Summary
Genetic silencing of leukemia-associated proteins with small interfering RNAs (siRNAs) is a straightforward way to delineate their functions. It can be very challenging to deliver siRNAs to leukemia-derived cells with high transfection efficiency and without compromising their viability. This protocol describes an efficient approach to silence oncogenic feline McDonough sarcoma (FMS)-like tyrosine kinase-3 in leukemia cells using siRNAs that are delivered by electroporation. The protocol maintains high cell viability and is generally useful to decrease RNAs encoding proteins of interest.
For complete details on the use and execution of this protocol, please refer to Beyer et al. (2022).
Subject areas: Cell Biology, Cell culture, Cell-based Assays, Cancer, Molecular Biology
Graphical abstract

Highlights
-
•
Transient knockdown of proteins in leukemic cells with survival rates around 80%
-
•
Technique demonstrated through genetic attenuation of the leukemogenic kinase FLT3-ITD
-
•
Applicability for various cell systems and RNAs/proteins of interest
-
•
Genetic reduction can be used as a comparison for inhibitor studies
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Genetic silencing of leukemia-associated proteins with small interfering RNAs (siRNAs) is a straightforward way to delineate their functions. It can be very challenging to deliver siRNAs to leukemia-derived cells with high transfection efficiency and without compromising their viability. This protocol describes an efficient approach to silence oncogenic feline McDonough sarcoma (FMS)-like tyrosine kinase-3 in leukemia cells using siRNAs that are delivered by electroporation. The protocol maintains high cell viability and is generally useful to decrease RNAs encoding proteins of interest.
Before you begin
Overview
This protocol explains how to culture and transfect leukemic cells with siRNAs against the oncogenic mutant FMS-like tyrosine kinase-3. This will knock-down but not eliminate this protein. The technique is also useful to decrease other proteins of interest in leukemic cells.
Experimental design and assessment of control conditions
In most solid tumor-derived cells, lipid-based (for example, Lipofectamine reagent) or polymer-based (for example, TurboFect reagent) transfection protocols efficiently transfer siRNAs. Unfortunately, this is not the case for most if not all leukemic cell lines. To prevent other researchers from failure and waste of money and resources, this protocol provides a possible option using electroporation.
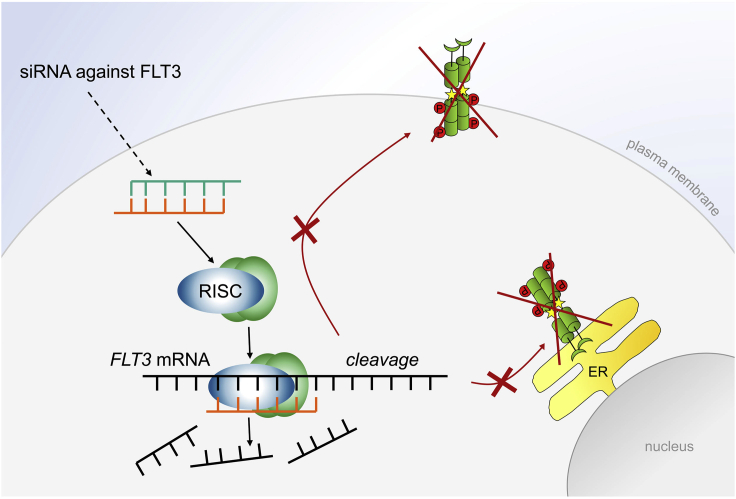
To succeed with the experiments described here, it is mandatory to carefully choose the right model cell line(s) and RNAi molecules. We describe the method based on MV4-11 cells which depend on the FLT3-ITD oncogene (Beyer et al., 2022). In Figure 1, a simplified scheme of the siRNA pathway illustrates how a gene of interest is silenced.
Figure 1.
Silencing of oncogenic FLT3 by RNA interference
Double-stranded siRNA enters the cytoplasm upon electroporation of the cells. After unwinding and cleavage into single-stranded RNA, one siRNA strand binds to the RNA-induced silencing complex (RISC). The activated RISC-siRNA complex binds to the selected mRNA (in our case FLT3 mRNA) which leads to its cleavage and a subsequent reduction of the protein of interest. In the case of mutant FLT3-ITD, FLT3-ITD that locates to the plasma membrane and to the endoplasmic reticulum (ER) is decreased. This attenuates FLT3-ITD-dependent proliferative signaling, and consequently causes cell cycle arrest and apoptotic cell death; P, phosphorylation; yellow star, mutation in the ITD of FLT3 (Beyer et al., 2022).
In advance of the experiment, it must be clarified that there is a power supply and enough space under a clean bench. This allows carrying out the electroporation under sterile conditions.
Before starting the experiment, make sure that the cells are growing exponentially. In the case of the AML cell line MV4-11, it is optimal to use cells that are in culture between 2 weeks and 2 months. It is not recommended to use cells that are freshly thawed or have been in culture for a long time. Make sure that the cells are not stressed due to growth in high density (more than 0.6 × 106 cells per mL) or a missing cell-cell contact (less than 0.1 × 106 cells per mL). This will lead to a cell cycle arrest in the G1-phase, low transfection efficiency, and poorly reproducible data.
In this protocol, the Neon™ Transfection system (100 μL-Kit) is used to attenuate FLT3-ITD, and the success of this approach is proven by immunoblot. Dependent on the research interest and the experiments that are necessary, it is also possible to use the NeonTM Transfection system 10 μL-Kit.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FLT3 (used at 1:250 dilution) | Abcam | Cat#ab245116 |
| GAPDH (used at 1:2,000 dilution) | Santa Cruz | Cat#sc-32233 |
| IRDye® 800CW Donkey anti-Mouse IgG Secondary Antibody (used at 1:10,000 dilution) | LI-COR | Cat#926-32212 |
| IRDye® 800CW Donkey anti-Rabbit IgG Secondary Antibody (used at 1:10,000 dilution) | LI-COR | Cat#926-32213 |
| Chemicals, peptides, and recombinant proteins | ||
| Phosphate buffered saline (PBS) | Bio&SELL | Cat#BS.L 182-50 |
| RPMI-1640 | Sigma | Cat#R8758 |
| Fetal Bovine Serum (FBS) Superior | Sigma | Cat#S0615 |
| Tris | Carl Roth | Cat#5429 |
| NaCl | Carl Roth | Cat#3957 |
| Glycerin | Carl Roth | Cat#3783 |
| EDTA | AppliChem | Cat#131669.1211 |
| SDS | Carl Roth | Cat#8029 |
| Bromophenol blue | Sigma | Cat#B0126 |
| HCl | Carl Roth | Cat#4625 |
| IGEPAL® CA-630 | Merck | Cat# 56741 |
| DTT | Carl Roth | Cat# 6908 |
| Powdered milk | Carl Roth | Cat#T145 |
| Tween® 20 | Carl Roth | Cat#9127 |
| cOmplete™ Protease Inhibitor Cocktail | Roche | Cat#11697498001 |
| Phosphatase Inhibitor Cocktail 2 | Sigma | Cat#P5726 |
| Critical commercial assays | ||
| Neon™ Transfection system 100 μL-Kit Contains:
|
Thermo Fisher Scientific | Cat#MPK10025 |
| Experimental models: Cell lines | ||
| Human: MV4-11, diploid cells from a 10-year-old male with biphenotypic B-myelomonocytic leukemia | DSMZ/ATCC | ACC 102/CRL-9591™ |
| Oligonucleotides | ||
| siRNA-A noncoding control | Santa Cruz | Cat#sc-37007 |
| ON-TARGETplus Human FLT3 (2322) siRNA - SMARTpool | Dharmacon | Cat#L-003137-00-0005 |
| Software and algorithms | ||
| Image Studio Lite Ver 5.0 | LI-COR | https://www.licor.com/bio/image-studio-lite/ |
| Prism6 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Excel | Microsoft | https://www.microsoft.com/de-de/microsoft-365/excel |
| Other | ||
| InvitrogenTM NeonTM Transfection System | Thermo Fisher Scientific | Cat#10090314 |
| Sonicator UP200Ht | Hielscher | https://si.vwr.com/store/product/14037364/ultrasonic-homogenisers-up200ht |
| Neubauer Counting Chamber (depth 0.1 mm) | Marienfeld Superior | Cat# 0640110 |
Materials and equipment
Complete RPMI medium
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI-1640 | n/a | 450 mL |
| Fetal bovine serum (FBS) | 10% | 50 mL |
| Total | n/a | 500 mL |
Store at 4°C and prewarm at 15°C up to 25°C before use.
CRITICAL: FBS (also termed fetal or cosmic calf serum, FCS) is a bovine-derived product that can contain animal pathogens. Touch only with gloves, also to avoid contaminating it. Human platelet lysate (hPL) can be used as a better controlled alternative to FBS, and hPL also allows electroporation of cells with siRNAs (Pons et al., 2019).
NET-N lysis buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 8.0 | 10 mM | 302.9 mg |
| NaCl | 100 mM | 1.5 g |
| Glycerin | 10% | 25 mL |
| EDTA | 1 mM | 93.1 mg |
| IGEPAL® CA-630 | 0.5% | 1.25 mL |
| Add ddH2O to | n/a | 250 mL |
Store at 4°C for 6 months.
Note: Dissolve Tris in deionized water. Adjust the correct pH value by adding HCl dropwise and use a pH meter as control.
CRITICAL: Add protease inhibitor to prevent protein degradation in lysates. One protease inhibitor from Roche is dissolved in 10 mL NET-N lysis buffer and can be stored at 4°C for up to 14 days. If protein phosphorylation is to be analyzed, add phosphatase inhibitor from Sigma at 1:100 dilution to the buffer containing protease inhibitor. 1 mM DTT is added freshly to the buffer. Due to its short half-life, it is advisable to prepare a stock solution of 1 M DTT, to aliquot in small volumes, and store them at – 20°C for maximally 6 months.
CRITICAL: Tris and the detergent IGEPAL® CA-630 can be irritating to skin and respiratory system. Avoid contact by wearing gloves and a face mask or weigh the substances under a hood. HCl is a strong acid and causes painful irritation on skin, eyes, etc. If you or someone else had contact with such substances, wash with plenty of clear water. Do not neutralize with alkaline solutions.
Alternatives: To block the activities of proteases and phosphatases during protein lysis, any other suitable inhibitors can be used.
6× sample buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 6.8 | 375 mM | 1.4 g |
| SDS | 12% | 3.6 mL |
| Glycerin | 30% | 9 mL |
| DTT | 500 mM | 2.3 g |
| Bromophenol blue | 0.01% | small spatula tip (solution becomes midnight blue) |
| Add ddH2O to | n/a | 30 mL |
Aliquot 1 mL buffer in 1.5 mL centrifugation tubes and can be stored at −20°C for up to 1 year.
Note: Dissolve Tris in deionized water. Adjust the correct pH value by adding HCl dropwise, control with a pH meter.
CRITICAL: Bromophenol blue is a potential carcinogen.
Alternatives: The 6× sample buffer can be prepared easily in the lab. It is also possible to use commercially available sample buffers or to use other recipes for sample buffers if desired.
CRITICAL: The usage of an electroporation device is essential for this experiment. In this protocol, the Invitrogen™ Neon™ Transfection System is mentioned (Figures 2A and 2B). For the detection of immunoblots, LI-COR fluorescence coupled secondary antibodies and the LI-COR Odyssey Classic imaging system were used.
Figure 2.
NeonTM Transfection system
(A and B) Contents of the transfection system are (A) a pipette and a pipette unit and (B) the NeonTM transfection device.
Tris-based buffer with Tween® 20 (TBS-T)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 7.6 | 20 mM | 2.4 g |
| NaCl | 140 mM | 8.2 g |
| Tween® 20 | 0.05% | 500 μL |
| Add ddH2O to | n/a | 1 L |
Store at 15°C up to 25°C for up to 1 year.
Note: Dissolve Tris in deionized water. Adjust the correct pH value by adding HCl dropwise, control with a pH meter.
Alternatives: For the electroporation of cells, other systems on the market can be used. The settings of every system must be optimized for each cell type. For the detection of proteins with immunoblotting, other possibilities like enhanced chemiluminescence (ECL) will also work.
Alternatives: In this protocol, a Neubauer counting chamber is used for determining cell numbers. Any other cell counting method can be used, too. Note that because of the small size of MV4-11 cells (and many other leukemic cells), machine-based counting methods may have difficulties to discriminate between intact cells and larger particles of debris or clumping dead cells.
Step-by-step method details
Cell seeding – 1st day
Timing: 1 h
This step is very important to equalize the number of cells in every sample. This will improve the reproducibility of the experiments. The following steps must be done under sterile conditions.
-
1.Resuspend the cells in the culture flask for equal distribution (due to gravity, these cells fall to the bottom of the cell culture flask but remain in suspension) and transfer them into a 50 mL Falcon tube.
-
a.Pipette a volume of 50 μL cell suspension into a 1.5 mL centrifugation tube.
-
b.Count the cells with a Neubauer counting chamber. Calculate the number of cells per mL.
-
a.
Note: Certain leukemic cells fall to the bottom of the flask and must be resuspended more vigorously (for example, we see this with erythroleukemic HEL cells). Solid tumor-derived cells usually attach to the bottoms of flasks and must be detached by trypsinization.
Note: Prepare a dilution of the cell suspension with PBS for more reliable cell counting results. For MV4-11 cells, a dilution of 1:2 works best (this step can be done under non-sterile conditions because the counted cells shall not go back to the main culture). Add 50 μL PBS to the cell suspension and make sure that the cells are resuspended thoroughly.
-
2.Seed 0.75 × 106 cells per mL in a final volume of 4 mL in a 6 well plate (final number of cells per sample: 3 × 106 cells in 4 mL medium). This will yield material for about two immunoblot analyses and one flow cytometry analysis.
-
a.Calculate for 3 samples: siRNA control, siRNA against FLT3, and one additional sample to facilitate accurate pipetting. In total, 9 × 106 cells are necessary.
-
b.Centrifuge the 9 × 106 cells in a 50 mL Falcon tube for 5 min at 200 × g and room temperature (15°C up to 25°C).
-
c.During the centrifugation step, prepare a 6 well plate with 3 mL fresh complete growth medium for the samples that will be electroporated and place it in the incubator to prewarm the media.
-
d.After centrifugation, aspirate the supernatant and resuspend the cells in 3 mL fresh complete growth medium thoroughly (1 mL for every sample) by pipetting.
-
e.Add 1 mL cell suspension to every well of the prewarmed media in the 6 well plate. Thus, you will have two wells with 3 × 106 cells each.
-
f.Let the cells adapt overnight (approximately 16 h), incubate them at 37°C, 5% CO2 in a humidified atmosphere.
-
a.
Note: The cell line MV4-11 should be cultured at a density between 0.3 × 10ˆ6 and 0.5 × 10ˆ6 cells per mL to achieve the best growth. This gives a volume of 18–30 mL of cell suspension, i.e., a volume that fits in a 50 mL Falcon tube.
Note: The optimal cell numbers for electroporation must be adjusted for every cell line. For different leukemia cell lines, cell numbers between 2 × 106 to 3 × 106 cells per sample usually gives good results.
CRITICAL: Make sure that the medium that is used for electroporation does not contain antibiotics. All buffers and media should have room temperature (15°C up to 25°C) to not stress the cells.
Transfection of cells – 2nd day
Timing: 1 h
The following part is the key step. The siRNA is transferred into the cells. All steps are done under sterile conditions.
-
3.
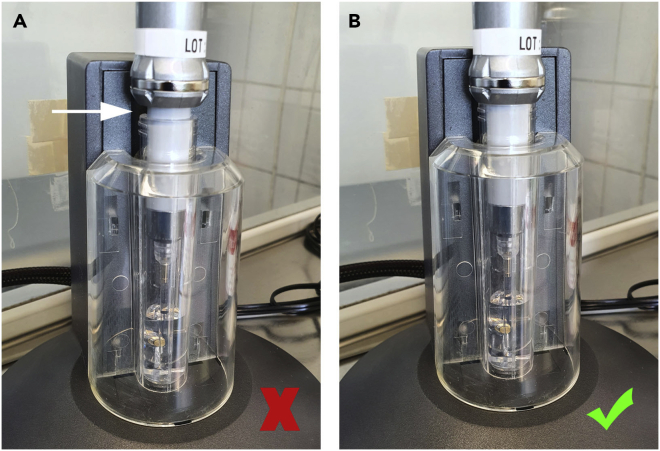
Prepare a NeonTM Electroporation tube with 3 mL Electrolytic Buffer E2 and place it in the transfection unit as shown in Figures 3A–3C.
Note: The NeonTM Electroporation tube can be used several times. It is highly advisable to renew the Electrolytic Buffer E2 between different siRNAs and when using different cell lines.
-
4.Prepare the cells for electroporation. Transfer the cells from the 6 well plate into 15 mL conical centrifugation tubes.
-
a.Resuspend the cells in the well thoroughly and transfer them with a 1 mL pipette into the centrifugation tube.
-
b.Wash each well with 1 mL PBS and transfer it into the tube as well.
-
c.Centrifuge for 5 min at 200 × g at room temperature (15°C up to 25°C).
-
d.Aspirate the supernatant with a vacuum pump and resuspend the cells in 2 mL PBS.
-
e.Centrifuge for 5 min at 200 × g at room temperature (15°C up to 25°C).
-
f.During centrifugation, prepare a 6 well plate with 2 mL complete media per sample and place it in the incubator to warm the media.
-
g.Aspirate the supernatant to remove remaining FBS and resuspend the cells in 100 μL resuspension buffer R from the Thermo Fisher transfection kit.
-
a.
Note: It is very important to aspirate the supernatant very thoroughly. This can be done with a 1 mL pipette if there is no vacuum pump available. A significant excess of PBS leads to strong dilution of the resuspension buffer R. The NeonTM Electroporation Pipette is adjusted for exactly 100 μL; excess of PBS leads to a loss of cells that can be electroporated.
-
5.
Transfer the cells in the electroporation buffer into a 1.5 mL conical centrifugation tube.
-
6.
Add 100 pmol siRNA to the cells. A stock concentration of 10 μM is used for siRNAs (recommended by the manufacturer), so add 10 μL to the cells.
Note: This concentration of siRNA is useful to decrease several proteins in our hands. To transfect cells by electroporation, higher amounts of siRNA are necessary than for liquid-/polymer-based transfection protocols. It is necessary to adjust the concentration for every siRNA of interest and for each cell type.
-
7.
Use the Neon Electroporation Pipette and place the cells in the electroporation unit as shown in Figures 4A and 4B (see also troubleshooting, problem 1). Use the following settings for electroporation of MV4-11 cells.
| Voltage: | 1,350 V |
|---|---|
| Width: | 35 ms |
| Pulses: | 1 pulse |
Note: The Neon™ Tips can be used several times. Change the tips between different cell lines and different siRNAs. It is possible to use the same tip for siRNA control first and siRNA of interest second. It is necessary to adjust the correct settings for every cell line. Thermo Fisher provides a database with protocols of selected cell lines (https://www.thermofisher.com/de/de/home/life-science/cell-culture/transfection/neon-transfection-system/neon-transfection-system-cell-line-data.html). Even if the cell line of interest is not listed, the database can help to establish the correct settings.
CRITICAL: Other settings can be more useful for other leukemic cell types. We effectively decrease proteins by electroporation and RNAi in MOLM-13 acute myeloid leukemia cells (voltage: 1,600 V, width: 10 ms, 3 pulses), in RS4-11 B cell precursor leukemia cells (voltage: 1,750 V, width: 20 ms, 1 pulse), and in HEL erythroleukemia cells (voltage: 1,400 V, width: 20 ms, 2 pulses). For K562 chronic myeloid leukemia cells and NB4 promyelocytic leukemia cells, see our recent publications (Pons et al., 2019, 2021).
-
8.
Transfer the cells into the 6 well plate (as shown in Figure 5) with the prewarmed medium. Let the cells adapt overnight (approximately 16 h).
Figure 3.
Correct placement of NeonTM Electroporation tube
It is very important to place the electroporation tube correctly in the pipette unit.
(A) Prepare the electroporation tube with 3 mL Electrolytic Buffer E2.
(B) Place the tube in the pipette unit. Make sure that it is placed correctly. You can feel it click into place.
(C) The tube is correctly placed when there is no large gap under the tube in the unit.
Figure 4.
Correct placement of the NeonTM Pipette
The correct placement of the electroporation tip is required for the proper working of the electroporation device.
(A) There should be no gap between the electroporation tube and the pipette.
(B) If the pipette is placed correctly, you can feel it click into place.
Figure 5.
Transfer of electroporated cells into 6 well plate
Stimulation/incubation of cells – 3rd day
Timing: 1 h
After adaption time, count and reseed the cells as necessary for the experiment of interest. In this protocol, the cells are analyzed using immunoblot. The following steps are done under sterile conditions in a clean bench.
-
9.
Count the cells using a Neubauer counting chamber as described in step 1.
-
10.
Seed 0.2 × 106 cells per mL in a final volume of 5 mL in complete medium (final number of cells per sample: 1 × 106 cells in 5 mL medium). Pipette the volume of 1 × 106 cells in a 6 well plate and adjust a final volume of 5 mL with complete medium in every well.
CRITICAL: It is advisable not to stress the cells. Avoid any centrifugation steps because the cells are very sensitive after electroporation. It is recommended to test the viability of the cells after electroporation; for example, by flow cytometry using annexin V/propidium iodide staining for early and late apoptotic events (Beyer et al., 2022). The samples can be divided in the experiment and a small amount of 1 mL cell suspension can be used to evaluate apoptosis or cell cycle distribution. The control transfected samples typically show viability of over 80%. If the cells show a high apoptosis rate see troubleshooting, problem 2.
-
11.
Let the cells adapt for at least 1 h before stimulation (e.g., with drugs if necessary) or let the cells incubate overnight (approximately 16 h) at 37°C, 5% CO2 in a humidified atmosphere before harvesting.
Note: For MV4-11 cells, incubation of 48 h after electroporation gives the best results regarding knock-down efficiency. Using another cell line, the ideal time for gene silencing must be identified.
Note: This protocol is not only working for siRNA. It can also be used for plasmids and the CRISPR-Cas9 technology. It can likewise be used to decrease RNAs of interest, e.g., miRNAs or long non-coding RNAs with known or to be identified biological functions.
Harvesting and lysis of cells – 4th day
Timing: 2 h
In this protocol, the cells are analyzed by immunoblot. The following steps can be done under non-sterile conditions and must be performed on ice if indicated.
-
12.Cell harvest.
-
a.Transfer the cells from the 6 well plate into conical 15 mL centrifugation tubes.
-
b.Centrifuge for 5 min at 800 × g at room temperature (15°C up to 25°C).
-
c.Aspirate the supernatants with a vacuum pump and a Pasteur pipette or with a 1 mL pipette. Resuspend the cells in 1 mL PBS and transfer the cells into 1.5 mL centrifugation tubes.
-
d.Centrifuge for 5 min at 800 × g at 4°C.
-
e.Aspirate the supernatants thoroughly.
-
a.
Note: Make sure that there is as little PBS left as possible after aspiration. A high excess of PBS will dilute the lysis buffer and lead to incomplete cell lysis.
-
13.Cell lysis.
-
a.Add an appropriate amount of complete NET-N lysis buffer (i.e., containing protease/phosphatase inhibitors and DTT) to the cells. As rule of thumb add around 100 μL of complete NET-N lysis buffer to 1 × 106 cells.
-
b.Lyse the cells for 30 min on ice.
-
c.Sonicate the samples for 10 s and an amplitude of 40%. This shears DNA and thereby facilitates the loading of protein samples into the SDS-PAGE gel. Benzonase might alternatively be used to shear DNA.
-
d.Centrifuge the tubes for 25 min at 17,000 × g at 4°C.
-
a.
-
14.Determination of protein concentration and final sample preparation.
-
a.Transfer the supernatants into new 1.5 mL centrifugation tubes and determine protein concentrations (e.g., Bradford or BCA assays).
-
b.Equalize the amount of protein in every sample and add 6× sample buffer to a final concentration of 1× buffer.
-
c.Denature the proteins by boiling for 5 min at 95°C.
-
a.
Note: Certain proteins do not tolerate boiling for 5 min at 95°C. If necessary, reduce boiling time and/or temperature.
Note: If cytosolic fractions are collected, shearing/degradation of genomic DNA is not necessary. Together with nuclear proteins, it will remain in the centrifuged nuclear fraction; see (Göder et al., 2021) for further details.
Pause point: After preparing the lysates and boiling, it is possible to continue the next day. You can freeze the samples at – 20°C for several weeks, until running the immunoblot.
-
15.
Run immunoblot and analyze the proteins of interest. For complete details on the use and execution of performing an immunoblot, please refer to (Beyer et al., 2017).
Note: Primary antibodies are prepared in TBS-T buffer with 2% skim milk powder; dilutions of antibodies are listed in the key resources table. Secondary antibodies are prepared in TBS-T buffer.
Alternatives: The high starting numbers of cells for this experiment provide the ability to perform other experiments of interest after gene silencing.
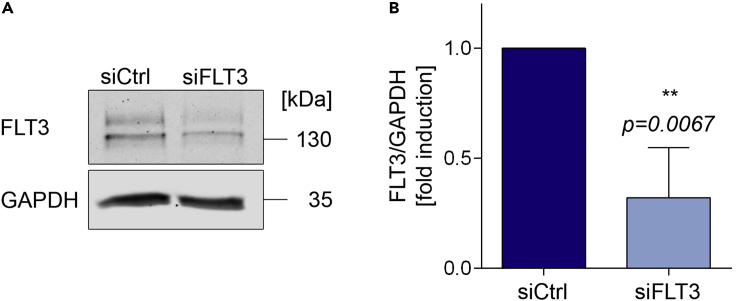
Expected outcomes
Samples obtained and analyzed with this protocol show a strong reduction in the protein expression of FLT3, as shown in Figures 6A and 6B. If the usage of the protocol failed, see troubleshooting, problem 3, problem 4, and problem 5.
Note: This experiment helps to understand the impact of FLT3 in AML cells. For example, it allows to investigate different cell signaling pathways that depend on FLT3 expression by immunoblot. The observed effects can be controlled with specific inhibitors of FLT3 (Beyer et al., 2022).
Figure 6.
Expected outcome of electroporation
(A) After analyzing the lysates by immunoblot, there should be a clear reduction of the protein of interest.
(B) Quantification of three independent blots (n=3; student’s one-tailed t-test; ∗∗p≤0.01). The control samples were set to 1 in each of the three experiments and the siFLT3 samples were set relative to these values of 1.
Quantification and statistical analysis
It is possible to quantify the reduction of proteins that are detected with immunoblot by densitometry. In this protocol, the LI-COR imaging software is used for quantification. Draw a rectangle around the bands of interest (see Figure 7) and copy the values shown into an Excel sheet. In the table below, calculation of signal intensity is explained. The test for statistical significance was done with GraphPad Prism 6.
Calculation of signal intensity
| Sample | Signal FLT3 | Signal GAPDH | Normalized signal FLT3 | Normalized signal GAPDH | Signal of FLT3 normalized to GAPDH |
|---|---|---|---|---|---|
| siCtrl | X | A | = X/X | =A/A | = (set as 1) |
| siFLT3 | Y | B | =Y/X | =B/A | = |
The LI-COR imaging software gives three different measurements. These are background, area, and signal. Each signal is calculated by subtracting the background signal in the measured area.
Alternatives: Here, LI-COR imaging software is used for quantification and Microsoft Excel for quantification of immunoblots, but any other suitable software (for example, ImageJ) can be used for densitometry. The calculation of significance can be done with any other working software (for example Excel or SPSS).
Figure 7.

Example of densitometric analysis of an immunoblot
Picture from Image Studio Lite Ver 5.0 exemplifies how to quantify an immunoblot. The program gives the intensity of the signal (background signal and size of the rectangle included in the signal calculation).
Limitations
Silencing a gene with siRNAs and the procedure of electroporation can stress cells extremely. This can lead to high induction of apoptosis and/or changes in cell cycle distribution, for example, G1-phase arrest. If this occurs with control siRNA transfections, it can falsify results. On the other hand, an induction of cell death with the specific siRNA can indicate that the desired effect was reached. For example, in the case of a FLT3-ITD knock-down in MV4-11 cells, apoptosis is a consequence; this mirrors the effects of FLT3 inhibitors (Beyer et al., 2022). Nonetheless, cell death should not occur rapidly after electroporation and/or non-specifically with the control siRNAs. If this happens, consider replacing such controls and modify the protocol.
The described method of gene silencing is a temporary effect and results obtained with this method must be repeated at least three times.
It can be very challenging to adjust the correct settings for electroporation, especially when using hard-to-handle cells. There are some variables to determine: Correct cell numbers, ideal electroporation settings, and incubation time before analyzing the cells.
The outcome of the experiment is highly dependent on the available siRNAs. Sometimes it is necessary to test several siRNAs to find the one that is working perfectly.
This protocol shows a possibility to silence a gene. However, there is still a residual activity of the gene and protein of interest. Using siRNA, it is not possible to create a complete and permanent knockout.
The use of independent siRNAs can corroborate the data that are obtained. This can also diminish results that stem from off-target effects. A mixture of siRNAs that targets multiple sites of the mRNA often increases the silencing. This also holds for mixtures of shRNAs that are encoded on plasmids that can be transfected into cells. Note that plasmids can generally be transfected less effectively into cells. This is due to their much larger size than siRNAs and the higher purity of the chemically produced siRNAs.
To further control the observed effects of siRNAs, siRNA-resistant molecules can be transfected with the siRNAs. These are often called rescue mutants (Krämer et al., 2006). This technique requires the creation of plasmids with silent mutations in the siRNA-binding sites. This can be troublesome if the providers of commercially available siRNAs do not disclose the exact sequences. Alternatively, siRNAs can be targeted to the untranslated regions (5′-UTR or 3′-UTR) of the mRNAs that are to be decreased and plasmids with the coding sequence lacking the UTRs can be transfected. Furthermore, siRNAs against a human mRNA can be transfected with rescue mutants from another species (often from Mus musculus). Depending on sequence conservation, this approach can be successful. Care should though be taken if the human and murine proteins do not have identical functions, complex formation behavior, or other differences.
Troubleshooting
Problem 1
When placing the NeonTM Electroporation Pipette in the transfection unit, mistakes can be done. If the pipette is placed correctly, there is a click noise. Other error messages may though appear in the display of the NeonTM Electroporation device (step 7).
Potential solution
Make sure that there are no air bubbles in the pipette tip. If there is a loss of volume during the transfer of the cells from the 15 mL tube to the 1.5 mL centrifugation tube, add some resuspension buffer R to the sample so that no air bubbles occur. Make sure that the Electroporation Pipette is placed perfectly in the transfection unit and that the connections have contact (see Figures 8A–8D).
Figure 8.
Correct placement and usage of a pipette tip on the NeonTM Transfection Pipette
To make sure that no error message appears, it is very important to use the transfection tip properly.
(A) Place the pipette tip on the pipette.
(B) Make sure that the tip fits correctly. Try to move the metallic electrode in the middle of the tip. If it does not move, replace the tip.
(C) Soak the cells in the resuspension buffer without air bubbles. This will lead to an error message.
(D) Air-bubble-free cells are in the resuspension buffer in the electroporation tip.
Problem 2
Cells show low viability (steps 9 and 10).
Potential solution
Check if the cells are growing exponentially and optimize the settings for transfection. Using wrong voltages or pulses can lead to high apoptosis rates. In this case, transfect the cells of interest with non-coding siRNA and try several settings. In case of hard-to-handle cells, it can be helpful to increase the percentage of FBS in the growth media. Not only the technical settings are important, also the density of the cells. Adjust the cell number for transfection thoroughly.
Problem 3
Using this protocol, no reduction of the targeted protein is detected (section expected outcomes).
Potential solution
If no reduction of the protein of interest is detectable, it may be necessary to repeat the experiment and change some conditions or to order another siRNA. Not every siRNA is working in every cell line, due to different mutations and splice variants (see Figure 1 for method details). A literature search is necessary to carefully choose the siRNAs. Most companies like Dharmacon or Thermo Fisher Scientific provide such tools and a look at the literature can be helpful (Khvorova and Watts, 2017; Li and Zamore, 2019). If you find siRNAs that are frequently and successfully used, they will likely be helpful. Additionally consider that some mRNAs are more abundant than others and this can lead to a slower decline of the proteins that they encode. Moreover, some proteins are very stable in cells and even a strong reduction of their mRNAs can cause no discernable reduction of such proteins. This can often be solved with multiple rounds of electroporations over several days. One must also consider that antibodies sometimes do not bind the protein that they are expected to. Other antibodies bind related proteins that are not affected by the siRNA. Both can be mistakenly interpreted as poor RNAi efficacy. In case of such issues and to ensure that your siRNA targets the mRNA of interest, you can additionally perform qPCR analyses to verify the reduction of this mRNA.
Always check if the cells are growing normally. Some cell lines can differentiate during culturing or acquire different features over time (e.g., increased chromosomal instability, senescence markers). In this case, thaw a new vial of cells or try to obtain cells from another source.
Problem 4
The targeted protein is reduced, but there is no biological effect or another type of biological effect than with an inhibitor for the protein (section expected outcomes).
Potential solution
Genetic silencing of a druggable protein, followed by analyses for biochemical alterations and cell fate decisions, can inform whether drug-induced effects are specific. Selecting the cell line of choice is key for the success of such an approach. If you aim to do research on mutant FLT3, you should use cell lines with FLT3-ITD or other FLT3 mutants (for example, AML cells such as MV4-11 cells or MOLM-13 cells). If you are interested in other kinases such as BCR-ABL, you should choose other cell types (for example, chronic myeloid leukemia cells such as K562 cells or LAMA cells). In other words, your protein of interest should be present in your cell type of interest. In addition, the protein must have a key biological function that can be revealed by its depletion with RNAi.
Also consider that effects of a pharmacological inhibitor of a protein and genetic elimination of it may be different, because the inhibition and the removal of a protein can trigger common and disparate effects in cells. Moreover, inhibitors usually act immediately, while RNAi-mediated protein reduction is a slower process. This can lead to a requirement for different time points to analyze inhibitor- and RNAi-induced effects.
Problem 5
Immunoblot is not able to detect the protein of interest (section expected outcomes).
Potential solution
This can have multiple reasons, such as low expression levels of the protein of interest in the chosen cell type, poor antibody affinity, too low concentrations of antibodies, too little amount of protein lysate loaded, inappropriate blocking solutions, transfer problems (this occurs especially with very short and large proteins), etc. Solutions for these and further issues with immunoblot are discussed in more detail in (Beyer et al., 2017, 2022).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Oliver H. Krämer (okraemer@uni-mainz.de).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
M.B. is funded by the German Research Foundation/Deutsche Forschungsgemeinschaft (DFG) KR2291/9-1, project number 427404172 (to O.H.K.) and intramural funding to M.B. Work done in the group of O.H.K. is also supported by DFG KR2291/12-1, project number 445785155; KR2291/14-1, project number 469954457; KR2291/15-1, project number 495271833; KR2291/16-1, project number 496927074; KR2291/17-1, project number 502534123; the Deutsche Forschungsgemeinschaft DFG-project number 393547839 - SFB 1361, sub-project 11; the Wilhelm Sander-Stiftung (2019.086.1); the DAAD Egypt/Germany; and the Brigitte und Dr. Konstanze Wegener-Stiftung (Projekt 65).
Author contributions
Both authors contributed to the writing, editing, and approval of this work.
Declaration of interests
M.B. and O.H.K. declare the patent “Synthesis, pharmacology, and use of new and selective FMS-like tyrosine kinase 3 (FLT3) inhibitors, WO2019/034538,” and O.H.K. declares the patent “Novel HDAC6 inhibitors and their uses, WO2016020369A1”. The first patent covers substance classes that are effective against mutant FLT3, but none of the patents deals with electroporation techniques or other improvements for cell transfection.
Contributor Information
Mandy Beyer, Email: manbeyer@uni-mainz.de.
Oliver H. Krämer, Email: okraemer@uni-mainz.de.
Data and code availability
This study did not generate/analyze new datasets or code.
References
- Beyer M., Henninger S.J., Haehnel P.S., Mustafa A.H.M., Gurdal E., Schubert B., Christmann M., Sellmer A., Mahboobi S., Drube S., et al. Identification of a highly efficient dual type I/II FMS-like tyrosine kinase inhibitor that disrupts the growth of leukemic cells. Cell Chem. Biol. 2022;29:398–411.e4. doi: 10.1016/j.chembiol.2021.10.011. [DOI] [PubMed] [Google Scholar]
- Beyer M., Kiweler N., Mahboobi S., Krämer O.H. How to distinguish between the activity of HDAC1-3 and HDAC6 with western blot. Methods Mol. Biol. 2017;1510:355–364. doi: 10.1007/978-1-4939-6527-4_26. [DOI] [PubMed] [Google Scholar]
- Göder A., Ginter T., Heinzel T., Stroh S., Fahrer J., Henke A., Krämer O.H. STAT1 N-terminal domain discriminatively controls type I and type II IFN signaling. Cytokine. 2021;144:155552. doi: 10.1016/j.cyto.2021.155552. [DOI] [PubMed] [Google Scholar]
- Khvorova A., Watts J.K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017;35:238–248. doi: 10.1038/nbt.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer O.H., Baus D., Knauer S.K., Stein S., Jäger E., Stauber R.H., Grez M., Pfitzner E., Heinzel T. Acetylation of Stat1 modulates NF-kappaB activity. Genes Dev. 2006;20:473–485. doi: 10.1101/gad.364306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zamore P.D. RNA interference and small RNA analysis. Cold Spring Harb. Protoc. 2019 doi: 10.1101/pdb.top097436. pdb.top097436. [DOI] [PubMed] [Google Scholar]
- Pons M., Nagel G., Zeyn Y., Beyer M., Laguna T., Brachetti T., Sellmer A., Mahboobi S., Conradi R., Butter F., Krämer O.H. Human platelet lysate as validated replacement for animal serum to assess chemosensitivity. ALTEX. 2019;36:277–288. doi: 10.14573/altex.1809211. [DOI] [PubMed] [Google Scholar]
- Pons M., Zeyn Y., Zahn S., Mahendrarajah N., Page B.D.G., Gunning P.T., Moriggl R., Brenner W., Butter F., Krämer O.H. Oncogenic kinase cascades induce molecular mechanisms that protect leukemic cell models from lethal effects of de novo dNTP synthesis inhibition. Cancers. 2021;13:3464. doi: 10.3390/cancers13143464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze new datasets or code.