Abstract
Many transcription factors are multifunctional and also influence DNA replication. So far, their mechanism of action has remained elusive. Here we show that a DNA-binding protein could rely on the same biochemical activity that activates transcription to stimulate replication from the yeast chromosomal ARS1 origin. Unexpectedly, the ability to stimulate replication from this origin was not restricted to polymerase II transcription factors, but was a property shared by polymerase III factors. Furthermore, activation of replication did not depend on the process of transcription, but rather on the ability of DNA-binding transcription factors to remodel chromatin. The natural ARS1 activator Abf1 and the other transcription factors that stimulated replication remodeled chromatin in a very similar manner. Moreover, the presence of a histone H3 mutant that was previously shown to generally increase transcription also facilitated replication from ARS1 and partially compensated for the absence of a transcription factor. We propose that multifunctional transcription factors work by influencing the chromatin architecture at replication origins so as to generate a structure that is favorable to the initiation of replication.
INTRODUCTION
Initiation of DNA replication is limited to a specific time during the cell cycle and to specific locations on the chromosome. One level of regulation of DNA replication might be provided by the interplay between gene expression and replication, which is thought to facilitate the temporal control of both processes during the cell cycle and integrate origin selection with the gene transcription program (1–9).
In the yeast Saccharomyces cerevisiae, origins of replication are defined sequences that often harbor binding sites for transcription factors (10). For example, the so-called autonomously replicating sequence 1 (ARS1) is one of the best-characterized origins of replication and contains four genetically defined cis-acting A and B elements. The A element is the binding site for the origin recognition complex (ORC) (11) and is absolutely essential for ARS1 function, whereas individual B elements contribute positively to the efficiency of replication (12). The B1 element provides an additional binding site for the ORC (13,14), whereas the B2 element has been suggested to facilitate DNA unwinding (15). The B3 element is a binding site for the transcription factor Abf1 (16). The function of the B3 element of ARS1 can be replaced by binding sites for other yeast transcription factors such as Rap1 and Gal4 (12). In addition, activating domains from mammalian transcription factors such as VP16 and p53 can stimulate DNA replication in yeast when tethered to the ARS1 sequence (17). These results taken together show that at least some transcription factors are multifunctional and can also directly influence the initiation of DNA replication (18,19).
It has been postulated that involvement of the same transcription factors in the activation of promoter and origin functions requires that these proteins are either endowed with two different biochemical activities or that they have one single activity that stimulates two distinct processes (12,17). The results of in vitro experiments have suggested that at least some transcription factors might indeed possess two distinct biochemical activities. In these experiments, transcription factors capable of stimulating viral DNA replication were shown to interact not only with components of the RNA polymerase II (pol II) complex, but also with a replication factor, replication protein A (20,21). However, the results of plasmid stability assays from our laboratory indicate that one biochemical activity of transcription factors might be sufficient to stimulate both transcription from a promoter and replication from an origin site in yeast. In these experiments, a single protein–protein interaction established between the DNA-binding domain of Gal4 and the RNA pol II holoenzyme component Gal11P was shown to cause stimulation of plasmid replication in yeast (22). Since the same interaction also activates gene transcription by recruiting the pol II holoenzyme to DNA (23–25), it is most likely that the same mechanism causes stimulation of DNA replication by this DNA-binding protein. The question about which activity of the recruited pol II transcription complex could stimulate DNA replication was left open in our published work. According to one of the several hypotheses that have been put forward, recruitment of the transcription complex to DNA could remodel chromatin structures that act as general repressors of origin function (22). Such putative remodeling could indirectly facilitate the assembly of an active DNA replication complex at the ARS1 site. In agreement with this notion, biochemical studies with reconstituted chromatin templates have shown that some transcription factors activate viral DNA replication by counteracting the inhibitory effect of nucleosomes (26,27). Moreover, expression of the human breast cancer protein BRCA1 fused to the DNA-binding domain of Gal4, which can activate transcription in yeast, caused chromatin remodeling and stimulated DNA replication from a modified ARS1, where the B elements were replaced with binding sites for the fusion protein. Mutant forms of BRCA1 that did not stimulate replication were found to be unable to remodel chromatin (28). However, these experiments still did not clarify whether chromatin remodeling and stimulation of replication by the transcription factor were the consequence of the same activity that also activated transcription or of a different activity, e.g. interaction and recruitment of replication factors that would be specific for its function at this origin site.
Here we show that the defined Gal4–Gal11P interaction that can activate transcription by recruiting the pol II complex to DNA (23–25) can also stimulate replication from a chromosomal ARS1 site. We also demonstrate that stimulation of chromosomal DNA replication could be induced by factors involved in RNA polymerase III (pol III) transcription. Furthermore, we show that activation of replication did not depend on the process of transcription, but rather on the ability of DNA-binding transcription factors to remodel chromatin. Abf1, the natural ARS1 activator, also remodeled chromatin at the ARS1 site in a manner that was very similar to that of the other transcription factors capable of stimulating replication. In support of the idea that initiation of replication can be regulated by changes in chromatin structure, we also show that the presence of a mutant histone H3, which has been characterized for its ability to cause increased levels of transcription of several genes (29), facilitated replication. Thus, we suggest that DNA-binding transcription factors able to remodel chromatin in a manner that is favorable to the function of an origin site are capable of stimulating replication from that site.
MATERIALS AND METHODS
Strains and plasmids
Strains RL1 and RL5 have been described by Hu et al. (28), while the strains used for the experiments shown in Figure 6 have been described by Marahrens and Stillman (30). Strains RMY200 and RMY247 used for the experiments shown in Figure 3 have been described by Mann and Grunstein (29). Strains YKE1 and YKE2 are derivatives of RL1 in which the Gal4-binding sites in the modified ARS1 locus were replaced by the B-SNR6 and UASG-SNR6 cassettes by ‘pop-in and pop-out’ integration of plasmids pKE5 and pKE6, respectively. pKE5 was constructed by insertion of a 430 bp PCR product containing part of the TRP1 gene and an oligomer containing a mutated B1 element of ARS1 into a derivative of the pARS1/–B23/G24 plasmid (17) containing the SNR6 gene with the B block (B-SNR6) replacing the Gal4-binding site. CEN4 was then removed by EcoRI + PvuII digestion and religation. pKE6 was as pKE5 except that it contained the UASG-SNR6 cassette. Plasmid pKE11 was used to replace the endogenous SNR6 by the shorter SNR6-6 gene. To construct pKE11, a 580 bp fragment containing part of the SNR6 gene bearing two deletions of 3 bp each (SNR6-6) was amplified and inserted into YIplac211 that had been digested with XbaI and SphI. Centromeric plasmids expressing Gal11P or Gal11 protein (pKE9 and pKE10) are derivatives of pSO32 and pSO23 (24), respectively. The sequence encoding Gal4(1–100) was cloned into a yeast expression vector carrying the inducible CUP1 promoter (17). Gal4(1–94) and Gal4–CTF expression cassettes, a kind gift of Rong Li (31), were also under the control of the CUP1 promoter. Expression of the proteins was induced by growing yeast in the presence of 100 µM copper sulfate.
Figure 6.
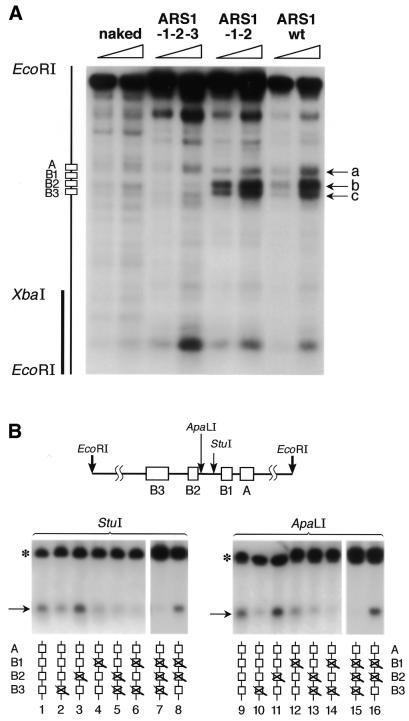
Abf1 remodels chromatin at the native ARS1 locus and causes structural changes that are very similar to those caused by the other transcription factors. (A) Indirect end-labeling was used to analyze the micrococcal nuclease digestion pattern around the ARS1 region. Two different concentrations of micrococcal nuclease were used (open triangles). The radioactive probe is indicated by a vertical bar (left). The approximate positions of the four elements of ARS1 are indicated at left. The cutting sites in chromatin were compared to those in deproteinized DNA (naked) from the ARS1–1–2–3 strain. The arrows (a–c) indicate the bands whose intensity was most significantly affected by the presence of a functional (Abf1-binding) B3 element (ARS1–1–2 and ARS1 wt). (B) Different restriction nuclease enzymes were used to analyze the accessibility of sites around ARS1 (not drawn to scale). The asterisk marks the fragment obtained by complete EcoRI digestion and the arrows point to fragments obtained by analytical restriction nuclease digestion.
Figure 3.
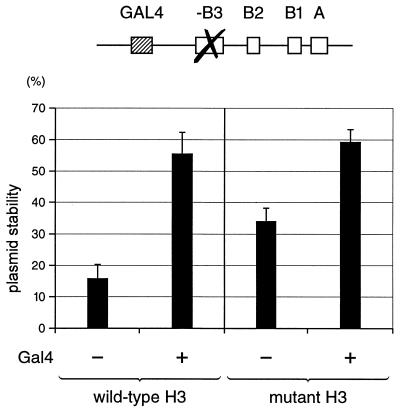
Histone H3 N-terminal mutations reduce the dependence on Gal4 for DNA replication from a ARS1 plasmid. Yeast cells containing the wild-type or mutant H3 were transformed with a centromeric plasmid bearing the modified ARS1 sequence shown in the diagram at the top. Cells were either grown on galactose to induce the activating function of Gal4 (+) or on glucose to repress Gal4 (–). Stability of the test plasmid, which reflects the efficiency of replication, is expressed as the percentage of yeast cells that still retained the plasmid after non-selective growth for 14 generations. The results are an average of three independent experiments.
Plasmid stability assays
Plasmid stability assays were performed as described (12). Gal4 expression was induced by growth in galactose-containing medium.
Two-dimensional gel electrophoresis
Two-dimensional gel electrophoresis of the genomic NcoI restriction fragment containing the ARS1 locus was performed as described before (28), except that a 2410 bp PCR product corresponding to the 5′ NcoI–EcoRV fragment of this region was used as probe.
Primer extension assays
Primer extension assays were performed by mixing 5–10 µg total RNA with 1 pmol 32P-labeled oligo C61G (5′-gcaggggaactcctgatcatctctg) specific for the SNR6 transcript. This mix was first denatured at 95°C, incubated for hybridization at 55°C for 1.5 h, precipitated and resuspended in 10 µl of water. The samples were then processed according to the standard protocol included in the Omniscript Reverse Transcription Kit (Qiagen).
Genomic chromatin analysis
Micrococcal nuclease and restriction nuclease digestion assays were performed as described (32,33). Genomic DNA was digested with EcoRI, resolved by gel electrophoresis and probed with a radiolabeled 186 bp oligonucleotide sequence corresponding to the EcoRI–XbaI fragment from the ARS1 locus. The size marker was a DNA ladder consisting of multiples of 256 bp.
RESULTS
A single biochemical activity of a DNA-binding protein that activates transcription also stimulates chromosomal DNA replication
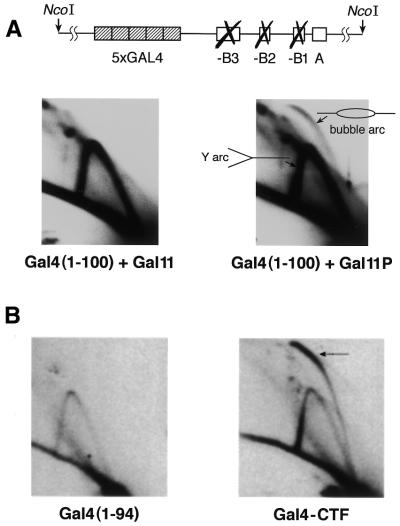
Our previous results have shown that the same, defined interaction between Gal4(1–100) and Gal11P, which activates transcription by recruitment of the pol II transcription complex to DNA, could also stimulate replication from a plasmid ARS1 in yeast (22). Expression of wild-type Gal11, which does not interact with Gal4(1–100), or the combination of Gal11P with Gal4(1–94), which lacks an essential part of the Gal11P-interacting domain (23), did not stimulate plasmid replication in these assays (22). To analyze whether the Gal4–Gal11P interaction can also stimulate replication from the chromosomal ARS1 site, we used the yeast strain RL1 (a kind gift from Rong Li) that carries a modified ARS1 in which the B elements are mutated and five Gal4-binding sites are inserted nearby (28). A vector expressing the Gal4(1–100) protein was integrated into the genome of RL1. The resulting strain was further transformed with centromeric plasmids that express either wild-type Gal11 or Gal11P (Gal11P is dominant over the endogenous wild-type Gal11) (23). Two-dimensional gel electrophoretic assays were performed to monitor chromosomal DNA replication. Two types of replication intermediates that encompassed the ARS1 origin could be distinguished: those that initiated from ARS1 and traced a characteristic bubble arc and those that resulted from passive replication of this region by a replication fork that initiated at neighboring origins and traced a so-called Y arc (Fig. 1A). Replication from the RL1 ARS1 site was stimulated only in the presence of both Gal11P and Gal4(1–100). Indeed, neither a combination of wild-type Gal11 with Gal4(1–100) nor of Gal11P with Gal4(1–94) was able to activate replication (Fig. 1A and data not shown). Moreover, the presence of a functional ORC-binding A element was absolutely required for initiation of replication stimulated by the Gal4–Gal11P interaction (data not shown).
Figure 1.
Recruitment of the RNA pol II holoenzyme component Gal11P to DNA stimulates replication from a chromosomal ARS1 site. (A) Two-dimensional gel analysis of chromosomal ARS1 replication intermediates generated in the RL1 yeast strain expressing Gal4(1–100) together with either Gal11 or Gal11P. The schematic structure of the modified ARS1 locus located in the 4.7 kb genomic NcoI restriction fragment is shown at the top. The indicated bubble arc is formed by replication intermediates that initiated from ARS1, while the Y arc is mostly generated by a replication fork that initiated at neighboring origins (see text for more details). (B) Two-dimensional gel analysis of chromosomal ARS1 replication intermediates generated in the RL1 yeast strain expressing Gal4(1–94) or Gal4–CTF (the Gal4 DNA-binding domain fused to the activation domain of CTF). The position of the bubble arc is indicated by an arrow.
In control experiments, we used the same technique to monitor DNA replication stimulated by Gal4–CTF, a fusion protein bearing the DNA-binding domain of Gal4 fused to the proline-rich activation domain of CTF that is known to stimulate replication from a chromosomal ARS1 site (31). Figure 1B shows that expression of this transcriptional activator stimulated replication from the ARS1 site of strain RL1, while Gal4(1–94)p, which bears the DNA-binding domain but lacks the activating function, was not able to stimulate replication. Stimulation of replication by Gal4–CTF was stronger than activation by the Gal4–Gal11P interaction. The expression of Gal4 derivatives used in these experiments was verified by electrophoretic mobility shift assays and by gene activation experiments in which both activation of a lacZ reporter gene and growth on galactose were monitored (data not shown).
These results show that the well-defined Gal4–Gal11P interaction, which activates transcription by recruitment of the pol II transcription complex to DNA and can stimulate replication of a plasmid, can also stimulate DNA replication from a chromosomal origin site.
RNA pol III transcription factors can also stimulate DNA replication
The transcription factors that have been shown to stimulate DNA replication all belong to the RNA pol II class of transcription factors (12,17,22,28). In order to investigate whether the ability to stimulate DNA replication is an exclusive property of pol II transcription factors, we tested stimulation of DNA replication by RNA pol III factors that are involved in activation of the yeast SNR6 gene. This gene, which encodes the U6 snRNA, is controlled by four distinct cis-regulatory elements that bind pol III transcription factors. These elements are the TATA sequence at position –30, a proximal sequence called PSE at –60, two overlapping intragenic A blocks at +21/+29 and a downstream B block at +234 bp from the initiation site (34,35). Mutations of the B block that abolish SNR6 transcription in vivo have been described (35).
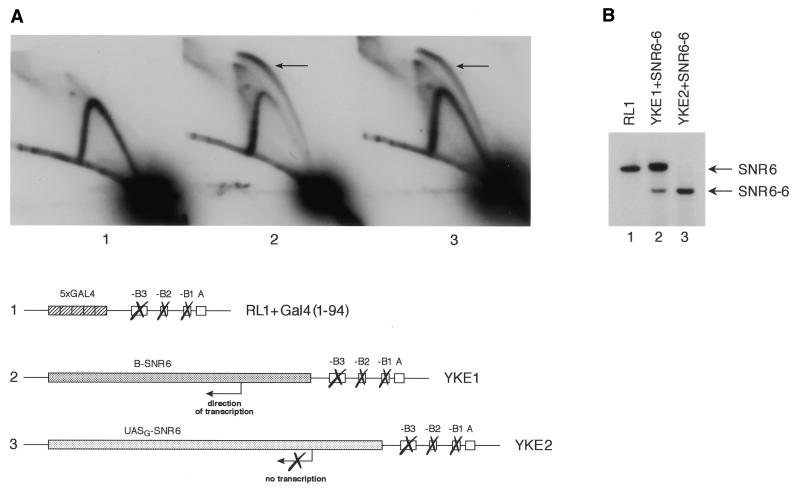
To test the potential for activating DNA replication by RNA pol III transcription factors, we substituted the Gal4-binding sites near the ARS1 locus of strain RL1 with either a SNR6 gene cassette bearing all the natural promoter elements (B-SNR6 in the YKE1 strain), or with the UASG-SNR6 gene cassette, in which a Gal4-binding UASG sequence is substituted for the B block (35) (UASG-SNR6 in the YKE2 strain). Both gene cassettes were inserted near the ARS1 locus so that, if active, they would be transcribed in the opposite direction to the replication origin. Two-dimensional electrophoretic analysis of these modified ARS1 sequences showed that the presence of both versions of the SNR6 gene near the modified ARS1 caused stimulation of DNA replication from this origin, as compared to strain RL1, in which ARS1 is inactive in the absence of DNA-tethered activating domains (Fig. 2A). The natural B-SNR6 gene showed a slightly higher stimulatory activity on DNA replication originating from the modified ARS1 than the UASG-SNR6 gene, as quantitated by comparative ImageQuaNT analysis of bubble arc signals with Y arc signals (Fig. 2A).
Figure 2.
DNA sequences that recruit RNA pol III transcription factors can also stimulate replication from a chromosomal ARS1 site. (A) Two-dimensional gel analysis of chromosomal ARS1 replication intermediates generated in the following yeast strains: (1) RL1 expressing Gal4(1–94); (2) YKE1, a derivative of RL1 in which the Gal4-binding sites near ARS1 have been replaced by the pol III B-SNR6 gene cassette; (3) YKE2, which contains the UASG-SNR6 cassette. The ARS1 regions of these strains (1–3) are schematically drawn to scale. The bubble arc is indicated by an arrow and is formed by replication intermediates that initiated from ARS1. (B) Primer extension analysis of the B-SNR6 and UASG-SNR6 gene products reveals that stimulation of replication by these pol III gene cassettes is not dependent on the transcription process. The endogenous wild-type SNR6 gene of both the YKE1 and YKE2 strains was replaced by a 6 bp shorter SNR6 sequence (SNR6-6) to allow the detection of transcripts specifically synthesized from the pol III gene cassettes near the ARS1 locus. Primer extension analysis of the RL1 endogenous SNR6 gene products served as a positive control for expression and size of the full-length U6 RNA.
The UASG-SNR6 gene is expected to be silent (35), while the B-SNR6 gene should be transcribed by RNA pol III. In such a case, one would conclude that stimulation of replication in these assays would not depend on the transcription process per se. To test this possibility, we used the primer extension technique to monitor transcription levels from the B-SNR6 and the UASG-SNR6 gene cassettes of strains YKE1 and YKE2, respectively. To distinguish between transcripts synthesized from the SNR6 locus and those from the B-SNR6 and the UASG-SNR6 gene cassettes near the ARS1 locus, we substituted the SNR6 gene at the original locus with a 6 bp shorter, yet fully functional, version of the SNR6 gene (SNR6-6 gene; M.Petrascheck and A.Barberis, unpublished results). Figure 2B shows that the B-SNR6 gene next to the modified ARS1 was actively transcribed, whereas no transcript from the UASG-SNR6 gene could be detected. The shorter SNR6-6 gene was present and transcribed in both strains YKE1 and YKE2. Strain RL1 served as a positive control in these assays to assess the expression of full-length U6 snRNA.
These results show for the first time that the ability to stimulate replication from ARS1 is not restricted to pol II transcription factors, but is also a property shared by RNA pol III factors. Moreover, the act of transcription by RNA pol III appears to be unnecessary for stimulation of DNA replication in our assays.
N-terminal mutations of histone H3 facilitate replication and reduce the dependence on transcription factor(s) for efficient initiation
Since DNA replication from the ARS1 origin can be stimulated by transcription factors involved in either RNA pol II or pol III transcription, it is reasonable to postulate the existence of a mechanism for activating replication that must be common to these different classes of transcription factors. Such a common mechanism might be chromatin remodeling, which could create a chromatin structure that favors initiation of the replication process. Based on this idea, we tested the possibility that a histone H3 mutant, which has been characterized for its ability to cause an increased level of transcription of several pol II and pol III genes, might increase the initiation rate from a replication origin site. This mutant H3 protein bears the amino acid substitutions K9G, K14G and K18G that knock out acetylation sites within its N-terminus. It has been suggested that this increased expression of pol II and pol III genes in the presence of the H3 mutant is due to a chromatin structure that is less constrained and perhaps more accessible to the transcriptional machinery (29,36). It is therefore possible that this mutant H3 protein might increase the efficiency of replication initiation by inducing a chromatin structure that is more accessible to the replication machinery.
DNA replication in yeast strains carrying either mutant or wild-type H3 was monitored by plasmid stability assay. For these experiments, we used a centromeric plasmid bearing a modified ARS1 sequence in which a Gal4-binding site had been inserted next to a mutated B3 element, which no longer binds the transcription factor Abf1. Replication efficiency of this plasmid was reflected in the percentage of yeast cells that still retained the plasmid after 14 generations of non-selective growth (12). Figure 3 shows that, as expected, Gal4 stimulated the replication efficiency of the reporter plasmid in the wild-type H3 strain, as indicated by increased plasmid stability in the presence of this transcription factor. In the mutant H3 strain, plasmid stability was higher than in the wild-type H3 strain in the absence of Gal4. This transcription factor further stimulated replication in the mutant H3 strain, but the overall stimulation was significantly lower than in the wild-type strain. Thus, the presence of the mutant H3 facilitated replication and partially overcame the requirement for the Gal4 transcription factor for more efficient replication of the reporter plasmid.
Activation of DNA replication by pol II and pol III transcription factors correlates with their ability to remodel chromatin
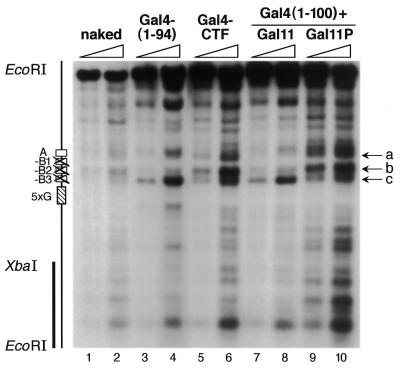
The preceding results are consistent with the idea that the primary role of transcription factors in stimulating replication is to remodel chromatin so as to create a structure that favors function of the replication machinery in initiating DNA replication at an origin site. To assess whether activation of replication from ARS1 by all the different transcription factors tested in this work correlates with chromatin remodeling at this site, we examined chromatin structures by micrococcal and restriction nuclease assay. Figure 4 shows that expression of Gal4–CTF, which is able to stimulate DNA replication, caused prominent changes in the micrococcal nuclease digestion pattern at the replication origin (lanes 5 and 6) relative to the pattern observed in the presence of Gal4(1–94) (lanes 3 and 4), which binds DNA but is unable to stimulate replication. The same types of changes in digestion pattern were also caused by expression of Gal4(1–100) together with the interacting Gal11P (lanes 9 and 10), which activates replication, as compared to expression of the same DNA-binding protein in the presence of the non-interacting wild-type Gal11 (lanes 7 and 8). In particular, a replication stimulatory function added to otherwise silent DNA-binding Gal4 derivatives enhanced the sensitivity of the ARS1 chromatin to micrococcal nuclease at two sites (indicated by arrows a and b), while maintaining the sensitivity of another site (indicated by arrow c).
Figure 4.
Activation of replication induces a specific alteration of the chromatin structure at the chromosomal ARS1 replication origin. Indirect end-labeling was used to analyze the micrococcal nuclease digestion pattern around the ARS1 region. Two different concentrations of micrococcal nuclease were used (open triangles). The radioactive probe is indicated by a vertical bar (left). The approximate positions of the Gal4-binding sites and the four elements of ARS1 are indicated at left. The cutting sites in chromatin were compared to those in deproteinized DNA (naked) from the RL1 strain. The arrows (a–c) indicate the bands whose intensity was most significantly affected by the stimulatory transcription factors Gal4–CTF and Gal4(1–100) + Gal11P.
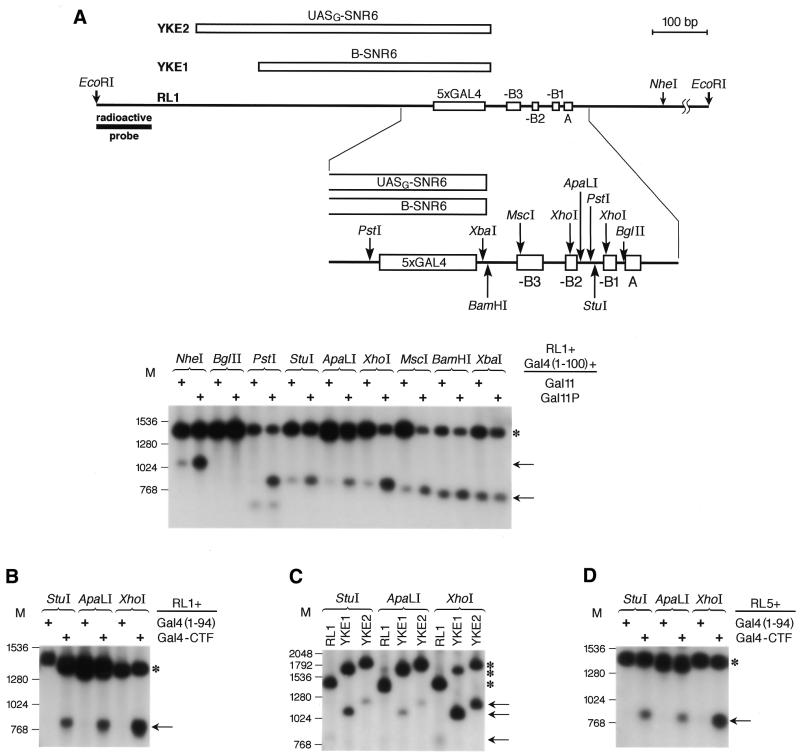
To better quantitate and map the changes in chromatin structures within and near the ARS1 site, we measured relative DNA accessibility by restriction nuclease assay. We first assessed chromatin accessibility in a RL1-derived yeast strain expressing Gal4(1–100) together with either wild-type Gal11 or Gal11P, which interacts with Gal4(1–100) and stimulates replication from the modified ARS1 (see above). The accessibility of most restriction sites present within and near the RL1 ARS1 locus was strongly enhanced by the presence of the replication-stimulating Gal4(1–100) + Gal11P pair, as compared to the degree of accessibility in the presence of the silent Gal4(1–100) + Gal11 pair (Fig. 5A). The most prominent changes in restriction nuclease sensitivity were present in the region of the ARS1 sequence in which the micrococcal nuclease hypersensitive sites were localized (compare Fig. 4 with 5A). The accessibility of the region surrounding the Gal4-binding sites seemed to be constitutive and was relatively high, while that of the A element region appeared to be constitutively very low, as expected for the continuous presence of the ORC protecting this region from nucleases (37). Quantitation of the levels of restriction site accessibility by scanning densitometry analysis gave the following results (increased accessibility in the presence of Gal11P versus Gal11): NheI, 6-fold increase; BglII, undetectable; PstI upper band, 52-fold; PstI lower band, unchanged; StuI, 5-fold; ApaLI, 10-fold, XhoI, 28-fold; MscI, 18-fold; BamHI, 1.7-fold; XbaI, 1.9-fold.
Figure 5.
Activators of the ARS1 function cause increased DNA accessibility to restriction nucleases over the chromosomal chromatin region of the replication origin. (A) Different restriction nuclease enzymes were used to analyze the accessibility of sites at and around ARS1. Strains, restriction nuclease sites and the radioactive probe used are depicted at the top (drawn to scale). The asterisk marks the fragment obtained by complete EcoRI digestion and the arrows point to fragments obtained by analytical restriction nuclease digestion (the stronger the band, the more accessible the site). Nuclease restriction sites, in particular those localized over the B1–B2 region (PstI, StuI, ApaLI and XhoI), are more accessible in the RL1 strain expressing the activating Gal4p(1–100) + Gal11P pair than in the RL1 strain expressing the non-activating Gal4p(1–100) + Gal11 pair. (B) Restriction sites localized over the B1–B2 region are more accessible in the presence of Gal4(1–94)–CTF than in the presence of Gal4(1–94)p. The asterisk marks the fragment obtained by complete EcoRI digestion and the arrow points to fragments obtained by analytical restriction nuclease digestion. (C) The accessibility of restriction nuclease sites at the ARS1 locus of strains YKE1 and YKE2 is compared to the accessibility of the same sites in strain RL1 expressing non-activating Gal4(1–94). In both strain YKE1 and YKE2 chromatin is remodeled, although somewhat more efficiently in strain YKE1, which correlates with the slightly higher efficiency of replication stimulation in YKE1. The asterisk marks the fragment obtained by complete EcoRI digestion and the arrows point to fragments obtained by analytical restriction nuclease digestion. Fragment sizes vary between strains RL1, YKE1 and YKE2 because of the different inserts flanking the B3 element. (D) Chromatin remodeling is not a consequence of replication. Accessibility of the restriction sites in the (–A–B1–B2–B3) ARS1 of strain RL5 expressing Gal4(1–94) was compared to the accessibility of sites in the same strain expressing the activating Gal4–CTF fusion protein.
We also measured chromatin accessibility to restriction nucleases in strains in which replication from the ARS1 site was activated by either Gal4–CTF (Fig. 5B) or by RNA pol III transcription factors (Fig. 5C). Both types of transcription factors changed the accessibility of chromatin to restriction nucleases in a similar manner (compare Fig. 5B and C). The pattern of changes caused by Gal4–CTF and by RNA pol III transcription factors was also similar to that caused by the Gal4(1–100) + Gal11P pair (Fig. 5A–C and data not shown). Moreover, the relative accessibility of these sites to restriction nucleases (Fig. 5) roughly correlated with the extent of stimulation of replication brought about by the various transcription factors (see Figs 1 and 2). These results show that different types of pol II activators and pol III transcription factors that can stimulate replication from a nearby origin also cause a similar pattern of changes in the chromatin structure at the replication origin site.
It has been shown that chromatin remodeling by a Gal4–BRCA1 hybrid also occurs at a mutated ARS1 site that no longer sustains initiation of replication (28). For these experiments, the authors constructed the yeast strain RL5, a derivative of RL1 containing a point mutation in the ARS1 A element that completely abolishes replication initiation (17,30,38), yet does not affect chromatin remodeling caused by the presence of Gal4–BRCA1 (28). These results led the authors to conclude that chromatin remodeling is likely to be a cause, rather than an effect, of activated replication (28). We used the restriction nuclease assay to quantitate changes in chromatin structures caused by transcription factors in the absence of replication from the RL5 ARS1, which was confirmed (data not shown). Figure 5D shows that Gal4–CTF, but not Gal4(1–94), was able to enhance accessibility of chromatin even in the absence of replication initiation. These results confirm that the observed chromatin remodeling is not a consequence of DNA replication and support the idea that chromatin remodeling is the mechanism for activating replication that is common to all the different transcription factors tested so far.
Abf1 binding to the ARS1 B3 element causes chromatin changes that are very similar to those caused by other transcription factors
The analysis of chromatin remodeling and stimulation of DNA replication presented so far has been limited to pol II and pol III transcription factors that are not natural activators of the replication origins under study. It is therefore very important to analyze the effect on chromatin structures of a natural activator of origin function, such as the transcription factor Abf1. It has been shown that an ARS1 locus bearing a mutated B3 element unable to bind Abf1 displays a different chromatin structure from ARS1 sequences harboring an intact B3 element (28,39,40). We wanted to test the hypothesis that Abf1 may cause changes in chromatin structure at the modified ARS1 site that are similar to those caused by the pol II and pol III transcription factors analyzed previously. To this end, we analyzed chromatin accessibility to nucleases at the ARS1 locus of strains containing wild-type B elements (ARS1 wt) or mutated B1 and B2 elements, such as those present in strain RL1, in addition to either a mutated B3 element (ARS1-1-2-3) or a wild-type B3 element that can bind Abf1 (ARS1-1-2). DNA replication cannot efficiently initiate from ARS1-1-2-3, while it is stimulated by Abf1 from ARS1-1-2 (30). Figure 6A shows that the presence of the Abf1-binding element B3 in both the ARS1 wt and ARS1-1-2 sequences enhanced sensitivity to micrococcal nuclease in a region of the origin sequence that was much less sensitive in the absence of this element (ARS1-1-2-3). The enhanced sensitivity caused by the presence of the B3 element was qualitatively and quantitatively similar between ARS1 wt and ARS1-1-2. Most importantly, the micrococcal nuclease digestion pattern caused by the Abf1-binding element B3 strongly resembled the digestion pattern of the RL1 ARS1 site in the presence of the replication-stimulating factors Gal4–CTF and Gal4(1–100) + Gal11P (compare bands a–c of Fig. 6A to those shown in Fig. 4). Therefore, efficient DNA replication from this ARS1 site requires chromatin remodeling so as to create a structure that is specifically favorable to the assembly of a functional replication machinery. Bands a and b shown in both Figures 4 and 6A are diagnostic for such a replication-favorable chromatin structure at this region around the B1 and B2 elements of the ARS1 origin.
Restriction nuclease analysis of ARS1-1-2-3 and ARS1-1-2 (Fig. 6B) confirmed that Abf1 binding to the B3 element enhanced DNA accessibility to a degree that strongly resembled that observed with the RL1 ARS1 site in the presence of replication-stimulating pol II and pol III transcription factors (compare Fig. 6B with 5). Indeed, the region between the B1 and B2 elements was much more accessible to the restriction nucleases ApaLI and StuI in the presence of the B3 element than in its absence. A very similar result was obtained with the pol II and pol III transcription factors, as shown in Figure 5. Since the pattern of chromatin remodeling caused by a natural activator of ARS1 and that caused by the pol II and pol III ARS1 activators tested here are very similar, we conclude that remodeling of chromatin at a specific region of ARS1 is the most likely mechanism by which the endogenous factor Abf1 stimulates replication from ARS1.
Figure 6B also shows the contribution of the individual B elements and of combinations of them to the chromatin structure of this region of the ARS1 origin. Compared to wild-type ARS1, mutation of the B2 element alone did not change DNA accessibility to ApaLI and StuI. In contrast, mutations of either the B1 or the B3 element partially reduced such accessibility. The combination of mutated B1 and B3 elements caused a marked protection of this ARS1 region against these restriction nucleases. Thus, in addition to the Abf1-binding B3 element, the B1 element also plays a role in determining the chromatin structure of this region of ARS1. This is consistent with the function of B1 in providing a binding site for ORC in addition to that provided by the essential A element (13,14), and it is also consistent with a recent report that the ORC has the ability to position nucleosomes (41).
DISCUSSION
The results of our analysis of the factors that can regulate DNA replication show that the function of the endogenous transcription factor Abf1 in stimulating DNA replication from the chromosomal ARS1 origin can be replaced by a specific and unique interaction between a DNA-binding protein [Gal4(1–100)] and a component of the pol II holoenzyme (Gal11P). This particular interaction is known to activate gene transcription from a promoter by recruiting the holoenzyme to DNA (23–25,42). Similarly, the stimulatory function of Abf1 can be replaced by DNA sequences (SNR6 gene cassettes) that tether pol III transcription factors near the ARS1 origin site. In this case, we also show that transcription is not required for stimulation of DNA replication (see Figs 1 and 2). This set of results indicates that: (i) DNA-binding proteins can use the same biochemical activity that activates transcription from a gene promoter to stimulate DNA replication from the chromosomal ARS1 sequence bearing its recognition site; (ii) the ability to stimulate replication from this origin is not a prerogative of pol II transcription factors and does not require the process of transcription. To our knowledge, this is the first report indicating that pol III transcription factors binding near an origin site can stimulate DNA replication. As yet, no replication origin has been characterized in yeast or in metazoans that is functionally linked to a nearby pol III gene. However, a database search indicated that some S.cerevisiae ARS elements are located close to RNA pol III genes. The yeast ribosomal ARS elements are also located near 5S genes transcribed by RNA pol III (43) and might therefore be among candidates of interest for investigating the potential effects of pol III genes on the stimulation of replication.
We have tested the hypothesis that chromatin remodeling might be a mechanism for stimulating DNA replication that is common to all the different transcription factors analyzed thus far. The idea is that various transcription factors use their ability to remodel chromatin, which might be inherent in their function of activating transcription, to create a chromatin structure that is favorable to the process of replication initiation. Several published results provide support for this idea. For example, many activators of pol II genes have been shown to cause chromatin remodeling (44). In the specific case of activation by the Gal4–Gal11P interaction, it has been shown that recruitment of the RNA pol II holoenzyme by this interaction can reposition nucleosomes at a promoter, even in the absence of transcription (45). Similarly, recruitment of the RNA pol III complex to the SNR6 gene is known to cause chromatin remodeling (36). Therefore, chromatin remodeling as a general mechanism provides the simplest explanation for how such a variety of activators, as well as the recruitment of the pol II and pol III transcription complexes, can all activate the same process. Our results show that, indeed, the transcription factors capable of stimulating replication in our experiments, including the natural activator Abf1 that binds the B3 element, are all also capable of remodeling chromatin at the ARS1 origin site. Most importantly, the pattern of micrococcal and restriction nuclease digestion at the replication origin is very similar for all tested transcription factors, thus revealing the presence of a chromatin structure that is common to ARS1 origin sites activated by different transcription factors. Therefore, the results of our chromatin analysis by micrococcal and restriction nuclease assays provide evidence in support of chromatin remodeling as the relevant mechanism for stimulation of replication by transcription factors.
One additional line of evidence in support of the chromatin remodeling hypothesis is provided by the results showing that the presence of a histone H3 mutant in yeast facilitates replication and reduces the dependence of a plasmid ARS1 on a transcription factor for efficient replication (see Fig. 3). This particular H3 mutant, which bears the acetylation site substitutions K9G, K14G and K18G within the N-terminus, has been characterized for its ability to cause increased levels of transcription of several pol II genes, including Gal4-regulated genes (29), as well as to enhance transcription of mutated SNR6 genes (36). Such an increased expression level of pol II as well as pol III genes in the presence of this H3 mutant has been interpreted as the consequence of a perturbed or more relaxed structure of nucleosomes containing this mutant histone, which could generally facilitate assembly of transcription complexes on the DNA and subsequent transcription (29,36). In our assays, this H3 mutant might promote a relaxed, more accessible chromatin structure and thus stimulate replication. An alternative explanation might be suggested by the reported interaction between Mcm2 and histone H3 (46): structural changes in H3, which might be caused by the amino acid substitutions described above, could enhance binding of Mcm2 to this histone protein and promote recruitment of other Mcm proteins to the replication origin. However, the fact that these mutations in H3 cause stimulation of both transcription and replication makes it unlikely that they have such a general effect by changing the interactions with factors that are specific for the replication process.
The experiments presented in this paper do not address the question of the identity of the activities that actually remodel chromatin. Clearly, DNA binding per se is not sufficient to remodel chromatin to create a structure that is favorable to the initiation of replication. This conclusion, which has already been drawn by other authors (28), is particularly evident in the case of stimulation of replication by the Gal4(1–100)–Gal11P interaction. In this case, in the presence of non-interacting wild-type Gal11, the same DNA-binding protein Gal4(1–100) is unable to induce a chromatin structure that allows, or stimulates, efficient initiation of replication. The following possibilities for the chromatin remodeling function of transcription factors can be imagined. Chromatin remodeling might be a consequence of some of the activities of the recruited pol II and pol III transcription complexes. Our results show, however, that transcription is not required for stimulation of replication, thus suggesting that the polymerase activity itself is not involved in the process. Alternatively, chromatin remodeling might be carried out by machineries specialized for the disruption or repositioning of nucleosomes. While it is not yet known whether chromatin remodeling machineries associate with pol III transcription factors in yeast, the human TFIIIC complex has been shown to contain a histone acetyltransferase activity (47). In contrast to the small amount of information regarding pol III-associated remodeling machineries, a large body of evidence has established that several specialized machineries work with pol II transcription factors to remodel chromatin. For example, the chromatin remodeling Swi–Snf complex can be directly contacted by sequence-specific pol II transcription factors and can also be indirectly recruited through its association with the pol II holoenzyme (48,49). Similarly, various histone acetylase activities that can modify the structure of chromatin have been shown to be direct targets of transcription factors and also to be associated with the pol II transcription complex that is recruited to DNA (50–52). Thus, these data suggest how sequence-specific pol II transcription factors that either directly contact chromatin remodeling machineries or bind the pol II transcription complex, as is clearly the case for the Gal4(1–100)–Gal11P interaction, can cause changes in the nearby chromatin structure. In support of the involvement of specialized chromatin remodeling machineries in stimulating DNA replication, recent results of plasmid stability assays have suggested that the Swi/Snf complex can play a role not only in transcription but also in replication (53). Nevertheless, it is known that chromatin can be remodeled by other activities in addition to Swi/Snf and histone acetylases (54,55). Considering the results showing that replication can be stimulated by a variety of pol II and pol III transcription factors, it is also likely that several of these activities can influence the chromatin architecture at replication origins so as to create a structure that is favorable to the assembly of an active replication complex.
The idea that chromatin structures are very important components of the regulatory process of DNA replication has already been suggested by several studies, which have correlated the presence of nucleosomes over an origin with a loss of origin function (39,40,56). In particular, it has been shown that forcing a nucleosome over the A and B1 elements of ARS1 eliminates initiation (56). Furthermore, in support of the relevance of chromatin structure in the regulation of replication initiation, recent results show that the ORC, which binds the A and B1 elements, is necessary to maintain the nucleosomal configuration adjacent to ARS1 (41). It has also been suggested that the nucleosome positioned by the ORC proximal to the origin is not only kept from invading the origin sequence, but might even play a positive role in stimulating initiation of replication. Our restriction nuclease analysis of ARS1 shows that the B1 element is also required to maintain an open (accessible) chromatin structure over the ARS1 regulatory elements. It is therefore possible that the ORC remodels chromatin on both sides of its recognition sequence, thereby positioning a positively acting nucleosome adjacent to the A element of the origin and interfering with formation of an inhibitory structure over the origin regulatory sequences. In this scenario, the transcription factor Abf1, as well as the other pol II and pol III transcription factors tested in this work, could work together with the ORC to maintain an open chromatin structure over the origin sequence that is readily accessible to additional replication factors.
Analysis of the mechanisms by which transcription factors can modulate the efficiency of origin function is very important for an understanding of regulation of replication not only in the yeast S.cerevisiae, where most characterized replication origins carry binding sites for known transcription factors such as Abf1 and Rap1, but also in other organisms. Indeed, it has recently been shown that many replication origins in Schizosaccharomyces pombe (6) and in mammals (1) are located in close proximity to gene promoters and/or enhancers. It is plausible that while replication origins in S.cerevisiae have evolved to carry their own regulatory transcription factor-binding sites, S.pombe and mammalian replication origins have been selected for their localization near enhancer/promoter sequences; these may represent two strategies to achieve the same goal, i.e. regulated and efficient initiation of replication through an interplay between transcription factors, chromatin and the DNA replication machinery.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs F. Thoma and J. Sogo for advice on chromatin analysis and two-dimensional gel assays, and for critical comments on the manuscript. We also thank L. Badi, U. Hübscher, L. Martin, M. Petrascheck and I. Stagljar for comments on the manuscript and R. Li, B. Stillman and W. Hörz for materials and protocols. This work was supported by grants from the Swiss National Science Foundation (31-49485.96), the Roche Research Foundation and the Helmut Horten Foundation.
REFERENCES
- 1.Delgado S., Gomez,M., Bird,A. and Antequera,F. (1998) Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J., 17, 2426–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benard M., Pallotta,D. and Pierron,G. (1992) Structure and identity of a late-replicating and transcriptionally active gene. Exp. Cell Res., 201, 506–513. [DOI] [PubMed] [Google Scholar]
- 3.de Stanchina E., Gabellini,D., Norio,P., Giacca,M., Peverali,F.A., Riva,S., Falaschi,A. and Biamonti,G. (2000) Selection of homeotic proteins for binding to a human DNA replication origin. J. Mol. Biol., 299, 667–680. [DOI] [PubMed] [Google Scholar]
- 4.DePamphilis M.L. (1999) Replication origins in metazoan chromosomes: fact or fiction? Bioessays, 21, 5–16. [DOI] [PubMed] [Google Scholar]
- 5.Ferguson B.M. and Fangman,W.L. (1992) A position effect on the time of replication origin activation in yeast. Cell, 68, 333–339. [DOI] [PubMed] [Google Scholar]
- 6.Gomez M. and Antequera,F. (1999) Organization of DNA replication origins in the fission yeast genome. EMBO J., 18, 5683–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatton K.S., Dhar,V., Brown,E.H., Iqbal,M.A., Stuart,S., Didamo,V.T. and Schildkraut,C.L. (1988) Replication program of active and inactive multigene families in mammalian cells. Mol. Cell. Biol., 8, 2149–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heintz N.H. (1996) DNA replication in mammals. In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 983–1004.
- 9.Ten Hagen K.G., Gilbert,D.M., Willard,H.F. and Cohen,S.N. (1990) Replication timing of DNA sequences associated with human centromeres and telomeres. Mol. Cell. Biol., 10, 6348–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newlon C.S. and Theis,J.F. (1993) The structure and function of yeast ARS elements. Curr. Opin. Genet. Dev., 3, 752–758. [DOI] [PubMed] [Google Scholar]
- 11.Bell S.P. and Stillman,B. (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature, 357, 128–134. [DOI] [PubMed] [Google Scholar]
- 12.Marahrens Y. and Stillman,B. (1992) A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science, 255, 817–823. [DOI] [PubMed] [Google Scholar]
- 13.Rao H. and Stillman,B. (1995) The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc. Natl Acad. Sci. USA, 92, 2224–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowley A., Cocker,J.H., Harwood,J. and Diffley,J.F. (1995) Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J., 14, 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin S. and Kowalski,D. (1997) Functional equivalency and diversity of cis-acting elements among yeast replication origins. Mol. Cell. Biol., 17, 5473–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diffley J.F. and Stillman,B. (1988) Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc. Natl Acad. Sci. USA, 85, 2120–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li R., Yu,D.S., Tanaka,M., Zheng,L., Berger,S.L. and Stillman,B. (1998) Activation of chromosomal DNA replication in Saccharomyces cerevisiae by acidic transcriptional activation domains. Mol. Cell. Biol., 18, 1296–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stucki M., Stagljar,I., Jonsson,Z.O. and Hubscher,U. (2000) A coordinated interplay: proteins with multiple functions in DNA replication, DNA repair, cell cycle/checkpoint control and transcription. Prog. Nucleic Acid Res. Mol. Biol., 65, 261–298. [DOI] [PubMed] [Google Scholar]
- 19.van der Vliet P.C. (1996) Roles of transcription factors in DNA replication. In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 87–118.
- 20.He Z., Brinton,B.T., Greenblatt,J., Hassell,J.A. and Ingles,C.J. (1993) The transactivator proteins VP16 and GAL4 bind replication factor A. Cell, 73, 1223–1232. [DOI] [PubMed] [Google Scholar]
- 21.Li R. and Botchan,M.R. (1993) The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell, 73, 1207–1221. [DOI] [PubMed] [Google Scholar]
- 22.Stagljar I., Hubscher,U. and Barberis,A. (1999) Activation of DNA replication in yeast by recruitment of the RNA polymerase II transcription complex. Biol. Chem., 380, 525–530. [DOI] [PubMed] [Google Scholar]
- 23.Barberis A., Pearlberg,J., Simkovich,N., Farrell,S., Reinagel,P., Bamdad,C., Sigal,G. and Ptashne,M. (1995) Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell, 81, 359–368. [DOI] [PubMed] [Google Scholar]
- 24.Farrell S., Simkovich,N., Wu,Y., Barberis,A. and Ptashne,M. (1996) Gene activation by recruitment of the RNA polymerase II holoenzyme. Genes Dev., 10, 2359–2367. [DOI] [PubMed] [Google Scholar]
- 25.Gaudreau L., Adam,M. and Ptashne,M. (1998) Activation of transcription in vitro by recruitment of the yeast RNA polymerase II holoenzyme. Mol. Cell, 1, 913–916. [DOI] [PubMed] [Google Scholar]
- 26.Cheng L. and Kelly,T.J. (1989) Transcriptional activator nuclear factor I stimulates the replication of SV40 minichromosomes in vivo and in vitro. Cell, 59, 541–551. [DOI] [PubMed] [Google Scholar]
- 27.Li R. and Botchan,M.R. (1994) Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc. Natl Acad. Sci. USA, 91, 7051–7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y.F., Hao,Z.L. and Li,R. (1999) Chromatin remodeling and activation of chromosomal DNA replication by an acidic transcriptional activation domain from BRCA1. Genes Dev., 13, 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mann R.K. and Grunstein,M. (1992) Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J., 11, 3297–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marahrens Y. and Stillman,B. (1994) Replicator dominance in a eukaryotic chromosome. EMBO J., 13, 3395–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li R. (1999) Stimulation of DNA replication in Saccharomyces cerevisiae by a glutamine- and proline-rich transcriptional activation domain. J. Biol. Chem., 274, 30310–30314. [DOI] [PubMed] [Google Scholar]
- 32.Gregory P.D., Barbaric,S. and Horz,W. (1999) Restriction nucleases as probes for chromatin structure. Methods Mol. Biol., 119, 417–425. [DOI] [PubMed]
- 33.Livingstone-Zatchej M. and Thoma,F. (1999) Mapping of nucleosome positions in yeast. Methods Mol. Biol., 119, 363–378. [DOI] [PubMed] [Google Scholar]
- 34.Brow D.A. and Guthrie,C. (1990) Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev., 4, 1345–1356. [DOI] [PubMed] [Google Scholar]
- 35.Marsolier M.C., Chaussivert,N., Lefebvre,O., Conesa,C., Werner,M. and Sentenac,A. (1994) Directing transcription of an RNA polymerase III gene via GAL4 sites. Proc. Natl Acad. Sci. USA, 91, 11938–11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marsolier M.C., Tanaka,S., Livingstone-Zatchej,M., Grunstein,M., Thoma,F. and Sentenac,A. (1995) Reciprocal interferences between nucleosomal organization and transcriptional activity of the yeast SNR6 gene. Genes Dev., 9, 410–422. [DOI] [PubMed] [Google Scholar]
- 37.Diffley J.F., Cocker,J.H., Dowell,S.J. and Rowley,A. (1994) Two steps in the assembly of complexes at yeast replication origins in vivo. Cell, 78, 303–316. [DOI] [PubMed] [Google Scholar]
- 38.Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- 39.Thoma F., Bergman,L.W. and Simpson,R.T. (1984) Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J. Mol. Biol., 177, 715–733. [DOI] [PubMed] [Google Scholar]
- 40.Venditti P., Costanzo,G., Negri,R. and Camilloni,G. (1994) ABFI contributes to the chromatin organization of Saccharomyces cerevisiae ARS1 B-domain. Biochim. Biophys. Acta, 1219, 677–689. [DOI] [PubMed] [Google Scholar]
- 41.Lipford J.R. and Bell,S.P. (2001) Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell, 7, 21–30. [DOI] [PubMed] [Google Scholar]
- 42.Barberis A. and Gaudreau,L. (1998) Recruitment of the RNA polymerase II holoenzyme and its implications in gene regulation. Biol. Chem., 379, 1397–1405. [DOI] [PubMed] [Google Scholar]
- 43.Banditt M., Koller,T. and Sogo,J.M. (1999) Transcriptional activity and chromatin structure of enhancer-deleted rRNA genes in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 4953–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kadonaga J.T. (1998) Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell, 92, 307–313. [DOI] [PubMed] [Google Scholar]
- 45.Gaudreau L., Schmid,A., Blaschke,D., Ptashne,M. and Horz,W. (1997) RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell, 89, 55–62. [DOI] [PubMed] [Google Scholar]
- 46.Ishimi Y., Komamura,Y., You,Z. and Kimura,H. (1998) Biochemical function of mouse minichromosome maintenance 2 protein. J. Biol. Chem., 273, 8369–8375. [DOI] [PubMed] [Google Scholar]
- 47.Kundu T.K., Wang,Z. and Roeder,R.G. (1999) Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol. Cell. Biol., 19, 1605–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallberg A.E., Neely,K.E., Hassan,A.H., Gustafsson,J.A., Workman,J.L. and Wright,A.P. (2000) Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor tau1 activation domain. Mol. Cell. Biol., 20, 2004–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson C.J., Chao,D.M., Imbalzano,A.N., Schnitzler,G.R., Kingston,R.E. and Young,R.A. (1996) RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell, 84, 235–244. [DOI] [PubMed] [Google Scholar]
- 50.Brown C.E., Lechner,T., Howe,L. and Workman,J.L. (2000) The many HATs of transcription coactivators. Trends Biochem. Sci., 25, 15–19. [DOI] [PubMed] [Google Scholar]
- 51.Peterson C.L. and Workman,J.L. (2000) Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev., 10, 187–192. [DOI] [PubMed] [Google Scholar]
- 52.Sudarsanam P. and Winston,F. (2000) The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet., 16, 345–351. [DOI] [PubMed] [Google Scholar]
- 53.Flanagan J.F. and Peterson,C.L. (1999) A role for the yeast SWI/SNF complex in DNA replication. Nucleic Acids Res., 27, 2022–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belotserkovskaya R. and Berger,S.L. (1999) Interplay between chromatin modifying and remodeling complexes in transcriptional regulation. Crit. Rev. Eukaryot. Gene Expr., 9, 221–230. [DOI] [PubMed] [Google Scholar]
- 55.Kornberg R.D. and Lorch,Y. (1999) Chromatin-modifying and -remodeling complexes. Curr. Opin. Genet. Dev., 9, 148–151. [DOI] [PubMed] [Google Scholar]
- 56.Simpson R.T. (1990) Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature, 343, 387–389. [DOI] [PubMed] [Google Scholar]