Abstract
We examined the clinical features and pathology, and identified the causative mutation, of osteogenesis imperfecta in a 2-mo-old kitten with growth retardation and abnormal gait. Blood and radiographic examinations were performed on presentation. Radiographs revealed decreased opacity of numerous bones. Fractures were observed in some long bones, including femur and tibia. Histologic examination of the tibia showed decreased osteoid and osteoblasts at the primary spongiosa extending from the growth plate. The periosteum was thickened, and cortical bone and osteoblasts were decreased. Consequently, osteogenesis imperfecta was diagnosed. Genomic DNA and total RNA were extracted from the skin and used for PCR. Whole-genome sequencing identified a 2-bp deletion (c.370_371delTG; p.C124fs), which resulted in a homozygous frameshift mutation on exon 3 of CREB3L1. This mutation introduced a premature stop codon, suggesting production of the truncated protein without a functional domain as a transcription factor for expression of COL1A1 mRNA. This error may have affected collagen fibril formation, leading to the development of osteogenesis imperfecta.
Keywords: cats, CREB3L1, mutation, osteogenesis imperfecta, osteopenia
Osteogenesis imperfecta (OI) is a congenital connective tissue disease characterized by generalized bone fragility. In humans, OI is classified into 4 groups based on clinical features and putative inheritance. 7 Patients with type I have blue sclera and moderate bone fragility; type II is lethal in the perinatal period; type III patients have white sclera and severe progressive deformity; and type IV patients have white sclera and variable bone fragility. 7 There are >20 categories of OI based on molecular, biochemical, and/or radiographic criteria. 7 Mutations have been identified in numerous genes that are related to the type I collagen molecule.4,7 The mode of inheritance is different in each type of OI. 2
Collagen fiber abnormalities are common in each OI type. Type I collagen comprises one α1 chain and two α2 chains, which form a triple-helix structure. Type I collagen is a major component of bones, tendons, and most connective tissues. 21 In patients with OI, the basic bone construct based on type I collagen cannot be formed as a result of collagen fibril abnormalities or deficiency. 15
OI or suspected OI has been reported in many animal species, including dogs, 6 cattle, 4 sheep, 1 and cats.8,11,23 Previous studies have reported causative mutations in COL1A1 (dog, cattle) and COL1A2 (dog).4,6,14,19 In another type of canine OI with spontaneous fractures of bones and teeth, joint hyperlaxity SERPINH1 mutations have been identified.10,16 In cattle, severe bone and dental fragility and blue sclera have been reported, with evidence of fractures in utero. 18 In sheep, OI has been reported to be lethal, and clinical features included multiple intrauterine fractures, subcutaneous edema, joint laxity, blue sclera, and pink teeth. 1 In cats, several studies have reported bone fragility,8,11,23 which was radiographically or microscopically diagnosed as OI. Mature cortical and trabecular bone was decreased, similar to observations in human patients with OI. 11 However, to our knowledge, causative mutations have not been reported in feline OI.
We present here the case of a domestic cat with growth retardation and bone fragility that was radiographically and histologically diagnosed as OI. Moreover, we identified a 2-bp deletion mutation in CREB3L1, which is involved in COL1A1 expression, suggesting a causative mutation in our case.
An ~8-wk-old male kitten was brought to the Nasunogahara Animal Clinic (Japan) because of lameness. The kitten was born to a stray cat and had obvious stunting and an abnormal gait. Blood and urine samples were obtained for hematologic and urine tests, and radiographic examination was performed. The owner abandoned the kitten because of a suspected congenital anomaly, and it was adopted by the author (M. Takanosu).
However, the kitten died 34 d after it was presented to our animal clinic. An autopsy was performed after the kitten’s death, and the whole body was immersed in 10% neutral-buffered formalin for fixation. After bone demineralization (Plank-Rychlo solution, Decalx; Jokoh), tissues were processed routinely, and 4-µm sections were stained with H&E.
A sample of skin was excised from the abdominal wall with sterilized scissors just after death of the kitten, immersed immediately in RNA Save (Biological Industries), and stored at −20°C for DNA extraction. Genomic DNA (gDNA) was extracted from the skin (High Pure PCR template preparation kit; Roche) according to the manufacturer’s instructions. DNA concentration was measured using a spectrophotometer (NanoDrop 1000; Thermo Fisher) and adjusted to 10 µg/µL with 10 mM Tris-HCl, pH 8.0. Total RNA was extracted from the skin (NucleoSpin TriPrep; Macherey-Nagel) following the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized from 1 µg of total RNA (ReverTra Ace qPCR RT master mix; Toyobo Life Science) containing oligo dT and random hexamer primers, and then stored at −20°C until use.
For whole-genome sequencing (WGS), 1 µg of gDNA was used for preparation of the library (TruSeq DNA PCR-free library prep; Illumina). The average insert size was 350 bp. Next-generation sequencing was performed (NovaSeq 6000; Illumina) with a standard 150-bp paired-end read protocol on the HiSeq X Ten platform (Illumina). The total read data was 90 Gb, and average alignment depth was 33. 5 Phred quality scores of 20 and 30 were calculated as 98.8% and 95.6%, respectively. FastQC (v.0.11.6) was used for the quality check for sequencing data (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Burrows-Wheeler Aligner (v.0.7.17) was used for aligning reads to reference sequences (http://bio-bwa.sourceforge.net). Picard (v.2.17.2) was used to examine aligned records in the supplied sequence alignment map (SAM) or binary format for the SAM file to locate duplicate molecules (http://broadinstitute.github.io/picard/). SAMTools was used to identify variants (http://samtools.sourceforge.net). SnpEff was used for annotating the possible effects of variants (http://snpeff.sourceforge.net/). The feline genome obtained from a database (https://www.ncbi.nlm.nih.gov/assembly/GCF_000181335.3/) was used as a reference. All steps for WGS and data analysis were performed at Macrogen Japan (https://www.macrogen-japan.co.jp). The data obtained were visualized using Genome Browser (https://genome.ucsc.edu). Single nucleotide variants were analyzed using a prediction software of functional effects (PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/).
The forward and reverse primers for detection of a CREB3L1 mutation in gDNA were as follows: 5′-AGTCCCCT GAGCCTGGTC-3′ and 5′-CATGGCTGAGGAGGCGG-3′, respectively. The primers for reverse-transcription PCR (RT-PCR) were as follows: TTTCTTTGACGACCCTGTGCT-3′ and 5′-CTCCTGCTTCACCATGACGG-3′. PCR was performed in a total volume of 10 µL containing 1 µL of 10 × PCR buffer, 1 µL of 2 mM deoxynucleotide triphosphates, 1 µL of each forward and reverse primer (2 µM), and 0.5 units of Taq DNA polymerase (Blend Taq plus; Toyobo Life Science). The PCR cycling conditions were as follows: initial denaturation at 95°C for 60 s, 35 cycles of 94°C for 20 s, 60°C for 20 s, and 72°C for 20 s. The PCR products were analyzed using a capillary electrophoresis system with a high-resolution cartridge (Qsep100; BiOptic). For sequencing analysis, these products were treated (ExoSAP-IT; Thermo Fisher) and analyzed on a DNA sequencer (ABI 3730xl DNA analyzer; Thermo Fisher) at Macrogen.
Genotyping of the CREB3L1 mutation was performed on the PCR allelic competitive extension (PACE) system (3CR Bioscience) using 136 random feline blood samples obtained for blood examination for diagnosis or medical check-up. We used 2 allele-specific primers for the wild-type (5′-GAAGGTGAC CAAGTTCATGCTGGGTGCTGGGACACAAACTGT-3′) and mutant (5′-GAAGGTCGGAGTCAACGGATTGGGT GCTGGGACACAAACTGC-3′), as well as one common reverse primer (5′-CTCCGGGCTCTGCTCCTGC-3′). PCR was performed in a total volume of 8 µL containing 1× PACE master mix containing 10 ng of the gDNA, 0.1 µL of each allele-specific primer (12 µM), and 0.1 µL of the reverse primer (30 µM) on a real-time PCR instrument (MyGo Mini; IT-IS Life Science). The PCR conditions were as follows: after initial denaturation at 94°C for 15 min, 10 cycles of 94°C for 15 s, 65°C–62°C with −0.3°C in each cycle, 30 cycles of 94°C for 15 s, and 62°C for 40 s.
The body weight of the kitten was 600 g, which was obviously lower than that of a littermate (1.1 kg). The kitten had reduced agility and toddled (Suppl. Video). All teeth were normal in appearance, and the color of sclera was also normal. Joint laxity and skin fragility were not observed. Although complete blood count and serum chemistry findings were within RIs, the parathyroid hormone (PTH) concentration was slightly lower than the RI, and the creatinine concentration was low, which may have been a result of poor body condition (Suppl. Table 1). Calcium, ionized-calcium, and phosphorus concentrations were within their RIs. Although the kitten had a normal appetite and activity, it experienced difficulty in defecating because of pelvic stenosis. Constipation was not improved by 0.2 mL of lactulose administration PO q12h. An enema with water at 37°C was performed to prompt defecation. Although defecating was improved slightly, constipation continued. Bacterial peritonitis developed as a result of colon perforation, resulting in spontaneous death on day 34.
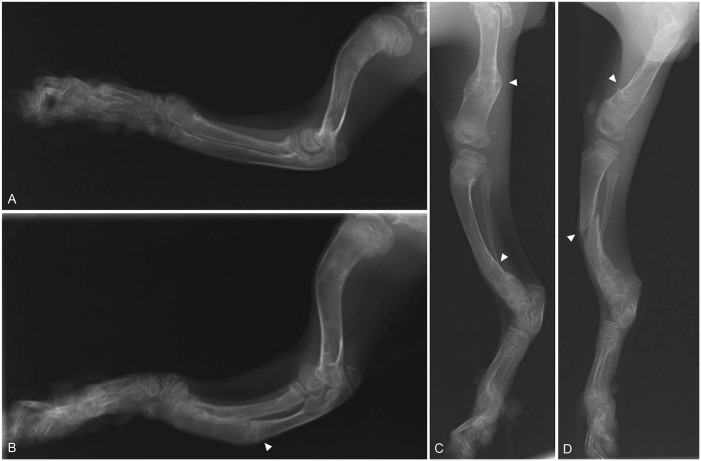
There was increased radiolucency of bones throughout the body (Fig. 1). The pelvis was extremely narrow. Healing or healed fractures were observed in the ribs and the diaphysis of several long bones, including the femurs, tibias, and ulnas. Cortices of long bones were thinned. The right and left humeri were bent forward at the proximal end and laterally at the distal diaphysis (Fig. 2A, 2B). The left ulna was fractured in the middle of the diaphysis. The left radius and ulna were bent caudally with respect to the right radius. The digits were not deformed; however, they had increased radiolucency. Similar to the forelimbs, the hindlimbs had abnormalities, specifically cortical bone thinning as well as increased radiolucency and fractures (Fig. 2C, 2D). These radiographic observations were strongly suggestive of OI or fibrous osteodystrophy.
Figure 1.
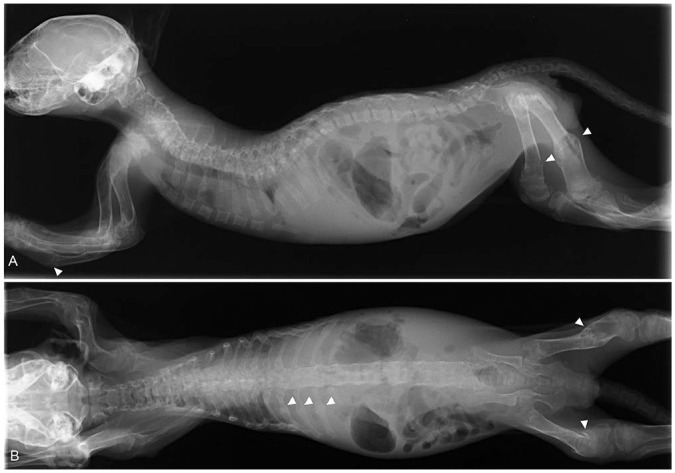
Whole-body radiographic images of a kitten with severe osteogenesis imperfecta. A. Lateral view. B. Ventral-dorsal view. There is increased bone radiolucency in the whole body. Arrowheads indicate bone fractures.
Figure 2.
Radiographic images of the forelimbs and hindlimbs of a kitten with severe osteogenesis imperfecta. A. Lateral view of the right forelimb. B. Lateral view of the left forelimb. The radius and ulna are curved with respect to the right. A fracture can be seen in the ulna (arrowhead). C. Lateral view of the right hindlimb. D. Lateral view of the left hindlimb. There are bilateral femoral and tibial fractures (arrowheads).
Autopsy examination of long bones revealed a remarkable decrease in cancellous bone in the diaphysis and metaphysis. Histologic findings revealed that long bones were affected mainly, but not the tendons, teeth, or alveolar, mandibular, or cranial bones. Growth plate morphology was normal, and chondrocytes were regularly aligned with normal differentiation. The primary spongiosa was thinner than that in a normal kitten (Fig. 3A, 3B). Osteoid was remarkedly decreased at the primary spongiosa compared with normal (Fig. 3C, 3D). Osteoblasts were not observed on trabecular surfaces in the primary spongiosa. Secondary spongiosa was absent in the metaphyses. The cortex of long bones was thinner, and osteoid was remarkably decreased compared with a normal kitten; a small amount of cartilage was present in the cortical bone, and a few osteoblasts were observed at the thickened outer periosteum (Fig. 3E, 3F). Abnormalities were not evident in the teeth (Fig. 3G). Dentin and alveolar bone appeared normal. Although the morphology of the vertebral growth plates was normal, the primary spongiosa was thin and obvious osteoblasts were not observed on trabecular surfaces in the primary spongiosa. No morphologic abnormalities were observed in the patellar tendons or the sclera.
Figure 3.
Histologic sections of the tibia of a kitten with severe osteogenesis imperfecta. A, B. The growth plate of the case (A) is intact; however, the amount of osteoid at the primary spongiosa is decreased compared with the control normal kitten (B). C. High magnification of the primary spongiosa of the case. A tiny amount of osteoid is observed on trabecular surfaces in the primary spongiosa, and a few flattened and abnormally shaped osteoblasts are present (arrowhead). D. The primary spongiosa of the control normal kitten. Osteoblasts align in a simple layer and osteoid can be seen (double arrowheads). E. Cortical bone of the case is decreased, and osteoblasts are not observed at the periosteum (po). The cambium layer (C) is prominent. F. The cortical bone of the control normal kitten is thick, and osteoblasts can be seen at the periosteum (po). G. A part of the tooth and alveolar bone of the case. The dentin (d) and alveolar bone (al) are normal. H&E. A, B: bar = 100 µm; C, D: bar = 25 µm; E–G: bar = 200 µm.
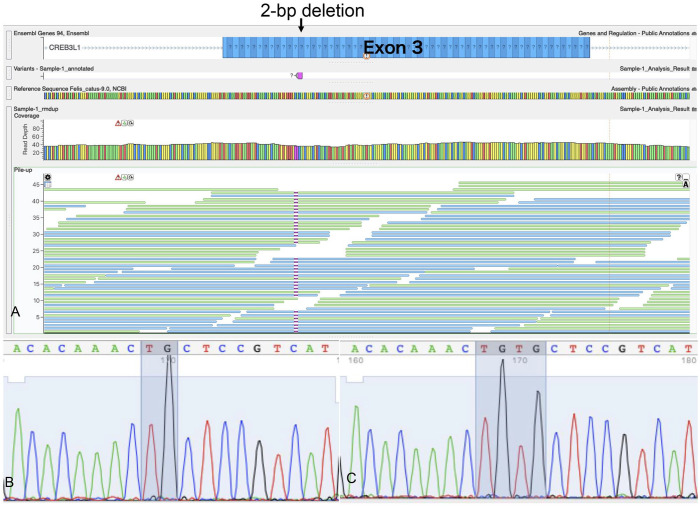
Using WGS, genes responsible for human OI were examined for variants. Although several missense mutations were found on P3H1, PPIB, FKBP10, TMEM38B, and MBTPS2, all were ruled out as a mutation that might be responsible for our case based on PolyPhen-2 analysis of the altered amino acid sequence. WGS revealed a 2-bp deletion in exon 3 of CREB3L1 (c.370_371delTG). This mutation was detected in all sequenced contigs except for the vector end, which indicated homozygosity (Fig. 4A). The 2-bp deletion mutation caused a frameshift and induced a premature stop codon (p.C124fs). Sequencing of the PCR product from RT-PCR revealed a 2-bp deletion in the cDNA of CREB3L1 (Fig. 4B, 4C) as well. All of the 136 cats who underwent genotyping of the 2-bp deletion of CREB3L1 were wild-type (Suppl. Fig. 1; Suppl. Table 2).
Figure 4.
A. Whole-genome sequencing of DNA obtained from our case of feline osteogenesis imperfecta (OI) with a 2-bp deletion on exon 3 of CREB3L1. All contigs except vector ends show a 2-bp deletion, which suggests that the mutation is homozygous. B, C. Sanger sequencing of RT-PCR products from the patient and a control cat. B. cDNA sequence of a part of the CREB3L1 exon 3 amplified from the cat with OI showing deletion TG (shaded) compared with that from a control cat. C. cDNA sequence of the control cat. TGTG is shaded.
Radiographically, the long bones of this kitten were deformed, and cortices were thin. There were several fractures, including new, healing, or healed fractures, suggesting OI or fibrous osteodystrophy, with secondary hyperparathyroidism as a possible diagnosis. Although PTH was slightly decreased from the RI in our case, this may be an artifact because the sample spent a few days in transportation to an external laboratory. 3 The appetite of the kitten was good; commercial cat food was being fed. Primary and secondary hyperparathyroidism or nutritional osteoporosis were ruled out. OI was the most likely diagnosis.
Histologic examination revealed impairment of both membranous and endochondral ossification; lesions noted resembled those reported for canine or feline OI.11,19,20 We observed no abnormalities in the teeth, sclera, or tendon, which differed from other cases. Our case corresponds most closely with type III human OI, which is characterized by white sclera with severe progressive deformity, and includes CREB3L1 as a causative mutation. 7 However, there are few reports in regard to feline OI; therefore, further studies are needed for the classification of OI in cats.
Human OI16 is caused by a CREB3L1 mutation. 22 Patients with OI16 are characterized by bone fragility and a high risk of fractures. 12 Three OI16 mutation types have been identified in humans: 3-bp deletion (p.L312del), 13 nonsense mutation (p.Y428X), 15 and missense mutation (p.A304V). 12 The basic-leucin zipper domain that plays a role in binding to DNA is located at the 291–381 amino acid position. 9 Therefore, p.L312del and p.A304V disrupt the functional domain.12,13 This disruption has a negative effect on osteoblast differentiation and COL1A1 transcription. Although p.Y428X is outside the basic-leucin zipper domain, the truncated molecule loses the function as a transcription factor for COL1A1 in the osteoblast. 15 Similarly, Creb3l1 knockout mice have growth retardation, osteopenia, and a high risk of fractures in homozygous mutants. 17 Moreover, Creb3l1 is critical for bone formation through transcription factor binding to the promoter region of Col1a1. 17 Human patients with OI16 and knockout mice with CREB3L1 have the homozygous mutation, which renders the functional protein completely absent.15,17 In our case, c.370_371delTG on CREB3L1 exon 3, which was homozygous, as in humans and mice, may lose the basic-leucin zipper domain because of the premature stop codon. Therefore, we suggest that our kitten had decreased COL1A1 mRNA expression and osteoblast differentiation. Taken together with histologic findings, we concluded that the 2-bp deletion in CREB3L1 was the mutation responsible for OI in our case.
Our study was limited by the lack of trio analysis to confirm whether the parents of the kitten carried the mutation. Our kitten had a homozygous mutation, which indicates an autosomal recessive trait. To confirm the inheritance of the disease as an autosomal recessive trait, the genotype of the parents should have been analyzed. Therefore, we cannot conclude that this disease is inherited. If the disease is autosomal recessive, the gene frequency of the mutation is extremely low, given that genotyping in 136 random cats did not reveal the mutation. A stray cat moves outdoors freely during pregnancy; the dam of this kitten might have been exposed to mutagenic substances. We cannot rule out the possibility that the c.370_371delTG is a de novo mutation.
Supplemental Material
Supplemental material, sj-pdf-1-vdi-10.1177_10406387221081227 for Severe osteogenesis imperfecta caused by CREB3L1 mutation in a cat by Masamine Takanosu and Yumiko Kagawa in Journal of Veterinary Diagnostic Investigation
Footnotes
Declarations of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Masamine Takanosu  https://orcid.org/0000-0002-5500-7811
https://orcid.org/0000-0002-5500-7811
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Masamine Takanosu, Nasunogahara Animal Clinic, Ohtawara, Tochigi, Japan (Takanosu).
Yumiko Kagawa, North Lab, Hondori, Sapporo, Hokkaido, Japan (Kagawa).
References
- 1. Arthur DG, et al. Lethal osteogenesis imperfecta and skin fragility in newborn New Zealand Romney lambs. N Z Vet J 1992;40:112–116. [DOI] [PubMed] [Google Scholar]
- 2. Balasubramanian M, et al. Osteogenesis imperfecta: ultrastructural and histological findings on examination of skin revealing novel insights into genotype-phenotype correlation. Ultrastruct Pathol 2016;40:71–76. [DOI] [PubMed] [Google Scholar]
- 3. Barber PJ. Disorders of the parathyroid glands. J Feline Med Surg 2004;6:259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bourneuf E, et al. Rapid discovery of de novo deleterious mutations in cattle enhances the value of livestock as model species. Sci Rep 2017;7:11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Byers PH, et al. Osteogenesis imperfecta: translation of mutation to phenotype. J Med Genet 1991;28:433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Campbell BG, et al. Canine COL1A2 mutation resulting in C-terminal truncation of pro-alpha2(I) and severe osteogenesis imperfecta. J Bone Miner Res 2001;16:1147–1153. [DOI] [PubMed] [Google Scholar]
- 7. Chetty M, et al. The evolution of the nosology of osteogenesis imperfecta. Clin Genet 2021;99:42–52. [DOI] [PubMed] [Google Scholar]
- 8. Cohn LA, Meuten DJ. Bone fragility in a kitten: an osteogenesis imperfecta-like syndrome. J Am Vet Med Assoc 1990;197:98–100. [PubMed] [Google Scholar]
- 9. Denard B, et al. The membrane-bound transcription factor CREB3L1 is activated in response to virus infection to inhibit proliferation of virus-infected cells. Cell Host Microbe 2011;10:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drögemüller C, et al. A missense mutation in the SERPINH1 gene in Dachshunds with osteogenesis imperfecta. PLoS Genet 2009;5:e1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evason MD, et al. Suspect osteogenesis imperfecta in a male kitten. Can Vet J 2007;48:296–298. [PMC free article] [PubMed] [Google Scholar]
- 12. Guillemyn B, et al. A homozygous pathogenic missense variant broadens the phenotypic and mutational spectrum of CREB3L1-related osteogenesis imperfecta. Human Mol Genet 2019;28:1801–1809. [DOI] [PubMed] [Google Scholar]
- 13. Keller RB, et al. Monoallelic and biallelic CREB3L1 variant causes mild and severe osteogenesis imperfecta, respectively. Genet Med 2018;20:411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Letko A, et al. A de novo in-frame duplication in the COL1A2 gene in a Lagotto Romagnolo dog with osteogenesis imperfecta. Anim Genet 2019;50:786–787. [DOI] [PubMed] [Google Scholar]
- 15. Lindahl K, et al. Homozygosity for CREB3L1 premature stop codon in first case of recessive osteogenesis imperfecta associated with OASIS-deficiency to survive infancy. Bone 2018;114:268–277. [DOI] [PubMed] [Google Scholar]
- 16. Lindert U, et al. Molecular consequences of the SERPINH1/HSP47 mutation in the Dachshund natural model of osteogenesis imperfecta. J Biol Chem 2015;290:17679–17689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murakami T, et al. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol 2009;11:1205–1211. [DOI] [PubMed] [Google Scholar]
- 18. Petersen JL, et al. Evidence for a de novo, dominant germ-line mutation causative of osteogenesis imperfecta in two Red Angus calves. Mamm Genome 2019;30:81–87. [DOI] [PubMed] [Google Scholar]
- 19. Quist EM, et al. Identification of a candidate mutation in the COL1A2 gene of a Chow Chow with osteogenesis imperfecta. J Hered 2018;109:308–314. [DOI] [PubMed] [Google Scholar]
- 20. Seelinger F, et al. Osteogenesis imperfecta in two litters of dachshunds. Vet Pathol 2003;40:530–539. [DOI] [PubMed] [Google Scholar]
- 21. Steiner RD, Basel D. COL1A1/2 osteogenesis imperfecta. Rev. 2021 May 6. In: Adam MP, ed. Gene Reviews [Internet]. University of Washington. https://www.ncbi.nlm.nih.gov/books/NBK1295/ [Google Scholar]
- 22. Symoens S, et al. Deficiency for the ER-stress transducer OASIS causes severe recessive osteogenesis imperfecta in humans. Orphanet J Rare Dis 2013;8:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Won S, et al. Clinical application of quantitative computed tomography in osteogenesis imperfecta-suspected cat. J Vet Sci 2017;18:415–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-vdi-10.1177_10406387221081227 for Severe osteogenesis imperfecta caused by CREB3L1 mutation in a cat by Masamine Takanosu and Yumiko Kagawa in Journal of Veterinary Diagnostic Investigation