Abstract
The protozoan parasite Giardia lamblia synthesizes a diverse and surprisingly abundant array of sterile transcripts unable to code for proteins. Random sampling of cDNAs from two evolutionarily divergent Giardia strains indicates that ∼20% of cDNAs in the libraries represent polyadenylated sterile transcripts. RNase protection analysis and northern blot hybridization of three sterile transcript loci demonstrated that both the sterile transcript and a complementary mRNA were made in each case, further categorizing these sterile transcripts as antisense transcripts. Investigation of the genomic loci for these same three sterile antisense transcripts showed typical transcription units for the sense transcripts, but still failed to reveal a usable open reading frame for the sterile antisense transcripts. 5′-RACE mapped the transcription start site for one of the sterile antisense transcripts to an AT-rich region, as is typical for Giardia. It is unclear whether these sterile transcripts represent errors in transcription or whether they have regulatory functions within the cell, although preliminary investigations failed to reveal evidence for a role in developmental gene regulation. In either case, the presence of such a large pool of sterile antisense transcripts is dramatic evidence of the unusual molecular machinery of the early diverging protist G.lamblia.
INTRODUCTION
Giardia lamblia is a protozoan parasite of medical and evolutionary importance (1,2). Giardia lamblia differentiates from an infective cyst into a trophozoite in the lumen of the small intestine, where its presence may result in gastrointestinal symptoms. The parasite differentiates back into the cyst and is excreted in this form (3,4). There is substantial literature addressing the early divergence of the parasite on the eukaryotic branch of evolution (5–9) and available evidence suggests that it exhibits unusual molecular genetics for a eukaryotic cell, like many other early diverging parasitic protists. The parasite is thought to be asexual based on clonal populations in the wild (10), although recent evidence of the existence of reverse transcriptase genes in Giardia suggests the possibility of a sexual stage (11). Additional evidence suggests that Giardia is tetraploid (12), with an exceedingly compact genome (13,14) and two equivalent nuclei (15). The compact genome is reflected in the fact that introns have not yet been identified, intergenic regions are small (often <25 bp) and the untranslated regions (UTRs) of mRNAs analyzed so far are typically <20 nt at the 5′-end and <50 nt at the 3′-end (reviewed in 16). The longest reported 5′-UTR is 146 nt (17), while the longest reported 3′-UTR is 288 nt (18). In addition, promoter analysis indicates that a very short region (<50 bp) of proximal upstream G.lamblia DNA is all that is required to drive expression of a reporter gene in transfected parasites (17,19–22). Sequence alignment of several promoter regions fails to reveal any highly conserved sequences, suggesting that these promoter sequences are also highly degenerate (23,24).
‘Sense’ transcripts are RNA messages that can code for a protein, while ‘antisense’ transcripts are RNAs that are complementary to the sense transcript. Natural antisense transcripts are produced normally by a cell and are typically sterile RNAs, lacking an open reading frame (ORF) and therefore unable to code for a protein. Multiple examples of natural antisense transcripts have been documented in both prokaryotic and eukaryotic cells, as well as from RNA and DNA viruses. Despite numerous specific examples, the overall abundance of antisense transcripts in a cell is assumed to be exceedingly low. In prokaryotic cells antisense transcripts are typically <100 nt with a conserved stem–loop structure that is essential for their roles in the negative regulation of gene expression, plasmid copy number, transposition and bacteriophage replication (reviewed in 25). The roles of eukaryotic antisense messages and their mechanisms of action are less clear. In eukaryotes, antisense transcripts appear as either short (≤100 nt) transcripts, partially complementary to the sense message and derived from a different locus, or as longer (≥100 nt) transcripts, complementary to the sense message, and usually derived from the same locus (reviewed in 26–29). Good evidence exists for involvement of the shorter transcripts in pre-mRNA splicing (30) and rRNA maturation (31), as well as a role in regulating gene expression (32). The function of many of the longer antisense transcripts in gene regulation is less well defined, but several examples strongly suggest a possible role for antisense transcripts in developmental gene regulation (33–40). Various mechanisms have been proposed for antisense transcript control of gene expression, including mRNA stability, transcriptional control, alternative splicing and mRNA truncation (26,27).
Our investigation of transcripts in G.lamblia has led us to the surprising discovery that the molecular uniqueness of G.lamblia extends to a profusion of natural antisense RNAs transcribed by the parasite. Random sampling of cDNA libraries has revealed the presence of ∼20% sterile, antisense transcripts. Importantly, this abundance is the result of low levels of transcription from many different loci, not simply high levels of transcription from a few loci. Detailed characterization of three loci confirm that both sense and antisense transcripts are synthesized and that their relative abundance is unchanged throughout the developmental cycle of Giardia.
MATERIALS AND METHODS
Database accession
All sequence data have been deposited in GenBank. Accession numbers for the three complete genomic loci are: cysteine protease, AF399008; NAD(P)H oxidoreductase (NOR), AF399009; DEAD RNA helicase, AF399010. Accession numbers for the cDNAs are; WB cDNAs 1–17, AF398985–AF399001; GS cDNAs 1–6, AF399002–AF399007.
Materials
Unless specified otherwise, all enzymes were obtained from New England Biolabs (Beverly, MA).
Cultures
Trophozoites of two G.lamblia isolates, GS clone GS/H7 (ATCC no. 50581) and WB clone WB/1267 (ATCC no. 50582), were cultured as described previously (41). Briefly, cells were maintained anaerobically in glass culture tubes at 37°C in Keister’s modified TYI-S-33 medium supplemented with antibiotics. Induction of encystation was performed as previously described (42) and encysting cells were harvested 24 h after initiation of encystation.
Library construction and cDNA sampling
A previously described WB directional cDNA library (43) was used for cDNA sampling. Briefly, this library was constructed in λgt22A using SuperScript II reverse transcriptase (Life Technologies, Gaithersburg, MD) and polyadenylated RNA from encysting parasites. A directional cDNA library in λZAPII was made separately from the GS/H7 isolate of G.lamblia starting with 5 µg poly(A)+ RNA from trophozoites as directed by the manufacturer (Stratagene, La Jolla, CA) for use in parallel studies. Both libraries contained polyadenylated cDNAs as required by the method of construction.
Inserts from the WB library were amplified by PCR using λgt11 F and R primers (New England Biolabs) and Taq polymerase (Perkin Elmer, Burlingame, CA) or Taq Precision Plus (Stratagene) under the following conditions: 2 min at 95°C; 30 cycles of 30 s at 95°C, 1 min at 65°C, 3 min at 72°C; 4 min at 72°C. Inserts from the GS library were amplified with T3 and T7 primers (Stratagene) as above. Agarose gel electrophoresis was used to estimate cDNA insert size.
For subcloning, PCR products from the WB library were purified by ethanol precipitation and digested with SalI and NotI to release the cDNA insert. pBluescript II SK– (Stratagene) vector was prepared by digestion with SalI and NotI, dephosphorylation with calf intestinal phosphatase, and gel purification (Geneclean II; Bio101, Vista, CA). PCR products and vector were ligated and recombinant plasmids identified by diagnostic restriction digestion.
Alternatively, PCR products from the WB and GS libraries were prepared for direct sequencing in one of two ways: (i) directly purified (Wizard PCR Preps DNA Purification System; Promega, Madison WI) with or without prior treatment with shrimp alkaline phosphatase and exonuclease III (Amersham Pharmacia, Piscataway, NJ); (ii) gel purified (Geneclean II).
An ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer, Foster City, CA) and the same primers used for PCR were used to sequence cDNAs. Reactions were run on an ABI377 automated sequencer (Perkin-Elmer). Internal oligonucleotide primers were designed as needed to complete sequencing of the entire cDNA. Sequences were analyzed for ORFs using DNA Strider 1.2 (CEA, France). BLASTX and BLASTP searches were performed on the cDNA sequence or predicted protein sequence to search for similarities to known genes in GenBank.
Identification and sequencing of genomic clones
To generate probes to be used in genomic library screening, sterile cDNA inserts 1–3 from the WB library (sense transcript designation: 1, cysteine protease; 2, NOR; 3, DEAD RNA helicase), subcloned into pBluescript II SK– as described above, were digested with SalI and NotI and inserts were purified from agarose gels (Geneclean II). The DNA was labeled with [α-2P]dCTP by random priming (Ready to Go; Amersham Pharmacia). Approximately 60 000 plaques from a G.lamblia WB genomic DNA λFIXII library (44) were screened for each probe and several positive plaques were purified to homogeneity. Lambda phage DNA was prepared (Lofstrand Laboratories, Gaithersburg, MD) and sequenced directly as described above using 5 µg λ DNA per sequencing reaction. Internal oligonucleotide primers were designed as needed to complete sequencing of the full loci on both strands.
RNA purification
Polyadenylated RNA was isolated from either trophozoites or 24 h encysting cells using a Quik-Prep poly(A)+ RNA kit (Amersham Pharmacia) and quantitated by absorbance at 260 nm.
RNase protection assays
To generate RNA probes for RNase protection assays, sterile cDNA inserts 1–3 from the WB library (sense transcript designation as above), subcloned into pBluescript II SK– as described above, were linearized with BamHI (cysteine protease, antisense), XmnI (cysteine protease, sense), EcoNI (NOR, antisense), PstI (NOR, sense) or AflIII (DEAD helicase, antisense and sense). Linearized plasmids were purified by proteinase K digestion, phenol/chloroform extraction and ethanol precipitation. Probes were synthesized using 500 ng linearized plasmid, either T3 (sense probes) or T7 (antisense probes) primers, RNA polymerase (Maxiscript; Ambion, Austin, TX) and [32P]dUTP (Amersham Pharmacia). Predicted probe sizes are cysteine protease antisense 286 nt, cysteine protease sense 344 nt, NOR antisense 264 nt, NOR sense 185 nt, DEAD helicase antisense 209 nt and DEAD helicase sense 430 nt. All antisense probes have 22 nt of vector DNA and all sense probes have 20 nt of vector DNA which should be removed during RNase digestion. Probes were purified by DNase I digestion, G-25 chromatography and electrophoresis on 5% acrylamide, 1× TBE, 7 M urea gels.
RNase protection assays were carried out using 50 000 c.p.m. of each probe and ethanol precipitated with 5 µg yeast RNA alone or with 0.25 µg G.lamblia WB poly(A)+ RNA. Probes were resuspended in annealing buffer (RPAII; Ambion) at 42°C overnight. Single-stranded RNA was then digested with a mixture of RNase A and RNase T1 (RPAII) diluted 1:100 at 37°C for 60 min, except for the NOR antisense probe, which was digested with a 1:1000 dilution of RNase A and RNase T1 for 30 min at 37°C. One set of yeast reactions was left undigested to demonstrate riboprobe size on subsequent gel analysis. RNase digestion was stopped by precipitation (RPAII) and samples were electrophoresed on 5% acrylamide, 1× TBE, 7 M urea gels and exposed to film overnight at –80°C. The autoradiogram was digitized and imported into Adobe Photoshop 3.0 for editing and labeling. Only 10% of the no RNase control samples were loaded onto gels. A 100 nt ladder of RNA molecules was synthesized from a Century template set (Ambion). Since the amounts of Giardia RNA were not titrated, results are not quantitative.
Rapid amplification of cDNA ends (RACE)
5′- and 3′-RACE were performed according to the manufacturer’s instructions using 1 µg polyadenylated RNA and gene-specific primers (Life Technologies). Nested PCR products were gel purified (Geneclean II) and directly sequenced as described above.
Northern blot analysis
An aliquot of 10 µg WB trophozoite or cyst polyadenylated RNA was loaded per gel lane, separated on a 1.2% formaldehyde–agarose gel in MOPS buffer and transferred to a nylon filter (Nytran TurboBlot; Schleicher & Schull, Keene, NH). Oligonucleotide probes complementary to either the cysteine protease sense (5′-GCCTTCGTCTCCATGCCGATGAAGA-3′) or antisense (5′-GTTCATGACGTTCCGCTCCCACTCG-3′) transcript or to the NOR sense (5′-CCTCGCTGACCTGCTCAGGGGTCAT-3′) or antisense (5′-ATGACCCCTGAGCAGGTCAGCGAGG-3′) transcript were end-labeled with [γ-32P]ATP; labeling efficiencies were determined by scintillation counting. Alternatively, double-stranded probes against ADP-ribosylation factor (ARF), immunoglobulin-binding protein (BiP) and cyst wall protein 2 (CWP2) were amplified by PCR using Taq Precision Plus (Stratagene) under the following conditions: 2 min at 95°C; 34 cycles of 30 s at 95°C, 1 min at 50°C, 3 min at 72°C; 4 min at 72°C. Oligonucleotides used for PCR were: ARF(F), 5′-ATGGGCCAAGGCGCATCAAAGATC-3′; ARF(R), 5′-CTTGTCGAAGATGTAGTCACTAAG-3′; BIP(F), 5′-ATGCTCGCTCTTGTCTTTGCCGC-3′; BIP(R), 5′-TTAGAGTTCATCTTTTTCTGC-3′; CWP2(F), 5′-ATGATCGCAGCCCTTGTTCTAGG-3′; CWP2(R), 5′-TCACCTTCTGCGGACAATAGGC-3′. PCR products were gel purified (Geneclean II) and labeled with [α-32P]dCTP by random priming (Ready-to-Go). Filters were hybridized to probes in 5× SSPE, 5× Denhardt’s, 1% SDS, 100 µg/ml denatured salmon sperm DNA at 42°C for oligonucleotide probes or 65°C for double-stranded PCR product probes. After washing at 55°C in 2× SSC, 0.1% SDS, filters were exposed to film or phosphorimager cassettes overnight, which were then scanned (Storm 560 PhosphorImager; Molecular Dynamics) and images were transferred to Adobe Photoshop 3.0 for editing and labeling.
RESULTS
Sterile antisense transcripts are highly abundant in G.lamblia
To examine mRNA transcripts in Giardia, we chose to use two highly divergent isolates of Giardia with obvious genetic differences: WB, a member of Group 1 (Assemblage A), and GS, a member of Group 3 (Assemblage B) (16,45). The evolutionary distance between the two isolates has been previously demonstrated through molecular analyses, which revealed an 18% nucleotide divergence for the triosephosphate isomerase gene (46), with no similarity in non-coding regions (47,48), and a 13% nucleotide divergence for the ARF gene (49). Polyadenylated RNA for library construction was isolated from two different stages of the parasite life cycle from the two isolates, encysting cells for WB and trophozoites for GS. Directional cDNA libraries (cloned using SalI at the 5′-end and NotI at the polyadenylated 3′-end of the original mRNAs for the WB library and cloned using EcoRI at the 5′-end and XhoI at the polyadenylated 3′-end of the original mRNAs for the GS library) were synthesized independently for these two strains, using poly(dT) primers to further ensure that incorporated transcripts were polyadenylated and directionally incorporated. Comparisons of results between the two libraries should therefore have significant implications for the biology of G.lamblia independent of isolate, life cycle stage and library.
During an initial screening of the G.lamblia WB cDNA library using a panel of monoclonal antibodies, three sterile antisense transcripts were detected (Fig. 1, WB cDNAs 1–3). In this screen nine cDNAs were isolated from the library and fully sequenced on both strands. Of these, five cDNAs were later judged to react with the monoclonal antibody used in their isolation, one contained a non-reactive but coding sense transcript and the final three appeared to represent sterile, antisense transcripts (see below for a further description). The finding of three antisense transcripts in Giardia from such a small sample suggested that antisense transcripts might be quite common in Giardia, in contrast to the rarity of such transcripts in other cells.
Figure 1.
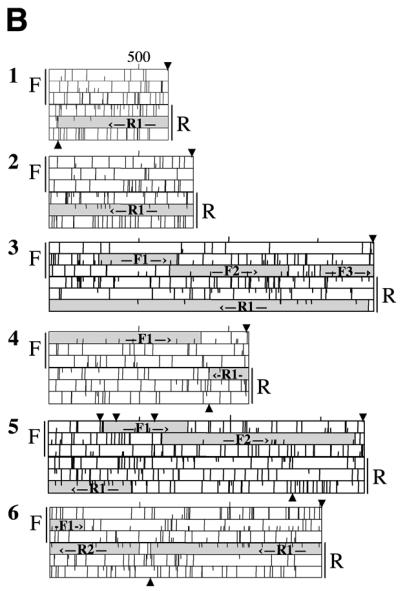
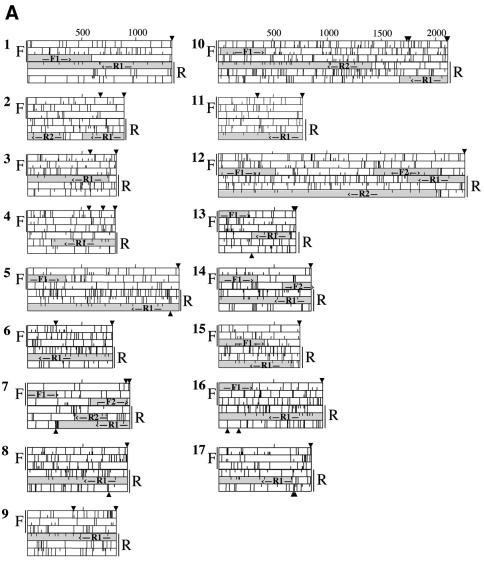
Sterile antisense cDNAs in G.lamblia. Individual plaques were picked from cDNA libraries from two strains of Giardia, WB and GS. The cDNAs were amplified by PCR and fully sequenced on one strand. Open reading frames were analyzed in all six reading frames using DNA Strider 1.2. The sequences have been translated in all six reading frames, with F and R indicating forward and reverse strands, respectively. Half lines indicate ATG codons and full lines indicate stop codons. Possible ORFs longer than 300 nt are indicated by hatched boxes and labeled F# and R#. Arrows above and below the plots indicate the location of consensus polyadenylation signal sequences. (A) Sterile antisense cDNAs from the WB library. (B) Sterile antisense cDNAs from the GS library.
To assess the relative percentage of sense and antisense transcripts in Giardia, we randomly picked plaques from each directional cDNA library (91 plaques from the WB library and 50 plaques from the GS library), amplified inserts by PCR and sequenced the full insert on at least one strand. Empty plaques and aberrantly incorporated cDNAs (e.g. internal cloning sites within the cDNA or multiple cDNA inserts) were eliminated from consideration, leaving 54 inserts from the WB library and 37 inserts from the GS library for further analysis. All cDNAs examined had putative Giardia polyadenylation signals (A/TGTPuAA) and were polyadenylated. Forty of the 54 WB cDNAs and 31 of the 37 GS cDNAs had ORFs in the forward direction and were therefore judged to be coding transcripts (data not shown). The remaining 14 of the 54 WB transcripts (26%) and six of 37 GS transcripts (16%) did not have usable ORFs in the forward direction (Fig. 1, WB cDNAs 4–17 and GS cDNAs 1–6), indicative of their sterility. Each, however, did have at least one ORF on the reverse strand, potentially making them antisense transcripts. These data indicate a phenomenal abundance of sterile antisense transcripts in Giardia, with ∼20% of the sampled cDNAs being non-coding. It is evident that the unprecedented frequency of sterile antisense transcripts is not simply a peculiarity of one isolate of G.lamblia or encysting cells or a library artifact, but is instead a basic feature of the molecular biology of the parasite.
It is important to understand the unique genomic organization of G.lamblia and our process of sequence analysis that permitted us to identify these transcripts as sterile. We have analyzed all ORFs longer than 300 bp in all six reading frames of the cDNAs. Although it is possible that ORFs shorter than 300 bp may code for proteins, these smaller ORFs would in turn simply have longer UTRs and would therefore again be unlikely to be translated. Sense transcripts are defined as having an ORF in the forward (SalI→NotI or EcoRI→XhoI) direction, regardless of similarities to known proteins, and are polyadenylated. In contrast, sterile antisense transcripts are polyadenylated and have an ORF (sometimes not fully contained within the length of the transcript) in the reverse direction (NotI→SalI or XhoI→EcoRI), yet do not appear to contain a usable ORF in the forward direction. The term ‘usable’ refers to the fact that both 5′- and 3′-UTRs are exceptionally short in Giardia transcripts, typically <20 nt at the 5′HUTR and <50 nt at the 3′-UTR. With the longest identified 5′-UTR at 146 nt and the longest 3′-UTR at 288 nt, it is reasonable to predict that cDNAs with 5′- or 3′-UTRs >300 nt will not be competent for translation; in particular, it is impractical to suggest that our results are simply 23 examples of UTRs of unprecedented length. We have therefore chosen 300 nt as the upper limit on permissible UTRs for identification of ORFs, and potential ORFs with UTRs longer than 300 nt are considered sterile. Because the quality of the cDNA libraries are important in this analysis, we emphasize that the average cDNA insert size in the libraries is ∼1 kb; sequencing of six independent sense transcripts indicated that the cDNA clones extended to within 100 bp of the 5′-end of the gene. It is also important to note that there is no evidence for introns in Giardia, making it highly unlikely that all 23 cDNA clones represent splicing intermediates.
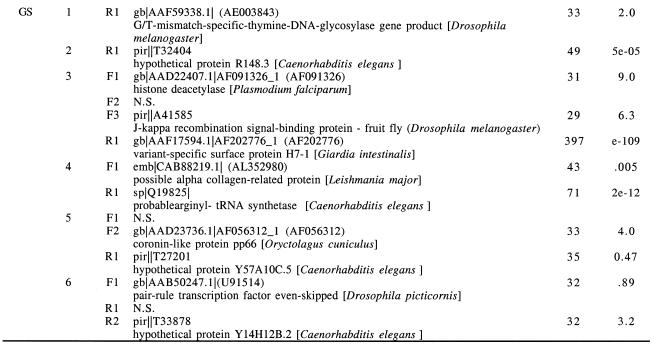
Predicted translation products of the ORFs from these putative sterile, antisense cDNAs were subjected to BLASTP searches (Table 1) with the result that only one of the 18 ORFs on the forward strands had similarity to known proteins (using an arbitrary cut-off of an E-value <0.5), while 18 of the 28 ORFs on the reverse strands had similarity to known proteins (P < 0.005, χ2 test). In addition, five of the seven reverse strand ORFs which contained the stop codon for the ORF also had polyadenylation signals immediately following these stop codons. To further look for the possibility of coding regions in the forward direction, all cDNA sequences were subjected to BLASTX searches which translate the sequence in all six reading frames for comparison with protein databases. No further protein similarities were noted for the forward direction. Although indirect evidence, the BLAST searches further corroborate the conclusion that the transcripts are sterile and antisense. We do not conclude that all antisense transcripts must be sterile or that all sterile transcripts must be antisense; our data only address the general population of nonsense transcripts in Giardia.
Table 1. Significant sequence similarities of possible ORFs >300 bp.
[CRC Table-insert tagged image=gke606t01]Random cDNA inserts from two different directional libraries were sequenced. In the WB library results, the first transcripts listed were the result of a preliminary screening; the remaining 14 transcripts were observed in an analysis of 54 cDNAs. In the GS library results, the six transcripts listed were observed in an analysis of 37 cDNAs. ORFs were identified using DNA Strider 1.2 and translation products of all ORFs were used for BLASTP searches of GenBank (http://www.ncbi.nl.nih.gov/blast/blast.cgi?Jform=1) (54). The most similar protein for each ORF is listed along with the BLAST score and E values.
Characterization of three loci producing sterile antisense transcripts
To further study these sterile transcripts, genomic clones of the first three loci initially identified in screening the WB library (nos 1–3) were isolated and completely sequenced on both strands. Maps of the genomic loci are annotated to indicate possible ORFs and polyadenylation signals on both strands (Fig. 2). The sterile cDNAs on the forward strand are indicated, along with the full-length transcripts predicted by northern blot analysis for these cDNAs (Fig. 5 and data not shown). The corresponding transcripts on the complementary strand are indicated and labeled by similarity to known proteins (cysteine protease, NOR and RNA helicase). The NOR gene has been further analyzed and the recombinant protein found to have functional NOR activity (50). The genomic sequences fully corroborated the cDNA sequences, excluding the possibility that the original cDNA clones were merely sequencing or library artifacts and, therefore, confirming the validity of the directional library as a means for detecting antisense transcripts. Furthermore, extending the genomic sequence further upstream of the cDNA clones revealed only small ORFs without significant similarities to sequences in GenBank for the NOR and cysteine protease antisense transcripts. These transcripts are therefore either sterile or represent unprecedented lengths of 5′- and 3′-UTRs in Giardia. The RNA helicase antisense transcript clearly cannot code for protein nor did we see evidence for potential protein coding abilities in several other antisense transcripts subsequently analyzed (see above). Thus, while we cannot exclude the possibility of coding regions within the ‘sterile’ transcripts, we find it highly unlikely since these transcripts would again represent enormous and unprecedented lengths of untranslated mRNA compared to all other known Giardia transcripts (reviewed in 1 and numerous other examples).
Figure 2.
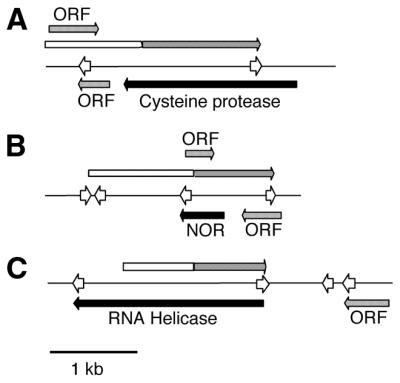
Transcriptional maps of loci expressing sense and antisense RNAs. Genomic clones for the three cDNAs sequenced in Figure 1 were isolated from a Giardia strain WB genomic DNA library (44) and the loci sequenced. The sequences corresponding to the original cDNAs are indicated by long gray arrows preceded by open boxes. The transcription initiation sites were mapped by 5′-RACE and are indicated by the end of the black boxes (sense transcripts) or open boxes (antisense transcripts). Polyadenylation signals are indicated by open arrowheads. All other open reading frames greater than 100 amino acids and beginning with a methionine are indicated by short gray arrows and are labeled ‘ORF’s. These open reading frames have no similarity to known proteins and are labeled simply ORF. (A) Cysteine protease. (B) NOR. (C) ATP-dependent RNA helicase.
Figure 5.
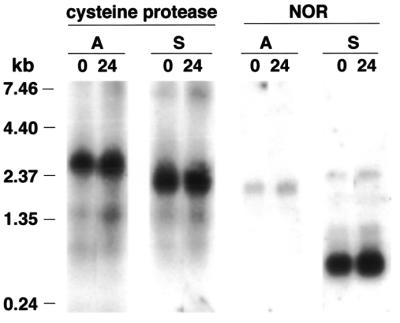
Northern blot analysis of sense and antisense transcripts. Oligonucleotides complementary to the cysteine protease antisense (A) or sense (S) transcripts or to the NOR antisense or sense transcripts were end-labeled with [γ-32P]ATP to equivalent specific activities and hybridized to either WB trophozoite (0 h) or encysting cell (24 h) polyadenylated RNA.
To examine transcription start sites and polyadenylation sites, the 5′- and 3′-ends of both the sense and antisense transcripts from all three loci were identified using 5′- and 3′-RACE. The complete sequence for the genomic locus of clone 2 (NOR) from the WB library is shown in Figure 3. The coding region for the NOR gene is underlined (to maintain the forward and reverse nomenclature, the NOR gene is actually on the complementary strand to that shown here) and transcription start sites and polyadenylation sites for the NOR sense and antisense transcripts are boxed. The results demonstrate that the 5′-flanking DNA of both transcripts is AT-rich, as is typical in Giardia promoter regions, and that polyadenylation signals appear slightly degenerate using the sequence A/TGTPyAA. As is typical for Giardia genes, the start and stop codons of the NOR gene are coincident with the transcription start sites and polyadenylation signals. Similar results were also seen for the cysteine protease and RNA helicase sense and antisense transcripts (data not shown), although the 5′-ends of the antisense transcripts for the cysteine protease and RNA helicase loci could not be unambiguously determined because multiple start sites appeared to be used. However, based on the sizes of the PCR products and of transcripts detected by northern analysis (Fig. 5 and data not shown), the approximate transcriptional start sites were identified and were within the region of sequenced genomic DNA, as shown in Figure 2.
Figure 3.
Sequence of the genomic locus for NOR. The sequence is shown as a single strand encoding the ‘forward’ direction; hence the original mRNA (the sterile, antisense transcript) would appear on this strand. The sequence complementary to the NOR gene is underlined. The AT-rich sequences surrounding the transcription start sites and the polyadenylation signals for both the sense and antisense transcripts are boxed and are as follows: transcription start site for sense transcript (AAAAAATG at 2040, start codon underlined); polyadenylation signal for sense transcript (AGTAAA at 1545, stop codon underlined); transcription start site for antisense transcript (AAAA at 510); polyadenylation signal for antisense transcript (TGTAAA at 2610).
Confirmation of the presence of sense and antisense transcripts from three loci
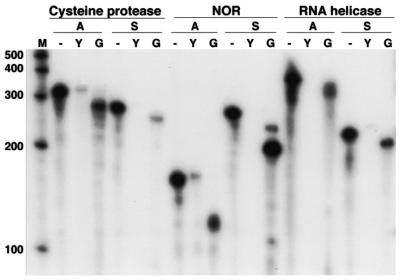
During the RACE analyses described above, products were not obtained when reverse transcriptase was omitted from the reactions, confirming their presence as RNA transcripts in populations of Giardia. RNase protection assays were also used to independently confirm the presence of both the antisense and sense transcripts. For all three examples unique protected RNA fragments of the expected sizes were detected for both the sense and antisense messages only with Giardia polyadenylated RNA (Fig. 4). No protected fragment was detected by hybridization with yeast RNA. Although a small amount of undigested probe remains in the cysteine proteinase and NOR antisense probe samples, these cannot represent protected RNA since the probes contain 20 nt of sequence derived from the Bluescript cloning vector which was not removed. The remaining probe is therefore the result of incomplete digestion. This then clearly establishes that for these three examples both the sense and antisense messages are present in a population of Giardia trophozoites.
Figure 4.
RNase protection analysis of Giardia RNA. Lane M shows a 100 nt RNA ladder. The cDNAs analyzed in Figure 1 were used to generate 32P-labeled riboprobes. Riboprobes complementary to the antisense (A) or sense (S) strands of the cysteine protease, NOR or ATP-dependent RNA helicase loci were added to yeast RNA and left undigested as an indication of riboprobe size (-). To detect the presence of the antisense and sense transcripts, the riboprobes were added to yeast (Y) RNA or G.lamblia (G) polyadenylated trophozoite RNA and digested with a 1:100 dilution of RNase A + RNase T1, except for the NOR probe complementary to the sense strand, for which a 1:1000 dilution was used. Results are not quantitative since titrations of RNA were not used.
Apparent absence of developmental gene regulation
Examples from other eukaryotic cells suggest that the antisense transcripts could serve a role in the regulation of developmental gene expression. In Giardia this would correspond to a role in the developmental cycle between the trophozoite and cyst. To begin to address this question, we therefore performed northern blot analysis using polyadenylated RNA isolated from either trophozoites or encysting cells with oligonucleotide probes to allow strand-specific detection of transcripts from the cysteine protease and NOR loci. Probes complementary to both the sense and antisense strands detected transcripts of a size predicted by the 5′- and 3′-RACE experiments, validating the specificity of this approach and confirming the presence of the transcripts as previously demonstrated by RNase protection (Fig. 5). We added equivalent amounts of labeled sense and antisense oligonucleotide probes to permit quantitative analysis of these northern blots. Probes for the NOR locus were complementary to each other and would therefore also have had the same hybridization kinetics. If these antisense transcripts did have a role in regulating gene expression during development, then one might expect the absolute levels of sense RNA to differ between trophozoites and encysting cells or that the ratio of sense to antisense RNA might differ between developmental stages. However, the amounts of sense and antisense RNAs do not vary between trophozoites (0 h time points) and encysting cells (24 h time points) and the ratio of antisense (lanes A) to sense (lanes S) RNA did not change, indicating that antisense transcripts of these two loci probably do not play a role in regulating sense transcript levels during the developmental cycle. Translational or post-translational mechanisms have been observed to regulate the developmental expression of BiP in Giardia (51), and we cannot rule out roles for antisense RNAs in this process. However, using double-stranded probes for northern analysis, we have been unable to detect antisense transcripts for BiP, CWP2 (43) or ARF (49), three genes known to be developmentally regulated in Giardia (data not shown). In contrast, full-length double-stranded probes against the NOR gene did detect both sense and antisense transcripts (data not shown).
A role for these antisense transcripts in gene regulation also seems unlikely because the affected loci cannot be simply classified as ones involved in development or, for that matter, any other single cellular process. Instead, the loci include a number of basic housekeeping genes which would not be expected to be developmentally regulated (Table 1). Furthermore, in circumstances where either natural or artificially introduced antisense transcripts have been proven to have a regulatory function, the amount of antisense transcript often far exceeded the amount of sense transcript that was being down-regulated. The antisense transcripts we have analyzed in detail in Giardia, however, each appear to be of equal or significantly lower abundance as judged by northern analysis than the corresponding sense transcript (Fig. 5). They are therefore unlikely to have a major effect on expression of the corresponding sense transcript via mechanisms which require an excess of antisense RNA. So although we cannot argue against a regulatory role for any specific transcript, the available evidence indicates that the bulk of the antisense transcripts do not serve a regulatory function.
DISCUSSION
In the work presented here we show that G.lamblia synthesizes an inordinately large number of sterile, antisense transcripts. We analyzed a total of 100 cDNA clones from two directional libraries and found 23 clones which we believe represent sterile transcripts. While small ORFs were occasionally found on the forward strands of these transcripts, we believe that they are functionally sterile since they would all represent UTRs of unprecedented length for Giardia and because only one of 18 has a homolog in GenBank; a much lower frequency than the homologs found for 18 of 28 ORFs on the complementary strands. Identification of the initial three antisense transcripts was confirmed by several independent means, including sequencing of the relevant genomic loci (Fig. 2), RNA-dependent 5′- and 3′-RACE (Fig. 3), RNase protection assays (Fig. 4) and strand-specific northern blot analysis (Fig. 5). In each instance we have shown that both the sense and antisense transcripts are synthesized by a population of cells.
Based on the survey of inserts in cDNA libraries we estimate that one in five transcripts in Giardia are sterile. These transcripts could interfere with the synthesis, stability or translational capability of the corresponding sense transcripts. However, while it would be impossible to prove that no antisense transcript in Giardia has a role in regulating gene expression, we have been unable to find any evidence that would support such a role. The abundance of antisense molecules is generated by transcription from many different loci, rather than a higher level of transcription from a few loci. The identity of the corresponding sense transcripts is quite diverse, resisting easy categorization into groups such as ‘developmentally important genes’, and indeed several of the genes, such as the two different tRNA synthetases, are typically constitutively expressed in cells (Table 1). Taken together, these data imply that if the transcripts were to have a regulatory role, the effect would be almost global for a very large number of G.lamblia genes, regardless of any apparent need for regulation. It is difficult to imagine the purposeful production of these antisense molecules for such ill-defined control. Furthermore, strand-specific northern blot analysis on two of the antisense/sense transcript pairs in G.lamblia shows that the ratio of sense to antisense transcripts does not vary during the course of development (Fig. 5).
We propose that the sterile transcripts are the result of a loose molecular mechanism controlling transcription. Recent work from our laboratory indicates that transcription of a reporter gene in Giardia can be driven by several different AT-rich initiator sequences as short as 8 bp inserted just upstream of the ATG translation initiation site (22), and previous studies comparing proximal upstream regions in Giardia have found limited sequence conservation (23,24). It is therefore likely that cryptic promoters are abundant in Giardia, simply because the sequences necessary to initiate transcription are short and not highly specific. Indeed, the sequences immediately upstream of the 5′-end of the NOR antisense transcript are typical of the short AT-rich initiator elements (Fig. 2). Additionally, 5′-RACE analyses of the three tested sense and antisense transcripts indicated the presence of multiple transcription start sites for these antisense transcripts (data not shown). Subsequent rounds of nested PCR resulted in a single clearly dominant product which was used to examine the nucleotide identity at the major transcriptional start site of NOR. However, the presence of multiple sizes of RNA molecules from a single locus again supports the presence of functional cryptic promoters in Giardia. The presence of multiple cryptic promoters would lead to the production of several overlapping sterile transcripts with different transcriptional start sites. Transcription would continue until the first common polyadenylation signal was reached, and indeed the first polyadenylation signal in each sequence is found at the 3′-end of each RNA (Fig. 2).
Why then should the sterile messages be antisense? This might be explained simply by the extraordinarily tight gene packing in the Giardia genome. Intergenic regions are typically quite short (52). The inference from this is that in most instances in Giardia one or the other strand of DNA usually contains an ORF. There is simply not much ‘junk’ DNA in the genome and therefore a sterile transcript will often be, by definition, transcribed from a region opposite a gene. Notably, not all sense transcripts appear to have a corresponding antisense transcript (data not shown).
It should be noted that two examples suggest that genes might actually exist on overlapping antisense transcripts in G.lamblia (17,53). In the first example an ORF exists on a transcript complementary to the rRNA and there is evidence that a protein is synthesized from this transcript (53). The other example is an ORF with no known homologs on a transcript antisense to glucosamine 6-phosphate isomerase. This antisense transcript contains very large 5′- and 3′-UTRs and there is no evidence yet that it codes for a protein (17). Further work will be needed to determine if this RNA codes for a protein or is another sterile antisense transcript.
While the data clearly show the presence of abundant antisense transcripts in populations of G.lamblia, they do not address what might be happening in individual cells. It remains to be investigated whether every cell in a population makes the same subset of sterile transcripts or indeed whether each of the two nuclei within Giardia makes the same subset of sterile transcripts. It therefore remains possible that individual cells express copious amounts of one or a few antisense messages which do regulate gene expression. This is unlikely, however, since the particular locus involved would have to be different in each cell in the population. It will also be interesting to define the half-lives and subcellular localizations of these antisense transcripts. In particular, potential base pairing with the corresponding sense transcripts in vivo and whether such double-stranded RNA molecules might induce RNA interference mechanisms will require further investigations.
The abundance of sterile antisense transcripts in Giardia is surprising and does not fit current models in other eukaryotic systems involving antisense transcripts in gene regulation. It will indeed be important to discover whether any antisense transcript in Giardia serves such a regulatory function. More important, however, will be to determine what unique selective pressures have driven Giardia to adopt such molecular mechanics and what other implications this might have on parasite biology.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Sara Davis-Hayman, Kirk Deitsch and Tom Templeton for helpful discussions and suggestions throughout this work.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed at present address. Tel: +1 202 687 9883; Fax: +1 202 687 5662; Email: hge@georgetown.edu Present address:Heidi G. Elmendorf and Steven M. Singer, Department of Biology, 306A Reiss Building, 37th and O Streets NW, Georgetown University, Washington, DC 20057, USA AF398985–AF399010
REFERENCES
- 1.Adam R.D. (1991) The biology of Giardia spp. Microbiol. Rev., 55, 706–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson R.C., Reynoldson,J.A. and Mendis,A.H. (1993) Giardia and giardiasis. Adv. Parasitol., 32, 71–160. [DOI] [PubMed] [Google Scholar]
- 3.Lujan H.D., Mowatt,M.R. and Nash,T.E. (1997) Mechanisms of Giardia lamblia differentiation into cysts. Microbiol. Mol. Biol. Rev., 61, 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillin F.D., Reiner,D.S. and McCaffery,J.M. (1996) Cell biology of the primitive eukaryote Giardia lamblia. Annu. Rev. Microbiol., 50, 679–705. [DOI] [PubMed] [Google Scholar]
- 5.Sogin M.L., Gunderson,J.H., Elwood,H.J., Alonso,R.A. and Peattie,D.A. (1989) Phylogenetic meaning of the kingdom concept: an unusual ribosomal RNA from Giardia lamblia. Science, 243, 75–77. [DOI] [PubMed] [Google Scholar]
- 6.Cavalier-Smith T. and Chao,E.E. (1996) Molecular phylogeny of the free-living archezoan Trepomonas agilis and the nature of the first eukaryote. J. Mol. Evol., 43, 551–562. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto T., Nakamura,Y., Nakamura,F., Shirakura,T., Adachi,J., Goto,N., Okamoto,K. and Hasegawa,M. (1994) Protein phylogeny gives a robust estimation for early divergences of eukaryotes: phylogenetic place of a mitochondria-lacking protozoan, Giardia lamblia. Mol. Biol. Evol., 11, 65–71. [DOI] [PubMed] [Google Scholar]
- 8.Hashimoto T., Nakamura,Y., Kamaishi,T., Nakamura,F., Adachi,J., Okamoto,K. and Hasegawa,M. (1995) Phylogenetic place of mitochondrion-lacking protozoan, Giardia lamblia, inferred from amino acid sequences of elongation factor 2. Mol. Biol. Evol., 12, 782–793. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez L.B., Hashimoto,T. and Muller,M. (1996) Sequence of a malic enzyme gene of Giardia lamblia. Mol. Biochem. Parasitol., 82, 145–151. [DOI] [PubMed] [Google Scholar]
- 10.Tibayrenc M., Kjellberg,F. and Ayala,F.J. (1990) A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas and Trypanosoma and their medical and taxonomical consequences. Proc. Natl Acad. Sci. USA, 87, 2414–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arkhipova I. and Meselson,M. (2000) Transposable elements in sexual and ancient asexual taxa. Proc. Natl Acad. Sci. USA, 97, 14473–14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y. and Adam,R.D. (1994) Allele-specific expression of a variant-specific surface protein (VSP) of Giardia lamblia. Nucleic Acids Res., 22, 2102–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Blancq S.M. and Adam,R.D. (1998) Structural basis of karyotype heterogeneity in Giardia lamblia. Mol. Biochem. Parasitol., 97, 199–208. [DOI] [PubMed] [Google Scholar]
- 14.Lanzer M., Fischer,K. and Le Blancq,S.M. (1995) Parasitism and chromosome dynamics in protozoan parasites: is there a connection? Mol. Biochem. Parasitol., 70, 1–8. [DOI] [PubMed] [Google Scholar]
- 15.Kabnick K.S. and Peattie,D.A. (1990) In situ analyses reveal that the two nuclei of Giardia lamblia are equivalent. J. Cell Sci., 95, 353–360. [DOI] [PubMed] [Google Scholar]
- 16.Adam R.D. (2000) The Giardia lamblia genome. Int. J. Parasitol., 30, 475–484. [DOI] [PubMed] [Google Scholar]
- 17.Knodler L.A., Svard,S.G., Silberman,J.D., Davids,B.J. and Gillin,F.D. (1999) Developmental gene regulation in Giardia lamblia: first evidence for an encystation-specific promoter and differential 5′ mRNA processing. Mol. Microbiol., 34, 327–340. [DOI] [PubMed] [Google Scholar]
- 18.Que X., Svard,S.G., Meng,T.C., Hetsko,M.L., Aley,S.B. and Gillin,F.D. (1996) Developmentally regulated transcripts and evidence of differential mRNA processing in Giardia lamblia. Mol. Biochem. Parasitol., 81, 101–110. [DOI] [PubMed] [Google Scholar]
- 19.Singer S.M., Yee,J. and Nash,T.E. (1998) Episomal and integrated maintenance of foreign DNA in Giardia lamblia. Mol. Biochem. Parasitol., 92, 59–69. [DOI] [PubMed] [Google Scholar]
- 20.Sun C.H. and Tai,J.H. (1999) Identification and characterization of a ran gene promoter in the protozoan pathogen Giardia lamblia. J. Biol. Chem., 274, 19699–19706. [DOI] [PubMed] [Google Scholar]
- 21.Yee J., Mowatt,M.R., Dennis,P.P. and Nash,T.E. (2000) Transcriptional analysis of the glutamate dehydrogenase gene in the primitive eukaryote, Giardia lamblia. Identification of a primordial gene promoter. J. Biol. Chem., 275, 11432–11439. [DOI] [PubMed] [Google Scholar]
- 22.Elmendorf H.G., Singer,S.M., Pierce,J., Cowan,J. and Nash,T.E. (2001) Initiator and upstream elements in the alpha-2 tubulin promoter of Giardia lamblia. Mol. Biochem. Parasitol., 113, 157–169. [DOI] [PubMed] [Google Scholar]
- 23.Holberton D.V. and Marshall,J. (1995) Analysis of consensus sequence patterns in Giardia cytoskeleton gene promoters. Nucleic Acids Res., 23, 2945–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yee J. and Dennis,P.D. (1994) The NADP-dependent glutamate dehydrogenase of Giardia lamblia: a study of function, gene structure and expression. Syst. Appl. Microbiol., 16, 759–767. [Google Scholar]
- 25.Wagner E.G. and Simons,R.W. (1994) Antisense RNA control in bacteria, phages and plasmids. Annu. Rev. Microbiol., 48, 713–742. [DOI] [PubMed] [Google Scholar]
- 26.Kumar M. and Carmichael,G.G. (1998) Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev., 62, 1415–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanhee-Brossollet C. and Vaquero,C. (1998) Do natural antisense transcripts make sense in eukaryotes? Gene, 211, 1–9. [DOI] [PubMed] [Google Scholar]
- 28.Knee R. and Murphy,P.R. (1997) Regulation of gene expression by natural antisense RNA transcripts. Neurochem. Int., 31, 379–392. [DOI] [PubMed] [Google Scholar]
- 29.Nellen W. and Sczakiel,G. (1996) In vitro and in vivo action of antisense RNA. Mol. Biotechnol., 6, 7–15. [DOI] [PubMed] [Google Scholar]
- 30.Kramer A. (1996) The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 367–409. [DOI] [PubMed] [Google Scholar]
- 31.Smith C.M. and Steitz,J.A. (1997) Sno storm in the nucleolus: new roles for myriad small RNPs. Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- 32.Austin J. and Kenyon,C. (1994) Developmental timekeeping. Marking time with antisense. Curr. Biol., 4, 366–369. [DOI] [PubMed] [Google Scholar]
- 33.Kimelman D. and Kirschner,M.W. (1989) An antisense mRNA directs the covalent modification of the transcript encoding fibroblast growth factor in Xenopus oocytes. Cell, 59, 687–696. [DOI] [PubMed] [Google Scholar]
- 34.Li A.W., Seyoum,G., Shiu,R.P. and Murphy,P.R. (1996) Expression of the rat BFGF antisense RNA transcript is tissue-specific and developmentally regulated. Mol. Cell. Endocrinol., 118, 113–123. [DOI] [PubMed] [Google Scholar]
- 35.Volk R., Koster,M., Poting,A., Hartmann,L. and Knochel,W. (1989) An antisense transcript from the Xenopus laevis bFGF gene coding for an evolutionarily conserved 24 kd protein. EMBO J., 8, 2983–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lazar M.A., Hodin,R.A., Darling,D.S. and Chin,W.W. (1989) A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA alpha transcriptional unit. Mol. Cell. Biol., 9, 1128–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noguchi M., Miyamoto,S., Silverman,T.A. and Safer,B. (1994) Characterization of an antisense Inr element in the eIF-2 alpha gene. J. Biol. Chem., 269, 29161–29167. [PubMed] [Google Scholar]
- 38.Farrell C.M. and Lukens,L.N. (1995) Naturally occurring antisense transcripts are present in chick embryo chondrocytes simultaneously with the down-regulation of the alpha 1 (I) collagen gene. J. Biol. Chem., 270, 3400–3408. [DOI] [PubMed] [Google Scholar]
- 39.Hildebrandt M. and Nellen,W. (1992) Differential antisense transcription from the Dictyostelium EB4 gene locus: implications on antisense-mediated regulation of mRNA stability. Cell, 69, 197–204. [DOI] [PubMed] [Google Scholar]
- 40.Cock J.M., Swarup,R. and Dumas,C. (1997) Natural antisense transcripts of the S locus receptor kinase gene and related sequences in Brassica oleracea. Mol. Gen. Genet., 255, 514–524. [DOI] [PubMed] [Google Scholar]
- 41.Keister D.B. (1983) Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R. Soc. Trop. Med. Hyg., 77,487–488. [DOI] [PubMed] [Google Scholar]
- 42.Boucher S.E. and Gillin,F.D. (1990) Excystation of in vitro-derived Giardia lamblia cysts. Infect. Immun., 58, 3516–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lujan H.D., Mowatt,M.R., Conrad,J.T., Bowers,B. and Nash,T.E. (1995) Identification of a novel Giardia lamblia cyst wall protein with leucine-rich repeats. Implications for secretory granule formation and protein assembly into the cyst wall. J. Biol. Chem., 270, 29307–29313. [DOI] [PubMed] [Google Scholar]
- 44.Mowatt M.R., Aggarwal,A. and Nash,T.E. (1991) Carboxy-terminal sequence conservation among variant-specific surface proteins of Giardia lamblia. Mol. Biochem. Parasitol., 49, 215–227. [DOI] [PubMed] [Google Scholar]
- 45.Nash T.E. and Mowatt,M.R. (1992) Identification and characterization of a Giardia lamblia group-specific gene. Exp. Parasitol., 75, 369–378. [DOI] [PubMed] [Google Scholar]
- 46.Mowatt M.R., Weinbach,E.C., Howard,T.C. and Nash,T.E. (1994) Complementation of an Escherichia coli glycolysis mutant by Giardia lamblia triosephosphate isomerase. Exp. Parasitol., 78, 85–92. [DOI] [PubMed] [Google Scholar]
- 47.Lu S.Q., Baruch,A.C. and Adam,R.D. (1998) Molecular comparison of Giardia lamblia isolates. Int. J. Parasitol., 28, 1341–1345. [DOI] [PubMed] [Google Scholar]
- 48.Baruch A.C., Isaac-Renton,J. and Adam,R.D. (1996) The molecular epidemiology of Giardia lamblia: a sequence-based approach. J. Infect. Dis., 174, 233–236. [DOI] [PubMed] [Google Scholar]
- 49.Murtagh J.J.,Jr, Mowatt,M.R., Lee,C.M., Lee,F.J., Mishima,K., Nash,T.E., Moss,J. and Vaughan,M. (1992) Guanine nucleotide-binding proteins in the intestinal parasite Giardia lamblia. Isolation of a gene encoding an approximately 20-kDa ADP-ribosylation factor. J. Biol. Chem., 267, 9654–9662. [PubMed] [Google Scholar]
- 50.Sanchez L.B., Elmendorf,H., Nash,T.E. and Muller,M. (2001) NAD(P)H:menadione oxidoreductase of the amitochondriate eukaryote Giardia lamblia: a simpler homologue of the vertebrate enzyme. Microbiology, 147, 561–570. [DOI] [PubMed] [Google Scholar]
- 51.Lujan H.D., Mowatt,M.R., Conrad,J.T. and Nash,T.E. (1996) Increased expression of the molecular chaperone BiP/GRP78 during the differentiation of a primitive eukaryote. Biol. Cell, 86, 11–18. [DOI] [PubMed] [Google Scholar]
- 52.McArthur A.G., Morrison,H.G., Nixon,J.E., Passamaneck,N.Q., Kim,U., Hinkle,G., Crocker,M.K., Holder,M.E., Farr,R., Reich,C.I., Olsen,G.E., Aley,S.B., Adam,R.D., Gillin,F.D. and Sogin,M.L. (2000) The Giardia genome project database. FEMS Microbiol. Lett., 189, 271–273. [DOI] [PubMed] [Google Scholar]
- 53.Upcroft J.A., Healey,A., Mitchell,R., Boreham,P.F. and Upcroft,P. (1990) Antigen expression from the ribosomal DNA repeat unit of Giardia intestinalis. Nucleic Acids Res., 18, 7077–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]