Abstract
The stemness of cancer cells contributes to tumorigenesis, the heterogeneity of malignancies, cancer metastasis, and therapeutic resistance. However, the roles and regulatory mechanisms maintaining stemness among breast cancer subtypes remain elusive. Our previous studies have demonstrated that ectopic expression and dynamic alteration of the mesenchymal transcription factor forkhead box F2 (FOXF2) differentially regulates breast cancer progression and metastasis organotropism in a cell subtype-specific manner. Here, we reveal the underlying mechanism by which FOXF2 enhances stemness in luminal breast cancer cells but suppresses that in basal-like breast cancer (BLBC) cells. We show that luminal breast cancer and BLBC cells with FOXF2-regulated stemness exhibit partial mesenchymal stem cell properties that toward osteogenic differentiation and myogenic differentiation, respectively. Furthermore, we show that FOXF2 activates the Wnt signaling pathway in luminal breast cancer cells but represses this pathway in BLBC cells by recruiting nuclear receptor coactivator 3 (NCoA3) and nuclear receptor corepressor 1 (NCoR1) to the promoters of Wnt family member 2B (WNT2B) and frizzled class receptor 1 (FZD1) genes to activate and repress their transcription, respectively. We propose that targeting the Wnt signaling pathway is a promising strategy for the treatment of breast cancers with dysregulated expression of FOXF2.
Keywords: FOXF2, Wnt signaling pathway, WNT2B, FZD1, NCoR1, NCoA3, stemness, luminal breast cancer, basal-like breast cancer, metastasis
Abbreviations: BLBC, basal-like breast cancers; cDNA, complementary DNA; ChIP, chromatin immunoprecipitation; CSC, cancer stem cell; DMFS, distant metastasis-free survival; EMT, epithelial–mesenchymal transition; EMyoT, epithelial–myogenic transition; EOT, epithelial–osteogenic transition; ER, estrogen receptor; FBS, fetal bovine serum; FCM, flow cytometric; qPCR, quantitative PCR; TF, transcription factor; TNBC, triple-negative breast cancers
Breast cancer is a highly heterogeneous disease that can be classified into several intrinsic subtypes with distinct behavior and clinical implications based on molecular profiles (1, 2). Breast cancers most commonly metastasize to the bone, lung, and liver. The organ preference of metastasis is different among breast cancer subtypes. Luminal breast cancers are prone to metastasize to bone at advanced stage, while basal-like breast cancers (BLBC)/triple-negative breast cancers (TNBC) tend to develop visceral metastasis at early stage (3, 4). Epithelial–mesenchymal transition (EMT) is a crucial cellular process for conferring cell plasticity and enabling the invasion-metastasis cascade. EMT involves multiple transition stages that cause cells to gradually progress from epithelial to completely mesenchymal. Cells in different stages of EMT display differences in differentiation status and metastatic potential (5, 6, 7), which is tightly linked with the balance between the maintenance of stemness properties and the regulation of differentiation.
Forkhead box F2 (FOXF2), a mesenchymal transcription factor (TF), plays critical roles in the maintenance of tissue homeostasis by promoting the differentiation of mesenchymal cells and controlling the mesenchymal transformation of adjacent epithelial cells (8). Our previous studies have demonstrated that ectopic expression and dynamic alteration of FOXF2 differentially regulates breast cancer progression and metastasis organotropism in a cell subtype-specific manner. FOXF2 overexpression promotes EMT/epithelial–osteogenic transition (EOT) and bone metastasis by pleiotropically transactivating bone-related genes in luminal breast cancer and BLBC cells (9) but represses EMT/epithelial–myogenic transition (EMyoT) and visceral metastasis by pleiotropically transrepressing genes encoding EMT-inducing TFs (10, 11, 12), TGF-βs, and miRNA182 (13) in BLBC cells. However, whether the mesenchymal stemness and differentiation direction of breast cancer cells that have undergone FOXF2-regulated EMT determine metastasis organotropism in a cell subtype-specific manner remain to be further explored. In addition, although the nuclear receptor corepressor 1 (NCoR1)–mediated transrepression function of FOXF2 in BLBC cells has been found in our previous studies (11, 13), the mechanism underlying the transactivation function of FOXF2 in luminal breast cancer cells is still unknown.
The Wnt signaling pathway is responsible for stemness and plasticity maintenance and promotes metastatic colonization in the bone (14, 15, 16) and visceral organs (17, 18, 19) in luminal breast cancer and BLBC cells. The canonical Wnt signaling pathway requires the binding of Wnt ligands to frizzled receptors and low-density lipoprotein receptor-related protein 5/6 (LRP5/6) coreceptors to transduce intracellular signaling via the β-catenin–T-cell-specific transcription factor/lymphoid enhancer–binding factor (TCF/LEF) signaling cascade. Dysregulation of Wnt ligands and frizzled receptors has been linked with cancer stemness properties, carcinogenesis, and metastasis (20), which are mediated by the transcriptional regulation of multiple genes by the Wnt/β-catenin signaling cascade, such as MYC (21), MMP9 (22) and VEGF (23). FOXF2 has been found to act as an upstream inhibitor of Wnt signaling in mesenchymal cells (24, 25). However, whether and how FOXF2 regulates the Wnt/β-catenin signaling pathway in luminal breast cancer and BLBC cells remains undefined.
In this study, we investigated the role of FOXF2 in regulating mesenchymal stemness and differentiation direction in luminal breast cancer and BLBC cells as well as the underlying mechanisms by which FOXF2 regulates the Wnt ligand and frizzled receptor of the Wnt/β-catenin signaling pathway. We further explored the transactivating and transrepressive functions of FOXF2 for its target genes of Wnt ligand and frizzled receptor in luminal breast cancer and BLBC cells and the underlying mechanisms by which FOXF2 recruits nuclear receptor coactivator and nuclear receptor corepressor, respectively. We also tested the therapeutic effects of Wnt signaling pathway inhibitors on breast cancers with FOXF2 dysregulation-driven metastasis.
Results
FOXF2 promotes stemness in luminal breast cancer cells but suppresses that in BLBC cells
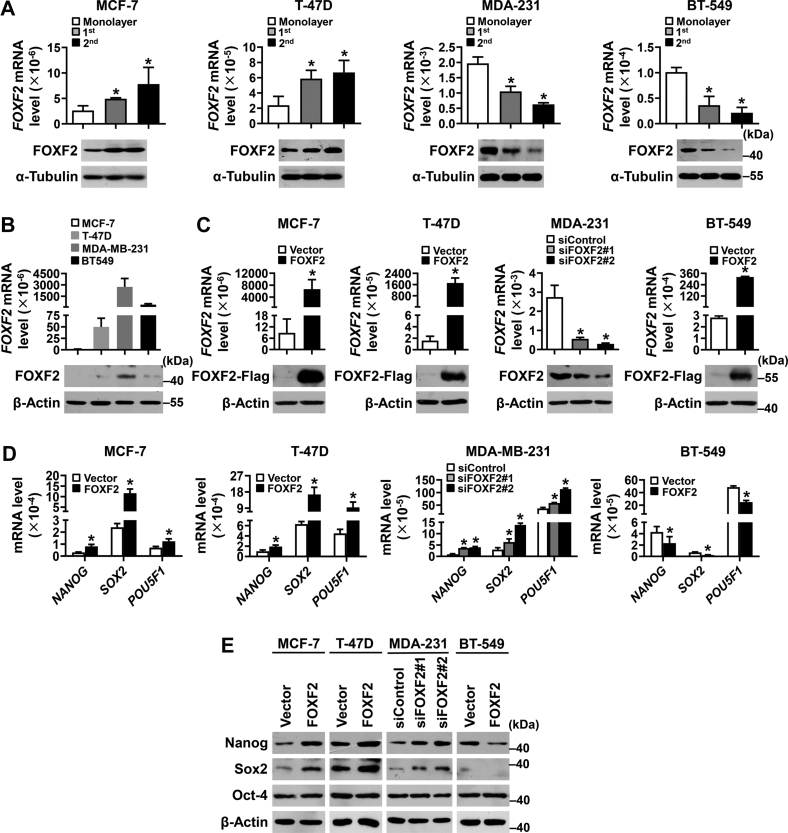
To investigate the role of FOXF2 in regulating the stemness of breast cancer cells, human luminal breast cancer cell lines MCF-7 and T-47D as well as BLBC cell lines MDA-MB-231 and BT-549 were subjected to sphere culture. FOXF2 mRNA and protein levels were shown to be upregulated in luminal breast cancer cell mammospheres but downregulated in BLBC cell mammospheres (Fig. 1A). Then, FOXF2 expression was forced in MCF-7, T-47D (with no or less FOXF2 expression), and BT-549 (with low FOXF2 expression) cells, and FOXF2 expression was knocked down in MDA-MB-231 cells (with higher FOXF2 expression; Fig. 1, B and C). The stem cell markers nanog homeobox (NANOG) and sex-determining region Y-box 2 (SOX2) but not POU class 5 homeobox 1 (POU5F1)/octamer-binding transcription factor 4 (OCT4) were positively regulated by FOXF2 at the mRNA and protein expression levels in luminal breast cancer cells and negatively regulated by FOXF2 in BLBC cells (Fig. 1, D and E). These results suggest that FOXF2 promotes the stemness of luminal breast cancer cells but suppresses the stemness of BLBC cells.
Figure 1.
FOXF2 enhances the stemness of luminal breast cancer cells but inhibits that of BLBC cells.A–C, the mRNA and protein levels of FOXF2 in the indicated cells were detected by RT-qPCR and immunoblot. 1st: first-generation mammospheres; 2nd: second-generation mammospheres. D and E, the mRNA (D) and protein (E) expression levels of stem cell–related markers in the indicated cells were detected by RT-qPCR and immunoblot. ∗, p < 0.05 compared with control cells. BLBC, basal-like breast cancers; RT-qPCR, reverse transcription-quantitative PCR.
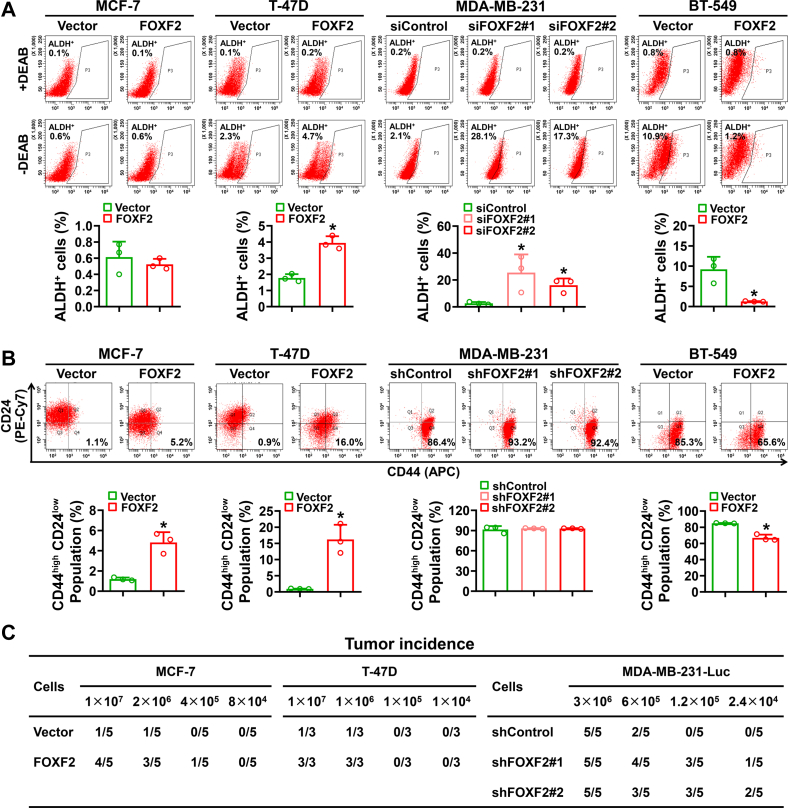
To investigate the cancer stem cell (CSC) phenotype of FOXF2-regulated luminal breast cancer and BLBC cells, the aldehyde dehydrogenase–positive (ALDH+) cell population and CD44high/CD24low cell population were assessed by flow cytometric (FCM) analysis. The results showed that FOXF2 positively regulated the ratio of ALDH+ and/or CD44high/CD24low populations in MCF-7 and T-47D luminal breast cancer cells but negatively regulated that in MDA-MB-231 and BT-549 BLBC cells (Fig. 2, A and B). The tumorigenesis ability assessed by an in vivo limiting dilution assay revealed that FOXF2 promoted the tumor-initiation capability of MCF-7 and T-47D cells in NOD/SCID mice and suppressed that of MDA-MB-231 cells in nude mice (Figs. 2C and S1). These results indicate that FOXF2 oppositely regulates stemness in luminal breast cancer and BLBC subtypes.
Figure 2.
FOXF2 positively regulates ALDH+and/or CD44high/CD24lowpopulations and tumor-initiating capacity in luminal breast cancer cells but negatively regulates those in BLBC cells.A and B, cell populations of ALDH+ (A) and CD44high/CD24low (B) were assessed by FCM analysis. ∗, p < 0.05 compared with control cells. C, the tumor-initiating capability of the indicated cells was assessed by an in vivo limiting dilution tumor formation assay. Table showing the incidences of xenograft tumors formed by inoculating gradient concentrations of the indicated cells in NOD/SCID mice or nude mice. BLBC, basal-like breast cancers; FCM, flow cytometric.
Luminal breast cancer and BLBC cells with FOXF2-regulated stemness exhibit partial mesenchymal stem cell properties that toward different directions of differentiation
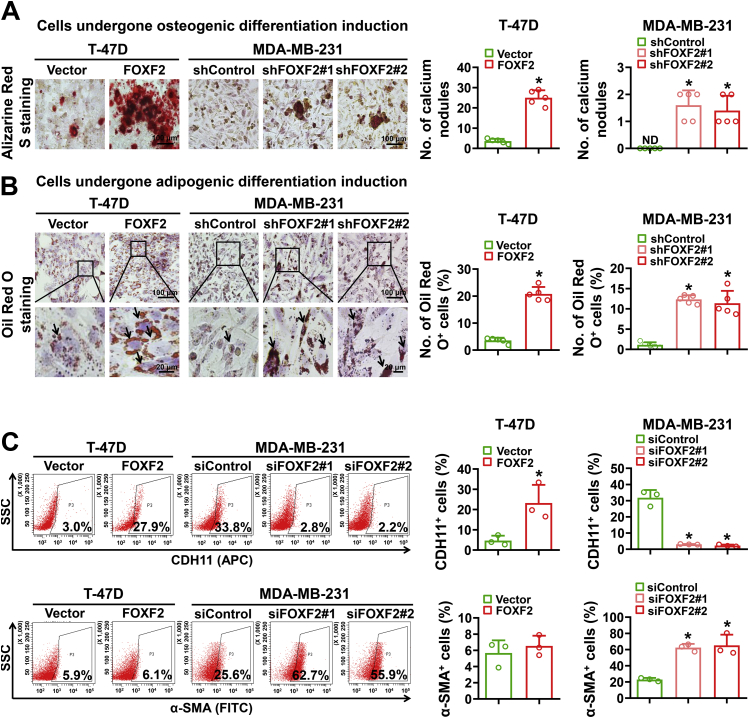
To further investigate whether breast cancer cells with FOXF2-regulated stemness have mesenchymal stem cell properties, T-47D cells with forced FOXF2 expression and MDA-MB-231 cells with FOXF2 knockdown as well as their control cells were induced toward osteogenic differentiation and adipogenic differentiation. The results showed that the differentiation capabilities toward osteogenesis and adipogenesis were significantly enhanced by forced FOXF2 expression in T-47D cells and by FOXF2 knockdown in MDA-MB-231 cells (Fig. 3, A and B). It is worth noting that the number and size of calcium nodules formed by T-47D cells with forced FOXF2 expression were greatly more than that formed by MDA-MB-231 cells with FOXF2 knockdown. Considering that ectopic overexpression of FOXF2 lead to EOT in luminal breast cancer and BLBC cells and FOXF2 deficiency results in EMyoT in BLBC cells, we detected the osteogenic marker cadherin 11 (CDH11) and the myofibroblastic marker actin alpha 2, smooth muscle/alpha smooth muscle actin (ACTA2/α-SMA) of these cells by FCM analysis. The results showed that FOXF2 positively regulated CDH11 expression in both T-47D and MDA-MB-231 cells, while negatively regulated α-SMA expression in MDA-MB-231 cells but not in T-47D cells (Fig. 3C). This result is consistent with our previous reports that both luminal breast cancer and BLBC cells with high FOXF2 expression present pro-osteogenic features and have a tendency to metastasize to bone (9), while FOXF2-deficient BLBC cells have a myofibroblast phenotype and preferentially metastasize to visceral organs (13). These results indicate that FOXF2 differentially regulates the transition of luminal breast cancer and BLBC cells into partial mesenchymal stem-like cells that toward different differentiation directions.
Figure 3.
Luminal breast cancer and BLBC cells regulated by FOXF2 exhibit partial mesenchymal stem cell properties toward osteogenic differentiation and myogenic differentiation, respectively.A and B, the osteogenic differentiation (A) and adipogenic differentiation (B) capabilities of the indicated cells were assessed by induction with osteogenic differentiation media for 21 days and adipogenic differentiation media for 25 days and then staining with Alizarin Red S and Oil Red O, respectively. The number of calcium nodules was counted and the percentage of Oil Red O+ cells was calculated. Arrow points to lipid droplet. C, CDH11+ and α-SMA+ cell populations were detected by FCM analysis. ND: not detected. ∗, p < 0.05 compared with control cells. BLBC, basal-like breast cancers; FCM, flow cytometric.
FOXF2 actives the Wnt signaling pathway in luminal breast cancer cells but represses that in BLBC cells through differential regulation of WNT2B and FZD1 transcription
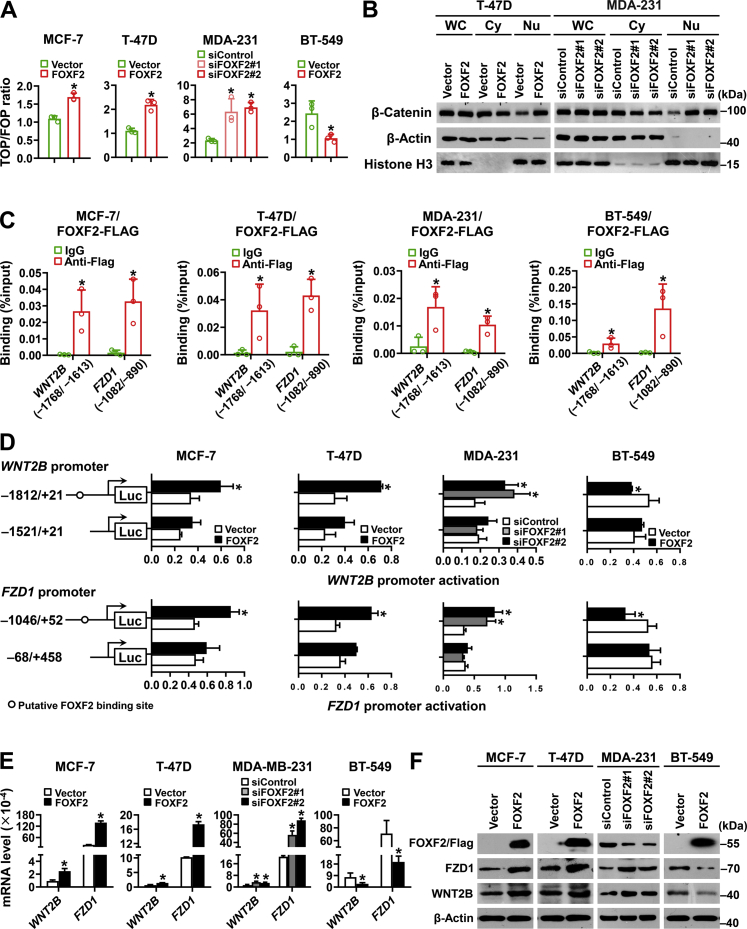
To investigate the mechanism by which FOXF2 differentially regulates stemness in luminal breast cancer and BLBC cells, Wnt signaling pathway activity regulated by FOXF2 was assessed in luminal breast cancer and BLBC cells using TOPflash/FOPflash luciferase reporter assays. The results revealed that FOXF2 positively regulated Wnt signaling pathway activity in MCF-7 and T-47D luminal breast cancer cells but negatively regulated that in MDA-MB-231 and BT-549 BLBC cells (Fig. 4A). Consistently, the nuclear distribution of β-catenin was significantly increased by forced expression of FOXF2 in T-47D luminal breast cancer cells but increased by FOXF2 knockdown in MDA-MB-231 BLBC cells (Fig. 4B). To further investigate the mechanism by which FOXF2 regulates the Wnt/β-catenin signaling pathway, putative FOXF2-binding sequences in the proximal promoter regions of genes encoding Wnt ligands and receptors were analyzed to identify candidate target genes. WNT2B and FZD1 were found to be candidate target genes of FOXF2 (Fig. S2A). Chromatin immunoprecipitation (ChIP)-PCR and ChIP-quantitative PCR (qPCR) assays confirmed that FOXF2 could bind to the proximal promoters of WNT2B and FZD1 containing the FOXF2-binding elements (Figs. S2B and 4C). Dual-luciferase reporter assays validated that FOXF2 positively regulated the activity of the WNT2B and FZD1 promoters containing the FOXF2-binding elements in luminal breast cancer cells and negatively regulated that in BLBC cells (Fig. 4D). Consistently, FOXF2 positively regulated the expression of WNT2B and FZD1 as well as their encoding proteins in luminal breast cancer cells and negatively regulated that in BLBC cells (Fig. 4, E and F). Wnt signaling pathway activity regulated positively by FOXF2 in luminal breast cancer cells and regulated negatively by FOXF2 in BLBC cells could be rescued through the knockdown or overexpression of WNT2B and FZD1 (Fig. S2, C and D). These results demonstrated that FOXF2 oppositely regulates Wnt signaling pathway activity through differential regulation of WNT2B and FZD1 transcription in luminal breast cancer and BLBC cells.
Figure 4.
FOXF2 activates the Wnt signaling pathway in luminal breast cancer cells but inhibits that in BLBC cells by directly targeting WNT2B and FZD1.A, the indicated cells were cotransfected with TOP/FOP flash and Renilla pRL-TK plasmids for 48 h and then subjected to a dual-luciferase reporter assay to detect Wnt signaling pathway activity. The ratio of TOPflash to FOPflash was analyzed. B, nuclear translocation of β-catenin in the indicated cells was analyzed by immunoblot. WC: whole-cell extracts; Cy: cytoplasmic extracts; Nu: nuclear extracts. C, the binding of FOXF2 to the WNT2B and FZD1 promoters containing the putative binding sites in the indicated cells transfected with FOXF2-FLAG plasmids was assessed by ChIP-qPCR assays. D, the transcriptional activity of the WNT2B and FZD1 promoters in the indicated cells was evaluated by a dual-luciferase reporter assay. pGL3-WNT2B or pGL3-FZD1 promoter luciferase reporter construct containing or lacking the FOXF2-binding element was transfected into the indicated cells. E and F, the mRNA (E) and protein (F) levels of WNT2B and FZD1 in the indicated cells were detected by RT-qPCR and immunoblot. ∗, p < 0.05 compared with control cells. BLBC, basal-like breast cancers; ChIP-qPCR, chromatin immunoprecipitation-quantitative PCR; RT-qPCR, reverse transcription-quantitative PCR.
FOXF2 oppositely regulates WNT2B and FZD1 transcription in luminal breast cancer and BLBC cells by recruiting NCoA3 and NCoR1
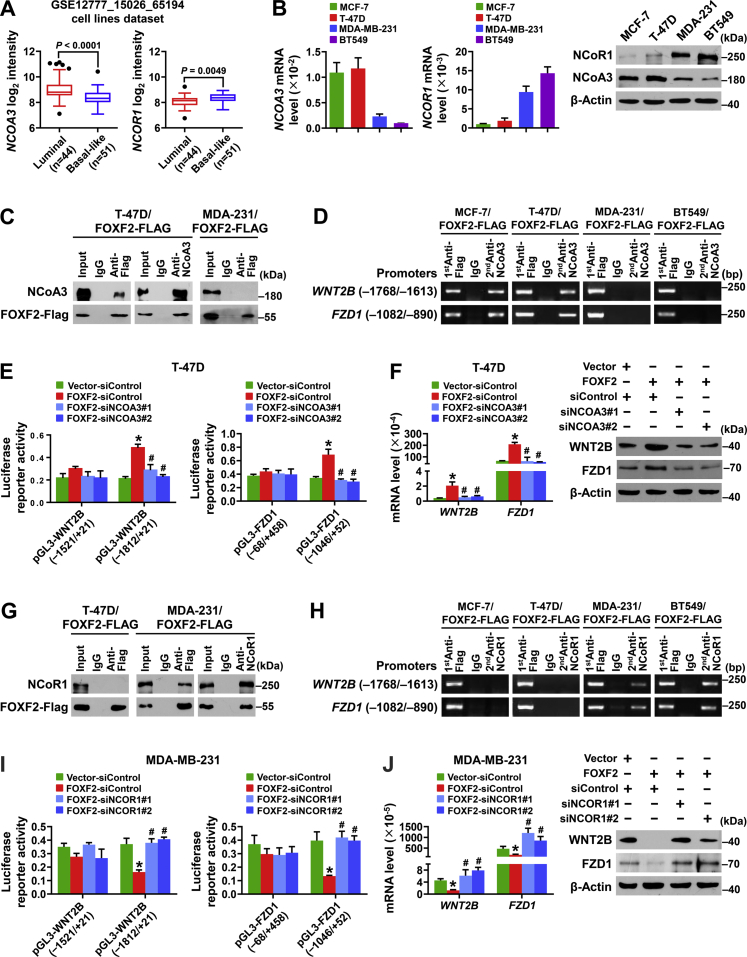
To investigate the mechanism by which FOXF2 differentially regulates WNT2B and FZD1 transcription in luminal breast cancer and BLBC cells, the expression patterns of NCoR- and NCoA-coding genes in luminal breast cancer and BLBC cells were analyzed based on the pooled gene expression profiling dataset GSE12777-GSE15026-GSE65194, which includes 44 luminal subtype cell lines and 51 basal-like subtype cell lines. The results showed that NCOA3 expression was significantly higher in luminal breast cancer cells than in BLBC cells and that NCOR1 expression was significantly higher in BLBC cells than in luminal breast cancer cells (Fig. 5A). The mRNA and protein expression levels of NCoA3 and NCoR1 in MCF-7, T-47D, MDA-MB-231, and BT-549 cell lines were detected by reverse transcription-qPCR and immunoblot. Consistently, NCoA3 expression was significantly higher in MCF-7 and T-47D luminal breast cancer cells than in MDA-MB-231 and BT-549 BLBC cells and NCoR1 expression was significantly higher in MDA-MB-231 and BT-549 cells than in MCF-7 and T-47D cells (Fig. 5B). These results suggest that NCoA3 and NCoR1 contribute to the transactivating function of FOXF2 in luminal breast cancer cells and the transrepressive function of FOXF2 in BLBC cells, respectively.
Figure 5.
FOXF2 oppositely regulates the transcription of WNT2B and FZD1 in luminal breast cancer and BLBC cells by recruiting NCoA3 and NCoR1, respectively.A, the expression levels of NCOR1 and NCOA3 in luminal and basal-like subtypes of breast cancer cells were compared. The data were mined from the combined gene-expression profiling dataset GSE12777_GSE15026_GSE65194 of the human breast cancer cell lines (n = 95). B, the mRNA and protein expression levels of NCOR1 and NCOA3 were detected by RT-qPCR and immunoblot. C and G, the interaction between FOXF2 and NCoA3 (C) or NCoR1 (G) in the indicated cells was assessed by Co-IP assay using anti-FLAG and anti-NCoA3 or anti-NCoR1 antibodies. D and H, the enrichment of the WNT2B and FZD1 promoter regions containing FOXF2-binding sites in the indicated cells transfected with FOXF2-FLAG plasmids was detected by ChIP-PCR and Re-ChIP-PCR analyses. The promoter region was first enriched with anti-FLAG antibody and then with anti-NCoA3 (D) or anti-NCoR1 (H) antibody. E and I, the transcriptional activities of the WNT2B and FZD1 promoters in the indicated cells were assessed by dual-luciferase reporter assays. F and J, the mRNA and protein levels of WNT2B and FZD1 in the indicated cells were detected by RT-qPCR and immunoblot. ∗, p < 0.05 compared with control cells; #, p < 0.05 compared with FOXF2-overexpressing cells. Co-IP, co-immunoprecipitation; RT-qPCR, reverse transcription-quantitative PCR.
To validate the mediated role of NCoA3 and NCoR1 in FOXF2 differentially regulating WNT2B and FZD1 transcription in luminal breast cancer and BLBC cells, protein immunoprecipitation, ChIP, and dual-luciferase reporter assays were performed in the breast cancer cell lines. The results showed that FOXF2 recruited NCoA3 to form a complex (Fig. 5C) that bound to the WNT2B and FZD1 promoters in luminal breast cancer cells but not in BLBC cells (Fig. 5D). FOXF2 enhanced the transcriptional activities of WNT2B and FZD1 promoters containing FOXF2-binding elements and NCoA3 mediated FOXF2-regulated WNT2B and FZD1 transcription and translation (Fig. 5, E and F) in luminal breast cancer cells. Conversely, FOXF2 recruited NCoR1 to the promoters of WNT2B and FZD1 in BLBC cells but not in luminal breast cancer cells (Fig. 5, G and H). FOXF2 repressed the transcriptional activities of the WNT2B and FZD1 promoters containing FOXF2-binding elements in BLBC cells, and NCoR1 mediated FOXF2-regulated WNT2B and FZD1 transcription and translation in BLBC cells (Fig. 5, I and J). Thus, FOXF2 oppositely regulates WNT2B and FZD1 transcription in luminal breast cancer and BLBC cells by recruiting NCoA3 and NCoR1, respectively.
Breast cancer cells with FOXF2-regulated stemness are sensitive to Wnt signaling pathway inhibitors
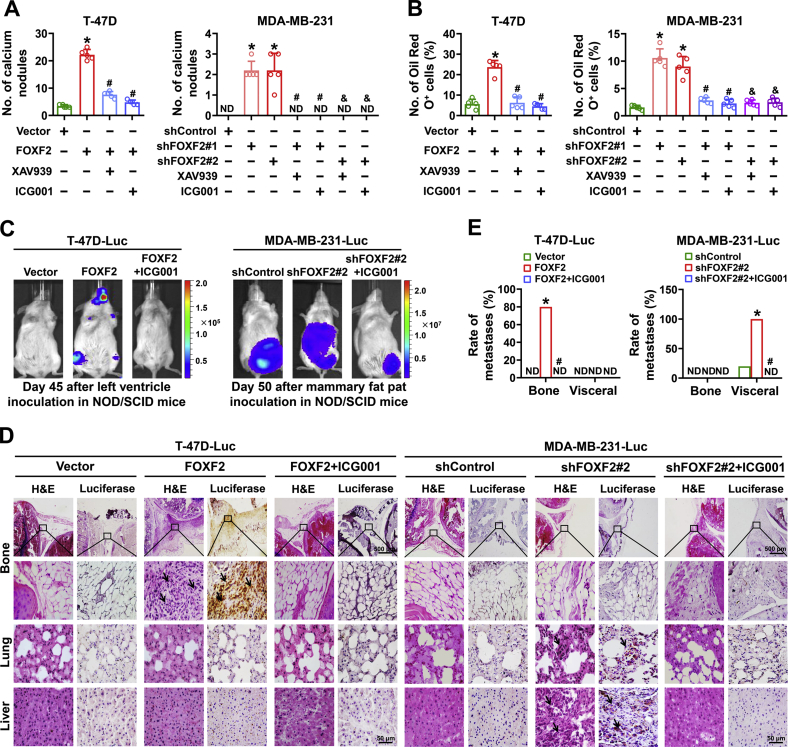
To investigate the role of the Wnt signaling pathway in FOXF2-regulated stemness in luminal breast cancer and BLBC cells, the Wnt signaling pathway inhibitors XAV939 and ICG001 were used to block the Wnt signaling pathway in the above T-47D and MDA-MB-231 cells in vitro. XAV939 and ICG001 treatment significantly inhibited the osteogenic differentiation (Fig. 6A) and adipogenic differentiation capacities (Fig. 6B) of FOXF2-regulated cells. The in vivo assays with ventricular injections of T-47D-FOXF2-Luc cells and T-47D-Vector-Luc cells in NOD/SCID mice showed that the forced expression of FOXF2 promoted bone metastasis and that ICG001 treatment repressed bone metastasis of T-47D-FOXF2-Luc cells (Fig. 6C–E). The in vivo assays with mammary fat pad inoculations of MDA-MB-231-Luc-shFOXF2 cells and MDA-MB-231-Luc-shControl cells in NOD/SCID mice showed that FOXF2 depletion significantly promoted visceral organ metastasis of MDA-MB-231-Luc cells. ICG001 treatment repressed visceral organ metastasis of MDA-MB-231-Luc-shFOXF2 cells (Figs. 6C–E and S3A–C). The increased protein levels of c-Myc, MMP9, and VEGF, the Wnt signaling pathway downstream targets, in tumor tissues formed by MDA-MB-231-Luc-shFOXF2 cells were also significantly inhibited by ICG001 (Fig. S3D). These results indicate that targeting the Wnt signaling pathway is a promising strategy for the treatment of breast cancers with abnormal expression of FOXF2.
Figure 6.
Breast cancer cells with FOXF2-regulated stemness sensitize to Wnt signaling pathway inhibitor.A and B, Wnt signaling pathway inhibitor XAV939 (10 μM) or ICG001 (10 μM) was used to inhibit the Wnt signaling pathway in the indicated cells. A and B, the characteristics of mesenchymal stem cells which differentiate into osteoclasts (A) and adipocytes (B) were evaluated by osteogenic and adipogenic differentiation assays, and the number of calcium nodules and the ratio of Oil Red O+ cells were calculated. C–E, a total of 1 × 107 T-47D-FOXF2-Luc or T-47D-Vector-Luc cells were injected into the left ventricle of female NOD/SCID mice (n = 5 per group). A total of 3 × 106 MDA-MB-231-Luc-shFOXF2 or MDA-MB-231-Luc-shControl cells were inoculated into abdominal mammary fat pad of female NOD/SCID mice (n = 5 each group). The mice-bearing tumors formed by T-47D-FOXF2-Luc or MDA-MB-231-Luc-shFOXF2 cells were treated with ICG001 (20 mg/kg) by i.p. injection three times a week for 7 weeks starting on the 11th day after initial inoculation. Bioluminescence images of xenograft mice on day 45 (T-47D-Luc) or 50 (MDA-MB-231-Luc) after indicated cell inoculation (C). The bone and visceral metastases in xenograft mice injected with indicated cells was identified by H&E staining and luciferase immunohistochemical staining (D). The rate of bone and visceral metastases in each group was analyzed (E). ND: not detected. ∗, p < 0.05 compared with control cells or control mice. # and &, p < 0.05 compared with FOXF2-overexpressing or FOXF2-depleted cells (# for shFOXF2#1 and & for shFOXF2#2) and the mice-bearing tumors from T-47D-FOXF2-Luc or MDA-MB-231-Luc-shFOXF2 cells, respectively. Arrow points to metastatic foci.
Discussion
In this study, we demonstrated that the ectopic high expression of FOXF2 in luminal breast cancer cells and deficiency of FOXF2 expression in BLBC cells lead to enhancement of stemness that was characterized by elevated expression of stem cell markers, increased populations of ALDH+ and CD44high/CD24low, and enhanced capacities of tumor initiation. We also found that FOXF2 differentially regulated the transition of luminal breast cancer and BLBC cells into partial mesenchymal stem-like cells with osteogenic and myogenic differentiation directions, respectively. This is consistent with our previous reports that breast cancer cells with high FOXF2 expression present an osteogenic feature and have a tendency to metastasize to bone, while FOXF2-deficient BLBC cells appear to have a myogenic phenotype and properly metastasize to visceral organs (9, 13). Thus, we further reveal the mechanism of FOXF2-regulated metastasis organotropism of breast cancer cells by which FOXF2 differentially regulates the transitions of luminal breast cancer and BLBC cells into partial mesenchymal stem cell properties that toward different directions of differentiation.
The canonical Wnt signaling pathway plays an important role in cell fate determination including tumorigenesis and metastasis. WNT2B gene, also designated WNT13, encodes two WNT2B protein isoforms from its alternative promoters and splicing variants. WNT2B1 and WNT2B2 are secreted-type and transmembrane-type glycoproteins, respectively. WNT2B2, a major WNT2B transcript, is a canonical Wnt signal and is expressed in multiple human cancer types and some tissues with chronic inflammation (20, 26). FOX-binding sites within the WNT2B promoter have been identified through refined integrative genomic analyses (27). In this study, we verified the binding of FOXF2 to the WNT2B promoter. Importantly, we demonstrate for the first time that FOXF2 oppositely regulates WNT2B transcription in luminal breast cancer and BLBC cells. Cell subtype-specific regulation by FOXF2 consistently appeared in FZD1 transcription. FZD1 has been found to be upregulated in various cancers and mediates metastasis and chemoresistance by the activity of the Wnt/β-catenin pathways and maintenance of the stemness of CSCs (28, 29, 30, 31). FZD1 is a target gene activated by the oncoprotein YB-1 (31) but repressed by tumor suppressors (32). In this study, we identified that FOXF2 acts as a direct regulator of FZD1 and oppositely regulates its transcription in luminal breast cancer and BLBC cells.
We also analyzed the correlation between FOXF2 and ALDH1A1, ALDH1A2, CD44, WNT2B, or FZD1 expression levels in breast cancer tissues based on The Cancer Genome Atlas (TCGA) dataset. Significant positive correlations occurred in luminal subtype tumors but weak positive correlations or no correlations appeared in TNBC subtype tumors (Fig. S4, A and B). Extensive intertumor and intratumor heterogeneity have been identified in TNBC tumors than that in luminal tumors (33, 34, 35), which may be the underlying reason for the inconsistency between cell lines and clinical tissues in BLBC/TNBC subtype. On the other hand, FOXF2 and its target genes also lie on the downstream of multiple regulators. The role and mechanism of FOXF2 in regulating its downstream target genes in cells (e.g., immune cells and fibroblasts) in tumor microenvironment needs to be studied in the future. Furthermore, we analyzed the relationship between combined expression levels of FOXF2/WNT2B or FOXF2/FZD1 in breast cancer tissues and distant metastasis-free survival (DMFS) of patients based on Kaplan–Meier Plotter dataset. As expected, patients bearing tumor of FOXF2high/WNT2Bhigh or FOXF2high/FZD1high had worse DMFS than the patients bearing tumor of FOXF2low/WNT2Blow or FOXF2low/FZD1low in luminal subtype cases. While the patients bearing tumor of FOXF2low/WNT2Bhigh had worse DMFS than the patients bearing tumor of FOXF2high/WNT2Blow in TNBC subtype cases (Fig. S4C). Aforementioned evidence implies that FOXF2-WNT2B and/or FOXF2-FZD1 regulatory axis drives and controls the distant metastasis in luminal breast cancer and BLBC, respectively.
The transactivating or transrepressive function of TFs is dependent on epigenetic modification and chromatin remodeling through the recruitment of cofactors. Our previous studies have demonstrated that FOXF2 reprograms EMT/EOT by pleiotropically transactivating BMP4, SMAD1, and bone-related genes in breast cancer cells of both luminal and basal-like subtypes (9) but represses EMT/EMyoT by pleiotropically transrepressing genes encoding EMT-inducing TFs (10, 11, 12), TGF-βs and miRNA182 (13) in only BLBC cells. In this study, we found that FOXF2 selectively transactivates WNT2B and FZD1 in luminal breast cancer cells and transrepresses that in BLBC cells. Mechanistically, FOXF2 acts as a transrepressor in a BLBC cell subtype-specific manner, which has been reported by our group (10, 11, 12, 13, 36). FOXF2 recruits NCoR1 to form a complex with histone deacetylase 3 (HDAC3) in BLBC cells but not in luminal breast cancer cells (11). In this study, we identified that the transrepression of WNT2B and FZD1 by FOXF2 in BLBC cells followed this mechanism. Importantly, we found, for the first time, that FOXF2 acts as a transactivator for WNT2B and FZD1 in a luminal breast cancer cell subtype-specific manner. FOXF2 recruits NCoA3 to perform transactivating function. NCoA3, also known as amplified in breast cancer 1 (AIB1)/steroid receptor coactivator-3 (SRC-3), has histone acetyltransferase activity and functions as a primary coactivator in an estrogen receptor (ER)–dependent fashion. NCoA3 is amplified and contributes to tamoxifen resistance in human epidermal growth factor receptor–positive and ER-positive breast cancer (37, 38, 39). In this study, we found that NCoA3 was more highly expressed in luminal breast cancer cells than in BLBC cells and conferred transactivating function on FOXF2 in luminal breast cancer cells but not in BLBC cells. We speculate that the interaction of FOXF2 with NCoA3 may be mediated by ER or other coactivators existing in luminal breast cancer cells but not in BLBC cells.
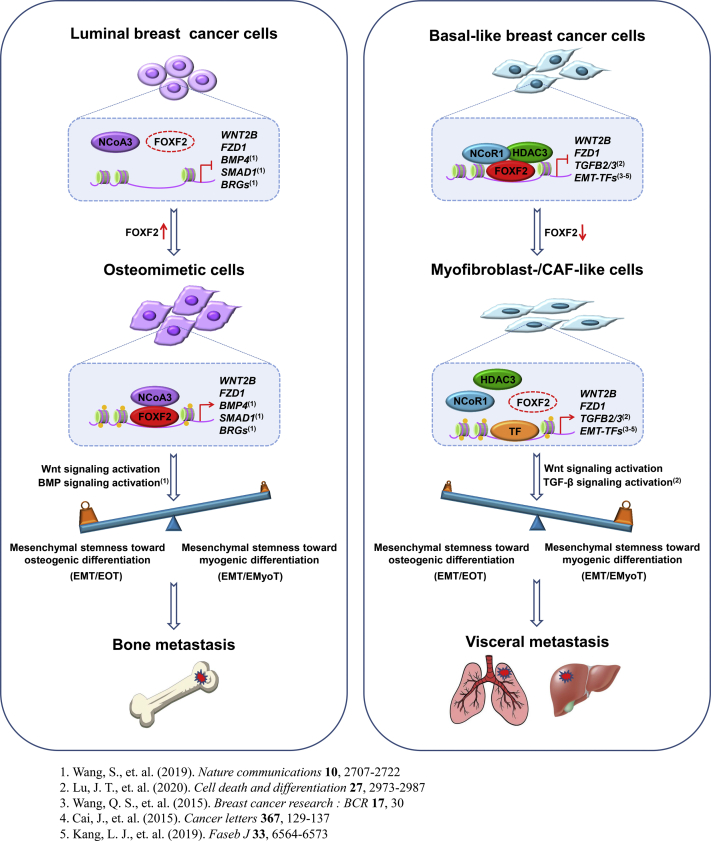
There is growing evidence that the Wnt/β-catenin signaling pathway plays a crucial role in metastasis to bone and visceral in breast cancer cells of both luminal and basal-like subtypes (14, 15, 16, 17, 18, 19). Although the tumor-derived Wnt signaling inhibitor dickkopf-1 (Dkk1) is associated with breast cancer bone-specific metastasis (40, 41, 42), Dkk1 is a target of the Wnt/β-catenin signaling pathway, and its aberrant expression in breast tumors and bone metastases may be caused by dysregulation of the Wnt/β-catenin signaling pathway (41). Consistently, our research indicated that the Wnt/β-catenin signaling pathway mediates both FOXF2 deficiency–accelerated visceral metastasis in BLBC and FOXF2 overexpression-driven bone metastasis in luminal breast cancer. Combined with our previous reports (9, 13), FOXF2 regulates metastasis organotropism of breast cancer in a cell subtype-specific manner depending on the network formed by the BMP, TGF-β, and Wnt/β-catenin signaling pathways (Fig. 7).
Figure 7.
Diagram of FOXF2 regulating metastasis organotropism of breast cancer in a cell subtype-specific manner.
Considering the critical roles of the Wnt/β-catenin pathway in stemness maintenance, chemoresistance, and multiorgan metastases, various small molecule and monoclonal antibody drugs targeting this pathway have been developed for cancer treatment (19, 43, 44, 45, 46). Based on the factors that FOXF2 regulates Wnt/β-catenin signaling pathway activation and stemness in a breast cancer cell subtype context, we treated FOXF2-dysregulated breast cancer cells with Wnt/β-catenin pathway inhibitors. We found that breast cancer cells with FOXF2-regulated stemness were sensitive to Wnt signaling pathway inhibitors.
In summary, this study identifies that FOXF2 oppositely regulates stemness in luminal breast cancer and BLBC cells through the Wnt/β-catenin signaling pathway. Luminal breast cancer and BLBC cells with FOXF2-regulated stemness exhibit partial mesenchymal stem cell properties that toward osteogenic differentiation and myogenic differentiation, respectively. This finding provides further evidence to explain the roles of FOXF2 in differentially regulating breast cancer progression and metastasis organotropism in a cell subtype-specific manner. This study also identified WNT2B and FZD1 as transcriptional target genes of FOXF2, and NCoA3 and NCoR1 as a coactivator and corepressor of FOXF2 confer the functions of transactivating and transrepressing on FOXF2 in luminal breast cancer and BLBC cells, respectively. This study suggests that targeting Wnt/β-catenin signaling pathway is a promising strategy for the treatment of breast cancers with dysregulated expression of FOXF2.
Experimental procedures
Cell culture and treatment
The human breast cancer cell lines MCF-7, T-47D, MDA-MB-231, and BT-549 were obtained from American Type Culture Collection. MDA-MB-231-Luc-D3H2LN (MDA-MB-231-Luc) cells were obtained from Caliper Life Sciences. All cell lines were authenticated by short tandem repeat profiling and tested for mycoplasma contamination. Cells were grown in RPMI1640 or Dulbecco's modified Eagle’s medium (DMEM; Thermo Fisher Scientific/Gibco) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific/Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin. For mammosphere culture, 2 × 104 cells were cultured in serum-free DMEM/F12 medium supplemented with B27 (Life Technologies/Gibco), 20 mg/ml epidermal growth factor (Peprotech), and 20 mg/ml basic fibroblast growth factor (Peprotech) in an ultralow attachment 6-well plate (Corning Inc) for 7 days (first-generation) and another 7 days (second-generation). For Wnt signaling pathway inhibition, cells were cultured in medium supplemented with 10 μM ICG001 (Selleck; catalog no. S2662) or 10 μM XAV939 (Sigma–Aldrich; catalog no. X3004).
Induction of osteogenic differentiation and adipogenic differentiation
For induction of osteogenic differentiation, cells were plated into 6-well culture dishes at a concentration of 1 × 103 cells/well and cultured in DMEM with 10% FBS, 50 μM ascorbic acid, 10 nM dexamethasone, and 10 mM β-glycerophosphate for 21 days. Calcium depositions were visualized with 1% Alizarin Red S staining for 10 min at room temperature (RT) and the total number of calcium nodules was counted under a microscope. For induction of adipogenic differentiation, cells were cultured in DMEM with 10% FBS, 0.5 mM 3-isobutyl 1-methylxanthine, l mM dexamethasone, 100 μM indomethacin, and 10 μg/ml insulin for 25 days. Oil droplets were stained with Oil Red O for 20 min at RT. The number of Oil Red O positive (Oil Red O+) cells in five randomly selected fields was counted under a microscope at 200× magnification. The percentage of Oil Red O+ cells in the total number of cells was calculated.
Lentiviral infection
Recombinant lentiviruses carrying FLAG- or Luc-tagged full-length human FOXF2 complementary DNA (cDNA) (NM_001452; FOXF2-FLAG, FOXF2-Luc) or empty vectors as well as recombinant lentiviruses carrying shRNAs targeting distinct sequences of human FOXF2 mRNA (shFOXF2#1 and shFOXF2#2) or negative control (shControl) were constructed by Shanghai Genechem Co, Ltd for the infection of breast cancer cells to stably silence endogenous FOXF2 or express exogenous FOXF2 that were selected in 1.0 μg/ml puromycin for 2 weeks. The shFOXF2 sequences are listed in Table S1.
Transfection of plasmids and siRNA
FLAG-tagged full-length human FOXF2 cDNA expression plasmid (FOXF2-FLAG), full-length human WNT2B and FZD1 cDNA expression plasmids (Genewiz), siRNAs targeting distinct sequences of human FOXF2 mRNA (siFOXF2#1 and siFOXF2#2), WNT2B mRNA (siWNT2B#1 and siWNT2B#2), FZD1 mRNA (siFZD1#1 and siFZD1#2), NCOR1 mRNA (siNCOR1#1 and siNCOR1#2), NCOA3 mRNA (siNCOA3#1 and siNCOA3#2), and the corresponding controls (RiboBio Co, Ltd) were transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The sequences of siRNAs are listed in Table S1.
Reverse transcription-qPCR
Total RNA extraction, reverse transcription, qPCR, and quantification of target gene expression were performed as described previously (47). The Platinum Quantitative PCR SuperMix-UDG system (Invitrogen) and SYBR Premix DimerEraser system (TaKaRa) were used for qPCR. All primers and probes were synthesized by Sangon Biological Engineering Technology & Services Co, Ltd and are listed in Table S2.
Immunoblot and immunohistochemistry assays
Immunoblot and immunohistochemistry assays were performed as previously described (48). For immunoblot, whole-cell, cytoplasmic and nuclear extracts were prepared using the Nuclear Protein Extraction kit (Active Motif) according to the manufacturer's instructions. β-Actin and histone H3 were used as internal references for total/cytoplasmic and nuclear extracts, respectively. All antibodies and their dilutions are listed in Table S3.
Flow cytometric analysis
Cells were harvested and resuspended in PBS containing 5% FBS and then doubly labeled with anti-CD44-APC (eBioscience) and anti-CD24-PE-Cyanine7 (BioLegend) antibodies or singly labeled with antihuman CDH11-APC (BioLegend) or antihuman α-SMA-FITC (Abcam) antibody at 4°C for 30 min. The identification of ALDH+ cell population was performed using the ALDEFLUOR kit (StemCell Technologies) according to the manufacturer’s instructions. Flow cytometric analysis was performed using a BD FACSCantoII flow cytometer (BD Biosciences).
ChIP-PCR, ChIP-qPCR, and Re-ChIP-PCR
The FZD1 and WNT2B promoter regions containing putative FOXF2-binding elements were enriched using an anti-FLAG antibody in cells transfected with FOXF2-FLAG plasmids. The anti-FLAG-enriched promoter regions of the target genes were used to perform Re-ChIP by anti-NCoA3 or anti-NCoR1 antibody, as previously described (11). ChIP-PCR assays were performed as previously described (10). The quantity of ChIP-enriched DNA fragments was calculated according to the percent input. All antibodies and their dilutions are listed in Table S3. All sequences of primers for ChIP-PCR, ChIP-qPCR, and Re-ChIP-PCR are listed in Table S4.
Protein immunoprecipitation
Cells transfected with FOXF2-FLAG plasmids were lysed with radioimmunoprecipitation assay lysis buffer containing a protease inhibitor cocktail (Thermo Fisher Scientific). The detailed procedure of protein immunoprecipitation was performed as described previously (11).
Dual-luciferase reporter assays
Dual-luciferase reporter assays were performed as previously described (10). To assess the status of the Wnt signaling pathway, cells growing in a 24-well plate were transiently cotransfected with 0.5 μg of TOPflash luciferase reporter plasmid (Millipore) that contains two sets of three copies of the WT TCF-binding regions and 0.025 μg of Renilla luciferase pRL-TK plasmid. FOPflash plasmid containing mutated TCF-binding regions was used as a negative control. TOPflash and FOPflash activities were measured using a dual-luciferase reporter system and normalized to Renilla luciferase activity. All sequences of primers for the construction of luciferase reporters are listed in Table S4.
Xenograft tumor assays
For in vivo limiting dilution tumor formation assay, MDA-MB-231-Luc-shFOXF2 or MDA-MB-231-Luc-shControl cells were inoculated into the abdominal mammary fat pads of 6-week-old female nude mice (GemPharmatech Co, Ltd) at gradient concentrations ranging from 2.4 × 104 to 3 × 106 (n = 5, each group). T-47D-FOXF2-Luc or T-47D-Vector-Luc cells were inoculated at gradient concentrations ranging from 1 × 104 to 1 × 107 (n = 3, each group). MCF-7-FOXF2-Luc or MCF-7-Vector-Luc cells mixed with Matrigel (BD Biosciences) in 1: 1 were inoculated at gradient concentrations ranging from 8 × 104 to 1 × 107 (n = 5, each group). Mice bearing MCF-7 cells were subcutaneously injected with estrogen (Selleck; 2 mg/kg, once a week for 10 weeks) from 1 week before inoculation. Tumor incidence was identified by anatomy at the appropriate time after cell inoculation. For drug treatment, 3 × 106 MDA-MB-231-Luc-shFOXF2 or MDA-MB-231-Luc-shControl cells were inoculated into the abdominal mammary fat pads of NOD/SCID mice, and 1 × 107 T-47D-FOXF2-Luc or T-47D-Vector-Luc cells were injected into the left ventricle of NOD/SCID mice. From the 11th day after the initial inoculation, the mice were intraperitoneally injected with ICG001 (20 mg/kg, three times a week for 7 weeks). The primary tumor and metastases were visualized with bioluminescence assessment and validated by H&E and immunohistochemistry staining of paraffin-embedded tissue sections. Animals of similar age and weight were randomly selected and grouped. Histological evaluation was performed in a blinded fashion. All experimental procedures were approved by the Laboratory Animal Ethics Committee at Tianjin Medical University Cancer Institute and Hospital.
Gene expression profiling datasets of breast cancer cell lines and clinical specimens
The breast cancer cell line gene expression profiling datasets GSE12777 (n = 51), GSE15026 (n = 30), and GSE65194 (n = 14) were pooled into a dataset (GSE12777-GSE15026-GSE65194) including 44 luminal subtype cell lines and 51 basal-like subtype cell lines. The expression levels of transcriptional coactivators and corepressors in luminal and basal-like subtype cell lines were compared. Gene expression profiling of 580 luminal subtype tumor tissues and 159 TNBC subtype tumor tissues in TCGA dataset were downloaded for analysis of the correlation between genes. The data were converted to transcripts per million and normalized as log2 (transcripts per million +1). Kaplan–Meier Plotter online tool (kmplot.com) was used to assess the effect of the genes on breast cancer prognosis. The cutoff values of each gene expression level for separating patients into high- and low-expression groups with distinct metastasis statuses were determined by the auto select best cutoff parament.
Statistical analysis
All in vitro experiments were performed at least two independent times each in triplicate, and the data are presented as the mean ± SD. Student’s t test was used to compare the differences between the experimental and control groups and differences in gene expression levels between the luminal and basal-like groups. For in vivo experiments, Fisher's exact test was used to compare the differences in the incidence of metastasis among different xenograft-bearing mice. Differences with p < 0.05 were considered statistically significant. All analyses were performed using GraphPad Prism 6.0 Software (GraphPad).
Data availability
The data supporting this study are available from the corresponding author upon request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 82173349, 81872403, 81672894, 81472680, 81272357).
Author contributions
Y. M. F. conceptualization; X. Z., R. Z., R. H., T. H. Z., and Q. L. Z. formal analysis; X. Z., R. Z., C. H., and R. H. investigation; Y. M. F. writing–original draft; Y. M. F., X. Z., Q. S. W., X. Q. L., and R. Z. writing–review & editing; X. Z. visualization; Y. M. F. supervision; Y. M. F. project administration; Y. M. F. funding acquisition.
Edited by Eric Fearon
Supporting information
References
- 1.Perou C.M., Sorlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rakha E.A., Chan S. Metastatic triple-negative breast cancer. Clin. Oncol. (R Coll. Radiol.) 2011;23:587–600. doi: 10.1016/j.clon.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Pinilla S.M., Sarrio D., Honrado E., Hardisson D., Calero F., Benitez J., et al. Prognostic significance of basal-like phenotype and fascin expression in node-negative invasive breast carcinomas. Clin. Cancer Res. 2006;12:1533–1539. doi: 10.1158/1078-0432.CCR-05-2281. [DOI] [PubMed] [Google Scholar]
- 5.Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 6.Pastushenko I., Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biology. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Lu W., Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev. Cell. 2019;49:361–374. doi: 10.1016/j.devcel.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aitola M., Carlsson P., Mahlapuu M., Enerback S., Pelto-Huikko M. Forkhead transcription factor FoxF2 is expressed in mesodermal tissues involved in epithelio-mesenchymal interactions. Dev. Dyn. 2000;218:136–149. doi: 10.1002/(SICI)1097-0177(200005)218:1<136::AID-DVDY12>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 9.Wang S., Li G.X., Tan C.C., He R., Kang L.J., Lu J.T., et al. FOXF2 reprograms breast cancer cells into bone metastasis seeds. Nat. Commun. 2019;10:2707. doi: 10.1038/s41467-019-10379-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q.S., Kong P.Z., Li X.Q., Yang F., Feng Y.M. FOXF2 deficiency promotes epithelial-mesenchymal transition and metastasis of basal-like breast cancer. Breast Cancer Res. 2015;17:30. doi: 10.1186/s13058-015-0531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang L.J., Yu Z.H., Cai J., He R., Lu J.T., Hou C., et al. Reciprocal transrepression between FOXF2 and FOXQ1 controls basal-like breast cancer aggressiveness. Faseb J. 2019;33:6564–6573. doi: 10.1096/fj.201801916R. [DOI] [PubMed] [Google Scholar]
- 12.Cai J., Tian A.X., Wang Q.S., Kong P.Z., Du X., Li X.Q., et al. FOXF2 suppresses the FOXC2-mediated epithelial-mesenchymal transition and multidrug resistance of basal-like breast cancer. Cancer Lett. 2015;367:129–137. doi: 10.1016/j.canlet.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Lu J.T., Tan C.C., Wu X.R., He R., Zhang X., Wang Q.S., et al. FOXF2 deficiency accelerates the visceral metastasis of basal-like breast cancer by unrestrictedly increasing TGF-beta and miR-182-5p. Cell Death Differ. 2020;27:2973–2987. doi: 10.1038/s41418-020-0555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eyre R., Alferez D.G., Santiago-Gomez A., Spence K., McConnell J.C., Hart C., et al. Microenvironmental IL1beta promotes breast cancer metastatic colonisation in the bone via activation of Wnt signalling. Nat. Commun. 2019;10:5016. doi: 10.1038/s41467-019-12807-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kar S., Jasuja H., Katti D.R., Katti K.S. Wnt/beta-Catenin signaling pathway regulates osteogenesis for breast cancer bone metastasis: experiments in an in vitro nanoclay scaffold cancer testbed. ACS Biomater. Sci. Eng. 2020;6:2600–2611. doi: 10.1021/acsbiomaterials.9b00923. [DOI] [PubMed] [Google Scholar]
- 16.Previdi S., Maroni P., Matteucci E., Broggini M., Bendinelli P., Desiderio M.A. Interaction between human-breast cancer metastasis and bone microenvironment through activated hepatocyte growth factor/Met and beta-catenin/Wnt pathways. Eur. J. Cancer. 2010;46:1679–1691. doi: 10.1016/j.ejca.2010.02.036. [DOI] [PubMed] [Google Scholar]
- 17.Malanchi I., Santamaria-Martinez A., Susanto E., Peng H., Lehr H.A., Delaloye J.F., et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 18.DiMeo T.A., Anderson K., Phadke P., Fan C., Perou C.M., Naber S., et al. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009;69:5364–5373. doi: 10.1158/0008-5472.CAN-08-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fatima I., El-Ayachi I., Playa H.C., Alva-Ornelas J.A., Khalid A.B., Kuenzinger W.L., et al. Simultaneous multi-organ metastases from chemo-resistant triple-negative breast cancer are prevented by interfering with WNT-signaling. Cancers (Basel) 2019;11:2039. doi: 10.3390/cancers11122039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh M. WNT2B: comparative integromics and clinical applications (Review) Int. J. Mol. Med. 2005;16:1103–1108. [PubMed] [Google Scholar]
- 21.He T.C., Sparks A.B., Rago C., Hermeking H., Zawel L., da Costa L.T., et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 22.Papathanasiou I., Malizos K.N., Tsezou A. Bone morphogenetic protein-2-induced Wnt/beta-catenin signaling pathway activation through enhanced low-density-lipoprotein receptor-related protein 5 catabolic activity contributes to hypertrophy in osteoarthritic chondrocytes. Arthritis Res. Ther. 2012;14:R82. doi: 10.1186/ar3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Gaspard J.P., Chung D.C. Regulation of vascular endothelial growth factor by the Wnt and K-ras pathways in colonic neoplasia. Cancer Res. 2001;61:6050–6054. [PubMed] [Google Scholar]
- 24.Ormestad M., Astorga J., Landgren H., Wang T., Johansson B.R., Miura N., et al. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development. 2006;133:833–843. doi: 10.1242/dev.02252. [DOI] [PubMed] [Google Scholar]
- 25.Nik A.M., Reyahi A., Ponten F., Carlsson P. Foxf2 in intestinal fibroblasts reduces numbers of Lgr5(+) stem cells and adenoma formation by inhibiting Wnt signaling. Gastroenterology. 2013;144:1001–1011. doi: 10.1053/j.gastro.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 26.Katoh M., Hirai M., Sugimura T., Terada M. Cloning, expression and chromosomal localization of Wnt-13, a novel member of the Wnt gene family. Oncogene. 1996;13:873–876. [PubMed] [Google Scholar]
- 27.Katoh M. Transcriptional regulation of WNT2B based on the balance of Hedgehog, Notch, BMP and WNT signals. Int. J. Oncol. 2009;34:1411–1415. [PubMed] [Google Scholar]
- 28.Milovanovic T., Planutis K., Nguyen A., Marsh J.L., Lin F., Hope C., et al. Expression of Wnt genes and frizzled 1 and 2 receptors in normal breast epithelium and infiltrating breast carcinoma. Int. J. Oncol. 2004;25:1337–1342. [PubMed] [Google Scholar]
- 29.Peng Q., Wang L., Zhao D., Lv Y., Wang H., Chen G., et al. Overexpression of FZD1 is associated with a good prognosis and resistance of Sunitinib in clear cell renal cell carcinoma. J. Cancer. 2019;10:1237–1251. doi: 10.7150/jca.28662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L., Yang Z., Li D., Liu Z., Zou Q., Yuan Y., et al. Overexpression of FZD1 and CAIX are associated with invasion, metastasis, and poor-prognosis of the pancreatic ductal adenocarcinoma. Pathol. Oncol. Res. 2018;24:899–906. doi: 10.1007/s12253-017-0284-5. [DOI] [PubMed] [Google Scholar]
- 31.Yang F., Cui P., Lu Y., Zhang X. Requirement of the transcription factor YB-1 for maintaining the stemness of cancer stem cells and reverting differentiated cancer cells into cancer stem cells. Stem Cel. Res. Ther. 2019;10:233. doi: 10.1186/s13287-019-1360-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dang S., Zhou J., Chen Y., Chen P., Ji M., Shi B., et al. Dynamic expression of ZNF382 and its tumor-suppressor role in hepatitis B virus-related hepatocellular carcinogenesis. Oncogene. 2019;38:4804–4819. doi: 10.1038/s41388-019-0759-9. [DOI] [PubMed] [Google Scholar]
- 33.Karaayvaz M., Cristea S., Gillespie S.M., Patel A.P., Mylvaganam R., Luo C.C., et al. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat. Commun. 2018;9:3588. doi: 10.1038/s41467-018-06052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung W., Eum H.H., Lee H.O., Lee K.M., Lee H.B., Kim K.T., et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costa A., Kieffer Y., Scholer-Dahirel A., Pelon F., Bourachot B., Cardon M., et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33:463–479.e10. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 36.Wang Q.S., He R., Yang F., Kang L.J., Li X.Q., Fu L., et al. FOXF2 deficiency permits basal-like breast cancer cells to form lymphangiogenic mimicry by enhancing the response of VEGF-C/VEGFR3 signaling pathway. Cancer Lett. 2018;420:116–126. doi: 10.1016/j.canlet.2018.01.069. [DOI] [PubMed] [Google Scholar]
- 37.Osborne C.K., Bardou V., Hopp T.A., Chamness G.C., Hilsenbeck S.G., Fuqua S.A., et al. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J. Natl. Cancer Inst. 2003;95:353–361. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- 38.Oh J.H., Lee J.Y., Kim K.H., Kim C.Y., Jeong D.S., Cho Y., et al. Elevated GCN5 expression confers tamoxifen resistance by upregulating AIB1 expression in ER-positive breast cancer. Cancer Lett. 2020;495:145–155. doi: 10.1016/j.canlet.2020.09.017. [DOI] [PubMed] [Google Scholar]
- 39.Yi P., Wang Z., Feng Q., Chou C.K., Pintilie G.D., Shen H., et al. Structural and functional impacts of ER coactivator sequential recruitment. Mol. Cell. 2017;67:733–743.e4. doi: 10.1016/j.molcel.2017.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhuang X., Zhang H., Li X., Cong M., Peng F., Yu J., et al. Differential effects on lung and bone metastasis of breast cancer by Wnt signalling inhibitor DKK1. Nat. Cell Biol. 2017;19:1274–1285. doi: 10.1038/ncb3613. [DOI] [PubMed] [Google Scholar]
- 41.Kasoha M., Bohle R.M., Seibold A., Gerlinger C., Juhasz-Boss I., Solomayer E.F. Dickkopf-1 (Dkk1) protein expression in breast cancer with special reference to bone metastases. Clin. Exp. Metastasis. 2018;35:763–775. doi: 10.1007/s10585-018-9937-3. [DOI] [PubMed] [Google Scholar]
- 42.Mariz K., Ingolf J.B., Daniel H., Teresa N.J., Erich-Franz S. The Wnt inhibitor dickkopf-1: A link between breast cancer and bone metastases. Clin. Exp. metastasis. 2015;32:857–866. doi: 10.1007/s10585-015-9750-1. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z.M., Wu J.F., Luo Q.C., Liu Q.F., Wu Q.W., Ye G.D., et al. Pygo2 activates MDR1 expression and mediates chemoresistance in breast cancer via the Wnt/beta-catenin pathway. Oncogene. 2016;35:4787–4797. doi: 10.1038/onc.2016.10. [DOI] [PubMed] [Google Scholar]
- 44.Anastas J.N., Moon R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 45.Mukherjee N., Panda C.K. Wnt/beta-Catenin signaling pathway as chemotherapeutic target in breast cancer: an update on pros and cons. Clin. Breast Cancer. 2020;20:361–370. doi: 10.1016/j.clbc.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 46.Xi Y., Chen Y. Wnt signaling pathway: implications for therapy in lung cancer and bone metastasis. Cancer Lett. 2014;353:8–16. doi: 10.1016/j.canlet.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 47.Kong P.Z., Yang F., Li L., Li X.Q., Feng Y.M. Decreased FOXF2 mRNA expression indicates early-onset metastasis and poor prognosis for breast cancer patients with histological grade II tumor. PloS One. 2013;8 doi: 10.1371/journal.pone.0061591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Y., Xiao C.H., Tan L.D., Wang Q.S., Li X.Q., Feng Y.M. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of breast cancer cells through paracrine TGF-beta signalling. Br. J. Cancer. 2014;110:724–732. doi: 10.1038/bjc.2013.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this study are available from the corresponding author upon request.