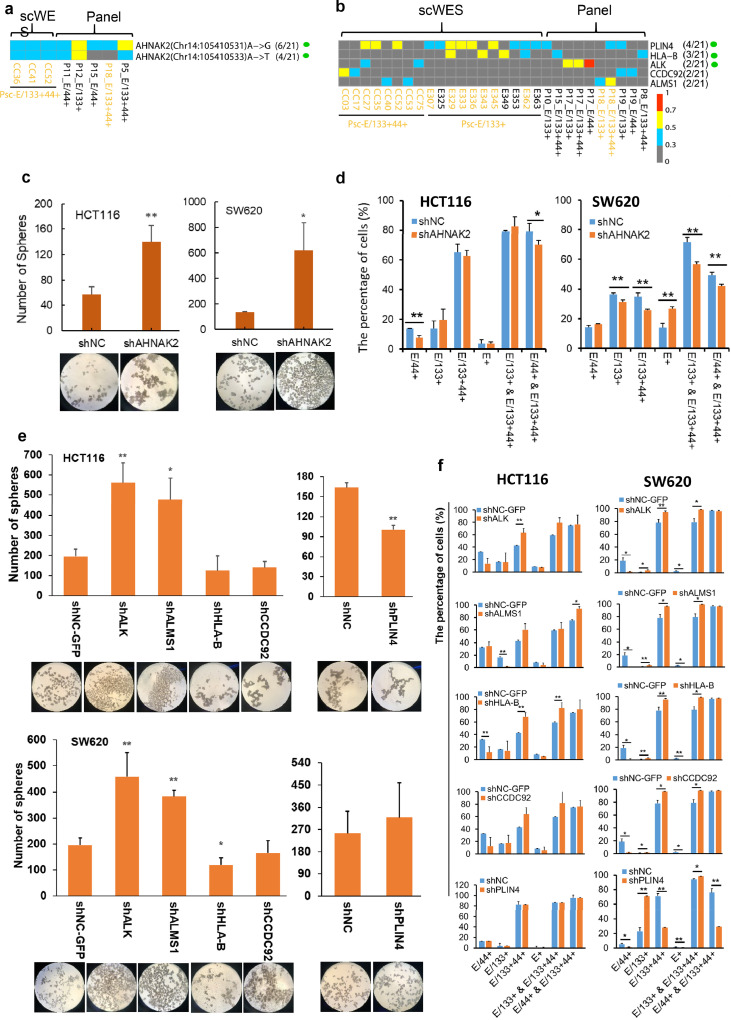
Figure 5.
The same CRCICs mutational sites or mutated genes found in scWES data and targeted sequencing data, and the knock down expression analysis of these mutated genes. a. The same CRCICs mutational sites found in both data. 6/21 and 4/21 means the corresponding patients with the mutation vs. total 21 patients. b. The same CRCICs mutant genes found in both data. Px means the serial number of each CRC patient for targeted sequencing data. Psc means the patient for scWES data. Green solid circle means that the subcellular location of the gene is at the cell surface. ‘x’ in (x/21) means how many patients has this mutation, while “21” in (x/21) means there are totally 21 patients including the patient for scWES and the 20 patients for validation. KRAS-mutant cells are marked with yellow fonts. a, b. Genes with variant allele frequency (VAF) of ‘≥ 0.7 to 1’, ‘≥ 0.5 to < 0.7’, ‘≥ 0.3 to < 0.5’, ‘0 to <0.3’ were labeled with squares of red, yellow, blue, and grew, respectively. c. Sphere formation of AHNAK2 knock-down HCT116 and SW620 cells. Magnification: 40 X. d. Compared with shNC cells, the percentage changes of each CRCIC or CRCC group in AHNAK2 knock-down HCT116 and SW620 cells by FACS assays. e. Sphere formation of other five genes knock-down HCT116 and SW620 cells. Magnification: 40 X. f. Compared with shNC cells, the percentage changes of each CRCIC or CRCC group in other five genes knock-down HCT116 and SW620 cells by FACS assays, n=3. Student t-test was used. Data are presented as mean ± SD. * P < 0.05; ** P < 0.01. shNC and shNC-GFP cells are negative control cells transfected with lentivirus-packing plasmids.