pair group method with mathematical average (UPGMA). Bootstrap values were based on 100 replicates.
Recovery of bands from DGGE gels and sequence analysis.
Prominent DGGE bands were selected and used for excision and nucleotide sequence determination (Fig. 1). For each band selected, only the middle portion was excised with a sterile razor, and slices (approximately 30 mg [wet weight]) were placed in 2-ml screw-cap polypropylene tubes containing 0.1 g of glass beads (diameter, 0.1 mm; BioSpec Products) and 0.1 ml of TE buffer. The tubes were shaken at 5,000 rpm for 30 s in a minibeadbeater (BioSpec Products), frozen at −20°C for 15 min, and shaken a second time for 60 s. The tubes were then incubated overnight at 37°C. After centrifugation at 5,000 × g for 1 min, 1 ml of buffer containing DNA was used as the template for a PCR performed under the conditions described above for environmental samples. A 5-μl sample of each PCR product was subjected to agarose gel electrophoresis to confirm product recovery and to estimate product concentration. Five microliters of each reaction mixture was also subjected to DGGE analysis, as described above for environmental samples, to check the purity and to confirm the melting behavior of the band recovered. Some DNA samples still contained mixed products, as shown by multiple DGGE bands. In each of these cases the intended band was excised from the recovered pattern. The remaining PCR product (40 μl) was purified by using Wizard PCR Preps (Promega), and 1 μg of DNA per sample was used as a template for each of two double-stranded cycle sequencing reactions. The reactions were performed with Cy 5-labeled primers 968f (without a GC clamp) and 1401r by using a Thermo Sequenase primer-labeled kit (Amersham Pharmacia Biotech) according to the protocol provided by the manufacturer. Long Read ReproGels were run on an ALFexpress II automated sequencing system (Amersham Pharmacia Biotech). For sequence comparisons with the database we used the FASTA program (31).
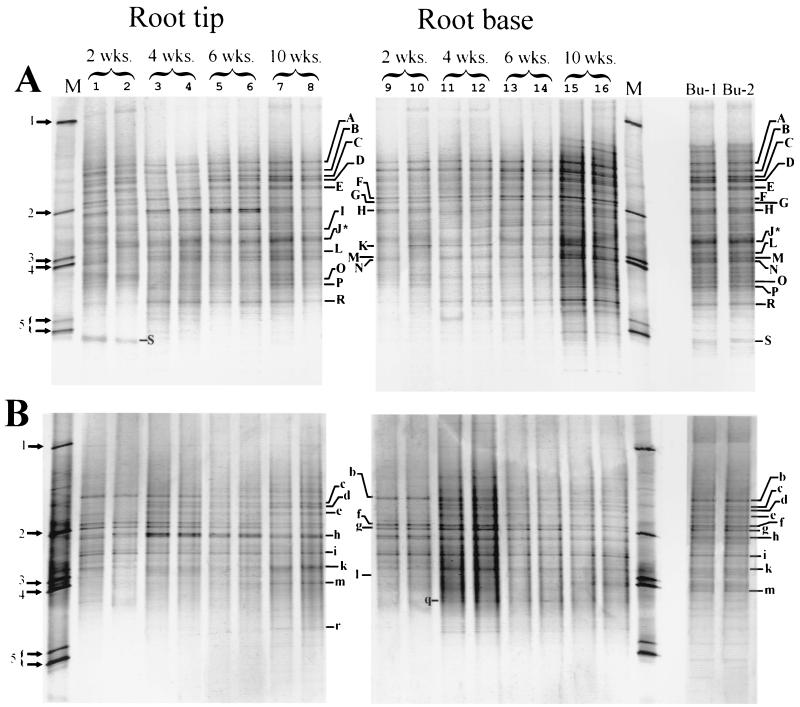
FIG. 1.
DGGE patterns produced from 16S rDNA (A) and 16S rRNA (B) templates isolated from chrysanthemum rhizosphere samples. Samples were taken from either root tips (lanes 1 through 8) or root bases (lanes 9 through 16), from plants harvested at the ages indicated at the top. Two samples from each developmental stage and root system location were analyzed and were numbered sequentially as shown above the lanes. Lanes, Bu-1, and Bu-2, contained samples from bulk soil. Lanes M contained a DGGE marker, consisting of a mixture of 16S rDNA fragments. The marker bands in the marker lanes are as follows: 1, Enterobacter cloacae BE1; 2, Listeria innocua ALM105; 3, Rhizobium leguminosarum subsp. trifolii; 4, Arthrobacter sp.; 5, Pseudomonas cepacia P2. Uppercase letters indicate bands from the DNA-derived DGGE patterns that were excised for sequence analysis, while lowercase letters indicate bands from the RNA-derived patterns with identical sequences (Table 1). The asterisks indicate that excised band J produced an ambiguous sequence, probably due to the presence of multiple DNA fragments within this band.
Nucleotide sequence accession numbers.
Band sequences determined in this study have been deposited in the EMBL database under accession numbers AJ86726 to AJ86742.
RESULTS
All chrysanthemum rhizosphere samples, as well as the two bulk soil samples, produced positive PCR and RT-PCR results with the 968f-GC and 1401r primers. DGGE of recovered PCR products based on amplified rhizosphere DNA produced between 22 and 30 detectable bands ranging in mobility from approximately 48 to 62% denaturant. The bulk samples each contained 34 detectable bands. Duplicate PCR yielded nearly indistinguishable DGGE results, as did multiple DNA isolations from a single rhizosphere sample (results not shown), suggesting that there was a high level of reproducibility in the DNA isolation, PCR, and DGGE procedures. The DGGE profiles derived from RT-PCR of rhizosphere 16S rRNA templates contained 10 to 18 visible bands per sample. Bands were visible at approximately 50 to 60% denaturant. The bulk samples produced an average of 21 detectable bands. Duplicate RT-PCR amplifications and RNA extractions also resulted in nearly identical DGGE patterns. The DGGE patterns recovered from the bulk soil remained constant over the course of the 10-week experiment (8; results not shown).
To gain insight into the identities of major bacterial populations, prominent DGGE bands from both the DNA- and RNA-derived rhizosphere profiles and bands derived from bulk soil were excised and used for nucleotide sequence analysis (Fig. 1). Most of the excised bands produced legible DNA sequences; the only exception was band J, and this rather diffuse band may have contained DNA from more than one bacterial species. DGGE bands in a single gel that appeared to be identical based on mobility did indeed produce identical nucleotide sequences (a total of 10 duplicate bands were sequenced [data not shown]). Furthermore, DGGE bands that appeared to be identical in the DNA- and RNA-derived profiles also produced identical sequences, as indicated by the upper- and lowercase letter designations used in Fig. 1 and Table 1. The majority of the DGGE bands showed the highest levels of identity to clones recovered from soil environments or sequences obtained from strains isolated from soils. The highest level of identity between a DGGE band and a previously defined sequence was the level of identity observed for bands B and b (Table 1). These bands showed 99.5% identity to a sequence recovered from a grassland soil (12) and 97.2% identity to a previously isolated Bacillus strain (accession number AJ132749). The band sequences that were least similar to previously recovered sequences were the sequences obtained from bands F and f, G and g, O, and P, which showed between 88.4 and 91.5% identity to the most closely related database sequences. The sequence obtained from bands E and e had the lowest level of identity with sequences of cultured bacterial species. Although this sequence was 94.9% identical to a grassland clone characterized by Felske et al. (12), it displayed only 80.7% similarity with the sequence of Verrucomicrobium spinosum, the mostly closely matching cultured strain (42).
TABLE 1.
Sequence analysis of bands excised from DGGE gels derived from bacterial 16S rDNA and 16S rRNA extracted from rhizosphere samples of chrysanthemum plants grown in a loamy sand soil.
| Band (s)a | Most closely related bacterial sequenceb | % Identity | Accession no. | Reference |
|---|---|---|---|---|
| A | Unidentified bacterium | 94.5 | AJ000983 | 12 |
| Bacillus lentus | 94.3 | D16272 | T. Suzuki, unpublished data | |
| B, b | Unidentified bacterium | 99.5 | Y07574 | 12 |
| Bacillus species | 97.2 | AJ132749 | A. Felske, unpublished data | |
| C, c | Unidentified bacterium | 98.5 | AJ001222 | 12 |
| Bacillus cohnii | 95.8 | X76437 | 30 | |
| D, d | Unidentified bacterium | 96.8 | AJ001222 | 12 |
| Bacillus pseudomegaterium | 96.2 | X77791 | Lafay et al.,unpublished data | |
| E, e | Unidentified bacterium | 94.9 | Y07575 | 12 |
| Verrucomicrobium spinosum | 80.7 | X90515 | 42 | |
| F, f | Acidobacterium capsulatum | 91.5 | D26171 | A. Hiraishi, unpublished data |
| G, g | Acidobacterium capsulatum | 90.8 | D26171 | A. Hiraishi, unpublished data |
| H, h | Unidentified bacterium | 93.3 | Y07582 | 12 |
| Pseudomonas carboxydohydrogen | 92.5 | Ab021393 | Anzai et al., unpublished data | |
| I, i | Variovorax paradoxus | 93.3 | Ab008000 | T. Hamada, unpublished data |
| K, k | Unidentified γ-proteobacterium | 95.5 | Ab015547 | Kato and Li, unpublished data |
| Pseudomonas graminis sp. nov. | 95.3 | Y11150 | 2 | |
| L, l | Comamonas testosteroni | 94.5 | Ab007997 | T. Hamada, unpublished data |
| M, m | Unidentified bacterium | 96.5 | Y12598 | A. Felske, unpublished data |
| Afripia genospecies 2 | 89.8 | U87765 | A. M. Whitney, unpublished data | |
| N | Unidentified bacterium | 96.3 | Y12596 | 12 |
| Azospirillum brasilense | 91.1 | X79739 | 10 | |
| O | Unidentified bacterium | 88.9 | Y07583 | 12 |
| Acetobacter sp. | 87.1 | Ab016865 | Ueda et al., unpublished data | |
| P | Unidentified bacterium | 89.9 | Y0758 | 12 |
| Acetobacter sp. | 88.4 | Ab016865 | Ueda et al., unpublished data | |
| q | Pseudomonas lundensis | 93.5 | Ab021395 | Anzai et al., unpublished data |
| R, r | Arthrobacter oxydans | 90.4 | X83408 | 22 |
| S | Ralstonia eutropha | 96.0 | Ab015605 | 19 |
The uppercase letters indicate bands derived from 16S rDNA, and the lowercase letters indicate bands derived from 16S rRNA.
The database entry with the highest level of identity is shown. When the most similar sequence was the sequence of an unidentified bacterium or environmental clone, the value for the most closely related identified bacterium is also given.
Most of the bands examined for the RNA-derived community profiles were also detected in the DNA-derived DGGE patterns; the only exception was band q. Thus, for the most part the RNA-derived bands were a subset of the bands detected by DNA isolation and PCR. In the DNA-derived profiles, approximately 11 bands were detected for all samples. A total of seven of the RNA-derived bands were present in all samples, and these seven bands were also all present in all of the DNA profiles. Most of the bands detected were observed for more than a single experimental treatment (i.e., harvest time and root location). An exception was band S; this Ralstonia-like band was detected only in the DNA of 2-week-old root tip samples.
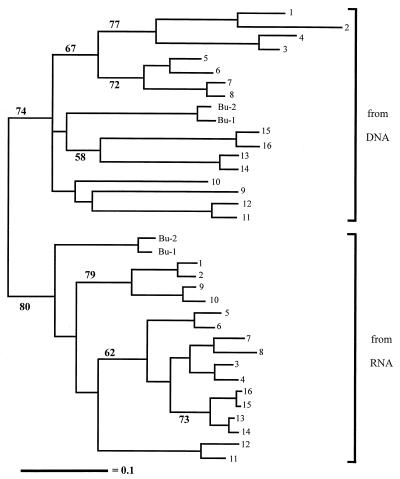
In order to compare DGGE patterns, Pearson's indices were determined for comparisons of all profiles, and UPGMA was used to create a dendrogram describing pattern similarities (Fig. 2). This analysis clearly distinguished between the DNA-and RNA-derived DGGE patterns. Most of the duplicated samples, which were collected under the same conditions, produced DGGE patterns that grouped together as most similar to each other; the only exceptions were DNA samples 9 and 10 (2-week-old root base samples). The grouping based on DNA-derived patterns clustered all root tip samples together separate from the root base samples. Within the root tip cluster, samples from the first 4 weeks of the experiment grouped together, as did samples from the 6- and 10-week sampling dates. The RNA-derived profiles showed a different pattern of clustering. In this case, all 2-week samples grouped together regardless of whether they were obtained from root tips or root bases, and no clear separation between root tip and root base samples was observed.
FIG. 2.
UPGMA tree representing the genetic similarity of the microbial community profiles obtained by PCR-DGGE and RT-PCR–DGGE. Samples 1 through 16, Bu-1, and Bu-2 refer to the lane numbers in Fig. 1 (DNA-derived patterns in Fig. 1A and RNA-derived patterns in Fig. 1B); and Bu-1 and Bu-2 were samples from bulk soil. Bootstrap values greater than 50 are given at nodes only for clusters containing more than two samples.
DISCUSSION
Plant roots are thought to differentially influence the survival, growth, and activity of microorganisms in the rhizosphere, depending on the plant species or cultivar, developmental stage of the plant, and root part examined. The latter two parameters were examined in this study, which was designed to monitor the presence and activity of dominant bacterial populations in the rhizosphere of developing chrysanthemum seedlings. To the best of our knowledge, this study was the first attempt to use culture-independent methods to monitor both the presence and the activity of dominant bacterial populations in relation to plant and root development. While the so-called “rhizosphere effect” has been observed in several culture-based studies (3, 6, 23), such studies addressed only the small fraction of the bacterial community that is amenable to pure-culture isolation.
There was a high level of similarity between the DGGE profiles obtained after PCR amplification of rhizosphere 16S rDNA, regardless of the root region (root tip versus root base) or developmental stage of the plant. The patterns obtained for all rhizosphere samples also showed high degrees of similarity to DGGE patterns obtained from rDNA amplified from the bulk soil, although the latter patterns contained a number of additional bands and a higher level of background signal. Thus, plant root effects did not cause complete shift in the bacterial community but rather caused subtle changes, which is in agreement with previous culture-independent attempts to characterize bacterial communities in the rhizosphere (8, 11, 12, 20). Nevertheless, the DGGE profiles derived from DNA isolated from root tip samples were found to be more similar to each other than to profiles derived from root base DNA extraction. This result is in agreement with the results of related culture-based studies in which stable bacterial communities in root tip rhizosphere samples were examined (9; B. M. Duineveld, J. Postma, and J. A. Van Veen, submitted for publication). The similarity of bulk and rhizosphere samples is influenced by the amount of soil adhering to the roots, which we used to define the rhizosphere, and soil adhesion to roots appeared to be constant over the entire growth period except for the very beginning (9; Duineveld et al., submitted).
The root tip PCR-DGGE profiles obtained for the first 4 weeks of the experiment were more similar to each other than to the profiles obtained for later harvest dates in the experiment. Thus, although the total-community compositions for the different root regions or developmental plant stages remained rather similar, these factors did seem to influence some aspects of the community profiles obtained from the rhizosphere samples. Samples obtained from the rhizospheres of different individual plants that received the same treatment (i.e., the same location and the same plant age) almost always produced DGGE profiles that were nearest neighbors in the dendrogram analysis. This suggests that bacterial community development was a reproducible process, at least for the dominant groups of bacteria detected by PCR-DGGE.
In earlier work (8), it was concluded that much of the rhizosphere effect might be masked by the continued presence of dominant groups already present in soil. Many dominant bacterial groups might be dormant under particular rhizosphere conditions, but their presence would still be detected by DNA-based analyses. We therefore also targeted 16S rRNA, thought to give a first approximation of total cell activity (41), by performing RT-PCR analyses and compared the resulting DGGE profiles with those obtained by the DNA-targeted analysis. The band patterns produced when 16S rRNA templates were targeted were less complex than the DNA-derived profiles. The smaller number of bands in the RNA-based profiles suggests that several groups predominantly present in the rhizosphere were not dominant actively metabolizing groups in the rhizosphere, although differences due to inefficiency of reverse transcription cannot be ruled out. The RNA-derived bands were essentially a subset of the bands detected in the DNA-based analysis, suggesting that the bacterial populations that were responsible for the most activity also were populations that were numerically dominant (i.e., they accounted for ≥ 0.1 to 1% of the total community PCR product as determined by the methods of Heuer and Small [20] and Muyzer et al. [27]).
As observed with the rDNA-derived profiles, no dramatic shifts in the total DGGE profiles of the active bacterial populations were observed, although DGGE pattern comparisons did reveal some trends with respect to plant growth. The DGGE profiles derived from bulk soil RNA were also quite similar to the rhizosphere profiles, although they exhibited a higher background level and contained several additional bands. In contrast to the DNA-derived profiles, there was no clear clustering of root tip samples. However, there was clustering of all samples from the plants harvested after 2 weeks, regardless of the root region examined. This result was similar to that observed in a previous culture-based analysis in which it was suggested that young plants contained bacterial communities that were distinct from the bacterial communities contained by plants at other developmental stages (9). These results imply that a certain degree of succession occurs in the bacterial community of the rhizosphere during plant development.
In total, the results described above are only in partial agreement with the general hypotheses concerning bacterial dynamics in the rhizosphere. Young roots are known to excrete more organic material than older roots, which can result in different specific bacterial populations (4, 6, 23). Young roots might be suitable for so-called r-strategists, bacteria which in this case may be characterized by high growth rates and success under less crowded conditions. The niches provided by the roots of older plants may be occupied by so-called K-strategists, bacteria with lower growth rates that can compete in communities under crowded, substrate-limited conditions (1). Interestingly, the RNA-derived DGGE patterns were more similar to each other than the DNA-derived patterns were to each other (compare the horizontal distances in Fig. 2). One might expect the responses at the rRNA level to be more rapid and to have greater amplitude than those at the rDNA level, thus resulting in greater variation among rRNA-derived profiles. However, the opposite was observed, apparently due to the similar responses of specific bacterial populations in the rhizosphere. The somewhat tighter clustering of RNA-derived profiles may, however, be an artifact caused by the generally simplified nature of the profiles. In any case, the data do not support the notion that totally different bacterial populations are activated at different stages of chrysanthemum development or in different root regions. It may be that soil not only has a great influence on the dominant bacterial populations present in the rhizosphere (14) but also affects which bacteria become most active in the rhizosphere. Gelsomino et al. (14) recently found soil type to be an important determinant of bacterial community structure as evidenced by PCR-DGGE. However, these authors presented evidence obtained with only a single plant species, and it remains to be seen if other plants produce a more varied pattern of bacterial stimulation. The present study was also conducted with only a single plant species and a controlled pot cultivation system. It is not yet known how well such simple conditions mimic rhizosphere conditions in complex field situations, where roots from several plant species may be intertwined.
Sequencing of DGGE bands revealed that the majority of the dominant populations detected had 16S rDNA sequences that were most closely related to those of previously described soil bacteria (i.e., Acetobacter, Bacillus, Pseudomonas, and Arthrobacter) or unidentified bacteria detected as environmental clones. However, no sequence recovered was an exact match with a previously recovered sequence. Our results suggest that the activity of bacteria within the rhizosphere is rather strain or ecotype specific instead of determined at the genus level or a higher taxonomic level. For example, several Bacillus-like sequences were detected in the DNA-derived profiles, but only some of these sequences appeared to be derived from populations that were metabolically active in the rhizosphere. Furthermore, the taxonomic affiliations of the sequences detected by the RT-PCR approach covered a broad phylogenetic range. The high levels of detection of Bacillus-like sequences were in agreement with the results of Felske et al. (12), who found that Bacillus was the most predominantly active bacterium in grassland soils. Major taxonomic shifts were not observed in our study with respect to plant parameters or type of nucleic acid targeted. Rather, most profile differences were due the relative presence or activity of different bacterial populations in a particular genus.
One of the initial goals of this study was to gain insight into potential natural antagonists of Pythium, a major disease-causing oomycete in chrysanthemum. Identification of bacterial populations that are predominantly present and active in the chrysanthemum rhizosphere, particularly in the early developmental stages when Pythium is most damaging (25), might help focus future biocontrol efforts. In addition to the more recognized bacterial groups that have been identified as dominantly active in the chrysanthemum rhizosphere, populations related to Acidobacterium capsulatum were detected as some of the few predominantly active bacterial populations in young root samples. The Holophaga-Acidobacterium group was also detected in this study and has been shown previously to be active in grassland soils (12, 39). Actinomycete-related sequences were detected only at the DNA level, suggesting that they were not highly active in this system. Although the methods used proved to be highly reproducible, potential biases inherent in nucleic acid extraction and PCR amplification must kept in mind (5, 32, 35, 40). Thus, although this study helped provide insight into the dynamics of bacterial communities in the rhizosphere, it is clearly the combination of data such as the data which we obtained with data obtained by other approaches, including continued isolation of potentially antagonistic bacteria, that will lead to future advances in the biological defense against plant pathogens.
ACKNOWLEDGMENTS
We thank Klaas Vrieling, Dirk Passchie, and Bertie Joan van Heuven from the Institute of Evolutionary and Ecological Sciences for assistance with the DGGE band sequencing.
This project was sponsored by the Leiden University Institutes of Evolutionary and Ecological Sciences and of Molecular Plant Sciences and by Plant Research International, Wageningen, The Netherlands.
Footnotes
NIOO publication number 2706.
REFERENCES
- 1.Andrews J H, Harris R F. r- and K-selection and microbial ecology. In: Marshall K C, editor. Advances in microbial ecology. Vol. 9. New York, N.Y: Plenum Press; 1986. pp. 99–147. [Google Scholar]
- 2.Behrendt U, Ulrich A, Schumann P, Erler W, Burghardt J, Seyfarth W. A taxonomic study of bacteria isolated from grasses: a proposed new species, Pseudomonas graminis sp. nov. Int J Syst Bacteriol. 1999;49:297–308. doi: 10.1099/00207713-49-1-297. [DOI] [PubMed] [Google Scholar]
- 3.Bolton H, Jr, Frederickson J K, Elliott L F. Microbial ecology of the rhizosphere. In: Metting F B, editor. Soil microbial ecology. New York, N.Y: Marcel Dekker Inc.; 1992. pp. 27–63. [Google Scholar]
- 4.Bowen G D, Rovira A D. The rhizosphere, the hidden half. In: Waisel Y, Eshel A, Kafkafi U, editors. Plant roots—the hidden half. New York, N.Y: Marcel Dekker Inc.; 1991. pp. 641–649. [Google Scholar]
- 5.Chang Y-J, Stephen J R, Richter A P, Venosa A D, Bruggemann J, Macnaughton S J, Kowalchuk G A, Haines J R, Kline E, White D C. Phylogenetic analysis of aerobic freshwater and marine enrichment cultures efficient in hydrocarbon degradation: effect of profiling method. J Microbiol Methods. 2000;40:19–31. doi: 10.1016/s0167-7012(99)00134-7. [DOI] [PubMed] [Google Scholar]
- 6.Curl E A, Truelove B. The rhizosphere. Berlin, Germany: Springer Verlag; 1986. [Google Scholar]
- 7.Duarte G F, Rosado A S, Sedin L, Keijzer-Wolters A C, van Elsas J-D. Extraction of ribosomal RNA and genomic DNA from soil for studying the diversity of the indigenous bacterial community. J Microbiol Methods. 1998;32:21–29. [Google Scholar]
- 8.Duineveld B M, Rosado A S, van Elsas J D, van Veen J A. Analysis of the dynamics of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns. Appl Environ Microbiol. 1998;64:4950–4957. doi: 10.1128/aem.64.12.4950-4957.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duineveld B M, Van Veen J A. The number of bacteria in the rhizosphere during plant development: relating colony-forming units to different reference units. Biol Fertil Soils. 1999;28:285–291. [Google Scholar]
- 10.Fani R, Bandi C, Bazzicalupo M, Ceccherini M T, Fancelli S, Gallori E, Gerace L, Grifoni A, Miclaus N, Damiani G. Phylogeny of the genus Azospirillum based on 16S rDNA sequences. FEMS Microbiol Lett. 1995;129:195–200. doi: 10.1111/j.1574-6968.1995.tb07579.x. [DOI] [PubMed] [Google Scholar]
- 11.Felske A, Rheims H, Wolterink A, Stackebrandt E, Akkermans A D L. Ribosome analysis reveals prominent activity of an uncultured member of the class actinobacteria in grassland soils. Microbiology. 1997;143:2983–2989. doi: 10.1099/00221287-143-9-2983. [DOI] [PubMed] [Google Scholar]
- 12.Felske A, Wolterink A, Van Lis R, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferris M J, Muyzer G, Ward D J. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelsomino A C, Keijzer-Wolters A C, Gacco G, Van Elsas J-D. Assessment of bacterial community structure in soil by polymerase chain reaction and denaturing gradient gel electrophoresis. J Microbiol Methods. 1999;38:1–15. doi: 10.1016/s0167-7012(99)00054-8. [DOI] [PubMed] [Google Scholar]
- 15.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 16.Grayston S J, Wang S Q, Campbell C D, Edwards A C. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol Biochem. 1998;30:369–378. [Google Scholar]
- 17.Griffiths B S, Ritz K, Ebblewhite N, Dobson G. Soil microbial community structure: effects of substrate loading rates. Soil Biol Biochem. 1999;31:145–153. [Google Scholar]
- 18.Hale M G, Moore L D, Griffin G J. Root exudates and exudation. In: Dommergues Y R, Krupa S V, editors. Interactions between non-pathogen soil microorganisms and plants. Amsterdam, The Netherlands: Elsevier; 1978. pp. 163–203. [Google Scholar]
- 19.Hanada S, Shigematsu T, Shibuya K, Eguchi M, Hasegawa T, Suda Y, Kamagata Y, Kanagawa T, Kurare R. Phylogenetic analysis of trichloroethylene-degrading strains newly isolated from polluted soil with the contaminant. J Ferment Bioeng. 1998;80:539–544. [Google Scholar]
- 20.Heuer H, Smalla K. Application of denaturing gradient gel electrophoresis and temperature gradient gel electrophoresis for studying soil microbial communities. In: Van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker Inc.; 1997. pp. 353–373. [Google Scholar]
- 21.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:6793–6774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch C, Klatte S, Kroppenstedt R, Schumann P, Stackebrandt E. Reclassification of Arthrobacter picolinophilus as Rhodococcus erythropolis. Int J Syst Bacteriol. 1995;45:837–839. doi: 10.1099/00207713-45-4-837. [DOI] [PubMed] [Google Scholar]
- 23.Liljeroth E, Burgers S L G E, Van Veen J A. Changes in bacterial populations along roots of wheat seedlings. Biol Fertil Soils. 1991;10:276–280. [Google Scholar]
- 24.Lynch J M, Whipps J M. Substrate flow in the rhizosphere. Plant Soil. 1990;129:1–10. [Google Scholar]
- 25.Martin F N. Pythium. In: Kohmoto K, Singh U S, Singh R P, editors. Pathogenesis and host specificity in plant diseases. Vol. 2. Oxford, United Kingdom: Pergamon Press; 1995. pp. 17–36. [Google Scholar]
- 26.Moran M A, Torsvik V L, Torsvik T, Hodson R E. Direct extraction and purification of rRNA for ecological studies. Appl Environ Microbiol. 1993;59:915–918. doi: 10.1128/aem.59.3.915-918.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neal J R, Jr, Atkinson T G, Larson R I. Changes in the rhizosphere microflora of spring wheat induced by disomic substitution of a chromosome. Can J Microbiol. 1970;16:153–158. doi: 10.1139/m70-027. [DOI] [PubMed] [Google Scholar]
- 29.Neal J R, Jr, Larson R I, Atkinson T G. Changes in rhizosphere populations of selected physiological groups of bacteria related to substitution of specific pairs of chromosomes in spring wheat. Plant Soil. 1973;39:209–212. [Google Scholar]
- 30.Nielsen P, Rainey F A, Outtrop H, Priest F G, Fritze D. Comparative 16S rDNA sequence analysis of some alkaliphilic bacilli and the establishment of a sixth rRNA group within the genus Bacillus. FEMS Microbiol Lett. 1994;117:61–66. [Google Scholar]
- 31.Pearson W R, Lipman D J. Improved tools for biological sequence analysis. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polz M F, Cavanaugh C M. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosado A S, Duarte G R, Seldin L, Van Elsas J D. Genetic diversity of nifH gene sequences in Paenibacillus azotofixans strains and soil samples analyzed by denaturing gradient gel electrophoresis of PCR-amplified gene fragments. Appl Environ Microbiol. 1998;64:2770–2779. doi: 10.1128/aem.64.8.2770-2779.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozak D B, Colwell R R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1978;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swinnen J, Van Veen J A, Merckx R. 14-C pulse labeling of field-grown spring wheat: an evaluation of its use in rhizosphere carbon budget estimations. Soil Biol Biochem. 1994;26:161–170. [Google Scholar]
- 37.Torsvik V, Salte K, Sorheim R, Goksoyr J. Comparison of phenotypic diversity and DNA heterogenity in a population of soil bacteria. Appl Environ Microbiol. 1990;56:776–781. doi: 10.1128/aem.56.3.776-781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker Inc.; 1997. [Google Scholar]
- 39.Van Veen J A, Van Overbeek L S, Van Elsas J D. Fate and activity of microorganisms introduced in soil. Microbiol Mol Biol Rev. 1997;61:121–135. doi: 10.1128/mmbr.61.2.121-135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Von Wintzingerode F, Goebel E B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 41.Wagner R. The regulation of ribosomal RNA synthesis and bacterial cell growth. Arch Microbiol. 1994;161:100–106. doi: 10.1007/BF00276469. [DOI] [PubMed] [Google Scholar]
- 42.Ward-Rainey N, Rainey F A, Schlesner H, Stackebrandt E. Assignment of hitherto unidentified 16S rDNA species to a main line of descent within the domain Bacteria. Microbiology. 1995;141:3247–3250. [Google Scholar]