Abstract
In this study, we utilized unsupervised machine learning techniques to examine the relationship between different symptoms in cases who died of COVID-19 and cases who recovered from it. First, our data was cleared of redundancies, and the ten most important variables were selected using a filter-based technique (extra-tree classifier). Next, we calculated the Silhouette, Davis Boldin (DB), and the mean intra-cluster distance measures to select the optimal number of clusters, then clustered the data using both the K-means and hierarchical clustering based on Self Organizing Map (SOM) neural network. Our results revealed that patients who died of COVID-19 had high mean values in different symptoms, but not all patients with this characteristic necessarily died. Besides, our result indicated that the patient's age is directly related to the hospital duration, and elderly patients are more likely to be assigned to the intensive care unit (ICU). However, the patient's sex has the same distribution in different groups and does not correlate with other symptoms. In conclusion, our results confirmed past studies. Also, this research helps physicians improve medical services by considering other important factors for treating different groups of COVID-19 patients.
Keywords: COVID-19, Unsupervised machine learning, Clustering, Self-organizing map, Neural network
1. Introduction
In late 2019, the world faced a major dilemma that affected all aspects of human life. A new type of coronavirus has emerged in Wuhan, China [1]. The infectious virus targets the human respiratory system and is rapidly transmitted from person to person [2]. Its high rate of transmission over several days led to the reporting of the first samples of the virus in different countries. Finally, with the spread of the virus in 216 countries, the World Health Organization on March 11, 2020, declared the outbreak of novel coronavirus as a deadly pandemic [3].
There are several types of coronaviruses in the world. The most important of these viruses are acute respiratory syndrome and middle east respiratory syndrome [4]. The newly identified virus is called SARS-COV-2 and is the cause of COVID-19 disease [4]. Like other families of coronaviruses, COVID-19 is a deadly infectious disease that targets the lungs in an infected person [4].
Several effective coronavirus vaccines have been developed by reputable companies around the world so far. But, adherence to government guidelines on safety precaution measures and social distancing are still the two most efficient measures in preventing COVID-19 [4].
Numerous studies have reported various neurological, pathological, and recurrence-based SARS-COV-2 features. Although novel coronavirus does not directly affect the central nervous system, delirium and septic encephalopathy are the most important neurological symptoms in critically ill patients. In young and female patients, smell dysfunction and headache are the predominant symptoms in most patients, and muscle pain is a common symptom in both severe and mild patients [5]. Infection with the new coronavirus, on the other hand, may result in pathological changes in some organs including the heart, brain, lung, or kidney. Generally, pathological observations may not be a reliable way to diagnose, but they can reveal pathological changes associated with SARS- COV-2 infection and the cause of death in patients [6]. Besides, several studies have shown that some of the pathological changes in the COVID-19 cases are similar to those
Seen in SARS and MERS cases [6]. Additionally, feature extraction from different genomic sequences of SARS-COV-2 using nonlinear methods such as recurrence quantification analysis (RQA) can provide valuable information about both virus mutations and vaccine development. The RQA-related features make it possible to compare genomic sequences of SARS-COV-2 [7]. However, researchers in computer science have tried to study various aspects of the new found virus and help handle the epidemic. These methods, which are focused on diagnosing the disease or predicting the pattern of pandemic outbreaks, include machine learning [8,9], deep learning [10,11], mathematical modeling [12,13], and social network analysis (SNA) [14,15] techniques. With the advent of deep learning approaches in recent years, supervised learning methods have been replaced as much as possible with unsupervised methods [16]. Deep learning approaches with several parameters and hierarchies require a large number of data samples for training, so deep learning methods with supervised fashion require labeling a large number of samples, which is a very time-consuming process [16]. Therefore, paradigms that require less or no labeled samples are preferred for training deep learning models [16].
In this study, we used two unsupervised learning techniques to discover possible hidden patterns among the various neurological and pathological features of the COVID-19 cases. We set a target to assess the relationship between the different characteristics of recovered COVID-19 cases and patients who died of COVID-19 in different age groups. Our methods include clustering the COVID-19 cases by the K-means and SOM neural network methods. We tried to find out what is the relationship between patients' characteristics in a similar group? What is the difference between patients' characteristics in dissimilar groups? What characteristics play an important role in determining the outcome of COVID-19 cases (recovery or death)? What is the relationship between patients' characteristics and their outcomes?
Our result showed that the patient's age is directly related to the hospitalization, and elderly patients are more likely to be allocated to the ICU, but the patient's sex has the identical distribution in diverse groups and does not associate with other symptoms. Also, our outcomes demonstrated that patients who died of COVID-19 had high mean values in various symptoms, but not all patients with this attribute necessarily passed. To conclude, our results proved past research. Besides, this research assists physicians enhance medical services by evaluating other elements for treating diverse groups of COVID-19 cases.
In the next section of the article, we first summarize some related works then describe the study area and how to collect data. Second, we display statistical characteristics of the research data through various visualization plots. Next, we describe the data preprocessing and implemented models in the learning subsections and define research methods in detail. The research findings are displayed in the results section. In the discussion section, the results of different techniques and research applications are explained. Finally, in the conclusion section, the purpose of the research and our findings are reviewed, and the limitations of the study are stated to do more future work.
1.1. Related works
According to the latest statistics, 7.1% of the machine learning approaches proposed to deal with the COVID-19 are unsupervised [17]. These approaches often involve a clustering approach to detect hidden patterns among different disease symptoms [17].
Julian et al. [18] clustered the COVID-19 cases and distinguished severity subgroups among patients using the X-mean method. The researchers used four different tests C-reactive protein (CRP), the serum levels of aspartate transaminase (AST), the number of neutrophils, lactate dehydrogenase (LDH) as the main variables to assign items to three separate clusters. The results showed that the X-mean clustering method can well identify the severity subgroups of the COVID-19 patients through applied tests.
Chaudhary and Singh [19] used the principal component analysis (PCA) technique to reduce the size of the COVID-19 patient data set, then identified hidden communities across different countries using the K-means clustering method. The results of the study revealed the communities around the world that play an important role in transmitting the disease to other regions.
In another study, researchers used an unsupervised method called the biterm topic model (BTM) to statistically and geographically identify user-generated content related to the COVID-19. To do this, the researchers grouped users' tweets with the symptom, test, and recovery keywords associated with the COVID-19 into five different clusters with specific keywords. The researchers in this study found that the group of people who reported the symptoms of COVID 19 disease and did not test to manifest the disease was more dominant than the other groups. Therefore, it is never possible to report the exact number of confirmed cases of the COVID-19 [20].
Xue, Jia et al. [21] used an unsupervised approach, latent dirichlet allocation (LDA), to analyze tweets posted by users and identify content related to family violence during the COVID-19 epidemic. Researchers have extracted nine various themes related to family violence from identified tweets, and claim that their findings could define appropriate policies to reduce family violence in future outbreaks.
Similarly, researchers used the LDA method to analyze COVID-19-related tweets to find out the emotions and concerns of users. The researchers categorized different tweets into several groups with specific keywords and found that the predominant sentiments associated with the new coronavirus epidemic among users are related to mixed feelings of fear, trust, and anger [22,23].
Karadayi et al. [24] designed a hybrid deep learning-based architecture to identify an unsupervised anomaly in multivariate spatio-temporal data of COVID-19 patients. In this study, the network is composed of a three-dimensional convolutional neural network (CNN) encoder and a convolutional long short-term memory (CLSTM) decoder that is trained with an unsupervised paradigm to detect the anomaly. The authors claimed that the proposed method has significantly better performance in detecting anomalies than the state-of-the-art methods.
In [25], researchers used K-means clustering to separate different countries into five different groups in the first and second surges of the epidemic. In this study, researchers used the susceptible-exposed-infected-recovered (SEIR) model to estimate the effective reproductive number (t) of each country, then used the t > 1 to t < 1 duration to identify high-risk communities at the global level and define different clusters.
Mingxiang et al. [26] presented a hybrid unsupervised and supervised approach for clustering and classifying tweets related to the COVID-19 symptoms, respectively. First, relevant tweets are extracted and clustered using the BTM method. Then, tweets in various clusters are classified as different data classes using a deep learning model. The results demonstrated that the deep learning model has a better performance than other models developed in this study.
Haupt, Michael Robert et al. [27] used unsupervised machine learning and social network analysis techniques to analyze conversations on Twitter related to protest movements against the COVID-19 epidemic policies. In this study, researchers used an unsupervised fashion using natural language processing to extract and cluster topics and used the SNA technique to analyze the re-tweet network. Cluster analysis in this study showed that the number of tweets concerning the protest movement was more than the opposition, and the protest movement is the dominant phenomenon. In addition, supporters of the protest movement are more likely to re-tweet users than non-supporters.
As another example, researchers in Ref. [28] clustered 155 countries based on air pollution, health services, socio-economic empowerment variables using the K-means method with a different number of clusters. Next, the clusters were compared based on the number of confirmed cases, recovered cases, and deaths of COVID-19 infection using a one-way ANOVA test. The results confirmed the best model for clustering countries with three PCA parameters and five clusters. In addition, the results revealed that the K-means algorithm with five clusters can well stratify countries based on the number of confirmed cases [28].
Finally, researchers in Ref. [29] investigated the relationship between the COVID-19 mortality and Bacille Calmette-Guerin (BCG) vaccination program. In this study, researchers used the K-means clustering method to cluster the countries that followed the BCG vaccination, then compared the mortality rates of countries in the same cluster with samples belonging to other clusters. The results showed that there is a significant relationship between mortality rate and the BCG vaccination, but the relationship between the COVID-19 mortality rate and other vaccination programs was not significant. The government that implemented the BCG vaccination policy for at least the last 15 years has recorded a lower mortality rate. In addition, the results showed that with an increase in population over 65 years, the mortality rate increases sharply.
2. Methodology
Fig. 1 demonstrates the block diagram of the proposed methodology. First, the research data is described, and a brief description is provided of COVID-19 patients' characteristics, then data samples are visualized with two well-known visualization methods. Visualization helps to reveal hidden statistical properties among data samples. In the third step, the data are pre-processed using various data cleaning and transformation methods then important patient characteristics are selected. In the processing stage, the number of optimal clusters is calculated using the DB and Silhouette indexes then the samples are clustered by the K-means and SOM methods. Finally, the statistical characteristics of the generated clusters are calculated, and the results are compared and analyzed.
Fig. 1.
The block diagram of the research methodology.
2.1. Experiment data
The research data has been collected from a set of clinical symptoms of patients with the COVID-19 in Saveh Hospital in Iran through questionnaires, interviews, and medical records of patients between August 2020 and November 2020. Research data includes 1142 cases with 39 variables per patient. The number of recovered cases and deaths in the data set are 1131 and 111 samples, respectively. Also, the most important variables include age, sex, hospitalized ward, cough, fever, various types of underlying diseases, hospital duration, smoking, intubation, and the outcome of patients (recovery or death). Table 1 provides a detailed description and possible values of all the important variables in our data set.
Table 1.
Description and possible values of important variables in this research.
| Variable | Description |
|---|---|
| Age | Patient's age |
| Gender | 0; female, 1; male |
| Taken to the hospital | 1; no, 2; yes |
| Section of hospital | the ward where the patient has been hospitalized. 1; regular ward, 2; intensive care unit, 3; no hospitalization |
| Contact coronavirus | 0; no history of contact with COVID-19 patients, 1; history of contact with COVID-19 patients. |
| Result PCR | 0; negative for COVID-19, 1; positive for COVID-19, 3; test result is pending. |
| Fever, cough, headache, chest pain | 0 stands for absence of symptom, and 1 stands for presence of the symptom |
| Shortness of breath | 0 stands for absence of symptom, and 1 stands for presence of the symptom |
| CT scan manifestation | 1; CT scan results for COVID-19 are negative, 2; CT scan results for COVID-19 are positive |
| Rate of partial pressure of oxygen, Po2 | 0; PO2 levels are greater than 93, 2; PO2 levels are less than 93 |
| Condition entering the hospital | 0; sever, 1; mild |
| Presence of underlying diseases | 0 stands for absence of underlying diseases, and 1 stands for presence of the underlying diseases. |
| Hospital duration | number of hospitalization days |
| Pregnancy | 0; no, 1; yes |
| Intubation | 1; patient has undergone intubation, 2; patient has not undergone intubation |
| Death | no; patient has recovered, yes; patient has died. |
2.2. Data visualization
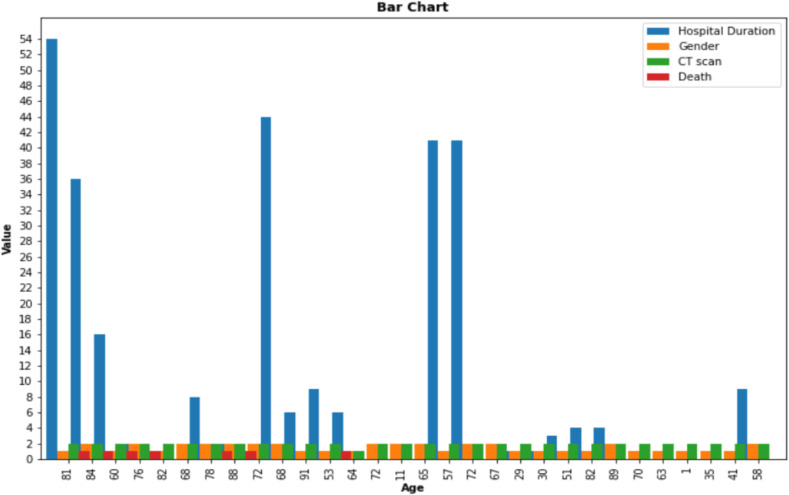
The purpose of data visualization is to reveal hidden characteristics within the data that are not statistically visible. We visualize the data with two visualization plots. A useful way to examine the different characteristics of patients relative to each other is to display data with a bar plot [30]. The bar diagram shows how to assign a variety of features to a specific sample. Besides, it is a convenient way to compare the variable values of one case with another case based on a particular variable [30]. For example, in Fig. 2 , we show the gender, hospital duration, CT scan result, and class label (death or recovery) variables of several patients toward their age. The horizontal axis in Fig. 2 represents the age of patients and the vertical axis shows the values of gender, hospital duration, CT scan, and class label variables corresponding to the age of each particular patient. The value of the hospital duration variable indicates the number of days the patient has been hospitalized. Values 1 and 2 for the “CT-scan” variable indicate negative and positive CT-scan manifestation, respectively, and for the “gender” variable denote female and male patients, respectively. According to Fig. 2, most male patients over 50 years old were hospitalized longer than younger cases, and the CT-scan result has been positive for most patients of both sexes.
Fig. 2.
Bar plot of sex, length of hospitalization, CT-scan result, and the class label variables to their age.
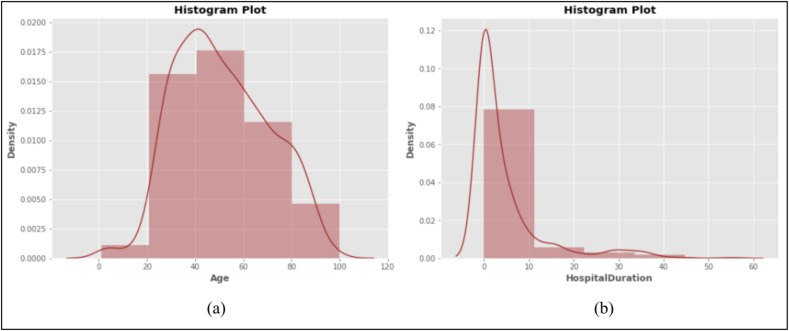
As the latest visualization plot, we have used a histogram plot to show the distribution of a particular feature in different patients. The histogram shows how the values of a variable are assigned to different intervals. We plotted two histograms to examine the distribution of two features that play an important role in the results of this study. Fig. 3 shows histogram plots of age and hospital duration variables in this study. The horizontal axis represents the set of variable values in different intervals, and the vertical axis shows the density of values distribution in the defined intervals. According to Fig. 3 a, the density of patients’ age is higher in the range of 40–60 years. It means that the age of the patients has the highest frequency in this period. Similarly, the density of the hospitalization in the range of 0–10 days is maximum (Fig. 3b). It indicates that most patients were hospitalized for less than ten days.
Fig. 3.
Histogram plot with distribution density of age (a) and hospital duration (b) variables of the COVID-19 cases in this study.
2.3. Data pre-processing
In the real world, the data collected may be distorted by environmental conditions such as noise or human mistakes and may not be sufficiently valid for training the learning model. The purpose of data preprocessing is the preparation of data as much as possible for the processing and learning phase [31]. Data preprocessing strongly affects the performance of learning models and ultimately leads to improving model performance. This stage includes steps to clear, transform, integrate, and reduce the data [31]. In general, the output of the preprocessing phase is a decremental set of redundancies, null values, and outliers. Besides, depending on the learning method, data preprocessing may involve data sampling and normalization.
In this research, we have applied various preprocessing techniques to the data. Null values in a particular attribute are filled with the average value of that attribute. This ensures that the learning model does not incline to a specific value in a feature. Moreover, we used the K-means clustering method to detect outliers in the data. Based on the K-means clustering method, our data lacked outliers. It is due to accurate methods such as patients' medical records and questionnaires in collecting data for this study.
2.3.1. Feature selection
Feature selection is a preprocessing step to reduce data volume. The goal is to select an optimal subset of features and eliminate unrelated variables in the research data. So, it leads to improving the learning model metrics and reduces computational costs [32]. The most well-known methods for feature selection are Filter, Wrappers, and Embedded approaches [32]. The filter approach uses feature ranking independent of the learning algorithm. Feature ranking is interpreted as the degree of importance of a feature in distinguishing between data classes [32]. Techniques based on the wrapper approach are implemented along with the learning algorithm [32]. These methods repeatedly invoke the learning algorithm on different sets of features and use the model performance results to select the best subset. Finally, the embedded techniques are made from a combination of filter and wrapper methods [32]. In this study, we used the extra tree classification to select the ten most influential features in diagnosing the outcome of COVID-19 cases (recovery or death). The extra tree algorithm is a filter-based technique that is applied independently of the learning algorithm to select the optimal features on the data set. This algorithm is an ensemble and majority-vote-based method similar to the random forest method that uses a set of decision trees to identify class labels [33].
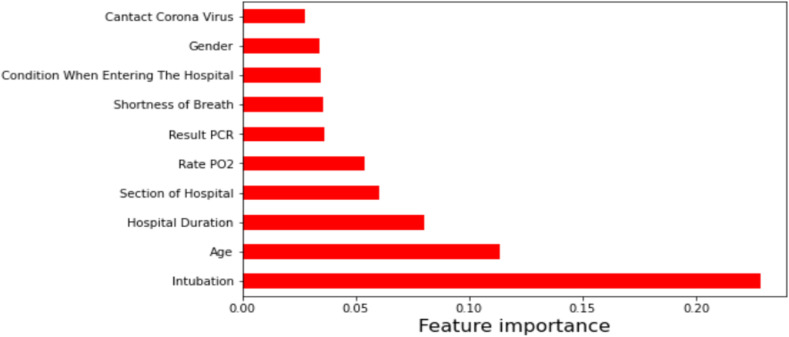
As mentioned earlier, our aim in this study was to investigate the relationship between the different characteristics of patients who died of COVID-19 or recovered from it. Therefore, we only examine the relationship between the variables that have the most impact on determining the outcome of patients infected with the disease (class labels). This action increases the validity of the results and prevents biases that may be caused by considering some irrelevant features. Since our techniques in this study are unsupervised, we removed class labels from the data set after the feature selection stage. The most important variables filtered by a filter-based method (extra tree) are presented in Fig. 4 . In the processing phase, the features selected in Fig. 4 are used by learning methods. According to this figure, intubation, age, and hospital duration are the most relevant characteristics to class labels, respectively.
Fig. 4.
Top 10 important features selected using the filter-based method.
2.4. Processing
The processing step is the main step in machine learning techniques. In the processing stage, the learning models are applied to preprocessed data. In machine learning, various learning methods have been proposed for different applications in the literature. These methods are mainly based on supervised, unsupervised, or semi-supervised feature learning [31].
2.4.1. Unsupervised learning algorithms
We have used the data clustering methods in unsupervised learning. The purpose of clustering is to separate data samples into different groups so that the samples belonging to a group have the most similarity with the members of the same group and have the least similarity with the members of other groups. We implemented the K-means clustering and neural network-based SOM methods for unsupervised learning to identify possible hidden patterns in the symptoms of patients. The identified patterns help us understand the similarities between the same symptoms of patients in the same group and understand the difference between the symptoms of these patients and patients assigned to other groups.
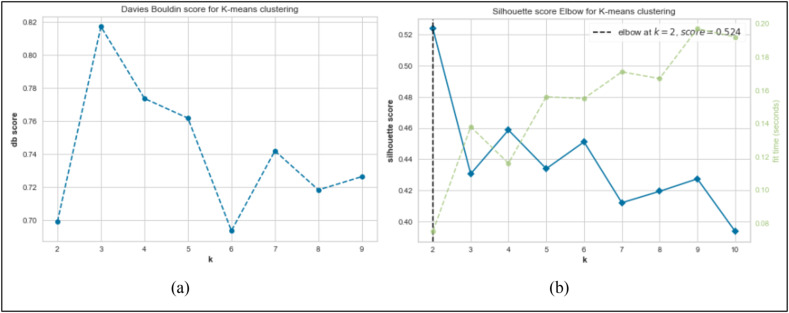
To reduce the cost of computations, we first calculated the number of optimal clusters for the research data using the clustering quality evaluation indexes, then clustered the data to this number optimally. We used Davies-Bouldin (DB) and Silhouette indexes to calculate the number of optimal clusters. DB and Silhouette indexes are two internal measures for clustering evaluation [34]. Internal measures do not depend on external information (prior knowledge) because they directly evaluate the clustering structure from the original data. External indexes, on the other hand, require information about the clustering problem [34]. The DB index shows the ratio of inter-cluster differentiation to intra-cluster similarity. So, a lower value in the DB index indicates better performance of the clustering algorithm [34]. But the silhouette index depends on the cohesion within the clusters as well as their degree of separability. The silhouette index for each point measures its correlation to its cluster relative to the adjacent cluster(s). The value of silhouette is between +1 and −1, so a value close to 1 indicates a good match between the point and its cluster. If the silhouette measure is close to 1 for all cases within the clusters, the clustering operation is performed correctly [34]. Otherwise, it indicates poor clustering results, which may also be due to improper selection of the number of clusters (k). Fig. 5 reveals the calculated DB and silhouette indexes for the different number of clusters. According to the DB metric, the best number of clusters for clustering our research data is two clusters (Fig. 5a). Similarly, the Silhouette index has determined the number of two clusters as the optimal number of clusters (Fig. 5b). The DB and Silhouette indices are calculated through Equations (1), (2)), respectively.
| DB = k∑i = 1Max {Δ(Ci)+Δ(Cj)}, i ≠ j | (1) |
| Silhouette = Max{Δ(Ci), y} | (2) |
Fig. 5.
The calculated Davies-Bouldin and Silhouette indices for the different number of clusters(k).
Where Δ(Ck) stands for the average intra-cluster distance within the cluster Ck, δ(Ci, Cj) is the inter-cluster distance between the cluster Ci and Cj, and y represents the smallest average inter-cluster distance between all clusters.
2.4.2. K-means algorithm
K-means algorithm takes the parameters k from the input and divides the n sample into k clusters so that the internal similarity of the clusters is high and the external similarity of the clusters is low. The degree of similarity in each cluster is measured by the average distance of the samples within that cluster. In fact, this algorithm is a heuristic method for minimizing the square error measure given in Equation (3) [16].
| (3) |
In this relation, E stands for the sum of squared errors for all data samples, p represents a data sample that belongs to cluster Ci, and mi is the mean of samples in cluster Ci.
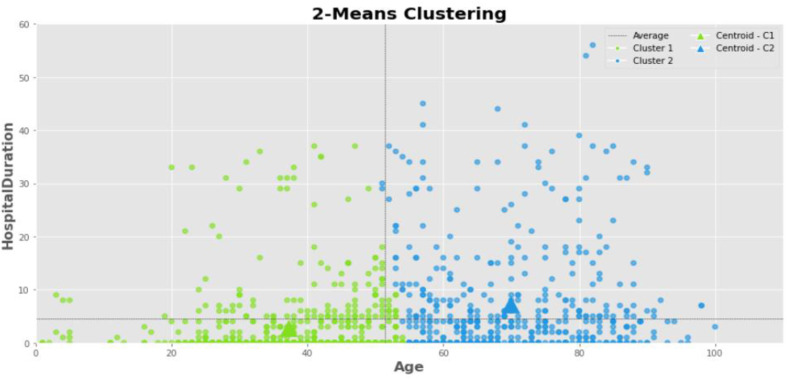
Fig. 6 shows the K-means clustering of the COVID-19 cases into two clusters. In this figure, the clusters distinguish by different colors and triangle symbols represent the cluster centers. The horizontal axis corresponds to the age of the patients, and the vertical axis shows the hospital duration. Also, the centers of the clusters were calculated approximately at points [22,38,70]. So, most of the values of the age and hospital duration variables are close to these points since the average distance between cases and these centers is minimal. Besides, the cases belonging to the two clusters 1 and 2 are in the age range of lower and higher than 58 years, respectively. Also, the distribution of samples based on the number of hospitalization days in both clusters is almost the same, but the cases with longer duration in cluster 2 with older cases are more than cluster 1.
Fig. 6.
The COVID-19 cases belonging to clusters 1 and 2 in the 2-means clustering based on age and hospital duration characteristics.
2.4.3. Self-organizing map (SOM)
The SOM method is based on artificial neural networks and is a powerful tool to reduce data size for exhibiting multidimensional data in two dimensions [35]. The SOM is also a network of neurons (nodes) that maintains a spatial connection between data samples by matching the neighbors of the winning neuron [35]. Therefore, in addition to clustering the data into separate areas, similar areas are usually placed next to each other. Unlike traditional neural networks, the SOM technique does not require a target vector to learn the features [35]. In SOM, each neuron has a specific spatial position and has weights of the same dimensions as the input vector [35]. In addition, unlike conventional neural networks, there is no connection in a SOM network, and the drawing lines only indicate the proximity between nodes [35]. The connection between network nodes refers to.
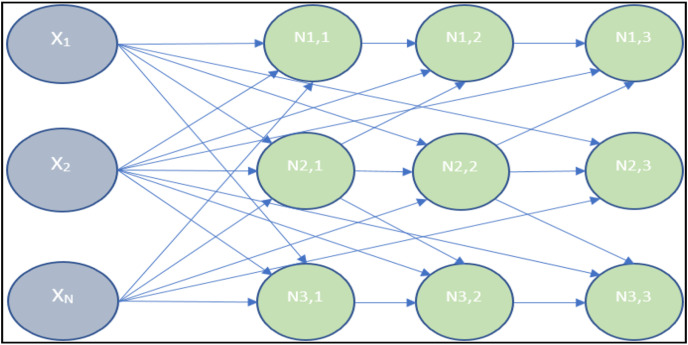
The existence of a path between them to exchange data samples. In a SOM network, all nodes are connected to the network input at the same time [35]. Therefore, the samples assigned to the nodes are permanently stored, and no data samples are exchanged between the nodes [35]. Fig. 7 presents a 3 * 3 SOM with an input layer and a middle layer.
Fig. 7.
A 3 × 3 self-organizing map neural network with an n-dimensional input vector.
Initially, the SOM randomly assigns a weight vector to all neurons equal to the dimension of the input vector. Each input sample is assigned to a node that is more similar to its weight [35]. The winning neuron, which receives the sample, modifies its weight to match the values of the input vector. In addition, the winning neuron simultaneously corrects the weight of the nodes present in its neighborhood radius [35]. Besides, the weight of the neuron that is closer to the winning neuron changes more. In subsequent iterations, with the arrival of new samples, SOM tends to map with stable areas. The measure for assessing the similarity of the input vector with the weight of network neurons is usually the Euclidean distance. The Euclidean distance between two vectors is equal to Equation (4), where X is the input vector, and W is the weight vector.
As mentioned earlier, after determining the winning neuron, the weight of the neighboring neurons changes. To do this, we must first calculate its neighborhood radius, then identify the nodes within the radius. Over time, with the arrival of each new sample, the neighborhood of neurons becomes smaller by decreasing the neighborhood radius. For this action, the exponential reduction function in Equation (5) is used.
| (4) |
| (5) |
where σ stands for the network radius at the moment t0, γ is a time constant, and variable t shows the current time step. The value of γ depends on the number of rounds selected to run the algorithm. Now suppose that t is a time step, L is an indicator called learning rate, W stands for the weight vector of the neurons, and X represents the input vector, then the weight of the nodes in the neighborhood radius is corrected as Equation (6).
| W(t + 1) = W(t) + L(t) (X(t) − W(t)) | (6) |
Also, the learning rate decreases as follows in each cycle (Equation (7)). Initially, the learning rate is a fixed value (L) and gradually tends to zero.
| (7) |
In Equation (6), not only the learning rate should be reduced over time, but the learning effect should be proportional to the distance of one neuron from the winning neuron. The learning effect at a higher radius should be less than the lower radius and close to the winning neuron [35]. Therefore, Equation (6) must be changed to Equation (8), in which θ is calculated by Equation (9) and represents the effect of distance from the winning neuron on the learning of the neighbor node. In Equation (9), the variable d is the distance between the neighbor node and the winning node, and σ indicates the neighborhood radius.
| W(t + 1) = W(t) + θ(t)L(t) (X(t) − W(t)) | (8) |
| (9) |
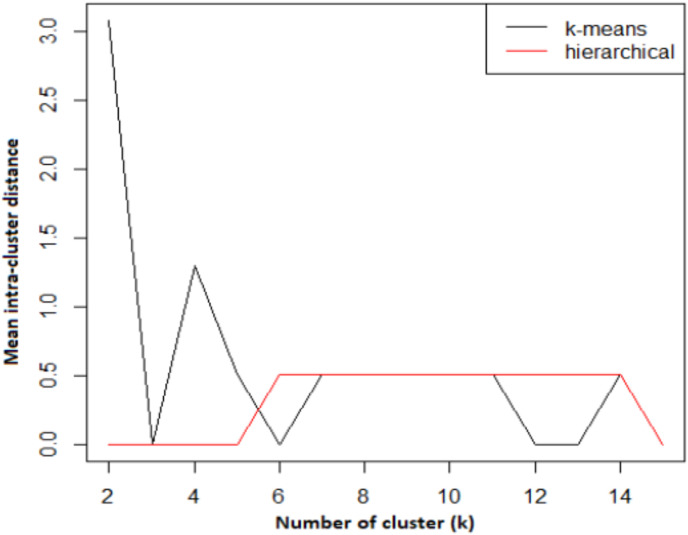
In practice, how to determine the dimensions of the network for different applications is different. It is achieved by preference so that the designed network has no empty neurons. We used a 3 × 3 network to design the SOM and allocate the COVID-19 cases. Fig. 8 a reveals how the primary weights (codes vector) are assigned to the neurons. This figure shows what samples each neuron receives for allocation. Similarly, Fig. 8 b displays the number of samples allocated to the neurons with different colors in the same network. In this figure, red neurons have the least number of data samples, and the lower the color intensity, the greater the number of samples assigned to the neuron. Before clustering network nodes, we must calculate the number of optimal clusters by clustering measure. As mentioned earlier, in clustering, the goal is to cluster the data so that the samples in the same group have the least distance (most similarity) with each other. Since in a SOM the allocation of samples and the neighborhood radius are determined based on the distance measure, so a suitable way for determining the number of clusters in a SOM is to calculate the average intra-cluster distance in the different number of clusters (k). We calculated the mean intra-cluster distance for the two K-mean and hierarchical clustering methods for different numbers of k (Fig. 9 ). Hierarchical clustering uses a tree structure to cluster data samples [31]. This technique is usually based on either Agglomerative or Divisive approaches [31]. In the agglomerative method, each case initially forms a cluster, and clusters are repeatedly combined until the stop condition is met or only one cluster is available. On the other hand, in the divisive approach, all the samples form a single cluster, then they are repeatedly divided into smaller clusters until each sample forms a cluster [31].
Fig. 8.
(a). The initial weight vectors (codes vector) of SOM neurons. (b). The number of COVID-19 cases assigned to SOM neurons with a different color. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 9.
The number of optimal clusters based on the average intra-cluster distance for the two K-mean and hierarchical clustering methods.
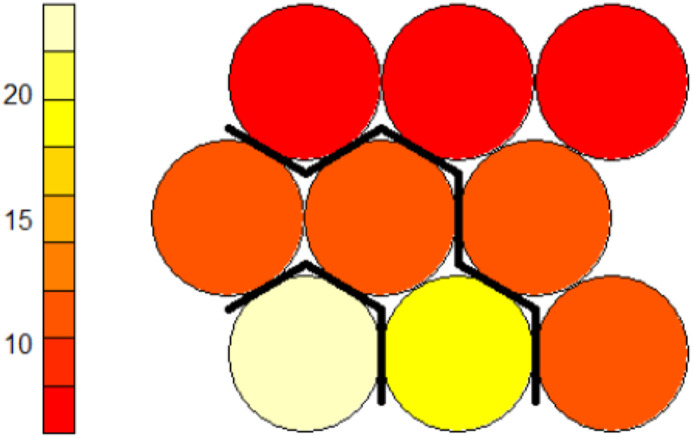
According to the average intra-cluster, 3 clusters is the optimal number of clusters for COVID-19 cases. Besides, Fig. 10 presents the SOM neurons colored based on the average distance between neurons whose clusters (k = 3) are separated by hierarchical clustering. In this network, more distance (less similarity) indicates a lighter color, and less distance (more similarity) is darker in color. In the results section, we examine what knowledge the SOM clustering provides about the COVID-19 cases.
Fig. 10.
Hierarchical clustering (k = 3) of colored SOM based on the average distance of each neuron from its neighbors (higher color intensity indicates less distance and more similarity). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.5. Analysis environment
We used a system with a CPU 2.3Ghz (five-core), 6 gigabytes of RAM, and one terabyte of disk space to implement various algorithms in this study. We implemented visualization, preprocessing, and K-means clustering stages with the Python programing language version 3.5. Also, R programming language version 3.5.1 was used to implement SOM neural network due to its ability to visualize results.
3. Result
In this section, our results for different machine learning methods are presented separately, and the data clusters in different techniques are compared with each other.
3.1. K-means clustering
In Section 2.4.1 we estimated that the best number of clusters for clustering our data is 2 clusters. We have given some statistical characteristics (count, mean) of each cluster for data classes (recovery or death) based on some main features in Table 2 . Samples of the two clusters are essentially patients who have recovered from the disease. It shows that there is not much difference between the symptoms of patients in different classes. The similarity of patients who died of coronavirus infection to the cases belonging to clusters zero and one is 1.9% and 7.8%, respectively. The noteworthy point in Table 2 is that the average of features in cluster zero is generally lower than the average of the corresponding features in cluster one. Because fewer deaths are assigned to cluster zero, it proves that patients who recovered from the disease have a lower mean age, are not admitted to the ICU, require less intubation, and have been hospitalized for shorter periods. The results also show that the patient's sex is equally distributed in the two clusters. In addition, the results indicate that the average of the mentioned features improved in some patients in cluster number one, with a higher number of similar deaths. In general, the clustering results confirm the principle that the recovery of infected patients is not greatly affected by their features, but a high average of the characteristics can effectively increase risk of death in them. Finally, the accuracy measure for K-means clustering was 62%, which indicates how well the clustering algorithm is able to group samples with the same class in the same cluster.
Table 2.
Statistical characteristics of clusters in K-means clustering.
| Class | Cluster | Count (%) | Age | Gender | Intubation |
Result PCR | Hospital Duration | Hospital Section |
|---|---|---|---|---|---|---|---|---|
| Mean | ||||||||
| Recovery | 0 | 54.7 | 37.1 | 1.5 | 1.3 | 0.3 | 2.5 | 1.3 |
| 1 | 35.4 | 68.6 | 1.4 | 1.5 | 0.6 | 6.8 | 1.5 | |
| Death | 0 | 1.9 | 40.4 | 1.3 | 1.9 | 0.7 | 6.0 | 1.5 |
| 1 | 7.8 | 76.0 | 1.4 | 1.9 | 0.7 | 8.1 | 1.6 |
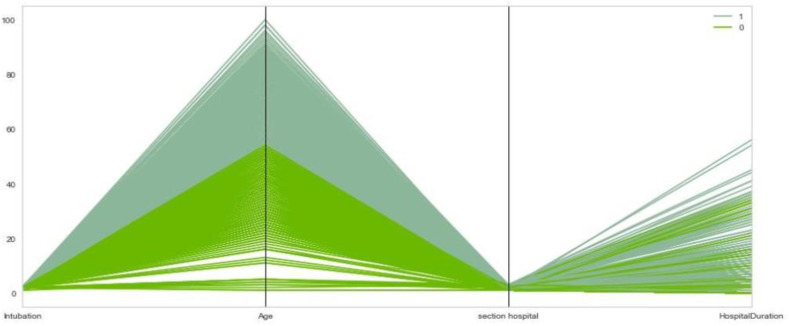
We have used the parallel coordinates plot to visualize the samples belonging to each cluster (Fig. 11 ). For more clarity in Fig. 11, we consider only four features (age, intubation, hospital section, hospital duration) for each case. Also, the samples were separated into different clusters with different colors. Similar to Table 2, Fig. 11 reveals that cluster number zero, which includes more recovered cases, is at a lower level of the plot, and cluster one, which has the highest number of deaths, is at a higher level.
Fig. 11.
Parallel Coordinates diagram of age, intubation, hospital duration, and hospital section variables for cases belong to clusters 0 and 1.
3.2. SOM clustering
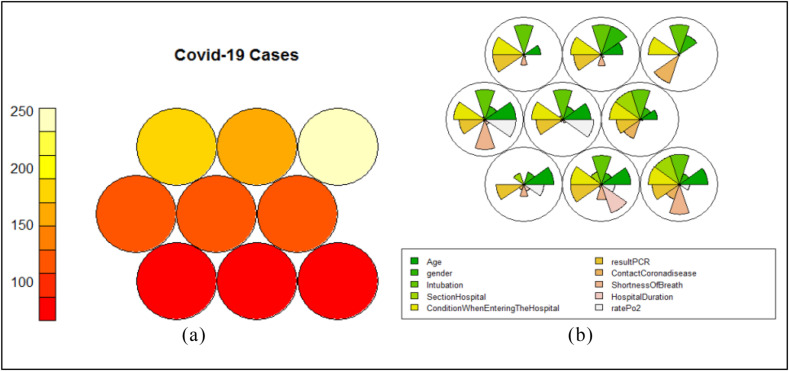
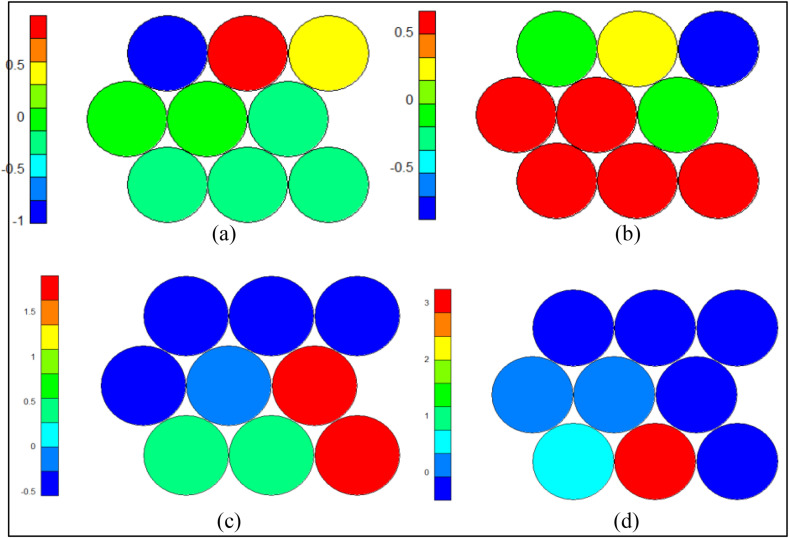
We divided the SOM nodes into 3 clusters using hierarchical clustering (Section 2.4.3). As shown, the first cluster (cluster zero) consists of nodes (3.1), the nodes in position (2,1), (2,2) and (3,2) form the second cluster (cluster one), and finally, the last cluster (cluster two) includes all of the remaining nodes in the SOM network (Fig. 10). In this section, we take a closer look at the clustering of the SOM nodes. We examine how the distribution of different features in the samples belonging to each cluster regardless of their class label through the heat map diagram. Fig. 12 presents the distribution of age, sex, hospital duration, and hospital section variables of COVID-19 cases using the heat map diagram.
Fig. 12.
Distribution of age (a), sex (b), hospital duration (c), and hospital section (d) features of COVID-19 cases in 3*3 SOM network.
The average age of the cases in the first and second clusters is higher than the cases assigned to the third cluster (Fig. 12a). On the other hand, the average sex of patients in clusters one and two is almost zero, so clusters zero and one are composed of the same distribution of male and female patients (Fig. 12b). This shows that patients' age and sex are not related to each other. In addition, although the patients' sex in clusters 0 and 1 tends to be more female (light green), the COVID-19 cases are not generally affected by their sex.
Another significant analysis is the study of the SOM nodes from the hospital duration perspective (Fig. 12c). The length of hospitalization is another remarkable feature that was considered to examine its relationship with other characteristics. A cursory glance at Fig. 12 shows that clusters 0 and 1, which accounted for more elderly COVID-19 cases, were hospitalized longer. On the other hand, younger patients are discharged early (due to recovery or death) and spend the same time in the hospital. Therefore, clustering results prove that the patients' age is directly related to the number of days that they spend in hospital.
The hospital section feature is the latest feature of the COVID-19 cases which its distribution has been studied in this subsection (Fig. 12d). In this study, hospital sections are divided into usual ward (common symptoms) and special ward (severe symptoms) based on the deterioration of patients. A higher number describes the regular section, and a lower number represents the special ward. Fig. 12 displays that all items in clusters zero and one are hospitalized in the special wards, and there is the same distribution of both units in cluster two. It indicates that although the hospital wards do not generally depend on the patients' age and hospital duration, the elderly who have a long time of hospital duration make up most of the patients in the ICU.
(c) (d)
4. Discussion
It has been more than a year since the emergence of the new coronavirus, and the world is still in the face of a human catastrophe. The novel coronavirus kills many people every day around the world. Although widespread vaccination against COVID-19 disease has begun in most countries, health experts say it is still a long way from the end of the pandemic. Accordingly, it is a public duty for everyone to do their best to help solve this global dilemma. One way is to look at the different features in the COVID-19 patients and discover the hidden aspects of the disease by analyzing the relationship between the various symptoms.
In this study, we used different machine learning techniques to assess the clinical symptoms of patients who died of COVID-19 infection or recovered from it. Machine learning techniques in this research mainly include unsupervised methods. We implemented the two K-means and self-organizing map clustering methods to examine the symptoms in different groups of the COVID-19 cases using unsupervised features learning. The K-means clustering is a usual method in unsupervised learning that does not have a high computational cost. We used this method to determine the statistical characteristics of cases belong to different clusters. Before that, we calculated the number of optimal clusters using the Davis & Bolding and Silhouette clustering evaluation measures. These indices calculated 2 clusters as an optimal clustering of our data (Fig. 5). The K-means clustering results showed that patients who died of infection generally have a higher mean age and are hospitalized more in special wards (ICU) than usual wards (Table 2). Recovered COVID-19 patients, on the other hand, have a relatively lower average age and fewer hospitalization days. However, the opposite is not necessarily true because several patients have a high average of features but have recovered. In addition, the patients' sex does not affect their outcome because there is the same distribution of male and female patients in both clusters.
The SOM approach is another way for the proper clustering of data. It is another unsupervised technique we used in this study to determine the correlation and relationship between several symptoms in clusters. We first calculated the number of optimal clusters for hierarchical clustering. This clustering method proposed 3 clusters as the optimal number of clusters (Fig. 9). We assigned different samples in 10 replications to a 3 * 3 SOM and divided its nodes into three clusters by hierarchical clustering (Fig. 10). The advantage of using SOM is the adjustment of the nodes with the nodes within their neighborhood radius. This advantage provides a unique overview of various features and enables visual analysis of data. We used the heat map diagram to show the average values of 4 attributes in different nodes by different colors. Next, we examined which variables are directly related to each other in different clusters (Fig. 12). From the SOM network point of view, our results revealed a direct relationship between age and length of hospital stay attributes. It means that the older the patients, the more hospital duration and vice versa. In addition, the results show that the hospitalization of patients in the special wards (ICU) is directly related to older age and more hospitalization days. However, the patients' sex is not related to other symptoms.
Our results confirmed the research presented in the past. Most studies have identified patients' age as an essential factor in the outcome of COVID-19 infection in patients. According to the latest research, more than 70% of deaths in Iran are among people over 60 years old. In addition, the underlying disease is another important factor that increases the risk of pulmonary involvement. However, in this study, the underlying diseases were identified as secondary symptoms and were not placed as the ten most efficient variables in our study. The reason is the threshold (top ten effective features) set for selecting the features in this research. We considered this threshold value to reduce the cost of computations and increase the reliability of clustering results. Finally, our results provide new information to health professionals. For example, specialists can better understand the relationship between different symptoms of patients.
4.1. Significant statement
As mentioned in the previous section, our results confirm past research and present new facts. The results of this study help health specialists consider other factors to improve services to the COVID-19 patients in addition to characteristics such as age and underlying diseases. Specialists can take various measures to treat different groups of patients by measuring the factors that have been recognized as effective. Also, they should take into account the relationship found between variables in this study. Our results provide an acceptable estimate of the outcome of COVID-19 cases (recovery or death) based on different conditions, so physicians can focus more on patients who are more likely to die. In simple terms, it ultimately leads to reducing in coronavirus mortality.
To sum up, the main findings of this research are as follows:
-
1.
Patients who died of COVID-19 have higher mean values in symptoms, but the opposite is not necessarily true. It means that not all patients with these characteristics necessarily die.
-
2.
There is a direct relationship between patients' age and length of their hospitalization, and the older the patients, the more they are admitted to the ICU than regular wards.
-
3.
The patient's sex has the same distribution in different groups and does not correlate with other symptoms.
4.2. Limitations
This research faced some limitations. First, a large proportion of our samples formed patients who recovered from COVID-19 infection. It leads to learning algorithms more being influenced by recovered cases. One solution is to increase the number of samples belonging to death class to create an acceptable balance of both classes in the data set, which increases the reliability of the results. Second, we considered ten features as the most determinative features to decrease computational costs and increase reliability. Therefore, by raising this threshold, more features will be examined. Third, our data cannot be analyzed by new machine learning methods such as deep artificial neural networks. Because the architecture of deep neural network models is basically suitable for unstructured data or complex data types. In addition, they require a large amount of data for high-level performance. One solution is to implement deep artificial neural networks using a Big Data platform by increasing the diversity of data and integrating existing data with other data types such as CT-scan images and ECG signals. However, it greatly increases the computational complexity of the operations.
5. Conclusion
We investigated different aspects of the new coronavirus using unsupervised machine learning methods. After data preprocessing, we developed the K-means and SOM neural network techniques to cluster the COVID-19 cases. Our results revealed that patients who died of COVID-19 mostly had high mean values in age, intubation, length of hospitalization, and special ward (ICU) features. However, the opposite is not necessarily true. It means though the patients who died of coronavirus infection have a higher mean in their characteristics, there are patients whose average values of their symptoms are high but have recovered. In addition, we developed a SOM neural network for hierarchical clustering of the COVID-19 cases. In this way, we examine the relationship between different features in different groups of patients uniquely. The results indicated that the patients' age was directly related to the length of hospital stay, and elderly patients were admitted to the ICU more than usual wards. Also, the patients' sex has the same distribution in different groups and is not related to other characteristics.
5.1. Future works
Raising the feature selection threshold in this research can be a basis for further study in the future. In addition, in the clustering section, we considered only four characteristics (age, sex, hospital duration, hospital section) to examine the type of their correlation. So, more features such as blood oxygen level, intubation, and shortness of breath can take into account for further analysis. Furthermore, more statistical characteristics can be calculated on clusters to find out more useful knowledge.
Increasing data types and providing the variety of Big Data to take advantage of the capabilities of a Big Data platform such as Apache Spark to provide a framework for implementing deep learning techniques could be another suggestion for future researches.
Finally, an effective way to compare the results of this study is to solve the class imbalance problem for clustering the COVID-19 cases. The class imbalance problem is a major problem in the supervised learning paradigm, and its solution prevents learning algorithms from biasing towards a particular class label [31]. Three over-sampling, under-sampling, and synthetic approaches have been introduced in the literature to solve the class imbalance problem [31]. In the under-sampling technique, some samples of the majority class are removed, in the over-sampling approach, some cases of the minority class are replicated, and in the synthetic method, some cases of the minority class are combined to produce new samples [31].
Data availability
The data utilized for finding the outcomes of this research have been taken through questionnaires and patients' medical records in Saveh Hospital, Iran. Research data was approved by the SMC in Iran and was provided by figshare repository with unique identifier “http://doi.org/10.6084/m9. fihshare.12,446,120. v1″ and under “Attribution 4.0 (CC BY 4.0)" license.
Funding statement
The authors received no financial support for the research and/or authorship of this article.
Author contributions
S.I. collected research data, developed machine learning methods, provided the ideas of study, performed the statistical analysis, and wrote the article. A.A. and E.A. conceived the study, reviewed the various sections of the paper, conceptualized the results, and approved the final version of the manuscript.
Ethical approval
Our research was confirmed by the Institutional Review Board of Department of Information Technology Engineering, Industrial and System Engineering Faculty, Tarbiat Modares University. Ethical review and approval were waived for this study due to the data samples lacked the participants' personal information, and our study did not violate participants' privacy.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to express our full thanks to Saveh Medical Center for providing medical data.
References
- 1.Chen N., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus 37 pneumonia in Wuhan, China: a descriptive study. The lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoosefi Lebni J., et al. How the COVID-19 pandemic effected economic, social, political, and cultural factors: a lesson from Iran. Int J Soc Psychiatr. 2020 doi: 10.1177/0020764020939984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Health Organization "World. World Health Organization; Geneva: 2020. Coronavirus disease (COVID-19) pandemic. [Google Scholar]
- 4.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orsucci Daniele, et al. Neurological features of COVID-19 and their treatment: a review. Drugs in context. 2020;9 doi: 10.7573/dic.2020-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabary Mohammadreza, et al. Pathologic features of COVID-19: a concise review. Pathol Res Pract. 2020 doi: 10.1016/j.prp.2020.153097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olyaee, Hossein Mohammad, et al. RCOVID19: recurrence-based SARS-CoV-2 features using chaos 10 game representation. Data Brief. 2020;32 doi: 10.1016/j.dib.2020.106144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ilbeigipour Sadegh, Albadvi Amir. Supervised learning of COVID-19 patients’ characteristics to discover symptom patterns and improve patient outcome prediction. Inform Med Unlocked. 2022;30 doi: 10.1016/j.imu.2022.100933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuli S., Tuli S., Tuli R., Gill S.S. Predicting the growth and trend of COVID-19 pandemic using machine learning and cloud computing. Internet of Things. 2020;11 doi: 10.1016/j.iot.2020.100222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva Pedro, et al. COVID-19 detection in CT images with deep learning: a voting-based scheme and cross-datasets analysis. Inform Med Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Islam Md Zabirul, Islam Md Milon, Asraf Amanullah. A combined deep CNN-LSTM network for 18 the detection of novel coronavirus (COVID-19) using X-ray images. Inform Med Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ndaïrou F., Area I., Nieto J.J., Torres D.F. Mathematical modeling of COVID-19 transmission dynamics with a case study of Wuhan. Chaos, Solit Fractals. 2020;135 doi: 10.1016/j.chaos.2020.109846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H.M., Junior L.L., Castro F.F.M., Yang A.C. Mathematical model describing COVID-19 in Sao Paulo, Brazil–evaluating isolation as control mechanism and forecasting epidemiological scenarios of release. Epidemiol Infect. 2020;148 doi: 10.1017/S0950268820001600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saraswathi S., Mukhopadhyay A., Shah H., Ranganath T. Social network analysis of COVID-19 transmission in Karnataka, India. Epidemiol Infect. 2020;148 doi: 10.1017/S095026882000223X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pascual-Ferra P., Alperstein N., Barnett D.J. Social network analysis of COVID-19 public discourse on twitter: implications for risk communication. Disaster Med Public Health Prep. 2020:1–9. doi: 10.1017/dmp.2020.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Károly, Artúr István, Fullér Róbert, Galambos Péter. Unsupervised clustering for deep learning: a tutorial survey. Acta Polytechnica Hungarica. 2018;15(8):29–53. [Google Scholar]
- 17.Kwekha-Rashid AmeerSardar, Abduljabbar Heamn N., BilalAlhayani Coronavirus disease (COVID-19) cases analysis using machine-learning applications. Appl Nanosci. 2021:1–13. doi: 10.1007/s13204-021-01868-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benito-Leon Julián, et al. Using unsupervised machine learning to identify age-and sex-independent severity subgroups among patients with COVID-19: observational longitudinal study. J Med Internet Res. 2021;23(5) doi: 10.2196/25988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaudhary Laxmi, Singh Buddha. Community detection using unsupervised machine learning techniques on COVID-19 dataset. Social Network Analysis and Mining. 2021;11(1):1–9. doi: 10.1007/s13278-021-00734-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackey Tim, et al. Machine learning to detect self-reporting of symptoms, testing access, and recovery associated with COVID-19 on Twitter: retrospective big data infoveillance study. JMIR public health and surveillance. 2020;6(2) doi: 10.2196/19509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue Jia, et al. The hidden pandemic of family violence during COVID-19: unsupervised learning of tweets. J Med Internet Res. 2020;22(11) doi: 10.2196/24361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue Jia, et al. Twitter discussions and emotions about the COVID-19 pandemic: machine learning approach. J Med Internet Res. 2020;22(11) doi: 10.2196/20550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue Jia, et al. Public discourse and sentiment during the COVID 19 pandemic: using latent dirichlet allocation for topic modeling on twitter. PLoS One. 2020;15(9) doi: 10.1371/journal.pone.0239441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karadayi Yildiz, Aydin Mehmet N., Selçuk Öǧrenci Arif. Unsupervised anomaly detection in multivariate Spatio-Temporal data using deep learning: early detection of COVID-19 outbreak in Italy. IEEE Access. 2020;8:164155–164177. doi: 10.1109/ACCESS.2020.3022366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Wei-Chun, et al. Journalofthe Formosan MedicalAssociation; 2021. Classification of community-acquired outbreaks for the global transmission of COVID-19:Machine learningand statisticalmodelanalysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai Mingxiang, et al. IEEE international conference on communications workshops (ICC workshops. IEEE; 2021. Evaluation of hybrid unsupervised and supervised machine learning approach to detect self-reporting of COVID-19 symptoms on twitter. 2021. [Google Scholar]
- 27.Haupt Michael Robert, et al. Characterizing twitter user topics and communication network dynamics of the “liberate” movement during COVID-19 using unsupervised machine learning and social network analysis. Online Social Networks and Media. 2021;21 [Google Scholar]
- 28.Carrillo-Larco, Rodrigo M., Castillo-Cara Manuel. Using country-level variables to classify countries according to the number of confirmed COVID-19 cases: an unsupervised machine learning approach. Wellcome Open Research. 2020;5 doi: 10.12688/wellcomeopenres.15819.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brooks Nathan A., et al. The association of Coronavirus Disease-19 mortality and prior bacille Calmette-Guerin vaccination: a robust ecological analysis using unsupervised machine learning. Sci Rep. 2021;11(1):1–9. doi: 10.1038/s41598-020-80787-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.https://chartio.com/learn/charts/bar-chart-complete-guide/
- 31.Han Jiawei, Pei Jian, Kamber Micheline. Elsevier; 2011. Data mining: concepts and techniques. [Google Scholar]
- 32.Chandrashekar G., Sahin F. A survey on feature selection methods. Comput Electr Eng. 2014;40(1):16–28. [Google Scholar]
- 33.Geurts P., Ernst D., Wehenkel L. Extremely randomized trees. Mach Learn. 2006;63(1):3–42. [Google Scholar]
- 34.Rendon E., Abundez I., Arizmendi A., Quiroz E.M. Internal versus external cluster validation indexes. International Journal of computers and communications. 2011;5(1):27–34. [Google Scholar]
- 35.Kohonen T. Essentials of the self-organizing map. Neural Network. 2013;37:52–65. doi: 10.1016/j.neunet.2012.09.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data utilized for finding the outcomes of this research have been taken through questionnaires and patients' medical records in Saveh Hospital, Iran. Research data was approved by the SMC in Iran and was provided by figshare repository with unique identifier “http://doi.org/10.6084/m9. fihshare.12,446,120. v1″ and under “Attribution 4.0 (CC BY 4.0)" license.