Abstract
Familial dysautonomia (FD), a hereditary sensory and autonomic neuropathy, is caused by a mutation in the Elongator complex protein 1 (ELP1) gene that lead to a tissue-specific reduction of ELP1 protein. Our work to generate a phenotypic mouse model for FD headed to the discovery that homozygous deletion of the mouse Elp1 gene leads to embryonic lethality prior to mid-gestation. Given that FD is caused by a reduction, not loss, of ELP1, we generated two new mouse models by introducing different copy numbers of the human FD ELP1 transgene into the Elp1 knockout mouse (Elp1−/−) and observed that human ELP1 expression rescues embryonic development in a dose dependent manner. We then conducted a comprehensive transcriptome analysis in mouse embryos to identify genes and pathways whose expression correlates with the amount of ELP1. We found that ELP1 is essential for the expression of genes responsible for nervous system development. Further, gene length analysis of the differentially expressed genes showed that the loss of Elp1 mainly impacts the expression of long genes and that by gradually restoring Elongator their expression is progressively rescued. Finally, through evaluation of co-expression modules, we identified gene sets with unique expression patterns that depended on ELP1 expression.
Introduction
Elongator is a highly conserved multiprotein complex composed of two copies of each of its six subunits, named Elongator complex proteins 1 to 6 (ELP1–6). Elongator subunits are evolutionarily highly conserved from yeast to humans both in their sequence and interaction with other subunits (Krogan and Greenblatt, 2001; Hawkes et al., 2002; Li et al., 2005; Chen et al., 2006; Chen et al., 2009; Kojic and Wainwright, 2016). Conserved function across all species has been clearly demonstrated using a variety of different cross-species rescue experiments (Li et al., 2005; Chen et al., 2006; Chen et al., 2009). Deletion of any of the genes encoding the six subunits confers almost identical biochemical phenotypes in yeast (Fellows et al., 2000; Winkler et al., 2001; Frohloff et al., 2003), suggesting that there is a tight functional association between the proteins comprising Elongator complex, and that the functional integrity of Elongator is compromised in the absence of any of its subunits (Frohloff et al., 2003; Huang et al., 2005). Both yeast and human Elongator have lysine acetyltransferase activity that is mediated by the catalytic subunit Elp3. Elp3 has two identified substrates: histone H3 and α-Tubulin (Otero et al., 1999; Wittschieben et al., 1999; Creppe et al., 2009; Tran et al., 2012). While the acetylation of histone H3 is linked to the role of the complex in transcriptional elongation (Otero et al., 1999; Wittschieben et al., 1999; Pokholok et al., 2005), cytosolic acetylation of α-Tubulin has been linked to its role in microtubule organization particularly in the context of cell migration (Creppe et al., 2009). Elongator was isolated as a complex that associates with chromatin and interacts with the elongating phosphorylated form of RNA polymerase II (RNAPII) both in yeast and human (Otero et al., 1999; Wittschieben et al., 1999; Kim et al., 2002). The catalytic subunit Elp3, by acetylating histone H3, facilitates RNAPII access to actively transcribed genes. In human cells, Elongator is required for the expression of several genes involved in migration and in the expression of HSP70 (Close et al., 2006; Han et al., 2007). In addition, accumulating evidence supports the role of this complex in maintaining translational fidelity through tRNA modifications. Specifically, Elongator is essential for the formation of 5-methoxycarbonylmethyl (mcm5) and 5-carbamoylmethyl (ncm5) groups on uridine nucleosides present at the wobble position of many tRNAs (Huang et al., 2005; Bauer and Hermand, 2012).
Several loss-of-function studies have demonstrated the key role of Elongator during development. Yeast Elp mutants are hypersensitive to high temperature and osmotic conditions, and they showed defects in exocytosis, telomeric gene silencing, DNA damage response and adaption to new growth medium (Wittschieben et al., 1999; Rahl et al., 2005; Li et al., 2009). In Arabidopsis thaliana, mutations in Elp subunits resulted in impaired root growth (Nelissen et al., 2005) and deletion of Elp3 in Drosophila melanogaster was lethal at the larval stage (Walker et al., 2011). Depletion of Elongator in Caenorhabditis elegans led to defects in neurodevelopment (Solinger et al., 2010). In mice, Elp1 knockout results in embryonic lethality due to failed neurulation and vascular system formation (Chen et al., 2009; Dietrich et al., 2011). Consistent with its crucial role during development, several human neurodevelopmental disorders have been associated with mutations in Elongator complex subunits. Familial dysautonomia (FD) is caused by a splicing mutation in ELP1 (Anderson et al., 2001; Slaugenhaupt et al., 2001; Cuajungco et al., 2003) that reduces the amount of functional protein in a tissue specific manner, missense mutations in ELP2 have been found in individuals with severe intellectual disability (ID) (Najmabadi et al., 2011; Cohen et al., 2015), variants of ELP3 have been associated with amyotrophic lateral sclerosis (ALS) (Simpson et al., 2009), ELP4 variants have been implicated in autism spectrum disorder and Rolandic epilepsy (RE) (Strug et al., 2009; Addis et al., 2015) and a mutation in Elp6 causes Purkinjie neuron degeneration and ataxia-like phenotypes in mice (Kojic et al., 2018).
FD is a neurodevelopmental disorder characterized by widespread sensory and autonomic dysfunction and by central nervous system (CNS) pathology (Axelrod et al. 2010; Mendoza-Santiesteban et al. 2012; Mahloudji et al., 1970; Pearson, 1979; Ochoa, 2003; Mendoza-Santiesteban et al., 2017). The major mutation in FD is a splicing mutation in ELP1 intron 20 that leads to variable skipping of exon 20 and to a reduction of ELP1 mostly in the nervous system (Slaugenhaupt, 2002; Cuajungco et al., 2003). In 2009 we generated a knockout (KO) Elp1 mouse, Elp1−/− and showed that complete ablation of Elp1 resulted in early embryonic lethality (Chen et al., 2009; Dietrich et al., 2011). To gain a better understanding of how reduction of ELP1 leads to FD, we generated two new mouse models by introducing different copy numbers of the human ELP1 transgene with the major FD mutation (Hims et al., 2007), TgFD1 (one copy) and TgFD9 (nine copies), into the Elp1−/− mouse. Although the human FD transgene did not rescue embryonic lethality of the Elp1−/− mouse, its expression rescues embryonic development in a dose dependent manner in TgFD1; Elp1−/− and TgFD9; Elp1−/− embryos. In order to understand the gene regulatory networks that are dependent on ELP1 expression, we conducted a comprehensive transcriptome analysis in these mouse embryos.
Results
Generation of mice expressing an increasing amount of ELP1.
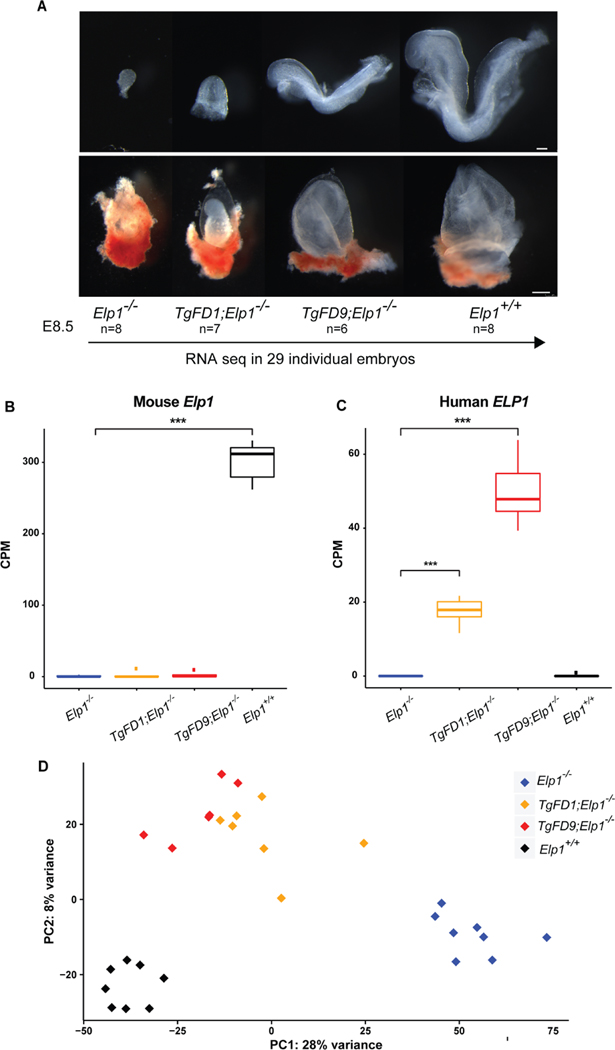
We previously demonstrated that homozygous deletion of the mouse Elp1 gene leads to embryonic lethality prior to mid-gestation (Chen et al., 2009). Detailed phenotypic characterization of Elp1−/−KO embryos at early developmental stages revealed several abnormalities, including a dramatic reduction in size, disruption of the extraembryonic vascular networks, failure of germ layer inversion, and interruption of cephalic neural-tube closure (Chen et al., 2009; Dietrich et al., 2011). In an effort to understand the molecular mechanisms that characterize FD we have generated several transgenic mouse lines carrying the wild-type (WT) and FD human ELP1 gene that differ by the copy number of the transgene (Hims et al., 2007). The murine Elp1 protein is 80% identical to human ELP1 and by introducing the human WT ELP1 transgene into the Elp1−/− mouse, we completely rescued development and mice were born alive and healthy, confirming ELP1 functional conservation between human and mouse (Chen et al., 2009). To test whether the abnormalities caused by ablation of mouse Elp1 could be improved by the human FD transgene, heterozygote mice carrying different copy numbers of the FD ELP1 transgene (TgFD1; Elp1+/−or TgFD9; Elp1+/−) were crossed with mice heterozygous for the Elp1 knockout allele (Elp1+/−). Mice were collected at either E8.5 or P0 and genotyped using genomic DNA from the visceral yolk sac. Although neither TgFD1 nor TgFD9 rescued embryonic lethality in the Elp1+/− mice (Table 1), the development of the FD1/KO (TgFD1; Elp1−/−) and FD9/KO (TgFD9; Elp1−/−) embryos progressed further as human ELP1 expression increased (Fig. 1A). The KO embryos degenerate by E12.5 while KO/FD1 and KO/FD9 embryos degenerate by E14.5 (Fig. S1) (Chen et al., 2009). Because familial dysautonomia is caused by a developmental reduction of ELP1 protein, we conducted a comprehensive transcriptome analysis to identify gene expression changes that correlate with the observed developmental delay. We collected 29 individual C57BL/6 mouse embryos at E8.5 (n = 8 KO, n = 7 FD1/KO, n = 6 FD9/KO, n = 8 WT) and total RNA was extracted from each single embryo (Fig. 1A). KO and WT embryos were obtained from the same breeders and the correct staging of each litter was confirmed by counting the somite number of the control littermates (EMA Anatomy Atlas of Mouse Development). We specifically performed the transcriptome analysis at E8.5 because we previously published that Elp1 is required in early embryogenesis and the Elp1−/− KO embryos degenerate after E10.5 (Chen et al., 2009). Further, FD affects the peripheral nervous system (PNS) and E8.5 is a critical time for PNS development. At E8.5 the neural crest, which will give rise to most of the PNS, including the DRG and ganglia of the autonomic nervous system, has acquired its identity and migration has begun (Labosky and Kaestner, 1998; Teng et al., 2008). Finally, E8.5 was the earliest stage at which we could collect sufficient RNA from individual embryos to generate RNA-Seq libraries. As expected, KO embryos do not express any WT Elp1 (Fig. 1B), whereas FD1/KO and FD9/KO embryos express increasing amounts of WT ELP1 with FD9/KO embryos expressing three times more ELP1 than FD1/KO embryos (Fig. 1C). Normalized gene count comparisons revealed that expression of human ELP1 in FD1/KO embryos is ~6% of WT Elp1 while in the FD9/KO human ELP1 expression is ~16% of WT Elp1 (Fig. 1C). Interestingly, the expression of the other Elongator subunits (Elp2–6) was not affected by Elp1 loss (Fig. S2). Principal component analysis revealed that the four genotypes exhibited distinct expression profiles with PC1 explaining 28% of the total expression variance across samples, suggesting that ELP1 dosage plays a critical role in embryonic transcriptome regulation (Fig. 1D).
Table 1.
Genotype ratios of offspring generated by TgFD; Elp1+/−X Elp1+/− pairings. The time of conception is estimated to be E0.5 day prior to the observation of a vaginal plug. P0, postnatal day zero. Expected mendelian ratio is indicated below the genotype, percentage of embryos of each genotype in parentheses.
| TgFD; Elp1+/− X Elp1+/− | Time | Number of animals | Total | |||||
|---|---|---|---|---|---|---|---|---|
| Elp1+/+ 12.5% | Elp1+/− 25% | Elp1−/− 12.5% | Tg; Elp1+/+ 12.5% | Tg; Elp1+/− 25% | Tg; Elp1−/− 12.5% | |||
| TgFD1 | E8.5 | 17(11%) | 37(25%) | 19(13%) | 17(11%) | 43(29%) | 17(11%) | 150 |
| TgFD1 | P0 | 43(19%) | 88(39%) | 0(0%) | 29(13%) | 63(28%) | 0(0%) | 223 |
| TgFD9 | E8.5 | 10(7%) | 30(21%) | 16(11%) | 25(17%) | 45(31%) | 17(11%) | 143 |
| TgFD9 | P0 | 24(23%) | 42(40%) | 0(0%) | 13(12%) | 27(25%) | 0(0%) | 106 |
Fig. 1.
Generating mouse embryos expressing increasing amount of ELP1. A: Morphological phenotypes of Elp1−/−, TgFD1; Elp1−/−, TgFD9; Elp1−/− and Elp1+/+ embryos (top) and extraembryonic components (bottom) at E8.5. RNA-seq experiment was performed using total RNA extracted by individual embryo. B: Expression of the endogenous WT Elp1 across different embryos. The median for each group is shown. C: Expression of the human WT ELP1 across different embryos. The median for each group is shown. D: Principal component analysis of all the embryos colored by genotype. In box-and-whisker plots in B and C, each box extends to 1.5 times inter-quartile range (IQR) from upper and lower hinges, respectively. Outliers are not shown. Only comparisons with significant difference are marked by stars (two-tailed, unpaired Welch’s t test with Bonferroni correction). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Scale bars, 100 μm (A, top); 250 μm (A, bottom).
Major transcriptome changes in Elp1 KO embryos.
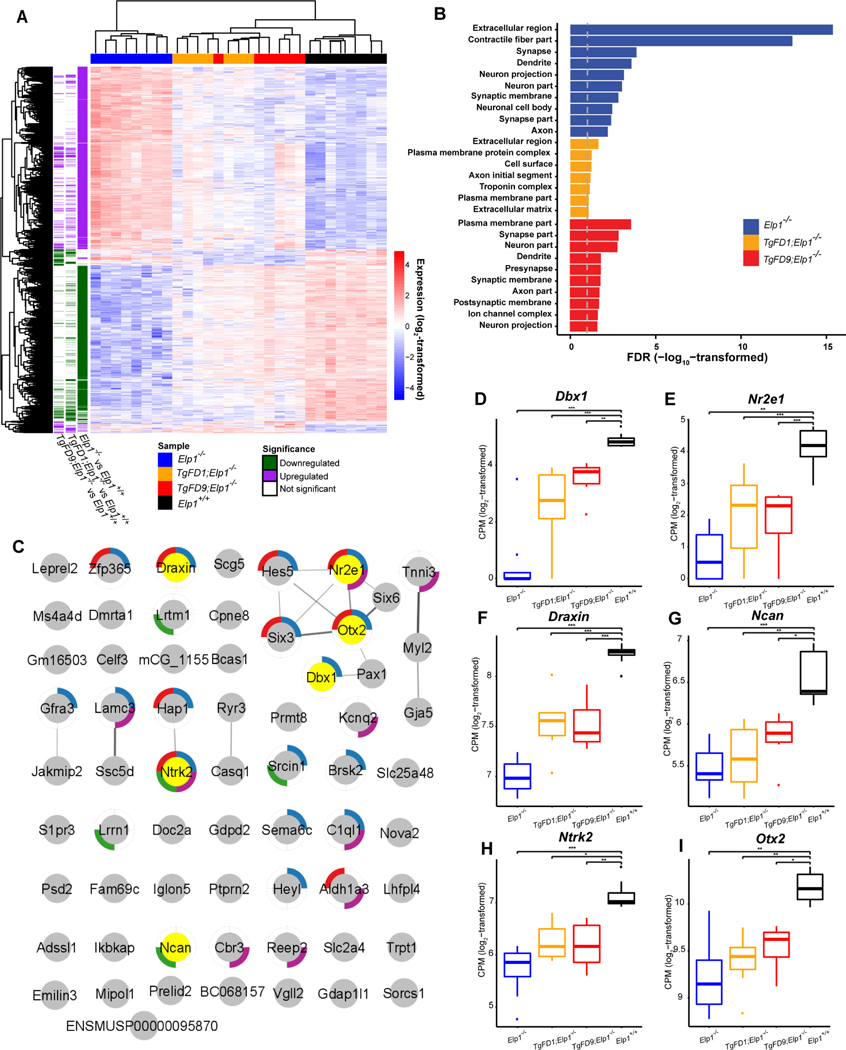
Transcriptome profiling in embryos expressing an increasing amount of ELP1 show that the number of differentially expressed genes (DEGs), (False Discovery Rate or FDR < 0.1; and Fold Change or FC ≥ 1.5), proportionally declines as ELP1 expression increases. In KO embryos 2399 out of 19,619 (12.23%) genes were differentially expressed when compared with WT embryos (Fig. 2A, Table S1). Strikingly, in FD1/KO embryos the DEGs were only 601 (3.06%), while in FD9/KO embryos there were 494 DEGs (2.52%) (Fig. 2A; Table S1), demonstrating that a minimal increase in ELP1 is sufficient to rescue the expression of ~80% of all DEGs. Gene Ontology (GO) analysis of the down-regulated genes (FDR < 0.1) in KO, FD1/KO and FD9/KO embryos highlighted pathways important for nervous system development including synapse formation, neuron projection and axon growth (Fig. 2B; Table S2). These findings are consistent with the body of work supporting the role of ELP1 during early development in target tissue innervation and with the fact that neuronal loss in FD is mostly due to failure of innervation (Close et al., 2006; Johansen et al., 2008; Cheishvili et al., 2011; George et al., 2013; Abashidze et al., 2014; Jackson et al., 2014; Ohlen et al., 2017). Notably, of the 71 genes that were significantly downregulated in all 3 KO genotypes (Table S3), 24 of them (~33%) have a critical role in nervous system or brain development (Fig. 2C). STRING analysis of these genes revealed an enrichment for protein-protein interactions (PPI enrichment = 2.9E-4 according to STRING v11) (Fig. 2C) (Szklarczyk et al., 2019). Among the neuronal genes, Dbx1 and Nr2e1 were the two most down-regulated genes across all 3 KO genotypes (Fig. 2D and 2E). Dbx1, also known as Developing Brain Homeobox 1, is expressed in a regionally restricted pattern in the developing mouse CNS and encodes for a transcription factor that plays a pivotal role in interneuron differentiation in the ventral spinal cord (Lu et al., 1996). In vertebrates, spinal interneurons modulate the motor neuron activity elicited by incoming sensory information and, by relaying the proprioceptive data to the brain, play a critical role in locomotor coordination (Lanuza et al., 2004). Nr2e1 is a transcription factor that regulates the expression of genes essential for retinal development (Yu et al., 2000). Loss of Nr2e1 in mice has been shown to cause severe early onset retinal degeneration with death of various retinal cells including retinal ganglion cells (RGCs) (Miyawaki et al., 2004; Abrahams et al., 2005; Zhang et al., 2006). Interestingly, the list of neuronal genes that were significantly downregulated in all three KO genotypes, KO, FD1/KO and FD9/KO, also included the chemorepulsive axon guidance protein draxin, the neuronal adhesion protein involved in neurite growth neurocan (Ncan), the brain-derived neurotrophic factor BDNF-receptor TrkB (Ntrk2) and the homeobox protein involved in brain and sensory organ development otx2 (Fig. 2F–I). GO pathways associated with up-regulated genes in the KO embryos included several terms related to apical plasma membrane, specifically associated with brush border glucose transport and lipoprotein metabolism (Fig. S3; Table S2). The activation of these metabolic pathways might represent a compensatory response to the failure of the embryos to proceed through development.
Fig. 2.
Transcriptome profiling of embryos expressing increasing amount of ELP1. A: Heatmap of the 2619 differentially expressed genes for each genotype compared to WT Elp1+/+. B: Gene ontology analysis of the downregulated genes in Elp1−/−, TgFD1; Elp1−/−, TgFD9; Elp1−/−embryos. The graph shows FDR values for the most significant specific GO terms (also see in Table S2). C: STRING interaction map for the 71 proteins that were significantly downregulated in all 3 KO genotypes (also see in Table S3). Each pie indicates a protein. Note, Ikbkap is the alternative name of Elp1. The pies surrounded by colored edges indicate the 24 proteins that have a critical role in neurogenesis (GO:0022008, blue), brain development (GO:0007420, red), regulation of synapse organization (GO:0050807, green), and nervous system process (GO:0050877, purple), respectively. The pies in yellow indicate the proteins whose expression are highlighted in panels D–I. D–I: box-and-whisker plots showing the expression of key genes for nervous system development that are significantly downregulated across all three KO genotypes: Dbx1 (D), Nr2e1 (E), Draxin (F), Ncan (G), Ntrk2 (H) and Otx2 (I). *, P <0.05; **, P <0.01 and ***, P < 0.001, Welch’s t test.
Long neuronal genes require Elongator activity for their expression.
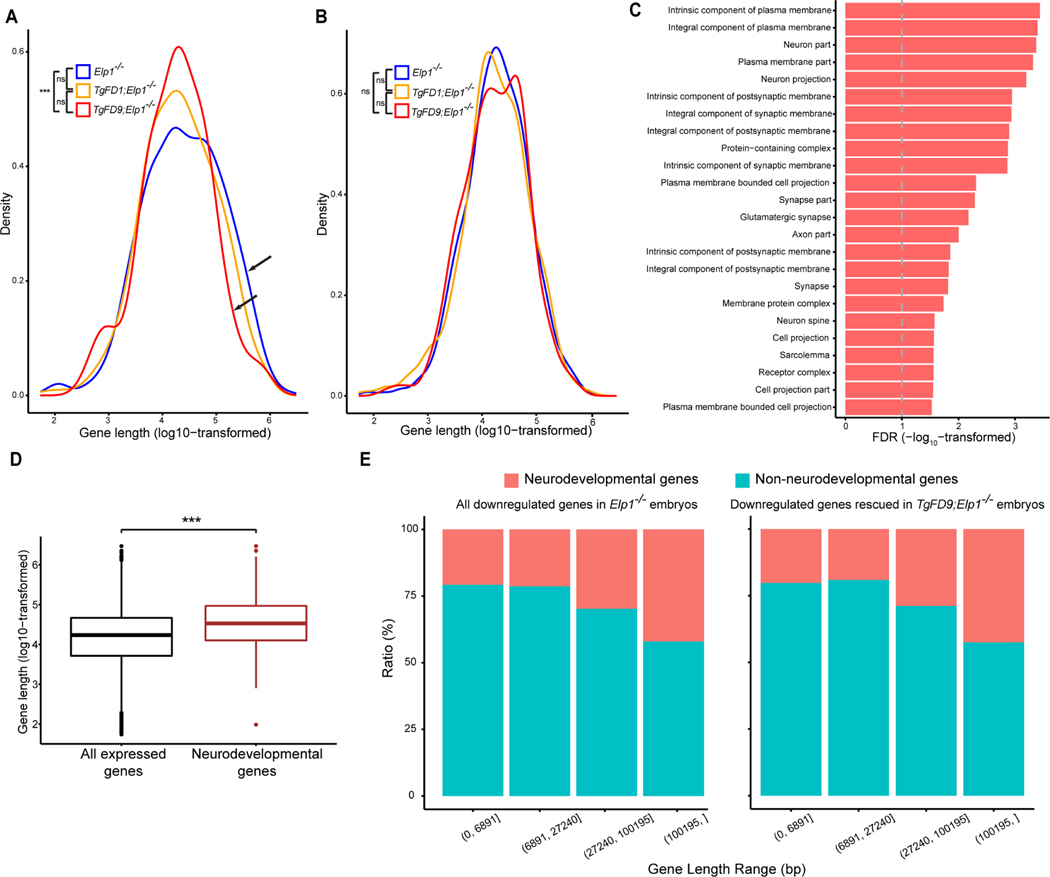
Although Elongator plays a number of roles in the cell, in the nucleus this complex directly interacts with RNAPII and facilitates transcriptional elongation through altering chromatin structure (Otero et al., 1999; Wittschieben et al., 1999; Hawkes et al., 2002; Kim et al., 2002). Elongator has histone acetyltransferase activity via its ELP3 subunit and regulates the accessibility of RNAPII to the chromatin. Using Chromatin immunoprecipitation (ChIP) we have previously shown that in the absence of ELP1, histone H3 acetylation was significantly reduced at the 3′ ends of genes (Chen et al., 2009). Moreover, Close et al demonstrated that upon reduction of ELP1 there was progressively lower RNAPII density at the 3′ end of target genes than in the promoter region (Close et al., 2006), supporting the role of Elongator in transcriptional elongation. To examine if the downregulation in gene expression observed in the KO embryos might be due to a failure in transcriptional elongation, we compared the length distribution of the DEGs among embryos expressing an increasing amount of ELP1. We discovered that long genes, especially those longer than 100 kb, were downregulated significantly more often in the KO embryos than in the FD9/KO embryos (FDR < 0.01, Fig. 3A) suggesting that Elongator loss has a more pronounced effect on the expression of longer genes. Importantly, ELP1 restoration efficiently rescued their expression (Fig. 3A). In contrast, there was no difference in gene length in the upregulated genes between the different genotypes (Fig. 3B). Of the 257 long genes (> 100 kb) that were downregulated in KO embryos, 216 (84.05%) were rescued in the FD1/KO embryos and 247 (96.11%) were rescued in FD9/KO embryos (Fig. S4; Table S4). GO analysis of these genes highlighted pathways important for synapse formation, neuron projection and axon growth (Fig. 3C; Table S5). Since long downregulated genes were enriched for pathways important for nervous system development, we compared the average length of neurodevelopmental genes with the average length of all expressed genes (see Material and Methods) and we observed that neurodevelopmental genes (GO:0048666) were significantly longer (Fig. 3D). We then investigated whether the ELP1-dependent gene regulation was driven by length or if neuronal genes were more likely to require ELP1 for efficient transcription. We divided all downregulated genes in KO embryos into four gene-length categories and examined the proportion of neurodevelopmental genes (GO:0048666) in each category (Fig. 3E left panel). If neurodevelopmental genes were more likely to require ELP1 for efficient transcription, we would expect to see more rescue with higher ELP1 dosage. Given that the proportion of neurodevelopmental genes rescued in the FD9/KO embryos in all gene-length categories was similar to those of the downregulated genes in the KO embryos (Fig. 3E right panel), we concluded that neurodevelopmental genes are more susceptible to ELP1-loss simply because they are longer than the non-neurodevelopmental genes.
Fig. 3.
Gene length distribution in embryos expressing different ELP1 amounts. A: Length distribution of the down-regulated genes in Elp1−/−, TgFD1; Elp1−/− and TgFD9; Elp1−/− embryos. Arrows indicate the comparison between Elp1−/− and TgFD9; Elp1−/− distribution. B: Length distribution of the up-regulated genes in Elp1−/−, TgFD1; Elp1−/− and TgFD9; Elp1−/− embryos. In both (A) and (B), K-S test was applied. C: Gene ontology analysis of the 247 long genes (> 100 kb) downregulated in Elp1−/− embryos and rescued at TgFD9; Elp1−/−. The graph shows FDR values for each specific GO term (also see in Table S5). D: Comparison between length distributions of all expressed genes in RNA-Seq with the length distribution of neuronal genes (GO:0048666 in Gene Ontology database) via two-tailed, unpaired Welch’s t test. E: Ratio of neurodevelopmental and non-neurodevelopmental genes in different gene-length ranges. The left panel represents all downregulated genes in Elp1−/− embryos, while the right panel represents genes which expression was rescued in TgFD9; Elp1−/− embryos. The gene-length ranges were determined in order to have an equal number of genes in each range. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Identification of genes whose expression correlates with amount of ELP1.
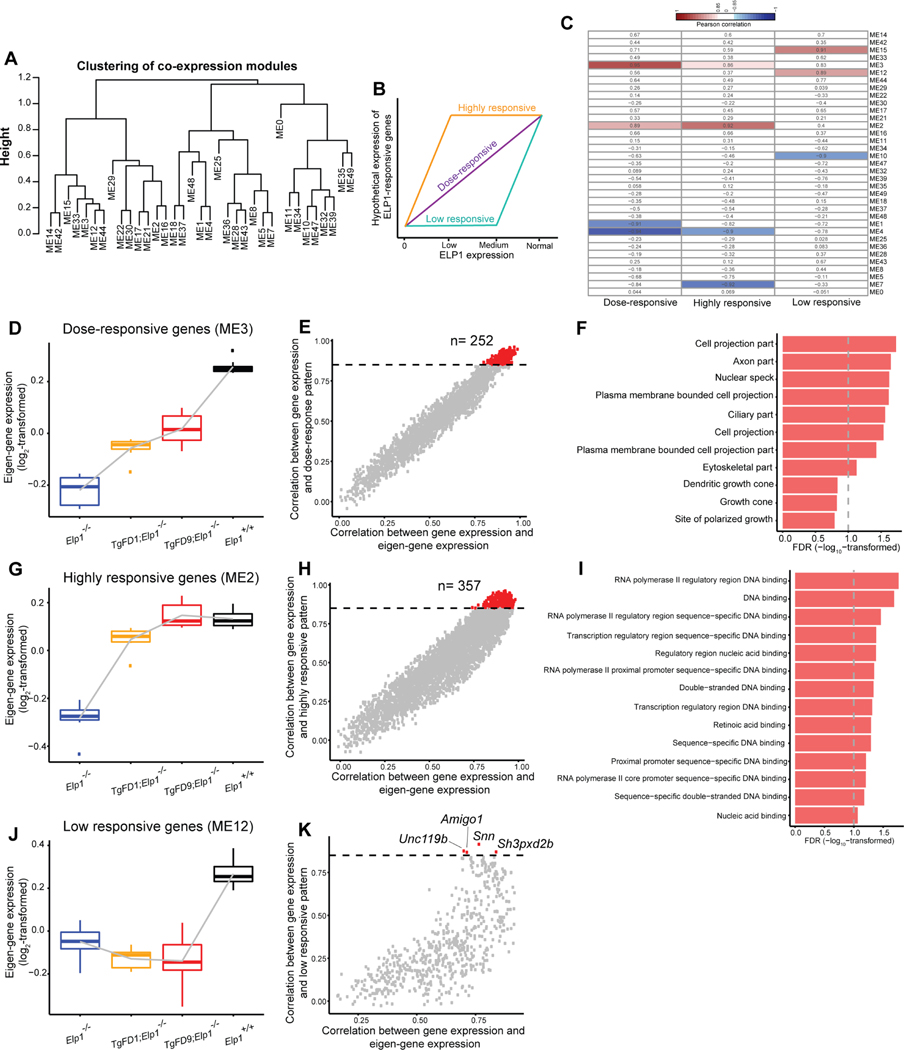
Elongator has been linked to transcriptional regulation (Close et al., 2006; Han et al., 2007; Li et al., 2011). Our unique mouse models provide, for the first time, the ability to perform a comprehensive transcriptome analysis to identify genes whose expression depends on the amount of ELP1. This study is highly relevant to better understanding FD pathogenesis, as the disease is caused by a reduction, not loss, of ELP1 primarily in the nervous system. We built the gene co-expression network across embryonic RNA-Seq data for all genotypes (Materials and Methods). We identified 35 co-expression distinct Modules Eigengenes (MEs) (Fig. 4A). We then postulated that genes whose expression relies on ELP1 expression would be grouped into three major categories (Fig. 4B): (1) genes whose expression changes as a monotonic function of ELP1, referred as “dose-responsive genes”; (2) genes whose expression is completely rescued with low ELP1 expression, referred as “highly responsive genes” and (3) genes whose expression is restored only when ELP1 is expressed at WT levels, referred as “low responsive genes”. Among the 35 MEs identified, ME3, ME2 and M12 had the highest positive correlation with these hypothesized gene patterns (Pearson correlation ≥ 0.85, FDR < 0.1, Fig. 4C). ME3 included the dose-responsive genes (Fig. 4D); ME2 included the highly responsive genes (Fig. 4G) and M12 contained the low responsive genes (Fig. 4J). Interestingly, ME4, ME7 and ME10 had the highest negative correlation with our hypothesized patterns (Fig. S5).
Fig. 4.
Identification of ELP1-responsive gene patterns. A: Modules of distinct Eigengenes (ME) analysis across embryos expressing increasing amount of ELP1 identified 35 distinct MEs (see also Materials and Methods). B: Hypothetical expression trajectories of ELP1-responsive genes. C: Heatmap displays the Pearson correlation between the eigengene of each co-expression module and the eigengene of each hypothetical expression trajectory. The blue domain indicates a negative correlation while the red domain indicates a positive correlation. The number in each grid demonstrates the Pearson correlation coefficient. D: Boxplot displays the eigengene expression of ME3 at each genotype. E: Module membership of each gene in ME3. Each dot represents a gene. The X-axis demonstrates the Pearson correlation between the expression of genes in ME3 and the module eigengenes of ME3 while the Y-axis demonstrates the Pearson correlation between gene expression and the eigengene of each hypothetical expression trajectory. The horizontal dashed line shows a correlation coefficient of 0.85. F: Gene ontology analysis of the 252 dose-responsive genes which expression strictly increase as monotonic function of ELP1. The graph shows FDR values for each specific GO term (also see in Table S6). G: Boxplot displays the eigengene expression of ME2 at each genotype. H: Module membership of each gene in ME2. I: Gene ontology analysis of the 357 highly responsive genes whose expression is completely restored in FD9/KO embryos. The graph shows FDR values for each specific GO term (also see in Table S6). J: Boxplot displays the eigengene expression of ME12 at each genotype. K: Module membership of each gene in ME12. *, P < 0.05; **, P < 0.01; ***, P < 0.001, Welch’s t test.
Overall, we identified 252 dose-responsive genes whose expression strictly increases as a monotonic function of ELP1 (Pearson correlation ≥ 0.85, FDR < 0.1, Fig. 4E). The GO analysis of these genes highlighted pathways important for axon and cell projection formation (Fig. 4F; Table S6). This result supports, once again, the role of ELP1 in the expression of genes important for target tissue innervation and is consistent with the innervation failure observed in FD (Close et al., 2006; Johansen et al., 2008; Cheishvili et al., 2011; George et al., 2013; Abashidze et al., 2014; Jackson et al., 2014; Ohlen et al., 2017). We found 357 highly responsive genes whose expression was completely restored in FD9/KO embryos (Pearson correlation ≥ 0.85, FDR < 0.1, Fig. 4H). GO analysis of these genes highlighted pathways associated with transcriptional regulation (Fig. 4I; Table S6) suggesting that a small increase of functional Elongator is enough to restore normal expression of important transcriptional regulators. This finding might explain the dramatic phenotypic improvement observed in FD9/KO embryos when compared with FD1/KO and KO. Finally, we identified four low responsive genes whose expression was rescued solely in WT embryos: Amigo1, Snn, Unc119b and Sh3pxd2b (Pearson correlation ≥ 0.85, FDR < 0.1, Fig. 4K). Interestingly, Amigo1 is a cell adhesion molecule that promotes attachment and neurite outgrowth in embryonic hippocampal neurons (Kuja-Panula et al., 2003). Snn is a transmembrane protein that localizes in the mitochondria and other vesicular organelles and mediates neuronal cell apoptosis induced by trimethyltin chloride (Buck-Koehntop et al., 2005; Billingsley et al., 2006). Unc119b is required for ciliary trafficking and null mutations in the Unc119 gene family are associated with nephronophthisis and retinal degeneration (Constantine et al., 2012). Lastly, Sh3pxd2b encodes Tks4, a scaffold protein involved in podosome organization that is important for adult bone homeostasis (Vas et al., 2019).
In the negatively correlated patterns ME4, ME7 and ME10 (Pearson correlation ≥ 0.85, FDR < 0.1) we did not find any enrichment for neuronal terms (Fig. S5; Table S7). Genes in ME4 highlighted pathways involved in mitochondrial and respiratory chain activity (Fig. S5; Table S7) while genes in ME7 and ME10 did not show any significant enrichment for GO terms (FDR < 0.1). These results show that the expression of many genes that are involved in transcriptional regulation and nervous system development positively correlate with ELP1 expression.
Discussion
Variants in Elongator subunits are associated with various human neurodevelopmental disorders, including FD (ELP1), ID (ELP2), ALS (ELP3), autism spectrum disorder and Rolandic epilepsy (ELP4) (Anderson et al., 2001; Slaugenhaupt et al., 2001; Cuajungco et al., 2003; Simpson et al., 2009; Strug et al., 2009; Najmabadi et al., 2011; Addis et al., 2015; Cohen et al., 2015; Kojic et al., 2018). To gain a better understanding the pathogenesis of FD, we generated two new mouse models expressing an increasing amount of ELP1, FD1/KO and FD9/KO. Although the human FD transgene did not rescue embryonic lethality in the ELP1 KO mouse, its expression improved embryonic development in a dose dependent manner. To identify genes and pathways whose expression is highly correlated with ELP1 and that are ultimately essential for embryonic development, we conducted a comprehensive transcriptome analysis in KO, FD1/KO, FD9/KO and WT embryos. We found that even a minimal increase in ELP1 has a dramatic effect on overall gene expression with the majority of the KO DEGs being completely rescued in the FD1/KO embryos, which only expresses an amount of ELP1 that is approximately 6% of the Elp1 amount expressed in the WT embryos. A significant portion of the down-regulated genes across the different genotypes KO, FD1/KO and FD9/KO, have a crucial role in nervous system development. Among these neuronal genes, Dbx1 and Nr2e1 were the two most down-regulated genes in all three genotypes with absent or reduced expression of ELP1.
Dbx1, also known as Developing Brain Homeobox 1, is expressed in a regionally restricted pattern in the developing mouse CNS and encodes for a transcription factor that has pivotal role in interneuron differentiation in the ventral spinal cord (Lu et al., 1996; Pierani et al., 2001). Dbx1 mutant mice exhibit marked changes in motor coordination, supporting the role of Dbx1-dependent interneurons as key components of the spinal locomotor circuits that control stepping movements in mammals (Lanuza et al., 2004). Interestingly, one of the most characteristic features of FD is poor locomotor coordination and both patients as well as a phenotypic mouse model of FD showed progressive impairment in coordination that leads to severe gait ataxia (Macefield et al., 2011; Macefield et al., 2013; Morini et al., 2019).
Nr2e1, or nuclear receptor subfamily 2 group E member 1, encodes a highly conserved transcription factor known to be a key stem cell fate determinant in both the developing mouse forebrain and retina (Pignoni et al., 1990; Yu et al., 1994; Monaghan et al., 1995; Monaghan et al., 1997; Jackson et al., 1998; Young et al., 2002; Miyawaki et al., 2004; Li et al., 2008; Schmouth et al., 2012). During development, Nr2e1 null mice display an increase in apoptotic levels of RGCs in the ganglion cell layer (GCL), which results in a marked reduction in thickness of the distinct layers in the adult retina and optic nerve dystrophy (Young et al., 2002; Miyawaki et al., 2004; Zhang et al., 2006). Intriguingly, degeneration of RGCs is observed in two different Elp1 conditional knock-out mice (Ueki et al., 2016; Ueki et al., 2018). In addition, patients with FD show RGCs loss with reduction in the thickness of the retinal nerve fiber layer (RNFL) and progressive vision loss (Mendoza-Santiesteban et al. 2012; Mendoza-Santiesteban et al. 2014; Mendoza-Santiesteban et al., 2017). Although further studies will be necessary to determine the link between ELP1 reduction and downregulation of specific key neurodevelopmental genes, the identified ELP1-dependent transcriptome profiles constitute an excellent foundational resource for understanding Elongator biology and also help to shed light into the molecular pathways that underlie diseases caused by disruption of Elongator activity.
In the nucleus, Elongator facilitates transcriptional elongation through altering chromatin structure (Otero et al., 1999; Wittschieben et al., 1999; Hawkes et al., 2002; Chen et al., 2009). Therefore, we analyzed the length distribution of DEGs among embryos expressing different ELP1 amounts. Our data clearly showed that long genes are more affected by the loss of functional Elongator compared with shorter genes and, by gradually increasing ELP1 amount, we were able to progressively restore their expression. Moreover, the observation that neuronal genes are significantly longer than all expressed genes, offers a possible explanation about why the nervous system is the tissue that most relies on functional Elongator during embryonic development. In the future it would be interesting to investigate the role of ELP1 in transcriptional elongation for each identified target, in order to identify direct regulatory effects.
Several genes that require Elongator to be efficiently expressed have been identified using either cell lines or conditional KO mouse tissues with reduced Elp1 expression (Boone et al. 2012; Cohen-Kupiec et al. 2011; Close et al., 2006; Abashidze et al., 2014; Zeltner et al., 2016; Goffena et al., 2018). In addition, RNA microarray analysis in post-mortem FD tissues has shown that a subset of genes involved in myelination require ELP1 for efficient transcription (Cheishvili et al., 2007). In the current study we have identified gene patterns whose expression varies as a function of ELP1 amount: (1) genes whose expression changes as a monotonic function of ELP1; (2) genes whose expression is completely restored with low amount of ELP1 and (3) genes whose expression is restored only when ELP1 is expressed at WT levels (Fig. 4C). It is interesting that distinctive groups of genes display different Elongator dependence. Genes important for axon formation and cell projection responded in a dose-dependent manner to functional Elongator amounts. This is consistent with the observation that, even though Elongator is a ubiquitously expressed protein complex, variants affecting different Elongator subunits all lead to neurodevelopmental diseases (Anderson et al., 2001; Slaugenhaupt et al., 2001; Cuajungco et al., 2003; Simpson et al., 2009; Strug et al., 2009; Najmabadi et al., 2011; Addis et al., 2015; Cohen et al., 2015; Kojic et al., 2018). On the other hand, the expression of important transcriptional regulators is already restored in FD9/KO embryos, suggesting that only a small increase of functional Elongator is necessary to rescue their expression. This might underlie the dramatic phenotypic improvement observed in FD9/KO embryos when compared with FD1/KO and KO. We then identified sets of genes whose expression negatively correlated with ELP1. GO analysis of these genes highlighted pathways involved in mitochondrial and respiratory chain activity.
In conclusion, our study is the first to assess the in vivo dose-dependent effect of ELP1 in early development using transcriptome analysis. We demonstrated that even a minimal increase in ELP1 can have a dramatic effect on both mouse embryonic development and global gene expression. Although loss of ELP1 compromised the expression of many genes, this study shows that neuronal genes are more sensitive to ELP1 reduction. Further studies will be necessary to determine which of the identified genes are directly regulated by ELP1, which would suggest that the expression changes result from a failure in transcriptional elongation, versus those that are modified indirectly, potentially due to the role of Elongator in translation. Previous studies using ChIP have demonstrated that ELP1 associates with the coding region of the genes whose expression is affected by its loss, and that histone H3 acetylation is reduced across these genes, supporting its direct role in transcriptional elongation (Close et al., 2006). However, we cannot exclude that some of the observed gene expression changes might be downstream effects due to the role of Elp1 in tRNA modification. It is worth noting, however, that even if the observed gene expression changes are not directly mediated by ELP1, they are certainly the result of ELP1 dosage.
The transcriptome-wide identification of gene networks and biological pathways that are regulated by ELP1 dosage described in this study is highly relevant to better understand the pathogenesis of FD, as well as other neurodevelopmental diseases caused by Elongator deficiency. The data presented here will help to identify potential biomarkers for future clinical studies and new targetable pathways for therapy.
Materials and Methods
Generation of FD1/KO, FD9/KO mouse models and genotyping
The detailed description of the original strategy to generate the Elp1 knockout mouse line has been previously published (Chen et al., 2009). A detailed description of the generation the TgFD transgenic lines carrying different copy number of the human ELP1 gene with the IVS20+6T>C mutation can be found in our previous manuscript by Hims et al. (Hims et al., 2007). To create the TgFD1; Elp1−/− and TgFD9; Elp1−/− mouse, we crossed the previously generated TgFD1 or TgFD9 transgenic mouse line with the mouse line heterozygous for the null allele Elp1−/+. The resulting progeny was genotyped to detect the presence of the TgFD1 or TgFD9 transgene and of the null allele Elp1−/−. As expected, the TgFD1 and TgFD9 transgene segregated independently from the null allele; therefore, around one-fourth of the F1 mice carried both the TgFD transgene and null Elp1 alleles (TgFD1; Elp1+/−or TgFD9; Elp1+/−). Subsequently, we crossed the TgFD1; Elp1+/− and TgFD9; Elp1+/− mice with the mouse line heterozygous for the null Elp1 allele (Elp1+/−). The resulting progeny was genotyped to detect the presence of the TgFD1 or TgFD9 transgene as well as the null allele in homozygosis (Elp1−/−). The Elp1−/−, TgFD1; Elp1−/−, TgFD9; Elp1−/− and Elp1+/+ embryos were produced by crossing heterozygote mice carrying different copy numbers of the FD ELP1 transgene (TgFD1; Elp1+/− or TgFD9; Elp1+/−) with heterozygote mice (Elp1−/+). The day of vaginal-plug discovery was designated E 0.5. The mice used for this study were housed in the animal facility of Massachusetts General Hospital (Boston, MA), provided with constant access to a standard diet of food and water, maintained on a 12-hour light/dark cycle, and all experimental protocols were approved by the Subcommittee on Research Animal Care at the Massachusetts General Hospital.
The genotypes of animals and embryos were determined by PCR analysis of genomic DNA from tail biopses and from embryos and/or visceral yolk sacs, respectively. The primer sets used were as follows: for determining the wild-type Elp1 allele, 5′-ACCCTCAGGCAGTTTGATTG-3′ and 5′-CATGGCTCCATAAAACAAACAC-3′; for detecting the knockout allele, 5′-ACCCTCAGGCAGTTTGATTG-3 ′ and 5 ′-GGCTACCGGCTAAAACTTGA-3 ′; and for determining the human TgFD transgenes, TgProbe1F 5 ′-GCCATTGTACTGTTTGCGACT-3 ′ and TgProbe1R 5′-TGAGTGTCACGATTCTTTCTGC-3 ′.
Morphological analysis of embryos
Photographs of visceral yolk sacs and embryos were taken with a digital camera Leica DFC7000 T mounted on a Leica M205 FCA dissection microscope. LAS X software (Leica) was used for image processing.
RNA-Seq experiment
RNA was extracted from 8 Elp1−/−, 7 TgFD1; Elp1−/−, 6 TgFD9; Elp1−/− and 8 Elp1+/+ individual embryos at the embryonic stage of E8.5 using the QIAzol Reagent and following the manufacturer’s instructions. RNAseq Libraries were prepared using TruSeq® Stranded mRNA Library Prep kit (Illumina 20020594), using 100 ng total RNA as input. Libraries were evaluated for final concentration and size distribution by Agilent 2200 TapeStation and/or qPCR, using Library Quantification Kit (KK4854, Kapa Biosystems), and multiplexed by pooling equimolar amounts of each library prior to sequencing. Pooled libraries were 50 base pair paired-end sequenced on Illumina HiSeq 2500 across multiple lanes. Real time image analysis and base calling were performed on the HiSeq 2500 instrument using HiSeq Sequencing Control Software (HCS) and FASTQ files demultiplexed using CASAVA software version 1.8.
A synthesized transcriptome reference was generated, by artificially adding the sequence of human ELP1 gene from the Ensembl human transcriptome reference GRCh37.75 to the Ensembl mouse transcriptome reference GRCm38.83 as an independent chromosome. RNA-Seq reads were mapped to this synthesized transcriptome reference 3 by STAR v2.5.2b, allowing only uniquely mapped reads with 5% mismatch (Dobin et al., 2013).
Differential gene expression analysis
Gene counts were generated by HTSeq-count v0.11.2 with “-s reverse” option to be compatible with Illumina TruSeq reads, according to the gene annotations of the synthesized transcriptome reference. Four genotypes were defined as “FD0”, “FD1”, “FD9” and WT to reflect the amounts of human and mouse ELP1. Gene counts across samples were filtered so that only genes whose median expression amounts were no less than 0.1 counts-per-million-reads in at least one genotype were kept for the downstream analysis. The R (v3.6.1) package “SVA” v3.32.1 was implemented on the filtered gene expression matrix to estimate surrogated variables (SVs) among samples. A generalized linear model was built by the R package “DESeq2” v1.24.0 to correlate gene expression to genotypes reflecting human ELP1 and mouse Elp1 amounts, together with all the estimated SVs. During the differential gene expression analysis, the FC and FDR of each genotype were estimated per gene. PCA analysis was implemented based on the top 500 most variable genes across samples, from the filtered gene expression matrix with SVs corrected.
GO analysis
GO analysis was done by GOrilla (Eden et al., 2009) (http://cbl-gorilla.cs.technion.ac.il/). Organism was set as Mus musculus. Two unranked lists of genes were used for each GO analysis. The GO analysis for DEGs used the following two lists: 1) the list of DEGs for a genotype and 2) the list of all genes in the filtered gene expression matrix. The GO analysis for co-expression modules used the following two lists: 1) the list of genes highly correlated with a hypothetical pattern in a co-expression module and 2) the list of all genes in the filtered gene expression matrix. In the figures related to the GO analysis for DEGs, ten significant terms in the ontology of Cellular Component were selected to be plotted in Fig. 2B. The complete list of DEG GO terms for each genotype can be found in Supplementary Table 2.
Mouse neuronal genes
Mouse neuronal genes were extracted and downloaded via AmiGO2 from geneontology.org, using the key word ‘neuron’ and restricting organism as ‘Mus musculus’. Totally 3,442 unique neuronal genes were found.
Co-expression modules analysis
R package “WGCNA” (Zhang and Horvath, 2005) v1.68 was implemented to the filtered gene expression matrix with SVs corrected. The soft-thresholding power was determined to be 5. The minimal module size was set to 30. The raw modules were merged using a dis-similarity cutoff of 0.15.
Correlation between co-expression modules and hypothetical patterns
Dummy expression data were generated to mimic three hypothetical patterns, namely the monotonic increase pattern, the monotonic decrease pattern, and the saturated pattern. The eigen vector representing each co-expression module was then correlated with each of the hypothetical patterns using Pearson correlation.
Statistical Analysis
All raw P values in this study, if multiple tests were involved, were corrected by the Benjamini-Hochberg Procedure and converted to FDR values. Wald test was applied in the differential gene expression analysis. A significant DEG of a genotype, compared to WT, was defined as FDR < 0.1 and the FC ≥ 1.5 for that genotype. Fisher’s exact test was used for GO analysis, where a significant enrichment for a GO term was defined as FDR < 0.1. Kolmogorov-Smirnov test (K-S test) was applied to compare the gene length distribution of genes from different groups. A significant difference in length distribution between two groups was defined as FDR < 0.1. A significant correlation throughout this study was defined as the Pearson correlation coefficient ≥ 0.85 and FDR < 0.1. For all box plots, the middle lines inside boxes indicated the medians. The lower and upper hinges corresponded to the first and third quartiles. Each box extended to 1.5 times inter-quartile range (IQR) from upper and lower hinges respectively. The symbols *, ** and ***, if appeared in the figures, indicated FDR < 0.1, < 0.01 and < 0.001, respectively.
Data availability
The RNA sequencing datasets generated during the current study are available in the GEO database: GSE186465
Any other relevant data are available from the authors upon reasonable request.
Supplementary Material
Acknoledgments
We thank Dr. Lucy Norcliffe-Kaufmann and Dr. Horacio Kaufmann of the Dysautonomia Treatment and Evaluation Center at New York University Medical School for their long-standing collaboration and helpful discussions. We are also grateful to Dr. Frances Lefcort for her comments on the manuscript. This work was supported by National Institutes of Health (NIH) grants (R37NS095640 to S.A.S.) and the Francis Crick Institute (to PC and JQS).
Funding: Research support from PTC Therapeutics, Inc. (S.A.S.).
Personal financial interests: Susan A. Slaugenhaupt is a paid consultant to PTC Therapeutics and is an inventor on several U.S. and foreign patents and patent applications assigned to the Massachusetts General Hospital, including U.S Patents 8,729,025 and 9,265,766, both entitled “Methods for altering mRNA splicing and treating familial dysautonomia by administering benzyladenine,” filed on August 31, 2012 and May 19, 2014 and related to use of kinetin; and U.S. Patent 10,675,475 entitled, “Compounds for improving mRNA splicing” filed on July 14, 2017 and related to use of BPN-15477. Elisabetta Morini, Dadi Gao, Michael E. Talkowski and Susan A. Slaugenhaupt are inventors on an International Patent Application Number PCT/US2021/012103, assigned to Massachusetts General Hospital and entitled “ RNA Splicing Modulation” related to use of BPN-15477 in modulating splicing.
Footnotes
CRediT authorship contribution statement
Elisabetta Morini: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - Original Draft. Dadi Gao: Conceptualization, Methodology, Software, Formal analysis, Writing - Original Draft. Emily Logan: Investigation. Monica Salani: Investigation, Writing - Review & Editing. Aram Krauson: Data Curation, Writing - Review & Editing. Anil Chekuri: Data Curation, Writing - Review & Editing. Yei-Tsung Chen: Investigation, Writing - Review & Editing. Ashok Ragavendran: Methodology, Software. Probir Chakravarty: Data Curation. Serkan Erdin: Methodology, Software, Writing - Review & Editing. Alexei Stortchevoi: Investigation. Jesper Svejstrup: Data Curation, Writing - Review & Editing. Michael Talkowski: Conceptualization, Supervision, Writing - Review & Editing. Susan Slaugenhaupt: Conceptualization, Supervision, Writing - Review & Editing, Funding acquisition.
Conflict of Interest
The authors declare competing financial interests.
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abashidze A, Gold V, Anavi Y, Greenspan H,Weil M, 2014. Involvement of IKAP in peripheral target innervation and in specific JNK and NGF signaling in developing PNS neurons. PLoS One 9, e113428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams BS, Kwok MC, Trinh E, Budaghzadeh S, Hossain SM,Simpson EM, 2005. Pathological aggression in “fierce” mice corrected by human nuclear receptor 2E1. J. Neurosci. 25, 6263–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addis L, Ahn JW, Dobson R, Dixit A, Ogilvie CM, Pinto D, Vaags AK, Coon H, Chaste P, Wilson S, et al. , 2015. Microdeletions of ELP4 Are Associated with Language Impairment, Autism Spectrum Disorder, and Mental Retardation. Hum Mutat 36, 842–850. [DOI] [PubMed] [Google Scholar]
- Anderson SL, Coli R, Daly IW, Kichula EA, Rork MJ, Volpi SA, Ekstein J,Rubin BY, 2001. Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 68, 753–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod FB, Hilz MJ, Berlin D, Yau PL, Javier D, Sweat V, Bruehl H,Convit A, 2010. Neuroimaging supports central pathology in familial dysautonomia. J. Neurol. 257, 198–206. [DOI] [PubMed] [Google Scholar]
- Bauer F,Hermand D, 2012. A coordinated codon-dependent regulation of translation by Elongator. Cell Cycle 11, 4524–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billingsley ML, Yun J, Reese BE, Davidson CE, Buck-Koehntop BA,Veglia G, 2006. Functional and structural properties of stannin: roles in cellular growth, selective toxicity, and mitochondrial responses to injury. J. Cell. Biochem. 98, 243–250. [DOI] [PubMed] [Google Scholar]
- Boone N, Bergon A, Loriod B, Deveze A, Nguyen C, Axelrod FB,Ibrahim EC, 2012. Genome-wide analysis of familial dysautonomia and kinetin target genes with patient olfactory ecto-mesenchymal stem cells. Hum. Mutat. 33, 530–540. [DOI] [PubMed] [Google Scholar]
- Buck-Koehntop BA, Mascioni A, Buffy JJ,Veglia G, 2005. Structure, dynamics, and membrane topology of stannin: a mediator of neuronal cell apoptosis induced by trimethyltin chloride. J. Mol. Biol. 354, 652–665. [DOI] [PubMed] [Google Scholar]
- Cheishvili D, Maayan C, Cohen-Kupiec R, Lefler S, Weil M, Ast G,Razin A, 2011. IKAP/Elp1 involvement in cytoskeleton regulation and implication for familial dysautonomia. Hum. Mol. Genet. 20, 1585–1594. [DOI] [PubMed] [Google Scholar]
- Cheishvili D, Maayan C, Smith Y, Ast G,Razin A, 2007. IKAP/hELP1 deficiency in the cerebrum of familial dysautonomia patients results in down regulation of genes involved in oligodendrocyte differentiation and in myelination. Hum. Mol. Genet. 16, 2097–2104. [DOI] [PubMed] [Google Scholar]
- Chen YT, Hims MM, Shetty RS, Mull J, Liu L, Leyne M,Slaugenhaupt SA, 2009. Loss of mouse Ikbkap, a subunit of elongator, leads to transcriptional deficits and embryonic lethality that can be rescued by human IKBKAP. Mol. Cell. Biol. 29, 736–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhang H, Jablonowski D, Zhou X, Ren X, Hong X, Schaffrath R, Zhu JK,Gong Z, 2006. Mutations in ABO1/ELO2, a subunit of holo-Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol. Cell. Biol. 26, 6902–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close P, Hawkes N, Cornez I, Creppe C, Lambert CA, Rogister B, Siebenlist U, Merville MP, Slaugenhaupt SA, Bours V, et al. , 2006. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol. Cell. 22, 521–531. [DOI] [PubMed] [Google Scholar]
- Cohen JS, Srivastava S, Farwell KD, Lu HM, Zeng W, Lu H, Chao EC,Fatemi A, 2015. ELP2 is a novel gene implicated in neurodevelopmental disabilities. Am. J. Med. Genet. A 167, 1391–1395. [DOI] [PubMed] [Google Scholar]
- Cohen-Kupiec R, Pasmanik-Chor M, Oron-Karni V,Weil M, 2011. Effects of IKAP/hELP1 deficiency on gene expression in differentiating neuroblastoma cells: implications for familial dysautonomia. PLoS One 6, e19147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantine R, Zhang H, Gerstner CD, Frederick JM,Baehr W, 2012. Uncoordinated (UNC)119: coordinating the trafficking of myristoylated proteins. Vision Res. 75, 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O, Laguesse S, Cornez I, Rahmouni S, Ormenese S, et al. , 2009. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell 136, 551–564. [DOI] [PubMed] [Google Scholar]
- Cuajungco MP, Leyne M, Mull J, Gill SP, Lu W, Zagzag D, Axelrod FB, Maayan C, Gusella JF,Slaugenhaupt SA, 2003. Tissue-specific reduction in splicing efficiency of IKBKAP due to the major mutation associated with familial dysautonomia. Am. J. Hum. Genet. 72, 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich P, Yue J, E, Dragatsis IS, 2011. Deletion of exon 20 of the Familial Dysautonomia gene Ikbkap in mice causes developmental delay, cardiovascular defects, and early embryonic lethality. PLoS One 6, e27015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M,Gingeras TR, 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D,Yakhini Z, 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellows J, Erdjument-Bromage H, Tempst P,Svejstrup JQ, 2000. The Elp2 subunit of elongator and elongating RNA polymerase II holoenzyme is a WD40 repeat protein. J. Biol. Chem. 275, 12896–12899. [DOI] [PubMed] [Google Scholar]
- Frohloff F, Jablonowski D, Fichtner L,Schaffrath R, 2003. Subunit communications crucial for the functional integrity of the yeast RNA polymerase II elongator (gamma-toxin target (TOT)) complex. J. Biol. Chem. 278, 956–961. [DOI] [PubMed] [Google Scholar]
- George L, Chaverra M, Wolfe L, Thorne J, Close-Davis M, Eibs A, Riojas V, Grindeland A, Orr M, Carlson GA, et al. , 2013. Familial dysautonomia model reveals Ikbkap deletion causes apoptosis of Pax3+ progenitors and peripheral neurons. Proc. Natl. Acad. Sci. U S A 110, 18698–18703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffena J, Lefcort F, Zhang Y, Lehrmann E, Chaverra M, Felig J, Walters J, Buksch R, Becker KG,George L, 2018. Elongator and codon bias regulate protein levels in mammalian peripheral neurons. Nature communications 9, 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q, Hou X, Su D, Pan L, Duan J, Cui L, Huang B,Lu J, 2007. hELP3 subunit of the Elongator complex regulates the transcription of HSP70 gene in human cells. Acta Biochim. Biophys. Sin. (Shanghai) 39, 453–461. [DOI] [PubMed] [Google Scholar]
- Hawkes NA, Otero G, Winkler GS, Marshall N, Dahmus ME, Krappmann D, Scheidereit C, Thomas CL, Schiavo G, Erdjument-Bromage H, et al. , 2002. Purification and characterization of the human elongator complex. J. Biol. Chem. 277, 3047–3052. [DOI] [PubMed] [Google Scholar]
- Hims MM, Shetty RS, Pickel J, Mull J, Leyne M, Liu L, Gusella JF,Slaugenhaupt SA, 2007. A humanized IKBKAP transgenic mouse models a tissue-specific human splicing defect. Genomics 90, 389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Johansson MJ,Bystrom AS, 2005. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11, 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A, Panayiotidis P,Foroni L, 1998. The human homologue of the Drosophila tailless gene (TLX): characterization and mapping to a region of common deletion in human lymphoid leukemia on chromosome 6q21. Genomics 50, 34–43. [DOI] [PubMed] [Google Scholar]
- Jackson MZ, Gruner KA, Qin C,Tourtellotte WG, 2014. A neuron autonomous role for the familial dysautonomia gene ELP1 in sympathetic and sensory target tissue innervation. Development 141, 2452–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen LD, Naumanen T, Knudsen A, Westerlund N, Gromova I, Junttila M, Nielsen C, Bottzauw T, Tolkovsky A, Westermarck J, et al. , 2008. IKAP localizes to membrane ruffles with filamin A and regulates actin cytoskeleton organization and cell migration. J. Cell. Sci. 121, 854–864. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lane WS,Reinberg D, 2002. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc Natl Acad Sci U S A 99, 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic M, Gaik M, Kiska B, Salerno-Kochan A, Hunt S, Tedoldi A, Mureev S, Jones A, Whittle B, Genovesi LA, et al. , 2018. Elongator mutation in mice induces neurodegeneration and ataxia-like behavior. Nature communications 9, 3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic M,Wainwright B, 2016. The Many Faces of Elongator in Neurodevelopment and Disease. Front. Mol. Neurosci. 9, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ,Greenblatt JF, 2001. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 8203–8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuja-Panula J, Kiiltomaki M, Yamashiro T, Rouhiainen A,Rauvala H, 2003. AMIGO, a transmembrane protein implicated in axon tract development, defines a novel protein family with leucine-rich repeats. J. Cell. Biol. 160, 963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labosky PA,Kaestner KH, 1998. The winged helix transcription factor Hfh2 is expressed in neural crest and spinal cord during mouse development. Mech. Dev. 76, 185–190. [DOI] [PubMed] [Google Scholar]
- Lanuza GM, Gosgnach S, Pierani A, Jessell TM,Goulding M, 2004. Genetic identification of spinal interneurons that coordinate left-right locomotor activity necessary for walking movements. Neuron 42, 375–386. [DOI] [PubMed] [Google Scholar]
- Li F, Lu J, Han Q, Zhang G,Huang B, 2005. The Elp3 subunit of human Elongator complex is functionally similar to its counterpart in yeast. Mol Genet Genomics 273, 264–272. [DOI] [PubMed] [Google Scholar]
- Li F, Ma J, Ma Y, Hu Y, Tian S, White RE,Han G, 2011. hElp3 directly modulates the expression of HSP70 gene in HeLa cells via HAT activity. PLoS One 6, e29303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Fazly AM, Zhou H, Huang S, Zhang Z,Stillman B, 2009. The elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet. 5, e1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Sun G, Yang S, Qu Q, Nakashima K,Shi Y, 2008. Nuclear receptor TLX regulates cell cycle progression in neural stem cells of the developing brain. Mol Endocrinol 22, 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Shashikant CS,Ruddle FH, 1996. Separate cis-acting elements determine the expression of mouse Dbx gene in multiple spatial domains of the central nervous system. Mech. Dev. 58, 193–202. [DOI] [PubMed] [Google Scholar]
- Macefield VG, Norcliffe-Kaufmann L, Gutierrez J, Axelrod FB,Kaufmann H, 2011. Can loss of muscle spindle afferents explain the ataxic gait in Riley-Day syndrome? Brain 134, 3198–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macefield VG, Norcliffe-Kaufmann LJ, Axelrod FB,Kaufmann H, 2013. Relationship between proprioception at the knee joint and gait ataxia in HSAN III. Mov. Disord. 28, 823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahloudji M, Brunt PW,McKusick VA, 1970. Clinical neurological aspects of familial dysautonomia. J. Neurol. Sci. 11, 383–395. [DOI] [PubMed] [Google Scholar]
- Mendoza-Santiesteban CE, Hedges Iii TR, Norcliffe-Kaufmann L, Axelrod F,Kaufmann H, 2014. Selective retinal ganglion cell loss in familial dysautonomia. J Neurol 261, 702–709. [DOI] [PubMed] [Google Scholar]
- Mendoza-Santiesteban CE, Hedges TR 3rd, Norcliffe-Kaufmann L, Warren F, Reddy S, Axelrod FB,Kaufmann H, 2012. Clinical neuro-ophthalmic findings in familial dysautonomia. J. Neuroophthalmol. 32, 23–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Santiesteban CE, Palma JA, Hedges TR 3rd, Laver NV, Farhat N, Norcliffe-Kaufmann L,Kaufmann H, 2017. Pathological Confirmation of Optic Neuropathy in Familial Dysautonomia. J. Neuropathol. Exp. Neurol. 76, 238–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki T, Uemura A, Dezawa M, Yu RT, Ide C, Nishikawa S, Honda Y, Tanabe Y,Tanabe T, 2004. Tlx, an orphan nuclear receptor, regulates cell numbers and astrocyte development in the developing retina. J. Neurosci. 24, 8124–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan AP, Bock D, Gass P, Schwager A, Wolfer DP, Lipp HP,Schutz G, 1997. Defective limbic system in mice lacking the tailless gene. Nature 390, 515–517. [DOI] [PubMed] [Google Scholar]
- Monaghan AP, Grau E, Bock D,Schutz G, 1995. The mouse homolog of the orphan nuclear receptor tailless is expressed in the developing forebrain. Development 121, 839–853. [DOI] [PubMed] [Google Scholar]
- Morini E, Gao D, Montgomery CM, Salani M, Mazzasette C, Krussig TA, Swain B, Dietrich P, Narasimhan J, Gabbeta V, et al. , 2019. ELP1 Splicing Correction Reverses Proprioceptive Sensory Loss in Familial Dysautonomia. Am. J. Hum. Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, Hosseini M, Behjati F, Haas S, Jamali P, et al. , 2011. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478, 57–63. [DOI] [PubMed] [Google Scholar]
- Nelissen H, Fleury D, Bruno L, Robles P, De Veylder L, Traas J, Micol JL, Van Montagu M, Inze D,Van Lijsebettens M, 2005. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc. Natl. Acad. Sci. U S A 102, 7754–7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa JG, 2003. Familial dysautonomia (Riley-Day syndrome) may be associated with epilepsy. Epilepsia 44, 472. [DOI] [PubMed] [Google Scholar]
- Ohlen SB, Russell ML, Brownstein MJ,Lefcort F, 2017. BGP-15 prevents the death of neurons in a mouse model of familial dysautonomia. Proc Natl Acad Sci U S A 114, 5035–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P,Svejstrup JQ, 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell. 3, 109–118. [DOI] [PubMed] [Google Scholar]
- Pearson J, 1979. Familial dysautonomia (a brief review). J. Auton. Nerv. Syst. 1, 119–126. [DOI] [PubMed] [Google Scholar]
- Pierani A, Moran-Rivard L, Sunshine MJ, Littman DR, Goulding M,Jessell TM, 2001. Control of interneuron fate in the developing spinal cord by the progenitor homeodomain protein Dbx1. Neuron 29, 367–384. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Baldarelli RM, Steingrimsson E, Diaz RJ, Patapoutian A, Merriam JR,Lengyel JA, 1990. The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell 62, 151–163. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. , 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Chen CZ,Collins RN, 2005. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol. Cell. 17, 841–853. [DOI] [PubMed] [Google Scholar]
- Schmouth JF, Banks KG, Mathelier A, Gregory-Evans CY, Castellarin M, Holt RA, Gregory-Evans K, Wasserman WW,Simpson EM, 2012. Retina restored and brain abnormalities ameliorated by single-copy knock-in of human NR2E1 in null mice. Mol. Cell. Biol. 32, 1296–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CL, Lemmens R, Miskiewicz K, Broom WJ, Hansen VK, van Vught PW, Landers JE, Sapp P, Van Den Bosch L, Knight J, et al. , 2009. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet. 18, 472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaugenhaupt SA, 2002. Genetics of familial dysautonomia. Tissue-specific expression of a splicing mutation in the IKBKAP gene. Clin. Auton. Res. 12 Suppl. 1, I15–19. [DOI] [PubMed] [Google Scholar]
- Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, Cuajungco MP, Liebert CB, Chadwick B, Idelson M, Reznik L, et al. , 2001. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 68, 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Paolinelli R, Kloss H, Scorza FB, Marchesi S, Sauder U, Mitsushima D, Capuani F, Sturzenbaum SR,Cassata G, 2010. The Caenorhabditis elegans Elongator complex regulates neuronal alpha-tubulin acetylation. PLoS Genet. 6, e1000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strug LJ, Clarke T, Chiang T, Chien M, Baskurt Z, Li W, Dorfman R, Bali B, Wirrell E, Kugler SL, et al. , 2009. Centrotemporal sharp wave EEG trait in rolandic epilepsy maps to Elongator Protein Complex 4 (ELP4). Eur. J. Hum. Genet. 17, 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. , 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic. Acids. Res. 47, D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng L, Mundell NA, Frist AY, Wang Q,Labosky PA, 2008. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development 135, 1615–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HT, Nimick M, Uhrig RG, Templeton G, Morrice N, Gourlay R, DeLong A,Moorhead GB, 2012. Arabidopsis thaliana histone deacetylase 14 (HDA14) is an alpha-tubulin deacetylase that associates with PP2A and enriches in the microtubule fraction with the putative histone acetyltransferase ELP3. Plant. J. 71, 263–272. [DOI] [PubMed] [Google Scholar]
- Ueki Y, Ramirez G, Salcedo E, Stabio ME,Lefcort F, 2016. Loss of Ikbkap Causes Slow, Progressive Retinal Degeneration in a Mouse Model of Familial Dysautonomia. eNeuro 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki Y, Shchepetkina V,Lefcort F, 2018. Retina-specific loss of Ikbkap/Elp1 causes mitochondrial dysfunction that leads to selective retinal ganglion cell degeneration in a mouse model of familial dysautonomia. Dis. Model. Mech. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vas V, Kovacs T, Kormendi S, Brody A, Kudlik G, Szeder B, Mezo D, Kallai D, Koprivanacz K, Mero BL, et al. , 2019. Significance of the Tks4 scaffold protein in bone tissue homeostasis. Sci. Rep. 9, 5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J, Kwon SY, Badenhorst P, East P, McNeill H,Svejstrup JQ, 2011. Role of elongator subunit Elp3 in Drosophila melanogaster larval development and immunity. Genetics 187, 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler GS, Petrakis TG, Ethelberg S, Tokunaga M, Erdjument-Bromage H, Tempst P,Svejstrup JQ, 2001. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 276, 32743–32749. [DOI] [PubMed] [Google Scholar]
- Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P,Svejstrup JQ, 1999. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell. 4, 123–128. [DOI] [PubMed] [Google Scholar]
- Young KA, Berry ML, Mahaffey CL, Saionz JR, Hawes NL, Chang B, Zheng QY, Smith RS, Bronson RT, Nelson RJ, et al. , 2002. Fierce: a new mouse deletion of Nr2e1; violent behaviour and ocular abnormalities are background-dependent. Behav. Brain. Res. 132, 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu RT, Chiang MY, Tanabe T, Kobayashi M, Yasuda K, Evans RM,Umesono K, 2000. The orphan nuclear receptor Tlx regulates Pax2 and is essential for vision. Proc. Natl. Acad. Sci. U S A 97, 2621–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu RT, McKeown M, Evans RM,Umesono K, 1994. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature 370, 375–379. [DOI] [PubMed] [Google Scholar]
- Zeltner N, Fattahi F, Dubois NC, Saurat N, Lafaille F, Shang L, Zimmer B, Tchieu J, Soliman MA, Lee G, et al. , 2016. Capturing the biology of disease severity in a PSC-based model of familial dysautonomia. Nat. Med. 22, 1421–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B,Horvath S, 2005. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, Article17. [DOI] [PubMed] [Google Scholar]
- Zhang CL, Zou Y, Yu RT, Gage FH,Evans RM, 2006. Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes. Dev. 20, 1308–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing datasets generated during the current study are available in the GEO database: GSE186465
Any other relevant data are available from the authors upon reasonable request.